Significance
Reactivating sleep-like quiescent cells in the body (most notably stem cells) to divide is fundamental to tissue homeostasis and regeneration. Like sleep having shallow and deep stages, quiescence exhibits graded depths with different speeds and rates responding to stimulation signals. Understanding how quiescence depth is regulated is fundamental to our understanding of tissue repair and regeneration. Here we show that quiescence depth is regulated like a “dimmer switch” by lysosomes (cell organelles that break down and recycle biomolecules), through the lysosomal function of reducing oxidative stress. We developed a gene signature to predict quiescence depth and show that quiescence deepening likely represents a transition path from cell division to irreversibly arrested senescence, related to the aging process.
Keywords: quiescence, dormancy, senescence, aging, lysosome
Abstract
The reactivation of quiescent cells to proliferate is fundamental to tissue repair and homeostasis in the body. Often referred to as the G0 state, quiescence is, however, not a uniform state but with graded depth. Shallow quiescent cells exhibit a higher tendency to revert to proliferation than deep quiescent cells, while deep quiescent cells are still fully reversible under physiological conditions, distinct from senescent cells. Cellular mechanisms underlying the control of quiescence depth and the connection between quiescence and senescence are poorly characterized, representing a missing link in our understanding of tissue homeostasis and regeneration. Here we measured transcriptome changes as rat embryonic fibroblasts moved from shallow to deep quiescence over time in the absence of growth signals. We found that lysosomal gene expression was significantly up-regulated in deep quiescence, and partially compensated for gradually reduced autophagy flux. Reducing lysosomal function drove cells progressively deeper into quiescence and eventually into a senescence-like irreversibly arrested state; increasing lysosomal function, by lowering oxidative stress, progressively pushed cells into shallower quiescence. That is, lysosomal function modulates graded quiescence depth between proliferation and senescence as a dimmer switch. Finally, we found that a gene-expression signature developed by comparing deep and shallow quiescence in fibroblasts can correctly classify a wide array of senescent and aging cell types in vitro and in vivo, suggesting that while quiescence is generally considered to protect cells from irreversible arrest of senescence, quiescence deepening likely represents a common transition path from cell proliferation to senescence, related to aging.
Cell proliferation in multicellular organisms is tightly regulated, and the vast majority of cells in the body stay dormant and out of the cell cycle at any given moment. The dormant state, when reversible upon growth signals, is referred to as cellular quiescence. Quiescence protects cells against stress and irreversible arrest, such as senescence; it is fundamental to many physiological phenomena, such as stem cell homeostasis and tissue repair (1–4). Consequently, dysregulation of cellular quiescence can lead to a range of hyper- and hypoproliferative diseases, including cancer and aging (5–7).
It becomes increasingly recognized that quiescence is a heterogeneous state with graded depth. For example, following injury, muscle and neural stem cells at noninjury sites in the body move adaptively into shallower quiescence (alert or primed quiescent phase), positioning cells to more quickly respond to injury and reenter the cell cycle when needed (8, 9). In other cases, cells move into deep quiescence and require stronger growth stimulation and longer times to reenter the cell cycle, as seen in hepatocytes upon partial hepatectomy in older rats than in younger ones (10, 11). Similarly, the quiescent state deepens in cells that are cultured longer under quiescence-inducing signals, such as contact inhibition (12, 13) and serum starvation (14). Deep quiescent cells can still revert to proliferation under physiological conditions, appearing distinct from irreversibly arrested senescent cells (SI Appendix, Fig. S1A). A graded quiescence depth indicates graded potentials for tissue repair and regeneration; yet, how quiescence depth is regulated in the cell and the connection between deep quiescence and senescence are not well understood.
In this study, we investigate what regulates quiescence depth in rat embryonic fibroblast (REF) cells. We identified sequential transcriptome changes as cells move progressively deeper into quiescence under longer-term serum starvation. In particular, we found that lysosomal gene expression and lysosomal mass (indicated by the intensity of lysosomal staining and the number of lysosomes per cell) continuously increase as quiescence deepens; however, autophagy flux decreases. Lysosomes are hydrolytic enzyme-filled organelles in the cell that break down many types of biomolecules; lysosomal function, primarily through processes of autophagy and endocytosis, has been shown to prevent irreversible cellular states, such as senescence, terminal differentiation, and apoptosis (15–18). Here we found that the increased lysosomal mass in deep quiescent cells is, in part, due to decreased autophagy flux, which is partially but not fully compensated by increased lysosomal gene expression and lysosomal biogenesis. We show that lysosomal function, like a dimmer switch, continuously regulates quiescence depth and thus the proliferative potential of quiescent cells by reducing the accumulation of reactive oxygen species (ROS). We found that a set of “senescence core signature” genes (19) show similar expression patterns in deep quiescence as in senescence, and that a gene-expression signature we developed to indicate cellular quiescence depth by comparing deep and shallow quiescent REF cells is able to correctly classify senescent and aging cells in a wide array of cell lines in vitro (20, 21) and tissues in vivo (22, 23), suggesting that deep quiescence may serve as a common transition path from cell proliferation to senescence, related to aging.
Results
Dynamic Transcriptome Changes during Quiescence Deepening.
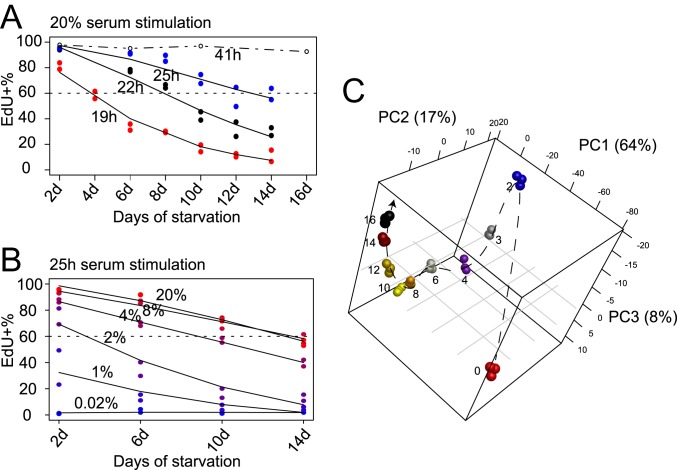
Similar to our previous observation (14), REF cells moved into deeper quiescence progressively with longer-term serum starvation. After 2-d serum starvation, the entire cell population entered quiescence, as demonstrated by their negative DNA incorporation of 5-ethynyl-2′-deoxyuridine (EdU) and a complete shut-off of E2f1 expression (SI Appendix, Fig. S1B). E2f1 is a member of the E2f family of transcription factors; it up-regulates a large battery of genes involved in DNA replication and cell cycle progression, acting as the key effector of an Rb-E2f bistable switch that controls the all-or-none transition from quiescence to proliferation (24, 25). With increasingly longer serum starvation, cells moved into deeper quiescence, requiring a longer time and a greater serum stimulation strength (concentration) to reenter the cell cycle than cells in shallower quiescence. For example, when the duration of serum starvation increased gradually from 2 to 14 d, the cell cycle reentry time upon 20% serum stimulation increased from <19 h to ∼25 h (to achieve ∼60% EdU+ rate) (Fig. 1A and SI Appendix, Fig. S1C), and the serum concentration required for cell cycle reentry increased from <2% to ∼8% (to achieve ∼60% EdU+ measured at the 25th hour of serum stimulation) (Fig. 1B). This result is consistent with our earlier findings that the time and serum threshold required for cell cycle reentry are 2 related features that both reflect the same biological state, the quiescence depth (14). Importantly, deep quiescent cells were not irreversibly arrested. For example, upon 20% serum stimulation, 92.6% of 16-d serum-starved deep quiescent REF cells were able to reenter the cell cycle (EdU+) within 41 h (SI Appendix, Fig. S1B).
Fig. 1.
Sequential quiescence deepening and transcriptome change in serum-starved REF cells over time. (A and B) EdU+% of 2- to 14-d serum-starved cells upon stimulation. Cells in duplicated wells (n = 2) were harvested 19, 22, and 25 h after 20% serum stimulation (A), or 25 h after stimulation with serum at indicated concentrations (0.02 to 20%) (B). The EdU+% at 41 h after 20% serum stimulation (from SI Appendix, Fig. S1B) is shown in A for comparison. Lines were fitted using the smooth.spline function in R. (C) Principal component (PC) analysis of proliferating and 2- to 16-d serum-starved cells based on 2,736 differentially expressed genes in RNA-seq time course. Days of serum starvation are indicated next to the sample (triplicates per condition). Dashed arrow is plotted for clarity. Percentage of the variation that each principal component accounts for is shown on the axis.
