The Sonic Hedgehog (Shh) protein is essential for embryonic patterning, but can contribute to various cancers when the underlying signaling pathway is aberrantly activated. Here, the structure of the human Shh signaling domain is presented in complex with octahedrally coordinated Zn2+ and Mg2+ ions at 1.43 Å resolution that give rise to localized changes. The modes of ion coordination are compared with previous structures determined in isolation or in complex with binding partners, which reveals significant plasticity in the modes of metal coordination that has implications for the design of peptide drugs to repress the signaling pathway.
Keywords: Sonic Hedgehog protein, protein–protein interactions, divalent ion coordination, cell signaling, drug design, peptide inhibitors, Mg2+ ions, Zn2+ ions
Abstract
The Hedgehog pathway is an essential cell-signaling paradigm implicated in cancer tumorigenesis and the developmental disorder holoprosencephaly, making it an attractive target for therapeutic design. The N-terminal domain of the Sonic Hedgehog protein (Shh-N) is the essential signaling molecule in the Hedgehog pathway. In this role Shh-N interacts with its cognate membrane receptor Patched, as well as the regulatory proteins HHIP and CDO, by utilizing interfaces harboring one or more divalent ions. Here, the crystal structure of human Shh-N is presented at 1.43 Å resolution, representing a landmark in the characterization of this protein. The structure reveals that the conserved Zn2+-binding site adopts an atypical octahedral coordination geometry, whereas an adjacent binding site, normally occupied by binuclear Ca2+, has been supplanted by a single octahedrally bound Mg2+. Both divalent sites are compared with those in previous Shh-N structures, which demonstrates a significant degree of plasticity of the Shh-N protein in terms of divalent ion binding. The presence of a high Mg2+ concentration in the crystallization medium appears to have influenced metal loading at both metal ion-binding sites. These observations have technical and design implications for efforts focused on the development of inhibitors that target Shh-N-mediated protein–protein interactions.
1. Introduction
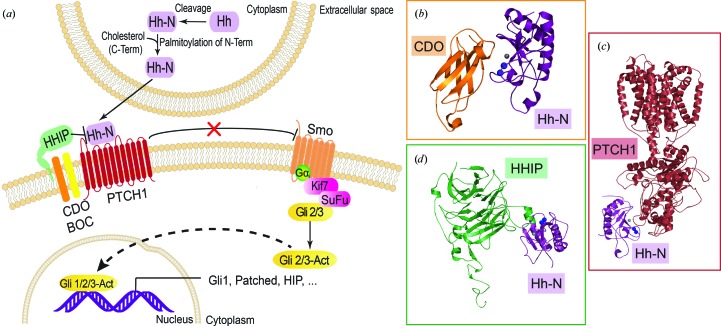
The Hedgehog (Hh) signaling pathway (Fig. 1 ▸ a) has been ascribed many roles in embryonic development, including cell growth, differentiation, tissue patterning and organogenesis (Chiang et al., 1996 ▸; Ingham & McMahon, 2001 ▸; Ingham & Placzek, 2006 ▸; Heussler et al., 2002 ▸). Chemical disruption of Hh signaling was initially discovered to lead to birth defects. One notorious event was the development of cyclopia in Idaho sheep that fed on corn lily, which was subsequently traced to the natural product cyclopamine, a steroidal alkaloid that disrupts Hh signaling through inhibition of the Smoothened receptor (Heretsch et al., 2010 ▸). Pathological dysregulation of the Hedgehog pathway has also been implicated in cancer malignancies in multiple tissues (Rubin & de Sauvage, 2006 ▸; Theunissen & de Sauvage, 2009 ▸; Taipale & Beachy, 2001 ▸; Berman et al., 2003 ▸), making Hedgehog molecular recognition an important entry point for the development of anticancer therapies (Ng & Curran, 2011 ▸; Dlugosz & Talpaz, 2009 ▸). In mammals three Hedgehog (Hh) proteins are known: Indian Hedgehog (Ihh), Desert Hedgehog (Dhh) and Sonic Hedgehog (Shh) (Ingham & McMahon, 2001 ▸). Hedgehog proteins undergo multiple cleavage and modification reactions to yield secreted forms that act in a dose-dependent manner (Ingham & McMahon, 2001 ▸; Briscoe & Thérond, 2013 ▸). Processing entails proteolytic removal of the N-terminal signal sequence and autocleavage mediated by the C-terminal domain, yielding a 19 kDa N-terminal domain (Mann & Beachy, 2004 ▸). This domain is modified by palmitoylation at the N-terminus and cholesterol addition to the C-terminal amino acid (Porter et al., 1996 ▸; Buglino & Resh, 2008 ▸), resulting in the mature signaling protein Shh-N. Similar processing of other Hedgehog N-terminal domains yields the additional functional, secreted signaling ligands Ihh-N and Dhh-N (Lee et al., 1994 ▸; Bürglin, 2008 ▸; Varjosalo & Taipale, 2007 ▸).
Figure 1.
Schematic overview of the Hedgehog signaling pathway and depiction of key regulatory interfaces. (a) The Hedgehog (Hh) protein N-terminal domain is cleaved by the C-terminal domain, yielding Hh-N products (Shh-N, Dhh-N or Ihh-N signified by the Hh protein), inside Hh-producing cells. Addition of cholesterol to the C-terminus and palmitoylation of the N-terminus produces the active, secreted signaling molecule. Shh-N binding to PTCH1 and the co-receptors CDO and BOC alleviates the inhibition of Smoothened (Smo), allowing the dissociation of Gli proteins from the Kif7 and SuFu proteins. Subsequent activation and transport to the nucleus allows the Gli proteins to carry out activation or repression of transcription, including the upregulation of Hh pathway proteins. (b) Cartoon diagram of the quaternary complex between the FNIII domain of co-receptor CDO, a human homologue of Interference Hedgehog (Ihog), bound to Shh-N (PDB entry 3d1m; McLellan et al., 2008 ▸). (c) Cartoon diagram of the quaternary complex of the PTCH1 transmembrane receptor bound to Shh-N (PDB entry 6dmy; Gong et al., 2018 ▸). (d) Cartoon diagram of the quaternary complex of the HHIP membrane protein bound to Shh-N (PDB entry 2wfx; Bishop et al., 2009 ▸).
Hedgehog ligands are well characterized in terms of their binding to the extracellular domain of the transmembrane receptor Patched (PTCH1), as well as the co-receptors CDO and BOC, which are the human homologs of Interference Hedgehog (Ihog; Figs. 1 ▸ b and 1 ▸ c; Carpenter et al., 1998 ▸; Tenzen et al., 2006 ▸). Binding to PTCH1 by an Hh-N ligand leads to a sequence of molecular-signaling events that give rise to an intracellular cascade (Ingham & Placzek, 2006 ▸; Jiang & Hui, 2008 ▸; Wong & Reiter, 2008 ▸). Hh-N binding to PTCH1 results in de-repression of the Smoothened (SMO) receptor and the dissociation of Gli proteins from Kif7 and SuFu (Taipale et al., 2002 ▸; Zhao et al., 2007 ▸; Liem et al., 2009 ▸; Svärd et al., 2006 ▸). Upon activation of the Hh pathway, Gli proteins are differentially phosphorylated as transcriptional activators and repressors controlling the expression of Hh-regulated genes (Fig. 1 ▸ a). Among the activated genes are several Hh pathway genes that include PTCH1 as part of a negative-feedback loop regulating the pathway (Humke et al., 2010 ▸). Mammalian Hedgehog signaling can be regulated negatively by the membrane protein human Hedgehog interacting protein (HHIP), which binds to Hh-N ligands, preventing their interaction with PTCH1 (Fig. 1 ▸ d; Richards & Degnan, 2009 ▸). The latter molecular-recognition event led to the development of macrocyclic peptides that inhibit the Hh pathway (Owens et al., 2017 ▸).
