Abstract
Gastro-intestinal nematodes, especially Haemonchus contortus, are widespread pathogenic parasites of small ruminants. Studying their spatial genetic structure is as important as studying host genetic structure to fully understand host-parasite interactions and transmission patterns. For parasites having a simple life cycle (e.g., monoxenous parasites), gene flow and spatial genetic structure are expected to strongly rely on the socio-spatial behavior of their hosts. Based on five microsatellite loci, we tested this hypothesis for H. contortus sampled in a wild Mediterranean mouflon population (Ovis gmelini musimon × Ovis sp.) in which species- and environment-related characteristics have been found to generate socio-spatial units. We nevertheless found that their parasites had no spatial genetic structure, suggesting that mouflon behavior was not enough to limit parasite dispersal in this study area and/or that other ecological and biological factors were involved in this process, for example other hosts, the parasite life cycle, or the study area history.
Keywords: Ovis gmelini musimon × Ovis sp., Host-parasite co-structure, Population genetics, Nematode, Mouflon, Haemonchus contortus
Abstract
Les nématodes gastro-intestinaux, et plus particulièrement Haemonchus contortus, sont cosmopolites et pathogènes chez les petits ruminants. Étudier leur structure génétique spatiale est aussi important que d’étudier celle des hôtes pour pleinement comprendre les interactions hôtes-parasites et les processus de transmission. Pour les parasites ayant des cycles de vie simples (par exemple, les parasites monoxènes), on s’attend à ce que les flux de gènes et la structure génétique spatiale dépendent fortement du comportement socio-spatial de leurs hôtes. En utilisant cinq loci microsatellites, nous avons testé cette hypothèse pour des H. contortus échantillonnés dans une population sauvage de mouflons méditerranéens (Ovis gmelini musimon × Ovis sp.) dans laquelle les caractéristiques de l’espèce et de l’environnement génèrent des unités socio-spatiales. Nous avons néanmoins mis en évidence que leurs parasites ne présentent pas de structure génétique spatiale, ce qui suggère que le comportement des mouflons ne restreint pas la dispersion des parasites dans cette aire d’étude et/ou que d’autres facteurs biologiques et écologiques tels que d’autres hôtes, le cycle de vie du parasite, ou l’histoire de l’aire d’étude jouent un rôle dans ce processus.
Introduction
Parasitism has been shown to impact numerous host characteristics (e.g., survival [45], body condition [20] and behavior [23]). Studying parasite population ecology is thus crucial to better understand and predict parasite impacts on host populations. Among the diverse ecological elements to be studied to have a complete picture of parasite population ecology, population genetics is among the most important since it makes it possible to identify ecological drivers of population structure, helping to gather information about processes often difficult to observe directly in parasite species (e.g., species, such as dispersal or demographic changes [34]). Due to their lifestyle, panmixia of a parasite population (and thus random spatial distribution of allelic frequencies) can be disrupted by the non-random transmission of parasites between hosts [34]. Spatial genetic structures of parasite populations are thus expected to rely on different hosts characteristics (e.g., spatial behavior and dispersal) [15, 41, 62].
Ruminants can host a large diversity of external and internal parasite species, especially in their digestive tract [71, 76]. Gastrointestinal parasites are major parasites of ruminants, due to their high prevalence and potential impact on their host fitness and population dynamics, in both domestic (e.g., cattle Bos taurus [11] and sheep Ovis aries [69]), and wild ruminants (e.g., Soay sheep Ovis aries L. [16, 40]). The most prevalent gastrointestinal parasites are often the Coccidia and nematodes (e.g., in roe deer Capreolus capreolus [2], Mediterranean mouflon Ovis gmelini musimon × Ovis sp [14], see also [44] for a review, African buffalo Syncerus caffer [29], zebra Equus quagga, springbok Antidorcas marsupialis, blue wildebeest Connochaetes taurinus, gemsbok Oryx gazella [44]). Among the diverse abomasal species of parasites identified, the nematode Haemonchus contortus is a widespread and pathogenic parasitic worm of small domestic and wild ruminants (e.g., mouflon [44, 52, 65, 82], chamois Rupicapra r. rupicapra, roe deer, Alpine ibex Capra ibex ibex, domestic goat Capra hircus and sheep [9], African buffalo [6]). Studying population ecology of this parasite, including population genetics, is of prime importance for a better understanding and management of its impact on host populations. While most studies on the genetic structures of H. contortus populations were performed on livestock (see [9, 72]), revealing low levels of genetic differentiation even at large (e.g., state or country) spatial scales (reviewed by [33]), scarce knowledge is available in wild populations [15]. In such populations, the behavioral ecology of the host species (e.g., philopatry, sexual segregation, dispersal or migration) and landscape structure and connectivity influence host movements (e.g., [42, 83]) and may generate marked socio-spatial structures at small intra population scales (e.g., [53, 64]). However, how these host populations’ structures influence the gene flow of their parasites is still an open question for numerous populations of wild ruminants [15].