To identify molecular mechanisms regulating quiescence depth at the transcriptional level, we performed RNA-sequencing (RNA-seq) analysis of 0- to 16-d serum-starved cells (26). As expected, the expression of well-characterized proliferation genes—such as E2f1, Cdk2, and Cdk4 (SI Appendix, Fig. S2A)—E2f1 target genes (SI Appendix, Fig. S2B), and proliferation-related gene clusters (#7 and 9, SI Appendix, Fig. S2C) was quickly down-regulated within 2-d serum starvation. Conversely, the expression of growth inhibitory genes, such as Rb1, Cdkn1a (p21Cip1), and Cdkn1b (p27Kip1), was up-regulated upon serum starvation (SI Appendix, Fig. S2A). The expression of Cdkn2a (p16INK4A), a senescence marker (27), remained low as cells moved deeper in quiescence (3- to 16-d serum starvation) (SI Appendix, Fig. S2D), consistent with quiescence being a reversible state.
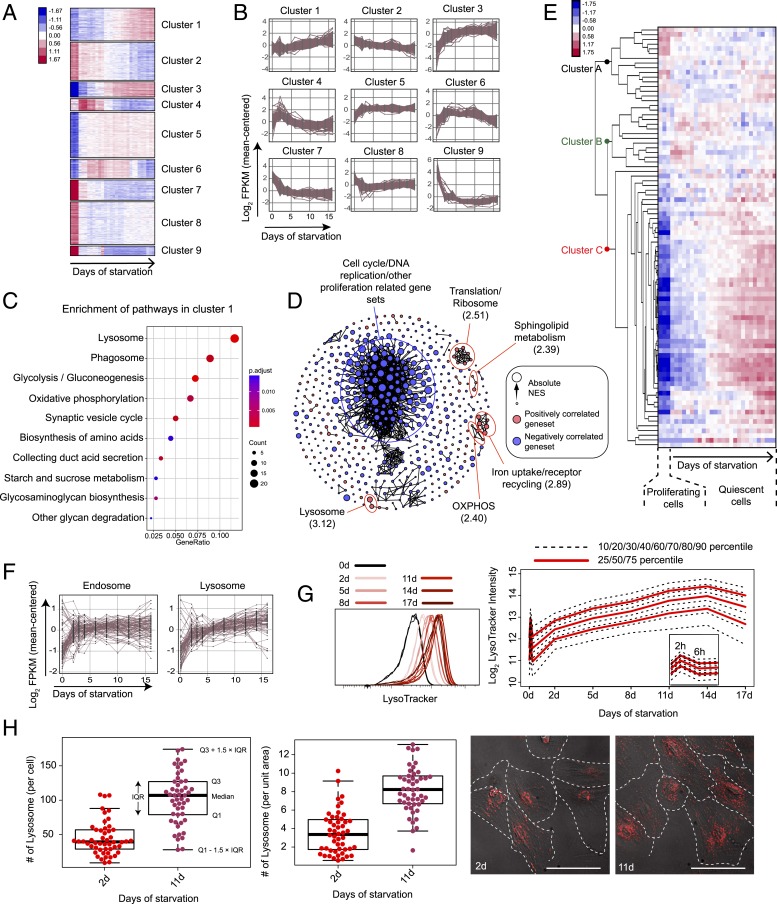
The global gene-expression profile changed drastically when cells transitioned from proliferation to quiescence (0- to 2-d serum starvation) and continued to change sequentially as cells moved from shallow to deep quiescence (2- to 16-d serum starvation) (Fig. 1C). This sequential change was reflected in 9 gene clusters that exhibited different temporal dynamic patterns (Fig. 2 A and B). In particular, the expression of 2 gene clusters appeared to be positively correlated with quiescence deepening and increased monotonically as cells moved from shallow to deep quiescence (clusters 1 and 3) (Fig. 2B). In cluster 1, multiple biological functions were enriched; among them, lysosome was overall the most statistically significant in Kyoto Encyclopedia of Genes and Genomes (KEGG) overrepresentation tests [Fig. 2C, using clusterProfiler (28); SI Appendix, Fig. S2C, using DAVID (29)]. In cluster 3, lysosome was the single function significantly enriched statistically (SI Appendix, Fig. S2C). Consistently, lysosomal genes also had the strongest positive correlation with deep quiescence in gene set enrichment analysis (GSEA) (30) (normalized enrichment score [NES] = 3.12) (Fig. 2D).
Fig. 2.
Lysosomal gene expression and lysosomal mass increase along quiescence deepening. (A) K-means clustering of 2,736 differentially expressed genes in RNA-seq analysis. Sample columns (0-, 2-, 3-, 4-, 6-, 8-, 10-, 12-, 14-, and 16-d serum starvation) are ordered chronologically with 3 replicates at each time point. Up- and down-regulation of genes are shown in red and blue, respectively; color gradient bar indicates the degree of change in gene expression (normalized and log-transformed; see Materials and Methods for details). (B) Gene-expression dynamics within each K-means cluster in A. Expression of each gene in a given cluster is shown in a time-course curve. (C) Pathways enriched (P < 0.05) in cluster 1. GeneRatio (x axis) and dot size indicate the fraction and number of genes involved in the indicated pathway in cluster 1, respectively. Color gradient bar indicates statistical significance of pathway enrichment in KEGG overrepresentation test based on P values adjusted by Benjamini-Hochberg correction. (D) Gene sets significantly correlated to quiescence depth (false-discovery rate < 0.1) in GSEA analysis. Gene sets are connected by edges based on their similarity (Jaccard index ≥ 0.5) when applicable. Gene sets positively and negatively correlated to quiescence deepening are shown in red and blue nodes, respectively. Node size represents the absolute NES in the GSEA result. Biological functions that are the most negatively and positively (top 5 in statistical significance) correlated with quiescence deepening are demarcated by blue and red circles, respectively; the maximum NES value of all enclosed nodes in a circle is shown in parenthesis. (E) Heat map of time-course expression of lysosomal genes in RNA-seq analysis (0- to 16-d serum starvation). Lysosomal genes (rows) are defined by the Mouse Genome Database (MGD) (83) and ordered by hierarchical clustering (with gene names shown in SI Appendix, Table S1). RNA-seq samples (columns) are ordered chronologically (as in A). Color gradient bar indicates the degree of change in gene expression as in A. (F) Time-course expression of differentially expressed endosomal and lysosomal genes in RNA-seq analysis (0- to 16-d serum starvation). Endosomal and lysosomal genes are defined by the MGD (83). (G) LysoTracker intensity of serum-starved REF cells. LysoTracker intensity distribution of each cell population (0- to 17-d serum starvation, triplicates of ∼10,000 cells each) is shown (Left); the log2-transformed agglomeration of the triplicates at each time point is plotted with the indicated percentiles (Right). (H) Number of lysosomes (LysoTracker foci) per cell and per unit area of cell in 2- and 11-d serum-starved cells (n = 50 each). Box plot: Q1 and Q3 refer to the first and third quantiles, respectively; IQR, interquartile range = Q3 − Q1; the same below unless otherwise noted. (Inset) Representative lysoTracker foci microscopy. Each red dot represents a LysoTracker-stained lysosome in the cell. (Scale bar, 50 µm.) Cell boundaries were manually determined using both POL and Cy5 filters.
Lysosomal Gene Expression and Lysosomal Mass Increase as Quiescence Deepens.
The vast majority of lysosomal genes, encoding for various lysosomal enzymes, activator proteins, membrane proteins, and ion channel proteins, increased their expression continuously as cells moved from shallow to deep quiescence (Fig. 2E [cluster C and the bottom half of cluster A], Fig. 2F, and SI Appendix, Fig. S3A). As a comparison, the expression of most genes associated with endosomes, another cellular organelle in the endosomal–lysosomal system, only increased significantly during the first 2 d of serum starvation but not afterward as quiescence continued to deepen (Fig. 2F and SI Appendix, Fig. S3 B and C).
Lysosomal mass also increased in deep quiescence: Following a brief pulsatile adaptive response (within the first 6 h) upon serum starvation, lysosomal mass continuously increased over the next 14 d, as seen from the stained LysoTracker intensity (Fig. 2G). The initial pulsatile response of lysosomal mass was likely due to an adaptive mammalian target of rapamycin (mTOR)-autophagy response to serum starvation, as previously reported (31). The continuously increased LysoTracker intensity in deep quiescence was consistent with an increased number of lysosomes, another indicator of lysosomal mass; for example, 11-d serum-starved cells exhibited significantly more lysosomal foci per cell and per unit area of cell than 2-d serum-starved cells (per cell, > 2.5-fold, P = 4.9 × e−14 in a 1-tailed t test; per unit area of cell, > 2.3-fold, P = 2.9 × e−15 in a 1-tailed t test) (Fig. 2H).
Lysosomal Function and Lysosome Destruction Decrease as Quiescence Deepens.
