Significance
Viruses need to package nucleic acid genomes so that the genetic material (either RNA or DNA) can survive outside of a cell and without the DNA repair that takes place within all cells. We have studied the structure of an icosahedral virus that infects a host living in nearly boiling acid. The outer capsid which covers the genome-containing membrane vesicle is composed of 2 proteins that have a variation of a previously observed fold. The genomic DNA is in the A-form first described for desiccated DNA, rather than the B-form more commonly found in biology. A-DNA may be the prevalent storage form of DNA in everything from bacterial spores to viruses inhabiting the most extreme environments.
Keywords: DNA structure, hyperthermophilic acidophiles, cryo-EM
Abstract
Studies on viruses infecting archaea living in the most extreme environments continue to show a remarkable diversity of structures, suggesting that the sampling continues to be very sparse. We have used electron cryo-microscopy to study at 3.7-Å resolution the structure of the Sulfolobus polyhedral virus 1 (SPV1), which was originally isolated from a hot, acidic spring in Beppu, Japan. The 2 capsid proteins with variant single jelly-roll folds form pentamers and hexamers which assemble into a T = 43 icosahedral shell. In contrast to tailed icosahedral double-stranded DNA (dsDNA) viruses infecting bacteria and archaea, and herpesviruses infecting animals and humans, where naked DNA is packed under very high pressure due to the repulsion between adjacent layers of DNA, the circular dsDNA in SPV1 is fully covered with a viral protein forming a nucleoprotein filament with attractive interactions between layers. Most strikingly, we have been able to show that the DNA is in an A-form, as it is in the filamentous viruses infecting hyperthermophilic acidophiles. Previous studies have suggested that DNA is in the B-form in bacteriophages, and our study is a direct visualization of the structure of DNA in an icosahedral virus.
The morphological diversity of DNA viruses which infect archaea thriving in extreme geothermal environments is astounding (1, 2). Along with common filamentous and spherical viruses, it includes viruses with diverse and unusual shapes resembling bottles, droplets, coils, and spindles. The morphogenesis of the majority of these viruses involves formation of a nucleoprotein filament—from the genomic DNA and capsid proteins—and its condensation into bodies of different shapes. In case of the Sulfolobus polyhedral virus 1 (SPV1) from the family Portogloboviridae, such a nucleoprotein filament is condensed into the globular core, which is surrounded by a lipid envelope and further encased by an outer polyhedral capsid (3). The general design of the virion superficially resembles that of the icosahedral bacterial and archaeal viruses with an inner lipid envelope (families Tectiviridae, Corticoviridae, Sphaerolipoviridae, and Turriviridae), in which, however, DNA is present in the naked form (4–9).
We have used electron cryo-microscopy (cryo-EM) to study in detail the structure of SPV1. We have been able to determine structures for the 2 outer capsid proteins (VP4 and VP10), visualize subcapsomer proteins linking the outer capsid to the icosahedral inner membrane, determine a structure for the VP1-wrapped circular double-stranded DNA (dsDNA) genome, and generate a model for the packing of the nucleoprotein core into 5 concentric shells within the membrane.
Results
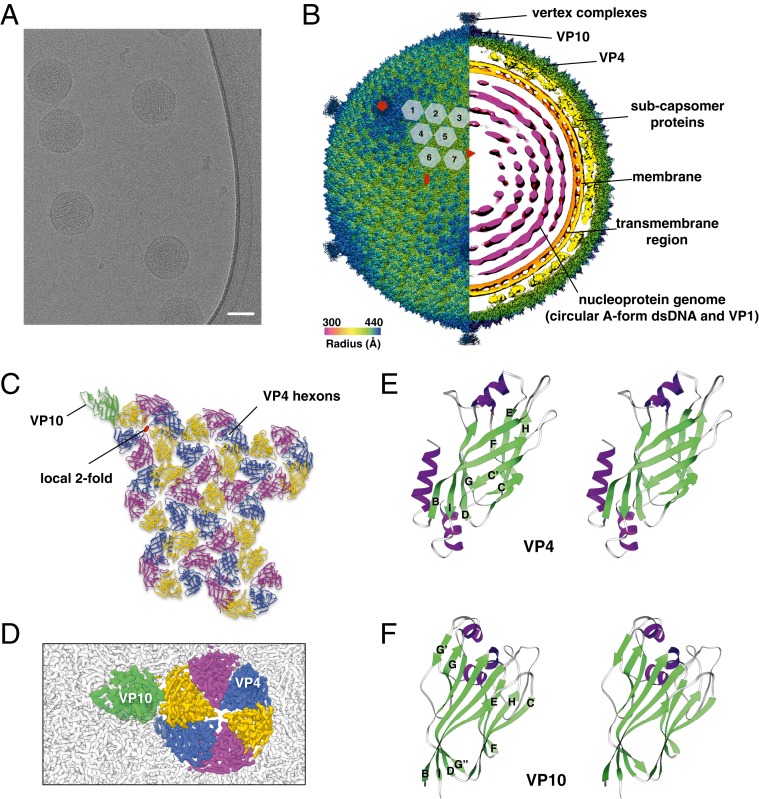
To obtain an atomic description of the SPV1 capsid, we used cryo-EM imaging (Fig. 1A). A 3.7-Å resolution icosahedrally averaged reconstruction of SPV1 was calculated with RELION (10) combined with Ewald sphere correction (11), using 19,092 images of virion particles. The reconstructed virion (Fig. 1B) is 87 nm in diameter and has a T = 43 surface lattice, where T is the triangulation number. There will thus be 420 hexamers and 12 pentamers in this icosahedral surface, and the icosahedral asymmetric unit (IAU) contains 7 hexamers and 1 subunit from the pentamer (Fig. 1C). Such a capsid arrangement has not been previously described in viruses. A comparison of icosahedral capsid size versus triangulation number shows that SPV1 is consistent with this relation for β-barrel fold capsids, while being significantly smaller in diameter than an HK97-fold virus having a similar T value (SI Appendix, Fig. S1).