The importance of Hedgehog signaling pathways in human health and disease has stimulated efforts to manipulate Shh-N ligand–receptor interactions (Stanton et al., 2009 ▸; Owens et al., 2017 ▸). Many cancers exhibit abnormal activation by Hh ligands (Heretsch et al., 2010 ▸). As such, efforts to elucidate the three-dimensional structures of Hh ligands continue to be of interest, including the isolated Shh-N domains from mouse and human (Pepinsky et al., 2000 ▸; Hall et al., 1995 ▸), as well as of these ligands in complex with binding partners such as the ectodomain of the PTCH1 cell-surface receptor (Qi, Schmiege, Coutavas, Wang et al., 2018 ▸; Gong et al., 2018 ▸; Qi, Schmiege, Coutavas & Li, 2018 ▸), the conserved, vertebrate-specific inhibitor of Hedgehog signaling, HHIP (Bishop et al., 2009 ▸; Bosanac et al., 2009 ▸), and the third fibronectin type III (FNIII) domain of the cell-surface receptor CDO (McLellan et al., 2008 ▸) (Fig. 1 ▸). An exceptional aspect of the latter interfaces is that they feature direct and indirect contributions from divalent metal ions (McLellan et al., 2008 ▸; Bosanac et al., 2009 ▸). Indeed, Shh-N features a metalloprotease-like fold in which Zn2+ occupies a pseudo-active site with nearby binuclear Ca2+ coordination. These ion sites are located on a protein surface that is accessible to interacting extracellular binding partners but is distal to the modified termini (Bosanac et al., 2009 ▸). The correct protein loading and participation of divalent ions in Shh-N recognition of its cognate ectodomains has implications for the design and screening of molecules to disrupt these interactions and modulate Shh signaling (Owens et al., 2017 ▸).
Here, we report the crystal structure of human Shh-N at 1.43 Å resolution with R work and R free values of 12.4% and 13.9%, respectively, which represents the highest reported resolution for this protein to our knowledge. The overall fold is comparable to prior human and mouse structures, although differences are observed in two loops near the metal ion-coordination sites. Despite the crystallization of Shh-N in the presence of a cyclic peptide designed to bind at the tetrahedral Zn2+-coordination site of the protein (Owens et al., 2017 ▸), we did not observe this inhibitor in the structure. Instead, the Zn2+ ion exhibits octahedral geometry similar to that of Shh-N in complex with CDO (McLellan et al., 2008 ▸), but appears to be distinct from the tetrahedral geometry observed in other isolated Shh-N structures (Pepinsky et al., 2000 ▸; Bishop et al., 2009 ▸; Bosanac et al., 2009 ▸; Hall et al., 1995 ▸). We also observed that the binuclear Ca2+ site was replaced by a single, octahedrally coordinated Mg2+ that has not been described elsewhere. Here, we compare the observed modes of Zn2+ and Mg2+ coordination with prior Zn2+ and Ca2+ ligation by Shh-N in the context of unbound and receptor-bound Shh-N structures. Our observations reveal plasticity at both ion-binding sites, which has implications for the structural analysis of this important molecular-recognition interface and drug design.
2. Materials and methods
2.1. Crystallization and X-ray data collection
Human Shh-N protein was subcloned into pET-42b (Novagen) and expressed as a His6-GST fusion protein by Rosetta(DE3) bacterial overexpression. Recombinant cells were grown in LB medium with kanamycin (50 µg ml−1) and chloramphenicol (34 µg ml−1). At an OD600 of 0.6, the cells were induced with 0.25 mM IPTG and grown for 18 h at 18°C. Following immobilized nickel-affinity chromatography (Invitrogen), the GST-Shh was diluted with buffer (50 mM Tris pH 7.5, 300 mM NaCl, 20 mM imidazole, 0.5 mM EDTA, 1 mM TCEP) to a concentration of 1 mg ml−1. The His6-GST tag was cleaved at 4°C for 16 h by incubation with TEV protease (Sigma) using a 1:200 enzyme:substrate molar ratio. The His6-GST tag and TEV protease were removed by cation-exchange chromatography (GE Healthcare). The resulting protein was exchanged into a buffer comprising 10 mM HEPES pH 7.5, 100 mM NaCl, 0.25 mM TCEP, followed by concentration to 1 mM using centrifugal filtration (Amicon Ultra). The purity of the protein was confirmed by SDS–PAGE. The sample was subjected to crystallization trials against JBScreen JCSG++ (Jena Bioscience GmbH) at 5°C using a Mosquito robot (TTP Labtech). Manual optimization of the initial hits from 24-well plates yielded cubic crystals from hanging-drop vapor-diffusion experiments comprising equal volumes (1 µl) of protein and well solution. The mother liquor comprised 24%(w/v) polyethylene glycol 8000, 0.10 M Tris pH 7.0, 0.24 M MgCl2 and 1 mM L2-m5 peptide (Owens et al., 2017 ▸) dissolved in DMSO. The crystals reached maximum dimensions of 80 × 60 × 60 µm after three weeks and were cryoprotected by 3 min soaks in synthetic mother liquor containing an additional 5–20%(v/v) glycerol. Single cubes were captured in thin nylon loops mounted on 16 mm copper pins (Hampton Research) and plunged into liquid N2 for shipping to the Stanford Synchrotron Radiation Lightsource (SSRL). X-ray data collection was conducted remotely on beamline 12-2 using the Blu-Ice software and the Stanford Auto-Mounter (Soltis et al., 2008 ▸; McPhillips et al., 2002 ▸; Cohen et al., 2002 ▸). The wavelength was 0.9537 Å, Δφ was 0.2° and the exposure time was 0.2 s per frame; diffraction data were recorded at a distance of 300 mm for 900 images on a PILATUS 6M detector (Dectris). X-ray fluorescence scans were conducted at the Zn K edge and were analyzed by CHOOCH (Evans & Pettifer, 2001 ▸). The data-collection strategy was generated using Web-Ice (Soltis et al., 2008 ▸). Diffraction data were reduced with the autoxds script (Gonzalez & Tsai, 2010 ▸) using XDS, POINTLESS, AIMLESS and TRUNCATE to generate intensities and structure-factor amplitudes (Winn et al., 2011 ▸; Kabsch, 2010 ▸). Representative data-collection statistics are provided in Table 1 ▸.
Table 1. Shh-N X-ray data-collection and refinement statistics.
Values in parentheses are for the outer shell.
| Data collection | |
| Diffraction source | Beamline 12-2, SSRL |
| Resolution range (Å) | 37.55–1.43 (1.45–1.43) |
| Wavelength (Å) | 0.9537 |
| Space group | I23 |
| a = b = c (Å) | 118.7 |
| α = β = γ (°) | 90.0 |
| Completeness (%) | 99.9 (99.4) |
| Multiplicity | 18.4 (8.9) |
| R p.i.m. (%) | 2.1 (46.2) |
| R merge (%) | 9.0 (129.6) |
| 〈I/σ(I)〉 | 20.0 (1.5) |
| CC1/2 (%) | 99.9 (59.3) |
| Refinement | |
| No. of reflections, working set | 51261 |
| No. of reflections, test set | 2297 |
| R work/R free | 0.1237/0.1391 |
| No. of non-H atoms | |
| Protein | 1257 |
| Ligand | 23 |
| Water | 256 |
| Total | 1536 |
| Maximum-likelihood coordinate error (Å) | 0.12 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Angles (°) | 0.96 |
| Average B factors (Å2) | |
| Protein | 18 |
| Metal-ion ligands† | 13 |
| DMSO | 68 |
| Glycerol | 71 |
| Cl− | 24 |
| Zn2+/Mg2+ site‡ | 17/12 |
| Mg2+ site | 15 |
| Waters | 33 |
| Ramachandran plot | |
| Most favored (%) | 99 |
| Allowed (%) | 1 |
Includes atoms from amino-acid side chains that coordinate ions at the Zn2+ and Mg2+ sites, as well as inner-sphere waters.
