Abstract
Background
Sjögren's syndrome (SS) is one of the most common autoimmune disorders leading to exocrine gland dysfunction. Both immune-dependent processes – like Type I Interferon (IFN) signaling and immune-independent processes – such as calcium signaling in epithelial cells – contribute to disease pathophysiology. However, a mechanistic link between these processes has not been demonstrated.
Methods
Primary human salivary gland cells were used to evaluate the differential expression of miRNAs with smRNA-seq in primary epithelial cells culture and digital PCR was conducted in SS human salivary glands (SG) biopsies to verify the results. With siRNA screening and pull-down assays to establish the role of miRNA in IFN activation.
Findings
Activation of IFN-β by miR-1248 is through the direct association with both RIG-I and AGO2. Further functional studies establish a unique dual functional role of miR-1248 in phSG cells: i) activation of the RIG-I pathway by acting as ligand of this sensor leading to IFN production and ii) regulation of the expression of mRNAs through the canonical microRNA function. Importantly, ITPR3, a key component of calcium signaling in epithelial cells, that has previously shown to be downregulated in SS SG, was directly targeted and downregulated by miR-1248, inducing the same functional calcium signaling changes as observed in SS SGs.
Interpretation
Identification of the first endogenous mammalian microRNA that binds to RIG-I inducing IFN production but also demonstrate a novel pathophysiological underlying mechanism in which miR-1248 overexpression links two major pathways associated with SS, namely activation of IFN production with modulation of calcium signaling. Together, these findings suggest a unifying hypothesis for the immune-independent and -dependent processes contributing to the pathogenesis of SS.
Fund
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Dental and Craniofacial Research (NIDCR).
Keywords: Sjögren's syndrome, miR-1248, Interferon, RIG-I and ITPR3
Research in context.
Evidence before this study
Sjögren's syndrome is one of the most common autoimmune disorders leading to exocrine gland dysfunction. Many studies reported recently indicate both immune-dependent processes – like Type I Interferon (IFN) signaling – and immune-independent processes – such as calcium signaling in epithelial cells – contribute to disease pathophysiology. More than 50% of SS patients show an IFN signature elevation. However, a mechanistic link between these processes has not been demonstrated.
Added value of this study
Our study identify the role of endogenous mammalian miRNA that not only could initiate IFN signaling but also demonstrate a novel pathophysiological underlying mechanism in which miR-1248 overexpression links two major pathways associated with SS, namely activation of IFN production with modulation of calcium signaling. These findings suggest a unifying hypothesis for the immune-independent and –dependent processes contributing to the pathogenesis of SS.
Implications of all the available evidence
The role of miR-1248 in IFN induction and signaling in salivary epithelial cells identifies new mechanistic pathways and potential target for therapies.
Alt-text: Unlabelled Box
1. Introduction
Sjögren's syndrome (SS) is an autoimmune disorder that targets exocrine glands, primarily the salivary and lacrimal, resulting in dry mouth (xerostomia) and dry eyes (xeropthalmia). In the salivary glands (SGs), the reduction or complete loss of saliva secretion has been attributed to acinar cell dysfunction [1,2]. Numerous hypotheses have been proposed for the pathogenesis of salivary epithelial cell dysfunction in SS, including both immune-independent and immune-mediated mechanisms [3,4].
Emerging studies have shown alterations in interferon (IFN) signaling pathways, especially the upregulation of IFN-inducible genes (IFN signature) in salivary glands and peripheral blood in subgroups of SS patients [[5], [6], [7], [8], [9]]. IFNs have been shown to regulate more than 2000 coding and non-coding RNA transcripts, in a highly coordinated manner, depending on subtype, timing, dosage, cell types, and pathophysiological status [10]. An IFN signature has been shown to be present in peripheral blood in 55% of SS patients and has been linked to higher disease activity, presence of anti-Ro/SSA and anti-La/SSB antibodies and hypergammaglobulinemia [11]. It is likely that IFN promotes adaptive immune responses through the activation of immune cells and induction of cytokine production [[12], [13], [14]]. However, it still remains unclear what initiates the IFN response in SS patients.
MicroRNAs (miRNAs) are small non-coding RNAs that bind to target transcripts and block translation. Studies have shown that miRNAs regulate a wide range of biological processes [[15], [16], [17], [18], [19]], including innate immune response and viral infection. In the context of autoimmune diseases, particularly systemic lupus erythematosus (SLE), rheumatoid arthritis (RA) and Sjögren's syndrome [[20], [21], [22]], miRNAs have been shown to modulate IFN signaling by targeting key genes [23,24]. However, in SS patients, these same miRNAs can show apparently conflicting or paradoxical effects on IFN [25].
There is growing evidence that salivary dysfunction in SS may not be tightly linked to the autoimmune response. Bookman et al. [26] found that characteristic clinical measures of SS, such as stimulated salivary flow, correlate poorly with inflammation (r2 < 0.11). In our own studies, we have shown that SGs and SG epithelial cells from patients with SS display a marked reduction in ITPR3, a key protein involved in store-operated calcium entry (SOCE) that triggers fluid secretion in SG acini [27].
We previously established primary human SG epithelial cell cultures (phSG) that retain highly proliferative growth in low calcium medium (0.05 mM) but display differentiated acinar-like phenotype in high calcium (1.2 mM) medium [28]. We performed small RNA profiling using smRNA-seq to identify calcium-regulated miRNAs that may be involved in the change in phenotype of the phSG cells. One of these miRNAs (miR-1248) was found in parallel Whole Transcriptome profiling experiments of SS SG as significantly up-regulated in SS and a high degree of correlation among endogenous miR-1248 copy number and IFN signature in SS.
We pursued mechanistic studies to decipher the functional role of this microRNA in SG. We document the link between miR-1248 and the activation of IFN signaling pathway and confirmed the secretion of IFN-β in miR-1248-transfected phSG cells. Further, through investigation with siRNA screening and pull-down assays, coupled with transcriptome sequencing analysis, we confirmed that miR-1248 directly binds to both AGO2 acting as a canonical miRNA, and RIG-I (DDX58) activating directly the IFN pathway. Importantly, we also identified DAK1, a negative regulator of IFN-β synthesis, and ITPR3, as target transcripts of miR-1248.
2. Materials and methods
2.1. Human samples
Human subjects research was carried out in accordance with approved National Institute of Health (NIH) guidelines conforming to the standards of the Declaration of Helsinki.All participants provided informed consent prior to the initiation of any study procedures. Human samples were obtained from NIH Institutional Review Board approved protocols (ClinicalTrials.gov Identifiers: NCT00001196, NCT00001390) in the Sjögren's Syndrome Clinic at the National Institute of Dental and Craniofacial Research (NIDCR) at the NIH in Bethesda, MD.
2.2. Plasmid construction
A 0.5 kb DNA fragment encompassed precursor miR-1248 (pre-miR-1248) region was amplified by PCR using human genomic DNA as a template and cloned into pAC-LTR-EF-1α expression vector to generate pAC-PremiR1248 plasmid (detail see supplement). The luciferase reporter constructs contained either DAK 3’-UTR (2.25 kb, named pLuc-DAK-3’UTR) or ITPR3 coding region (8.0 kb, named pLuc-ITPR3-CDS) were obtained from GeneCopeia. Luciferase reporter plasmids of Cignal IRF1 Reporter (CCS-7040 L) and Cignal ISRE Reporter (STAT1/STAT2, CCS-008 L) were used to monitor type I IFN-induced signal transduction pathways (Qiagen). Expression construct of RIG-I (pEF-Bos-FLAG-RIG-I) was a generous gifted from Dr. M. Gale, Jr. (University of Washington).
2.3. Cell transfection, quantitative real-time PCR and luciferase assays
Indicated concentration of miRNAs, siRNAs and luciferase plasmids were transfected with either Lipofectamine RNAiMAX (Invitrogen) or Attractene transfectant (Qiagen) (see supplement). The synthetic miRNAs (mimic) of has-miR-1248 and All Star negative control miRNA (miR-N.C.) were from Qiagen. The siRNAs of IFNAR1, IFNAR2, AGO2, DAK, HMGB1, MAVS, MYD88, LGP2, RIG-1, ITPR3 were obtained from Ambion. Firefly and renilla luciferase activities were determined by Dual-Luciferase Reagent Assay System (Promega) with Fluostar Omega microplate reader (BMG Labtech). Firefly luciferase activity was normalized with renilla luciferase activity and expressed as fold change over control (see supplement).
2.4. Co-immunoprecipitation of protein and miRNA complex
The cell pellets of phSG cells transfected transfected with miR-1248 mimic or transduced with viral pAC-Pre-miR1248 plasmid were collected following the protocol of RNA ChIP-IT assay (Active Motif) (see supplement). The binding complexes were pull-downed with antibodies against either RIG-I, AGO2 or rabbit IgG (control, all from Cell Signaling), or GFP (control, Ab290, Abcam) and the total RNAs were obtained and subjected to real-time PCR (RT-qPCR) using a TaqMan probe of miR-1248. (see supplement).
