Abstract
Ensuring correct radiograph view labeling is important for machine learning algorithm development and quality control of studies obtained from multiple facilities. The purpose of this study was to develop and test the performance of a deep convolutional neural network (DCNN) for the automated classification of frontal chest radiographs (CXRs) into anteroposterior (AP) or posteroanterior (PA) views. We obtained 112,120 CXRs from the NIH ChestX-ray14 database, a publicly available CXR database performed in adult (106,179 (95%)) and pediatric (5941 (5%)) patients consisting of 44,810 (40%) AP and 67,310 (60%) PA views. CXRs were used to train, validate, and test the ResNet-18 DCNN for classification of radiographs into anteroposterior and posteroanterior views. A second DCNN was developed in the same manner using only the pediatric CXRs (2885 (49%) AP and 3056 (51%) PA). Receiver operating characteristic (ROC) curves with area under the curve (AUC) and standard diagnostic measures were used to evaluate the DCNN’s performance on the test dataset. The DCNNs trained on the entire CXR dataset and pediatric CXR dataset had AUCs of 1.0 and 0.997, respectively, and accuracy of 99.6% and 98%, respectively, for distinguishing between AP and PA CXR. Sensitivity and specificity were 99.6% and 99.5%, respectively, for the DCNN trained on the entire dataset and 98% for both sensitivity and specificity for the DCNN trained on the pediatric dataset. The observed difference in performance between the two algorithms was not statistically significant (p = 0.17). Our DCNNs have high accuracy for classifying AP/PA orientation of frontal CXRs, with only slight reduction in performance when the training dataset was reduced by 95%. Rapid classification of CXRs by the DCNN can facilitate annotation of large image datasets for machine learning and quality assurance purposes.
Keywords: Deep learning, Deep convoluted neural networks, Artificial intelligence, PACS
Introduction
The application of artificial intelligence (AI) in medicine is gaining much momentum across several medical specialties, particularly for automated classification of clinical images [1–5]. Within radiology, AI has been met with much enthusiasm and optimism, especially with the recent development of deep learning (DL), which has proven to be useful in automated image classification across multiple image modalities and disease states [4, 6, 7].
One potential application for DL in radiology is automated annotation of radiographic view for the purposes of machine learning database curation. The accuracy of DL algorithms depends on the number of training images and validity of “groundtruth” labels, which may be limited by occasionally mislabeled metadata in medical imaging [8]. In clinical practice, such study descriptions, such as radiographic view, are stored in the digital imaging and communications in medicine (DICOM) metadata and displayed as overlaid annotations on a standard picture archiving and communication system (PACS) viewer. Aakre et al. previously found general errors in plain radiograph labels to be as high as 2.4%, demonstrating the need for quality assurance tools [9]. An automated method for radiographic view semantic labeling could facilitate the curation of large databases for medical image machine learning, as well as facilitate radiologists’ workflow in the interpretation of studies from outside facilities, the labels of which are heterogeneous, occasionally inaccurate, and, therefore, not always reliable.
Although radiologists traditionally utilize anatomical markers such as position of the scapulae and relative size of the cardiac silhouette to determine the projection of a CXR [10], deep learning could facilitate the automated classification of CXRs by radiographic view. Prior work by Rajkomar et al. demonstrated 99% accuracy of a DL system in automatically classifying CXRs into frontal (anteroposterior (AP) and posteroanterior (PA)) vs. lateral views [8], which suggests a similar approach could be applied towards classifying AP vs. PA CXRs.
The purpose of this study was to develop and test the performance of a deep convolutional neural network (DCNN) for the rapid automated classification of frontal CXRs into AP or PA views.
Methods
Datasets
All patient data were de-identified and compliant with the Health Insurance Portability and Accountability Act (HIPAA). This retrospective study was approved by the Institutional Review Board. We obtained CXRs from the publicly available NIH ChestX-ray14 database [4, 6], comprised of 112,120 frontal CXRs (44,810 (40%) AP, 67,310 (60%) PA) from 30,805 patients. We created a second database only comprised of CXRs from pediatric patients < 18 years old (2885 (49%) AP and 3056 (51%) PA) (Table 1). We also obtained an external test dataset comprised of CXRs from Shenzhen, China, and Montgomery County, USA, which were composed of 800 PA CXRs [11]. In order to test our algorithm’s generalizability to radiographs obtained from a different hospital system, 200 de-identified CXRs (100 (50%) AP and 100 (50%) PA) were obtained from Institution Name and were used for further testing (Institution city and state). All images were saved using lossless Portable Network Graphics (PNG) format and then resized to a 256 × 256 matrix from a native resolution of 1024 × 1024.