This site was modeled with partial occupancy comprising 0.4 Zn2+ ions and 0.2 Mg2+ions.
2.2. Structure determination and refinement
The Shh-N structure was determined by molecular replacement using Phenix (Adams et al., 2010 ▸; McCoy et al., 2007 ▸) starting from PDB entry 1vhh as a search model (Hall et al., 1995 ▸), which yielded a top translation-function Z-score of 63 and a top log-likelihood gain of 3823. This is the first time that Shh-N has been crystallized in space group I23 to our knowledge. The initial Shh-N model was modified by phenix.autobuild, followed by manual adjustments in Coot (Emsley et al., 2010 ▸) with intervening cycles of phenix.refine (Adams et al., 2010 ▸). This approach converged on R work and R free values of 12.4% and 13.9% at 1.43 Å resolution. Reduced-bias OMIT maps demonstrated atomistic features that helped to define the placement of the divalent metal ions and inner-sphere ligands. Although crystallization occurred in the presence of the L2-m5 peptide, this inhibitor was not observed in the Shh-N crystal structure. In the later stages of refinement, we modeled residual nonprotein electron density that appeared to be consistent with DMSO, glycerol and chloride ions bound at the protein surface. The corresponding temperature factors are higher than those of nearby side chains (Table 1 ▸), indicating that these atoms do not bind with full occupancy. Cα superposition analysis was performed in CCP4i2 (Krissinel, 2012 ▸; Potterton et al., 2018 ▸). All cartoons and schematic diagrams derived from coordinates were generated in PyMOL (Schrödinger). Schematic molecular diagrams were produced in ChemDraw Professional (PerkinElmer). The final refinement statistics are provided in Table 1 ▸. Coordinates and structure-factor amplitudes were deposited in the Protein Data Bank as entry 6pjv.
2.3. Occupancy of the Zn2+ site and supporting analysis
Negative electron density in reduced-bias 2mF o − DF c and mF o − DF c maps at the Zn2+ site supported an ion model with reduced occupancy. This site was modeled best with occupancies of 0.4 for Zn2+ and 0.2 for Mg2+ (discussed below). The resulting ion-occupancy scheme used here was based on multiple refinement trials in which the Zn2+ and Mg2+ ions summed to various partial occupancies at the Zn2+ site. The deciding factor in the choice of occupancies was the identification of B factors for both ions that were comparable with the values observed for the amino-acid ligands (Table 1 ▸), as well as mF o − DF c maps devoid of appreciable positive or negative electron density. Although there are examples of octahedrally coordinated Zn2+ ions in other Shh-N structures with 0.5 occupancy, we considered the prospect that the Zn2+ site comprises a mixture of Zn2+ and Mg2+, since the latter ion was present at 0.24 M concentration in the mother liquor used for flash-cooling. To assess the compatibility of the ligands with both ions, we searched the Cambridge Structural Database (CSD). The results demonstrated that octahedrally coordinated Mg2+ is feasible for a constellation of inner-sphere ligands that includes water, carboxylate and imidazolyl groups. Such complexes were observed in CSD refcodes DEZING, ODENAN, PUNREV, PUHPUE, UDIVEJ and WEZQUO, which were used to calculate ion-coordination distances and angles. Comparable entries retrieved for octahedrally coordinated Zn2+ with water, carboxylate and imidazolyl ligands include BIBBAQ, BIBBEU, COVPOU and DAHREK. Based on our refinement, we conclude that mixed loading of Zn2+ and Mg2+ into the Zn2+ site provides the most parsimonious explanation of why the site exhibits well ordered ligand side chains while accounting for only partial Zn2+ occupancy. Inductively coupled plasma mass spectrometric analysis of human Shh-N samples revealed that they contained substoichiometric levels of Zn2+ (0.4–0.5 equivalents per Shh-N protein) prior to crystallization. No effort was made to reload Zn2+ prior to or during crystallization.
2.4. Isothermal titration calorimetry
Pure Shh-N protein was dialyzed at 4°C using a 3.5 kDa molecular-weight cutoff Slide-A-Lyzer (Thermo) against 4 l buffer comprising 10 mM HEPES pH 7.4, 100 mM NaCl, 5 mM EGTA, 0.5 mM ZnCl2, which was exchanged for fresh buffer after 4 h and allowed to dialyze for an additional 2 h. A second round of dialysis was completed against 4 l buffer comprising 10 mM HEPES pH 7.4, 100 mM NaCl, which was exchanged for fresh buffer after 4 h and allowed to dialyze overnight. Stocks of CaCl2 and MgCl2 were prepared at 1.0 M concentrations in water and 0.2 µm filtered. Divalent cation stocks were diluted using the final (used) dialysis buffer. Measurements were conducted at 20°C on a PEAQ-ITC (Malvern Panalytical). The divalent ion in the syringe was injected into Shh-N located in the sample cell using 19 2.0 µl injections spaced 150 s apart, except for the technical injection of 0.4 µl. Thermograms were analyzed with MicroCal PEAQ-ITC Analysis Software using a one-set-of-sites binding model with a stoichiometry of 2:1. Two identical Ca2+ sites were assumed based on prior Shh-N structures and were used as a constraint in fitting. Experiments were conducted with cell concentrations ranging from 20 to 50 µM Shh-N and syringe concentrations of divalent ions of between 1 and 50 mM. Reproducible results were obtained for Ca2+ at a concentration of 8 mM with a cell concentration of 40–55 µM. Titrations with Mg2+ under comparable conditions yielded no significant heats of binding.