We aimed here to help answer this question by studying the spatial genetic structure of a H. contortus population parasitizing an isolated wild Mediterranean mouflon population. In this population, males and females have been shown to be spatially structured (Supplementary Data 1 and [32, 57, 66]) and to have stable home ranges from year to year (see [54], Appendix S2 in [55]). Because H. contortus has a direct life cycle (monoxenous parasite), for which free-living infesting larvae remain close to the host feces [59], we expected the socio-spatial behavior to generate a spatial genetic signature in the population of H. contortus.
Materials and methods
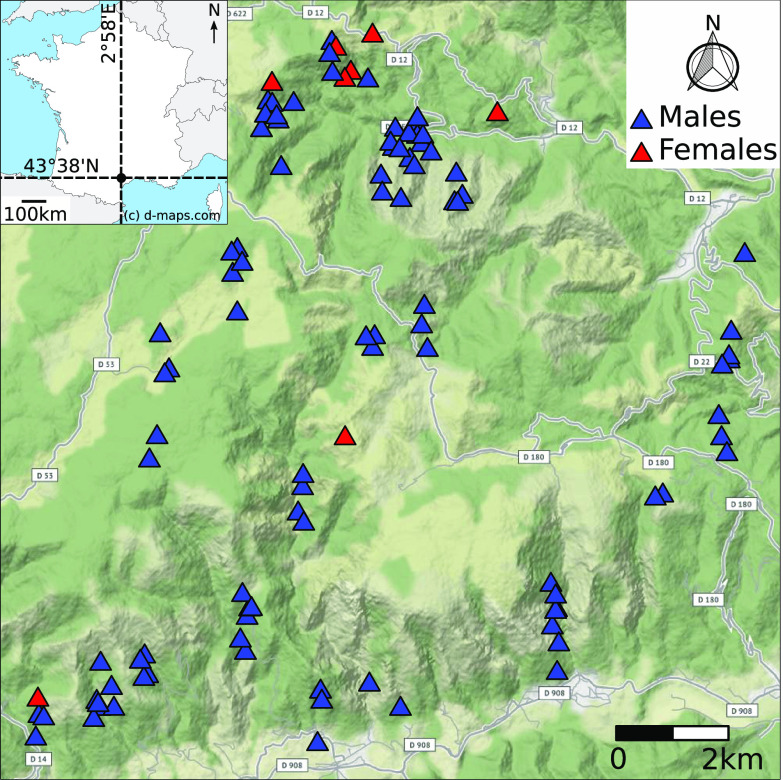
Parasites were sampled from the abomasum of 85 Mediterranean mouflon harvested between September 2011 and February 2012 in the Caroux-Espinouse massif (43°38′N, 2°58′E, 17,000 ha, 130–1124 m a.s.l, southern France, Fig. 1). Morphologically identified H. contortus [74] were present in 70.5% of individuals (based on an aliquot of the abomasum content: estimated median number of worms by parasitized individuals = 39, min = 3, max = 1100). For genetic analyses, a total of 115 adult H. contortus (107 males and 8 females, Fig. 1) were sampled from 43 mouflon (33 males and 10 females, all but one older than 4 years, i.e., adult individuals having a fixed home range [24, 25, 27]) so that a mean of 2.67 H. contortus were sampled by host (min = 1, max = 7).
Figure 1.
Map of the Caroux-Espinouse massif and locations of the Haemonchus contortus sampled (blue triangles: males, red triangles: females). Random spatial noise of a few meters using a uniform distribution was introduced to avoid obtaining duplicate coordinates for parasites sampled in a same host. The map was generated with the ggmap R package [49]. France country map© d-maps.com (https://d-maps.com/carte.php?num_car=2818&lang=en).
DNA was extracted from 5 mm of the body of each individual, sampled in the head extremity, and avoiding female genital cords and hence egg DNA contamination. We used the EZ-10 Spin Column genomic DNA Minipreps Biobasic kit (ref BS628). Following supplier recommendations but adjusting volumes to the small size of samples, the lysis was performed in 100 μL of ACL buffer and 7 μL of proteinase K. Samples were incubated for 1 h at 55 °C under agitation (400 rpm). Purification and the two washing steps were performed using 150 μL of AB solution and 200 μL of buffer for each washing. Elution was done using 50 μL of EB buffer.
