Abstract
Objectives: Epigenetic markers, and in particular DNA methylation, have come to the fore as new tools in the personalization of the treatment of obesity and its comorbidities. The objectives of the current investigation were to identify epigenetic biomarkers that might be predictive of response to a weight-loss intervention, and to better understand the influence of certain nutrients (particularly antioxidants) on the epigenome.
Methods: Global DNA (LINE-1) methylation levels were assessed in peripheral blood mononuclear cells (PBMCs) from 96 obese volunteers of the Metabolic Syndrome Reduction in Navarra study, using a methylation-sensitive high resolution melting approach after bisulfite modification.
Results: Baseline LINE-1 DNA methylation levels were significantly higher (5.41%) in high responders (>8% of weight loss) as compared to low responders (<8%) to the energy-restricted treatment. Indeed, a LINE-1 methylation higher than 84.15% may be predictive of a high response to the hypocaloric diet. Statistically significant correlations were found between LINE-1 baseline DNA methylation levels and the response to the treatment involving total fat mass and body weight. Furthermore, LINE-1 baseline methylation levels positively correlated with baseline dietary total antioxidant capacity (TAC).
Discussion: LINE-1 methylation levels in PBMCs might be used to predict response to a dietary weight-loss intervention, and seem to be related to the dietary TAC.
Trial Registration: www.clinicaltrials.gov: NCT01087086.
Keywords: Antioxidants, Biomarkers, Epigenetic, Energy-restriction, LINE-1, Methylation, Obesity
Background
Obesity, which is defined as the excessive accumulation of adiposity in relation to body mass, occurs as a result of an imbalance in energy homeostasis between energy intake and energy expenditure.1 Given the steady increase in the prevalence of obesity,2 there is an urgent need to find new robust biomarkers that could lead to better prevention of the disorder and a more personalized form of treatment.3 In recent years, different epigenetic biomarkers have been identified in relation to the personalized response to different dietary approaches to treat obesity.4,5
Epigenetics is defined as the heritable changes in gene activity and expression that occur without abnormalities in DNA sequences, and is involved in the onset and progression of obesity and obesity-related diseases.6 At a general level, DNA methylation is usually assessed in peripheral blood mononuclear cells (PBMCs) by measuring repeat interspersed regions, such as long interspersed nucleotide element 1 (LINE-1) or Alu regions, a kind of short interspersed nucleotide element (SINE). PBMCs, which have the longest life span of all blood cells, are considered to best represent the biological changes related to environmental exposures or modifications to lifestyle and behavior.7 On the other hand, LINE-1 and SINE sequences are retrotransposons, mobile sequences that use a ‘copy-and-paste’ mechanism to change their location and their copy number using an RNA intermediate.8 In the case of obesity, one trial reported that a high or low birth weight, as well as premature birth, was associated with significantly lower LINE-1 methylation levels.9 Similarly, Perng et al. found that LINE-1 hypomethylation was related to the development of adiposity in boys.10
Although nutrition plays a role in disease prevention, the interactions between the epigenome (including overall DNA methylation) and different dietary bioactive compounds and food combinations have not been the focus of much research. For example, it has been reported that some antioxidants, including genistein, zinc, selenium, and vitamin A, could affect DNA methylation,11 whereas dietary compounds and interventions could modify LINE-1 DNA methylation.12
Hence, the objectives of the current research were to study whether overall DNA methylation levels (such as LINE-1 methylation at baseline) could be a predictive biomarker of response to a hypocaloric diet (by comparing low and high responders to the dietary treatment) and to elucidate the influence of certain nutrients, particularly antioxidants, on overall DNA methylation.
Methods
Subjects and study protocol
The current analysis was conducted within the RESMENA (Metabolic Syndrome Reduction in Navarra) project, a randomized nutritionally controlled trial.13 The sample for this study consisted of 96 adults with MetS as defined by International Diabetes Federation criteria. All those subjects who did not meet the previously detailed inclusion criteria13 or for whom relevant information was lacking were excluded. Subjects presenting chronic diseases related to nutrient metabolism, those following special diets and those experiencing body weight changes in the last 3 months were also excluded from the trial. Participants underwent two energy-restricted dietary patterns for 8 weeks, both with an energy restriction of −30% of the studied requirements.13 The control diet was based on the American Heart Association (AHA) guidelines, including 3–5 meals/day, a macronutrient distribution of 55% total caloric value from carbohydrates, 15% from proteins, and 30% from lipids. In contrast, the RESMENA diet was designed with a higher meal frequency, consisting of 7 meals/day, and a macronutrient distribution of 40% total caloric value from carbohydrates, 30% from proteins and 30% from lipids, as described elsewhere.13 As no differences in anthropometric and biochemical variables were found between both dietary intervention groups, the two subject groups were merged for further analyses.14 Body composition was specifically measured using a dual-energy X-ray absorptiometry (DXA Lunar Prodigy, GE Medical Systems, Madison, WI, USA). Diet composition was analyzed using the DIAL software (Alce Ingenieria, Madrid, Spain), and dietary total antioxidant capacity (TAC) was calculated using the Carlsen et al. values.15 TAC was determined using the ferric reducing ability of plasma (FRAP) assay, which was able to estimate the antioxidant power of antioxidants such as ascorbic acid, uric acid, bilirubin, trolox (a water-soluble analog of vitamin E), alpha-tocopherol, and albumin in plasma and in mixtures of these antioxidants.16
The study was approved by the Ethics Committee of the University of Navarra (065/2009) and appropriately registered at www.clinicaltrials.gov (NCT01087086). All the participants provided written informed consent for participation in agreement with the Declaration of Helsinki. The study was performed following the CONSORT 2010 guidelines.17
Sample collection and DNA isolation
A blood sample was collected from each volunteer and transferred into a 10 mL EDTA tube at baseline and after 8 weeks of treatment. Blood was immediately placed on ice, centrifuged within 30 minutes to isolate the buffy coat, and stored at −20°C until use. DNA was isolated from the PBMCs using the Master Pure DNA Purification Kit for Blood Version II kit (Epicenter, Madison, WI, USA) according to the instructions provided by the manufacturer. Purified DNA was stored at −20°C until use. Purified DNA was quantified by PicoGreen dsDNA Quantitation Reagent (Invitrogen, Carlsbad, CA, USA).
