ABSTRACT
Toll-like receptor 3 (TLR3) is a viral sensor that induces apoptosis in response to double-stranded RNA (dsRNA). Common genetic changes in the TLR3 gene may influence breast cancer susceptibility and development. However, all of the polymorphisms in the previous study were only markers of the TLR3 gene, not causative polymorphisms. In this study, we performed a case-control study focusing on the relationship between rs5743305 (−926T>A), a single nucleotide polymorphism (SNP) in the promoter region of TLR3, and breast cancer. We found that the genetic variant rs5743305 increased the risk of breast cancer under the dominant and codominant models (dominant model: AT+AA vs TT.: OR = 1.3023, 95%CI: 1.0778–1.5736, P = .0062; codominant model: AA vs. TT: OR = 1.3919, 95%CI: 1.0177–1.9036, P = .0384; AT vs. TT: OR = 1.2799, 95%CI: 1.0475–1.5639, P = .0158) but not under the recessive model (TT vs. AT+AA, OR = 1.2387, 95%CI: 0.9197–1.6682, P = .1588). The same trends were found in the age-adjusted logistic regression study and stage 2 study. Furthermore, the electrophoretic mobility shift assay (EMSA) and luciferase reporter assay showed that rs5743305 decreased the transcriptional activity of TLR3. There was consistently reduced TLR3 mRNA and protein expression in human breast cancer samples from patients with TLR3 − 926A. Therefore, TLR3 rs5743305 increases the risk of breast cancer by decreasing the transcriptional activity of TLR3. This study may provide a better understanding of the genetic architecture underlying disease susceptibility and may advance the potential for preclinical prediction in future genetic testing.
KEYWORDS: Toll-like receptor 3 (TLR3), breast cancer, single nucleotide polymorphism
Introduction
Breast cancer is the most frequently diagnosed type of cancer and the second leading cause of cancer death in women.1,2 Previous studies have shown that both the innate and the adaptive immune system play a role in preventing relapse in women with breast cancer.3 Toll-like receptors (TLRs) are a class of proteins that play a key role in immune responses.One of the ten TLRs in the human genome, Toll-like receptor 3 (TLR3),4,5 is a viral sensor that induces apoptosis in response to double-stranded RNA (dsRNA), which is produced by most viruses at some point during their replication.6 Activation of TLR3 induces the activation of NF-kB and drives cellularapoptosis.7,8
In our previous study, we found that common genetic changes in the TLR3 gene may influence breast cancer susceptibility and development.9 TLR3 plays a negative regulatory role in the initiation and progression of human breast cancer cells, at least in part by downregulating the EGFR/PI3K/AKT pathway.9 However, all the single nucleotide polymorphisms (SNPs) in our previous study were only markers of the TLR3 gene, not causative polymorphisms.
In this study, we investigated a single nucleotide polymorphism, rs5743305 (−926T>A), which is in the promoter region of TLR3. We performed a case-control study focusing on the relationship between TLR3 − 926T>A and breast cancer. Furthermore, the function of this SNP was studied to investigate whether it is a causative polymorphism or just a marker of the TLR3 gene.
Results
The genetic variant rs5743305 is associated with the risk of breast cancer
A case–control study often compares the prevalence of a specific disease among persons with normal alleles and persons with variant alleles, which generates an odds ratio (OR). In this study, the single-nucleotide polymorphism consists of a major allele (T) and a minor allele (A). Thus, the genotype can be a major allele homozygote (TT), a heterozygote (TA) or a minor allele homozygote (AA). Odds are given for each genotype, and a pair of odds generates an OR. The dominant model compares AT + AA versus TT, the recessive model compares AA versus TT + AT and the codominant models compares AT versus TT and AA versus TT.
We first analyzed the odds ratio (OR) for rs5743305 in different models (Figure 1(A)). Univariate analysis revealed that rs5743305 increased the risk of breast cancer under the dominant and codominant models (dominant model: AT+AA vs. TT: OR = 1.3023, 95%CI: 1.0778–1.5736, P = .0062; codominant model: AA vs. TT: OR = 1.3919, 95%CI: 1.0177–1.9036, P = .0384; AT vs. TT: OR = 1.2799, 95%CI: 1.0475–1.5639, P = .0158) but not under the recessive model (TT vs. AT+AA, OR = 1.2387, 95%CI: 0.9197–1.6682, P = .1588). In this study, the A allele is dominant over T, causing heterozygous offspring to have a higher risk of expressing the same phenotype as their homozygous dominant parent.
