Abstract
Gastric cancer (GC) is a serious health problem worldwide. The potential involvement of long noncoding RNAs in GC progression remains largely unexplored. Here, we identified a novel long noncoding RNA referred to as onclncRNA-626 (oncogenic lncRNA RP11-626H12.3), which was highly upregulated in GC tissues. The high expression levels of onclncRNA-626 in GC patients predicted poor prognoses. Functional assays indicated that onclncRNA-626 could promote the proliferation and metastasis of GC cells in vitro and in vivo. In exploring the molecular mechanisms guiding these functions, we found that onclncRNA-626 specifically interacted with serine- and arginine-rich splicing factor 1 (SRSF1) and increased its stability. SRSF1 was upregulated in GC tissues and correlated with onclncRNA-626 expression and patient survival. Furthermore, RNA-seq data revealed that onclncRNA-626 affected multiple signaling pathways, including the p53 signaling pathway. Rescue experiments showed that onclncRNA-626 probably performed its biological function through SRSF1 mediation of the p53 pathway. Together, our findings demonstrate that onclncRNA-626 promotes GC progression by binding SRSF1; further, this lncRNA is a potential prognostic biomarker for GC patients.
Keywords: OnclncRNA-626, SRSF1, proliferation, metastasis, gastric cancer
Introduction
Gastric cancer is one of the most common malignant tumors in humans, with the third highest mortality rate worldwide [1,2]. Every year, nearly 1 million patients are diagnosed with gastric cancer worldwide, half of which occur in China [3,4]. Over the past decades, despite the improvement of surgical techniques and the development of targeted therapies and immunotherapies, the 5-year survival rate has still been unsatisfactory [5]. Several studies have indicated that drug resistance, angiogenesis and immune escape might contribute to treatment failure, and the underlying mechanisms for these processes are still poorly understood and need to be further clarified.
Recently, lncRNAs have attracted great attention due to their variable roles in tumorigenesis [6]. LncRNAs are long noncoding RNAs more than 200 bp in length with no or limited protein coding abilities [7]. LncRNAs perform their biological functions through complex mechanisms and can regulate gene expression at epigenetic, transcriptional and posttranscriptional levels [8,9]. Recently, mounting evidence has demonstrated that deregulated lncRNAs are involved in proliferation, metastasis, apoptosis, genomic instability and stem cell-like characteristics in cancer [10]. For example, the lncRNA MetaLnc9 promotes metastasis in lung cancer by specifically interacting with PGK1 and NONO and activating the AKT/mTOR pathway [11]. The lncRNA TLSN8 acts as a tumor suppressor and inactivates IL-6/STAT3 in HCC [12]. The lncRNA TROJAN promotes triple-negative breast cancer progression via ZMYND8 degradation [13]. However, the potential role of lncRNAs in gastric cancer remains largely unknown and needs to be elucidated.
In the present study, we identified a novel lncRNA in gastric cancer termed onclncRNA-626 (oncogenic LncRNA RP11-626H12.3). OnclncRNA-626 was significantly upregulated in gastric cancer tissues and was associated with poor outcome. In loss- and gain-of-function experiments, onclncRNA-626 promoted proliferation and metastasis in vitro and in vivo. Mechanistically, onclncRNA-626 probably performs its function by specifically binding to SRSF1, which affects the downstream p53 pathway; thus, onclncRNA-626 is a potential therapeutic target for GC.
Materials and methods
GC cell lines and reagent
Human gastric cancer cell lines BGC-823, MKN-45, MKN-28, AGS, MGC-803, HGC-27 and normal gastric epithelial cell line GES-1 were purchased from Shanghai Cell Bank Type Culture Collection Committee (CBTCCC, Shanghai, China). All the cell lines were authenticated by short tandem repeat (STR) analysis and were found to be mycoplasma-free. Cells were cultured in DMEM medium (HyClone, Logan, UT, USA) with 10% fetal bovine serum (Biological Industries, Israel) at 37°C under a humidified atmosphere with 5% CO2 as described previously [14]. Cycloheximide (CHX) was purchased from MedChemExpress Company (USA), and rapamycin was obtained from CNSpharm (Shanghai, China).
Human GC tissues
A total of 97 paired gastric cancerous and adjacent normal specimens were obtained from the Biobank at Fudan University Shanghai Cancer Center (FUSCC). Each patient underwent gastrectomy from 2008 to 2010 in FUSCC and was diagnosed with gastric cancer. None of the patients had received chemotherapy or radiation before surgery. The overall survival (OS) time was defined as the time from the surgery date to death, and disease-free survival (DFS) time was defined as the time from surgery date to first recurrence. The clinicopathological features of the patients are shown in Table S1 and are described according to the 7th version American Joint Committee on Cancer staging system (AJCC). All procedures were approved by the ethics committee of Fudan University Shanghai Cancer Center.
RNA isolation, reverse transcription PCR and quantitative real-time PCR
Total RNA from tissue specimens and cells was extracted using TRIzol reagent (Invitrogen) as described previously. For each sample, 500 ng RNA was reverse-transcribed into cDNA using the PrimeScriptTM RT reagent kit (Takara, Dalian, China). For quantification, real-time PCR was performed with SYBR Premix Ex Taq (Takara, Dalian, China) using the Applied Biosystems Prism 7900 system (Applied Biosystems, Life Technologies, USA). The relative expression of detected genes was calculated using the comparative CT (2-ΔΔCT) method, and 18S or β-actin expression functioned as the endogenous control. The sequences of the primers are listed in Supplementary Data 1.
Subcellular fractionation
For nuclear/cytoplasmic fractionation, MKN-45, MGC-803 and HGC-27 cells (1×107 of each) were collected and suspended in cell fraction buffer on ice for 10, 20 and 20 min, respectively. The cell fraction buffer contained the following: 20 mM pH 7.5 Tris-cl, 10 mM NaCl, 0.1% NP4O, and 2 mM MgCl2. Then, RNA was extracted, and qRT-PCR was performed. β-actin served as the cytoplasmic endogenous control, and U2 small nuclear RNA served as the nuclear endogenous control.
RNA scope
The RNA ISH was measured with the RNAscope 2.5 HD Detection Kit (Brown) and HybEZTM system (Advanced Cell Diagnostics) according to the manufacturer’s instructions [15]. In brief, FFPE gastric cancer tissues were treated with H2O2 buffer for 10 min and then with the 1× target retrieval reagent for 20 min at 100°C; finally, tissues were digested in protease buffer plus for 30 min. According to the instructions, the tissue slides were moved into a HybEZ oven (Advanced Cell Diagnostics) for hybridization with probes and signal amplification and detection. The specific onclncRNA-626 probe and corresponding positive/negative probes were synthesized by Advanced Cell Diagnostics. The onclncRNA-626 probe targets 38-1298 of NR_120529.1 and does not cross react with NR_120530.1.