2.5. RNA library preparation and sequencing
Total RNA was isolated from cultured cells or from ten 20 μm sections of minor salivary gland tissue in OCT using the miRCURY™ RNA Isolation Kit - Cell & Plant (Exiqon). Total RNA (200 ng – 500 ng) was rRNA depleted with the RiboMinus Eukaryotic Kit v2 (Ambion) and were subject to library preparation using the Ion Total RNA-Seq kit v2 (Thermo Fisher Scientific) according to manufacturer protocols for small RNA or total RNA libraries as indicated. The barcoded cDNA library was quantified a Bioanalyzer 2100 (Agilent) and input for template preparation using the Ion PI™ Hi-Q™ Chef Kit and the Ion Chef instrument, followed by sequencing on the Ion Proton sequencer (Thermo Fisher Scientific).
2.6. RNA-Seq data analysis
Following sequencing, the data were separated by barcode, aligned with TMAP, and exported as part of the Ion Torrent Suite 4.2.1 pipeline (Thermo Fisher Scientific). For small RNA sequencing (smRNA-Seq) in cell culture experiments, aligned BAM files for each barcode were used as input into Partek Genomics Suite (version 6.15.1016) and features were summarized by FPKM using miRBase 20 annotations. Differential expression was computed as the ratio of FPKM values between low and high calcium samples. For whole RNA sequencing in SG tissue, features were quantified using the featureCounts software [29] from the aligned BAM files and analyzed for differential expression using the standard workflow of the DESeq2 package [30] for the R statistical programming language. Reported p-values from RNA-Seq data are from a Wald test adjusted with the Benjamini-Hochberg correction for multiple hypothesis testing. The total RNA-Seq data were normalized using the regularized log transformation from the DESeq2 package before computing IFN scores. A scoring system described previously [31] was used to calculate scores using normalized values for each sample. The score is the average number of standard deviations above the mean expression in healthy samples of a preselected set of genes. Genes were selected for IFN-alpha (IFIT1, IFI44, EIF2AK2), beta (ISG15, OAS1, IFIH1, RSAD2, IFI6), and gamma (IRF1, GBP1, SERPING1) [32].
2.7. Live cell calcium imaging using confocal microscopy
HSG cells were loaded with Fluo-4 AM (10 μM; Invitrogen) in its growth medium for 20 min at 37 °C; they were then washed for 20 min in their growth medium without Fluo-4 AM. Standard extracellular solution without calcium was used to perform the experiments. The cells were stimulated with 100 μM Carbachol (CCh, Sigma, St. Louis, MO, U.S.A.); and by subsequent addition of CaCl2 (1 mM). Cells were imaged using a FluoView 1000 (Olympus) confocal microscope; images were acquired every 1.5`s. Fluo-4 AM was excited with the 488-nm line of an argon ion laser with emission at 510 nm. Regions of interest (ROI) were selected and fluorescence intensity was determined as a function of time and expressed relative to the initial fluorescence.
2.8. Digital PCR quantification
To estimate the copy number of miR-1248 in salivary gland, total RNAs were isolated from 14 biopsy samples (6 health controls and 8 SS patients) with miRCURY RNA Isolation Kit (Exiqon). Fifty nanogram of total RNA was reversed transcribed with the TaqMan miRNA Reverse Transcriptase kit (Thermo Fisher Scientific) and the PCR reaction was loaded on chips (two per sample) on the ProFlex PCR system. End-point detection of the hsa-miR-1248 TaqMan Assay was performed on the QuantStudio 3D Digital PCR System (Thermo Fisher Scientific) and the data were analyzed by QuantStudio 3D software for copy number and quality control calculations.
2.9. Statistical analysis
Statistical significance was evaluated using Student's t-test analysis, unless otherwise specified. The level of significance was set p < .05 for all analyses.
2.10. RNA sequencing data
The data analyzed in this study are deposited in database dbGaP under the study accession phs001842.v1.p1 and study name “RNAseq of Sjögren's Syndrome and Healthy Volunteers' Salivary Glands”.
3. Results
3.1. Differential expression of miRNAs in SS and in phSG cells
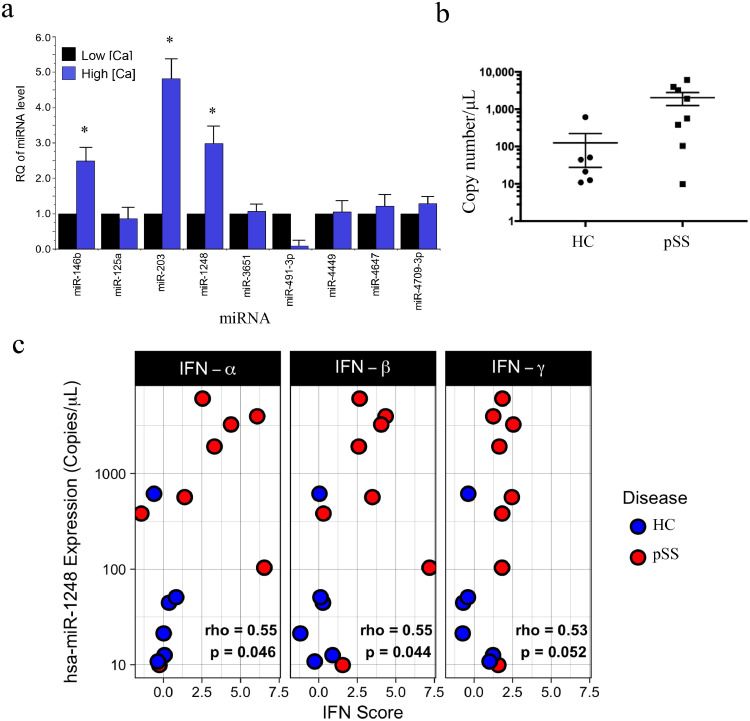
Alterations in calcium signaling have been shown to be associated with SS and salivary gland dysfunction [27,33] In an attempt to understand the biological pathways and processes that underlie this phenomenon, we performed a pilot experiment and compared miRNA expression profiles using small-RNA deep-sequencing (smRNA-Seq) in phSG cells maintained in keratinocyte growth medium (KGM) containing either low (0.05 mM, KGM-L) or high (1.2 mM, KGM-H) calcium. Thirty miRNAs were differentially expressed with >10-fold up-regulation in phSG cells grown in KGM-H vs. in KGM-L condition. Among those, miR-125A, miR-664, miR-3651, miR-4647 and miR-4709 were only expressed in phSG cells maintained in KGM-H (Table S1). We used real-time RT-PCR experiments to validate these results, and as shown in Fig. 1A, the most notable differentially expressed miRNAs were miR-146b, miR-203, and miR-1248, which were up-regulated by 2.5-, 5- and 3-fold, respectively (p < .05), whereas miR-491-3p was down-regulated about 8-fold (p < .05). Of these calcium-regulated miRNAs, we also found that miR-1248 was significantly more abundant in minor SGs of SS patients using digital PCR (Fig. 1B). Based on the above, we further examined the biological role of miR-1248 in SS using phSG cells as a model for salivary acinar cells.
Fig. 1.
Validation of differentially expressed miRNAs in phSG cells and in SS patients. (a) Total RNAs were isolated from phSG cells maintained either in KGM-L or KGM-H medium and used for RT-qPCR with indicated TaqMan probes. Data are average of three separate experiments with mean ± S.E. and presented as a fold change over each respective low and high calcium sample (* p < .05, Student's t-test). (b) Expression level of miR-1248 in MSG of Healthy Controls (n = 6, circle) and SS subjects (n = 8, square) measured by digital PCR as copies/μl. (c) Total RNA-seq and digital PCR assays were conducted with total RNAs from healthy control (n = 6, blue) and SS patients (n = 8, red) biopsies for whole transcriptome analysis and miR-1248 copy number determination, respectively. Data are presented as the copy number of miR-1248 vs. interferon score of type I and II IFN. The interferon score is the average number of standard deviations from the mean expression level of IFN stimulated genes in healthy controls (IFN-α: IFIT1, IFI44, EIF2AK2; IFN-β: ISG15, OAS1, IFIH1, RSAD2, IFI6; IFN-γ: IRF1, GBP1, SERPING1). Each panel is labeled with the Spearman's rho value for the correlation between copy number and IFN score of all samples in the panel and the p-value from a Spearman rank test. HC: healthy control; pSS: Sjögren's syndrome patient. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Correlation of miR-1248 expression, IFN signaling and SS
To investigate whether there is a correlation between elevated miR-1248 expression and IFN signaling in SS patients (Table S2), total RNA-seq and digital PCR assays were used to evaluate the expression of interferon-stimulated genes (ISGs) and the copy number of miR-1248, respectively. Expression of ISGs was summarized by a scoring method (see Methods and Materials) that yields increasingly positive values as expression of one or more ISGs is increased in a sample compared to the average of the healthy volunteer samples, and increasingly negative values if they are decreased. As shown in Fig. 1C, using the interferon score to represent the type I and type II IFN signaling, not only did the copy number of miR-1248 correlate positively with interferon score, but the expression level of miR-1248 was also significantly higher in most of the SS patient samples than in healthy controls (Welch's t-test p value = .045). These data demonstrate that miR-1248 expression is positively correlated to a modest degree (average Spearman rho of 0.5), with the degree of IFN activation in salivary gland tissue of SS patients.
