Abstract
Background
Metastasis is the main cause for gastric cancer (GC)-related deaths. Better understanding of GC metastatic mechanism would provide novel diagnostic markers and therapeutic targets. Though it has been reported that mammalian sterile-20-like kinase 4 (MST4) exerts the oncogenic role in other tumors, the prognostic value and biological role of MST4 in GC are still unknown.
Methods
The expression level of MST4 in GC was analyzed by using TCGA database. Then, Western blot and polymerase chain reaction (PCR) were used to determine the MST4 expression in GC tissues and cell lines. Immunohistochemistry was performed to investigate the expression of proteins in human GC tissues, and its correlation with clinicopathologic parameters as well as the prognosis for patients with GC was analyzed. In addition, the biological function and its molecular mechanism of MST4 in GC were investigated by in vitro and in vivo assays.
Results
It demonstrated that MST4 expression was significantly upregulated in GC tissues and cell lines. High expression of MST4 was correlated with aggressive clinicopathological parameters such as lymph node metastasis, lymphovascular invasion (all P < 0.05). GC patients with high MST4 expression had both shorter overall survival (OS) and disease-free survival (DFS) than those with low MST4 expression (all P < 0.05). MST4 expression was an independent and significant risk factor for OS and DFS of GC patients (all P < 0.05). Results of functional experiments showed that MST4 could promote GC cells migration, invasion in vitro and metastasis in vivo. In terms of mechanism, MST4 promoted metastasis by facilitating epithelial–mesenchymal transition (EMT) through activating Ezrin pathway in GC. Further studies indicate that down-regulated miR-124-3p expression contributes to upregulated MST4 expression in GC.
Conclusion
Our data showed that MST4 predicts poor prognosis and promotes metastasis by facilitating epithelial–mesenchymal transition in GC. Therefore, our study suggests that MST4 can be used as a valuable prognostic biomarker and a potential therapeutic target in GC.
Keywords: MST4, gastric cancer, prognosis, epithelial-mesenchymal transition, metastasis
Introduction
Gastric cancer (GC) remains a lethal cancer worldwide as it the fifth most frequently diagnosed cancer and the third leading cause of cancer death.1 Although greatly improved in the treatment for GC patients including the surgical technique, chemotherapy, etc., the clinical outcome of GC patients is still unfavorable due to tumor metastasis.2 Tumor metastasis after curative resection is the main obstacle for the overall survival of patients.2 Metastasis is a multistage cascade reaction that involves many tumor suppressor gene inactivation and oncogene activation.3 Therefore, a better understanding of GC metastatic mechanism would provide novel diagnostic markers and therapeutic targets.
The epithelial–mesenchymal transition (EMT) process has been observed to underlie embryo development and tissue repair.4 However, many evidence have proven EMT plays a critical role in the genesis, invasion and metastasis of many tumors,5,6 including GC.7 EMT process involves distinct phenotypic changes detected by a series of EMT biomarkers, such as epithelial marker E-cadherin and mesenchymal marker vimentin.8 Multiple complex signaling pathways are required for the induction of EMT as epithelial cells undergoing EMT must accompany by upregulation and downregulation of critical genes.9 Therefore, it is necessary to further clarify the underlying mechanism of EMT in invasion and metastasis of GC.
Mammalian sterile-20-like kinase 4 (MST4), also named Serine/Threonine Kinase 26 (STK26), is a member of the germinal center kinase (GCK) group III family of kinases, which are a subset of the Ste20-like kinases.10 MST4 is first cloned and characterized by using a screen for Raf-interacting proteins and composes a C-terminal regulatory domain and an N-terminal kinase domain, which are required for full activation of the kinase.11 Due to its kinase activity, there are many biological functions for MST4 in normal physiological conditions and diseases. It has been reported that MST4 could regulate cell polarity and cytoskeleton.12,13 In tumors, MST4 is frequently upregulated in pancreatic cancer, prostate cancer and hepatocellular carcinoma and such expression is significantly correlated with patient prognosis.14–16 MST4 can promote proliferation and tumorigenesis of prostate cancer.17 It also has been demonstrated that MST4 can facilitate the proliferation and migration of pancreatic cancer.14 In addition, MST4 could promote EMT and metastasis of hepatocellular carcinoma by activating ERK pathway.16 However, little is known about the function and clinical significance of MST4 in GC.
In the present study, MST4 expression in GC was first analyzed by using TCGA database. Combined with our own analysis, we found MST4 expression is upregulated in GC tissues (GCT) compared with corresponding adjacent noncancerous gastric mucosae (ANGM). High MST4 expression is associated with aggressive clinicopathological parameters and poor prognosis of GC patients. In vitro and in vivo experiments demonstrated that MST4 promotes GC cell cytoskeleton rearrangements, EMT, invasion, and metastasis. Mechanically, the studies showed that MST4 interacted with Ezrin and enhanced the Ezrin activity by promoting its phosphorylation. Furthermore, we found post-translational regulation by miR-124-3p downregulation is the one important cause for MST4 upregulation in GC patients.
Materials And Methods
GC Patients And Tissue Samples
Fifty pairs of fresh frozen GC tissues (GCT) and matched adjacent noncancerous gastric mucosae (ANGM) were collected following laparoscopic or robotic radical distal gastrectomy at the Department of General Surgery, the First Affiliated Hospital of Nanchang University between January 2017 and December 2017. Formalin-fixed, paraffin-embedded paired GCT and ANGM samples obtained from 150 patients undergoing open radical distal gastrectomy at the Department of General Surgery the First Affiliated Hospital of Nanchang University between January 2008 and December 2011 were designated as the training cohort. Another cohort containing 120 samples, including matched GCT and ANGM, from patients who underwent laparoscopic radical distal gastrectomy between January 2011 and December 2012 at the Department of General Surgery, the First Affiliated Hospital of Nanchang University was designated as the validation cohort. The tumour was located in the gastric antrum. None of the patients received preoperative treatment. Pathological diagnosis was identified by at least two pathologists. The present study was approved by the Ethics Committee of the Institutional Review Boards of the First Affiliated Hospital of Nanchang University (Nanchang, China). Prior informed consent was obtained from all participants, and it was conducted in accordance with the Declaration of Helsinki and current ethical guidelines.
Follow-Up And Prognostic Study
All patients were regularly followed-up by trained and experienced researchers. Patient mortality from other causes was treated as censored cases. Disease-free survival (DFS) was defined as the length of time following resection during which a patient survived without signs of recurrence or metastasis. Overall survival (OS) was defined as the interval between the date of surgery and mortality date, or between surgery date and the last observation date for surviving patients. Research protocols followed the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) recommendations for reporting prognostic biomarkers in cancer.18
Cell Lines And Cell Culture
Immortalized human gastric mucosal epithelial cell line GES-1 and four human GC cell lines (AGS, SGC-7901, HGC27 and MKN45) were purchased from the American Type Culture Collection ((ATCC, Manassas, VA)) and maintained in RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS (GE Healthcare Life Sciences, Logan, UT) and 1% penicillin/streptomycin (Thermo Fisher Scientific) in a humidified incubator at 37°C with 5% CO2. Short tandem repeat (STR) DNA fingerprinting was used to authenticate all cell lines prior to the commencement of the study.
Vector Construction And Transfection
The lentiviral vector (LV) encoding short hairpin RNAs (shRNAs) for MST4 knockdown and the LV encoding MST4 gene ORF were purchased from GeneChem Technologies (GeneChem, Shanghai, China). The procedures of lentivirus transinfection were performed as described previously.19 The shRNA target sequences for human MST4 were as follows: shMST4-1: 5ʹ- GCTGCCAATGTCTTGCTCTCA-3ʹ; shMST4-2: 5ʹ-GCTGGTCAGCTGACAGATACA-3ʹ; shMST4-3: 5ʹ-GGCAGAAGGACACAGTGATGA-3ʹ. AGS cells were transfected with lentiviral vectors encoding the shRNAs, and MKN45 cells were transfected with lentiviral vectors encoding the human MST4 gene. An empty vector was used as the negative control and was designated as LV-control. The lentiviral vectors were transfected into the GC cells with a proper multiplicity of infection (MOI). At 48 h after transfection, 3.0 μg/mL puromycin (OriGene) was added, and the cells were incubated for 2 weeks to select stable transfected cells. Overexpression or down-regulated expression of MST4 was confirmed by qPCR and Western blot. The inhibitory efficiency of three shRNAs was validated and the MST4-shRNA-3 (named shMST4 in the figures) was adopted for subsequent study because of highly effective inhibition of MST4 expression in AGS cells. In addition, the plasmids pcDNA3.1+HA, pcDNA3.1+HA-Ezrin and pcDNA3.1+HA-Ezrin Thr567 (mutant T567D Ezrin, a constitutively active phosphomimetic Ezrin) were constructed and transfected into corresponding GC cells using Lipofectamine 2000. G418 (500 µg/mL) was used to select stable transfected cells. For Ezrin knockdown, the shRNA target sequences were as follows: shEzrin-1: 5′-CCTGGAAATGTATGGAATCAA-3′; shEzrin-2: 5′-GGGTCCTACGCTGTGCAGGCC-3′; shEzrin-3: 5′-AACAGCTGGAAACAGAGAAGAAA-3′. The inhibitory efficiency of three shRNAs was validated and the Ezrin-shRNA-3 was adopted for subsequent study because of highly effective inhibition of Ezrin expression.
