We uncover a phage- and antibiotic-induced stress response in the clinically important opportunistic pathogen Pseudomonas aeruginosa. Phage-infected P. aeruginosa subpopulations are isolated from uninfected subpopulations by the production of a stress-induced signal. Activation of the stress response by antibiotics causes P. aeruginosa to physically be repelled from the area containing antibiotics altogether, consistent with a mechanism of antibiotic evasion. The stress response observed here could increase P. aeruginosa resilience against antibiotic treatment and phage therapy in health care settings, as well as provide a simple evolutionary strategy to avoid areas containing stress.
KEYWORDS: antibiotics, bacteriophage, quorum sensing, stress response, swarming
ABSTRACT
We investigate the effect of bacteriophage infection and antibiotic treatment on the coordination of swarming, a collective form of flagellum- and pilus-mediated motility in bacteria. We show that phage infection of the opportunistic bacterial pathogen Pseudomonas aeruginosa abolishes swarming motility in the infected subpopulation and induces the release of the Pseudomonas quinolone signaling molecule PQS, which repulses uninfected subpopulations from approaching the infected area. These mechanisms have the overall effect of limiting the infection to a subpopulation, which promotes the survival of the overall population. Antibiotic treatment of P. aeruginosa elicits the same response, abolishing swarming motility and repulsing approaching swarms away from the antibiotic-treated area through a PQS-dependent mechanism. Swarms are entirely repelled from the zone of antibiotic-treated P. aeruginosa, consistent with a form of antibiotic evasion, and are not repelled by antibiotics alone. PQS has multiple functions, including serving as a quorum-sensing molecule, activating an oxidative stress response, and regulating the release of virulence and host-modifying factors. We show that PQS serves additionally as a stress warning signal that causes the greater population to physically avoid cell stress. The stress response at the collective level observed here in P. aeruginosa is consistent with a mechanism that promotes the survival of bacterial populations.
IMPORTANCE We uncover a phage- and antibiotic-induced stress response in the clinically important opportunistic pathogen Pseudomonas aeruginosa. Phage-infected P. aeruginosa subpopulations are isolated from uninfected subpopulations by the production of a stress-induced signal. Activation of the stress response by antibiotics causes P. aeruginosa to physically be repelled from the area containing antibiotics altogether, consistent with a mechanism of antibiotic evasion. The stress response observed here could increase P. aeruginosa resilience against antibiotic treatment and phage therapy in health care settings, as well as provide a simple evolutionary strategy to avoid areas containing stress.
INTRODUCTION
Stress responses enable individual bacteria to adapt to environmental stresses such as low pH, low ion concentrations, and low nutrient availability (1–4). At the population level, stress responses also promote survival of the group. Bacteria use cell-to-cell signaling, known as quorum sensing (QS), to activate phage defense mechanisms at high cell density when phage could otherwise rapidly spread throughout the bacterial population (5–8). In addition, the emergence of phenotypic heterogeneity in bacterial populations gives rise to persister and antibiotic-tolerant cells that are transiently resistant to antibiotics (9–11). In natural and host environments, bacterial populations are spatially heterogeneous, giving rise to multiple subpopulations of the same species (12, 13). Coordinated spatial stress responses between subpopulations could facilitate the survival of the species.
Swarming is a collective form of bacterial motility driven by flagella and pili that promotes antibiotic resistance and pathogenesis in humans and animals (14–19). High-cell-density bacterial populations swarm on semisolid surfaces, which have physical characteristics similar to those of mucous layers surrounding epithelial membranes (20, 21). In particular, semisolid agar media and mucus are both non-Newtonian fluids that share overlapping ranges of viscosities (22–24).
P. aeruginosa is an opportunistic bacterial pathogen that swarms and is responsible for a range of illnesses, including lung infection in cystic fibrosis patients, hospital-acquired infections, sepsis, and disease in immunocompromised patients (25). Strains of P. aeruginosa have been identified that are resistant to all classes of antibiotics, making the development of new therapeutics an important priority (26). The spatial organization and dynamics of P. aeruginosa swarms are controlled by the secretion of rhamnolipids and 3-(3-hydroxyalkanoyloxy) alkanoic acid (HAAs) (Fig. 1A, left, and Fig. 1Bi), which modulate the repulsion and attraction between different swarming subpopulations (27, 28). Whether and how these signals are coordinated in response to stress are unknown. In particular, threats to the collective in the form of phage and antibiotics could have a significant impact on P. aeruginosa group behaviors.
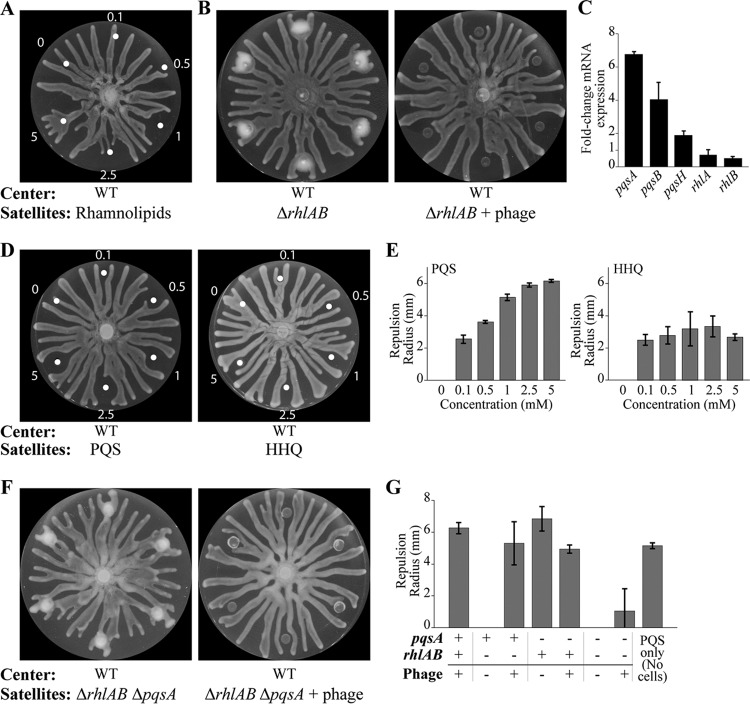
FIG 1.

Infection of P. aeruginosa by bacteriophage inhibits motility and induces repulsion of healthy swarms. (A) Schematic depicting the release of rhamnolipids (RLs) and HAAs (arrows) during swarming (left) and the regulation of rhamnolipids, HAAs, HHQ, and PQS by stress and QS (right). (B to D) Swarm agar assays after 16 to 18 h of growth at 37°C. Wild-type (WT) P. aeruginosa strain PA14 without phage (i) or with phage (ii) or the ΔrhlAB strain without phage (iii) was spotted at the center (B), on the right (C), or at concentric satellite positions surrounding the center (D). Wild-type P. aeruginosa without phage is spotted on the left in panel C and at the center in panel D. Graphics below the images indicate the initial spot positions and corresponding cultures. Petri dishes are 9 cm in diameter.
P. aeruginosa regulates expression of virulence genes and group behaviors through a hierarchical QS network (29). The Pseudomonas quinolone signal 2-heptyl-3-hydroxy-4-quinolone (PQS) is a molecule secreted by P. aeruginosa that has diverse roles, including mediating cell-to-cell signaling through QS, regulating virulence factor expression, iron acquisition, inducing both oxidative stress and an antioxidative response, and modulating host immune responses (30, 31). PQS is found in the lungs of cystic fibrosis patients, indicating the important role of this molecule in long-term persistence of P. aeruginosa infection (32). The synthesis of PQS is regulated by an intricate network of QS regulators, including the LasI/R, RhlI/R, and IQS QS systems (33, 34) (Fig. 1A, right), and is additionally enhanced in response to nutrient starvation, antibiotics, and phage infection (33, 35–37). PQS is synthesized from anthranilate by enzymes encoded by the pqsABCD operon and pqsH (38–41). The multifunctional role of the molecule in signaling and stress responses suggests that it coordinates diverse functions in a collective.
