Diatom ALB3b is required for insertion of Fx-Chl-binding proteins in thylakoid membranes and has a novel conserved domain implying that its interaction partners differ from those in plants/green algae.
Abstract
The family of chloroplast ALBINO3 (ALB3) proteins function in the insertion and assembly of thylakoid membrane protein complexes. Loss of ALB3b in the marine diatom Phaeodactylum tricornutum leads to a striking change of cell color from the normal brown to green. A 75% decrease of the main fucoxanthin-chlorophyll a/c-binding proteins was identified in the alb3b strains as the cause of changes in the spectral properties of the mutant cells. The alb3b lines exhibit a truncated light-harvesting antenna phenotype with reduced amounts of light-harvesting pigments and require a higher light intensity for saturation of photosynthesis. Accumulation of photoprotective pigments and light-harvesting complex stress-related proteins was not negatively affected in the mutant strains, but still the capacity for nonphotochemical quenching was lower compared with the wild type. In plants and green algae, ALB3 proteins interact with members of the chloroplast signal recognition particle pathway through a Lys-rich C-terminal domain. A novel conserved C-terminal domain was identified in diatoms and other stramenopiles, questioning if ALB3b proteins have the same interaction partners as their plant/green algae homologs.
Diatoms (Bacillariophyceae) are a major group of eukaryotic phytoplankton belonging to the phylum Heterokont that evolved through a secondary endosymbiotic event around 200 to 180 million years ago (Brown and Sorhannus, 2010). Diatoms are key primary producers in the marine food chain. They account for 40% of the total carbon fixation in oceans and 25% of the total global oxygen production (Falkowski et al., 1998). Diatom plastids differ substantially from those in green algae and land plants due to their peculiar inheritance and evolution (Oudot-Le Secq et al., 2007). Because of secondary endosymbiotic events, four membranes surround the diatom chloroplast. The outer envelope, known as chloroplast endoplasmic reticulum, is a continuum with the nuclear envelope. The diatom thylakoids are organized in stacked bands of three membranes, also known as girdle lamellae, spanning along the entire length of the plastid. This configuration differs substantially from the classic grana stacks and interconnecting stroma-exposed thylakoid organization found in higher plant chloroplasts (Austin and Staehelin, 2011). Light-harvesting complexes (LHCs) are embedded in the thylakoid membrane of the chloroplast and surround the photosynthetic reaction centers of the photosystems.
In contrast to land plants, where specific LHCs serve either PSI or PSII, diatoms are characterized by a peripheral fucoxanthin (Fx)-chlorophyll (Chl) a/c antenna complex believed to deliver excitation energy to both photosystems, in addition to having a PSI-associated antenna (Lepetit et al., 2010; Büchel, 2015). Proteins of the peripheral Fx-Chl a/c antenna complex in diatoms belong to the LHC superfamily (Durnford et al., 1996) but are often referred to as Fx-Chl a/c-binding proteins (FCPs) in order to distinguish them from the LHCs of the green lineages (Falkowski and Raven, 2007). In addition to the light-harvesting pigments, FCPs also bind diadinoxanthin (Ddx) and diatoxanthin (Dtx), photoprotective pigments essential during light stress conditions (Wang et al., 2019). The FCPs belong to three major LHC classes: the LHCFs, including the main Fx-Chl a/c-binding proteins, the red alga-like LHCRs, and the LHCXs, related to the LhcSRs in Chlamydomonas reinhardtii (Büchel, 2015). The latter has been shown to play a central role in dissipating excessively absorbed energy through nonphotochemical quenching (NPQ) in cooperation with photoprotective pigments (Bailleul et al., 2010; Taddei et al., 2016, 2018; Lepetit et al., 2017).
LHC proteins and certain photosystem core proteins are known to be integrated into the thylakoid membrane of land plants and green microalgae through the posttranslational or cotranslational part of the chloroplast signal recognition particle (CpSRP) assembly pathway (Sundberg et al., 1997; Schuenemann et al., 1998; Bellafiore et al., 2002; Gerdes et al., 2006; Kirst et al., 2012a, 2012b; Kirst and Melis, 2014). The plant/green algae CpSRP pathway includes the LHC-specific chaperone CpSRP43, the GTPase CpSRP54, the signal recognition receptor CpFTSY, and the ALBINO3 insertase (ALB3; Bellafiore et al., 2002; Kirst and Melis, 2014). Homologs of CpSRP54, CpFTSY, and ALB3 can be identified in diatom genomes (Armbrust et al., 2004; Bowler et al., 2008; Mock et al., 2017), whereas no homologs for the molecular chaperone CpSRP43 have been identified (Träger et al., 2012). CpSRP43 orthologs appear to be restricted to plants and green algae; however, distantly related ankyrin repeat proteins can be found in Haptophyceae. Diatom CpSRP54 knockout (KO) mutants have been shown to be light sensitive (Nymark et al., 2016), but no further information exists about CpSRP54’s role, or the role of any other members of the CpSRP pathway, in the integration and assembly of thylakoid membrane proteins in diatoms. It has been shown, however, that efficient integration of FCPs depends on stromal factors and on the presence of GTP (Lang and Kroth, 2001).
In land plants and green microalgae, members of the CpSRP pathway guide certain chloroplast proteins to the thylakoid membranes where ALB3 mediates protein insertion in the developing thylakoids. ALB3 belongs to the YidC/Oxa1/Alb3 family of proteins that function in folding, insertion, and assembly of membrane protein complexes in bacteria and in certain eukaryotic organelles, such as mitochondria and chloroplasts (Hennon et al., 2015). The homologs within each subfamily have different C-terminal domains (CTDs) that are crucial for their function and protein-protein interaction. Two homologs belonging to this protein family are found in the chloroplasts of Arabidopsis (Arabidopsis thaliana), ALB3 and ALB4 (Sundberg et al., 1997; Gerdes et al., 2006), and C. reinhardtii, ALB3.1 and ALB3.2 (Bellafiore et al., 2002). ALB3 mutants of Arabidopsis have a severe phenotype. They are characterized by white/pale-yellow leaves, are defective in thylakoid membrane development, have strongly decreased pigment content, and are unable to survive phototrophically beyond the seedling stage when grown on soil (Sundberg et al., 1997). The Arabidopsis ALB3 insertase is essential for insertion of LHC proteins through the posttranslational CpSRP pathway and seems to be involved in cotranslational assembly of certain chloroplast-encoded membrane proteins (Sundberg et al., 1997; Moore et al., 2000; Kugelmann et al., 2013). Functional data exist also for the two C. reinhardtii ALB3 homologs, ALB3.1 and ALB3.2 (Bellafiore et al., 2002; Ossenbühl et al., 2004; Göhre et al., 2006). ALB3.1 of C. reinhardtii has been shown to be crucial for insertion of LHC proteins into the developing thylakoid membrane and to play a role in the assembly of D1 reaction center protein into PSII (Bellafiore et al., 2002; Ossenbühl et al., 2004). In contrast to the Arabidopsis ALB3 mutants, C. reinhardtii cells lacking ALB3.1 are still capable of phototrophic growth. The other C. reinhardtii ALB3 homolog, ALB3.2, is however essential for cell survival and is believed to be associated with the assembly and maintenance of the photosystems (Göhre et al., 2006).
Important differences have been identified between the functions of the ALB3 homologs of organisms within the green lineage. We therefore hypothesized that characterization of diatom ALB3 insertases has the potential to uncover other and unique functional features connected to this protein family. Using a reverse genetics approach, we applied the clustered, regulary interspaced, short palindromic repeat (CRISPR)/CRISPR-associated Protein9 (Cas9) technology to knock out ALB3b, encoding one of the two ALB3 proteins present in the diatom Phaeodactylum tricornutum. We demonstrate here that ALB3b’s primary functional role pertains to the insertion of light-harvesting antenna proteins in the developing thylakoid membrane. This, however, does not include antenna proteins functioning in photoprotection. Reduced levels of light-harvesting antenna proteins resulted in changes in the spectral properties, pigment content, growth rate, and photosynthetic performance of the cells.
RESULTS
Two homologs of the ALB3 insertase were identified in P. tricornutum and in all other stramenopiles where sequence data are available (Supplemental Fig. S1). Phylogenetic analyses showed that ALB3 proteins in plants/green algae and ALB3 proteins from stramenopiles were clearly divided into two distinct groups (Supplemental Fig. S1). Sequence similarity with the two ALB3 proteins with known functions in the green alga C. reinhardtii could therefore not be used to predict the individual functions of the two P. tricornutum ALB3 proteins (ALB3a and ALB3b). The ALB3a paralog has a basic Lys-rich CTD with similarities to CTDs in ALB3 proteins in plants and green algae (Supplemental Fig. S2). In Arabidopsis, this domain has been reported to interact directly with CpSRP43 and CpSRP54-CpFTSY complexes (Falk et al., 2010; Falk and Sinning, 2010; Lewis et al., 2010; Dünschede et al., 2011; Chandrasekar and Shan, 2017). ALB3b proteins in stramenopiles, however, do not contain the Lys-rich CTD but have instead a unique conserved domain (Fig. 1). Both P. tricornutum ALB3 genes (ALB3a [Phatr2_43657] and ALB3b [Phatr2_46411]) were targeted for CRISPR/Cas9-mediated disruption, but we were only able to generate viable KO lines for the ALB3b gene. Three independent alb3b KO lines (alb3b-14, alb3b-16, and alb3b-19) with large insertions of different sizes toward the 5′ end of the gene (Supplemental Fig. S3) were identified and cultured from single cells. All insertions consisted of fragments of the vectors used for transformation and caused premature stop codons at the N-terminal part of the protein (Fig. 1B). To verify that both alleles were mutated and that no wild-type sequence was present, allele-specific PCR was performed. Both alleles could be amplified in the wild type whereas only one allele could be amplified in the mutant strains, indicating larger insertion or deletion events that prevent amplification of the other mutated allele by PCR (Supplemental Fig. S4). Complementation of all three alb3b KO mutants with a codon-modified ALB3b (to avoid gene editing) was performed to confirm that the phenotype described below was the result of a lack of a functional ALB3B insertase.
Figure 1.
Presentation of intact and truncated ALB3b protein. A, The area of the ALB3b protein corresponding to the 20-bp target region for CRISPR/Cas9-based gene editing is located toward the N-terminal part of the protein (blue highlighting), with the protospacer adjacent motif (PAM) target site located at the reverse DNA strand (green highlighting). CTD, C-terminal domain; CTP, chloroplast-targeting peptide; 60 kD IMP, 60-kD inner membrane protein domain. B, Overview of amino acid sequences resulting from CRISPR/Cas9-induced inserts in the three alb3b KO lines causing premature stop codons and truncated ALB3b proteins. Color coding is as follows: blue, wild-type (WT) target sequence; green, amino acid corresponding to the PAM site; red, insert. Asterisks indicate premature stops. C, Protein alignment based on the CTD of ALB3b proteins in diatoms.
Spectral Properties of the Wild Type and alb3b Mutants
Previous studies on green algae and plants showed that mutations causing a reduction in the size of the light-harvesting antenna result in a pale green color of the chloroplasts (Sundberg et al., 1997; Bellafiore et al., 2002; Polle et al., 2003; Kirst et al., 2012a, 2012b; Oey et al., 2013; Gu et al., 2017). The diatom FCP complexes contain, in addition to Chl a and c, high amounts of Fx responsible for the golden-brown coloration of the diatom cells (Gundermann and Büchel, 2014; Büchel, 2015; Wang et al., 2019). The absorption properties of Fx are strongly dependent on the protein environment and undergo extreme bathochromic shifts upon protein binding, dividing the different Fx molecules into more red-, green-, and blue-absorbing complexes (Premvardhan et al., 2009, 2010; Gundermann and Büchel, 2014; Wang et al., 2019). We therefore hypothesized that a distortion of the normal antenna size/structure of P. tricornutum could result in a visible change in cell coloration. Disruption of the gene encoding the ALB3B insertase did indeed cause a change in coloration from the normal golden brown of the wild-type cells to a green coloration, suggesting structural changes of the light-harvesting antenna in the alb3b KO mutants (Fig. 2A).
Figure 2.
Color differences and spectral characteristics of wild type (WT) and alb3b mutants. A, Visual representation of the alb3b phenotype compared with the wild type at low light (LL; 35 µmol photons m−2 s−1; left side) and medium light (ML; 200 µmol photons m−2 s−1; right side). For comparison and visualization of the color differences, all cultures were adjusted to equal cell densities (3 × 107 cells mL−1). B and C, Absorbance spectra (B) and in vivo fluorescence excitation spectra (C) of cultures acclimated to ML. Isolated intact thylakoid membranes were used for recording of the absorption spectra to avoid scattering. Fluorescence emission was measured at 730 nm to ensure origin from the reaction center II Chl a. Insets are as follows: difference spectra between the absorbance of the wild type and alb3b KO lines (B) and excitation energy transfer in the blue-green region of the in vivo fluorescence excitation spectra (C). Wild-type data are presented as averages of three biological replicates; alb3b data are presented as averages of the three alb3b KO lines 14, 16, and 19 ± sd for all data points indicated by the gray area around the graphs. Three biological replicates were measured for each line.
