Abstract
Biocontrol offers a promising alternative to synthetic fungicides for the control of a variety of pre- and post-harvest diseases of crops. Black rot, which is caused by the pathogenic fungus Ceratocytis fimbriata, is the most destructive post-harvest disease of sweet potato, but little is currently known about potential biocontrol agents for this fungus. Here, we isolated several microorganisms from the tuberous roots and shoots of field-grown sweet potato plants, and analyzed their ribosomal RNA gene sequences. The microorganisms belonging to the genus Pantoea made up a major portion of the microbes residing within the sweet potato plants, and fluorescence microscopy showed these microbes colonized the intercellular spaces of the vascular tissue in the sweet potato stems. Four P. dispersa strains strongly inhibited C. fimbriata mycelium growth and spore germination, and altered the morphology of the fungal hyphae. The detection of dead C. fimbriata cells using Evans blue staining suggested that these P. dispersa strains have fungicidal rather than fungistatic activity. Furthermore, P. dispersa strains significantly inhibited C. fimbriata growth on the leaves and tuberous roots of a susceptible sweet potato cultivar (“Yulmi”). These findings suggest that P. dispersa strains could inhibit black rot in sweet potato plants, highlighting their potential as biocontrol agents.
Subject terms: Biotic, Microbe
Introduction
Endophytes are microorganisms that reside within plant tissues without causing any apparent disease symptoms in their hosts1–4. These organisms interact with plants in a variety of ways, affecting all stages of development through to the end of the plant life cycle5. Some endophytes promote plant growth, the mechanisms behind which have been extensively studied and include the facilitation of nutrient uptake (e.g., phosphorus), nitrogen fixation for plant use, the sequestration of iron for plants by siderophores, the production of plant hormones [e.g., gibberellins (GAs), indoleacetic acid (IAA), and cytokines], the elimination of ethylene from plants, reduction of the amount of iron available to plant pathogens in the rhizosphere, the synthesis of fungal cell wall-degrading enzymes, and competition with several phytopathogens6–9.
Many biotic and abiotic factors influence the structure and function of the bacterial communities in plants–for example, climate change, pesticide treatment, soil type, plant health, and developmental stage10–14. In addition, the composition of the root exudates varies between plants and affects the relative abundance of microorganisms near the root15. Not only do plants provide nutrients to microorganisms, but some plant species also contain antimicrobial metabolites that are unique to their exudates16,17. Therefore, the compounds that are secreted by the roots act as a signal, attracting or repelling microorganisms to the plants18,19, and thus regulating the interaction between the roots and soil microorganisms.
Sweet potato [Ipomoea batatas (L.)] is one of the most important cultivated crops in the world, with an annual production area of 8.0 million hectares and a total global production of 106,569 million tons20. Sweet potato is also a valuable medicinal plant due to its anti-cancer, anti-diabetic, anti-oxidant, and anti-inflammatory activities21–23. However, sweet potato plants experience many post-harvest diseases during transportation and storage. Black rot, which is caused by Ceratocystis fimbriata, is one of the most devastating, causing serious economic and resource losses worldwide24–26. These pathogenic fungi are carried in the tuberous roots and cause disease in the progeny, and cannot currently be controlled by fungicides or other chemicals. Therefore, the control of black rot in sweet potato is a serious challenge, irrespective of the reproduction, planting, and storage techniques that are used, due to the extensive and intractable nature of this disease24,26,27. It has recently been shown that an endophytic bacterium of Arabidopsis thaliana known as Rhodococcus sp. KB6 inhibits black rot disease caused by C. fimbriata in sweet potato leaves25. However, there are still few reports of methods for controlling this disease.
A number of endophytic bacteria have been isolated from sweet potato at different production stages and from a range of tissues and plant genotypes28. In particular, bacteria in the genus Bacillus have been shown to be highly enriched in the tuber rhizosphere of many genotypes of sweet potato, while other genera exhibit a plant genotype dependent affluence28,29. In particular, the genera Brucella, Sphingobium, Comamonas, Methylophilus, Pantoea, Acinetobacter, Pseudomonas, Stenotrophomonas, Chryseobacterium, and Sphingobacterium are abundant in the tuber rhizosphere of the IPB-137 genotype28. Some of the yellow-pigmented, gram-negative bacteria in the genus Pantoea (family Erwiniaceae) produce antimicrobials and have been developed into commercial biocontrol products to help control fire blight on apple and pear trees, such as BlightBan C9-1 and BloomtimeTM biological30–32, while others have bioremediation potential, with the ability to degrade herbicides without generating toxic products33. However, while the endophyte P. agglomerans has been isolated from the stem of sweet potato34, it remains unknown whether bacteria in this genus can counteract the growth of sweet potato fungal pathogens.
In this study, we isolated microorganisms from the shoots and tuberous roots of sweet potato plants growing in the field and examined their inhibition activity against the fungal pathogen C. fimbriata. We found that four P. dispersa strains (RO-18, RO-20, RO-21, and SO-13) showed strong inhibition activity against C. fimbriata, with cell-free culture supernatant inhibiting spore germination and causing cellular changes in the hyphal morphology, including hyphal swelling, distortion, and cytoplasmic aggregation. Furthermore, we confirmed that these bacteria exhibit fungicidal rather than fungistatic activity. These results indicate the value of P. dispersa strains for reducing black rot infection in sweet potato and decreasing the use of agricultural chemicals.
Results
Molecular identification and phylogenetic analysis of the microbial communities in sweet potato plants
We collected a total of 75 species of microorganisms from the tuberous roots (RO) and shoots (SH) of field-grown sweet potato plants and the bulk soil (SO). These isolates could be identified to the genus and species level with a sequence similarity of 97–100% based on 16S or 5.8S rRNA gene sequences in the NCBI database.
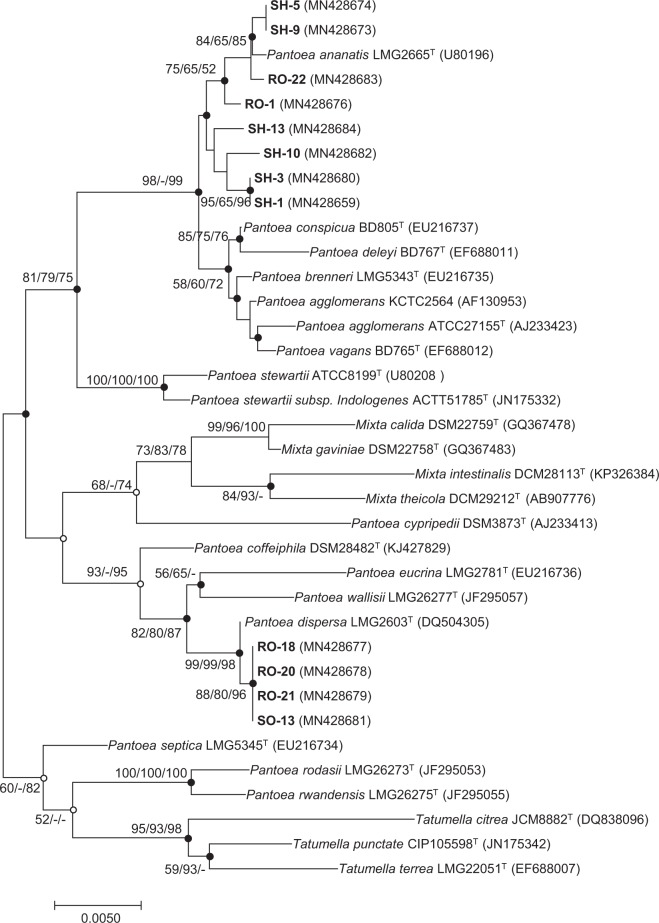
The isolates were identified as bacteria belonging to 15 genera: Arthrobacter, Bacillus, Bacterium, Burkholderia, Cupriavidus, Enterobacter, Leclercia, Lysinibacillus, Microbacteriaceae, Microbacterium, Pantoea, Pseudomonas, Psychrobacillus, Serratia, and Streptomyces; fungi belong to 17 genera: Aspergillus, Cladosporium, Cryptococcus, Fusarium, Gongronella, Mortierella, Mucor, Neurospora, Papiliotrema, Penicillium, Phoma, Rhizomucor, Rhodosporidium, Rhodotorula, Sakaguchia, Torula, and Mucor. Results revealed that 75 species endophytic microorganisms were belonging to 32 genera. At the genus level, the samples were predominantly populated with variable numbers of individuals in the genera Pantoea and Bacillus (Supplementary Tables S2–S4). The phylogenetic tree based on the 16S rRNA, gyrB and rpoB gene which generated from NJ, ML and ME algorithm showed that all species of the genus Pantoeas divided into multiple clades, while Pantoea isolates in this study fell into two clades, P. dispersa, and P. ananatis (Fig. 1; Supplementary Figs S1 and S2). The isolates RO-18, RO-20, RO-21, and SO-13 were most closely related to the type strain Pantoea dispersa LMG2603T, while isolates RO-1, RO-22, SH-5, SH-9, SH-1, SH-10, SH-13, and SH-3 were most closely related to the type strain Pantoea ananatis LMG2665T. These independent monophyletic clades clearly determined the species name of Pantoea collected from sweet potato tissue.