Fig. 1.
Cryo-EM of SPV1 and its major capsid proteins. (A) Cryo-EM image of SPV1. (Scale bar, 50 nm.) (B) Overall cryo-EM density map of SPV1, rendered to display the outer capsid (left half) and a central slice showing the interior of the virus (right half). The virus is colored by radius. (C) The icosahedral asymmetric unit of SPV1, with VP10 protein and VP4 hexons labeled. (D) Zoomed in cryo-EM map of a capsid region showing VP10 (green) and VP4 hexons (yellow, blue, and magenta). The surrounding capsid is colored gray. (E and F) “Wall-eyed” stereo views of atomic models of VP4 (E) and VP10 (F) show variations of the jelly-roll fold.
The cryo-EM reconstruction clearly revealed 3 layers of SPV1: the capsid, the subcapsomer proteins and membrane, and the genome wrapped inside the membrane (Fig. 1B). At this resolution (SI Appendix, Fig. S2 A and B), we were able to build de novo an atomic model of the SPV1 major capsid protein (MCP) VP4 (Fig. 1 B–D and SI Appendix, Fig. S3) into the hexameric densities. The VP4 proteins arrange into hexons which have a weak 6-fold symmetry and strong pseudo–2-fold symmetry. This is due to the fact that the VP4 subunits are not strictly related to each other by a 6-fold symmetry, as well as by the fact that here are some minor variations when the 6 subunits in the same hexon are aligned. The nonequivalence likely has the following 2 causes: 1) the interactions between different VP4s and the subcapsomer protein array are not equivalent and 2) the interactions between a VP4 and adjacent hexons are not equivalent as well, due to the curvature of the capsid and the location of the pentons. It was clear that the VP4 atomic model could not explain the pentameric densities located at the 5-fold axes in the virion. We therefore searched for other proteins encoded in the SPV1 genome (3) and found VP10 (open reading frame 27), not previously identified as a structural protein due to its low abundance (the stoichiometry of VP4:VP10 is 42:1). As with VP4, we were able to build de novo an atomic model for the pentamers formed by VP10 (Fig. 1 D and F). Despite the limited sequence identity between VP4 and VP10 (∼15%), they share a high degree of structural similarity with a Z-score of 6.9, and both display a variation of the “jelly-roll” fold (12), a prevalent fold in viral capsid proteins (13). Unlike the classical jelly-roll fold, first observed in tomato bushy stunt virus (14), which contains 2, 4-stranded β-sheets (BIDG and CHEF), VP4 and VP10 have additional β-strands inserted next to the C-strand (C′) and G-strand (G′, G′′), respectively (Fig. 1 E and F and SI Appendix, Fig. S4).
Vertex complexes were observed above all VP10 pentamers (SI Appendix, Fig. S5 A and B), even when icosahedral symmetry was not applied (eliminating the possibility that some pentamers did not contain this spike). Most of the vertex complex was not as ordered as the major capsid proteins and thus had a very limited resolution. However, in the middle of the VP10 pentamers there are extra helices extending from the vertex complex that make interactions with VP10 and thus anchor the spikes in the pentamers (SI Appendix, Fig. S5 C and D). Pentamers that function as adaptors for spikes appear to be a general theme among dsDNA viruses with β-barrel MCPs (4, 6, 15, 16). In addition, 5 of the 6 jumbophages (tailed bacteriophages with genomes larger than 200 kbp) studied had HK97 folds with extra density above the pentons (17).