RESULTS
Phage-infected P. aeruginosa colonies repel uninfected P. aeruginosa swarms.
We characterized the effect of virulent phage infection on P. aeruginosa UCBPP-PA14 swarms using the phage DMS3vir, which is an engineered lytic form of the DMS3 phage that was isolated previously from clinical human samples (42). Stationary-phase P. aeruginosa cultures were mixed with phage, spotted on semisolid agar swarming plates, and cultured for 16 to 18 h at 37°C. Phage were prepared by diluting the stock lysate 1:100 to establish a concentration of phage (1012 PFU/ml) that did not cause the complete lysis of the P. aeruginosa population.
Whereas the uninfected wild-type strain swarms (Fig. 1Bi), phage infection inhibited swarming motility (Fig. 1Bii; see also Movie S1 in the supplemental material) comparable to that of the swarming-defective ΔrhlAB strain (Fig. 1Biii). The ΔrhlAB strain does not produce rhamnolipids and HAAs, which are required for swarming (Fig. 1A). The swarming defect due to phage infection is consistent with a previous report in which DMS3 lysogenization of P. aeruginosa inhibited swarming motility through a mechanism dependent on the CRISPR-Cas (clustered regularly interspaced short palindromic repeat [CRISPR]–CRISPR-associated) adaptive immune defense (43).
We investigated the ability of the phage infection to spread to neighboring swarms by spotting both uninfected and infected P. aeruginosa colonies on the same plate (Fig. 1C and Movie S1) or in six-way swarming assays in which uninfected P. aeruginosa was spotted at the center of the swarming plate and P. aeruginosa was spotted at satellite positions surrounding the center (Fig. 1D and Movie S2). Uninfected P. aeruginosa spotted at the satellite positions exhibited swarming motility and repulsed the tendrils of the center swarm (Fig. 1Ci and Di and Movie S2), which is consistent with the ability of HAAs to repel swarms (44). In contrast, phage-infected satellite colonies were inhibited for swarming motility and repulsed uninfected swarms from the vicinity of the infection (Fig. 1Cii and Dii). The inhibition of swarming motility in phage-infected wild-type cells was comparable to that in a ΔrhlAB strain, which is defective in swarming motility due to the lack of HAA and rhamnolipid production (Fig. 1Biii). As expected, the uninfected ΔrhlAB strain did not repulse swarms from the center (Fig. 1Ciii and Diii), consistent with the requirement of HAAs to repel swarms (44).
The ability of phage-infected wild-type cells to repulse center swarms appeared to require active infection, as phage lysate alone did not induce repulsion of the uninfected swarms (Fig. S1A). Either the factors required for repulsion were not present in the phage lysate or the concentration of these factors was insufficient to trigger repulsion. Together, these results suggest that phage-infected cells are deficient in swarming, which would serve to constrain the phage infection locally.
P. aeruginosa organisms that survive phage infection are heterogeneous in response to subsequent infection.
To determine whether the swarm deficiency and repulsion phenotypes were inherited in descendants of phage-infected P. aeruginosa, individual colonies were isolated from phage-infected satellite colonies and tested for the ability to be reinfected using the cross-streaking method, in which cells are streaked past a line of phage. The ability of bacteria to grow beyond the phage line indicates that the strains are not susceptible to phage killing. We observed that two out of the four colonies that were isolated from the phage-infected satellite colonies were not susceptible to phage killing (Fig. 2A). Phage-resistant strains that were remixed with phage were not inhibited for swarming motility (Fig. 2B), suggesting that these isolates are spontaneous surface mutants and/or have acquired CRISPR-Cas-dependent adaptive immunity against the phage. In contrast, reinfection of phage-sensitive strains inhibited motility and caused repulsion of the uninfected swarm (Fig. 2C), which was phenotypically identical to the P. aeruginosa organisms that were initially infected (Fig. 1Cii and Dii). These results indicate that the dual phenotype of swarming inhibition and repulsion of uninfected swarms requires an active infection by phage. We expected the majority of P. aeruginosa organisms that survive phage infection would be phage resistant. However, only half of the surviving P. aeruginosa isolates were phage resistant (Fig. 2A). This observation suggests that a transient phage defense mechanism is associated with the swarming deficiency and repulsion phenotypes.
FIG 2.
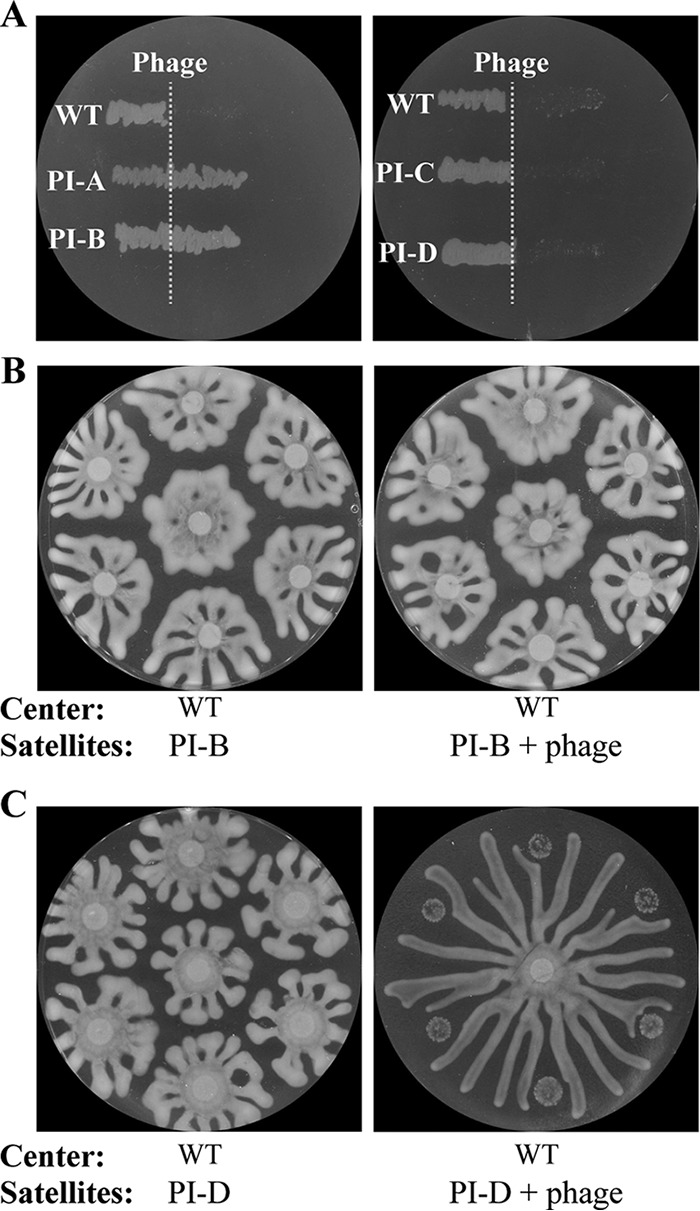
Phage resistance, inhibition of motility, and repulsion in isolates that survive phage treatment. (A) Cross-streak assay in which phage (dashed line) is applied along a line on the dish and phage-infected isolates (PI) of P. aeruginosa from satellite colonies are streaked from left to right across the phage line. Growth beyond the phage line is observed in PI-A and PI-B, indicating resistance to phage, whereas no growth is observed in PI-C and PI-D, indicating sensitivity. (B and C) PI-B (B) or PI-D (C) on swarm agar petri dishes in the absence or presence of phage at the satellite positions. Uninfected wild-type P. aeruginosa is spotted at the center. Inhibition of motility and repulsion are observed in PI-D, which is phage sensitive, in the presence of phage.
Phage-infected colonies repel uninfected swarms independently of rhamnolipids and HAAs.
