Summary
To overcome nitrogen deficiencies in the soil, legumes enter symbioses with rhizobial bacteria that convert atmospheric nitrogen into ammonium. Rhizobia are accommodated as endosymbionts within lateral root organs called nodules that initiate from the inner layers of Medicago truncatula roots in response to rhizobial perception. In contrast, lateral roots emerge from predefined founder cells as an adaptive response to environmental stimuli, including water and nutrient availability. CYTOKININ RESPONSE 1 (CRE1)-mediated signaling in the pericycle and in the cortex is necessary and sufficient for nodulation, whereas cytokinin is antagonistic to lateral root development, with cre1 showing increased lateral root emergence and decreased nodulation. To better understand the relatedness between nodule and lateral root development, we undertook a comparative analysis of these two root developmental programs. Here, we demonstrate that despite differential induction, lateral roots and nodules share overlapping developmental programs, with mutants in LOB-DOMAIN PROTEIN 16 (LBD16) showing equivalent defects in nodule and lateral root initiation. The cytokinin-inducible transcription factor NODULE INCEPTION (NIN) allows induction of this program during nodulation through activation of LBD16 that promotes auxin biosynthesis via transcriptional induction of STYLISH (STY) and YUCCAs (YUC). We conclude that cytokinin facilitates local auxin accumulation through NIN promotion of LBD16, which activates a nodule developmental program overlapping with that induced during lateral root initiation.
Keywords: nitrogen, endosymbiosis, rhizobia, lateral root/nodule organogenesis, NODULE INCEPTION, CYTOKININ RESPONSE FACTOR, LATERAL ORGAN BOUNDARIES DOMAIN, YUCCA, auxin, Medicago truncatula
Graphical Abstract
Highlights
-
•
Lateral roots and nodules show extensive overlap in organogenesis and transcription
-
•
Nodule and lateral root initiation converges at an auxin maximum and LBD16
-
•
Cytokinin promotes auxin biosynthesis via NIN and LBD16 during nodule initiation
To overcome growth-limiting nitrogen deficiency in soils, legumes host nitrogen-fixing bacteria in symbiotic root nodules. Schiessl et al. show that despite different environmental stimuli and organ function, nodule and lateral root initiation converges on a local accumulation of the plant hormone auxin and a set of auxin-responsive regulators.
Introduction
Nodules initiate as lateral root organs in response to the perception of rhizobial bacteria at the root surface. Rhizobial nodulation (Nod) factors activate symbiosis signaling in root epidermal cells, which in turn activates cytokinin signaling in the root cortex and pericycle [1, 2, 3, 4]. CYTOKININ RESPONSE 1 (CRE1)-mediated signaling in inner-root tissues leads to the induction of the symbiosis-specific transcription factor NODULE INCEPTION (NIN), and both CRE1 and NIN are indispensable for nodule initiation [5, 6, 7, 8, 9, 10]. While cytokinin signaling is both necessary and sufficient for the induction of nodules [7, 8], cytokinin suppresses lateral root development [11, 12, 13, 14], with cre1 mutants showing increased lateral root emergence and decreased nodulation [13, 15].
Nodules and lateral roots initiate from pericycle, endodermal, and inner-cortical cells as a function of local auxin accumulation [16, 17]. Accompanying both lateral root and nodule development is upregulation of auxin-responsive WOX5 and PLETHORAs at the initiation site of both organs [18, 19]. While lateral roots form from founder cells that are proposed to be primed by periodically oscillating auxin maxima, there is no evidence that nodules originate from such predefined founder cells [17, 20, 21]. Rather, the initiation of an auxin maxima during nodulation has been proposed to result from suppression of rootward polar auxin transport below the site of rhizobial recognition as a function of cytokinin recognition by CRE1 [3, 22, 23, 24]. Pharmacological suppression of auxin transport promotes nodule organogenesis [22, 25], while genetic suppression of auxin transporters blocks nodule organogenesis [26, 27]. This work, together with computational modeling [28], has led to the proposition that symbiotic induction of cytokinin signaling blocks polar auxin transport below the site of rhizobial recognition, leading to a localized auxin maximum that coordinates nodule organogenesis.
These previous studies indicate that a number of parallels can be drawn between nodule and lateral root development, but their modes of initiation differ significantly. In this study, we directly compared lateral root and nodule development with high spatial and temporal resolution to identify the commonalities and differences that underlie their development. We demonstrate that lateral roots and nodules share overlapping developmental programs that converge on the formation and interpretation of an auxin maximum. This is exemplified by our finding that auxin-responsive LOB-DOMAIN PROTEIN 16 (LBD16) is required for formation of both nodule and lateral root primordia. NIN and LBD16 are necessary for cytokinin promotion of the auxin biosynthesis regulators STYLISH (STY) and YUCCAs (YUC), suggesting that the recruitment of NIN and LBD16 into a symbiotic response allows cytokinin promotion of a root developmental program during nodulation.
Results
Lateral Roots and Nodules Show Extensive Overlap in Development and Transcription
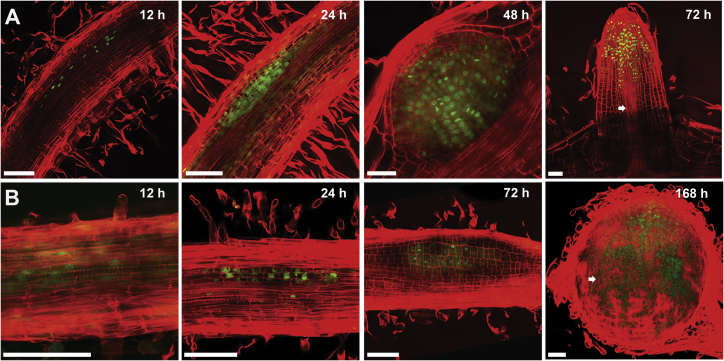
To better understand the commonalities and differences in lateral root and nodule development, we compared their organogenesis and correlated this with changes in gene expression. To initiate lateral roots in Medicago truncatula, we turned 2-day-old seedlings 135° to create a bend in the root [29], while nodules were induced with droplets of Sinorhizobium meliloti culture applied on the root susceptibility zone (Figures S1A and S1B). For both organs, we observed initial cell-cycle activation 12 h post induction (hpi) using the fluorescently labeled nucleotide analog 5-ethynyl-2-deoxyuridine (EdU). Anticlinal cell divisions initiated at 16 hpi and by 24 hpi primordia of both organs consisted of several cell layers, implying multiple rounds of periclinal cell divisions (Figures 1A and 1B). Lateral root primordia developed consistently faster than nodules, with cone shaped primordia at 48 hpi and fully emerged lateral roots at 72 hpi, whereas nodules developed as flat and concealed primordia up to 72 hpi, with nodule emergence occurring between 120–168 hpi (Figures 1A and 1B).
Figure 1.
Lateral Roots and Nodules Show Overlapping Development
(A and B) (A) Optical sections of lateral roots and (B) nodules hours (h) post induction. Red propidium iodide demarks cell walls and green EdU-labeled nuclei DNA replication. Arrowheads indicate vascular strands that in lateral roots are apparent by 72 hpi compared to nodules at 120–168 hpi. Scale bars:100 μm.
See also Figure S1.
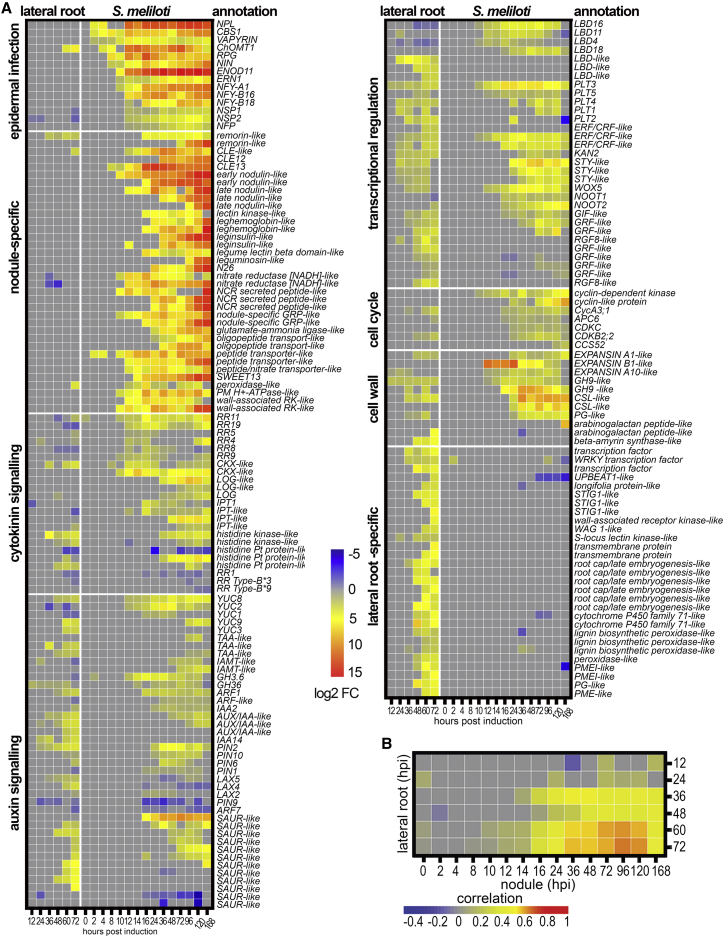
To correlate these developmental processes with gene expression changes, we performed RNA sequencing (RNA-seq) on 2- to 3-mm segments of gravitropically stimulated and non-stimulated roots at six time points between 12 and 72 hpi and of S. meliloti or mock spot-inoculated roots at 15 time points from 0 to 168 hpi (Figures 2A, and S1D–S1F). This revealed a high overlap in gene expression changes: 74% of upregulated genes and 81% of downregulated genes in lateral root development were similarly responsive in nodulation (Figures 2A, 2B, and S2; Data S1).
Figure 2.