3.3. Induction of IFN signaling pathway by miR-1248 mimic in phSG cells
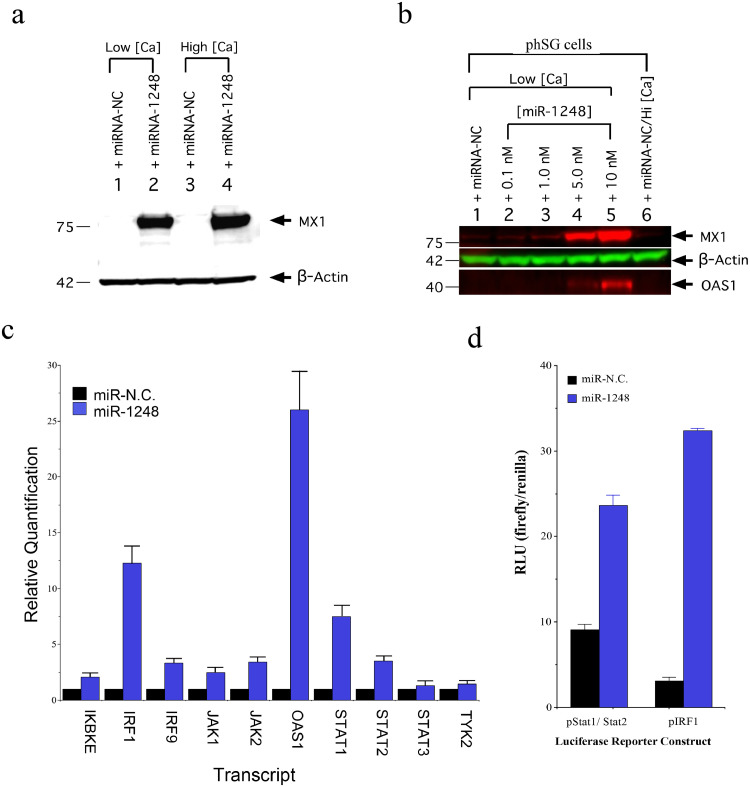
To identify systems-level alterations correlated with this miRNA, whole transcriptome profiling of miR-1248-transfected phSG cells was compared to negative control (miR-NC) transfected samples (Table S3). Differentially expressed transcripts were analyzed by Ingenuity Pathway Analysis (IPA) and revealed an enrichment for genes involved in canonical IFN signaling pathways. We selected MX1 and OAS1, two ISGs commonly used as markers for Type I Interferon activation [34], to monitor the induction of IFN. As shown in Fig. 2A, MX1 protein was markedly increased in miR-1248-transfected phSG cells in both KGM-L and KGM-H conditions. Importantly, although endogenous miR-1248 levels increased three-fold in KGM-H (Fig. 1A), no MX1 expression was detected in miR-NC-transfected cells (Fig. 2A, lane 3). Interestingly, when miR-1248 was transfected into primary fibroblasts, HeLa, HEK293 or human salivary gland (HSG) cell line, no MX1 was detected (Fig. S1), indicating that miR-1248-induced MX1 expression is cell type-specific. To further confirm the specificity of ISGs induced by miR-1248, when increasing concentrations of miR-1248 were used for transfection both MX1 and OAS1 levels increased in a dose-dependent manner (Fig. 2B), confirming that miR-1248 is responsible for the up-regulation of ISGs in phSG cells.
Fig. 2.
Induction of MX1 by miR-1248 in phSG cells. (a) Transfection of miR-1248 in phSG cells induces MX1 protein expression under both high and low calcium conditions. (b) The induction of MX1and OAS1 due to miR-1248 were dose-dependent (β-actin was used as loading control in both a and b). (c) Total RNAs from phSG cells transfected either with miR-N.C. (control) or miR-1248 were subjected to RT-qPCR. Data are shown as a mean ± S.E. from three independent experiments and presented as a relative quantification to each respective miR-N.C. control sample. The error bars represent a 95% Confidence Interval. (d) Indicated luciferase reporter plasmid was transfected into phSG cells 24 h prior to the transfection of indicate miRNAs for 48 h. Cellular lysates were used for both firefly and renilla luciferase activity. Data are shown as mean ± S.E. of RLU from three independent experiments, after normalized with renilla luciferase activity.
The up-regulation of several critical IFN signaling components was further analyzed by quantitative RT-PCR. Compared to the miR-NC-treated control, the expression of TYK2 and STAT3 was mildly up-regulated in miR-1248-transfected phSG cells (Fig. 2C). However, the expression of IKBKE, IRF1, IRF9, JAK1, JAK2, STAT1, STAT2 displayed more robust increases ranging from 2- to 12-fold, and OAS1 increased >25-fold in the presence of miR-1248 (Fig. 2C). Furthermore, when miR-1248 was co-transfected together with the luciferase reporter constructs containing either the STAT1/STAT2-responsive motif or the IRF1-binding motif in the promoter region, luciferase activity showed a 2.5- and 10-fold increase, respectively, compared to miR-NC-transfected controls (Fig. 2D). This verifies that the increase of ISGs expression in miR-1248 transfected phSG cells was mediated by the IFN/STAT/JAK signaling.
3.4. MiR-1248 leads to secretion of IFN-β
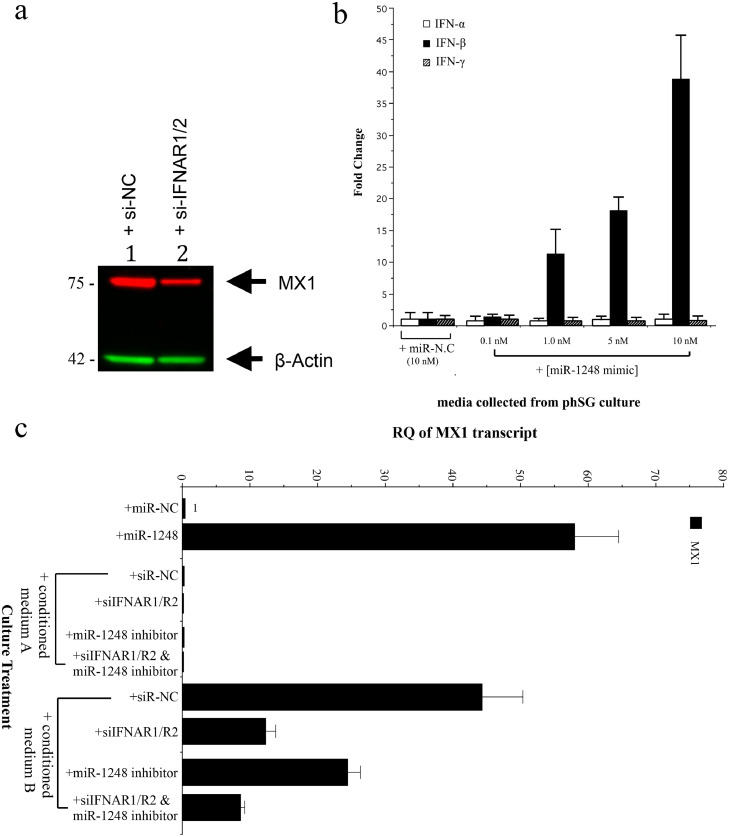
Next, we studied whether induction of the IFN signaling pathway by miR-1248 is responsible for the activation of the STAT/JAK signaling pathway. A mixture of siRNAs against type I IFN-α/β receptor (IFNAR1 and IFNAR2) markedly reduced the expression of MX1 protein in miR-1248-transfected phSG cells (Fig. 3A), indicating that the activation of STAT/JAK signaling pathway by miR-1248 is dependent upon the presence of IFNAR1/R2 receptors in phSG cells. To identify if, and which, IFN cytokines were produced, phSG cells were transfected with various concentrations of miR-1248 and the extracellular media were collected for ELISA assays. As shown in Fig. 3B, only IFN-β, and not IFN-α or IFN-γ, was produced and secreted into the culture media in a miR-1248 dose-dependent manner. To evaluate the biologic activity of secreted IFN-β, phSG cells were cultured with the conditioned media from miR-1248-transfected phSG cultures. In these cultures, MX1 expression was significantly increased (Fig. 3C). However, this elevation was reduced with siRNA-mediated knockdown of IFNAR1 and IFNAR2 or treatment with an miR-1248 inhibitor prior to the addition of conditioned medium (Fig. 3C). Taken together, these data demonstrate that miR-1248 induces the production and secretion of IFN-β, subsequently activates STAT/JAK signaling pathway and leads to upregulation of MX1, OAS1 and ISG15 (see Fig. 4A and S2) expression in phSG cells.