Figure 1.
Neighbor-joining phylogenetic tree based on 16S rRNA showing the relative positions of the Pantoea isolates and other Pantoea type strains. Evolutionary analyses were conducted in MEGA7.0 and bootstrap values (based on 1,000 replicates) greater than 50 are indicated at the branch nodes. Filled circles on the nodes indicate that the relationships were also recovered by ML and ME algorithms, whereas open circles indicate nodes recovered by either the ML or the ME algorithm. The bar represents 0.005 substitutions per nucleotide position.
Colonization of Pantoea in sweet potato plant tissues
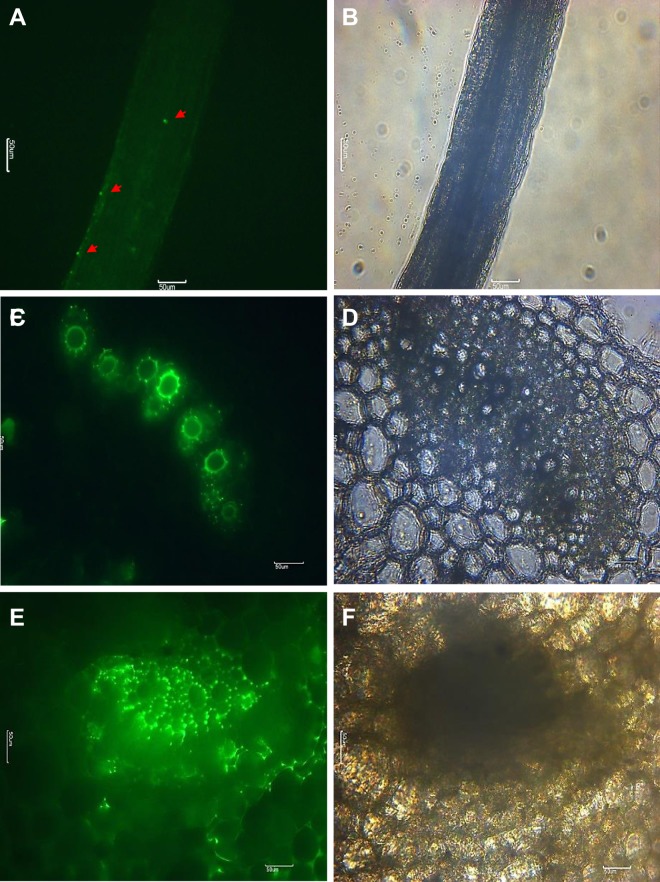
To visualize the location of Pantoea in sweet potato plants by live-cell imaging, we inoculated the roots of the sweet potato cultivar “Yulmi” with a suspension of GFP-labeled P. dispersa RO-21. Individual colonies of GFP-expressing RO-21 were observed in the roots 18 h after inoculation (Fig. 2A,B; Supplementary Fig. S3). After 48 h, bacterial aggregates were observed in horizontal sections of stems (Fig. 2C–F), and cells were effectively observed residing in the intercellular spaces of the outer cortex and there was extensive colonization in the cellular pits of the xylem tracheids in the stems (Fig. 2C–F). After 7 d, GFP-expressing macro-colonies of RO-21 was found in the leaf petioles (Supplementary Fig. S4), and it was able to recover by selective medium (Supplementary Fig. S5). This result suggests that P. dispersa effectively colonizes sweet potato tissues as an endophytic bacterium.
Figure 2.
Visualization of green fluorescent protein (GFP)-expressing Pantoea in sweet potato (Ipomoea batatas “Yulmi”) plant tissues. Sweet potato plants were inoculated with GFP-labeled P. dispersa RO-21 at 108 colony-forming units (CFU) mL−1. The plant roots and vascular system were then examined under an epifluorescence microscope. (A) Single colonies were observed in the roots at 18 h after inoculation. (C,E) Multiple colonies were observed inside the plant’s vascular system at 48 h after inoculation. (B,D,F) Transmission images of plant cells are shown. Green fluorescence and transmission images were taken at 20× (A,B) and 40× (C–F) magnification. Each arrow indicates the position of single colonies of GFP-expressing Pantoea. Scale bars are 50 μm.
Evaluation of in vitro antagonism
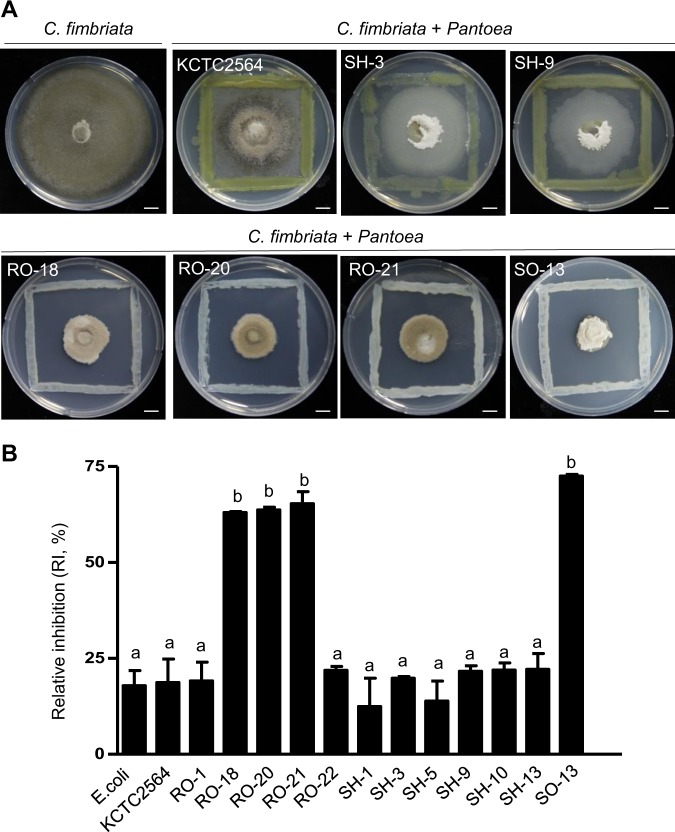
To determine the potential of endophytic Pantoea to control C. fimbriata, Pantoea isolates were screened for their inhibitory ability using an in vitro dual culture assay on PDA media. Mycelial plugs of C. fimbriata (5-mm diameter) were placed at the center of PDA plates and bacterial colonies were streaked around the borders of the plates (Fig. 3A; Supplementary Fig. S6). The antagonistic activity of the Pantoea strains was then estimated by comparing the fungal growth with C. fimbriata alone. Since P. agglomerans was previously identified from sweet potato31, we introduced P. agglomerans KCTC2564 (=ATCC27155) as additional Pantoea strain for this test. Fungal growth was monitored by measuring the diameter of the mycelium until day 16 at 28 °C (Fig. 3A; Supplementary Fig. S6A and S6B). This approach allowed the relative growth of C. fimbriata mycelia to be quantitatively assessed (Fig. 3B).
