Abstract
Many genes and mutations have been reported for colorectal cancer (CRC); however, very few have been associated with colorectal cancer liver metastasis (CRLM). We performed gene expression profiling experiments to identify genetic markers for CRLM and elucidate the molecular mechanisms. Microarray experiments were performed on CRC primary tumor samples with or without liver metastasis (LM) using the Affymetrix U133 plus 2.0 GeneChip Array. A new identified gene-scinderin (SCIN) was overexpressed with synchronous LM at both the RNA level evaluated with quantitative real-time PCR and protein level evaluated with immunohistochemistry and also with short overall survival analyzed with Kaplan-Meier method. With multivariate analysis indicated that SCIN served as an independent poor prognostic predictor for CRC patients. Disease-free survival was also significantly lower in SCIN overexpressing CRC patients with metachronous LM. In addition, SCIN knockdown significantly reduced cell proliferation, induced cell cycle arrest, and promoted the expression of some cell cycle apoptosis-related protein. Moreover, the DIAPH1, STAT3, CDK2, CDK4, and EGFR levels were downregulated, whereas CDKN2B and COL4A1 were upregulated in DLD-1-shSCIN cells by microarray analysis compared with DLD-1 shCon cells. These findings revealed that SCIN may serve as an important predictor of CRLM and poor outcome for CRC patients. SCIN may be a potential therapeutic target in human CRC. However, translation of its roles into clinical practice will require further investigation and additional experimental validation.
Keywords: scinderin, colorectal cancer, liver metastasis, genome-wide association study, prognosis
Introduction
Even in developed countries, colorectal cancer (CRC) is the third most common malignancy diagnosed in both men and women and is the second leading cause of cancer deaths (Siegel et al., 2013). Approximately 25% of patients with CRC present with liver metastasis (LM) at initial diagnosis and almost 50% will develop LM. Approximately, 25%–50% of patients with surgically resected colorectal cancer liver metastasis (CRLM) survive 5 or more years (Manfredi et al., 2006; Robertson et al., 2009). Even after radical resection, relapse can occur in 75% of patients, generally occurring within the first 2 years after surgery, and 50% of relapses are in the liver (Petrelli, 2008). Therefore, identifying biomarkers for early CRLM diagnosis and those with a prognostic value for CRC after resection may help patients choose a suitable follow-up interval and subsequent therapy.
Scinderin (SCIN), or adseverin, was initially identified as a calcium-regulated, actin depolymerizing agent involved in secretion; it was named in response to its original isolation from the adrenal medulla. SCIN possesses the ability to sever actin filaments and is a member of the gelsolin family of actin-binding proteins (Maekawa and Sakai, 1990; Rodriguez Del Castillo et al., 1990); In addition, it can bind actin monomers and is present in secretory cells (Rodriguez Del Castillo et al., 1990). SCIN comprises six homologous domains (A1–A6) that share 60% identity to the six domains from gelsolin (G1–G6), which can inhibit mitochondrial apoptosis by closing voltage-dependent anion channels (VDACs) (Kusano et al., 2000). Miura et al. (2012) reported the interaction between SCIN and VDAC, particularly in acisplatin-resistant human bladder cancer cell line overexpressing SCIN, and this binding was suggested to contribute to cisplatin resistance via the inhibition of mitochondria-mediated apoptosis.
Recently, studies found that SCIN functioning in the development and progression of some human cancers. SCIN was highly expressed in gastric cancer tissues and the level of expression associated with the depth of tumor invasion, lymph node metastasis, and poor overall survival. SCIN knockdown inhibited the invasion and metastasis of gastric cancer cells and restrained the filopodium formation and Cdc42 expression (Liu et al., 2016). In hepatocellular carcinoma, SCIN knockdown sensitized cancer cells to chemotherapy and inhibited tumor growth in vivo. Consistently, SCIN overexpression protected cells from apoptosis, promoted xenografted tumor cell growth (Qiao et al., 2018). In prostate cancer, SCIN knockdown significantly downregulated the protein expression of epidermal growth factor receptor (EGFR), impaired cell proliferation-mediated by epidermal growth factor, and inhibited the signaling pathway activation of the downstream mitogen-activated protein kinase (MEK) and extracellular signal-regulated kinase (ERK). SCIN knockdown promoted prostate cell apoptosis by inhibition of B-cell lymphoma-extra-large (Bcl-xl) expression and caspase signaling (Lai et al., 2018). However, the clinical significance and molecular mechanism for SCIN in CRC remain unknown.
Based on the whole-genome expression profiling of CRC, we observed that SCIN, a newly recognized biomarker, was significantly upregulated in CRC patients with synchronous liver metastasis (SLM) and a poor prognosis. Thus, we further evaluated the potential molecular mechanisms responsible for these effects of SCIN in CRC cells.
Materials and Methods
Selection of Patient Material
Tumor specimens with patient clinical and follow-up data were selected from our tumor bank. We divided the samples into two groups according to the presence of LM. The samples were obtained from patients with no family history of CRC or secondary malignancy, and these patients had not received radiation or chemotherapy before surgery. The patients in the nonmetastatic group had a minimum of 3 years disease-free survival (DFS) after surgery. For additional details related to patient materials, see Table 1 and Supplementary Tables 1 and 3.
Table 1.
Clinicopathologic variables and SCIN expression in 300 CRCs.
| Variables | SCIN expression | P-value | |
|---|---|---|---|
| High | Low | ||
| Age (years) | |||
| =60 | 172 | 32 | 0.507 |
| >60 | 78 | 18 | |
| Gender | |||
| Male | 119 | 23 | 0.836 |
| Female | 131 | 27 | |
| Tumor location | |||
| Colon | 160 | 28 | 0.286 |
| Rectum | 90 | 22 | |
| Tumor size | |||
| =5 cm | 184 | 37 | 0.953 |
| >5 cm | 66 | 13 | |
| Gross appearance | |||
| Ulcerative | 190 | 37 | 0.764 |
| Exophytic | 60 | 13 | |
| Histological type | |||
| Adenocarcinoma | 220 | 43 | 0.695 |
| Mucinous adenocarcinoma | 30 | 7 | |
| Tumor differentiation | |||
| Well or moderate | 80 | 18 | 0.582 |
| Poor or other | 170 | 32 | |
| Depth of invasion | |||
| T1,2 | 13 | 36 | <0.001 |
| T3,4 | 237 | 14 | |
| Lymph node metastasis | |||
| Absent | 107 | 30 | 0.026 |
| Present | 143 | 20 | |
| CRC and LM | |||
| CRC without LM | 78 | 22 | 0.021a |
| CRC with SLM | 90 | 10 | 0.103b |
| CRC with MLM | 82 | 18 | 0.480c |
a, CRC without LM vs. CRC with SLM P = 0.021.
b, CRC with SLM vs. CRC with MLM P = 0.480.
c, CRC with MLM vs. CRC without LM P = 0.103.