5’ and 3’ RACE
According to the manufacturer’s instructions, a SMARTer RACE cDNA Amplification kit (Clontech, California, USA) was used to perform 5’ and 3’ RACE experiments [16]. The sequences of the PCR primers in the RACE analysis are listed in Supplementary Data 1.
Vector constructs, siRNA and transfection
The overexpression plasmid and lentivirus for onclncRNA-626 (NR_120529.1) were constructed and packaged by Hanyin Biotechnology Company (Shanghai, China). The SRSF1 overexpression plasmid and the corresponding negative control plasmid were purchased from Vigene Biosciences (Shandong, China). The siRNAs for onclncRNA-626 and SRSF1 were synthesized by RiboBio (Guangzhou, China). The sequences of the above siRNAs are listed in Supplementary Data 1. For transient transfection, Lipofectamine 2000 (Invitrogen) was used to transfect the above plasmids and siRNAs into cells according to the manufacturer’s instructions, as described previously.
Proliferation and colony formation assay
Cells with the indicated treatments were seeded into a 96-well plate at a density of 2000/100 μl. On the next day, cell viability was measured using Cell Counting Kit-8 (CCK-8) assays (Dojindo, Japan). Each experiment was performed in triplicate, and cells were measured continuously for 5 days. For the colony formation assay, 1000 cells were seeded into a 6-well plate with complete medium and were grown for approximately two weeks. Finally, visible colonies were fixed with 4% paraformaldehyde, stained with 1% crystal violet and counted.
Migration and invasion assay
For the migration assay, 4×104 transfected HGC-27 or MGC-803 cells and 8×104 MKN-45 or BGC-823 cells were suspended in 200 μl FBS-free DMEM medium and seeded into the upper chamber of a transwell insert (24-well insert; pore size, 8 µM; BD Biosciences), and 600 μl 10% FBS DMEM medium was added into the lower chamber. For the invasion assay, Matrigel (1:10) (BD Biosciences) was diluted and polymerized in the transwell insert for 30 min at 37°C, and then the above cells were seeded in the top chamber. After incubation for 24 h, the cells that migrated into the opposite side of the chamber were fixed with 4% paraformaldehyde, stained with 1% crystal violet and counted.
In vivo assay
Four-week-old male BLAB/c nude mice were purchased from the Shanghai SLAC Laboratory Animal Center of the Chinese Academy of Sciences (Shanghai, China) and randomly divided into two groups (n=6/group). To investigate the potential effect of onclncRNA-626 on metastasis in vivo, 6×106 BGC-823 cells (stably expressing the control vector or the onclncRNA-626) in 200 μl PBS were injected into the tail vein of mice. After 8 weeks, the mice were anesthetized and killed. The metastatic loci in the lungs were analyzed and statistically calculated by two independent pathologists after hematoxylin and eosin staining.
RNA pull down
The RNA pull down was performed using the PierceTM Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher, USA) according to the manufacturer’s instructions. In brief, biotin RNA labeling Mix and T7 RNA polymerase (Roche) was used to transcribe the onclncRNA-626 (RP11-626H12.3) sequence in vitro. Then, the biotinylated RNA was incubated with streptavidin-linked magnetic beads and HGC-27 cell lysates at room temperature for 2 h. After washing with 1× binding buffer four times, the bead-RNA-protein complexes were precipitated and diluted in protein lysis buffer. The proteins were finally separated on an SDS-PAGE gel for mass spectrometry or western blot analysis. The primers used for in vitro transcription are listed in Supplementary Data 1.
RNA immunoprecipitation (RIP)
The RIP assay was performed with the Magna RIP RNA-binding Protein Immunoprecipitation kit (Millipore, USA) according to the manufacturer’s instructions. In brief, 5×107 HGC-27 cells grown in 10 cm dishes were harvested and lysed in 500 μl lysis buffer on ice for 5 min, and the lysate was centrifuged at 12000 rpm for 30 min. The supernatants were incubated with beads at room temperature for 20 min. Then, the indicated antibodies (SRSF1 or Normal mouse IgG) were added to the supernatants for incubation at 4°C overnight. Finally, the beads were washed three times with wash buffer, and the RNA was extracted. For the RIP assay of deletion mutants, plasmids with FLAG-tagged full-length and truncated SRSF1 were transiently transfected into HGC-27 cells. Then, the cells were lysed, and the RIP assay was performed as described above with FLAG antibody.
RNA sequencing
RNA-seq was performed to investigate the mRNA expression profiles after upregulating onclncRNA-626 in BGC-823 cells. Three biological replicates were carried out at the Genergy Biology Company (Shanghai, China) using HiSeq 3000 (Illumina, USA). Fold change >1.3 and <0.8 were set as the cutoff values to select differentially expressed genes (Supplementary Data 2), and gene set enrichment analysis (GSEA) analysis was applied for pathway enrichment with P<0.05.
Western blot
Western blots were performed as previously described [17,18]. The following primary antibodies were used: β-actin (1:2000; Cell Signaling Technology), Flag (1:5000; Sigma-Aldrich, Merck), SRSF1 (1:1000; Santa Cruz Biotechnology), BCL-AF1 (1:1000; Santa Cruz Biotechnology), FBP1 (1:1000; Santa Cruz Biotechnology), hnRPC1/2 (1:1000; Santa Cruz Biotechnology), YBX-1 (1:1000; Santa Cruz Biotechnology), and p53 (1:1000; Santa Cruz Biotechnology). The following secondary antibodies were used: goat anti-rabbit or anti-mouse IgG (1:10000 each; Jackson ImmunoResearch Laboratories).
Immunohistochemistry (IHC)
A total of 90 paired gastric TMAs with overall survival follow-up data was purchased from Outdo Biotech Co., LTD. (Cat: HStmAde180Sur04, Shanghai, China), and immunohistochemistry was performed as previously reported. In brief, paraffin sections were deparaffinized and then rehydrated. After antigen retrieval in a microwave, endogenous peroxidase activity was blocked by incubation with 3% hydrogen peroxide. Nonspecific binding sites were blocked with PBS containing 10% normal goat serum. The sections were then incubated with the primary antibody against SRSF1 (1:50) overnight at 4°C. Finally, the TMA immunostaining score was assessed independently by two pathologists.
