Abstract
Background
Melanoma is a heterogeneous tumour, but the impact of this heterogeneity upon therapeutic response is not well understood.
Methods
Single cell mRNA analysis was used to define the transcriptional heterogeneity of melanoma and its dynamic response to BRAF inhibitor therapy and treatment holidays. Discrete transcriptional states were defined in cell lines and melanoma patient specimens that predicted initial sensitivity to BRAF inhibition and the potential for effective re-challenge following resistance. A mathematical model was developed to maintain competition between the drug-sensitive and resistant states, which was validated in vivo.
Findings
Our analyses showed melanoma cell lines and patient specimens to be composed of >3 transcriptionally distinct states. The cell state composition was dynamically regulated in response to BRAF inhibitor therapy and drug holidays. Transcriptional state composition predicted for therapy response. The differences in fitness between the different transcriptional states were leveraged to develop a mathematical model that optimized therapy schedules to retain the drug sensitive population. In vivo validation demonstrated that the personalized adaptive dosing schedules outperformed continuous or fixed intermittent BRAF inhibitor schedules.
Interpretation
Our study provides the first evidence that transcriptional heterogeneity at the single cell level predicts for initial BRAF inhibitor sensitivity. We further demonstrate that manipulating transcriptional heterogeneity through personalized adaptive therapy schedules can delay the time to resistance.
Funding
This work was funded by the National Institutes of Health. The funder played no role in assembly of the manuscript.
Keywords: Melanoma, MITF, Resistance, Heterogeneity, Mathematical modelling
Research in context.
Evidence before this study
The majority of patients with BRAF-mutant melanoma are not cured through BRAF-MEK inhibitor therapy. Current targeted therapy paradigms for melanoma are based upon continuous BRAF-MEK inhibitor treatment. This almost invariably leads to therapy failure that results from the selection of resistance-conferring genetic mutations or the adoption of drug resistance- associated transcriptional programs. Some melanoma patients who failed BRAF inhibitor therapy can be successfully re-challenged, through mechanisms that remain poorly understood. Recent work has demonstrated that melanomas are transcriptionally heterogeneous and may be comprised of multiple populations of cells with differing levels of drug resistance. It is therefore likely that drug schedules could be personalized to maintain the balance between sensitive and resistant cell states, improving the durations of response. There is already some preclinical evidence that intermittent BRAF inhibitor dosing, using fixed on/off schedules can delay the time to resistance in some melanoma xenograft models. In the present study, we sought to understand how transcriptional diversity at baseline predicted for BRAF inhibitor sensitivity and the likelihood of a successful re-challenge after therapeutic escape. This data was then leveraged to define mathematical model-driven personalized dosing schedules that improved therapeutic responses in vivo.
Added value of this study
Little is known about how transcriptional heterogeneity impacts the response of BRAF-mutant melanoma to BRAF inhibitor therapy. The present study used single cell mRNA analyses to define the transcriptional heterogeneity of melanoma at baseline, following treatment and then following the withdrawal of BRAF inhibitor therapy. These patterns of heterogeneity, which were predictive of both baseline therapy response and sensitivity to re-challenge, were mathematically modelled and leveraged to define personalized on/off drug schedules in real-time that led to improved therapeutic responses in in vivo melanoma models. Our work provides the first preclinical evidence that transcriptional heterogeneity at the single cell level predicts for the initial sensitivity to BRAF inhibitor therapy, and the potential for re-challenge following therapy failure. We further demonstrate that manipulating transcriptional heterogeneity through personalized adaptive therapy schedules can delay the time to resistance.
Implications of all of the available evidence
The cumulative data suggest that melanomas are transcriptionally diverse and can adopt phenotypes with a wide range of behaviours and drug sensitivities. It is likely that the transcriptional composition of melanomas at baseline is predictive of the depth of the initial response to therapy and whether patients will respond to subsequent rounds of treatment following the onset of resistance. Personalizing drug dosing schedules to account for the dynamics of transcriptional heterogeneity could be one strategy of improving outcomes for melanoma patients using existing FDA-approved therapies.
Alt-text: Unlabelled Box
1. Introduction
Continuous MAPK pathway inhibition in BRAF-mutant melanomas typically leads to transcriptional reprogramming and a switch to an epithelial-to-mesenchymal transition (EMT)-like phenotype characterized by upregulated receptor tyrosine kinase (RTK) expression [[1], [2], [3]], increased c-JUN expression and decreased expression of the melanocyte lineage factor microphthalmia (MITF) [[4], [5], [6], [7], [8], [9]]. Transcriptional reprogramming is one of the most common responses to BRAF inhibitor therapy, occurring in up to 94% of cases [10]. This switch in transcriptional state results in the emergence of phenotypically dedifferentiated, invasive and drug-tolerant tumour cells [10]. At this time, it is not known whether all of the melanoma cells in a tumour respond to BRAF inhibition in a similar manner, if there is any heterogeneity in the transcriptional response at the single cell level, or what these differences could mean for therapeutic response.
Current clinical management for stage IV melanoma dictates that most patients, irrespective of BRAF mutational status, will receive immune checkpoint therapy as their frontline treatment. While this is usually undertaken with the hope of a curative response, only ~30% of patients are likely to respond [11,12]. Among patients with advanced BRAF-mutant melanomas who receive BRAF inhibitors and the BRAF-MEK inhibitor combination, three patterns of response are typically seen: 1) a majority initially respond to therapy and then later relapse, 2) a percentage experience very long-term responses to therapy, 3) a small percentage of patients show little or no response to therapy [13]. Among those who initially respond to BRAF inhibitor therapy and then progress, resistance can sometimes be reversible. A recent clinical trial showed that up to 34% of patients who failed prior BRAF inhibitor therapy could be successfully retreated, suggesting that drug-sensitive melanoma cells are retained in these cases [14]. At this time, little is known about the molecular characteristics of the BRAF-mutant melanomas that exhibit outstanding responses to BRAF-MEK inhibitor therapy, or those that may benefit from drug holidays followed by retreatment.
Current targeted therapy paradigms are based upon continuous BRAF-MEK inhibitor treatment, which almost invariably leads to resistance [10,15,16]. A different approach, pioneered by our group, exploits the fact that tumours are phenotypically heterogeneous and that drug-resistant cells show reduced fitness in the absence of drug. Under this paradigm, short bursts of intermittent therapy to reduce tumour volume are followed by appropriately timed drug holidays that allow residual sensitive cells to regrow, restoring drug sensitivity and prolonging responses [[17], [18], [19]]. Although this strategy has shown promise in early-stage clinical trials [[17], [18], [19]], little is currently known about its molecular basis. In the present study, we performed single cell transcriptional analysis of melanoma cells on/off BRAF inhibitor therapy and defined how the transcriptional states were modulated by the therapy. We identified melanomas in which the initial drug sensitive state was retained under chronic BRAF inhibitor therapy, which allowed for successful therapy re-challenge. This data led to the development of a mathematical model that defined real-time personalized BRAF inhibitor on/off schedules that led to improved tumour control in vivo.
2. Materials and methods
2.1. Cell culture, tumour tissues and reagents
The 1205Lu, WM9, WM793, WM164, WM983A, WM39 and 451Lu melanoma cell lines were a generous gift from Dr. Meenhard Herlyn (The Wistar Institute, Philadelphia, PA, USA). SK-Mel-28 and A375 melanoma cell lines were purchased from ATCC (Manassas, VA). M229 and M233 cell lines were a generous gift from Dr. Antoni Ribas (UCLA Medical Center, Santa Monica, CA, USA). All cell lines were authenticated using short tandem repeat validation analysis (BioSynthesis Inc., Lewisville, TX, USA). WM164R and 1205LuR cell lines were generated through chronic treatment with vemurafenib (2 μM and 3 μM, respectively) [20]. WM164R-DW and 1205LuR-DW cell lines were generated by withdrawing chronic BRAF inhibitor treatment from WM164R and 1205LuR lines for 10+ weeks. Cell lines were maintained in 5% FBS/RPMI-1640, with the addition of vemurafenib for resistant cell lines, and routinely tested for mycoplasma contamination.
2.2. Single cell gene expression analysis
One million cells were plated in 10 cm tissue culture plates and allowed to attach overnight. Cell culture media was changed to fresh media and cells were allowed to grow an additional 24 h. Cells were detached through trypsinization and loaded onto the Fluidigm C1 IFC, for single, live cell annotation, lysis, reverse transcription and pre-amplification according to manufacturer's protocol. cDNA products were split up evenly among ten Dynamic Array IFCs, including pooled cDNA and multiple other positive and negative controls to monitor amplification and plate-to-plate effects. IFCs were analysed on the Fluidigm Biomark HD using a panel of 88 Delta Gene Assays (Supplemental Table 1). This was a custom-built gene panel chosen to represent the key signalling molecules and transcription factors known to be involved in melanoma biology and BRAF inhibitor resistance.