Figure 3.
Inhibition effects of Pantoea isolates against the phytopathogenic fungus C. fimbriata. (A) Dual culture assay of C. fimbriata and Pantoea isolates at 16 d. Agar plugs of C. fimbriata (5-mm diameter) were co-incubated with Pantoea strains SH-3, SH-9, RO-18, RO-20, RO-21, and SO-13 for 16 d at 28 °C. Escherichia coli and P. agglomerans KCTC2564 were used as controls (scale bar = 1 cm). (B) The relative inhibition (RI) rate of each Pantoea strain against C. fimbrata. The RI was calculated using the formula RI (%) = [(radial growth in control − radial growth in sample)/radial growth in control] × 100%. The bars indicate the standard errors of triplicate samples. Values followed by a different letter are significantly different (P ≤ 0.05).
Pantoea strains RO-18, RO-20, RO-21, and SO-13, which were identified as P. dispersa, had the highest inhibition rates, reaching >63% at 16 d after co-incubation (Fig. 3B). Among these, RO-21 and SO-13 showed the best inhibition activity, decreasing mycelial growth by up to 72% and significantly reducing the diameter of the fungal disks from 7.3 cm with the control treatment to 2.4–2.6 cm (Supplementary Fig. S6B). All four of these strains exhibited a clear inhibition halo (Fig. 3A). Some of the other bacterial strains also showed weak inhibition activity on C. fimbriata growth (19–34%), including Escherichia coli DH5 α, P. agglomerans KCTC2564, and P. ananatis strains SH-5, SH-9, SH-1, RO-22, SH-3, and RO-1 (Fig. 3A,B; Supplementary Fig. S6A); however, there was no statistically significant reduction in the diameter of the fungal disks with these treatments and the negative control (P ≥ 0.05) (Fig. 3B). These results indicate that P. dispersa strains RO-18, RO-20, RO-21, and SO-13 are good candidates as potential biocontrol agents of C. fimbriata.
Effect of cell-free culture supernatant of Pantoea on spore germination and hyphal morphology
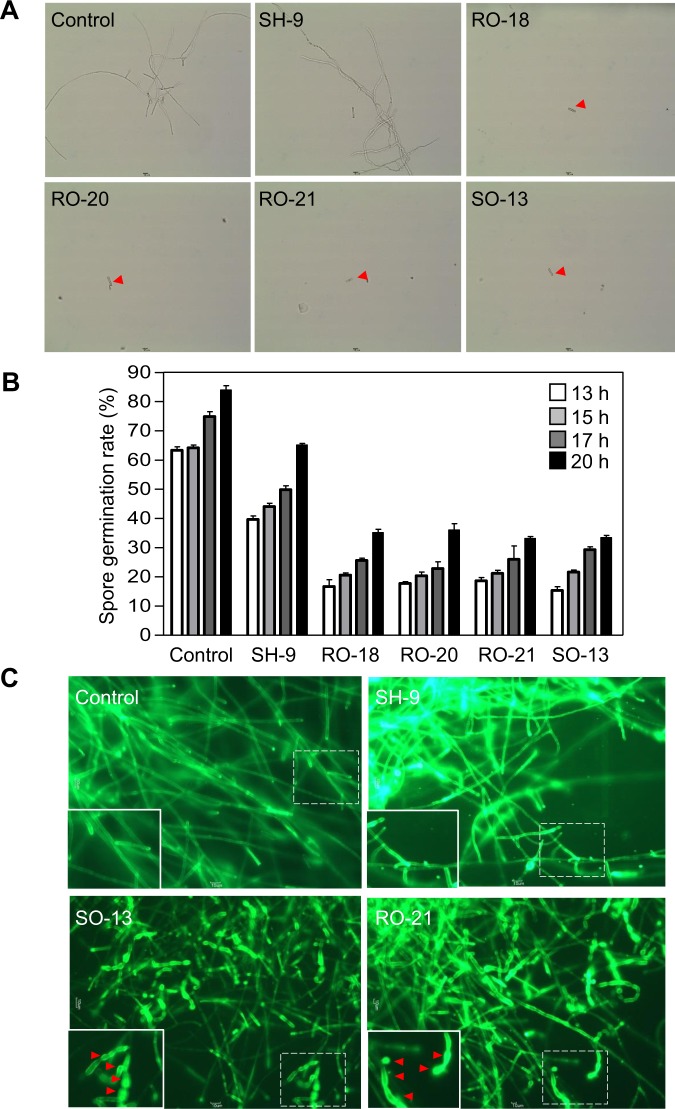
To determine whether the extracellular compounds that are produced by P. dispersa have antifungal activity, we observed the spore germination rates and morphologies of C. fimbriata following co–culture of the spores (105 CFU mL−1) with 900 μL of cell–free culture supernatant of Pantoea strains SH-9, RO-18, RO-20, RO-21, and SO-13. Light microscopy was then used to evaluate the effect of extracellular metabolites in the culture filtrates on the spore germination rates. Incubation with the cell-free culture supernatant of SH-9 did not significantly affect the C. fimbriata germination rates compared with the control after 20 h [germination rates = Control (84.0%) and SH-9 (65.1%), respectively]. However, the germination rates of the spores were inhibited by incubation with the cell-free supernatant of RO-18 (35.1%), RO-20 (36.0%), RO-21 (33.1%), and SO-13 (33.4%) after 20 h (Fig. 4A,B). Staining with FITC-WGA further showed that extracellular metabolites in the culture filtrates of P. dispersa strains RO-21 and SO-13 caused cellular changes in the hyphal morphology of C. fimbriata, including hyphal swelling, distortion, and cytoplasmic aggregation, while incubation with the cell-free culture supernatant of P. ananatis strain SH-9 did not cause any changes in hyphal morphology (Fig. 4C). Together, these findings suggest that the extracellular metabolites of P. dispersa have antifungal activity against C. fimbriata.
Figure 4.
Effects of the cell-free supernatant of Pantoea isolates on the C. fimbriata spore germination rates and hyphal morphology. (A) Effects of various Pantoea strains on the C. fimbriata spore germination rates. Strains RO-18, RO-20, RO-21, and SO-13 strongly inhibited spore germination, whereas strain SH-9 exhibited only weak antifungal activity and the negative control (no treatment) showed no inhibition. The experiment was repeated twice with similar results. (B) Effect of the spore germination rates on the co-culturing time. (C) Alteration of the fungal cell wall by cell-free culture supernatant of Pantoea isolates. Mycelia were stained with fluorescein isothiocyanate-labeled wheat germ agglutinin (FITC-WGA). Insets show enlarged views of the boxed areas. Arrows indicate hyphal swelling, distortion, and cytoplasmic aggregation.
