Fusobacterium spp. are emerging pathogens that contribute to mammalian and human diseases, including colorectal cancer. Despite a validated connection with disease, few proteins have been characterized that define a direct molecular mechanism for Fusobacterium pathogenesis. We report a comprehensive examination of virulence-associated protein families in multiple Fusobacterium species and show that complete genomes facilitate the correction and identification of multiple, large type 5a secreted autotransporter genes in previously misannotated or fragmented genomes. In addition, we use protein sequence similarity networks and human cell interaction experiments to show that previously predicted noninvasive strains can indeed bind to and potentially invade human cells and that this could be due to the expansion of specific virulence proteins that drive Fusobacterium infections and disease.
KEYWORDS: Fusobacterium nucleatum, Fusobacterium necrophorum, virulence factor, autotransporter, type 5 secretion, FadA, T5SS, Fap2, colorectal cancer, adhesin, host-pathogen, Lemierre’s syndrome, Fusobacterium
ABSTRACT
Fusobacterium spp. are Gram-negative, anaerobic, opportunistic pathogens involved in multiple diseases, including a link between the oral pathogen Fusobacterium nucleatum and the progression and severity of colorectal cancer. The identification and characterization of virulence factors in the genus Fusobacterium has been greatly hindered by a lack of properly assembled and annotated genomes. Using newly completed genomes from nine strains and seven species of Fusobacterium, we report the identification and corrected annotation of verified and potential virulence factors from the type 5 secreted autotransporter, FadA, and MORN2 protein families, with a focus on the genetically tractable strain F. nucleatum subsp. nucleatum ATCC 23726 and type strain F. nucleatum subsp. nucleatum ATCC 25586. Within the autotransporters, we used sequence similarity networks to identify protein subsets and show a clear differentiation between the prediction of outer membrane adhesins, serine proteases, and proteins with unknown function. These data have identified unique subsets of type 5a autotransporters, which are key proteins associated with virulence in F. nucleatum. However, we coupled our bioinformatic data with bacterial binding assays to show that a predicted weakly invasive strain of F. necrophorum that lacks a Fap2 autotransporter adhesin strongly binds human colonocytes. These analyses confirm a gap in our understanding of how autotransporters, MORN2 domain proteins, and FadA adhesins contribute to host interactions and invasion. In summary, we identify candidate virulence genes in Fusobacterium, and caution that experimental validation of host-microbe interactions should complement bioinformatic predictions to increase our understanding of virulence protein contributions in Fusobacterium infections and disease.
IMPORTANCE Fusobacterium spp. are emerging pathogens that contribute to mammalian and human diseases, including colorectal cancer. Despite a validated connection with disease, few proteins have been characterized that define a direct molecular mechanism for Fusobacterium pathogenesis. We report a comprehensive examination of virulence-associated protein families in multiple Fusobacterium species and show that complete genomes facilitate the correction and identification of multiple, large type 5a secreted autotransporter genes in previously misannotated or fragmented genomes. In addition, we use protein sequence similarity networks and human cell interaction experiments to show that previously predicted noninvasive strains can indeed bind to and potentially invade human cells and that this could be due to the expansion of specific virulence proteins that drive Fusobacterium infections and disease.
INTRODUCTION
Bacterial pathogens use a repertoire of diverse virulence proteins to establish infection and confer long-term survival in their respective hosts and environments. A central theme to these virulent phenotypes is the expression of surface-exposed and secreted proteins that interact with a variety of macromolecule receptors on host cells (1–4). In addition, these proteins can be deployed by intracellular bacteria within the host cytoplasm or niche-specific vacuole to confer survival, replication, and dissemination. These phenotypes in Gram-negative bacteria are frequently achieved by using large, multiprotein secretion systems, or nanomachines, divided into six categories (secretion systems for types 1 to 6 [T1SS to T6SS]) (5). While these are the most common systems to introduce virulence factors into the host or competing bacteria, Fusobacterium cells are unique in that they lack all of the aforementioned multiprotein secretion systems except for the type 5 secretion system (T5SS) (6). This system is unique in that it is not a large nanomachine but is divided into five distinct categories (T5aSS to T5eSS) that are composed of only one (5a, 5c, 5d, and 5e) or two proteins (T5bSS). These subtypes can be divided into monomeric autotransporters (5a and 5d) (7, 8), two-partner secretion systems (5b) (9), homotrimeric autotransporters (5c) (10), and intimins (5e) (11, 12). The majority of characterized autotransporters are large adhesins or proteases of the T5aSS, or homotrimeric T5cSS adhesins that include YadA from Yersinia species (13). A large-scale bioinformatic analysis showed that 100% of Fusobacterium genomes encode T5aSS proteins, the highest percentage in all Gram-negative bacteria tested (14).
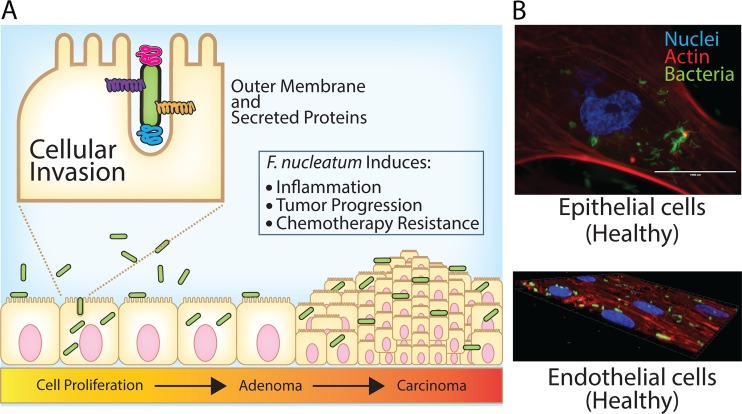
Fusobacterium spp. are Gram-negative, nonmotile, anaerobic bacteria generally isolated from the human oral cavity, but they can also infect other higher mammals, including cattle and sheep (15–17). A strong correlation has been established between the presence of F. nucleatum in colorectal cancer (CRC) tumors and a direct induction of increased tumor size, frequency, and stimulation of a proinflammatory tumor microenvironment (Fig. 1) (18–20). The interaction of this bacterium with host cells also induces chemoresistance by blocking apoptosis (21), and viable bacteria have been shown to travel within metastatic cells to the liver (22). In addition, increased F. nucleatum loads within patient sampled tumors correlate with decreased human life expectancy (23).
FIG 1.
F. nucleatum is an invasive opportunistic pathogen capable of multitissue colonization and infections, with a validated role in colorectal cancer progression. (A) Overview of F. nucleatum cellular invasion, bioinformatically or experimentally characterized proteins that participate in this phenotype, and consequences of infection within localized tissue niches. (B) Fluorescence microscopy images showing F. nucleatum subsp. nucleatum ATCC 23726 invasion of human epithelial and endothelial cells.
In a recent large-scale bioinformatic study, Fusobacterium species were divided into actively invading species that drive their own cellular entry (F. nucleatum, F. periodonticum, F. varium, and F. ulcerans), passively invading species that require a compromised epithelial barrier for cellular entry (F. necrophorum and F. gonidiaformans), and those with unknown invasive potential (F. mortiferum) (24). However, it was previously experimentally shown that multiple F. necrophorum and F. mortiferum strains were significantly more invasive than F. nucleatum strains into keratinocytes, which is in direct conflict with bioinformatic reports that place these species in the noninvasive or passively invasive category (24, 25). While host cell binding and invasion have been deemed critical for F. nucleatum pathogenesis in connection with colorectal cancer, the role of invasion in non-F. nucleatum species remains understudied. For instance, it is possible that invasive F. necrophorum and F. mortiferum strains could induce similar host cell signaling changes seen in CRC, and yet they might not be associated with specific diseases due to their verified low abundance compared to that of F. nucleatum. However, this hypothesis has not been tested experimentally, and F. necrophorum and F. mortiferum have not been associated with CRC. A recent study reported that F. nucleatum is able to induce chemoresistance by interacting with surface-exposed toll-like receptors (21). However, we note that the strain used in that study was highly invasive (F. nucleatum 25586), while the amount of intracellular bacteria was not determined. Since previously characterized mutants with decreased invasion into human cell lines are also deficient in cellular binding, it could be that there is a complex phenotype that bridges the need for initial host cell docking, and subsequent intracellular modulation of cell signaling. In summary, despite extensive phylogenetic analyses of Fusobacterium, our knowledge of the specific virulence mechanisms of this bacterial genus remains limited.
There is a clear gap in our understanding of invasion and virulence in the genus Fusobacterium and the proteins that are involved in driving diverse phenotypes in hosts from humans to cattle and sheep. Most of our knowledge comes from a limited number of F. nucleatum strains and a small sampling of outer membrane adhesins that have been experimentally validated as critical for oral interactions, preterm birth, and colorectal cancer. The reason for this lack of molecular studies in Fusobacterium is its well-known genetic recalcitrance, with only four strains of F. nucleatum yielding chromosomal modification (15, 26–28). To aid in our understanding of virulence at the genetic and molecular level, we recently completed nine Fusobacterium genomes and created the FusoPortal database that includes detailed genomic and bioinformatic analysis of this emerging pathogen (29, 30). These genomes were used here to identify and correct protein families of the autotransporter, FadA, and MORN2 domain-containing proteins, all of which are predicted to play key roles in cellular binding and invasion. Among the virulence associated proteins that have been experimentally characterized is Fap2, a large (3,786 amino acids [aa] in F. nucleatum 23726), dual-function, autotransporter adhesin that binds to the natural killer receptor TIGIT to inhibit tumor cell clearance (40) and initiates host cell docking and altered signaling through the sugar Gal-GalNAc on the surfaces of colorectal cancer cells (43).
FadA is a small (∼125 aa) adhesin that multimerizes on the surface of F. nucleatum and has been shown to directly bind to E-cadherin, where it induces β-catenin signaling in human cancer xenografts in mice (31). For multiple F. nucleatum genomes, we highlight the identification of two homologues of FadA (FadA2 and FadA3), with multiple identical copies of the FadA3 gene being identified and verified throughout each genome (28). FadA is directly involved in host cell binding and invasion in the strain F. nucleatum subsp. polymorphum 12230 and yet, likely due to genetic restraints, it has not been characterized in other strains.