Methylation standards and bisulfite conversion
Cells-to-CpG™ Methylated gDNA Control Kit (Life Technologies, Waltham, MA, USA) and DNA from placenta cells (D3160, Sigma Aldrich, St. Louis, MO, USA) were used as methylated and non-methylated DNA standards, respectively. To generate a range of methylated and unmethylated DNA standards, the two control DNA standards were mixed in 0, 20, 40, 60, 80, and 100% methylated to unmethylated template ratios.
One microgram of each standard and sample DNA was bisulfite-converted (BSC) by using the Epitect Fast Bisulfite Conversion Kit (Qiagen, Venlo, Limburg, The Netherlands) according to the manufacturer's instructions, thus converting non-methylated cytosines into uracil. All BSC DNAs were diluted to 5 ng/μL for use in PCR.
Methylation-sensitive high resolution melting (MS-HRM) analysis
Primers were designed against the completely methylated sense strand sequence and according to the recommendations of Wojdacz and Dobrovic18 in order to minimize PCR bias. The promoter region of the consensus LINE-1 sequence (GenBank: X58075) was used to design primer sets with the Primer3 website.19 The primers were the followings: forward-GCGAGGTATTGTTTTATTTGGGA; reverse-CGCCGTTTCTTAAACC. It screened eight CpGs in an amplicon length of 141 bp.
PCR amplification of the DNA was carried out using a 7900HT Fast Real-Time PCR System (Life Technologies, Waltham, MA, USA) equipped with the SDS Software (Version 2.4.1, Life Technologies, Waltham, MA, USA). PCR was performed in a 10-μL reaction volume, and 5 ng of BSC DNA templates for LINE-1 assay were added to each well, which contained 1 × MeltDoctor™ HRM Master Mix (HRM) (Life Technologies, Waltham, MA, USA) and 0.2 μM of each primer. The cycling protocol conditions included a single enzyme activation step of 10 minutes at 95°C followed by 40 cycles of the following steps: denaturation 95°C, 15 seconds, and annealing 60°C, 1 minute. The MS-HRM step was performed after 40 cycles of amplification and the MS-HRM analysis was initiated by denaturing all products at 95°C for 1 minute, followed by annealing at 55°C for 1 minute. Samples were slowly warmed to 95°C at 0.1°C/second.
The High ReSolution Melt Software v2.0 (Life Technologies, Waltham, MA, USA) was employed for end-product analysis. This algorithm allowed the raw melt curves to be normalized for fluorescence intensity, and a temperature shift was applied to align the normalized melt curves, which facilitated the analysis of samples with varying crossing threshold (Ct) values. A difference curve was then derived from the first derivative of the melt curves. Data for the difference melt curves were exported to Excel (Office 2007; Microsoft Corp., Redmond, WA) for further analyses. Graphs were plotted and inverted vertically. Both peak-height and area-under-the-curve from the normalized, temperature-shifted, difference curves were used to generate a standard curve and determine the degree of methylation of each DNA sample. All participant DNA samples were analyzed on a 384-well plate, which included a no-template control (NTC) and a set of reference methylation standards. Reference methylation standard curves and experimental samples were tested in triplicate.
Statistical analyses and graph representation
This ancillary study could be considered as observational, but this kind of approach is considered as valid as the initially designed interventional approach.20 Different statistical models were applied to analyze the methylation results. All data were examined using parametric tests after confirming normal distribution with the Shapiro-Wilk test. The baseline methylation differences between low and high responders to the nutritional intervention were analyzed using ANCOVA. This approach allowed prediction of a response to a diet according to a specific methylation pattern. Pearson's correlations were fitted to evaluate the potential correlations of the methylation values with biochemical and anthropometric variables, and also with the dietary components. Receiver operating characteristic (ROC) analysis was used to evaluate the sensitivity and specificity of DNA methylation levels of LINE-1 for predicting a high response to a hypocaloric diet.
A P < 0.05 was considered statistically significant. SPSS 15.0 software (SPSS Inc., Chicago, IL, USA) for Windows XP (Microsoft) was used for all the statistical analyses.
To show graphically and visually the results, graphs, and figures were created with GraphPad Prism® version 6.0C (La Jolla, CA, USA).