RNA Extraction And qRT-PCR Analysis
Total RNA was extracted from cell lines or fresh frozen tumor specimens using TRIzol reagent (Thermo Fisher Scientific). After synthesizing the first-strand cDNA via the universal cDNA synthesis kit (Toyobo, Osaka, Japan), qRT-PCR was performed in triplicate using SYBR®-Green Realtime PCR Master Mix assay kit (Toyobo). The primer sequences for MST4 were as follows: forward primer, 5ʹ- TTCGAGCTGGTCCATTTGATG-3ʹ and reverse primer, 5ʹ- TGAATGCAGATAGTCCAGACCT-3ʹ. The primer sequences for E-cadherin were as follows: forward primer, 5ʹ- ATTTTTCCCTCGACACCCGAT-3ʹ and reverse primer, 5ʹ- TCCCAGGCGTAGACCAAGA-3ʹ. The primer sequences for vimentin were as follows: 5ʹ- AGTCCACTGAGTACCGGAGAC-3ʹ and reverse primer, 5ʹ- CATTTCACGCATCTGGCGTTC-3ʹ. GAPDH was used as a control using the following primers: forward primer: 5ʹ-GCACCGTCAAGGCTGAGAAC-3ʹ; reverse primer: 5ʹ-TGGTGAAGACGCCAGTGGA-3ʹ. The experimental conditions were followed by the manufacturer’s protocol.
Western Blot Analysis
Total proteins were extracted with RIPA lysis buffer. Protein concentration was then determined using the bicinchoninic acid method. Equal protein was separated by SDS-PAGE and then transferred to the PVDF membrane (EMD Millipore, Bedford, MA). The membrane was blocked with 5% skimmed milk and incubated with the appropriate antibody. The antigen-antibody complex on the membrane was detected with enhanced chemiluminescence reagents (Thermo Fisher Scientific). Antibody used in this study included mouse anti-β-actin antibody (EMD Millipore), mouse anti-MST4 antibody (Santa Cruz Biotechnology, Dallas, TX), Rabbit anti-Ezrin antibody (CST, Danvers, MA), Rabbit anti-phospho-Ezrin antibody (Abcam, Cambridge, MA), mouse anti-E-cadherin antibody (CST), mouse anti-vimentin antibody (CST).
Cell Proliferation And Colony Formation Assays
Methyl thiazolyl tetrazolium (MTT) assay was used to determine the level of cell proliferation and had been described previously.19 For colony formation assays, 500 cells were seeded into 35mm dishes (Corning Costar Corp, Corning, NY) and cultured for 2 weeks at 37°C. The numbers of colonies per dish were counted after staining with crystal violet. Only positive colonies (diameter > 40 µm) in the dishes were counted and compared. These experiments were performed in triplicate.
Transwell Migration And Invasion Assay
Transwell migration and invasion assay were used to determine the GC cell motility and invasion, respectively. The upper chamber of the insert plated with Matrigel (BD Biosciences, Franklin Lakes, NJ) was used for Transwell invasion assay, while the insert without Matrigel was used to Transwell migration assay. Briefly, the tumor cells were preincubated with Mitomycin-C (10 µg/mL) for 1 hr at 37°C to suppress cell proliferation. After 24 hrs of incubation at 37°C, we removed the cells remaining in the upper chamber. The cells adhered to the lower membrane of the inserts were fixed by methanol and counted after staining with a solution containing 0.1% crystal violet (Beyotime Institute of Biotechnology, Shanghai, China). For each experimental group, the assays were performed in triplicate, and three randomly fields in each replicate were chosen for the cell number quantification.
Adhesion Assay
Cell-ECM adhesion assay and cell-cell adhesion assay were performed as described previously.19 For the cell-ECM adhesion assay, a 96-well plate was coated with fibronectin at 37°C for 1 hr and washed twice with washing buffer. The plates were blocked in blocking buffer at 37°C for 60 min. The cells (100 µL) at a density of 1x105/mL were added into each well and cultured at 37°C. Five wells for each group were detected at 60, 90 or 120 min. After removing unbound cells, the bounded cells were detected by MTT assay. For the cell-cell adhesion assay, single cells (100 µL) at density of 1x105/mL were added into a 96-well plate with a fully confluent single cell layer, and cultured at 37°C. After removing unbound cells, the remaining cells were collected and quantified. The adhesion rate was determined by counting representative aliquots from each sample on a hematocytometer. The percentage of adhesion was quantified at 60, 90 or 120 min as: (N0-Nt)/N0 x 100, where Nt is the total number of unbound cells at the indicated incubation time, and N0 is the total number of cells.
Cellular Cytoskeleton Analysis
The cells grown on cover slides were fixed by formaldehyde, and then washed and incubated with rhodamine-conjugated phalloidin (Solarbio Science and Technology, Beijing, China). Following staining with DAPI (Beyotime Institute of Biotechnology), images of the slides were captured using an inverted fluorescence TE-2000S microscope (Nikon, Tokyo, Japan).
Immunohistochemistry (IHC)
Formalin-fixed paraffin sections of clinical samples were stained using the streptavidin-peroxidase system (ZSGB-BIO, Beijing, China). Briefly, 5μm tissue sections were deparaffinized in xylene and rehydrated using a series of graded alcohols. Slides were blocked with 10% goat serum before incubating with the primary antibody. The samples were incubated overnight with a primary antibody, and subsequent secondary antibody followed by DAB labeled secondary antibody. The protein expression was scored based on staining intensity (SI) and percentage of positive cells (PP) using the immunostaining score (IS) as described previously.19 IS = SI × PP. The SI was classified into four grades: 0, negative; 1, weak; 2, moderate; 3, strong. The PP was defined into five categories: 0, 0% positive cells; 1, 0–25% positive cells; 2, 25–50% positive cells; 3, 50–75% positive cells, and 4, 75–100% positive cells. Therefore, the value range of IS was 0~12 points. We defined 0 as negative (-), 1~4 as weakly positive (+), 5~8 as positive (++) and 9~12 as strongly positive (+++). The protein expression in GC specimens was also divided into a low expression group (- and +) and a high expression group (++ and +++) for further analysis.
In Vivo Metastatic Assay
BALB/c-nu/nu mice were performed and housed in the Animal Institute of Nanchang University (Nanchang, China) according to the protocols approved by the Medical Experimental Animal Care Commission of Nanchang University (Nanchang). Ethical and legal approval was obtained prior to the commencement of the study. Mice were housed under specific pathogen-free conditions: Temperature, 25°C; relative humidity, ~40%; lighting, 10 h/day with fluorescent lights. The mice received ad libitum access to sterilized food and water. The experiments were performed according to the protocols approved by the Medical Experimental Animal Care Commission. For experimental metastasis assays, the nude mice were injected with 1 × 106 cells (resuspended in PBS) via the tail vein. All and lungs were fixed with 10% phosphate-buffered neutral formalin, sectioned serially and stained with hematoxylin and eosin (H&E) for histological examination.
Statistical Analysis
All data were analyzed using the statistical software SPSS 18.0 (SPSS Inc., Chicago, IL). The differences between the two groups were analyzed by Student’s t test when the variance is homogeneous. If the variance is not homogeneous, the differences between two groups were analyzed by Mann–Whitney U-test. χ2 analysis was used to analyze the correlation between the expression of MST4 and clinicopathologic parameters. Survival curves were constructed with the Kaplan–Meier method and compared using the log rank test. The Cox proportional hazards regression model was established to identify independent factors for OS and DFS rates of patients. A two-tailed P value of less than 0.05 was considered as statistical significance.