In Fusobacterium necrophorum, the leukotoxin LktA (lktBAC operon) is secreted by the type 5b two-partner secretion system and has been characterized in cattle and sheep infections causing liver abscesses, spontaneous abortion, and foot rot (F. necrophorum subsp. necrophorum) (32, 33). In humans, F. necrophorum subsp. funduliforme is the predominant F. necrophorum subspecies and a native inhabitant of the human oropharynx. This strain is leukotoxin positive and causes infections of the throat and jugular vein in the form of the potentially fatal Lemierre’s syndrome (62). Despite our knowledge that LktA induces immune cell toxicity, the mechanisms by which these opportunistic subspecies become invasive and establish infection in different organisms are poorly understood. To our knowledge, we have sequenced, annotated, and bioinformatically characterized the only two complete F. necrophorum genomes in F. necrophorum subsp. necrophorum ATCC 25286 (34) and F. necrophorum subsp. funduliforme 1_1_36S (29), which are highlighted through experimental characterization of host cell binding in this study.
Of the protein families characterized in this study, the most frequent and extensively enriched are MORN2 (membrane ontology and recognition nexus type 2) domain-containing proteins that can have 1 to 27 repeats of this ∼20-amino-acid domain. This expansion of MORN2 domain proteins is highly specific to Fusobacterium (24), with the exception of multiple genes found in Helicobacter bilis, which is involved in colitis and hepatitis, as well as infectious abortions in sheep (35, 36). No known function has been assigned to the MORN2 domain-containing proteins; however, most contain signal sequences allowing for export into the periplasmic space and potential further export to the outer membrane or secretion into the extracellular environment. As was previously shown, MORN2 domain proteins are enriched in invasive F. nucleatum species and are frequently present in genomic regions with type 5a autotransporter genes.
With the recent development of a selectable gene knockout system in F. nucleatum 23726 (27), the need to identify accurate gene boundaries and surrounding gene clusters is critical for identifying and characterizing virulence proteins used by these bacteria to establish infection. We believe this study will provide a critical tool to drive the improvement of genetic manipulation, therefore allowing previously difficult or impossible cloning and recombinant expression of proteins for the development of antibodies and protein structure-function studies. In addition, these data will ultimately help us answer the question of how a genus of Gram-negative bacteria that lack multiprotein secretion machinery has evolved an alternative mechanism of driving host cell interactions and colonization through an abundance of outer membrane presented adhesins and enzymes. In summary, this study provides a foundation for an increased understanding of how Fusobacterium spp. infect a diverse range of host tissues.
RESULTS
Fusobacterium open reading frame predictions and comparison with previous database annotations.
As shown in Fig. S1 in the supplemental material, the previously completed F. nucleatum subsp. nucleatum ATCC 25586 genome (37) and database depositions of T5aSS proteins were improperly annotated through shortcomings in annotation software and not errors in the genome. We show that 15 of 15 T5aSS autotransporters, including Fap2 (NP_604343.1; 3,165 aa) were previously misannotated (KEGG, GenBank, and UniProt databases) as determined by greatly increased open reading frame length using Prokka (38) and Prodigal (39), and the subsequent identification of the required secretory (SEC) signal sequence for inner membrane translocation. Reannotation of the F. nucleatum 25586 genome in a previous publication also showed a correction of T5aSS open reading frame annotations (6). To add experimental evidence to our annotations, previous work showed that gene interruption of radD in F. nucleatum 23726 resulted in a missing protein band, as seen by SDS-PAGE, at ∼370 kDa in F. nucleatum 23726, which matches well with our annotated size of this protein at 3,461 amino acids. However, the RadD protein was previously annotated as 2,143 amino acids in F. nucleatum 25586, and our new annotation of 3,472 aa matches much better with this experimental data validating a protein of ∼370 kDa.
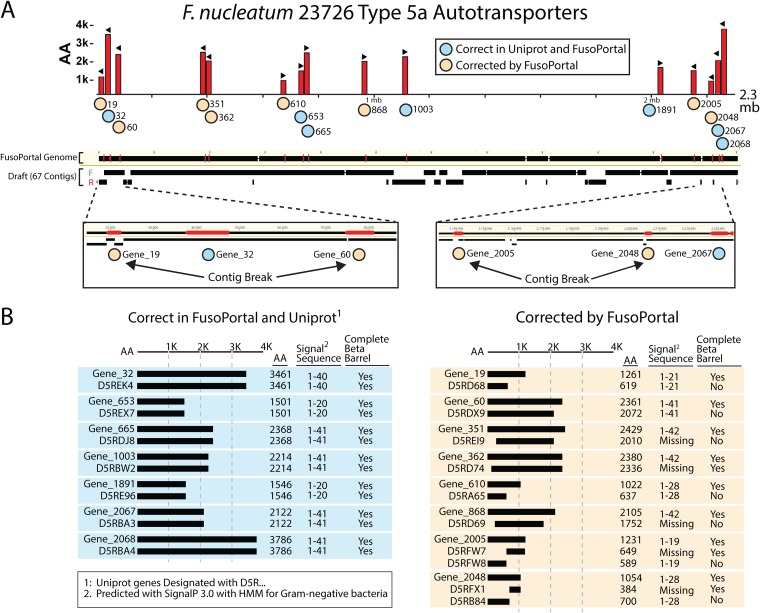
This initial open reading frame correction study led us to reannotate additional publicly available Fusobacterium genomes to determine if this was a common occurrence. We discovered that multiple genomes suffered from large proteins either being misannotated due to software limitations or that these large genes spanned the broken boundaries of multicontig genomes. For example, upon our resequencing and completion of the F. nucleatum subsp. nucleatum ATCC 23726 genome and comparison to the previous 67 contig draft (Fig. 2A), we show that 8 of 15 T5aSS open reading frames were incorrectly annotated, with four of these being due to contig breaks resulting in fragmented genes. These errors are only partially due to the large gene length (>11 kb for fap2 [FN1449]), since the other errors were annotation driven (Fig. 2B). Many of these proteins are predicted surface adhesins with homology to Fap2, a key protein involved in F. nucleatum-induced colorectal cancer modulation. We subsequently sequenced and completed a total of nine Fusobacterium genomes and previously showed that genes larger than 3 kb had a high error rate, as validated in this study for the T5aSS proteins (29, 30, 34). A comparison of all annotations of T5aSS to T5dSS autotransporters for strains F. nucleatum 25586 and F. nucleatum 23726 can be found in Table S1 in the supplemental material. In addition, we provide files containing open reading frames for all virulence proteins discussed in this paper (T5SS, FadA, and MORN2) in FASTA format (OSF repository), and Table S2 contains a list of all open reading frames with InterPro domains and N-terminal signal sequence validation.
FIG 2.
Comparison of T5aSS autotransporter gene annotations from incomplete and complete F. nucleatum 23726 genomes. (A) Locations (red bars) and directions (arrowheads) of T5aSS open reading frames in the complete F. nucleatum 23726 genome (NCBI, ASM301978v1) from FusoPortal. Contigs from the draft genome (NCBI, ASM17889v1) were aligned on the complete genome, and contig breaks that affected gene annotation are highlighted. (B) Autotransporters that were previously correct (blue) and corrected (tan) are highlighted and show that annotation errors were a combination of N-terminal (signal sequence) and C-terminal (outer membrane embedding β-barrel) truncations.
Phylogenetic and protein sequence similarity network analysis of seven Fusobacterium species.
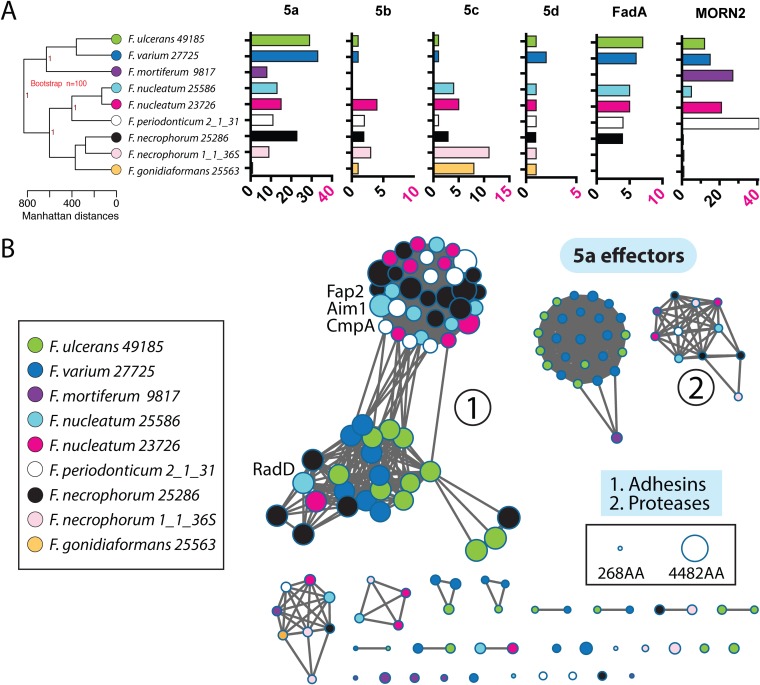
Our full-genome phylogenetic analysis of nine Fusobacterium genomes spanning seven species (Fig. 3A) shows a clear lineage, where F. ulcerans, F. varium, and F. mortiferum are distinct from F. nucleatum. Previous phylogenetic analysis predicted that F. necrophorum and F. nucleatum were more distantly related than indicated by our current analysis (24). In the bar graphs in Fig. 3A, we highlight groups of virulence proteins and the number of genes in each family, as predicted by hidden Markov models (HMM) and InterPro analysis, and confirm that each contains a Sec-dependent signal sequence. Upon examination of both F. necrophorum genomes, we discovered that F. necrophorum 25286 has an expansion of T5aSS autotransporter subsets found in F. nucleatum that share homology with Fap2, whereas F. necroporum 1_1_36S has far fewer T5aSS genes, and none of them cluster with the Fap2 gene and its homologues (Fig. 3B). Both F. necrophorum species nearly lack MORN2 (membrane ontology and recognition nexus type 2) domain-containing proteins. In addition, F. necrophorum 1_1_36S does not have any fadA family genes, whereas the invasive strains F. nucleatum 23726 and F. necrophorum 25286 have five and four, respectively. However, we do note that a potential limitation of this analysis lies in using seven species of Fusobacterium, whereas five of these species are only represented by a single genome. Since we saw drastic difference in F. necrophorum gene profiles, it will be crucial to expand future analyses to include enough genomes to determine interspecies differences that may be apparent at the subspecies level.
FIG 3.
Virulence factor analysis of Fusobacterium. (A) Full-genome phylogenetic tree of seven Fusobacterium species encompassing nine strains. (B) Sequence similarity networks of all T5aSS autotransporters (E value, 10−125) as colored by their designated genomes. Each node represents a single protein (142 total), and the node size represents the protein size in amino acids.