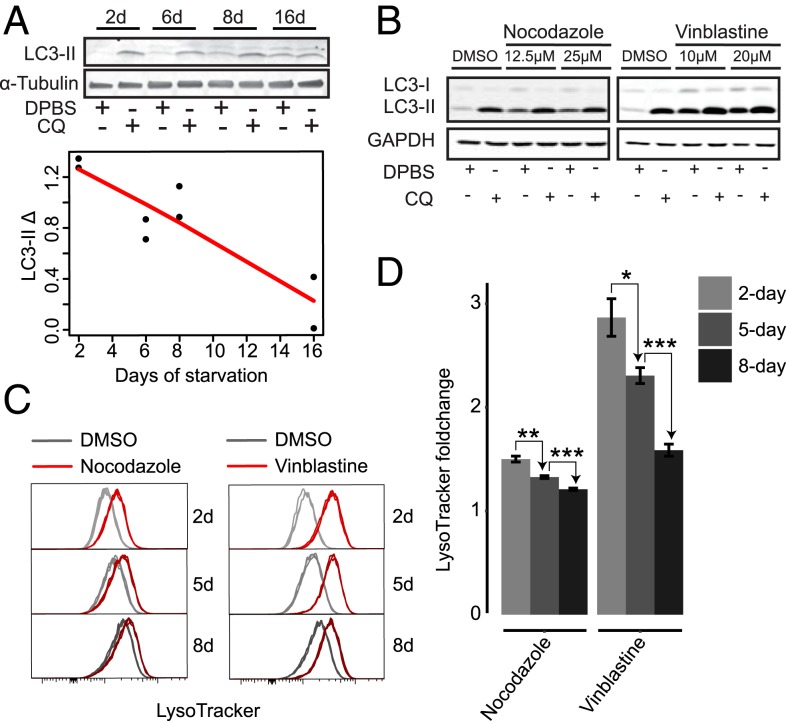
We thought the increases of both lysosomal gene expression and lysosomal mass as quiescence deepens would lead to an increase of lysosomal function. Instead, we found that autophagy flux, an indicator for the primary lysosomal function, did not increase but gradually declined as cells moved deeper into quiescence, as seen in an LC3-II turnover assay (32) (Fig. 3A). This result posed 2 immediate questions: 1) What is the relationship between increased lysosomal mass and gene expression to decreased lysosomal function? 2) Does declined lysosomal function cause quiescence deepening and, if so, how?
Fig. 3.
Lysosomal–autophagic function and lysosome destruction decline as quiescence deepens. (A) LC3-II turnover assay in 2-, 6-, 8-, and 16-d serum-starved cells. Quantified LC3-II Δ (the difference between LC3-II signal intensity normalized against α-Tubulin control in CQ-treated and nontreated cells) in duplicate samples is shown with a linear fit (red line). (Inset) A representative immunoblot image. (B) LC3-II turnover assay in 2-d serum-starved cells treated with nocodazole, vinblastine, or vehicle control for 24 h at indicated doses. (C) LysoTracker intensity of 2-, 5-, and 8-d serum-starved cells treated with 25 μM nocodazole, 20 μM vinblastine, or vehicle control for 24 h (triplicates). (D) Relative LysoTracker intensity of treated samples over vehicle control in C at the indicated serum starvation time. Error bar, SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 (1-tailed t test).
An increase of lysosomal mass can be caused by an increase of lysosome biogenesis or a decrease of lysosome destruction or both. Since lysosome itself is destructed via an autophagic process named lysophagy (33, 34), the declined autophagy flux we observed in cells with prolonged serum starvation (Fig. 3A) suggests that the rate of lysosome destruction likely declines accordingly in deep quiescence. If this is the case, when we inhibit autophagy/losophagy, we expect that 1) lysosomal mass will increase in cells, and 2) the degree of increase (fold-change) will be smaller in deep than shallow quiescent cells. Indeed, when we treated 2-, 5-, and 8-d serum-starved quiescent cells with an autophagy inhibitor (nocodazole or vinblastine) (35) to repress autophagy/lysophagy (as seen by decreased autophagy flux in LC3 turnover assay) (Fig. 3B), we found that the relative increase of LysoTracker intensity was smaller in cells under prolonged starvation (Fig. 3 C and D). Although the result should be taken with caveat since nocodazole and vinblastine are general autophagy inhibitors but not specific lysophagy inhibitors (which are lacking in the field to the best of our knowledge), and thus may cause other side effects, this result, together with decreased autophagy (Fig. 3A), suggested that lysosome destruction decreased as quiescence deepened. Insufficient lysosome destruction by autophagy/lysophagy can lead to an accumulation of damaged lysosomes in the cell, which in turn interferes with lysosomal homeostasis and function (34).
Interestingly, while blocking autophagy/lysophagy by either nocodazole or vinblastine increased the LysoTracker intensity in quiescent REF cells, it did not increase the number of lysosomes (SI Appendix, Fig. S4 A and C) and even decreased the lysosomal number over time (SI Appendix, Fig. S4 B and D). This result is different from the increased lysosomal number we observed, along with quiescence deepening (Fig. 2H); it suggests that decreased lysosome destruction was not the only factor contributing to the increase of lysosomal mass in deep quiescent cells. Indeed, increased lysosome biogenesis (consistent with the increased lysosomal gene expression, as seen in Fig. 2 E and F) also played a role in this process, as we will explain further in the Discussion. Altogether, our study suggests that increased lysosomal mass (containing damaged lysosomes) is both partially caused by and contributes to decreased lysosomal function and autophagic clearance in deep quiescence.
Inhibiting Lysosomal Function Deepens Cellular Quiescence.
Our next question concerns whether the decreased lysosomal function is causal to, or merely a consequence of, deep quiescence. If decreased lysosomal function causes deep quiescence, we expect that inhibiting lysosomal function in quiescent cells would further deepen quiescence and that increasing lysosomal function would lead to shallow quiescence.
To test this idea, we first performed pharmacological inhibition of lysosomal function and measured corresponding changes in quiescence depth (Fig. 4A). As seen in Fig. 1 A and B, deep quiescent cells under longer-term serum starvation both require a higher serum concentration and take a longer time to reenter the cell cycle. This result is consistent with our earlier study with targeted perturbations (although the relative degree of required increases in serum concentration and time may vary depending on the specific quiescence-deepening factors) (14). Here we used at least 1 consistent indicator, the minimum serum concentration required for cell cycle reentry, to measure quiescence depth throughout this study.
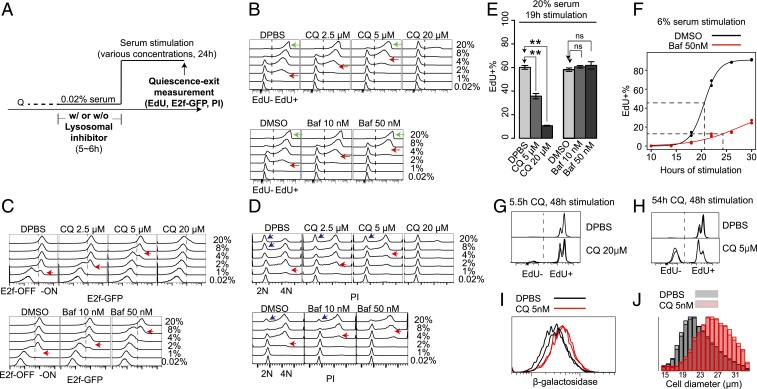
Fig. 4.
Lysosomal inhibition pushes cells into deeper quiescence and senescent-like state. (A) Experimental scheme of the quiescence-depth assays in B–D with lysosomal inhibitor treatment. Q, quiescent state. (B–D) Readouts of EdU (B), E2f-GFP (C), and PI staining (D) in quiescence-depth assay of 2-d serum-starved cells (∼10,000 cells per sample, with the highest frequency set to 100% at the y axis of each histogram). Cells at EdU− and EdU+ (B), E2f-ON and -OFF states (C), and G0/G1 (2N), S (2-4N), and G2/M phase (4N) (D) are as indicated. Red arrows in B–D, serum concentrations that activated E2f1 or initiated DNA replication in approximately half of the cell population. Green arrows in B, cell cycle reentry (EdU+) of the indicated whole-cell populations. Blue arrows in D, cell populations that contained a subset of recently divided cells following strong serum stimulation. Quantifications of cell death and irreversible arrest percentages at treatment conditions in B–D are shown in SI Appendix, Table S3. (E and F) Cell cycle reentry (EdU+) of 2-d serum-starved cells treated with vehicle, CQ, or Baf for 6 h (as in A), followed by serum stimulation at 20% (E) or 6% (F). Cells were harvested at the indicated time points (19 h, E; 10 to 30 h, F). Error bar in E, SEM (n = 2); **P < 0.01, and ns, insignificance, P > 0.05 (1-tailed t test). Dotted lines in F indicate the estimated median cell cycle reentry time (50 nM Baf-treated = 24.3 h; DMSO control = 20.6 h) of the quiescence-exited cells by the 30th hour (n = 2 at each time point). (G and H) EdU profiles of 2-d serum-starved cells treated with 20 μM CQ for 5.5 h (G) or 5 μM CQ for 54 h (H), followed by 20% serum stimulation for 48 h (n = 2). (I and J) Cells were treated as in H (5 μM CQ or vehicle control for 54 h, n = 2), then either measured for β-galactosidase activity (I), or stimulated with 20% serum for 48 h followed by cell size measurement (J).
