Abstract
RNAs, particularly noncoding RNAs (ncRNAs), are becoming increasingly important therapeutic targets, because they are causative and antagonists of human disease. Indeed, aberrant RNA structural elements and expression deregulate biological processes. In this review, we describe methodologies to discover and optimize small molecules interacting with RNA (SMIRNAs), including the evaluation of direct target engagement and the rescue of RNA-mediated phenotypes in vitro and in vivo. Such studies are essential to fully characterize the mode of action of SMIRNAs and advance our understanding of rationally and efficiently drugging RNAs for therapeutic benefit.
Introduction
Most drug discovery campaigns focus on modulating defective or aberrantly expressed protein targets, enabled by technological advancements in high-throughput screening (HTS), protein-biased small-molecule compound libraries, assessment of direct target engagement in cells, and structure determination. By and large, these technologies are less developed for RNA targets. The identification of RNAs as key pathological agents in many diseases and the serendipitous discovery that small molecules identified from phenotypic screens act on the RNA [1] or premRNA and the RNA/spliceosome interface [2,3] support the notion of targeting RNA for therapeutic benefit. Indeed, advances in structural determination of RNA and RNA-protein and/or small-molecule complexes [4] have revealed that RNAs exhibit well-defined druggable pockets thought to be reserved for protein targets. Alternatively, targetable sites in RNA could be highly flexible or dynamic [5], providing a transient structure that could accommodate or be stabilized by small molecules, akin to an induced-fit model used to describe some protein–small-molecule interactions.
Until recently, few RNAs had been investigated as potential drug targets, such as bacterial ribosomes, RNAs found in pathogenic yeast and/or fungi, and viral RNAs, such as HIV-1 TAR [6,7]. Recent breakthroughs showed that various disorders, such as Huntington’s disease (HD), myotonic dystrophy type 1 (DM1), amyotrophic lateral sclerosis (ALS), and frontotemporal dementia (FTD), are triggered by RNA repeat expansions, such as r(CAG)exp, r(CUG)exp, and r(G4C2)exp, respectively. These feature well-defined RNA secondary structures, and their contribution to disease emergence have been thoroughly investigated, substantiating even further the necessity of considering RNA as a bona fide drug target. Yet, these examples represent a tiny fraction of the underexplored and therapeutically relevant RNA biological space. Indeed, the results of the Encyclopedia of DNA Elements (ENCODE) project revealed that most of the transcriptome does not translate into proteins and, therefore, is noncoding [8]. Recent developments in RNA sequencing [9], prediction of RNA secondary structure from sequence [10], as well as determination of RNA structures in cells [11,12] are adding more evidence of the structural and functional roles of ncRNAs, such as miRNAs and long ncRNAs (lncRNAs) in the progression of various human diseases. Indeed, increasing amounts of evidence [13–16] emphasize that biological activity is controlled by various RNA secondary structure elements (RNA motifs) within ncRNAs, the modulation of which with small molecules might lead to the development of novel lead medicines.
Herein, we review developments of small molecules and chemical probes [17–19] targeting key RNA structural elements from HIV-1, pathogenic yeasts, and microsatellite repeat expansions as well as emerging ncRNAs, such as miRNAs and lncRNAs. We focus on the key screening technologies and/or methodologies that successfully identified small molecules targeting the disease-causing RNA structural elements. Whenever applicable, we describe the optimization process of the initial hit together with the mode of binding of the small molecule to the target RNA. In addition, we emphasize the importance of translating the in vitro results to the physiologically relevant cellular models of the disease-causing ncRNA by applying recently developed target engagement techniques. Confirming the RNA-centric mode of action of the small molecules is vital to develop selective chemical probes and tools that efficiently modulate ncRNA function in vitro and in vivo. Therefore, we further discuss recent advances in the modular assembly of inhibitors in situ templated by RNA structural elements as well as small molecules that cleave RNA targets. Collectively, these efforts have the potential to open new avenues in drug discovery by providing novel therapeutic opportunities for diseases, the pathology of which is triggered by disease-causing RNAs.
Structure-based design strategies
Characterization of RNA–protein macromolecular complexes represents a rich source of structural information, fostering an in-depth understanding of the molecular recognition events and their impact on biological processes. High-resolution structures can reveal atomistic details of the RNA–protein binding interface, showing the key amino acids that drive the macromolecular complex formation. Therefore, mimicking these interactions can represent the basis for the rational design of peptides, peptidomimetics, and macrocycles as RNA binders.
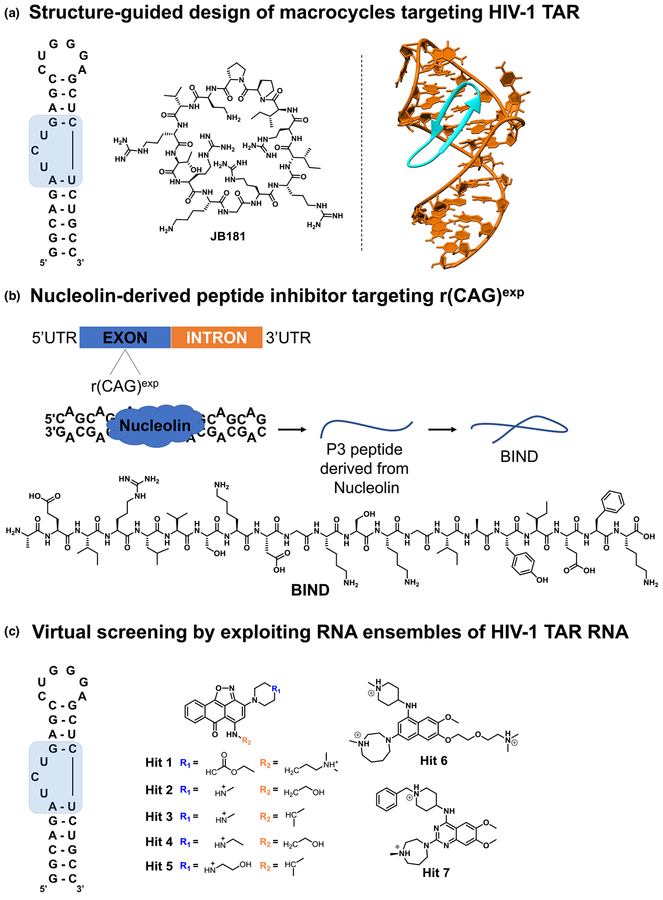
Structure-guided design of macrocycles targeting HIV-1 TAR
The trans-activator protein (Tat) and the trans-activation response element (TAR) RNA interaction is crucial for the viral replication of HIV-1 and, therefore, is steadily pursued for the development of novel antiviral agents. Shortridge et al. [20] reported the structural-guided optimization of L22, a conformationally constrained β-hairpin-containing peptide derived from the arginine rich motif (ARM) of the Bovine Immunodeficiency Virus (BIV) Tat–TAR complex. A focused positional scanning strategy with noncanonical amino acids was applied to the BIV ARM-derived peptide L22, which yielded the macrocyclic peptide JB181 (Fig. 1a), a picomolar affinity and selective binder of TAR RNA even in large excess of tRNA competitor. The strong interaction yielded a 1.4Å resolution NMR structure that confirmed the key residues driving the peptide-RNA interaction (Fig. 1a). Despite the > 100-fold higher in vitro binding affinity of JB181 to the RNA target relative to the starting peptide L22, no significant difference in bioactivity was observed in various cellular assays. This discrepancy suggests a complex interplay of the conformational changes or stabilization occurring upon the protein–RNA interaction in vitro and their relevance in the assembly of multicomponent macromolecular complexes in cells. In conclusion, the report highlighted the potential of structure-guided optimization of cyclic peptides to generate subnanomolar binders of TAR RNA with good selectivity, which will aid efforts to develop novel and potent antivirals.
FIGURE 1.
Graphical representations of the structure-based design strategies used to identify small molecules interacting with RNA. (a) Macrocycle JB181 was generated from L22 peptide via a focused positional scanning strategy with noncanonical amino acids. The RNA motif targeted by JB181 is highlighted in blue. (b) The binding interface of the Nucleolin–r(CAG)exp complex was used to generate peptide P3, which was subsequently optimized to adopt a b-hairpin conformation as in Beta-structured Inhibitor for Neurodegenerative Diseases (BIND). (c) Chemical structures of small-molecule hit compounds identified by a Tat-peptide displacement assay and confirmed by virtual screening using an ensemble-based virtual screening strategy.
Nucleolin-derived peptide inhibitor targeting r(CAG)exp
Numerous neurodegenerative disorders are triggered by trinucleo-tide repeat expansions [r(CAG)exp], including Huntington’s disease (HD), spinocerebellar ataxia type 2 (SCA2), and spinocerebellar ataxia type 3 (SCA3) [21]. Long stretches of r(CAG) exp form a hairpin structure featuring 1 × 1 AA internal loops that sequester various RNA-binding proteins such, as muscleblind-like protein 1 (MBNL1) and nucleolin (NCL) [21], which is in part responsible for the disease etiology. The interaction between NCL and r(CAG)exp was previously characterized and is mediated by the RNA recognition motifs (RRMs) 2 and 3of NCL. Therefore, by analyzing the binding interface of the r(CAG)exp and RRM regions, truncated peptides can be derived as starting points for the design of novel r(CAG)exp binders. However, compared with the protein domains they were derived from, peptides tend to be weaker binders and additionally suffer from various liabilities, such as cellular stability and permeability. Therefore, downstream optimization strategies, such as secondary structure rigidification, are further applied to circumvent these drawbacks.