2.3. Analysis of heterogeneity and transcriptional state landscape
The SinCHet MATLAB toolbox was utilized for the analysis of heterogeneity and to identify the transcriptional states present in each sample, as described in [4]. Briefly, unsupervised hierarchical cluster analysis using Ward linkage was used to cluster 292 cells among four cell lines into distinct subpopulations whose transcriptional states are significantly different, based on single cell gene expression similarities. Shannon Profile (SP) consists of the Shannon index, a quantitative measure of how many different transcriptional states exist in the dataset and their proportional abundance, calculated at all possible resolutions (from 1 to 292 for this dataset) to evaluate overall transcriptional heterogeneity for all 292 cells. The profile of the differences of Shannon index, PSD, was calculated as the Shannon index differences at the same resolution as SP across all possible clustering resolutions and was used to characterize the heterogeneity differences. The D statistic, defined as the differences of the areas under the PSD between two conditions, was calculated to evaluate heterogeneity difference between pair-wise conditions or cell lines. Its statistical significance was estimated using 1000 permutations. Change points of the PSD between two conditions were detected using multivariate adaptive regression splines (MARS) model as previously reported [25]. In this study, there are total 6 pair-wise comparisons across the four cell lines. The number of identified transcriptional states was determined using the minimum change point of the PSDs (Supplemental Fig. 1A) with significant heterogeneity differences (p < .05, Supplemental Fig. 1B). A snapshot of the transcriptional states was summarized using a pie chart and heatmap (Fig. 1F, Supplemental Fig. 1C and Supplemental Table 2). For biomarker comparison across phenotypic states (Supplemental Figs. 2–3), a Kruskal-Wallis test was first performed for overall p-value, while pair-wise p-values were generated to adjust for multiple comparisons. False discovery rate (FDR) was estimated using the Benjamin and Hochberg method to adjust for the multiple hypotheses testing (across genes). Interactome analysis of top 15 differentially expressed genes was performed using Metacore (Clarivate Analytics, Philadelphia, PA, USA) and visualized using Cytoscape (cytoscape.org).
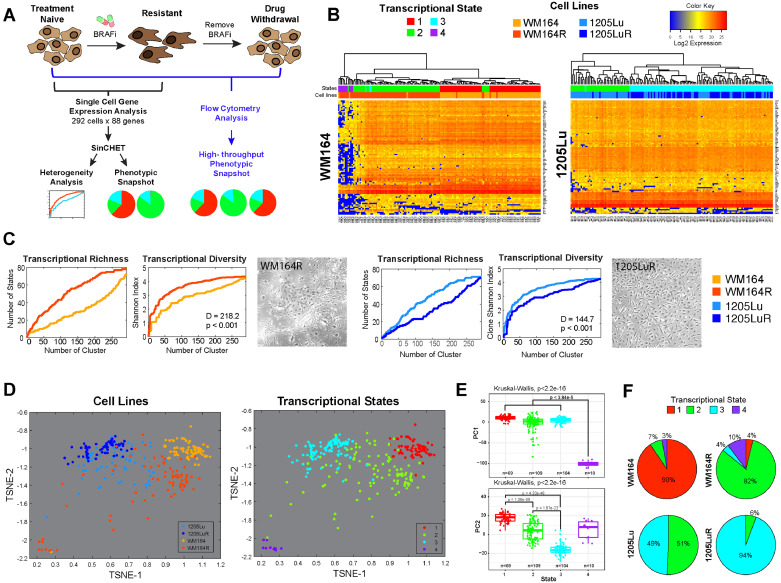
Fig. 1.
Defining the transcriptional diversity of melanoma. A. Overview of the experimental workflow. B. Single cell mRNA analysis of BRAF inhibitor naïve and treated WM164 and 1205Lu melanoma cells identifies 4 different transcriptional subpopulations. Heatmaps of gene expression data of 88 genes (y-axis) for individual cells (x-axis) of treatment-naive WM164 and 1205Lu and their drug-resistant (R) counterparts. Colour-coding under the dendogram depicts the transcriptional state and the cell line each cell belongs to. C. BRAF inhibition leads to either an increase or a decrease in transcriptional diversity. Shannon heterogeneity analyses of the single cell data from Fig. 1B reveals WM164 to become more diverse on therapy and the 1205Lu cells to become less diverse. Transcriptional richness is a measure of distinct transcriptional states present. Graphs show the number of distinct transcriptional states, as well as the Shannon Diversity Index, at different heights along the gene expression dendogram from Fig. 1B. D. t-SNE analysis demonstrates the relationship between drug-naïve and treated melanoma cell lines and the 4 transcriptional states. Data points show the individual cells and their orientation in transcriptional space. Note that drug-resistant (WM164R) cells are the most transcriptionally diverse. States #1, #3 and #4 are quite distinct with #2 showing overlap with #1 and #3. E. Four transcriptional states are statistically distinct. Principal component analysis and Kruskal-Wallis test performed comparing all cells in each transcriptional state highlights strong differences in gene expression among the four states. F. WM164 and 1205Lu cells show different transcriptional landscapes, which change under chronic BRAF inhibitor treatment. Pie charts are derived from data shown in Fig. 1B using SinCHet software analysis and show the percentage of cells in each transcriptional state.
2.4. Western blot
Western blot analysis was performed as previously described [21]. Antibodies for pERK (Thr202/Tyr204, #9101) and ERK (#9102) were purchased from Cell Signalling Technology (Danvers, MA, USA).
2.5. Patient single cell RNAseq analysis
A previously published melanoma single cell RNAseq dataset was downloaded through Gene Expression Omnibus (accession number GSE72056) [22]. A total of 1159 tumour cells (from the original 4645 single cells, which included non-malignant cells) from 14 patients were included in this analysis. To identify the potential corresponding phenotypic states in these patient samples using the observed states in our cell line data, the overlapping 88 genes were first harmonized and de-batched using COMBAT [23]. The de-batched expression dataset was then used in the consecutive analysis. To investigate the expression similarity of each patient cell with respect to the 4 transcriptional states identified in cell line data (N = 292 cells), we modified the unsupervised clustering algorithm as follows: First, the centroid of each transcriptional state from the cell line data (i.e., 69 cells in state 1, 109 cells in state 2, 104 cells in state 3 and 10 cells in state 4) was calculated and assigned to represent the original identified state (Supplemental Table 2). Then, unsupervised hierarchical cluster analysis of a total of 1163 cell groups was performed. The 1163 cell groups were consisted of the 4 pre-identified clusters of cells from cell lines and 1159 tumour cells from patient samples. The weight of each cell group, when calculating distance between cell groups using Ward linkage, is the number of cells in each cell group. That is, 69 was the weight for the cell group with 69 cells in state 1, and 109 as the weight for the cell group with 109 cells in state 2, 104 for those in state 3, and 10 for those in state 4 while 1 was used as the weight for each tumour cell in patient samples. After the hierarchical cluster analysis was performed, the final number of transcriptional states was determined using the minimum number of clusters while the originally identified 4 transcriptional states remained identified as separated states. Therefore, the final number of states would be ≥4 depending on the expression similarity between patient tumour cells and cells in cell lines.
2.6. Flow cytometry
One million cells were plated in 10 cm tissue culture plates and allowed to attach overnight. Cell culture media was changed to fresh media and cells were allowed to grow an additional 24 h, then detached using trypsinization. Cells were fixed using 4% paraformaldehyde for 10 min. Cells were permeabilized with 100% ice-cold methanol and stained using Axl conjugated to Alexa Fluor 647, ERBB3 conjugated to Alexa Fluor 594 (R&D, Minneapolis, MN, USA), Ki67 conjugated to BV786, pERK conjugated to BV421 (BD, Franklin Lakes, NJ, USA) and MITF conjugated to Alexa Fluor 488 (Abcam, Cambridge, UK). Staining data was captured on the BD LSR II instrument and analysed using FCS Express software (De Novo Software, Glendale, CA, USA). The same cell state gating strategy (Supplemental Fig. 4) was use on all samples. For transcriptional state analysis following intermittent drug treatment, 3 μM vemurafenib was used. One million WM164 cells were plated in 10-cm cell culture dishes and allowed to attach overnight. Then cells were treated according to different treatment schedules: 4 days on, 10 days on, 4 days on then 4 days off, and 10 days on then 4 days off. Cells were then harvested and analysed as above. WM164R cultured under chronic vemurafenib (2 μM) and treatment-naïve WM164 were used as controls.
2.7. Cell growth assays
For short-term growth analyses, cells were plated at 100,000 cells/well in 6-well cell culture plates and allowed to adhere overnight. Cells in each well were then counted using the Countess Automated Cell Counter (Invitrogen, Carlsbad, CA, USA) over the course of 4–5 days until confluency. Doubling time was calculated based on Td = (t2-t1)*((log(2)/log(q2/q1)), where Td is doubling time, t1 is the first day of measurement, t2 is the last day of measurement, q1 is the number of cells on the first day of measurement and q2 is the number of cells on the last day of measurement. For long-term growth analyses, one million WM164 or 1205Lu cells were plated into T75 flask and allowed to attach overnight. Cells were then treated chronically with 2 μM (WM164) or 3 μM (1205Lu) vemurafenib. Cells are counted at confluency and re-plated at one million cells per T75 flask for 72 days. The projected total cell number, had the cells not been split, was calculated based on cell counts at each passage.
