Significance
It is recognized that an optimized mixture of endo- and exo-glucanases, beta-glucosidases, and accessory enzymes need to work cooperatively to result in effective cellulose hydrolysis. However, a key factor that impacts hydrolysis is the effectiveness of enzyme access to the cellulose. A quantitative method based on fluorescence-tagged CBMs and CLSM was developed to assess the cellulose structure, quantify its accessibility, and elucidate how this might influence the efficiency of cellulose hydrolysis. This work provides insights into the substrate–enzyme interactions that occur during enzyme-mediated hydrolysis of cellulose.
Keywords: carbohydrate-binding modules, enzyme accessibility, supramolecular cellulose structure, confocal laser scanning microscopy, quantitative image analysis
Abstract
Two fluorescence-tagged carbohydrate-binding modules (CBMs), which specifically bind to crystalline (CBM2a-RRedX) and paracrystalline (CBM17-FITC) cellulose, were used to differentiate the supramolecular cellulose structures in bleached softwood Kraft fibers during enzyme-mediated hydrolysis. Differences in CBM adsorption were elucidated using confocal laser scanning microscopy (CLSM), and the structural changes occurring during enzyme-mediated deconstruction were quantified via the relative fluorescence intensities of the respective probes. It was apparent that a high degree of order (i.e., crystalline cellulose) occurred at the cellulose fiber surface, which was interspersed by zones of lower structural organization and increased cellulose accessibility. Quantitative image analysis, supported by 13C NMR, scanning electron microscopy (SEM) imaging, and fiber length distribution analysis, showed that enzymatic degradation predominates at these zones during the initial phase of the reaction, resulting in rapid fiber fragmentation and an increase in cellulose surface crystallinity. By applying this method to elucidate the differences in the enzyme-mediated deconstruction mechanisms, this work further demonstrated that drying decreased the accessibility of enzymes to these disorganized zones, resulting in a delayed onset of degradation and fragmentation. The use of fluorescence-tagged CBMs with specific recognition sites provided a quantitative way to elucidate supramolecular substructures of cellulose and their impact on enzyme accessibility. By designing a quantitative method to analyze the cellulose ultrastructure and accessibility, this study gives insights into the degradation mechanism of cellulosic substrates.
It is largely acknowledged that feedstock recalcitrance to enzymatic deconstruction is one of the major limitations in establishing effective biomass-based biorefinery processes (1, 2). The effectiveness of the cellulases, a mixture of endo- and exo-glucanases, beta-glucosidases, and accessory enzymes on cellulose hydrolysis, is influenced by a variety of factors. These include the nature of the lignin–carbohydrate complex, particle size, porosity, and surface area (3, 4), with the limited accessibility of the enzymes to the cellulose recognized as being a major impediment (5, 6). As accessibility of cellulose to enzymes is mainly influenced by the cellulose structure (5, 6), studies made use of methods, such as XRD, NMR, and GPC (7, 8), which provided useful information on overall cellulose structural characteristics, such as crystallinity and the degree of polymerization. One of the parameters most often used to describe the cellulose structure is the crystallinity index (CI), which categorizes cellulose into either crystalline or paracrystalline substructures (6, 7, 9). However, the structure of cellulose cannot be strictly categorized into paracrystalline or crystalline (10), and enzyme accessibility has been shown to be affected by the various levels of cellulose organization, ranging from the nano- to micrometer scale (4, 11).
Previous work has suggested that enzyme-mediated hydrolysis is predominantly initiated at more accessible regions within the cellulose matrix (12–17), with these regions often referred to as dislocation zones, slip planes, microcompressions, or kinks (18, 19). These regions in the fiber cell wall have been shown to be mechanically weak points in the cellulose fiber (20–22), consequently making them of ongoing interest to the pulp and paper sector (18). Methods such as polarized light microscopy (13, 15, 20–24) or SEM (17, 21, 24) were used to better elucidate these disruptions. The disruptions have been shown to vary in severity, from microcracks at the fiber surface to buckling and folding of the S1 and S2 layers (i.e., at the primary and secondary cell wall). However, the exact nature of these disruptions is less clear. Polarized light microscopy studies have indicated that these regions are birefringent, with some researchers interpreting this as increased crystallinity (15) and that the microfibrils continue unbroken through these regions (22). As a result, it has been suggested that the shear-failures at these zones occur because of weaker or misplaced intermicrofibrillar bonds rather than intramicrofibrillar deformations or weaknesses (22, 25). Related work has further shown that these localized areas of morphological changes are predominantly paracrystalline, based on their CI (26) and susceptibility to acid (12, 27) and enzymatic (12–17) hydrolysis. In more recent work, the bending stress exhibited on microfibrils has been shown to result in the breakage of single glucan chains, with the resulting defects enhancing the accessibility to processive cellulases (28). However, an ongoing challenge has been to effectively monitor changes to the supramolecular structure of cellulose as enzyme-mediated deconstruction proceeds.
Carbohydrate-binding modules (CBMs) are the noncatalytic part of cellulolytic enzymes and are thought to assist with enzyme synergism via a proximity and targeting effect (29). Several CBMs have been successfully used to assess the topology of cellulosic substrates during enzymatic hydrolysis (30). More than 80 CBM families have been categorized into 3 types according to their substrate affinity (31), with type A predominantly binding to crystalline cellulose, type B to the single glucan chains that are prevalent in paracrystalline cellulose, and type C to soluble carbohydrates (32). Based on their specific adsorption toward different carbohydrate substructures (31), fluorescence-tagged CBMs have been successfully used to assess the glyco-architecture of plant cell walls when combined with confocal laser scanning microscopy (CLSM) (33).
Related studies, using either type A or B CBMs, have suggested that these CBMs predominantly bind to the more disorganized cellulose regions of the cellulose fibers, providing valuable information on the role of these defects on enzyme accessibility (15, 33–37). As CBM binding depends on the respective binding mechanism (29, 32, 36, 38), they have the potential to be used to both evaluate cellulose morphology as well as its impact on enzyme accessibility.
In the work described below, 2 fluorescence-tagged CBMs, a type A (CBM2a) and a type B (CBM17), were combined with CLSM to better elucidate the cellulose ultrastructure and assess how key substrate characteristics might influence enzyme-mediated deconstruction of cellulose. In contrast to previous studies, the CBM/CLSM method provided information on the spatial distribution of the carbohydrate fine structures within the fiber and allowed us to monitor changes at the fiber surface over time. This provided localized and quantitative information of where hydrolysis predominated in a time-resolved manner.