Microarray Sample Preparation and Gene Expression Analysis
RNA isolation and microarray procedures were performed according to the manufacturer’s instructions, as previously described (Watanabe et al., 2006). Briefly, for sample preparation, snap-frozen tumor samples were cryosectioned (5 µm), and tumor tissues (~1 mm2) were collected using laser-capture microdissection. RNA was isolated from the microdissected material using Trizol (California Carlsbad, Invitrogen, USA) extraction, following the manufacturer’s guidelines. Microarray profiling was performed using human U133 Plus 2.0 GeneChip(r) (Affymetrix, Santa Clara, CA) arrays according to the manufacturer’s protocol.
Cell Lines, Plasmid Construction, and Transfection
The cell lines were obtained from the Chinese Academy of Sciences and cultured in 1640 medium with 10% fetal bovine serum (Logan Utah, HyClone, USA). The pGC-LV/GFP expression vector was purchased from Shanghai Genechem Ltd. SCIN cDNA was obtained from the RZPD clone bank (Germany). The interfering oligonucleotide designed with a short hairpin structure targeting SCIN and a scrambled shRNA as a control were cloned into the pGC-LV/GFP vector. The recombinant vector pGC-LV/GFP/-shRNA SCIN was confirmed by DNA sequencing and enzyme digestion analysis. DLD-1 and SW480 cells were transfected with pGC-LV/GFP/-shRNA SCIN and pGC-LV/GFP/-control using Lipofectamine2000 (Invitrogen), as recommended by the manufacturer.
Quantitative Real-Time PCR
For quantitative real-time PCR (qRT-PCR) to measure the levels of SCIN RNA (Fritzmann et al., 2009), tumor cryosections of 60 CRC samples, including those without LM (n = 20), with SLM (n = 20), and with metachronous LM (MLM) (n = 20), were processed, and RNA was examined as previously described (Fritzmann et al., 2009). Gene-specific primers for PCR products were designed using PPRIMER5 software with information from GenBank (NCBI). The primer sequences are shown in Supplementary Table 7.
Immunohistochemistry and Staining Evaluation
For SCIN protein expression analysis, formalin-fixed paraffin-embedded tissues from 300 patients (18–75 years old) including paired normal mucosa were used for immunohistochemistry (IHC) as previously described (Spano et al., 2005) (rabbit polyclonal anti-SCIN; Sigma Corporation, Cat#: HPA020518, USA).
The intensity of the IHC staining of SCIN was scored using the semiquantitative immunoreactivity scoring (IRS) system (Weichert et al., 2008). The percentage of labeled cells was graded as follows: grade 0, no positive cells; grade 1, 1%~25% labeled tumor cells; grade 2, 26%~50% labeled tumor cells; grade 3, 51%~75% labeled tumor cells; and grade 4, >75% positive tumor cells. The intensity of peroxidase deposits, ranging from light beige to dark brown, was assessed visually in the tumor cell cytoplasm and scored as 0 (negative), 1 (weak), 2 (moderate), or 3 (strong). A composite score, potentially ranging from 0 to 12, was obtained by multiplying the grade by the intensity. An IRS value between 0 and 6 was considered low expression, and IRS value >6 was considered high expression. All specimens were pathologically reassessed independently by two gastroenterology pathologists blinded to the clinical data.
MTT Assay
The proliferation of transfected cells was measured using an MTT assay. A total of 10 µl of MTT (5 mg/ml; Sigma) was added to each well for a final volume of 100 µl of culture medium containing viable cells. After an additional incubation of 4 h, the resulting formazan was dissolved in 100 µl of isopropanol with 40 mM hydrochloric acid. Spectrophotometric absorbance at 570 nm (for formazan dye) was measured with absorbance at 630 nm as a reference.
Western Blotting
Cells were harvested and scraped in RIPA buffer containing 10% protease inhibitor cocktail to obtain the total protein content. The BCA method was used to measure the protein concentration. Each sample was fractionated using 10% SDS-PAGE and blotted onto PVDF membranes. The membranes were incubated in 5% nonfat dry milk to block nonspecific binding and then blotted with a primary antibody overnight at 4°C. After washes with TBST and incubation with anti-rabbit horseradish peroxidase-conjugated secondary antibody (Biosynthesis Biotechnology, China) for 2 h at room temperature, the immunocomplexes were visualized using chemiluminescence (GE, USA) according to the manufacturer’s protocol (Sun et al., 2008).
Flow Cytometry Analysis
Cell cycle progression was analyzed by flow cytometry. In brief, 4 days after lentivirus infection, cells were collected, washed by PBS, and fixed by 75% ethanol. Cells were stained with propidium iodide (PI) and RNase overnight at 4°C. Samples were then analyzed.
Gene Expression Analysis Between DLD-1-Shscin and DLD-1-Shcon Cells
Total RNA was isolated from DLD-1-shSCIN and DLD-1-shCon cells. The RNA samples were converted to biotinylatedc RNA and hybridized to the GeneChip, as described above. Then, gene cluster analysis, gene ontology (GO) analysis (including molecular function, biological process and cellular component), and pathway analysis were applied between the differently expressed genes, and the discrepancy in gene expression was confirmed using qRT-PCR, as described above (Fritzmann et al., 2009).
The Significance Analysis of Microarrays
The popular statistical technique of significance analysis of microarrays (SAM) applying to detect the differentially expressed genes was first introduced by Tusher et al. in 2001 (Tusher et al., 2001). SAM identifies gene by assimilating a set of gene-specific t-tests. Each gene is allocated a score on account of its change in gene expression associated to the standard deviation of repeated detection for that gene. Gene with score greater than an assumed threshold is considered potentially significant. The percentage of the gene identified by probability is the false discovery rate (FDR). The different number of genes can be identified by adjusting the threshold, and FDRs can be calculated for each set (Tusher et al., 2001; Rieger and Chu, 2004; Del Rio et al., 2007; Cescon et al., 2015; Steinberg et al., 2017; Feng et al., 2018; Hao et al., 2019).