2.8. Growth inhibition assay
MTT growth inhibition assays were carried out as previously described [24] using vemurafenib. IC50 values were calculated by non-linear regression analysis of log(inhibitor) vs. response using GraphPad Prism Software (La Jolla, CA, USA).
2.9. Apoptosis assay
One million cells were plated in 10 cm dishes and allowed to attach overnight. Cells were then treated with vehicle control or 3 μM vemurafenib for 72 h. Cells were trypsinized, stained using tetramethylrhodamine methyl ester (TMRM) and analysed by flow cytometry.
2.10. Mouse xenografts
Seven-week-old female NSG mice (The Jackson Laboratory, Bar Harbor, ME, USA) were subcutaneously injected with 5 × 105 WM164 cells per mouse. Tumours were allowed to establish over 3 days. Mice were randomly separated into treatment cohorts using GraphPad's random treatment group assignment (graphpad.com), consisting of 11 mice per cohort. Mice received D10001 control chow or AIN-76A 417 mg/kg PLX4720-formulated chow (Research Diets, New Brunswick, NJ, USA) daily. Tumour volumes (½ × L(length) × W(width)2) were measured every 2–3 days. All animal experiments were carried out in compliance with ethical regulations and protocols approved by the University of South Florida Institutional Animal Care and Use Committee.
2.11. Mathematical model for personalized adaptive xenograft treatment
To describe tumour volume response to BRAF inhibitor treatment, a two-compartment Ordinary Differential Equation model was developed consisting of a sensitive (S) and a resistant (R) compartment, , where gS, R indicate growth rate of S and R, respectively, and δ is the death rate of S. The two compartments share a carrying capacity K, the maximum capacity of the tumour either due to nutrient or space constraints. We also allow transition between the two states (α and β are the transition rates). To predict an effective fixed intermittent inhibitor therapy schedule, the model was calibrated to xenograft tumour growth dynamics on no-treatment, continuous, 2-day on/6-day off, 7-day on/7-day off and 14-day on/14-day off schedules. These estimated parameters were later used as initial ranges for parameterizing the mathematical model to determine the individual mouse-specific treatment schedules. To predict adaptive therapy for each mouse, we trained the model to reproduce all previous tumour volume changes, and then used it to forecast the expected tumour volumes (both on and off) at future time points. The mathematical model was calibrated with each mouse tumour volume change data every 2–3 days. We used an optimization algorithm called implicit filtering, a steepest descent algorithm for problems with bound constraints, to determine the parameters (H) that minimised the difference between predicted normalized tumour volume (V(t; H), V(0, H) = 1) and mouse tumour volume (D(t)/D(0), normalized tumour volume) [25]. The mathematical definition of our problem was:
where the goal is to minimize the objective function f subject to the condition that H ∈ ℝN is in the feasible region Ω. Estimated parameters produced fitted curves and small root-mean-squared errors (average error = 0.34, minimum error = 0.09, and maximal error = 0.79). Then, a critical tumour volume (Fig. 4D, yellow asterisk) was calculated that produces the most impact when switching treatment off (c.f., Fig. 4D below dotted line, off volume < on volume). Using the estimated parameters for each individual mouse, we simulated the impact of treatment both on and off for two or three days, resulting in multiple potential tumour volume trajectories. We then compared the simulated tumour volumes of these two groups (on and off treatment). We examine the predicted tumour volume increase in comparison to the previous treatment decision (either on or off) time point. If this predicts that the tumour volume on (V on) > tumour volume off (V off) at the next time point, we name this volume the “critical volume”. Next, we compared the actual tumour volume to the critical value and recommended whether to restart or hold drug; if for example, actual tumour volume < critical tumour volume, the model suggests to stop treatment, by calculating the probability of success on/off drug. To accommodate individual tumour dynamics, both the critical volume and the treatment on/off decisions were personalized for each mouse.
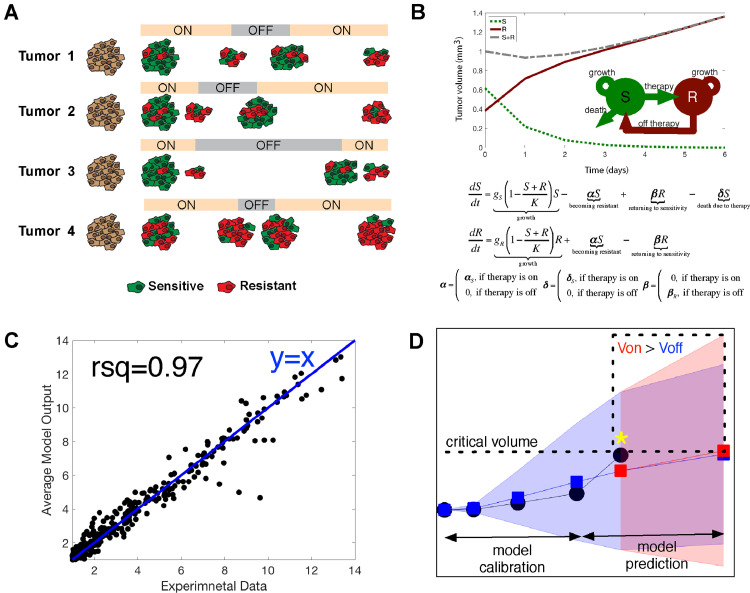
Fig. 4.
Development of a mathematical model to maintain drug sensitive cell states through adaptive drug-dosing. A. Model showing the basis for using personalized intermittent (adaptive) therapy to control transcriptional heterogeneity. It is assumed that melanomas are composed of cells with transcriptional states that either convey drug sensitivity (red) or resistance (green). Drug holidays are associated with the recovery of cells with drug sensitivity. B. Scheme showing projected temporal changes in tumour growth in a two-compartment model consisting of a sensitive (S) and a resistant (R) compartment (top). Mathematical expression for the model, where gS, R indicate growth rate of S and R, respectively; K is a carrying capacity; δ is death rate of S; α & β are transition rates between two states (bottom). C. Linear regression curve shows the model parameters generated tumour dynamics that matched with experimental data (R-squared = 0.97 and average relative error = 0.35, 35%). D. Modelling the effects of intermittent/continuous BRAF inhibitor treatment in human melanoma mouse xenografts. Model prediction was updated in real-time using the compartment model and direct tumour volume sampling. Tumour volume trajectory was then calculated for which the effects of drug off was greater than drug on (Voff < Von, dotted box). A critical tumour volume was then determined (VC, indicated by yellow asterisk) that satisfied the condition Voff < Von. The decision to treat was determined by comparing the critical tumour volume with experimentally measured tumour volume (VE). If VE was less than VC, the mouse was taken off treatment. Otherwise, treatment continued. Experimental data is depicted in black, model predicted volume changes during treatment-off is blue, model predicted tumour volume during treatment-on is red. The yellow asterisk indicates a critical tumour volume, which is simulated tumour volume at the current time point that makes volume on (V on) > tumour volume off (V off) at the next time point. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Results
3.1. Defining the transcriptional diversity of melanoma
To better understand the role of transcriptional heterogeneity in melanoma drug response, we developed a single cell analysis workflow to simultaneously quantify the expression of 88 genes per cell (Fig. 1A–B, Supplemental Tables 1–3). This gene panel was chosen to represent the key signalling molecules and transcription factors involved in melanoma biology and the response of melanoma cells to BRAF inhibitor therapy. The goal was to define the number of distinct cell states in melanoma cultures, and to determine how these cell states were regulated by BRAF inhibitor therapy. We used isogenic cell line pairs that were either drug naïve or had been continuously treated with BRAF inhibitor for >6 months until resistance was acquired [3,26]. Single cell gene expression data was analysed using the Single Cell Heterogeneity (SinCHet) MATLAB toolbox developed by our group, which allows different, coexistent transcriptional states to be identified [27]. Using Shannon diversity index metrics that quantify species richness (a measure of the number of distinct transcriptional states present) and evenness, and the D statistic that quantifies an overall difference of Shannon index between two populations [27], we found some melanomas exhibited an increase in diversity/heterogeneity after chronic drug treatment while others showed decreased diversity (Fig. 1C). These transcriptional changes were also mirrored by changes in the diversity of cell morphologies seen (Fig. 1C, Supplemental Fig. 5). Our SinCHet analysis, which used the minimum change point at the lowest cluster level (Supplemental Fig. 1), initially identified four distinct transcriptional states (Fig. 1D–F). An examination of the relationship between the transcriptional states using T-distributed stochastic neighbour embedding (t-SNE) analysis, showed #1, #3 and #4 to be more distinct, with state #2 being highly diverse and occupying the most transcriptional space (Fig. 1D). Overlay of the cell line identity data demonstrated the commonality of the states between the two cell lines and highlighted the increased diversity of the WM164 cells and the reduced diversity of the 1205Lu cells following chronic BRAF inhibition (Fig. 1D). Principal component analysis and Kruskal-Wallis test demonstrated that the four transcriptional states were statistically distinct (Fig. 1E). Phenotypic snapshots generated by our SinCHet software showed melanoma cell lines with BRAF inhibitor sensitivity (such as WM164) to be composed of a dominant population of transcriptional State #1 and minor populations of transcriptional States #2 and #4; this changed following chronic BRAF inhibition, with State #1 declining and States #2, #3 and #4 expanding (Fig. 1F). In contrast, some melanomas with low BRAF inhibitor sensitivity (such as 1205Lu) lacked cell state #1 and instead consisted of transcriptional states #2 and #3 at baseline, with transcriptional state #3 being enriched following long-term drug treatment (Fig. 1F). Both cell lines showed an equivalent initial level of ERK inhibition following BRAF inhibitor treatment (Supplemental Fig. 6), suggesting that the different patterns of transcriptional heterogeneity at baseline did not impact the level of inhibition in the MAPK signalling pathway.