Lateral Roots and Nodules Show Overlap in Gene Expression
(A) Heatmap showing selected genes induced during lateral root and nodule development with fold changes ≥±1.5; p < 0.05. Expression depicts log2 fold changes.
(B) Correlation heatmap depicting the overlap between genes differentially expressed during lateral root and nodule organogenesis over a time course of development.
See also Figures S1 and S2 and Data S1.
The earliest-responding genes to S. meliloti inoculation at 2 hpi were genes previously described as activated in the root epidermis [30]: NPL, CBS1, VAPYRIN, ChOMT, RPG, NIN, and NF-YA1 (Figures 2A and S2; Data S1). Reporters for cytokinin signaling were induced with the onset of cell-cycle activation in the root cortex and pericycle at 10–12 hpi, and these showed little or no response during lateral root initiation (Figures 2A and S2; Data S1). Genes associated with auxin biosynthesis and signaling, together with a set of auxin-responsive transcriptional regulators, were among the earliest-upregulated genes with overlapping expression during lateral root and nodule development: YUC, PLETHORAs (PLT), and STY, previously shown to regulate auxin metabolism, transport, and signaling during lateral root development in Arabidopsis [31, 32, 33, 34]. The upregulation of auxin-related genes coincides with the first significant auxin responses that occurred 12 h post S. meliloti inoculation, as evidenced by the auxin-response reporter DIRECT REPEAT5 driving green fluorescent protein (DR5-GFP) (Figure S1C).
LBD16 Represents a Point of Convergence between Nodule and Lateral Root Development
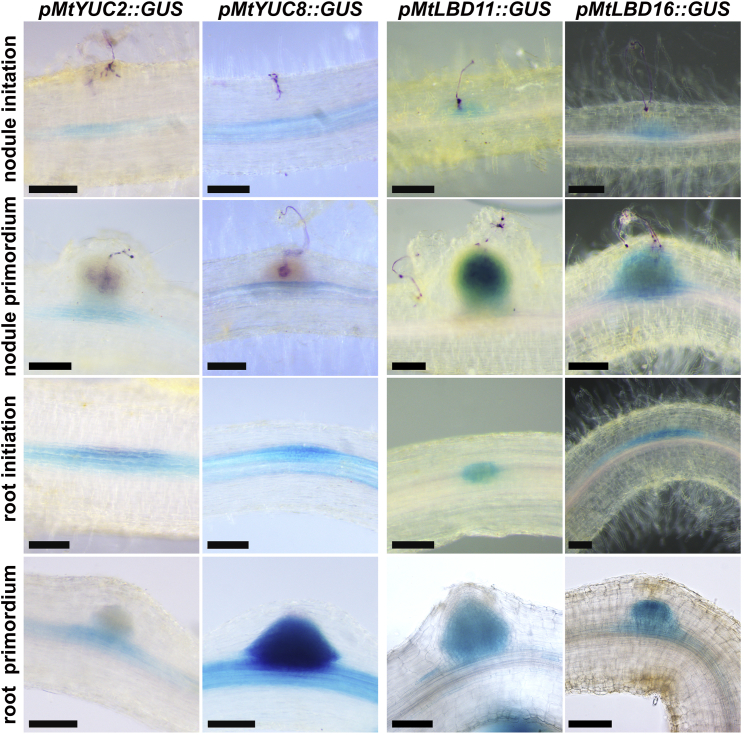
During lateral root initiation in Arabidopsis, YUC and LBDs are activated and associated with the regulation and response to auxin [35, 36]. In our transcriptional profiling, we found YUC2, YUC8, LBD11, and LBD16 induction within 12 h in response to rhizobial treatment, which was coordinated with the overall auxin response (Figures 2A, S1C, S2, and S3A; Data S2A and S2B). Promoter-β-glucuronidase (GUS) analysis of these four genes revealed that all were induced at the sites of lateral root and nodule initiation very early during primordia development (Figure 3).
Figure 3.
LATERAL ORGAN BOUNDARIES and YUCCAs Are Expressed during Root and Nodule Primordium Initiation and Development
Expression patterns of YUC2, YUC8, LBD11, and LBD16 during nodule and lateral root development visualized by GUS staining (blue). Rhizobial-expressed LacZ is stained magenta. Scale bars: 100 μm.
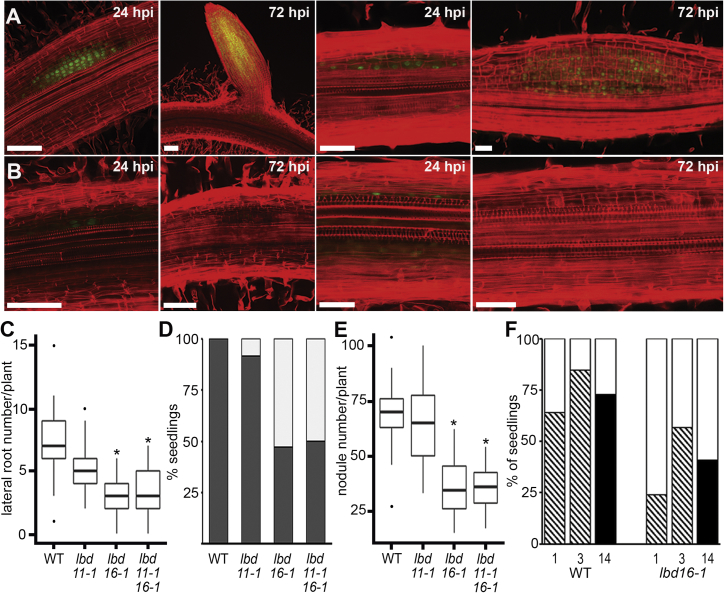
To further assess the importance of LBDs, we identified mutants in LBD11 (lbd11-1) and LBD16 (lbd16-1, lbd16-2) (Figures S3B–S3G). Mutants in lbd16 and lbd11lbd16 showed significant defects in root organogenesis, with 50% reductions in the number of emerged lateral roots and nodules (Figures 4C–4F, S3D–S3G, and S4). This reduction is associated with early defects in cell division during primordium formation. In the most severe cases, root sections with no primordia emerging 72 hpi appeared to have terminated development after a few divisions (Figures 4A and 4B). From this, we infer that LBD16 represents a convergence point between lateral root and nodule development, being important for the promotion of cell proliferation during organ initiation and early primordium development. Consistent with an early role in primordium formation, lbd16 and lbd11lbd16 mutants showed dramatic reductions in nodule-associated gene expression: 93% of rhizobial-induced genes at 24 hpi are LBD16 dependent (Figures 5A, 5B, and S2; Data S1). This includes auxin-signaling genes, auxin regulators such as STYLISH and YUCCAs, and cell-wall-remodeling genes, all of which have overlapping expression in nodules and lateral roots.
Figure 4.
Lateral Root and Nodule Number Are Reduced in lbd16
(A and B) Optical sections of lateral roots and nodules in (A) wild-type (WT) and (B) lbd16-1 at 24 or 72 hpi. Scale bars: 50 μm.
(C) Lateral root number in 14-day-old seedlings. Boxplots show median (thick line), second to third quartiles (box), minimum and maximum ranges (lines), and outliers (single points). A one-way Kruskal-Wallis rank-sum test showed that lateral root number is dependent on genotype; asterisks indicate significantly different (95% confidence) means compared with WT. n = 56 (WT), 58 (lbd11-1), 64 (lbd16-1), and 66 (lbd11lbd16).
(D) Percentage of gravi-stimulated seedlings with ≥1 lateral roots (dark gray) or 0 lateral roots (white) in the bend 5 dpi in WT (n = 25), lbd11-1 (n = 60), lbd16-1 (n = 53), and lbd11lbd16 (n = 60) showing significant reduction in the number of emerging lateral roots in lbd16-1 and lbd11lbd16 compared to WT. p = 0.166, 7.401e−07, and 1.413e−06 respectively; Fisher’s exact test.
(E) Nodule number 21 days post S. meliloti inoculation. n = 13 (WT), 15 (lbd11-1), 14 (lbd16-1), and 14 (lbd11lbd16). A normal distribution allowed a one-way ANOVA test revealing the mean number of nodules differed significantly between genotypes; asterisks indicate significantly different (95% confidence) means compared to WT.
(F) Percentage of seedlings with ≥1 primordia (hashed), ≥1 emerged nodule (black), or no structure (white) developing at the spot inoculation site at 1, 3, and 14 days post inoculation (dpi; n = 116 WT and 142 lbd16-1). lbd16-1 showed significantly different rates of initiation of primordia or nodules at all time points: p = 0.003 (1 dpi), 0.022 (3 dpi), and 2.084e−04 (14 dpi); Fisher’s exact test.
See also Figures S3 and S4 and Data S2A.
Figure 5.
The Impact of CRE1, NIN, and LBD16 on Nodulation-Associated Gene Expression
(A) Heatmap of selected genes (as in Figure 2; see also Figure S2 for gene identifiers and Data S1) in WT (jemalong), cre1-1, and nin-1 root sections at 12 and 24 h and WT (R108), lbd16-1 (ls), and lbd11lbd16 (ld) at 24 h post S. meliloti spot inoculation; response to LBD11 (11) and LBD16 (16) overexpression in 3-week-old hairy roots compared to control roots and during lateral root induction. Expression represents log2 fold changes.
(B) Pairwise comparisons of all differentially expressed genes dependent on cre1-1 (blue), nin-1 (red), and lbd16-1 (purple). cre1-1 and nin-1 comparisons were to WT jemalong and lbd16-1 to WT R108.