Fig. 3.
Secretion of IFN-β in miR-1248 transfected phSG cells. (a) Western blot analysis showed reduced miR-1248-induced MX1 expression after knockdown of IFNAR1/R2 receptors in phSG cells. (b) Quantification of secreted interferons by ELISA showed a dose-dependent increase of IFN-β. Error bars represent standard error from three independent experiments with duplicated samples. (c) Conditioned media collected after phSG cells were transfected with miR-N.C. (medium A) or miR-1248 mimic (medium B) for 24 h was incubated with phSG cells previously transfected with indicated siRNAs or miR-1248 inhibitor for 24 h before exposure to the conditioned media for another 24 h. MX1 transcript level was shown to be induced by Medium B only. Data are shown as RQ (relative quantification) over the phSG cells transfected with miR-N.C. The error bars represent a 95% Confidence Interval. RQ: relative quantification.
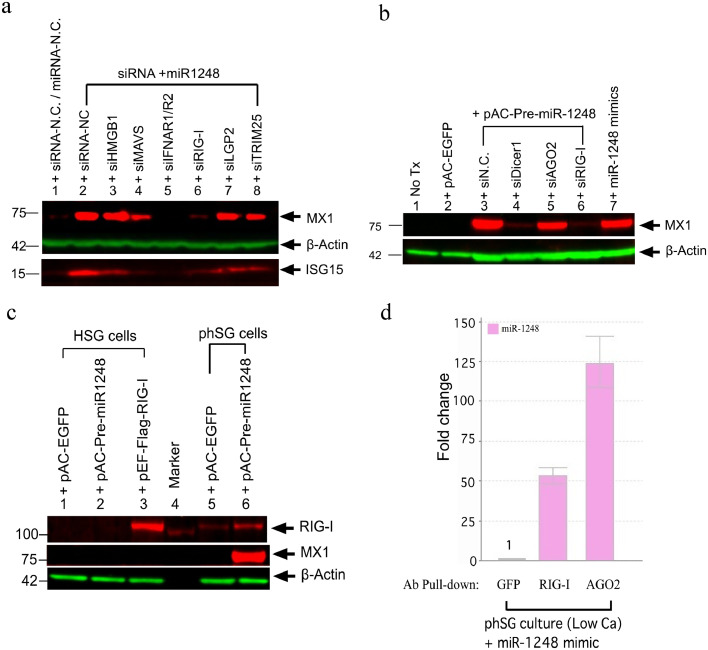
Fig. 4.
RIG-I plays a key role in MX1 expression induced by miR-1248 in phSG cells. (a) Western blot analyses of MX1 and ISG15 in phSG cells transfected with the indicated siRNAs together with miR-1248 mimic showed that only knockdown of RIG-I and IFNAR1/R2 abolished MX1 induction (lanes 5 and 6). Knockdown of MAVS attenuates MX1 induction (lane 4). (b) Transduction of pAC-Pre-miR-1248 also induced MX1 expression and this effect is abolished upon knockdown of DICER1 and RIG-I. pAC-EGFP was used as viral transduction control. (c) Western blot analysis of MX1 and RIG-I indicates that transduction of pAC-Pre-miR-1248 induced expression of both proteins in phSG cells (lanes 6), but not in the HSG cell line (lane 2). pAC-EGFP (lane 1, 5) served as a negative control for viral transduction and a RIG-I expression construct (pEF-Flag-RIG-I, lane 3) was used as a positive control. β-actin was used for loading control (A, B). (d) Protein-miRNAs complex was immunoprecipitated by GFP, RIG-I or AGO2 antibody and the level of miR-1248 was quantified by RT-qPCR, showing direct interaction of miR-1248 with both RIG-I and AGO2. Data are shown as fold change (mean ± S.E., average from three independent experiments) over the GFP antibody control. Tx: transfection.
3.5. Activation of RIG-I-mediated IFN signaling by miR-1248
To identify members of the IFN signaling pathway involved in the miR-1248-mediated induction of IFN-β in phSG cells, we used siRNAs targeting these members prior to the treatment of miR-1248. First, to determine whether endosomal TLRs are involved, siRNAs were also used to knockdown either MYD88, IRF7 or TLR3. No effect was observed on the miR-1248-induced MX1 expression (Fig. S2), showing that endosomal TLRs were not responsible for the immune response. Knockdown of HMGB1 (Fig. 4A, lane 3) also did not affect miR-1248-induced MX1 expression compared to the siRNA-NC control (Fig. 4A, lane 2). However, knockdown of MAVS (Fig. 4A, lane 4), LGP2 (Fig. 4A, lane 7) or TRIM25 (Fig. 4A, lane 8) reduced MX1 expression by 60%, 30% and 40%, respectively. Knockdown of either IFN-α/β receptors (INFAR1/R2, Fig. 4A, lane 5) or retinoic acid-inducible gene-I (RIG-I, Fig. 4A, lane 6) completely abolished MX1 expression, not only confirming the importance of the role of type I IFN receptors, but also revealing that RIG-I plays a critical role in miR-1248 mediated IFN-β production.
To rule out possible artifacts caused by the modification used to stabilize the synthetic microRNAs, we generated a viral expression construct containing pre-miR-1248 DNA sequences (pAC-Pre-miR-1248) and transduced it into phSG cells. The increase of the mature miR-1248 was confirmed by RT-qPCR (Fig. S4). As shown in Fig. 4B, viral transduction on its own (pAC-EGFP, as a negative control) did not induce MX1 expression (lane 2), indicating that the viral transduction alone was not sufficient to activate IFN signaling. However, as expected, expression of pAC-Pre-miR-1248 resulted in an induction of MX1 in phSG cells (lane 3). Knockdown of the endoribonuclease Dicer1 (lane 4) and RIG-I (lane 6) significantly diminished MX1 expression, indicating that processing by Dicer and generation of the mature miR-1248 are required to up-regulate MX1 expression through RIG-I-mediated activation of IFN signaling. Interestingly, knockdown of AGO2 (lane 5), which is a critical component of the RNA-induced silencing complex (RISC) and normal miRNA function, led to only a 25% reduction of MX1 expression, suggesting that miR-1248 might play an additional role in IFN signaling activation through canonical miRNA function. In summary, these data demonstrate that the viral-transduced miR-1248 precursor is processed by Dicer into its mature form in phSG cells and results in up-regulation of MX1 expression similar to that we observed using miR-1248 mimics. In addition, the decrease in MX1 in the absence of AGO2 strongly suggests a role of this miRNA on IFN signaling by targeting other key players in the IFN generation pathway as a post-transcriptional regulator.
3.6. Upregulation of RIG-I expression by miR-1248 in phSG cells
It has been reported that RIG-I expression is up-regulated by IFN signaling [35,36], so we examined whether expression of RIG-I was regulated by miR-1248 in epithelial cells. Transduction of pAC-Pre-miR-1248 in HSG cells resulted in neither an increase of MX-1 expression, nor a detectable RIG-I protein band (Fig. 4C, lane 2). Trace amounts of RIG-I with no MX1 induction were observed in phSG cells transduced with a pAC-EGFP construct (lane 5), indicating low levels of endogenous RIG-I in phSG cells. Expression of miR-1248 precursor showed markedly increase of both RIG-I and MX1 expression (lane 6). These observations were further supported by RT-qPCR in which the level of RIG-I and MX1 transcripts displayed a similar pattern after miR-1248 transfection (Fig. S3A). Compared to the miR-NC-transfected control, the expression of RIG-I shows a 17-, 35- and 24-fold increase at 24 h, 48 h and 72 h, respectively. Similarly, the expression of MX1 shows an 80-, 190-, and 100-fold increase at 24 h, 48 h and 72 h, respectively. These observations were further supported by an increase in both RIG-I and MX1 transcript levels in phSG cells treated with IFN-β cytokine directly (Fig. S3B). Together, these data demonstrate that expression of RIG-I was up-regulated by miR-1248-mediated activation of IFN-β in phSG cells.