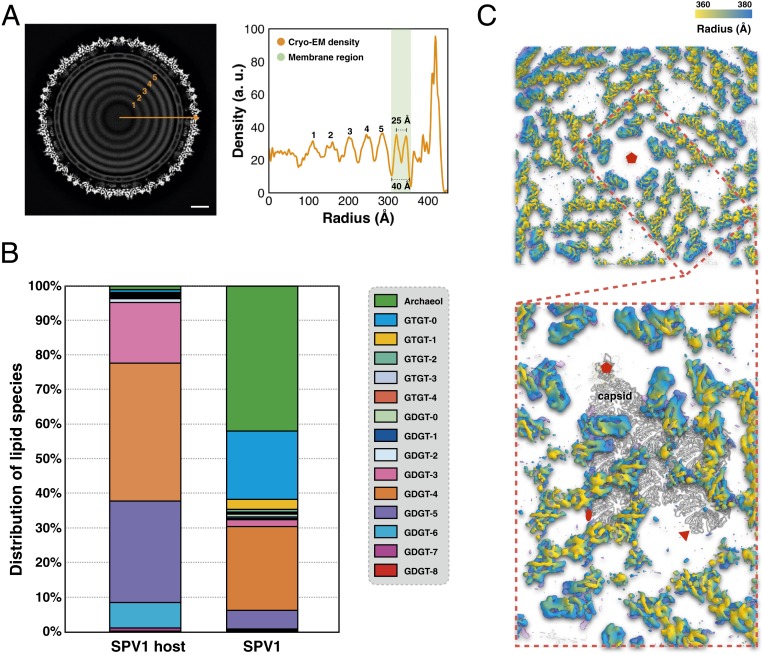
The icosahedral reconstruction of the SPV1 virus shows that the membrane has an inner and outer density peak (Figs. 1B and 2A) due to the lipid headgroups, as seen in many bilayer structures. The thickness of the membrane is comparable with bilayers in other icosahedral viruses (8, 18): the distance between the inner and outer peaks is ∼25 Å, and the total thickness is about 40 Å (Fig. 2A). The lipid composition of the SPV1 membrane and its host has been analyzed by liquid chromatography with tandem mass spectrometry (Fig. 2B). Interestingly, the distribution of the lipid species in SPV1 virions is very different from that in the host. The host membrane is dominated (97%) by membrane-spanning C40 glycerol dibiphytanyl glycerol tetraether (GDGT) species, especially GDGT-3, GDGT-4, GDGT-5, and GDGT-6. In contrast, the virion membrane is specifically enriched in the archaeol (42%) and GDGT-0 (20%), which together represent less than 2% of the host lipid. Such abnormal selection of archaeol or GDGT-0 was also observed in 2 helical viruses, SFV1 (19) and AFV1 (20), and a spindle-shaped virus, SSV1 (21), living in similar environmental settings and infecting similar Sulfolobales strains.
Fig. 2.
Subcapsomer architecture. (A) Central slice of the icosahedral cryo-EM reconstruction (Left) and its radial density profile (Right). (Scale bar, 100 Å.) (B) Distribution of the lipid species identified in Sulfolobus S38A (SPV1 host) and SPV1 virions. (C) Protein array connecting capsid and membrane. These proteins are colored by radial distance from the center of the virion.
As with several other icosahedral viruses (6, 8, 18, 22), the SPV1 membrane displays an icosahedral shape, not a spherical one, presumably due to the presence of the subcapsomer protein array that not only inserts into the membrane bilayer but also spans the space between the membrane and outer capsid (Fig. 2C). As described previously (3), VP1–VP3 reside mostly within the SPV1 membrane, whereas VP4–VP10 are found outside the membrane. Predictions of transmembrane regions using the TMHMM server (23) suggest with high confidence that VP3, VP5, VP6, and VP7 have transmembrane helices (SI Appendix, Fig. S6). Since VP3 is a small protein (119 residues) predicted to be nearly all α-helical and the protein abundance is the same in particles with or without the outer shell (3), it is very likely that VP3 is an integral membrane protein. On the other hand, VP5, VP6, and VP7 have similar length (∼300 residues), but their secondary structures are predicted to be quite different (SI Appendix, Fig. S7). This is consistent with the observed subcapsomer array, in which there are clearly several different proteins within the IAU. Unfortunately, given the limited resolution in this area (∼7 Å), those densities cannot be segmented unambiguously to build atomic models. This also indicates that the subcapsomer is not as ordered as the capsid region after icosahedral averaging.
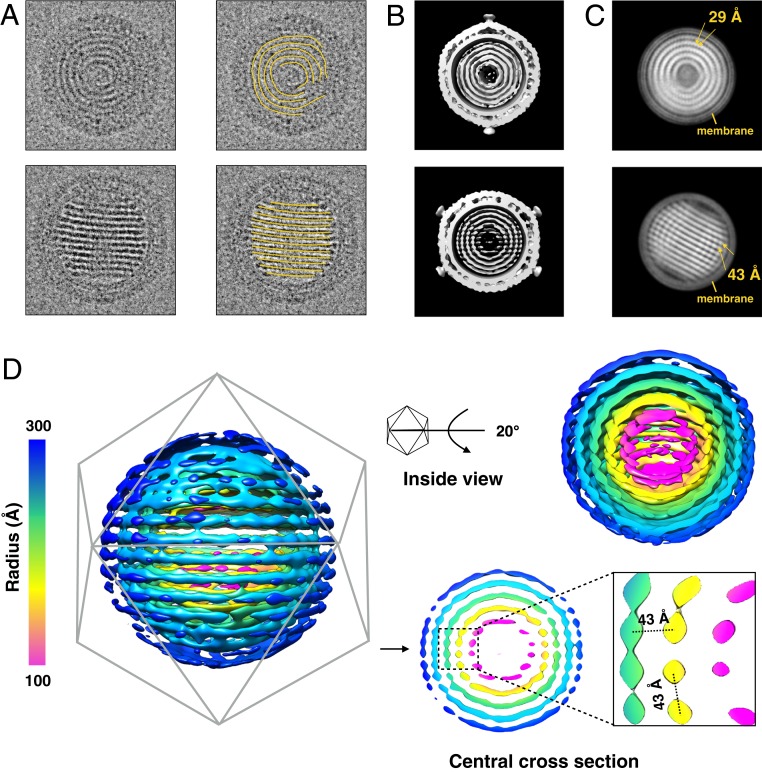
In the reconstruction of SPV1, the genome was seen as 5 discrete concentric spheres after applying icosahedral symmetry (Fig. 2A and SI Appendix, Fig. S8). Interestingly, 2 special views were frequently observed in the raw electron microscopy (EM) micrographs (Fig. 3A): one with discontinuous ring-like features and the other with parallel striations, which we ascribe to being top and side views, respectively. To better visualize the SPV1 genome, asymmetrical reconstructions were done using both RELION (10) and JSPR (24), starting with the icosahedrally averaged volume as an initial reference but only allowing it to move among 1 of the 60 possible orientations related by icosahedral symmetry. Both software packages achieved similar results with ∼35-Å resolution (Fig. 3B). The central portion of the asymmetrical reconstruction shows similar patterns as the “top view” and the “side view” (Fig. 3B). Using a 700-Å diameter spherical mask excluding most of the virion outside the membrane, 2 RELION class averages representing the top view and the side view were obtained (Fig. 3C). In the top view class average, the rings are not uniform: The rings are smeared in some regions, while in the remaining