Table 1.
Datasets used for deep convolutional neural network training, validation, and testing
| DCNN | Radiograph type | Train | Validate | Test |
|---|---|---|---|---|
| All patients | AP | 31,367 | 4481 | 8962 |
| PA | 47,117 | 6731 | 13,462 | |
| Total | 78,784 | 11,212 | 22,424 | |
| Pediatric | AP | 2020 | 288 | 577 |
| PA | 2139 | 306 | 611 | |
| Total | 2020 | 594 | 1188 |
AP anteroposterior, PA posteroanterior
Computer Hardware and Software Specifications
All DCNN development and testing was performed using PyTorch framework (https://pytorch.org) on a 2.5 GHz Intel Haswell dual socket (12-core processors) (Intel, Santa Clara, CA) with 128 GB of RAM and 2 NVIDIA K80 GPUs (NVIDIA Corporation, Santa Clara, CA).
DLS Development
Following typical DL methodology [12], we randomly assigned 70% of the data into the “training” dataset, 10% of the data into the “validation” dataset, and 20% into the “testing” dataset, ensuring no overlap in images between these datasets (Table 1). Briefly, the training phase utilizes the majority of the available data to train DCNNs to classify images into predefined categories by identifying image features specific to each category. The validation phase utilizes a smaller proportion of available data to test the DCNNs trained in the training phase and select the highest-performing algorithms. The final testing phase consists of assessing the diagnostic performance of the best-performing algorithm(s) on a dataset that was not utilized in either the training or validation phase.
We utilized the ResNet-18 [13] DCNN pretrained on 1.2 million color images of everyday objects from ImageNet (http://www.image-net.org/) prior to training on the CXRs. This technique is known as transfer learning and allows for modification of pretrained neural network architectures to be used for classification of different datasets not used in training of the original network [3, 4, 14]. Transfer learning has previously demonstrated superior performance in medical image classification compared with the use of untrained networks [3, 4, 14]. The solver parameters used for our DCNN training were as follows: 50 training epochs; stochastic gradient descent (SGD) with a learning rate of 0.001, momentum of 0.9, and weight decay of 1 × 105. During each training epoch, each image was augmented by a random rotation between − 5 and 5°, random cropping, and horizontal flipping.
Experimental Setup
Two separate DCNNs were trained, one using the entire dataset and another using only pediatric CXRs. Each DCNN was tested on a unique testing dataset not used for training or validation.
The DCNN trained on the entire dataset was further externally tested on 800 PA radiographs from Shenzhen, China, and Montgomery County, USA, performed previously for tuberculosis screening [11]. Two hundred de-identified radiographs from randomly selected patients from the Johns Hopkins Hospital (Baltimore, MD) were used to test the algorithm for further validation in a hospital system.
Testing time was divided by the number of testing cases to determine the rate of classification in number of images per second. To identify the distinguishing image features used by the DCNN for classification, we created heatmaps via class activation mapping (CAM) [15].
Statistical Analysis
For each testing dataset, receiver operating characteristic (ROC) curves with area under the curve (AUC) were generated and statistically compared between DCNNs using the DeLong parametric method, along with standard measures of diagnostic performance (sensitivity, specificity, accuracy, positive predictive value, negative predictive value) at optimal diagnostic thresholds chosen using Youden’s J statistic [16, 17]. AP images were considered to be positive and PA negative for statistical purposes.