Zhang et al. [22] reported the structure-guided optimization of a previously identified 13-amino acid peptide (P3) derived from the RRM2 domain of NCL. Given that P3 was predicted to adopt a flexible conformation without a defined secondary structure, optimization efforts led to a 21-mer peptide termed ‘Beta-structured Inhibitor for Neurodegenerative Diseases’ (BIND) (Fig. 1b) that folds into a b-hairpin-containing secondary structure. An alanine scan strategy throughout its sequence highlighted Glu2, Lys13, Gly14, Ile18, Glu19, and Phe20 as the key amino acids involved in the binding to r(CAG)exp. To facilitate the translocation across the cellular membrane, the BIND peptide was conjugated with the 11-amino acid cell-penetrating transactivator of transcription (TAT) peptide. The TAT-BIND peptide rescued the cellular toxicity in r(CAG)78-transfected HEK293 cells and reduced nucleolar stress by restoring the normal localization of NCL and nucleophosmin (NPM; B23) in the nucleoli. Interestingly, in Drosophila and a mouse striatal cell model of HD, the binding potency of the BIND peptide increased with the CAG repeat size and suppressed neurodegeneration in vivo. In conclusion, the NCL-r(CAG)exp interaction represents an excellent starting point in structure-guided campaigns to generate peptides binding r(CAG)exp.
Virtual screening by exploiting RNA ensembles of HIV-1 TAR RNA
Structure-based (SB) approaches are emerging as essential tools that can cost- and time-efficiently accelerate drug discovery campaigns for RNA targets. Such strategies include the prediction, identification, and optimization of small molecules toward an RNA target of interest using the 3D structural information obtained by X-ray crystallography and/or NMR spectroscopy. Within SB drug discovery (SBDD) strategies, virtual screening (VS) enables docking of virtual chemical libraries of small molecules using a well-defined binding pocket or region of interest within an RNA structure. This process usually yields a set of possible binding poses as well as an estimation of their corresponding binding free energy. Therefore, VS represents an alternative to HTS campaigns, allowing a straightforward exploration of the chemical space at reduced costs. However, VS applied to even well-characterized RNA targets, such as HIV-1 TAR, is currently elusive and is mainly attributed to the inability to select the biologically relevant RNA conformation(s) [23] that the chemical libraries should be screened against. Therefore, the results of the screening campaign might vary depending on the quality of the RNA model as well as on the structural determination methodology used for generating it, hampering a clear-cut distinction of the true hits that can be further pursued experimentally.
Target-based approaches
Ganser et al. [24] reported a hybrid experimental–computational approach using RNA dynamic ensembles derived from HIV-1 TAR RNA. The authors employed a training set comprising 177 experimentally validated hit molecules, seven out of which were novel (Fig. 1c) and 103 349 nonbinders, as determined by HTS via a Tat-peptide displacement assay. VS was then performed on a dynamic ensemble of 20 equally populated HIV-1 TAR conformations obtained by NMR spectroscopy and molecular dynamics (MD) simulations. This ensemble-based VS (EBVS) approach overcomes the shortcomings of identifying experimentally validated hits because of either the number of individual conformations or poorly represented RNA ensembles lacking experimental confirmation in solution. True hits were enriched, and their enrichment correlated with the accuracy of the generated RNA ensemble as well as its size. Moreover, the ligand was placed in the proximity of the RNA binding motif as experimentally determined by NMR spectroscopy. In conclusion, the study emphasizes the importance of using RNA dynamic ensembles in VS campaigns to reduce the gap between in silico predictions and experimental validation.
Ligand-based approaches
A chemical-similarity search was used by Parkesh et al. [25] to identify r(CUG)exp binders using Hoechst 33258 and pentamidine as queries using National Cancer Institute (NCI; 250 000 compounds) and eMolecules (8 000 000 molecules) databases. A robust 3D alignment software termed ‘Rapid Overlay of Chemical Structure’ (ROCS) straightforwardly sampled the two databases for novel derivatives that were chemically similar to the initially selected lead molecules. The most promising candidates were tested in various binding and functional assays in vitro and in a DM1 cellular model, respectively. These efforts yielded derivative H1 that disrupted r(CUG)exp nuclear foci and improved DM1 splicing defects both in vitro and in vivo.
Using a pharmacophore-based VS search, Angelbello et al. [26] identified novel compounds that target r(CUG)exp. The authors built a pharmacophore model using a training set of ten known small-molecule binders of r(CUG)exp. Then, novel chemotypes were identified within ~4.3 million compounds with drug-like properties part of the ZINC database [27,28]. The selected candidates were validated to bind r(CUG)exp in vitro and correct various phenotypes in cellular models of DM1. For example, derivative p7 stimulated the nucleocytoplasmic transport of r(CUG)exp retained in the nucleus and improved pre-mRNA splicing defects associated with the sequestration of MBNL1.
In conclusion, these studies reveal important features of targeting RNA with small molecules. First, previously reported small molecules binding r(CUG)exp represent excellent starting points for pharmacophore-based VS strategies to explore chemical space for novel chemotypes even in the absence of a thoroughly characterized RNA target. Second, integrating VS in silico with robust binding assays in a high-throughput format can rapidly delineate true hits that can subsequently be tested in physiologically relevant cell lines that best represent the disease-causing RNA under investigation.
Fluorescence-based screening strategies
HTS of compound libraries represents a well-established paradigm for the identification of small-molecule modulators of various biological targets. Among the possible readout formats, measuring fluorescence change informs the conformational changes in the vicinity of a fluorescently labeled RNA or RNA-binding ligands[29]. The ability to examine such processes in solution in a homogenous plate-format, at relatively low cost, across multiple time points or in an end-point fashion, makes the fluorescence readout readily applicable in target-based drug discovery HTS campaigns. For example, HTS of commercially available compound collections is routinely used as preliminary screen against fluorescently labeled RNA, using various formats, such as fluorescence polarization (FP), Förster resonance energy transfer (FRET), or in combination with fluorescent dyes, as in differential scanning fluorimetry (DSF) [30].
Inhibitors of group II intron via fluorescence-based enzymatic assays
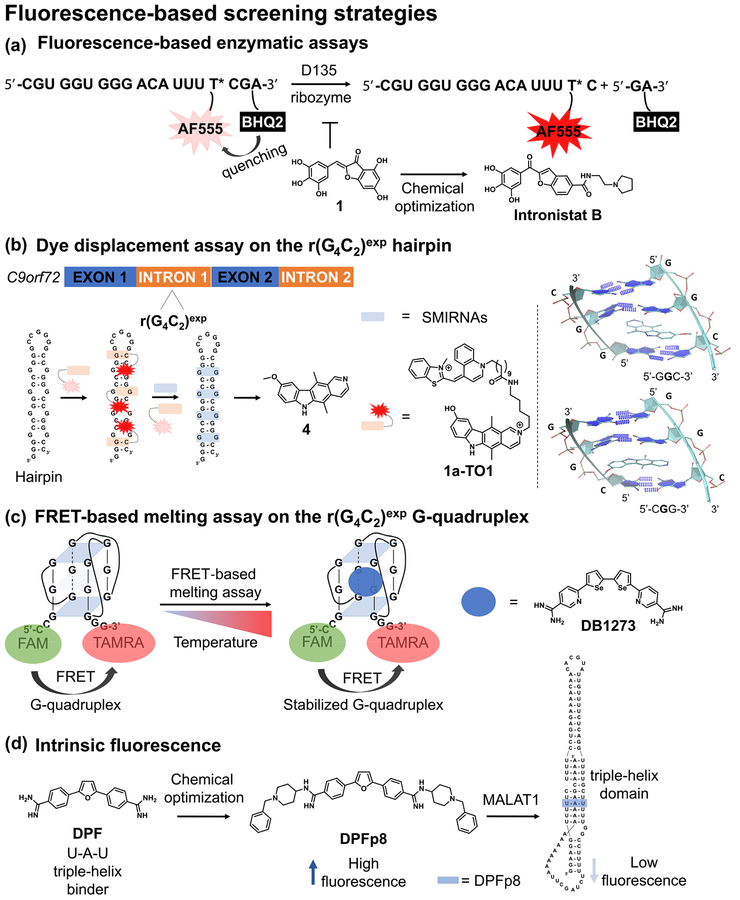
An assay set-up readily applicable for enzymatic reactions relies on the turn-on fluorescence signal of a fluorescently quenched construct. For example, an oligonucleotide containing an Alexa555 fluorophore proximal to the Black Hole Quencher 2 (BHQ2) is fluorescently inactive because of the latter, which absorbs the excitation energy from the adjacent fluorophore [31]. However, the removal of this moiety from the oligonucleotide substrate induces an increase in the (turned-on) fluorescent signal, which makes this assay set-up well suited for the interrogation of RNA molecules with enzymatic activity, such as group II ribozymes. These represent intriguing self-splicing introns, the splicing of which is exclusively mediated by rearrangement of large RNA (sub)domains without the assistance of proteins [32]. These are unique RNA targets present within the mitochondrial genome of pathogenic yeasts, and their inhibition can directly impact the fungal metabolism. This is therapeutically appealing, given that pathogenic yeasts represent a serious threat to patients with compromised immunity.
Starting from the ai5γ II intron of Saccharomyces cerevisiae, Fedorova et al. [31] engineered a multiturnover ribozyme termed ‘D135’ capable of cleaving fluorescently quenched oligonucleotide RNA substrates that resemble the parental 5′ splice site. A fluorescence-based HTS was performed on 10 000 molecules from commercially available libraries, including NCI, US Food and Drug Administration (FDA)-approved drugs, ENZO, ChemBridge, and ChemDiv. The initially identified benzylidene-benzofuran lead molecule was optimized via medicinal chemistry approaches unraveling structure–activity relationships (SAR) that ultimately led to Intronistat B (Fig. 2a). This derivative inhibited growth of Saccharomyces cerevisiae and Candida parapsilosis in the same bioactivity range as the established antifungal agent amphotericin B, by disrupting the splicing of the mitochondrially encoded cyto-chrome c oxidase I (COX1) gene, and was well tolerated in mammalian cells. In conclusion, the study emphasizes that commercially available libraries of small molecules contain derivatives that target ribozymes derived from pathogenic yeasts. Further studies in elucidating the exact binding site might offer novel opportunities to identify the conformational changes that block the activity of the ribozyme.