To address limitations in predicting the virulence potential of a bacterial strain using whole genomes, we implemented protein sequence similarity networks to determine whether any strains and species shared similarity with previously characterized T5aSS virulence proteins from F. nucleatum (e.g., Fap2 and AimI) (Fig. 3B). To our knowledge, within the Fusobacterium autotransporters, no protein sequence similarity networks have previously been reported. In addition, we provide a full phylogenetic tree of all 142 T5aSS autotransproters in Fig. S2. We analyzed all T5aSS autotransporters from nine genomes encompassing seven species of Fusobacterium and show distinct subsets of predicted functions that include surface exposed adhesins, serine proteases, and proteins of unknown function. Within the large adhesins, there is a distinct divide between two subsets of T5aSS adhesins at stringent clustering conditions (10−125 via EFI-EST) (58). We show that while previous autotransporter analysis broadly associated RadD with all other type 5a monomeric autotransporters, this protein clusters with a subsection of autotransporters that are more commonly found in multiple copies in F. varium and F. ulcerans. When we compared strains of F. nucleatum and F. varium, there were not significant differences in the numbers of autotransporter, FadA, and MORN2 genes. However, it is worth noting that F. varium lacks homologous Fap2 adhesins that are critical for colorectal cancer pathogenesis. Oddly, F. periodonticum has an abundance of these Fap2-like adhesins, yet it has no proteins from the RadD family. Finally, to add additional complexity to predicting the function of autotransporters, recent analysis showed that CmpA (Gene_60 in F. nucleatum 23726 [FN0254]), which clusters with the host binding adhesin Fap2, is a crucial adhesin that binds to Streptococcus gordonii during oral biofilm formation.
Comparison of colorectal cancer cell interactions between F. nucleatum and F. necrophorum.
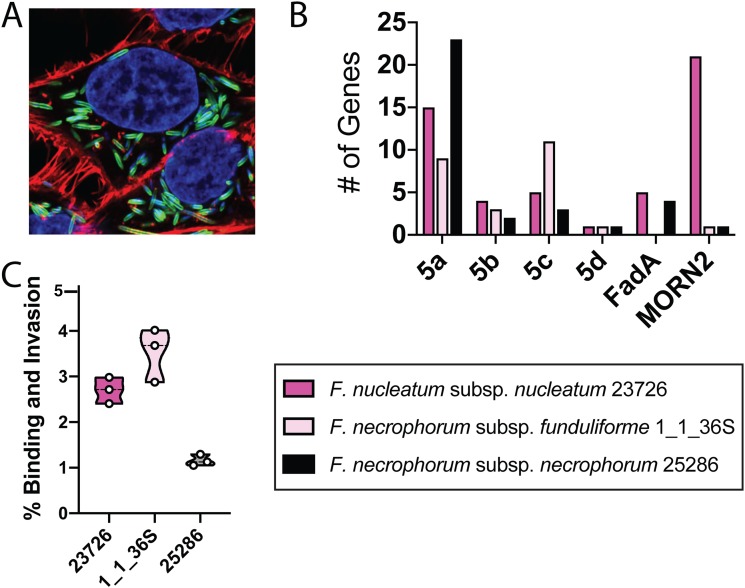
F. nucleatum 23726 is a highly invasive strain of Fusobacterium, and we show surface-bound and intracellular bacteria in HCT116 CRC cells via fluorescence microscopy in Fig. 4A. Because the potential virulence proteins between F. necrophorum subsp. necrophorum 25286, F. necrophorum subsp. funduliforme 1_1_36S, and F. nucleatum 23726 were strikingly different (Fig. 4B), we tested each strain for the ability to bind HCT116 cells. Using 30-min bacterial binding assays at a low multiplicity of infection (MOI; 1:40 bacterial/human cells) with monolayers of HCT116 cells (Fig. 4C), we show that the predicted low-binding strain F. necrophorum 1_1_36S surprisingly binds with higher efficiency than the well-characterized and highly invasive strain F. nucleatum 23726. However, in spite of a genetic expansion of predicted F. nucleatum-like T5aSS adhesins, F. necrophorum 25286 has far lower binding potential than the previously described strains. When we compared the virulence protein families between these three strains, we noted that F. necrophorum 1_1_36S contains far more type 5c trimeric autotransporter adhesins than F. nucleatum 23726 and F. necrophorum 25286, and yet it lacks the type 5a adhesin Fap2, has no FadA genes, and has only one MORN2 domain protein.
FIG 4.
Quantitation of cell-associated F. nucleatum and F. necrophorum to HCT116 cancerous colonocytes. (A) Superresolution imaging of surface bound and intracellular F. nucleatum 23726 in HCT116 CRC cells. (B) Quantitation of genes in six potential virulence gene families in F. nucleatum 23726, F. necrophorum 25286, and F. necrophorum 1_1_36S. (C) HCT116 binding assays where percent invasion is determined as the number of bacteria recovered as live CFU after incubation and washing compared to the number of input bacteria.
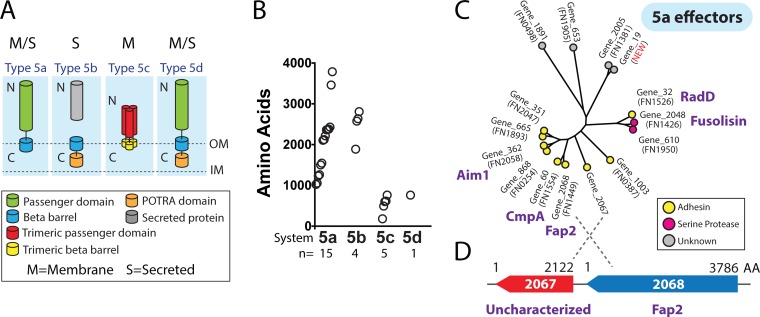
A detailed view of T5SS autotransporters in the genetically tractable strain F. nucleatum 23726.
Autotransporters are proteins specific to Gram-negative bacteria and constitute the largest family of secreted proteins in bacteria (41). Increasing interest in F. nucleatum 23726 has occurred due to several key studies on fap2 gene interruptions (40, 42) and the recent development of a double-crossover, markerless gene deletion system for this strain (27). In Fig. 5A, we highlight the basic domain structures of the four classes of autotransporter proteins (T5aSS to T5dSS) that are encoded in Fusobacterium genomes. F. nucleatum 23726 has 25 total autotransporters (Fig. 5B), ranging in size up to 3,786 aa (Fap2), with the majority falling in the classic monomeric T5aSS proteins. Phylogenetic analysis of the T5aSS autotransporters (Fig. 5C) from F. nucleatum 23726 fits well with the multigenome analysis found in Fig. 3B and Fig. S2. We have bioinformatically divided these proteins into three categories based on the predicted function and nearest neighbors: adhesins, serine proteases, and proteins of unknown function. We observed that the previously characterized adhesin RadD from F. nucleatum 23726 does not strictly cluster near verified adhesins such as Fap2 and AimI, but near the serine proteases that include fusolisin (FN1426) (63). As shown in Fig. 5D, we have also identified another T5aSS autotransporter adhesin directly upstream in the F. nucleatum 23726 genome, of which there has been no characterization.
FIG 5.
Analysis of T5aSS autotransporters in F. nucleatum 23726. (A) Domain structure of the Gram-negative-bacterium-specific, outer-membrane-embedded or secreted autotransporter protein family. Type 5e (intimins) is not represented because Fusobacterium spp. do not contain intimin genes. (B) Quantities of each family in F. nucleatum 23726, as well as the size of each protein. (C) Phylogenetic tree showing all 15 T5aSS proteins divided by node color into predicted protein functions. (D) Highlighted diagram of an uncharacterized Fap2 homologue that is just upstream of the well-characterized virulence protein Fap2.
Our complete F. nucleatum 23726 genome also uncovered a new T5aSS autotransporter (Gene_19) missing in the previous draft assembly. This protein shares strong homology with Gene_2005, previously named FN1381 using the F. nucleatum 25586 naming system. We identified that Gene_19 and Gene_2005 (FN1381) are potential gene duplications that have diverged because the C-terminal ∼700 amino acids share 100% sequence identity.
Discovery and renaming of T5bSS and T5cSS genes in F. nucleatum 23726.
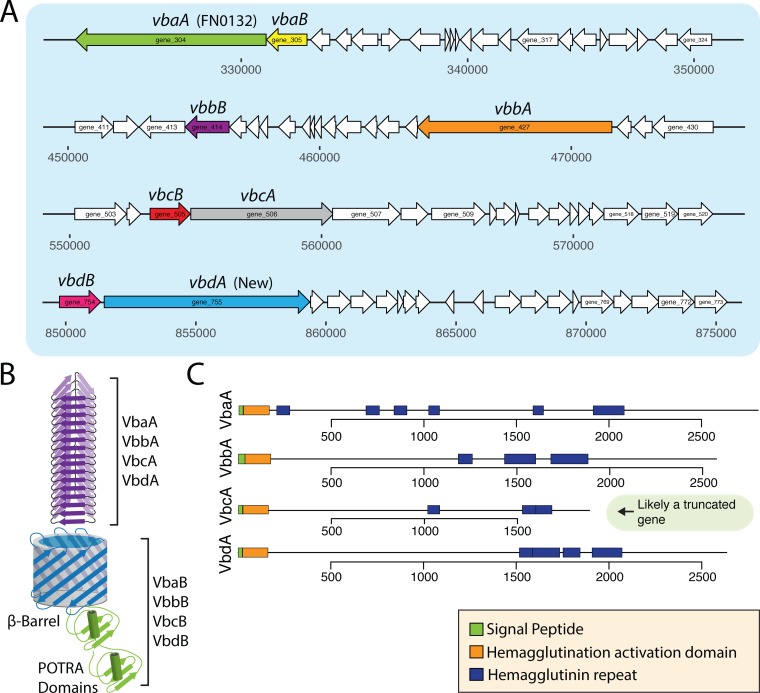
As shown in Fig. 6 and Table S1, we have identified four full open reading frames for T5bSS effectors (two-partner secretion [TPS]) in F. nucleatum 23726, and the full repertoire of these effectors from the nine Fusobacterium genomes characterized in this study can be analyzed in a phylogenetic tree in Fig. S3. We renamed the secreted effector genes type Vb TpsA (vbaA, vbbA, vbcA, and vbdA) (Fig. 6A). The paired genes encoding for β-barrel translocation proteins were renamed type Vb TpsB (vbaB, vbbB, vbcB, and vbdB), and a schematic of TPS architecture is shown in Fig. 6B. The previous draft genome of F. nucleatum 23726 had the VbaA and VbbA proteins properly annotated, but the vbcA gene was fragmented into two open reading frames. vbdA is a newly identified gene encoding a 2,634-aa protein.