3. Results
3.1. Shh-N structural quality, fold and comparison with other Shh-N domains
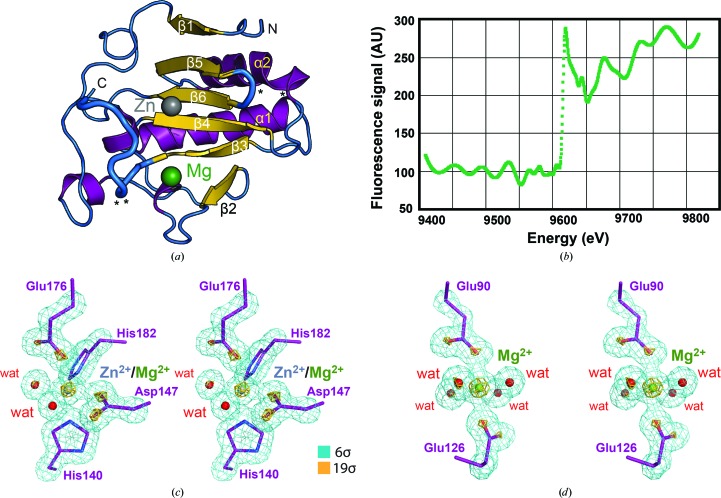
The 1.43 Å resolution structure of Shh-N reported here represents an advance in resolution compared with previous structures of human Shh-N, for which the best observed diffraction resolution (d min) was 1.83 Å (Maun et al., 2010 ▸). The quality of the Shh-N model is indicated by R cryst, R work and R free values of 12.4%, 12.4% and 13.9%, respectively, satisfactory geometry (r.m.s.d.s for bonds and angles of 0.007 Å and 0.96°) and 99% of the backbone dihedral angles in allowed regions of the Ramachandran plot, with no outliers (Table 1 ▸). The global Shh-N fold concurs with previous analyses and can be described as an α/β structure comprising a mixed, six-stranded β-sheet flanked by two major α-helices, two short 310-helices and multiple joining loops and turns (Fig. 2 ▸ a). A comparison with previously characterized human and murine Shh-N models [PDB entries 2wfx, 2wg4 (Bishop et al., 2009 ▸), 3d1m (McLellan et al., 2008 ▸), 3ho5 (Bosanac et al., 2009 ▸), 3m1n (Pepinsky et al., 2000 ▸), 3mxw (Maun et al., 2010 ▸), 1vhh (Hall et al., 1995 ▸), 4c4n (Whalen et al., 2013 ▸), 6oev (Qi, Schmiege, Coutavas & Li, 2018 ▸) and 6dmy (Gong et al., 2018 ▸)], as well as an Ihh-N model (PDB entry 3k7g; Y.-X. He, Y. Kang, W. J. Zhang, J. Yu, G. Ma & C.-Z. Zhou, unpublished work), reveals that the new Shh-N structure here varies at distinct locations from previous models based on r.m.s. Cα backbone deviations, which fall within a range from 0.5 to 1.2 Å, significantly higher than the average atomic coordinate error of 0.12 Å (Table 1 ▸). Notable deviations are observed at Glu130 and Asp131, which lie on the surface of the protein and make crystal contacts with the side chains of Lys178′ and Arg155′ in the context of the unique cubic crystal-packing environment (Fig. 2 ▸ a). Three loop regions that show significant variation between known Shh-N structures that cannot be explained solely by crystal contacts include Gly127–Asp129, Gly132–Glu137 and Val173–Cys183 (β5 and β6; Fig. 2 ▸ a), which flank the metal-binding sites. Variation in the latter ‘pseudo-active-site’ region confirms the known flexibility of the structural elements that surround and compose the metal-binding sites on the Shh-N surface.
Figure 2.
Overall fold of human Sonic Hedgehog N-terminal domain (Shh-N) and evidence for divalent-ion coordination. (a) Ribbon diagram depicting the Shh-N α/β fold with the surface ion-binding pockets flanked by a series of β-sheets (yellow), helices (purple) and loops (blue). Spheres represent Zn2+ (gray) and Mg2+ (green) binding sites within the pseudo-metalloprotease active site. Loops with higher Cα r.m.s.d.s compared with previous Shh-N structures are depicted with thicker diameters. The side chains of Glu130 and Asp131 and of Lys178 and Arg155 are involved in crystal contacts that affect the loop conformation and are denoted by asterisks (*). (b) Plot of an X-ray fluorescence scan from a single crystal indicating an absorption edge at 1.2823 Å (9667 eV), consistent with the presence of Zn2+. (c) Stereoview of a reduced-bias simulated-annealing OMIT map at contour levels of 6σ (teal) and 19σ (gold) that depicts the final model for the Shh-N amino-acid ligands (purple), coordinating waters (red), Mg2+ (green) and Zn2+ (gray). These coordinates were removed from the electron-density calculation to demonstrate the existence of these features in the experimental data, while corroborating the octahedral coordination geometry at the Zn2+ site. (d) Stereoview of a reduced-bias simulated-annealing OMIT map at contour levels of 6σ (teal) and 19σ (gold) depicting the final model of the Shh-N amino-acid ligands (purple), coordinating waters (red) and Mg2+ (green). These coordinates were removed from the electron-density calculation to demonstrate the existence of these features in the experimental data, while corroborating the octahedral ion-coordination geometry and the placement of a single Mg2+ ion at a site typically occupied by binuclear Ca2+.
3.2. Octahedral coordination of Zn2+ on the Shh-N surface
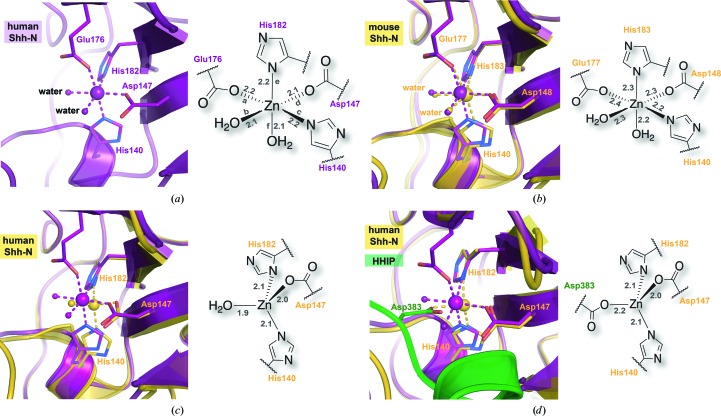
The available human and mouse Shh-N structures show coordination to Zn2+ at a site located adjacent to strands β4 and β6 and the β3–β4 loop (Fig. 2 ▸ a). X-ray fluorescence scans of crystals indicated an absorption edge at 1.2823 Å (9667 eV), consistent with Zn2+ (Fig. 2 ▸ b; Xie et al., 2004 ▸). However, efforts to fit a full-occupancy Zn2+ led to negative electron density in Fourier maps, suggesting that the ion is not bound with full occupancy. This point is corroborated by the poor quality of anomalous difference maps and inductively coupled plasma mass spectrometry. Reduced-bias simulated-annealing OMIT maps calculated in the absence of an ion and ligands of the inner sphere support the presence of an ion model and corroborate its octahedral coordination geometry (Fig. 2 ▸ c). This map shows electron density at contour levels exceeding 27σ, in which the largest peak is centered on the ion. Accordingly, we modeled a Zn2+ ion with a partial occupancy of 0.4, as well as an Mg2+ ion with a partial occupancy of 0.2, consistent with the observed Zn2+ anomalous signal and the high levels of MgCl2 in the crystallization mother liquor. The mixed-occupancy Zn2+/Mg2+ ion exhibits regular octahedral coordination geometry based on bond distance and angle measurements to inner-sphere ligands, which produced average values of 2.2 ± 0.1 Å and 89.9 ± 4.2° (Fig. 3 ▸ a).
Figure 3.
Ball-and-stick and schematic depictions of ion coordination at the Zn2+ site of Shh-N crystal structures. (a) The octahedral coordination geometry of the current study depicting inner-sphere Zn2+/Mg2+ coordination by the side chains of His140, Asp147, Glu176 and His182 and two water molecules. The distances between each ligand and divalent ion are shown in Å. The corresponding line segments between ligands and ions are labeled with letters to describe the associated bond angles: ∠ab = 85.8°, ∠ad = 92.6°, ∠ae = 90.8°, ∠af = 87.4°, ∠bc = 89.8°, ∠be = 85.0°, ∠bf = 84.4°, ∠cd = 91.7°, ∠ce = 89.6°, ∠cf = 91.4°, ∠de = 99.9° and ∠df = 90.7°. Here and elsewhere, schematic diagrams of the coordination sphere are depicted on the right. (b) Octahedral Zn2+-coordination geometry from a previously described co-crystal structure of mouse Shh-N (residue number +1 compared with human structures) in complex with the third fibronectin-type III domain of CDO (gold; PDB entry 3d1m; McLellan et al., 2008 ▸) superimposed (Cα r.m.s.d. of 0.54 Å) on the current model (purple). (c) Tetrahedral Zn2+ coordination of a previously reported human Shh-N structure (gold; PDB 3mxw; Maun et al., 2010 ▸) with a water molecule as the fourth ligand in the tetrahedral coordination sphere superimposed (Cα r.m.s.d. of 0.64 Å) on the current model (purple). (d) Tetrahedral Zn2+ coordination of a previously reported human Shh-N structure (gold; PDB entry 3ho5; Bosanac et al., 2009 ▸) in complex with HHIP (green) that contributes residue Asp383 to complete the tetrahedral coordination sphere superimposed (Cα r.m.s.d. of 0.63 Å) on the current model (purple).