We applied 2 lysosomal inhibitors, bafilomycin A1 (Baf) and chloroquine (CQ), that prevent lysosomal acidification (36). A short exposure (6 h) to either drug was sufficient to inhibit lysosomal function in REF cells, evident by the impaired proteolytic degradation within the lysosomal compartment (SI Appendix, Fig. S5A). When quiescent cells were treated with either of these 2 lysosomal inhibitors, increasingly higher serum concentrations were required to activate cells to reenter the cell cycle, in a drug dose-dependent manner (Fig. 4 B–D; red arrow points to serum concentration for activating ∼50% of cells, judged based on EdU incorporation, E2f-GFP reporter, and propidium iodide [PI] staining, respectively). Correspondingly, at a given serum concentration (e.g., 4% serum), a decreasing percentage of cells were able to reenter the cell cycle (EdU+) with an increasing dose of either lysosomal inhibitor, Baf or CQ (Fig. 4B). The lysosomal inhibition-caused quiescence deepening occurred regardless of the preceding quiescence depth before drug treatment: As shown in SI Appendix, Fig. S5B, a higher serum concentration was required to activate drug-treated (blue curve) than nontreated (red curve) cells at all test conditions (serum-starvation days).
Consistently, cells took longer time to reenter the cell cycle under the treatments of lysosomal inhibitors Baf and CQ. For example, with either the treatment of 5 µM CQ or control (Dulbecco’s phosphate-buffered saline, DPBS), although nearly all cells were able to reenter the cell cycle (EdU+) by the 24th hour of serum stimulation (20%) (green arrow in Fig. 4 B, Upper), the EdU+ percentage of cells was significantly smaller with 5 µM CQ than with DPBS at the 19th hour (Fig. 4E), indicating a delayed cell cycle entry with CQ treatment. The entry–delay effect of Baf at the tested dose range was weaker than that of CQ: Under strong serum stimulation (20%), the EdU+ percentage of cells with either the treatment of 50 nM Baf or DMSO control did not show a significant difference at either the 19th hour (Fig. 4E) or 24th hour (green arrow in Fig. 4 B, Lower); however, under reduced serum stimulation strength (6%) and longer observation time window, the effect of Baf becomes clear in causing a delayed half-activation time of the quiescence-exit cell population compared to the DMSO control (Fig. 4F).
As seen in Fig. 4B, a short but strong inhibition of lysosomal function by CQ treatment (6 h, 20 μM) deepened quiescence; the vast majority of cells were still EdU− at the 24th hour after 20% serum stimulation. However, these cells were reversible and became EdU+ by the 48th hour (Fig. 4G). In comparison, a prolonged lysosomal inhibition by CQ treatment at a lower dose (54 h, 5 μM) drove a significant subpopulation of quiescent cells into a more irreversible state: As seen in Fig. 4H, 41.4 ± 2.9% such treated cells remained EdU− after 48 h of 20% serum stimulation; in comparison, <2.5% of nontreated control cells remained EdU− (Fig. 4H). This result in quiescent cells may be related to earlier findings that a repeated or long exposure to sublethal stresses can effectively induce proliferating cells to senescence (37, 38). In addition to the irreversible phenotype, we found that long-term CQ exposed quiescent cells exhibited other hallmarks of senescence: A significantly increased β-galactosidase activity (P = 0.011 in 1-tailed t test) (Fig. 4I) and hypertrophy with a significantly increased cell size upon serum stimulation (P = 0.003 in 1-tailed t test) (Fig. 4J). Taken together, our data suggest that an impairment of lysosomal function in quiescent cells can lead to deep quiescence and potentially a senescence-like state.
Enhancing Lysosomal Function Pushes Cells toward Shallower Quiescence.
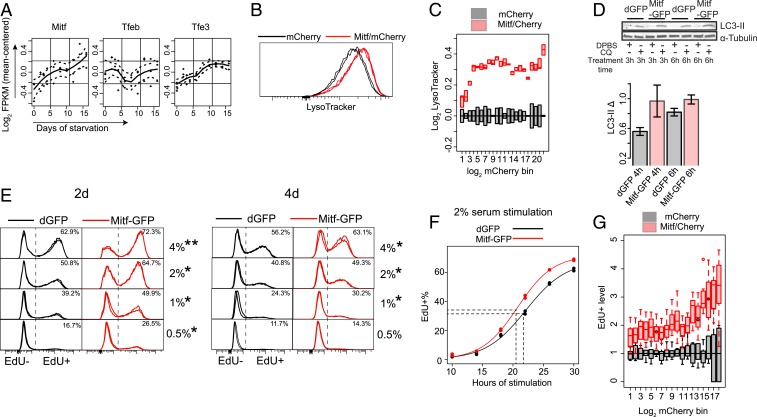
If the impaired lysosomal function is responsible for quiescence deepening, enhancing such function will likely counteract this trend and push cells into shallower quiescence. Consistent with this hypothesis, we observed that nutrient starvation, a known inducer of autophagy, pushed cells into shallower quiescence: Serum-starved REF cells treated with PBS (i.e., under additional nutrient starvation) for 320 min showed significantly higher E2f-ON% (SI Appendix, Fig. S6A) and EdU+% (SI Appendix, Fig. S6B) upon serum stimulation than treated with control (i.e., without nutrient starvation). This result is similar to what was previously observed in quiescent neural stem cells (NSCs) (39). However, nutrient starvation induces a range of cellular responses (e.g., AMPK activation and mTOR inhibition) in addition to autophagy. To increase lysosomal function directly, we sought to enhance lysosomal gene expression and biogenesis in REF cells. Lysosomal gene expression and biogenesis can be up-regulated by a MiT/TFE family of transcription factors Tfeb, Mitf, and Tfe3, which bind to a CLEAR-box sequence upstream of many lysosomal genes and are known as the master regulator of lysosome biogenesis (40–43). Remarkably, the expression of Mitf and Tfe3 but not Tfeb increased significantly in deep quiescence (Fig. 5A) (P = 0.028, 0.032, and 0.827 for Mitf, Tfe3, and Tfeb, respectively, in a 1-tailed t test comparing 16-d and 2-d serum-starved cells); meanwhile, Mitf but not Tfeb and Tfe3 showed a high degree of coexpression with lysosomal genes in quiescence (SI Appendix, Fig. S6C). Together, these results suggested a unique role for Mitf in regulating lysosomal function in the quiescent REF cell model. Indeed, we found that ectopic Mitf expression in quiescent REF cells (arrow in SI Appendix, Fig. S6D) significantly increased both the LysoTracker intensity (P = 0.017 in a 1-tailed t test) (Fig. 5 B and C) and lysosomal number (SI Appendix, Fig. S6E) in the cell, as well as enhanced autophagy flux (higher LC3-II Δ) (Fig. 5D), demonstrating the ability of Mitf to increase lysosomal biogenesis and function. Consistently, ectopic Mitf expression pushed cells into shallower quiescence, as seen in both a higher EdU+% upon serum stimulation (Fig. 5E) and a shorter time to reenter the cell cycle (Fig. 5F). Notably, there was a monotonic correlation between the level of introduced Mitf expression vector (indicated by mCherry intensity) (x axis in Fig. 5G) and the EdU+ level upon serum stimulation (normalized to mCherry control) (y axis in Fig. 5G), suggesting that cells can be continuously driven to shallower quiescence by enhancing lysosomal function.
Fig. 5.
Enhancing lysosomal function pushes cells toward shallower quiescence. (A) Time-course expression of MiT/TFE family members in RNA-seq analysis. Dashed line, SEM of the fitted line. (B and C) Lysosomal mass indicated by LysoTracker intensity. Cells were cotransfected with Mitf-GFP and mCherry expression vectors (Mitf/mCherry, triplicates) or mCherry control alone (mCherry, duplicates) and induced to quiescence by 4-d serum starvation. (B) Distribution of LysoTracker intensity in ∼10,000 cells per sample, with the highest frequency set at 100% (y axis). (C) Binned LysoTracker intensity (log-transformed with the median of mCherry control set at 0; bottom and top edges of each box indicate the first and third quartiles, respectively). Cells were grouped according to their mCherry intensity (log-transformed) into 22 even-width bins (bins with cell number <40 or with background fluorescence level similar to nontransfected cells were filtered out). (D) LC3-II turnover assay. Cells were transfected with Mitf-GFP expression vector or dGFP control and induced to quiescence by 4-d serum starvation. (Upper) Representative immunoblot image. (Lower) Quantified LC3-II Δ (between CQ-treated and nontreated cells as in Fig. 3A, duplicates). Error bar, SEM. (E) Quiescence-depth assay by EdU profile (n = 2). Cells were transfected with Mitf-GFP expression vector or dGFP control and induced to quiescence by 2- or 4-d serum starvation, followed by serum stimulation at indicated concentrations (0.5 to 4%) for 24 h. *P < 0.05 and **P < 0.01 (1-tailed t test). (F) Cell cycle reentry (EdU+) of 2-d serum-starved cells transfected with Mitf-GFP vector or dGFP control, followed by 2% serum stimulation for the indicated durations. Dotted lines indicate the estimated median cell cycle reentry time (Mitf-GFP = 20.5 h; dGFP = 21.7 h) of the quiescence-exited cells by the 30th hour (n = 2 at each time point). (G) Quiescence-depth assay in mCherry or Mitf/mCherry transfected cells. Transfected cells were induced to quiescence by 4-d serum starvation and stimulated with 0.5% serum. EdU+% was measured after 24 h of serum stimulation (in 7 and 9 replicates of mCherry and mCherry/Mitf transfection, respectively; ∼10,000 cells each). Transfected cells were binned according to mCherry intensity as in C. EdU+% of mCherry/Mitf cells was normalized to that of mCherry control cells in each bin. The resultant “EdU+ level” (y axis) indicates relative fold increase of quiescence exit (EdU+) associated with ectopic Mitf level (indicated by mCherry intensity, x axis) over corresponding mCherry control. (B, C, and G) Cotransfected mCherry was used as a proxy to quantify Mitf-GFP expression (see SI Appendix, Fig. S6F for verification). Using GFP for quantification otherwise may be interefered by the E2f-GFP reporter signal in the cell; in addition, GFP fluorescence is quenched by the Click-iT reaction in EdU assay and thus cannot be used for quantification.