FIG 6.
Type 5b secreted proteins (T5bSS) in F. nucleatum 23726. (A) Genomic location and open reading frame sizes for four T5bSS effectors with their respective β-barrel translocation proteins. (B) Schematic of the secreted VbaA, VbbA, VbcA, and VbdA effector proteins and their respective outer membrane translocation proteins VbaB, VbbB, VbcB, and VbdB. (C) Predicted domains of each secreted protein.
In strain F. nucleatum 25586, a previous study revealed that the N-terminal Sec signal sequence was out of frame in the T5bSS β-barrel translocation protein, but the presence of a poly(A) stretch that bridged this region in the T5bSS β-barrel translocation proteins led the authors to propose that slipped-strand translation could occur during phase variation, therefore translating these signal sequence-lacking open reading frames into fully functional proteins capable of translocating a T5bSS effector. Our analysis also identified β-barrel translocation proteins without signal sequences in F. nucleatum 25586. However, in the closely related F. nucleatum 23726, all open reading frames for the secreted protein and translocation machinery are in frame and are predicted to allow for protein translation. Because of this discrepancy, all F. nucleatum 25586 T5bSS systems do not appear functional, and therefore we have not included them in our analysis until biochemical characterization proves the slipped-strand translation and expression of these proteins. We have highlighted all potential F. nucleatum 25586 T5bSS genes in Tables S1 and S2 in the supplemental material, but note that these genes suffer from shifted or in some cases split reading frames.
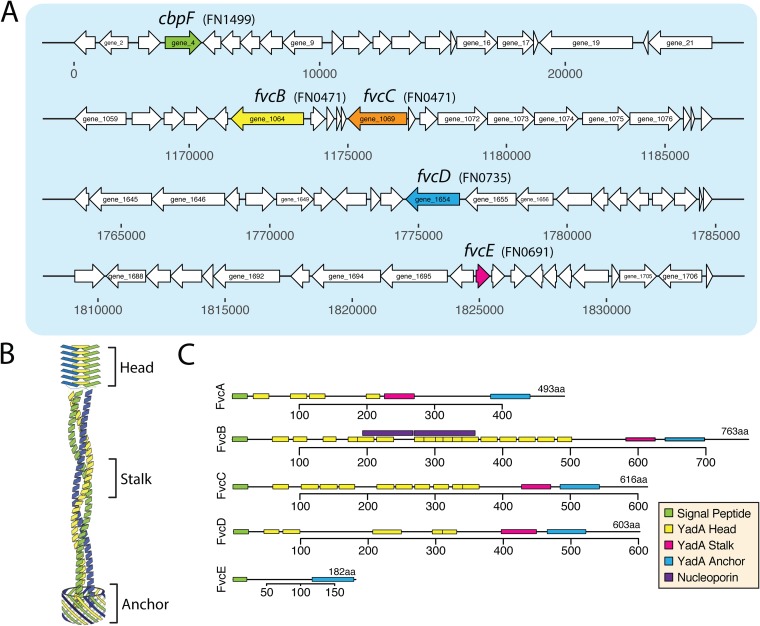
We report the presence of five T5cSS trimeric autotransporter adhesin (TAA) genes in F. nucleatum 23726 as shown in Fig. 7A, and we have renamed them Fusobacterium type Vc protein genes (fvcA, fvcB, fvcC, fvcD, and fvcE). Three of the five open reading frames were previously misannotated, with that of FvcC being the most extreme (new, 615 aa; old, 456 aa). In addition to the classic head-stalk-anchor architecture (Fig. 7B), several other domains have been characterized that differentiate the role of TAAs (44). As shown in Fig. 7C, TAAs can have multiple head domains, and the stalk domains are α-helical coiled coils. FvcE differs in that it is quite small at 181 aa and contains only the required Sec signal sequence and anchor domain. We hypothesized that FvcE could be a nonfunctioning T5cSS adhesin. In Fig. S4, phylogenetic analysis of TAAs from nine Fusobacterium genomes shows that proteins from highly invasive F. nucleatum strains cluster away from those of strains predicted to be less invasive.
FIG 7.
Type 5c secreted trimeric autotransporter proteins (T5cSS) in F. nucleatum 23726. (A) Genomic location and open reading frame sizes for five T5cSS trimeric autotransporter proteins. (B) Schematic of the head stalk and anchor domains. Multiple head domains can be present in one protein. (C) Predicted domains of each secreted protein.
Analysis of the newly expanded FadA protein family.
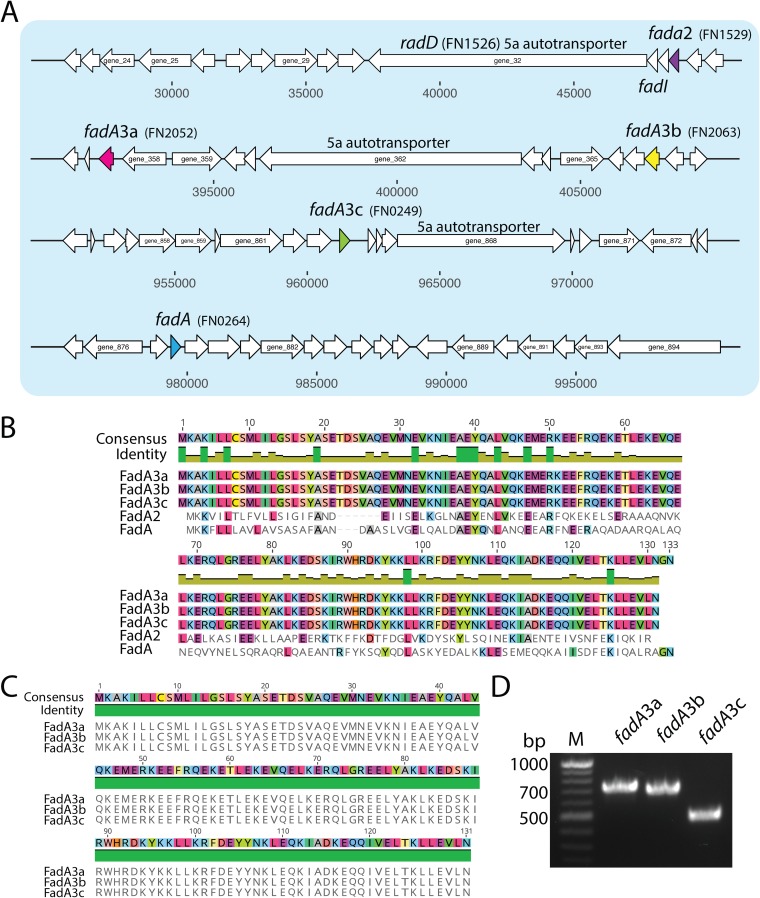
In addition to the type 5 autotransporters, this present study highlights the unique FadA-like adhesins of Fusobacterium (Fig. 8A and B). Our phylogenetic data (Fig. S6) show that multiple fadA-like adhesin genes are present across Fusobacterium species, where invasive strains possess a higher number and diversity of the corresponding proteins. In contrast, the predicted passive invaders F. necrophorum 1_1_36S and F. gonadiaformans 25563 lack the entire FadA protein family. We report newly found FadA proteins that we have named FadA, FadA2, FadA3a, FadA3b, and FadA3c, with the FadA3 proteins being complete gene triplications that are 100% identical at the amino acid level (Fig. 8C). The FadA3a to c genes were also found in F. nucleatum 25586, as shown by phylogenetic analysis in Fig. S6, but were not found in triplicate in the other seven Fusobacterium strains analyzed. We validated that these genes were in fact correct annotations and not genome errors using PCR with primers designed for unique upstream regions of each gene in F. nucleatum 23726 (Fig. 8D; Fig. S7).
FIG 8.
FadA family proteins in F. nucleatum 23726. (A) Genomic location and open reading frame sizes for five FadA family proteins. (B) CLUSTAL alignment of FadA proteins from F. nucleatum 23726. (C) CLUSTAL alignment of FadA3a, FadA3b, and FadA3c showing 100% sequence identity at the amino acid level. (D) PCR to confirm the presence of the fadA3a, fadA3b, and fadA3c genes at distinct locations in the genome of F. nucleatum 23726.
Identifying and characterizing MORN2 domain proteins in Fusobacterium.
Genomic studies have greatly detailed the expansion of MORN2 domain-containing proteins in Fusobacterium, with the greatest enrichment in the bioinformatically predicted active invaders (24). In Fig. S8 we generated a protein tree for the 120 MORN2 domain-containing proteins from nine Fusobacterium genomes. We show in Fig. 3A that F. periodonticum contains more than 40 MORN2 domain proteins, which is twice as many as the two experimentally validated invasive F. nucleatum strains. Since there are no experiments showing that these MORN2 domain proteins are involved in invasion, we address the potential roles of MORN2 domain containing proteins in Fusobacterium virulence in the Discussion.
DISCUSSION
Fusobacterium spp. are opportunistic pathogens that cause diverse infections and show strong connections and contributions to multiple diseases in humans and higher mammals (15). F. nucleatum has garnered significant attention as an “oncobacterium” that contributes to the progression and severity of colorectal cancer and has also been implicated in malignant oral leukoplakia (45), oral squamous cell carcinoma (46, 47), and pancreatic cancer (48, 49). Because F. nucleatum is an oncomicrobe, F. nucleatum infection studies have shown that administration of the antibiotic metronidazole reduces the tumor burden in human tumor-generated xenografts in mice (22). Since antibiotic therapy to treat disease could result in altered gut flora that changes the efficacy of chemotherapy drugs, a new paradigm would be to control infections at the disease site without antibiotics or to block this pathogen from leaving its native human oral cavity. For F. necrophorum strains, mostly of the subspecies funduliforme, infections of the jugular vein and subsequent progression to Lemierre’s syndrome can be fatal in humans. F. necrophorum subsp. necrophorum is the primary causative agent of bovine hoof rot and liver abscess, causing severe monetary loss in the livestock industry. The prediction of Fusobacterium virulence overall has been complicated and speculative, especially with respect to species outside of F. nucleatum . For instance, F. varium has been identified in the colonic mucosa of ulcerative colitis patients, and supernatants from these colonic mucosa samples were able to induce ulcerative colitis in mice (50, 51). However, F. varium is not associated with colorectal cancer, and this species is not predicted to be highly invasive like F. nucleatum. Since ulcerative colitis is defined as inflammation of the intestines and since we generally associate Fusobacterium cellular invasion with host-induced inflammation, there could be an alternative molecular mechanism other than cellular entry that allows F. varium to experimentally induce ulcerative colitis. As discussed below, there is a vast difference in our predicted virulence factors between F. nucleatum and F. varium, which still needs to be experimentally validated.