In this study, a set of genes with a FDR < 0.05 were then selected and an absolute value (fold-change cutoff) of 2.7 was applied to reduce the number of potential probes between CRC patients with and without LM. Statistical analyses were performed using the open source software R version 2.13.1.
Statistical Analyses
Continuous data were measured using a t-test. For categorical data, chi-squared analysis or Fisher’s exact test was used. The survival rate was analyzed with the Kaplan-Meier method, and differences in survival rates were assessed with the log-rank test. A Cox proportional hazards model was used for multivariate analysis. All statistical analyses were performed using SPSS 16.0 software (SPSS, Chicago, IL, USA). Two-sided P-values were calculated, and P < 0.05 was considered significant.
Results
SCIN Is Significantly Upregulated in SLM
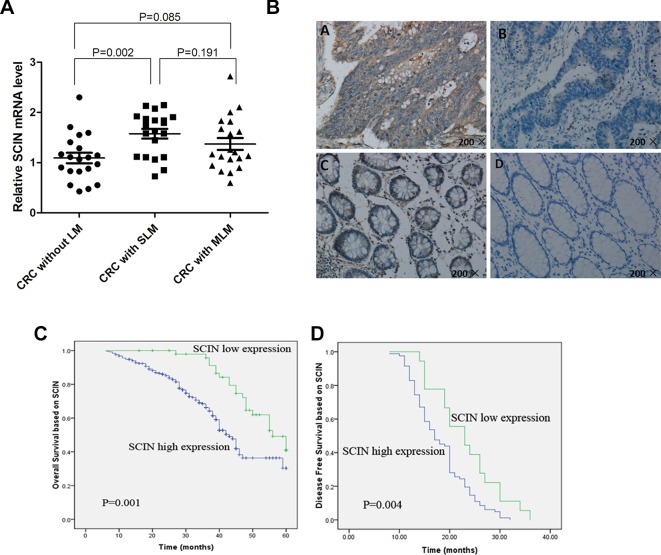
Microarray profiling was performed using human U133 Plus 2.0 GeneChip(r) in primary tumors of CRLM patients (n = 5) and CRC patients without LM (n = 6). We applied the SAM to analyze gene expression differences (for scores with an absolute value ≥2.7) and identified a list of 110 genes differentially expressed at significant levels in patients with and without LM (with different absolute value, different numbers of genes can be obtained; the baseline data of the GeneChip patients and differentially 110 genes were in Supplementary Tables 1 and 2, respectively). Of these, 60 genes (including the SCIN) were shown to have higher expression levels in patients with LM, and 50 genes were shown to have lower expression levels. Based on literature search, bioinformatics analysis, and patent database of cancer key gene, after excluding some Star Genes that have been studied, we focusing on 24 genes (SCIN is one of them). Then, we examined SCIN using a qRT-PCR method based on SYBR green for CRC primary tumors without LM (n = 20), with SLM (n = 20), and with MLM (n = 20) (The baseline data of the 60 patients examined by qRT-PCR was in Supplementary Table 3). We found that the SCIN was expressed at significantly higher level in CRC tumor samples with SLM compared to patients without LM (P = 0.002), no significantly different was found between patients without LM and MLM (P = 0.085), and no significantly different was found between SLM and MLM patients (P = 0.191) (Figure 1A). Then, we used IHC to examine SCIN protein expression levels in 300 CRC patients, including those without LM (n = 100), with SLM (n = 100), and with MLM (n = 100). We observed brown SCIN staining in the cancer cell cytoplasm, and very few paracancerous samples showed low expression levels (Figure 1B). In addition, we found that high SCIN expression levels in cancerous tissues were significantly associated with SLM compared to CRC patients without LM (P = 0.021) (Table 1).
Figure 1.
SCIN expression in CRC tissue samples and Kaplan-Meier analysis with OS and DFS. (A) qRT-PCR was used to detect SCIN mRNA expression levels in CRC primary tumor tissues without LM, with SLM, or with MLM. (B) Representative immunostains of SCIN expression in CRC clinical samples. Positive cells are stained brown high-intensity SCIN expression in cancerous tissues (upper left). Low-intensity SCIN expression in cancerous tissues (upper right). Positive SCIN staining in paracancerous tissue (below left). Negative SCIN staining in paracancerous tissue (below right). (C) Kaplan-Meier analysis of OS related to SCIN expression in patients with CRC (n = 300; P = 0.001). (D) DFS (P = 0.004) related to SCIN expression in MLM patients (n = 100).
The Correlation Between SCIN Expression and Clinicopathologic Parameters
According to the extent of SCIN staining in the cytoplasm, the experimental samples were divided into two groups [the high SCIN expression group (n = 250) and the low SCIN expression group (n = 50)] to investigate SCIN expression in association with clinicopathologic variables. We found high SCIN expression was related to the depth of invasion and lymph node metastasis (P < 0.05), but was not related to age, gender, tumor location, tumor size, gross appearance, histological type, and tumor differentiation (P > 0.05; Table 1).
SCIN Expression and Prognosis of CRC Patients
We applied Kaplan-Meier survival analysis to study the relationship between SCIN expression and overall survival (OS) among the 300 patients previously described. OS was significantly lower among SCIN-overexpressing patients (P = 0.001; Figure 1C). In addition to SCIN expression, univariate analysis also identified poor tumor differentiation, depth of invasion infiltration (T3-4), lymph node metastasis, and distant metastasis (SLM and MLM) as significantly associated with poor OS (Table 2). Other clinicopathologic features such as age, gender, tumor location, tumor size, gross appearance, and histological type were not significant prognostic factors (Table 2). Table 2 also shows the results of multivariate analysis in the final model through the use of a multivariate Cox proportional hazard regression model, which included SCIN expression, tumor differentiation, depth of invasion infiltration, lymph node metastasis, and distant metastasis. In this model, the variables of high SCIN expression and distant metastasis were independent prognostic predictors for CRC patients. We also analyzed DFS among MLM patients using Kaplan-Meier survival analysis and found that DFS was significantly lower in patients with SCIN overexpression (P = 0.004; Figure 1D).
Table 2.