3.7. Direct binding of miR-1248 with RIG-I and AGO2 in phSG cells
It has been known that RIG-I acts as an RNA sensor to mediate Toll-like receptor (TLRs)-independent IFN-α/β induction in the presence of replicating RNA viruses [37,38]. As shown above, knockdown of RIG-I markedly abolished miR-1248-induced MX1 expression in phSG cells, and given its function as an RNA sensor, it is likely that RIG-I binds miR-1248 directly to trigger activation of the IFN-β signaling pathway. To study this, RIG-I/miR-1248 complexes were immunoprecipitated (IP) with a monoclonal antibody against RIG-I from extracts of either pAC-Pre-miR-1248-transduced or miR-1248 mimic-transfected phSG cells. Co-immunoprecipitation of endogenous RIG-I was very efficient and the majority of RIG-I protein was recovered from cell lysates (Fig. S5A). In addition, the association between miR-1248 and RIG-I was observed in either pAC-Pre-miR-1248-transduced or miR-1248 mimic-transfected phSG cells (Fig. S5B). Due to the possible dual role of miR-1248, the co-IP was repeated with antibodies against RIG-I, AGO2 and GFP. The amount of miR-1248 associated with RIG-I and AGO2 was enriched 50- and 125-fold, respectively, compared to that of GFP antibody pull-down control (Fig. 4D). These data demonstrate that miR-1248 can interact with either RIG-I or AGO2 in phSG cells, providing direct evidence of the dual role of miR-1248 as an inducer of IFN signaling initiated by the binding with RIG-I and as a canonical miRNA associated with AGO2.
3.8. MiR-1248 directly targets DAK and ITPR3 transcripts
To confirm the canonical miRNA function of miR-1248 and to identify its direct mRNA targets, we used the RNA22 miRNA target prediction database [39,40]. The genes ITPR3 and DAK were of interest since the role of ITPR3 in SS salivary gland dysfunction has been documented [27] and DAK plays a role as a repressor for type I IFN signaling pathway [41]. Therefore, we sought to further explore the effects of miR-1248 on these two target genes.
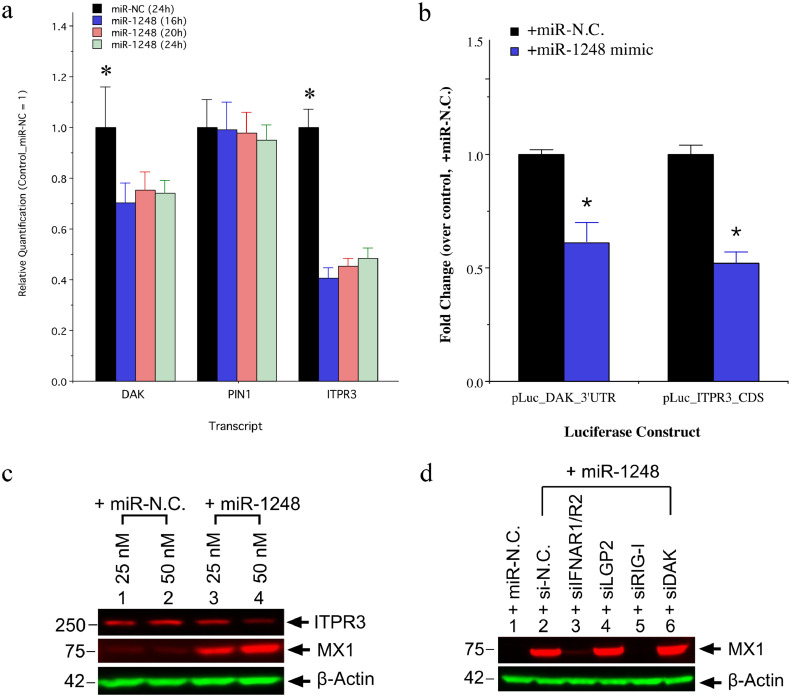
To confirm direct targeting of the ITPR3 and DAK transcripts by miR-1248, we measured their expression over time in miR-1248 mimic-transfected phSG cells. As shown in Fig. 5A, the level of DAK and ITPR3 transcript decreased about 30% and 55%, respectively, beginning as early as 16 h post-transfection compared to the miR-NC-transfected control sample. No reduction was observed in PIN1 transcript, which is another repressor for Type I IFN signaling pathway and was used as a control. Target prediction by RNA22 revealed two regions in the coding sequence of ITPR3 and one in DAK's 3′-UTR as potential miR-1248 binding sites. In addition, as shown in Fig. 5B, not only was the induction of MX1 expression dose-dependent (lane 3 and 4), but a reduction of ITPR3 protein level was also observed after miR-1248 mimic transfection (lane 4). To confirm these target sites, luciferase reporter constructs and, as shown in Fig. 5C, miR-1248 mimic down-regulated luciferase activities of the DAK-3′UTR and the ITPR3-CDS plasmids by 40% and 50%, respectively, compared to the miR-N.C.-transfected controls confirming functionally relevant binding of miR-1248 to both the ITPR3 and the DAK transcript.
Fig. 5.
Canonical targets of miR-1248 in phSG cells. (a) Down-regulation of DAK and ITPR3 expression was observed in the presence of miR-1248 by RT-qPCR. Relative quantification over the control (miR-NC) is presented as the mean from three independent experiments (*, p < .05, Student's t-test). (b) Decrease of ITPR3 in the presence of miR-1248 was also seen in the protein level, with a concomitant increase in the MX1 levels. The most significant decrease was observed with 50 nM of miR-1248. (c) Luciferase constructs containing the 3’-UTR of DAK or full-length CDS of ITPR3 were co-transfected with the indicated miRNAs into phSG cells. Luciferase activities were measured and the data are shown as fold change over each respective control (miR-N.C.-transfected). Data are presented as the mean from three independent experiments (mean ± S.E., *, p < .05, Student's t-test). (d) Knockdown of DAK enhances the induction of MX1 by miR-1248. Indicated siRNAs were transfected into phSG cells 24 h prior to the transfection of miR-1248. Cellular extracts were analyzed by Western blots for MX1 expression.
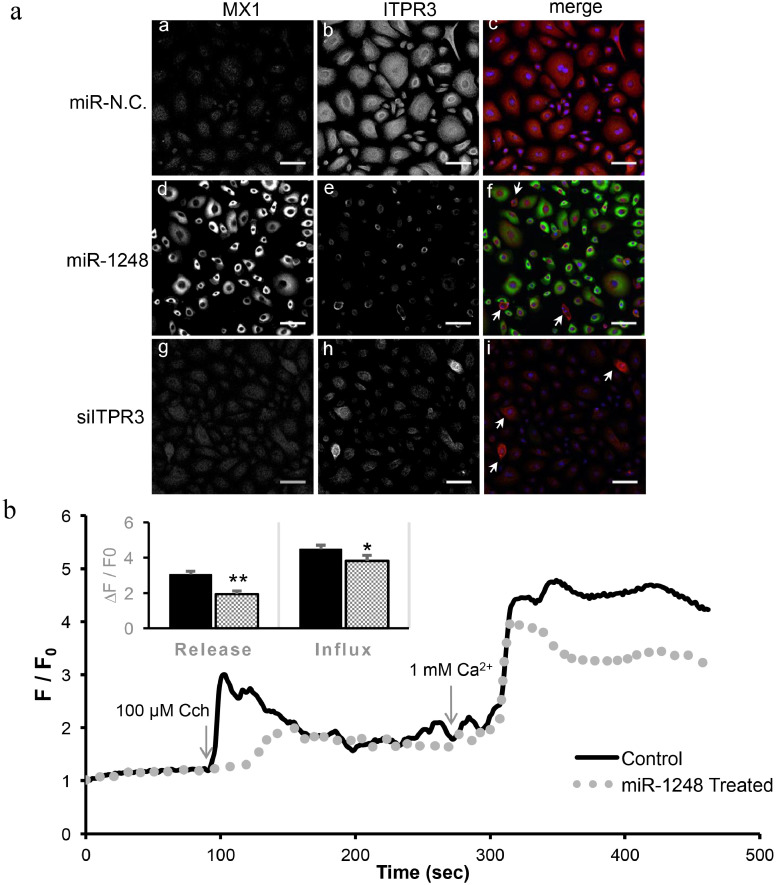
As an orthogonal approach to confirming the effect on ITPR3, double immunofluorescence staining was performed in miRNA-transfected phSG cells for ITPR3 and for MX1 which was used as an indicator for successful transfection. As shown in Fig. 6A, the control miR-N.C.-transfected cells (a-c) show faint staining of MX1 (a) and strong staining of ITPR3 (b) with a distribution in the cytoplasmic region. In contrast, the miR-1248 mimic-transfected cells display strong MX1 staining (d) and weak ITPR3 staining (e). There were few cells displaying strong ITPR3 signal with very weak or no MX1 staining (f, arrow) suggesting that these cells were most likely untransfected and that MX1 and ITPR3 staining were mutually exclusive. The siITPR3-transfected phSG cells used as a control, show background staining of MX1 (g) and relative weak ITPR3 (h) staining. Few untransfected cells exhibited strong ITPR3 staining (i, arrow).