Results
Characteristics of the study population
After the dietary intervention, the main anthropometric and biochemical measurements significantly decreased (P < 0.001) (data not shown). For example, body mass index (BMI) values were reduced by the energy-restricted program (from 35.8 [95% CI: 34.9–36.8] kg/m2 to 33.4 [95% CI: 32.4–34.3] kg/m2, P < 0.001), thus improving the adiposity status from obesity class II to obesity class I. No differences were found between either dietary intervention groups, neither in the anthropometric nor in the biochemical variables (Table 1). Moreover, there were no significant differences between the two dietary intervention groups for the two variables of interest (baseline LINE-1 methylation levels and baseline TAC; P = 0.794 and P = 0.905, respectively). Therefore, the two groups were merged and subsequently analyzed together as a single experimental group, as previously published.14,21 Such merging increased the statistical power of the study as a result of the larger population.
Table 1. Anthropometric and biochemical changes after 8 weeks of treatment depending on the diet.
| Variable | AHA (n = 48) | RESMENA (n = 48) | P |
|---|---|---|---|
| Sex (%) | ns | ||
| Male | 56.3 | 50.0 | |
| Female | 43.7 | 50.0 | |
| ΔWeight (kg) | −6.7 (−7.6 to −5.8) | −7.1 (−7.9 to −6.3) | 0.555 |
| ΔBMI (kg/m2) | −2.4 (−2.7 to −2.1) | −2.5 (−2.8 to −2.2) | 0.732 |
| ΔWaist (cm) | −6.8 (−8.2 to −5.4) | −7.5 (−8.6 to −6.4) | 0.432 |
| ΔTotal fat mass by DXA (kg) | −4.6 (−5.3 to −3.9) | −5.2 (−5.9 to −4.5) | 0.208 |
| ΔTrunk mass by DXA (kg) | −2.6 (−4.1 to −1.1) | −3.7 (−4.3 to −3.1) | 0.183 |
| ΔSBP (mmHg) | −11 (−16 to −6) | −13 (−19 to −7) | 0.570 |
| ΔDBP (mmHg) | −6 (−9 to −3) | −8 (−11 to −5) | 0.371 |
| ΔTotal cholesterol (mg/dl) | −16.9 (−26.6 to −7.2) | −16.3 (−29.9 to −2.7) | 0.943 |
| ΔHDL-c (mg/dl) | −4.0 (−6.1 to −1.9) | −2.0 (−3.9 to −0.1) | 0.158 |
| ΔGlucose (mg/dl) | −13.0 (−21.8 to −4.2) | −13.6 (−24.6 to −2.6) | 0.939 |
| ΔTG (mg/dl) | −30.9 (−51.5 to −10.3) | −43.5 (−66.6 to −20.4) | 0.421 |
Data expressed as mean (95% CI). Abbreviations: BMI, body mass index; DXA, dual-energy X-ray absorptiometry; SBP, systolic blood pressure; DBP, diastolic blood pressure; HDL-c, high density lipoprotein cholesterol; TG, triglycerides.
Anthropometric and biochemical measurements were recorded at baseline and after 8 weeks of the energy-restricted dietary program, and the volunteers were classified according to the response to the weight-loss intervention (Table 2). According to the diogenes study inclusion criteria,22 subjects were considered as ‘high responders’ (HR) when weight loss was ≥8% after 8 weeks of energy-restricted program (n = 38) and ‘low responders’ (LR) when weight loss was <8% (n = 58), respectively. A total of 39.6% of the population was considered as high responders. No statistical differences were found at baseline between high and low responders, except for triglyceride levels. As expected, participants in the HR group showed greater reductions in body weight, BMI, waist circumference, total fat mass, and truncal fat mass than those in the LR group (Table 2).
Table 2. Characteristics of the population classified by the response to the energy-restricted dietary program.
| Variable | Low responders (n = 58) | High responders (n = 38) | P† | ||
|---|---|---|---|---|---|
| Baseline | 8 weeks | Baseline | 8 weeks | ||
| Age (years) | 48 (45.4–50.6) | 48 (45.4–50.6) | 51 (48.0–54.0) | 51 (48.0–54.0) | 0.133 |
| Sex (%) | 0.938 | ||||
| Male | 53.4 | 53.4 | 52.6 | 52.6 | |
| Female | 46.6 | 46.6 | 47.5 | 47.5 | |
| Weight (kg) | 101.2 (96.0–106.2) | 96.0 (91.2–100.8)*** | 97.4 (92.4–102.4) | 88.0 (83.5–92.5)*** | <0.001 |
| BMI (kg/m2) | 36.0 (34.8–37.2) | 34.2 (33.1–35.3)*** | 35.5 (33.9–37.1) | 32.1 (30.6–33.6)*** | <0.001 |