FIGURE 2.
Graphical representations of the fluorescence-based screening strategies used to identify small molecules interacting with RNA. (a) A fluorescently inactive oligonucleotide embedding AF555 fluorophore and BHQ2 quencher was designed to mimic the natural substrate cleaved by the group II self-splicing introns. Upon cleavage by the engineered D135 ribozyme, BHQ is released, leading to turn-on fluorescence signal. Initial hit compound 1 was optimized via medicinal chemistry approaches to the more potent derivative Intronistat B. (b) The fluorescently inactive 1a-TO1 construct exhibits turn-on fluorescent signal when bound to the hairpin form of r(G4C2)8, which is quenched upon displacement with small molecules interacting with the RNA. The bound structures of compound 4 to 50-GGC-30 and 50-CGG-30 sites were obtained by combining 2D NOESY NMR experiments with computational modeling (G represents the G residue within the 1 × 1 GG internal loops). (c) The degree of Förster resonance energy transfer (FRET) from 50-FAM to 30-TAMRA within the dual-labeled r(G4C2)4 construct is monitored as a function of temperature in the presence and absence of small-molecule ligands. Derivative DB1273 stabilizes the G-quadruplex form of r(G4C2)4 and exhibits FRET at higher temperatures relative to the vehicle control, i.e. higher melting temperature (Tm). (d) Binding of DPFp8 to the triple helix region of MALAT1 can be monitored by measuring the quenching of compound intrinsic fluorescence upon interaction with the nucleic acid.
Dye displacement assay yields bioactive derivatives of the r (G4C2)exp hairpin
Fluorescent intercalator displacement (FID) [33–35] uses turn-on fluorophores that exhibit enhanced fluorescence properties upon binding a macromolecular target. For example, thiazole orange (TO)-based dyes exhibit low fluorescent properties when freely diffused in solution, but the fluorescence is dramatically enhanced upon binding RNA because of lowering of the degrees of rotation. Therefore, SMIRNAs can be identified upon fluorescence quenching as the bound dye is displaced from the RNA target [36]. TO dyes can also be covalently linked to known RNA binders, whereby the small molecule will bring TO into the vicinity of the target RNA, yielding a fluorescent signal.
Intronic r(G4C2)exp hexanucleotide repeat expansions within the C9orf72 gene are the most common cause for genetically defined ALS and FTD, collectively termed ‘c9ALS/FTD’ [37,38]. Key pathologies responsible for the clinical manifestations of c9ALS/FTD are caused by RNA gain-of-function mechanisms of r(G4C2)exp, such as the formation of RNA foci in the nucleus and the production of toxic dipeptide repeats (DPR) via repeat-associated non-ATG translation (RANT) in the cytosol, which ultimately lead to neuronal death. However, the precise RNA secondary structure(s) of r(G4C2)exp that are responsible for the disease phenotypes are not well understood. Literature reports show that r(G4C2)exp can form at least two structural folds in equilibrium [39–41], depending on the cations present in solution. Specifically, Na+ ions favor the hairpin form, comprising 1 × 1 GG internal loops, whereas the addition of K+ ions completely shifts the equilibrium towards the formation of a thermodynamically stable G-quadruplex structure.
Focusing on the hairpin form, Wang et al. [42] identified small molecules targeting c9ALS/FTD pathology by means of an in vitro dye displacement assay using the 1a-TO1 conjugate. This encompasses the previously reported binder of r(G4C2)exp, termed 1a, covalently attached to TO1 dye via a linker (Fig. 2b). By monitoring quenching of the 1a-TO1 signal upon displacement with a focused compound collection of 45 derivatives chemically similar to 1a, the authors identified the ellipticine derivative termed ‘compound 4’. This derivative more potently bound r(G4C2)8 over various RNA control sequences and reduced RANT in a r (G4C2)66 transfected HEK293T disease model to a greater extent than 1a, an antisense oligonucleotide, and several known G-quadruplex binders. In vitro, compound 4 potently disrupted a deleterious RNA–protein interaction characterized in c9ALS/FTD cellular models, such as r(G4C2) exp–hnRNP H, via a time-resolved FRET (TR-FRET) assay. Indeed, compound treatment released the sequestered hnRNP H protein from nuclear RNA foci in the r(G4C2)66-transfected HEK293T disease model. Various biophysical techniques showed that the hairpin rather than the G-quadruplex conformation undergoes RANT and that compound 4 stabilizes the 1 × 1 GG internal loops via stacking interactions (Fig. 2b). Mechanistically, the small molecule hinders the assembly of polysomes on the toxic transcript r(G4C2)66 inhibiting RANT without affecting mRNA levels. In conclusion, the study indicates that the stabilization of the r(G4C2) exp hairpin form is a therapeutically viable solution to develop novel lead medicines for c9ALS/FTD.
FRET-based melting assay yields bioactive derivatives of the r(G4C2) exp G-quadruplex form
A third popular fluorescence-based screening approach relies on the FRET between two fluorescently active moieties. When found in proximity, the energy released upon exciting the donor fluorophore is transferred to the acceptor fluorophore, the emission of which is then recorded. Therefore, the lower the distance between the fluorophores, the stronger the energy transfer and the higher the emission recorded on the acceptor channel. Given that FRET reports changes in distances at the nanometer scale, it can be applied in various scenarios where conformational changes of the RNA molecule(s) under investigation are measured upon applying external stimuli, such as heat. Increasing the temperature over a wide range triggers the denaturation of the RNA structure and consequently lowers or abolishes the FRET signal.
Focusing on the G-quadruplex form of r(G4C2)4, Simone et al.[43] used a FRET-based melting assay on 138 compounds to identify small-molecule stabilizers of this RNA fold. Using the FRET signal arising from the 5′-FAM and 3′-TAMRA dual-labeled r(G4C2)4 construct to measure the melting temperature (Tm) in the presence of the derivatives, the authors identified three structurally related small molecules, DB1246, DB1247, and DB1273, (Fig. 2c) that stabilized r(G4C2)4 over d(G4C2)4. The most promising derivative, DB1273, successfully reduced the number of nuclear RNA foci in patient-derived induced pluripotent stem cell (iPSC)-derived motor and cortical neurons and lowered the dipeptide repeats poly(GP) levels with no cytotoxicity. In addition, compound treatment improved the survival of a c9ALS/FTD Drosophila model at different developmental stages by lowering the levels of poly(GP) and poly(GR) without affecting the RNA transcript, indicating an RNA-based mode of action.
In conclusion, both studies emphasize the importance of pursuing structurally and mechanistically well-characterized disease-causing RNAs in compound-screening campaigns. The combination of target-based screening assays, such as dye-displacement, FRET, and TR-FRET, and bioactivity testing in physiologically relevant disease models can accelerate the development of c9ALS/FTD therapeutics in a more straightforward manner.
Exploiting intrinsic fluorescence for the identification of ligands against MALAT1
An additional fluorescence-based screening approach is based on monitoring the fluorescent properties of SMIRNAs. In this case, the signal of intrinsically fluorescent active ligands in solution is quenched upon interaction with the RNA due to various factors, such as conformational changes of the bound ligand or stacking with the internal nucleotides. This approach is particularly appealing because it does not need chemical derivatization of the ligand to attach a fluorescently active unit. The identification of a position suited for functionalization that will not perturb binding activity to the intended RNA target usually implies additional synthetic steps, which can often be cumbersome depending on the molecular complexity of the ligand. Therefore, intrinsically fluorescent small molecules can directly be applied in screening campaigns for assessing RNA binding and selectivity.
The lncRNA MALAT1 is overexpressed in numerous cancer models and its ablation was reported to decrease invasion and tumor growth [44,45]. Recent studies showed that MALAT1 transcript levels are governed by a specific region within the 3΄-end that folds into a U-A-U triple helix structure, recently characterized by X-ray crystallography [46]. Destabilizing this region with small molecules represents a novel strategy to reduce the cellular stability of MALAT1, leading to its rapid degradation in cancers where this transcript is overexpressed [47]. Using the propensity of the diphenylfuran derivative furamidine (DPF) to bind 5′-AU/3′-UA base pairs [48], Donlic et al. [49] reported its medicinal chemistry optimization yielding DPFp8 (Fig. 2d). This interacted with the triple helix domain of MALAT1 in vitro more potently than control RNA and DNA sequences, as determined by the quenching of the intrinsic fluorescence of the compound. Interestingly, DPFp8 exhibited the most pronounced rod-like character among the entire series of synthesized derivatives, exhibiting a nearly planar conformation and dihedral angles that preclude intramolecular interactions between the functional groups and the aromatic rings. In conclusion, the report highlights the optimization of a previously reported triplex binder by combining lead optimization with 3D shape and conformational analysis that led to high affinity and selectivity towards the desired RNA target in vitro. Further studies are required to establish whether the optimized candidate DPFp8 modulates the transcript levels of MALAT1 by the proposed mechanism in disease-relevant cellular models. Interestingly, DPF was previously reported to potently bind r(AUUCU)exp in vitro by targeting the 5′-AU/3′-UA base pairs[48]. An azide-functionalized DPF derivative was then used to generate a dimeric compound that potently corrected various phenotypes associated with spinocerebellar ataxia type 10 (SCA10) in patient-derived cells. Therefore, DPF is a versatile building block for designing and optimizing small molecules targeting various RNA structural motifs containing 5′-AU/3′-UA base pairs and U-A-U triple helices.