For each sample, seven microsatellites (Hcms25, Hcms27, Hcms33, Hcms36, Hcms40, Hcms22co3 and Hcms8a20, see [63, 68], three multiplexes, see Supplementary Table 1) were amplified through polymerase chain reaction (PCR) in a final volume of 15 μL composed of QIAGEN Multiplex PCR kit Mastermix (ref. 206145), 40 nM of each primer, and 2 μL of DNA solution. PCR cycles consisted of 15 min of activation (95 °C), followed by 40 cycles of denaturation (30 s, 94 °C), annealing (1 min 30 s at primer-specific annealing temperature, see Supplementary Data 2) and extension (1 min, 72 °C). Cycles were followed by a final extension step (30 min, 60 °C). PCR products were resolved on a capillary sequencer ABI 3730XL (Thermo Fisher Scientific) by the Genoscreen laboratory (Lille, France). The electropherograms obtained were analyzed using GENEMAPPER software (Applied Biosystems/Life Technologies) and read by two independent analysts to determine allele sizes for each individual and markers. This microsatellite panel is known to produce unambiguous genotypes and to be highly polymorphic, and has demonstrated its relevance in previous H. contortus genetic structure studies (e.g., [68]).
Genotyping errors were tracked using MICROCHECKER v.2.2.3 software [78]. Using FSTAT v.2.9.3.2 software [35, 36], we determined genetic diversity indices (see Table 1) and tested for departures from Hardy–Weinberg (HW) equilibrium and linkage disequilibrium between pairs of loci (none detected, results not shown). Observed heterozygosity (Ho) was determined using R software (R core team 2016), and the hierfstat package [37].
Table 1.
Number of alleles sampled (N a), allelic richness (A R), observed heterozygosity (Ho), expected heterozygosity (He) and Fis values (bold values are significantly different from zero, adjusted Bonferroni nominal levels: 0.01 [3]) for the five loci included in the population genetics analyses of the Haemonchus contortus sampled in the Caroux-Espinouse massif.
The spatial population genetic structure of H. contortus was first investigated using a sPCA (spatial Principal Component Analysis, library adegenet of R software) [47, 48, 60]. We performed eigenvalue tests (n = 9999) to assess the significance of the local and global spatial structures [60]. The connection network was set using the inverse of the Euclidean pairwise distances between individuals. We then ran 10 independent runs of the MCMC simulations implemented in GENELAND v.4.0.8 software [39], using the correlated allele frequencies and the null allele models (see Results) to test for K varying from 1 to 10, with 1,000,000 iterations, a thinning of 100, and a burn-in of 1000. All analyses involving R packages were conducted with R 3.3.2 (R core team 2016).
Results
Of the seven loci selected for genotyping, one (Hcms8a20, Supplementary Data 2) failed to amplify and was thus not included in the dataset. MICROCHECKER indicated higher than 0.05 null allele frequencies for the loci Hcms22Co3, Hcms27 and Hcms40 (f = 0.18, 0.29 and 0.20, respectively, Van Oosterhout et al.’s estimator, [78]). In addition, 28% of the sampled individuals failed to be genotyped at the Hcms27 locus which was thus excluded from the dataset. The five remaining loci considered in subsequent analyses showed a relatively high level of genetic diversity (Table 1). For all loci, the Fis value was 0.16 and significant (p = 0.01), suggesting overall deviation from Hardy–Weinberg equilibrium likely attributed to the high null allele frequencies observed for some loci [19].
The sPCA revealed no significant global (p = 0.97) or local (p = 0.32) spatial genetic structures when considering the first positive and negative axes (Supplementary Data 3). Accordingly, in the 10 independent GENELAND runs, the maximum posterior density was obtained for K = 1 (Supplementary Data 3), indicating an absence of spatial genetic structure in the study area.
Discussion
In the present study, we hypothesized the socio-spatial behavior of Mediterranean mouflon (Supplementary Data 1, [32, 57, 66]) to limit H. contortus gene flow, resulting in a significant genetic structure in the parasite population. We, however, did not evidence any spatial patterns in the distribution of parasite genetic variability. An absence of genetic differentiation, even at large spatial scales, has already been described for nematodes in wild host populations [1, 72]. Several explanations can be proposed to explain such a result in the study area.
First, gene flow of parasite species such as H. contortus having an environmental phase in their life cycle might be maximized even if their hosts are only slightly mobile, because this allows for parasite exchanges without a need for the hosts to encounter one another, but just to share feeding areas. In addition, male mouflon perform reproductive excursions during the rutting period (Marchand et al., unpublished data [66]) and, even though this is not systematic, young males might disperse [26] and act as super-spreaders [51, 79] due to higher shedding rates than adults (e.g., Bourgoin et al. unpublished data, [80]). Finally, although Mediterranean mouflon are spatially structured, some overlap persists between socio-spatial unit home ranges (see Fig. 1 in [56], Supporting information G in [66]). Taken together, these host behavioral characteristics might favor step by step parasite exchanges between sub-populations of hosts, and be sufficient to ensure parasite gene flow across the entire study area.