| Waist circumference (cm) | 111.7 (108.2–115.2) | 106.2 (103.1–109.3)*** | 110.2 (106.2–114.2) | 100.6 (96.9–104.3)*** | <0.001 |
| Total fat mass (kg)‡ | 43.3 (40.9–45.7) | 39.4 (37.0–41.8)** | 40.9 (37.5–44.3) | 34.3 (31.2–37.4)*** | <0.001 |
| Trunk mass (kg)‡ | 25.5 (23.8–27.2) | 23.4 (21.7–25.1)** | 24.9 (22.8–27.0) | 20.1 (18.2–22.0)*** | 0.001 |
| SBP (mmHg) | 147 (143–151) | 138 (134–142)*** | 153 (145–161) | 136 (130–142)*** | <0.05 |
| DBP (mmHg) | 85 (83–87) | 79 (77–81)*** | 85 (81–89) | 77 (74–80)*** | 0.122 |
| TC (mg/dl) | 214.2 (202.4–226) | 204.3 (193.1–214.9) | 229.1 (215.8–242.4) | 202.1 (187.5–216.7)*** | <0.05 |
| HDL-c (mg/dl) | 44.4 (41.6–47.2) | 42.4 (39.7–45–1) | 45.1 (42.2–48.0) | 40.7 (38.3–43.1)** | 0.126 |
| Glucose (mg/dl)§ | 123.9 (113.6–134.2) | 111.0 (105.9–116.1)** | 120.2 (110.2–130.2) | 106.4 (98.9–113.9)*** | 0.945 |
| TG (mg/dl)§ | 180.8 (151.9–209.7) | 154.0 (132.8–175.2)** | 191.7 (156.6–226.8) | 137.1 (106.7–167.5)*** | 0.797 |
| Energy intake (kcal) | 2111 (1970–2252) | 1383 (1297–1469)*** | 2311 (2158–2464) | 1291 (1224–1358)*** | <0.01 |
| Dietary TAC (mmol/day) | 7.5 (6.7–8.3) | 9.8 (7.9–9.7)*** | 7.0 (6.0–8.0) | 11.2 (10.0–12.4)*** | 0.159 |
Data expressed as mean (95% CI). Abbreviations: BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; TC, total cholesterol; HDL-c, high density lipoprotein cholesterol; TG, triglycerides; TAC, total antioxidant capacity. High responders categorized by 8% body weight loss from baseline.
†P of variable changes between high responders and low responders.
*P < 0.05 between baseline and 8 weeks after.
**P < 0.01 between baseline and 8 weeks after.
***P < 0.001 between baseline and 8 weeks after.
‡Measurement by dual-energy X-ray absorptiometry (DXA).
§Non-normally distributed variables were transformed for the model analysis.
Association between the baseline DNA methylation levels and the response to the energy-restricted dietary program
Baseline LINE-1 DNA methylation levels were significantly higher in HR (mean: 85.49; 95% CI: 83.45–87.53) as compared to LR (mean: 80.08; 95% CI: 79.33–81.84) (P < 0.001) (Fig. 1A). Statistically significant correlations (P < 0.05) were observed between baseline LINE-1 methylation levels and the loss of certain anthropometric parameters such as total fat mass and body weight (Fig. 1B and C). Moreover, baseline LINE-1 methylation levels were also associated with changes in waist circumference (r2 = 0.319, P = 0.003; r2 = 0.279, P = 0.026; respectively) and with changes in triglyceride levels (r2 = 0.317, P = 0.016).
Figure 1.
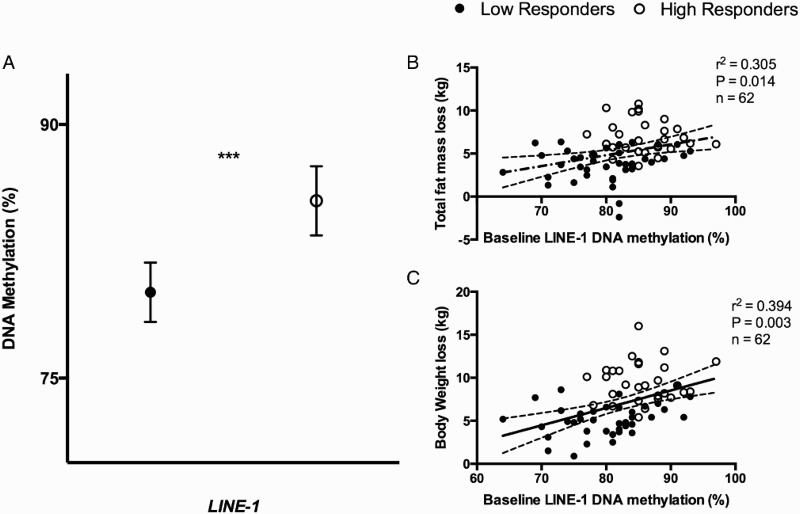
Baseline DNA methylation levels of low and high responders to the intervention. (A) Baseline DNA methylation levels (%) of low responders and high responders adjusted for age, gender, and number of plate. P from ANCOVA test: *0.05 < P < 0.01; ***P < 0.001. (B) Pearson's correlation between total fat mass loss (kg) and baseline DNA methylation levels (%) of LINE-1; (C) Pearson's correlation between body weight loss (as kg) and baseline DNA methylation levels (%) of LINE-1. r2 represents Pearson correlation coefficient and P is the stringency of the correlation. A P < 0.05 was considered statistically significant. Full circles: LR; empty circles: HR.
Influence of dietary total antioxidant capacity on DNA methylation levels
Pearson's correlation analyses were performed to assess the possible association between TAC and LINE-1 methylation levels. Baseline TAC was associated (P < 0.01) with LINE-1 methylation levels at baseline (Fig. 2). By using a linear regression model, we observed that total fat mass and TAC explained about 22% of the variation of the baseline DNA methylation of LINE-1 (Table 3).
Figure 2.