The CLSM images were complemented by SEM imaging, 13C NMR analysis, and fiber length quantification, and together the results showed that disorganized zones occur at regular intervals on the fiber surface. These zones are composed of more accessible crystalline and paracrystalline cellulose structures, and, as a result, enzymes can bind to and degrade these areas more efficiently, leading to fiber fragmentation and the production shorter fiber fragments.
Results and Discussion
The Use of Fluorescence-Tagged CBMs and CLSM to Assess the Supramolecular Structure of Northern Bleached Softwood Kraft Pulp during Enzymatic Hydrolysis.
In earlier work, photoactivated localization microscopy was successfully used to determine the selectivity of binding of 6 CBMs, including CBM2a and CBM17, to various cellulosic substrates (29). Over a selectivity range of 0.16 (paracrystalline) to 0.39 (crystalline), CBM17 and CBM2a showed an affinity of 0.21 and 0.27, respectively, suggesting that CBM2a bound to areas with a higher degree of structural organization than did the CBM17. However, CBM2a also showed a greater degree of binding promiscuity. Although it primarily targeted the hydrophobic [110] faces of the cellulose crystal lattice (according to Miller indices), it also bound to less ordered paracrystalline regions (29, 38, 39).
In the work reported here, CBM2a (type A) and CBM17 (type B), which had been covalently bound to Rhodamine-RedX (red) and fluorescein isothiocyanate (FITC) (green), respectively, were used to assess the substructures of northern bleached softwood Kraft (NBSK) fibers. In the initial phase of the hydrolysis reaction (Fig. 1A), the majority of the NBSK fibers were predominantly red (CBM2a-RRedX) and interspersed at regular intervals by discrete zones of green (CBM17-FITC). Earlier work had suggested that, at these less organized regions (green), enzymatic activity was most pronounced, resulting in their rapid degradation (9, 12, 13, 15). As described below, the CBM/CLSM results supported these earlier observations as this method allowed us to elucidate the nature and the spatial distribution of localized supramolecular cellulose structures and to quantitatively assess changes over hydrolysis time. Also, the CBM/CLSM method allowed the cellulose substructures to be analyzed before changes to the overall fiber structure (i.e., fiber fragmentation) occurred.
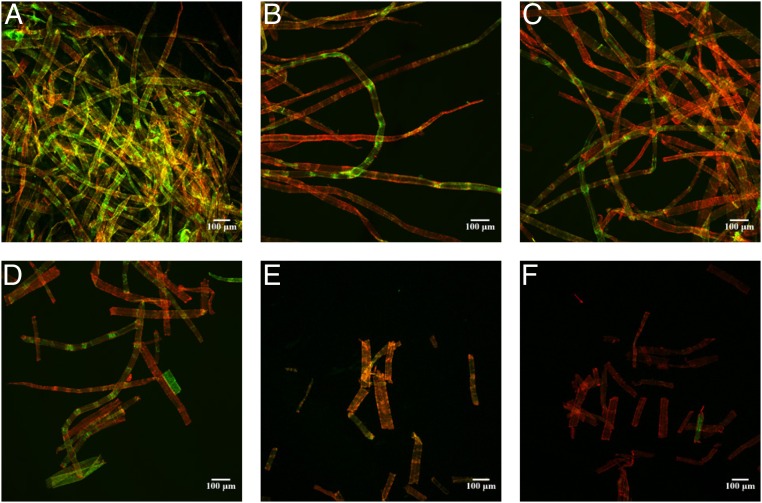
Fig. 1.
Changes in the morphology of NBSK pulp fibers after 0 h (A), 0.5 h (B), 1 h (C), 2 h (D), 4 h (E), and 6 h (F) of hydrolysis, visualized using CBM17-FITC and CBM2a-RRedX binding with CLSM. (Scale bars: 100 µm.)
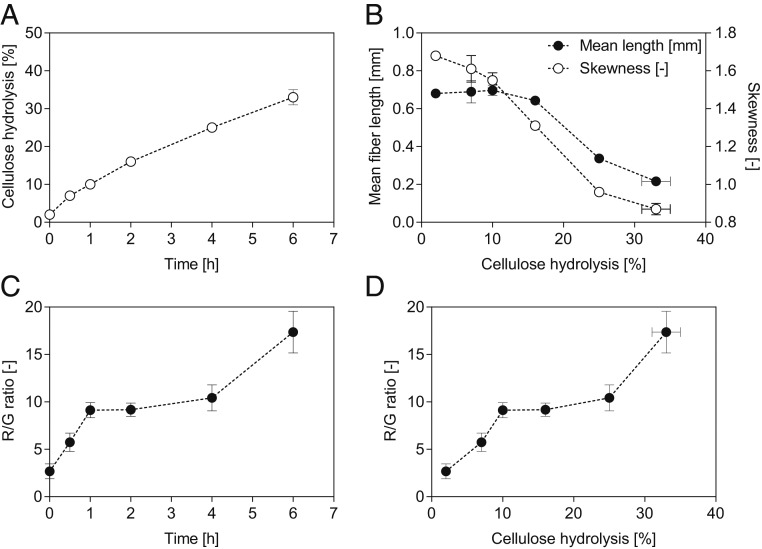
A 1% (wt/vol) concentration of NBSK was hydrolyzed by Ctec3 at an enzyme loading of 10 filter paper units (FPU)⋅g−1, and the resulting CLSM images after 0.5, 1, 2, 4, and 6 h of hydrolysis are depicted in Fig. 1 B–F. The corresponding hydrolysis profile is summarized in Fig. 2A. It was apparent that the fiber morphology changed only slightly before 2 h of hydrolysis, after which fiber fragmentation occurred (Fig. 1 D–F). At the same time the “red” fluorescence (indicating binding of CBM2a-RRedX to highly ordered cellulose) increased in predominance as hydrolysis proceeded while the green fluorescence intensity (from the paracrystalline binding CBM17-FITC) decreased (Fig. 1). Previous work (12, 13, 16, 21, 27) has reported similar fiber fragmentation patterns, suggesting that the increased susceptibility of disordered zones to acid and enzymes likely resulted in their more rapid degradation.