The presence of mixed-occupancy Zn2+/Mg2+ at the Zn2+ site of Shh-N best explains the observed octahedral coordination geometry and electron density. Precedents for octahedral Mg2+ coordinated by carboxylate and imidazole ligands come from small-molecule structures (Gryz et al., 2007 ▸; Stadie et al., 2007 ▸; Liu & Liu, 2010 ▸; Ye et al., 1998 ▸; Srinivasan et al., 2007 ▸). Likewise, small-molecule examples are available for octahedral Zn2+ coordination arising from two imidazole and two carboxylate ligands (Carballo et al., 2004 ▸; Li et al., 2014 ▸; Shimizu et al., 2004 ▸). Remarkably, these Mg2+ and Zn2+ ions exhibit nearly identical octahedral coordination distances of 2.12 ± 0.06 and 2.12 ± 0.05 Å, with average bond angles of 89.5 and 89.8°. Given the compatibility of the ligands and the geometry with both Mg2+and Zn2+, it is reasonable that the well ordered electron density observed here for Shh-N is the result of a mixture of these ions. Precedents exist for mixed-ion loading in Zn2+ enzymes that could not be defined solely by crystallography (Wedekind et al., 1995 ▸; Ruzicka et al., 1995 ▸).
The Zn2+ site receives coordination from the side chains of His140, Asp147, Glu176 and His182 and two water molecules (Fig. 3 ▸ a). Precedents for octahedral Zn2+ coordination geometry in Shh-N come from the previously described co-crystal structure of murine Shh-N in complex with the third fibronectin type III domain of CDO (Fig. 3 ▸ b; PDB entry 3d1m; McLellan et al., 2008 ▸). Like the structure reported here, the Shh-N–CDO complex was also modeled with a partially occupied Zn2+ (0.5), although no Mg2+ was present in the mother liquor. The Shh-N domain of the latter complex superimposes well on the human Shh-N structure reported here (Cα r.m.s.d. of 0.54 Å), especially considering that the murine Shh-N protein was crystallized in a quaternary complex with CDO, whereas the human Shh-N protein is in the unbound state. Analysis of other known Shh-N structures indicates that the Zn2+ site coordination adopts a distorted trigonal planar (not shown; Pepinsky et al., 2000 ▸) or tetrahedral (Figs. 3 ▸ c and 3 ▸ d) geometry, consistent with its description as a pseudo-metalloprotease active site (Bosanac et al., 2009 ▸). In all cases of tetrahedral geometry that we have examined, His140, Asp147 and His182 contribute three of the coordinating residues from Shh-N, whereas the fourth coordination site is occupied by a water molecule in structures of the isolated Shh-N domain (Fig. 3 ▸ c). The plasticity of the Zn2+-coordination site is underscored not only by its ability to adopt multiple coordination geometries, which appear to bind other ions, but also by its ability to integrate ligands from other proteins into its coordination sphere. In this respect, the bound water provides a nexus for displacement and engagement by residues contributed by binding partners that coordinate directly to Zn2+ (Bosanac et al., 2009 ▸; Pepinsky et al., 2000 ▸; Maun et al., 2010 ▸). For example, HHIP contributes Asp383 to supplant water, yielding tetrahedral coordination in the quaternary complex (Fig. 3 ▸ d). Despite a defined role in protein–protein interactions in some complexes, other interfaces do not utilize Zn2+ coordination, and Zn2+ removal has no effect in terms of structure or stability (Bosanac et al., 2009 ▸).
3.3. Octahedral coordination by Mg2+ replaces binuclear Ca2+ on the Shh-N surface
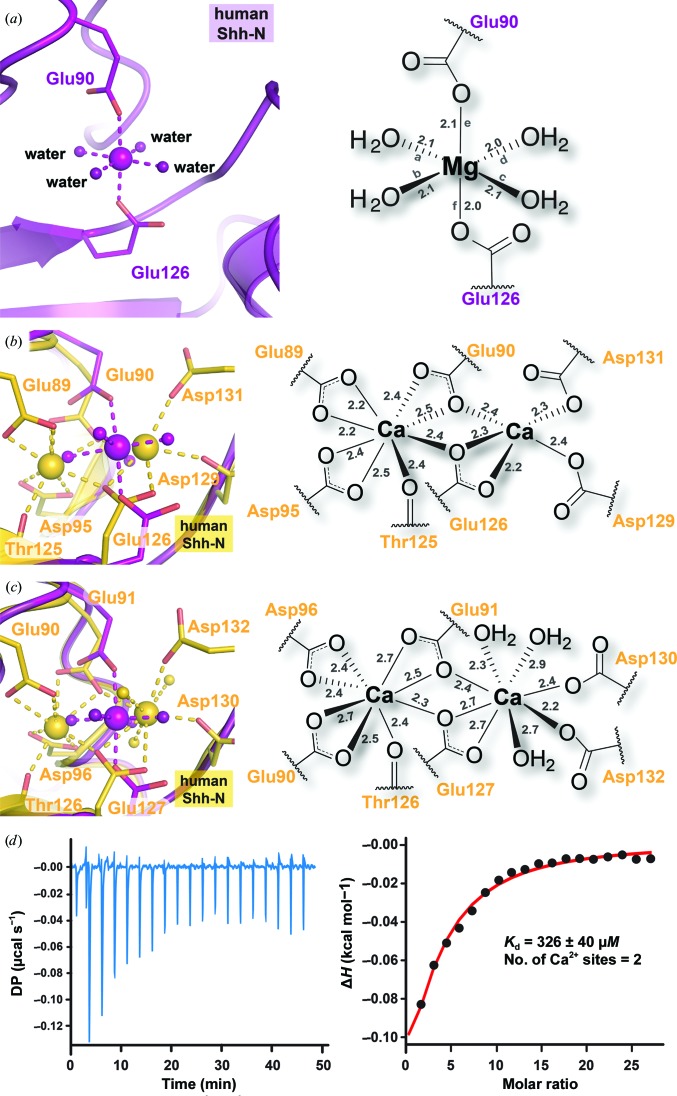
A second metal-binding site lies adjacent to the Zn2+ ion site in the pseudo active site between the β2–α1 and β3–β4 loops (Fig. 2 ▸ a). Owing to the high concentration of MgCl2 in the crystallization medium, the octahedral geometry of the ion, the hardness of the ligands and coordination distances, this site is consistent with Mg2+. Reduced-bias simulated-annealing OMIT maps calculated in the absence of the metal and inner-sphere coordination model corroborate the placement of the ion (Fig. 2 ▸ d). These maps further reveal that the Mg2+ ion is detectable at contour levels exceeding 31σ. Coordination is achieved by two axial carboxylate groups derived from Glu90 and Glu126 with four equatorial water molecules that complete the coordination sphere (Fig. 4 ▸ a). The coordination distances range from 2.0 to 2.1 Å, consistent with the oxygen–Mg2+ distances of 2.064 ± 0.025 Å observed in high-resolution small-molecule structures (Wedekind, 2011 ▸). The bond angles to adjacent ligands show an average value of 90.0 ± 2.0°, consistent with regular octahedral geometry (Fig. 4 ▸ a). The Mg2+ ion is centered in the Ca2+-binding pocket, although only a single Mg2+ ion occupies an otherwise binuclear metal-coordination site (Figs. 4 ▸ b and 4 ▸ c). In this manner, the carboxylate groups used to coordinate Mg2+ continue to point inwards toward a cavity on the Shh-N surface, maintaining the side-chain rotamers in conformations comparable to those used to coordinate the two Ca2+ ions.