To further explore the visual changes in spectral properties in the alb3b mutants compared with wild-type cultures, we recorded the in vivo absorbance (Fig. 2B) and fluorescence excitation spectra (Fig. 2C) for ML-acclimated cultures. The spectra showed that less light energy in the blue-green region is absorbed and available for photosynthesis in cultures lacking the ALB3b insertase. In vivo fluorescence excitation spectra were used to indicate the pigments’ relative energy transfer efficiency to Chl a in the reaction center of PSII. The differences in the in vivo fluorescence excitation spectra between the wild type and alb3b mutants (Fig. 2C, inset) strongly resembled the absorption characteristics of Chl c (peak at 462 nm) and Fx (peak at 520 nm; Bricaud et al., 2004; Premvardhan et al., 2009; Gundermann and Büchel, 2014), implying a substantially lower contribution in energy transfer from Chl c and Fx to the reaction center of PSII in the alb3b KO mutants. Smaller differences between the wild type and mutant strains are expected for the absorption spectra, as these spectra will also include pigments associated with PSI and non-protein-bound carotenoids dissolved in the thylakoid membrane that do not transfer absorbed energy to PSII (Lepetit et al., 2010). Even so, the difference in the peak profile for the absorption spectra (Fig. 2B, inset) matches the difference in the in vivo fluorescence excitation spectra, confirming a reduction of Chl c and Fx in the mutants.
Low-temperature (77 K) fluorescence measurements were performed to clarify the distribution of excitation energy between PSII and PSI in the wild type compared with alb3b mutant cultures (Fig. 3). The same samples were excited with either 435 nm (targeting Chl a absorption maxima; Fig. 3A) or 470 nm (targeting antenna pigments [Chl c and carotenoids]; Fig. 3B). The 77 K emission spectra recorded from ML-acclimated samples revealed fluorescence emission maxima at 688 and 710 nm, which are traditionally attributed to PSII and PSI, respectively (Ikeda et al., 2008; Yamagishi et al., 2010; Juhas and Büchel, 2012). In addition, an increase in fluorescence at 710 nm (F710) emission at the expense of F687 was observed in P. tricornutum cells that were in a state of high NPQ (Lavaud and Lepetit, 2013). In wild-type samples, the chosen excitation wavelengths caused a preferential energy transfer to PSII, displaying a relative amplitude of PSII fluorescence emission that was 2.5-fold (435 nm) or 3.3-fold (470 nm) higher than the PSI emission (F687/F710). In contrast, the average F687/F710 observed in the alb3b mutants were F687/F710 = 1.3 (435 nm) or 1.4 (470 nm), implying that excitation energy transfer to PSII was relatively more affected than energy transfer to PSI.
Figure 3.
77 K fluorescence emission spectra of wild-type (WT) and alb3b KO samples acclimated to ML. Samples were excited at either 435 nm (A) or 470 nm (B). The emission spectra were normalized at their 710-nm maximum. Data for alb3b are averages of the three alb3b KO lines 14, 16, and 19 ± sd for all data points indicated by the gray area around the graphs. Three biological replicates were measured for each line, including the wild type.
Effect of Lack of ALB3b Insertase on the Organization of the Photochemical Apparatus
The green color of the alb3b KO mutants and the combined results from the absorbance, fluorescence excitation, and emission spectra suggested that these mutants have an altered functional light-harvesting antenna size. To investigate this in more detail, the wild type and the alb3b KO lines were analyzed using an absorbance difference spectrophotometer (Melis, 1989). The rate of light absorption per second by PSII and PSI was measured by using low-intensity actinic light selected by cutoff and interference filters to selectively excite Fx (533 nm) or Chl a (670 nm), respectively (Table 1). When exiting Fx, the rate of light utilization by the photosystems revealed a severe decrease in the absorption cross section both for PSII and PSI in the alb3b mutant lines compared with the wild type (Table 1). The functional Chl a antenna sizes of PSII and PSI in the mutants were less affected because of the Chl a molecules bound to the photosystem core subunits (Ben-Shem et al., 2003; Nelson and Yocum, 2006; Ago et al., 2016; Table 1). In accordance with the 77 K data, these data also suggest a more severe decrease of the antenna size of PSII compared with PSI (Table 1).
Table 1. Photosystem absorption cross section and Chl a content per P700 in alb3b mutants compared with wild-type cells.
Photosystem absorption cross section was measured as rate of 533-nm (Fx) or 670-nm (Chl a) photons absorbed by the functional thylakoid membranes. The actinic light intensity was adjusted to I670 = 2.1 µmol photons m−2 s−1 and I533 = 12 µmol photons m−2 s−1. Rates of light absorption and utilization are given in photons per second ± sd. P700 quantification was measured from the light-induced ΔA700 with 670-nm (Chl a) actinic illumination.
| Photosystem | Parameter | Wild Type | alb3b-14 | alb3b-16 | alb3b-19 | Average alb3b | alb3b/Wild Type |
|---|---|---|---|---|---|---|---|
| PSI | (Fx) 533 nm | 2.61 ± 0.40 | 1.10 ± 0.08 | 1.17 ± 0.10 | 1.09 ± 0.00 | 1.10 ± 0.06 | 42% |
| (Chl) 670 nm | 1.93 ± 0.11 | 1.43 ± 0.05 | 1.39 ± 0.15 | 1.39 ± 0.14 | 1.40 ± 0.01 | 72.5% | |
| PSII | (Fx) 533 nm | 32.30 ± 0.7 | 13.92 ± 1.96 | 8.10 ± 1.16 | 8.58 ± 0.93 | 10.17 ± 3.24 | 35% |
| (Chl) 670 nm | 12.62 ± 2.69 | 7.21 ± 0.00 | 7.08 ± 0.90 | 6.71 ± 1.36 | 7.00 ± 0.26 | 55% | |
| Chl a/P700 | 663% ± 9%:1 | 466% ± 11% | 414% ± 9% | 394% ± 11% | 425:1 | 64% | |
Organization of the photochemical apparatus was further studied by quantification of PSI (P700) relative to the Chl a content of the cells. P700 content was measured from the light-induced ΔA700 absorbance change at 700 nm attributed to photooxidation of P700. On a P700 basis, there was a substantially lower number of Chl a molecules in alb3b (i.e. from 663 Chl a/P700 in the wild type down to an average of 425 Chl a/P700 in the mutants; Table 1). This directly reflects the lowering of Chl a pigments per electron transport chain (i.e. per P700) in the alb3b mutants relative to the wild type.
Western blot was used for examination of the role of the ALB3b insertase in incorporating proteins in the thylakoid membrane. Antibodies specific for antenna proteins (LHCFs and LHCXs) and photosystem subunits (D1, D2, and PsaC) were used, and an antibody against AtpB was employed as a loading control. The level of LHCF proteins in the alb3b mutants was assessed by an antibody binding to a highly conserved epitope of the LHCF1 to LHCF11 proteins (Juhas et al., 2014) and found to be lowered to about 25% of wild-type levels in cells grown under both LL and ML conditions (Fig. 4A). The relative decline of LHCF proteins is in good agreement with the smaller functional antenna size of PSII, as estimated from the kinetic spectrophotometric measurements using Fx excitation (Table 1). The relative gene expression levels of four LHCF genes (LHCF1, LHCF2, LHCF5, and LHCF8) were examined to determine if the low content of LHCF proteins in the alb3b lines could be explained by a strong down-regulation of the expression of these genes. Our data showed high gene expression levels (low cycle threshold values) of the examined LHCFs in all lines (Supplemental Table S1). Of the examined LHCF genes, only LHCF8 was significantly, but moderately, down-regulated in all alb3b lines (Supplemental Fig. S5). No antibodies are available for the detection of LHCR proteins constituting the main LHC protein fraction associated with the PSI antenna (Lepetit et al., 2010; Grouneva et al., 2011; Gundermann and Büchel, 2014). However, the smaller functional PSI antenna size in the mutant lines implied that ALB3b plays a vital role also in the insertion of LHCR proteins. An antibody (anti-FCP6) against an LHCX (FCP6) of Cyclotella meneghiniana, which also cross-reacts with the P. tricornutum LHCX proteins (Juhas et al., 2014), was used for comparison of the relative content of these photoprotective proteins. LHCX1 is crucial for NPQ to take place, whereas LHCX2 and LHCX3 can provide additional NPQ capacity during high-light (HL) stress (Bailleul et al., 2010; Taddei et al., 2016, 2018; Lepetit et al., 2017). LHCX1 and LHCX3 are of highly similar size (21.9 and 22.8 kD, respectively); therefore, complete separation by western-blot analysis is challenging. Based on the expression pattern of the LHCX isoforms known from the literature, we interpret the proteins detected under both LL and ML conditions to be a mix of LHCX1 and LHCX3, with the major contribution coming from LHCX1 under these conditions (Taddei et al., 2016, 2018). The relative content of the LHCX1+3 proteins in the mutants compared with the wild type seemed to be unaffected (slightly reduced levels of LHCX1+3 in alb3b-14) in both light conditions (Fig. 4A). The LHCX2 protein (24.7 kD) was detected at similar levels in the wild type and alb3b lines after 6 h of ML exposure (Supplemental Fig. S6B), but it was not detectable in LL- or ML-acclimated samples (Fig. 4A). The strong band of ∼22 kD detected in the wild type and alb3b lines 6 h after the shift from LL to ML (Supplemental Fig. S6B) is likely to contain large amounts of LHCX3 in addition to LHCX1 (Taddei et al., 2016, 2018). Based on western-blot analyses performed on PSI/II core proteins, the lack of a functional ALB3b insertase does not seem to have a negative impact on the incorporation of chloroplast-encoded photosystem subunits (Fig. 4B).
Figure 4.
Western-blot analysis of thylakoid membrane proteins from the wild type (WT) and alb3b mutant lines acclimated to LL or ML conditions. A, Abundance of LHC proteins belonging to the LHCF group was evaluated using an antibody recognizing LHCF1-11, whereas the LHCX proteins were recognized by anti-FCP6 (an LHCX family member of C. meneghiniana). A dilution series of the wild-type samples was used to assess the level of LHC proteins in alb3b mutants compared with the wild type. B, Protein expression of PSII and PSI core proteins was evaluated with antibodies against the D1 (PSII), D2 (PSII), and PsaC (PSI) core subunits. A dilution series of the alb3b samples was used to assess the level of photosystem subunits in alb3b mutants compared with the wild type. An antibody recognizing the β-subunit of ATP synthase (AtpB) was used as a loading control on each of the individual blots. Lanes marked with 100% contain 10 µg (20 µg for analysis of LHCX levels) of protein extracts. Images have been cropped.
Preliminary analysis with transmission electron microscopy showed a lower number of thylakoid membranes per chloroplast, but no obvious difference in the thylakoid architecture could be observed in the alb3b-14 mutant line acclimated to LL (Supplemental Fig. S7).
Functional Properties of the alb3b KO Mutants
To study the capability of the alb3b mutant to respond to a shift in light conditions, LL-acclimated cells (0 h) were shifted to ML conditions and sampled after 0.5, 6, 24, 48, and 168 h. The pigment content (Fig. 5) and photosynthetic performance (Figs. 6 and 7) of the acclimating cells were analyzed.
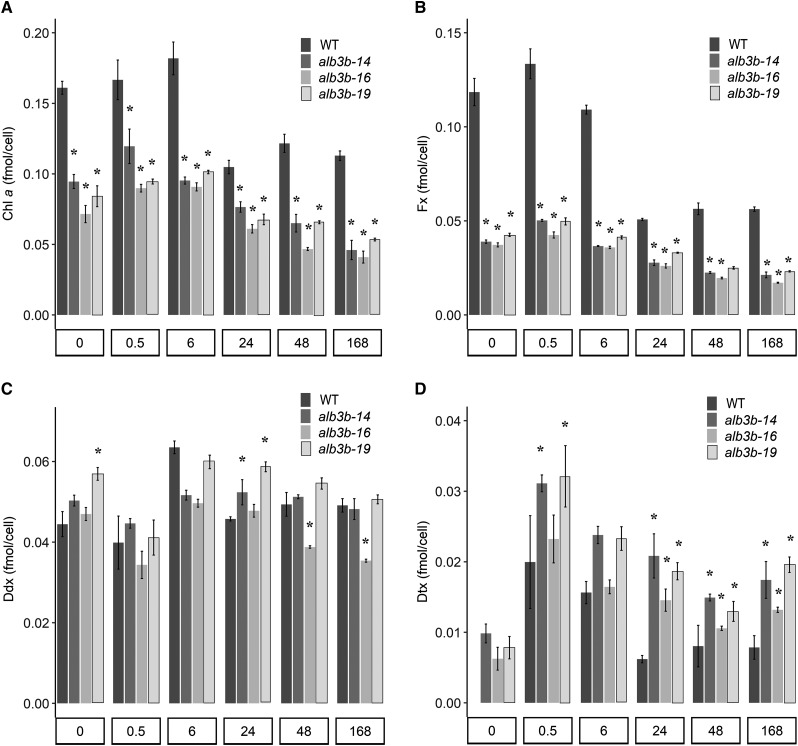
Figure 5.