Second, H. contortus is a generalist parasite of small ruminants [75], and roe deer (Capreolus capreolus) inhabit our study area. Although roe deer are present at much lower densities than those for Mediterranean mouflon [4], they may contribute to H. contortus gene flow by having different socio-spatial behaviors such as territoriality [43], attraction for forest edges [70], or marked dispersal abilities [21], linked to their parasitic infestation [22]. In addition, while currently only low numbers of domestic ruminants (~300 sheep and 300 cattle, French Ministry of Agriculture and Food 2000) are reared in the massif, mainly in the outlying areas of the mouflon range [5, 17, 18], domestic sheep and cattle were historically present at high densities in the study area before Mediterranean mouflon introduction (60 years ago). H. contortus might thus also have been present before the mouflon introduction, and the current spatial genetic structure of H. contortus could result from the historical and contemporary interactions between these three different hosts’ spatial structures and behaviors.
We encountered methodological issues with null alleles that may also raise questions about our statistical power for detecting spatial genetic structure in the parasite population. However, null alleles tend generally to induce an overestimation of genetic structure rather than the opposite, and only interfere slightly with assignments of individuals to genetic clusters [7, 10]. Since we observed no genetic structure, we were therefore confident that the presence of null alleles here led to conservative results. In addition, the sPCA uses allele frequencies as variables [47] and numerous alleles were identified in the five loci studied here (see Table 1), consequently increasing the statistical power to detect genetic structure, even though only a limited number of loci are involved. The relatively high null allele frequencies observed reflected the high genetic diversity reported in H. contortus worldwide and in the present study, in which the number of alleles by locus is even higher than the number observed in other populations (see e.g., [68, 81]). Null alleles have frequently been reported in other studies on H. contortus (e.g., [12, 13, 46, 68]) and more generally on parasitic nematodes (e.g., [38, 73]). This can be explained by the high effective population sizes (one host can carry thousands of worms) characterizing parasitic worms, favoring rapid evolution of DNA sequences and thus mutation in the flanking region of microsatellite loci [10, 63].
Conclusions and perspectives
Contrary to our expectations, we did not detect any spatial genetic structure in H. contortus parasitizing Mediterranean mouflon of the Caroux-Espinouse massif. This result highlights that studying both sides of host-parasite interactions is crucial to fully understand and predict the sanitary evolution of populations, since parasite dispersal is often the result of more than one ecological factor [58]. It also reveals that results about the impacts of host spatial behavior on parasite transmission (e.g., [41]) might be difficult to generalize to diverse host-parasite systems. Specific studies on given host-parasite systems are thus needed to conclude about parasite population ecology and impacts of host ecology on their evolution. It is especially important for parasites having an indirect transmission process such as H. contortus or for wild hosts being in contact with domestic animals since these biological characteristics might interact with host ecology and increase the spatial scale at which parasite transmission occurs (e.g., [8]). In the current global context of habitat fragmentation [30], which has the potential to impact host population structures, genetic diversity and thus fitness [31, 67], the results of the present study also demonstrate how the overall impact of changes on populations should be assessed at the community level, since different species, such as hosts and parasites, might be differently impacted. To go further in the understanding of H. contortus population ecology, an interesting perspective could be to resample H. contortus in the different host species (mouflon, roe deer and domestic ruminants) present in the study area to determine whether the panmictic population we detected in Mediterranean mouflon also extends to other sympatric species. Such a comparison would help us to understand how parasites spread in the Caroux-Espinouse massif and give supplementary information to wildlife managers when defining management and conservation planning.
Acknowledgments
We warmly thank all the professionals from the Office National de la Chasse et de la Faune Sauvage, the Office National des Forêts and the Groupement d’Intérêt Environnemental et Cynégétique of Caroux-Espinouse (Patricia Michel and all the hunters who allowed us to collect digestive tracts of mouflon). We thank all the technical personnel of LBBE for helping with the laboratory steps of the study and Julien Maerten and Guillaume Pratviel for parasitological analyses of mouflon. This work was supported financially by Vetagro Sup, the LBBE-UMR CNRS 5558 and the Office National de la Chasse et de la Faune Sauvage. The authors declare they have no conflict of interest. The authors are grateful to the two anonymous reviewers for their help in improving the manuscript. G. Bourgoin has received the support of the EU in the framework of the Marie-Curie FP7 COFUND People Programme, through the award of an AgreenSkills+ fellowship (under grant agreement n°609398).
Cite this article as: Portanier E, Garel M, Devillard S, Duhayer J, Poirel M-T, Henri H, Régis C, Maillard D, Redman E, Itty C, Michel P & Bourgoin G. 2019. Does host socio-spatial behavior lead to a fine-scale spatial genetic structure in its associated parasites? Parasite 26, 64.