Rhamnolipids and HAAs modulate the repulsion and attraction between swarming tendrils and are required for swarming motility (27, 28). The secretion of rhamnolipids and HAAs by P. aeruginosa into the vicinity of phage-infected cells was the most likely explanation for the observed repulsion (Fig. 1Cii and Dii). Indeed, purified rhamnolipid fractions spotted at the satellite positions induced repulsion of approaching swarms at intermediate concentrations (Fig. 3A). However, higher concentrations of rhamnolipid fractions attracted the swarms (Fig. 3A and Movie S3A), demonstrating that the effects of attraction and repulsion by rhamnolipids are concentration dependent. To test the hypothesis that phage infection causes repulsion through the production of rhamnolipids and HAAs by P. aeruginosa, ΔrhlAB mutant cells, which are defective in the production of rhamnolipids and HAAs (44), were spotted at the satellite positions. Swarming motility was inhibited in the ΔrhlAB satellite colonies, consistent with the requirement for the production of rhamnolipids and HAAs for swarming motility (27). Importantly, uninfected center swarm tendrils that approached the uninfected ΔrhlAB satellite colonies were not repulsed (Fig. 1Ciii and Diii, 3B and G, and S1B and Movie S4A), consistent with the role of HAAs as a repellant (44). However, phage-infected ΔrhlAB satellite colonies repelled the uninfected center swarm tendrils (Fig. 3B and G and Movie S4B), indicating that the production of rhamnolipids and HAAs was not responsible for the phage-induced repulsion.
FIG 3.
Role of rhamnolipids, PQS, and HHQ in mediating the repulsion response to phage infection. (A) Swarming assay in which water and increasing concentrations (millimolar) of rhamnolipids are spotted at the satellite positions. (B) Spotting of the ΔrhlAB strain at the satellite positions without phage (left) or with phage (right). (C) Fold change in relative mRNA transcript levels of pqsA, pqsB, pqsH, rhlA, and rhlB, normalized by 5S expression, in phage-infected wild-type P. aeruginosa at the satellite positions and compared to levels for uninfected cells at the center of the swarm dish, determined through qRT-PCR. (D and E) Swarming assays, and quantification thereof, in which dimethyl sulfoxide and increasing concentrations (millimolar) of PQS and HHQ are spotted at the satellite positions. (F) Spotting of the ΔrhlAB ΔpqsA strain at the satellite positions without (left) or with (right) phage. (G) Quantification of the repulsion radii at the satellite positions for different strain backgrounds and with or without phage. The repulsion induced by spotting 1 mM PQS is included as a reference. White dots indicate the centers of positions where rhamnolipids, PQS, or HHQ was spotted. Bars in panel C are the averages from at least 2 independent experiments (n = 2). Bars in panel E for PQS and HHQ are the averages from 2 or 4 satellite colonies (n = 2 or 4), respectively, and bars in panel G are the averages for at least 6 satellite colonies (n = 6). Error bars indicate standard deviations. Uninfected wild-type P. aeruginosa is spotted at the center of all assays. The repulsion radius quantification is described in Materials and Methods.
Importantly, these results indicate that the repulsion mechanism observed during phage infection (Fig. 1Cii and Dii) is distinct from the repulsion by uninfected wild-type cells (Fig. 1Ci and Di), the latter of which relies on the production of HAAs. Furthermore, the phage infection of strains defective in the QS master regulator LasR, the QS regulator RhlR, the combination of both, or the CRISPR-Cas system caused repulsion of the center swarm (Fig. S1C to F). These results suggest that phage infection induces the secretion of a molecule that repulses approaching uninfected swarms using a pathway that is activated independently of the master regulators of QS and CRISPR-Cas.
Phage infection activates PQS quorum-sensing signaling.
A recent preprint suggested that genes required for production of the QS molecule (PQS) are upregulated in response to phage infection (37). We verified that the expression of genes responsible for the production of PQS, pqsA and pqsB (38–40), was significantly increased in phage-infected satellite colonies compared to that of the uninfected center swarm (P < 0.05) (Fig. 3C), suggesting that PQS is involved in the repulsion mechanism. The relative transcript levels of the rhamnolipid production genes rhlA and rhlB were slightly decreased in phage-infected cells, but the decrease was not statistically significant (P > 0.1).
We investigated whether PQS alone induces repulsion of swarms. Indeed, spotting increasing concentrations of PQS over the same range of rhamnolipids at the satellite positions produced a monotonic increase in repulsion radius (Fig. 3D and E and Movie S3B). Spotting of 2-heptyl-4-quinolone (HHQ), the precursor molecule to PQS, at the satellite positions caused swarm repulsion to a lesser extent (Fig. 3D and E and Movie S3C), indicating that PQS has greater repelling capacity than HHQ.
Given the ability of PQS to repulse the center swarm tendrils, we hypothesized that PQS is responsible for the repulsion observed during phage infection. Deletion of the pqsA gene, which encodes the synthase that produces the early precursor to PQS, abolished swarming of uninfected satellite colonies (Fig. S2A). The lack of swarming is consistent with the requirement of PQS production for swarming (45, 46). The ΔpqsA strain retained the capacity to repel the wild-type center swarm (Fig. 3G and Fig. S2A), which may be attributed to the production of rhamnolipids and HAAs in this strain. Infection of the ΔpqsA strain decreased the repulsion of the wild-type center swarm (Fig. 3G and Fig. S2A), which suggests that phage infection decreases the production of non-PQS repulsion molecules. The repulsion of the wild-type center swarm was significantly decreased in the phage-infected ΔrhlAB ΔpqsA strain (Fig. 3F and G and Movie S4D) which does not produce rhamnolipids, HAAs, or PQS (40). We observed that the center swarm collided with 60% of the phage-infected ΔrhlAB ΔpqsA satellite colonies, which induced a change in the optical properties of the newly infected tendril population, consistent with phage-mediated lysis (Fig. 3F and Movie S4D). The minimal repulsion by the infected ΔrhlAB ΔpqsA strain may be due to the infection of the approaching swarm by phage that diffuse out of the satellite colonies, which triggers repulsion itself, or due to unidentified secreted factors.
PQS is produced during stationary-phase growth (47). However, the repulsion effects observed here are not due to PQS in the inoculum, as the sterile-filtered supernatant from the ΔrhlAB inoculum did not cause repulsion of the wild-type center swarm (Fig. S2B). Furthermore, the effect is not due to PQS contained within cells in the inoculum, as the sonication and subsequent sterile filtration of the ΔrhlAB inoculum did not cause repulsion (Fig. S2B). Together, these results suggest that phage infection upregulates PQS production, which is largely responsible for the repulsion of uninfected swarms.
Antibiotic-stressed colonies repulse swarms.
PQS promotes the survival of the fittest cells within well-mixed P. aeruginosa populations by sensitizing cells to oxidative stress and other stresses (31). We reasoned that the PQS-mediated repulsion of healthy swarms promotes the survival of the bacterial population by directing the spatial organization of the population. The application of cell stress through other means, including treatment by antibiotics, may also direct the spatial organization of the population to promote overall survival. To test this hypothesis, wild-type P. aeruginosa was mixed with gentamicin, which inhibits growth through stalling ribosomal tRNA translocation (48, 49), and immediately spotted at satellite positions. Minimal growth of P. aeruginosa was observed at the satellite positions (Fig. 4A), indicating that the effective concentration of gentamicin was below the MIC due to the diffusion of the antibiotic through the agar. The slow growth of antibiotic-treated P. aeruginosa (Fig. 4A and Movie S5A) was phenotypically comparable to that due to phage infection (Fig. 1Dii and Movie S2B). Notably, gentamicin treatment inhibited swarming motility at the satellite positions and caused repulsion of untreated center swarm tendrils (Fig. 4A and B and Movie S5A) comparable to that observed in phage-infected P. aeruginosa. The swarm repulsion was not due to the presence of gentamicin alone, as untreated swarms passed through the P. aeruginosa-free gentamicin spots (Fig. 4A and B). Gentamicin-induced swarm repulsion was observed in the ΔrhlAB strain but was significantly reduced in the ΔpqsA strain and was not observed in the ΔrhlAB ΔpqsA strain (Fig. 4A and B and Movie S5B and C). The repulsion effect was abolished in the strain deleted for pqsH, which encodes the synthase that converts HHQ into PQS, and in the ΔrhlAB ΔpqsH strain (Fig. S3C and D), suggesting that PQS is the dominant molecule responsible for inducing repulsion. Together, these data demonstrate the requirement of PQS production for antibiotic-induced repulsion.