Pigment concentrations per cell for the wild type (WT) and alb3b mutant lines as a function of ML exposure time. Cellular pigment concentrations are shown for Chl a (A), Fx (B), Ddx (C), and Dtx (D) in wild-type and alb3b mutant cells as a function of time following a shift from LL conditions (0 h; 35 µmol photons m−2 s−1) to ML conditions (200 µmol photons m−2 s−1) for 0.5, 6, 24, 48, and 168 h. Results are presented as means of three biological replicates ± sd. Asterisks indicate the results of two-tailed Student’s t tests: *, P < 0.05.
Figure 6.
DES index and NPQ capacity of the wild type (WT) and alb3b mutants. A, DES index [DES = Dtx/(Dtx + Ddx)] calculated from the HPLC pigment data from LL-acclimated (0 h) wild-type and alb3b cultures exposed to ML for 0.5, 6, 24, 48, and 168 h. B and C, Capacity for NPQ calculated from rapid light curves derived from LL-acclimated cells approximately 2 months after isolation of mutated single cells (B) and after being maintained in culture for one more year (C). NPQ = (Fm′max/Fm′) – 1. Fm′max replaces the commonly used Fm, since Fm′ values frequently occur that are higher than the Fm from dark-treated diatom samples (Serôdio et al., 2006). Results are presented as means of three biological replicates ± sd.
Figure 7.
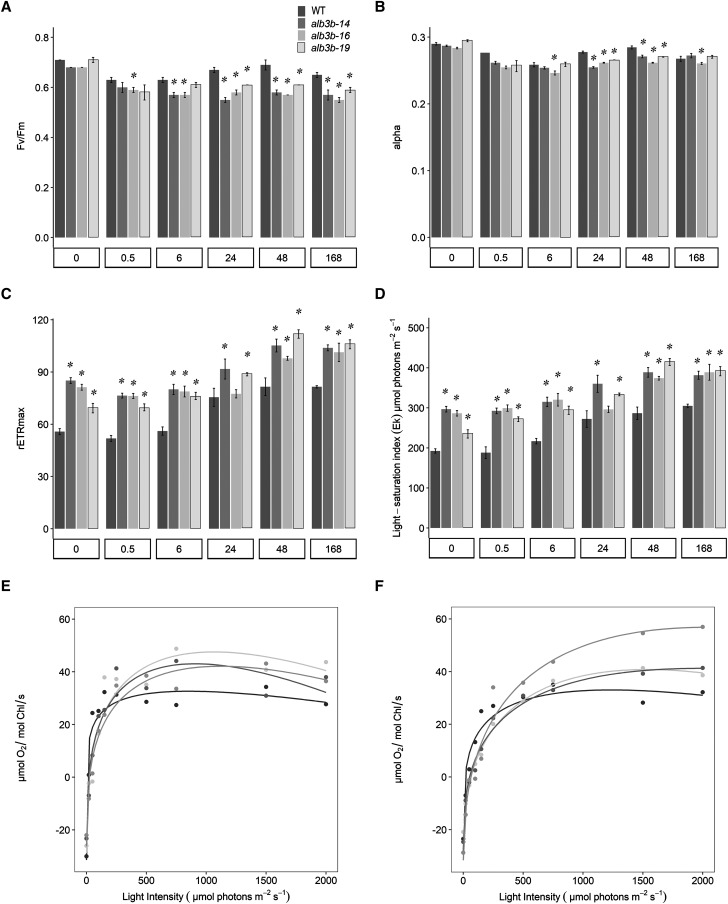
Photo-physiological responses of the wild type (WT) and alb3b mutant lines. A to D, In vivo Chl a fluorescence kinetics (PAM) were used to estimate Fv/Fm (A), α (B), rETRmax (C), and Ek (D) in LL (0 h)-acclimated wild-type and alb3b KO lines as a function of ML exposure time (0.5–168 h). Values are presented with sd bars. Asterisks indicate the results of two-tailed Student’s t tests: *, P < 0.05. E and F, Light-saturation curves of photosynthesis based on oxygen evolution were produced for LL-acclimated (E) and ML-acclimated (F) wild type and alb3b KO lines. The oxygen concentration was normalized on a per-Chl basis. The results were fit with curves based on a polynomial regression using R. All values are presented as averages of three biological replicates for each line, and sd for each value can be found in Supplemental Table S3.
Capacity for Photoacclimation and Photoprotection
As expected from the changed coloration and spectroscopic analyses, the alb3b KO mutants had a significantly lower content of light-harvesting pigments (LHPs) per cell compared with the wild type (Fig. 5). Even though the content of LHPs in LL-acclimated alb3b mutants was already lower than in ML-acclimated wild-type cells, the LHP concentration in the mutants decreased further as a response to the ML treatment (Fig. 5, A and B). This observation implies that the mechanisms controlling the down-regulation of the LHPs in response to an increase in available light are independent of the actual pigment concentration in the cells. The alb3b mutant lines contained ∼40% to 60% less Chl a and ∼60% to 65% less Fx in response to the light treatment (Fig. 5, A and B; Supplemental Table S2).
The smaller antenna size of the mutant lines had no negative impact on the cell content of the xanthophyll cycle carotenoids Ddx and Dtx (Fig. 5, C and D). Both the wild type and alb3b mutant lines showed the expected photoprotective response to a shift to a higher light intensity (Nymark et al., 2009), which could be observed as an immediate rise in Dtx concentration inversely to a decrease in Ddx concentration. The conversion of Ddx to Dtx peaked at the 0.5-h time point, as evident by the deepoxidation state (DES) index (Fig. 6A). The DES index decreased and stabilized at a lower level after prolonged exposure to ML, indicating that the algae were acclimating to the new light condition. Although changes in the DES index for both the wild type and mutants followed the same pattern after the shift to higher light intensities, the DES index was higher in the mutants than in wild-type cultures at all time points. The NPQ capacity of the alb3b mutants was initially (approximately 2 months after the isolation of mutated single cells) found to be lowered to around half of that in the wild type at irradiance levels greater than 400 µmol m−2 s−1 (Fig. 6B), but when the same experiment was repeated after the cells had been maintained in culture for an additional year (approximately 100–150 generations), the differences between the wild type and mutants had declined for all lines (Fig. 6C). Measurements of time-dependent NPQ development in alb3b mutants and the wild type produced highly similar results to those found when calculating NPQ from rapid light curves (Supplemental Fig. S8). The NPQ of alb3b-16 was closer to wild-type levels, whereas a lower NPQ was observed in the two other alb3b lines. The smaller differences in NPQ capacity between alb3b lines and the wild type led us to also reanalyze the relative LHCF protein content, pigment levels, and photosynthetic parameters in LL-acclimated alb3b and wild-type cultures after one more year of growth (Supplemental Figs. S6A, S9, and S10). No major changes were observed for the alb3b lines relative to wild-type cells compared with the initial analyses of these parameters.
Photosynthetic Performance
Variable Chl a fluorescence (pulse amplitude modulated fluorescence measurements) was used to calculate the photosynthetic (PSII) efficiency (Fv/Fm) of wild-type and mutant lines during the light experiment. In LL-acclimated cells, the Fv/Fm was ∼0.7 for all lines (Fig. 7A), which is around the maximum value expected for algal cells under optimal growth conditions (Falkowski and Raven, 2007). After 0.5 h of ML exposure, both wild-type and mutant cells showed a modest decrease in Fv/Fm (Fig. 7A). The Fv/Fm in the mutant cultures stabilized close to ∼0.6 in ML, whereas Fv/Fm in wild-type cultures increased after prolonged exposure to ML. The maximum relative electron transport rate (rETRmax) and light saturation index (Ek) values increased as a function of ML exposure time in all cultures (Fig. 7, C and D), as the photoacclimation mechanisms enabled the cells to utilize the increased amount of light energy available for photosynthesis (Nymark et al., 2009). However, the alb3b mutants displayed, on average, ∼30% to 40% higher rETRmax and Ek compared with wild-type cultures, showing the largest differences during the first part of the light experiment before the cells had been able to downsize the photosynthetic apparatus in response to the increased light intensities. Less pronounced differences in rETRmax and Ek were found between wild-type and alb3b cultures at the 24-h time point due to a more rapid change in photoacclimation status in wild-type cells, probably because of a higher cell division rate as described below. To further investigate the apparent increased photosynthetic performance of the alb3b KO lines indicated by the PAM measurement, light-saturation curves of photosynthesis based on oxygen evolution were measured for the wild type and alb3b KO lines acclimated to either LL (Fig. 7E) or ML (Fig. 7F). The maximum photosynthetic rate (Pmax [µmol oxygen mol−1 Chl s−1]), the maximum light utilization coefficient (α), and the saturation intensity (Es) of photosynthesis (Pmax/α [µmol photons m−2 s−1]) were calculated from the light-saturation curves of photosynthesis (Table 2; Powles and Critchley, 1980). When normalized to Chl a, the mutant lines showed a typical truncated light-harvesting antenna (TLA) mutant phenotype with higher Pmax and Es and slightly lower α compared with the wild type due to the lower functional absorption cross section caused by the smaller antenna (Kirst et al., 2014). Thus, it should be noted that these results do not indicate a higher photosynthetic performance per cell. In fact, when oxygen evolution was normalized per cell, the mutant lines showed a Pmax similar to the wild type (Supplemental Fig. S11). Also, the light saturation curves of the alb3b KO lines acclimated to LL showed a tendency of declining photosynthetic activity at light intensity greater than 1,000 µmol photons m−2 s−1 (Fig. 7E).
Table 2. Photosynthesis and respiration properties of the wild type and the alb3b KO lines.
Parameters are calculated from the light-saturation curves of photosynthesis based on oxygen evolution of the wild type and the alb3b KO lines (LL, Fig. 7E; ML, Fig. 7F). Data for alb3b are presented as averages of the three independent alb3b KO lines (alb3b-14, alb3b-16, and alb3b-19) ± sd. A minimum of three biological replicates were measured for each independent line.
| Property | LL | ML | ||
|---|---|---|---|---|
| Wild Type | alb3b | Wild Type | alb3b | |
| Respiration (μmol oxygen mol−1 Chl s−1) | 30.0 ± 13.6 | 23.8 ± 1.7 | 23.5 ± 5.9 | 24.9 ± 3.2 |
| Pmax (μmol oxygen mol−1 Chl s−1) | 57.7 ± 11.5 | 63.2 ± 3.1 | 55.7 ± 4.9 | 71.8 ± 7.6 |
| Es (µmol photons m−2 s−1) | 96.5 | 250 | 170 | > 400 |
| α | 0.35 | 0.32 | 0.29 | 0.25 |
Effect of Light Intensity on Cell Growth
Growth parameters were calculated from the exponential phase in batch cultures of LL- and ML-acclimated cultures (Table 3; Supplemental Fig. S12) to investigate how the changes in antenna size and composition affected the cell division rate. The results showed that wild-type cells grew faster than alb3b KO mutants at both light conditions, but a shift from LL to ML intensities diminished that growth rate gap between the alb3b KO mutants and the wild type (Table 3), as recently observed in other TLA mutants (Kirst et al., 2014; Formighieri and Melis, 2017). At ML conditions, the wild-type cells already divided at a maximum rate slightly above two cell divisions per day (Fawley, 1984). We hypothesized that if the slower growth rate of the alb3b mutants was caused by a lower ability to capture light energy, increasing the light intensities should have a positive effect on growth of the mutant cells. To investigate if a further increase in light intensity could close the growth rate gap, mutant and wild-type cells were acclimated to HL conditions (480 µmol photons m−2 s−1). The growth temperature was set to 23°C, which supports the highest cell division rate in P. tricornutum (Fawley, 1984). During the HL acclimation period (2 weeks), the majority of cells in one of the alb3b lines (alb3b-16) changed from the fusiform morphotype to a rounded phenotype. The rounded cells showed a tendency for aggregation, making accurate counting necessary for growth rate calculations difficult. The attempt to acclimate alb3b-16 to HL was repeated after the discovery of the strongly increased NPQ capacity in cells that had been maintained in culture for 1 year after isolation of single cells, but the HL treatment induced the same change in morphotype as previously observed. The two other alb3b lines did not show a change in morphotype during the HL acclimation period or during the following growth rate experiments, but prolonged HL treatment (months), including periods in stationary phase, induced the formation of the rounded cell type also in the two other alb3b lines. The same treatment did not provoke the formation of round cells in wild-type cultures. Growth curves are included in Supplemental Figure S13A. The growth rate calculations from the exponential part of the curve showed that the wild-type cells still divided twice per day in HL, whereas the average maximal growth rate of the alb3b mutants dropped from 1.2 in ML to 0.8 divisions per day under HL (Table 3). The physiological status of the cells, measured as Fv/Fm, was monitored during the length of the growth experiment (Supplemental Fig. S13B). The average Fv/Fm in wild-type cultures during the period of maximal growth was found to be 0.63. In contrast, the corresponding Fv/Fm value in the alb3b mutants was 0.41, pointing to a higher degree of photodamage. In order to investigate the presence of oxidative damage, levels of lipid peroxidation were measured for HL-acclimated wild-type and mutant cells (alb3b-14 and alb3b-19). The mutant lines did not show higher levels of lipid peroxidation compared with the wild type (Supplemental Fig. S14). Similar levels of xanthophyll pigments in the mutant compared with the wild type could explain these results, considering their role in the stabilization and protection of the thylakoid membrane lipids from peroxidation (Havaux et al., 2007).