Lysosomal Function Prevents Quiescence Deepening via ROS Reduction.
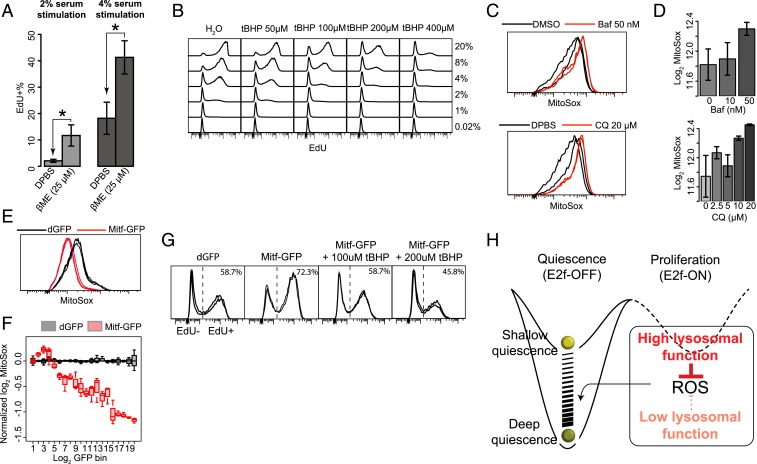
Lysosomes are known to play an antioxidative role in quiescent stem cells (15, 16). Thus, here we tested whether lysosomal function potentially prevents quiescence deepening by antioxidation. If it does, we reasoned that increasing antioxidation in the cell would reduce quiescence depth. Indeed, we found that supplementing antioxidant 2-mercaptoethanol (βME) drove quiescent cells into a shallower state, from which cells became more sensitive to growth signals (higher EdU+% upon serum stimulation) (Fig. 6A); conversely, supplementing an oxidative reagent tert-butyl hydroperoxide (tBHP) drove cells into deeper quiescence, shown by the lower EdU+% (Fig. 6B) and lower E2f-ON% (SI Appendix, Fig. S7A) upon stimulation at a given serum concentration with increasing tBHP concentration. These results indicate that the degree of oxidative stress is positively correlated with quiescence depth.
Fig. 6.
Lysosomal function reduces oxidative stress and prevents quiescence deepening. (A) Two-day serum-starved cells were further cultured in starvation medium containing βME for 4 d and stimulated with 2% or 4% serum for 24 h, followed by EdU assay. *P < 0.05, 1-tailed t test statistical significance and ns, insignificance, (triplicates with ∼10,000 cells each). Error bar, SEM. (B) Two-day serum-starved cells were treated with tBHP at indicated concentrations for 1 h and stimulated with various concentrations of serum as indicated for 24 h, followed by EdU assay. (A and B) Quantifications of cell death and irreversible arrest percentages at treatment conditions in A and B are shown in SI Appendix, Table S3. (C and D) Intracellular ROS level in quiescent REF cells treated with lysosomal inhibitors. Two-day serum-starved cells were treated with Baf or CQ at indicated concentrations for 6 h and subjected to MitoSox measurement (duplicates with ∼10,000 cells each). MitoSox signal distribution with the highest frequency set at 100% (y axis, C) and its median (log-transformed, D) are shown. Error bar, SEM. (E and F) Intracellular ROS level in quiescent cells transfected with Mitf expression vector. Cells were transfected with Mitf-GFP or dGFP control vectors and induced to quiescence by 4-d serum starvation, followed by MitoSox measurement (triplicates with ∼10,000 cells each). REF cells without the E2F-GFP reporter were used to avoid interfering Mitf-GFP quantification by the reporter signal. (E) MitoSox signal distribution with the highest frequency set at 100% (y axis). (F) Binned MitoSox intensity (log-transformed and normalized with the median of dGFP control set at 0). Cells were grouped according to their log-transformed GFP intensity into 20 even-width bins. (G) Quiescence-depth assay of Mitf transfected cells cotreated with tBHP (n = 2). Cells were transfected with Mitf-GFP expression vector or dGFP control, induced to quiescence by 2-d serum starvation. Mitf-transfected cells were further treated with tBHP at indicated concentration for 1 h. Cells were subsequently stimulated with 4% serum for 24 h and subjected to EdU assay. (H) Model of quiescence-depth regulation by lysosomal function.
Next, we found that inhibiting lysosomal function by Baf and CQ, which deepened quiescence, increased mitochondrial ROS level (higher MitoSox intensity) (Fig. 6 C and D). Conversely, enhancing lysosomal function by ectopic Mitf expression, which reduced quiescence depth, suppressed mitochondrial ROS (lower MitoSox intensity) (Fig. 6E) in a dose-dependent manner (Fig. 6F). Consistently, tBHP blocked the effect of Mitf on reducing quiescence depth: While the ectopic Mitf expression increased the EdU+% over control, additional tBHP treatment in Mitf-transfected cells decreased the EdU+% in a tBHP dose-dependent manner (Fig. 6G). We also considered that lysosomes may prevent quiescence deepening by increasing energy production, another known role of lysosomes (18). However, treatment with metabolites known to increase ATP production, such as methyl pyruvate (MPy), did not reduce quiescence depth with statistical significance (SI Appendix, Fig. S7B); it actually increased quiescence depth at high concentrations (SI Appendix, Fig. S7C), indicating that ATP generation is likely not responsible for how lysosomal function prevents quiescence deepening in the tested conditions, reminiscent of similar findings in yeasts (44). Taken together, our findings suggest that: 1) Increasing oxidative stress deepens quiescence, and reducing oxidative stress prevents quiescence from deepening; 2) reducing lysosomal function increases ROS, the mediator of oxidative stress, and increasing lysosomal function decreases ROS; and 3) increasing or decreasing lysosomal function continuously modulates quiescence depth, acting like a dimmer switch (Fig. 6H).
Quiescence Deepening Is Linked to Cellular Senescence and Aging.
Finally, we examined whether and how deep quiescence in REF cells is related to the quiescent states of other cell types (case Q) and other nongrowth cellular states, such as senescence (case S). To this end, we first developed a gene-expression signature indicating the quiescence depth in REF cells by performing a linear regression analysis of the time-course RNA-seq data (see Materials and Methods for details). To identify a signature that corresponds to a deepening of quiescence as opposed to simply halting the cell cycle, we omitted the proliferating sample (0 d) from the analysis and used samples corresponding to 2- to 16-d serum starvation (i.e., shallow to deep quiescence) to build the regression model. We next applied this quiescence-depth signature to other publicly available RNA-seq datasets related to cases Q and S, above.