Figure 1.
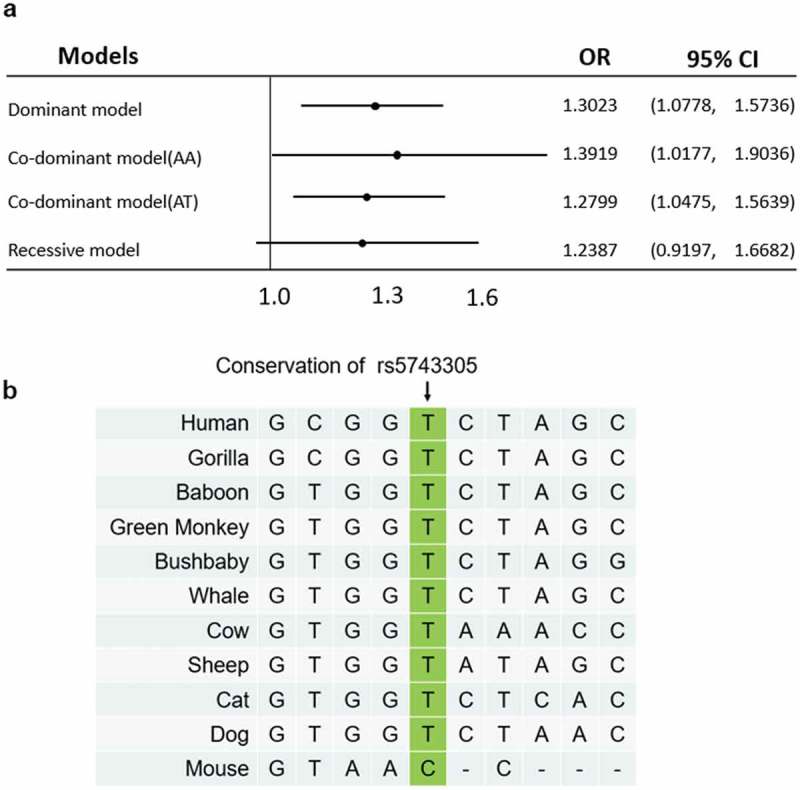
Odds ratio plot and homology study of rs5743305. (A) The odds ratio plot shows the allelic effects of rs5743305 in different models. Dominant model: TT vs. AT+AA; codominant model: AA vs. TT, and AT vs. TT; recessive model (TT vs. AT+AA). The error bars represent the 95% confidence intervals of the odds ratios.OR = odds ratio. (B) The homology study showed that the rs5743305 position of the TLR3 gene is relatively conserved in most animals. Most animals have a T at this position, whereas mouse has a C at this position.
Moreover, we added age as a multivariate factor and performed the logistic regression study. The adjusted OR of the dominant model (AA + AT vs. TT) is 1.307 (95% CI: 1.028–1.585), the OR of the codominant model (AA vs. TT) is 1.406 (95% CI: 1.019–1.792), the OR of the co-dominant model (AT vs. TT) is 1.340 (95% CI: 1.033–1.646), and the OR of the recessive model (AA vs. AT + TT) is 1.296 (95% CI: 0.922, 1.669). (Table 1)
Table 1.
Allelic effects of rs5743305 in different models in stage 1.
| Breast cancer n =1,031 Control n = 1,272 | OR (95% CI) Without adjusting | p | OR (95% CI) Adjusted by age | p |
|---|---|---|---|---|
| Dominant model(AA+AT vs. TT) | 1.302 (1.078, 1.574) | 0.006 | 1.307 (1.028-1.585) | 0.013 |
| Co-dominant model(AA vs. TT) | 1.392 (1.018, 1.904) | 0.038 | 1.406 (1.019-1.792) | 0.008 |
| Co-dominant model(AT vs. TT) | 1.280 (1.048, 1.564) | 0.016 | 1.340 (1.033-1.646) | 0.023 |
| Recessive model(AA vs. AT+TT) | 1.239 (0.920, 1.668) | 0.159 | 1.296 (0.922, 1.669) | 0.200 |
OR = odds ratio
95% CI = 95% Confidence interval
A homologous study of rs5743305 showed that the promoter of the TLR3 gene is relatively conserved in most animals. Most animals have a T at this position. Only mice have a C at this position (Figure 1(B)).