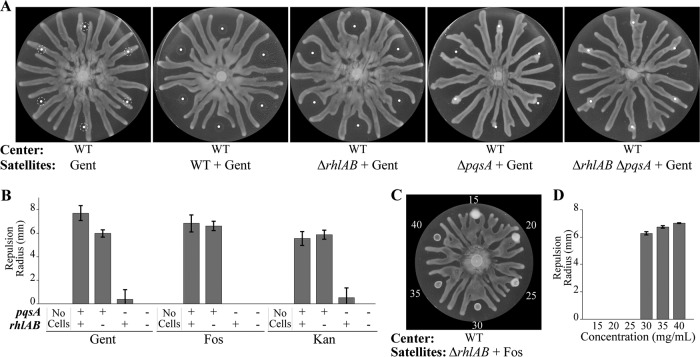
FIG 4.
Antibiotics inhibit motility and induce the repulsion response. (A) Swarming assays in which satellite positions are spotted with 0.5 mg/ml gentamicin (Gent) and wild-type, ΔrhlAB, ΔpqsA, and ΔrhlAB ΔpqsA strains of P. aeruginosa. The dashed lines indicate the boundaries of the initial spots. (B) Repulsion radii of wild-type and mutant P. aeruginosa strains that were spotted with 0.5 mg/ml gentamicin (Gent), 40 mg/ml fosfomycin (Fos), or 25 mg/ml kanamycin (Kan). (C and D) Swarming assay (C) and corresponding repulsion radii (D) in which the initial culture of the ΔrhlAB strain was spotted with fosfomycin (concentrations in mg/ml). White dots indicate the centers of positions where antibiotics, cells, or the combination of antibiotics and cells was spotted. Bars in panels B and D indicate averages from at least 6 or 2 independent experiments (n = 6 or 2), respectively. Error bars indicate standard deviations. The swarming assays for the kanamycin and fosfomycin treatments depicted in panel B are shown in Fig. S3A and B in the supplemental material. Overnight cultures were mixed with antibiotics to the indicated concentrations, and 6-μl aliquots of the mixtures were spotted at satellite positions on antibiotic-free swarming media.
The inhibition of motility and induction of repulsion were similarly observed through treatment of P. aeruginosa at satellite positions using the antibiotics kanamycin (Fig. 4B and Fig. S3A), which inhibits tRNA translocation similarly to gentamicin (48, 49), and fosfomycin (Fig. 4B to D and Fig. S3B), which inhibits the synthesis of the cell wall component peptidoglycan (50). In particular, the repulsion radius at satellite positions clearly increased with increasing concentrations of antibiotics (Fig. 4C and D and Fig. S3E and F). We note that repulsion is triggered sharply by a small change in fosfomycin concentration between 25 and 30 mg/ml and is concomitant with the inhibition of growth of the satellite colonies. In contrast, continuously increasing repulsion is observed using synthetic PQS (Fig. 3D and E). These results suggest that the activation of PQS in response to antibiotic stress is switch-like rather than graded.
The effects of other antibiotics, including carbenicillin, cefsulodin, ciprofloxacin, and tetracycline, were assessed on repulsing center swarms. However, the presence of these antibiotics alone (without P. aeruginosa) inhibited the motility of center swarms at lower concentrations than required to inhibit growth of the satellite colonies (Fig. S4C). Thus, we were unable to assess whether these antibiotics could cause P. aeruginosa to repel swarms.
Antibiotic stress increases PQS signaling.
Phage infection increased the relative abundance of PQS synthesis transcripts (Fig. 3C). We reasoned that antibiotic-induced cell stress may promote a similar effect. Indeed, the treatment of wild-type P. aeruginosa with gentamicin at the satellite positions significantly enhanced transcript levels of PQS synthesis genes (over 100-fold for pqsA) (Fig. 5A). These results support the model that antibiotic stress increases PQS synthesis and inhibits swarming motility. Furthermore, the long-range diffusion of PQS was confirmed by performing mass spectrometry analysis of agar extracts in the zone of repulsion outside of the antibiotic-treated P. aeruginosa satellite colony. PQS was detected in the agar surrounding the gentamicin-treated or fosfomycin-treated ΔrhlAB strain but not in that of the treated ΔrhlAB ΔpqsA strain (Fig. 5B and Fig. S5A). Together, these results indicate that the induction of stress through inhibition of tRNA translocation or cell wall synthesis inhibits swarming motility and induces the long-range dispersion of PQS, which repulses swarms from entering the area containing the stress-inducing agent.
FIG 5.
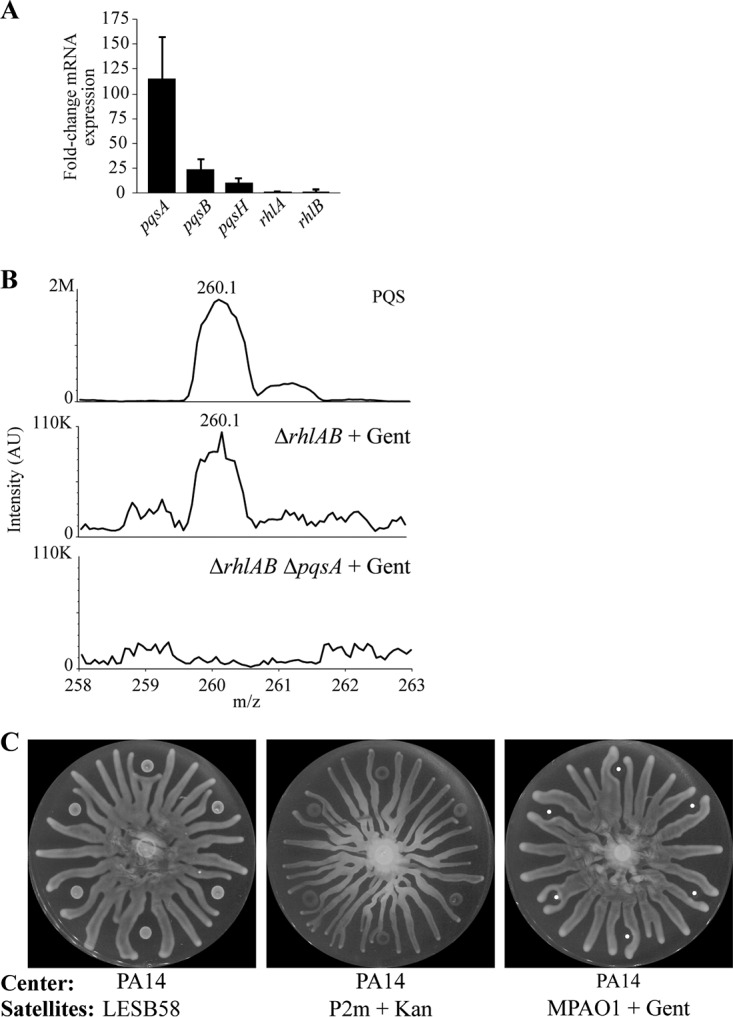
Antibiotics induce production of PQS, which is detected outside the area containing cells. (A) Fold change in mRNA transcripts, as determined by qRT-PCR, of gentamicin-treated wild-type P. aeruginosa on swarming dishes compared to that of untreated cells at the center of the swarming dish. (B) Liquid chromatography-mass spectrometry of 10 μM PQS and agar extracts of the zone of repulsion surrounding ΔrhlAB and ΔrhlAB ΔpqsA strains of P. aeruginosa that were treated with 0.5 mg/ml gentamicin. The area containing cells is excluded. AU, arbitrary units. (C) Swarming assay in which the untreated hypervirulent P. aeruginosa strain LESB58, the mucoid P. aeruginosa cystic fibrosis isolate P2m treated with 25 mg/ml of kanamycin, or MPAO1 treated with 0.5 mg/ml of gentamicin was spotted at the satellite positions. Bars in panel A indicate averages from at least 2 independent experiments. Error bars indicate standard deviations. Overnight cultures were mixed with antibiotics to the indicated concentrations, and 6-μl aliquots of the mixtures were spotted on antibiotic-free swarming media.