Small molecule microarray strategies
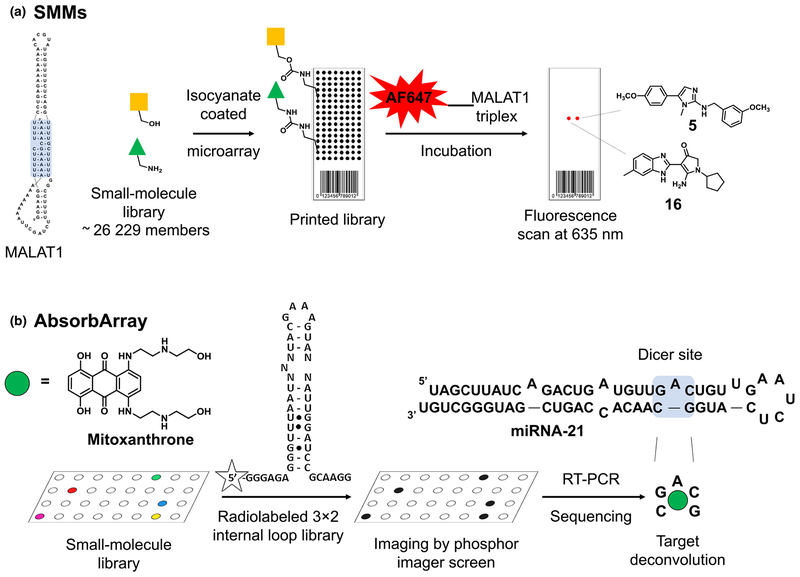
Small molecule microarrays (SMMS) represent a pivotal technology with major applications in target-based drug discovery campaigns. The principle relies on the covalent or noncovalent immobilization of small molecules on microchips depending on the chosen screening strategy. Thousands of small-molecule spots can be deposited in an array format on microarray slides, which are subsequently screened against fluorescently labeled biomacromolecules. Hit compounds can easily be identified by deconvoluting the spots where the target molecule is highly enriched.
Small-molecule microarray yields bioactive compounds targeting MALAT1
Using a SMM strategy, Abulwerdi et al. [50] reported the discovery of two structurally unrelated derivatives that target the triplex region of MALAT1 derived from mouse. Briefly, isocyanate-coated glass slides were reacted with the amino or hydroxy functional group of 26 229 commercially available derivatives, yielding smallmolecule libraries immobilized on chip. These were incubated with the fluorescently labeled triple helix region of MALAT1 and spots exhibiting a high fluorescent signal were identified as hit compounds. Compound 5 and compound 16 (Fig. 3a) emerged as potential lead molecules after passing various selection criteria, including internal SMM screening campaigns against various hairpins, riboswitches, and others. The selected compounds reduced the expression levels of MALAT1 and the degree of branching morphogenesis in an organoid mouse model of mammary cancer. In conclusion, this study reports the robust application of SMMs against MALAT1, which yielded two structurally dissimilar small molecules biologically active in mouse organoid tumor models. Further studies are required to validate the RNA-centric model of action of these compounds, which will increase our understanding of how to rationally design small molecules that affect MALAT1 biology in relevant disease models.
FIGURE 3.
Graphical representations of the small-molecule microarray strategies used to identify small molecules interacting with RNA. (a) Small molecules are covalently attached to the surface of a microchip using straightforward chemistries, followed by incubation with the fluorescently labeled triple helix region of MALAT1. After subsequent washing steps, the highly fluorescent spots are identified and hit molecules are deconvoluted. (b) Small-molecule libraries are absorbed on the agarose-coated surface of a chip. This is then incubated with a 32P-radiolabeled RNA library of 3 × 2 internal loops, followed by extensive washing steps. Afterwards, the radioactive spots are isolated and the RNA targets are identified via RT-PCR and sequencing. This method provides the RNA motif–small molecule pair.
Drug repurposing via library versus library screening (AbsorbArray)
A novel microarray-based screening technology was recently developed, termed ‘AbsorbArray’ (Fig. 3b). In this case, small-molecule compounds from commercially available libraries are noncovalently adhered on the surface of agarose-coated micro-arrays and screened against RNA motif libraries commonly found in ncRNAs. The lack of covalent attachment on the microarray slides is particularly appealing, because readily available compound libraries of small molecules can be applied on the chip without further derivatization, which might affect the binding mode to the RNA target(s). Compared with the traditional micro-array approach, where each RNA target of interest is screened in a stepwise fashion, the RNA motif library versus small-molecule library screening format of AbsorbArray, termed 2D combinatorial screening (2DCS), probes an even greater number of interactions in a single step that will ultimately yield the preferred RNA motif for each individual small molecule. Therefore, compounds targeting RNA structural element(s) that are part of disease-causing RNAs can straightforwardly be identified together with their relative affinity to the entire RNA motif library.
The half-life of ncRNAs is orchestrated by a fine-tuned balance between synthesis and degradation and is correlated with various human diseases. In particular, miRNA-21 upregulation has been shown to contribute to several cancer pathologies, including lung carcinoma [51] and pancreatic ductal adenocarcinoma [52]. Therefore, lowering miRNA-21 levels by blocking its processing and subsequent maturation shuts down the activation of various downstream biological pathways modulated by this miRNA.
Velagapudi et al. [53] confirmed that the invasive phenotype of MDA-MB-231 triple-negative breast cancer cells can be ablated by the chemotherapeutic agent mitoxantrone by specifically targeting a key A-bulge within pre-miRNA-21, affecting its processing. AbsorbArray was performed on ~ 1000 compounds within the NIH Clinical Collection (NIH-CC), kinase- and RNA-focused libraries, and ~ 1024 RNA motifs comprising 3 × 2 asymmetric internal loops and bulges present in various cellular RNAs. The presence of competitor oligonucleotides eliminated nonspecific binders and enriched the selection of compounds binding strongly to defined RNA structural elements. The identification of highly confident RNA motif small-molecule pairs was revealed by sequencing RNAs bound to the microarray chip and deconvoluted via a stringent statistical analysis tool, termed ‘high-throughput structure-activity relationships through sequencing’ (HiT-StARTS). This strategy revealed that mitoxantrone reliably binds the A bulge in the Dicer site of pre-miRNA-21 (Fig. 3b) inhibiting Dicer processing in vitro and in MDA-MB-231 cells. As a result, the levels of the tumor suppressor phosphatase and tensin homolog (PTEN) increased, ultimately leading to the ablation of the invasive phenotype of MDA-MB-231. RNA target modulation by mitoxantrone was confirmed by over-expressing pre-miRNA-21, which abolished the biological activity of this compound, and by target engagement using the Chemical-Crosslinking and Isolation by Pull-Down (Chem-CLIP) procedure.
In conclusion, this study highlights important conclusions for the field of drugging RNA with small molecules. First, AbsorbArray furnishes high-confidence small molecules binding disease-causing RNA structural motif(s) in vitro, which is appealing in drug repurposing campaigns. Second, the invasive phenotype of MDAMB-231 can be ablated by mitoxantrone targeting a key RNA structural element of pre-miRNA-21, therefore validating an RNA-based mechanism of action in addition to inhibiting type II topoisomerase.
Database search strategies
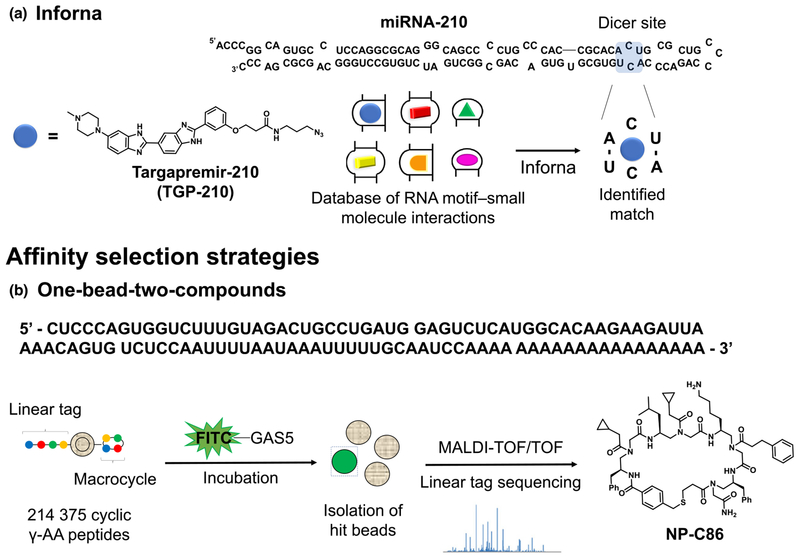
Mining Inforna yields an inhibitor of miRNA-210 processing
The recent technological advances in various research areas, such as screening methodologies, sequencing, computational power, and bioinformatics, have yielded a wealth of information about targeting RNA with small molecules freely accessible online. As a result, tremendous efforts have been invested into compiling all this information in databases that researchers can use as starting points for testing biological hypotheses related to drugging RNA with small molecules. The Inforna platform merges RNA secondary structure prediction with the results obtained via 2DCS [54]. Upon simultaneously probing millions of possible RNA small-molecule interactions, valuable data are then extracted by means of stringent statistical analysis which ultimately yields high-throughput SAR between RNA motifs and small molecules. That is, for each disease-causing RNA structural motif, Inforna can provide high-quality lead small molecule(s) as well as their relative affinity towards structurally similar and dissimilar secondary structures [54,55].