Fig. 6.
Effect of miR-1248 on calcium signaling. (a) Immunofluorescence staining of phSG cells transfected with miR-N.C. (a-c), miR-1248 (d-f) or siRNA against ITPR3 (g-i). Cells were fixed and stained with antibodies to MX1 (a, d, g) or ITPR3 (b, e, h). Images (c, f, i) are merged between MX1 (green) and ITPR3 (red). DAPI was used for staining nuclei (blue). Arrows indicate the non-transfected cells. Bar = 50 μm. MX1 increase and ITPR3 decrease in the presence of miR-1248 in phSG cells. (b) HSG cells (control, solid line; miR-1248-treated, dotted line) were loaded with Fluo-4 and the mobility of calcium ion was monitored with CCh stimulation. Values are averages from 3 experiments in each group (minimum of 50 ROIs). Quantitation of first peak increase in fluorescence (representing internal Ca2+ release) for control HSG (3.045 ± 0.1882) and for miR-1248-treated cells (1.937 ± 0.1889). ** indicates significant difference (p < .0001, Student's t-test). Quantitation of second peak of fluorescence increase (due to Ca2+ entry) for control HSG cells (4.483 ± 0.2274) and for miR-1248-treated cells (3.827 ± 0.3112). * indicates a significant difference p < .05 (Student's t-test). Statistically significant differences were observed for both calcium release and influx in miR-1248 transfected cells. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.9. Canonical miRNA function of miR-1248 has functional effects on both Ca2+ and IFN signaling
To establish the functional alterations caused by the down-regulation of ITPR3 in the presence of miR-1248 and to separate the canonical and non-canonical effects of miR-1248, we examined CCh-induced [Ca2+]i increases in HSG cells by measuring changes in Fluo-4 fluorescence. Agonist-stimulated intracellular Ca2+ release was measured by administering 100 μM CCh in the absence of extracellular calcium, measured as a transient increase in fluorescence. Subsequent addition of 1 mM calcium to the external medium triggered a second increase in fluorescence due to Ca2+ entry. As shown in Fig. 6B, when compared to controls (HSG cells transfected with miR-N.C.) there is a 36% and 14% reduction in calcium release and calcium entry, respectively, in miR-1248-treated cells. Together, these data demonstrate that the reduction of ITPR3 by miR-1248 impairs the SOCE signaling pathway in HSG cells.
Besides the direct binding with RIG-I, we also studied the role of miR-1248 as a canonical miRNA involved in the IFN signaling pathway. Knockdown of AGO2 and RIG-I resulted in a 50% and 90% MX1 reduction, respectively, compared to control in pAC-Pre-miR-1248-transduced phSG cells (Fig. S6) and this reduction was also observed at the protein level (Fig. 4B). Furthermore, knockdown of DAK in miR-1248 mimic-transfected phSG cells resulted in a modest increase of MX1 expression (Fig. 5D, lane 6) compared to the control (lane 2), confirming the negative role of DAK in IFN induction. Together, these data confirm that miR-1248 targets the DAK transcript, contributing to the regulation of IFN signaling in phSG cells by regulating this negative regulator of IFN signaling as a canonical miRNA.
4. Discussion
SS pathobiology remains elusive. To understand the biological processes that underlie the immunological progression of SS, we applied deep sequencing and analysis based on well-characterized cellular alterations to identify key players and their function in the pathophysiology of salivary glands in SS. As miRNAs are considered as master regulators of cellular functions, we examined their functional role in SS in a systematic way.
Whole transcriptome profiling of phSG cells identified the calcium-regulated increase of a miRNA, miR-1248. Extensive whole transcriptome experiments of minor SGs of SS patients and healthy controls also identified miR-1248 to be significantly elevated in SS. Interestingly, the increased levels of this microRNA correlated with an increase in the expression of IFN signature genes, therefore we proceeded to further characterize the role of this miRNA in SS. In silico miRNA target prediction and exhaustive experiments with the phSG cell culture model confirmed two direct targets of miR-1248: DAK, a negative regulator of type I IFN, and ITPR3, a key component of calcium signaling. The use of whole tissue lysates does not does not account for the underlying glandular cell-type heterogeneity and this was a limitation that led us to examine the expression and effect of this miRNA in various types of cell lines.
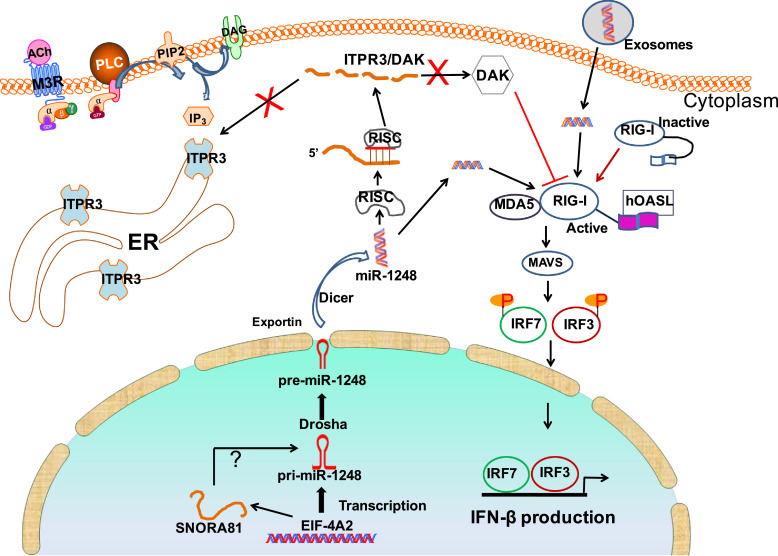
The down-regulation of DAK caused by miR-1248 synergistically enhances the activation of the IFN response, while down-regulation of ITPR3 provides a functional mechanism for a previously reported observation in SS salivary glands that has been shown to be a key player in SS-associated SG dysfunction [27]. Collectively, our data provide strong evidence toward a multifaceted role for miR-1248, presented as a graphical representation in Fig. 7. By modulating both IFN and calcium signaling in SS, miR-1248 affects two important functions of a target organ in this disease, and therefore, we propose that it serves as an underlying molecular mechanism involved in this autoimmune syndrome.
Fig. 7.
Schematic diagram of the proposed mechanisms in SS MSG. The miR-1248 plays a dual role by (1) associating with RIG-I and activating IFN-β synthesis and (2) down-regulating both DAK and ITPR3 expression in a canonical microRNA pathway. Reduction of DAK leads to an increase of IFN-β production, while reduction of ITPR3 impairs SOCE signaling and results in decreased salivary flow. The endogenous level of miR-1248 in acinar cells can be elevated from exogenous miR-1248 transported by the exosomes secreted from infiltrated lymphocytes. ER: endoplasmic reticulum; PLC: phospholipase C. M3R: muscarinic receptor 3.
Few studies have examined biological roles of miR-1248. Hooten et al., reported that miR-1248 increases in serum and could be used as a biomarker for aging [42]. In our study, the copy number of miR-1248 is poorly correlated with age (Spearman's rho = 0.2, p = .49), and shows moderate but non-significant correlation with focus score (rho = 0.43, p = .13), unstimulated salivary flow (rho = −0.47, p = .09), and stimulated salivary flow (rho = −0.48, p = .08). Copy number is significantly associated with presence of SSA and SSB autoantibodies (Kruskal Wallis p = .04), but not with C3 (p = .49) or C4 (p = .1). Differential expression of miR-1248 was found in serum of asthmatics patients and involved in up-regulation of IL-5 expression [43].
The genomic region containing miR-1248 is a complex locus with several overlapping annotated features in the RefSeq database. The annotated 5′-end of the miRNA precursor is only three bases upstream of the small nucleolar RNA (snoRNA), SNORA81 and both map to intron 7 of the protein-coding gene EIF4A2. This genomic organization, where the precursor sequence is almost completely nested within the snoRNA, poses a challenge for RNA-Seq studies. The consensus recommendation for summarizing gene counts in RNA-Seq data calls for discarding reads that fall within such an overlapping locus [29,44], since the true source of a read fragment is indistinguishable given the short-read sequencing strategy employed by most NGS platforms. Therefore, it is important to note that despite the high copy number we observed in patients using digital PCR, the upregulation of miR-1248 would not have been detectable using standard analysis techniques in whole transcriptome RNA-Seq expression comparisons between SS patients and healthy volunteers. Numerous studies have reported on the processing of snoRNAs into miRNAs, or snoRNAs performing miRNA-like function [[45], [46], [47]], but it remains unclear whether miR-1248 is a processed product of SNORA81 or if it is independently transcribed. Nevertheless, the elevation of miR-1248 in differentiated phSG cells and in SGs of SS patients suggests its expression is a calcium-dependent and disease-related event.