Virulent P. aeruginosa strains repulse healthy swarms.
The importance of PQS in host pathogenesis (30–32) suggests a role for the stress response observed here during human infection. We investigated the potential role of the PQS-mediated repulsion response in the Liverpool epidemic strain LESB58, which is a hypervirulent isolate of P. aeruginosa (51). The strain was spotted without antibiotics or phage at the satellite positions, while wild-type PA14 was spotted in the center. LESB58 grew slowly, did not swarm, repulsed PA14 swarms (Fig. 5C), and produced elevated levels of PQS in the surrounding agar (Fig. S5B). Thus, the hypervirulent LESB58 strain exhibited a phenotype that is consistent with a constitutively active cell stress response, even though no external stress was present. In addition, the repulsion response was characterized in the mucoid P. aeruginosa cystic fibrosis isolate P2m (52) and in the strain MPAO1 (53). Center swarm tendrils contacted untreated satellite colonies but were repulsed by antibiotic-treated colonies (Fig. 5C and Fig. S4A and B). Together, these data indicate that the stress response is present in clinical isolates and suggest a role for the stress response in pathogenesis and treatment tolerance.
DISCUSSION
Stress responses enable bacteria to respond appropriately to stimuli that threaten their survival. We demonstrate that the detection of stress by a P. aeruginosa subpopulation induces the production of PQS, which repulses approaching swarms far beyond the area containing the stress and thereby serves as a long-range signal (Fig. 6). This long-range stress response complements the short-range kin lysis stress system previously reported in P. aeruginosa (54). PQS activates heightened stress responses in P. aeruginosa, including the formation of outer membrane vesicles, which interfere with cytokine production and deliver toxins to target host cells (55–57). Through activation of the PqsR receptor, PQS also induces pqsE expression, which synthesizes a QS molecule that activates RhlR and is thereby responsible for regulation of key virulence factors that kill plants and animals, including hydrogen cyanide and elastase (41, 58, 59). Our data suggest that PQS functions additionally as a coordinator of spatial organization during swarming. Importantly, this work shows that PQS repulses healthy populations from the area containing stress. Given the ability of a stress response to impact swarming, which is a collective behavior, we refer to this response as the collective stress response.
FIG 6.
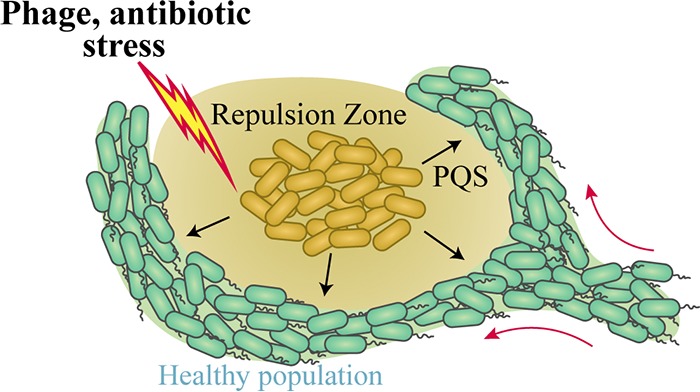
Schematic of the collective stress response. Antibiotics and bacteriophage inhibit swarming motility and induce the production and release of PQS. The release of PQS repels swarms such that untreated bacteria do not approach the infected/treated area. The stress response potentially reduces the spread of phage and the exposure of P. aeruginosa to antibiotics.
The collective stress response is activated by diverse types of stress and may provide the overall population with protection from the stress. In the context of bacteriophage infection, the stress response physically quarantines the infected population by repelling healthy (uninfected) swarms. This response may serve as a warning system to direct the healthy population to stay clear of the zone of danger. The overall effect is that the healthy P. aeruginosa population is protected from infection by the phage-infected population. Thus, the repulsion of uninfected P. aeruginosa swarms by the infected subpopulation reduces the potential spread of the infection to other parts of the population. In the ΔrhlAB ΔpqsA strain, which is defective in the repulsion response, uninfected wild-type swarms approached and contacted infected populations, which enabled the infection to spread to the healthy population (Fig. 3F).
In natural environments, the stress response could mitigate the spread of phage infection throughout a population and thereby serve as a bacterial defense mechanism against phage to complement other defenses, including the CRISPR-Cas system (60). In particular, membrane vesicles function as phage decoys, and their production is upregulated in response to PQS in P. aeruginosa (56, 57, 61). Thus, the production of PQS and membrane vesicles in response to phage infection could function as a mechanism to increase transient immunity to phage. We note that bacteria that survive phage infection are typically resistant to further infection. However, only half of the surviving phage-infected P. aeruginosa organisms were resistant (Fig. 2), which suggests that P. aeruginosa employs a protection mechanism that enables survival against phage infection without acquiring heritable phage resistance. This mechanism is consistent with the upregulation of membrane vesicle production by PQS in response to phage infection.
The collective stress response to antibiotics induces P. aeruginosa populations to secrete PQS into the surrounding environment. Healthy swarms are repulsed from the area of antibiotic-treated cells altogether, directing the swarming tendril toward areas free of antibiotics. In natural environments, this could promote the evasion of P. aeruginosa from competing microbes that produce antibiotics, enabling the greater population to avoid a close encounter with the microbe. In the context of antibiotic use to treat human disease, the collective stress response could have a role in lowering the efficacy of antibiotic treatment by guiding the bacterium away from areas of high local concentrations of antibiotics. Together, our results show that the collective stress response repels P. aeruginosa populations from environments containing phage or antibiotics, which promotes the survival of the overall population.
The ability of rhamnolipids/HAAs and PQS to repulse swarms raises the question of why PQS is produced under stressful conditions. It is possible that PQS is produced as an alternative repulsive molecule because rhamnolipids/HAAs cannot be produced by stressed cells or because rhamnolipids/HAAs do not guarantee repulsion, as high concentrations of rhamnolipids attract swarms (Fig. 3A). Alternatively, the production of PQS provides additional functions that could benefit P. aeruginosa, including signaling other P. aeruginosa organisms in the vicinity of the presence of stress, the production of an oxidative environment (31) that could protect against invasion by other microbes, and the production of membrane vesicles, which could kill invading host cells and serve as phage decoys (56, 62).
Future work will address the mechanisms by which antibiotics and phage infection activate the production of PQS and the impacts of the stress signal on other P. aeruginosa organisms, microbes, and host cells in the vicinity. Our results suggest that the regulation of PQS in response to stress is not achieved through the canonical quorum-sensing regulators LasR and RhlR. In addition, stress-mediated PQS production of outer membrane vesicles could have a significant role in bacterium-host interactions and impact the severity of bacterial infections.
MATERIALS AND METHODS
Growth conditions.
Strains were streaked from frozen stocks, which were maintained at –80°C, onto LB broth, Miller (Becton, Dickinson, Franklin Lakes, NJ)-supplemented petri dishes containing 1.5 to 2% Bacto agar (Becton, Dickinson) and grown at 37°C overnight. Single colonies were inoculated into sterilized LB medium and grown to saturation for 16 to 18 h in a shaker at 225 rpm or in a roller drum. For strain construction and plasmid maintenance, gentamicin and carbenicillin were used at 30 μg/ml and 200 μg/ml, respectively. DMS3vir lysate was prepared using standard phage preparation procedures (63) using the ΔCRISPR Δcas strain as a host. Phage were diluted into LB to a concentration of 1012 PFU/ml and stored at 4°C. All strains and plasmids used in this study are described in Table S1 in the supplemental material. All P. aeruginosa PA14 strains used in this study were derived from PA14 UCBPP-PA14, obtained from the O’Toole laboratory.
Strain construction.
A markerless strain of PA14 that constitutively expresses mCherry was constructed by flipping out the aacC1 gene, which is responsible for gentamicin resistance, by transforming AFS27E with pFLP2, yielding AFS27E.1.