Figure 4.
Ball-and-stick and schematic depictions of Mg2+ and Ca2+ ion coordination in Shh-N crystal structures. (a) The observed Mg2+ octahedral coordination geometry occurs in the context of the binuclear Ca2+-binding site. The new structure (purple) coordinates to Mg2+ by the side chains of Glu90 and Glu126 and four water molecules. Distances between each ligand and divalent ion are shown in Å. The corresponding line segments between ligand atoms and ions are labeled with letters to describe the associated bond angles: ∠ab = 89.9°, ∠ad = 87.0°, ∠ae = 88.3°, ∠af = 89.2°, ∠bc = 91.2°, ∠be = 89.9°, ∠bf = 86.4°, ∠cd = 91.9°, ∠ce = 92.6°, ∠cf = 90.0°, ∠de = 92.1° and ∠df = 91.5°. (b) The Ca2+ binuclear coordination site observed in the previously reported human Shh-N structure (gold; PDB entry 3ho5; Bosanac et al., 2009 ▸) superimposed (Cα r.m.s.d. of 0.63 Å) on the current Mg2+ coordination site (purple). Coordinating residues are indicated, but water molecules were not modeled owing to limited resolution. (c) The Ca2+ binuclear coordination site of a previously reported mouse (residue number +1 compared with human structures) Shh-N structure (gold; PDB entry 3d1m; McLellan et al., 2008 ▸) superimposed (Cα r.m.s.d. of 0.54 Å) on the current Mg2+ coordination site. Coordinating residues are indicated along with water molecules to complete each Ca2+ coordination sphere. (d) Binding analysis of Ca2+ by ITC using a one set of sites binding model with a stoichiometry of 2:1 provides an estimate of affinity assuming two identical ions.
The location and mode of Mg2+ ion coordination differ from previously determined Shh-N structures, in which two Ca2+ ions coordinate most frequently to six carboxylate groups provided by Glu89, Glu90, Asp95, Glu126, Asp129 and Asp131, as well as the carbonyl of Thr125, located in a negatively charged pocket on the protein surface. Water molecules complete the Ca2+ coordination spheres. In previously published Shh-N structures in complex with HHIP (PDB entry 3ho5) and the FNIII domain of CDO (PDB entry 3d1m) the traditionally observed Ca2+ coordination is present (Figs. 4 ▸ b and 4 ▸ c; McLellan et al., 2008 ▸; Bosanac et al., 2009 ▸). Despite their presence in Shh-N complex structures, the Ca2+ ions do not directly contribute to quaternary complex interactions. The apparent K d of Ca2+ dissociation from Shh-N was estimated to be >100 µM (McLellan et al., 2008 ▸). Using isothermal titration calorimetry, we estimated an average K d value of 357 ± 43 µM, assuming two identical Ca2+ binding sites per human Shh-N domain, which provided the best binding model for the titration data (Fig. 4 ▸ d). In contrast, a comparable analysis of Mg2+ showed no significant heats of binding, implying that Mg2+ coordinates with an appreciably lower affinity; this is in agreement with the structure, in which only two amino acids serve as ligands (Fig. 4 ▸ a). The weak binding of Ca2+ provides a plausible explanation for the heterogeneous modes of Ca2+ binding among Shh-N structures, as well as the observation that Ca2+ readily dissociates during protein purification. Previous structures lacking Ca2+ showed that the structure remains well ordered without these ions and that coordination residues such as Glu90 point outwards into the solvent (Bishop et al., 2009 ▸). These observations are consistent with the finding here that Mg2+ can be introduced into the Ca2+ pocket at millimolar concentrations (i.e. a level tenfold greater than the K d), which alters the local loop conformations and the positions of metal ligands (Figs. 4 ▸ b and 4 ▸ c) without changing the overall fold.
4. Discussion
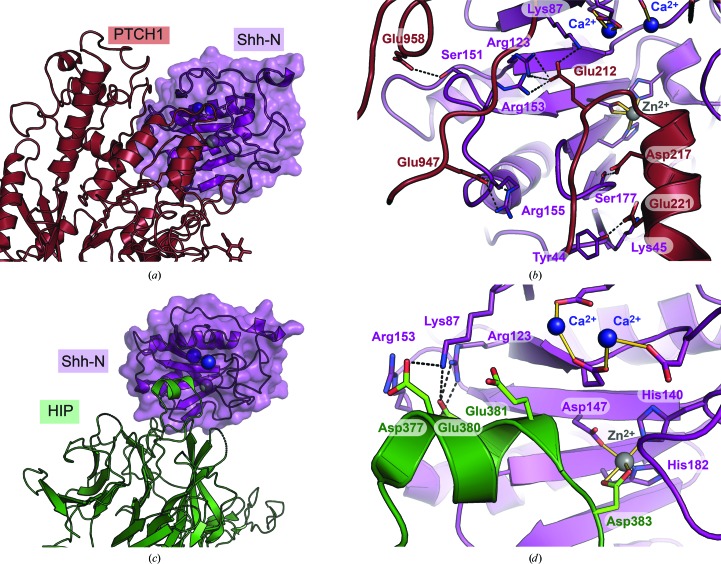
Understanding the Shh-N metal-loading requirements and the plasticity of the divalent-ion coordination sites is significant in the context of understanding the underlying basis of protein–protein interactions that are relevant for therapeutic development (Heretsch et al., 2010 ▸). Indeed, Shh-N has multiple interacting partners with various modes of binding to its surface (Fig. 1 ▸). This diversity in protein-binding sites provides an opportunity to generate inhibitors that target Shh-N, which is tethered on the outer membrane of the cell as a consequence of the cholesterol modification (Jeong & McMahon, 2002 ▸). Structural information derived from the current and previous structures provides a deeper understanding of Shh-N binding to ions and has implications for the design and screening of inhibitors of this protein. Previous structures of Shh-N in complex with the surface receptor Patched 1 (PTCH1) and Hedgehog interaction protein (HHIP; Figs. 1 ▸ c and 1 ▸ d) revealed key information about the inhibitory function of HHIP (Gong et al., 2018 ▸; Bishop et al., 2009 ▸; Qian et al., 2019 ▸). Notably, the structure of Shh-N in complex with PTCH1 shows an extensive quaternary interface that is supported by a series of specific interactions between acidic and basic residues contributed by PTCH1 and Shh-N (Figs. 5 ▸ a and 5 ▸ b). Here, Ca2+ appears to prevent electrostatic repulsion between the two proteins. Despite this quaternary interaction, many accessible pockets remain on the surface of Shh-N near the metal-binding sites. In contrast, the Shh-N inhibitor HHIP exhibits a much less extensive quaternary interface that binds by inserting a β-propeller loop into the Zn2+-binding cavity of Shh-N (Fig. 5 ▸ c). The interface utilizes Asp383 of HHIP to make an inner-sphere contact with Zn2+ of Shh-N to complete its coordination sphere (Fig. 5 ▸ d). In this manner, the coordination of HHIP residues deep within the metal-binding pocket provides a means of accessing the Shh-N surface and stably disrupting PTCH1 binding by steric overlap. Notably, mutation or chelation of the Ca2+ site resulted in a tenfold decrease in Shh-N binding despite the lack of direct interaction between HHIP and the Ca2+ ions, once again demonstrating the importance of both Shh-N metal-coordination sites as determinants of molecular recognition (McLellan et al., 2008 ▸).
Figure 5.