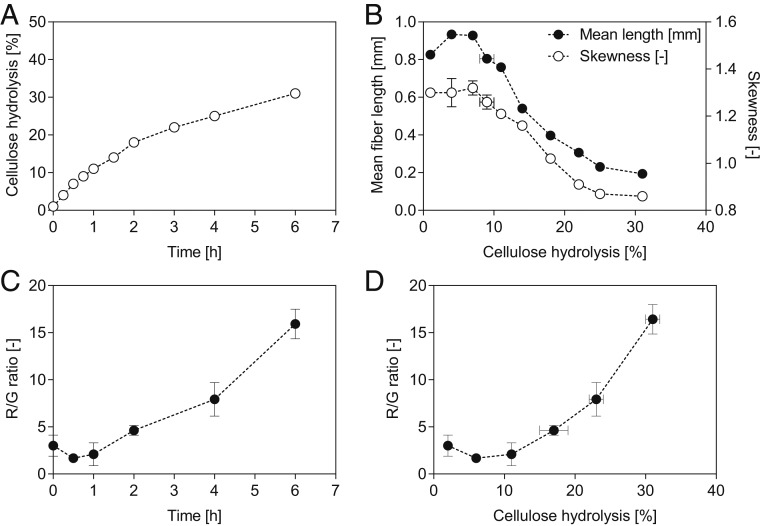
Fig. 2.
Enzymatic hydrolysis of NBSK. Shown are (A) hydrolysis time course, (B) change in mean fiber length (filled circles) and skewness (empty circles), (C) change in R/G ratio over time, and (D) change in R/G ratio with increasing cellulose hydrolysis. Duplicate experiments were performed. Data in A and B show mean values and the spread. Data in C and D were derived from 8 to 10 image frames, and mean values and the SDs are shown.
The Influence of Enzyme-Mediated Cellulose Hydrolysis on Fiber Size.
When the mean length and skewness of the NBSK fibers during hydrolysis was assessed (Fig. 2B), it was apparent that the mean fiber length increased initially. As the skewness plateaued in the same time frame, it can be concluded that, in this phase, the fines are degraded at a faster rate than fiber fragmentation occurred. Fines are defined by the lower boundary of the FQA, below 0.07 mm in length. Both the mean fiber length and the skewness decreased, confirming the fragmentation of the long fibers into shorter more homogenous fragments (Fig. 2B). Once 25% to 30% of the cellulose had been hydrolyzed, the mean length of the fibers leveled out at ∼0.2 mm (Fig. 2B). Previous enzyme-mediated hydrolysis of softwood Kraft pulp found similar changes to the mean fiber length (13, 33). The CLSM images additionally indicated that the fiber fragments had approximately the same distance between major structural defects (“green zones,” Fig. 1A), as reported previously (12, 13, 16, 21, 27). This strongly suggested that fiber fragmentation had occurred at the more disorganized zones.
Quantifying Changes in the Supramolecular Cellulose Structure during Hydrolysis Time Using the CBM/CLSM Method and Computer-Based Image Analysis.
When the crystallinity of the pulp was assessed using 13C NMR, the initial NBSK was shown to have a CI of ∼0.52 (calculated via the NMR spectra) (SI Appendix, Fig. S1), which is on a par with Avicel (10). When the accessibility of the NBSK substrates was assessed using the CBM adsorption assay (Table 1), the CBM2aH6 to CBM17 binding ratio was 10, supporting the notion of a greater accessibility of the CBM2aH6, as compared to the CBM17. It should be noted that the adsorption data reflected accessibility to all of the binding sites throughout the substrate whereas the CBM/CLSM method was able to quantify the relative binding of the CBMs to the fiber surface, representing the optical section of the CLSM. One of the advantages of the CBM/CLSM method over the CBM adsorption and the NMR analysis is that it provided an estimate of the topography of the substrate, as well as spatial distribution of the cellulose morphologies, helping to better elucidate the nature and the spatial distribution of localized supramolecular structures.
Table 1.
The change of CBM17 and CBM2aH6 adsorption on NBSK over hydrolysis time
| Time, h | CBM2aH6, mgprotein⋅gcellulose−1 | CBM17, mgprotein⋅gcellulose−1 |
| 0 | 24.9 ± 0.5 | 2.4 ± 0.0 |
| 0.5 | 21.6 ± 1.5 | 2.3 ± 0.0 |
| 1 | 21.0 ± 2.0 | 1.9 ± 0.0 |
| 2 | 18.2 ± 1.3 | 1.5 ± 0.0 |
| 6 | 15.2 ± 0.9 | 1.6 ± 0.0 |
Depicted are mean values and the SD from triplicate experiments.
The ratio of red (from CBM2a-RRedX fluorescence) and green (from CBM17-FITC fluorescence) was then used to quantify changes in cellulose accessibility (Fig. 2C). When the red/green (R/G) ratios were plotted over hydrolysis time, the initial ratio (0 to 1 h) was constant and continuously increased afterward. Over prolonged hydrolysis, an increase in variance was observed (Figs. 2 and 3 and SI Appendix, Fig. S3), likely due to the development of different populations showing varying degrees of degradation (as exemplified in Fig. 1D). Over 6 h of hydrolysis, which resulted in ∼30% NBSK hydrolysis, the R/G ratio increased 5.3-fold. This increase in the R/G ratio was likely due to a loss in CBM17-FITC binding (Fig. 1), with the CBM adsorption data in Table 1 also showing a faster decline in CBM17 binding as compared to CBM2aH6 binding. As an increase in the R/G ratio implied an increase in surface crystallinity, 13C NMR analysis was used to assess the CI of the various samples. As summarized in SI Appendix, Fig. S1, the crystallinity had indeed increased from 0.529 to 0.543 within the first 2 h of hydrolysis, supporting the proposed increase in the overall crystallinity of the fiber. After 2 h of hydrolysis, it was apparent that the structure of the fibers had changed significantly, in both their size and supramolecular organization. Thus, it was difficult to ascribe changes in the 13C NMR spectra to just changes to the CI.