regions there is a strong 29-Å periodicity along the genome filament. On the other hand, the separation in the side view is uniformly quite sharp, with a spacing of ∼43 Å between striations. This 43-Å separation corresponds to the separation between filaments, which can also be clearly observed in the central cross-section of the asymmetrical reconstruction (Fig. 3D) from the following 2 different directions: 1) between axial layers (e.g., yellow to yellow) as also shown in Fig. 3C and 2) between radial shells (e.g., green to yellow, or yellow to magenta). In contrast to the known naked dsDNA genome in many icosahedral viruses where the spacing is 25 to 29 Å between shells (25–28), the 43-Å spacing within the SPV1 genome strongly suggests that the genome is actually a nucleoprotein filament formed by VP1 copies bound to dsDNA. This is supported by several lines of evidence: 1) Several helical viruses (SIRV2, AFV1, and SFV1) with a nucleoprotein genome also have similar 43-Å interfilament distances (19, 20, 29). 2) Within the SPV1 membrane, there is a large amount of soluble VP1 protein present, even when compared with the MCP VP4 (3). (3) Recombinant VP1 expressed and purified from Escherichia coli binds dsDNA in vitro by a gel shift assay at temperatures of 65 to 85 °C (SI Appendix, Fig. S9). 4) The thickness of the filaments in the asymmetrical reconstruction appears too large to be naked dsDNA (Fig. 3D).
Fig. 3.
Asymmetrical reconstruction of SPV1. (A) Two distinctive views of SPV1 from the raw EMs, showing concentric rings (Top) and striations (Bottom). (B) The central portion of the asymmetrical reconstruction of SPV1 filtered to 40 Å in the corresponding orientations shown in A. (C) Two-dimensional class averages of masked SPV1 (excluding the outer capsid) match both views seen in A. In the Top average, there is a strong helical periodicity along the nucleoprotein filaments with a pitch of 29 Å. In the Bottom average, the 43-Å spacing between layers is evident. (D) The same EM map in B showing the genome region only, colored by radius. A central section (Bottom Right) highlights the conserved 43-Å spacing between layers and between filaments in the same layer.
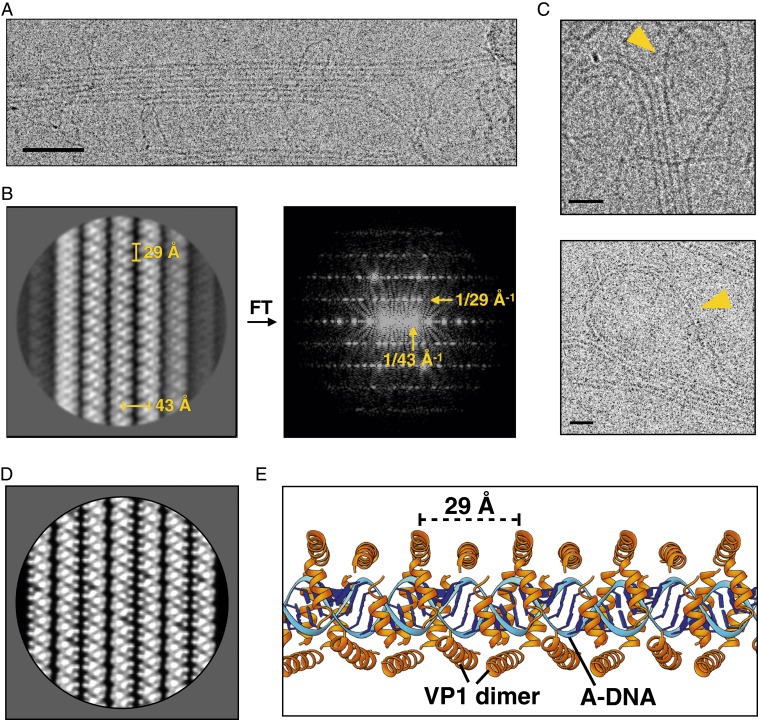
To further understand the SPV1 genome packing, the virions were dissociated by multiple cycles of freezing and thawing, which result in the disruption of capsids and the majority of enveloped cores. By this means the genome was released from the particles. Electron microscopic examination of the extracted genome revealed 4 types of filaments: naked dsDNA, single nucleoprotein filaments, supercoiling bundles of nucleoprotein filaments, and 2-dimensional (2D) rafts of nucleoprotein filaments (Fig. 4A and SI Appendix, Fig. S10). A 2D class average was generated after manually picking ∼6,000 raft-like regions (Fig. 4B). In this class average, the 29-Å periodicity within each filament is clearly seen. The canonical model (30) for A-form DNA has 11 base pairs (bp) per turn, with a rise of 2.6 Å/bp, generating a pitch of 29 Å. It has previously been shown for the filamentous viruses AFV1 (20), SIRV2 (29), and SFV1 (19) that the twist values of A-form DNA are 10.8, 11.2, and 11.3 bp per turn, respectively, all bracketing 11.0 bp/turn. We suggest that the twist of A-form DNA in SPV1 is 11.0 bp/turn, which explains why no helical features are seen and every turn of the 29-Å pitch filament is identical in projection. The interfilament distance in this class average is 43 Å, consistent with the spacings in the asymmetrical reconstruction. In addition to this, in the class average each filament is a mirror image of an adjacent filament, showing that each is rotated azimuthally by 180° from the neighboring filament. This indicates that the interactions between filaments are quite specific, and not the result of random aggregation. All evidence suggests that the interactions within these rafts are the same as within the intact membrane-enveloped genome, and attractive interactions between filaments must hold these rafts together after the membrane is ruptured, in stark contrast to the strongly repulsive interactions that exist between neighboring helices in capsids containing naked dsDNA (25).