Global and close-up views of key protein–protein interfaces involved in Hedgehog signaling. (a) Quaternary structure of the extracellular domain of PTCH1 engaged at the metal-bound surface of Shh-N (PDB entry 6dmy; Gong et al., 2018 ▸). (b) Close-up view of (a) showing human Shh-N interactions with PTCH1. Amino acids involved in formation of the complex are depicted as sticks (oxygen, red; nitrogen, blue) with putative hydrogen bonds indicated by dashed lines. Interactions with metal ions are indicated by solid gold lines. (c) Quaternary structure of HHIP engaged at the metal-bound surface of Shh-N (PDB entry 2wfx; Bishop et al., 2009 ▸). (d) Close-up view of (c) depicting mouse Shh-N (residue numbering adjusted by −1 for ease of comparison with human structures) interactions with HHIP. Amino acids involved in complex formation are depicted as sticks (oxygen, red; nitrogen, blue) with putative hydrogen bonds indicated by broken lines.
At present, macrocyclic peptides are being developed that mimic the inhibitory function of HHIP (Owens et al., 2017 ▸). The information gained by the current Shh-N structure reveals how the plasticity of the metal-binding site can be used in inhibitor design and screening. Because the binding of HHIP depends on coordination to Zn2+, HHIP peptide analogs have been designed to exploit the tetrahedral coordination geometry at the protein–protein interface (see, for example, Fig. 5 ▸ d; Owens et al., 2017 ▸). Based on the present findings, we envision the development of peptide mimics that can utilize the expanded octahedral coordination capacity of Zn2+ observed here (Figs. 3 ▸ a and 3 ▸ b). The observed plasticity of the Ca2+-binding site also suggests that additional ligands for Ca2+ could be installed into HHIP peptide mimics to potentially enhance affinity and/or specificity (for example, at a position equivalent to Glu381; Fig. 5 ▸ d). On a technical note, the results here demonstrate that Shh-N can benefit from Zn2+ and Ca2+ supplementation after purification, especially if these ions are desired for inhibitor-binding studies, screening experiments or crystallization trials. Notably, the millimolar concentrations of multivalent ions present in typical commercial broad screens (Fazio et al., 2014 ▸) can supplant Ca2+ or can co-occupy substoichiometrically bound ion sites, as observed here for Mg2+. These effects can lead to an interaction interface that differs from the physiologically relevant one.
5. Conclusions
The high-resolution structure of Shh-N reported here provides insight into the plasticity of Shh-N divalent ion-binding sites. This variation is exemplified by the ability of metal ligands to adopt myriad geometries that accommodate multiple types of metals. Documenting both the atypical octahedral coordination state of Zn2+ (and its mixed occupancy) as well as the presence of Mg2+ in the Ca2+-binding pocket contributes further to our understanding of the Shh-N binding surface. The diversity of quaternary complexes formed with Shh-N provides potential avenues for therapeutic targeting of Hedgehog signaling. The detailed view of human Shh-N gained here and in previous structural studies builds a foundation for the design and development of chemical agents useful for interfering with Shh-N-mediated signaling.
Supplementary Material
PDB reference: human Sonic Hedgehog N-terminal signaling domain, 6pjv
Acknowledgments
We thank members of the Wedekind laboratory for technical assistance and Professor Dirk Bohmann for helpful discussions. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences.
Funding Statement
This work was funded by National Institutes of Health, National Institute of General Medical Sciences grant R01GM123864 to Joseph Wedekind. National Cancer Institute grant R21CA187502 to Rudi Fasan.
References
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. [DOI] [PMC free article] [PubMed]
- Berman, D. M., Karhadkar, S. S., Maitra, A., Montes De Oca, R., Gerstenblith, M. R., Briggs, K., Parker, A. R., Shimada, Y., Eshleman, J. R., Watkins, D. N. & Beachy, P. A. (2003). Nature (London), 425, 846–851. [DOI] [PubMed]
- Bishop, B., Aricescu, A. R., Harlos, K., O’Callaghan, C. A., Jones, E. Y. & Siebold, C. (2009). Nature Struct. Mol. Biol. 16, 698–703. [DOI] [PMC free article] [PubMed]
- Bosanac, I., Maun, H. R., Scales, S. J., Wen, X., Lingel, A., Bazan, J. F., de Sauvage, F. J., Hymowitz, S. G. & Lazarus, R. A. (2009). Nature Struct. Mol. Biol. 16, 691–697. [DOI] [PubMed]
- Briscoe, J. & Thérond, P. P. (2013). Nature Rev. Mol. Cell Biol. 14, 416–429. [DOI] [PubMed]
- Buglino, J. A. & Resh, M. D. (2008). J. Biol. Chem. 283, 22076–22088. [DOI] [PMC free article] [PubMed]
- Bürglin, T. R. (2008). Genome Biol. 9, 241. [DOI] [PMC free article] [PubMed]
- Carballo, R., Castiñeiras, A., Covelo, B., García-Martínez, E., Niclós, J. & Vázquez-López, E. M. (2004). Polyhedron, 23, 1505–1518.
- Carpenter, D., Stone, D. M., Brush, J., Ryan, A., Armanini, M., Frantz, G., Rosenthal, A. & de Sauvage, F. J. (1998). Proc. Natl Acad. Sci. USA, 95, 13630–13634. [DOI] [PMC free article] [PubMed]
- Chiang, C., Litingtung, Y., Lee, E., Young, K. E., Corden, J. L., Westphal, H. & Beachy, P. A. (1996). Nature (London), 383, 407–413. [DOI] [PubMed]
- Cohen, A. E., Ellis, P. J., Miller, M. D., Deacon, A. M. & Phizackerley, R. P. (2002). J. Appl. Cryst. 35, 720–726. [DOI] [PMC free article] [PubMed]
- Dlugosz, A. A. & Talpaz, M. (2009). N. Engl. J. Med. 361, 1202–1205. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, G. & Pettifer, R. F. (2001). J. Appl. Cryst. 34, 82–86.
- Fazio, V. J., Peat, T. S. & Newman, J. (2014). Acta Cryst. F70, 1303–1311. [DOI] [PMC free article] [PubMed]
- Gong, X., Qian, H., Cao, P., Zhao, X., Zhou, Q., Lei, J. & Yan, N. (2018). Science, 361, eaas8935. [DOI] [PubMed]
- Gonzalez, A. & Tsai, Y. (2010). autoxds. http://smb.slac.stanford.edu/facilities/software/xds/#autoxds_script.
- Gryz, M., Starosta, W. & Leciejewicz, J. (2007). J. Coord. Chem. 60, 539–546.
- Hall, T. M., Porter, J. A., Beachy, P. A. & Leahy, D. J. (1995). Nature (London), 378, 212–216. [DOI] [PubMed]
- Heretsch, P., Tzagkaroulaki, L. & Giannis, A. (2010). Angew. Chem. Int. Ed. 49, 3418–3427. [DOI] [PubMed]
- Heussler, H. S., Suri, M., Young, I. D. & Muenke, M. (2002). Arch. Dis. Child. 86, 293–296. [DOI] [PMC free article] [PubMed]
- Humke, E. W., Dorn, K. V., Milenkovic, L., Scott, M. P. & Rohatgi, R. (2010). Genes Dev. 24, 670–682. [DOI] [PMC free article] [PubMed]
- Ingham, P. W. & McMahon, A. P. (2001). Genes Dev. 15, 3059–3087. [DOI] [PubMed]
- Ingham, P. W. & Placzek, M. (2006). Nature Rev. Genet. 7, 841–850. [DOI] [PubMed]
- Jeong, J. & McMahon, A. P. (2002). J. Clin. Invest. 110, 591–596. [DOI] [PMC free article] [PubMed]
- Jiang, J. & Hui, C.-C. (2008). Dev. Cell, 15, 801–812. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Krissinel, E. (2012). J. Mol. Biochem. 1, 76–85. [PMC free article] [PubMed]
- Lee, J. J., Ekker, S. C., von Kessler, D. P., Porter, J. A., Sun, B. I. & Beachy, P. A. (1994). Science, 266, 1528–1537. [DOI] [PubMed]
- Li, B., Fan, L.-M., Song, W.-K., Liu, X.-Z. & Zhang, X.-T. (2014). Jiegou Huaxue, 33, 1311.