Table 3. Growth rates of the wild type and alb3b mutant lines acclimated to different light intensities.
Maximum cell division per day was calculated from three biological replicates of the wild type and alb3b KO lines acclimated to LL (35 µmol photons m−2 s−1), ML (200 µmol photons m−2 s−1), or HL (480 µmol photons m−2 s−1). Values are presented ± sd. n/a, Growth rate for the alb3b-16 mutant in HL was not calculated because of cell aggregation.
| Light Intensity | Wild Type | alb3b-14 | alb3b-16 | alb3b-19 | alb3b Average |
|---|---|---|---|---|---|
| LL | 1.6 ± 0.23 | 0.4 ± 0.02 | 0.6 ± 0.02 | 0.6 ± 0.03 | 0.5 ± 0.09 |
| ML | 2.2 ± 0.03 | 1.1 ± 0.01 | 1.2 ± 0.03 | 1.4 ± 0.05 | 1.2 ± 0.13 |
| HL | 2.0 ± 0.05 | 0.8 ± 0.17 | n/a | 0.9 ± 0.25 | 0.8 ± 0.19 |
Complementation Studies of alb3b Mutants
A plasmid containing the codon-modified ALB3b under the control of its native promoter was introduced to the three alb3b lines by biolistic bombardment. As a result, 70 of 75 transformed colonies regained their brown coloration. Six brown colonies (two colonies derived from each of the three complemented lines) were randomly picked and subjected to PCR analysis followed by sequencing. The introduction of the modified ALB3b gene and the absence of wild-type sequence were confirmed (Supplemental Fig. S15). Three brown colonies (representing each of the three complemented mutant lines) were cultured for analyses of pigment and LHCF content. The results showed that the wild-type phenotype was recovered by introduction of the modified ALB3b gene (Fig. 8).
Figure 8.
Culture color, LHCF protein level, and pigment concentration in complemented alb3b lines compared with the wild type (WT). A, Wild type and complemented alb3b KO lines (alb3b-14C, alb3b-16C, and alb3b-19C) were acclimated to LL and ML conditions. All cultures were concentrated and adjusted to equal cell densities (3 × 107 cells mL−1) for comparison. B, Western-blot analysis of LHCF proteins in the wild type and complemented alb3b mutant lines acclimated to LL and ML conditions. LHCF protein levels were evaluated using LHCF1-11 antibody. An antibody recognizing the β-subunit of ATP synthase was used as a loading control. Ten micrograms of total protein from cell lysates was loaded onto the gel. C, Cellular pigment concentrations of Chl a and Fx in LL conditions. Results are presented as means of three biological replicates ± sd. Asterisks indicate the results of two-tailed Student t tests: *, P < 0.05.
DISCUSSION
Effects of Loss of the P. tricornutum ALB3b Insertase
The substantially lower level of antenna proteins belonging to the LHCF group (Fig. 4A) indicate that the primary role of the P. tricornutum ALB3b insertase is the efficient integration of the main LHC proteins into the thylakoid membrane. However, a small functional antenna size is still assembled, implying a phenotype where some LHC proteins can be inserted through other thylakoid membrane insertion pathways or that some functional redundancy exists between ALB3b and the uncharacterized diatom homolog ALB3a. The mainly unaffected levels of photoprotective LHCX proteins found in alb3b mutants (Fig. 4A) clearly indicate the presence of other integration pathway(s) for antenna proteins. The lower level of LHPs and smaller functional antenna size, the changed spectral properties, and the increased light saturation level can be seen as effects of the lower amount of antenna proteins causing a truncated light-harvesting antenna. The phenotypic traits listed above are characteristic of TLA-phenotype mutants, previously generated in cyanobacteria, green microalgae, and land plants (Polle et al., 2003; Kirst et al., 2012a, 2012b, 2014, 2017, 2018; Formighieri and Melis, 2017; Gu et al., 2017). TLA mutants have been shown to grow at relatively similar rates to the wild type when enough light energy is available (Bellafiore et al., 2002; Polle et al., 2003; Kirst et al., 2014; Gu et al., 2017).
The slow growth of the alb3b mutants compared with wild-type cells might be partially explained by a reduced ability to capture light energy, since an increase in light intensity from 35 (LL) to 200 µmol photons m−2 s−1 (ML) diminished the difference in growth rate between the wild type and mutants by a factor of 2. If the smaller antenna size of the mutants was the sole reason for the slow growth rate, a further increase in irradiance should further diminish the difference in growth between the wild type and mutants. Instead, analyses of algae cultures acclimated to HL (∼480 µmol photons m−2 s−1) revealed a negative effect on cell division rate, photodamage of the alb3b mutants, and induction of a round cell phenotype. The round or oval cell shape has previously been reported to be associated with prolonged exposure to abiotic stress (De Martino et al., 2007, 2011; Herbstová et al., 2017). The apparent increased photosynthetic capacity estimated for alb3b mutants at both LL and ML light conditions seems counterintuitive if the alb3b mutants are HL sensitive. However, these data are calculated from light-response curves where the algae are subjected to HL intensities for relatively short periods of time (minutes). The HL experienced by the algae during the generation of light-response curves might be too short for extensive photodamage to occur. However, mutants acclimated to LL conditions did show signs of photoinhibition, observed as a decrease in oxygen production when exposed to light intensities greater than 1,000 µmol photons m−2 s−1 (Fig. 7E).
NPQ is an important photoprotective mechanism providing the ability to dissipate excessively absorbed energy harmlessly as heat during HL exposure. In the alb3b mutants, the NPQ capacity was reduced compared with wild-type levels (Fig. 6, B and C; Supplemental Fig. S7), suggesting a reduced capability to handle prolonged HL exposure. Several studies show a convincing relationship between the amount of both LHCX and Dtx and the capacity for NPQ, and the presence of LHCX proteins and the conversion of protein-bound Ddx to Dtx has been found to be essential for NPQ to take place (Lavaud et al., 2002; Bailleul et al., 2010; Lepetit et al., 2012, 2013, 2017; Taddei et al., 2018). The level of LHCX proteins and the content of the xanthophyll cycle pigments (Ddx + Dtx) were not negatively affected by the lack of ALB3b insertase. However, Ddx and Dtx are found in three different pools in diatoms, one located in a lipid shield around the FCPs and two that are bound to antenna proteins connected to PSI or the peripheral FCP antenna, respectively (Lepetit et al., 2010). Only the protein-bound fraction of the peripheral antenna contributes to NPQ after the conversion of Ddx to Dtx (Lepetit et al., 2010). Because of the potential to store xanthophyll cycle pigments in the lipid phase of the thylakoid membrane, the amount of accumulated Ddx + Dtx that is protein bound might still be reduced even though the cell concentrations in the alb3b lines are similar to or higher than in the wild type. The molecular role of LHCX and Dtx in NPQ is still elusive, and no data exist about the precise localization of FCPs or the LHCX proteins. The latest models for NPQ in diatoms suggest that there are two quenching sites (Q1 and Q2) present in the diatom thylakoids (Miloslavina et al., 2009; Büchel, 2014; Lavaud and Goss, 2014; Goss and Lepetit, 2015; Giovagnetti and Ruban, 2017). NPQ at Q1 is believed to involve physical detachment of FCP oligomers from PSII that in P. tricornutum can be measured as an increase in 77 K emission at 710 nm and as a decrease of PSII cross section (Lavaud and Lepetit, 2013; Giovagnetti and Ruban, 2017), whereas Q2 seems to take place in FCPs functionally connected to PSII and involves antenna reorganization and aggregation of LHC trimers (Miloslavina et al., 2009; Büchel, 2014; Lavaud and Goss, 2014; Giovagnetti and Ruban, 2017). Q2 is suggested to be dependent on the presence of protein-bound Dtx and provides a much higher level of NPQ compared with Q1 (Giovagnetti and Ruban, 2017). Despite the comparable content of photoprotective antenna proteins and pigments in the wild type and alb3b mutants, the strong decrease in alb3b antenna size might disturb crucial protein-pigment or protein-protein (e.g. LHCF-LHCX) interactions potentially necessary for effective antenna aggregation (Q2) and lower the pool of detachable antenna involved in Q1. This might lead to the lower NPQ capacity observed in the alb3b mutants. However, the difference in NPQ capacity between alb3b lines and the wild type decreased after the alb3b lines had been maintained in culture for an additional 1 year (approximately 100–150 generations). The increase in NPQ compared with the wild type was especially prominent for alb3b-16. No major differences in pigment or LHCF content between the individual alb3b lines or changes in the pigment or LHCF ratios between alb3b and the wild type were observed that could explain the changes in NPQ capacity over time. The different NPQ levels in the mutants and the general increase in NPQ over time in the alb3b lines compared with wild-type levels therefore cannot be explained by changes in antenna size over time. Giovagnetti and Ruban (2017) showed that the amount of antenna detached is not proportional to the level of NPQ and that the NPQ can continue to increase without a further reduction of the PSII cross section. We therefore suggest that the increase in NPQ over time is caused not by a larger pool of detachable antenna but that the alb3b lines, over many generations, have been able to increase their capacity for NPQ at Q2 through an unknown mechanism.
Role of Diatom ALB3b in the Integration of Nucleus- and Plastid-Encoded Proteins Compared with ALB3 in Green Algae and Plants
P. tricornutum ALB3b showed functional similarities to the C. reinhardtii homolog ALB3.1 (Bellafiore et al., 2002; Ossenbühl et al., 2004). Both the diatom ALB3b and the green algae ALB3.1 play a role in the insertion of LHC proteins into the thylakoid membrane (Bellafiore et al., 2002; Kirst and Melis, 2014), and loss of the insertase causes a notably smaller antenna size (Bellafiore et al., 2002). In addition, C. reinhardtii cells lacking ALB3.1 contain a substantially increased fraction of highly stable membrane inserted but unassembled D1 protein (Ossenbühl et al., 2004). The D1 content in C. reinhardtii alb3.1 mutants was half that of wild-type cells. Based on the above-described findings, an additional role in the assembly of D1 into PSII was identified in green microalgae (Bellafiore et al., 2002; Ossenbühl et al., 2004). Subunits of PSI (PsaC), PSII (D1 and D2), and the ATP synthase complex (AtpB) were not negatively affected by the absence of the ALB3b insertase in diatom cells (Fig. 4B), but our analyses does not discriminate between unassembled proteins in the thylakoid membrane and proteins that are incorporated into photosynthetic complexes. More extensive protein analyses would be necessary to rule out a role of the diatom ALB3b insertase in the integration/assembly of chloroplast-encoded thylakoid membrane proteins. Assembled PSII complexes are fully functional in both C. reinhardtii (Ossenbühl et al., 2004) and P. tricornutum alb3b mutants (Fig. 7A). We detected no differences in photosynthetic efficiency in LL-acclimated cells between the wild type and mutants. This implies that even though the alb3b KO lines have a truncated antenna size, there is no difference in the probability of the trapped excitation energy being used for photochemistry between the wild type and mutants. However, a less efficient repair of PSII from photodamage (Guenther and Melis, 1990) and an associated slower replacement of damaged D1 could explain the on average ∼12% to 14% lower Fv/Fm measured in alb3b mutants during prolonged ML exposure and the on average ∼36% lower Fv/Fm observed in HL-acclimated mutant cells. An efficient PSII-repair mechanism including a more frequent replacement of photodamaged D1 is required under such conditions (Baroli and Melis, 1996; Theis and Schroda, 2016). Alternatively (or additionally), the PSII of the alb3b mutants might be more susceptible to photodamage because of the altered light-harvesting antenna disturbing the normally efficient NPQ mechanism (Fig. 6, B and C) functioning in this alga (Lavaud and Goss, 2014). However, the transformation of the normally fusiform alb3b-16 line into the rounded morphotype in HL regardless of having a lower (Fig. 6B) or more similar (Fig. 6C; Supplemental Fig. S7) NPQ capacity than the wild type indicates that there are other reasons for why alb3b mutants are sensitive to HL.