Transfection of miRNAs or siRNAs leading to an unexpected IFN induction has also been documented [[48], [49], [50]]. The unintended results in some cases were due to the use of liposome for delivery into cells [48]. Reynolds et al., observed that dsRNA sequences longer than 23 bp transfected in concentrations of 10–100 nM induced IFN response in a cell type-dependent manner [51]. However, Zhao et al., showed that miR-136, which is a 23mer miRNA, could activate the RIG-I signaling pathway, possibly through a direct association with RIG-I, but the precise binding mechanism between miR-136 and RIG-I was not clarified [50]. To exclude non-specific effects, we implemented the following specific strategies. First, based on the dose-response study of miR-1248 on MX1 expression, we used low concentrations (≤5 nM) of miR-1248 in most of the experiments, levels which have not been associated with non-specific activation of IFN. Second and most importantly, we expressed the pre-miR-1248 DNA construct and confirmed miR-1248-mediated IFN activation., The miR-1248-mediated IFN-β induction seems to be cell type-dependent since such IFN response was observed only in primary epithelial, but not in primary fibroblasts or in immortal cell lines like HSG, HeLa and HEK293 (Fig. S1).
It has been reported that transfection a 27mer dsRNA induces more IFN-response genes in HeLa S3 than in HEK293 cells [51]. This correlates with the degree of IFN induction on the expression of Toll-like receptor 3 (TLR3) and RNA-dependent protein kinase R (PRKR). However, in this study we suspect that the reason for the lack of induction of IFN-response in miR-1248-transfected cell lines is due to either low level or undetectable RIG-I protein as shown in the case of HSG cells (Fig. 4B, lane 1 and 2). It has been documented that RIG-I preferentially associates with short dsRNA, or a 5′-triphosphate group at the end of ssRNA longer than 19 mers with rich in U residues [52,53]. Also, some siRNAs or miRNAs contain immunostimulatory sequences (GUCCUUCAA or GU-rich) and can activate IFN pathways in certain cell types [[54], [55], [56]]. This motif is not present in miR-1248 – although the seed sequence, CCUUCUUG, comes close – so the precise nature the binding of miR-1248 to RIG-I remains unclear.
The length and sequence of viral RNAs or miRNAs acting as a ligand to RIG-I have been studied extensively, though no study has reported on the threshold level of RIG-I ligand needed to induce IFN signaling. Even as a three-fold increase of miR-1248 was observed in phSG grown in KGM-H (Fig. 1A), no MX1 induction was detected (Fig. 2A). However, under different experimental conditions, transfection as low as 1 nM of miR-1248 mimic into phSG cells was sufficient to detect MX1 induction after 48 h (Fig. 3B). Therefore, it is possible that miR-1248 expression needs to reach a certain threshold level in order to induce IFN signaling in phSG cells. Further studies are needed to identify the causes of the increased levels of this microRNA in the salivary epithelial cells. Animal experiments tracking the trafficking of exosomes enriched in miR-1248 between cells and tissues would dissect the impact of exogenous vs endogenous produce miR-1248 and which events occur first and drive the observed IFN release.
In summary, our findings identify an intriguing mechanism of the pathogenesis of SS and the ensuing SG dysfunction as they reveal a dual role of miR-1248 in human SG acinar cells; the activation of the IFN pathway by binding to RIG-I and the dysregulation of IFN signaling and calcium signaling by down-regulating DAK1 and ITPR3, respectively, through its canonical miRNA function. This provides important insights into the pathogenesis and potential therapeutic approaches for the protection of acinar cells from dysfunction in SS patients.
Funding
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Dental and Craniofacial Research (NIDCR).
Author contributions
Author's contribution: SJ: Designed study, collected data, analyzed and interpreted data, generated figures, wrote manuscript. MT: Collected data, analyzed and interpreted data, generated figures, wrote manuscript. LT: Collected data, analyzed data, generated figures, reviewed manuscript. CYZ: Collected data, analyzed data, reviewed manuscript. BMW: Collected data, generated table, provided feedback on the results, reviewed/edited the manuscript. IA: Designed study, analyzed and interpreted data, wrote manuscript.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, NIDCR. The authors acknowledge the work of the NIDCR/NIH Sjögren's Syndrome research nurses, Lolita Bebris and Eileen Pelayo, and patient coordinator, Donna Kelly for their valuable assistance in data collection and ensuring data integrity. The authors also acknowledge the work of Mohammad Hadavand and Hiba Mohiuddin for generating the digital PCR data.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.09.010.
Appendix A. Supplementary data
Supplementary material
References
- 1.Alamanos Y., Tsifetaki N., Voulgari P.V., Venetsanopoulou A.I., Siozos C., Drosos A.A. Epidemiology of primary Sjogren's syndrome in north-west Greece, 1982-2003. Rheumatology. 2006;45(2):187–191. doi: 10.1093/rheumatology/kei107. [DOI] [PubMed] [Google Scholar]
- 2.Helmick C.G., Felson D.T., Lawrence R.C., Gabriel S., Hirsch R., Kwoh C.K. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Arthritis Rheum. 2008;58(1):15–25. doi: 10.1002/art.23177. [DOI] [PubMed] [Google Scholar]
- 3.Billings M., Dye B.A., Iafolla T., Baer A.N., Grisius M., Alevizos I. Significance and implications of patient-reported xerostomia in Sjogren's syndrome: findings from the National Institutes of Health Cohort. EBioMedicine. 2016;12:270–279. doi: 10.1016/j.ebiom.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallo A., Jang S.I., Ong H.L., Perez P., Tandon M., Ambudkar I. Targeting the Ca(2+) sensor STIM1 by exosomal transfer of Ebv-miR-BART13-3p is associated with Sjogren's syndrome. EBioMedicine. 2016;10:216–226. doi: 10.1016/j.ebiom.2016.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bave U., Nordmark G., Lovgren T., Ronnelid J., Cajander S., Eloranta M.L. Activation of the type I interferon system in primary Sjogren's syndrome - a possible etiopathogenic mechanism. Arthritis Rheum. 2005;52(4):1185–1195. doi: 10.1002/art.20998. [DOI] [PubMed] [Google Scholar]
- 6.Emamian E.S., Leon J.M., Lessard C.J., Grandits M., Baechler E.C., Gaffney P.M. Peripheral blood gene expression profiling in Sjogren's syndrome. Genes Immun. 2009;10(4):285–296. doi: 10.1038/gene.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottenberg J.E., Busson M., Loiseau P., Cohen-Solal J., Lepage V., Charron D. TGF-beta 1 and TNF-alpha polymorphisms are associated with anti-SSB(La) ntibodies secretion in primary Sjogren's syndrome. Arthritis Rheum. 2003;48 doi: 10.1002/art.11103. [9):S258-S] [DOI] [PubMed] [Google Scholar]
- 8.Hjelmervik TO PK, Jonassen I., Jonsson R., Bolstad A.I. Gene expression profiling of minor salivary glands clearly distinguishes primary Sjögren's syndrome patients from healthy control subjects. Arthritis Rheum. 2005;52(5):1534–1544. doi: 10.1002/art.21006. [DOI] [PubMed] [Google Scholar]
- 9.Perez P., Anaya J.M., Aguilera S., Urzua U., Munroe D., Molina C. Gene expression and chromosomal location for susceptibility to Sjogren's syndrome. J Autoimmun. 2009;33(2):99–108. doi: 10.1016/j.jaut.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H. INTERFEROME v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41(D1) doi: 10.1093/nar/gks1215. (D1040-D6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brkic Z., Maria N.I., van Helden-Meeuwsen C.G., van de Merwe J.P., van Daele P.L., Dalm V.A. Prevalence of interferon type I signature in CD14 monocytes of patients with Sjogren's syndrome and association with disease activity and BAFF gene expression. Ann Rheum Dis. 2013;72(5):728–735. doi: 10.1136/annrheumdis-2012-201381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ambrosi A., Wahren-Herlenius M. Update on the immunobiology of Sjogren's syndrome. Curr Opin Rheumatol. 2015;27(5):468–475. doi: 10.1097/BOR.0000000000000195. [DOI] [PubMed] [Google Scholar]
- 13.Espinosa A., Dardalhon V., Brauner S., Ambrosi A., Higgs R., Quintana F.J. Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL-23-Th17 pathway. J Exp Med. 2009;206(8):1661–1671. doi: 10.1084/jem.20090585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theofilopoulos A.N., Baccala R., Beutler B., Kono D.H. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 15.Callis T.E., Chen J.F., Wan D.Z. MicroRNAs in skeletal and cardiac muscle development. DNA Cell Biol. 2007;26(4):219–225. doi: 10.1089/dna.2006.0556. [DOI] [PubMed] [Google Scholar]