Statistics analysis
Data are presented as the mean ± standard error of the mean (SEM) from three independent experiments. The Student’s t-test was employed to compare the differences between two groups or among more than two groups. Fisher’s exact test was applied to analyze the relationship between clinicopathological features and onclncRNA-626 expression. Pearson’s correlation was performed to analyze the link between SRSF1 and onclncRNA-626 expression in GC tissues. P<0.05 was considered statistically significant. All data were analyzed using GraphPad Prism 7.0.
Results
OnclncRNA-626 was aberrantly upregulated in GC patients
By examining 97 paired GC specimens from the Fudan University Shanghai Cancer Center (FUSCC) from 2008 to 2010, we found that onclncRNA-626 expression was markedly higher in GC than in normal tissue specimens (P=0.0011, Figure 1A, 1B), and survival analysis showed that high onclncRNA-626 expression was significantly correlated with poor disease-free survival (P=0.0038, Figure 1C) and overall survival (P=0.0088, Figure 1D). We also investigated the expression of onclncRNA-626 in the TCGA data portal. The results from GEPIA (http://gepia.cancer-pku.cn) showed that patients with higher onclncRNA-626 levels exhibited shorter disease-free survival (P=0.0033, Figure 1E) and worse overall survival (P=0.02, Figure 1F) than patients with lower onclncRNA-626 levels. In addition, we analyzed the relationship between onclncRNA-626 expression and clinicopathological features. As shown in Table S1, onclncRNA-626 correlated with pT4 stage (P=0.0473) and pTNM stage (P=0.0279). These findings suggest that onc3lncRNA-626 might play a critical role in GC progression.
Figure 1.
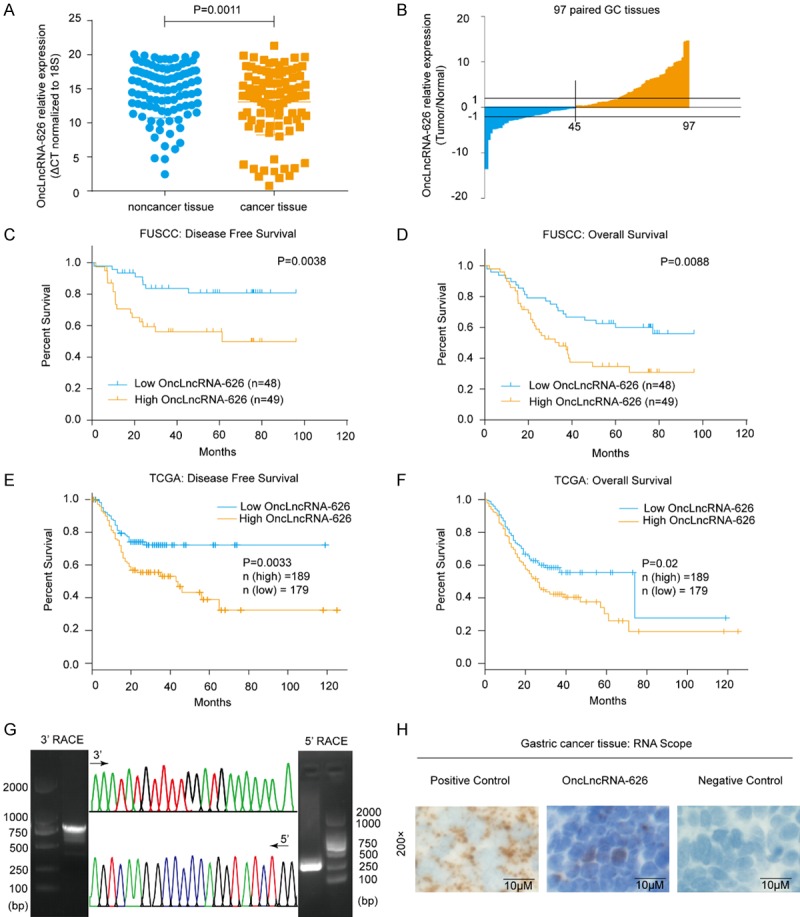
A. OnclncRNA-626 expression levels were quantified in 97 pairs of GC tissues and adjacent normal tissues using qPCR. B. Fold-changes in onclncRNA-626 expression in GC tissues. C, D. Analysis of the correlation between onclncRNA-626 RNA levels and disease-free survival as well as overall survival in 97 GC patients. E, F. The disease-free survival and overall survival time in relation to onclncRNA-626 expression in the TCGA cohort (http://gepia.cancer-pku.cn). G. Representative images of PCR products from the 3’ RACE and 5’ RACE (left and right: images of agarose gel following electrophoresis; middle, sequencing of RACE products). H. RNAscope in situ hybridization images showing the distributions of onclncRNA-626 in GC tissues (left and right: images of positive and negative control; middle: images of onclncRNA-626). Bars: 10 μM.
OnclncRNA-626 is located on chromosome 11 and has two annotated transcripts in the National Center for Biotechnology Information database (NCBI) (Figure S1A). The results of quantitative real-time PCR (qPCR) (Figure S1B) and 5’ and 3’ rapid amplification of the cDNA ends (RACE) (Figure 1G) revealed that the 1755-bp onclncRNA-626 was the predominant transcript in the GC cell lines and the GC tissues. The sequence of the full-length onclncRNA-626 is shown in Figure S1C. The Coding Potential Calculator (CPC), Coding Potential Assessment Tool (CPAT) and PhyloCSF codon substitution frequency analysis indicated that onclncRNA-626 is a noncoding RNA (Figure S1D-F). Moreover, the results of nuclear/cytoplasmic RNA fractionation from the subcellular distribution assay demonstrated that onclncRNA-626 was located in both the cytoplasm and the nucleus of MGC-803, HGC-27 and MKN-45 cell lines (Figure S1G). In addition, the RNAscope assay confirmed that onclncRNA-626 was consistently located in the cytoplasm and nucleus in GC tissues (Figure 1H). Finally, expression levels of onclncRNA-626 in GC cell lines were determined, and onclncRNA-626 was found to be upregulated to varying degrees in comparison with GES-1 cells (Figure S1H). Taken together, the above results suggest that onclncRNA-626 plays an oncogenic role in GC.