Results
MST4 Expression Was Significantly Upregulated In GC
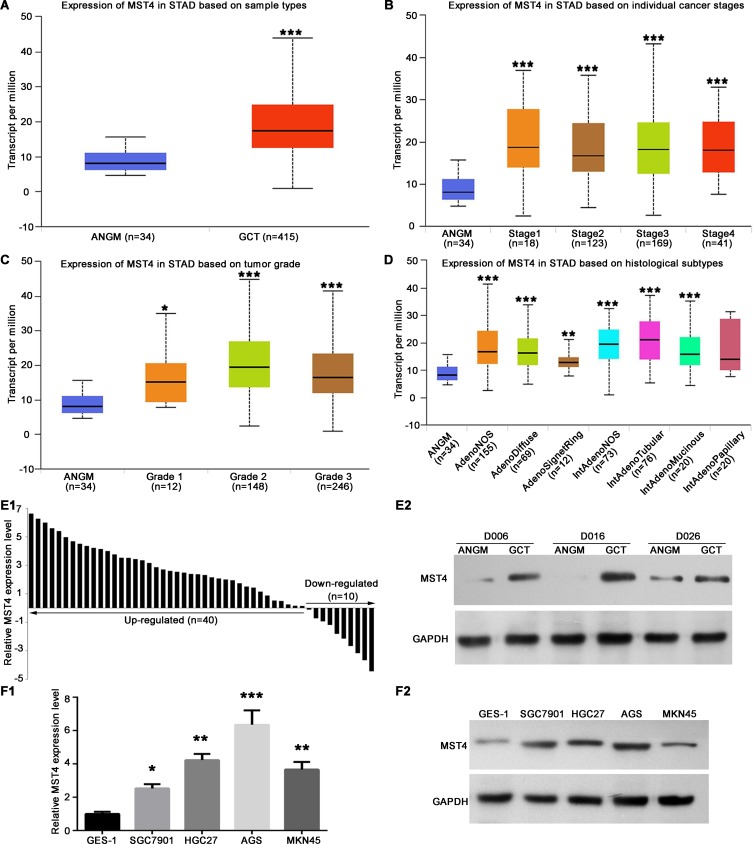
In order to investigate the expression level of MST4 in GC, we first examined mRNA expression of MST4 in TCGA gastric database by using UALCAN (www.ualcan.path.uab.edu). MST4 mRNA expression level in GCT was significantly higher than ANGM (Figure 1A). Further analysis based on cancer stages (Figure 1B) and tumor grade (Figure 1C) showed MST4 expression level in all tumor stages and tumor grade of NCT was significantly higher than in ANGM, but there are no significant differences between each tumor stages, indicating MST4 upregulation is an early event during GC progression. In addition, except for papillary intestinal adenocarcinoma, all histological subtypes of GCT had a higher MST4 expression than ANGM (Figure 1D). Next, we determined the expression level of MST4 in GC and cell lines by qRT‐PCR and Western blot. Compared with their corresponding ANGM, qRT‐PCR showed MST4 mRNA was upregulated in GCT (40/50, 80%; Figure 1E1), and Western blot also showed MST4 protein expression level was upregulated in GCT (Figure 1E2). Similarly, MST4 mRNA and protein expression level in gastric cell lines including AGS, SGC-7901, HGC27 and MKN45 was higher than normal gastric epithelial cell line GES-1 (Figure 1F1 and F2).
Figure 1.
MST4 expression was significantly upregulated in gastric cancer (GC). (A) The mRNA expression profile of MST4 in GCTs and ANGMs was analyzed by UALCAN program using TCGA data. GCT, GC tissue; ANGM, adjacent noncancerous gastric mucosa. (B) Expression of MST4 in TCGA database based on tumor stages. (C) Expression of MST4 in TCGA database based on tumor grade. (D) Expression of MST4 in TCGA database based on histological subtypes. (E) MST4 expression was significantly up-regulated in GCT. (E1) qRT-PCR was used to analyze MST4 mRNA expression in GCTs (n=50) and ANGMs (n=50). (E2) Western blot results showed that MST4 protein expression was higher in GCTs than in ANGMs. (F1 and F2) MST4 expression was significantly up-regulated in GC cell lines. (F1) Quantitative real-time PCR (qRT-PCR) analysis of MST4 mRNA showed, compared with normal gastric epithelial cell line GES-1, MST4 mRNA expression elevated in GC cell lines including AGS, SGC-7901, HGC27 and MKN45 cell line. (F2) Western blot results showed MST4 protein was overexpressed in AGS, SGC-7901, HGC27 and MKN45 cell line relative to GES-1 cell line. *P < 0.05; **P < 0.01; ***P < 0.001.
MST4 Upregulation Is Significantly Associated With Aggressive Clinicopathological Parameters Of GC Patients
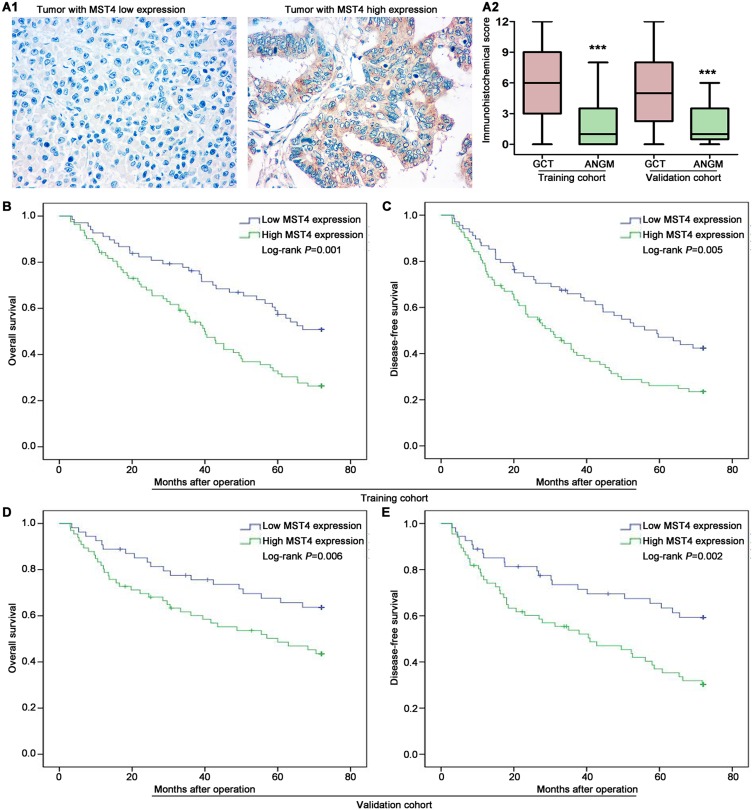
To explore the potential clinical significance of MST4 expression in GC patients, the relationship between MST4 protein expression and clinicopathologic parameters was first analyzed. The clinicopathologic parameters of GC in training cohort and validation cohort are shown in Table 1. IHC was adopted to stain MST4 in training and validation cohort patients (Figure 2A1). According to IHC score, we found MST4 expression score in GCT is significantly higher than that in ANGM (Figure 2A2). Combined with clinical data, we further found high MST4 expression was positively associated with aggressive clinicopathological parameters such as lymph node metastasis (P<0.001, Table 2), lymphovascular invasion (P=0.004, Table 2) in training cohort. Likewise, MST4 upregulation was also associated with aggressive clinicopathological parameters such as lymph node metastasis (P<0.001, Table 3), lymphovascular invasion (P=0.001, Table 3) in validation cohort. These results also indicated MST4 might play a crucial role in gastric cancer invasion and metastasis.
Table 1.