Fig. 4.
Cryo-EM of the disrupted SPV1 nucleoprotein genome. (A) Electron micrograph of extracted SPV1 genome shows filaments bundled into 2D rafts. (Scale bar, 500 Å.) (B) Two-dimensional class average of rafts (Left) and its power spectrum (Right) shows the 29-Å VP1-dimer axial repeat and the 43-Å interfilament spacing. (C) Electron micrographs show the adjacent filaments within the same raft connected by loops. (Scale bars, 200 Å.) (D) Two-dimensional raft projection generated from a VP1-DNA filament model, filtered to a 10-Å resolution. (E) A model for the nucleoprotein filament showing A-form DNA (blue) and VP1 dimers (orange) wrapped around the DNA.
To reconstruct the nucleoprotein filament, segments boxed from both the rafts and free single filaments were combined. The helical symmetry was determined to be a 29 Å rise with a 0.0° twist (1.0 subunits per turn), but a helical reconstruction could never be obtained via Iterative Helical Real Space Reconstruction (31) that had better than ∼30-Å resolution. The simplest explanation was that the dataset was dominated by raft images which provide only one unique projection of the filament (the 180° projection is simply the mirror image of the unique view), and the free filaments failed to adequately cover other views. However, the reconstruction showed clear dihedral symmetry (i.e., the filaments are bipolar), meaning that there must be 2 copies of VP1 within the 29-Å asymmetrical unit forming a symmetrical dimer. The A-form DNA will also have dihedral symmetry when the sequence is ignored. The secondary structure of VP1 was predicted to be 4 α-helices with similar length (SI Appendix, Fig. S8). We therefore attempted to build a model using the ∼7-Å resolution 2D class average as a constraint. Both A-form DNA and the VP1 helices can be built into a model (Fig. 4E), the projection of which (Fig. 4D) matches the class average (Fig. 4B) extremely well. The sequence of VP1 is enriched in arginine, lysine, and asparagine (22% in total), and this is significantly higher than the other 9 SPV1 virion proteins (ranging from 8 to 13%). Those 3 types of residues can form strong polar contacts with the backbone phosphate groups in the DNA, and such enrichments were also seen in the nucleoprotein complexes in the helical virions SIRV2 (29), SFV1 (19), and AFV1 (20).
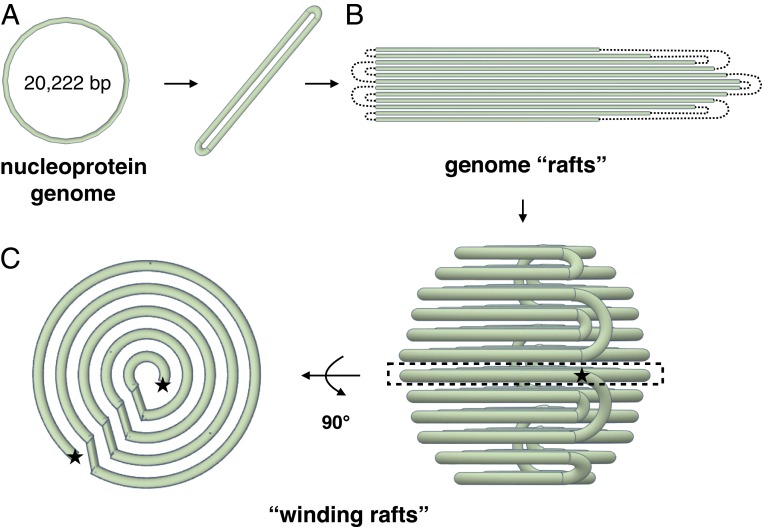
As described previously, the SPV1 genome is a single circular dsDNA molecule. Given this, an interesting question is how the filaments within the rafts are connected to each other. Fortunately, such connections were captured in micrographs showing that the adjacent filaments in the rafts can be connected to each other by loops (Fig. 4C). This has allowed us to combine a number of pieces of information to build a model for the overall packing of the circular genome in the virion (Fig. 5). The circular DNA is best thought of as a long linear double filament with loops at each end (Fig. 5A). This double filament then folds to form a 2D raft with loops connecting adjacent filaments (Fig. 5B). This raft is then spooled into concentric shells (Fig. 5C), giving rise to the 2 characteristic views: the concentric rings, seen in the top view, and the striations, seen in the side view. Since there are crossings between shells, the top views (Fig. 3 A and C) do not show complete concentric rings, but the side views (Fig. 3 A and C) do show sharp and uniform striations.