Results
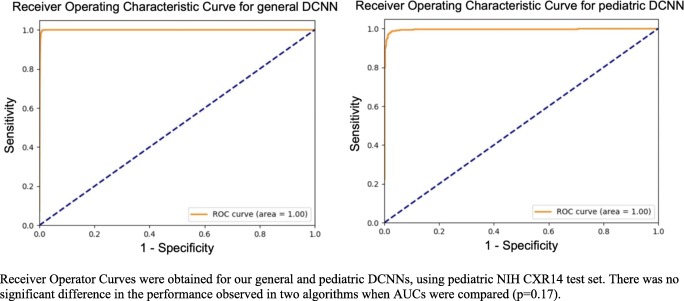
DCNNs trained using the entire CXR dataset and pediatric CXR dataset achieved AUCs of 1.0 and 0.997, respectively (p = 0.17), and accuracy of 99.6% and 98%, respectively, for distinguishing between AP and PA CXR in the testing dataset. A representative ROC curve is depicted in Fig. 1. Sensitivity and specificity were 99.6% and 99.5%, respectively, for the DCNN trained on the entire dataset and 98% for both sensitivity and specificity for the DCNN trained on the pediatric dataset. Positive predictive values were 1 and 0.98 for general and pediatric algorithms respectively, and negative predictive values were 0.99 for both algorithms (Table 2).
Fig. 1.
Receiver operator curves for general and pediatric classifier. Receiver operator curves were obtained for our general and pediatric DCNNs, using pediatric NIH CXR14 test set. There was no significant difference in the performance observed in two algorithms when AUCs were compared (p = 0.17)
Table 2.
Performance of deep convolutional networks (DCNNs) for radiograph orientation
| DCNN | Testing set | AUC | Sensitivity | Specificity | PPV | NPV | Accuracy (%) |
|---|---|---|---|---|---|---|---|
| General | NIH CXR14 | 1 | 1 | 0.99 | 1 | 0.99 | 99.6 |
| Shenzhen, China and Montgomery County, USA | 0.999 | N/A | 0.99 | N/A | N/A | 99.3 | |
| Peds NIH CXR14 | 0.999 | 0.99 | 0.99 | 1 | 0.99 | 99.4 | |
| Johns Hopkins Hospital (Baltimore, MD) | 0.985 | 0.98 | 0.96 | 0.96 | 0.98 | 96.0 | |
| Pediatrics | Peds NIH CXR14 | 0.997 | 0.98 | 0.98 | 0.98 | 0.98 | 98.0 |
AUC area under the receiver operating characteristic (ROC) curve, PPV positive predictive value, NPV negative predictive value. Positive refers to AP radiographs while negative refers to PA radiographs
The DCNN trained using all images was tested using 800 PA radiographs from Montgomery County, USA, and Shenzhen, China, for external generalizability, and achieved an AUC of 0.999 and accuracy of 99.3%. When deployed for radiographs retrieved from Institution Abbreviation, our algorithm reached an AUC of 0.985, accuracy of 96% with sensitivity of 98% and specificity of 97%. Overall, classification of the CXRs occurred at a rate of 33 radiographs per second.
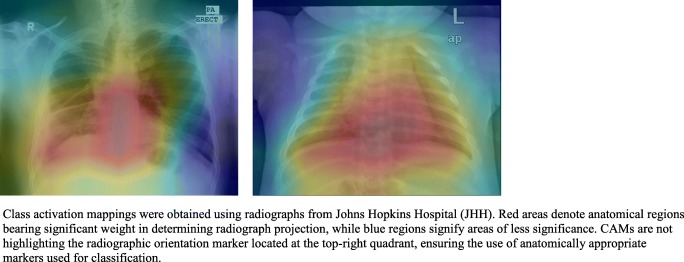
Representative heatmaps showing distinguishing features weighted most heavily by the DCNNs are demonstrated in Fig. 2 for both pediatric and general DCNNs. The DCNNs most frequently highlighted the scapulae, heart, stomach, and diaphragm. Although some radiographs contained radiopaque labels indicating “AP” or “PA,” none of the CAMs highlighted the region containing the labels.
Fig. 2.