Supplementary material
Supplementary materials are available at https://www.parasite-journal.org/10.1051/parasite/2019062/olm
References
- 1.Archie EA, Ezenwa VO. 2011. Population genetic structure and history of a generalist parasite infecting multiple sympatric host species. International Journal for Parasitology, 41, 89–98. [DOI] [PubMed] [Google Scholar]
- 2.Body G, Ferté H, Gaillard JM, Delorme D, Klein F, Gilot-Fromont E. 2011. Population density and phenotypic attributes influence the level of nematode parasitism in roe deer. Oecologia, 167, 635–646. [DOI] [PubMed] [Google Scholar]
- 3.Bonferroni CE. 1936. Teoria statistica delle classi e calcolo delle probabilità. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze, 8, 3–62. [Google Scholar]
- 4.Bourgoin G, Marchand P, Hewison AJM, Ruckstuhl KE, Garel M. 2018. Social behaviour as a predominant driver of sexual, age-dependent and reproductive segregation in Mediterranean mouflon. Animal Behaviour, 136, 87–100. [Google Scholar]
- 5.Bousquel V. 1999. Analyse historique de l’évolution de la végétation sur le massif du Caroux-Espinouse. Rapport Life “Gestion conservatoire des pelouses et landes en région méditerranéenne”.
- 6.Budischak SA, O’Neal D, Jolles AE, Ezenwa VO. 2018. Differential host responses to parasitism shape divergent fitness costs of infection. Functional Ecology, 32, 324–333. [Google Scholar]
- 7.Carlsson J. 2008. Effects of microsatellite null alleles on assignment testing. Journal of Heredity, 99, 616–623. [DOI] [PubMed] [Google Scholar]
- 8.Cassirer EF, Manlove KR, Almberg ES, Kamath PL, Cox M, Wolff P, Roug A, Shannon J, Robinson R, Harris RB, Gonzales BJ, Plowright RK, Hudson PJ, Cross PC, Dobson A, Besser TE. 2018. Pneumonia in Bighorn sheep: risk and resilience. Journal of Wildlife Management, 82, 32–45. [Google Scholar]
- 9.Cerutti MC, Citterio CV, Bazzocchi C, Epis S, D’Amelio S, Ferrari N, Lanfranchi P. 2010. Genetic variability of Haemonchus contortus (Nematoda: Trichostrongyloidea) in alpine ruminant host species. Journal of Helminthology, 84, 276–283. [DOI] [PubMed] [Google Scholar]
- 10.Chapuis MP, Estoup A. 2007. Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution, 24, 621–631. [DOI] [PubMed] [Google Scholar]
- 11.Charlier J, Höglund J, von Samson-Himmelstjerna G, Dorny P, Vercruysse J. 2009. Gastrointestinal nematode infections in adult dairy cattle: impact on production, diagnosis and control. Veterinary Parasitology, 164, 70–79. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhry U, Redman EM, Raman M, Gilleard JS. 2015. Genetic evidence for the spread of a benzimidazole resistance mutation across southern India from a single origin in the parasitic nematode Haemonchus contortus. International Journal for Parasitology, 45, 721–728. [DOI] [PubMed] [Google Scholar]
- 13.Chaudhry U, Redman EM, Ashraf K, Shabbir MZ, Rashid MI, Ashraf S, Gilleard JS. 2016. Microsatellite marker analysis of Haemonchus contortus populations from Pakistan suggests that frequent benzimidazole drug treatment does not result in a reduction of overall genetic diversity. Parasites & Vectors, 9, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cockenpot A. 2013. Étude des facteurs de variation de l’excrétion parasitaire mesurée par analyse coproscopique chez le mouflon Méditerranéen (Ovis gmelini musimon × Ovis sp.) dans le massif du Caroux-Espinouse. France: Veterinary dissertation, VetAgro Sup, Campus vétérinaire de Lyon, Université de Lyon. [Google Scholar]
- 15.Cole R, Viney M. 2018. The population genetics of parasitic nematodes of wild animals. Parasites & Vectors, 11, 590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig BH, Pilkington JG, Pemberton JM. 2006. Gastrointestinal nematode species burdens and host mortality in a feral sheep population. Parasitology, 133, 485–496. [DOI] [PubMed] [Google Scholar]