OnclncRNA-626 promoted proliferation and metastasis in GC
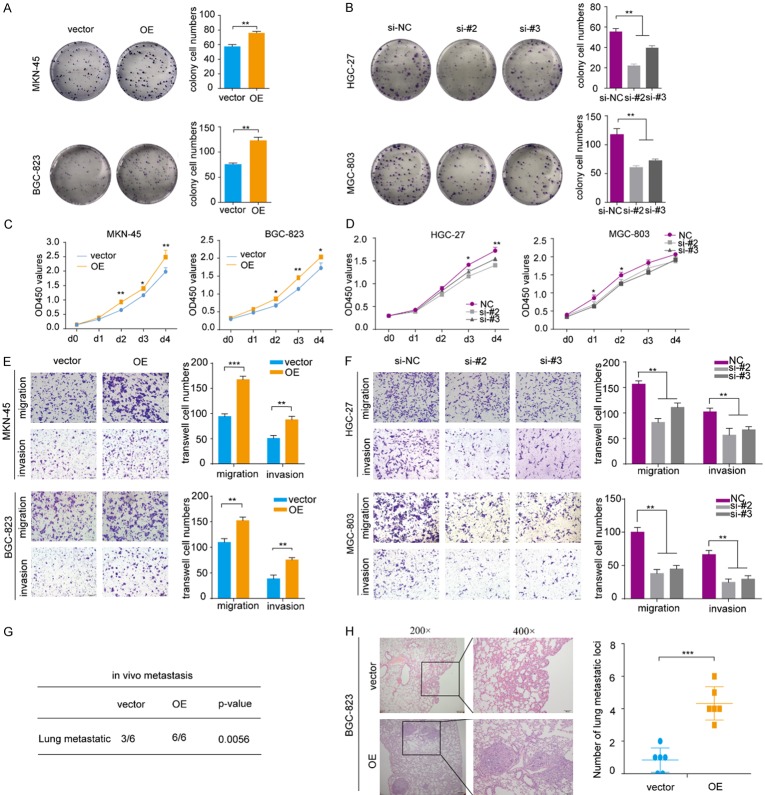
To investigate the biological roles of onclncRNA-626 in GC, plasmids were used to exogenously overexpress onclncRNA-626 in both MKN-45 and BGC-823 cells (Figure S2A). The results showed that forced overexpression of onclncRNA-626 strongly increased colony formation in MKN-45 and BGC-823 cells (Figure 2A); in addition, the silencing of onclncRNA-626 by specific small interfering RNAs (siRNAs) (Figure S2B) significantly inhibited colony formation in MGC-803 and HGC-27 cells (Figure 2B). Consistent with these results, cell proliferation assays showed that onclncRNA-626 overexpression accelerated the proliferation rates of MKN-45 and BGC-823 cells (Figure 2C), whereas onclncRNA-626 siRNA treatment decreased the proliferation of MGC-803 and HGC-27 cells. Additionally, transwell assays showed that onclncRNA-626 overexpression significantly promoted the migration and invasion capacities of MKN-45 and BGC-823 cells (Figure 2E), whereas onclncRNA-626 knockdown inhibited the migration and invasion of MGC-803 and HGC-27 cells (Figure 2F). Furthermore, we assessed the impact of onclncRNA-626 on metastasis in vivo using a lung metastasis mouse model. Hematoxylin-eosin staining showed that onclncRNA-626 overexpression dramatically increased the number of metastatic foci in the lung sections (Figure 2G, 2H). Taken together, these data indicated a vital role for onclncRNA-626 in promoting proliferation and metastasis of GC.
Figure 2.
(A, B) Colony formation assays with representative images of MKN-45 and BGC-823 cells infected with the lentivirus expressing onclncRNA-626, as well as HGC-27 and MGC-803 cells transfected with two independent onclncRNA-626 siRNAs. (C, D) CCK-8 assays of MKN-45 and BGC-823 cells infected with the lentivirus expressing onclncRNA-626 and HGC-27 and MGC-803 cells transfected with two independent onclncRNA-626 siRNAs. (E, F) Transwell migration and invasion assays of MKN-45 and BGC-823 cells as well as in HGC-27 and MGC-803 cells after manipulating onclncRNA-626 expression. (G) Hematoxylin-eosin staining of lung sample sections with metastatic nodules obtained from nude mice after injection with BGC-823 cells infected with the lentivirus expressing onclncRNA-626. (H) Statistics analysis of the metastatic foci in the lung detected by hematoxylin-eosin staining. Data are shown as the mean ± SEM, n=3 in (A, B, E and F), n=6 in (H). *P<0.05, **P<0.01, ***P<0.001. Bars: 100 μM.
OnclncRNA-626 physically interacted with SRSF1 in GC
To explore the potential molecular mechanism underlying how onclncRNA-626 promoted proliferation and metastasis in GC cells, we performed RNA pull-down assays using a biotin-labeled RNA probe to identify potential proteins associated with onclncRNA-626 in HGC-27 cells. After SDS-PAGE electrophoresis and silver staining, the sense and antisense samples were subjected to mass spectrometry (Figure 3A, Supplementary Data 3, Figure S3A). After independent confirmation by western blot, SRSF1 was identified as a binding target of onclncRNA-626 (Figures 3B, S3B). The specificity of this interaction was further confirmed through RNA immunoprecipitation (RIP) assays (Figure 3C). Notably, deletion-mapping analysis was performed based on the secondary structure of onclncRNA-626 to map the precise binding regions for SRSF1 (Figure 3D). The results revealed that the 5’-end segment (156-440 nt) of onclncRNA-626 mediates the interaction with SRSF1 (Figure 3E). On the other hand, RIP assays with full-length and truncated SRSF1 showed that the RRM2 domain of SRSF1 is responsible for their interaction with onclncRNA-626 (Figure 3F, 3G). The above results indicated that onclncRNA-626 probably performed its function by specifically binding to SRSF1.
Figure 3.