3.2. The effects of BRAFi therapy upon transcriptional heterogeneity
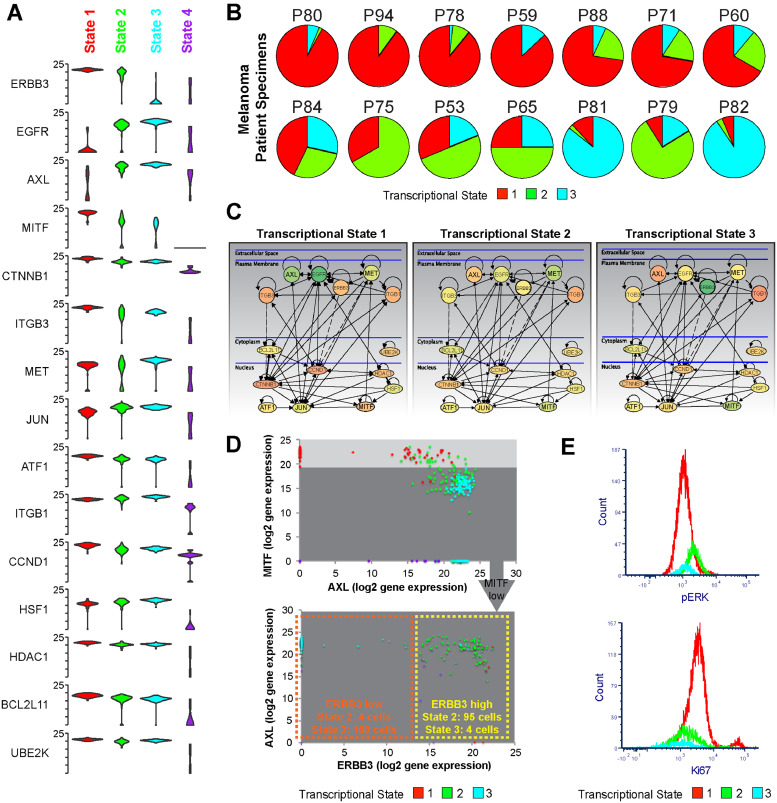
We interrogated the transcriptional data and represented the top 15 genes using Violin plots to show the diversity of the gene expression across the population at the single cell level (Fig. 2A shows the top genes, Supplemental Fig. 2 shows the entire panel). These data showed State #1 to be characterized by high expression of cyclin D1, ERBB3, STAT3/5, ATF1, ATF4, MITF and β-catenin and low expression of c-JUN and RTKs such as Axl and EGFR (Fig. 2A and Supplemental Figs. 2–3). State #2 was characterized by high expression of ERBB3, Axl and the transcription factor c-JUN. Decreased levels of MET, RELB, E2F1, BIM, ULK1, SMAD1/9 and XIAP were also observed. State #3 was characterized by high Axl, c-JUN, E2F1, WEE1, c-MET and EGFR expression and lower expression of MITF, ERBB3 and SMAD9. Increased integrin β1 expression and lower E-cadherin expression was also seen, characteristic of an EMT-like state. State #4 had low MITF/RTK expression and a generally suppressed gene expression profile associated with cell death; this state was excluded from further analyses. The same cellular states were also conserved in melanoma clinical specimens. SinCHet Analysis of single cell RNAseq data from 14 human melanoma specimens [22] detected all three transcriptional states, with each melanoma showing a unique transcriptional makeup (Fig. 2B). Grouping these changes into network maps highlights the differences between the RTK signalling, cell cycle regulation and transcription factor dependencies of each of the three major cellular states (Fig. 2C). In light of previous studies demonstrating that MITF and Axl are anti-correlated [5,28], we next looked at the expression of these two genes at the single cell level. It was noted that although some cells were MITF high and Axl low, a population of cells (~17%) were identified that were both Axl-high and MITF-high (Fig. 2D). This population had a gene expression profile that was intermediate between cells that were either Axl-high or Axl-low and clustered with both transcriptional states #1 and #2 (Fig. 2D, Supplemental Fig. 3). Key genes that distinguished the MITF-high/Axl-high and the MITF-high/Axl-low were increased expression of CDH2 (N-cadherin), CEBPA, EGFR, FOXO3, RELA, JUN, Vimentin and TP53 and decreased expression of Integrin αV, ETS1/2, c-MET and WEE1 (Supplemental Fig. 3). It therefore seemed that the MITF-High/Axl-low and MITF-low/Axl-high expression signatures were not binary states, and that the melanoma cells could potentially adopt a range of intermediary phenotypes. An analysis of the phenotypic behaviours of the transcriptional states demonstrated State #1 to express high levels of Ki67 (consistent with the highest cyclin D1 and E2F1 expression), whereas State #2 had the highest MAPK pathway signalling as shown by increased phospho-ERK levels (Fig. 2E). Together these integrated transcriptional and functional data predicted that State #1 would be proliferative, but sensitive to BRAF inhibition, State #2 to be less proliferative with higher baseline MAPK signalling and that State #3, with its enrichment for EGFR, c-JUN and Axl, would potentially show BRAF inhibitor resistance.
Fig. 2.
Gene level analysis identifies unique transcriptional profiles for each transcriptional state. A. Violin plots showing expression of each of the top 15 differentially expressed mRNAs clustered by transcriptional state. Note higher Axl and c-JUN in States #2 and #3, Cyclin D1 (CCND1), MITF in State #1, ERBB3 in States #1 and #2 B. Melanoma patient samples exhibit a diverse array of transcriptional state compositions. Data shows heterogeneity analysis of single cell RNA-Seq data from ref. (22) of melanoma patient specimens, based upon the 88 genes from our panel. C. major signalling differences among the three main transcriptional states. Metacore interactome analysis of the top 15 differentially expressed genes among the three transcriptional states illustrates rewiring of major signalling among the subpopulations. Node colour denotes mean level of mRNA expression detected (green = lower expression, red = higher expression). D. MITF-High/Axl-low and MITF-low/Axl-high expression signatures are not binary states. Dot plots showing the relative expression of MITF, AXL and ERBB3 in all cells derived from data in Fig. 1B show the varied distribution of MITF/AXL expression and the need to examine additional markers (such as ERBB3) for a more complete analysis of the transcriptionally heterogeneous subpopulations. E. Transcriptional State #2 has the highest pERK levels and Transcriptional State #1 shows the highest proliferative capacity. Comparison of pERK and Ki67 staining by flow cytometry between transcriptional States #1, 2 and 3. Data shows expression of phospho-ERK in each transcriptional state resolved by the gating shown in Supplemental Fig. 4 in WM164 and WM164R cells. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Transcriptional state composition dictates response to BRAF inhibition
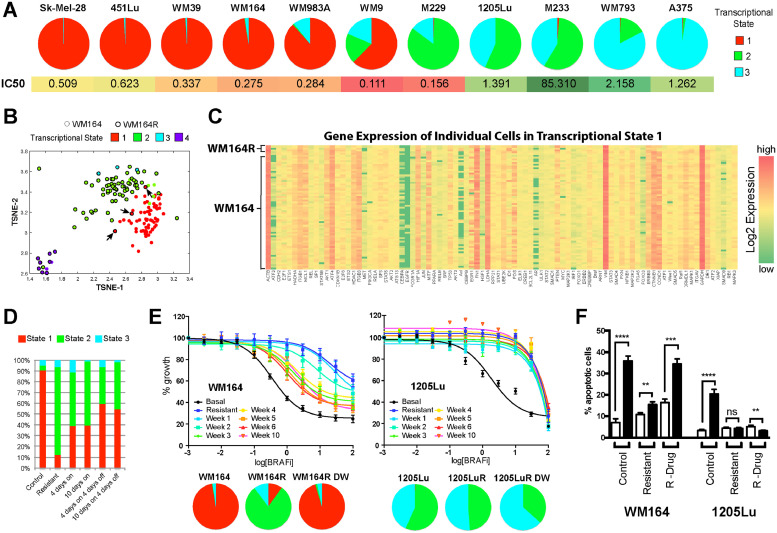
To more easily quantify melanoma heterogeneity across multiple samples, we defined three markers that allowed the cell state compositions to be delineated flow cytometry (Supplemental Fig. 4). Although this methodology relies on protein-level expression as opposed to mRNA level expression and the identification of a small number of MITF high/AXL high cells from State # 2 may be missed, the proportions of the transcriptional state subpopulations identified by flow cytometry were analogous to those defined by the single cell analysis approach. Application of these methods to a larger cell line panel revealed an association between a high percentage of cells in transcriptional State #1 and a lower vemurafenib IC50 (two sample t-test: t = −2.02, p = .07; Fig. 3A and Supplemental Fig. 7), suggesting that this represents a more drug sensitive state. Only one cell line, the M229, exhibited high sensitivity to vemurafenib but did not appear to harbor many cells in State #1. We next determined the influence of cell-state composition at baseline and the time to BRAF inhibitor resistance. Here, the cell lines were treated with vemurafenib and the growth dynamics under chronic drug treatment were measured. We found melanomas lacking State #1 became resistant more rapidly than those in which State #1 was the dominant population (Supplemental Fig. 8). As evolution-based studies have demonstrated drug resistance to be associated with a fitness cost, we next asked whether the transcriptional state composition determined growth fitness in the absence or presence of drug. It was noted that melanomas with a greater growth fitness (shortest doubling times) in the absence of drug experienced a reduction in growth fitness after the acquisition of resistance (Supplemental Table 4). In contrast, treatment-naïve melanomas that lacked the drug-sensitive State #1 already exhibited reduced growth fitness at baseline (Supplemental Table 4, doubling time 1205Lu > WM164) and experienced little change in their fitness after the acquisition of drug resistance (Supplemental Table 4, doubling time 1205Lu~1205LuR).