Univariate and multivariate analysis of clinicopathological factors for OS in 300 CRC patients.
| Characteristics | OS | ||||
|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | ||||
| χ2 | P | Exp(B) | 95%CI for Exp(B) | P | |
| Age | 0.289 | 0.591 | |||
| Gender | 3.632 | 0.057 | |||
| Tumor location | 0.569 | 0.450 | |||
| Tumor size | 0.946 | 0.331 | |||
| Gross appearance | 0.036 | 0.849 | |||
| Histological type | 0.300 | 0.584 | |||
| Tumor differentiation | 5.620 | 0.018 | 0.918 | 0.590–1.428 | 0.704 |
| Depth of invasion | 4.745 | 0.029 | 0.826 | 0.457–1.492 | 0.526 |
| Lymph node metastasis | 11.118 | 0.001 | 0.715 | 0.475–1.077 | 0.109 |
| With distant metastasis | 79.905 | <0.001 | 8.511 | 5.032–14.394 | <0.001 |
| SCIN expression | 11.097 | 0.001 | 0.343 | 0.185–0.635 | 0.001 |
SCIN Knockdown Inhibits Cell Proliferation and Induces Cell Cycle Arrest as Well as Cell Apoptosis
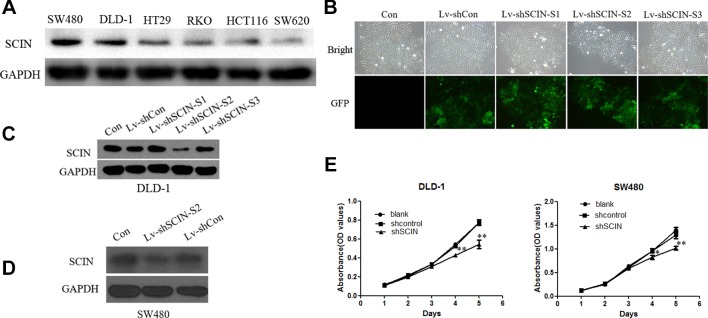
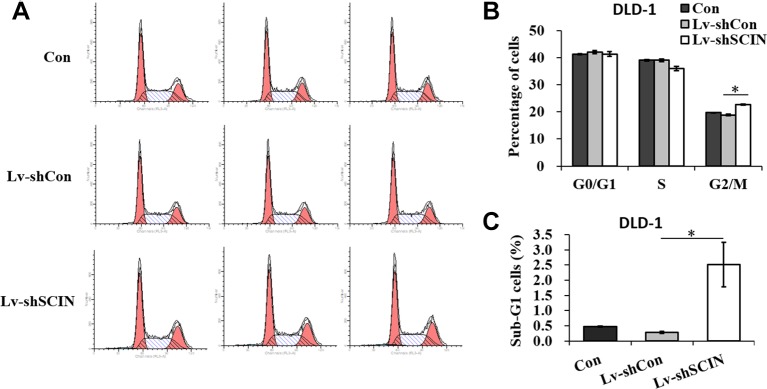
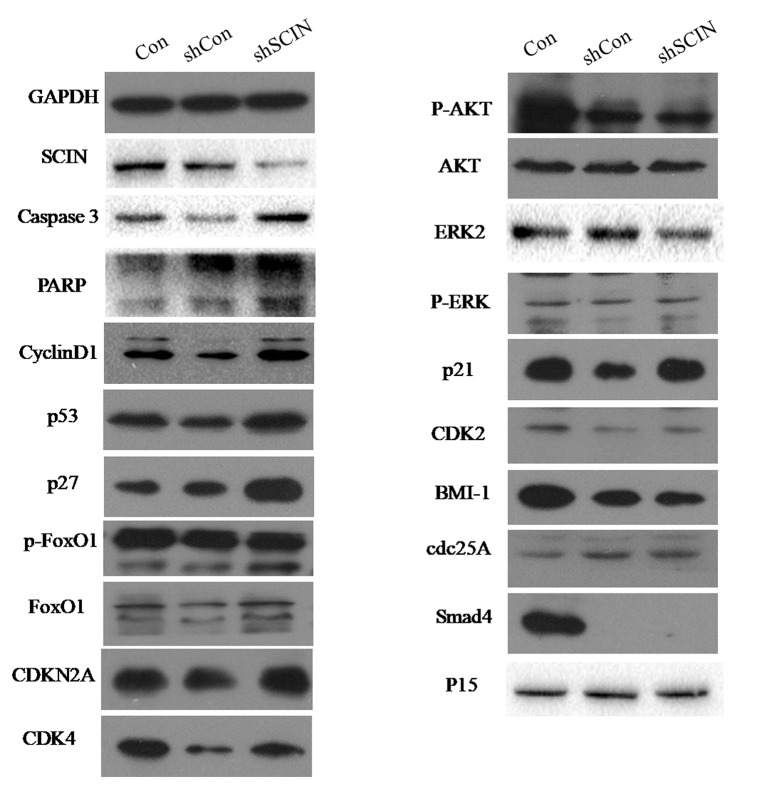
Among six CRC cell lines, SCIN protein was highly expressed in SW480 and DLD-1 cell lines. So, we used DLD-1 and SW480 cell lines to study the biological function of SCIN in cancer cell growth. In order to rule out the potential off-target effect, we also introduced Lv-shSCIN-S1, Lv-shSCIN-S2, and Lv-shSCIN-S3 targeting SCIN into DLD-1 cell, and only Lv-shSCIN-S2 inhibited the SCIN expression significantly (Figures 2A–D). We selected specific SCIN-targeting shRNA (S2) for the follow-up experiments, lentivirus mediated RNA interference in DLD-1 and SW480 cell lines were performed. The roles of SCIN in cell proliferation was investigated in the DLD-1 and SW480 cell lines using an MTT assay, we found that SCIN knockdown significantly reduced cell proliferation (Figure 2E). Flow cytometry analysis was used to study the mechanisms by which SCIN knockdown induced cell apoptosis. After Lv-shSCIN infection, the DLD-1 cell percentage in the G2/M phase was significantly increased (Figure 3). SCIN knockdown led to the DLD-1 cell accumulation in the sub-G1 phase, which represents apoptotic cells. The cell cycle markers expression were also increased, including CyclinD1, P53, P27, p-FoxO1, caspase 3, and PARP (Figure 4). These results suggested that knockdown of SCIN in CRC cells blocked cell cycle progression via upregulation of CyclinD1, P53, P27, p-FoxO1, caspase 3, and PARP.
Figure 2.
Depletion of SCIN in CRC cells by Lv-shSCIN infection. (A) Expression of SCIN in CRC cell lines (SW480, DLD-1, HT29, RKO, HCT116, and SW620), as revealed using Western blotting. (B) SCIN expression levels in DLD-1 cells transduced with Lv-shSCIN. Infection efficiencies were evaluated by GFP fluorescence in DLD-1 cells after lenti-virus infection. (C) The expression of SCIN was significantly inhibited after Lv-shSCIN-S2 was successfully transfected into DLD-1 cell line. (D) The expression of SCIN was significantly inhibited after Lv-shSCIN-S2 was successfully transfected into SW480 cell line. (E) SCIN knockdown significantly reduced cell proliferation in the cell line of DLD-1 and SW480, as determined using an MTT assay. (*P < 0.05, **P < 0.01).