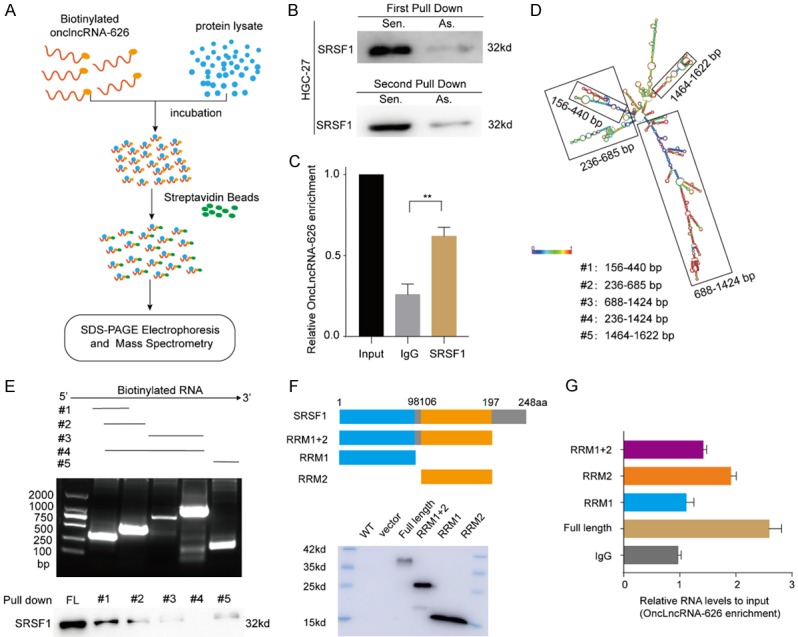
(A) Schematic of the RNA pull-down procedures for the identification of onclncRNA-626-associated proteins. (B) Immunoblotting for specific associations of SRSF1 with onclncRNA-626 from two independent RNA pull-down assays. (C) RNA immunoprecipitation (RIP) assays using antibodies against SRSF1, and qPCR for detecting onclncRNA-626 enrichment. (D) Graphic illustration of predicted onclncRNA-626 secondary structure analyzed by LNCipedia (http://www.lncipedia.org) and the truncated fragments of onclncRNA-626 according to the stem-loop structure. (E) Immunoblotting for SRSF1 in samples pulled down with biotinylated full-length onclncRNA-626 or biotinylated truncated onclncRNA-626 RNA motifs (#1: 156-440 nt; #2: 236-685 nt; #3: 688-1424 nt; #4: 236-1424 nt; #5: 1464-1622 nt). (F, G) Deletion mapping to identify the domains of SRSF1 that bind to onclncRNA-626. RIP analysis for onclncRNA-626 enrichment in cells transiently transfected with full-length or truncated FLAG-tagged constructs. Data are shown as the mean ± SEM, n=3 in (C and G). *P<0.05, **P<0.01.
P53 was involved in onclncRNA-626 mediated downstream pathway
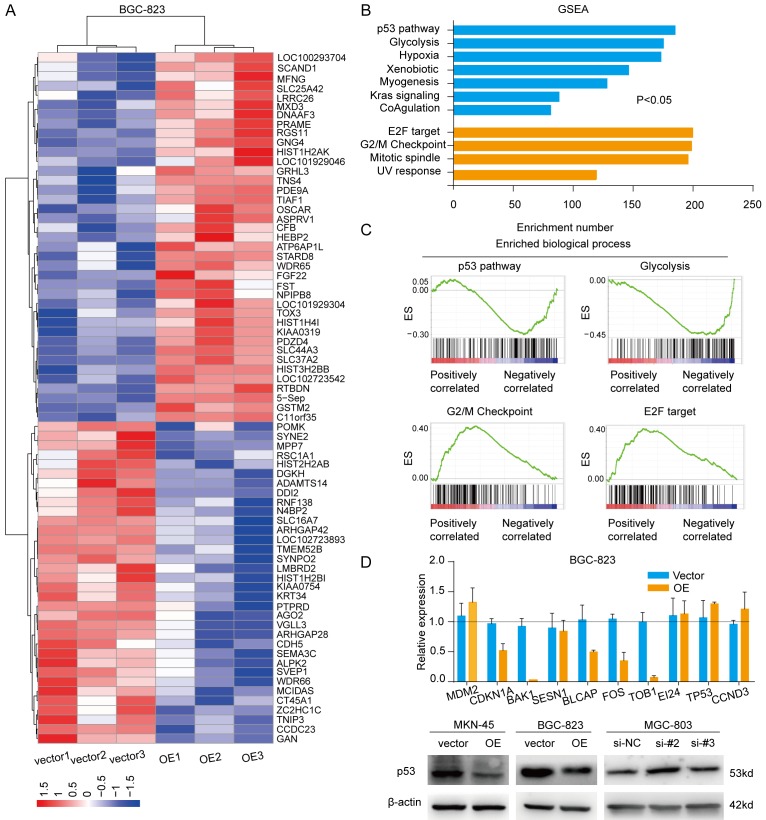
To better understand the molecular mechanisms of onclncRNA-626 as they pertain to GC cell proliferation and metastasis, we next performed RNA-sequencing analysis to obtain the transcriptional mRNA profiles of BGC-823 cells after overexpressing onclncRNA-626 (Figure 4A, Supplementary Data 2). Then, gene set enrichment analysis (GSEA) revealed that onclncRNA-626 primarily affected the p53 signaling pathway, glycolysis, the G2/M checkpoint and E2F targeting (Figure 4B, 4C). Since several studies have identified a relationship between SRSF1 and p53, we attempted to investigate onclncRNA-626’s effect on p53. The mRNA levels of selected genes (CDKN1A, BAK1, BLCAP, FOS, TOB1) in the p53 signaling pathway were consistently affected by onclncRNA-626 in the GC cells (Figure 4D). However, the mRNA levels of p53 were not changed after overexpressing onclncRNA-626. We then examined p53 protein levels after modulating onclncRNA-626 levels. As shown in Figure 4D, p53 protein levels were dramatically decreased after overexpressing onclncRNA-626, and the reverse was true following the knockdown of onclncRNA-626 expression. Taken together, the above results suggest that onclncRNA-626 probably exerts its biological function through the inactivated p53 pathway.
Figure 4.
(A) Heat-map representation of RNA-sequencing results for the gene expression profiles of BGC-823 cells overexpressing onclncRNA-626. Genes are shaded with blue, white, or red in the heat map to indicate low, intermediate, or high expression, respectively. (B) GSEA enrichment focused on a set of signaling pathways and biological processes after onclncRNA-626 overexpression, and the results are summarized based on the enrichment number. (C) GSEA enrichment of the subsets of genes involved in the p53 pathway, glycolysis, the G2/M checkpoint and E2F targeting after onclncRNA-626 overexpression. (D) Upper part: qPCR assays for the selected genes from the p53 pathway in BGC-823 cells overexpressing onclncRNA-626. Lower part: immunoblotting for the protein levels of p53 after overexpressing onclncRNA-626 in MKN-45 and BGC-823 cells or silencing onclncRNA-626 in MGC-803 cells. β-actin served as the internal control. Data are shown as the mean ± SEM, n=3 in (D).