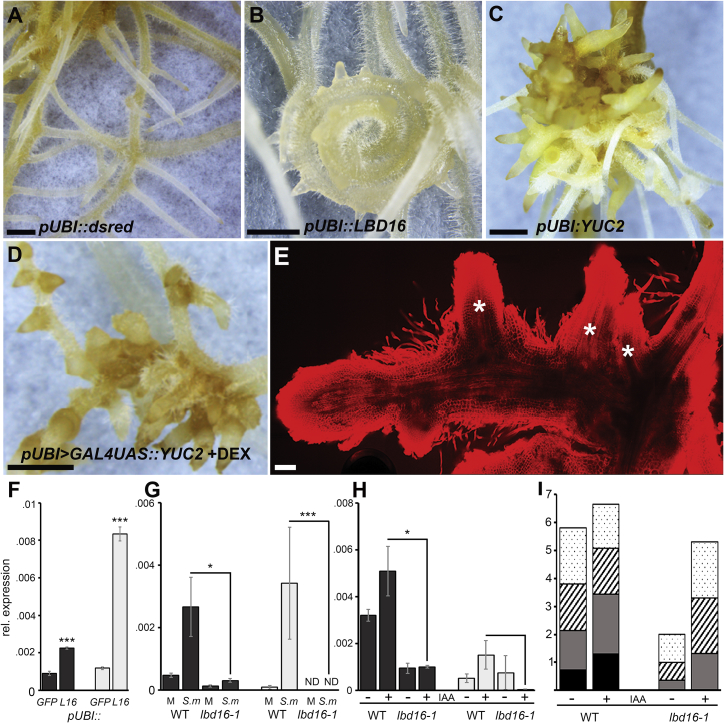
Overexpression of LBD16 or YUC2 Is Sufficient to Promote Root Primordia Formation
LBD16 in Arabidopsis is induced by a localized auxin maximum and controls initial cell divisions during lateral root formation [32, 36, 37]. Our LBD16 mutants in M. truncatula imply a similar function in this species. To further understand the role that LBD16 plays during root organogenesis, we overexpressed LBD16 in M. truncatula using the Lotus japonicus UBIQUITIN1 (LjUBI) promoter. Roots overexpressing LBD16 showed extensive curling with ectopic root primordia initiation (Figures 6A, 6B, S5A, and S5B). LBD16 overexpression was associated with the constitutive induction of two STY-like transcription factors and these same STY genes were dependent on LBD16 for their induction by rhizobia (Figures 5A, 6F, 6G, S2, and S5K). In Arabidopsis, STY control the regulation of the YUC auxin biosynthesis genes [33]. Expression of YUC2 from the constitutive LjUBI promoter or induced through dexamethasone regulation resulted in initiation and emergence of ectopic root primordia (Figures 6C–6E, and S5C–S5G). From this, we conclude that LBD16, most likely through the regulation of auxin biosynthesis via STY, controls root organ initiation. However, the overexpression of LBD16 led primarily to the expression of auxin-associated genes, but not the induction of nodule or lateral root-specific genes (Figure 5A). We conclude that induction of LBD16 or promotion of auxin biosynthesis is sufficient to activate root primordia formation, but additional factors need to be coordinately induced to give rise to nodule or lateral root identity.
Figure 6.
Overexpression of LBD16 or YUC2 Is Sufficient to Promote Root Primordia Formation
(A–E) Constitutive expression of (A) dsred (control), (B) LBD16, or (C) YUC2 under control of the LjUBI promoter in hairy roots. 44 out of 103 and 53 out of 57 transformed plants expressing pLjUBI:LBD16 and pLjUBI:YUC2, respectively, showed similar phenotypes as depicted in (B) and (C).
(D and E) (D) Bright field image and (E) optical sections of propidium iodide-stained root structures expressing dexamethasone (Dex)-inducible YUC2 under control of the LjUBI promoter (pLjUBI > GAL4UAS::MtYUC2). 82 out of 167 plants transformed with pLjUBI > GAL4UAS::MtYUC2 showed ectopic primordium induction 2 weeks post Dex treatment as depicted in (D) and (E) and Figures S5C–S5G compared to 3 out of 147 Dex treated plants transformed with pLjUBI > GAL4UAS::GFP. Ectopic primordia in (E) are indicated with asterisks. Scale bars: (A–D) 1 mm and (E) 50 μm.
(F–H). (F) Quantification of transcript levels by qRT-PCR of STY-like (Medtr1g023320) (dark gray bars) and STY-like (Medtr8g076620) (light gray bars) in hairy roots constitutively expressing LBD16 or GFP (pUBI::LBD16 or pUBI::GFP), (G) S. meliloti and mock spot inoculation, and (H) IAA (+) and mock (−) treatment in WT and lbd16-1 root sections at 24 hpi. Expression levels were measured by qRT-PCR and normalized to HH3. Statistical comparisons were performed as indicated. Values are the mean of 3 biological replicates ± SEM (Student’s t test; asterisks indicate statistical significance; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
(I) Number of lateral root primordia in 3-day-old WT and lbd16-1 seedlings 24 h post IAA or mock treatment represented as means (n ≥ 11). The different stages of primordia development are indicated: dotted, stages I–II; striped, stage III; light gray, stages IV–V; black, emerged. Mock-treated lbd16-1 seedlings had significantly fewer primordia compared to WT (Student’s t test; p < 0.001). IAA treatment significantly increased primordia number in lbd16-1 seedlings (Student’s t test; p < 0.001), but not in WT. More stages IV–V and emerged primordia developed in WT than in lbd16-1 in both treatments (Student’s t test; p < 0.001).
See also Figures S5 and S6.
Similar to what has been reported in Arabidopsis [36], we found that indole-3-acetic acid (IAA) treatment of M. truncatula roots activates LBD16 expression (Figure S5H), revealing that LBD16 is responsive to auxin accumulation. IAA treatment of lbd16-1 partially rescued the lateral root phenotype, enhancing the number of root primordia that formed. However, primordia emergence was still defective in lbd16-1 (Figure 6I). Despite this partial rescue of the mutant phenotype by IAA treatment, we observed that the induction by both rhizobia and IAA of STY, PLT3, and a cell-wall-modifying POLYGALACTURONASE-like gene was abolished in lbd16-1 (Figures 6H, S5I, and S5J). We conclude that LBD16 is both responsive to auxin and responsible for the promotion of auxin, implying that LBD16 has a function in the amplification of auxin accumulation.
Cytokinin Promotion of STY and YUC Is a Function of NIN and LBD16
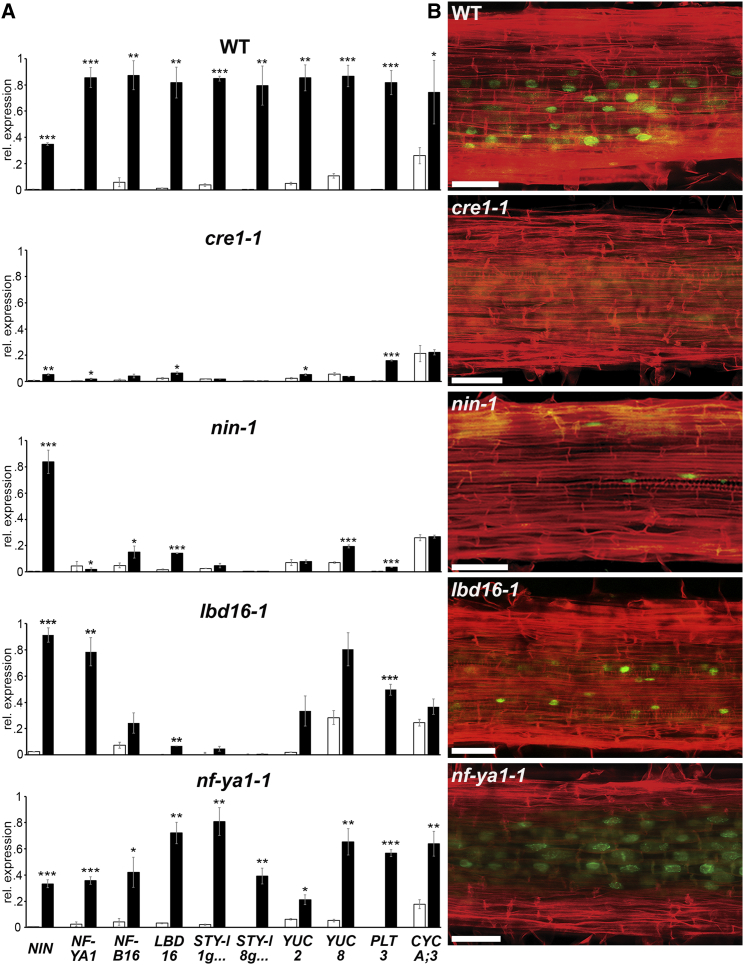
Induction of nodules in response to rhizobial bacteria is dependent on the cytokinin receptor CRE1 and the cytokinin-inducible transcription factor NIN [4, 5, 9, 13]. Consistent with this, RNA-seq on S. meliloti spot-inoculated root segments of the cre1-1 or nin-1 mutants at 24 hpi revealed that 98.7% and 95.7% of S. meliloti-responsive genes are dependent on CRE1 or NIN, respectively. Genes dependent on LBD16 showed almost complete overlap with CRE1 and NIN dependencies with the exception of infection-associated genes in the root epidermis and some nodule-specific genes (Figures 5A, 5B, and S2). Such a large overlap between NIN- and LBD16-dependent gene expression suggested the possibility that LBD16 may function downstream of NIN and may be required for NIN induction of nodule initiation. Consistent with such a hypothesis, we observed that overexpression of NIN is sufficient to activate LBD16 expression along with its known target NF-YA1 [10] (Figure S5L).