Figure 3.
SCIN knockdown induces cell cycle arrest. (A) Flow cytometry analysis of cell cycle in DLD-1 cells, DLD-1-shSCIN, and DLD-1-shCon cells. (B) Statistic results of cell percentages in G0/G1, S, and G2/M phases in DLD-1 cells, DLD-1-shSCIN, and DLD-1-shCon cells. (C) Statistic results of cell percentages in the sub-G1 phase in DLD-1 cells, DLD-1-shSCIN, and DLD-1-shCon cells (*P < 0.05).
Figure 4.
SCIN knockdown promoted the expression of some cell cycle apoptosis-related protein. The expression of CyclinD1, P53, P27, p-FoxO1, caspase 3, and PARP in CRC cells was increased by Western blotting analysis. GAPDH was used as a control protein.
Other Potential Molecular Mechanisms of SCIN Functioning in CRC Cells Analyzed by Microarray Analysis
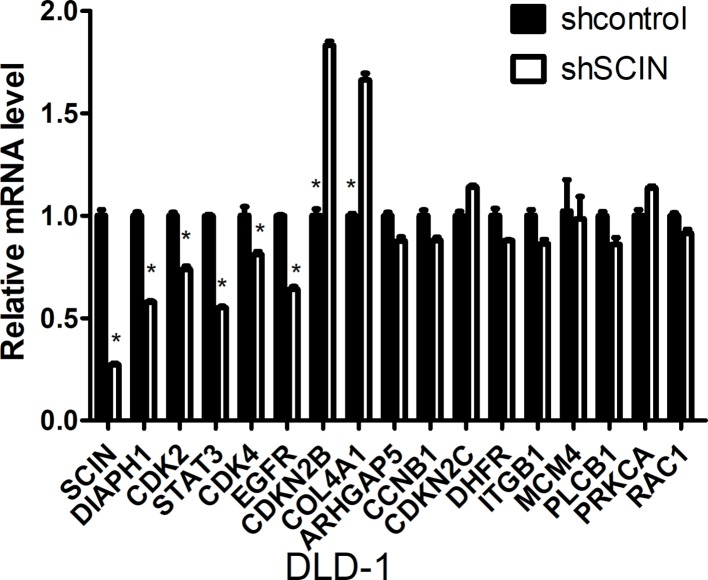
Microarray analysis was performed using the Affymetrix GeneChip for DLD-1-shSCIN and DLD-1-shCon cells (the results are listed in Supplementary Table 4). Then, we performed gene cluster analysis, GO analysis (molecular function, biological process, and cellular component of MGI GO Slim) and pathway analysis (http://www.broadinstitute.org/gsea/msigdb/annotate.jsp), and the results are listed in Supplementary Tables 5 and 6. After comprehensive analysis, we identified 16 genes (CDKN2C, CDKN2B, DIAPH1, MCM4, STAT3, EGFR, RAC1, DHFR, CDK2, COL4A1, PRKCA, CDK4, CCNB1, ARHGAP5, ITGB1, and PLCB1) that were significantly correlated with SCIN expression. Then, we further examined these differentially expressed genes using qRT-PCR (the primer sequences are shown in Supplementary Table 7). We found that between DLD-1-shSCIN and DLD-1-shCon cells, DIAPH1, CDK2, STAT3, CDK4, and EGFR RNA were significantly downregulated (P < 0.05), whereas CDKN2B and COL4A1 RNA were significantly upregulated (P < 0.05; Figure 5).
FIGURE 5.
qRT-PCR validation of candidate genes between DLD-1-shSCIN and DLD-1-shCon cells by microarray analysis. Between DLD-1-shSCIN and DLD-1-shCon cells, DIAPH1, CDK2, STAT3, CDK4, and EGFR RNA were significantly downregulated, whereas CDKN2B and COL4A1 RNA were significantly upregulated. The data shown represent the mean ± s.e.m of the experiment performed in triplicate (*P < 0.05).
Discussion
CRC is one of the most common malignancies, with an increasing global incidence. Whereas approximately one-half of patients can be cured through surgical resection and adjuvant therapy for well-confined primary tumors, metastatic disease (with the liver as the most common site of CRC metastasis) is largely incurable and is the most frequent cause of mortality due to its systemic nature and the resistance of disseminated tumor cells to existing therapeutic agents (Stein et al., 2009; Valastyan and Weinberg, 2011). The early molecular mechanisms underlying the dynamic and intricate process of metastasis, which are crucial for diagnosis, remain poorly understood (Valastyan and Weinberg, 2011; Budhu and Wang, 2012). Currently, clinical and histopathological findings and tissue-based molecular markers are insufficient for the early identification of individuals at high risk for LM (Duffy et al., 2007).
Very few gene expression profiling studies on CRC have been performed using microarray technology, and these studies have mainly focused on the carcinogenesis process, prognosis prediction, and treatment response prediction (Wang et al., 2004; Arango et al., 2005; Stein et al., 2009), rather than LM in CRC. Gene expression profiling is an innovative and promising approach to investigate underlying molecular mechanisms. Stein et al. (2009) identified a novel gene termed metastasis-associated in colon cancer-1 (MACC1), which was found to be associated with colon cancer through genome-wide expression analysis in primary and metastatic carcinomas. The authors demonstrated that MACC1 expression in tumor specimens served as an independent prognostic indicator of metastasis formation and metastasis-free survival. Wang et al. (2004) used the Affymetrix U133a GeneChip to evaluate RNA samples from 74 patients (31 patients developed tumor relapse in less than 3 years, whereas 43 patients remained disease-free for more than 3 years after surgery). These authors identified a 23-gene signature that predicted recurrence in Dukes’ B patients, which indicated that patients with this 23-gene expression pattern should be upstaged to receive adjuvant therapy, similar to Dukes’ C patients. Arango et al. (2005) performed microarray analysis in a unique set of fresh-frozen tumor samples from Dukes’ C patients who had received surgery as the only form of treatment and found that a high-density oligonucleotide microarray analysis accurately distinguished patients with good and poor prognosis after surgery; in particular, the Ras homologue family member A (RHOA) was shown to identify a subset of patients with poor prognosis who could benefit from more aggressive treatment. In the current study, our goal was to identify genes that could predict LM. We applied genome-wide expression analyses in primary tumors with or without LM and identified SCIN for the first time. Our study results further suggested that SCIN may serve as an important predictor of both LM and poor outcome in CRC.