OnclncRNA-626 promoted GC progression via the SRSF1-mediated p53 pathway
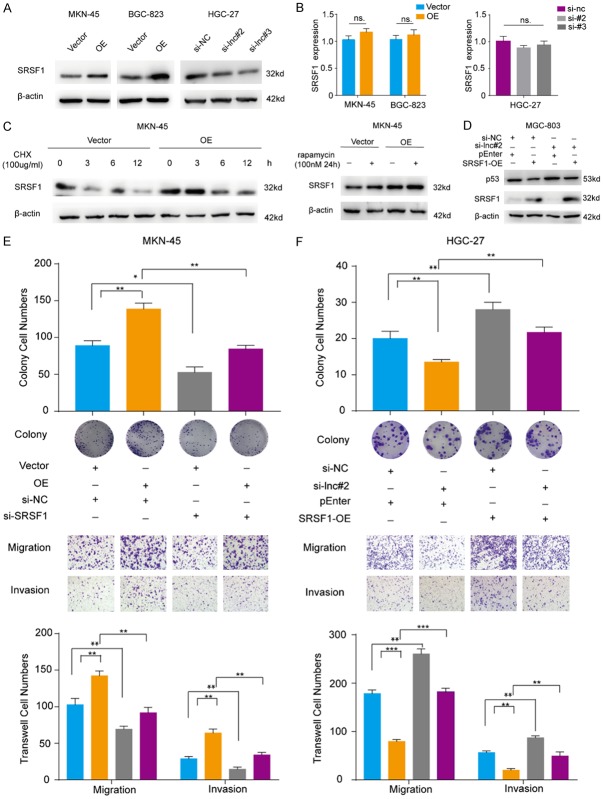
Next, we detected the expression levels of SRSF1 in GC cells after manipulating onclncRNA-626 levels. As shown in Figure 5A, SRSF1 protein levels were dramatically increased when onclncRNA-626 was overexpressed in MKN-45 and BGC-823 cells, and the protein levels were reduced when onclncRNA-626 was silenced in HGC-27 cells. However, no significant changes in the mRNA levels of SRSF1 were observed after modulating onclncRNA-626 expression (Figure 5B). In addition, overexpression of onclncRNA-626 could increase the half-life of SRSF1 in MKN-45 cells after treatment with the protein synthesis inhibitor cycloheximide (CHX). In addition, after treating the cells with rapamycin, we found that the protein level of SRSF1 was increased (Figure 5C). This suggested that onclncRNA-626 inhibited the degradation of SRSF1 by lysosomal mechanisms. Several studies have demonstrated that SRSF1 affects p53 expression. We hypothesize that onclncRNA-626 exerts its function through SRSF1-mediated p53 degradation. To test this hypothesis, we performed rescue experiments, which revealed that when SRSF1 was overexpressed in MGC-803/onclncRNA-626 knockdown cells, p53 expression was partially restored (Figure 5D). Moreover, when endogenous SRSF1 was effectively silenced in onclncRNA-626-overexpressing MKN-45 cells, the colony formation, migration and invasion abilities were remarkably reduced (Figure 5E). Unsurprisingly, the decreased cell growth and mobility in onclncRNA-626-silenced HGC-27 cells was partially restored by SRSF1 overexpression (Figure 5F). Taken together, our results indicate that onclncRNA-626 probably performs its biological function through the SRSF1 pathway.
Figure 5.
(A) Immunoblotting for the protein levels of SRSF1 after overexpressing onclncRNA-626 in MKN-45 and BGC-823 cells or silencing its expression in HGC-27 cells. β-actin served as the internal control. (B) Quantification of SRSF1 mRNA levels by qPCR after overexpressing onclncRNA-626 in MKN-45 and BGC-823 cells or silencing onclncRNA-626 in HGC-27 cells. (C) MKN-45 cells overexpressing onclncRNA-626, while control cells were treated with cycloheximide (CHX, 100 μg/ml) for the indicated times or with rapamycin (100 nM) for 24 h. Immunoblotting for determining SRSF1 levels in whole-cell extracts. (D) Immunoblotting for the protein levels of p53 after overexpressing SRSF1 and silencing onclncRNA-626 in MGC-803 cell rescue assays. (E) Rescue assay analysis using transwell migration and invasion assays and colony formation assays after overexpressing onclncRNA-626 and silencing SRSF1 in MKN-45 cells. (F) Rescue assay analysis using transwell migration and invasion assays and colony formation assays after overexpressing SRSF1 and silencing onclncRNA-626 in HGC-27 cells. Data are shown as the mean ± SEM, n=3 in (B, E and F).
SRSF1 was upregulated and associated with poor outcome in GC tissue
To assess the clinical significance of SRSF1, we evaluated the protein levels of SRSF1 in 90 paired GC and NCT samples using IHC (Figures 6A, 6B, S4). Interestingly, the IHC staining results showed that high SRSF1 expression was significantly correlated with poor overall survival (P=0.0245, Figure 6C). Furthermore, the expression of onclncRNA-626 and SRSF1 in 72 paired GC tissues displayed a positive correlation (r2=0.6285, P<0.0001, Figure 6D). Taken together, as shown in Figure 6E, we found that onclncRNA-626 was expressed at a high level in GC and could directly bind to SRSF1 to stabilize it, which inactivated the p53 pathway to promote malignancies in GC.
Figure 6.
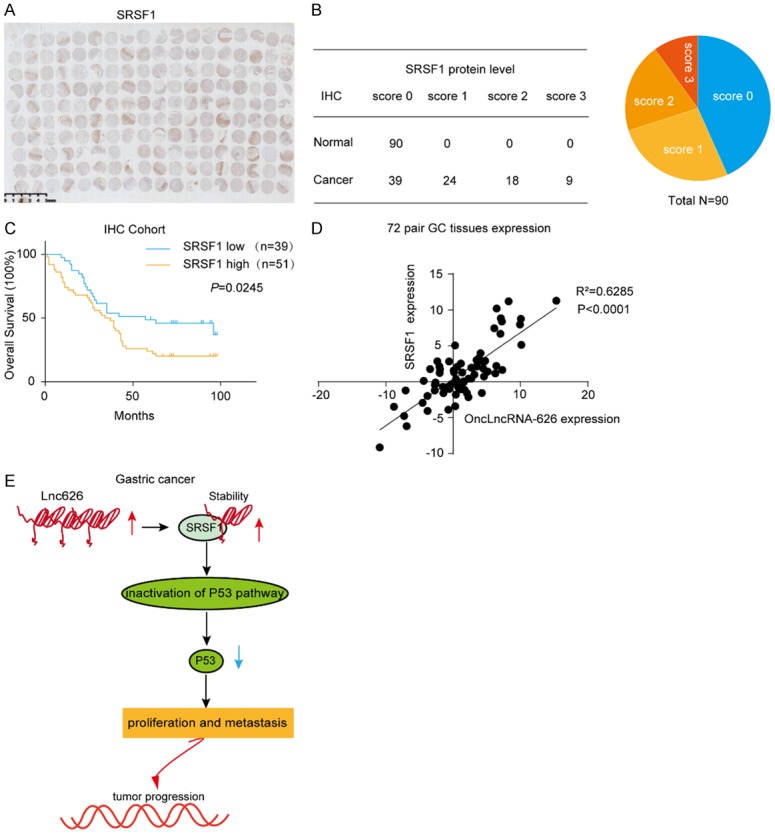
A. IHC staining of the SRSF1 protein in a 90 paired gastric cancer tissue microarray. Scale bar: 5 mm. B, C. Expression levels and Kaplan-Meier analysis of the correlation between SRSF1 protein and overall survival in 90 GC patients. D. qPCR to determine the expression of SRSF1 and onclncRNA-626 in 72 pairs of GC tissues and adjacent normal tissues. E. Diagram of the biological mechanism of onclncRNA-626 in GC.