Fig. 3.
Transcriptional state composition determines BRAF inhibitor sensitivity. A. BRAF-mutant melanoma cell lines with a high percentage of Transcriptional State #1 exhibit a reduced vemurafenib IC50. Transcriptional state composition was measured in 11 cell lines by flow cytometry based on Axl, MITF and ERBB3 (top); Vemurafenib IC50s were calculated by MTT assay (bottom). B. t-SNE analysis showing that Transcriptional State 1 is the same in both the drug sensitive WM164 and resistant WM164R cell lines. C Gene expression heatmap comparing cells in State #1 from drug sensitive (WM164) and drug resistant (WM164R) melanoma cells. D. Drug holidays allow for the recovery of Transcriptional State #1. Transcriptional state distribution was measured in WM164 cells using flow cytometry (as above) following different treatment/drug holiday schedules. E. Resistant melanomas that retain drug-sensitive Transcriptional State #1 return to sensitivity following drug holidays. Data show responses of drug-naïve WM164 and 1205Lu, drug-resistant WM164R and 1205LuR and drug-resistant WM164R and 1205LuR with vemurafenib removed from cell culture media for increasing periods of time (1–10 weeks, top) by MTT assay. Transcriptional state composition was measured by flow cytometry. F. Return of melanoma cell lines to sensitivity following drug removal. Flow cytometry-based apoptosis assay shows the percentage of apoptotic (TMRM-) cells in control, resistant and resistant cultures following drug withdrawal treated with vemurafenib (72 h, 3 μM).
One surprising finding from the single cell mRNA analyses was the observation that the drug sensitive State #1 was not eradicated from the population, even after >6 months of drug treatment (Fig. 1F), with small numbers of cells from drug-resistant WM164 cells remaining clustered with the drug-naïve WM164 cells (Fig. 3B). An analysis of the gene expression profiles showed a very strong overlap between State #1 populations from the two cell lines (Fig. 3B,C). We reasoned that this minor population of sensitive cells would exhibit greater fitness during a drug holiday and may have the potential to outgrow the resistant cells. Analysis of the cell state dynamics showed that treatment of drug-naïve WM164 cells with BRAFi led to a rapid decline in the proportion of cells in State #1 by day 4, but this partly recovered following removal of drug (Fig. 3D). Importantly, the recovery of State #1 was also seen in cultures that had been chronically treated with BRAF inhibitor therapy. Removal of drug from BRAF-inhibitor resistant WM164R cells led to a slow return to drug sensitivity, that was associated with an expansion of transcriptional State #1 and the regression of the other states (Fig. 3E). This return to sensitivity was also associated with the restoration of the apoptotic response to vemurafenib in the drug-withdrawn cultures (Fig. 3F). By contrast, a cell line that lacked the drug-sensitive transcriptional State #1 at baseline did not return to sensitivity when drug was removed for up to 10 weeks (Fig. 3E,F). It thus seemed likely that the initial presence of cellular State #1 was one determinant of both initial drug sensitivity and also future response to drug re-challenge following initial relapse. The potential clinical significance of these findings was indicated by a retrospective chart review that identified 34 BRAF-mutant melanoma patients treated with BRAF and BRAF-MEK inhibitor therapy, at least 23 of whom displayed a second clinical response upon drug holiday and re-treatment (Supplemental Table 5). It is thus possible that the secondary responses to BRAF and BRAF-MEK inhibitor therapy observed in melanoma patients could reflect a drug sensitive population of cells being retained even following the acquisition of overt resistance.
3.4. Development of a mathematical model to maintain drug sensitive cell states through adaptive drug-dosing
The analysis of our heterogeneity data predicted that some melanomas were composed of a transcriptional state with a high level of drug sensitivity that declined on therapy and then recovered when drug was withdrawn. We next determined whether the differences in fitness between the sensitive and resistant transcriptional states could be leveraged to improve tumour control through dosing schedules that maximized the number of sensitive, drug responsive cells in the tumour (Schematic in Fig. 4A). We reasoned that the decrease in tumour volume on drug and then the rate of re-growth in the absence of drug in vivo was a reflection of the balance between sensitive (#1) and resistant (#2 and #3) transcriptional states. To better understand this, we developed a two-compartment mathematical (ODE: ordinary differential equation) model that described the competition between sensitive (S: State #1) and resistant (R: States #2 and #3) cell growth dynamics (Fig. 4B). The model allowed for transition between the sensitive and resistant cell types and was calibrated using tumour growth dynamics from melanoma WM164 xenografts grown under vehicle, continuous, 2-day on/6-day off, 7-day on/7-day off and 14-day on/14-day off treatments with the BRAF inhibitor PLX4720 (Supplemental Fig. 9). The estimated parameters generated tumour dynamics that matched with experimental data (R-squared = 0.97 and average relative error = 0.35, 35%, Fig. 4C). The estimated parameter set served as an initial insight into designing the personalized adaptive dosing schedule, in which each mouse had a 2-week lead-in of PLX4720 followed by mathematical model-driven treatment decisions. The model was re-calibrated in real-time using individual animal tumour growth dynamics three times per week, which then determined whether drug should be held or reinitiated for each mouse (Fig. 4D). It is known that clinical responses are only seen to BRAF inhibitor therapy when tumour pERK levels are decreased by >80% [29]. With this in mind, we retained the standard dose of PLX4720 and instead altered the schedule to ensure that the drug sensitive population of cells was retained.