Fig. 5.
“Spooling raft” model of the SPV1 genome packing. (A) The dsDNA nucleoprotein genome of SPV1 is circular, but the packing involves a long double filament with connecting loops at each end. (B) Rafts of SPV1 filaments with dotted lines indicating the connecting loops. (C) Spooling raft model of SPV1 genome packing. (Left) A central slice is shown from a top view, where one can see both the concentric shells and the crossings between shells. (Right) A side view giving rise to the sharp striations seen in projection (Fig. 3A).
Discussion
We have determined the structure of SPV1, the sole representative of the family Portogloboviridae. We find that the 2 capsid proteins, VP4 and VP10, forming the external icosahedral capsid have a variant of the β-barrel fold, widely known as the single jelly roll, which is one of the most common structural motifs in capsid proteins of viruses with either RNA or DNA genomes (12). Previous structural studies have revealed 2 major ways in which jelly-roll capsid proteins are used to build icosahedral shells. In small RNA and single-stranded DNA viruses, the axis of the β-barrel is parallel to the capsid surface, whereas, in larger dsDNA viruses, the capsid proteins are oriented so that the axis of the β-barrel is roughly perpendicular to the capsid (32). It has been suggested that during evolution capsid proteins in the 2 virus groups have been recruited from cellular ancestors independently of each other (12). Furthermore, 2 variations in bacterial and archaeal dsDNA viruses with internal membranes and jelly-roll MCPs have been described previously: 1) viruses with hexagonal capsomers formed from a trimer of double jelly-roll MCPs, where 2 jelly-roll motifs are fused into a single polypeptide chain (families Corticoviridae, Tectiviridae and Turriviridae) and 2) viruses with hexagonal capsomers formed from heterohexamers of 2 different single jelly-roll MCPs (family Sphaerolipoviridae). In contrast, the capsid proteins of SPV1 are arranged in a manner different from that observed in these other viruses. The SPV1 virion is formed from a single MCP which forms homohexamers and, unlike in other known viruses, the axis of the β-barrel in SPV1 MCP is tilted 30° with respect to the normal to the capsid surface. Tailed dsDNA viruses have the HK97 capsid protein fold, and these subunits can be organized into both hexamers and pentamers. But dsDNA viruses with β-barrel folds—adenovirus, PRD1, and others—have trimeric “hexons” that feature 3 pairs of β-barrels and separately encoded penton subunits featuring a single β-barrel. SPV1 seems to share features of both the HK97-tailed capsid proteins with a pentamer–hexamer organization and the adenovirus capsid proteins (β-barrel, separate pentamer, and hexamer proteins), but the hexamer subunit has only one β-barrel. A comparison of the SPV1 capsid protein organization with other single and double jelly-roll proteins is shown in SI Appendix, Fig. S11.
The 2 capsid proteins, VP4 and VP10, display the same fold, but show very low pairwise sequence identity (∼15%). A similar observation has been made in the case of the filamentous virus SFV1 (19) where the sequences of the 2 MCPs were found to have no significant similarity to each other or to any other proteins in all existing databases using BLAST (33). However, structural studies revealed that the 2 proteins each had the same overall fold, and both were clearly homologs of proteins characterized in 2 other filamentous viruses, SIRV2 (29) and AFV1 (20). From the same environmental sample, by metagenomic sequencing, we were able to recover another filamentous virus, SBFV2, with capsid proteins that are homologous to those of SFV1 (34). We expect that greatly increased sampling of icosahedral viruses in these environments should reveal structural homologs of VP4 and VP10.
Many studies have explored how naked dsDNA is packaged under pressures of 10s of atmospheres within bacterial and eukaryotic viruses (25, 35–41). This pressure arises both from the mutual repulsion of the negatively charged phosphate backbone of the DNA and from the need to tightly bend the semiflexible DNA. The spacings between helices of naked DNA that have been seen (26, 42) in such tight packaging are typically 25 to 29 Å. While it has never been directly visualized by high-resolution EM that the dsDNA is in a B-form, other techniques have suggested that this is the case (43–46). In contrast, we have shown that VP1-covered dsDNA within the SPV1 virion forms shells that are spaced 43 Å apart, and within these shells filaments are also spaced laterally by 43 Å. The interactions between adjacent helices is attractive, as judged by the fact that they persist when the core is disrupted. Strikingly, the spacing between helical turns in 3 filamentous viruses (SIRV2, AFV1, and SFV1) that infect hyperthermophilic acidophiles is also ∼43 Å, and in these viruses the DNA is completely wrapped by dimers formed from either 1 (SIRV2) or 2 (AFV1, SFV1) capsid proteins (19, 20, 29). We have been able to build a model for the nucleoprotein filament using a 2D average as a strong constraint, and all evidence suggests that the DNA is in A-form with 11 bp per 29-Å pitch turn. The A-form DNA that is present in SPV1, with a rise of 2.6 Å per base pair (in contrast to B-DNA, with ∼10.4 bp per turn and a rise of 3.4 Å) matches the parameters of A-form DNA in crystals almost exactly (47).