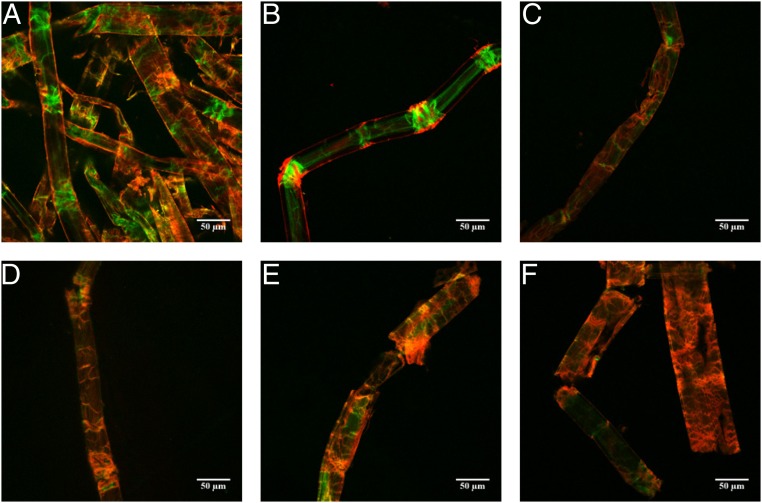
Fig. 3.
Visualization of fiber breakage of NBSK pulp fibers hydrolyzed for 0 h (A), 2 h (B–D), and 4 h (E and F) using CBM17-FITC and CBM2a-RRedX binding with CLSM. (Scale bars: 50 µm.)
During Initial Hydrolysis, the Disorganized Zones Are Predominantly Hydrolyzed, as Evidenced by CLSM and SEM Imaging.
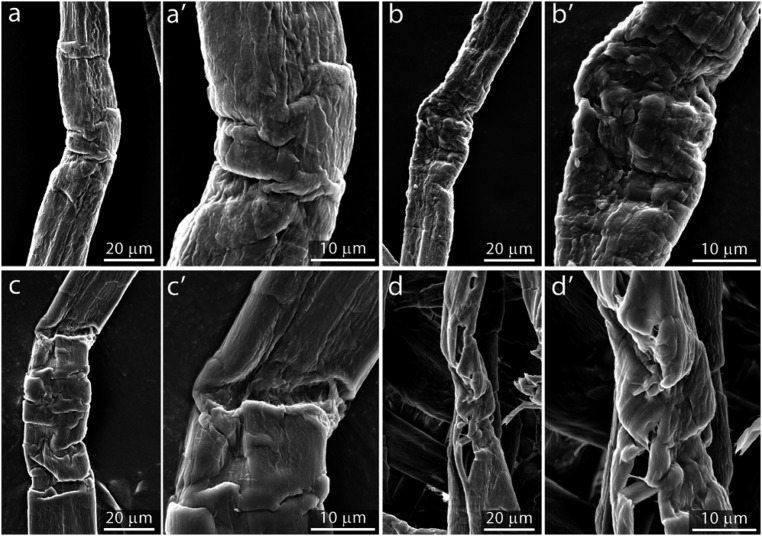
Higher magnification imaging by CLSM (Fig. 3) and SEM (Fig. 4) was employed to investigate localized morphological changes that resulted from the initial phase of digestion. CLSM images from the beginning of the reaction (Fig. 3 A and B) (0 to 1 h of hydrolysis time) suggested that enzymatic degradation was most extensive at the disorganized zones. With increasing hydrolysis time (Fig. 3 C–F), the disorganized zones increased in size, resulting in fiber shearing and breakage. When SEM was used to support these observations (Fig. 4), it appeared that those areas that had shown significant deformation (buckling of the S1/S2 layer) had developed into larger cracks and crevices (Fig. 4 A–C′), leading to the fragmentation and disruption of the fiber (Fig. 4 D and D′).
Fig. 4.
Visualization (SEM) of disorganized zones after 0.5 h (A and A′), 1 h (B and B′), 2 h (C and C′), and 4 h (D and D′) of incubation. (Magnification: A–D, 2,000×; A′–D′, 5,000×.)
Previous work that looked at the thermodynamics involved in enzymatic hydrolysis showed that the energy required for degradation of crystalline cellulose is much higher than the energy needed to hydrolyze a single glucan chain within paracrystalline cellulose (40). Thus, as indicated by the data presented in Figs. 1–4, it was likely that the disorganized zones were more quickly hydrolyzed, resulting in the observed fiber fragmentation.
Elucidating Differences in Enzyme- and Substrate-Induced Changes during Enzyme-Mediated Cellulose Hydrolysis.
Drying has been shown to result in hornification, an effect that leads to internal fiber volume shrinkage and the inability to regain the original state when resuspended in water. This collapse of the porous structure in turn has been shown to restrict enzyme accessibility to cellulose, negatively influencing hydrolyzability (41, 42). We next used the CBM/CLSM method to evaluate the effect drying might have on enzyme accessibility (CLSM images are shown in SI Appendix, Fig. S2). Although the initial hydrolysis profile of the never-dried pulp (Fig. 5A) was very similar to that of the dried NBSK pulp (Fig. 2A), the mean fiber length and skewness (Fig. 5B) and the R/G ratio (Fig. 5 C and D) differed significantly. In contrast to the initial increase in mean fiber length observed with the dried pulp (Fig. 2B), the mean fiber length of the never-dried pulp did not change until more than 15% of the cellulose had been hydrolyzed. In contrast, the skewness decreased at the onset of hydrolysis, implying that fragmentation occurred at the start of the reaction at the same time as the fines were being hydrolyzed. This indicated that, from the beginning of the hydrolysis reaction, the enzymes were able to access and degrade the disorganized zones within the never-dried pulp. This was supported by the R/G ratio profiles (Fig. 5 C and D) where an immediate and pronounced increase in the R/G ratio was observed for the same phase (0 to 1 h or 0% to 10% cellulose conversion) (Fig. 5 C and D) that showed stagnant R/G ratios when dried NBSK was the substrate (Fig. 2 C and D). In agreement with previous observations (41, 42), it is likely that the never-dried fibers had a higher accessibility to the enzymes due to the porous architecture of the cell wall. This is supported by previous CBM adsorption studies using CBM17 and CBM2aH6 adsorption to never-dried pulp (cf. MFC0 in ref. 11) where higher adsorption values were obtained as compared to the dried NBSK (Table 1). In contrast to the dried NBSK, the R/G ratio of the never-dried fiber plateaued between 1 and 4 h of hydrolysis (10% to 30% of cellulose conversion) (Fig. 5 C and D), signifying a similar change in binding for both CBMs. After 4 to 6 h, the R/G ratio increased, reaching a similar value as the dried NBSK (Fig. 2 C and D). Interestingly, over the same time frame, the mean fiber length and skewness showed a rapid decrease (Fig. 5B), suggesting that the fines were already degraded and enzyme action on the longer fibers resulted in their rapid fragmentation. It is possible that the static R/G ratio indicated that, due to fiber fragmentation, the accessibility of both CBMs was temporarily increased. However, after fragmentation, it is possible that the enzymes can act on the disordered regions at the ends of the fiber fragments, resulting in a drop in CBM17-FITC binding and the observed final rise in the R/G ratio.