Discussion
Recently, numerous studies have demonstrated the vital roles of long noncoding RNAs in the progression of various human cancers. Due to advances in next-generation sequencing and public bioinformatic databases, an increasing number of lncRNAs have been identified. However, a large number of lncRNAs remain largely unknown and need to be further described. In the current study, we identified a novel oncogenic lncRNA termed onclncRNA-626, which is located on chromosome 11. OnclncRNA-626 was overexpressed in GC tissues, and its increased expression was associated with poor survival time in GC patients, indicating a potential oncogenic role in GC. Further functional analysis revealed that onclncRNA-626 promoted proliferation and metastasis in vitro and in vivo. Mechanistic investigation showed that onclncRNA-626 probably performed its function by specifically binding to SRSF1, which affected the downstream p53 pathway.
LncRNAs can regulate gene expression in various ways, including through chromatin modification, transcriptional regulation and posttranscriptional processing. In addition, lncRNAs can act as guides to promote transcription, or as enhancers, decoys, or scaffolds to interact with various proteins to exert diversity activities [19]. Among these, interacting with specific RNA binding proteins (RBPs) is one of the most important functions. Here, through RNA pull-down and RIP assays, SRSF1 was identified as one of the binding proteins of onclncRNA-626. SRSF1 (also known as SF2/ASF) is one of the members of the SR family and plays an oncogenic role in cancer development [20]. The molecular structure shows that SRSF1 has two domains: RRM1 and RRM2 [21]. Due to its specific structure, SRSF1 has diverse biological functions, such as providing mRNA stability, nuclear export, and translation; further, it interacts with diverse proteins [22]. In the current study, a truncation assay revealed that the RRM2 domain of SRSF1 was the dominant binding site for onclncRNA-626.
SRSF1 is frequently upregulated in various cancers. For example, SRSF1 was overexpressed in breast cancer and promoted mammary epithelial cell transformation [23]. SRSF1 was upregulated in small cell lung cancer (SCLC) and may play a key role in DNA repair and chemo-sensitivity [24]. Here, the IHC results of TMA revealed that SRSF1 was elevated in paired GC tissues and was associated with poor survival. However, to our knowledge, investigation on the interaction between SRSF1 and lncRNAs is limited. Only the lncRNAs MALT1 with SRSF1 was reported in HCC [25,26]. In the study, we found onclncRNA-626 could regulate SRSF1 expression at the protein level and increase the half-life time of SRSF1. In addition, after treated with rapamycin, the protein level of SRSF1 in onclncRNA-626 overexpressed gastric cancer cells was increased, indicating that onclncRNA-626 probably inhibited the degradation of SRSF1 by a lysosomal mechanism.
Various lncRNAs have been shown to regulate pathways that orchestrate metastasis and progression in numerous cancers [27,28]. Through RNA-seq and subsequent GESA analysis, we found that onclncRNA-626 regulated p53 and p53 target genes to play a role in promoting proliferation and metastasis. The tumor suppressor protein p53 (TP53) is a critical regulator of cellular homeostasis. Many lncRNAs are capable of directly or indirectly regulating p53. For example, the lncRNA PANDAR competitively interacted with p53 and thus blocked CDKN1A transcription in gastric cancer [29]. The lncRNA MEG3 stimulated p53-mediated transactivation to inhibit cell proliferation, meaning that it acted as a tumor suppressor [30].
In the study, onclncRNA-626 could affect the p53 pathway and specifically interact with SRSF1. Interestingly, several studies have described a link between SRSF1 and p53. For example, SRSF1 could promote proliferation through the Δ133p53/EGR1/KLF5 pathway in vascular smooth muscle cell [31]. Shipra Das et al found that the oncogenic role of SRSF1 was dependent upon the inactivation of the p53 pathway [20]. In line with these data, our rescue experiment also revealed that when SRSF1 was overexpressed in MGC-803/onclncRNA626 knockdown cells, p53 expression was partially restored, indicating that onclncRNA-626 probably exerted its biological function through SRSF1 mediation of the p53 pathway. Based on this conclusion, we inferred that onclncRNA-626 was upregulated in GC and would directly bind SRSF1, making SRSF1 more stable; thus, the p53 pathway was affected, which would lead to increases in GC malignancies.
In summary, we identified a novel lncRNA in gastric cancer termed onclncRNA-626, which was upregulated and played an oncogenic role in GC. Moreover, the underlying mechanism of onclncRNA-626 probably involved the SRSF1-mediated p53 pathway, suggesting that onclncRNA-626 is a prospective therapeutic target in GC.
Acknowledgements
We would like to take our special thanks to Dr. Wang Xiaoling from Shanghai Jiao Tong University Ruijin Hospital for her kindly constructing the SRSF1 truncated plasmids. This work was supported by funds from the National Natural Science Foundation of China (81772576). The GC samples from the biobank of FUSCC were approved by the Ethics Committee of FUSCC, and informed consent was obtained from all the participants. In addition, the study complied with the Animal Care guidelines of FUSCC.
Disclosure of conflict of interest
None.