During nodulation, auxin accumulation and auxin signaling are activated by cytokinin at the site of rhizobial infection in a CRE1-dependent manner [22, 38], and we found that auxin accumulation in spot-inoculated root sections was CRE1- and NIN-dependent (Figure S6D). Wild-type M. truncatula roots respond to cytokinin treatments with upregulation of nodule-associated transcriptional regulators including NIN, NF-YA1, and NF-YB16 and auxin-associated genes including LBD16, PLT3, STY, and YUC (Figures 7A and S6B). Furthermore, such cytokinin treatment is sufficient to promote cell-cycle activation in the root cortex, as evidenced by EdU staining (Figures 7B, S6A, and S6C). Such cytokinin responses are abolished in cre-1-1 and nin-1, consistent with their role in cytokinin signaling and response. While cytokinin treatment of lbd16-1 resulted in NIN and NF-YA1 induction, we found that the induction of STY and YUC by cytokinin was significantly reduced in lbd16-1, as was the induction of CYCLINA;3. By contrast, loss of NF-YA1-1 function appeared to have little effect on gene regulation and cell-cycle activation by cytokinin. We conclude that cytokinin promotion of auxin biosynthesis and cell division, which appears to uniquely occur in roots during nodulation, is the function of NIN and LBD16.
Figure 7.
NIN and LBD16 Mediate Auxin Regulators and Cell-Cycle Activation in Response to Cytokinin
(A) Expression profiling on root segments treated with 100 nM (6-Benzylaminopurine) BAP for 24 h by qRT-PCR normalized to HH3. Statistical comparisons were performed between mock (white bars) and BAP (black bars). Values are the mean ΔCt values of three biological replicates normalized to the maximum value obtained for that gene within the ecotype. Data are presented ± SEM (Student’s t test; ∗p < 0.05; ∗∗p < 0.01, ∗∗∗p < 0.001).
(B) Representative optical sections (≥20 roots analyzed) in root segments (susceptibility zone) treated with 100 nM BAP. Red, cell walls; green, cell-cycle activation. Scale bars: 50 μm.
See also Figures S6A–S6C.
Discussion
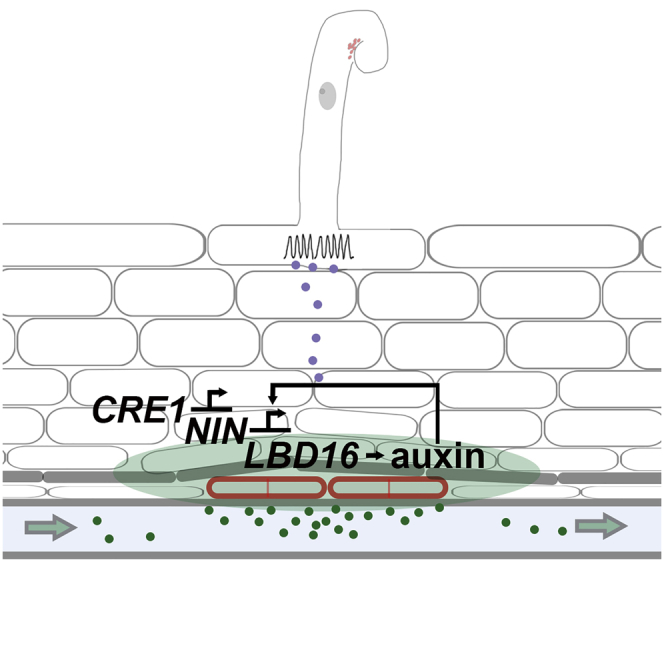
Our work demonstrates that lateral root and nodule development converge on a core root developmental program primarily associated with the formation and interpretation of a localized auxin maximum. The induction of this core developmental program is in part dependent on LBD16, which shows similar patterns of expression during nodule and lateral root development. LBD16 has been shown to promote root organogenesis in response to an array of environmental stimuli, including hydropatterning and wounding [39, 40], and our work further supports the essential role that LBD16 plays as a key integrator in the adaptation of root system architecture. We propose that the integration of LBD16 into the symbiotic response to rhizobial bacteria is responsible for the recruitment of a core root developmental program and represents a point of convergence between nodule and lateral root development.
Nodulation is restricted to a group of plants in the so-called “nitrogen-fixing clade” and is specifically associated with the accommodation of nitrogen-fixing bacteria. A diversity of species within this clade show nodulation, and recent studies imply a single origin for the emergence of nodulation within this clade [41, 42]. Actinorhizal species (nodulating species outside the legumes) show nodules that possess a centralized vasculature [43], similar in morphology to lateral roots. Considering the new insights in nodule evolution, it seems likely that the unique architecture of the legume nodule is a derived state from the more primitive structures shown by actinorhizal plants. This is consistent with our studies that demonstrate a 75% overlap in the gene expression changes induced in lateral roots and nodules. We suggest that the initial stages of nodule evolution involved the recruitment of lateral root organogenesis into a symbiotic program, with later adaptations that created the unique features of the nodule.
The overlap between nodules and lateral roots appears to primarily converge on the response to an auxin maxima at the site of nodule and lateral root initiation. The auxin maxima during nodule formation was thought to be primarily caused by cytokinin and/or flavonoid-derived suppression of polar auxin transport [3, 22, 23]. However, here, we show that YUCs are activated below the site of the developing nodule primordia, and this precedes marker genes for auxin responses. We suggest that it is a combination of localized auxin biosynthesis, coupled with changes in polar auxin transport, that both contribute to the auxin maxima that forms in response to rhizobial recognition. Localized increases of intracellular auxin levels, mediated by YUC, have been proposed to promote further auxin accumulation in Arabidopsis leaves via reorientation of PIN-FORMED (PIN) proteins [44]. A similar situation could exist during nodulation whereby initial activation of local auxin biosynthesis below the site of rhizobial perception could modulate polar auxin transport to further promote auxin accumulation [24, 28].
Upregulation of LBD16 upon rhizobial spot inoculation coincides with the formation of the auxin maxima and precedes activation of the cell cycle, implying an early role for LBD16 in this process. Loss-of-function analysis showed that LBD16 is necessary for the appropriate initiation of root and nodule primordia, and ectopic expression of LBD16 showed promotion of root primordia and induction of many regulators of root development that are activated during both nodule and lateral root development. Previous work in Arabidopsis has shown that LBD16 is required for the transition from root-founder cell identity to primordium cell identity, including activation of cell proliferation [40, 45, 46]. Expanding such observations into nodule initiation, we suggest that in the absence of founder cells, promotion of nodules involves activation of root organogenesis via cytokinin, which provides a novel route into the induction of LBD16 that then promotes primordial cell identity and proliferation. Additional factors must also be induced that promote organ-specific differentiation into lateral roots or nodules.
While nodulating and non-nodulating species of plants respond to auxin treatment with emergence of nodule-like structures [47], only nodulating legumes do so in response to cytokinin [48]. Our work suggests that cytokinin promotion of NIN activates LBD16 that then promotes local auxin biosynthesis through the induction of STY and YUC. This suggests that modifications to the NIN, LBD16 regulon in legumes allowed a novel root developmental response to cytokinin that is critical for the promotion of nitrogen-fixing nodules. In L. japonicus, two STY genes are induced during nodulation in a manner dependent on NF-YA1 [49], a direct target of NIN. However, in our work, we see LBD16 dependence for cytokinin activation of STY, but not a dependence on NF-YA1. NIN-like proteins function in the adaptive response of roots to nitrogen availability [50, 51], and it is striking that NIN has been specifically recruited during the evolution of nodulation with no role in promoting lateral root development (Figure S6E) [41, 42]. The high degree of overlap in nodule and lateral root development implies that engineering efforts to transfer nodule organogenesis to species that lack this trait should focus on the cytokinin induction of root organogenesis, as well as on the functional characterization of components that specifically promote nodule organ identity, rather than the necessity to engineer the entire nodulation developmental program.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Sinorhizobium meliloti strain 2011 | [52] | N/A |
| Sinorhizobium meliloti strain 2011 pXLGD4 lacZ strain | [52] | N/A |
| Agrobacterium rhizogenes strain AR1193 | [53] | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| buffered nodulation media (BMN) | [54] | N/A |
| AVG-Cl (Aminoethoxyvinyl glycine hydrochloride) | ABCAM | ab145382, CAS: 55720-26-8 |
| Luteolin | Sigma-Aldrich | L9283,CAS: 491-70-3 |
| Terragreen | Oil-DriCompany | 72111537 |
| RNeasy Micro Kit | QIAGEN | Cat 74004 |
| RNase free DNase Kit | QIAGEN | Cat 79254 |
| Indole-3-acetic acid | Sigma-Aldrich | I5148, CAS: 6505-45-9 |
| 5-Bromo-4-chloro-3-indolyl-β-D-glucuronic acid, sodium salt trihydrate | Melford Laboratories | CAS:12954-41-9 |
| Magenta-5-Bromo-6-chloro-3-indolyl-β-D-galactopyranoside (magenta-x-gal) | Melford Laboratories | CAS: 93863-88-8 |
| Propidium iodide | Sigma-Aldrich | P4170, CAS:25535-16-4 |
| 5-ethynyl-2’-deoxyuridine (EdU) | Invitrogen | A10044 |
| Alexa Fluor™ 488 5-Carboxamido-(6-Azidohexanyl), Bis(Triethylammonium Salt)), 5-isomer | Invitrogen | A10266 |
| α-Amylase from Bacillus licheniformis | Sigma-Aldrich | A3403, CAS: 9000-85-5 |
| RNA Transcriptor First Strand cDNA Synthesis Kit | Roche Diagnostics | 04379012001 |
| LightCycler 480 SYBR green I master | Roche Diagnostics | 04707516001 |
| 6-Benzylaminopurine | Sigma-Aldrich | B3408, CAS:1214-39-7 |
| Dexamethasone | Sigma-Aldrich | D4902, CAS:50-02-2 |
| Critical Commercial Assays | ||
| IAA and iP quantification by liquid chromatography-tandem mass spectrometry | [55, 56] | N/A |
| Illumina TruSeq Stranded mRNA HT kit | Illumina, performed by IMGM Laboratories | 20020594 |
| BsaI-HF | New England BioLabs | R3535 |
| BpiI | Thermo Fisher Scientific | ER1011 |
| T4 DNA Ligase 2,000,000 units/mL | New England BioLabs | M0202T |
| Deposited Data | ||
| Short read sequencing data | this manuscript | GEO: GSE133612 |
| Experimental Models: Organisms/Strains | ||
| Medicago truncatula cultivar jemalong | Heritage Seeds Pty, Adelaide, AU | jemalong |
| Medicago truncatula ecotype R108 incl tnt insertion lines NF20768 (lbd16-1), NF15962 (lbd16-2), NF20919 (lbd11-1) originally obtained from Nobel Research institute LLC, Ardmore, USA (Cheng et al., 2014) | this manuscript | lbd16-1, lbd16-2, lbd11-1 |
| Medicago truncatula ecotype A17 mutant cre1-1 | [3] | cre1-1 |
| Medicago truncatula ecotype A17 mutant nin-1 | [5] | nin-1 |
| Medicago truncatula ecotype A17 mutant nfya1-1 | [57] | nfya1-1 |
| Oligonucleotides | ||
| For qRTPCR and genotyping oligos see Table S1 | N/A | N/A |
| Recombinant DNA | ||
| Golden Gate Level 0, distributed via https://www.ensa.ac.uk | GeneArt, Thermo Fisher Scientific | N/A |
| Binary plasmids generated using Golden Gate Cloning [58], details see Table S2 | this manuscript | N/A |
| Dex inducible system (GVG and 6xGAL4UAS) | [59] | N/A |
| Software and Algorithms | ||
| Sequence info M. truncatula Mt4.0v1 genome retrieved Phytozome https://phytozome.jgi.doe.gov | [60, 61] | N/A |
| R package STAR | [62] | N/A |
| Feature Counts in R package Rsubread | [63] | N/A |
| R package edgeR | [64] | N/A |
| R package DESeq2 | [65] | N/A |
| Synonym locus ID matches obtained from Uniprot | [66, 67] | N/A |
| fluorescence quantification software in FIJI | [68] | N/A |
| HMMER3.1b2 HMMSEARCH, HMMALIGN | http://hmmer.org/ | N/A |
| Lotus genome annotation retrieved | ftp://ftp.kazusa.or.jp/pub/lotus/ | N/A |
| Aligning sequences with MAFFTv72712 | [69] | N/A |
| Extacting alignments with Jalview | [70] | N/A |
| Interactive Tree of Life Visualization Tool iToL | [71] | N/A |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by The Lead Contact, Giles E.D. Oldroyd (gedo2@cam.ac.uk), subject to material transfer agreements. This study did not generate unique reagents.