There have been several bioinformatic studies of Fusobacterium, but none of these used databases that were entirely populated with complete genomes. This lack of complete genomes led us to sequence, assemble, annotate, and create the FusoPortal database (29) of Fusobacterium genomes to aid in the bioinformatic and molecular experiments found in this study. These data provide highly accurate gene boundaries and therefore will greatly facilitate the design and production of recombinant proteins for structural and functional studies, an area that is severely lacking in the Fusobacterium field. Our data confirmed that Fusobacterium spp. are unique in that they lack functional protein secretion machinery of the type 1, 2, 3, 4, and 6 varieties, and yet they are still opportunistic pathogens. However, a recent proteomic analysis of Fusobacterium nucleatum revealed that secreted outer membrane vesicles contain proteins from each of the virulence factor families we analyzed in this study, potentially providing an additional mechanism for protein dispersal (52).
To build upon our understanding of Fusobacterium virulence, this study reports whole-genome phylogenetic analysis combined with protein sequence similarity networks and cellular invasion studies to probe complex interactions between the host and various Fusobacterium species. Although we know that Fusobacterium spp. contribute to several diseases, our understanding of the overarching molecular mechanisms driving the virulence of these bacteria remains limited. We focused our virulence factor analyses on the type 5 secreted autotransporters, FadA adhesins, and MORN2 domain protein families previously hypothesized to drive bacterial invasion into host cells. We show the importance of having accurate and complete genomes in identifying large proteins; the majority being type 5a autotransporters that have been characterized as virulence factors in a wide range of Gram-negative bacteria. Previous reports highlight the importance of the type 5a autotransporter protein Fap2 in F. nucleatum virulence. We highlight several additional observations from our bioinformatic data and note that a key contribution of this work is the identification of Fap2 homologues in multiple Fusobacterium genomes, and we report that F. necrophorum subsp. necrophorum 25286 has two close homologues of this adhesin. However, our CRC cell binding studies (Fig. 4) show that F. necrophorum 25286, despite having multiple Fap2 homologues, binds more weakly to host cells than does F. nucleatum 23726, which has a single copy of fap2. In addition, previous whole-genome phylogenetic studies hypothesized that strains of F. necrophorum are passive invaders that needed an injured epithelial barrier to induce entry into host cells. However, our experimental data show that F. necrophorum 1_1_36S is quite competent at binding human colonocytes, a phenotype shared with F. nucleatum 23726. Bioinformatic analysis hints that this adhesive phenotype in F. necrophorum 1_1_36S is not because of an overabundance of type 5a autotransporters, since it lacks a fap2 gene, but could be more adhesive due to 11 type 5c trimeric autotransporter adhesins (6 more than F. nucleatum 23726) that have been well characterized in other bacteria for cellular binding and invasion (53). We show that both F. necrophorum strains analyzed virtually lack MORN2 domain-containing proteins, whereas F. nucleatum strains are generally rich (>20 open reading frames) in this protein family, leading us to rethink the role of MORN2 domain proteins in cellular binding, invasion, and Fusobacterium virulence.
F. nucleatum in the oral cavity serves as a bridge for bacterium-bacterium aggregation and interactions with mammalian cells and inert tooth surfaces within the gingival pocket (64, 65). The T5aSS proteins RadD and CmpA are critical for interspecies adherence and the overall architecture of multispecies biofilms (66). Therefore, type 5a autotransporters play key roles in coaggregation, cell-cell interaction, and biofilm formation during healthy and pathogenic states. Our data suggest that newly identified subsets of T5aSS autotransporters are present in a variety of Fusobacterium species, and therefore new genetic and biochemical studies should be designed to characterize these proteins in virulence. Oddly, RadD from F. nucleatum clusters with the F. varium and F. ulcerans adhesins and not Fap2 in F. nucleatum. It remains to be determined whether RadD plays a critical role in cellular invasion or the progression of colorectal cancer associated with F. nucleatum. More evidence for this comes from the fact that RadD drives interactions with a diverse set of bacteria (67) and is present in a single copy in the two F. nucleatum strains analyzed. An alternative hypothesis is that RadD-like adhesins have unidentified functions that drive the inflammation seen in F. varium-induced ulcerative colitis but are not critical for direct cellular entry. In addition, F. varium and F. ulcerans share the most similar profile of T5aSS autotransporters, with an uncharacterized subset of proteins that are not found in the other seven Fusobacterium strains analyzed.
Although Fusobacterium have been reported as nonmotile, we sought to determine whether F. nucleatum T5aSS autotransporters share homology with IcsA from Shigella flexneri or with T5cSS proteins with BimA from Burkholderia species, since these proteins localize to a single bacterial pole (old pole) and coordinate intracellular actin-based motility (68, 69). Using our custom BLAST server that is built into the FusoPortal database, we show no homology to IcsA and low homology to the membrane-associated β-barrel of BimA. These data agree well with a lack of actin-based motility reported by all previous studies and our own observation of intracellular F. nucleatum. Although actin-based motility is not used for intracellular movement, another intriguing observation was that actin localizes to intracellular F. nucleatum in human keratinocytes and that inhibition of new actin synthesis blocked intracellular entry (25). The protein or proteins involved in this direct or indirect actin recruitment have yet to be identified. Determining whether intracellular F. nucleatum cloaks itself or a host vacuole in actin to evade host clearance through ubiquitination and xenophagy, as seen for Listeria monocytogenes and Chlamydia trachomatis (70, 71), could be a key piece in understanding the intracellular persistence and dissemination of this pathogen.
In addition, we provide the most thorough characterization of several understudied and potential virulence protein families in Fusobacterium, including the T5bSS (two-partner secretion), T5cSS (trimeric autotransporter adhesins), and MORN2 domain proteins. To our knowledge, two-partner secretion systems, or T5bSS effectors, have not been experimentally analyzed in any F. nucleatum studies. T5bSS analyses in multiple pathogenic bacteria report that these secreted effectors contain multiple hemagglutinin domains and function as cytolysins, hemolysins, adhesins, and proteins that initiate contact-dependent growth inhibition to fight off neighboring bacteria (72, 73). For example, a T5bSS filamentous hemagglutinin in Bordetella pertussis serves as an adhesin and is essential for colonization of tissues (74). Furthermore, ShlA in Serratia marcescens plays a cytotoxic role by contributing to the colonization of tissues (75). Thus, while it remains possible that type Vb autotransporters play a role in tissue colonization, they could alternatively be involved in survival and bacterial competition in oral and colorectal niche environments. Within the Fusobacterium species that we analyzed, there is a wide range of these uncharacterized proteins, with the largest number of functional genes being found in F. nucleatum 23726. Since no active released classic toxins have been reported for F. nucleatum, it could be that these proteins instead act as uncharacterized effectors on or within peripheral blood mononuclear cells (PBMCs), for which F. nucleatum has been shown to induce apoptosis (61, 76). Within the T5bSS toxin family, both F. necrophorum 25286 and F. necrophorum 1_1_36S express the LktA leukotoxin that induces the activation and apoptosis of leukocytes (77, 78). The lktBAC operon responsible for the production of this T5bSS secreted toxin is absent in all other strains of Fusobacterium analyzed and could explain the severe abscess phenotype induced by this species. Since F. necrophorum 25286 is associated with serious livestock infections, these data, combined with the potential to perform genetic manipulation in these strains, could lead to a more effective live, attenuated vaccine for bovine hoof rot and liver abscesses.
Recently the first F. nucleatum TAA CbpF (FN1499) was characterized and was shown to bind to CEACAM1 on human cells (86). The TAA protein family consists of important virulence factors in other pathogenic Gram-negative bacteria, including YadA from Yersinia pestis and Yersinia pseudotuberculosis (13, 53), Hia from Haemophilus influenzae (79), and SadA of Salmonella enterica (80). TAAs form long fibrous proteins that can extend more than 100 nm from the surfaces of the bacteria, thereby presenting the adhesive head domains to dock with host cells. YadA binds to the human extracellular matrix proteins fibronectin and collagen, drives invasion into epithelial and phagocytic cells, and inhibits activation of the serum complement (81). Previous bioinformatic analysis implied that a genetic expansion of T5cSS autotransporters in less-invasive strains indicates that these proteins are likely not used by Fusobacterium for cellular invasion, despite evidence of the TAAs being critical for virulence in multiple human-pathogenic Gram-negative bacteria. We propose that TAAs from pathogenic F. nucleatum could play a role in cellular invasion and that genetic and biochemical studies need to complement one another before we discount the TAAs as unimportant for Fusobacterium virulence.
Genetic and biochemical studies of FadA in F. nucleatum 12230 showed that this small (∼125 aa) adhesin multimerizes on the bacterial surface and subsequently binds to E-cadherin to modulate endothelial barrier permeability, signaling, and inflammatory responses in models of human cancer (31). This implicates FadA in the entry and exit of blood vessels to translocate F. nucleatum to the fetal-placental unit, where multiple studies support a role for this bacterium in preterm birth (82, 83). The identification of up to six FadA family proteins in a genome leads us to hypothesize that there is cooperativity among this protein family and that the FadA homologues, FadA2 and FadA3, could have similar functions as adhesins but have unidentified host receptor molecules. These include a gene triplication of fadA3 in F. nucleatum 23726 and F. nucleatum 25586. In F. nucleatum 12230, a ΔfadA mutant shows greatly reduced proliferation of human cancer cells (HCT116 and HT29), but this procarcinogenic phenotype was recovered by fadA complementation and the addition of purified active FadA to cell cultures. Bioinformatic analysis of the F. nucleatum 12230 genome (GenBank assembly accession no. GCA_003226385.1) revealed a single copy of fadA and a single copy of fadA3 that appears to encode a protein with N-terminal amino acids missing, hinting that FadA and not FadA3 drives cellular binding and cancer cell proliferation. In addition, FadA is also important for the colonization of F. nucleatum 12230 in mouse placenta (83). With five FadA family proteins in the genetically tractable F. nucleatum 27326, it will be crucial to delete multiple copies of these genes to determine whether they act synergistically during infection or whether FadA, FadA2, and FadA3 play distinct roles in virulence and colonization in diverse tissue niches, including the subgingival microbial community, human placenta, or the colon. Finally, we show that the greatest number of fadA genes are found in F. varium and F. ulcerans, two species with lower predicted invasive potential than the experimentally tested FadA positive strain F. nucleatum 12230. Since FadA was shown to drive host cell signaling in F. nucleatum 12230, it could be that this protein family plays a similar role in F. varium-induced ulcerative colitis.