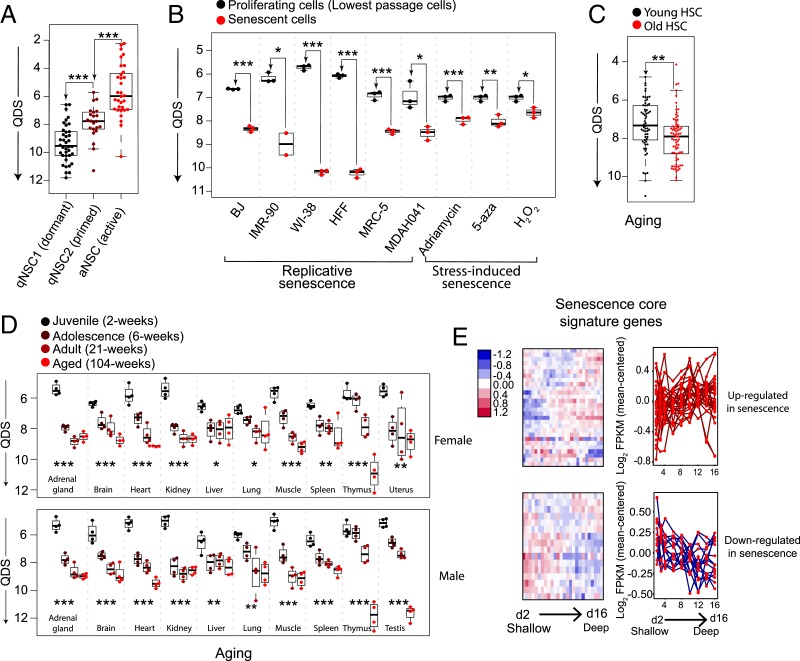
As an example of case Q, dormant NSCs in vivo transition into a shallower quiescent state called primed NSCs upon neural injury (9). When we applied the quiescence-depth gene signature of REF cells to the RNA-seq data corresponding to NSCs before and after neural injury, we obtained quiescence depth scores (QDS) from the linear regression model (see Materials and Methods for details) to indicate the relative quiescence depth of NSCs. We found that the QDS of primed NSCs after injury was significantly smaller (i.e., shallower quiescence) than that of dormant NSCs before injury (Fig. 7A). This result suggests that the quiescence-depth signature of REF cells in vitro can predict the relative quiescence depth of NSCs in vivo, indicating that this gene signature may reflect shared quiescence regulatory mechanisms across different cell types.
Fig. 7.
Quiescence deepening parallels cellular senescence in vitro and aging in vivo. (A) QDS of dormant, primed, and activated NSCs predicated based on the quiescence-depth gene expression signature developed in REF cells. Each dot represents a single cell sampled by single-cell RNA (scRNA)-seq (9). See Materials and Methods for details. *P < 0.05, **P < 0.01, and ***P < 0.001 (1-tailed t test, same below in B and C). (B) Predicted QDS of indicated cell lines at proliferation versus replicative senescence or stress-induced senescence (caused by DNA damaging agent Adriamycin, DNA demethylation agent 5-aza-2-deoxycytidine [5-aza], or oxidant H2O2). Each dot represents a cell population sampled by RNA-seq (20, 21). (C) Predicted QDS of hematopoietic stem cells (HSCs) collected from young (2 to 3 mo) or aged (20 to 25 mo) mice. Each dot represents a single cell sampled by scRNA-seq (22). (D) Predicted QDS of indicated organ types harvested from rats (female, Upper; male, Lower) at indicated age stages. *P < 0.05, **P < 0.01, and ***P < 0.001 (1-way ANOVA). Each dot represents a cell population sampled by RNA-seq (23). (E) Expression dynamics of “senescence core signature” genes in RNA-seq analysis of 2- to 16-d serum-starved cells, shown in a hierarchical clustering heatmap (Left) or an overlaid line plot (Right). “Senescence core signature” genes up- or down-regulated in senescent cells are grouped, Upper and Lower, respectively.
To test case S, we analyzed RNA-seq datasets associated with cellular senescence and aging by applying the QDS model with the quiescence-depth gene signature of REF cells. Strikingly, we found that QDS correctly predicted cellular senescence and aging in a wide array of cell lines in vitro (20, 21) and tissues in vivo (22, 23) (Fig. 7 B–D). In all 6 cell lines (BJ, IMR-90, WI-38, HFF, MRC-5, and MDAH041) studied under replicative senescence and in all 3 conditions (Adriamycin, 5-aza, and H2O2) studied under stress-induced senescence, QDS was significantly larger than that in proliferating controls (Fig. 7B). Notably, QDS was constructed without involving proliferating samples (by comparing shallow and deep quiescent cells under 2- to 16-d serum starvation) and thus likely reflects a common feature between deep quiescence and senescence. Consistently, a set of “senescence core signature” genes identified from metaanalysis (19) exhibit similar expression changes (i.e., up- or down-regulation) under both deep quiescence and senescence (Fig. 7E). Furthermore, QDS was found significantly larger in aged hematopoietic stem cells (Fig. 7C) and monotonically increasing in all 11 studied rat organs undergoing aging processes from juvenile to adolescence, adult, and aged stages (Fig. 7D). The success of QDS gene signature to predict both senescence in vitro and aging in vivo suggests that quiescence deepening may share a molecular mechanistic basis with and act as a transitional state toward senescence and aging (see more in Discussion). Consistently, it was recently reported that long-term cultured quiescent human fibroblasts eventually transit into senescence under physiological oxygen conditions (45).
Discussion
The lysosome–autophagy pathway is known to protect adult stem cells against irreversible states, such as senescence, apoptosis, and terminal differentiation (15, 16, 18). The present study shows that instead of a simple OFF–ON switch, lysosomes function as a dimmer switch to continuously modulate quiescence depth (Fig. 6H) and proliferative capacity of quiescent cells.
Our study suggests that as cells move deeper in quiescence, lysosomal gene expression and lysosomal mass increase, but lysosomal function and lysosome destruction decrease. Decreased lysosome destruction via autophagy/lysophagy contributes to an increased lysosomal mass and likely an accumulation of damaged lysosomes, which in turn contributes to a decreased lysosomal–autophagy function. Decreased lysosome destruction alone, however, does not fully explain the increased lysosomal mass (both lysoTracker intensity of lysosome number) observed in deep quiescent cells (Fig. 2 G and H), as blocking lysosome destruction increased LysoTracker intensity but not the number of lysosomes in REF cells (SI Appendix, Fig. S4); similarly, it was reported previously that lysosomal number did not change or even mildly decreased in autophagy-impaired fibroblasts with Atg4B and Atg7 mutations (46). The other contributing factor of increased lysosomal mass in deep quiescence appears to be lysosomal biogenesis: The expression of Mitf, a master regulator of lysosomal biogenesis, significantly increased the lysosomal number (SI Appendix, Fig. S6E) in quiescent REF cells. Consistently, we found that applying small chemical inhibitors to repress Mcoln1 (also known as TRPML1) that is crucial for lysosome biogenesis (47) significantly decreased lysosomal number in quiescent REF cells (SI Appendix, Fig. S8 A and B). The increase in lysosome biogenesis itself is likely compensatory to quiescence deepening: Mitf expression increases significantly along with quiescence deepening (Fig. 5A); relatedly, inhibiting lysosome biogenesis further deepens quiescence (lower EdU+% with Mcoln1 inhibitors than with DMSO control) (SI Appendix, Fig. S8C). The dynamic balance and relative contributions of lysosome biogenesis and degradation destruction in deep quiescence await further investigation.
REF cells in deep quiescence exhibited reduced autophagy flux (Fig. 3A), which may be relevant to the declined autophagy observed in aging (15, 48–50) and senescent cells (17, 51). It is unclear what resulted in declined autophagy in deep quiescence despite increased lysosomal gene expression (Fig. 2E) and biogenesis. Note that not every lysosomal gene increased expression with quiescence deepening: A small subset of lysosomal genes enriched for those that also function in the Golgi network showed a general decrease (see cluster B in Fig. 2E for enrichment analysis and SI Appendix, Table S2). As the Golgi network is critical to transporting lysosomal hydrolases from cytoplasmic ribosomes into lysosomal vesicles (52), the reduced expression of some of these related genes, therefore, may lead to a reduced lysosomal–autophagy function. Alternatively, the “net” decrease in lysosomal function may be adaptive and serve as a cellular counter for serum-deprivation duration (i.e., environmental growth restriction), which in turn determines how cautious the cell will get before resuming growth. The exact mechanisms dictating the relationship between lysosomal function and quiescence depth awaits further studies.
Our results suggest that lysosomal function prevents quiescence deepening via ROS reduction (Fig. 6). Previous studies have shown that the lysosome–autophagy pathway reduces ROS in several types of quiescent cells (15, 16) through mitophagy: That is, selective autophagic degradation of damaged mitochondria (15). It remains to be tested whether lysosomes prevent quiescence deepening in REF cells via mitophagy, but the reduced mitochondrial ROS levels resulting from enhanced lysosomal function (Fig. 6 E and F) is consistent with this model.
It was shown previously that mTORC1 can transform muscle stem cells into a shallower quiescent state, called GAlert, in which cells are sensitized to growth stimulation (8). Consistently, continuous mTORC1 activation led to depletion of certain types of quiescent stem cells in vivo by forcing cell cycle reentry (53, 54). It is well established that mTORC1 drives cell growth and inhibits the lysosome-autophagy pathway (43, 55). Accordingly, the effect of mTORC1 on quiescence depth is likely 2-sided: By promoting cell growth, mTORC1 facilitates quiescence exit and thus reduces quiescence depth; by inhibiting lysosomal function, mTORC1 drives cells deeper into quiescence. In the GAlert case, where mTORC1 is activated both before and during stimulation, the net result appears to be shallower quiescence. The exact bimodal effects of mTORC1 on quiescence depth may be complicated and condition-dependent, awaiting future studies.