Fig. 5.
Enzymatic hydrolysis of never-dried NBSK. Shown are (A) hydrolysis time course, (B) change in mean fiber length (filled circles) and skewness (empty circles), (C) change in R/G ratio over time, and (D) change in R/G ratio with increasing cellulose hydrolysis. Duplicate experiments were performed. Data in A and B show mean values and the spread. Data in C and D were derived from 8 to 10 image frames, and mean values and the SDs are shown.
Conclusions
Fluorescence-tagged CBMs were successfully combined with CLSM and quantitative image analysis to show that, during initial hydrolysis, enzyme-mediated deconstruction predominated at localized regions of cellulose showing lower degrees of organization. The higher concentration of the green fluorescent (caused by pronounced CBM17-FITC binding) suggested their more paracrystalline nature. When 13C NMR, SEM, and fiber characterization were combined with the CBM adsorption and CBM/CLSM data, it was apparent that fiber fragmentation and the preferential hydrolysis of the more disorganized fiber regions occurred at the same time. When the CBM/CLSM method was used to follow the hydrolysis of dried and never-dried Kraft pulp fibers, it was apparent that drying reduced cellulose accessibility, delaying the hydrolysis of the more accessible cellulose regions and the onset of fiber fragmentation.
Materials and Methods
Substrates and Chemicals Used.
Unless otherwise mentioned, all chemicals were obtained from Thermofisher Scientific (Massachusetts), and enzymes were obtained from Novozymes (Denmark). NBSK from handsheets and never-dried NSBK (MFC0) were used as substrates. NBSK handsheets were soaked in water at 1% consistency, and the fibers were redispersed nondisruptively with a standard disintegrator (Robert Mitchell Company Ltd, QC, Canada) for 15 min at 3,000 rpm (≤45,000 revolutions, TAPPI T205 standard) (43). The redispersed fibers were dewatered by filtration and washed in 2 L of water for 30 min under gentle stirring. Afterward, the fibers were again dewatered by filtration, washed with 2 L of water, and manually pressed out to a dry matter content of ∼40%. Conductivity measurements in the wash fractions confirmed the absence of soluble salts in the NBSK. The never-dried softwood Kraft pulp was prepared as described previously (cf. MFC0, ref. 11). The chemical composition of 2 substrates was determined following the protocol of the National Renewable Energy Laboratories (44). The cellulose, hemicellulose, and lignin content was, respectively, 75.6%, 8.2%, and 4.5% dry matter for NBSK and 80.4%, 8.7%, and 2.3% dry matter for the never-dried fiber.
Enzymatic Hydrolysis of NBSK and Never-Dried Pulp.
Hydrolysis reactions of NBSK were performed in 100-mL screw cap shake flasks with 25 g total reaction weight. Incubation was at 50 °C and 150 rpm in an incubator shaker (25 gIST-4075; GMI Inc., Minnesota). Never-dried pulp was hydrolyzed in 2-mL screw cap tubes with 1.5 g total reaction weight at 50 °C in a thermo block. The reaction was mixed by hourly vortexing. Parallel experiments showed that the change in agitation had no influence on either fiber fragmentation, hydrolysis efficiency, or the R/G ratio profiles (SI Appendix, Fig. S4). Due to the high viscosity of the reaction mixture, representative sampling was not possible. Thus, 2 individual reactions were prepared for each time point, and the total reaction mass was harvested. All of the reactions were performed in 20 mM sodium-acetate buffer (pH 4.8) at a substrate loading of 10 g dry mass⋅L−1. The commercial Ctec3 enzyme mixture was used (total cellulase and β-glucosidase activity of 205 FPU⋅mL−1 and 6,400 CBU⋅mL−1, respectively). The enzyme loading of all experiments was 10 FPU⋅g−1 dry mass, corresponding to 4.4 mg protein⋅mL−1. To stop the reaction, the mixtures were brought to 100 °C for 10 min and then stored at −20 °C for future analysis.
Analysis of Enzymatic Activities and Glucose Concentrations and Determination of Hydrolysis Yields.
Total cellulase activities (FPU) were measured as described by Ghose et al. (45). β-glucosidase activities were determined with p-nitro-phenyl β-d-1.4-glucopyranoside as substrate, following a previously established protocol (46). Released glucose was analyzed using a YSI Biochemistry Analyzer (2700 Select; Fisher Scientific, Massachusetts). The conversion of cellulose to glucose, Yg (% of theoretical maximum), was calculated by the following equation:
where Cg (g⋅L−1) is the concentration of solubilized glucose in the sample supernatant, Cg0 (g⋅L−1) is the initial glucose concentration, φG (-) is the molecular weight ratio of glucose to glucan monomer (φG = 180/162 = 1.11), Cis0 (g⋅L−1) is the initial concentration of insoluble solids, and xG0 (-) is the initial mass fraction of glucan in the insoluble solids. The equation assumes that: 1) the density of the hydrolysis slurry is 1,000 g⋅L−1, 2) the volume of the liquid equals that of the hydrolysis slurry, and 3) the volume of the liquid remains unchanged during the hydrolysis (47). These assumptions provide sufficient accuracy of the yield prediction at low solids loadings (47).
Analysis of the Supramolecular Structure of the NBSK Fibers Using the CBM/CLSM Method.
CBM production and preparation of the CBM-fluorescence dye conjugates.
CBM2a and CBM17 were sourced from Cellulomonas fimi and Clostridium cellulovorans, respectively (39, 48). The 2 CBMs, as well as a his-tagged version of CBM2a (CBM2aH6), were recombinantly expressed in Escherichia coli. The cultivation and purification strategy has been previously published elsewhere (11). In brief, a high cell density-fed batch cultivation with a carbon limited glycerol feed was run, typically achieving ∼100-g cell dry weight per liter. The cells were harvested by centrifugation and then disrupted chemically. Purification was performed by affinity chromatography using either cellulose (CBM2a, CBM17) or nickel beads (CBM2aH6) as carrier (11). The purity of CBMs was verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE).