Despite a great interest in the potential role of MORN2 domain proteins in cellular invasion due to their genomic proximity to T5aSS proteins and expansion in invasive F. nucleatum strains, no studies, to our knowledge, have shown a direct role for this hypothesis. MORN2 domain-containing proteins could enhance adhesive and active invasive traits since their genes clustered near those for FadA, RadD, and additional T5aSS autotransporter adhesins. Since this study, a role for these proteins in interspecies interactions has been suggested (15), as they are predicted surface-associated proteins, and some of them are part of the “FusoSecretome” (84). More recently, MORN2 domain-containing proteins were found to be secreted into F. nucleatum outer membrane vesicles (52). In addition, the fact that these proteins possess a YwqK domain suggests that they could be acting as toxin-antitoxin systems for interspecies competition or as bacterial abortive infection systems that limit viral replication and are activated by phage infection (85).
Though not analyzed here, Fad-I is an outer membrane protein that was shown to induce human beta defensin 2 (hBD-2) through a Toll-like receptor-mediated host response. The immune modulation induced by Fad-I in F. nucleatum strains 23726 and 25586 was far more potent than that seen in F. nucleatum 10953 (54). As with all protein families, this is a great example of how small sequence variations in key proteins could account for altered virulence between phylogenetically similar strains of Fusobacterium.
In conclusion, we provide bioinformatic identification and analysis of virulence factors, as well as host-pathogen infection studies to dissect the role of the role of potential virulence protein families in cellular binding and invasion. We hypothesize that autotransporters, FadA, and MORN2 proteins synergistically form a host cell docking and invasion network that confer the host, tissue, and disease mechanism of the diverse range of Fusobacterium species. These results show that a more detailed analysis of invasion using additional strains is warranted and could help in creating more accurate predictive models of invasive potential from genome sequences. This study will benefit from future work that expands upon our nine genome analysis to include additional Fusobacterium species and clinical strains, which will ultimately lead to a deeper understanding of individual proteins in disease. It is our hope that these genomes, bioinformatic analyses, and Fusobacterium invasion studies spark the discovery of new virulence factors and drive studies that further our understanding of virulence mechanisms at the molecular level in the diverse bacterial Fusobacterium genus.
MATERIALS AND METHODS
Use of genomic information for bioinformatic analysis of nine Fusobacterium strains.
Fusobacterium genomes and all associated data used for this study can be accessed under the NCBI BioProjects PRJNA433545 and PRJNA513186. Annotations for all genes were determined with Prodigal (39) or Prokka (38) and can be found on the FusoPortal website (http://fusoportal.org) or our Open Science Framework data repository (http://osf.io/2c8pv). In addition, NCBI annotated each genome using their Prokaryotic Genome Annotation Pipeline. Signal peptide prediction was done with SignalP 3.0 and the HMM option (55). Protein functions were predicted using InterPro analysis (56) within Blast2GO 5.2.5 software (57), and predictions for all virulence proteins used in this study can be found in Table S2 in the supplemental material.
Protein sequence similarity networks.
Networks were created by identifying all protein families of interest using the InterPro server within Blast2Go software. Protein hits were manually checked for accuracy and the presence of a Sec signal sequence. Qualifying proteins were then analyzed using the “all-vs-all” BLAST feature within the EFI-EST server (58), followed by cluster mapping in Cytoscape 3.0 (59). All raw files for proteins in fasta format, as well as Cytoscape networks, are posted in our Open Science Framework repository.
Phylogenetic analysis of Fusobacterium genomes.
Full genome phylogenetic analysis of nine Fusobacterium genomes (Fig. 3A) was performed using the MicroPan (60) R-package of pangenome analysis. All protein open reading frames for each genome were included in the analysis. Nodes for each protein were manually colored in Adobe Illustrator to match the genome key depicted in Fig. 3B.
Phylogenetic analysis of virulence factors.
Protein trees (Fig. S2, S3, S4, S5, S6, and S8) were built within Geneious 9.02 using the Geneious Tree Builder. Pairwise alignment was run using a Blosum62 cost matrix and neighbor-joining trees were built using a Jukes-Cantor genetic distance model. Nodes for each protein were manually colored in Adobe Illustrator to match the genome key depicted in Fig. 3B.
Quantitation of Fusobacterium binding to HCT116 cells.
A single colony was used to start overnight cultures of F. nucleatum 23726, F. necrophorum 25286, or F. necrophorum 1_1_36S. Stationary-phase cultures were back diluted to an optical density at 600 nm (OD600) of 0.01 in 5 ml of Columbia broth supplemented with hemin and vitamin K (CBHK) and grown to exponential phase (OD600 = 0.4). Fusobacterium cultures were then transferred to a 1.5 ml conical tube and centrifuged at a 14,000 relative centrifugal force for 3 min to pellet bacterial cells. The CBHK was removed, and the bacterial cell pellet was resuspended in McCoy’s 5A medium with no serum or antibiotics. Fusobacterium cells were added (MOI ∼ 1:40 bacterium/human cells; ∼10,000 bacteria per well based on plated colony counts with 4 × 105 HCT116 cells) to the media of confluent HCT116 human cell monolayer cultures in 12-well plates (for binding assays) or 1.5 coverslip bottom plates (fluorescence microscopy), followed by incubation for 30 min at 37°C in 5% CO2. After infection, epithelial cells were washed three times with 0.1 M MOPS (pH 7.2)–1 mM MgCl2. HCT116 monolayers were scraped and resuspended in warm deionized water before plating on CBHK agar for growth of CFU. Since Fusobacterium species vary significantly in the number of bacteria per OD600 unit, controls for the number of bacteria were determined by directly plating 10% of the bacterial input for infections and counting CFU. The percent binding and invasion (Fig. 4C) was calculated by dividing the number of recovered colonies from the HCT infection by the calculated number of bacteria input based on the CFU.
Fluorescence microscopy of intracellular F. nucleatum in FHC and Ca9-22 gingival cells.
In Fig. 1B (top panel), a confluent monolayer of healthy fetal human colonocytes (FHC; ATCC CRL-1381) were grown in Dulbecco modified Eagle medium (DMEM)–F-12 medium supplemented with 10% FBS (Atlanta Biologicals) in 1.5 coverslip glass-bottom plates. Cells were infected in antibiotic free DMEM–F-12 medium with a 10:1 MOI of F. nucleatum 23726 labeled with the fluorescent lipid FM 1-43FX were for 1 h at 37°C in 5% CO2. Extracellular bacteria were then killed with DMEM–F-12 medium supplemented with 100 IU/ml penicillin and 100 (μg/ml) streptomycin. After being washed with medium, the cells were fixed in wells with phosphate-buffered saline (PBS)–3.7% paraformaldehyde and then permeabilized with 1% Triton X-100. Cells infected with F. nucleatum 23726 were then stained with 1.0 μg/ml DAPI (4′,6′-diamidino-2-phenylindole; Invitrogen) to visualize DNA and Texas Red-X–phalloidin for F-actin filaments (Thermo Fisher). Images were acquired using a 63× oil objective on an EVOS FL fluorescence microscope. For Fig. 4A, HCT116 cancerous colonocytes were used for invasion with F. nucleatum 23726. Images were acquired using a 60× objective on a Zeiss 880 Airyscan-equipped fluorescence microscope.
Fluorescence microscopy of intracellular F. nucleatum HMVEC.
In Fig. 1B (bottom panel), adult human dermal microvascular endothelial cells (HMVEC) (Lonza) were cultured in a custom-built three-dimensional printed chamber where a 1.5 coverslip was adhered to the bottom of the printed well using biocompatible polydimethylsiloxane (PDMS; Sylgard 184 silicone elastomer kit). The CAD model of the microchamber was designed using Autodesk Inventor Professional 2018. The chamber was printed with PLA using MakerBot Ultimaker 2. Additionally, cell attachment factor (Cell Systems) and a thin lining of collagen (5 mg/ml) were used to coat the coverslip before seeding the host cells within the wells. HMVEC were grown in EGM MV-2 (Lonza) media at 37°C in 5% CO2. Cells were infected in antibiotic-free DMEM–F-12 medium with a 10:1 MOI of F. nucleatum 23726 labeled with the fluorescent lipid FM 1-43FX were for 1 h at 37°C in 5% CO2. Extracellular bacteria were then killed with DMEM–F-12 medium supplemented with 100 IU/ml penicillin and 100 (μg/ml) streptomycin. After being washed with medium, the cells were fixed in wells with a 10% formalin solution. The fixed cells were washed with PBS and blocked with a solution of PBS-X containing 10 ml of PBS, 5 μl of Triton X-100, and 200 mg of bovine serum albumin (Fisher BioReagents). After a rinse with PBS, the cells were stained with Alexa Fluor 568-phalloidin (Life Technologies) for F-actin filaments (1:100 dilution) and 1.0 μg/ml DAPI (Invitrogen) for the nucleus. Imaging was performed using a Zeiss LSM 800 confocal microscope. Representative images and z-stacks were obtained using a Zeiss 63× 1.8-numerical-aperture (NA) oil immersion lens with 4× averaging.
PCR to validate the presence of three separate genes for fadA3a to -c in F. nucleatum 23726.
Genomic DNA was extracted from exponential-phase F. nucleatum 23726. The primers (Fig. S7) are designated prDJSVT1006 to prDJSVT1009. The following 5′-to-3′ sequences correspond to each primer: prDJSVT1006, 5′-CTAAACTCTTCCTTTCTTTCCATTTCC-3′; prDJSVT1007, 5′-CCTATCAGGAAGTATGATAACTTTAACAAG-3′; prDJSVT1008, 5′-CCTATACCCTAGCAAACAATAACAAAGTC-3′; and prDJSVT1009, 5′-GAACATGAAATAAGACAAATATTTTAAGGATG-3′. PCR was run using Q5 polymerase (NEB) and a 50 to 62°C annealing temperature gradient. The products were visualized using 1% agarose gels stained with ethidium bromide.
Data availability.
All genomic data used for protein annotation can be found on NCBI under BioProject numbers PRJNA433545 and PRJNA513186. All associated raw data outside of the supplemental material, which includes the T5aSS autotransporter protein similarity network and a FASTA file with all virulence factor open reading frames, can be found on our Open Science Framework data repository (https://osf.io/vs3fd).