- 17.Cugnasse JM, Chiappin D. 1992. Statut ancien et actuel du chevreuil (Capreolus capreolus) dans le département du Gard, perspectives. Bulletin mensuel de l’Office. National de la Chasse, 164, 26–38. [Google Scholar]
- 18.Cugnasse JM, Houssin H. 1993. Acclimatation du mouflon en France : la contribution des réserves de l’Office national de la chasse. Bulletin mensuel de l’Office National de la Chasse, 183, 26–37. [Google Scholar]
- 19.De Meeûs T. 2018. Revisiting FIS, FST, Wahlund effects, and null alleles. Journal of Heredity, 109, 446–456. [DOI] [PubMed] [Google Scholar]
- 20.Debeffe L, Mcloughlin PD, Medill SA, Stewart K, Andres D, Shury T, Wagner B, Jenkins E, Gilleard JS, Poissant J. 2016. Negative covariance between parasite load and body condition in a population of feral horses. Parasitology, 143, 983–997. [DOI] [PubMed] [Google Scholar]
- 21.Debeffe L, Morellet N, Cargnelutti B, Lourter B, Bon R, Gaillard JM, Hewison AJM. 2012. Condition-dependent natal dispersal in a large herbivore: heavier animals show a greater propensity to disperse and travel further. Journal of Animal Ecology, 81, 1327–1337. [DOI] [PubMed] [Google Scholar]
- 22.Debeffe L, Morellet N, Verheyden H, Hoste H, Gaillard JM, Cargnelutti B, Picot D, Sevila J, Hewison AJM. 2014. Parasite abundance contributes to condition-dependent dispersal in a wild population of large herbivores. Oikos, 123, 1121–1125. [Google Scholar]
- 23.Dobson AP. 1988. The population biology of parasite-induced changes in host behaviour. Quarterly Review of Biology, 63, 139–165. [DOI] [PubMed] [Google Scholar]
- 24.Dubois M, Gerard J, Maublanc ML. 1992. Seasonal movements of females Corsican mouflon (Ovis ammon) in a Mediterranean mountain range, southern France. Behavioural Processes, 26, 155–166. [DOI] [PubMed] [Google Scholar]
- 25.Dubois M, Quenette P, Bideau E. 1993. Seasonal range use by European mouflon rams in medium altitude mountains. Acta Theriologica, 38, 185–198. [Google Scholar]
- 26.Dubois M, Khazraïe K, Guilhem C, Maublanc ML, Le Pendu Y. 1996. Philopatry in mouflon rams during the rutting season: psycho-ethological determinism and functional consequences. Behavioural Processes, 35, 93–100. [DOI] [PubMed] [Google Scholar]
- 27.Dupuis J, Badia J, Maublanc ML, Bon R. 2002. Survival and spatial fidelity of mouflon (Ovis gmelini): a Bayesian analysis of an age-dependent capture-recapture model. Journal of Agricultural, Biological, and Environmental Statistics, 7, 277–298. [Google Scholar]
- 28.El Mousadik A, Petit RJ. 1996. High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theoretical and Applied Genetics, 92, 832–839. [DOI] [PubMed] [Google Scholar]
- 29.Ezenwa V, Jolles A. 2008. Horns honestly advertise parasite infection in male and female African buffalo. Animal Behaviour, 75, 2013–2021. [Google Scholar]
- 30.Fahrig L. 2003. Effects of habitat fragmentation on biodiversity. Annual Review of Ecology, Evolution, and Systematics, 34, 487–515. [Google Scholar]
- 31.Frankham R, Ballou JD, Briscoe DA. 2004. A primer of conservation genetics. New York: Cambridge University Press. [Google Scholar]
- 32.Garel M, Cugnasse JM, Maillard D, Gaillard JM, Hewison AJM, Dubray D. 2007. Selective harvesting and habitat loss produce long-term life history changes in a mouflon population. Ecological Applications, 17, 1607–1618. [DOI] [PubMed] [Google Scholar]
- 33.Gilleard JS, Redman E. 2016. Chapter Two – Genetic diversity and population structure of Haemonchus contortus. Advances in Parasitology, 93, 31–68. [DOI] [PubMed] [Google Scholar]
- 34.Gorton MJ, Kasl AL, Detwiler JT, Criscione CD. 2012. Testing local-scale panmixia provides insights into the cryptic ecology, evolution, and epidemiology of metazoan animal parasites. Parasitology, 139, 981–997. [DOI] [PubMed] [Google Scholar]
- 35.Goudet J. 1995. FSTAT: a computer program to calculate F-statistics. Journal of Heredity, 86, 485–486. [Google Scholar]
- 36.Goudet J. 2001. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www2.unil.ch/popgen/softwares/fstat.htm. Updated from Goudet (1995).