The covering of the DNA by the largely α-helical VP1 dimers can be compared with the wrapping observed in the helical viruses SIRV2, AFV1, and SFV1, where a largely α-helical capsid protein forms a symmetrical dimer and wraps around the DNA. The level of arginine, lysine, and asparagine in the VP1 homodimer is 22%, which is also similar in the SIRV2 homodimer (19%), the AFV1 heterodimer (18 and 19%), and the SFV1 heterodimer (17 and 21%). However, the total length of the VP1 sequence (78 residues) is shorter than the DNA-binding core region in SIRV2, AFV1, and SFV1 (∼100 residues), suggesting that VP1 may not wrap the DNA as extensively as occurs in the helical viruses.
Collectively, our results reveal a way of building an icosahedral capsid from single jelly-roll MCPs, with a T = 43 lattice not previously observed in other viruses. The finding that the genomic dsDNA within the icosahedral capsid adopts an A-form, which has been previously described only in structurally related filamentous viruses (19, 20, 29), indicates that this might be a more general adaptation to extreme environments. Since life on earth may have arisen in extreme environments, it also raises the intriguing possibility that the ability of DNA to switch to an A-form may have been a critical selective force in the evolution of early life.
Methods
Detailed methods for SPV1 production and purification, cryo-EM and image processing, model building of the major capsid proteins, VP1 purification and gel shift assay, dissociation of viral particles, and analysis of both host and viral lipids are provided in SI Appendix.
Data Availability.
The atomic model of the SPV1 asymmetrical unit has been deposited in the Protein Data Bank (ID 6OJ0), and the reconstructed volume has been deposited in the Electron Microscopy Data Bank (accession no. EMD-20083). Other data are available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant GM122510 (to E.H.E.); the European Union’s Horizon 2020 Research and Innovation Program Grant 685778, project VIRUS X (to D.P.); and l’Agence Nationale de la Recherche Grant ANR-17-CE15-0005-01 (to M.K.). Imaging by the National Cancer Institute (NCI) National Cryo-EM Facility (Frederick National Laboratory for Cancer Research) was supported by contract HSSN261200800001E. We thank Dr. Ulrich Baxa and Dr. Thomas Edwards for their help in NCI data collection; the Ultrastructural BioImaging unit of Institut Pasteur for access to electron microscopes; and Dr. Wen Jiang (Purdue University) for very helpful discussions.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. W.K.O. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Electron Microscopy Data Bank, https://www.ebi.ac.uk/pdbe/emdb/ (accession no. 20083) and in the Protein Data Bank (ID 6OJ0).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908242116/-/DCSupplemental.
References
- 1.Prangishvili D., et al. , The enigmatic archaeal virosphere. Nat. Rev. Microbiol. 15, 724–739 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Munson-McGee J. H., Snyder J. C., Young M. J., Archaeal viruses from high-temperature environments. Genes (Basel) 9, E128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y., et al. , A novel type of polyhedral viruses infecting hyperthermophilic archaea. J. Virol. 91, e00589-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos-Pérez I., et al. , Structural basis for assembly of vertical single β-barrel viruses. Nat. Commun. 10, 1184 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Colibus L., et al. , Assembly of complex viruses exemplified by a halophilic euryarchaeal virus. Nat. Commun. 10, 1456 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veesler D., et al. , Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography. Proc. Natl. Acad. Sci. U.S.A. 110, 5504–5509 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abrescia N. G., et al. , Insights into virus evolution and membrane biogenesis from the structure of the marine lipid-containing bacteriophage PM2. Mol. Cell 31, 749–761 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Cockburn J. J., et al. , Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature 432, 122–125 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Rissanen I., et al. , Bacteriophage P23-77 capsid protein structures reveal the archetype of an ancient branch from a major virus lineage. Structure 21, 718–726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russo C. J., Henderson R., Ewald sphere correction using a single side-band image processing algorithm. Ultramicroscopy 187, 26–33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossmann M. G., Johnson J. E., Icosahedral RNA virus structure. Annu. Rev. Biochem. 58, 533–573 (1989). [DOI] [PubMed] [Google Scholar]
- 13.Krupovic M., Koonin E. V., Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. U.S.A. 114, E2401–E2410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison S. C., Olson A. J., Schutt C. E., Winkler F. K., Bricogne G., Tomato bushy stunt virus at 2.9 A resolution. Nature 276, 368–373 (1978). [DOI] [PubMed] [Google Scholar]