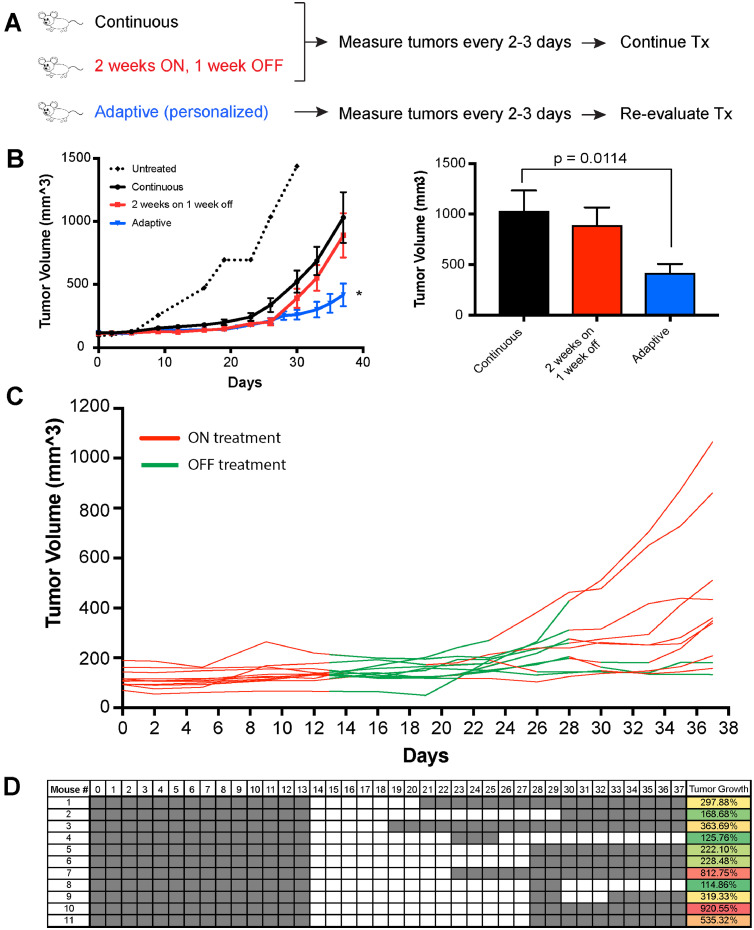
As the final step, we validated our mathematical model in vivo to determine whether personalized, adaptive dosing schedules would lead to improved anti-tumour responses. For the adaptive arm, mouse tumours were measured every 2–3 days. The tumour volumes were then entered into the mathematical model in real-time to predict whether the inhibition of tumour growth on that day would be better if drug was administered or held. Supplemental Fig. 10 shows the distribution of the estimated parameters for the 11 mice on the adaptive treatment arm. As our comparator arms, we used standard continuous dosing schedules (such as those used clinically) and a fixed intermittent dosing schedule, that has been postulated to outperform continuous dosing schedules in preclinical studies [30]. Control mice were treated with PLX4720 either continuously or on a fixed intermittent dosing schedule (2 weeks on/1 week off; Fig. 5A). The efficacy of PLX4720 compared to vehicle control has been well established previously by our group and others (Supplemental Fig. 11) [9,21,31]. The adaptive schedule outperformed both continuous treatment and a fixed drug-holiday schedule, and led to significantly improved tumour control (Fig. 5B). Intriguingly, the adaptive dosing schedules were quite different for each mouse, despite the same cell line being xenografted, suggesting the potential influence of host factors in regulating heterogeneity (Fig. 5C–D). Supplemental Fig. 12 shows model predictions of tumour volume change from day 0 to day 37 fit to the measured tumour volumes for each mouse and Supplementary Table 6 contains the 200 parameter sets for each mouse that generated the model prediction curves. An analysis of final tumour volume at the end of the experiment demonstrated significantly smaller tumour volumes for the adaptive dosing schedule compared to either the continuous drug dosing or the 2 week on/1 week off intermittent dosing (Fig. 5B).
Fig. 5.
Evolutionary-informed BRAF inhibitor dosing schedules outperform continuous or fixed intermittent dosing in vivo A. Treatment schema for mouse experiment comparing continuous and fixed intermittent schedule of BRAF inhibitor treatment to personalized, adaptive dosing using mathematical modelling of individual xenografted tumours. B. Evolutionary-informed adaptive dosing schedules are associated with better tumour control than either continuous or fixed intermittent drug dosing. Data show mean tumour volume data from WM164 melanoma xenografts between the three BRAF inhibitor treatment groups over time (left). Untreated “sentinel” mice receiving drug free chow is shown by the dotted line. Average tumour volumes are shown for each treatment group on the last day of the experiment (right). C. Mathematical modelling of tumour response dynamics under drug predicts individual dosing schedules for each mouse. Individual tumour volume and treatment data are shown from the adaptive treatment group over time (green line = off therapy, red line = on therapy). D. Chart shows individual dosing schedules for each mouse on the adaptive treatment arm of the xenograft experiment (grey: on therapy. White: off therapy), along with the increases in individual tumour volumes at the end of the experiment. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
The treatment landscape for advanced melanoma has changed dramatically over the past 7 years [12,[32], [33], [34]]. Although these new therapies, including the BRAF-MEK inhibitor combination and immune checkpoint inhibitors, have reduced death rates by ~30%, cures remain infrequent and most patients eventually fail therapy. For the patients with BRAF-mutant melanoma, a legitimate goal is to develop therapy schedules that prolong the duration of response and/or disease control. There is already clinical evidence that a sub-set of patients with BRAF-mutant melanoma can be successfully re-challenged with BRAF and BRAF-MEK inhibitor therapy following an initial round of response [14,35]. In the present study we have used innovative single cell heterogeneity analyses to define how the baseline mixture of cellular states predicts for initial BRAF inhibitor sensitivity. This data was then leveraged to develop personalized adaptive dosing schedules that accounted for the fitness of individual melanoma cells within heterogeneous tumours, with the goal of improving therapeutic responses in vivo.
Our single cell mRNA and heterogeneity analyses suggested that melanoma cell lines and patient specimens consisted of four transcriptional states. Among these, State #1 had a proliferative gene expression profile and showed a good level of BRAF inhibitor sensitivity [5,16,28]. Transcriptional State #3 showed increased expression of c-JUN and EGFR; two genes previously associated with BRAF inhibitor resistance [2,[6], [7], [8]]. These findings mirrored those of another recent single cell RNA-Seq analysis that identified multiple transcriptional states in melanoma Patient Derived Xenografts (PDXs) that were characterized as #1) high MITF expression #2) increased AP-1 expression (e.g. c-FOS and c-JUN) associated with an EMT-like state and #3) a neural-crest like state [36]. Our in-depth heterogeneity analysis demonstrated that the AP-1-high cell state could be divided into two distinct sub-states. One of these states, that we defined as State #2, was noted to be high in c-JUN and Axl but statistically distinct from States #1 and #3. Specific gene expression differences between State #2 and #3 included lower expression of Rel-B, c-MET and higher expression of ERBB3. Transcriptional State #2 also exhibited some overlap between both State #1 and #3, and appeared to be a diverse intermediate (or transitional) state that was frequently enriched for under drug treatment. The identification of intermediate cell states is not unprecedented. The development of single cell RNA-Seq technologies have allowed intermediate transcriptional states to be defined in other contexts, including the epithelial-to-mesenchymal transition (EMT). In this instance, multiple transitional transcriptional states have been defined during EMT that fall in between those states typically characterized as either epithelial and mesenchymal [37].
A prevailing paradigm in the melanoma field has been the role of the melanoma lineage marker MITF in the switch between a proliferative state (high MITF) and an invasive state (low MITF). Our single cell analyses demonstrated the co-existence of individual cells that were MITF- high/Axl-low and MITF-low/Axl-high in the majority of melanoma cell lines and patient specimens. These changes in MITF, which are often anti-correlated with expression of Axl, have been linked to melanoma drug sensitivity with MITF-high/Axl-low melanomas showing increased sensitivity to BRAF inhibitors and the MITF-low/Axl-high melanomas being intrinsically resistant [5,28]. At the same time, increased expression of MITF has also been reported to be associated with drug resistance [38]. One explanation for these seemingly contradictory findings is our observation that the MITF-high cells can cluster with distinct transcriptional states that are either drug-sensitive or drug resistant (e.g. States #1 and #2). At the single cell level, individual cells were identified that had high expression of MITF, Axl and cyclin D1 and increased expression of resistance conferring genes such as EGFR [2,39]. A further minor subset of cells was also identified that were MITF-low and Axl-low, which clustered with transcriptional State #1. Together these studies demonstrate that melanomas consist of cells that can adopt a range of phenotypic behaviours, some of these adopting hybrid or intermediate transcriptional profiles that fell outside of the well-defined MITF-high/Axl-low and MITF-low/Axl-high states. It is likely that this range of transcriptional profiles constitutes a reservoir of cells with differing levels of drug sensitivity and that some of these may ultimately drive resistance. There is already some evidence that these minor populations of cells have unique therapeutic vulnerabilities and that these can be targeted to prevent the onset of drug resistance [36]. Studies from our own group have shown that combined targeting of BRAF along with c-JUN transcriptional activity through HDAC8 inhibition can also suppress the onset of resistance [9].
Multiple transcriptional states were often present in treatment-naïve melanoma cell lines and melanoma specimens, confirming previous reports that pools of resistant cells may pre-exist in drug-naïve tumours [40] [36]. We conversely demonstrated that small numbers of drug-sensitive cells were also retained under drug resistance and, in some cases, these could then expand following drug holidays, permitting secondary therapeutic responses. Although the need to retain drug sensitive cells under a state of drug resistance is not immediately clear, there are examples from the ecology field in which heterogeneity within cell populations serves as a bet-hedging strategy against population extinction [[41], [42], [43]]. In organisms as diverse as yeast and bacteria, small numbers of phenotypically distinct cells are consistently identified in otherwise normal populations [43]. These cells typically show diminished fitness under normal conditions, but their reduced rates of growth and tolerance to harsh conditions provide an effective buffer against drastic environmental changes. Our data show the converse is also true and suggest that the retention of a drug sensitive cell population in drug-resistant melanoma could be leveraged as a strategy to improve therapeutic responsiveness. Of course, there may be other eco-evolutionary explanations that describe the retention of drug-sensitive cells in an otherwise drug resistant culture such as the social foraging theory, which may explain how heterogeneous, group-living subpopulations of cells may gain from persistent “social” bonds and lead to increased tolerance in competitive environments, including the producer-scrounger game [[44], [45], [46]].