Furthermore, we performed a stage 2 study in 468 breast cancer cases and 913 cases. (Table 2) The results showed that rs5743305 increased the risk of breast cancer under the dominant and codominant models [dominant model: AT + AA vs. TT: OR = 1.320 (95% CI: 1.055, 1.651), P = .015; codominant model: AA vs. TT: OR = 1.481 (95% CI: 1.020, 2.149), P = .039; AT vs. TT: OR = 1.282 (95% CI: 1.011, 1.625), P = .040)] but not under the recessive model [(TT vs. AT + AA, OR = 1.317 (95% CI: 0.924, 1.877), P = .1588]. (Table 3)
Table 2.
rs5743305 data in stage 2 study.
| Genotype |
|||
|---|---|---|---|
| AA | AT | TT | |
| Breast cancer cases | 57 | 207 | 204 |
| Controls | 87 | 365 | 461 |
Table 3.
Allelic effects of rs5743305 in different models in stage 2.
| Breast cancer n =468 Control n = 913 | OR (95% CI) Without adjusting | p | OR (95% CI) Adjusted by age | p |
|---|---|---|---|---|
| Dominant model(AA+AT vs. TT) | 1.320 (1.055, 1.651) | 0.015 | 1.402 (1.125, 1.678) | 0.012 |
| Co-dominant model(AA vs. TT) | 1.481 (1.020, 2.149) | 0.039 | 1.586 (1.016, 2.155) | 0.001 |
| Co-dominant model(AT vs. TT) | 1.282 (1.011, 1.625) | 0.040 | 1.327 (1.022, 1.632) | 0.026 |
| Recessive model(AA vs. AT+TT) | 1.317 (0.924, 1.877) | 0.128 | 1.343 (0.899, 1.786) | 0.086 |
OR = odds ratio
95% CI = 95% Confidence interval
The functional polymorphism rs5743305 decreased the transcriptional activity of TLR3
We next investigated the effect of the rs5743305 (TLR 3 − 926T>A) variant on the transcriptional activity of the TLR3 gene. Wildtype and −926T>A TLR3 plasmids were verified before use by direct sequencing (Figure 2(A)).
Figure 2.
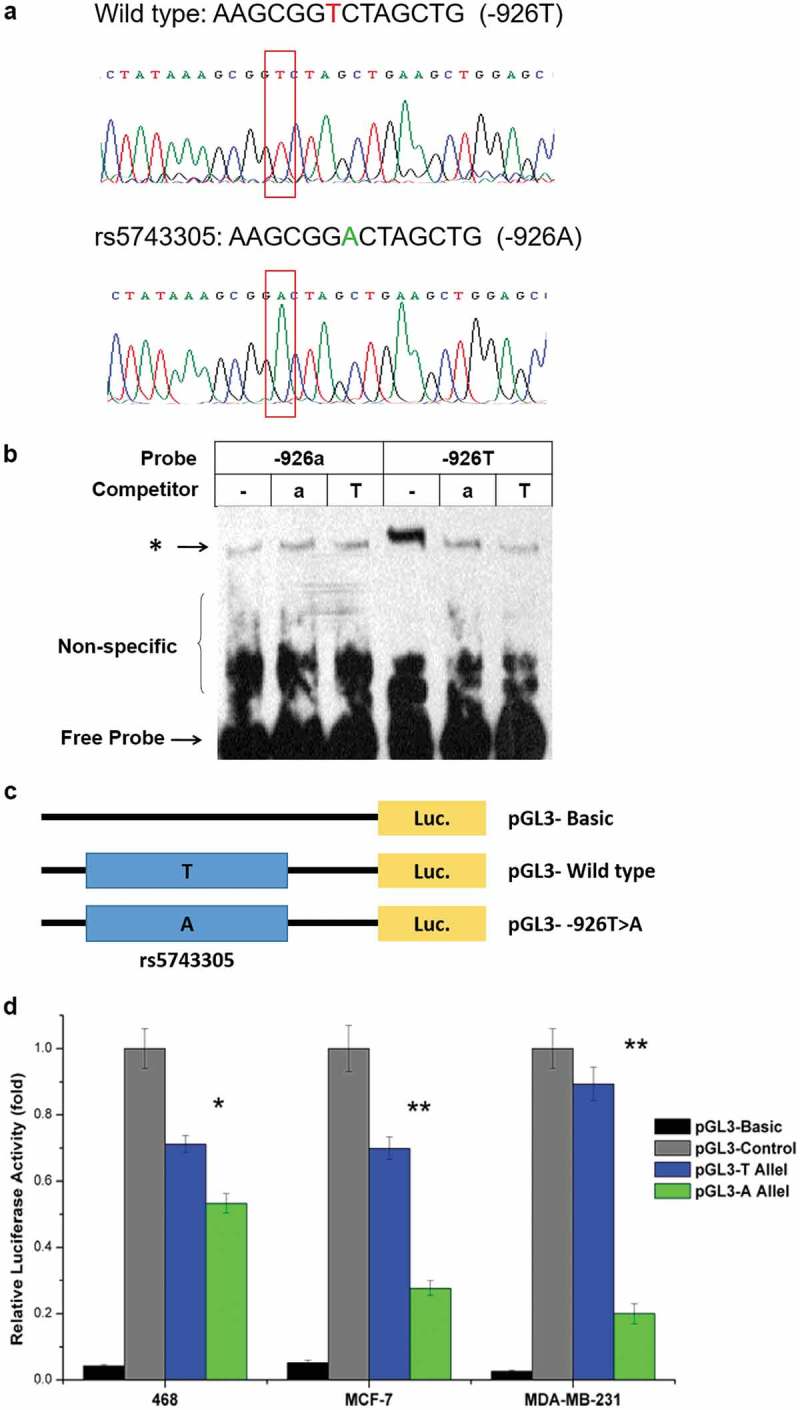