The Arabidopsis alb3p mutant has also been reported to be photosensitive. The mutant requires very low light intensities (12 µmol photons m−2 s−1) to produce detectable levels of photosynthetic complexes like LHC trimers and PSII monomers and dimers (Kugelmann et al., 2013). To explain the severe phenotype of the alb3p mutants, additional functions beyond the CpSRP pathway have been suggested for ALB3p (Kugelmann et al., 2013). Based on phenotypic similarities between alb3p and mutants defective in carotenoid synthesis, it has been speculated that ALB3p has a role in the integration and assembly of carotenoids into photosynthetic complexes (Kugelmann et al., 2013). The slow growth of the P. tricornutum alb3b mutants that cannot be compensated by increased light intensities, and the susceptibility to prolonged HL exposure, suggest additional roles for the ALB3b insertase. A future comparison with other types of P. tricornutum TLA mutants will be valuable for dissecting the primary effects of the absence of ALB3b from the secondary effects of having a truncated light-harvesting antenna size.
CONCLUSION
Our results show that ALB3b is essential for the assembly of a full-size light-harvesting antenna in diatoms. In land plants and green algae, ALB3 insertases are part of the CpSRP pathway, and the basic Lys-rich CTD is necessary for the interaction with other members of the pathway (Bellafiore et al., 2002; Chandrasekar and Shan, 2017). We also identified this domain within the ALB3a proteins of the stramenopiles but not in the ALB3b proteins, which have a unique CTD. The LHC-specific chaperone CpSRP43 is one of ALB3’s known interaction partners through its Lys-rich CTD, but neither we nor others (Träger et al., 2012) could identify this chaperone in diatoms or other stramenopiles. Also, the P. tricornutum CpSRP54 mutant was not reported to have a changed coloration, only to be light sensitive (Nymark et al., 2016). The different CTD in ALB3b proteins, the absence of CpSRP43, and the unchanged coloration of the diatom CpSRP54 mutant imply that the ALB3b proteins have distinct interaction partners from those of ALB3a and ALB3 of plants and green algae. A hypothetical model for the role of diatom ALB3 insertases is presented in Figure 9. For verification of the model, a more thorough investigation of the P. tricornutum CpSRP54 mutant and characterization of diatom FTSY mutants should be performed. This will clarify if ALB3b is part of the posttranslational CpSRP pathway or if diatom LHC proteins are guided to ALB3b through other mechanisms.
Figure 9.
Proposed model of the role of diatom ALB3 insertases in insertion/assembly of thylakoid membrane proteins. LHC proteins are synthesized on ribosomes on the chloroplast endoplasmic reticulum membrane (cERM), transported through the four membranes surrounding the secondary plastid of diatoms, and guided to ALB3b by an unknown protein complex before incorporation into the thylakoid membrane (left side). Chloroplast-encoded proteins are suggested to be integrated by the cotranslational cpSRP pathway including cpSRP54, FTSY, and ALB3a (right side). IEM, Plastid inner envelope membrane; OEM, plastid outer envelope membrane; PPM, periplastidial membrane.
MATERIALS AND METHODS
An axenic Phaeodactylum tricornutum culture originating from the sequenced clone Pt1 8.6 (CCMP2561) was obtained from the culture collection of the Provasoli-Guillard National Center for Marine Algae and Microbiota, Bigelow Laboratory for Ocean Sciences, in East Boothbay, Maine.
Experimental Conditions
Axenic culturing of P. tricornutum wild-type cells and the three alb3b KO lines (alb3b-14, alb3b-16, and alb3b-19) was performed as described previously unless otherwise stated (Nymark et al., 2009). Cell cultures were grown at 15°C under continuous cool-white fluorescent light at scalar irradiance ( photosynthetically active radiation [EPAR]) of ∼35 µmol photons m−2 s−1 (LL) or ∼200 µmol photons m−2 s−1 (ML). For the HL experiment, the wild type and the three independent alb3b KO lines were acclimated to 480 µmol m−2 s−1 and grown at 23°C in a Vötsch VB 1514 plant growth chamber (Vötsch Industrietechnik) equipped with metal halide lamps (Powerstar HQI-BT 400 W/D). The cultures were kept in the exponential growth phase for at least 3 weeks under these conditions to ensure that all cells were fully acclimated prior to conducting measurements.
For the spectrophotometric and kinetic analyses, cells were grown in F/2 enriched artificial seawater medium (Guillard and Ryther, 1962). To avoid carbon limitation during growth, the medium was supplemented with NaHCO3 (final concentration of 23.5 mm, pH 7.4). Cultures were grown at 25°C in 2-L glass bottles constantly stirred to ensure homogenous growth. Continuous illumination was provided by white fluorescent light tubes at ML. For the measurements, 80% to 85% of the total culture volume was harvested during the midexponential growth phase.
Growth Rates
Growth rates were estimated in batch cultures of the wild type and alb3b KO lines (three biological replicates) acclimated to LL, ML, or HL using a starting concentration of 100,000 (ML and HL) or 200,000 (LL) cells mL−1. Counting was performed either manually using a Bürker-Türk counting chamber after fixation with Lugol’s solution (LL samples) or with a BD Accuri C6 Flow Cytometer (BD Bioscience; ML and HL samples). For the latter, glutaraldehyde (2% [v/v] final solution) was used for the fixation of cells. Samples were excited by a 20-mW, 488-nm solid-state blue laser, and Chl fluorescence was measured by a greater than 670-nm optical filter (FL3). The average maximum growth rates (cell divisions d−1) were calculated by using a mean of the growth rates from the three biological replicates during the exponential phase.
Phylogenetic Analyses
ALB3 proteins in the National Center for Biotechnology Information protein database and from the iMicrobe transcriptome database (https://www.imicrobe.us/) were selected for phylogenetic analyses. Accession numbers for the protein sequences used in the analysis are listed in Supplemental Table S4. The analysis involved 47 ALB3 proteins from plants and algae, and each species was represented with two ALB3 paralogs (ALB3.1/ALB3.2 or ALB3a/ALB3b). The protein alignment was generated by using the ClustalX program (Thompson et al., 1997) and manually refined in GeneDoc 2.7.000 (Nicholas et al., 1997). The evolutionary relationships were estimated using the maximum likelihood method based on the Le-Gascuel model (Le and Gascuel, 2008) and the neighbor-joining method (Saitou and Nei, 1987). The initial trees for both maximum likelihood and neighbor-joining analyses were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances, estimated using a JTT model, and the trees with best topology were selected. For the maximum likelihood analyses, a discrete γ distribution was used to model evolutionary rate differences among sites (using five categories). All positions with less than 80% site coverage were eliminated. Tree branch confidence values were calculated by running 1,000 bootstrap replicates for neighbor joining and 100 replicates for maximum likelihood. The phylogenetic analyses were conducted in MEGA7 (Kumar et al., 2016).
CRISPR/Cas9 Gene Editing of the ALB3b Insertase
All steps for performing CRISPR/Cas9 editing of the ALB3b insertase gene (Phatr2_46411; XM_002180751), including selection of target site, ligation of adapter for target of interest into the pKS diaCas9-sgRNA plasmid (Nymark et al., 2016), transformation of diatom cells, and screening and identification of cells with biallelic mutations, were performed as described in the published protocol for CRISPR/Cas9 gene editing in P. tricornutum (Nymark et al., 2017). ALB3b-specific oligonucleotides for the creation of the adapter inserted into the single-guide RNA (sgRNA) cassette of the CRISPR/Cas9 vector, and primers used for the screening of cells with CRISPR/Cas9-mediated mutations, are presented in Supplemental Table S5. Three alb3b KO lines named alb3b-14, alb3b-16, and alb3b-19 were selected for functional characterization. These three selected lines were checked for off-target mutations by PCR amplification and sequencing of the regions containing the five most likely off-target sites. To identify potential off-target sites, a custom-made Perl-based script was used to search the genome for sites with high homology to the seed (protospacer adjacent motif-proximal) region of the target site. The script uses a string-based approach, which allows for up to three mismatches in the seed region. Off-targets are ranked by their similarity to the target site as well as the positions of the mismatches. No off-target mutations were found at any of the investigated sites. The Phatr2 identifiers for the genes containing the potential off-target sites and primers used for the screening process are listed in Supplemental Table S5.
Allele-Specific PCR
Allele-specific PCR was performed as an additional control as previously described (Serif et al., 2017). In short, primers for PCR were derived that include an allele-specific difference on the 3′ terminal base (see primers in Supplemental Table S5), thereby preventing polymerases without proofreading function from amplifying the respective other allele. Both alleles were amplified separately using HiDi polymerase (myPols) according to the manufacturer’s instructions.
Isolation of Thylakoid Membranes
Cells were harvested by centrifugation at 1,000g for 8 min at 4°C. The pelleted cells were resuspended in 50 mm Tricine-NaOH (pH 7.8) in ice-cold isolation buffer containing 300 mm Suc, 5 mm MgCl2, 10 mm NaCl, 2% (w/v) polyvinylpyrrolidone, 0.1% (w/v) BSA, and 5 mm ascorbic acid. The pellet was washed twice with the described buffer to remove residual salts from the growth medium. Cells were broken using a Branson 250 sonicator (pulse mode, 50% duty cycle, output power of 5) with a precooled tip for 45 s followed by 1 min of cooling in dim light. This process was repeated four times to ensure rupture of the majority of the cells. Unbroken cells were removed by centrifugation at 6,500 rpm for 10 min at 4°C. The thylakoid suspension was centrifuged at 75,000g for 45 min at 4°C using a Beckman Coulter ultracentrifuge. The thylakoid pellet was resuspended in 5 mL of ice-cold Tricine-NaOH (pH 7.8) buffer containing 10 mm NaCl and 5 mm MgCl2. Samples were measured immediately upon preparation.
Spectrophotometric and Kinetics Analyses
Photosystem kinetics and PSI quantitation analyses were performed using a laboratory-constructed absorbance difference spectrophotometer (Melis and Brown, 1980; Melis, 1989). The premise for this method is that, under light-limiting conditions, the rate of primary photochemistry is directly proportional to the light-harvesting antenna size (Melis, 1989). PSI (P700) content was measured from the light-induced ΔA700 using a differential extinction coefficient of 64 mm−1 cm−1 (Hiyama and Ke, 1972). Actinic excitation was provided in the red region of the spectrum using a transmittance interference 670-nm filter combined with a yellow cutoff filter (CS 3-69). The reaction mixture contained 50 to 100 µm Chl a, 0.02% (w/v) SDS, 250 µm methyl viologen, and 2.5 mm sodium ascorbate. The sample was illumined once prior to measuring to ensure the oxidation of cytochrome c6 and possibly cytochrome f. Two or three experimental replicates were measured, with at least three technical replicates taken. Chl a concentration in the samples was calculated after extraction in 90% (v/v) acetone for 30 min in the dark using the Jeffrey-Humphrey equation for diatoms (Jeffrey and Humphrey, 1975). Photocatalytic kinetics of the two photosystems were measured based on Chl a fluorescence induction for PSII and P700 oxidation for PSI (Melis, 1989). Actinic illumination was provided in the red and green regions of the spectrum using narrow interference filters with transmittance peaks at 670 and 533 nm. These filters were chosen after examination of the thylakoid absorbance spectra so that the 670-nm filter would excite predominantly Chl a, whereas the 533-nm filter would excite Fx and other carotenoids. Incident light intensity provided was 12 µmol photons m−2 s−1 in the green region and 2.1 µmol photons m−2 s−1 in the red region. The reaction mixture for the fluorescence kinetic measurements contained approximately 5 to 10 µm Chl a and 20 µm 3-(3,4-dichlorophenyl)-1,1-dimethylurea, and that for the P700 oxidation kinetics contained 100 to 200 µm Chl a, 250 µm methyl viologen, and 20 µm 3-(3,4-dichlorophenyl)-1,1-dimethylurea.
Absorbance Spectra
To avoid light scattering, absorption spectra were measured from thylakoid membrane extracts. Prior to measurement, the samples were placed in darkness in an ice bath to avoid thermal breakdown of thylakoid structure. Absorbance spectra of all extracts were scanned spectrophotometrically from 400 to 750 nm with a Shimadzu UV-1800 UV-visible spectrophotometer. The resuspension buffer was used as a blank and for baseline calibration.
In Vivo Fluorescence Excitation
In vivo fluorescence excitation spectra (400–700 nm) were measured as described previously using a Hitachi F-3000 spectrofluorometer (Nymark et al., 2013). Spectra were obtained by recording the Chl a fluorescence intensity (Chl a fluorescence from PSII) at 1 nm spectral resolution (5-nm bandwidth) at a fixed wavelength of emission (730 nm, 5-nm bandwidth). The emission of light was measured as a function of absorbed light at different wavelengths for ML-acclimated cultures. All spectra were normalized to the red emission maximum of Chl a of the wild-type cultures, so as to study the differences in excitation energy transfer efficiency by the main photosynthetic pigments Chl a, Chl c, and Fx in the blue-green part of the photosynthetically active radiation spectrum, where they exhibit their maximum absorption.