The ΔrhlAB strain was constructed through lambda red recombineering using the procedure described previously (64). All primers used for strain construction are given in Table S2. The upstream region of rhlA (rhlA′) and the downstream region of rhlB (′rhlB) were amplified using the primer pairs rhlA-lred-u1/rhlA-lred-l1 and rhlB-lred-u3/rhlB-lred-l3, respectively. The region containing FRT-aacC1-FRT was amplified from pAS03 using the primers rhlA-lred-u2 and rhlB-lred-l2. The rhlA’, FRT-aacC1-FRT, and ‘rhlB products were combined through isothermal assembly, amplified using rhlA-lred-u1 and rhlB-lred-l3, transformed into PA14/pUCP18-RedS, selected for gentamicin resistance, and cured of pUCP18-RedS by growing on sucrose, resulting in a PA14 ΔrhlAB::aacC1 strain. The resulting strain was transformed with pFLP2 to flip out the gentamicin resistance, yielding BR04.1.
The ΔpqsA strain was constructed by amplifying the upstream region of pqsA (pqsA′) by using the primers pqsA-lred-u1 and pqsA-lred-l1. The downstream region of pqsA (′pqsA) was amplified using the primers pqsA-lred-u3 and pqsA-lred-l3. The region containing FRT-aacC1-FRT was amplified from pAS03 using the primers pqsA-lred-u2 and pqsA-lred-l2. The pqsA′, FRT-aacC1-FRT, and ′pqsA products were combined through isothermal assembly, amplified using pqsA-lred-u1 and pqsA-lred-l3, transformed into AFS27E.1/pUCP18-RedS, selected for gentamicin resistance, and cured of pUCP18-RedS by growing on sucrose, resulting in an AFS27E.1 ΔpqsA::aacC1 strain. The resulting strain was transformed with pFLP2 to flip out the gentamicin resistance, yielding AFS79.1. The ΔrhlAB ΔpqsA strain was constructed by transforming the pqsA′-FRT-aacC1-FRT-′pqsA product used in the construction of AFS79.1 into BR04.1/pUCP18-RedS, selecting for gentamicin resistance, and curing of pUCP18-RedS by growing on sucrose, yielding AFS82.1.
The pqsH strain was constructed by amplifying the pqsH::Mar2xT7 allele from the P. aeruginosa ordered transposon library (65) using the primers pqsH-u1 and pqsH-l1, transforming the product into AFS27E.1/pUCP18-RedS, selecting for gentamicin resistance, and curing of pUCP18-RedS by growing on sucrose, yielding AFS77. The ΔrhlAB ΔpqsH strain was constructed by transforming the product into BR04.1/pUCP18-RedS, selecting for gentamicin resistance, and curing of pUCP18-RedS by growing on sucrose, yielding BR07.
Cross-streak assay.
A straight line of phage was introduced into LB petri dishes containing 1.5% agar by pressing on the agar with a sterilized straight edge and pipetting 25 μl of 1012 PFU/ml DMS3vir phage lysate down the line. Plates were dried until the line was no longer visibly wet. Single colonies were streaked across the plate perpendicular to the line of phage. Plates were incubated overnight at 37°C.
Swarming assay.
Swarming petri dishes (100 mm by 15 mm) contained 20 ml of M8 minimum medium supplemented with 1 mM MgSO4, 0.2% glucose, 0.5% Casamino Acids (Becton, Dickinson), and 0.5% agar (66). Petri dishes were dried in a single stack for 1 h on the bench and for an additional 30 to 60 min at room temperature with the petri dish lids off in a laminar flow hood at 300 cubic ft/min with approximately 40 to 50% ambient humidity. P. aeruginosa was cultured overnight (16 to 18 h) from single colonies to saturation in LB in a roller drum or shaker at 225 rpm at 37°C. Five microliters of culture was spotted in the center or at 6 equidistant satellite positions on a 5.8-cm-radius concentric circle around the center of the dish. Plates were incubated overnight at 37°C in a humidified chamber with a modified petri dish lid on an Epson photo scanner (Epson, Long Beach, CA). Images were acquired at 30-min intervals for 16 to 18 h and processed using ImageJ (NIH, Bethesda, MD).
For phage infection assays, 1 μl of 1012 PFU/ml of DMS3vir was mixed with 5 μl of overnight P. aeruginosa culture, and 6 μl of the resulting mixture was spotted at satellite positions. For the phage-only experiment, 1 μl containing 1012 PFU/ml was mixed with 5 μl water and spotted. For antibiotic treatments, overnight culture or water was mixed with antibiotics to final concentrations of 500 μg/ml of gentamicin (Sigma-Aldrich, St. Louis, MO), 25 mg/ml of kanamycin (Sigma-Aldrich), 40 mg/ml of fosfomycin (Tokyo Chemical Industry, Portland, OR), 0.05 mg/ml to 10 mg/ml of carbenicillin (Teknova, Hollister, CA), cefsulodin (Research Product International, Mount Prospect, IL), ciprofloxacin (Fisher Scientific, Hampton, NH), or tetracycline (Sigma-Aldrich), and 6 μl of the resulting mixture was spotted at satellite positions. For compound repulsion/attraction assays, 6 μl of rhamnolipids (R90-1G; AGAE Technologies, Corvallis, OR), PQS (94398-10MG; Sigma-Aldrich), or HHQ (SML0747-10MG; Sigma-Aldrich) was spotted at the satellite positions. For sonicated culture assays, strains were cultured overnight for 16 to 18 h to saturation and sonicated at 75% power using 15-s on/5-s off intervals for 1 min using a Sonic Dismembrator 500 sonicator (Fisher Scientific) and centrifuged at 21,000 × g for 5 min. The supernatant was filtered using a 0.2-μm filter and then filtered additionally using a 0.02-μm filter. Six microliters of the supernatant was spotted at satellite positions. For filtered culture assays, the sonication step was skipped.
Measurement of repulsion radius.
The radius of repulsion by each satellite colony was determined by identifying the concentric circle centered at the middle of the petri dish that passed through the center of the satellite colony (see Fig. S5C in the supplemental material). The distance from the center of each satellite colony to the two nearest swarming tendrils along a line tangent to the concentric circle was measured and averaged (Fig. S5C). If the tendril visibly contacted the initial zone of inoculation, which was determined by the first time-lapse image, or the distance between the center of the satellite colony to a tendril was less than or equal to the average radius of an initial spot (determined to be 1.99 mm with a standard deviation of 0.05 mm measured from 72 satellite colonies), the radius of repulsion was set to 0. Repulsion radii were averaged over at least six satellite colonies.
qRT-PCR.
P. aeruginosa cells were harvested from the center of the swarming petri dish or from satellite colonies after 18 to 20 h at 37°C. The agar was cut, placed in a separate petri dish, and washed with water. Cells were bound to a 0.22-μm filter membrane and resuspended in total lysis solution (10 mM Tris-HCl, 1 mM EDTA, 0.5 mg/ml lysozyme, 1% SDS, pH 8.0). RNA was harvested using hot phenol extraction as performed previously (67) and digested with DNase (Life Technologies, Carlsbad, CA). cDNA was synthesized using a reverse transcriptase kit (Applied Biosystems, Foster City, CA) and quantified using SsoAdvanced Universal SYBR green (Bio-Rad, Hercules, CA) on a CFX96 Touch real-time PCR detection system (Bio-Rad) quantitative PCR machine. All primers used for quantitative PCR are given in Table S2. The transcript abundance for each sample was normalized by 5S rRNA abundance, which was determined using the 5S_qPCR primer (8). The fold change in transcript abundance due to phage or antibiotic treatment was computed by dividing the average of the normalized transcript abundances in the satellite colonies by that of cells from the center swarm and using the resulting value (n) as the exponent in 2−n.
Mass spectrometry.