rs5743305 decreased the transcriptional activity of TLR3. (A) Wild type and −926T>A TLR3 plasmids were verified by direct sequencing. (B) Electrophoretic mobility shift assay (EMSA) showed that a much clearer DNA-protein complex was detected with the −926T probe than with the −926A probe. (C) Three luciferase reporter gene constructs were generated. They share identical backbone sequences except for the polymorphisms. (D) Significantly lower luciferase activity of the −926A haplotype was observed when compared with the wild type (−926T) haplotype vectors in the 468 cell line, MCF-7 cell line, and MDB-MB-231 breast cancer cell line.
* = P < .05; ** = P < .01.
We used an electrophoretic mobility shift assay (EMSA) to investigate whether the difference in activity between sequences containing −926T or −926A could be attributed to a different affinity of these two alleles in binding to transcription factors. As shown in Figure 2(B), a much clearer DNA-protein complex was detected with the −926T probe than with the −926A probe. Competition experiments revealed that the −926T band could be competed away with a 100-fold molar excess of the same unlabeled probe but could not compete with the same concentration of unlabeled −926A type probe. The corresponding results clearly showed a vastly greater affinity of −926T rather than −926A for transcription factors.
To assess the individual and cooperative effects of the two polymorphisms, we generated three luciferase reporter gene constructs that share identical backbone sequences except for the polymorphisms (Figure 2(C)). As shown in Figure 2(D), reporter gene expression driven by the −926T-containing TLR3 promoter (pGL3-T Allel) was greater (1.25-fold in the 468 cell line, 2.39-fold in the MCF-7 cell line, and 3.52-fold in the MDB-MB-231 cell line) than that driven by the-926A-containing counterpart (pGL3-T Allel) (P < .05). Significantly lower luciferase activity of the −926A haplotype was observed when compared with the wild type haplotype vectors in all three breast cancer cell lines.