- 15.Cao C., et al. , Conserved fiber-penton base interaction revealed by nearly atomic resolution cryo-electron microscopy of the structure of adenovirus provides insight into receptor interaction. J. Virol. 86, 12322–12329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huiskonen J. T., Manole V., Butcher S. J., Tale of two spikes in bacteriophage PRD1. Proc. Natl. Acad. Sci. U.S.A. 104, 6666–6671 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua J., et al. , Capsids and genomes of jumbo-sized bacteriophages reveal the evolutionary reach of the HK97 fold. MBio 8, e01579-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gil-Carton D., et al. , Insight into the assembly of viruses with vertical single β-barrel major capsid proteins. Structure 23, 1866–1877 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Liu Y., et al. , Structural conservation in a membrane-enveloped filamentous virus infecting a hyperthermophilic acidophile. Nat. Commun. 9, 3360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasson P., et al. , Model for a novel membrane envelope in a filamentous hyperthermophilic virus. eLife 6, e26268 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quemin E. R., et al. , Sulfolobus spindle-shaped virus 1 contains glycosylated capsid proteins, a cellular chromatin protein, and host-derived lipids. J. Virol. 89, 11681–11691 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laurinmäki P. A., Huiskonen J. T., Bamford D. H., Butcher S. J., Membrane proteins modulate the bilayer curvature in the bacterial virus Bam35. Structure 13, 1819–1828 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L., Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Guo F., Jiang W., Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol. Biol. 1117, 401–443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purohit P. K., Kondev J., Phillips R., Mechanics of DNA packaging in viruses. Proc. Natl. Acad. Sci. U.S.A. 100, 3173–3178 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerritelli M. E., et al. , Encapsidated conformation of bacteriophage T7 DNA. Cell 91, 271–280 (1997). [DOI] [PubMed] [Google Scholar]
- 27.North A. C., Rich A., X-ray diffraction studies of bacterial viruses. Nature 191, 1242–1245 (1961). [DOI] [PubMed] [Google Scholar]
- 28.Gellert M., Davies D. R., Organization of DNA in bacteriophage T4. J. Mol. Biol. 8, 341–347 (1964). [DOI] [PubMed] [Google Scholar]
- 29.DiMaio F., et al. , Virology. A virus that infects a hyperthermophile encapsidates A-form DNA. Science 348, 914–917 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franklin R. E., Gosling R. G., Molecular configuration in sodium thymonucleate.Nature 171, 740–741 (1953). [DOI] [PubMed] [Google Scholar]
- 31.Egelman E. H., A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Abrescia N. G., Bamford D. H., Grimes J. M., Stuart D. I., Structure unifies the viral universe. Annu. Rev. Biochem. 81, 795–822 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Altschul S. F., et al. , Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y., et al. , New archaeal viruses discovered by metagenomic analysis of viral communities in enrichment cultures. Environ. Microbiol. 21, 2002–2014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bauer D. W., et al. , Exploring the balance between DNA pressure and capsid stability in herpesviruses and phages. J. Virol. 89, 9288–9298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evilevitch A., et al. , Effects of salts on internal DNA pressure and mechanical properties of phage capsids. J. Mol. Biol. 405, 18–23 (2011). [DOI] [PubMed] [Google Scholar]
- 37.São-José C., de Frutos M., Raspaud E., Santos M. A., Tavares P., Pressure built by DNA packing inside virions: Enough to drive DNA ejection in vitro, largely insufficient for delivery into the bacterial cytoplasm. J. Mol. Biol. 374, 346–355 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Li Z., Wu J., Wang Z. G., Osmotic pressure and packaging structure of caged DNA. Biophys. J. 94, 737–746 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grayson P., Han L., Winther T., Phillips R., Real-time observations of single bacteriophage lambda DNA ejections in vitro. Proc. Natl. Acad. Sci. U.S.A. 104, 14652–14657 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González-Huici V., Salas M., Hermoso J. M., The push-pull mechanism of bacteriophage Ø29 DNA injection. Mol. Microbiol. 52, 529–540 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Tzlil S., Kindt J. T., Gelbart W. M., Ben-Shaul A., Forces and pressures in DNA packaging and release from viral capsids. Biophys. J. 84, 1616–1627 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Earnshaw W. C., Casjens S. R., DNA packaging by the double-stranded DNA bacteriophages. Cell 21, 319–331 (1980). [DOI] [PubMed] [Google Scholar]
- 43.Yu T. Y., Schaefer J., REDOR NMR characterization of DNA packaging in bacteriophage T4. J. Mol. Biol. 382, 1031–1042 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Overman S. A., et al. , Conformation and interactions of the packaged double-stranded DNA genome of bacteriophage T7. Biospectroscopy 4 (suppl. 5), S47–S56 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Earnshaw W., Casjens S., Harrison S. C., Assembly of the head of bacteriophage P22: X-ray diffraction from heads, proheads and related structures. J. Mol. Biol. 104, 387–410 (1976). [DOI] [PubMed] [Google Scholar]
- 46.Lerman L. S., Wilkerson L. S., Venable J. H. Jr, Robinson B. H., DNA packing in single crystals inferred from freeze-fracture-etch replicas. J. Mol. Biol. 108, 271–293 (1976). [DOI] [PubMed] [Google Scholar]
- 47.Vargason J. M., Henderson K., Ho P. S., A crystallographic map of the transition from B-DNA to A-DNA. Proc. Natl. Acad. Sci. U.S.A. 98, 7265–7270 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic model of the SPV1 asymmetrical unit has been deposited in the Protein Data Bank (ID 6OJ0), and the reconstructed volume has been deposited in the Electron Microscopy Data Bank (accession no. EMD-20083). Other data are available from the corresponding authors upon reasonable request.