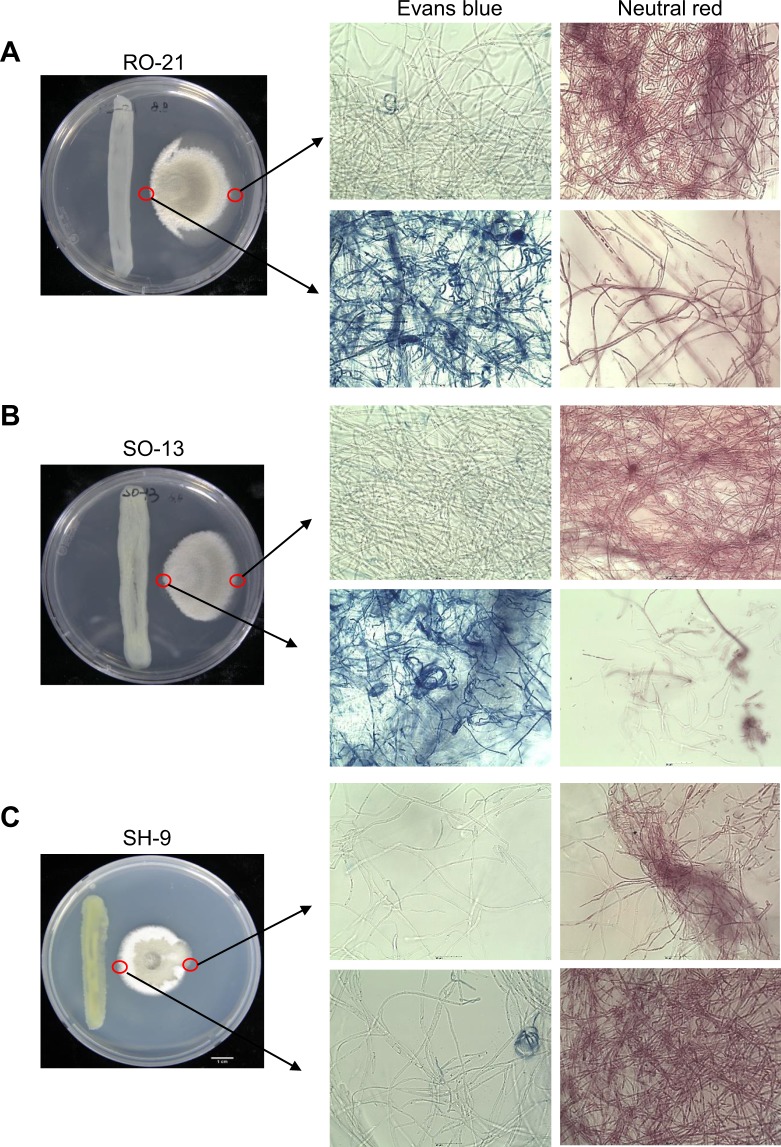
In vitro interaction between Pantoea isolates and C. fimbriata
To better understand the mode of action of the antifungal activity of P. dispersa strains, C. fimbriata was grown alongside Pantoea strains and the cell viability was visualized by staining with Evans blue, which stains dead cells blue, and Neutral red, which stains viable cells red. P. dispersa RO-21 and SO-13 appeared to cause the dramatic breakage of C. fimbriata hyphae in the contact zone, with the mycelia that were exposed to P. dispersa staining blue, while those in the control zone on the other side only exhibited faint blue staining (Fig. 5A,B). Similar results were obtained from RO-20 and RO-18 (Supplementary Fig. S7A and S7B). By contrast, in the presence of P. ananatis SH-9 and SH-3, faint blue staining and red staining were observed in both the contact and control zones (Fig. 5C; Supplementary Fig. 7C). These findings indicate that P. dispersa has fungicidal activity rather than fungistatic activity against C. fimbriata.
Figure 5.
Interaction effect of C. fimbriata with Pantoea RO-21, SO-13 and SH-9. C. fimbriata cells were stained with Evans blue (dead cells stain blue) and Neutral red (viable cells stain red) after co-cultivation with strain RO-21, SO-13 and SH-9 for 10 d. (A,B) Mycelia growing near to RO-21 and SO-13 stained blue, while those in the control zone on the other side only exhibited faint staining. (C) By contrast, mycelia that were co-cultured with SH-9 did not stain blue in any region. Scale bars are 50 μm.
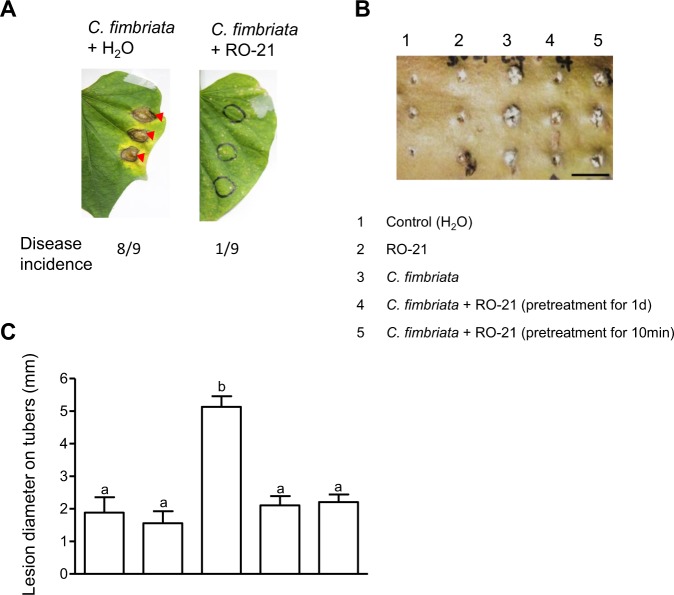
Inhibition of C. fimbriata infection in sweet potato leaves and tuberous roots
To assess the biocontrol efficiency of P. dispersa in the leaves and tuberous roots of the sweet potato cultivar “Yulmi”, which is susceptible to C. fimbrata infection23, samples were pre-treated with RO-21 or water (as a control) and then inoculated at the same site with droplets containing C. fimbriata spores. The disease incidence on the leaves and tuberous roots was then determined. Leaves that had been pre-treated with water showed disease symptoms at the inoculation site, with dark brown circular patches of 4–6 mm diameter appearing on the upper surface of the leaves and the surrounding areas turning yellow after 36 h. By contrast, the disease incidence was reduced in leaves that had been pre-treated with RO-21 (Fig. 6A). Disease symptoms were also dramatically reduced in tuberous roots that had been pre-treated with RO-21 cells for 1 d before C. fimbriata inoculation compared with those that had been pre-treated with water as a control (Fig. 6B,C). Furthermore, pre-treatment with RO-21 for only 10 min also reduced the size of the black lesions around the inoculation site compared with the water pre-treatment (Fig. 6C). Together, these findings indicate that P. dispersa could prevent the infection of sweet potato leaves and tuberous roots by the pathogenic fungus C. fimbriata.
Figure 6.
Effects of co-inoculation with Pantoea RO-21 and C. fimbriata on the detached leaves and tuberous roots of sweet potato (Ipomoea batatas “Yulmi”) plants. (A) Disease incidence in the leaves of sweet potato plants pre-treated with water (control; left) and RO-21 (right) at 36 h after co-incubation with a spores droplet of C. fimbriata. (B) Disease incidence in the tuberous roots at 36 h after inoculation following pre-treatment with RO-21 or water (control) at 10 d after co-incubation with a spores droplet of C. fimbriata (scale bar = 1 cm). (C) Diameter of the lesion region, as evaluated by ImageJ analysis. The values shown are the average of at least two independent experiment performed in triplicate. Error bars indicate the standard error of the means (SEM) values. Values followed by a different letter are significantly different (n = 3–6, P ≤ 0.05).
Discussion
The composition of any given plant-associated microbial community is likely to be determined by multiple factors, including the soil type, phytopathogen population12, plant age35, and plant genotype, as well as stochastic sampling factors36. In the present study, we identified 75 culturable microbes that were associated with field-grown sweet potato plants based on their colony morphologies, colors, and patterns of growth. These included members of the genera Bacillus, Pantoea, Enterobacter, Serratia, and Microbacterium (Supplementary Tables S2 and S3), indicating that these plants are associated with diverse bacterial communities. However, only a few species dominated these communities, with the genera Bacillus and Pantoea predominating in the shoots and tuberous roots (Supplementary Tables S2 and S3).
Bacillus species are considered attractive biocontrol agents due to their ability to produce hard, resistant endospores and antibiotics that control a broad range of plant fungal pathogens37. However, it has previously been found that B. subtilis does not effectively inhibit the growth of C. fimbriata38, which is consistent with the findings for the Bacillus strains isolated in the present study (data not shown). The bacterial genus Pantoea comprises many versatile species that have been isolated from a multitude of environments, such as mammals, agricultural areas, water, and particularly soil39. El Amraoui40 found that species showed stronger antifungal activity against Candida albicans CIP 48.72, Candida albicans CIP 884.65, and Cryptococcus neoformans CIP 960 than Bacillus sp., while Town41 showed that Pantoea sp. inhibits the potato pathogen Phytophthora infestans. In the present study, Pantoea strains RO-18, RO-20, RO-21, and SO-13, which were isolated from sweet potato, had a high homology with the type strain P. dispersa LGM 2603T and exhibited remarkable antifungal activity against C. fimbriata both in vitro and in vivo, with the in vitro dual culture assay showing that they inhibited C. fimbriata mycelium growth (Fig. 3A) and spore germination (Fig. 4A,B), appeared to cause dramatic breakage of the fungal hyphae (Fig. 4C), and killed the fungal hyphae, acting as a fungicidal agent (Fig. 5A,B). Therefore, genus Pantoea has a greater potential to control C. fimbriata than genus Bacillus.