Clinicopathologic Parameters Of Patients With GC In The Training Cohort And Validation Cohort
| Clinicopathologic Parameters | Training Cohort | Validation Cohort | P |
|---|---|---|---|
| Gender | |||
| Male | 95 | 84 | |
| Female | 55 | 36 | 0.300 |
| Age (y) | |||
| ≤60 | 71 | 64 | |
| > 60 | 79 | 56 | 0.391 |
| Tumor size(cm) | |||
| ≤5 | 64 | 46 | |
| >5 | 86 | 74 | 0.533 |
| Grade of differentiation | |||
| Well and moderate | 52 | 51 | |
| Poor and not | 98 | 69 | 0.208 |
| Lauren’s classification | |||
| Intestinal | 109 | 88 | |
| Diffuse | 41 | 32 | 0.902 |
| Lymph node metastasis | |||
| Negative | 54 | 45 | |
| Positive | 96 | 75 | 0.799 |
| Lymphovascular invasion | |||
| Negative | 105 | 89 | |
| Positive | 45 | 31 | 0.497 |
| TNM stage | |||
| I & II | 72 | 62 | |
| III | 78 | 58 | 0.624 |
Abbreviations: GC, gastric cancer; TNM, tumor node metastasis.
Figure 2.
High MST4 expression is significantly associated with poor prognosis of GC. (A1 and A2) MST4 protein is higher in GCTs than in corresponding ANGMs. (A1) Representative IHC images of low MST4 expression case and high MST4 expression case from training and validation cohort. (A2) IHC score showed GCTs exhibited higher MST4 protein expression than corresponding ANGMs. Box-plot analyzed the differential expression between GCTs and ANGMs from training and validation cohort. IHC, Immunohistochemistry. (B and C) Kaplan-Meier analysis of overall survival (B) and disease-free survival (C) based on MST4 expression in the training cohort. (D and E) Kaplan-Meier analysis of overall survival (D) and disease-free survival (E) based on MST4 expression in the validation cohort. ***P < 0.001.
Table 2.
Correlations Between Expression Of MST4 In GC Tissues And Clinicopathologic Parameters Of Patients With GC In The Training Cohort
| Clinicopathologic Parameters | n | MST4 Expression | P | |
|---|---|---|---|---|
| Low (68) | High (82) | |||
| Gender | ||||
| Male | 95 | 45 | 50 | |
| Female | 55 | 23 | 32 | 0.610 |
| Age (y) | ||||
| ≤60 | 71 | 30 | 41 | |
| > 60 | 79 | 38 | 41 | 0.514 |
| Tumor size(cm) | ||||
| ≤5 | 64 | 24 | 40 | |
| >5 | 86 | 44 | 42 | 0.101 |
| Grade of differentiation | ||||
| Well and moderate | 52 | 19 | 33 | |
| Poor and not | 98 | 49 | 49 | 0.125 |
| Lauren’s classification | ||||
| Intestinal | 109 | 54 | 55 | |
| Diffuse | 41 | 14 | 27 | 0.101 |
| Lymph node metastasis | ||||
| Negative | 54 | 37 | 17 | |
| Positive | 96 | 31 | 65 | <0.001 |
| Lymphovascular invasion | ||||
| Negative | 105 | 56 | 49 | |
| Positive | 45 | 12 | 33 | 0.004 |
| TNM stage | ||||
| I & II | 72 | 36 | 36 | |
| III | 78 | 32 | 46 | 0.270 |
Abbreviations: GC, gastric cancer; TNM, tumor node metastasis; MST4, mammalian sterile-20-like kinase 4.
Table 3.
Correlations Between Expression Of MST4 In GC Tissues And Clinicopathologic Parameters Of Patients With GC In The Validation Cohort
| Clinicopathologic Parameters | n | MST4 Expression | P | |
|---|---|---|---|---|
| Low (54) | High (66) | |||
| Gender | ||||
| Male | 84 | 39 | 45 | |
| Female | 36 | 15 | 21 | 0.692 |
| Age (y) | ||||
| ≤60 | 64 | 30 | 34 | |
| >60 | 56 | 24 | 32 | 0.715 |
| Tumor size (cm) | ||||
| ≤5 | 46 | 18 | 28 | |
| >5 | 74 | 36 | 38 | 0.349 |
| Grade of differentiation | ||||
| Well and moderate | 51 | 27 | 24 | |
| Poor and not | 69 | 27 | 42 | 0.133 |
| Lauren’s classification | ||||
| Intestinal | 88 | 44 | 44 | |
| Diffuse | 32 | 10 | 22 | 0.096 |
| Lymph node metastasis | ||||
| Negative | 45 | 31 | 14 | |
| Positive | 75 | 23 | 52 | <0.001 |
| Lymphovascular invasion | ||||
| Negative | 89 | 48 | 41 | |
| Positive | 31 | 6 | 25 | 0.001 |
| TNM stage | ||||
| I & II | 62 | 29 | 33 | |
| III | 58 | 25 | 33 | 0.688 |
Abbreviations: GC, gastric cancer; TNM, tumor node metastasis; MST4, mammalian sterile-20-like kinase 4.
High MST4 Expression Is Significantly Associated With Poor Prognosis Of GC Patients
To further determine the relationship between MST4 expression and prognosis of GC patients, we conducted survival analyses by Kaplan–Meier survival analysis and Cox proportional hazards analysis. In training cohort, GC patients with high MST4 expression had shorter overall survival (OS) (P=0.001; Figure 2B) and shorter disease-free survival (DFS) (P=0.005; Figure 2C) compared with those with low MST4 expression. Univariate and then multivariate Cox regression analysis showed high MST4 expression was an independent risk factor for OS (P=0.011; Table 4) and DFS (P=0.038; Table 5) for GC patients. Similarly, in validation cohort, Kaplan–Meier analysis indicated that GC patients with high MST4 expression level had a much worse OS (P=0.006; Figure 2D) and DFS (P=0.002; Figure 2E). Cox regression analysis also showed high MST4 expression was an independent prognostic factor for poor OS (P=0.023; Table 6) and DFS (P=0.021; Table 7) of GC patients.
Table 4.
Cox Proportional Hazard Regression Analyses For OS In The Training Cohort
| Parameters | Univariate Analysis | P | Multivariate Analysis | P |
|---|---|---|---|---|
| HR (95% CI) | HR (95% CI) | |||
| Gender (male vs female) | 1.017 (0.673–1.539) | 0.935 | NA | |
| Age (y, ≤60 vs >60) | 1.294 (0.853–1.962) | 0.225 | NA | |
| Tumor size (cm, >5 vs ≤5) |
1.412 (0.929–2.147) | 0.106 | NA | |
| Grade of differentiation (poor and not vs well and moderate) | 1.403 (0.920–2.140) | 0.116 | NA | |
| Lauren’s classification (intestinal vs diffuse) | 1.179 (0.777–1.789) | 0.440 | NA | |
| Lymph node metastasis (positive vs negative) | 2.127 (1.295–3.495) | 0.003 | 1.767 (1.165–2.681) | 0.017 |
| Lymphovascular invasion (positive vs negative) | 1.751 (1.151–2.664) | 0.009 | 1.328 (1.037–1.701) | 0.025 |
| TNM stage (III vs I & II) |
2.651 (1.418–4.957) | <0.001 | 2.208 (1.444–3.376) | 0.002 |
| MST4 expression (high vs low) | 2.033 (1.325–3.119) | 0.001 | 1.820 (1.149–2.880) | 0.011 |
Abbreviations: OS, overall survival; NA, not adopt; TNM, tumor node metastasis; MST4, mammalian sterile-20-like kinase 4.
Table 5.
Cox Proportional Hazard Regression Analyses For DFS In The Training Cohort
| Parameters | Univariate Analysis | P | Multivariate Analysis | P |
|---|---|---|---|---|
| HR (95% CI) | HR (95% CI) | |||
| Gender (male vs female) | 1.163 (0.782–1.728) | 0.456 | NA | |
| Age (y, ≤60 vs >60) | 1.475 (0.989–2.200) | 0.153 | NA | |
| Tumor size (cm, >5 vs ≤5) |
1.438 (0.965–2.143) | 0.180 | NA | |
| Grade of differentiation (poor and not vs well and moderate) | 1.337 (0.897–1.994) | 0. 274 | NA | |
| Lauren’s classification (intestinal vs diffuse) | 1.315 (0.881–1.963) | 0. 206 | NA | |
| Lymph node metastasis (positive vs negative) | 2.742 (1.428–5.268) | <0.001 | 2.052 (1.243–3.388) | 0.008 |
| Lymphovascular invasion (positive vs negative) | 1.910 (1.338–3.513) | 0.003 | 1.665 (1.085–2.555) | 0.031 |
| TNM stage (III vs I & II) | 2.154 (1.438–3.227) | 0.001 | 1.836 (1.233–2.733) | 0.012 |
| MST4 expression (high vs low) | 1.780 (1.184–2.674) | 0.005 | 1.587 (1.026–2.456) | 0.038 |
Abbreviations: DFS, disease-free survival; NA, not adopt; TNM, tumor node metastasis; MST4, mammalian sterile-20-like kinase 4.