Experimental Model and Subject Details
Plant material and S. meliloti strains including growth conditions
Medicago truncatula ecotypes jemalong, cultivar Jester, and ecotype R108 were used in this study. Jemalong was used to perform spot inoculation and hairy root transformations and as wild-type for comparisons to cre-1, nin-1, and nf-ya1-1, previously described [3, 5, 57]. All Tnt1 retrotransposon insertion lines described (NF20768 (lbd16-1), NF15962 (lbd16-2), and NF20919 (lbd11-1) were derivatives of the R108 ecotype and obtained from the Tnt1 Retrotransposon Mutant Collection (Noble Research Institute, Ardmore USA [72]; and as such R108 was used as the wild type for analysis of these mutants. Genotyping was performed using TntF and TntR oligos combined with the corresponding forward and reverse oligos encompassing the insertions (Table S1).
Seeds were scarified, surface sterilized with 10% (v/v) bleach solution, stratified for 3 days at 4°C and germinated on water agar plates. Plants were grown in sterile conditions in controlled environment rooms at 22°C (80% humidity, 16 h light/8 h dark, 300 μmol m2 s-1 light intensity) on filter paper-lined agar media in sealed plates unless otherwise specified. For spot inoculation seedlings were grown for 2 days on buffered nodulation medium (BNM) [54] supplemented with 1 μM aminoethoxyvinylglycine (AVG; Sigma-Aldrich Company Ltd, Darmstadt, Germany) at 22°C (16 h light/8 h dark, 300 μmol m-2 s-1 light intensity). Sinorhizobium meliloti strain 2011 [52] was grown in minimal medium supplemented with 3 μM luteolin (Sigma-Aldrich Company Ltd, Darmstadt, Germany) and diluted to a final concentration of 0.02 OD 600 nm using Fahraeus medium. The mock treatment consisted of Fahraeus medium with luteolin diluted to an equivalent concentration as the inoculum. Approximately 1 μL of S. meliloti suspension or mock treatment was inoculated onto the susceptibility zone (where the root hairs first appear) and marked by puncturing the filter paper alongside the site of inoculation. After periods ranging from 0 to 168 h, 2 mm sections of the root alongside the site of inoculation were harvested for RNA isolation or microscopy.
For spray inoculation of S. meliloti for plants grown on plates, seedlings were grown under similar conditions as described above. Roots of 1-day-old seedlings were covered with filter paper and sprayed with 2 mL S. meliloti of final concentration 0.02 OD 600 nm grown in minimal medium without luteolin. For inoculation of hairy roots, composite plants were transferred to terragreen:sharp sand mix (1:1) (Oil-DriCompany, Wisbech, UK) in P60 trays and left to grow for 7 days before inoculation with S. meliloti 2011 pXLGD4 (lacZ) (1.5 mL of overnight culture per plant diluted in liquid BNM to 0.02 OD 600 nm). Plants were grown for up to a further 4 weeks for nodule quantification and histochemical staining.
Bacterial strains
Agrobacterium rhizogenes strain AR1193 was used to introduce all binary vectors used in this study (Table S2) to M. truncatula jemalong seedlings following a previously published transient hairy root transformation protocol [53].
Method Details
Lateral root induction using gravi-stimulation
For lateral root induction [29], seedlings were grown for 2 days on modified Fahraeus medium plates and turned 135° for 12 h, then subsequently returned to their original orientation. Control plants were marked at the root tip at the time point of turning but left to grow straight. After periods ranging from 12 to 72 h, material was harvested for RNA isolation or microscopy or lateral roots were left to grow for 5 days before lateral root number was scored.
Construct production
The Golden Gate modular cloning system was used to prepare the plasmids [58]. This included a restriction digest-ligation protocol of 25 cycles of 3 min at 37°C and 4 min at 16°C using T4 DNA Ligase (New England BioLabs, Ipswich, UK) combined with the restriction enzymes BsaI-HF (New England BioLabs, Ipswich, UK) and BpiI (Thermo Fisher Scientific, Waltham, USA) for level1 and level2 assembly, respectively. All Level 0 s used in this study are held for distribution in the ENSA project core collection (https://www.ensa.ac.uk/) and are listed along with the binary plasmid details in Table S2. Sequences were domesticated, synthesized and cloned into pMS (GeneArt, Thermo Fisher Scientific, Waltham, USA). Sequence information for Medtr7g096530 (LBD16), Medtr4g060950 (LBD11) and Medtr6g086870 (YUC2) and Medtr7g099330 (YUC8) were obtained from the M. truncatula Mt4.0v1 genome via Phytozome (https://phytozome.jgi.doe.gov) [60]. The Dex-inducible system (GVG and 6xGAL4UAS) was adapted from the original system [59].
Hairy root transformation
Transformed Agrobacterium strains AR1193 [53] harboring the binary vectors (Table S2) including the AtUBI:dsred selection marker were cultured on LB medium plates supplemented with the corresponding antibiotics for 36 h at 28°C. For transformation, Agrobacteria were washed off the plates and resuspended using 1 mL of distilled water. The bacterial suspension was used to dip 1-day old seedlings after the root tip had been cut off (1/4 of total root length). Dipped seedlings were subsequently transferred to and grown on modified Fahraeus medium plates for 3 weeks. Selection was performed using a Leica M205FA stereo microscope with LED illumination and filters for dsred. Images of hairy root structures were obtained using a Leica DFC310FX color camera (Leica Microsystems, Wetzlar, Germany).
Hormone and chemical treatments
Dexamethasone (Dex; Sigma-Aldrich Company Ltd, Darmstadt, Germany), indole-3-acetic acid (IAA; Sigma-Aldrich Company Ltd, Darmstadt, Germany) and luteolin were dissolved in 70% ethanol. AVG and 6-Benzylaminopurine (BAP; Sigma-Aldrich Company Ltd, Darmstadt, Germany) were dissolved in water. Mock treatments were equal volumes of each solvent in the agar media. For BAP plate treatments (100 nM) and IAA plate treatments (100 nM) 2-day old seedlings were grown on BNM plates for 24 h with either BAP or IAA and supplemented with 1 μM AVG to replicate spot inoculation conditions. For the lateral root primordium assay 2-day old seedlings were transferred to modified Fahraeus medium supplemented with IAA (100 nm) or mock for 24 h. For Dex treatments, 3-week old plants with transformed roots were transferred to BNM agar plates supplemented with KNO3 (potassium nitrate, 2.5 mM) and either Dex (1 μM) or mock treatment, with a piece of filter paper placed over the root systems to ensure full contact with the additives.
Hormone quantification by liquid chromatography-tandem mass spectrometry
For extraction of auxin from M. truncatula root material, ∼10 mg of snap-frozen spot-inoculated root material was used. Tissue was ground to a fine powder using 3-mm stainless steel beads at 50 Hz for one minute in a TissueLyser LT (QIAGEN, Germantown, USA). Ground root samples were extracted with 1 mL of cold methanol containing [phenyl 13C6]-IAA (0.1 nmol/mL) as an internal standard in a 2-mL eppendorf tube. The tubes were vortexed and sonicated for 10 min in a Branson 3510 ultrasonic bath (Branson Ultrasonics, Eemnes, Netherlands) and placed overnight in orbital shaker at 4°C. The samples were centrifuged for 10 min at 11,500 rpm in a Heraeus Fresco 17 centrifuge (Thermo Fisher Scientific, Waltham, USA) at 4°C, after which the organic phase was transferred to a 4-mL glass vial. The pellets were re-extracted with another 1 mL of cold methanol. The combined methanol fractions were pooled and MeOH evaporated in a speed vacuum system (SPD121P, Thermo Savant, Hastings, UK) at room temperature. Residues were resuspended in 1 mL milliQ (1% formic acid) and then loaded on a 30mg/1cc Oasis MCX cartridge (Waters Corporation, USA). The cartridge was equilibrated with 1 mL of MeOH and 1 mL milliQ (1% formic acid) prior to sample loading. Subsequently the cartridge was washed with 1 mL milliQ (1% formic acid) and eluted with 1 mL of 100% MeOH. The MeOH was evaporated in a speed vacuum (SPD121P, Thermo Savant, Hastings, UK) at room temperature and the residue resuspended in 100 μl acetonitrile:water:formic acid (30:70:0.1, v/v/v). The sample was filtered through a 0.45 μm Minisart SRP4 filter (Sartorius, Goettingen, Germany) and measured on the same day. Auxin was analyzed on a Waters Xevo TQs tandem quadruple mass spectrometer as previously described [55].