Root-inoculated GFP-labeled RO-21 cells were initially visualized in the root surface and central vascular system of primary root in sweet potato plants within 18 h, following which a proliferation of cells was observed in the intercellular spaces between adjacent xylem tracheid cells in the caulosphere at 48 h after inoculation. The rapid spread of this strain from the root to the aerial tissues suggests that it uses the vascular system as a route for systemic colonization. Other endophytic bacteria are also located in the vascular system, with large fluorescent colonies having been clearly observed in the pits of the xylem cells in the plant cell wall42,43.
The mode of action of P. dispersa is similar to that of B. subtilis against Eutypa lata in grapes, whereby irregularities occur in the hyphal morphology, such as tip narrowing and vesicle formation44. B. subtilis has been shown to cause abnormalities in the hyphae of Fusarium oxysporum, causing cell wall lysis, breakage, granulation, and vacuolization45. Similar phenomenon was observed by P. dispersa isolates (Figs 4 and 5). Moreover, the leaves and tuberous roots of sweet potato plants had a significantly decreased incidence of disease symptoms when cultured in the presence of RO-21 (Fig. 6A,B), indicating that P. dispersa can inhibit the development of black rot disease in sweet potato. Moreover, strains RO-18, RO-20, RO-21, and SO-13 were initially isolated from the tuberous roots of agricultural sweet potato and soil, which can be explained by the fact that both P. dispersa and C. fimbriata are soil-borne microorganisms that primarily compete for the tuberous roots5.
Although P. ananatis strains SH-1, SH-3, SH-5, SH-9, SH-10, SH-13, RO-1, and RO-22 did not effectively inhibit the growth of C. fimbriata in this study, it has previously been shown that P. ananatis CPA-3 has strong antifungal activity against Penicillium expansum46. Similarly, P. ananatis 125NP12 have been found to protect tomato fruit against the grey mold fungus, Botrytis cinerea, by producing antifungal compounds47. In this respect, P. ananatis strains SH-1, SH-3, SH-5, SH-9, SH-10, SH-13, RO-1, and RO-22 might control other plant pathogens in sweet potato plants. In addition, the potential of P. ananatis SCB4789F-1 to promote plant growth has been demonstrated by its ability to solubilize phosphorus and zinc, produce siderophores, and synthesize IAA. P. ananatis B1-9 strain isolated from the rhizosphere of onions showed potential for promoting plant growth in peppers, cucumbers, and melons48. Therefore, further detailed studies are needed on the plant growth-promoting effect of P. ananatis isolates.
Several mechanisms have been reported for the biocontrol of plant pathogens, including competition for nutrients49, the induction of host resistance50,51, and the production of killer toxins52, degradable enzymes such as chitinase6, and antifungal metabolites53–55. Many Pantoea strains are strong environmental competitors that produce a variety of natural products with antibiotic activity, such as pantocins, microcins, and phenazines56–64. More recently, Pantoea Natural Product 1 (PNP-1), which has been isolated from P. ananatis BRT175, has been shown to have inhibitory activity against Erwinia amylovora and is likely to be similar in action to 4-formylaminooxyvinylglycine (FVG)65,66. In the present study, cell-free supernatant derived from P. dispersa cultures was found to have inhibitory effects on C. fimbriata spore germination and hyphae growth when added simultaneously or after some time, indicating that the antifungal activity of these strains may occur via similar mechanisms. Hence, further investigation is required to evaluate the potential bioactive compounds that may be of biocontrol importance in the metabolites of these isolates.
Together, our results indicate that P. dispersa strains represent useful biocontrol agents for protecting sweet potato plants from post-harvest infection by C. fimbriata and for decreasing the use of agricultural chemical.
Methods
Isolation of microorganisms
Tuberous roots and shoots were collected from healthy sweet potato (I. batatas) plants growing in a cultivation area in Jeongeup, Jeollabuk-do, Republic of Korea (35°30′29.8″N, 126° 50′13.9″E). To acquire plant extracts, the plant materials were extensively washed with sterile distilled water, and aliquot (100 μL) of the final rinse water was plated onto Laurie-Bertani (LB) and Potato Dextrose agar (PDA) media (Difco) to check the disinfection process. The plant tissues were then crushed with 50 mL of sterile phosphate-buffered saline (PBS: 100 mM Tris-HCl, pH 8.0; 150 mM NaCl) and filtered through four layers of sterile cheesecloth. The bacterial cells in the filtrate were serially diluted with PBS buffer, and 100 μL of each diluted sample was spread onto an LB and PDA media plates, and then incubated at 28 °C for 5–14 d.
The colony types in each sample were categorized according to their growth rate and phenotypic characteristics, such as sizes, colors, and colony morphology. Each type was then counted and a representative was taken for purification and identification. The following voucher specimens were deposited at the Korean Collection for Type Cultures (KCTC), Korea Research Institute of Bioscience & Biotechnology (KRIBB): KCTC 62154 (SO-13), KCTC 62153 (SH-13), KCTC 62152 (SH-10), KCTC 62151 (SH-9), KCTC 62150 (SH-5), KCTC 62149 (SH-3), KCTC 62148 (SH-1), KCTC 62147 (RO-22), KCTC 62146 (RO-21), KCTC 62145 (RO-20), KCTC 62144 (RO-18), and KCTC 62143 (RO-1).
Species identification and phylogenetic analysis using 16S and 5.8S rRNA gene sequences
The 16S, 5.8S rRNA and housekeeping genes (gyrB, and rpoB)67 of each species were amplified using the primers listed in Supplementary Table S1. 16S and 5.8S rRNA sequencing was carried out at BioFact Inc. (Daejeon, Korea), and sequence alignment was performed with the BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic trees were constructed using the neighbor-joining (NJ), minimum-evolution (ME), and maximum-likelihood (ML) method based on 16S, gyrB, and rpoB genes in MEGA (ver.7)68, and a bootstrap analysis of 1,000 replications was performed to evaluate the stability of the nodes.
Introduction of a plasmid-born green fluorescent protein (GFP) reporter into P. dispersa RO-21
To obtain GFP-expressing P. dispersa RO-21, the bacterial cells were transformed with the plasmid pGFPuvTM (Clontech Laboratories Inc., CA, USA). 1 μL of the plasmid (200 ng μL−1) was introduced into the cells by electroporation with the Gene Pulse XcellTM system (Bio-Rad, Hercules, CA, USA) in a 0.2-cm electroporation cuvette (2500 V cm−1, 25 μF, 200 Ω). The cells were allowed to recover in 1 mL of LB broth and then incubated for 30 min at 30 °C with shaking (150 rpm). The cells were then spread on a LB plate with ampicillin (100 μg mL−1) and incubated at 30 °C overnight to select the transformants, which were identified using ultraviolet (UV) light.
Monitoring bacterial colonization on sweet potato plants
Sweet potato plants that had been separated from their tuberous roots were placed in a continuous-floating hydroponic system for 5 d until their roots had reached a length of 4–5 cm. The roots of the plants were then inoculated with a suspension of GFP-expressing P. dispersa RO-21 [108 colony-forming units (CFU) mL−1]. After 15 min, the plant roots were washed three times with distilled water and then kept in the floating hydroponic system. GFP-expressing bacteria were detected in the root 18 h after inoculation. At 4 d and 7 d after inoculation, the stems of each sweet potato plants were cut into sections that were as thin as possible using a razor blade from 5 cm above the roots. Stem cross-sections and selected roots from each plant were then placed on a glass slide with distilled water, and the GFP-expressing bacteria were observed using an epifluorescence microscope with a GFP filter (Moticam Pro 205 A; Motic) at 20× or 40× magnification. To confirm the GFP signals were not due to the auto-fluorescence of the plant tissue itself, re-isolation of the Pantoea strain RO-21 were performed. Leaf petioles from the strain RO-21 inoculated plants for 7 d were collected, then surface-sterilized by using 1.05% sodium hypochlorite for 10 min and several repeats rinse in sterile distilled water. After homogenized with PBS buffer, samples were spread on LB agar plates supplementary with ampicillin (100 μg mL−1).