Agar was harvested from the zone of repulsion at satellite positions (outside the cell growth region) from swarming plates after 20 h of incubation at 37°C. Samples were prepared by excising the agar using the wide end of a 1-ml pipet tip (diameter, 7.5 mm), placing the agar in a microcentrifuge tube, adding ethyl acetate, vortexing for 30 s, and incubating for 10 min. The top layer was transferred and dried for 10 min in a speed vacuum at 50°C, resuspended in acetonitrile, and analyzed using an Acquity UPLC, Acquity QDa single-quadrupole detector (Waters, Milford, MA) with acetonitrile and 0.1% formic acid as column solvents. The peak at 1 to 2 s was used for analysis. For the LESB58 strain, supernatant from an overnight culture in LB was pelleted and extracted using ethyl acetate. Ten micromolars PQS in acetonitrile was used as a reference.
Statistical tests.
Bars indicate the means, and error bars indicate the standard deviations unless otherwise noted. The statistical significance of a change between two data sets was determined using two-tailed heteroscedastic Student's t test comparisons. Changes were deemed significant if the probability of the null hypothesis that two data sets originated from the same distribution was less than 0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bonnie Bassler, Allon Hochbaum, Shaun Lee, members of the Siryaporn laboratory, and members of the UCI Systems Microbiology group for helpful discussions and Felix Grun and Benjamin Katz from the UCI Mass Spectrometry Facility. The graphical schematic was designed by Robert Gant.
This work was supported by NIH award K22AI112816 and R21AI139968 grant to A.S. and by the University of California. N.M.H.-K. was supported by Lundbeck Fellowships R220-2016-860 and R251-2017-1070. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
All authors designed the experiments. J.-L.B., C.T., B.R., and A.S. performed the experiments and analyzed data. N.M.H.-K. and K.W. provided project direction. J.-L.B., C.T., N.M.H.-K., and A.S. wrote and edited the manuscript.
We have no competing interests to declare.
Footnotes
For a commentary on this article, see https://doi.org/10.1128/JB.00568-19.
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00383-19.
REFERENCES
- 1.Andrews SC, Robinson AK, Rodríguez-Quiñones F. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 2.Schaible UE, Kaufmann S. 2004. Iron and microbial infection. Nat Rev Microbiol 2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 3.Groisman EA, Hollands K, Kriner MA, Lee E-J, Park S-Y, Pontes MH. 2013. Bacterial Mg2+ homeostasis, transport, and virulence. Annu Rev Genet 47:625–646. doi: 10.1146/annurev-genet-051313-051025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Storz G, Hengge R (ed). 2011. Bacterial stress responses, 2nd ed American Society of Microbiology, Washington, DC. doi: 10.1128/9781555816841. [DOI] [Google Scholar]
- 5.Patterson AG, Jackson SA, Taylor C, Evans GB, Salmond GPC, Przybilski R, Staals RHJ, Fineran PC. 2016. Quorum sensing controls adaptive immunity through the regulation of multiple CRISPR-Cas systems. Mol Cell 64:1102–1108. doi: 10.1016/j.molcel.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan D, Svenningsen SL, Middelboe M. 2015. Quorum sensing determines the choice of antiphage defense strategy in Vibrio anguillarum. mBio 6:e00627. doi: 10.1128/mBio.00627-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Høyland-Kroghsbo NM, Maerkedahl RB, Svenningsen SL. 2013. A quorum-sensing-induced bacteriophage defense mechanism. mBio 4:e00362. doi: 10.1128/mBio.00362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Høyland-Kroghsbo NM, Paczkowski J, Mukherjee S, Broniewski J, Westra E, Bondy-Denomy J, Bassler BL. 2017. Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc Natl Acad Sci U S A 114:131–135. doi: 10.1073/pnas.1617415113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vega NM, Gore J. 2014. Collective antibiotic resistance: mechanisms and implications. Curr Opin Microbiol 21:28–34. doi: 10.1016/j.mib.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 11.Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Brien S, Williams D, Fothergill JL, Paterson S, Winstanley C, Brockhurst MA. 2017. High virulence sub-populations in Pseudomonas aeruginosa long-term cystic fibrosis airway infections. BMC Microbiol 17:30. doi: 10.1186/s12866-017-0941-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xavier JB. 2011. Social interaction in synthetic and natural microbial communities. Mol Syst Biol 7:483. doi: 10.1038/msb.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pezzlo M, Valter PJ, Burns MJ. 1979. Wound infection associated with Vibrio alginolyticus. Am J Clin Pathol 71:476–478. doi: 10.1093/ajcp/71.4.476. [DOI] [PubMed] [Google Scholar]
- 15.Coker C, Poore CA, Li X, Mobley HL. 2000. Pathogenesis of Proteus mirabilis urinary tract infection. Microbes Infect 2:1497–1505. doi: 10.1016/S1286-4579(00)01304-6. [DOI] [PubMed] [Google Scholar]
- 16.Overhage J, Bains M, Brazas MD, Hancock R. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J Bacteriol 190:2671–2679. doi: 10.1128/JB.01659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai S, Tremblay J, Déziel E. 2009. Swarming motility: a multicellular behaviour conferring antimicrobial resistance. Environ Microbiol 11:126–136. doi: 10.1111/j.1462-2920.2008.01747.x. [DOI] [PubMed] [Google Scholar]
- 18.Kim W, Killam T, Sood V, Surette MG. 2003. Swarm-cell differentiation in Salmonella enterica serovar typhimurium results in elevated resistance to multiple antibiotics. J Bacteriol 185:3111–3117. doi: 10.1128/jb.185.10.3111-3117.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler MT, Wang Q, Harshey RM. 2010. Cell density and mobility protect swarming bacteria against antibiotics. Proc Natl Acad Sci U S A 107:3776–3781. doi: 10.1073/pnas.0910934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeung ATY, Torfs ECW, Jamshidi F, Bains M, Wiegand I, Hancock REW, Overhage J. 2009. Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J Bacteriol 191:5592–5602. doi: 10.1128/JB.00157-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girod S, Zahm JM, Plotkowski C, Beck G, Puchelle E. 1992. Role of the physiochemical properties of mucus in the protection of the respiratory epithelium. Eur Respir J 5:477–487. [PubMed] [Google Scholar]
- 22.Lai SK, Wang Y-Y, Wirtz D, Hanes J. 2009. Micro- and macrorheology of mucus. Adv Drug Deliv Rev 61:86–100. doi: 10.1016/j.addr.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Che L, Zhou W, Chen XD. 2012. Rheological behavior of agar solution in relation to the making of instant edible bird’s nest products. Int J Food Eng 8:1556–3758. doi: 10.1515/1556-3758.2670. [DOI] [Google Scholar]
- 24.Robinson WB, Mealor AE, Stevens SE, Ospeck M. 2007. Measuring the force production of the hormogonia of Mastigocladus laminosus. Biophys J 93:699–703. doi: 10.1529/biophysj.107.104067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Driscoll JA, Brody SL, Kollef MH. 2007. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67:351–368. doi: 10.2165/00003495-200767030-00003. [DOI] [PubMed] [Google Scholar]
- 26.Lushniak BD. 2014. Antibiotic resistance: a public health crisis. Public Health Rep 129:314–316. doi: 10.1177/003335491412900402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caiazza NC, Shanks RMQ, O'Toole GA. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187:7351–7361. doi: 10.1128/JB.187.21.7351-7361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tremblay J, Richardson A-P, Lépine F, Déziel E. 2007. Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ Microbiol 9:2622–2630. doi: 10.1111/j.1462-2920.2007.01396.x. [DOI] [PubMed] [Google Scholar]