Supplementary Material
ACKNOWLEDGMENTS
This article has been supported by a Commonwealth Health Research Board Award (208-10-18, Slade), a National Science Foundation Career Award (CBET-1652112, Verbridge), the USDA National Institute of Food and Agriculture (Slade), and the Fralin Life Science Institute at Virginia Tech (Slade).
We thank Kristi Decourcy from the Fralin Life Sciences for assistance with microscopy.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00273-19.
REFERENCES
- 1.Casadevall A, Pirofski LA. 1999. Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect Immun 67:3703–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casadevall A, Pirofski LA. 2000. Host-pathogen interactions: basic concepts of microbial commensalism, colonization, infection, and disease. Infect Immun 68:6511–6518. doi: 10.1128/iai.68.12.6511-6518.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Didelot X, Walker AS, Peto TE, Crook DW, Wilson DJ. 2016. Within-host evolution of bacterial pathogens. Nat Rev Microbiol 14:150–162. doi: 10.1038/nrmicro.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ham H, Sreelatha A, Orth K. 2011. Manipulation of host membranes by bacterial effectors. Nat Rev Microbiol 9:635–646. doi: 10.1038/nrmicro2602. [DOI] [PubMed] [Google Scholar]
- 5.Costa TRD, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G. 2015. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 13:343–359. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- 6.Desvaux M, Khan A, Beatson SA, Scott-Tucker A, Henderson IR. 2005. Protein secretion systems in Fusobacterium nucleatum: genomic identification of type 4 piliation and complete type V pathways brings new insight into mechanisms of pathogenesis. Biochim Biophys Acta 1713:92–112. doi: 10.1016/j.bbamem.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan CW, Ma X, Paranjpe A, Jewett A, Lux R, Kinder-Haake S, Shi W. 2010. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect Immun 78:4773–4778. doi: 10.1128/IAI.00567-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casasanta MA, Yoo CC, Smith HB, Duncan AJ, Cochrane K, Varano AC, Allen-Vercoe E, Slade DJ. 2017. A chemical and biological toolbox for type Vd secretion: characterization of the phospholipase A1 autotransporter FplA from Fusobacterium nucleatum. J Biol Chem 292:20240–20254. doi: 10.1074/jbc.M117.819144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guérin J, Bigot S, Schneider R, Buchanan SK, Jacob-Dubuisson F. 2017. Two-partner secretion: combining efficiency and simplicity in the secretion of large proteins for bacterium-host and bacterium-bacterium interactions. Front Cell Infect Microbiol 7:148. doi: 10.3389/fcimb.2017.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leo JC, Grin I, Linke D. 2012. Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Philos Trans R Soc B 367:1088–1101. doi: 10.1098/rstb.2011.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins LM, Frankel G, Connerton I, Gonçalves NS, Dougan G, MacDonald TT. 1999. Role of bacterial intimin in colonic hyperplasia and inflammation. Science 285:588–591. doi: 10.1126/science.285.5427.588. [DOI] [PubMed] [Google Scholar]
- 12.Leo JC, Oberhettinger P, Schütz M, Linke D. 2015. The inverse autotransporter family: intimin, invasin and related proteins. Int J Med Microbiol 305:276–282. doi: 10.1016/j.ijmm.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 13.El Tahir Y, Skurnik M. 2001. YadA, the multifaceted Yersinia adhesin. Int J Med Microbiol 291:209–218. doi: 10.1078/1438-4221-00119. [DOI] [PubMed] [Google Scholar]
- 14.Abby SS, Cury J, Guglielmini J, Néron B, Touchon M, Rocha E. 2016. Identification of protein secretion systems in bacterial genomes. Sci Rep 6:23080. doi: 10.1038/srep23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brennan CA, Garrett WS. 2019. Fusobacterium nucleatum: symbiont, opportunist, and oncobacterium. Nat Rev Microbiol 17:156–166. doi: 10.1038/s41579-018-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gagnaire A, Nadel B, Raoult D, Neefjes J, Gorvel J-P. 2017. Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol 15:109–128. doi: 10.1038/nrmicro.2016.171. [DOI] [PubMed] [Google Scholar]
- 17.Han YW. 2015. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol 23:141–147. doi: 10.1016/j.mib.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. 2012. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. 2012. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. 2013. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang J-Y. 2017. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170:548–563.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, Chipashvili O, Hagan T, Walker M, Ramachandran A, Diosdado B, Serna G, Mulet N, Landolfi S, Ramon Y Cajal S, Fasani R, Aguirre AJ, Ng K, Élez E, Ogino S, Tabernero J, Fuchs CS, Hahn WC, Nuciforo P, Meyerson M. 2017. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science Dec 358:1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, Jenab M, Prehn JHM, Hughes DJ. 2014. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 33:1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 24.Manson McGuire A, Cochrane K, Griggs AD, Haas BJ, Abeel T, Zeng Q, Nice JB, MacDonald H, Birren BW, Berger BW, Allen-Vercoe E, Earl AM. 2014. Evolution of invasion in a diverse set of Fusobacterium species. mBio 5:e01864. doi: 10.1128/mBio.01864-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gursoy UK, Könönen E, Uitto V-J. 2008. Intracellular replication of fusobacteria requires new actin filament formation of epithelial cells. APMIS 116:1063–1070. doi: 10.1111/j.1600-0463.2008.00868.x. [DOI] [PubMed] [Google Scholar]
- 26.Karpathy SE, Qin X, Gioia J, Jiang H, Liu Y, Petrosino JF, Yerrapragada S, Fox GE, Haake SK, Weinstock GM, Highlander SK. 2007. Genome sequence of Fusobacterium nucleatum subspecies polymorphum: a genetically tractable fusobacterium. PLoS One 2:e659. doi: 10.1371/journal.pone.0000659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu C, Al Mamun AAM, Luong TT, Hu B, Gu J, Lee JH, D’Amore M, Das A, Ton-That H. 2018. Forward genetic dissection of biofilm development by Fusobacterium nucleatum: novel functions of cell division proteins FtsX and EnvC. mBio 9:e00360-18. doi: 10.1128/mBio.00360-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M, Sojar HT, Genco RJ, Kuramitsu HK, Deng CX. 2005. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol 187:5330–5340. doi: 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanders BE, Umana A, Lemkul JA, Slade DJ. 2018. FusoPortal: an interactive repository of hybrid MinION-sequenced Fusobacterium genomes improves gene identification and characterization. mSphere 3:e00228-18. doi: 10.1128/mSphere.00228-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Todd MS, Settlage RE, Lahmers KK, Slade DJ. 2018. Fusobacterium genomics using MinION and Illumina sequencing enables genome completion and correction. mSphere 3:e00269-18. doi: 10.1128/mSphere.00269-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. 2013. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oelke AM, Nagaraja TG, Wilkerson MJ, Stewart GC. 2005. The leukotoxin operon of Fusobacterium necrophorum is not present in other species of Fusobacterium. Anaerobe 11:123–129. doi: 10.1016/j.anaerobe.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Zhou H, Bennett G, Hickford J. 2009. Variation in Fusobacterium necrophorum strains present on the hooves of footrot infected sheep, goats, and cattle. Vet Microbiol 135:363–367. doi: 10.1016/j.vetmic.2008.09.084. [DOI] [PubMed] [Google Scholar]
- 34.Umaña A, Lemkul JA, Slade DJ. 2019. Complete genome sequence of Fusobacterium necrophorum subsp. necrophorum ATCC 25286. Microbiol Resour Announc 8:e00025-19. doi: 10.1128/MRA.00025-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gill J, Haydon TG, Rawdon TG, McFadden AMJ, Ha H-J, Shen Z, Feng Y, Pang J, Swennes AG, Paster BJ, Dewhirst FE, Fox JG, Spence RP. 2016. Helicobacter bilis and Helicobacter trogontum: infectious causes of abortion in sheep. J Vet Diagn Invest 28:225–234. doi: 10.1177/1040638716638704. [DOI] [PubMed] [Google Scholar]
- 36.Atherly T, Mosher C, Wang C, Hostetter J, Proctor A, Brand MW, Phillips GJ, Wannemuehler M, Jergens AE. 2016. Helicobacter bilis infection alters mucosal bacteria and modulates colitis development in defined microbiota mice. Inflamm Bowel Dis 22:2571–2581. doi: 10.1097/MIB.0000000000000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapatral V, Anderson I, Ivanova N, Reznik G, Los T, Lykidis A, Bhattacharyya A, Bartman A, Gardner W, Grechkin G, Zhu L, Vasieva O, Chu L, Kogan Y, Chaga O, Goltsman E, Bernal A, Larsen N, D’Souza M, Walunas T, Pusch G, Haselkorn R, Fonstein M, Kyrpides N, Overbeek R. 2002. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol 184:2005–2018. doi: 10.1128/JB.184.7.2005-2018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 39.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklić K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. 2015. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pallen MJ, Chaudhuri RR, Henderson IR. 2003. Genomic analysis of secretion systems. Curr Opin Microbiol 6:519–527. doi: 10.1016/j.mib.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Coppenhagen-Glazer S, Sol A, Abed J, Naor R, Zhang X, Han YW, Bachrach G. 2015. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect Immun 83:1104–1113. doi: 10.1128/IAI.02838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abed J, Emgård JEM, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, Mellul A, Chaushu S, Manson AL, Earl AM, Ou N, Brennan CA, Garrett WS, Bachrach G. 2016. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe 20:215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bassler J, Hernandez Alvarez B, Hartmann MD, Lupas AN. 2015. A domain dictionary of trimeric autotransporter adhesins. Int J Med Microbiol 305:265–275. doi: 10.1016/j.ijmm.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 45.Amer A, Galvin S, Healy CM, Moran GP. 2017. The microbiome of potentially malignant oral leukoplakia exhibits enrichment for Fusobacterium, Leptotrichia, Campylobacter, and Rothia species. Front Microbiol 8:2391. doi: 10.3389/fmicb.2017.02391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganly I, Yang L, Giese RA, Hao Y, Nossa CW, Morris LGT, Rosenthal M, Migliacci J, Kelly D, Tseng W, Hu J, Li H, Brown S, Pei Z. 2019. Periodontal pathogens are a risk factor of oral cavity squamous cell carcinoma, independent of tobacco and alcohol and human papillomavirus. Int J Cancer 145:775–784. doi: 10.1002/ijc.32152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Al-Hebshi NN, Nasher AT, Maryoud MY, Homeida HE, Chen T, Idris AM, Johnson NW. 2017. Inflammatory bacteriome featuring Fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. Sci Rep 7:1834. doi: 10.1038/s41598-017-02079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, Kanno S, Igarashi H, Naito T, Adachi Y, Tachibana M, Tanuma T, Maguchi H, Shinohara T, Hasegawa T, Imamura M, Kimura Y, Hirata K, Maruyama R, Suzuki H, Imai K, Yamamoto H, Shinomura Y. 2015. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget 6:7209–7220. doi: 10.18632/oncotarget.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaiser RA, Halimi A, Alkharaan H, Lu L, Davanian H, Healy K, Hugerth LW, Ateeb Z, Valente R, Fernández Moro C, Del Chiaro M, Sällberg Chen M. 2019. Enrichment of oral microbiota in early cystic precursors to invasive pancreatic cancer. Gut 2019:gutjnl-2018–317458. doi: 10.1136/gutjnl-2018-317458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohkusa T, Okayasu I, Ogihara T, Morita K, Ogawa M, Sato N. 2003. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut 52:79–83. doi: 10.1136/gut.52.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohkusa T, Sato N, Ogihara T, Morita K, Ogawa M, Okayasu I. 2002. Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J Gastroenterol Hepatol 17:849–853. doi: 10.1046/j.1440-1746.2002.02834.x. [DOI] [PubMed] [Google Scholar]