Table 6.
Cox Proportional Hazard Regression Analyses For OS In The Validation Cohort
| Parameters | Univariate Analysis | P | Multivariate Analysis | P |
|---|---|---|---|---|
| HR (95% CI) | HR (95% CI) | |||
| Gender (male vs female) | 1.298 (0.762–2.213) | 0.337 | NA | |
| Age (y, ≤60 vs >60) | 1.101 (0.647–1.872) | 0.723 | NA | |
| Tumor size (cm, >5 vs ≤5) |
1.647 (0.970–2.795) | 0.065 | 1.236 (0.668–2.287) | 0.500 |
| Grade of differentiation (poor and not vs well and moderate) | 1.456 (0.857–2.474) | 0.165 | NA | |
| Lauren’s classification (intestinal vs diffuse) | 1.080 (0.912–1.280) | 0.373 | NA | |
| Lymph node metastasis (positive vs negative) | 3.767 (1.509–9.403) | <0.001 | 2.502 (1.324–4.728) | 0.002 |
| Lymphovascular invasion (positive vs negative) | 1.908 (1.267–2.873) | 0.009 | 1.649 (1.097–2.479) | 0.028 |
| TNM stage (III vs I & II) | 2.633 (1.407–4.927) | 0.003 | 2.009 (1.149–3.513) | 0.014 |
| MST4 expression (high vs low) | 2.311 (1.304–4.096) | 0.006 | 1.862 (1.067–3.247) | 0.023 |
Abbreviations: OS, overall survival; NA, not adopt; TNM, tumor node metastasis; MST4, mammalian sterile-20-like kinase 4.
Table 7.
Cox Proportional Hazard Regression Analyses For DFS In The Validation Cohort
| Parameters | Univariate Analysis | P | Multivariate Analysis | P |
|---|---|---|---|---|
| HR (95% CI) | HR (95% CI) | |||
| Gender (male vs female) | 1.359 (0.831–2.221) | 0.221 | NA | |
| Age (y, ≤60 vs >60) | 1.088 (0.668–1.773) | 0.734 | NA | |
| Tumor size (cm, >5 vs ≤5) |
1.813 (1.002–3.280) | 0.046 | 1.613 (0.990–2.627) | 0.122 |
| Grade of differentiation (poor and not vs well and moderate) | 1.605 (0.983–2.618) | 0.158 | NA | |
| Lauren’s classification (intestinal vs diffuse) | 1.092 (0.929–1.284) | 0.285 | NA | |
| Lymph node metastasis (positive vs negative) | 2.907 (1.514–4.488) | <0.001 | 2.032 (1.409–2.930) | 0.007 |
| Lymphovascular invasion (positive vs negative) | 1.932 (1.095–3.410) | 0.010 | 1.795 (1.010–3.189) | 0.033 |
| TNM stage (III vs I & II) | 2.580 (1.569–4.243) | 0.001 | 1.902 (1.399–2.586) | 0.016 |
| MST4 expression (high vs low) | 2.329 (1.357–3.997) | 0.002 | 1.862 (1.211–2.863) | 0.021 |
Abbreviations: DFS, disease-free survival; NA, not adopt; TNM, tumor node metastasis; MST4, mammalian sterile-20-like kinase 4.
MST4 Promotes GC Cells Invasion And Metastasis
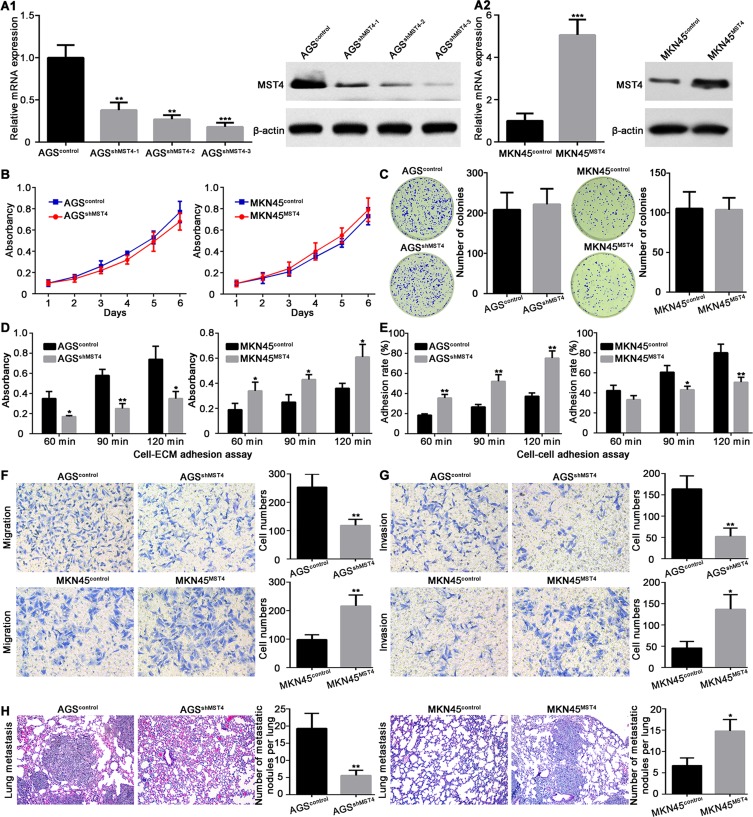
Considering the important clinical significance of MST4 in GC patients, we then asked MST4 biological functions in gastric cancer. In order to achieve this goal, we suppressed the expression of MST4 in AGS cells which expressed a relatively high level of MST4 and overexpressed MST4 expression in MKN45 cells which have relatively low MST4 expression. Then, the knockdown and overexpression efficiency of MST4 in AGS and MKN45 cells was confirmed by qRT-PCR and Western blot and met subsequent experimental requirements (Figure 3A1 and A2). MTT and colony formation assays were used to determine the role of MST4 in the proliferation of GC cells. Intriguingly, both results showed MST4 has no significant influence on the proliferation of GC cells (Figure 3B and C). Adhesion assay showed MST4 knockdown in AGS cells suppressed cell-ECM adhesion and enhanced cell-cell adhesion, whereas overexpression of MST4 in MKN45 cells increased cell-ECM adhesion and inhibited cell-cell adhesion (Figure 3D and E). Transwell migration assays showed that the downregulation of MST4 significantly decreased the motility capacity of AGS cells, while overexpression of MST4 significantly increased the motility of MKN45 cells (Figure 3F). Similar results were also observed in the Transwell invasion assays (Figure 3G). In addition, in vivo mouse tail vein injection experiment showed knockdown of MST4 in AGS cells significantly inhibited the formation of pulmonary metastasis focuses. MKN45 cells with MST4 overexpression had a more powerful capacity to form lung metastases than control (Figure 3H).
Figure 3.
MST4 promotes gastric cancer cells invasion and metastasis. (A1 and A2) Validation of the expression of MST4 in MKN45 and AGS cell line after transfection with knockdown (A1) and overexpression (A2) vectors, respectively. (B) Methyl thiazolyl tetrazolium (MTT) assay was performed to assess the effect of MST4 on GC cell proliferation. (C) Colony formation assay was used to determine the proliferation of GC cells. (D) MST4 promoted cell-ECM adhesion. (E) MST4 inhibited cell-cell adhesion. (F) Transwell migration assay showed MST4 promoted GC cell migration. (G) Transwell invasion assay showed MST4 promoted GC cell invasion. (H) Mice tail vein injection experiment showed MST4 significantly promoted the formation of pulmonary metastatic focuses. *P < 0.05; **P < 0.01; ***P < 0.001.