- 37.Goudet J. 2005. HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Molecular Ecology Notes, 5, 184–186. [Google Scholar]
- 38.Grillo V, Jackson F, Cabaret J, Gilleard JS. 2007. Population genetic analysis of the ovine parasitic nematode Teladorsagia circumcincta and evidence for a cryptic species. International Journal for Parasitology, 37, 435–447. [DOI] [PubMed] [Google Scholar]
- 39.Guillot G, Renaud S, Ledevin R, Michaux J, Claude J. 2012. A unifying model for the analysis of phenotypic, genetic and geographic data. Systematic Biology, 61, 897–911. [DOI] [PubMed] [Google Scholar]
- 40.Gulland FMD. 1992. The role of nematode parasites in Soay sheep (Ovis aries L.) mortality during a population crash. Parasitology, 105, 493–503. [DOI] [PubMed] [Google Scholar]
- 41.Gustafson KD, Newman RA, Rhen T, Tkach VV. 2018. Spatial and genetic structure of directly-transmitted parasites reflects the distribution of their specific amphibian hosts. Population Ecology, 60, 261–273. [Google Scholar]
- 42.Harris RJ, Reed JM. 2002. Behavioral barriers to non-migratory movements of birds. Annales Zoologici Fennici, 39, 275–290. [Google Scholar]
- 43.Hewison AJM, Vicent P, Reby D. 1998. Social organisation of European roe deer, in The European Roe Deer: The Biology of Success. Andersen R, Duncan P, Linnell JDC, Editors. Scandinavian University Press: Stockholm: p. 189–220. [Google Scholar]
- 44.Hille G. 2003. Untersuchungen zum Endoparasitenbefall des Muffelwildes in Thüringen. Germany: Inaugural-Dissertation, Universität Leipzig. [Google Scholar]
- 45.Hudson PJ, Dobson AP, Newborn D. 1998. Prevention of population cycles by parasite removal. Science, 282, 2256–2258. [DOI] [PubMed] [Google Scholar]
- 46.Hunt PW, Knox MR, Le Jambre LF, McNally J, Anderson LJ. 2008. Genetic and phenotypic differences between isolates of Haemonchus contortus in Australia. International Journal for Parasitology, 38, 885–900. [DOI] [PubMed] [Google Scholar]
- 47.Jombart T, Devillard S, Dufour AB, Pontier D. 2008. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity, 101, 92–103. [DOI] [PubMed] [Google Scholar]
- 48.Jombart T. 2008. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics, 24, 1403–1405. [DOI] [PubMed] [Google Scholar]
- 49.Kahle D, Wickham H. 2013. Ggmap: spatial visualization with ggplot2. The R Journal, 5, 144–161. [Google Scholar]
- 50.Kaufman L, Rousseeuw P. 1990. Finding groups in data: an introduction to cluster analysis. Hoboken: John Wiley & Sons Inc., Publication, Wiley-Interscience. [Google Scholar]
- 51.Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. 2005. Superspreading and the effect of individual variation on disease emergence. Nature, 438, 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maerten J. 2014. Détermination du parasitisme de l’abomasum chez le mouflon méditerranéen (Ovis gmelini musimon × Ovis sp.) dans le massif du Caroux-Espinouse. France: Veterinary dissertation, VetAgro Sup, Campus vétérinaire de Lyon, Université de Lyon. [Google Scholar]
- 53.Marchand P, Freycon P, Herbaux JP, Game Y, Toïgo C, Gilot-Fromont E, Rossi S, Hars J. 2017. Sociospatial structure explains marked variation in brucellosis seroprevalence in an Alpine ibex population. Scientific Reports, 7, 15592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marchand P, Garel M, Bourgoin G, Michel P, Maillard D, Loison A. 2014. Habitat-related variation in carcass mass of a large herbivore revealed by combining hunting and GPS data. Journal of Wildlife Management, 78, 657–670. [Google Scholar]
- 55.Marchand P, Garel M, Bourgoin G, Combining familiarity and landscape features helps break down the barriers between movements and home ranges in a non-territorial large herbivore. Journal of Animal Ecology, 86, 371–383. [DOI] [PubMed] [Google Scholar]
- 56.Marchand P, Garel M, Bourgoin G, Dubray D, Maillard D, Loison A. 2015. Sex-specific adjustments in habitat selection contribute to buffer mouflon against summer conditions. Behavioral Ecology, 26, 472–482. [Google Scholar]
- 57.Martins AG, Netto NT, Aulagnier S, Borges A, Dubois M, Vicente L, Gerard J-F, Maublanc ML. 2002. Population subdivision among mouflon sheep (Ovis gmelini) ewes and ranging behaviour of rams during the rut. Journal of Zoology, 258, 27–37. [Google Scholar]
- 58.Mazé-Guilmo E, Blanchet S, McCoy KD, Loot G. 2016. Host dispersal as the driver of parasite genetic structure: a paradigm lost? Ecology Letters, 19, 336–347. [DOI] [PubMed] [Google Scholar]
- 59.Molento MB, Buzatti A, Sprenger LK. 2016. Pasture larval count as a supporting method for parasite epidemiology, population dynamic and control in ruminants. Livestock Science, 192, 48–54. [Google Scholar]
- 60.Montano V, Jombart T. 2017. An Eigenvalue test for spatial principal component analysis. BMC Bioinformatics, 18, 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nei M. 1973. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences, 70, 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nieberding CM, Durette-Desset MC, Vanderpoorten A, Casanova JC, Ribas A, Deffontaine V, Feliu C, Morand S, Libois R, Michaux JR. 2008. Geography and host biogeography matter for understanding the phylogeography of a parasite. Molecular Phylogenetics and Evolution, 47, 538–554. [DOI] [PubMed] [Google Scholar]