Both EMSA and the luciferase reporter assay showed that rs5743305 is a functional polymorphism and that TLR3 − 926A leads to lower transcriptional activity than that of wild type (TLR3 − 926T).
Patients who carry rs5743305 had lower expression of TLR3
Furthermore, we tested the expression of TLR3 in breast cancer tissue of patients who carry rs5743305 and wild type. Real-time PCR (Figure 3(A)) showed that breast cancer patients carryingTLR3 − 926A had lower expression of TLR3 at the mRNA level (P = .0020). Western blotting (Figure 3(B,C)) showed that patients with TLR3 − 926A had lower TLR3 protein expression than that of TLR3 − 926T patients (P = .0015).
Figure 3.
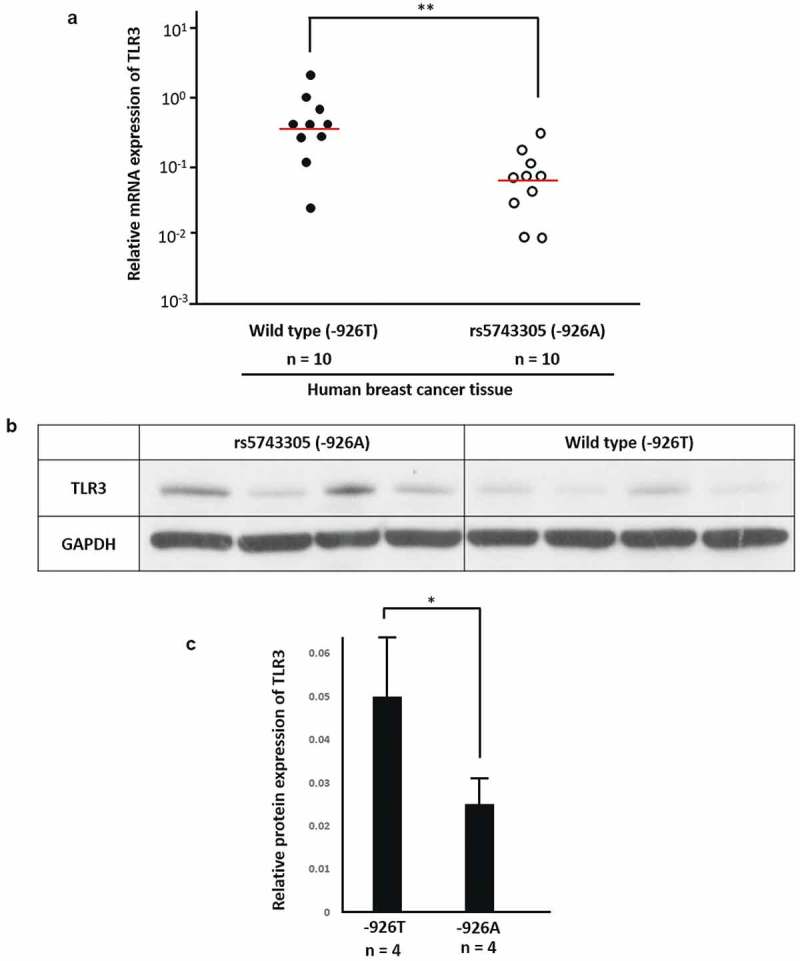
Patients with rs5743305 had lower expression of TLR3. (A) Real-time PCR showed that in breast cancer tissue of patients with TLR3 − 926A, TLR3 expression was lower at the mRNA level.(n = 10). (B) Western blot of breast cancer patients with TLR3 − 926A and wild type (−926T). (C) Tissue from patients with TLR3 − 926A had lower TLR3 protein expression than that from TLR3 − 926T patients. (n = 4).
** = P < .01.
Discussion
In this study, we performed a case-control study focusing on the relationship between rs5743305 (TLR 3 − 926T>A) and breast cancer and found that the genetic variant rs5743305 is associated with the risk of breast cancer. Moreover, we investigated the effect of this variant on the transcriptional activity of the TLR3 gene. Our data showed that rs5743305 is a functional polymorphism and that TLR3 − 926A leads to lower transcriptional activity than wild type.