MST4 Promotes GC Cell Metastasis By Facilitating EMT
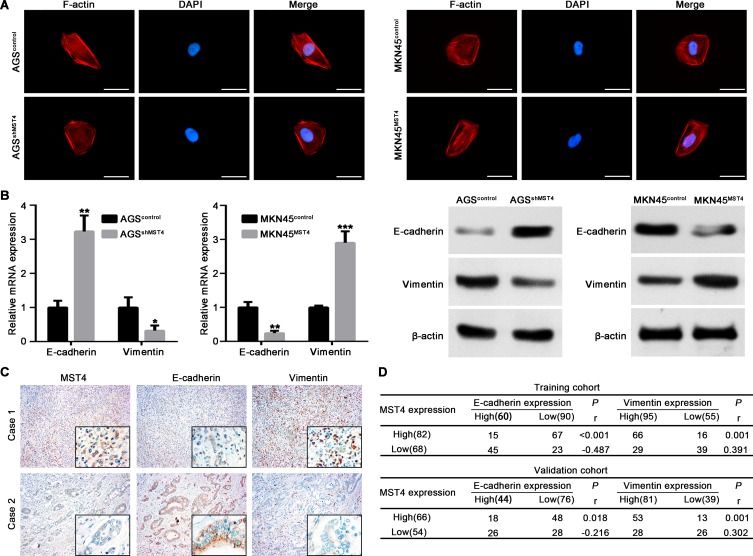
The above results showed MST4 played a crucial role in promoting invasion and metastasis of GC cells. EMT, by which GC cells gain cell motility and invasiveness, is defined by loss of epithelial cell polarity and E-cadherin expression, and by the acquisition of fibroblastic mesenchymal morphology and Vimentin expression.20 We detected the effect of MST4 on cell morphology through observation of actin cytoskeleton of GC cells by staining F-actin. The results showed AGS cells with MST4 downregulation exhibited a cobblestone shape and shrinkable F-actin fiber compared with control AGS cells. Inversely, MKN45 cells with MST4 upregulation presented an elongated cellular morphology and apparent F-actin fibers compared with control cells (Figure 4A). Then, qRT-PCR and Western blot were used to detect EMT markers, including epithelial marker E-E-cadherin and mesenchymal marker vimentin. The results showed knockdown MST4 expression in AGS cells increased E-cadherin expression and decreased vimentin expression, whereas overexpression of MST4 reduced expression of the epithelial marker, E-cadherin, and increased the mesenchymal marker, vimentin, in MKN45 cells (Figure 4B). IHC staining for serial sections of GCT showed tissues with high MST4 expression exhibited relatively high vimentin expression and low E-cadherin expression. However, tissues with low MST4 expression showed relatively low vimentin expression and high E-cadherin expression (Figure 4C). The spearman correlation analysis revealed that MST4 expression was positively correlative with vimentin and negatively correlative with E-cadherin expression in GCTs in both training and validation cohorts (Figure 4D). These results suggested that MST4 could promote HCC invasion and metastasis via EMT.
Figure 4.
MST4 promotes GC cell metastasis by facilitating EMT. (A) Representative images of cytoskeleton showed that MST4 affected the polymerization of F-actin in GC cells. (B) qRT-PCR and Western blot were used to determine the expression of E-cadherin and vimentin in GC cells with knockdown or overexpression of MST4. (C) Representative IHC images showed the expression of MST4, E-cadherin and vimentin expression in GCT with serial sections. The black frames in the lower right corner showed higher magnification of corresponding images. (D) Analysis of the relation of MST4 expression with E-cadherin and vimentin in GCT. Results showed a positive correlation of the expression of MST4 with the expression of Vimentin and a negative correlation of the expression of MST4 with the expression of E-cadherin in the GCT. *P < 0.05; **P < 0.01; ***P < 0.001.
MST4 Exerts The Function Through Activating Ezrin Signaling In GC
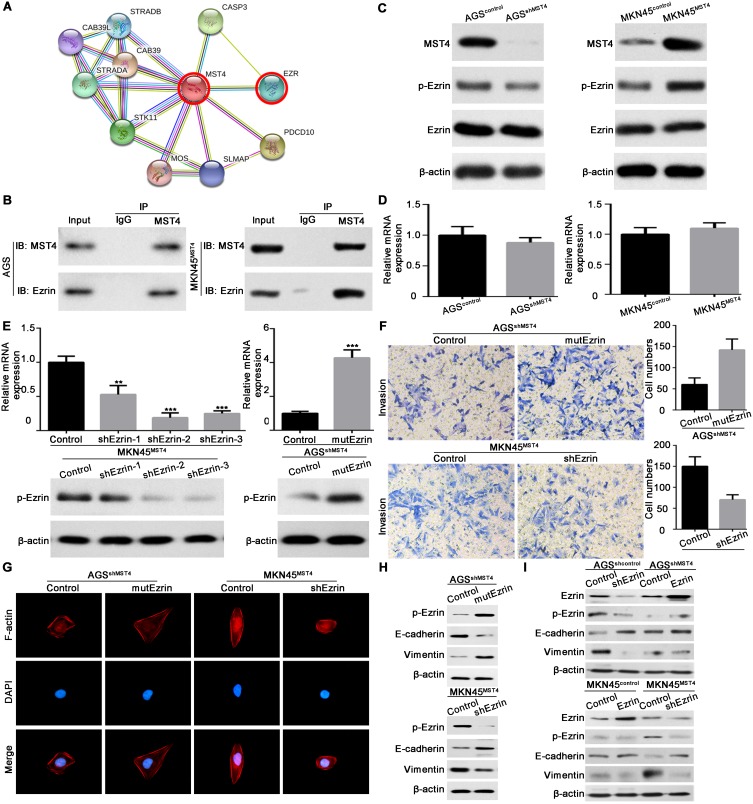
To determine the potential signaling by which MST4 exerts the function in GC, we screened the possible interactors of MST4 by STRING database (version 10.5, https://string-db.org) (Figure 5A).21 From these interactors, Ezrin (EZR) caught our attention, as Ezrin is frequently upregulated in GC tissues22,23 and its activation is associated with EMT process in many cancers.24,25 What is more, the interaction of MST4 and Ezrin participates in acid secretion of gastric parietal cells.26,27 Therefore, by using co-IP method, we revealed MST4 and Ezrin could interact with each other in GC cells (Figure 5B). Next, we found that silencing MST4 in AGS cells decreased phosphorylated Ezrin (p-Ezrin) protein expression, whereas overexpression of MST4 in MKN45 cells increased p-Ezrin expression (Figure 5C). Meanwhile, the total level of Ezrin was not altered by MST4 (Figure 5C). As expected, manipulating MST4 expression had little effect on Ezrin mRNA expression (Figure 5D). To further verify whether Ezrin mediates the function of MST4 in GC cells, we did the gain- and loss-of-function assays. The mutant T567D Ezrin (mutEzrin) is a constitutively active phosphomimetic Ezrin.28–30 We then transfected the exogenous mutEzrin expression plasmids into AGSshMST4 cells and Ezrin-shRNA plasmids into MKN45MST4 cells. The exogenous mutEzrin expression and silence efficacy of Ezrin were verified by qRT-PCR and Western blot (Figure 5E). The results showed p-Ezrin level was significantly elevated after exogenous mutEzrin transfected, while p-Ezrin level significantly descended after sharply knockdown of Ezrin (Figure 5E). Transwell invasion assay showed that exogenous mutEzrin restored its invasive capacity of AGSshMST4 cells, whereas sharply knockdown of Ezrin in MKN45MST4 cells counteracted the promoting effect of MST4 overexpression on cell invasion (Figure 5F). The cytoskeleton staining indicated that exogenous mutEzrin transfected into AGSshMST4 cells contributed to elongated cellular morphology and apparent F-actin fibers, whereas sharply knockdown of Ezrin in MKN45MST4 cells presented a cobblestone-like morphology with shrinkable F-actin fibers (Figure 5G). Likewise, Western blot results showed exogenous mutEzrin reversed the EMT phenotype of AGSshMST4 cells by E-cadherin downregulation and vimentin upregulation, while sharp downregulation of Ezrin expression caused MKN45MST4 cell E-cadherin upregulation and vimentin downregulation (Figure 5H). Our results also showed that the level of p-Ezrin was not affected by the changes of wild type Erzin expression in AGSshMST4 cells or MKN45control cells, indicating the activated Ezrin signaling was mainly dependent on phosphorylation role of MST4 in GC (Figure 5I). Taken together, these data indicate that MST4 exerts the function through activating Ezrin signaling in GC.
Figure 5.