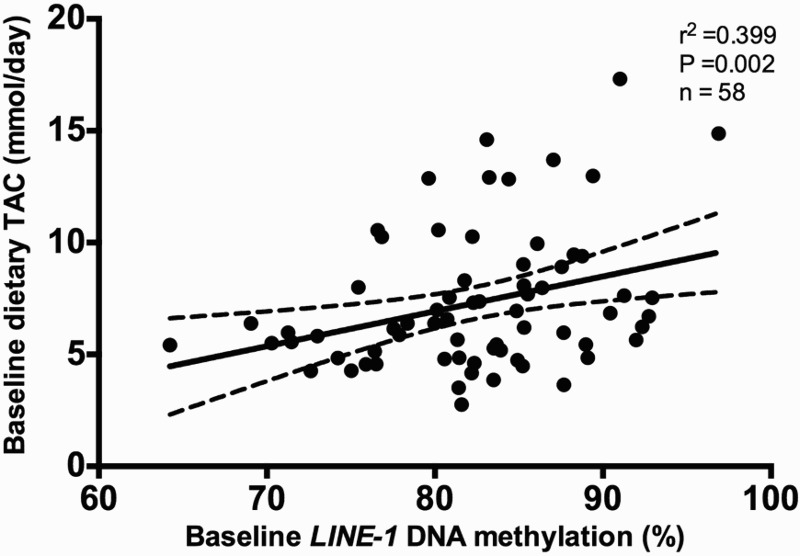
Pearson's correlation between baseline methylation levels (%) of LINE-1 and baseline dietary total antioxidant capacity (TAC; mmol/day), adjusted for age, sex, diet type, energy intake, and number of plate. r2 represents Pearson correlation coefficient.
Table 3. Multiple regression analyses showing the independent contributions of studied domains* to the variation of baseline DNA methylation of LINE-1.
| LINE-1 as dependent | B coefficient | 95% confidence interval | P |
|---|---|---|---|
| Total fat mass by DXA (%) | −0.379 | (−0.608; −0.167) | 0.001 |
| Dietary TAC (mmol/day) | 0.290 | (0.156; 1.053) | 0.007 |
| R2 = 0.22 (P < 0.001) | |||
Adjusted R2 and all independent variables included in each model are presented in the table. Italic style represents statistical significant P values.
*Domains are adiposity indicators (baseline total fat mass by DXA (%) and baseline dietary total antioxidant capacity (mmol/day)).
Discussion
DNA methylation in specific regions is a crucial step in the control and regulation of gene expression as it prevents or promotes the recruitment of regulatory proteins. As such it could be used as a biomarker of disease risk or to personalize treatment. For example, there is a long list of genes whose methylation status has been proposed as biomarkers for cancer diagnosis.23
The first objective of the present study was to determine if DNA methylation levels of LINE-1 at baseline could predict if a subject could be a low or a high responder to a dietary weight-loss program with a hypocaloric diet. It has been previously reported that the response to a weight-loss treatment could be predicted by analyzing the methylation levels of several genes at baseline.24 One of these studies24 found that five regions located in or near the genes AQP9, DUSP22, HIPK3, TNNT1, and TNNI3 were differentially methylated at baseline in HR and LR. Another study25 performed in obese adolescents who underwent 10 weeks of a multidisciplinary weight-loss intervention, found that DNA methylation levels in CpGs located in or near ATP10A, CD44, and WT1 showed statistically different methylation levels depending on the weight-loss outcome. The results of the present study show that baseline LINE-1 methylation levels were significantly different between HR and LR and thus confirm that methylation levels in PBMCs could be used as early biomarkers of weight-loss success with a hypocaloric diet. After performing a ROC curve (AUC = 0.75; 95% CI: 0.633–0.865), we concluded that a LINE-1 methylation value higher than 84.15% may be predictive of a higher response to a hypocaloric diet.