Although the correlation between SCIN and cancer has been revealed, gradually, the role of SCIN in tumor development and progression was controversial among different researches. SCIN expression in megakaryoblastic leukemia cells promoted cell apoptosis and impaired cell proliferation and tumor formation (Zunino et al., 2001), which is contrary to our study and others (Lai et al., 2018; Qiao et al., 2018). Another study found high levels of SCIN expression in gastric cancer tissue correlated with poor prognosis. Furthermore, SCIN promoted the invasion and metastasis of gastric cancer cells through activating the Cdc42 pathway to increase the formation of filopodia (Liu et al., 2016). We and other studies found changes of cell cycle distribution affected by SCIN (Wang et al., 2014; Liu et al., 2015), but Qiao et al. did not (Qiao et al., 2018). Qiao et al. (2018) and Miura et al. (2012) found that SCIN was identified as the cisplatin-resistant marker via interacting with VDAC and further inhibiting cell apoptosis in hepatocellular cancer cell and bladder cancer cell, respectively. SCIN comprises six homologous domains (A1–A6) that share 60% identity to the six domains from gelsolin (G1–G6), and gelsolin superfamily has been certified to take great role in cell apoptosis by modulating dynamic actin assembly (Kusano et al., 2000). Taken together, the results presented here demonstrated that SCIN plays an import role in mediating cytoskeleton reorganization and cell apoptosis.
Based on our preliminary CRC microarray analysis, SCIN may affect the diaphanous homolog (DIAPH1), cyclin-dependent kinases (CDK2), signal transducer and activator of transcription 3 (STAT3), cyclin-dependent kinases (CDK4), EGFR, cycline dependent kinase inhibitor 2B (CDKN2B), and collagen type IV alpha 1 (COL4A1) signaling pathways. DIAPH1 is a downstream effector of RhoA (Rho family of GTPases) and controls actin-dependent processes such as cytokinesis, serum response factor transcriptional activity, and cell motility (Narumiya et al., 2009). DIAPH1 dysregulation within the actin-cytoskeleton pathway may specifically promotes malignant transformation in inflammatory bowel disease-associated CRC (Kanaan et al., 2010). STAT3 is a transcriptional factor that is constitutively activated in many cancer types. STAT3 appears to play crucial roles in cell proliferation and survival, angiogenesis, tumor-promoting inflammation, and suppression of antitumor host immune responses in the tumor microenvironment (Yu et al., 2009). STAT3 activation in CRC is also associated with adverse clinical outcomes, supporting its potential role as a prognostic biomarker and a chemopreventive and/or therapeutic target (Morikawa et al., 2011); although, effectively inhibiting the activity of a transcriptional factor remains challenging due to their intracellular localization and lack of enzymatic activity. However, we found that inhibiting SCIN expression could downregulate STAT3 activity, which implies that anti-SCIN therapies may be an effective means to block STAT3 signaling in CRC. CDK2 and CDK4 are extremely important checkpoints of the cell cycle at the G1/S transition. CDK2 is differentially expressed during colorectal oncogenesis and cancer progression. CDK2 overexpression may facilitate lymph node metastasis of early cancer, and decreased CDK2 correlates with large tumor size, venous invasion, deep infiltration, hepatic metastasis, advanced stage, and poor prognosis (Li et al., 2001). CDK4 overexpression has been detected in intestinal adenomas and is associated with increased cell proliferative activity in premalignant neoplastic cells, which indicates that CDK4 may contribute to the malignant progression of adenomas (Zhang et al., 1997). EGFR belongs to the ERBB receptor tyrosine kinase network, and it plays an important role in CRC pathogenesis. EGFR is widely expressed in advanced CRC, with its expression ranging from 72% to 82% (Italiano et al., 2005). Many carcinoma types, including colon, breast, and lung, display heightened EGFR activity, which correlates with tumor recurrence and shorter patient survival (Yarden and Pines, 2012). CDKN2B is a cell-cycle inhibitor of INK4/ARF that binds CDK4 and CDK6 and induces an allosteric change to abrogate the binding of these kinases to D-type cyclins, thus inhibiting CDK4/6-mediated phosphorylation of retinoblastoma (Rb) family members. Studies have demonstrated that CDKN2B is deleted in a wide spectrum of tumors including melanoma, pancreatic adenocarcinoma, glioblastoma, certain leukemias, non-small cell lung cancer, and bladder carcinoma (Kim and Sharpless, 2006). In CRC, previous reports have suggested that CDKN2B methylation contribute to carcinogenesis along with p16 (INK4a) and that CDKN2B may serve as a prognostic factor in the early stages of disease (Ishiguro et al., 2006). Heterotrimers composed of COL4A1 and COL4A2 constituted the most abundant components of almost all basement membranes. Mutations in COL4A1 or COL4A2 were contribute to a broad spectrum of disorders, including glaucoma, myopathy, and hemorrhagic stroke (Kuo et al., 2012). COL4A1 was also identified as the potential therapeutic target genes in several human cancers including CRC (Jin et al., 2017).
Conclusion
Our studies indicate that SCIN may serve as an important predictor of CRLM and poor outcome for CRC patients. However, this is a preliminary study, to clarify the molecular mechanism of SCIN affection in cell growth and death, and further investigation and more experimental validation was required.
Ethics Statement
Clinical samples were collected from patients after obtaining written informed consent in accordance with a protocol approved by the Ethics Committee of Zhongshan Hospital of Fudan University (Shanghai, China).