77 K Chl Fluorescence Emission Measurements
Low-temperature fluorescence emission spectra were recorded for three biological replicates of ML-acclimated cell cultures using a custom-made 77 K fluorometer (Lamb et al., 2015). Monochromatic LEDs with an emission centered around either 435 nm (LED435-12-30; Roithner LaserTechnikor) or 470 nm (LED470; Roithner LaserTechnikor) were used as excitation wavelengths. Fluorescence emission spectra were recorded between 600 and 800 nm. Samples were adjusted to a Chl concentration of 1 µg mL−1, transferred to glass tubes, and frozen in liquid nitrogen before measuring the 77 K fluorescence emission. All spectra were normalized to the wild-type emission spectrum at 710 nm.
Protein Isolation, SDS-PAGE, and Western-Blot Analysis
Wild-type and alb3b mutant cultures acclimated to either LL or ML (three biological replicates for each line and light condition) were harvested by filtration (Durapore Membrane Filters, pore size 0.65 µm; Merck Millipore). Filters were transferred to 2-mL tubes (Sarstedt), and 1 mL of F/2 medium was added. The tubes were vortexed for 10 s for resuspension of the cells, before removal of filters and centrifugation of resuspended cells at 16,000g for 1 min at 15°C. The supernatant was removed, and the remaining pellet was flash frozen in liquid nitrogen and stored at −80°C. A 5-mm precooled stainless-steel bead (Qiagen) was added to each of the tubes with frozen cell pellets, and the cells were mechanically broken and homogenized in two steps using the TissueLyser system (Qiagen). The samples were first placed in a precooled (−80°C) adapter set followed by cell disruption for 2 min at 25 Hz. Before the second shaking step (8 min at 25 Hz), the samples were transferred to a room temperature (RT) adapter set, and 700 µL of lysis buffer (50 mm Tris, pH 6.8, and 2% [w/v] SDS) was added according to Juhas et al. (2014). Insoluble material was removed by centrifugation (100g for 30 min at 4°C). The supernatant was transferred to new tubes, and the protein concentration was determined using the DC Protein Assay kit (Bio-Rad) following the manufacturer’s instructions. In addition to the whole-cell extracts, lysates were also obtained from thylakoids isolated from cell cultures acclimated to either LL or ML conditions. Thylakoids were resuspended in lysis buffer (50 mm Tris, pH 6.8, and 2% [w/v] SDS), and protein extracts were obtained as above (the first step for cell breakage was omitted). Proteins were resolved on 12% or 15% SDS-PAGE gels, depending of the size of the protein of interest. Ten micrograms of the protein extracts was loaded onto the gel lanes. Western-blot analyses were performed on either total protein extracts (detection of LHCF and LHCX proteins) or thylakoid extracts (detection of D1, D2, and PsaC proteins). The PsaC antibodies produced a signal only when using thylakoid extracts, whereas the antibody recognizing LHCX proteins produced optimal results when using whole-cell extracts. LHC proteins and photosystem subunits were therefore analyzed in different extracts. The signal generated by AtpB polyclonal antibodies was used as loading controls on each blot, in addition to Coomassie Blue-stained gels that were run in parallel. Ten micrograms of the protein extracts was loaded onto the gels. Proteins were detected with the following antibodies: anti-D1 (AS05 084, Agrisera; 1:20,000), anti-D2 (AS06 146, Agrisera; 1:5,000), anti-PsaC (AS10 939, Agrisera; 1:1,000), anti-AtpB (AS05 085, Agrisera; 1:4,000), anti-LHCF1-11 (1:1,000), and anti-FCP6 (LHCX; 1:1,000; kind gifts from C. Büchel, University of Frankfurt [Juhas et al., 2014]). Primary antibody incubation was performed overnight at 4°C for all antibodies. Polyclonal Goat Anti-Rabbit Immunoglobulins/Biotinylated (Dako) was used as secondary antibody with an incubation time of 2 h at RT, followed by incubation with Horseradish Peroxidase Streptavidin (Vector Laboratories) for 1 h at RT. Protein-antibody cross-reactions were visualized with SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Scientific) and documented with a G:BOX ChemiXRQ gel doc system (Syngene).
Transmission Electron Microscopy
Electron microscopy was used to examine the status of the thylakoid architecture in the alb3b mutant lines. Wild-type and alb3b-14 cell cultures acclimated to LL were harvested by a light centrifugation step (4,000g for 10 min) and fixed overnight at RT in F/2 medium buffer containing 2.5% (v/v) glutaraldehyde and 2% (v/v) paraformaldehyde. Pellets were washed three times in F/2 medium buffer solution and embedded in a 5% (w/v) gelatin solution. After postfixation in 2% (w/v) osmium tetraoxide and 1.5% (w/v) kalium ferrocyanide, the samples were dehydrated in a gradient of ethanol. Samples were thereafter embedded with epoxy resins based on the protocol of Bozzola and Russell (1999) and sectioned with an ultramicrotome. Images were taken using a Tecnai 12 transmission electron microscope operating at 80 kV. Images were captured using a MORADA CCD camera.
Measurements of Malondialdehyde Content
The malondialdehyde (MDA) content was determined using the Lipid Peroxidation (MDA) assay kit (Sigma-Aldrich). The MDA concentration was measured based on its reaction with thiobarbituric acid and used as an index of lipid peroxidation. Wild-type and alb3b (alb3b-14 and alb3b-19) mutant cultures (three biological replicates for each line) acclimated to HL were harvested by filtration as described above. The cell pellet was resuspended in the MDA lysis buffer. To ensure complete lysis, the cells were briefly sonicated. Thereafter, the MDA content was determined based on the manufacturer’s instructions. In parallel, samples were collected and manually counted to determine cell concentration.
Isolation of Total RNA and Reverse Transcription Quantitative PCR
Three biological replicates of LL-acclimated wild-type and alb3b mutant cultures were harvested for isolation of total RNA in parallel to the samples harvested for protein analyses as described above. Total RNA isolation, quantification, and verification of RNA integrity were performed as described (Nymark et al., 2009). Reverse transcription of RNA was performed with the QuantiTect Reverse Transcrition kit (Qiagen) following the recommended protocol. One microgram of total RNA was used in each reaction. Reverse transcription quantitative PCR analysis was performed as described (Nymark et al., 2009) for calculation of relative expression ratios of four LHCF genes (LHCF1, LHCF2, LHCF5, and LHCF8). The geNorm module in the qBasePLUS software (Biogazelle) was used for determining the expression stability of the candidate reference gene. Based on the stability analysis, RPS5 (Phatr2_42848) and DLST (Phatr2_45557) were selected as reference genes (Nymark et al., 2013; Valle et al., 2014). LinRegPCR software (Ramakers et al., 2003; Ruijter et al., 2009) was used to calculate mean PCR efficiency per amplicon and cycle threshold values per sample. These data were imported into the qBasePLUS software (Biogazelle), which calculated relative expression ratios (given as calibrated normalized relative quantities) and performed statistical analyses on the results. The one-way ANOVA test integrated in the qBasePLUS software was used to evaluate the significance of the estimated relative expression ratios. Forward and reverse primers are listed in Supplemental Table S5.
Light-Shift Time-Series Experiments
LL-acclimated wild-type and alb3b KO lines were transferred to ML conditions and sampled after 0.5, 6, 24, 48, and 168 h following the shift in growth light intensity. LL samples (0 h) were harvested as controls. Three biological replicates were set up for each line and time point to reach a cell concentration of maximum 1 × 106 cells mL−1 at the day of harvesting. Samples were harvested for pigment analyses, monitoring of cell concentrations, variable in vivo Chl a fluorescence (PAM), and protein analyses.
Pigment Analyses
HPLC pigment analysis was performed according to Rodriguez et al. (2006) using a Hewlett-Packard HPLC 1100 Series system. Pigment values from the HPLC analysis were calculated as fmol of pigment per cell. Cell numbers were calculated from flow cytometer counts as described above.
Measurements of Photosynthetic Parameters
A PhytoPAM (System I) was used to measure variable Chl a fluorescence of the harvested samples. The photosynthesis versus irradiance relationship was obtained as described previously (Nymark et al., 2009). An additional step at 1,216 µmol photons m−2 s−1 was added for the samples that had been treated with ML for 1 week to ensure that light saturation levels were reached. The Fv/Fm, rETRmax, α, and Ek were calculated as described before (Nymark et al., 2009). The rETRmax is an estimate of the maximum photosynthetic capacity of the cells (∼Pmax), whereas the Ek (rETRmax/α) is a proxy for the threshold irradiance that separates light-limited and light-saturated photosynthesis (Genty et al., 1989; Sakshaug et al., 1997). Fm at low light intensities is commonly observed to be lower than the Fm′ level under low actinic light in diatoms (Serôdio et al., 2006; Cruz and Serôdio, 2008; Cruz et al., 2011). NPQ was therefore calculated from the light-response curve from LL-acclimated samples, using the maximum Fm′ level (Fm′max) instead of Fm as follows: NPQ = (Fm′max/Fm′) – 1 (Serôdio et al., 2006; Kalaji et al., 2017). NPQ development over time was additionally calculated from LL-acclimated cells exposed to 5 min of actinic light at an intensity setting of 832 µmol photons m−2 s−1. For the HL experiment, Fv/Fm was measured with an AquaPen-C (Photon System Instruments) at the end of a 30-min dark acclimation period to relax the fast-reversible component (qE) of NPQ so that only the photoinihibitory, slowly reversible quenching (qI), caused by damaged PSII reaction centers, would influence the Fv/Fm value.
Oxygen evolution was measured at 15°C using an S1 Clark-type polarographic oxygen electrode (Hansatech) increasingly illuminated with a 35-W cool-white spot LED. The measurements were done on cultures acclimated to both LL and ML. A 2-mL cell suspension from midexponential phase culture was added to a stirred chamber with temperature control and supplemented with sodium bicarbonate (30 µL of a 0.5 m solution) so that the oxygen production would not be limited by carbon availability. Prior to measuring, the Chl a concentration in the sample was adjusted to a concentration lower than 1.2 µm to avoid cell shading in the chamber. Simultaneously, cell concentration of the samples was determined by flow cytometry counting. Oxygen consumption in darkness was measured as a starting baseline; thereafter, the sample was exposed to gradually increasing light intensities and the oxygen evolution was measured continuously for at least 10 min. Each light intensity was adjusted by measuring the light intensity in the middle of the electrode chamber with a spherical US-SQS sensor (Walz).
Complementation of alb3b KO Lines
A modified version of the ALB3b gene was synthesized together with its native promoter by GeneArt Services Thermo Fisher Scientific (Supplemental Fig. S16). Modifications consisted of changes of the codon usage in the protospacer adjacent motif and target region of the ALB3b gene to avoid gene editing by the functional CRISPR/Cas9 system incorporated into the genome of the alb3b KO lines. MssI sites were included at the 5′ and 3′ ends of the module to facilitate blunt-end cloning into the pM9_4Compln vector from Madhuri et al. (2019) containing the bsr gene conferring resistance to blasticidin-S. Transformation of all three alb3b KO lines with the pM9_4Compln vector containing the synthesized ALB3b module was performed as described previously (Nymark et al., 2017). The algae were transferred to low-salt selection plates (25% [v/v] natural seawater supplemented with f/2-Si, 1% [w/v] agar, and 4 μg mL−1 blasticidin-S [Thermo Fisher Scientific]) ∼24 h after transformation. Transformed colonies appeared 3 to 4 weeks after transfer to selection plates. Colonies that had regained the normal brown color were randomly picked from the selection plates. PCR amplification of the ALB3b gene and subsequent sequencing were used to test for the presence of the modified version of the ALB3b gene and the absence of wild-type sequence. Primers used for both PCR amplification and sequencing were PtAlb3b-G1F and PtAlb3b-G1R (Supplemental Table S5). One complemented alb3b colony, resulting from each of the transformations performed with the alb3b KO lines, was cultivated for pigment and protein analyses, as described above.
Statistical Analyses
The one-way ANOVA test integrated in the qBasePLUS software (Biogazelle) was used to evaluate the significance of the estimated relative expression ratios of LHCF genes in alb3b mutants compared with wild-type cells. Two-tailed Student’s t tests were used to assess if there were significant differences in pigment concentration and photosynthetic parameters between alb3b mutants and the wild type.
Accession Numbers
Accession numbers for ALB protein sequences extracted from the GenBank National Center for Biotechnology Information, the iMicrobe database (Marine Microbial Eukaryote Transcriptome Sequencing Project), and the JGI genome portal are listed in Supplemental Table S4.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic relationship between members of the ALB3 family.