The lysosome–autophagy pathway plays an important role in cancer physiology and dormancy (5, 40, 41, 56–58). Activities of MiT/TFE family members, including Tfeb and Mitf, are up-regulated by overexpression or nuclear localization in multiple cancer types, including lung, pancreatic, and ovarian cancers (41, 59, 60), which is linked to poor prognosis and survival (59, 60). The lysosome–autophagy pathway appears to help maintain dormant cancer cells and facilitate survival and metastasis (5). Consistently, autophagy inhibition decreases the viability of dormant breast cancer cells and their metastatic recurrence, suggesting a promising treatment strategy (57). Future studies are needed to determine the optimal target and degree of lysosomal–autophagy inhibition in treatment to minimize disrupting the quiescence depth of normal cells.
Our study highlights that deep quiescent cells exhibit gene-expression signatures that are similar to senescent and aged cells (Fig. 7 B–E). The enriched up-regulation of lysosomal genes (KEGG_lysosome gene set) is shown in 10 of the 11 studied aging tissues (adrenal gland, brain, heart, kidney, liver, lung, spleen, thymus, uterus, testis, but not muscle); it is also shown in 5 of the 6 cell lines (BJ, IMR-90, WI-38, HFF, MRC-5, but not MDAH041) studied under replicative senescence, but not shown in stress (Adriamycin/5-aza/H2O2)-induced senescence in MDAH041 cells (SI Appendix, Table S4). This result is consistent with the findings that senescence and aging are partially driven by ROS and can be counteracted by lysosome–autophagy activities in many but not all conditions (6, 15, 17, 61–63), indicating the contributions of other potentially shared mechanisms underlying deep quiescence, senescence, and aging. Among them, DNA damage is a likely candidate, which can be caused by various stress signals (e.g., ROS and replication stress) and is associated with quiescence entry (64, 65) as well as cellular senescence (66, 67) and aging (68, 69). Furthermore, many cellular activities are up- or down-regulated as cells move deeper in quiescence (SI Appendix, Fig. S2C). Some of these cellular activities may be involved in the regulation of quiescence depth and potentially senescence and aging, with their detailed mechanisms awaiting further studies. In this regard, it has been shown recently in both NSCs and bacteria that the accumulation of protein aggregates is associated with quiescence deepening, and that the clearance of protein aggregates (by lysosomes in NSCs and by DnaK–ClpB complex in bacteria) drives cells to shallower quiescence and enhances their ability to reenter the cell cycle (39, 70).
Quiescence entry and exit are regulated by the Rb-E2f-Cyclin/Cdk gene network (71, 72) that functions as a bistable switch (25). How does the toggle switch-like nature of the Rb-E2f pathway fit with the lysosomal dimmer switch? We speculate that during quiescence exit, cells first move progressively into shallow quiescence and at a time point, the restriction point (73, 74), “flip” into the cell cycle by committing to proliferation; the whole process acts like adjusting a sliding dimmer before activating a toggle switch. In this regard, it will be important to figure out whether and how the lysosomal switch cross-talks with the Rb-E2f switch in controlling quiescence depth and exit in future studies.
Relatedly, recent in vivo studies on muscle and neural stem cells generally invoke 2 quiescence state: 1 shallow (e.g., GAlert and primed quiescence) and 1 deep. Our findings that the gene-expression signature we developed in the fibroblast model in vitro can correctly classify deep (dormant) and shallow (primed) NSCs in vivo (Fig. 7A) suggest a potential mechanistic link between the quiescent states in vitro and in vivo. As such, we predict that quiescence depth in vivo is also continuous instead of binary. It is possible that different levels of injuries can cause varying levels of quiescent states in vivo, which was not examined previously but would be interesting to test in future studies. Finally, in support of the continuous quiescence depth in vivo are the earlier findings that in the liver after partial hepatectomy and in the salivary gland after isoproterenol stimulation, cells in increasingly older rats took an increasingly longer time to initiate DNA synthesis, behaving like cells in increasing deeper quiescence (10, 75).
Quiescence is commonly believed to protect cells from irreversible arrest, such as senescence. Our work suggests that this protection declines in long-term quiescent cells and the gradual quiescence deepening likely represents a transition path from cell proliferation to senescence. Relatedly, the same Rb and p53 pathways appear to underlie both quiescence and senescence (76–79). Whether and how the lysosome–autophagy pathway interacts with Rb and p53 pathways to regulate the transition from deep quiescence to senescence, and whether such a transition is gradual following a continuum or abrupt as controlled by an ultrasensitive or bistable switch-like mechanism (80, 81), remain significant unanswered questions (SI Appendix, Fig. S1A).
Materials and Methods
Detailed descriptions of RNA-seq and downstream data analysis and modeling, modulation of lysosomal–autophagic function and lysosome biogenesis, assays for quiescence-depth, lysosomal mass and proteolytic activity, autophagy flux, mitochondrial ROS, β-galactosidase activity, cell size, and cytotoxicity are provided in SI Appendix.
Cell Culture and Quiescence Induction.
REFs used in this study are from a single-cell clone derived from REF52 cells and contain a stably integrated human E2F1 promoter-driven destabilized EGFP (E2f-GFP) reporter, as previously described [i.e., REF/E23 cells (25, 82)] unless otherwise noted. Cells were passaged every 2 to 3 d and maintained at subconfluency in growth medium: DMEM supplemented with 10% bovine growth serum (BGS; GE Healthcare, SH30541). To induce quiescence, cells were plated at ∼50% confluence in growth medium for a day, washed twice with DMEM, and cultured in serum-starvation medium (DMEM containing 0.02% BGS) for the indicated duration (≥2 d).
Quiescence-Depth Assay.
To assess quiescence depth, cells were switched from serum-starvation medium to serum-stimulation medium (DMEM containing BGS at indicated concentrations) and harvested at indicated time points by trypsinization. The cell fraction that reentered the cell cycle was quantified by assessing the profile of EdU incorporation (and the profiles of E2f-GFP reporter and PI DNA staining when indicated). Quiescence depth is determined by the serum threshold required to activate cells to reenter the cell cycle. At a given serum concentration, a smaller percentage of deeper quiescent cells are able to reenter the cell cycle by a given time than of shallower quiescent cells. See text for details.
Data Availability.
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE124109).
Supplementary Material
Acknowledgments
We thank Johnny Fares for valuable discussions and suggestions on lysosomal function and assays; Andrew Peak and Andrew Capaldi for critical reading and comments on the manuscript; and Haoxing Xu for providing Mcoln1 inhibitors. This work was supported by National Science Foundation Grants DMS-1463137 (to G.Y.), DMS-1418172 (to G.Y. and H.H.Z.), CCF-1740858 (to H.H.Z.), and DMS-1462049 (to J.X.); National Institutes of Health Grant R01DK119232 (to J.X.); Chinese National Science and Technology Major Project 2018ZX10302205 (to F.B.); and Guangdong Province Key Research and Development Program 2019B020226002 (to F.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE124109).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915905116/-/DCSupplemental.