MiRNA-210 is a therapeutically relevant ncRNA associated with poor prognosis in numerous breast [56], lung [57] and pancreatic cancers [58]. In normal conditions (normoxia), hypoxia inducible factor 1α (HIF-1α) is rapidly degraded by the proteasome via prolyl hydroxylase (PHD) upon ubiquitination and subsequent degradation [59]. However, under hypoxic conditions (hypoxia), the increased expression of miRNA-210 downregulates glycerol-3-phosphate dehydrogenase 1-like (GPD1L), which in turn represses PHD and prevents the proteolytic degradation of HIF-1α [60]. This mechanism promotes resistance to radiation treatments, and facilitates cell proliferation and survival in tumor environments, leading to tumor relapse [57]. Therefore, lowering the miRNA-210 levels by targeting its processing might represent a therapeutically viable solution.
Based on the Inforna mining, Costales et al. [61] identified the small molecule Targapremir-210 (TGP-210) as a high-confidence binder of the 1 × 1 CC internal loop within the Dicer site of pre miRNA-210, thus blocking its processing in vitro and in vivo (Fig. 4a). As a result, TGP-210 treatment upregulated GPD1L, decreased the stability of HIF-1α, and inhibited tumor growth in vivo in a NOD/SCID mouse model. By performing target engagement experiments by means of Chem-CLIP, the authors then demonstrated that the phenotype was the result of the modulation of pre-miRNA-210 levels. The results showed that TGP-210 recognizes the pre-miRNA-210 Dicer site selectively in vitro and can distinguish between RNA targets that feature the same binding motif as pre-miRNA-210 based on the different expression levels. Additionally, profiling studies by RT-qPCR highlighted that only miRNA-210 was significantly affected among hypoxia-related miRNAs. In summary, this study exploited the differential expression levels of miRNA-210 in hypoxic versus normoxic conditions to selectively modulate the Dicer processing of pre-miRNA-210. Moreover, important insights related to the druggability of ncRNAs with small molecules have been revealed. For example, the degree of RNA target occupancy in cells is driven not only by the affinity of the small molecule to a particular RNA structural motif, but also by the expression levels of the RNAs that feature this binding site.
FIGURE 4.
Graphical representations of the database search and affinity selection strategies used to identify small molecules interacting with RNA. (a) Inforna search reveals that Targapremir-210 (TGP-210) targets the Dicer site of miRNA-210 featuring a 1 × 1 CC internal loop; (b) A 214 375-member library on polymeric microbeads was spatially segregated allowing for spatial differentiation between cyclic γ-AA peptides (surface) and a corresponding linear tag (core). The beads were incubated with the fluorescently labeled oligonucleotide containing the UPF1-binding RNA sequence. Highly fluorescent beads were isolated, the linear tag was released from the beads, and its sequence deconvoluted via tandem mass spectrometry
Affinity selection strategies
One-bead-two-compounds (OBTC) strategy for disrupting GAS5-UPF1 interaction
Affinity selection techniques represent powerful tools in modern drug discovery. Among them, the split-and-pool synthesis of one-bead-one-compound (OBOC) and one-bead-two-compounds (OBTC) libraries yield unique and structurally diverse compounds attached as multiple copies onto solid support. The beads decorated with individual compounds are then incubated with fluorescently labeled macromolecules followed by subsequent isolation of the beads exhibiting the highest fluorescent signal. This is followed by chemical release of the hit(s) and subsequent sequence deconvolution by mass spectrometry (MS) techniques. Although the entire procedure involves multiple steps, the OBOC and OBTC approaches with structurally diverse combinatorial libraries are useful in targeting orphan macro-molecular targets.
Low levels of the lncRNA GAS5 in serum is considered as a potential biomarker for patients with type 2 diabetes mellitus (T2DM) [62]. Mechanistically, the GAS5 reduction blocks glucose uptake by regulating the expression levels of the insulin receptor in adipocytes. Therefore, increasing the GAS5 levels by inhibiting its degradation might stimulate insulin signaling. Its cellular levels are mediated by the regulator of nonsense transcripts 1 (UPF1) protein, a crucial component of the nonsense-mediated decay (NMD) pathway. Indeed, depletion of UPF1 led to GAS5 accumulation in cells, suggesting that interfering with the GAS5-UPF1 interaction is therapeutically viable.
Shi et al. [63] reported the first small-molecule disruptor of the GAS5-UPF1 lncRNA–protein interaction by applying an OBTC strategy. The authors generated a 214 375-member library that was incubated with the fluorescently labeled oligonucleotide containing the UPF1-binding RNA sequence of GAS5 in the presence of a ~ 1000-fold tRNA competitor. Highly fluorescent beads were isolated and sequence-deconvolution identified the macrocyclic derivative NP-C86 (Fig. 4b), exhibiting nanomolar binding affinity to lncRNA GAS5. Compound treatment restored the GAS5 levels in DM adipocytes without affecting the UPF1 levels. Consequently, this reinstated glucose uptake upon insulin stimulation in cells as well as in ex vivo diabetic adipose tissue (AT) explants. In conclusion, the OBTC strategy yielded the macrocycle NP-C86, which reduced the turnover of lncRNA GAS5 by blocking its interaction with UPF1, offering a therapeutically viable and novel approach to restore glucose homeostasis.
New modalities to tackle RNA disease biology
In situ inhibitor synthesis driven by RNA templated oligomerization
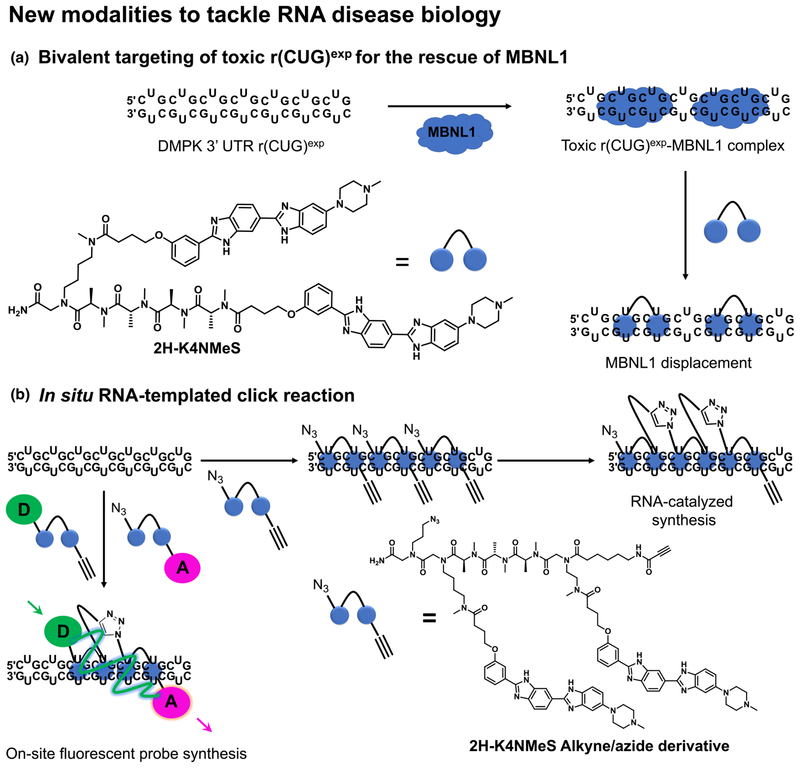
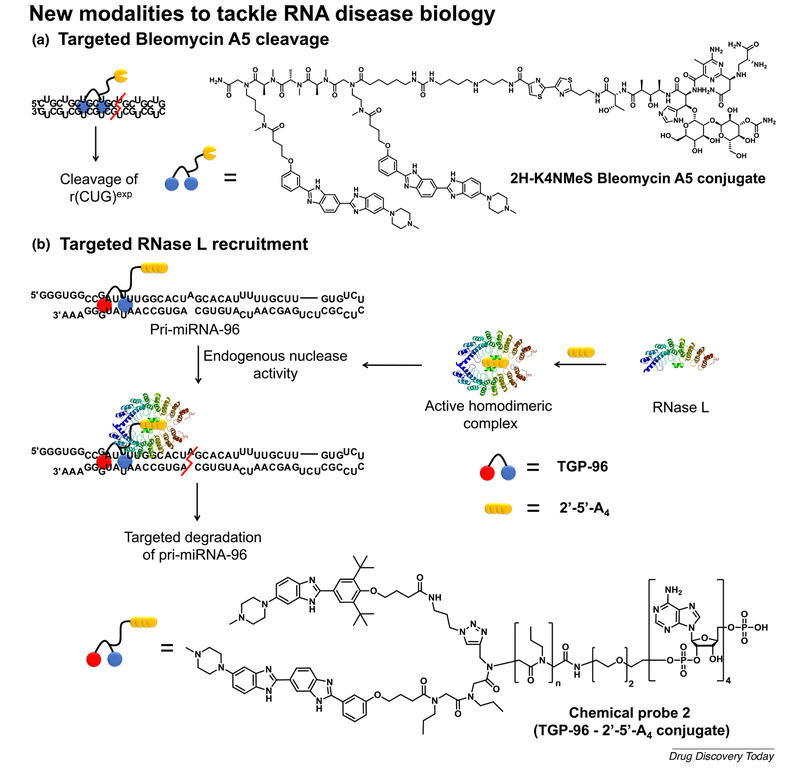
The RNA r(CUG)exp within the 3′-UTR of the dystrophia myotonica protein kinase (DMPK) mRNA transcript causes DM1. In patients with DM1, hundreds to thousands of such repeats form hairpin structures featuring 1 × 1 UU internal loops, which sequester various RNA-binding proteins, such as MBNL1. This RNA gain-of-function mechanism ultimately leads to dysregulation of the RNA splicing processes and defects in nucleocytoplasmic transport, causing muscle atrophy and myotonia along with numerous other multisystemic health issues. Significant resources were invested into developing chemical tools to probe and correct the dysfunction of r(CUG)exp by targeting the repetitive 1 × 1 UU internal loops with dimeric small molecules. A recent example published by Rzuczek et al. [64] describes 2H-K4NMeS, a dimeric chemical probe that potently targets r(CUG)exp and relieves various DM1-associated phenotypes (Fig. 5a). These effects are mediated by selectively engaging the r(CUG)exp in cells, as shown by the target engagement experiments via Chem-CLIP. Using this information, the initial 2HK4NMeS was equipped with alkyne and azide handles placed at optimal distances that are prone to in situ oligomerization by means of click chemistry when adjacently bound to RNA (Fig. 5b). As a result, the oligomeric product achieved 100-fold increased potency compared with the control probe. Interestingly, this RNA-catalyzed click approach also enabled the RNA templated synthesis of FRET sensors that not only confirmed target engagement, but also imaged the assembly process at the r(CUG)exp sites in cells (Fig. 5b). This work also described the development of small-molecule nucleic acid profiling by cleavage applied to RNA (Ribo-SNAP), which exploits the cleavage activity towards nucleic acids of the natural product Bleomycin A5. When covalently linked to 2H-K4NMeS, cleavage occurs at the RNA r(CUG)exp sites, depleting the RNA levels, which dramatically corrects the splicing defects in DM1 cellular models (Fig. 6a). Together, these synergistic chemical biology approaches can be applied to small molecules that potently and selectively bind repetitive sequences and could be applied to more than 30 microsatellite repeat expansions, many of which are still orphans of lead medicines.