- 63.Otsen M, Plas ME, Lenstra JA, Roos MH, Hoelstra R. 2000. Microsatellite diversity of isolates of the parasitic nematode Haemonchus contortus. Molecular Biochemical Parasitology, 110, 69–77. [DOI] [PubMed] [Google Scholar]
- 64.Podgórski T, Scandura M, Jedrzejewska B. 2014. Next of kin next door – philopatry and socio-genetic population structure in wild boar. Journal of Zoology, 294, 190–197. [Google Scholar]
- 65.Poglayen G, Urbani L, Modugno F, Scala A, Giannetto S, Rossi L. 2018. The Italian Mouflon (Ovis musimon): a brief history of its parasites in the last 45 years. Research Journal of Zoology, 1, 2. [Google Scholar]
- 66.Portanier E, Garel M, Devillard S, Marchand P, Andru J, Maillard D, Bourgoin G. 2017. Introduction history overrides social factors in explaining genetic structure of females in Mediterranean mouflon. Ecology and Evolution, 7, 9580–9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Portanier E, Garel M, Devillard S, Maillard D, Poissant J, Galan M, Benabed S, Poirel MT, Duhayer J, Itty C, Bourgoin G. 2019. Both candidate gene and neutral genetic diversity correlate with parasite resistance in female Mediterranean mouflon. BMC Ecology, 19, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Redman E, Packard E, Grillo V, Smith J, Jackson F, Gilleard JS. 2008. Microsatellite analysis reveals marked genetic differentiation between Haemonchus contortus laboratory isolates and provides a rapid system of genetic fingerprinting. International Journal for Parasitology, 38, 111–122. [DOI] [PubMed] [Google Scholar]
- 69.Roeber F, Jex AR, Gasser RB. 2013. Impact of gastrointestinal parasitic nematodes of sheep, and the role of advanced molecular tools for exploring epidemiology and drug resistance – an Australian perspective. Parasites & Vectors, 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saïd S, Servanty S. 2005. The influence of landscape structure on female roe deer home-range size. Landscape Ecology, 20, 1003–1012. [Google Scholar]
- 71.Samuel WM, Pybus MJ, Kocan AA. 2001. Parasitic diseases of wild mammals, 2nd edn Ames: Iowa State University Press. [Google Scholar]
- 72.Shen D, Wang J, Zhang D, Peng Z, Yang T, Wang Z, Bowman DD, Hou Z, Liu Z. 2017. Genetic diversity of Haemonchus contortus isolated from sympatric wild blue sheep (Pseudois nayaur) and sheep in Helan Mountains, China. Parasites & Vectors, 10, 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silvestre A, Sauve C, Cortet J, Cabaret J. 2009. Contrasting genetic structures of two parasitic nematodes, determined on the basis of neutral microsatellite markers and selected anthelmintic resistance markers. Molecular Ecology, 18, 5086–5100. [DOI] [PubMed] [Google Scholar]
- 74.Skryabin KI, Shikhobalova NP, Schulz RS, Popova TI, Boev SN, Delyamure SL. 1961. Key to parasitic nematodes, Vol. 3. Strongylata. Jerusalem: Israel Program for Scientific Translation. [Google Scholar]
- 75.Taylor M, Coop R, Wall R. 2016. Veterinary parasitology, 4th edn Sussex: Wiley Blackwell. [Google Scholar]
- 76.Taylor MA, Coop RL, Wall RL. 2014. Veterinary parasitology, 4th edn Sussex: Wiley Blackwell. [Google Scholar]
- 77.Turner WC, Getz WM. 2010. Seasonal and demographic factors influencing gastrointestinal parasitism in ungulates of Etosha national park. Journal of Wildlife Diseases, 46, 1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. 2004. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes, 4, 535–538. [Google Scholar]
- 79.VanderWaal KL, Ezenwa VO. 2016. Heterogeneity in pathogen transmission: mechanisms and methodology. Functional Ecology, 30, 1606–1622. [Google Scholar]
- 80.Wood ELD, Matthews JB, Stephenson S, Slote M, Nussey DH. 2013. Variation in fecal egg counts in horses managed for conservation purposes: individual egg shedding consistency, age effects and seasonal variation. Parasitology, 140, 115–128. [DOI] [PubMed] [Google Scholar]
- 81.Yin F, Gasser RB, Li F, Bao M, Huang W, Zou F, Zhao G, Wang C, Yang X, Zhou Y, Zhao J, Fang R, Hu M. 2016. Population structure of Haemonchus contortus from seven geographical regions in China, determined on the basis of microsatellite markers. Parasites & Vectors, 9, 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zaffaroni E, Manfredi MT, Citterio C, Sala M, Piccolo G, Lanfranchi P. 2000. Host specificity of abomasal nematodes in free ranging alpine ruminants. Veterinary Parasitology, 90, 221–230. [DOI] [PubMed] [Google Scholar]
- 83.Zimmermann B, Nelson L, Wabakken P, Sand H, Liberg O. 2014. Behavioral responses of wolves to roads: scale-dependent ambivalence. Behavioral Ecology, 25, 1353–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials are available at https://www.parasite-journal.org/10.1051/parasite/2019062/olm