After production and purification, the CBMs were concentrated to 10 g⋅L−1 in 10 mM bicarbonate buffer (pH 8.3), using sample concentrator tubes with a 3-kDa cutoff (Macrosep Advance Centrifugal Filter; Pall Laboratory, New York). Rhodamine red X N-hydroxysuccinimide (NHS) ester and Fluorescein NHS ester were each dissolved in DMSO to a concentration of 0.05 M. The dye solution was then mixed with the respective CBMs, resulting in a molar fluorescent dye to CBM ratio of 1.2. The reaction mixtures were incubated for 1 h at room temperature and for 12 h at 4 °C. The CBM-fluorescence dye conjugates were purified by size exclusion chromatography (HiPrep desalting 16/20 column; GE Healthcare Life Sciences, United Kingdom). SDS/PAGE was conducted to verify the correct sizes and purity of the CBM-fluorescence dye conjugates.
Sample preparation for CLSM imaging.
In samples taken from enzymatic hydrolyses, enzymes bound to the cellulose fibers were removed prior to CBM binding. For this, the fibers were washed 3 times by centrifuging (5,000 rpm, 10 min) and resuspending them in deionized water. After the last run, the fibers were taken up in SDS solution (SDS, 1% wt/vol) and incubated for 15 min at 85 °C. After cooling, the substrate samples were washed 2 times with absolute ethanol and 1 time with water. The washed samples were then suspended in PBS buffer (8 g⋅L−1 NaCl, 0.2 g⋅L−1 KCl, 1.44 g⋅L−1 Na2HPO4, 0.24 g⋅L−1 KH2PO4) and stored at 4 °C for CLSM analysis. Shortly before imaging, the fluorescence-tagged CBMs were mixed with the fibers, resulting in a protein loading of ∼5 μg of each CBM per gram of dry mass substrate. After a short incubation period (∼15 min, sufficient to ensure binding) (32, 39, 48), excess protein in the supernatant was removed by centrifuging to ensure minimal background fluorescence and high signal-to-noise ratio. The fibers were then gently resuspended in fresh PBS buffer. A small amount of sample was then mounted on objective glass, covered with a coverslip, and sealed with paraffin wax. Sample preparations and sample storage before CLSM imaging were conducted in the dark to prevent fluorescence quenching.
CLSM imaging.
Confocal laser scanning microscopy (CSLM) imaging was performed with an Olympus FV1000 (Olympus, Japan), using a 10× (numerical aperture [N.A.] 0.3 to 0.4) air objective, and a Zeiss 710 LSM (Carl Zeiss AG, Germany), using a 10× (N.A. 0.3 to 0.4) air objective and a 40× (N.A. 1.2) water objective. Following the specifications of the dyes, CBM17-FITC (green) was excited at 570 nm and emissions were detected at 590 to 650 nm. The excitation and emission of CBM2a-RRedX signal (red) was at 490 nm and 510 to 550 nm, respectively. Images were acquired in 5 to 6 µm (10×) and 0.4 to 0.5 µm (40×) thick optical sections. Bleed-through of fluorescence emission was tested and can be neglected for the presented image quantification method.
Quantitative CLSM image analysis.
Quantitative image analysis of the acquired CLSM images was performed to assess changes in concentrations of fluorophores and the distribution of structural features in the specimens with increasing cellulose conversion. High throughput could be achieved with a high degree of automation of the image analysis. The image analysis was performed in a Python environment (Build 2.7.10, Python Software Foundation) using open source image-processing toolboxes (49–52). A detailed description is given in SI Appendix. In brief, the red and green fluorescence layers of the acquired CLSM images were imported to the processing environment, the background values of the layers were subtracted from the fluorescence intensity data, and the data were scaled by the laser intensities. Point features (i.e., negative and saturated pixels) in the layers were removed from the corrected intensity datasets. Fibers were identified by semiautomated detection of the region of interest (ROI). In brief, a median filter with a 7 × 7 aperture was applied to reduce spatial noise in each layer. The filtered layers were segmented based on contrasting intensities between fibers and background by unsupervised thresholding using Otsu’s method (53) for the red layer and the triangle method (54) for the green layer. The triangle method was used for increased robustness for the latter because of the weaker bimodality of the intensity histogram. The ROI was defined as the union of the identified fiber subset of each layer. The ROI was used to mask the corrected fluorescence intensity layers, and the overall red to green ratio (R/G ratio) was calculated as the arithmetic mean of the spatially distributed ratios of the masked intensity layers.
Fiber Length Analysis.
The fiber length distributions of the hydrolysis samples were measured by a HiRes Fiber Quality Analyzer (LDA02-series; OpTest Equipment, ON, Canada). For this, fibers were dispersed in water and diluted to yield fiber counts of 30 to 50 s−1 in the analysis. The sample size was 10,000 fibers, and the data acquisition range was bounded by the lower detection limit, 0.07 mm (55), and 2.5 mm. Observations outside the data range were censored during data analysis. The arithmetic mean, median, and a nonparametric skewness (Pearson’s second skewness coefficient) of the fiber length distributions were derived from the resulting frequency tables (histograms). Data processing was performed in MATLAB R2017a (MathWorks Inc., Massachusetts), using the statistics and machine learning toolbox.
CBM Adsorption Analysis.
CBM adsorption onto NBSK cellulose fibers was quantified. For this, the fibers were washed with SDS to remove bound enzymes, as described above (Sample preparation for CLSM imaging). Afterward, a depletion assay using CBM2aH6 and CBM17 was performed as detailed elsewhere (11). In brief, CBMs (0.5 to 15 μM) were added to the NBSK (10 mg dry mass) in a final volume of 1 mL in PBS buffer. Samples were incubated for 30 min at room temperature, the supernatant was separated from the fibers by centrifugation, and the concentration of CBMs in the supernatant was quantified spectrophotometrically at 280 nm (Cary 50 Bio; Agilent Technologies, California). The molar extinction coefficients of CBM17 and CBM2aH6 have been described to be 31,010 M−1⋅cm−1 and 27,625 M−1⋅cm−1, respectively (32, 39).
NMR Analysis.
Solid-state NMR data were acquired on a Bruker 500-MHz instrument (Milton, Ontario, Canada). 13C CPMAS experiments were performed on a 2.5-mm HCN MAS probe, using RAMP-CP (56). Spinal-64 was used during the decoupling (57). All spectra were acquired with 10,240 scans, a recycle delay of 4 s, a 90° 1H pulse of 2.75 μs, and a cross-polarization time of 3 ms. The spectra were processed with a line broadening of 50 Hz and referenced to external adamantane. The assignments were as reported in ref. 10. The error of the integral can be assessed to be ±0.008, determined for the C4 peak of 3 consecutive runs.