FIGURE 5.
Graphical representation of emerging modalities that potently correct aberrant phenotypes triggered by disease-causing RNAs. (a) Dimeric compounds potently bind r(CUG)exp and release sequestered MBNL1. (b) Dimeric compounds equipped with azide and alkyne functional groups, respectively, undergo in situ oligomerization templated by the disease-causing RNA [e.g. r(CUG)exp]. This process can be followed in cells by means of a FRET sensor generated from dimeric compounds equipped with FRET-compatible fluorophores
FIGURE 6.
Graphical representation of emerging modalities that cleave disease-causing RNAs. (a) The dimeric molecule 2H-K4NMeS covalently linked with the nucleic acid cleavage moiety Bleomycin A5 efficiently cleaves r(CUG)exp. (b) The dimeric molecule Targaprimir-96 (TGP-96) covalently linked to the RNase L dimerization unit 2’−5’-A4 efficiently cleaves miRNA-96. This is achieved by the endogeneous nuclease activity of RNase L, which is placed in close proximity to miRNA-96 by the RNA-binding module TGP-96.
Cleavage of miRNA-96 by RNase L recruitment
MiRNA-96 is highly expressed in breast cancers and represents a crucial biological hub that represses the proapoptotic transcription factor Forkhead box protein O1 (FOXO1), leading to cancer cell proliferation and tumor progression [65]. Therefore, ablating the levels of this ncRNA might provide therapeutic benefit. Along these lines, Costales et al. [66] reported the development of a RIBOnuclease Targeting Chimera (RIBOTAC) strategy that harnesses ribonuclease L (RNase L) to specifically cleave miRNA-96 (Fig. 6b). RNase L is a crucial component of the antiviral immune response and is ubiquitous in all cells in an inactive monomeric state. Its activation occurs via dimerization when binding 2′–5′oligoadenylate [2′–5′poly(A), 2′–5′-A4], the levels of which are increased upon viral infection. Therefore, the 2′–5′-A4 levels dictate regardless of whether RNase L is active. The authors exploited this cellular mechanism using chemical probe 2 (Fig. 6b), comprised of the small molecule Targaprimir-96 (TGP-96) conjugated to 2′–5′-A4, the dimerization inducer of RNase L. The latter recruits endogenously present RNase L and, therefore, activates its nuclease activity proximal to the pri-miRNA-96 binding site of TGP-96 (Fig. 6b). Therefore, chemical probe 2 selectively decreases the levels of pri-miRNA-96 and, consequently, miRNA-96 in MDA-MB-231 cells, ablating its invasive phenotype. Finally, compound treatment significantly increased the levels of FOXO1 and selectively triggered apoptosis of the MDA-MB-231 cells in a miRNA-96-dependent manner. In conclusion, the study reports a novel strategy to cleave ncRNAs that harnesses the ribonucleic acid degradation machinery within cells. This approach promises the development of custom RNA degraders to direct the RNase L nuclease activity in the proximity of almost any small molecule that targets therapeutically relevant ncRNAs.
Concluding remarks
Major progress in SMIRNAs within the past few years has elevated RNA among the therapeutically relevant biological targets to include in drug discovery campaigns [17,67]. Indeed, the field has accelerated at a rapid pace because of a better understanding of RNA structure and biology, and technological advancements in areas, such as synthetic methodologies, commercial avail ability of compound libraries, computational power, and sequencing.
Including RNA among the myriad biologically relevant drug targets expands the possibilities to tackle human diseases triggered by aberrant function of RNA motifs with small molecules. However, one of the main challenges is the efficient exploration of the currently available chemical space that engage disease-causing RNA structural elements. Currently, commercially available chemical libraries are enriched with chemical probes and small molecules optimized against commonly drugged protein targets, such as G-protein-coupled receptors (GPCRs) and kinases. In addition, not only clinically approved drugs, but also derivatives that lacked clinical efficiency are continuously pursued as new leads in drug repurposing campaigns. Therefore, an immediate approach is to efficiently repurpose clinically relevant candidates towards diseases that are triggered by aberrant function of RNA. A recent study showed that approved drugs can modulate the oncogenic noncoding miRNA-21 for therapeutic gain [53], highlighting the AbsorbArray technique as a straightforward strategy to repurpose the already available chemical matter to novel therapeutic indications. In addition, because these chemotypes were optimized to bind protein targets, they represent a solid ground to apply medicinal chemistry approaches to repurpose their cellular profile towards modulating disease-causing RNAs while minimizing the off-target effects in a proteome- and transcriptome-wide fashion.
The main challenge for chemists and chemical biologists remains the de novo discovery of novel chemical matter that efficiently populates the biologically relevant chemical space interacting with RNA motifs, which willsteadily increase. The challenge will be the development of robust technologies to provide initial hits and assess the druggability of RNA motifs with smallmolecules. First, screening virtual chemical libraries allows a straightforward approach to assess how suitable the currently assembled and explored chemical space is for targeting the noncoding transcriptome. This process has remained elusive because of a lack of understanding of how small molecules affect RNA conformational changes and dynamics upon binding. As a result, currently applied methods lack consistency and robustness in providing candidates that can be experimentally confirmed. This drawback can be addressed by performing VS campaigns using dynamic ensembles of rigorously characterized RNA structures that better capture the conformational heterogeneity and dynamics of the RNA of interest, therefore providing high-quality hits. In addition, developing reliable force fields and molecular dynamics simulations of RNA structure(s) [68] will provide good starting points towards efficient VS campaigns where the RNA of interest lacks structural characterization. Second, various HTS techniques are currently applicable and well established to robustly identify SMIRNAs (Table S1 in the supplemental information online), including ligand displacement assays [29,34,35], NMR-based methods [69], catalytic enzyme-linked click chemistry assay (cat-ELCCA) [70,71], automated ligand identification system (ALIS) via affinity selection-mass spectrometry (AS-MS) [72,73], and microarrayformats, such as SMMs [74], 2DCS [75], and AbsorbArray [53]. For example, compound-immobilized chips can be used either in sequential RNA target-based screening, as in the case of SMMs, or in a massively parallel format with RNA motif libraries. Whereas in the former, lead molecules against a single target can be determined, the 2DCS and AbsorbArray simultaneously probe RNA motifs against small-molecule libraries. Such screening campaigns usually yield an enormous amount of data, for which the continuous improvement of bioinformatics tools, such as HiT-StARTS, Inforna [54,55], and Pattern Recognition of RNA by Small Molecules (PRRSM) [76], is crucial to provide chemotype centric hypotheses to efficiently and selectively target RNA motifs. These efforts will need to be followed by synthetic efforts towards optimization,generation of SAR studies, and, if possible, structural characterization of the RNA–small molecule interaction to better understand the underlying principles of the molecular recognition event as well as its impact on the stability and/or dynamics of the RNA–small molecule complex. In particular, various 1D- and 2D-NMR methods have been used to reveal atomistic details of the RNA–small molecule interaction [77–79].
These efforts must be accompanied by rigorous target engagement and profiling studies in cells to demonstrate the RNA-centric mechanism of action [80]. The chemical biology community has been actively addressing this issue by developing technologies, such as Chem-CLIP, Ribo-SNAP, and RIBOTAC, which are well suited for this purpose. Such chemical tools were recently used to map binding sites of small molecules to RNAs, thus correlating the data obtained in vitro and in vivo. Therefore, the interrogation of the biological effects of small molecules both transcriptome and proteome wide will provide a more in-depth understanding of their interaction with biological systems and clearly delineate a protein- and/or RNA-centric mode of action. When combined with RNA sequencing[81], these modalities could be applied to target identification campaigns following phenotypic screens with already existing drugs or denovo chemical matter, whereby no protein target was accurately assigned to the hit molecule(s). Although an exact mode of action is not necessary for clinical approval, the identification early on of possible transcriptome-wide RNA off-target effects can mitigate the risk of failure in more advanced stages of clinical trials.