The hypothesis of considering LINE-1 as a biomarker of obesity is not presumptuous. LINE-1 methylation levels have been previously proposed as biomarkers for several diseases, including melanoma, prostate adenocarcinoma, rectal cancer, or osteonecrosis.26 Furthermore, studies have reported that DNA methylation, including LINE-1 and Alu methylation, may play a role in chronic diseases, specifically, cardiovascular disease and diabetes.27,28 Indeed, recent epidemiological trials have also reported associations between LINE-1 methylation and obesity-related diseases.12,28 Although Zhu et al.29 found that LINE-1 and Alu methylation levels in white blood cells were not correlated with BMI, other studies have found that there was a significant positive association between BMI and LINE-1 methylation30 and that LINE-1 hypomethylation was related to the development of adiposity in boys.10 Newborns with low or high birthweight have been found to have significantly lower LINE-1 methylation levels in their cord blood compared to normal weight infants.9 In a prospective study of 470 child-bearing women, a lower degree of LINE-1 methylation in PBMC was associated with excess body weight,31 whereas women with higher BMI, a higher percentage of body fat or higher waist circumference had lower LINE-1 methylation levels. Another study observed lower white blood cell LINE-1 methylation levels in obese subjects (BMI ≥ 25 kg/m2) when compared with those of BMI ≤ 25 kg/m2.32
However, few studies have analyzed the relationship between DNA methylation and weight loss. Martin-Nunez et al.33 found a positive correlation between LINE-1 methylation changes in PBMC and weight change. A recent study has observed that weight loss over 12 months did not change LINE-1 DNA methylation levels,34 as we have corroborated in our data (data not shown). All these data are in agreement with the results of the present study, where baseline LINE-1 methylation levels were associated with changes in BMI, body weight, and total fat mass, suggesting that such levels could be used to predict the outcome of a weight-loss intervention. This predictor could be added to other epigenetic biomarkers previously identified as early predictors of body weight loss and maintenance of body weight after weight loss treatments.3
The effect of different bioactive food components on overall DNA methylation is only just starting to be understood. Studies with metabolites involved in one-carbon metabolism (vitamin B12, vitamin B6, vitamin B1, folate, methionine, choline, biotin), isothiocyanates, phytoestrogens, flavonoids and other phenolic compounds, lipoic acid, vitamin A, and metabolites of vitamin E have demonstrated an interaction between such substances and DNA methylation.11,35 Many of these compounds exert antioxidant activity. Thus, polyphenols may decrease DNA methylation by altering cellular SAM/SAH ratios and by indirectly inhibiting DNMT activity.35 Ascorbic acid is another antioxidant that has been reported to promote changes in the epigenome in cultured cells as it tends to oxidize 5-methylcytosine into 5-hydroxymethylcytosine in a demethylation pathway.36
TAC has been inversely associated with central adiposity measurements in healthy young adults, indicating that it could be useful in the assessment of the health benefits of cumulative antioxidant capacity from food intake.37 In line with this, an association between baseline LINE-1 methylation levels and baseline TAC intake was observed in the current study. This is in agreement with previous studies that have found an association between retinol levels and LINE-1 methylation status.38 The amount of fruits and vegetables that subjects declared in the 136-item food frequency questionnaire were the main factors that influenced TAC values, as these products have a much higher antioxidant content (mmol/serving).15 To our knowledge, this is the first study where overall LINE-1 methylation and TAC have been associated.
The main limitation of the current study is that we did not analyze LINE-1 methylation levels in adipose tissue, and it is possible that fat tissue displays a different tissue-specific methylation pattern than that observed in peripheral blood leukocytes. Nevertheless, previous studies have shown that DNA methylation patterns are largely conserved across tissues, and that leukocyte DNA methylation levels might serve as surrogate markers of DNA methylation in specific tissues.39 Furthermore, no blood cell count was carried out, resulting in a possible limitation in the interpretation of DNA methylation levels. Nevertheless, one of the aims of the present study was to use PBMCs as a source of easily isolated prognostic biomarkers as other studies have previously reported.25,40 In addition, taking into account that body composition was measured by DXA, and that the technique used to quantify the methylation levels was MS-HRM (a rapid and reliable approach to detect DNA methylation)41; the results obtained are very robust.
Although further studies with a larger sample size and a longer intervention period are required to confirm the influence of the dietary approach on methylation status, this study suggests that LINE-1 methylation levels in PBMCs may well be a biomarker of weight loss response and that they are influenced by diet composition, especially antioxidant content. This finding is of particular relevance in that it confirms that the epigenome plays a role in the development of obesity and in the individual response to the environmental/dietary factors, and represents a first step towards the personalization of weight-loss treatments.
Conclusion
Baseline LINE-1 methylation levels correlated with changes in anthropometric variables and could be used as a predictor of weight loss success by early differentiating between high and low responders to a specific treatment. Finally, LINE-1 methylation levels in PBMCs seem to be affected by dietary TAC.
Acknowledgments
We thank Ana Lorente for her laboratory assistance. This work was supported by Línea Especial about Nutrition, Obesity, and Health (University of Navarra LE/97), CIBERobn, MINECO (ref. AGL2013-45554-R) and the Health Department of the Government of Navarra (48/2009). MGL holds a grant from the Centre for Nutrition Research of the University of Navarra. MLM holds a Juan de la Cierva fellowship from the Spanish Ministry of Economy and Competitiveness. Dr Paul William Miller, lecturer in the Institute of Modern Languages of the University of Navarra, is acknowledged for his help in reviewing the English of the manuscript.
Disclaimer statements
Contributors MGL: field work, data collection, analysis, and writing of the manuscript; FIM: design, analysis, and editing of the manuscript; MAZ: design, field work, and data collection; JAM: project leader, scientific interpretation, financial management, and editing of the manuscript; MLM: design, field work, data collection, analysis, and writing of the manuscript. All authors read and approved the final manuscript.
Funding None.
Conflicts of interest The author(s) declare that they have no competing interests.
Ethics approval The study was approved by the Ethics Committee of the University of Navarra (065/2009) and appropriately registered at www.clinicaltrials.gov (NCT01087086). This work was performed following the CONSORT 2010 guidelines.