Supplemental Figure S2. CTDs of diatom ALB3a and ALB3b proteins.
Supplemental Figure S3. DNA sequences for the ALB3b wild-type gene and the inserts in the alb3b KO lines.
Supplemental Figure S4. Allele-specific amplification of the Cas9 target site within the ALB3b gene in the wild type and alb3b mutant strains.
Supplemental Figure S5. Relative expression levels of LHCF genes in alb3b lines compared with the wild type.
Supplemental Figure S6. Western-blot analysis of LHCF and LHCX proteins from the wild type and alb3b mutant lines.
Supplemental Figure S7. Transmission electron micrographs of wild-type and alb3b-14 mutant line cells.
Supplemental Figure S8. NPQ development over time in the wild type and alb3b lines.
Supplemental Figure S9. Reevaluation of pigment concentrations per cell for LL-acclimated wild-type and alb3b mutant lines.
Supplemental Figure S10. Reevaluation of photo-physiological responses of LL-acclimated wild-type and alb3b mutant lines.
Supplemental Figure S11. Light-saturation curves of photosynthesis for LL- and ML-acclimated wild-type and alb3b mutant lines presented as oxygen evolution per cell.
Supplemental Figure S12. Growth curves for the wild type and alb3b mutants.
Supplemental Figure S13. Growth curves and corresponding measurements of photosynthetic efficiency of the wild type and alb3b mutants in HL.
Supplemental Figure S14. MDA product of lipid peroxidation.
Supplemental Figure S15. PCR analysis and Sanger sequencing of PCR products from complemented alb3b lines.
Supplemental Figure S16. DNA sequence representing the synthetic ALB3b module used for complementation of the alb3b KO lines.
Supplemental Table S1. Cycle threshold values for LHCF and reference genes.
Supplemental Table S2. Fraction of Chl a and Fx content in alb3b mutant lines compared with the wild type in LL and after 0.5 to 168 h in ML.
Supplemental Table S3. Oxygen evolution values of the light-saturation curves of photosynthesis ± sd for LL- and ML-acclimated wild-type and alb3b mutant lines.
Supplemental Table S4. Accession numbers for ALB proteins included in the phylogenetic analyses.
Supplemental Table S5. Oligonucleotide and primer sequences.
ACKNOWLEDGMENTS
We thank Claudia Büchel for kindly providing LHCF and LHCX antibodies and Peter G. Kroth for the pM9_4Compln vector. We also thank Kjersti Andresen for assistance with the HPLC analyses, Geir Johnsen and Inga Aamot for access to and guidance on the use of the PhytoPAM, and Martin F. Hohmann-Marriot and Gunvor Røkke for training on how to produce and analyze 77 K data. We thank the Cellular and Molecular Imaging Core Facility (CMIC), Norwegian University of Science and Technology (NTNU), for guidance and help during the acquisition of the transmission electron microscopy images. CMIC is funded by the Faculty of Medicine at NTNU and the Central Norway Regional Health Authority.
Footnotes
This work was supported by the Research Council of Norway to A.M.B through funding of the project Downsizing Light-Harvesting Antenna to Scale Up Production Potential and Valorization from Cultivation of Marine Microalgae (267474) and to O.V. through funding of the project Microbially Produced Raw Materials for Aquafeed (project no. 239001), by a Peder Sather Foundation award to A.M. and A.M.B. (SRPSC4 1-50504-13618-44 ME1AM), and by the NTNU Enabling Technologies Program to P.W.
Articles can be viewed without a subscription.
References
- Ago H, Adachi H, Umena Y, Tashiro T, Kawakami K, Kamiya N, Tian L, Han G, Kuang T, Liu Z, et al. (2016) Novel features of eukaryotic photosystem II revealed by its crystal structure analysis from a red alga. J Biol Chem 291: 5676–5687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, et al. (2004) The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 306: 79–86 [DOI] [PubMed] [Google Scholar]
- Austin JR II, Staehelin LA (2011) Three-dimensional architecture of grana and stroma thylakoids of higher plants as determined by electron tomography. Plant Physiol 155: 1601–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul B, Rogato A, de Martino A, Coesel S, Cardol P, Bowler C, Falciatore A, Finazzi G (2010) An atypical member of the light-harvesting complex stress-related protein family modulates diatom responses to light. Proc Natl Acad Sci USA 107: 18214–18219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroli I, Melis A (1996) Photoinhibition and repair in Dunaliella salina acclimated to different growth irradiances. Planta 198: 640–646 [DOI] [PubMed] [Google Scholar]
- Bellafiore S, Ferris P, Naver H, Göhre V, Rochaix JD (2002) Loss of Albino3 leads to the specific depletion of the light-harvesting system. Plant Cell 14: 2303–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Frolow F, Nelson N (2003) Crystal structure of plant photosystem I. Nature 426: 630–635 [DOI] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, et al. (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456: 239–244 [DOI] [PubMed] [Google Scholar]
- Bozzola JJ, Russell LD (1999) Electron Microscopy: Principles and Techniques for Biologists, Ed 2 Jones and Bartlett, [Google Scholar]
- Bricaud A, Claustre H, Ras J, Oubelkheir K (2004) Natural variability of phytoplanktonic absorption in oceanic waters: Influence of the size structure of algal populations. J Geophys Res Oceans 109: C11010 [Google Scholar]
- Brown JW, Sorhannus U (2010) A molecular genetic timescale for the diversification of autotrophic stramenopiles (Ochrophyta): Substantive underestimation of putative fossil ages. PLoS ONE 5: e12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büchel C. (2014) Fucoxanthin-chlorophyll-proteins and non-photochemical fluorescence quenching of diatoms In Demmig-Adams B, Garab G, Adams W III, Govindjee, eds, Non-Photochemical Quenching and Energy Dissipation in Plants, Algae and Cyanobacteria. Advances in Photosynthesis and Respiration (Including Bioenergy and Related Processes), Vol 40 Springer, Dordrecht, The Netherlands, pp 259–275 [Google Scholar]
- Büchel C. (2015) Evolution and function of light harvesting proteins. J Plant Physiol 172: 62–75 [DOI] [PubMed] [Google Scholar]
- Chandrasekar S, Shan SO (2017) Anionic phospholipids and the Albino3 translocase activate signal recognition particle-receptor interaction during light-harvesting chlorophyll a/b-binding protein targeting. J Biol Chem 292: 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz S, Goss R, Wilhelm C, Leegood R, Horton P, Jakob T (2011) Impact of chlororespiration on non-photochemical quenching of chlorophyll fluorescence and on the regulation of the diadinoxanthin cycle in the diatom Thalassiosira pseudonana. J Exp Bot 62: 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz S, Serôdio J (2008) Relationship of rapid light curves of variable fluorescence to photoacclimation and non-photochemical quenching in a benthic diatom. Aquat Bot 88: 256–264 [Google Scholar]
- De Martino A, Bartual A, Willis A, Meichenin A, Villazán B, Maheswari U, Bowler C (2011) Physiological and molecular evidence that environmental changes elicit morphological interconversion in the model diatom Phaeodactylum tricornutum. Protist 162: 462–481 [DOI] [PubMed] [Google Scholar]
- De Martino A, Meichenin A, Shi J, Pan KH, Bowler C (2007) Genetic and phenotypic characterization of Phaeodactylum tricornutum (Bacillariophyceae) accessions. J Phycol 43: 992–1009 [Google Scholar]
- Dünschede B, Bals T, Funke S, Schünemann D (2011) Interaction studies between the chloroplast signal recognition particle subunit cpSRP43 and the full-length translocase Alb3 reveal a membrane-embedded binding region in Alb3 protein. J Biol Chem 286: 35187–35195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durnford DG, Aebersold R, Green BR (1996) The fucoxanthin-chlorophyll proteins from a chromophyte alga are part of a large multigene family: Structural and evolutionary relationships to other light harvesting antennae. Mol Gen Genet 253: 377–386 [DOI] [PubMed] [Google Scholar]
- Falk S, Ravaud S, Koch J, Sinning I (2010) The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J Biol Chem 285: 5954–5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S, Sinning I (2010) The C terminus of Alb3 interacts with the chromodomains 2 and 3 of cpSRP43. J Biol Chem 285: le25-le26, author reply le26–le28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowski PG, Barber RT, Smetacek V (1998) Biogeochemical controls and feedbacks on ocean primary production. Science 281: 200–207 [DOI] [PubMed] [Google Scholar]
- Falkowski PG, Raven JA (2007) Aquatic Photosynthesis, Ed 2 Princeton University Press, Princeton, NJ [Google Scholar]
- Fawley MW. (1984) Effects of light intensity and temperature interactions on growth characteristics of Phaeodactylum tricornutum (Bacillariophyceae). J Phycol 20: 67–72 [Google Scholar]
- Formighieri C, Melis A (2017) Heterologous synthesis of geranyllinalool, a diterpenol plant product, in the cyanobacterium Synechocystis. Appl Microbiol Biotechnol 101: 2791–2800 [DOI] [PubMed] [Google Scholar]
- Genty B, Briantais JM, Baker NR (1989) The relationship between the quantum yield of photosynthetic electron-transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta 990: 87–92 [Google Scholar]
- Gerdes L, Bals T, Klostermann E, Karl M, Philippar K, Hünken M, Soll J, Schünemann D (2006) A second thylakoid membrane-localized Alb3/OxaI/YidC homologue is involved in proper chloroplast biogenesis in Arabidopsis thaliana. J Biol Chem 281: 16632–16642 [DOI] [PubMed] [Google Scholar]
- Giovagnetti V, Ruban AV (2017) Detachment of the fucoxanthin chlorophyll a/c binding protein (FCP) antenna is not involved in the acclimative regulation of photoprotection in the pennate diatom Phaeodactylum tricornutum. Biochim Biophys Acta 1858: 218–230 [DOI] [PubMed] [Google Scholar]
- Göhre V, Ossenbühl F, Crèvecoeur M, Eichacker LA, Rochaix JD (2006) One of two alb3 proteins is essential for the assembly of the photosystems and for cell survival in Chlamydomonas. Plant Cell 18: 1454–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss R, Lepetit B (2015) Biodiversity of NPQ. J Plant Physiol 172: 13–32 [DOI] [PubMed] [Google Scholar]
- Grouneva I, Rokka A, Aro EM (2011) The thylakoid membrane proteome of two marine diatoms outlines both diatom-specific and species-specific features of the photosynthetic machinery. J Proteome Res 10: 5338–5353 [DOI] [PubMed] [Google Scholar]
- Gu J, Zhou Z, Li Z, Chen Y, Wang Z, Zhang H, Yang J (2017) Photosynthetic properties and potentials for improvement of photosynthesis in pale green leaf rice under high light conditions. Front Plant Sci 8: 1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther JE, Melis A (1990) The physiological significance of photosystem II heterogeneity in chloroplasts. Photosynth Res 23: 105–109 [DOI] [PubMed] [Google Scholar]
- Guillard RR, Ryther JH (1962) Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Can J Microbiol 8: 229–239 [DOI] [PubMed] [Google Scholar]
- Gundermann K, Büchel C (2014) Structure and functional heterogeneity of fucoxanthin-chlorophyll proteins in diatoms In Hohmann-Marriott M, ed, The Structural Basis of Biological Energy Generation. Advances in Photosynthesis and Respiration (Including Bioenergy and Related Processes), Vol 39 Springer, Dordrecht, The Netherlands, pp 21–37 [Google Scholar]
- Havaux M, Dall’Osto L, Bassi R (2007) Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiol 145: 1506–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennon SW, Soman R, Zhu L, Dalbey RE (2015) YidC/Alb3/Oxa1 family of insertases. J Biol Chem 290: 14866–14874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbstová M, Bína D, Kaňa R, Vácha F, Litvín R (2017) Red-light phenotype in a marine diatom involves a specialized oligomeric red-shifted antenna and altered cell morphology. Sci Rep 7: 11976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama T, Ke B (1972) Difference spectra and extinction coefficients of P 700. Biochim Biophys Acta 267: 160–171 [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Komura M, Watanabe M, Minami C, Koike H, Itoh S, Kashino Y, Satoh K (2008) Photosystem I complexes associated with fucoxanthin-chlorophyll-binding proteins from a marine centric diatom, Chaetoceros gracilis. Biochim Biophys Acta 1777: 351–361 [DOI] [PubMed] [Google Scholar]
- Jeffrey SW, Humphrey GF (1975) New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem Physiol Pflanz 167: 191–194 [Google Scholar]
- Juhas M, Büchel C (2012) Properties of photosystem I antenna protein complexes of the diatom Cyclotella meneghiniana. J Exp Bot 63: 3673–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, von Zadow A, Spexard M, Schmidt M, Kottke T, Büchel C (2014) A novel cryptochrome in the diatom Phaeodactylum tricornutum influences the regulation of light-harvesting protein levels. FEBS J 281: 2299–2311 [DOI] [PubMed] [Google Scholar]