Given that RNA structure is fundamentally different to protein structure, the principles of potently and selectively targeting disease causing RNAs might differ and new concepts along these lines are starting to emerge. For example, multipronged approaches to study RNA small molecule interactions will allow researchers to: (i) fully understand the molecular recognition event between the RNA motif and the small molecule; and (ii) identify the most relevant conformational changes that trigger a biological response. On the one hand, this will require a comprehensive understanding of the physicochemical properties of the RNA structural motif(s), such as electropositive and negative surfaces, impact of the closing base pairs, hydration, surrounding metal ions, or location of the motif within the RNA helix. More important is how these effects impact the conformational heterogeneity landscape (i.e., the interconversion and distribution between local and global minima states in the apo and ligand bound form). On the other hand, a good understanding of the physicochemical and conformational properties of the small molecule(s) is equally important. For example, compounds witha rod-like characterwere highlighted to exhibitaffinity and selectivity towards particular RNA targets [82]. In addition, conformational parameters that dictate small molecule preorganization and planarity were considered when optimizing the SMN2 splicing agent Risdiplam [83], indicating that SMIRNAs and/or pre-mRNA might occupy a privileged location of the chemical space that might or might not overlap with the chemical matter targeting the proteome. Moreover, all these efforts must be analyzed for each RNA structural motif–small molecule pair to better assess the druggability of RNA-binding site as well as the identification of the chemotypes that trigger biologically relevant conformational changes.
We are only at the beginning of truly understanding the structural prerequisities for developing potent and selective SMIRNAs, which might require a paradigm change of the drug-likeness concept [84,85]. Some of the prominent examples presented here, such as peptides, macrocycles, and dimeric molecules, showed efficient correction of the phenotype(s) triggered by the disease-causing RNA, but do not obey Lipinski’s rule of five [86,87]. Despite this, dimeric molecules exhibit improved binding and selectivity towards RNA repeat expansions compared to the monomeric units, therefore significantly lowering the dose needed for therapeutic gain. In a recent example, Angelbello et al. [88] showed that intraperitoneally delivered Cugamycin selectively exp cleaved r(CUG) in a DM1 preclinical mouse model. Therefore, future interrogation campaigns of the beyond rule of five chemical space as well as optimization efforts will reveal novel ways to address and overcome the physicochemical properties featured by these compound classes for therapeutic benefit [89–91].
Drugging RNA is not limited to the identification and characterization of SMIRNAs. The continuous need to fulfill unmet therapeutic needs revealed new chemical tools that comprise of RNA binders covalently linked with either natural products or recruiting units of cellular components of the RNA degradation machinery to manipulate RNA function and modulate biological processes. Recent development of PROTAC [92] and its fast-paced development towards in vivo studies [93] was followed within only a few years later by RIBOTAC [66] for targeted degradation of RNAs via RNase L recruitment, opening avenues to provide new and effective therapeutic modalities [94,95] arising from the beyond rule of five chemical space [96]. Together with Ribo-SNAP, these new modalities not only achieve a more effective therapeutic effect because the target RNA is ablated, but both can also be used to confirm target engagement, map binding sites, and perform profiling studies in vitro and in vivo in a transcriptome-wide fashion [88]. Given their modular assembly, the units responsible for RNA cleavage (Bleomycin A5 and the 2′ –5′-A4 part of Ribo-SNAP, and RIBOTAC respectively) can be attached to almost any small molecule, allowing the interrogation of target occupancy in a transcriptome-wide fashion. This will provide important insights into how target occupancy by various chemotypes targeting the same RNA motif correlates with quantitative functional pharmacological effects in vitro and in vivo. Ultimately, such studies will comprehensively assess the chemotype–RNA target–phenotype interplay, leading to an improved understanding of the pharmacokinetic and pharmaco-dynamic properties and, therefore, to a better projection of the dose to the clinic.
We are at the cusp of deciphering the principles of effective small-molecule modulation of RNA targets. This could improve the efficiency of existing treatments, extend therapeutic options for currently orphan diseases, and potentially improve the therapeutic portfolio of already existing disorders by targeting novel ncRNAs. Recent examples, including the sensitization of cell lines to already approved anticancer treatments [97], reprogramming oncogenic circuits [61], targeted degradation of ncRNAs [98], and the first examples of drugging lncRNAs [49,50,63] are setting the stage to a bright future towards more rational drug design and discovery campaigns against ncRNAs.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health for funding (RO1-GM097455–07, DP1-NS096898, and PO1-NS099114). A.U. acknowledges the financial support received from the Deutsche Forschungsgemeinschaft (DFG) and the ALS Association (ALSA).S.V-D. thanks the Fonds de Recherche du Québec-Nature et Technologie (FRQNT) for the Postdoctoral research scholarship. The authors are thankful to Jonathan L. Chen, Hafeez S. Haniff, Jessica L. Childs-Disney and the members of the Disney lab for the valuable input and helpful discussions that shaped the final version of the manuscript.
Footnotes
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.drudis.2019.06.019.
References
- 1.Howe JA et al. (2015) Selective small-molecule inhibition of an RNA structural element. Nature 526, 672. [DOI] [PubMed] [Google Scholar]
- 2.Naryshkin NA et al. (2014) SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science 345, 688–693 [DOI] [PubMed] [Google Scholar]
- 3.Palacino J et al. (2015) SMN2 splice modulators enhance U1–pre-mRNA association and rescue SMA mice. Nat. Chem. Biol 11, 511. [DOI] [PubMed] [Google Scholar]
- 4.Barnwal RP et al. (2017) Applications of NMR to structure determination of RNAs large and small. Arch. Biochem. Biophys 628, 42–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bevilacqua PC and Blose JM (2008) Structures, kinetics, thermodynamics, and biological functions of RNA hairpins. Annu. Rev. Phys. Chem 59, 79–103 [DOI] [PubMed] [Google Scholar]
- 6.Chavali SS et al. (2019) Face-time with TAR: Portraits of an HIV- 1 RNA with diverse modes of effector recognition relevant for drug discovery. J. Biol. Chem 294, 9326–9341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas JR and Hergenrother PJ (2008) Targeting RNA with small molecules. Chem. Rev 108, 1171–1224 [DOI] [PubMed] [Google Scholar]
- 8.The EPC et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozsolak F and Milos PM (2010) RNA sequencing: advances, challenges and opportunities. Nat. Rev.Genet 12, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrews RJ et al. (2017) RNAStructuromeDB: a genome-wide database for RNA structural inference. Sci. Rep 7, 17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubota M et al. (2015) Progress and challenges for chemical probing of RNA structure inside living cells. Nat. Chem. Biol 11, 933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathews DH et al. (2004) Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U. S. A 101, 7287–7292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun L et al. (2019) RNA structure maps across mammalian cellular compartments. Nat. Struct. Mol. Biol 26, 322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mustoe AM et al. (2018) Pervasive regulatory functions of mRNA structure revealed by high-resolution SHAPE probing. Cell 173, 181–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X and Bartel DP (2017) Widespread influence of 3′-end structures on mammalian mRNA processing and stability. Cell 169, 905–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langdon EM et al. (2018) mRNA structure determines specificity of a polyQ-driven phase separation. Science 360, 922–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Disney MD et al. (2018) Drugging the RNA world. Cold Spring Harb. Perspect. Biol 10 a034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donlic A and Hargrove AE (2018) Targeting RNA in mammalian systems with small molecules. Wiley Interdiscip. Rev. RNA 9, e1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizvi NF and Smith GF (2017) RNA as a small molecule druggable target. Bioorg. Med. Chem. Lett 27, 5083–5088 [DOI] [PubMed] [Google Scholar]
- 20.Shortridge MD et al. (2018) An ultra-high affinity ligand of HIV-1 TAR reveals the RNA structure recognized by P-TEFb. Nucleic Acids Res. 47, 1523–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adegbuyiro A et al. (2017) Proteins containing expanded polyglutamine tracts and neurodegenerative disease. Biochemistry 56, 1199–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q et al. (2016) Assessing a peptidylic inhibitor-based therapeutic approach that simultaneously suppresses polyglutamine RNA- and protein-mediated toxicities in patient cells and Drosophila. Dis. Mod. Mech 9, 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salmon L et al. (2014) Advances in the determination of nucleic acid conformational ensembles. Annu. Rev. Phys. Chem 65, 293–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganser LR et al. (2018) High-performance virtual screening by targeting a high-resolution RNA dynamic ensemble. Nat. Struct. Mol. Biol 25, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkesh R et al. (2012) Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif–ligand database and chemical similarity searching. J. Am. Chem. Soc 134, 4731–4742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angelbello AJ et al. (2016) Development of pharmacophore models for small molecules targeting RNA: application to the RNA repeat expansion in myotonic dystrophy type 1. Bioorg. Med. Chem. Lett 26, 5792–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irwin JJ et al. (2012) ZINC: a free tool to discover chemistry for biology. J. Chem. Inf. Model 52, 1757–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irwin JJ and Shoichet BK (2005) ZINC—a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model 45, 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patwardhan NN et al. (2019) Fluorescent peptide displacement as a general assay for screening small molecule libraries against RNA. Org. Biomol. Chem 17, 1778–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matarlo JS et al. (2019) The natural product butylcycloheptyl prodiginine binds pre-miR-21, inhibits dicer-mediated processing of Pre-miR–21, and blocks cellular proliferation. Cell Chem. Biol XX, YYY–ZZZ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fedorova O et al. (2018) Small molecules that target group II introns are potent antifungal agents. Nat. Chem. Biol 14, 1073–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambowitz AM and Zimmerly S (2011) Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb. Perspect. Biol 3 a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J et al. (2010) Fluorescent indicator displacement assay for ligand RNA interactions. J. Am. Chem. Soc 132, 3660–3661 [DOI] [PubMed] [Google Scholar]