ORCID
Fermin I. Milagro http://orcid.org/0000-0002-3228-9916
References
- 1.Haslam DW, James WP. Obesity. Lancet 2005;366(9492):1197–209. doi: 10.1016/S0140-6736(05)67483-1 [DOI] [PubMed] [Google Scholar]
- 2.Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C, et al. . Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014;384(9945):766–81. doi: 10.1016/S0140-6736(14)60460-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez JA, Milagro FI, Claycombe KJ, Schalinske KL. Epigenetics in adipose tissue, obesity, weight loss, and diabetes. Adv Nutr 2014;5(1):71–81. doi: 10.3945/an.113.004705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campion J, Milagro F, Goyenechea E, Martinez J. TNF-alpha promoter methylation as a predictive biomarker for weight-loss response. Obesity (Silver Spring) 2009;17:1293–7. [DOI] [PubMed] [Google Scholar]
- 5.Cordero P, Campion J, Milagro FI, Goyenechea E, Steemburgo T, Javierre BM, et al. . Leptin and TNF-alpha promoter methylation levels measured by MSP could predict the response to a low-calorie diet. J Physiol Biochem 2011;67(3):463–70. doi: 10.1007/s13105-011-0084-4 [DOI] [PubMed] [Google Scholar]
- 6.Milagro FI, Mansego ML, De Miguel C, Martinez JA. Dietary factors, epigenetic modifications and obesity outcomes: progresses and perspectives. Mol Aspects Med 2013;34(4):782–812. doi: 10.1016/j.mam.2012.06.010 [DOI] [PubMed] [Google Scholar]
- 7.Delgado-Cruzata L, Vin-Raviv N, Tehranifar P, Flom J, Reynolds D, Gonzalez K, et al. . Correlations in global DNA methylation measures in peripheral blood mononuclear cells and granulocytes. Epigenetics 2014;9(11):1504–10. doi: 10.4161/15592294.2014.983364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang CR, Burns KH, Boeke JD. Active transposition in genomes. Annu Rev Genet 2012;46:651–75. doi: 10.1146/annurev-genet-110711-155616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michels KB, Harris HR, Barault L. Birthweight, maternal weight trajectories and global DNA methylation of LINE-1 repetitive elements. PLoS One 2011;6(9):e25254. doi: 10.1371/journal.pone.0025254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perng W, Mora-Plazas M, Marin C, Rozek LS, Baylin A, Villamor E. A prospective study of LINE-1DNA methylation and development of adiposity in school-age children. PLoS One 2013;8(4):e62587. doi: 10.1371/journal.pone.0062587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tatarinova T, editor. Effects of dietary nutrients on DNA methylation and imprinting. DNA methylation – from genomics to technology. Rijeka, Croatia: InTech - Open Science Open Minds; 2012. p. 289–302. [Google Scholar]
- 12.Pearce MS, McConnell JC, Potter C, Barrett LM, Parker L, Mathers JC, et al. . Global LINE-1 DNA methylation is associated with blood glycaemic and lipid profiles. Int J Epidemiol 2012;41(1):210–7. doi: 10.1093/ije/dys020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zulet MA, Bondia-Pons I, Abete I, de la Iglesia R, Lopez-Legarrea P, Forga L, et al. . The reduction of the metabolyc syndrome in Navarra-Spain (RESMENA-S) study: a multidisciplinary strategy based on chrononutrition and nutritional education, together with dietetic and psychological control. Nutr Hosp 2011;26(1):16–26. [PubMed] [Google Scholar]
- 14.Perez-Cornago A, de la Iglesia R, Lopez-Legarrea P, Abete I, Navas-Carretero S, Lacunza CI, et al. . A decline in inflammation is associated with less depressive symptoms after a dietary intervention in metabolic syndrome patients: a longitudinal study. Nutr J 2014;13:36. doi: 10.1186/1475-2891-13-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlsen MH, Halvorsen BL, Holte K, Bohn SK, Dragland S, Sampson L, et al. . The total antioxidant content of more than 3100 foods, beverages, spices, herbs and supplements used worldwide. Nutr J 2010;9:3. doi: 10.1186/1475-2891-9-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benzie IF, Strain JJ. The ferric reducing ability of plasma (FRAP) as a measure of ‘antioxidant power’: the FRAP assay. Anal Biochem 1996;239(1):70–6. doi: 10.1006/abio.1996.0292 [DOI] [PubMed] [Google Scholar]
- 17.Schulz KF, Altman DG, Moher D. CONSORT 2010 statement: updated guidelines for reporting parallel group randomized trials. Ann Intern Med 2010;152(11):726–32. doi: 10.7326/0003-4819-152-11-201006010-00232 [DOI] [PubMed] [Google Scholar]
- 18.Wojdacz TK, Dobrovic A. Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res 2007;35(6):e41. doi: 10.1093/nar/gkm013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 2000;132:365–86. [DOI] [PubMed] [Google Scholar]
- 20.Concato J, Shah N, Horwitz RI. Randomized, controlled trials, observational studies, and the hierarchy of research designs. N Engl J Med 2000;342(25):1887–92. doi: 10.1056/NEJM200006223422507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Legarrea P, de la Iglesia R, Abete I, Navas-Carretero S, Martinez JA, Zulet MA. The protein type within a hypocaloric diet affects obesity-related inflammation: the RESMENA project. Nutrition 2014;30(4):424–9. doi: 10.1016/j.nut.2013.09.009 [DOI] [PubMed] [Google Scholar]