References
- 1.Rumman M., Dhawan J., Kassem M., Concise review: Quiescence in adult stem cells: Biological significance and relevance to tissue regeneration. Stem Cells 33, 2903–2912 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Cheung T. H., Rando T. A., Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 14, 329–340 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossi L., et al. , Less is more: Unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell 11, 302–317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coller H. A., Sang L., Roberts J. M., A new description of cellular quiescence. PLoS Biol. 4, e83 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sosa M. S., Bragado P., Aguirre-Ghiso J. A., Mechanisms of disseminated cancer cell dormancy: An awakening field. Nat. Rev. Cancer 14, 611–622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh J., Lee Y. D., Wagers A. J., Stem cell aging: Mechanisms, regulators and therapeutic opportunities. Nat. Med. 20, 870–880 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharpless N. E., DePinho R. A., How stem cells age and why this makes us grow old. Nat. Rev. Mol. Cell Biol. 8, 703–713 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Rodgers J. T., et al. , mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature 510, 393–396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llorens-Bobadilla E., et al. , Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17, 329–340 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Bucher N. L., Regeneration of mammalian liver. Int. Rev. Cytol. 15, 245–300 (1963). [DOI] [PubMed] [Google Scholar]
- 11.Roth G. S., Adelman R. C., Age-dependent regulation of mammalian DNA synthesis and cell division in vivo by glucocorticoids. Exp. Gerontol. 9, 27–31 (1974). [DOI] [PubMed] [Google Scholar]
- 12.Owen T. A., Soprano D. R., Soprano K. J., Analysis of the growth factor requirements for stimulation of WI-38 cells after extended periods of density-dependent growth arrest. J. Cell. Physiol. 139, 424–431 (1989). [DOI] [PubMed] [Google Scholar]
- 13.Augenlicht L. H., Baserga R., Changes in the G0 state of WI-38 fibroblasts at different times after confluence. Exp. Cell Res. 89, 255–262 (1974). [DOI] [PubMed] [Google Scholar]
- 14.Kwon J. S., et al. , Controlling depth of cellular quiescence by an Rb-E2F network switch. Cell Rep. 20, 3223–3235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Prat L., et al. , Autophagy maintains stemness by preventing senescence. Nature 529, 37–42 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Ho T. T., et al. , Autophagy maintains the metabolism and function of young and old stem cells. Nature 543, 205–210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang H. T., Lee K. B., Kim S. Y., Choi H. R., Park S. C., Autophagy impairment induces premature senescence in primary human fibroblasts. PLoS One 6, e23367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lum J. J., et al. , Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120, 237–248 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Hernandez-Segura A., et al. , Unmasking transcriptional heterogeneity in senescent cells. Curr. Biol. 27, 2652–2660.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell M., Kruger A., Tainsky M. A., Gene expression profiling of replicative and induced senescence. Cell Cycle 13, 3927–3937 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marthandan S., et al. , Conserved senescence associated genes and pathways in primary human fibroblasts detected by RNA-seq. PLoS One 11, e0154531 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grover A., et al. , Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat. Commun. 7, 11075 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Y., et al. , A rat RNA-seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat. Commun. 5, 3230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao G., Tan C., West M., Nevins J. R., You L., Origin of bistability underlying mammalian cell cycle entry. Mol. Syst. Biol. 7, 485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao G., Lee T. J., Mori S., Nevins J. R., You L., A bistable Rb-E2F switch underlies the restriction point. Nat. Cell Biol. 10, 476–482 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Fujimaki K., et al. , Graded regulation of cellular quiescence depth between proliferation and senescence by a lysosomal dimmer switch. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124109. Deposited 11 October 2019. [DOI] [PMC free article] [PubMed]
- 27.Sharpless N. E., Sherr C. J., Forging a signature of in vivo senescence. Nat. Rev. Cancer 15, 397–408 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nazio F., et al. , Fine-tuning of ULK1 mRNA and protein levels is required for autophagy oscillation. J. Cell Biol. 215, 841–856 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizushima N., Yoshimori T., Levine B., Methods in mammalian autophagy research. Cell 140, 313–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasegawa J., Maejima I., Iwamoto R., Yoshimori T., Selective autophagy: Lysophagy. Methods 75, 128–132 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Papadopoulos C., Meyer H., Detection and clearance of damaged lysosomes by the endo-lysosomal damage response and Lysophagy. Curr. Biol. 27, R1330–R1341 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Monastyrska I., Rieter E., Klionsky D. J., Reggiori F., Multiple roles of the cytoskeleton in autophagy. Biol. Rev. Camb. Philos. Soc. 84, 431–448 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klionsky D. J., et al. , Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Debacq-Chainiaux F., et al. , Screening of senescence-associated genes with specific DNA array reveals the role of IGFBP-3 in premature senescence of human diploid fibroblasts. Free Radic. Biol. Med. 44, 1817–1832 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Duan J., Duan J., Zhang Z., Tong T., Irreversible cellular senescence induced by prolonged exposure to H2O2 involves DNA-damage-and-repair genes and telomere shortening. Int. J. Biochem. Cell Biol. 37, 1407–1420 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Leeman D. S., et al. , Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359, 1277–1283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ploper D., et al. , MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc. Natl. Acad. Sci. U.S.A. 112, E420–E429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perera R. M., et al. , Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature 524, 361–365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouché V., et al. , Drosophila Mitf regulates the V-ATPase and the lysosomal-autophagic pathway. Autophagy 12, 484–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Settembre C., Fraldi A., Medina D. L., Ballabio A., Signals from the lysosome: A control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14, 283–296 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laporte D., et al. , Metabolic status rather than cell cycle signals control quiescence entry and exit. J. Cell Biol. 192, 949–957 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marthandan S., Priebe S., Hemmerich P., Klement K., Diekmann S., Long-term quiescent fibroblast cells transit into senescence. PLoS One 9, e115597 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maejima I., et al. , Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32, 2336–2347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller A., et al. , Mucolipidosis type IV protein TRPML1-dependent lysosome formation. Traffic 16, 284–297 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Vittorini S., et al. , The age-related accumulation of protein carbonyl in rat liver correlates with the age-related decline in liver proteolytic activities. J. Gerontol. A Biol. Sci. Med. Sci. 54, B318–B323 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Kaushik S., et al. , Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Rep. 13, 258–265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cuervo A. M., Dice J. F., Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 275, 31505–31513 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Han B. I., Hwang S. H., Lee M., A progressive reduction in autophagic capacity contributes to induction of replicative senescence in Hs68 cells. Int. J. Biochem. Cell Biol. 92, 18–25 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Luzio J. P., Pryor P. R., Bright N. A., Lysosomes: Fusion and function. Nat. Rev. Mol. Cell Biol. 8, 622–632 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Chen C., et al. , TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J. Exp. Med. 205, 2397–2408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adhikari D., et al. , Tsc/mTORC1 signaling in oocytes governs the quiescence and activation of primordial follicles. Hum. Mol. Genet. 19, 397–410 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laplante M., Sabatini D. M., mTOR signaling in growth control and disease. Cell 149, 274–293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang A., Kimmelman A. C., Inhibition of autophagy attenuates pancreatic cancer growth independent of TP53/TRP53 status. Autophagy 10, 1683–1684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vera-Ramirez L., Vodnala S. K., Nini R., Hunter K. W., Green J. E., Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat. Commun. 9, 1944 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mowers E. E., Sharifi M. N., Macleod K. F., Autophagy in cancer metastasis. Oncogene 36, 1619–1630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giatromanolaki A., et al. , Increased expression of transcription factor EB (TFEB) is associated with autophagy, migratory phenotype and poor prognosis in non-small cell lung cancer. Lung Cancer 90, 98–105 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Fu X., Zhang L., Dan L., Wang K., Xu Y., Expression of TFEB in epithelial ovarian cancer and its role in autophagy. Int. J. Clin. Exp. Pathol. 9, 10914–10928 (2016). [Google Scholar]
- 61.Rubinsztein C. D., Mariño G., Kroemer G., Autophagy and aging. Cell 146, 682–695 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Dou Z., et al. , Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Narita M., et al. , Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science 332, 966–970 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barr A. R., et al. , DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression. Nat. Commun. 8, 14728 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arora M., Moser J., Phadke H., Basha A. A., Spencer S. L., Endogenous replication stress in mother cells leads to quiescence of daughter cells. Cell Rep. 19, 1351–1364 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gire V., Roux P., Wynford-Thomas D., Brondello J. M., Dulic V., DNA damage checkpoint kinase Chk2 triggers replicative senescence. EMBO J. 23, 2554–2563 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.d’Adda di Fagagna F., Living on a break: Cellular senescence as a DNA-damage response. Nat. Rev. Cancer 8, 512–522 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Soares J. P., et al. , Aging and DNA damage in humans: A meta‐analysis study. Aging (Albany N.Y.) 6, 432–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beerman I., Seita J., Inlay M. A., Weissman I. L., Rossi D. J., Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell 15, 37–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pu Y., et al. , ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol. Cell 73, 143–156.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Spencer S. L., et al. , The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell 155, 369–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun D., Buttitta L., States of G0 and the proliferation-quiescence decision in cells, tissues and during development. Int. J. Dev. Biol. 61, 357–366 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Schwarz C., et al. , A precise Cdk activity threshold determines passage through the restriction point. Mol. Cell 69, 253–264.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matson J. P., Cook J. G., Cell cycle proliferation decisions: The impact of single cell analyses. FEBS J. 284, 362–375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adelman R. C., Stein G., Roth G. S., Englander D., Age-dependent regulation of mammalian DNA synthesis and cell proliferation in vivo. Mech. Ageing Dev. 1, 49–59 (1972). [Google Scholar]
- 76.Childs B. G., Durik M., Baker D. J., van Deursen J. M., Cellular senescence in aging and age-related disease: From mechanisms to therapy. Nat. Med. 21, 1424–1435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shay J. W., Pereira-Smith O. M., Wright W. E., A role for both RB and p53 in the regulation of human cellular senescence. Exp. Cell Res. 196, 33–39 (1991). [DOI] [PubMed] [Google Scholar]
- 78.Campisi J., d’Adda di Fagagna F., Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729–740 (2007). [DOI] [PubMed] [Google Scholar]
- 79.Bennett D. C., Human melanocyte senescence and melanoma susceptibility genes. Oncogene 22, 3063–3069 (2003). [DOI] [PubMed] [Google Scholar]
- 80.Tyson J. J., Chen K. C., Novak B., Sniffers, buzzers, toggles and blinkers: Dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol. 15, 221–231 (2003). [DOI] [PubMed] [Google Scholar]
- 81.Ferrell J. E. Jr, Ha S. H., Ultrasensitivity part III: Cascades, bistable switches, and oscillators. Trends Biochem. Sci. 39, 612–618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang X., et al. , Exit from quiescence displays a memory of cell growth and division. Nat. Commun. 8, 321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith C. L., Blake J. A., Kadin J. A., Richardson J. E., Bult C. J.; Mouse Genome Database Group , Mouse Genome Database (MGD)-2018: Knowledgebase for the laboratory mouse. Nucleic Acids Res. 46, D836–D842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE124109).