- 34.Wicks SL and Hargrove AE (2019) Fluorescent indicator displacement assays to identify and characterize small molecule interactions with RNA. Methods XX, YYY–ZZZ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asare-Okai PN and Chow CS (2011) A modified fluorescent intercalator displacement assay for RNA ligand discovery. Anal. Biochem 408, 269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran T and Disney MD (2012) Identifying the preferred RNA motifs and chemotypes that interact by probing millions of combinations. Nat. Commun 3, 1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mori K et al. (2013) The c9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338 [DOI] [PubMed] [Google Scholar]
- 38.DeJesus-Hernandez M et al. (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su Z et al. (2014) Discovery of a biomarker and lead small molecules to target r (GGGGCC)-associated defects in c9FTD/ALS. Neuron 83, 1043–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor JP (2014) G-quadruplex poses quadruple threat. Nature 507, 175. [DOI] [PubMed] [Google Scholar]
- 41.Haeusler AR et al. (2014) C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z-F et al. (2019) The hairpin form of r(G4C2) in c9ALS/FTD is repeat-associated non-ATG translated and a target for bioactive small molecules. Cell Chem. Biol 26, 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simone R et al. (2018) G-quadruplex-binding small molecules ameliorate c9orf72 FTD/ALS pathology in vitro and in vivo. EMBO Mol. Med 10, 22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gutschner T et al. (2013) The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 73, 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arun G et al. (2016) Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 30, 34–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown JA et al. (2014) Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol 21, 633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amodio N et al. (2018) MALAT1: a druggable long non-coding RNA for targeted anti–cancer approaches. J. Hematol. Oncol 11, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang W-Y et al. (2016) Design of a bioactive small molecule that targets r(AUUCU) repeats in spinocerebellar ataxia 10. Nat. Commun 7, 11647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donlic A et al. (2018) Discovery of small molecule ligands for MALAT1 by tuning an RNA-binding scaffold. Angew. Chem. Int. Ed 57, 13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abulwerdi FA et al. (2019) Selective small-molecule targeting of a triple helix encoded by the long noncoding RNA, MALAT1. ACS Chem. Biol 14, 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seike M et al. (2009) MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc. Natl. Acad. Sci. U. S. A 106, 12085–12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.du Rieu MC et al. (2010) MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clinical Chem. 56, 603–612 [DOI] [PubMed] [Google Scholar]
- 53.Velagapudi SP et al. (2018) Approved anti-cancer drugs target oncogenic non-coding RNAs. Cell Chem. Biol 25, 1086–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Velagapudi SP et al. (2014) Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat. Chem. Biol 10, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Disney MD et al. (2016) Inforna 2.0: a platform for the sequence-based design of small molecules targeting structured RNAs. ACS Chem. Biol 11, 1720–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foekens JA et al. (2008) Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc. Natl. Acad. Sci.U. S. A 105, 13021–13026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grosso S et al. (2013) MiR–210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis. 4, e544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greither T et al. (2010) Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int. J. Cancer 126, 73–80 [DOI] [PubMed] [Google Scholar]
- 59.Maxwell PH et al. (1999) The tumour suppressor protein VHL targets hypoxiainducible factors for oxygen-dependent proteolysis. Nature 399, 271. [DOI] [PubMed] [Google Scholar]
- 60.Kelly TJ et al. (2011) A hypoxia-induced positive feedback loop promotes hypoxiainducible factor 1a stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol. Cell. Biol 31, 2696–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Costales MG et al. (2017) Small molecule inhibition of microRNA-210 reprograms an oncogenic hypoxic circuit. J. Am. Chem. Soc 139, 3446–3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carter G et al. (2015) Circulating long noncoding RNA GAS5 levels are correlated to prevalence of type 2 diabetes mellitus. BBA Clin. 4, 102–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi Y et al. (2019) Stabilization of lncRNA GAS5 by a small molecule and its implications in diabetic adipocytes. Cell Chem. Biol 26, 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rzuczek SG et al. (2016) Precise small-molecule recognition of a toxic CUG RNA repeat expansion. Nat. Chem. Biol 13, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guttilla IK and White BA (2009) Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J. Biol. Chem 284, 23204–23216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Costales MG et al. (2018) Small molecule targeted recruitment of a nuclease to RNA. J. Am. Chem. Soc 140, 6741–6744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Connelly CM et al. (2016) The emerging role of RNA as a therapeutic target for small molecules. Cell Chem. Biol 23, 1077–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.poner J et al. (2018) RNA structural dynamics as captured by molecular simulations: a comprehensive overview. Chem. Rev 118, 4177–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garavís M et al. (2014) Discovery of selective ligands for telomeric RNA G-quadruplexes (TERRA) through 19F-NMR based fragment screening. ACS Chem. Biol 9, 1559–1566 [DOI] [PubMed] [Google Scholar]
- 70.Garner AL (2018) cat-ELCCA: catalysing drug discovery through click chemistry. Chem. Commun 54, 6531–6539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lorenz DA et al. (2018) Development and implementation of an HTS-compatible assay for the discovery of selective small-molecule ligands for pre-microRNAs. SLAS DISCOVERY: Adv. Life Sci. R&D 23, 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rizvi NF et al. (2018) Discovery of selective RNA-binding small molecules by affinity-selection mass spectrometry. ACS Chem. Biol 13, 820–831 [DOI] [PubMed] [Google Scholar]
- 73.Flusberg DA et al. (2019) Identification of G-quadruplex-binding inhibitors of Myc expression through affinity selection–mass spectrometry. SLAS DISCOVERY: Adv. Life Sci. R&D 24, 142–157 [DOI] [PubMed] [Google Scholar]
- 74.Connelly CM et al. (2017) Discovery of RNA binding small molecules using small molecule microarrays In Small Molecule Microarrays: Methods and Protocols Uttamchandani M and Yao SQ, eds), pp. 157–175, Springer; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tran T and Disney MD (2010) Two-dimensional combinatorial screening of a bacterial rRNA A-site-like motif library: defining privileged asymmetric internal loops that bind aminoglycosides. Biochemistry 49, 1833–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eubanks CS et al. (2019) Visualizing RNA conformational changes via pattern recognition of RNA by small molecules. J. Am. Chem. Soc 141, 5692–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thompson RD et al. (2019) NMR characterization of RNA small molecule interactions. Methods XX, YYY–ZZZ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rinnenthal J et al. (2011) Mapping the landscape of RNA dynamics with NMR spectroscopy. Acc. Chem. Res 44, 1292–1301 [DOI] [PubMed] [Google Scholar]
- 79.Mustoe AM et al. (2014) Hierarchy of RNA functional dynamics. Annu. Rev. Biochem 83, 441–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Disney MD (2019) Perspectives on targeting RNA with small molecules to capture opportunities at the intersection of chemistry, biology, and medicine. J. Am. Chem. Soc 141, 6776–6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ye C et al. (2018) DRUG-seq for miniaturized high-throughput transcriptome profiling in drug discovery. Nat. Commun 9, 4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morgan BS et al. (2017) Discovery of key physicochemical, structural, and spatial properties of RNA-targeted bioactive ligands. Angew. Chem. Int. Ed 56, 13498–13502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ratni H et al. (2018) Discovery of risdiplam, a selective survival of motor neuron-2 (SMN2) gene splicing modifier for the treatment of spinal muscular atrophy (SMA).J. Med. Chem 61, 6501–6517 [DOI] [PubMed] [Google Scholar]
- 84.Mignani S et al. (2018) Present drug-likeness filters in medicinal chemistry during the hit and lead optimization process: how far can they be simplified? Drug Discov. Today 23, 605–615 [DOI] [PubMed] [Google Scholar]
- 85.Shultz MD (2019) Two decades under the influence of the rule of five and the changing properties of approved oral drugs. J. Med. Chem 62, 1701–1714 [DOI] [PubMed] [Google Scholar]
- 86.Lipinski CA (2004) Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today: Technol 1, 337–341 [DOI] [PubMed] [Google Scholar]
- 87.Zhang M-Q and Wilkinson B (2007) Drug discovery beyond the ‘rule-of-five’. Curr. Opin. Biotechnol 18, 478–488 [DOI] [PubMed] [Google Scholar]
- 88.Angelbello AJ et al. (2019) Precise small-molecule cleavage of an r(CUG) repeat expansion in a myotonic dystrophy mouse model. Proc. Natl. Acad. Sci. U. S. A 116, 7799–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Doak Bradley C. et al. (2014) Oral druggable space beyond the rule of 5: insights from drugs and clinical candidates. Chem. Biol 21, 1115–1142 [DOI] [PubMed] [Google Scholar]
- 90.DeGoey DA et al. (2018) Beyond the rule of 5: lessons learned from AbbVie’s drugs and compound collection. J. Med. Chem 61, 2636–2651 [DOI] [PubMed] [Google Scholar]
- 91.Poongavanam V et al. (2018) Opportunities and guidelines for discovery of orally absorbed drugs in beyond rule of 5 space. Curr. Opin. Chem. Biol 44, 23–29 [DOI] [PubMed] [Google Scholar]
- 92.Cromm PM and Crews CM (2017) Targeted protein degradation: from chemical biology to drug discovery. Cell Chem. Biol 24, 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun X et al. (2019) A chemical approach for global protein knockdown from mice to non–human primates. Cell Discov. 5, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Valeur E et al. (2017) New modalities for challenging targets in drug discovery. Angew. Chem. Int. Ed 56, 10294–10323 [DOI] [PubMed] [Google Scholar]
- 95.Valeur E and Jimonet P (2018) New modalities, technologies, and partnerships in probe and lead generation: enabling a mode-of-action centric paradigm. J. Med. Chem 61, 9004–9029 [DOI] [PubMed] [Google Scholar]
- 96.Edmondson SD et al. (2019) Proteolysis targeting chimeras (PROTACs) in ‘beyond rule-of-five’ chemical space: recent progress and future challenges. Bioorg. Med. Chem. Lett 29, 1555–1564 [DOI] [PubMed] [Google Scholar]
- 97.Costales MG et al. (2019) A designed small molecule inhibitor of a non-coding RNA sensitizes HER2 negative cancers to Herceptin. J. Am. Chem. Soc 141, 2960–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Costales MG et al. (2019) Targeted degradation of a hypoxia-associated non-coding RNA enhances the selectivity of a small molecule interacting with RNA. Cell Chem. Biol XX, YYY–ZZZ [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.