- 22.Gogebakan O, Kohl A, Osterhoff MA, van Baak MA, Jebb SA, Papadaki A, et al. . Effects of weight loss and long-term weight maintenance with diets varying in protein and glycemic index on cardiovascular risk factors: the diet, obesity, and genes (DiOGenes) study: a randomized, controlled trial. Circulation 2011;124(25):2829–38. doi: 10.1161/CIRCULATIONAHA.111.033274 [DOI] [PubMed] [Google Scholar]
- 23.Tollefsbol TO, editor. DNA methylation alterations in human cancers. Epigenetics in human disease. London: Elsevier; 2012. p. 29–52. [Google Scholar]
- 24.Moleres A, Campion J, Milagro FI, Marcos A, Campoy C, Garagorri JM, et al. . Differential DNA methylation patterns between high and low responders to a weight loss intervention in overweight or obese adolescents: the EVASYON study. FASEB J 2013;27(6):2504–12. doi: 10.1096/fj.12-215566 [DOI] [PubMed] [Google Scholar]
- 25.Milagro FI, Campion J, Cordero P, Goyenechea E, Gomez-Uriz AM, Abete I, et al. . A dual epigenomic approach for the search of obesity biomarkers: DNA methylation in relation to diet-induced weight loss. FASEB J 2011;25(4):1378–89. doi: 10.1096/fj.10-170365 [DOI] [PubMed] [Google Scholar]
- 26.Benard A, van de Velde CJ, Lessard L, Putter H, Takeshima L, Kuppen PJ, et al. . Epigenetic status of LINE-1 predicts clinical outcome in early-stage rectal cancer. Br J Cancer 2013;109(12):3073–83. doi: 10.1038/bjc.2013.654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One 2010;5(3):e9692. doi: 10.1371/journal.pone.0009692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baccarelli A, Wright R, Bollati V, Litonjua A, Zanobetti A, Tarantini L, et al. . Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology 2010;21(6):819–28. doi: 10.1097/EDE.0b013e3181f20457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu ZZ, Hou L, Bollati V, Tarantini L, Marinelli B, Cantone L, et al. . Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol 2012;41(1):126–39. doi: 10.1093/ije/dyq154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cash HL, McGarvey ST, Houseman EA, Marsit CJ, Hawley NL, Lambert-Messerlian GM, et al. . Cardiovascular disease risk factors and DNA methylation at the LINE-1 repeat region in peripheral blood from Samoan Islanders. Epigenetics 2011;6(10):1257–64. doi: 10.4161/epi.6.10.17728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piyathilake CJ, Badiga S, Alvarez RD, Partridge EE, Johanning GL. A lower degree of PBMC L1 methylation is associated with excess body weight and higher HOMA-IR in the presence of lower concentrations of plasma folate. PLoS One 2013;8(1):e54544. doi: 10.1371/journal.pone.0054544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang FF, Santella RM, Wolff M, Kappil MA, Markowitz SB, Morabia A. White blood cell global methylation and IL-6 promoter methylation in association with diet and lifestyle risk factors in a cancer-free population. Epigenetics 2012;7(6):606–14. doi: 10.4161/epi.20236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin-Nunez GM, Cabrera-Mulero R, Rubio-Martin E, Rojo-Martinez G, Olveira G, Valdes S, et al. . Methylation levels of the SCD1 gene promoter and LINE-1 repeat region are associated with weight change: an intervention study. Mol Nutr Food Res 2014;58(7):1528–36. doi: 10.1002/mnfr.201400079 [DOI] [PubMed] [Google Scholar]
- 34.Duggan C, Xiao L, Terry MB, McTiernan A. No effect of weight loss on LINE-1 methylation levels in peripheral blood leukocytes from postmenopausal overweight women. Obesity (Silver Spring) 2014;22(9):2091–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramona G., Dumitrescu MV, editors. Diet, epigenetics, and cancer. Cancer epigenetics methods and protocols. Totowa, NJ: Humana Press; 2012. p. 377–94. [Google Scholar]
- 36.Yin R, Mao SQ, Zhao B, Chong Z, Yang Y, Zhao C, et al. . Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J Am Chem Soc 2013;135(28):10396–403. doi: 10.1021/ja4028346 [DOI] [PubMed] [Google Scholar]
- 37.Hermsdorff HH, Puchau B, Volp AC, Barbosa KB, Bressan J, Zulet MA, et al. . Dietary total antioxidant capacity is inversely related to central adiposity as well as to metabolic and oxidative stress markers in healthy young adults. Nutr Metab 2011;8:59. doi: 10.1186/1743-7075-8-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perng W, Rozek LS, Mora-Plazas M, Duchin O, Marin C, Forero Y, et al. . Micronutrient status and global DNA methylation in school-age children. Epigenetics 2012;7(10):1133–41. doi: 10.4161/epi.21915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma B, Wilker EH, Willis-Owen SA, Byun HM, Wong KC, Motta V, et al. . Predicting DNA methylation level across human tissues. Nucleic Acids Res 2014;42(6):3515–28. doi: 10.1093/nar/gkt1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friso S, Udali S, Guarini P, Pellegrini C, Pattini P, Moruzzi S, et al. . Global DNA hypomethylation in peripheral blood mononuclear cells as a biomarker of cancer risk. Cancer Epidemiol Biomarkers Prev 2013;22(3):348–55. doi: 10.1158/1055-9965.EPI-12-0859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mansego ML, Milagro FI, Campion J, Martinez JA. Techniques of DNA methylation analysis with nutritional applications. J Nutrigenet Nutrigenom 2013;6(2):83–96. doi: 10.1159/000350749 [DOI] [PubMed] [Google Scholar]