Gene expression analysis
For spot inoculation and lateral root induction time-course experiments, roots were dissected as 2- to 3-mm segments around the spot of inoculation or mock treatment. For BAP and IAA response experiments, segments were dissected around the susceptibility zone marked at the time of treatment. About 50 to 60 segments were pooled to obtain 1 biological replicate, with 3-6 biological replicates per treatment/genotype were analyzed. RNA was extracted using the RNeasy Micro Kit (QIAGEN, Germantown, USA) and the RNase free DNase kit (QIAGEN, Germantown, USA) was used to remove genomic DNA. For reverse transcription of 1 μg total RNA, Transcriptor First Strand cDNA Synthesis Kit was used according to the manufacturer’s instructions (Roche Diagnostics GmbH). Quantitative real-time polymerase chain reactions (qRT-PCR) were performed in technical triplicates in the LightCycler 480 System using LightCycler 480 SYBR green I master (04707516001, Roche Diagnostics GmbH, Mannheim, Germany) in a total reaction volume of 10 μl. The primer pairs used for gene expression analysis are listed in Table S1.
RNA-Seq
RNA sequencing (RNA-Seq) was performed by IMGM Laboratories (Martinsried, Germany). RNA-Seq libraries were prepared with the Illumina TruSeq Stranded mRNA HT kit and sequencing of the libraries was performed on the Illumina NextSeq500 next generation sequencing system using the high output mode with 1 × 75 bp single-end read and 2 x 150 bp paired-end read chemistry (Illumina, Cambridge, UK).
Histochemical assays and cellular stains
For GUS or X-Gal staining roots were washed in water and immediately fixed in 90% acetone on ice for 1 h. Subsequently, the acetone was replaced by a wash solution containing 50 mM phosphate buffer pH 7.2. The wash buffer was replaced by GUS staining buffer containing 50 mM phosphate buffer pH 7.2, 0.5 mM K3Fe(CN)6 (potassium ferricyanide), 0.5 mM K4Fe(CN)6 (potassium ferrocyanide) and 2 mM 5-bromo-4-chloro-3-indolyl-beta-D-glucuronide (X-Gluc, Melford Laboratories, Ipswich, UK), vacuum infiltrated for 15 min and incubated at 37°C overnight. For X-Gal staining, the tissue was washed in 50 mM phosphate buffer pH 7.2 and fixed in 2.5% glutaraldehyde by vacuum infiltration for 15 min and incubation at room temperature for 1 h. Tissue was washed 3X in Z-buffer containing 100 mM phosphate buffer pH 7, 10 mM KCl, 1 mM MgCl2 and incubated in X-Gal staining buffer (Z-buffer supplemented with 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6 and 0.08% Magenta-5-Bromo-6-chloro-3-indolyl-B-D-galactopyranoside; (X-Gal, Melford Laboratories, Ipswich, UK) at 28°C overnight and washed with water 3 times. Tissue was cleared, stored and imaged in chloralhydrate solution. Images were obtained using a Leica DM6000 compound microscope 20X air objective with bright field settings (Leica Microsystems, Wetzlar, Germany).
For combined 5-ethynyl-2-deoxyuridine (EdU; Invitrogen, Thermo Fisher Scientific, Waltham, USA) and modified pseudo-Schiff-propidium iodide (PI; Sigma-Aldrich Company, Darmstadt, Germany) staining, we modified the published method [56] for roots: root segments were transferred to growth medium supplemented with 10 μM EdU. At transfer, plants were covered with filter paper and sprayed with liquid BNM or modified Fahraeus medium supplemented with 20 μM EdU and incubated for 5 min before the filter paper was removed. After 4 additional h of growth on growth media supplemented with EdU, 1 cm root sections centered around the susceptibility zone were dissected and dehydrated for 15 min in an ethanol dilution series (15%, 30%, 50%, 70%, 85%, 95%, and 100% ethanol) and stored in 100% ethanol overnight. The samples were rehydrated through the same ethanol dilution series and incubated at 37°C overnight in 0.3 mg/mL alpha-amylase (Sigma-Aldrich Company, Darmstadt, Germany) in phosphate buffer (20 mM pH 7.0, 2 mM NaCl, 0.25 mM CaCl2). All Edu labeling and PI staining steps were performed at room temperature with gentle shaking. First, the root sections were rinsed in water and incubated in solution containing 10 mM Alexa 488-azide (Invitrogen, Thermo Fisher Scientific, Waltham, USA) and 100 mM Tris pH 8.5 for 1 h, followed by 30 min in solution containing 10 mM Alexa 488-azide, 100 mM Tris, 1 mM CuSO4, 100 mM ascorbic acid, pH 8.5. The roots were subsequently washed three times in water, treated in 1% periodic acid for 30 min, washed twice in water, and incubated in Schiff-PI reagent for 2 h. The samples were cleared with chloral hydrate solution (Sigma-Aldrich Company, Darmstadt, Germany) for 1 h and mounted in Hoyer’s medium [56]. Imaging was performed with a Zeiss 700 confocal scanning microscope with excitation at 488 nm and emission filters set to 572–625 nm for propidium iodide and 505–600 nm for EdU using a 20X air lens objective (Carl Zeiss AG, Oberkochen, Germany). Images were processed using FIJI [73].
DR5::GFP fluorescence quantification
Individual seedlings stably transformed with a DIRECT REPEAT5 element driving nuclear localized green fluorescent protein (DR5::GFP), were visualized at the site of droplet application using the Leica M205FA stereo microscope with standard microscopy settings (Leica Microsystems, Wetzlar, Germany). The progression of each seedling was tracked by keeping them undisturbed inside the sealed growth plates, with plates removed from the growth chamber only at the visualization time points. The images were analyzed for mean fluorescence inside regions of interest (ROIs, squares with side length equal to the width of the root and centered at the droplet treatment) using FIJI software, similarly to previous studies [73, 74]. At each time point, we quantified the mean fluorescence inside the inoculation spot ROI, and two identically sized ROIs, positioned 3-root widths above and below the site of inoculation. The ratio of inside to average outside fluorescence was then calculated (ROI[spot]/ ((ROI[above]+ROI[below])/2).
Phylogenetic analysis
LATERAL ORGAN BOUNDARY (LOB) proteins were detected in the Arabidopsis, Medicago and Lotus proteomes using HMMER3.1b2 HMMSEARCH (hmmer.org). The inputs to this program were the Pfam hidden Markov model (HMM), DUF260 (PF00271, Pfam release 30), and the protein datasets from the Arabidopsis (Araport11), Medicago (Phytozome, V10) and Lotus genome annotations (ftp://ftp.kazusa.or.jp/pub/lotus/). The protein sequences detected were aligned back to the HMM using HMMER3.1b2 HMMALIGN. Gap columns in the alignment were removed, sequences with less than 70% coverage across the alignment were removed and the longest sequence for each gene from the set of splice versions was used for phylogenetic analysis. Phylogenetic analysis was carried out using the MPI version of RAxML v8.2.9 with the following method parameters set: -f a, -x 12345, -p 12345, -# 100, -m PROTCATJTT. The tree was mid-point rooted and visualized using the Interactive Tree of Life (iToL) tool [68]. Clades that included MtLBD11 and MtLBD16 were pruned from the tree and are displayed in Data S2A.
A similar procedure was used to identify YUCCA genes from the same genomes (Data S2B). The HMM FMO-like (PF00743, Pfam release 30) was used with HMMSEARCH to obtain the proteins. Most proteins showed insufficient coverage across the domain after aligning with HMMALIGN, however most of them aligned well at the N-terminal end of this model. Therefore a new HMM was built by aligning the full lengths of these sequences with MAFFT v7.2712 [71]. The conserved region of the alignment was extracted using Jalview [69] and used to build the new HMM with HMMER3.1b2 HMMBUILD. Then the same procedure as described above for the LOB family was repeated using the new HMM, except that sequences with less than 50% coverage across the alignment were removed before running RAxML.
Quantification and Statistical Analysis
RNA-seq
Reads from the RNA-sequencing experiments provided as raw fastq data were quality controlled and mapped to the M. truncatula reference genome version 4.0 (Mt4.0v1) [70] using R package STAR [61]. The counts and RPKM (Reads per kilobase per million mapped reads) values were calculated with featureCounts in R package Rsubread [62]. Non-metric multidimensional scaling was exploited to account for outliers. At least 3 biological replicates were always included in the full analysis. Genes that showed low expression throughout all samples were removed by measuring CPM (counts per million) values using R package edgeR [63]. Differentially expressed genes (DEGs) were identified by pairwise comparisons of raw counts of mock treatment versus experimental treatment, using the R package DESeq2 [64] with the threshold of absolute fold change of over 1.5 and a false discovery rate (FDR) corrected p value more significant than 0.05. The heatmaps of differential expression were plotted with R package pheatmap. The synonyms for M. truncatula locus ids were obtained by manual curation and by retrieving names from the Uniprot database using BLAST matches [65, 66]. The descriptions for each genes were obtained from Phytozome (Data S1).
qRTPCR
Expression values of minimum three biological replicates in three technical replicates were analyzed using the Pfaffl method with histone H3 (HH3) as reference [67]. Statistical comparison was performed between WT and mutants or treatment and corresponding mock. Values depicted in bar charts are the mean of minimum 3 biological replicates ± SEM (Student’s t test; ∗ p < 0.05; ∗∗ p < 0.01, ∗∗∗ p < 0.001).