Analysis of Disaggregated Zones by SEM.
Hydrated samples were washed and freeze-dried to prevent dehydration artifacts prior to imaging. Samples were mounted on aluminum stubs (VWR) using double-stick carbon tape and sputter coated with 7 nm of iridium. SEM imaging was performed using an FEI Quanta 400 FEG with accelerating voltages ranging from of 10 to 15 keV.
Supplementary Material
Acknowledgments
We thank Dr. Carl Houtman (Forest Products Laboratory, US Department of Agriculture, Madison, WI) for providing the underlying python code for the quantitative image analysis and Dr. Claus Felby (Copenhagen University, Denmark) for insightful discussions on dislocation zones. We thank the UBC BioImaging Facility (University of British Columbia, Vancouver, BC, Canada) and the Newcomb Imaging Center (Department of Botany, University of Wisconsin-Madison, Madison, WI) for providing access to the CLSMs. The Natural Science and Engineering Research Council of Canada (NSERC) and the Austrian Science Fund (Project J 4062-B21) are acknowledged for funding this work.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912354116/-/DCSupplemental.
References
- 1.Sassner P., Galbe M., Zacchi G., Techno-economic evaluation of bioethanol production from three different lignocellulosic materials. Biomass Bioenergy 32, 422–430 (2008). [Google Scholar]
- 2.Olofsson J., Barta Z., Börjesson P., Wallberg O., Integrating enzyme fermentation in lignocellulosic ethanol production: Life-cycle assessment and techno-economic analysis. Biotechnol. Biofuels 10, 51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y. H. P., Lynd L. R., Toward an aggregated understanding of enzymatic hydrolysis of cellulose: Noncomplexed cellulase systems. Biotechnol. Bioeng. 88, 797–824 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Mansfield S. D., Mooney C., Saddler J. N., Substrate and enzyme characteristics that limit cellulose hydrolysis. Biotechnol. Prog. 15, 804–816 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Meng X., Ragauskas A. J., Recent advances in understanding the role of cellulose accessibility in enzymatic hydrolysis of lignocellulosic substrates. Curr. Opin. Biotechnol. 27, 150–158 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Leu S.-Y., Zhu J. Y., Substrate-related factors affecting enzymatic saccharification of lignocelluloses: Our recent understanding. BioEnergy Res. 6, 405–415 (2013). [Google Scholar]
- 7.Hall M., Bansal P., Lee J. H., Realff M. J., Bommarius A. S., Cellulose crystallinity–A key predictor of the enzymatic hydrolysis rate. FEBS J. 277, 1571–1582 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Bansal P., et al. , Elucidation of cellulose accessibility, hydrolysability and reactivity as the major limitations in the enzymatic hydrolysis of cellulose. Bioresour. Technol. 107, 243–250 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Fan L. T., Lee Y.-H., Beardmore D. H., Mechanism of the enzymatic hydrolysis of cellulose: Effects of major structural features of cellulose on enzymatic hydrolysis. Biotechnol. Bioeng. 22, 177–199 (1980). [Google Scholar]
- 10.Park S., Baker J. O., Himmel M. E., Parilla P. A., Johnson D. K., Cellulose crystallinity index: Measurement techniques and their impact on interpreting cellulase performance. Biotechnol. Biofuels 3, 10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aïssa K., Novy V., Nielsen F., Saddler J. N., Use of carbohydrate binding modules to elucidate the relationship between fibrillation, hydrolyzability, and accessibility of cellulosic substrates. ACS Sustain. Chem. Eng. 7, 1113–1119 (2019). [Google Scholar]
- 12.Ander P., Hildén L., Daniel G., Cleavage of softwood kraft pulp fibers by HCl and cellulose. BioResources 3, 477–490 (2008). [Google Scholar]
- 13.Clarke K., Li X., Li K., The mechanism of fiber cutting during enzymatic hydrolysis of wood biomass. Biomass Bioenergy 35, 3943–3950 (2011). [Google Scholar]
- 14.Thygesen L. G., Gierlinger N., The molecular structure within dislocations in Cannabis sativa fibres studied by polarised Raman microspectroscopy. J. Struct. Biol. 182, 219–225 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Thygesen L. G., Hidayat B. J., Johansen K. S., Felby C., Role of supramolecular cellulose structures in enzymatic hydrolysis of plant cell walls. J. Ind. Microbiol. Biotechnol. 38, 975–983 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Ander P., Daniel G., Lindgren C. G., Marklund A., Characterization of industrial and laboratory pulp fibres using HCl, cellulase and FiberMaster analysis. Nord. Pulp Paper Res. J. 20, 115–121 (2005). [Google Scholar]
- 17.Suchy M., et al. , Effects of commercial cellobiohydrolase treatment on fiber strength and morphology of bleached hardwood pulp. Holzforschung 63, 731–736 (2009). [Google Scholar]
- 18.Nyholm K., Ander P., Bardage S., Daniel G., Dislocations in pulp fibres–Their origin, characteristics and importance–A review. Nord. Pulp Paper Res. J. 16, 376–384 (2001). [Google Scholar]
- 19.Hidayat B. J., Felby C., Johansen K. S., Thygesen L. G., Cellulose is not just cellulose: A review of dislocations as reactive sites in the enzymatic hydrolysis of cellulose microfibrils. Cellulose 19, 1481–1493 (2012). [Google Scholar]
- 20.Eder M., Terziev N., Daniel G., Burgert I., The effect of (induced) dislocations on the tensile properties of individual Norway spruce fibres. Holzforschung 62, 77–81 (2008). [Google Scholar]
- 21.Terziev N., Daniel G., Marklund A., Dislocations in norway spruce fibres and their effect on properties of pulp and paper. Holzforschung 59, 163–169 (2005). [Google Scholar]
- 22.Thygesen L. G., Eder M., Burgert I., Dislocations in single hemp fibres—investigations into the relationship of structural distortions and tensile properties at the cell wall level. J. Mater. Sci. 42, 558–564 (2007). [Google Scholar]
- 23.Hernandez-Estrada A., Gusovius H.-J., Müssig J., Hughes M., Assessing the susceptibility of hemp fibre to the formation of dislocations during processing. Ind. Crops Prod. 85, 382–388 (2016). [Google Scholar]
- 24.Thygesen L. G., Bilde-Sørensen J. B., Hoffmeyer P., Visualisation of dislocations in hemp fibres: A comparison between scanning electron microscopy (SEM) and polarized light microscopy (PLM). Ind. Crops Prod. 24, 181–185 (2006). [Google Scholar]