- 52.Liu J, Hsieh C-L, Gelincik O, Devolder B, Sei S, Zhang S, Lipkin SM, Chang Y-F. 2019. Proteomic characterization of outer membrane vesicles from gut mucosa-derived Fusobacterium nucleatum. J Proteomics 195:125–137. doi: 10.1016/j.jprot.2018.12.029. [DOI] [PubMed] [Google Scholar]
- 53.Eitel J, Dersch P. 2002. The YadA protein of Yersinia pseudotuberculosis mediates high-efficiency uptake into human cells under environmental conditions in which invasin is repressed. Infect Immun 70:4880–4891. doi: 10.1128/IAI.70.9.4880-4891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhattacharyya S, Ghosh SK, Shokeen B, Eapan B, Lux R, Kiselar J, Nithianantham S, Young A, Pandiyan P, McCormick TS, Weinberg A. 2016. FAD-I, a Fusobacterium nucleatum cell wall-associated diacylated lipoprotein that mediates human beta defensin 2 induction through Toll-like receptor-1/2 (TLR-1/2) and TLR-2/6. Infect Immun 84:1446–1456. doi: 10.1128/IAI.01311-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 56.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang H-Y, Dosztányi Z, El-Gebali S, Fraser M, Gough J, Haft D, Holliday GL, Huang H, Huang X, Letunic I, Lopez R, Lu S, Marchler-Bauer A, Mi H, Mistry J, Natale DA, Necci M, Nuka G, Orengo CA, Park Y, Pesseat S, Piovesan D, Potter SC, Rawlings ND, Redaschi N, Richardson L, Rivoire C, Sangrador-Vegas A, Sigrist C, Sillitoe I, Smithers B, Squizzato S, Sutton G, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Xenarios I, Yeh L-S, Young S-Y, Mitchell AL. 2017. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res 45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, Whalen KL. 2015. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochim Biophys Acta 1854:1019–1037. doi: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Snipen L, Liland KH. 2015. micropan: an R-package for microbial pan-genomics. BMC Bioinformatics 16:79. doi: 10.1186/s12859-015-0517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huynh T, Kapur RV, Kaplan CW, Cacalano N, Kinder Haake S, Shi W, Sieling P, Jewett A. 2011. The role of aggregation in Fusobacterium nucleatum-induced immune cell death. J Endod 37:1531–1535. doi: 10.1016/j.joen.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 62.Kuppalli K, Livorsi D, Talati NJ, Osborn M. 2012. Lemierre’s syndrome due to Fusobacterium necrophorum. Lancet Infect Dis 12:808–815. doi: 10.1016/S1473-3099(12)70089-0. [DOI] [PubMed] [Google Scholar]
- 63.Doron L, Coppenhagen-Glazer S, Ibrahim Y, Eini A, Naor R, Rosen G, Bachrach G. 2014. Identification and characterization of fusolisin, the Fusobacterium nucleatum autotransporter serine protease. PLoS One 9:e111329. doi: 10.1371/journal.pone.0111329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Signat B, Roques C, Poulet P, Duffaut D. 2011. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol 13:25–36. [PubMed] [Google Scholar]
- 65.Thurnheer T, Karygianni L, Flury M, Belibasakis GN. 2019. Fusobacterium species and subspecies differentially affect the composition and architecture of supra- and subgingival biofilms models. Front Microbiol 10:1716. doi: 10.3389/fmicb.2019.01716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lima BP, Shi W, Lux R. 2017. Identification and characterization of a novel Fusobacterium nucleatum adhesin involved in physical interaction and biofilm formation with Streptococcus gordonii. Microbiologyopen 6:e444. doi: 10.1002/mbo3.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaplan CW, Lux R, Haake SK, Shi W. 2009. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol Microbiol 71:35–47. doi: 10.1111/j.1365-2958.2008.06503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goldberg MB, Theriot JA. 1995. Shigella flexneri surface protein IcsA is sufficient to direct actin-based motility. Proc Natl Acad Sci U S A 92:6572–6576. doi: 10.1073/pnas.92.14.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu Q, Xu Y, Yao Q, Niu M, Shao F. 2015. A polar-localized iron-binding protein determines the polar targeting of Burkholderia BimA autotransporter and actin tail formation. Cell Microbiol 17:408–424. doi: 10.1111/cmi.12376. [DOI] [PubMed] [Google Scholar]
- 70.Yoshikawa Y, Ogawa M, Hain T, Chakraborty T, Sasakawa C. 2009. Listeria monocytogenes ActA is a key player in evading autophagic recognition. Autophagy 5:1220–1221. doi: 10.4161/auto.5.8.10177. [DOI] [PubMed] [Google Scholar]
- 71.Jiwani S, Alvarado S, Ohr RJ, Romero A, Nguyen B, Jewett TJ. 2013. Chlamydia trachomatis Tarp harbors distinct G and F actin binding domains that bundle actin filaments. J Bacteriol 195:708–716. doi: 10.1128/JB.01768-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ruhe ZC, Low DA, Hayes CS. 2013. Bacterial contact-dependent growth inhibition. Trends Microbiol 21:230–237. doi: 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hayes CS, Koskiniemi S, Ruhe ZC, Poole SJ, Low DA. 2014. Mechanisms and biological roles of contact-dependent growth inhibition systems. Cold Spring Harb Perspect Med 4:a010025. doi: 10.1101/cshperspect.a010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Inatsuka CS, Julio SM, Cotter PA. 2005. Bordetella filamentous hemagglutinin plays a critical role in immunomodulation, suggesting a mechanism for host specificity. Proc Natl Acad Sci U S A 102:18578–18583. doi: 10.1073/pnas.0507910102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Di Venanzio G, Stepanenko TM, García Véscovi E. 2014. Serratia marcescens ShlA pore-forming toxin is responsible for early induction of autophagy in host cells and is transcriptionally regulated by RcsB. Infect Immun 82:3542–3554. doi: 10.1128/IAI.01682-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaplan CW, Lux R, Huynh T, Jewett A, Shi W, Haake SK. 2005. Fusobacterium nucleatum apoptosis-inducing outer membrane protein. J Dent Res 84:700–704. doi: 10.1177/154405910508400803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Narayanan S, Stewart GC, Chengappa MM, Willard L, Shuman W, Wilkerson M, Nagaraja TG. 2002. Fusobacterium necrophorum leukotoxin induces activation and apoptosis of bovine leukocytes. Infect Immun 70:4609–4620. doi: 10.1128/IAI.70.8.4609-4620.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tadepalli S, Stewart GC, Nagaraja TG, Narayanan SK. 2008. Human Fusobacterium necrophorum strains have a leukotoxin gene and exhibit leukotoxic activity. J Med Microbiol 57:225–231. doi: 10.1099/jmm.0.47598-0. [DOI] [PubMed] [Google Scholar]
- 79.St Geme JW III, Cutter D. 2000. The Haemophilus influenzae Hia adhesin is an autotransporter protein that remains uncleaved at the C terminus and fully cell associated. J Bacteriol 182:6005–6013. doi: 10.1128/JB.182.21.6005-6013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raghunathan D, Wells TJ, Morris FC, Shaw RK, Bobat S, Peters SE, Paterson GK, Jensen KT, Leyton DL, Blair JMA, Browning DF, Pravin J, Flores-Langarica A, Hitchcock JR, Moraes CTP, Piazza RMF, Maskell DJ, Webber MA, May RC, MacLennan CA, Piddock LJ, Cunningham AF, Henderson IR. 2011. SadA, a trimeric autotransporter from Salmonella enterica serovar Typhimurium, can promote biofilm formation and provides limited protection against infection. Infect Immun 79:4342–4352. doi: 10.1128/IAI.05592-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schindler MKH, Schütz MS, Mühlenkamp MC, Rooijakkers SHM, Hallström T, Zipfel PF, Autenrieth IB. 2012. Yersinia enterocolitica YadA mediates complement evasion by recruitment and inactivation of C3 products. J Immunol 189:4900–4908. doi: 10.4049/jimmunol.1201383. [DOI] [PubMed] [Google Scholar]
- 82.Han YW, Redline RW, Li M, Yin L, Hill GB, McCormick TS. 2004. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect Immun 72:2272–2279. doi: 10.1128/IAI.72.4.2272-2279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vander Haar EL, So J, Gyamfi-Bannerman C, Han YW. 2018. Fusobacterium nucleatum and adverse pregnancy outcomes: epidemiological and mechanistic evidence. Anaerobe 50:55–59. doi: 10.1016/j.anaerobe.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zanzoni A, Spinelli L, Braham S, Brun C. 2017. Perturbed human sub-networks by Fusobacterium nucleatum candidate virulence proteins. Microbiome 5:89. doi: 10.1186/s40168-017-0307-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dy RL, Przybilski R, Semeijn K, Salmond GPC, Fineran PC. 2014. A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism. Nucleic Acids Res 42:4590–4605. doi: 10.1093/nar/gkt1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brewer ML, Dymock D, Brady RL, Singer BB, Virji M, Hill DJ. 2019. Fusobacterium spp. target human CEACAM1 via the trimeric autotransporter adhesin CbpF. J Oral Microbiol 11:1565043. doi: 10.1080/20002297.2018.1565043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomic data used for protein annotation can be found on NCBI under BioProject numbers PRJNA433545 and PRJNA513186. All associated raw data outside of the supplemental material, which includes the T5aSS autotransporter protein similarity network and a FASTA file with all virulence factor open reading frames, can be found on our Open Science Framework data repository (https://osf.io/vs3fd).