Various mathematical and computational models have been used in personalized medicine. An ordinary differential equation (ODE) modelling approach, which we utilized here, is typically used to describe cell population-level responses to therapy or microenvironmental changes. Excellent studies utilizing this approach include models predicting the impact of a cytokine on survival and expansion of hematopoietic cells [47], as well as models of predicting adaptive therapy response [19,48]. Brady et al. developed a comprehensive quantitative framework to simulate prostate cancer dynamics during treatment [49]. In their study, >70 patients undergoing multiple intermittent androgen deprivation therapies were modelled and used to identify a key determinant resistance. A partial differential equation modelling approach has also been employed to describe individual patient brain tumour MRI changes during treatment [50]. Agent-based modelling has proven useful in describing the behaviour of individual cells as wells as their interaction with neighbours (cancer cells, stromal cells, immune cells). A recent review by Karolak et al. discussed various individual cell-based models that considered the spatial architecture of tumours and addressed tumour growth as well as treatment response [51].
The identification of drug-specific dynamics between the transcriptional states and the maintenance of a pool of sensitive cells following treatment offered the possibility of developing adaptive BRAF inhibitor dosing schedules that were tailored to the mix of transcriptional states in each tumour. To achieve this, we developed a mathematical model calibrated by the growth dynamics of melanomas in vivo on/off BRAF inhibitor therapy. The decisions to hold or initiate therapy were based on predicted tumour growth dynamics and the assumption that tumour growth was mostly dictated by the expansion and contraction of transcriptional State #1. Our goal was to achieve initial tumour shrinkage and then to maintain sensitive cells within the tumour, preventing the uncontrolled expansion of the more resistant cellular states. Personalization of the dosing to each mouse revealed a wide variation in schedules, with some tumours requiring a relatively short drug holiday and others showing improved responses following longer drug holidays. This variability was not entirely unexpected, as it is likely that environmental constraints help shape transcriptional diversity at baseline. It was found that tailoring the treatment schedules to the dynamics each tumour delivered more profound anti-tumour responses than either continuous or fixed intermittent therapy schedules. These results provided the first evidence that personalizing therapy to tumour response dynamics may be of utility in melanoma.
There is already clinical evidence that a subset of melanoma patients exist who can be successfully re-challenged following initial BRAF inhibitor therapy [14], and that the magnitude of the response at re-challenge is predicted by the depth of the initial therapeutic response [35]. This work offers the first mechanistic insights into these clinical observations and provides a framework to develop better, more personalized BRAF inhibitor dosing schedules. Our new data may also allow new biomarkers to be developed that allow patients to be stratified to receive BRAF-MEK inhibitor therapy as their frontline treatment (e.g. those with a very high percentage of cells in transcriptional State #1) in place of immunotherapy. This is particularly pertinent in light of recent long-term follow up data showing that 5-year overall survival in patients on BRAF-MEK inhibitor therapy can be up to 34% [13]. Our studies further provide the proof-of-concept that resistance can be delayed through adaptive scheduling of existing FDA-approved drugs, with the advantages of reduced drug exposure and toxicity to the patient. In future refinements of this approach and our mathematical model, we envision integration of our approach with more conventional diagnostic metrics (circulating DNA, imaging, and biomarkers) to further improve patient outcomes. Our group has recently initiated a phase I feasibility trial of adaptive BRAF-MEK inhibitor therapy to test this concept in patients with advanced BRAF-mutant melanoma (NCT03543969).
Funding sources
This work was supported by SPORE grant P50 CA168536, R21 CA198550, R21 CA216756 (to KSMS) K99 CA226679 (to IS), Moffitt Cancer Center PSOC, U54 CA193489 (EK & AA) and the Cancer Center Support Grant P30 CA076292 from the NIH (NCI/NIH). We also acknowledge the support Miami Center for AIDS Research (CFAR) at the University of Miami Miller School of Medicine funded by P30AI073961. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders played no role in study design, data collection, data analysis, interpretation, and writing of the report.
Author contributions
Inna Smalley: study design, data collection, data interpretation, and figure/manuscript preparation. Keiran Smalley: Supervised the study, study design, data interpretation and writing. Alexander R.A. Anderson: mathematical model development, simulation analysis, figures and writing. Lesley de Armas: data collection and analysis. Nalan Akgul Babacan: data collection. Y. Ann Chen: Study design, analysis, data interpretation and writing. Zeynep Eroglu: data collection. Robert Gatenby: data interpretation. Eunjung Kim: study design (mathematical model), data collection, analysis and writing. Jiannong Li: data analysis and figures. Jane Messina: pathology guidance and review of manuscript. Silvya S. Maria-Engler: assisted with the planning of the project and the writing and editing of the manuscript. Sion Llewelyn Williams: oversaw single cell capture and data generation. Vernon K. Sondak: data analysis and interpretation, writing and editing. Paige Spence: data collection. Clayton Wyatt: data collection.
Declaration of Competing Interest
Keiran Smalley and Inna Smalley report personal editorial fees from Elsevier, outside the submitted work. Vernon Sondak reports personal fees from Merck, personal fees from Bristol-Myers Squibb, personal fees from Novartis, personal fees from Regeneron, personal fees from Array, personal fees from Novartis, personal fees from Polynoma, personal fees from Pfizer, during the conduct of the study. Zeynep Eroglu reports a grant from Novartis and serves on the advisory board for Array and Regeneron.
Clayton Wyatt, Paige Spence, Sion Williams, Lesley DeArmas, Alexander Anderson, Robert Gatenby, Jane Messina, Silvya Maria-Engler, Eunjung Kim, Nalan Barbacan, Jiannong Li and Ann Chen have nothing to disclose.
Acknowledgements
We would like to thank the Flow Cytometry Core Facility at Moffitt Cancer Center for their assistance with the flow cytometry analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.09.023.
Contributor Information
Y. Ann Chen, Email: ann.chen@moffitt.org.
Alexander R.A. Anderson, Email: alexander.anderson@moffitt.org.
Keiran S.M. Smalley, Email: keiran.smalley@moffitt.org.
Appendix A. Supplementary materials
Supplemental Figures 1-12
Supplemental Tables 1-4
Supplemental Table 5
Supplemental Table 6
References
- 1.Nazarian R., Shi H., Wang Q., Kong X., Koya R.C., Lee H. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. doi nature09626 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun C., Wang L., Huang S., Heynen G.J., Prahallad A., Robert C. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508(7494):118–122. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 3.Paraiso K.H., Thakur M.D., Fang B., Koomen J.M., Fedorenko I.V., John J.K. Ligand-independent EPHA2 signaling drives the adoption of a targeted therapy-mediated metastatic melanoma phenotype. Cancer Discov. 2015;5(3):264–273. doi: 10.1158/2159-8290.CD-14-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zipser M.C., Eichhoff O.M., Widmer D.S., Schlegel N.C., Schoenewolf N.L., Stuart D. A proliferative melanoma cell phenotype is responsive to RAF/MEK inhibition independent of BRAF mutation status. Pigment Cell Melanoma Res. 2011;24(2):326–333. doi: 10.1111/j.1755-148X.2010.00823.x. [DOI] [PubMed] [Google Scholar]
- 5.Konieczkowski D.J., Johannessen C.M., Abudayyeh O., Kim J.W., Cooper Z.A., Piris A. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014;4(7):816–827. doi: 10.1158/2159-8290.CD-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramsdale R., Jorissen R.N., Li F.Z., Al-Obaidi S., Ward T., Sheppard K.E. The transcription cofactor c-JUN mediates phenotype switching and BRAF inhibitor resistance in melanoma. Sci Signal. 2015;8(390):ra82. doi: 10.1126/scisignal.aab1111. [DOI] [PubMed] [Google Scholar]
- 7.Fallahi-Sichani M., Moerke N.J., Niepel M., Zhang T., Gray N.S., Sorger P.K. Systematic analysis of BRAF(V600E) melanomas reveals a role for JNK/c-Jun pathway in adaptive resistance to drug-induced apoptosis. Mol Syst Biol. 2015;11(3):797. doi: 10.15252/msb.20145877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Titz B., Lomova A., Le A., Hugo W., Kong X., Ten Hoeve J. JUN dependency in distinct early and late BRAF inhibition adaptation states of melanoma. Cell Discov. 2016;2 doi: 10.1038/celldisc.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emmons M.F., Faiao-Flores F., Sharma R., Thapa R., Messina J.L., Becker J.C. HDAC8 regulates a stress response pathway in melanoma to mediate escape from BRAF inhibitor therapy. Cancer Res. 2019;79(11):2947–2961. doi: 10.1158/0008-5472.CAN-19-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hugo W., Shi H., Sun L., Piva M., Song C., Kong X. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell. 2015;162(6):1271–1285. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Postow M.A., Chesney J., Pavlick A.C., Robert C., Grossmann K., McDermott D. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med. 2015;372(21):2006–2017. doi: 10.1056/NEJMoa1414428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robert C., Long G.V., Brady B., Dutriaux C., Maio M., Mortier L. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372(4):320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 13.Robert C., Grob J.J., Stroyakovskiy D., Karaszewska B., Hauschild A., Levchenko E. Five-year outcomes with dabrafenib plus trametinib in metastatic melanoma. N Engl J Med. 2019 doi: 10.1056/NEJMoa1904059. [DOI] [PubMed] [Google Scholar]