- 25.Keckes J., et al. , Cell-wall recovery after irreversible deformation of wood. Nat. Mater. 2, 810–814 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Dai D., Fan M., Investigation of the dislocation of natural fibres by Fourier-transform infrared spectroscopy. Vib. Spectrosc. 55, 300–306 (2010). [Google Scholar]
- 27.Thygesen L. G., Quantification of dislocations in hemp fibers using acid hydrolysis and fiber segment length distributions. J. Mater. Sci. 43, 1311–1317 (2008). [Google Scholar]
- 28.Ciesielski P. N., et al. , Nanomechanics of cellulose deformation reveal molecular defects that facilitate natural deconstruction. Proc. Natl. Acad. Sci. U.S.A. 116, 9825–9830 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox J. M., et al. , A single-molecule analysis reveals morphological targets for cellulase synergy. Nat. Chem. Biol. 9, 356–361 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Yang F., et al. , A mini-review on the applications of cellulose-binding domains in lignocellulosic material utilizations. BioResources 10, 6081–6094 (2015). [Google Scholar]
- 31.Jamal-Talabani S., et al. , Ab initio structure determination and functional characterization of CBM36; a new family of calcium-dependent carbohydrate binding modules. Structure 12, 1177–1187 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J., Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gourlay K., Hu J., Arantes V., Penttilä M., Saddler J. N., The use of carbohydrate binding modules (CBMs) to monitor changes in fragmentation and cellulose fiber surface morphology during cellulase- and swollenin-induced deconstruction of lignocellulosic substrates. J. Biol. Chem. 290, 2938–2945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filonova L., et al. , Analysis of the surfaces of wood tissues and pulp fibers using carbohydrate-binding modules specific for crystalline cellulose and mannan. Biomacromolecules 8, 91–97 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Hildén L., Daniel G., Johansson G., Use of a fluorescence labelled, carbohydrate-binding module from Phanerochaete chrysosporium Cel7D for studying wood cell wall ultrastructure. Biotechnol. Lett. 25, 553–558 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Ding S. Y., et al. , Versatile derivatives of carbohydrate-binding modules for imaging of complex carbohydrates approaching the molecular level of resolution. Biotechniques 41, 435–436, 438, 440 passim (2006). [DOI] [PubMed] [Google Scholar]
- 37.Kawakubo T., et al. , Analysis of exposed cellulose surfaces in pretreated wood biomass using carbohydrate-binding module (CBM)-cyan fluorescent protein (CFP). Biotechnol. Bioeng. 105, 499–508 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Lehtiö J., et al. , The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules. Proc. Natl. Acad. Sci. U.S.A. 100, 484–489 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLean B. W., et al. , Carbohydrate-binding modules recognize fine substructures of cellulose. J. Biol. Chem. 277, 50245–50254 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Beckham G. T., et al. , Molecular-level origins of biomass recalcitrance: Decrystallization free energies for four common cellulose polymorphs. J. Phys. Chem. B 115, 4118–4127 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Esteghlalian A. R., Bilodeau M., Mansfield S. D., Saddler J. N., Do enzymatic hydrolyzability and Simons’ stain reflect the changes in the accessibility of lignocellulosic substrates to cellulase enzymes? Biotechnol. Prog. 17, 1049–1054 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Mansfield S. D., de Jong E., Stephens R. S., Saddler J. N., Physical characterization of enzymatically modified kraft pulp fibers. J. Biotechnol. 57, 205–216 (1997). [Google Scholar]
- 43.Tappi T., Forming Handsheets for Physical Tests of Pulp (Technical Association of the Pulp and Paper Industry, 2000). [Google Scholar]
- 44.Sluiter A., et al. , Determination of Structural Carbohydrates and Lignin in Biomass (National Renewable Energy Laboratory, Golden, CO, 2012). [Google Scholar]
- 45.Ghose T. K., Measurement of cellulase activities. Pure Appl. Chem. 59, 257–268 (1987). [Google Scholar]
- 46.Zhang Y. H. P., Hong J., Ye X., “Cellulase assays” in Biofuels: Methods and Protocols, Mielenz J. R., Ed. (Humana Press, 2009), pp. 213–231. [Google Scholar]
- 47.Zhu Y., Malten M., Torry-Smith M., McMillan J. D., Stickel J. J., Calculating sugar yields in high solids hydrolysis of biomass. Bioresour. Technol. 102, 2897–2903 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Notenboom V., et al. , Recognition of cello-oligosaccharides by a family 17 carbohydrate-binding module: An X-ray crystallographic, thermodynamic and mutagenic study. J. Mol. Biol. 314, 797–806 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Linkert M., et al. , Metadata matters: Access to image data in the real world. J. Cell Biol. 189, 777–782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamprecht M. R., Sabatini D. M., Carpenter A. E., CellProfiler: Free, versatile software for automated biological image analysis. Biotechniques 42, 71–75 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Clark A., et al. , The friendly PIL fork (Version 5.2.0, 2018) https://github.com/python-pillow/Pillow. Accessed 14 October 2019.
- 52.Bradsky G., Pisarevsky V., Itzees W. G., OpenCV (Version 3.4.3, 2018). https://opencv.org/. Accessed 14 October 2019.
- 53.Otsu N., A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9, 62–66 (1979). [Google Scholar]
- 54.Zack G. W., Rogers W. E., Latt S. A., Automatic measurement of sister chromatid exchange frequency. J. Histochem. Cytochem. 25, 741–753 (1977). [DOI] [PubMed] [Google Scholar]
- 55.Li B., Bandekar R., Zha Q., Alsaggaf A., Ni Y., Fiber quality analysis: OpTest fiber quality analyzer versus L&W fiber tester. Ind. Eng. Chem. Res. 50, 12572–12578 (2011). [Google Scholar]
- 56.Metz G., Wu X. L., Smith S. O., Ramped-amplitude cross polarization in magic-angle-spinning NMR. J. Magn. Reson. 110, 219–227 (1994). [Google Scholar]
- 57.Comellas G., Lopez J. J., Nieuwkoop A. J., Lemkau L. R., Rienstra C. M., Straightforward, effective calibration of SPINAL-64 decoupling results in the enhancement of sensitivity and resolution of biomolecular solid-state NMR. J. Magn. Reson. 209, 131–135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.