MST4 exerts the function through activating Ezrin signaling in GC. (A) STRING database showed the interaction of MST4 protein. (B) Protein expression of MST4, p-Ezrin and total Ezrin in GC cells with knockdown or overexpression of MST4. (C) Co-immunoprecipitation assays were performed to analyze the direct binding between MST4 and Ezrin in GC cells. (D) mRNA expression of Ezrin in GC cells with knockdown or overexpression of MST4. (E) Validate the expression of p-Ezrin in GC cell lines with overexpression of mutant T567D Ezrin or knockdown of Ezrin. (F) Transwell invasion assay was performed to determine Ezrin is a critical downstream effector in MST4-promoted invasion and metastasis in GC. (G) Immunofluorescence assays of cytoskeleton showed Ezrin is the key downstream effector of MST4-promoted F-actin filament rearrangement in GC cells. (H) Western blot tested the effect of Ezrin on EMT induced by MST4. (I) By knocking-in or -out of wild type Ezrin in corresponding GC cells, p-Ezrin and EMT markers were tested by Western blot. **P < 0.01; ***P < 0.001.
miR-124-3p Downregulation Contributes To MST4 Upregulation In GC
Recent studies had revealed that oncogene mRNA regulation resulted from miRNAs dysregulation is a critical reason for upregulation in cancers.31,32 By using an online bioinformatics database (TargetScan 7.2, http://www.targetscan.org), we identified that three miRNAs, miR-124-3p, miR-22-3p and miR-19-3p, might be putative negative regulators of MST4 in GC (Figure 6A). Then, we detected the expression of miR-124-3p, miR-22-3p and miR-19-3p in previous 50 GCT and matched ANGM. The results showed miR-124-3p was downregulated in GCT and had the most significant differences (Figure 6B). Next, we carried out luciferase reporter assay to validate whether miR-124-3p indeed regulated MST4 expression. The results showed that the relative luciferase activity was sharply inhibited after co-transfected with miR-124-3p mimic and wildtype MST4 3ʹ-UTR. What is more, the activity of MST4 3ʹ-UTR with a putative binding site mutated was not affected by miR-124-3p (Figure 6C). Western blot analysis confirmed that miR-124-3p suppressed the MST4 protein expression in GC cells (Figure 6D). Then, the MST4 protein expression was determined in GCT with high and low miR-124-3p expression according to the median miR-124-3p expression level by qRT-PCR results showed in Figure 6B. Typical IHC images for GCT showed that GCT with high miR-124-3p expression exhibited low MST4 protein expression, while GCT with low miR-124-3p expression exhibited high MST4 protein expression (Figure 6E). Above all, these data demonstrated that miR-124-3p downregulation contributes to MST4 upregulation in GC.
Figure 6.
miR-124-3p downregulation contributes to MST4 upregulation in GC. (A) Three microRNAs that target MST4 were screened according to a microRNA database TargetScan (www.targetscan.org). (B) qRT-PCR was used to determine the expression level of three miRNAs in GCTs and corresponding ANGMs. (C) Data of luciferase reporter assay. Relative luciferase activity was analyzed after MST4 wildtype or mutant reporter plasmids were co-transfected into 293T cells infected with miR-124-3p lentivirus or control lentivirus. (D) Western blot showed MST4 expression in AGS cells with miR-124-3p overexpression and MKN45 cells with miR-124-3p knockdown. (E) IHC assays showed miR-124-3p expression was negatively associated with MST4 expression in GC. Representative IHC pictures about MST4 expression in GCTs with high miR-124-3p expression and low miR-124-3p expression according to the median miR-124-3p expression level by qRT-PCR determined in (B). **P < 0.01; ***P < 0.001.
Discussion
Although GC incidence rate exhibits a steady decline trend in the last few decades, the overall survival of GC patients is still dismal.33 Metastasis remains the major threat for GC patients because of its systemic influence and the resistance of therapeutic agents. The underlying molecular mechanisms of metastasis have been well studied, but the exact molecular and cell-biological details are still ambiguous.34 Kinase is a type of protein enzyme that catalyzes the transfer of phosphate groups from high-energy, phosphate-donating molecules to specific substrates, which is critical in metabolism, cell signaling, cellular transport, secretory processes, and many other cellular pathways. MST4 is a kind of kinase, which is similar to Ste20.11,35 Previous studies have defined many biologic functions of MST4, such as cytoskeleton regulation, migration and proliferation.14–16 However, the role of MST4 in GC has not been studied by now. In this study, we try to elucidate the clinical and biologic significance and molecular mechanisms of MST4 in GC.
In this study, we first analyzed the expression level of MST4 in GC. Public database showed a high expression of MST4 in GC tissues compared with normal gastric tissues. By using GC clinical specimen and cell lines, we further confirmed the upregulation of MST4 in GCT and cell lines. Then, the clinical significances of MST4 in GC patients were further analyzed. Intriguingly, high MST4 expression was associated with clinicopathological parameters of cancer metastasis, such as lymphovascular invasion, lymph node metastasis. Multivariate Cox regression analysis revealed that MST4 was an independent prognostic factor for both OS and DFS in GC patients after radical gastrectomy. What is more, GC patients with high MST4 expression had shorter OS and DFS compared with those with low MST4 expression. Our results consisted of the previous studies that MST4 is frequently upregulated and negatively correlated with patient prognosis in other tumors.14–16 These results revealed the upregulation of MST4 in GC and suggested high MST4 expression may be related to GC metastasis.
To determine the role of MST4 in metastasis, we used a serial of in vitro and in vivo assays. The results showed that overexpression of MST4 in GC cells significantly promoted migration, invasion and metastasis, whereas knockdown of MST4 significantly inhibited GC cell migration, invasion and metastasis. However, MST4 had little effect on GC cell proliferation. Though controversial, EMT is deemed as a character of cancer and plays an important role in promoting cancer cell invasion and metastasis.36,37 Lin et al16 had found MST4 could promote HCC EMT process. As expected, our results showed MST4 could promote GC cell cytoskeleton rearrangements. In addition, MST4 promoted mesenchymal marker vimentin expression and decreased epithelial marker E-cadherin expression, indicating MST4 participate in GC cell EMT process. Therefore, these results confirmed the role of MST4 in promoting GC cell invasion and metastasis by EMT.
In this study, we further revealed the mechanism of MST4 in promoting GC progression. As a kinase, MST4 could activate ERK signaling pathway by direct phosphorylation or dependence on PDCD10.16,38,39 A recent study showed MST4 could disrupt the MST1-MOB1 complex and regulate Hippo signaling pathway by forming a complex with MOB4.14 In addition, PKA-MST4-ezrin and PKA-MST4-ACAP4 signaling cascade to polarized acid secretion in gastric parietal cells.26,27 By using STRING database, we screened the potential downstream effector and found Ezrin may be a target. Co-IP results showed MST4 could bind with Ezrin and confirmed our speculation. Further gain- and loss-of-function assays showed Ezrin was a critical factor to mediate MST4 function in GC cell invasion and metastasis. The previous study had demonstrated that MST4 could directly phosphorylate Ezrin in vitro, which further promotes brush border formation.12 Here, we also found the tumor-promoting role Ezrin signaling is dependent on the phosphorylation function of MST4 in GC.
miRNAs are small noncoding RNAs that base-pair with the 3ʹ untranslated regions (UTRs) of protein-encoding mRNAs, resulting in mRNA destabilization and/or translational inhibition.32 To figure out the possible cause for aberrant MST4 expression in GC, we analyzed potential miRNAs that bind to MST4 3ʹ-UTR region. The results found three potential miRNAs and miRNA-124-3p was more significantly down-regulated in GCT. The experiments determined miRNA-124-3p could bind to MST4 3ʹ-UTR region and inhibit MST4 expression, indicating miRNA-124-3p is a regulator of MST4. Therefore, these findings not only elucidated the down-stream effector of MST4 but also confirmed the up-stream regulator of MST4, which is critical to develop new targeted drugs to GC treatment.
In conclusion, the present study determined high MST4 expression in GC tissues and cell lines. In addition, high MST4 expression was associated with aggressive clinicopathological parameters and poor prognosis of GC patients. Functional assays showed MST4 significantly promotes GC cell invasion and metastasis by inducing EMT process. In terms of mechanism, miRNA-124-3p downregulation in GC leads to MST4 upregulation and the latter further binds to Ezrin and promotes its phosphorylation, which is critical for MST4 playing its roles. Therefore, our study suggests that MST4 can be used as a valuable prognostic biomarker and a potential therapeutic target in GC.