Dual culture assay
To test the activity of the bacterial strains against C. fimbriata, mycelial plugs of C. fimbriata (5-mm diameter) were placed in the center of PDA plates (pH 5.7) and the various bacterial strains were streaked in a square shape around each agar disk at a distance of 3 cm from the mycelial plug. In addition, mycelial plugs were placed on PDA plates without any bacterial strains as a negative control. Fungal growth was assessed by measuring the diameter (in centimeters) of the colony until 16 d at 25 °C. Each bacterial strain was tested in triplicate and the experiment was carried out twice. The relative inhibition (RI) was then calculated using the formula RI (%) = [(radial growth in control − radial growth in samples)/radial growth in control] × 100, as previously described69. Data were analyzed by one-way analysis of variance (ANOVA) using GraphPad Prism 4. Significant differences (P ≤ 0.05) between the means were determined by unpaired t-test and Tukey’s multiple comparison test.
Microscopic analysis
A fungal spore germination assay was performed using cell-free supernatant of the Pantoea strains. Bacterial cells were harvested from 6-d-old liquid cultures of SH-9, RO-18, RO-20, RO-21, and SO-13 by centrifugation for 20 min at 10,000×g. The resulting supernatant was then filtered through a 0.2 μm filter (WhatmanTM) to remove the cells. C. fimbriata spores were collected from a 2-week-old culture of fungi growing on PDA media and placed in 10 mL of sterile distilled water, following which they were passed through 25 μm sterile MiraclothTM (Millipore) to remove the hyphae and subsequently diluted to 105 spore mL−1 for the bioassay. Then, 104 spores were co-incubated with 900 μL of each of the cell-free supernatant of SH-9, RO-18, RO-20, RO-21, and SO-13 for 13, 15, 17, and 20 h in triplicate. Spore germination (%) was evaluated in each sample by placing the spores on a microscope slide and counting at least 300 spores per sample. Once the fungal spores had been left to germinate in the cell-free supernatant of the Pantoea strains for 36 h, the hyphae were stained with fluorescein isothiocyanate-labeled wheat germ agglutinin (FITC-WGA, 1 μg mL−1) for 30 min to investigate whether the cell-free supernatant of P. dispersa altered the fungal cell wall.
To evaluate the viability of this pathogenic fungus in the presence of Pantoea strains, C. fimbriata was co-cultivated alongside RO-18, RO-20, RO-21, SH-3 or SH-9 on PDA media at 25 °C for 10 d. The C. fimbriata mycelia that were adjacent to the bacteria, were then stained with the vitality stains Neutral red (0.1 mg mL−1; Dae Jung, Cat # 5603-4125) or Evans blue (0.5 mg mL−1; Alfa Aesar, Cat # A16774) by placing 10 μL of the solution on a slide, incubating the cells for 3–5 min at room temperature, and then washing 3–4 times with deionized water. Mycelia growing on the opposite sides from the bacteria were treated as a negative control. There were 3–4 replicates per treatment and photographs were taken under a light microscope (Nikon Eclipse Ci).
Disease incidence and symptoms
To evaluate the biocontrol effect of P. dispersa on C. fimbriata, the leaves (5–6 weeks old) and tuberous roots of plants of the sweet potato cultivar “Yulmi”, which is susceptible to C. fimbrata, were analyzed. The leaf surfaces were punched with a needle on the upper side, and then the punched sites were inoculated with 10 μL of P. dispersa cell suspension (107 CFU mL−1) pre-treatment for 1 d. The C. fimbriata spores after incubation for 7 d at 25 °C were scraped from well-grown PDA plates and filtered through 25 μm Miracloth in order to obtain spore suspensions. 5 μL of C. fimbriata suspension (106 CFU mL−1) were inoculated at the same site as used for pre-treatment. The inoculated leaves were covered with plastic wrap for 36 h, the yellow to black area of the punched leaves were recorded. All experiments were performed in triplicate, repeated at least twice. Surface of the tuberous roots was punched with a needle, then same treatment as described with leaves, unless inoculated with 10 μL of P. dispersa cell suspension (107 CFU mL−1) pre-treatment for 1 d and 10 min, then inoculated with 5 μL of C. fimbriata suspension at the same site as was used for pre-treatment, under moist conditions in the plastic box at 25 °C for 10 d. The disease incidence was recorded by observing the formation of the black spots of C. fimbriata and softening of the tissue in the collar region. All measurements were made in triplicate and repeated twice with similar results.
Supplementary information
Acknowledgements
This work was supported by the KRIBB Research Initiative Program (KGM5281913) and the Next-Generation BioGreen 21 Program (SSAC, Grant#: PJ01318604), Rural Development Administration, Republic of Korea. We thank Dr. Seung-Hwan Park (Biological Resources Center, KRIBB) for kindly sharing the vector pGFPuvTM.
Author contributions
S.W.K. and J.L. designed the research; L.J. carried out the experiment; J.-S.L., J.C.J., and D.-H.K. analyzed the results; J.L. and L.J. wrote the manuscript; J.M.P., J.-W.Y., M.H.L., S.H.C. and C.Y.K. modified the manuscript. All authors read and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Suk Weon Kim, Email: kimsw@kribb.re.kr.
Jiyoung Lee, Email: jiyoung1@kribb.re.kr.
Supplementary information
is available for this paper at 10.1038/s41598-019-52804-3.