- 29.Papenfort K, Bassler BL. 2016. Quorum sensing signal–response systems in Gram-negative bacteria. Nat Rev Microbiol 14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin J, Cheng J, Wang Y, Shen X. 2018. The Pseudomonas quinolone signal (PQS): not just for quorum sensing anymore. Front Cell Infect Microbiol 8:230. doi: 10.3389/fcimb.2018.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Häussler S, Becker T. 2008. The Pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog 4:e1000166. doi: 10.1371/journal.ppat.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collier DN, Anderson L, McKnight SL, Noah TL, Knowles M, Boucher R, Schwab U, Gilligan P, Pesci EC. 2002. A bacterial cell to cell signal in the lungs of cystic fibrosis patients. FEMS Microbiol Lett 215:41–46. doi: 10.1111/j.1574-6968.2002.tb11367.x. [DOI] [PubMed] [Google Scholar]
- 33.Lee J, Wu J, Deng Y, Wang J, Wang C, Wang J, Chang C, Dong Y, Williams P, Zhang L-H. 2013. A cell-cell communication signal integrates quorum sensing and stress response. Nat Chem Biol 9:339–343. doi: 10.1038/nchembio.1225. [DOI] [PubMed] [Google Scholar]
- 34.Lee J, Zhang L. 2015. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6:26–41. doi: 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schafhauser J, Lepine F, McKay G, Ahlgren HG, Khakimova M, Nguyen D. 2014. The stringent response modulates 4-hydroxy-2-alkylquinoline biosynthesis and quorum-sensing hierarchy in Pseudomonas aeruginosa. J Bacteriol 196:1641–1650. doi: 10.1128/JB.01086-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morales-Soto N, Dunham SJB, Baig NF, Ellis JF, Madukoma CS, Bohn PW, Sweedler JV, Shrout JD. 2018. Spatially dependent alkyl quinolone signaling responses to antibiotics in Pseudomonas aeruginosa swarms. J Biol Chem 293:9544–9552. doi: 10.1074/jbc.RA118.002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blasdel BG, Ceyssens P-J, Chevallereau A, Debarbieux L, Lavigne R. 2018. Comparative transcriptomics reveals a conserved bacterial adaptive phage response (BAPR) to viral predation. bioRxiv doi: 10.1101/248849. [DOI]
- 38.Dulcey CE, Dekimpe V, Fauvelle D-A, Milot S, Groleau M-C, Doucet N, Rahme LG, Lépine F, Déziel E. 2013. The end of an old hypothesis: the pseudomonas signaling molecules 4-hydroxy-2-alkylquinolines derive from fatty acids, not 3-ketofatty acids. Chem Biol 20:1481–1491. doi: 10.1016/j.chembiol.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Déziel E, Lépine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. 2002. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol 184:6472–6480. doi: 10.1128/jb.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mukherjee S, Moustafa DA, Stergioula V, Smith CD, Goldberg JB, Bassler BL. 2018. The PqsE and RhlR proteins are an autoinducer synthase-receptor pair that control virulence and biofilm development in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 115:E9411–E9418. doi: 10.1073/pnas.1814023115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O'Toole GA. 2012. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol 194:5728–5738. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O'Toole GA. 2009. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol 191:210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Déziel E, Lépine F, Milot S, Villemur R. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3–(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005–2013. doi: 10.1099/mic.0.26154-0. [DOI] [PubMed] [Google Scholar]
- 45.Ha D-G, Merritt JH, Hampton TH, Hodgkinson JT, Janecek M, Spring DR, Welch M, O'Toole GA. 2011. 2-Heptyl-4-quinolone, a precursor of the Pseudomonas quinolone signal molecule, modulates swarming motility in Pseudomonas aeruginosa. J Bacteriol 193:6770–6780. doi: 10.1128/JB.05929-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo Q, Kong W, Jin S, Chen L, Xu Y, Duan K. 2014. PqsR-dependent and PqsR-independent regulation of motility and biofilm formation by PQS in Pseudomonas aeruginosa PAO1. J Basic Microbiol 54:633–643. doi: 10.1002/jobm.201300091. [DOI] [PubMed] [Google Scholar]
- 47.Diggle SP, Winzer K, Chhabra SR, Worrall KE, Cámara M, Williams P. 2003. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol 50:29–43. doi: 10.1046/j.1365-2958.2003.03672.x. [DOI] [PubMed] [Google Scholar]
- 48.Cabañas MJ, Vázquez D, Modolell J. 1978. Inhibition of ribosomal translocation by aminoglycoside antibiotics. Biochem Biophys Res Commun 83:991–997. doi: 10.1016/0006-291x(78)91493-6. [DOI] [PubMed] [Google Scholar]
- 49.Feldman MB, Terry DS, Altman RB, Blanchard SC. 2010. Aminoglycoside activity observed on single pre-translocation ribosome complexes. Nat Chem Biol 6:54–62. doi: 10.1038/nchembio.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kahan FM, Kahan JS, Cassidy PJ, Kropp H. 1974. The mechanism of action of fosfomycin (phosphonomycin). Ann N Y Acad Sci 235:364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 51.Cheng K, Smyth RL, Govan JR, Doherty C, Winstanley C, Denning N, Heaf DP, van Saene H, Hart CA. 1996. Spread of beta-lactam-resistant Pseudomonas aeruginosa in a cystic fibrosis clinic. Lancet 348:639–642. doi: 10.1016/S0140-6736(96)05169-0. [DOI] [PubMed] [Google Scholar]
- 52.Quinn RA, Phelan VV, Whiteson KL, Garg N, Bailey BA, Lim YW, Conrad DJ, Dorrestein PC, Rohwer FL. 2016. Microbial, host and xenobiotic diversity in the cystic fibrosis sputum metabolome. ISME J 10:1483–1498. doi: 10.1038/ismej.2015.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LeRoux M, Kirkpatrick RL, Montauti EI, Tran BQ, Peterson SB, Harding BN, Whitney JC, Russell AB, Traxler B, Goo YA, Goodlett DR, Wiggins PA, Mougous JD. 2015. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. Elife 4:e05701. doi: 10.7554/eLife.05701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bomberger JM, MacEachran DP, Coutermarsh BA, Ye S, O'Toole GA, Stanton BA. 2009. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog 5:e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Florez C, Raab JE, Cooke AC, Schertzer JW. 2017. Membrane distribution of the Pseudomonas quinolone signal modulates outer membrane vesicle production in Pseudomonas aeruginosa. mBio 8:e01034-17. doi: 10.1128/mBio.01034-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reyes-Robles T, Dillard RS, Cairns LS, Silva-Valenzuela CA, Housman M, Ali A, Wright ER, Camilli A. 2018. Vibrio cholerae outer membrane vesicles inhibit bacteriophage infection. J Bacteriol 200:e00792-17. doi: 10.1128/JB.00792-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hazan R, He J, Xiao G, Dekimpe V, Apidianakis Y, Lesic B, Astrakas C, Déziel E, Lépine F, Rahme LG. 2010. Homeostatic interplay between bacterial cell-cell signaling and iron in virulence. PLoS Pathog 6:e1000810. doi: 10.1371/journal.ppat.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rampioni G, Pustelny C, Fletcher MP, Wright VJ, Bruce M, Rumbaugh KP, Heeb S, Cámara M, Williams P. 2010. Transcriptomic analysis reveals a global alkyl-quinolone-independent regulatory role for PqsE in facilitating the environmental adaptation of Pseudomonas aeruginosa to plant and animal hosts. Environ Microbiol 12:1659–1673. doi: 10.1111/j.1462-2920.2010.02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 61.Manning AJ, Kuehn MJ. 2011. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol 11:258. doi: 10.1186/1471-2180-11-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mashburn LM, Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 63.Malke H. 1993. Jeffrey H. Miller, A short course in bacterial genetics–a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor 1992. Cold Spring Harbor Laboratory Press. ISBN: 0–87969–349–5. J Basic Microbiol 33:278–278. doi: 10.1002/jobm.3620330412. [DOI] [Google Scholar]
- 64.Shen Y, Siryaporn A, Lecuyer S, Gitai Z, Stone HA. 2012. Flow directs surface-attached bacteria to twitch upstream. Biophys J 103:146–151. doi: 10.1016/j.bpj.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ha D-G, Kuchma SL, O’Toole GA. 2014. Plate-based assay for swarming motility in Pseudomonas aeruginosa, p 67–72. In Filloux A, Ramos J-L (ed), Pseudomonas methods and protocols, vol 1149 Springer, New York, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siryaporn A, Kuchma SL, O’Toole GA, Gitai Z. 2014. Surface attachment induces Pseudomonas aeruginosa virulence. Proc Natl Acad Sci U S A 111:16860–16865. doi: 10.1073/pnas.1415712111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.