- Kalaji HM, Schansker G, Brestic M, Bussotti F, Calatayud A, Ferroni L, Goltsev V, Guidi L, Jajoo A, Li P, et al. (2017) Frequently asked questions about chlorophyll fluorescence, the sequel. Photosynth Res 132: 13–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst H, Formighieri C, Melis A (2014) Maximizing photosynthetic efficiency and culture productivity in cyanobacteria upon minimizing the phycobilisome light-harvesting antenna size. Biochim Biophys Acta 1837: 1653–1664 [DOI] [PubMed] [Google Scholar]
- Kirst H, Gabilly ST, Niyogi KK, Lemaux PG, Melis A (2017) Photosynthetic antenna engineering to improve crop yields. Planta 245: 1009–1020 [DOI] [PubMed] [Google Scholar]
- Kirst H, García-Cerdán JG, Zurbriggen A, Melis A (2012a) Assembly of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii requires expression of the TLA2-CpFTSY gene. Plant Physiol 158: 930–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst H, Garcia-Cerdan JG, Zurbriggen A, Ruehle T, Melis A (2012b) Truncated photosystem chlorophyll antenna size in the green microalga Chlamydomonas reinhardtii upon deletion of the TLA3-CpSRP43 gene. Plant Physiol 160: 2251–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst H, Melis A (2014) The chloroplast signal recognition particle (CpSRP) pathway as a tool to minimize chlorophyll antenna size and maximize photosynthetic productivity. Biotechnol Adv 32: 66–72 [DOI] [PubMed] [Google Scholar]
- Kirst H, Shen Y, Vamvaka E, Betterle N, Xu D, Warek U, Strickland JA, Melis A (2018) Downregulation of the CpSRP43 gene expression confers a truncated light-harvesting antenna (TLA) and enhances biomass and leaf-to-stem ratio in Nicotiana tabacum canopies. Planta 248: 139–154 [DOI] [PubMed] [Google Scholar]
- Kugelmann M, Fausser A, Ossenbuhl F, Brennicke A (2013) Phenotypes of Alb3p and carotenoid synthesis mutants show similarities regarding light sensitivity, thylakoid structure and protein stability. Photosynthetica 51: 45–54 [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Forfang K, Hohmann-Marriott M (2015) A practical solution for 77 K fluorescence measurements based on LED excitation and CCD array detector. PLoS ONE 10: e0132258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M, Kroth PG (2001) Diatom fucoxanthin chlorophyll a/c-binding protein (FCP) and land plant light-harvesting proteins use a similar pathway for thylakoid membrane Insertion. J Biol Chem 276: 7985–7991 [DOI] [PubMed] [Google Scholar]
- Lavaud J, Goss R (2014) The peculiar features of non-photochemical fluorescence quenching in diatoms and brown algae In Demmig-Adams B, Garab G, Adams W III, Govindjee, eds, Non-Photochemical Quenching and Energy Dissipation in Plants, Algae and Cyanobacteria. Advances in Photosynthesis and Respiration (Including Bioenergy and Related Processes), Vol 40 Springer, Dordrecht, The Netherlands, pp 421–443 [Google Scholar]
- Lavaud J, Lepetit B (2013) An explanation for the inter-species variability of the photoprotective non-photochemical chlorophyll fluorescence quenching in diatoms. Biochim Biophys Acta 1827: 294–302 [DOI] [PubMed] [Google Scholar]
- Lavaud J, Rousseau B, van Gorkom HJ, Etienne AL (2002) Influence of the diadinoxanthin pool size on photoprotection in the marine planktonic diatom Phaeodactylum tricornutum. Plant Physiol 129: 1398–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25: 1307–1320 [DOI] [PubMed] [Google Scholar]
- Lepetit B, Gélin G, Lepetit M, Sturm S, Vugrinec S, Rogato A, Kroth PG, Falciatore A, Lavaud J (2017) The diatom Phaeodactylum tricornutum adjusts nonphotochemical fluorescence quenching capacity in response to dynamic light via fine-tuned Lhcx and xanthophyll cycle pigment synthesis. New Phytol 214: 205–218 [DOI] [PubMed] [Google Scholar]
- Lepetit B, Goss R, Jakob T, Wilhelm C (2012) Molecular dynamics of the diatom thylakoid membrane under different light conditions. Photosynth Res 111: 245–257 [DOI] [PubMed] [Google Scholar]
- Lepetit B, Sturm S, Rogato A, Gruber A, Sachse M, Falciatore A, Kroth PG, Lavaud J (2013) High light acclimation in the secondary plastids containing diatom Phaeodactylum tricornutum is triggered by the redox state of the plastoquinone pool. Plant Physiol 161: 853–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepetit B, Volke D, Gilbert M, Wilhelm C, Goss R (2010) Evidence for the existence of one antenna-associated, lipid-dissolved and two protein-bound pools of diadinoxanthin cycle pigments in diatoms. Plant Physiol 154: 1905–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NE, Marty NJ, Kathir KM, Rajalingam D, Kight AD, Daily A, Kumar TK, Henry RL, Goforth RL (2010) A dynamic cpSRP43-Albino3 interaction mediates translocase regulation of chloroplast signal recognition particle (cpSRP)-targeting components. J Biol Chem 285: 34220–34230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhuri S, Serif M, Rio Bartulos C, Lepetit B, Kroth PG (2019) A strategy to complement PtAUREO1a in TALEN knockout strains of Phaeodactylum tricornutum. Algal Res 39: 101469 [Google Scholar]
- Melis A. (1989) Spectroscopic methods in photosynthesis: Photosystem stoichiometry and chlorophyll antenna size. Philos Trans R Soc B 323: 397–409 [Google Scholar]
- Melis A, Brown JS (1980) Stoichiometry of system I and system II reaction centers and of plastoquinone in different photosynthetic membranes. Proc Natl Acad Sci USA 77: 4712–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miloslavina Y, Grouneva I, Lambrev PH, Lepetit B, Goss R, Wilhelm C, Holzwarth AR (2009) Ultrafast fluorescence study on the location and mechanism of non-photochemical quenching in diatoms. Biochim Biophys Acta 1787: 1189–1197 [DOI] [PubMed] [Google Scholar]
- Mock T, Otillar RP, Strauss J, McMullan M, Paajanen P, Schmutz J, Salamov A, Sanges R, Toseland A, Ward BJ, et al. (2017) Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature 541: 536–540 [DOI] [PubMed] [Google Scholar]
- Moore M, Harrison MS, Peterson EC, Henry R (2000) Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J Biol Chem 275: 1529–1532 [DOI] [PubMed] [Google Scholar]
- Nelson N, Yocum CF (2006) Structure and function of photosystems I and II. Annu Rev Plant Biol 57: 521–565 [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HBJ, Deerfield DWI (1997) GeneDoc: Analysis and visualization of genetic variation. EMBNEW News 4: 14 [Google Scholar]
- Nymark M, Sharma AK, Hafskjold MC, Sparstad T, Bones AM, Winge P (2017) CRISPR/Cas9 gene editing in the marine diatom Phaeodactylum tricornutum. Bio Protoc 7: e2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymark M, Sharma AK, Sparstad T, Bones AM, Winge P (2016) A CRISPR/Cas9 system adapted for gene editing in marine algae. Sci Rep 6: 24951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymark M, Valle KC, Brembu T, Hancke K, Winge P, Andresen K, Johnsen G, Bones AM (2009) An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS ONE 4: e7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymark M, Valle KC, Hancke K, Winge P, Andresen K, Johnsen G, Bones AM, Brembu T (2013) Molecular and photosynthetic responses to prolonged darkness and subsequent acclimation to re-illumination in the diatom Phaeodactylum tricornutum. PLoS ONE 8: e58722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oey M, Ross IL, Stephens E, Steinbeck J, Wolf J, Radzun KA, Kügler J, Ringsmuth AK, Kruse O, Hankamer B (2013) RNAi knock-down of LHCBM1, 2 and 3 increases photosynthetic H2 production efficiency of the green alga Chlamydomonas reinhardtii. PLoS ONE 8: e61375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenbühl F, Göhre V, Meurer J, Krieger-Liszkay A, Rochaix JD, Eichacker LA (2004) Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3. Plant Cell 16: 1790–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudot-Le Secq MP, Grimwood J, Shapiro H, Armbrust EV, Bowler C, Green BR (2007) Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: Comparison with other plastid genomes of the red lineage. Mol Genet Genomics 277: 427–439 [DOI] [PubMed] [Google Scholar]
- Polle JE, Kanakagiri SD, Melis A (2003) tla1, a DNA insertional transformant of the green alga Chlamydomonas reinhardtii with a truncated light-harvesting chlorophyll antenna size. Planta 217: 49–59 [DOI] [PubMed] [Google Scholar]
- Powles SB, Critchley C (1980) Effect of light intensity during growth on photoinhibition of intact attached bean leaflets. Plant Physiol 65: 1181–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premvardhan L, Bordes L, Beer A, Büchel C, Robert B (2009) Carotenoid structures and environments in trimeric and oligomeric fucoxanthin chlorophyll a/c2 proteins from resonance Raman spectroscopy. J Phys Chem B 113: 12565–12574 [DOI] [PubMed] [Google Scholar]
- Premvardhan L, Robert B, Beer A, Büchel C (2010) Pigment organization in fucoxanthin chlorophyll a/c2 proteins (FCP) based on resonance Raman spectroscopy and sequence analysis. Biochim Biophys Acta 1797: 1647–1656 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Rodriguez F, Chauton M, Johnsen G, Andresen K, Olsen LM, Zapata M (2006) Photoacclimation in phytoplankton: Implications for biomass estimates, pigment functionality and chemotaxonomy. Mar Biol 148: 963–971 [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF (2009) Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sakshaug E, Bricaud A, Dandonneau Y, Falkowski PG, Kiefer DA, Legendre L, Morel A, Parslow J, Takahashi M (1997) Parameters of photosynthesis: Definitions, theory and interpretation of results. J Plankton Res 19: 1637–1670 [Google Scholar]
- Schuenemann D, Gupta S, Persello-Cartieaux F, Klimyuk VI, Jones JD, Nussaume L, Hoffman NE (1998) A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proc Natl Acad Sci USA 95: 10312–10316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serif M, Lepetit B, Weißert K, Kroth PG, Rio Bartulos C (2017) A fast and reliable strategy to generate TALEN-mediated gene knockouts in the diatom Phaeodactylum tricornutum. Algal Res 23: 186–195 [Google Scholar]
- Serôdio J, Vieira S, Cruz S, Coelho H (2006) Rapid light-response curves of chlorophyll fluorescence in microalgae: Relationship to steady-state light curves and non-photochemical quenching in benthic diatom-dominated assemblages. Photosynth Res 90: 29–43 [DOI] [PubMed] [Google Scholar]
- Sundberg E, Slagter JG, Fridborg I, Cleary SP, Robinson C, Coupland G (1997) ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell 9: 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei L, Chukhutsina VU, Lepetit B, Stella GR, Bassi R, van Amerongen H, Bouly JP, Jaubert M, Finazzi G, Falciatore A (2018) Dynamic changes between two LHCX-related energy quenching sites control diatom photoacclimation. Plant Physiol 177: 953–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei L, Stella GR, Rogato A, Bailleul B, Fortunato AE, Annunziata R, Sanges R, Thaler M, Lepetit B, Lavaud J, et al. (2016) Multisignal control of expression of the LHCX protein family in the marine diatom Phaeodactylum tricornutum. J Exp Bot 67: 3939–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis J, Schroda M (2016) Revisiting the photosystem II repair cycle. Plant Signal Behav 11: e1218587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Träger C, Rosenblad MA, Ziehe D, Garcia-Petit C, Schrader L, Kock K, Richter CV, Klinkert B, Narberhaus F, Herrmann C, et al. (2012) Evolution from the prokaryotic to the higher plant chloroplast signal recognition particle: The signal recognition particle RNA is conserved in plastids of a wide range of photosynthetic organisms. Plant Cell 24: 4819–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle KC, Nymark M, Aamot I, Hancke K, Winge P, Andresen K, Johnsen G, Brembu T, Bones AM (2014) System responses to equal doses of photosynthetically usable radiation of blue, green, and red light in the marine diatom Phaeodactylum tricornutum. PLoS ONE 9: e114211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yu LJ, Xu C, Tomizaki T, Zhao S, Umena Y, Chen X, Qin X, Xin Y, Suga M, et al. (2019) Structural basis for blue-green light harvesting and energy dissipation in diatoms. Science 363: eaav0365. [DOI] [PubMed] [Google Scholar]
- Yamagishi A, Ikeda Y, Komura M, Koike H, Satoh K, Itoh S, Shibata Y (2010) Shallow sink in an antenna pigment system of photosystem I of a marine centric diatom, Chaetoceros gracilis, revealed by ultrafast fluorescence spectroscopy at 17 K. J Phys Chem B 114: 9031–9038 [DOI] [PubMed] [Google Scholar]