Class activation mappings extracted from general and pediatric DCNNs. Class activation mappings were obtained using radiographs from Johns Hopkins Hospital (JHH). Red areas denote anatomical regions bearing significant weight in determining radiograph projection, while blue regions signify areas of less significance. CAMs are not highlighting the radiographic orientation marker located at the top-right quadrant, ensuring the use of anatomically appropriate markers used for classification
Discussion
One application of deep learning is in the semantic labeling of radiographs, which could aid radiologist workflow in ensuring accuracy of metadata obtained from outside facilities and facilitate the creation of large datasets for medical machine learning projects. In our study, we developed a deep learning system to accurately distinguish the PA/AP orientation of chest radiographs. Our deep learning system achieved a high classification performance for both pediatric and adult populations, which also generalized well to an external dataset. Furthermore, we identified distinguishing features the algorithm used to come to its diagnosis.
Our deep learning systems achieved AUC of 1 and 0.997 for adult and pediatric populations respectively with no statistically significant difference observed in performance (p = 0.17), demonstrating strong diagnostic performance for diverse demographics. These findings are consistent with those reported by Rajkomar et al., who reported 100% accuracy for distinguishing between frontal and lateral chest radiographs, a similar yet slightly simpler task [8]. Similarly, a prior study by Cheng et al. demonstrated an accuracy of 77.9% in classifying abdominal ultrasound images into 11 different anatomical categories [18], which is also consistent with our findings demonstrating the feasibility of DCNN-classification of medical image semantic labeling. Altogether, these findings confirm the efficacy of applying deep learning using neural networks trained on everyday objects (i.e., ImageNet) towards medical imaging (i.e., transfer learning).
Unlike the prior two studies by Rajkomar et al. and Cheng et al. [8, 18], we did not artificially increase our training and validation dataset size through augmentation techniques (although we applied standard image augmentation during training and validation). Because of the multiple-fold difference in number of images available in our study (112,120), compared with 1885 images in Rajkomar et al. and 9298 images in Cheng et al., we hypothesized that it would be unnecessary to physically augment the images to increase our dataset. Our near-perfect accuracy supports general machine learning theory that increased size of dataset results in higher performance. Furthermore, the DCNN trained using a smaller subset of 5941 pediatric CXRs also achieved near-perfect diagnostic performance, which suggests that, for simple tasks, such as labeling of radiographic view, a large number of training images are not necessary for deep learning.
Current limitations of deep learning include the concern that neural networks act as “black boxes,” given the inability of a network to explain its diagnostic or decision-making reasoning. Class activation mapping is a visualization tool that visually represents the features of an image that are weighted the most heavily in DCNN’s decision. In the CAM analysis of our DCNNs, we demonstrated that our algorithm tended to emphasize the mediastinum, heart, diaphragm, and osseous structures to classify images into AP or PA views. Such anatomic landmarks are, interestingly, similar to those used by radiologists, such as position of diaphragm to signify adequacy of breath in a PA radiograph; however, we stress that these heatmaps merely show the areas most emphasized by the DCNNs for decision-making and do not explain exactly what it is about these areas that influences its decision-making.
We have demonstrated the rapid speed of our DCNN, classifying 33 CXRs per second. These findings are consistent with those shown by Rajkomar et al., who showed a classification of CXRs into frontal or lateral view at a rate of 38 images per second [8]. Assuming that a health system utilizes 100,000 CXRs a year, an entire decade of imaging data generated can be annotated in 8.4 h using our algorithm. Therefore, our DCNN is well-suited to tackle large databases and potentially can reach faster classification rate using CPUs with higher processing power.
Although we trained our DCNN with the aim of retrospective image database curation, integration of our DCNN into the workflow of radiology technicians could also automate the labeling process and decrease the human error that may occur with manual entry. A separate related challenge in building an accurate database for both PACS workflows and for machine learning is integrating image metadata from different equipment manufacturers. Even within standardized PACS, significant variations in metadata conventions exist among equipment manufacturers. Therefore, automated semantic labeling of medical images would be an efficient solution to this problem; our algorithm is well-suited for such rapid automated classification of large image databases.