Phenotyping
Data on number of lateral roots and nodules was depicted in boxplots which show the median (thick line), second to third quartiles (box), minimum and maximum ranges (lines), and outliers (single points). Normal distribution of data was tested using the Shapiro-Wilk normality test. For pairwise comparisons statistical analysis was performed using either unpaired Student’s t test, Wilcoxon test or Fisher’s exact test. For multiple comparisons, one-way analysis of variance (one-way ANOVA) or one-way Kruskal-Wallis rank sum test, followed by Tukey multiple comparisons of means or Dunn test. The R statistical package was used for these analyses. Sample size n is provided in the figure legends and refers to the number of individual plants. Statistical tests and significance levels are provided in the figure legends.
Data and Code Availability
The short-read sequencing data generated in this study have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus, with accession number GEO: GSE133612. Lists of differentially expressed genes are compiled in Data S1.
Acknowledgments
We thank Jane Langdale for help with Golden Gate design of the dexamethasone-inducible system; Kerstin Guhl for technical help with auxin extractions; Jian Feng for providing and preparing nin-1 seeds; Mandana Miri for help with hormone treatments; Raymond Wightman, Gareth Evans, Kim Findlay, Eva Wegel, Sergio Lopez, and Colleen Drapek for support with microscopy; and Chengwu Liu, Uta Paszkowski, and Sebastian Schornack for critical comments. This work was supported by the Bill and Melinda Gates Foundation as OPP1028264, the Biotechnology and Biological Sciences Research Council as BB/K003712/1, the Gatsby Foundation as GAT3395/GLH, the NWO as VENI863-15-010, the National Science Foundation, USA as DBI 0703285 and IOS-1127155, and the Noble Research Institute.
Author Contributions
K.S., J.L.S.L., T.L., and I.T. planned and performed the experiments and analyzed the data with assistance from A.T., J.L., M.D.C., and K.R.; W.K. performed the auxin measurements; P.C.B. performed the phylogenetic analysis; K.S.M. and J.W. provided Medicago mutants; G.E.D.O., S.A., and V.A.G. supervised the work; and K.S. and G.E.D.O. wrote the manuscript with input from V.A.G.
Declaration of Interests
The authors declare no competing interests.
Published: September 19, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2019.09.005.
Supplemental Information
(A) Phylogenetic analysis of the LBD gene family. The tree shows a clade of 62 proteins out of 139 LBD-like proteins from Arabidopsis thaliana, Lotus japonicus, and Medicago truncatula. (B) Phylogenetic analysis of the YUCCA gene family. The tree shows all YUCCA-like proteins identified from Arabidopsis thaliana, Lotus japonicus, and Medicago truncatula. Genes functionally characterized in this study are labeled in red. Green dots indicate bootstrap values > = 90%. Related to Figures 3 and 4.
References
- 1.Oldroyd G.E., Murray J.D., Poole P.S., Downie J.A. The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 2011;45:119–144. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- 2.Crespi M., Frugier F. De novo organ formation from differentiated cells: root nodule organogenesis. Sci. Signal. 2008;1:re11. doi: 10.1126/scisignal.149re11. [DOI] [PubMed] [Google Scholar]
- 3.Plet J., Wasson A., Ariel F., Le Signor C., Baker D., Mathesius U., Crespi M., Frugier F. MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 2011;65:622–633. doi: 10.1111/j.1365-313X.2010.04447.x. [DOI] [PubMed] [Google Scholar]
- 4.Liu J., Rutten L., Limpens E., van der Molen T., van Velzen R., Chen R., Chen Y., Geurts R., Kohlen W., Kulikova O. A Remote cis-Regulatory Region Is Required for NIN Expression in the Pericycle to Initiate Nodule Primordium Formation in Medicago truncatula. Plant Cell. 2019;31:68–83. doi: 10.1105/tpc.18.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marsh J.F., Rakocevic A., Mitra R.M., Brocard L., Sun J., Eschstruth A., Long S.R., Schultze M., Ratet P., Oldroyd G.E. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007;144:324–335. doi: 10.1104/pp.106.093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schauser L., Roussis A., Stiller J., Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- 7.Tirichine L., Sandal N., Madsen L.H., Radutoiu S., Albrektsen A.S., Sato S., Asamizu E., Tabata S., Stougaard J. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science. 2007;315:104–107. doi: 10.1126/science.1132397. [DOI] [PubMed] [Google Scholar]
- 8.Murray J.D., Karas B.J., Sato S., Tabata S., Amyot L., Szczyglowski K. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science. 2007;315:101–104. doi: 10.1126/science.1132514. [DOI] [PubMed] [Google Scholar]
- 9.Vernié T., Kim J., Frances L., Ding Y., Sun J., Guan D., Niebel A., Gifford M.L., de Carvalho-Niebel F., Oldroyd G.E. The NIN Transcription Factor Coordinates Diverse Nodulation Programs in Different Tissues of the Medicago truncatula Root. Plant Cell. 2015;27:3410–3424. doi: 10.1105/tpc.15.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soyano T., Kouchi H., Hirota A., Hayashi M. Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 2013;9:e1003352. doi: 10.1371/journal.pgen.1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bielach A., Podlesáková K., Marhavy P., Duclercq J., Cuesta C., Müller B., Grunewald W., Tarkowski P., Benková E. Spatiotemporal regulation of lateral root organogenesis in Arabidopsis by cytokinin. Plant Cell. 2012;24:3967–3981. doi: 10.1105/tpc.112.103044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laplaze L., Benkova E., Casimiro I., Maes L., Vanneste S., Swarup R., Weijers D., Calvo V., Parizot B., Herrera-Rodriguez M.B. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell. 2007;19:3889–3900. doi: 10.1105/tpc.107.055863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Rizzo S., Crespi M., Frugier F. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell. 2006;18:2680–2693. doi: 10.1105/tpc.106.043778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lohar D.P., Schaff J.E., Laskey J.G., Kieber J.J., Bilyeu K.D., Bird D.M. Cytokinins play opposite roles in lateral root formation, and nematode and Rhizobial symbioses. Plant J. 2004;38:203–214. doi: 10.1111/j.1365-313X.2004.02038.x. [DOI] [PubMed] [Google Scholar]
- 15.Herrbach V., Chirinos X., Rengel D., Agbevenou K., Vincent R., Pateyron S., Huguet S., Balzergue S., Pasha A., Provart N. Nod factors potentiate auxin signaling for transcriptional regulation and lateral root formation in Medicago truncatula. J. Exp. Bot. 2017;68:569–583. doi: 10.1093/jxb/erw474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao T.T., Schilderink S., Moling S., Deinum E.E., Kondorosi E., Franssen H., Kulikova O., Niebel A., Bisseling T. Fate map of Medicago truncatula root nodules. Development. 2014;141:3517–3528. doi: 10.1242/dev.110775. [DOI] [PubMed] [Google Scholar]
- 17.Herrbach V., Remblière C., Gough C., Bensmihen S. Lateral root formation and patterning in Medicago truncatula. J. Plant Physiol. 2014;171:301–310. doi: 10.1016/j.jplph.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Osipova M.A., Mortier V., Demchenko K.N., Tsyganov V.E., Tikhonovich I.A., Lutova L.A., Dolgikh E.A., Goormachtig S. Wuschel-related homeobox5 gene expression and interaction of CLE peptides with components of the systemic control add two pieces to the puzzle of autoregulation of nodulation. Plant Physiol. 2012;158:1329–1341. doi: 10.1104/pp.111.188078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franssen H.J., Xiao T.T., Kulikova O., Wan X., Bisseling T., Scheres B., Heidstra R. Root developmental programs shape the Medicago truncatula nodule meristem. Development. 2015;142:2941–2950. doi: 10.1242/dev.120774. [DOI] [PubMed] [Google Scholar]
- 20.Moreno-Risueno M.A., Van Norman J.M., Moreno A., Zhang J., Ahnert S.E., Benfey P.N. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science. 2010;329:1306–1311. doi: 10.1126/science.1191937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubrovsky J.G., Napsucialy-Mendivil S., Duclercq J., Cheng Y., Shishkova S., Ivanchenko M.G., Friml J., Murphy A.S., Benková E. Auxin minimum defines a developmental window for lateral root initiation. New Phytol. 2011;191:970–983. doi: 10.1111/j.1469-8137.2011.03757.x. [DOI] [PubMed] [Google Scholar]
- 22.Ng J.L.P., Hassan S., Truong T.T., Hocart C.H., Laffont C., Frugier F., Mathesius U. Flavonoids and Auxin Transport Inhibitors Rescue Symbiotic Nodulation in the Medicago truncatula Cytokinin Perception Mutant cre1. Plant Cell. 2015;27:2210–2226. doi: 10.1105/tpc.15.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wasson A.P., Pellerone F.I., Mathesius U. Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell. 2006;18:1617–1629. doi: 10.1105/tpc.105.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohlen W., Ng J.L.P., Deinum E.E., Mathesius U. Auxin transport, metabolism, and signalling during nodule initiation: indeterminate and determinate nodules. J. Exp. Bot. 2018;69:229–244. doi: 10.1093/jxb/erx308. [DOI] [PubMed] [Google Scholar]
- 25.Rightmyer A.P., Long S.R. Pseudonodule formation by wild-type and symbiotic mutant Medicago truncatula in response to auxin transport inhibitors. Mol. Plant Microbe Interact. 2011;24:1372–1384. doi: 10.1094/MPMI-04-11-0103. [DOI] [PubMed] [Google Scholar]