References
- 1.Rejeb KB, Abdelly C, Savouré A. How reactive oxygen species and proline face stress together. Plant Physiol. Biochem. 2014;80:278–284. doi: 10.1016/j.plaphy.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Walitang DI, et al. The influence of host genotype and salt stress on the seed endophytic community of salt-sensitive and salt-tolerant rice cultivars. BMC plant biology. 2018;18:51. doi: 10.1186/s12870-018-1261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gouda S, Das G, Sen SK, Shin H-S, Patra JK. Endophytes: a treasure house of bioactive compounds of medicinal importance. Front. microbiology. 2016;7:1538. doi: 10.3389/fmicb.2016.01538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mei C, Flinn BS. The use of beneficial microbial endophytes for plant biomass and stress tolerance improvement. Recent Patents on Biotechnol. 2010;4:81–95. doi: 10.2174/187220810790069523. [DOI] [PubMed] [Google Scholar]
- 5.Fouda AH, Hassan SE-D, Eid AM, Ewais EE-D. Biotechnological applications of fungal endophytes associated with medicinal plant Asclepias sinaica (bioss.) Annals Agric. Sci. 2015;60:95–104. doi: 10.1016/j.aoas.2015.04.001. [DOI] [Google Scholar]
- 6.Shehata HR, Ettinger CL, Eisen JA, Raizada MN. Genes required for the anti-fungal activity of a bacterial endophyte isolated from a corn landrace grown continuously by subsistence farmers since 1000 BC. Front. microbiology. 2016;7:1548. doi: 10.3389/fmicb.2016.01548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shehata HR, Lyons EM, Jordan KS, Raizada MN. Relevance of in vitro agar based screens to characterize the anti-fungal activities of bacterial endophyte communities. BMC microbiology. 2016;16:8. doi: 10.1186/s12866-016-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassan SE-D. Plant growth-promoting activities for bacterial and fungal endophytes isolated from medicinal plant of Teucrium polium L. J. advanced research. 2017;8:687–695. doi: 10.1016/j.jare.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kandel SL, et al. An in vitro study of bio-control and plant growth promotion potential of Salicaceae endophytes. Front. microbiology. 2017;8:386. doi: 10.3389/fmicb.2017.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemanceau P, et al. Effect of two plant species, flax (Linum usitatissinum L.) and tomato (Lycopersicon esculentum Mill.), on the diversity of soilborne populations of fluorescent pseudomonads. Appl. Environ. Microbiol. 1995;61:1004–1012. doi: 10.1128/aem.61.3.1004-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siciliano SD, et al. Selection of specific endophytic bacterial genotypes by plants in response to soil contamination. Appl. Environ. Microbiol. 2001;67:2469–2475. doi: 10.1128/AEM.67.6.2469-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granér G, Persson P, Meijer J, Alström S. A study on microbial diversity in different cultivars of Brassica napus in relation to its wilt pathogen, Verticillium longisporum. FEMS microbiology letters. 2003;224:269–276. doi: 10.1016/S0378-1097(03)00449-X. [DOI] [PubMed] [Google Scholar]
- 13.Jousset A, et al. Plants respond to pathogen infection by enhancing the antifungal gene expression of root-associated bacteria. Mol. plant-microbe interactions. 2011;24:352–358. doi: 10.1094/MPMI-09-10-0208. [DOI] [PubMed] [Google Scholar]
- 14.Rasche F, Trondl R, Naglreiter C, Reichenauer TG, Sessitsch A. Chilling and cultivar type affect the diversity of bacterial endophytes colonizing sweet pepper (Capsicum anuum L.) Can. journal microbiology. 2006;52:1036–1045. doi: 10.1139/w06-059. [DOI] [PubMed] [Google Scholar]
- 15.Somers E, Vanderleyden J, Srinivasan M. Rhizosphere bacterial signalling: a love parade beneath our feet. Critical reviews microbiology. 2004;30:205–240. doi: 10.1080/10408410490468786. [DOI] [PubMed] [Google Scholar]
- 16.Bakker PA, Berendsen RL, Doornbos RF, Wintermans PC, Pieterse CM. The rhizosphere revisited: root microbiomics. Front. plant science. 2013;4:165. doi: 10.3389/fpls.2013.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lladó S, Baldrian P. Community-level physiological profiling analyses show potential to identify the copiotrophic bacteria present in soil environments. PLoS One. 2017;12:e0171638. doi: 10.1371/journal.pone.0171638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakshmanan V, Selvaraj G, Bais HP. Functional soil microbiome: belowground solutions to an aboveground problem. Plant physiology. 2014;166:689–700. doi: 10.1104/pp.114.245811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mommer L, Hinsinger P, Prigent-Combaret C, Visser EJ. Advances in the rhizosphere: stretching the interface of life. Plant Soil. 2016;407:1–8. doi: 10.1007/s11104-016-3040-9. [DOI] [Google Scholar]
- 20.Nedunchezhiyan M, Byju G, Jata SK. Sweet potato agronomy. Fruit, Veg. Sereal Sci. Biotechnol. 2012;6:1–10. [Google Scholar]
- 21.Bovell-Benjamin AC. Sweet potato: a review of its past, present, and future role in human nutrition. Adv. food nutrition research. 2007;52:1–59. doi: 10.1016/S1043-4526(06)52001-7. [DOI] [PubMed] [Google Scholar]
- 22.El Sheikha AF, Ray RC. Potential impacts of bioprocessing of sweet potato. Critical reviews food science nutrition. 2017;57:455–471. doi: 10.1080/10408398.2014.960909. [DOI] [PubMed] [Google Scholar]
- 23.Paul NC, Nam S-S, Kachroo A, Kim Y-H, Yang J-W. Characterization and pathogenicity of sweet potato (Ipomoea batatas) black rot caused by Ceratocystis fimbriata in Korea. Eur. J. Plant Pathol. 2018;152:833–840. doi: 10.1007/s10658-018-1522-8. [DOI] [Google Scholar]
- 24.Muramoto N, et al. Transgenic sweet potato expressing thionin from barley gives resistance to black rot disease caused by Ceratocystis fimbriata in leaves and storage roots. Plant cell reports. 2012;31:987–997. doi: 10.1007/s00299-011-1217-5. [DOI] [PubMed] [Google Scholar]
- 25.Hong CE, et al. A leaf-inhabiting endophytic bacterium, Rhodococcus sp. KB6, enhances sweet potato resistance to black rot disease caused by Ceratocystis fimbriata. J Microbiol Biotechnol. 2016;26:488–492. doi: 10.4014/jmb.1511.11039. [DOI] [PubMed] [Google Scholar]
- 26.Okada, Y., Kobayashi, A., Tabuchi, H. & Kuranouchi, T. Review of major sweetpotato pests in Japan, with information on resistance breeding programs. Breed. science 16145 (2017). [DOI] [PMC free article] [PubMed]
- 27.Zhang M, et al. Perillaldehyde controls postharvest black rot caused by Ceratocystis fimbriata in sweet potatoes. Front. microbiology. 2018;9:1102. doi: 10.3389/fmicb.2018.01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marques JM, et al. Plant age and genotype affect the bacterial community composition in the tuber rhizosphere of field-grown sweet potato plants. FEMS microbiology ecology. 2014;88:424–435. doi: 10.1111/1574-6941.12313. [DOI] [PubMed] [Google Scholar]
- 29.Marques JM, et al. Bacterial endophytes of sweet potato tuberous roots affected by the plant genotype and growth stage. Appl. Soil Ecol. 2015;96:273–281. doi: 10.1016/j.apsoil.2015.08.020. [DOI] [Google Scholar]
- 30.Johnson K, et al. Effect of antagonistic bacteria on establishment of honey bee-dispersed Erwinia amylovora in pear blossoms and on fire blight control. Phytopathol. (USA) 1993;83:995–1002. doi: 10.1094/Phyto-83-995. [DOI] [Google Scholar]
- 31.Johnson K, Stockwell V. Management of fire blight: a case study in microbial ecology. Annu. review phytopathology. 1998;36:227–248. doi: 10.1146/annurev.phyto.36.1.227. [DOI] [PubMed] [Google Scholar]
- 32.Johnson K, Stockwell V, Sawyer T, Sugar D. Assessment of environmental factors influencing growth and spread of Pantoea agglomerans on and among blossoms of pear and apple. Phytopathol. 2000;90:1285–1294. doi: 10.1094/PHYTO.2000.90.11.1285. [DOI] [PubMed] [Google Scholar]
- 33.Pileggi M, et al. Isolation of mesotrione-degrading bacteria from aquatic environments in Brazil. Chemosphere. 2012;86:1127–1132. doi: 10.1016/j.chemosphere.2011.12.041. [DOI] [PubMed] [Google Scholar]
- 34.Asis C, Jr, Adachi K. Isolation of endophytic diazotroph Pantoea agglomerans and nondiazotroph Enterobacter asburiae from sweetpotato stem in Japan. Lett. Appl. Microbiol. 2004;38:19–23. doi: 10.1046/j.1472-765X.2003.01434.x. [DOI] [PubMed] [Google Scholar]