Our study should be viewed in light of a number of limitations. We caution that our findings are specific for the task of classifying chest radiographs into AP vs. PA and as such, may not be generalizable towards other more difficult tasks. However, our results do demonstrate the potential power of machine learning to perform tasks with accuracy and speed for image preprocessing and radiograph quality assurance purposes. Additionally, we utilized a single DCNN, as opposed to multiple DCNNs, as done in prior studies [4, 14]. Combining multiple pretrained models may result in higher performance, although the utility of using additional DCNNs is questionable for our aims, given the very high AUCs achieved by the ResNet-18 DCNN used in our study. Finally, DCNNs are limited by the inability to know precisely what our network is utilizing to make its decision. Nevertheless, visualization techniques, such as CAM, are able to give some insight into the areas that are most important for decision-making.
In conclusion, we were able to train a neural network with high accuracy for automated classification of frontal CXRs into AP or PA view. Although further clinical validation in a prospective manner is warranted for routine clinical use, our findings suggest that transfer learning can be successfully applied towards semantic labeling of medical imaging. If applied on a large scale and utilized towards other imaging modalities and views, our networks could facilitate curation of large medical imaging databases for machine learning purposes and ensure metadata quality for radiographs obtained from different sources.
Compliance with Ethical Standards
All patient data were de-identified and compliant with the Health Insurance Portability and Accountability Act (HIPAA). This retrospective study was approved by the Institutional Review Board.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yi PH, Hui FK, Ting DS. Artificial intelligence and radiology: collaboration is key. J Am Coll Radiol. 2018;15:781–783. doi: 10.1016/j.jacr.2017.12.037. [DOI] [PubMed] [Google Scholar]
- 2.Litjens G, Kooi T, Bejnordi BE, Setio AA, Ciompi F, Ghafoorian M, van der Laak JA, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Kim DH, MacKinnon T. Artificial intelligence in fracture detection: transfer learning from deep convolutional neural networks. Clin Radiol. 2018;73:439–445. doi: 10.1016/j.crad.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 4.Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284:574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 5.Wong TY, Bressler NM. Artificial intelligence with deep learning technology looks into diabetic retinopathy screening. JAMA. 2016;316:2366. doi: 10.1001/jama.2016.17563. [DOI] [PubMed] [Google Scholar]
- 6.Prevedello LM, Erdal BS, Ryu JL, Little KJ, Demirer M, Qian S, White RD. Automated critical test findings identification and online notification system using artificial intelligence in imaging. Radiology. 2017;285:923–931. doi: 10.1148/radiol.2017162664. [DOI] [PubMed] [Google Scholar]
- 7.Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP. Performance of a deep-learning neural network model in assessing skeletal maturity on pediatric hand radiographs. Radiology. 2017;287:313–322. doi: 10.1148/radiol.2017170236. [DOI] [PubMed] [Google Scholar]
- 8.Rajkomar A, Lingam S, Taylor AG, Blum M, Mongan J. High-throughput classification of radiographs using deep convolutional neural networks. J Digit Imaging. 2017;30:95–101. doi: 10.1007/s10278-016-9914-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aakre KT, Johnson CD. Plain-radiographic image labeling: a process to improve clinical outcomes. J Am Coll Radiol. 2006;3:949–953. doi: 10.1016/j.jacr.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Goodman LR: Felson’s principles of chest Roentgenology, a programmed text, 4th edition. Saunders, 2014
- 11.Jaeger S, Candemir S, Antani S, Wang Y, Lu PX, Thoma G. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant Imaging Med Surg. 2014;4:475–477. doi: 10.3978/j.issn.2223-4292.2014.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM: ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings - IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 3462–3471, 2017
- 13.He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition. arXiv preprint arXiv:1512.03385, 2015
- 14.Lakhani P. Deep convolutional neural networks for endotracheal tube position and X-ray image classification: challenges and opportunities. J Digit Imaging. 2017;30:460–468. doi: 10.1007/s10278-017-9980-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A: Learning deep features for discriminative localization. In Proceedings - IEEE Conference on Computer Vision and Pattern Recognition (CVPR): 2921–2929, 2016
- 16.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 17.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::AID-CNCR2820030106>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Cheng PM, Malhi HS. Transfer learning with convolutional neural networks for classification of abdominal ultrasound images. J Digit Imaging. 2017;30:234–243. doi: 10.1007/s10278-016-9929-2. [DOI] [PMC free article] [PubMed] [Google Scholar]