Toll-like receptors (TLRs) are important regulators in both the adaptive and innate immune responses.10–12 They are capable of inducing antitumor mediators and are highly expressed on antigen-presenting cells. TLRs selectively recognize a variety of conserved molecular structures in invading pathogens. For example, TLR1 recognizes various bacterial components and initiates complex downstream NF-κB and MAPK pathways.13–15 TLRs may lead to immune tolerance and cancer progression.2 Some TLR agonists have been studied in tumor therapy in attempts to change immune tolerance into antitumor immunity.16,17
TLR3 is predominantly a breast tumor suppresser. It inhibits tumor development through effects on cell proliferation and survival and plays important roles in breast cancer development and progression.18 TLR3 recognizes double-stranded RNA (dsRNA) and induces apoptosis in human breast cancer cells.19 Treatment with dsRNA has been found to be associated with a significant decrease in the risk of relapse in TLR3-positive breast cancer.20Polyinosinic-polycytidylic acid (poly I:C), a ligand of TLR3, improves the antitumor effects of chimeric antigen receptor modified T (CAR-T) cells.21 Ultimo et al. reported that poly(I:C)-conjugated nanoparticles efficiently targeted breast cancer cells due to the dsRNA-TLR3 interaction.19 Bernardo et al. found that retinoic acid and poly(I:C) cotreatment activates TLR3; induces the production of type I IFN autocrine signaling, caspase-8 and caspase-3 activation, as well as tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) signaling; blocks breast cancer cell proliferation; and leads to the apoptosis of breast cancer cells. Salaun et al. described that dsRNA mediates its therapeutic effect through TLR3 expressed on breast tumor cells, and TLR3 is a biomarker for the therapeutic efficacy of double-stranded RNA.20
TLR3 polymorphisms are associated with an increased risk of breast cancer. In this study, we found that the genetic variant rs5743305 of TLR3 is associated with an increased risk of breast cancer. The genetic association between the TLR3 variants and breast cancer has been reported in previous studies. We found that the T-allele of rs5743312 and the T-allele of rs3775296 conferred an increased risk of breast cancer incidence.9Chen et al. reported that the SNP rs3775291 in TLR3 may influence breast cancer patient outcome.2 However, Etokebe et al. reported that polymorphisms in the TLR3 gene were not likely to be associated with an increased risk for developing breast cancer.22 This negative result was most likely because they only tested 74 women with breast cancer. It is difficult to obtain positive resultsin an association study with such a low case number. We tested 1,031 patients with breast cancer and 1,272 female cancer-free control subjects.A high patient number makes the positive association results more convincing.
The association of polymorphisms and genes has two potential explanations. One is that the SNP is just a marker of the gene. Another possibility is that the SNP influences the function of this gene. However, previous studies only suggested an association between TLR3 and breast cancer. None of them studied how the variants influence the expression or function of TLR3.
In this study, we found that rs5743305 is a functional polymorphism, and its polymorphism (−926A) leads to lower transcriptional activity than its wild type (−926T). The variant rs5743305 is in the promoter region of TLR3. SNPs in the promoter region that overlap transcription factor binding sites alter transcription factor binding affinity and influence gene expression at the RNA level. In this study, EMSA showed a lower binding affinity of −926A than −926T to transcription factors. The luciferase reporter gene showed that TLR3 − 926A leads to lower TLR3 gene expression than wild type.
Furthermore, we found that breast cancer patients carrying rs5743305 had lower expression of TLR3 at both the mRNA and protein levels in their breast cancer tissue. These results confirm the above transcriptional activity studies. Previous studies have found that TLR3 directly triggers apoptosis in human cancer cells.7 We analyzed TLR3 expression in breast cancer and normal tissue in Caucasians in The Cancer Genomic Atlas (TCGA) database and found that TLR3 expression was lower in breast cancer samples than expression in controls (P < .05). (Supplementary Figure 1)
Subgroup analysis of TLR3 in the different types of breast cancer was performed and revealed that patients with high expression of TLR3 had a better relapse-free survival (RFS) rate than the rate in those with lower TLR3 expression. (Supplementary Figure 2) These results were in accordance with our findings that decreased transcriptional activity leads to an increased risk of breast cancer. The effect of TLR3 is not dependent of the breast molecular subtype. However, we did not record the survival clinical data in our experiments and cannot analyze whether this polymorphism affects survival when breast cancer occurs. We will analyze the effect of this polymorphism on breast cancer survival in our future experiments.