Author Contributions
QL, JL, YW, and JX conceived the idea and designed the experiments. QL, JL, DZ, and ZN carried out all the experiments and co-wrote the manuscript. XP, PX, and MJ helped with cell culture, PCR, WB, IHC, and FCM. YW and JX supervised the project. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (81272390, 81372315, and 81472228), the Shanghai Science and Technology Committee Project (13JC1401601 and 134119a4800), the Shanghai Science and Technology Committee Talent Program (12XD1401900), and the Outstanding Academic Leaders Project of the Health System in Shanghai (XBR2011031).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2019.01183/full#supplementary-material
References
- Arango D., Laiho P., Kokko A., Alhopuro P., Sammalkorpi H., Salovaara R., et al. (2005). Gene-expression profiling predicts recurrence in Dukes’ C colorectal cancer. Gastroenterology 129 (3), 874–884. 10.1053/j.gastro.2005.06.066 [DOI] [PubMed] [Google Scholar]
- Budhu A., Wang X. W. (2012). Transforming the microenvironment: a trick of the metastatic cancer cell. Cancer Cell 22 (3), 279–280. 10.1016/j.ccr.2012.08.018 [DOI] [PubMed] [Google Scholar]
- Cescon D. W., She D., Sakashita S., Zhu C. Q., Pintilie M., Shepherd F. A., et al. (2015). NRF2 Pathway activation and adjuvant chemotherapy benefit in lung squamous cell carcinoma. Clin. Cancer Res. 21 (11), 2499–2505. 10.1158/1078-0432.CCR-14-2206 [DOI] [PubMed] [Google Scholar]
- Del Rio M., Molina F., Bascoul-Mollevi C., Copois V., Bibeau F., Chalbos P., et al. (2007). Gene expression signature in advanced colorectal cancer patients select drugs and response for the use of leucovorin, fluorouracil, and irinotecan. J. Clin. Oncol. 25 (7), 773–780. 10.1200/JCO.2006.07.4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy M. J., van Dalen A., Haglund C., Hansson L., Holinski-Feder E., Klapdor R., et al. (2007). Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur. J. Cancer 43 (9), 1348–1360. 10.1016/j.ejca.2007.03.021 [DOI] [PubMed] [Google Scholar]
- Feng S., Bucuvalas J. C., Demetris A. J., Burrell B. E., Spain K. M., Kanaparthi S., et al. (2018). Evidence of chronic allograft injury in liver biopsies from long-term pediatric recipients of liver transplants. Gastroenterology 1551838-1851 (6), e1837. 10.1053/j.gastro.2018.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzmann J., Morkel M., Besser D., Budczies J., Kosel F., Brembeck F. H., et al. (2009). A colorectal cancer expression profile that includes transforming growth factor beta inhibitor BAMBI predicts metastatic potential. Gastroenterology 137 (1), 165–175. 10.1053/j.gastro.2009.03.041 [DOI] [PubMed] [Google Scholar]
- Hao M., Barlogie B., Tricot G., Liu L., Qiu L., Shaughnessy J. D., Jr., et al. (2019). Gene expression profiling reveals aberrant T-cell marker expression on tumor cells of Waldenstrom’s macroglobulinemia. Clin. Cancer Res. 25 (1), 201–209. 10.1158/1078-0432.CCR-18-1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro A., Takahata T., Saito M., Yoshiya G., Tamura Y., Sasaki M., et al. (2006). Influence of methylated p15 and p16 genes on clinicopathological features in colorectal cancer. J. Gastroenterol. Hepatol. 21 (8), 1334–1339. 10.1111/j.1440-1746.2006.04137.x [DOI] [PubMed] [Google Scholar]
- Italiano A., Saint-Paul M. C., Caroli-Bosc F. X., Francois E., Bourgeon A., Benchimol D., et al. (2005). Epidermal growth factor receptor (EGFR) status in primary colorectal tumors correlates with EGFR expression in related metastatic sites: biological and clinical implications. Ann. Oncol. 16 (9), 1503–1507. 10.1093/annonc/mdi282 [DOI] [PubMed] [Google Scholar]
- Jin R., Shen J., Zhang T., Liu Q., Liao C., Ma H., et al. (2017). The highly expressed COL4A1 genes contributes to the proliferation and migration of the invasive ductal carcinomas. Oncotarget 8 (35), 58172–58183. 10.18632/oncotarget.17345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaan Z., Qadan M., Eichenberger M. R., Galandiuk S. (2010). The actin-cytoskeleton pathway and its potential role in inflammatory bowel disease-associated human colorectal cancer. Genet. Test. Mol. Biomarkers 14 (3), 347–353. 10.1089/gtmb.2009.0197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. Y., Sharpless N. E. (2006). The regulation of INK4/ARF in cancer and aging. Cell 127 (2), 265–275. 10.1016/j.cell.2006.10.003 [DOI] [PubMed] [Google Scholar]
- Kuo D. S., Labelle-Dumais C., Gould D. B. (2012). COL4A1 and COL4A2 mutations and disease: insights into pathogenic mechanisms and potential therapeutic targets. Hum. Mol. Genet. 21 (R1), R97–110. 10.1093/hmg/dds346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano H., Shimizu S., Koya R. C., Fujita H., Kamada S., Kuzumaki N., et al. (2000). Human gelsolin prevents apoptosis by inhibiting apoptotic mitochondrial changes via closing VDAC. Oncogene 19 (42), 4807–4814. 10.1038/sj.onc.1203868 [DOI] [PubMed] [Google Scholar]
- Lai X., Su W., Zhao H., Yang S., Zeng T., Wu W., et al. (2018). Loss of scinderin decreased expression of epidermal growth factor receptor and promoted apoptosis of castration-resistant prostate cancer cells. FEBS Open Bio. 8 (5), 743–750. 10.1002/2211-5463.12412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Q., Miki H., Ohmori M., Wu F., Funamoto Y. (2001). Expression of cyclin E and cyclin-dependent kinase 2 correlates with metastasis and prognosis in colorectal carcinoma. Hum. Pathol. 32 (9), 945–953. 10.1053/hupa.2001.27116 [DOI] [PubMed] [Google Scholar]
- Liu H., Shi D., Liu T., Yu Z., Zhou C. (2015). Lentivirus-mediated silencing of SCIN inhibits proliferation of human lung carcinoma cells. Gene 554 (1), 32–39. 10.1016/j.gene.2014.10.013 [DOI] [PubMed] [Google Scholar]
- Liu J. J., Liu J. Y., Chen J., Wu Y. X., Yan P., Ji C. D., et al. (2016). Scinderin promotes the invasion and metastasis of gastric cancer cells and predicts the outcome of patients. Cancer Lett. 376 (1), 110–117. 10.1016/j.canlet.2016.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S., Sakai H. (1990). Inhibition of actin regulatory activity of the 74-kDa protein from bovine adrenal medulla (adseverin) by some phospholipids. J. Biol. Chem. 265 (19), 10940–10942. [PubMed] [Google Scholar]