- 26.Roy S., Robson F., Lilley J., Liu C.-W., Cheng X., Wen J., Walker S., Sun J., Cousins D., Bone C. MtLAX2, a Functional Homologue of the Arabidopsis Auxin Influx Transporter AUX1, Is Required for Nodule Organogenesis. Plant Physiol. 2017;174:326–338. doi: 10.1104/pp.16.01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huo X., Schnabel E., Hughes K., Frugoli J. RNAi Phenotypes and the Localization of a Protein:GUS Fusion Imply a Role for Medicago truncatula PIN Genes in Nodulation. J. Plant Growth Regul. 2006;25:156–165. doi: 10.1007/s00344-005-0106-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deinum E.E., Kohlen W., Geurts R. Quantitative modelling of legume root nodule primordium induction by a diffusive signal of epidermal origin that inhibits auxin efflux. BMC Plant Biol. 2016;16:254. doi: 10.1186/s12870-016-0935-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ditengou F.A., Teale W.D., Kochersperger P., Flittner K.A., Kneuper I., van der Graaff E., Nziengui H., Pinosa F., Li X., Nitschke R. Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2008;105:18818–18823. doi: 10.1073/pnas.0807814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breakspear A., Liu C., Roy S., Stacey N., Rogers C., Trick M., Morieri G., Mysore K.S., Wen J., Oldroyd G.E. The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell. 2014;26:4680–4701. doi: 10.1105/tpc.114.133496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinon V., Prasad K., Grigg S.P., Sanchez-Perez G.F., Scheres B. Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2013;110:1107–1112. doi: 10.1073/pnas.1213497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng Z., Zhu J., Du X., Cui X. Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana. Planta. 2012;236:1227–1237. doi: 10.1007/s00425-012-1673-3. [DOI] [PubMed] [Google Scholar]
- 33.Eklund D.M., Ståldal V., Valsecchi I., Cierlik I., Eriksson C., Hiratsu K., Ohme-Takagi M., Sundström J.F., Thelander M., Ezcurra I., Sundberg E. The Arabidopsis thaliana STYLISH1 protein acts as a transcriptional activator regulating auxin biosynthesis. Plant Cell. 2010;22:349–363. doi: 10.1105/tpc.108.064816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith D.L., Fedoroff N.V. LRP1, a gene expressed in lateral and adventitious root primordia of arabidopsis. Plant Cell. 1995;7:735–745. doi: 10.1105/tpc.7.6.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang L.P., Zhou C., Wang S.S., Yuan J., Zhang X.S., Su Y.H. FUSCA3 interacting with LEAFY COTYLEDON2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana. New Phytol. 2017;213:1740–1754. doi: 10.1111/nph.14313. [DOI] [PubMed] [Google Scholar]
- 36.Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goh T., Joi S., Mimura T., Fukaki H. The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development. 2012;139:883–893. doi: 10.1242/dev.071928. [DOI] [PubMed] [Google Scholar]
- 38.Suzaki T., Yano K., Ito M., Umehara Y., Suganuma N., Kawaguchi M. Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development. 2012;139:3997–4006. doi: 10.1242/dev.084079. [DOI] [PubMed] [Google Scholar]
- 39.Orosa-Puente B., Leftley N., von Wangenheim D., Banda J., Srivastava A.K., Hill K., Truskina J., Bhosale R., Morris E., Srivastava M. Root branching toward water involves posttranslational modification of transcription factor ARF7. Science. 2018;362:1407–1410. doi: 10.1126/science.aau3956. [DOI] [PubMed] [Google Scholar]
- 40.Liu W., Yu J., Ge Y., Qin P., Xu L. Pivotal role of LBD16 in root and root-like organ initiation. Cell. Mol. Life Sci. 2018;75:3329–3338. doi: 10.1007/s00018-018-2861-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griesmann M., Chang Y., Liu X., Song Y., Haberer G., Crook M.B., Billault-Penneteau B., Lauressergues D., Keller J., Imanishi L. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science. 2018;361 doi: 10.1126/science.aat1743. [DOI] [PubMed] [Google Scholar]
- 42.van Velzen R., Holmer R., Bu F., Rutten L., van Zeijl A., Liu W., Santuari L., Cao Q., Sharma T., Shen D. Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc. Natl. Acad. Sci. USA. 2018;115:E4700–E4709. doi: 10.1073/pnas.1721395115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pawlowski K., Bisseling T. Rhizobial and Actinorhizal Symbioses: What Are the Shared Features? Plant Cell. 1996;8:1899–1913. doi: 10.1105/tpc.8.10.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abley K., Sauret-Güeto S., Marée A.F.M., Coen E. Formation of polarity convergences underlying shoot outgrowths. eLife. 2016;5:e18165. doi: 10.7554/eLife.18165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu X., Xu L. Transcription Factors WOX11/12 Directly Activate WOX5/7 to Promote Root Primordia Initiation and Organogenesis. Plant Physiol. 2016;172:2363–2373. doi: 10.1104/pp.16.01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J., Hu X., Qin P., Prasad K., Hu Y., Xu L. The WOX11-LBD16 Pathway Promotes Pluripotency Acquisition in Callus Cells During De Novo Shoot Regeneration in Tissue Culture. Plant Cell Physiol. 2018;59:734–743. doi: 10.1093/pcp/pcy010. [DOI] [PubMed] [Google Scholar]
- 47.Hiltenbrand R., Thomas J., McCarthy H., Dykema K.J., Spurr A., Newhart H., Winn M.E., Mukherjee A. A Developmental and Molecular View of Formation of Auxin-Induced Nodule-Like Structures in Land Plants. Front. Plant Sci. 2016;7:1692. doi: 10.3389/fpls.2016.01692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gauthier-Coles C., White R.G., Mathesius U. Nodulating Legumes Are Distinguished by a Sensitivity to Cytokinin in the Root Cortex Leading to Pseudonodule Development. Front. Plant Sci. 2019;9:1901. doi: 10.3389/fpls.2018.01901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hossain M.S., Shrestha A., Zhong S., Miri M., Austin R.S., Sato S., Ross L., Huebert T., Tromas A., Torres-Jerez I. Lotus japonicus NF-YA1 Plays an Essential Role During Nodule Differentiation and Targets Members of the SHI/STY Gene Family. Mol. Plant Microbe Interact. 2016;29:950–964. doi: 10.1094/MPMI-10-16-0206-R. [DOI] [PubMed] [Google Scholar]
- 50.Castaings L., Camargo A., Pocholle D., Gaudon V., Texier Y., Boutet-Mercey S., Taconnat L., Renou J.-P., Daniel-Vedele F., Fernandez E. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009;57:426–435. doi: 10.1111/j.1365-313X.2008.03695.x. [DOI] [PubMed] [Google Scholar]
- 51.Lin J.S., Li X., Luo Z., Mysore K.S., Wen J., Xie F. NIN interacts with NLPs to mediate nitrate inhibition of nodulation in Medicago truncatula. Nat. Plants. 2018;4:942–952. doi: 10.1038/s41477-018-0261-3. [DOI] [PubMed] [Google Scholar]
- 52.Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J.C., Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 53.Boisson-Dernier A., Chabaud M., Garcia F., Bécard G., Rosenberg C., Barker D.G. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 2001;14:695–700. doi: 10.1094/MPMI.2001.14.6.695. [DOI] [PubMed] [Google Scholar]
- 54.Ehrhardt D.W., Atkinson E.M., Long S.R. Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science. 1992;256:998–1000. doi: 10.1126/science.10744524. [DOI] [PubMed] [Google Scholar]
- 55.Ruyter-Spira C., Kohlen W., Charnikhova T., van Zeijl A., van Bezouwen L., de Ruijter N., Cardoso C., Lopez-Raez J.A., Matusova R., Bours R. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol. 2011;155:721–734. doi: 10.1104/pp.110.166645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schiessl K., Kausika S., Southam P., Bush M., Sablowski R. JAGGED controls growth anisotropyand coordination between cell sizeand cell cycle during plant organogenesis. Curr. Biol. 2012;22:1739–1746. doi: 10.1016/j.cub.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laporte P., Lepage A., Fournier J., Catrice O., Moreau S., Jardinaud M.-F., Mun J.-H., Larrainzar E., Cook D.R., Gamas P., Niebel A. The CCAAT box-binding transcription factor NF-YA1 controls rhizobial infection. J. Exp. Bot. 2014;65:481–494. doi: 10.1093/jxb/ert392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. PLoS ONE. 2011;6:e16765. doi: 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aoyama T., Chua N.-H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- 60.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 63.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Consortium T.U., UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 67.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Letunic I., Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang H., Krishnakumar V., Bidwell S., Rosen B., Chan A., Zhou S., Gentzbittel L., Childs K.L., Yandell M., Gundlach H. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics. 2014;15:312. doi: 10.1186/1471-2164-15-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng X., Wang M., Lee H.-K., Tadege M., Ratet P., Udvardi M., Mysore K.S., Wen J. An efficient reverse genetics platform in the model legume Medicago truncatula. New Phytol. 2014;201:1065–1076. doi: 10.1111/nph.12575. [DOI] [PubMed] [Google Scholar]
- 73.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pacios-Bras C., Schlaman H.R., Boot K., Admiraal P., Langerak J.M., Stougaard J., Spaink H.P. Auxin distribution in Lotus japonicus during root nodule development. Plant Mol. Biol. 2003;52:1169–1180. doi: 10.1023/b:plan.0000004308.78057.f5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Phylogenetic analysis of the LBD gene family. The tree shows a clade of 62 proteins out of 139 LBD-like proteins from Arabidopsis thaliana, Lotus japonicus, and Medicago truncatula. (B) Phylogenetic analysis of the YUCCA gene family. The tree shows all YUCCA-like proteins identified from Arabidopsis thaliana, Lotus japonicus, and Medicago truncatula. Genes functionally characterized in this study are labeled in red. Green dots indicate bootstrap values > = 90%. Related to Figures 3 and 4.
Data Availability Statement
The short-read sequencing data generated in this study have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus, with accession number GEO: GSE133612. Lists of differentially expressed genes are compiled in Data S1.