- Liem, K. F. Jr, He, M., Ocbina, P. J. & Anderson, K. V. (2009). Proc. Natl Acad. Sci. USA, 106, 13377–13382. [DOI] [PMC free article] [PubMed]
- Liu, X.-Y. & Liu, L.-H. (2010). Acta Cryst. E66, m305.
- Mann, R. K. & Beachy, P. A. (2004). Annu. Rev. Biochem. 73, 891–923. [DOI] [PubMed]
- Maun, H. R., Wen, X., Lingel, A., de Sauvage, F. J., Lazarus, R. A., Scales, S. J. & Hymowitz, S. G. (2010). J. Biol. Chem. 285, 26570–26580. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McLellan, J. S., Zheng, X., Hauk, G., Ghirlando, R., Beachy, P. A. & Leahy, D. J. (2008). Nature (London), 455, 979–983. [DOI] [PMC free article] [PubMed]
- McPhillips, T. M., McPhillips, S. E., Chiu, H.-J., Cohen, A. E., Deacon, A. M., Ellis, P. J., Garman, E., Gonzalez, A., Sauter, N. K., Phizackerley, R. P., Soltis, S. M. & Kuhn, P. (2002). J. Synchrotron Rad. 9, 401–406. [DOI] [PubMed]
- Ng, J. M. & Curran, T. (2011). Nature Rev. Cancer, 11, 493–501. [DOI] [PMC free article] [PubMed]
- Owens, A. E., de Paola, I., Hansen, W. A., Liu, Y. W., Khare, S. D. & Fasan, R. (2017). J. Am. Chem. Soc. 139, 12559–12568. [DOI] [PMC free article] [PubMed]
- Pepinsky, R. B., Rayhorn, P., Day, E. S., Dergay, A., Williams, K. P., Galdes, A., Taylor, F. R., Boriack-Sjodin, P. A. & Garber, E. A. (2000). J. Biol. Chem. 275, 10995–11001. [DOI] [PubMed]
- Porter, J. A., Young, K. E. & Beachy, P. A. (1996). Science, 274, 255–259. [DOI] [PubMed]
- Potterton, L., Agirre, J., Ballard, C., Cowtan, K., Dodson, E., Evans, P. R., Jenkins, H. T., Keegan, R., Krissinel, E., Stevenson, K., Lebedev, A., McNicholas, S. J., Nicholls, R. A., Noble, M., Pannu, N. S., Roth, C., Sheldrick, G., Skubak, P., Turkenburg, J., Uski, V., von Delft, F., Waterman, D., Wilson, K., Winn, M. & Wojdyr, M. (2018). Acta Cryst. D74, 68–84. [DOI] [PMC free article] [PubMed]
- Qi, X., Schmiege, P., Coutavas, E. & Li, X. (2018). Science, 362, eaas8843. [DOI] [PMC free article] [PubMed]
- Qi, X., Schmiege, P., Coutavas, E., Wang, J. & Li, X. (2018). Nature (London), 560, 128–132. [DOI] [PMC free article] [PubMed]
- Qian, H., Cao, P., Hu, M., Gao, S., Yan, N. & Gong, X. (2019). Nature Commun. 10, 2320. [DOI] [PMC free article] [PubMed]
- Richards, G. S. & Degnan, B. M. (2009). Cold Spring Harb. Symp. Quant. Biol. 74, 81–90. [DOI] [PubMed]
- Rubin, L. L. & de Sauvage, F. J. (2006). Nature Rev. Drug Discov. 5, 1026–1033. [DOI] [PubMed]
- Ruzicka, F. J., Wedekind, J. E., Kim, J., Rayment, I. & Frey, P. A. (1995). Biochemistry, 34, 5610–5617. [DOI] [PubMed]
- Shimizu, E., Kondo, M., Fuwa, Y., Sarker, R. P., Miyazawa, M., Ueno, M., Naito, T., Maeda, K. & Uchida, F. (2004). Inorg. Chem. Commun. 7, 1191–1194.
- Soltis, S. M., Cohen, A. E., Deacon, A., Eriksson, T., González, A., McPhillips, S., Chui, H., Dunten, P., Hollenbeck, M., Mathews, I., Miller, M., Moorhead, P., Phizackerley, R. P., Smith, C., Song, J., van dem Bedem, H., Ellis, P., Kuhn, P., McPhillips, T., Sauter, N., Sharp, K., Tsyba, I. & Wolf, G. (2008). Acta Cryst. D64, 1210–1221. [DOI] [PMC free article] [PubMed]
- Srinivasan, B. R., Sawant, J. V., Näther, C. & Bensch, W. (2007). J. Chem. Sci. 119, 243–252.
- Stadie, N. P., Sanchez-Smith, R. & Groy, T. L. (2007). Acta Cryst. E63, m2153–m2154.
- Stanton, B. Z., Peng, L. F., Maloof, N., Nakai, K., Wang, X., Duffner, J. L., Taveras, K. M., Hyman, J. M., Lee, S. W., Koehler, A. N., Chen, J. K., Fox, J. L., Mandinova, A. & Schreiber, S. L. (2009). Nature Chem. Biol. 5, 154–156. [DOI] [PMC free article] [PubMed]
- Svärd, J., Heby-Henricson, K., Henricson, K. H., Persson-Lek, M., Rozell, B., Lauth, M., Bergström, A., Ericson, J., Toftgård, R. & Teglund, S. (2006). Dev. Cell, 10, 187–197. [DOI] [PubMed]
- Taipale, J. & Beachy, P. A. (2001). Nature (London), 411, 349–354. [DOI] [PubMed]
- Taipale, J., Cooper, M. K., Maiti, T. & Beachy, P. A. (2002). Nature (London), 418, 892–897. [DOI] [PubMed]
- Tenzen, T., Allen, B. L., Cole, F., Kang, J.-S., Krauss, R. S. & McMahon, A. P. (2006). Dev. Cell, 10, 647–656. [DOI] [PubMed]
- Theunissen, J. W. & de Sauvage, F. J. (2009). Cancer Res. 69, 6007–6010. [DOI] [PubMed]
- Varjosalo, M. & Taipale, J. (2007). J. Cell Sci. 120, 3–6. [DOI] [PubMed]
- Wedekind, J. E. (2011). Met. Ions Life Sci. 9, 299–345. [PubMed]
- Wedekind, J. E., Frey, P. A. & Rayment, I. (1995). Biochemistry, 34, 11049–11061. [DOI] [PubMed]
- Whalen, D. M., Malinauskas, T., Gilbert, R. J. C. & Siebold, C. (2013). Proc. Natl Acad. Sci. USA, 110, 16420–16425. [DOI] [PMC free article] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
- Wong, S. Y. & Reiter, J. F. (2008). Curr. Top. Dev. Biol. 85, 225–260. [DOI] [PMC free article] [PubMed]
- Xie, K., Sowden, M. P., Dance, G. S., Torelli, A. T., Smith, H. C. & Wedekind, J. E. (2004). Proc. Natl Acad. Sci. USA, 101, 8114–8119. [DOI] [PMC free article] [PubMed]
- Ye, B.-H., Mak, T., Williams, I. D. & Li, X. (1998). J. Chem. Soc. Dalton Trans., pp. 1935–1936.
- Zhao, Y., Tong, C. & Jiang, J. (2007). Nature (London), 450, 252–258. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: human Sonic Hedgehog N-terminal signaling domain, 6pjv