- Manfredi S., Lepage C., Hatem C., Coatmeur O., Faivre J., Bouvier A. M. (2006). Epidemiology and management of liver metastases from colorectal cancer. Ann. Surg. 244 (2), 254–259. 10.1097/01.sla.0000217629.94941.cf [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura N., Takemori N., Kikugawa T., Tanji N., Higashiyama S., Yokoyama M. (2012). Adseverin: a novel cisplatin-resistant marker in the human bladder cancer cell line HT1376 identified by quantitative proteomic analysis. Mol. Oncol. 6 (3), 311–322. 10.1016/j.molonc.2011.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa T., Baba Y., Yamauchi M., Kuchiba A., Nosho K., Shima K., et al. (2011). STAT3 expression, molecular features, inflammation patterns, and prognosis in a database of 724 colorectal cancers. Clin. Cancer Res. 17 (6), 1452–1462. 10.1158/1078-0432.CCR-10-2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narumiya S., Tanji M., Ishizaki T. (2009). Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 28 (1-2), 65–76. 10.1007/s10555-008-9170-7 [DOI] [PubMed] [Google Scholar]
- Petrelli N. J. (2008). Perioperative or adjuvant therapy for resectable colorectal hepatic metastases. J. Clin. Oncol. 26 (30), 4862–4863. 10.1200/JCO.2008.18.5868 [DOI] [PubMed] [Google Scholar]
- Qiao X., Zhou Y., Xie W., Wang Y., Zhang Y., Tian T., et al. (2018). Scinderin is a novel transcriptional target of BRMS1 involved in regulation of hepatocellular carcinoma cell apoptosis. Am. J. Cancer Res. 8 (6), 1008–1018. [PMC free article] [PubMed] [Google Scholar]
- Rieger K. E., Chu G. (2004). Portrait of transcriptional responses to ultraviolet and ionizing radiation in human cells. Nucleic Acids Res. 32 (16), 4786–4803. 10.1093/nar/gkh783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D. J., Stukel T. A., Gottlieb D. J., Sutherland J. M., Fisher E. S. (2009). Survival after hepatic resection of colorectal cancer metastases: a national experience. Cancer 115 (4), 752–759. 10.1002/cncr.24081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Del Castillo A., Lemaire S., Tchakarov L., Jeyapragasan M., Doucet J. P., Vitale M. L., et al. (1990). Chromaffin cell scinderin, a novel calcium-dependent actin filament-severing protein. EMBO J. 9 (1), 43–52. 10.1002/j.1460-2075.1990.tb08078.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R., Naishadham D., Jemal A. (2013). Cancer statistics, 2013. CA Cancer J. Clin. 63 (1), 11–30. 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- Spano J. P., Lagorce C., Atlan D., Milano G., Domont J., Benamouzig R., et al. (2005). Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann. Oncol. 16 (1), 102–108. 10.1093/annonc/mdi006 [DOI] [PubMed] [Google Scholar]
- Stein U., Walther W., Arlt F., Schwabe H., Smith J., Fichtner I., et al. (2009). MACC1, a newly identified key regulator of HGF-MET signaling, predicts colon cancer metastasis. Nat. Med. 15 (1), 59–67. 10.1038/nm.1889 [DOI] [PubMed] [Google Scholar]
- Steinberg S. M., Shabaneh T. B., Zhang P., Martyanov V., Li Z., Malik B. T., et al. (2017). Myeloid cells that impair immunotherapy are restored in melanomas with acquired resistance to BRAF inhibitors. Cancer Res. 77 (7), 1599–1610. 10.1158/0008-5472.CAN-16-1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B. S., Dong Q. Z., Ye Q. H., Sun H. J., Jia H. L., Zhu X. Q., et al. (2008). Lentiviral-mediated miRNA against osteopontin suppresses tumor growth and metastasis of human hepatocellular carcinoma. Hepatology 48 (6), 1834–1842. 10.1002/hep.22531 [DOI] [PubMed] [Google Scholar]
- Tusher V. G., Tibshirani R., Chu G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad Sci. U. S. A. 98 (9), 5116–5121. 10.1073/pnas.091062498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S., Weinberg R. A. (2011). Tumor metastasis: molecular insights and evolving paradigms. Cell 147 (2), 275–292. 10.1016/j.cell.2011.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Sun S. Q., Yu Y. H., Wu W. Z., Yang S. L., Tan J. M. (2014). Suppression of SCIN inhibits human prostate cancer cell proliferation and induces G0/G1 phase arrest. Int. J. Oncol. 44 (1), 161–166. 10.3892/ijo.2013.2170 [DOI] [PubMed] [Google Scholar]
- Wang Y., Jatkoe T., Zhang Y., Mutch M. G., Talantov D., Jiang J., et al. (2004). Gene expression profiles and molecular markers to predict recurrence of Dukes’ B colon cancer. J. Clin. Oncol. 22 (9), 1564–1571. 10.1200/JCO.2004.08.186 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Komuro Y., Kiyomatsu T., Kanazawa T., Kazama Y., Tanaka J., et al. (2006). Prediction of sensitivity of rectal cancer cells in response to preoperative radiotherapy by DNA microarray analysis of gene expression profiles. Cancer Res. 66 (7), 3370–3374. 10.1158/0008-5472.CAN-05-3834 [DOI] [PubMed] [Google Scholar]
- Weichert W., Roske A., Niesporek S., Noske A., Buckendahl A. C., Dietel M., et al. (2008). Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: specific role of class I histone deacetylases in vitro and in vivo. Clin. Cancer Res. 14 (6), 1669–1677. 10.1158/1078-0432.CCR-07-0990 [DOI] [PubMed] [Google Scholar]
- Yarden Y., Pines G. (2012). The ERBB network: at last, cancer therapy meets systems biology. Nat. Rev. Cancer 12 (8), 553–563. 10.1038/nrc3309 [DOI] [PubMed] [Google Scholar]
- Yu H., Pardoll D., Jove R. (2009). STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer 9 (11), 798–809. 10.1038/nrc2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Nanney L. B., Luongo C., Lamps L., Heppner K. J., DuBois R. N., et al. (1997). Concurrent overexpression of cyclin D1 and cyclin-dependent kinase 4 (Cdk4) in intestinal adenomas from multiple intestinal neoplasia (Min) mice and human familial adenomatous polyposis patients. Cancer Res. 57 (1), 169–175. [PubMed] [Google Scholar]
- Zunino R., Li Q., Rose S. D., Romero-Benitez M. M., Lejen T., Brandan N. C., et al. (2001). Expression of scinderin in megakaryoblastic leukemia cells induces differentiation, maturation, and apoptosis with release of plateletlike particles and inhibits proliferation and tumorigenesis. Blood 98 (7), 2210–2219. 10.1182/blood.V98.7.2210 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.