- 14.Schreuer M., Jansen Y., Planken S., Chevolet I., Seremet T., Kruse V. Combination of dabrafenib plus trametinib for BRAF and MEK inhibitor pretreated patients with advanced BRAF(V600)-mutant melanoma: an open-label, single arm, dual-centre, phase 2 clinical trial. Lancet Oncol. 2017;18(4):464–472. doi: 10.1016/S1470-2045(17)30171-7. [DOI] [PubMed] [Google Scholar]
- 15.Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hugo W., Zaretsky J.M., Sun L., Song C., Moreno B.H., Hu-Lieskovan S. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva A.S., Kam Y., Khin Z.P., Minton S.E., Gillies R.J., Gatenby R.A. Evolutionary approaches to prolong progression-free survival in breast cancer. Cancer Res. 2012;72(24):6362–6370. doi: 10.1158/0008-5472.CAN-12-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enriquez-Navas P.M., Kam Y., Das T., Hassan S., Silva A., Foroutan P. Exploiting evolutionary principles to prolong tumor control in preclinical models of breast cancer. Sci Transl Med. 2016;8(327):327ra24. doi: 10.1126/scitranslmed.aad7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J., Cunningham J.J., Brown J.S., Gatenby R.A. Integrating evolutionary dynamics into treatment of metastatic castrate-resistant prostate cancer. Nat Commun. 2017;8(1):1816. doi: 10.1038/s41467-017-01968-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paraiso K.H., Haarberg H.E., Wood E., Rebecca V.W., Chen Y.A., Xiang Y. The HSP90 inhibitor XL888 overcomes BRAF inhibitor resistance mediated through diverse mechanisms. Clin Cancer Res. 2012;18(9):2502–2514. doi: 10.1158/1078-0432.CCR-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fedorenko I.V., Abel E.V., Koomen J.M., Fang B., Wood E.R., Chen Y.A. Fibronectin induction abrogates the BRAF inhibitor response of BRAF V600E/PTEN-null melanoma cells. Oncogene. 2016;35(10):1225–1235. doi: 10.1038/onc.2015.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., 2nd, Treacy D., Trombetta J.J. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science (New York, NY) 2016;352(6282):189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostat (Oxford, Engl) 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 24.Fedorenko I.V., Abel E.V., Koomen J.M., Fang B., Wood E.R., Chen Y.A. Fibronectin induction abrogates the BRAF inhibitor response of BRAF V600E/PTEN-null melanoma cells. Oncogene. 2015 doi: 10.1038/onc.2015.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelley C.T. Implicit filtering. Soc Ind Appl Math. 2011;184 [Google Scholar]
- 26.Smyth T., Paraiso K.H., Hearn K., Rodriguez-Lopez A.M., Munck J.M., Haarberg H.E. Inhibition of HSP90 by AT13387 delays the emergence of resistance to BRAF inhibitors and overcomes resistance to dual BRAF and MEK inhibition in melanoma models. Mol Cancer Ther. 2014;13(12):2793–2804. doi: 10.1158/1535-7163.MCT-14-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Smalley I., Schell M.J., Smalley K.S.M., Chen Y.A. SinCHet: a MATLAB toolbox for single cell heterogeneity analysis in cancer. Bioinformatics. 2017;33(18):2951–2953. doi: 10.1093/bioinformatics/btx297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller J., Krijgsman O., Tsoi J., Robert L., Hugo W., Song C. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun. 2014;5:5712. doi: 10.1038/ncomms6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bollag G., Hirth P., Tsai J., Zhang J., Ibrahim P.N., Cho H. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. doi nature09454 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das Thakur M., Salangsang F., Landman A.S., Sellers W.R., Pryer N.K., Levesque M.P. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494(7436):251–255. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phadke M.S., Sini P., Smalley K.S. The novel ATP-competitive MEK/Aurora kinase inhibitor BI-847325 overcomes acquired BRAF inhibitor resistance through suppression of Mcl-1 and MEK expression. Mol Cancer Ther. 2015;14(6):1354–1364. doi: 10.1158/1535-7163.MCT-14-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larkin J., Ascierto P.A., Dreno B., Atkinson V., Liszkay G., Maio M. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med. 2014;371(20):1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 33.Larkin J., Chiarion-Sileni V., Gonzalez R., Grob J.J., Cowey C.L., Lao C.D. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373(1):23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robert C., Karaszewska B., Schachter J., Rutkowski P., Mackiewicz A., Stroiakovski D. Improved overall survival in melanoma with combined dabrafenib and trametinib. New Engl J Med. 2015;372(1):30–39. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 35.Tietze J.K., Forschner A., Loquai C., Mitzel-Rink H., Zimmer L., Meiss F. The efficacy of re-challenge with BRAF inhibitors after previous progression to BRAF inhibitors in melanoma: a retrospective multicenter study. Oncotarget. 2018;9(76):34336–34346. doi: 10.18632/oncotarget.26149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rambow F., Rogiers A., Marin-Bejar O., Aibar S., Femel J., Dewaele M. Toward minimal residual disease-directed therapy in melanoma. Cell. 2018;174(4):843–855. doi: 10.1016/j.cell.2018.06.025. e19. [DOI] [PubMed] [Google Scholar]
- 37.Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S. Identification of the tumour transition states occurring during EMT. Nature. 2018;556(7702):463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 38.Smith M.P., Brunton H., Rowling E.J., Ferguson J., Arozarena I., Miskolczi Z. Inhibiting drivers of non-mutational drug tolerance is a salvage strategy for targeted melanoma therapy. Cancer Cell. 2016;29(3):270–284. doi: 10.1016/j.ccell.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smalley K.S., Lioni M., Palma M.D., Xiao M., Desai B., Egyhazi S. Increased cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF V600E-mutated melanomas. Mol Cancer Ther. 2008;7(9):2876–2883. doi: 10.1158/1535-7163.MCT-08-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaffer S.M., Dunagin M.C., Torborg S.R., Torre E.A., Emert B., Krepler C. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature. 2017;546(7658):431–435. doi: 10.1038/nature22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levy S.F. Cellular heterogeneity: benefits besides bet-hedging. Curr Biol. 2016;26(9) doi: 10.1016/j.cub.2016.03.034. R355-R7. [DOI] [PubMed] [Google Scholar]
- 42.Martins B.M.C., Locke J.O.W. Microbial individuality: how single-cell heterogeneity enables population level strategies. Curr Opin Microbiol. 2015;24:104–112. doi: 10.1016/j.mib.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 43.Gasch A.P., Yu F.B., Hose J., Escalante L.E., Place M., Bacher R. Single-cell RNA sequencing reveals intrinsic and extrinsic regulatory heterogeneity in yeast responding to stress. PLoS Biol. 2017;15 doi: 10.1371/journal.pbio.2004050. (12) doi ARTN e2004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.L-AGT Caraco. Princeton University Press; 2000. Social foraging theory. [Google Scholar]
- 45.Morand-Ferron J., Giraldeau L.-A., Lefebvre L. Wild Carib grackles play a producer–scrounger game. Behav Ecol. 2007;18(5):916–921. [Google Scholar]
- 46.Harten L., Matalon Y., Galli N., Navon H., Dor R., Yovel Y. Persistent producer-scrounger relationships in bats. Sci Adv. 2018;4(2) doi: 10.1126/sciadv.1603293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gullo F., van der Garde M., Russo G., Pennisi M., Motta S., Pappalardo F. Computational modeling of the expansion of human cord blood CD133+ hematopoietic stem/progenitor cells with different cytokine combinations. Bioinformatics (Oxford, England) 2015;31(15):2514–2522. doi: 10.1093/bioinformatics/btv172. [DOI] [PubMed] [Google Scholar]
- 48.West J.B., Dinh M.N., Brown J.S., Zhang J., Anderson A.R., Gatenby R.A. Multidrug cancer therapy in metastatic castrate-resistant prostate cancer: an evolution-based strategy. Clin Cancer Res. 2019;25(14):4413–4421. doi: 10.1158/1078-0432.CCR-19-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brady R., Nagy J.D., Gerke T.A., Zhang T., Wang A.Z., Zhang J. Prostate-specific antigen dynamics predict individual responses to intermittent androgen deprivation. bioRxiv. 2019 doi: 10.1038/s41467-020-15424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jackson P.R., Juliano J., Hawkins-Daarud A., Rockne R.C., Swanson K.R. Patient-specific mathematical neuro-oncology: using a simple proliferation and invasion tumor model to inform clinical practice. Bull Math Biol. 2015;77(5):846–856. doi: 10.1007/s11538-015-0067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karolak A., Markov D.A., McCawley L.J., Rejniak K.A. Towards personalized computational oncology: from spatial models of tumour spheroids, to organoids, to tissues. J R Soc Interface. 2018;15(138) doi: 10.1098/rsif.2017.0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures 1-12
Supplemental Tables 1-4
Supplemental Table 5
Supplemental Table 6