Table S1, Figures S1-S4
Supplementary Data 1
Supplementary Data 2
Supplementary Data 3
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Qin S, Xu J, Xiong J, Wu C, Bai Y, Liu W, Tong J, Liu Y, Xu R, Wang Z, Wang Q, Ouyang X, Yang Y, Ba Y, Liang J, Lin X, Luo D, Zheng R, Wang X, Sun G, Wang L, Zheng L, Guo H, Wu J, Xu N, Yang J, Zhang H, Cheng Y, Wang N, Chen L, Fan Z, Sun P, Yu H. Randomized, double-blind, placebo-controlled phase III trial of apatinib in patients with chemotherapy-refractory advanced or metastatic adenocarcinoma of the stomach or gastroesophageal junction. J. Clin. Oncol. 2016;34:1448–1454. doi: 10.1200/JCO.2015.63.5995. [DOI] [PubMed] [Google Scholar]
- 5.Zong L, Abe M, Seto Y, Ji J. The challenge of screening for early gastric cancer in China. Lancet. 2016;388:2606. doi: 10.1016/S0140-6736(16)32226-7. [DOI] [PubMed] [Google Scholar]
- 6.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 11.Yu T, Zhao Y, Hu Z, Li J, Chu D, Zhang J, Li Z, Chen B, Zhang X, Pan H, Li S, Lin H, Liu L, Yan M, He X, Yao M. MetaLnc9 facilitates lung cancer metastasis via a PGK1-activated AKT/mTOR pathway. Cancer Res. 2017;77:5782–5794. doi: 10.1158/0008-5472.CAN-17-0671. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Li Z, Liu L, Wang Q, Li S, Chen D, Hu Z, Yu T, Ding J, Li J, Yao M, Huang S, Zhao Y, He X. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology. 2018;67:171–187. doi: 10.1002/hep.29405. [DOI] [PubMed] [Google Scholar]
- 13.Jin X, Xu XE, Jiang YZ, Liu YR, Sun W, Guo YJ, Ren YX, Zuo WJ, Hu X, Huang SL, Shen HJ, Lan F, He YF, Hu GH, Di GH, He XH, Li DQ, Liu S, Yu KD, Shao ZM. The endogenous retrovirus-derived long noncoding RNA TROJAN promotes triple-negative breast cancer progression via ZMYND8 degradation. Sci Adv. 2019;5:eaat9820. doi: 10.1126/sciadv.aat9820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Wang C, Zhang X, Hua R, Gan L, Huang M, Zhao L, Ni S, Guo W. Bmi-1 regulates stem cell-like properties of gastric cancer cells via modulating miRNAs. J Hematol Oncol. 2016;9:90. doi: 10.1186/s13045-016-0323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Zhang J, Liu X, Li S, Wang Q, Di C, Hu Z, Yu T, Ding J, Li J, Yao M, Fan J, Huang S, Gao Q, Zhao Y, He X. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat Commun. 2018;9:1572. doi: 10.1038/s41467-018-04006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu ZH, Lin C, Liu CC, Jiang WW, Huang MZ, Liu X, Guo WJ. MiR-616-3p promotes angiogenesis and EMT in gastric cancer via the PTEN/AKT/mTOR pathway. Biochem Biophys Res Commun. 2018;501:1068–1073. doi: 10.1016/j.bbrc.2018.05.109. [DOI] [PubMed] [Google Scholar]
- 18.Wu ZH, Tao ZH, Zhang J, Li T, Ni C, Xie J, Zhang JF, Hu XC. MiRNA-21 induces epithelial to mesenchymal transition and gemcitabine resistance via the PTEN/AKT pathway in breast cancer. Tumour Biol. 2016;37:7245–7254. doi: 10.1007/s13277-015-4604-7. [DOI] [PubMed] [Google Scholar]
- 19.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 20.Das S, Krainer AR. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol Cancer Res. 2014;12:1195–1204. doi: 10.1158/1541-7786.MCR-14-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimoni-Sebag A, Lebenthal-Loinger I, Zender L, Karni R. RRM1 domain of the splicing oncoprotein SRSF1 is required for MEK1-MAPK-ERK activation and cellular transformation. Carcinogenesis. 2013;34:2498–2504. doi: 10.1093/carcin/bgt247. [DOI] [PubMed] [Google Scholar]
- 22.Das S, Anczukow O, Akerman M, Krainer AR. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 2012;1:110–117. doi: 10.1016/j.celrep.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anczukow O, Akerman M, Clery A, Wu J, Shen C, Shirole NH, Raimer A, Sun S, Jensen MA, Hua Y, Allain FH, Krainer AR. SRSF1-regulated alternative splicing in breast cancer. Mol Cell. 2015;60:105–117. doi: 10.1016/j.molcel.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang L, Huang J, Higgs BW, Hu Z, Xiao Z, Yao X, Conley S, Zhong H, Liu Z, Brohawn P, Shen D, Wu S, Ge X, Jiang Y, Zhao Y, Lou Y, Morehouse C, Zhu W, Sebastian Y, Czapiga M, Oganesyan V, Fu H, Niu Y, Zhang W, Streicher K, Tice D, Zhao H, Zhu M, Xu L, Herbst R, Su X, Gu Y, Li S, Huang L, Gu J, Han B, Jallal B, Shen H, Yao Y. Genomic landscape survey identifies SRSF1 as a key oncodriver in small cell lung cancer. PLoS Genet. 2016;12:e1005895. doi: 10.1371/journal.pgen.1005895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malakar P, Shilo A, Mogilevsky A, Stein I, Pikarsky E, Nevo Y, Benyamini H, Elgavish S, Zong X, Prasanth KV, Karni R. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 upregulation and mTOR activation. Cancer Res. 2017;77:1155–1167. doi: 10.1158/0008-5472.CAN-16-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Wang H, Zhang Y, Zhen N, Zhang L, Qiao Y, Weng W, Liu X, Ma L, Xiao W, Yu W, Chu Q, Pan Q, Sun F. Mutual inhibition between YAP and SRSF1 maintains long non-coding RNA, Malat1-induced tumourigenesis in liver cancer. Cell Signal. 2014;26:1048–1059. doi: 10.1016/j.cellsig.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 27.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Ben Q, Lu E, He X, Yang X, Ma J, Zhang W, Wang Z, Liu T, Zhang J, Wang H. Long noncoding RNA PANDAR blocks CDKN1A gene transcription by competitive interaction with p53 protein in gastric cancer. Cell Death Dis. 2018;9:168. doi: 10.1038/s41419-017-0246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei GH, Wang X. lncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:3850–3856. [PubMed] [Google Scholar]
- 31.Xie N, Chen M, Dai R, Zhang Y, Zhao H, Song Z, Zhang L, Li Z, Feng Y, Gao H, Wang L, Zhang T, Xiao RP, Wu J, Cao CM. SRSF1 promotes vascular smooth muscle cell proliferation through a Delta133p53/EGR1/KLF5 pathway. Nat Commun. 2017;8:16016. doi: 10.1038/ncomms16016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.