- 35.Islam SMA, et al. Effect of plant age on endophytic bacterial diversity of balloon flower (Platycodon grandiflorum) root and their antimicrobial activities. Curr. microbiology. 2010;61:346–356. doi: 10.1007/s00284-010-9618-1. [DOI] [PubMed] [Google Scholar]
- 36.Hardoim PR, van Overbeek LS, van Elsas JD. Properties of bacterial endophytes and their proposed role in plant growth. Trends microbiology. 2008;16:463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Cavaglieri L, Orlando J, Rodriguez M, Chulze S, Etcheverry M. Biocontrol of Bacillus subtilis against Fusarium verticillioides in vitro and at the maize root level. Res. Microbiol. 2005;156:748–754. doi: 10.1016/j.resmic.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Sonyal S, Ravichandra N, Reddy B. Efficacy of bioagents on Ceratocystis fimbriata and Meloidogyne incognita wilt complex in pomegranate. ecialise Sp. 2015;49:350–354. [Google Scholar]
- 39.Walterson AM, Stavrinides J. Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 2015;39:968–984. doi: 10.1093/femsre/fuv027. [DOI] [PubMed] [Google Scholar]
- 40.El Amraoui B, El Amraoui M, Cohen N, Fassouane A. Anti-Candida and anti-Cryptococcus antifungal produced by marine microorganisms. J. de mycologie medicale. 2014;24:e149–e153. doi: 10.1016/j.mycmed.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Town J, Audy P, Boyetchko SM, Dumonceaux TJ. High-quality draft genome sequence of biocontrol strain Pantoea sp. oxwo6b1. Genome Announc. 2016;4:e00582–16. doi: 10.1128/genomeA.00582-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Germaine K, et al. Colonisation of poplar trees by gfp expressing bacterial endophytes. FEMS Microbiol. Ecol. 2004;48:109–118. doi: 10.1016/j.femsec.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 43.Akbari DLA, Akbari LF. Detection of gfp expression and colonization of wheat by two endophytic bacteria tagged with gfp gene. Int. J. Pure App. Biosci. 2017;5:844–848. doi: 10.18782/2320-7051.2476. [DOI] [Google Scholar]
- 44.Ferreira J, et al. Biological control of Eutypa lata on grapevine by an antagonistic strain of Bacillus subtilis. Phytopathol. 1991;81:283–287. doi: 10.1094/Phyto-81-283. [DOI] [Google Scholar]
- 45.Chaurasia B, et al. Diffusible and volatile compounds produced by an antagonistic Bacillus subtilis strain cause structural deformations in pathogenic fungi in vitro. Microbiol. research. 2005;160:75–81. doi: 10.1016/j.micres.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 46.Torres R, Teixido N, Usall J, Abadias M, Vinas I. Post-harvest control of Penicillium expansum on pome fruits by the bacterium Pantoea ananatis CPA-3. The J. Hortic. Sci. Biotechnol. 2005;80:75–81. doi: 10.1080/14620316.2005.11511895. [DOI] [Google Scholar]
- 47.Enya J, et al. Culturable leaf-associated bacteria on tomato plants and their potential as biological control agents. Microb. ecology. 2007;53:524–536. doi: 10.1007/s00248-006-9085-1. [DOI] [PubMed] [Google Scholar]
- 48.Kim S-N, Cho W-K, Kim W-I, Jee H-J, Park C-S. Growth promotion of pepper plants by Pantoea ananatis b1-9 and its efficient endophytic colonization capacity in plant tissues. The Plant Pathol. J. 2012;28:270–281. doi: 10.5423/PPJ.OA.02.2012.0026. [DOI] [Google Scholar]
- 49.Janisiewicz W, Tworkoski T, Sharer C. Characterizing the mechanism of biological control of postharvest diseases on fruits with a simple method to study competition for nutrients. Phytopathol. 2000;90:1196–1200. doi: 10.1094/PHYTO.2000.90.11.1196. [DOI] [PubMed] [Google Scholar]
- 50.Droby S, et al. Induction of resistance to Penicillium digitatum in grapefruit by the yeast biocontrol agent Candida oleophila. Phytopathol. 2002;92:393–399. doi: 10.1094/PHYTO.2002.92.4.393. [DOI] [PubMed] [Google Scholar]
- 51.El Ghaouth A, Wilson CL, Wisniewski M. Control of postharvest decay of apple fruit with Candida saitoana and induction of defense responses. Phytopathol. 2003;93:344–348. doi: 10.1094/PHYTO.2003.93.3.344. [DOI] [PubMed] [Google Scholar]
- 52.Rosa MM, Tauk-Tornisielo SM, Rampazzo PE, Ceccato-Antonini SR. Evaluation of the biological control by the yeast Torulaspora globosa against Colletotrichum sublineolum in sorghum. World J. Microbiol. Biotechnol. 2010;26:1491–1502. doi: 10.1007/s11274-010-0324-8. [DOI] [Google Scholar]
- 53.Ertani A, Pizzeghello D, Francioso O, Tinti A, Nardi S. Biological activity of vegetal extracts containing phenols on plant metabolism. Mol. 2016;21:205. doi: 10.3390/molecules21020205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Veras FF, Correa APF, Welke JE, Brandelli A. Inhibition of mycotoxin-producing fungi by Bacillus strains isolated from fish intestines. Int. journal food microbiology. 2016;238:23–32. doi: 10.1016/j.ijfoodmicro.2016.08.035. [DOI] [PubMed] [Google Scholar]
- 55.Slimene IB, et al. Isolation of a chitinolytic Bacillus licheniformis S213 strain exerting a biological control against Phoma medicaginis infection. Appl. biochemistry biotechnology. 2015;175:3494–3506. doi: 10.1007/s12010-015-1520-7. [DOI] [PubMed] [Google Scholar]
- 56.Vanneste J, Yu J, Reglinski T, Allan A. Biocontrol agent Pantoea agglomerans strain NZ501 induces a resistance-like response in kiwifruit and tobacco cells. IX International Workshop on Fire Blight. 2001;590:279–283. [Google Scholar]
- 57.Vanneste J, Cornish D, Yu J, Voyle M. The peptide antibiotic produced by Pantoea agglomerans Eh252 is a microcin. IX International Workshop on Fire Blight. 2001;590:285–290. [Google Scholar]
- 58.Vanneste JL, et al. Biological control of sapstain fungi with natural products and biological control agents: a review of the work carried out in new zealand. Mycol. Res. 2002;106:228–232. doi: 10.1017/S0953756201005020. [DOI] [Google Scholar]
- 59.Giddens SR, Feng Y, Mahanty HK. Characterization of a novel phenazine antibiotic gene cluster in Erwinia herbicola eh1087. Mol. microbiology. 2002;45:769–783. doi: 10.1046/j.1365-2958.2002.03048.x. [DOI] [PubMed] [Google Scholar]
- 60.Smits TH, Rezzonico F, Duffy B. Evolutionary insights from Erwinia amylovora genomics. J. biotechnology. 2011;155:34–39. doi: 10.1016/j.jbiotec.2010.10.075. [DOI] [PubMed] [Google Scholar]
- 61.Smits TH, et al. Complete genome sequence of the fire blight pathogen Erwinia amylovora cfbp 1430 and comparison to other Erwinia spp. Mol. Plant-Microbe Interactions. 2010;23:384–393. doi: 10.1094/MPMI-23-4-0384. [DOI] [PubMed] [Google Scholar]
- 62.Vanneste J, et al. Isolation of copper and streptomycin resistant phytopathogenic Pseudomonas syringae from lakes and rivers in the central North Island of New Zealand. New Zealand Plant Prot. 2008;61:80–85. doi: 10.30843/nzpp.2008.61.6822. [DOI] [Google Scholar]
- 63.Vanneste, J. L. Fire blight: the disease and its causative agent, Erwinia amylovora (CABI, 2000).
- 64.Walterson AM, Smith DD, Stavrinides J. Identification of a Pantoea biosynthetic cluster that directs the synthesis of an antimicrobial natural product. PloS one. 2014;9:e96208. doi: 10.1371/journal.pone.0096208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trippe K, McPhail K, Armstrong D, Azevedo M, Banowetz G. Pseudomonas fluorescens SBW25 produces furanomycin, a non-proteinogenic amino acid with selective antimicrobial properties. BMC microbiology. 2013;13:111–121. doi: 10.1186/1471-2180-13-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Halgren A, et al. Genetics of germination-arrest factor (GAF) production by Pseudomonas fluorescens WH6: identification of a gene cluster essential for GAF biosynthesis. Microbiol. 2013;159:36–45. doi: 10.1099/mic.0.062166-0. [DOI] [PubMed] [Google Scholar]
- 67.Delétoile A, et al. Phylogeny and identification of Pantoea species and typing of Pantoea agglomerans strains by multilocus gene sequencing. J. clinical microbiology. 2009;47:300–310. doi: 10.1128/JCM.01916-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. biology evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nascimento S, Lima A, Borges B, De Souza C. Endophytic bacteria from Piper tuberculatum Jacq.: isolation, molecular characterization, and in vitro screening for the control of Fusarium solani f. sp piperis, the causal agent of root rot disease in black pepper (Piper nigrum L.) Genet. Mol. Res. 2015;14:7567–7577. doi: 10.4238/2015.July.3.32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.