- 16.Chen C.Z., Li L., Lodish H.F., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 17.Fabbri M., Ivan M., Cimmino A., Negrini M., Calin G.A. Regulatory mechanisms of microRNAs involvement in cancer: the strange case of Dr Jekyll and Mr Hyde. Expert Opin Biol Th. 2007;7(7):1009–1019. doi: 10.1517/14712598.7.7.1009. [DOI] [PubMed] [Google Scholar]
- 18.Mestdagh P., Feys T., Bernard N., Guenther S., Chen C., Speleman F. High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res. 2008;36(21):e143. doi: 10.1093/nar/gkn725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams A.E., Moschos S.A., Perry M.M., Barnes P.J., Lindsay M.A. Maternally imprinted microRNAs are differentially expressed during mouse and human lung development. Dev Dyn. 2007;236(2):572–580. doi: 10.1002/dvdy.21047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gourzi V.C., Kapsogeorgou E.K., Kyriakidis N.C., Tzioufas A.G. Study of microRNAs (miRNAs) that are predicted to target the autoantigens Ro/SSA and La/SSB in primary Sjogren's Syndrome. Clin Exp Immunol. 2015;182(1):14–22. doi: 10.1111/cei.12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapsogeorgou E.K., Gourzi V.C., Manoussakis M.N., Moutsopoulos H.M., Tzioufas A.G. Cellular microRNAs (miRNAs) and Sjogren's syndrome: candidate regulators of autoimmune response and autoantigen expression. J Autoimmun. 2011;37(2):129–135. doi: 10.1016/j.jaut.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y., Peng L., Ma W., Yi F., Zhang Z., Chen H. Autoantigen-targeting microRNAs in Sjogren's syndrome. Clin Rheumatol. 2016;35(4):911–917. doi: 10.1007/s10067-016-3203-3. [DOI] [PubMed] [Google Scholar]
- 23.Singh R.P., Massachi I., Manickavel S., Singh S., Rao N.P., Hasan S. The role of miRNA in inflammation and autoimmunity. Autoimmun Rev. 2013;12(12):1160–1165. doi: 10.1016/j.autrev.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Chan V.S., Nie Y.J., Shen N., Yan S., Mok M.Y., Lau C.S. Distinct roles of myeloid and plasmacytoid dendritic cells in systemic lupus erythematosus. Autoimmun Rev. 2012;11(12):890–897. doi: 10.1016/j.autrev.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Xu W.D., Lu M.M., Pan H.F., Ye D.Q. Association of MicroRNA-146a with autoimmune diseases. Inflammation. 2012;35(4):1525–1529. doi: 10.1007/s10753-012-9467-0. [DOI] [PubMed] [Google Scholar]
- 26.Bookman A.A.M., Shen H., Cook R.J., Bailey D., McComb R.J., Rutka J.A. Whole stimulated salivary flow correlation with the pathology of inflammation and damage in minor salivary gland biopsy specimens from patients with primary Sjogren's syndrome but not patients with sicca. Arthritis Rheum. 2011;63(7):2014–2020. doi: 10.1002/art.30295. [DOI] [PubMed] [Google Scholar]
- 27.Teos L., Zhang J., Hak J., Swaim W., Ambrus J., Grisius M. IP3R deficit in acinar cells underlies loss of salivary gland fluid secretion in the autoimmune Exocrinopathy, Sjogren's syndrome. Scand J Immunol. 2015;81(5):359–360. [Google Scholar]
- 28.Jang S.I., Ong H.L., Gallo A., Liu X., Illei G., Alevizos I. Establishment of functional acinar-like cultures from human salivary glands. J Dent Res. 2015;94(2):304–311. doi: 10.1177/0022034514559251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 30.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirou K.A., Lee C., George S., Louca K., Papagiannis I.G., Peterson M.G. Coordinate overexpression of interferon-alpha-induced genes in systemic lupus erythematosus. Arthritis Rheum. 2004;50(12):3958–3967. doi: 10.1002/art.20798. [DOI] [PubMed] [Google Scholar]
- 32.Samarajiwa S.A., Forster S., Auchettl K., Hertzog P.J. INTERFEROME: the database of interferon regulated genes. Nucleic Acids Res. 2009;37(Database issue):D852–D857. doi: 10.1093/nar/gkn732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ambudkar I.S. Calcium signalling in salivary gland physiology and dysfunction. J Physiol-London. 2016;594(11):2813–2824. doi: 10.1113/JP271143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honda K., Yanai H., Takaoka A., Taniguchi T. Regulation of the type I IFN induction: a current view. Int Immunol. 2005;17(11):1367–1378. doi: 10.1093/intimm/dxh318. [DOI] [PubMed] [Google Scholar]
- 36.Oshiumi H., Sakai K., Matsumoto M., Seya T. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. Eur J Immunol. 2010;40(4):940–948. doi: 10.1002/eji.200940203. [DOI] [PubMed] [Google Scholar]
- 37.Kawai T., Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7(2):131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 38.Meylan E., Tschopp J., Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442(7098):39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 39.Loher P., Rigoutsos I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics. 2012;28(24):3322–3323. doi: 10.1093/bioinformatics/bts615. [DOI] [PubMed] [Google Scholar]
- 40.Miranda K.C., Huynh T., Tay Y., Ang Y.S., Tam W.L., Thomson A.M. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 41.Diao F.C., Li S., Tian Y., Zhang M., Xu L.G., Zhang Y. Negative regulation of MDA5-but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. P Natl Acad Sci USA. 2007;104(28):11706–11711. doi: 10.1073/pnas.0700544104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hooten N.N., Fitzpatrick M., Wood W.H., De S., Ejiogu N., Zhang Y.Q. Age-related changes in microRNA levels in serum. Aging-Us. 2013;5(10):725–740. doi: 10.18632/aging.100603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panganiban RP1 P.M., Maru S.Y., Jefferson S.J., Roff A.N., Ishmael F.T. Differential microRNA epression in asthma and the role of miR-1248 in regulation of IL-5. Am J Clin Exp Immunol. 2012:154–165. (Nov 15;1(2) [PMC free article] [PubMed] [Google Scholar]
- 44.Anders S., Pyl P.T., Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai B.Y., Yegnasubramanian S., Wheelan S.J., Laiho M. RNA-Seq of the nucleolus reveals abundant SNORD44-derived small RNAs. Plos One. 2014;9(9) doi: 10.1371/journal.pone.0107519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ender C., Krek A., Friedlander M.R., Beitzinger M., Weinmann L., Chen W. A human snoRNA with MicroRNA-like functions. Mol Cell. 2008;32(4):519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 47.Scott M.S., Ono M. From snoRNA to miRNA: dual function regulatory non-coding RNAs. Biochimie. 2011;93(11):1987–1992. doi: 10.1016/j.biochi.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karlsen T.A., Brinchmann J.E. Liposome delivery of MicroRNA-145 to mesenchymal stem cells leads to immunological off-target effects mediated by RIG-I. Mol Ther. 2013;21(6):1169–1181. doi: 10.1038/mt.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olejniczak M., Galka P., Krzyzosiak W.J. Sequence-non-specific effects of RNA interference triggers and microRNA regulators. Nucleic Acids Res. 2010;38(1):1–16. doi: 10.1093/nar/gkp829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao L.Z., Zhu J.P., Zhou H.B., Zhao Z.Z., Zou Z., Liu X.K. Identification of cellular microRNA-136 as a dual regulator of RIG-I-mediated innate immunity that antagonizes H5N1 IAV replication in A549 cells. Sci Rep-Uk. 2015;5 doi: 10.1038/srep14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reynolds A., Anderson E.M., Vermeulen A., Fedorov Y., Robinson K., Leake D. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. Rna. 2006;12(6):988–993. doi: 10.1261/rna.2340906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baum A., Sachidanandam R., Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A. 2010;107(37):16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ranjith-Kumar C.T., Murali A., Dong W., Srisathiyanarayanan D., Vaughan R., Ortiz-Alacantara J. Agonist and antagonist recognition by RIG-I, a cytoplasmic innate immunity receptor. J Biol Chem. 2009;284(2):1155–1165. doi: 10.1074/jbc.M806219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hornung V., Ellegast J., Kim S., Brzozka K., Jung A., Kato H. 5 '-triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 55.Hornung V., Guenthner-Biller M., Bourquin C., Ablasser A., Schlee M., Uematsu S. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11(3):263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 56.Judge A.D., Sood V., Shaw J.R., Fang D., McClintock K., MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23(4):457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material