Acknowledgement
The authors would like to thank Dr. Jian Lei (Department of Pathology, Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University) for providing technical support for histology.
Funding Statement
This study was supported by the Natural Science Foundation of Jiangxi, China (grant no. 20181BAB215025), National Natural Science Foundation of China (grant no. 81702922), National Health Commission Foundation of Jiang Xi (grant no. 20191016), Natural Science Fund of Education Department of Jiang Xi (grant no. GJJ170007).
Disclosure
The authors confirm that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.v68.6 [DOI] [PubMed] [Google Scholar]
- 2.Deng JY, Liang H. Clinical significance of lymph node metastasis in gastric cancer. World J Gastroenterol. 2014;20:3967–3975. doi: 10.3748/wjg.v20.i14.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rankin EB, Giaccia AJ. Hypoxic control of metastasis. SCIENCE. 2016;352:175–180. doi: 10.1126/science.aaf4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 5.Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118 [DOI] [PubMed] [Google Scholar]
- 6.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447 [DOI] [PubMed] [Google Scholar]
- 7.Yoo YA, Kang MH, Lee HJ, et al. Sonic hedgehog pathway promotes metastasis and lymphangiogenesis via activation of Akt, EMT, and MMP-9 pathway in gastric cancer. Cancer Res. 2011;71:7061–7070. doi: 10.1158/0008-5472.CAN-11-1338 [DOI] [PubMed] [Google Scholar]
- 8.Peng Z, Wang CX, Fang EH, Wang GB, Tong Q. Role of epithelial-mesenchymal transition in gastric cancer initiation and progression. World J Gastroenterol. 2014;20:5403–5410. doi: 10.3748/wjg.v20.i18.5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwatsuki M, Mimori K, Yokobori T, et al. Epithelial-mesenchymal transition in cancer development and its clinical significance. Cancer Sci. 2010;101:293–299. doi: 10.1111/j.1349-7006.2009.01419.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ling P, Lu TJ, Yuan CJ, Lai MD. Biosignaling of mammalian Ste20-related kinases. Cell Signal. 2008;20:1237–1247. doi: 10.1016/j.cellsig.2007.12.019 [DOI] [PubMed] [Google Scholar]
- 11.Qian Z, Lin C, Espinosa R, LeBeau M, Rosner MR. Cloning and characterization of MST4, a novel Ste20-like kinase. J Biol Chem. 2001;276:22439–22445. doi: 10.1074/jbc.M009323200 [DOI] [PubMed] [Google Scholar]
- 12.Ten KJ, Jansen M, Yuan J, et al. Mst4 and Ezrin induce brush borders downstream of the Lkb1/Strad/Mo25 polarization complex. Dev Cell. 2009;16:551–562. doi: 10.1016/j.devcel.2009.01.016 [DOI] [PubMed] [Google Scholar]
- 13.Madsen CD, Hooper S, Tozluoglu M, et al. STRIPAK components determine mode of cancer cell migration and metastasis. Nat Cell Biol. 2015;17:68–80. doi: 10.1038/ncb3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen M, Zhang H, Shi Z, et al. The MST4-MOB4 complex disrupts the MST1-MOB1 complex in the Hippo-YAP pathway and plays a pro-oncogenic role in pancreatic cancer. J Biol Chem. 2018;293:14455–14469. doi: 10.1074/jbc.RA118.003279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Ma X, Peng S, Nan X, Zhao H. Differential expression of MST4, STK25 and PDCD10 between benign prostatic hyperplasia and prostate cancer. Int J Clin Exp Pathol. 2014;7:8105–8111. [PMC free article] [PubMed] [Google Scholar]
- 16.Lin ZH, Wang L, Zhang JB, et al. MST4 promotes hepatocellular carcinoma epithelial-mesenchymal transition and metastasis via activation of the p-ERK pathway. Int J Oncol. 2014;45:629–640. doi: 10.3892/ijo.2014.2455 [DOI] [PubMed] [Google Scholar]
- 17.Sung V, Luo W, Qian D, Lee I, Jallal B, Gishizky M. The Ste20 kinase MST4 plays a role in prostate cancer progression. Cancer Res. 2003;63:3356–3363. [PubMed] [Google Scholar]
- 18.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji237 [DOI] [PubMed] [Google Scholar]
- 19.Lei X, Deng L, Liu D, et al. ARHGEF7 promotes metastasis of colorectal adenocarcinoma by regulating the motility of cancer cells. Int J Oncol. 2018;53:1980–1996. doi: 10.3892/ijo.2018.4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001 [DOI] [PubMed] [Google Scholar]
- 21.Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Wang YY, Zhao ZS, Ma J. Ezrin is associated with gastric cancer progression and prognosis. Pathol Oncol Res. 2011;17:909–915. doi: 10.1007/s12253-011-9402-y [DOI] [PubMed] [Google Scholar]
- 23.Jin J, Jin T, Quan M, Piao Y, Lin Z. Ezrin overexpression predicts the poor prognosis of gastric adenocarcinoma. Diagn Pathol. 2012;7:135. doi: 10.1186/1746-1596-7-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Lin Z, Chen B, et al. Ezrin/NF-kB activation regulates epithelial- mesenchymal transition induced by EGF and promotes metastasis of colorectal cancer. Biomed Pharmacother. 2017;92:140–148. doi: 10.1016/j.biopha.2017.05.058 [DOI] [PubMed] [Google Scholar]
- 25.Chen MJ, Gao XJ, Xu LN, Liu TF, Liu XH, Liu LX. Ezrin is required for epithelial-mesenchymal transition induced by TGF-beta1 in A549 cells. Int J Oncol. 2014;45:1515–1522. doi: 10.3892/ijo.2014.2554 [DOI] [PubMed] [Google Scholar]
- 26.Yuan X, Yao PY, Jiang J, et al. MST4 kinase phosphorylates ACAP4 protein to orchestrate apical membrane remodeling during gastric acid secretion. J Biol Chem. 2017;292:16174–16187. doi: 10.1074/jbc.M117.808212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang H, Wang W, Zhang Y, et al. Cell polarity kinase MST4 cooperates with cAMP-dependent kinase to orchestrate histamine-stimulated acid secretion in gastric parietal cells. J Biol Chem. 2015;290:28272–28285. doi: 10.1074/jbc.M115.668855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie YH, Li LY, He JZ, et al. Heat shock protein family B member 1 facilitates ezrin activation to control cell migration in esophageal squamous cell carcinoma. Int J Biochem Cell Biol. 2019;112:79–87. doi: 10.1016/j.biocel.2019.05.005 [DOI] [PubMed] [Google Scholar]
- 29.Zhou J, Feng Y, Tao K, et al. The expression and phosphorylation of ezrin and merlin in human pancreatic cancer. Int J Oncol. 2014;44:2059–2067. doi: 10.3892/ijo.2014.2381 [DOI] [PubMed] [Google Scholar]
- 30.Ren L, Hong SH, Chen QR, et al. Dysregulation of ezrin phosphorylation prevents metastasis and alters cellular metabolism in osteosarcoma. Cancer Res. 2012;72:1001–1012. doi: 10.1158/0008-5472.CAN-11-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology. 2012;143:35–47. doi: 10.1053/j.gastro.2012.05.003 [DOI] [PubMed] [Google Scholar]
- 32.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee YC, Chiang TH, Chou CK, et al. Association between helicobacter pylori eradication and gastric cancer incidence: a systematic review and meta-analysis. Gastroenterology. 2016;150:1113–1124. doi: 10.1053/j.gastro.2016.01.028 [DOI] [PubMed] [Google Scholar]
- 34.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/S0962-8924(01)01980-8 [DOI] [PubMed] [Google Scholar]
- 36.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 37.Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036 [DOI] [PubMed] [Google Scholar]
- 38.Ma X, Zhao H, Shan J, et al. PDCD10 interacts with Ste20-related kinase MST4 to promote cell growth and transformation via modulation of the ERK pathway. Mol Biol Cell. 2007;18:1965–1978. doi: 10.1091/mbc.e06-07-0608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin JL, Chen HC, Fang HI, Robinson D, Kung HJ, Shih HM. MST4, a new Ste20-related kinase that mediates cell growth and transformation via modulating ERK pathway. Oncogene. 2001;20:6559–6569. doi: 10.1038/sj.onc.1204818 [DOI] [PubMed] [Google Scholar]