In summary, TLR3 rs5743305 (TLR3 − 926T>A) increases the risk of breast cancer. The mechanism is that the TLR3 − 926A polymorphism leads to lower transcriptional activity than its wild type. Women carrying this SNP have lower expression of TLR3 on their breast cell surface. Consequently, reduced activation of TLR3 may play the most important role in the increased risk of breast cancer (Figure 4). This study may provide a better understanding of the genetic architecture underlying disease susceptibility and may advance the potential for preclinical prediction in future genetic testing.
Figure 4.
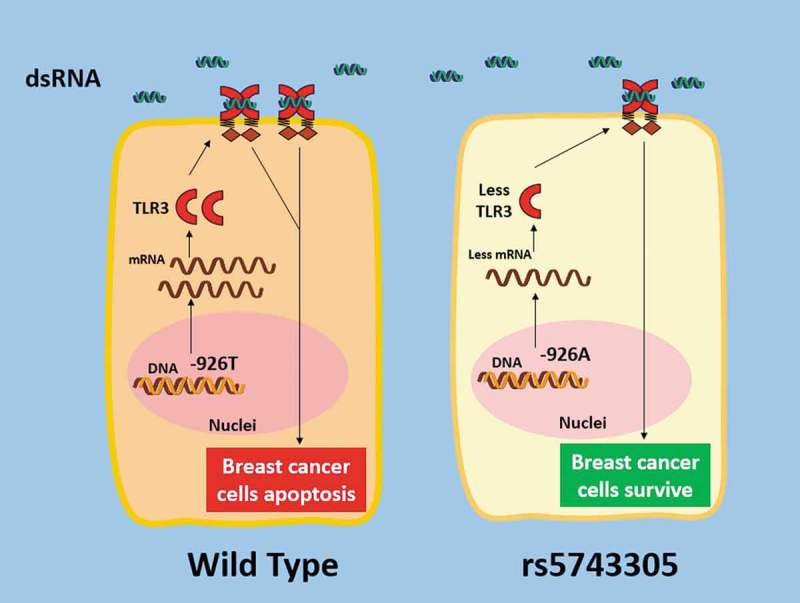
Schematic illustration of how the rs5743305 (TLR3 − 926T>A) variant increased the risk of breast cancer. TheTLR3 − 926A polymorphism leads to lower transcriptional activity than its wild type (−926T). Women carrying this SNP have lower expression of TLR3. Consequently, reduced activation of TLR3 may play a role in the increased risk of breast cancer.
Materials and methods
Ethics statement
This study was approved by the Ethical Committee of the Fudan University Shanghai Cancer Center. Each participant signed an informed consent document.
Study subjects and data collection
A total of 1,272 female cancer-free control subjects and 1,031 patients with breast cancer were identified as genetically unrelated Chinese in Shanghai City and its surrounding regions. Each participant was personally interviewed by doctors either in the outpatient department or in the inpatient department to obtain epidemiological and clinicopathological information. These subjects were recruited between January 2012 and June 2015 from the Department of Breast Surgery, Fudan University Shanghai Cancer Center. Patients with a previous history of other cancers (not breast cancer) were excluded. Primary ductal carcinoma in situ (DCIS) or infiltrating ductal carcinoma of the breast was pathologically confirmed. The control subjects were chosen from women who had come to our department for the purpose of breast cancer screening. The control subjects selected were proven to be free of breast cancer by a complete physical examination, ultrasonography, bilateral mammography, and biopsy when necessary. Women who had a previous history of cancer were also excluded. The controls were matched to the case patients on the basis of geographical area and age. All study subjects provided a 3- to 5-ml venous blood sample. All of the data collected were entered into a computerized database established by the Department of Breast Surgery of the Fudan University Shanghai Cancer Center.
SNP genotyping
Genomic DNA was extracted from peripheral blood lymphocytes using a GentraPureGene DNA Purification Kit (Gentra Systems, USA). The samples were stored at −20°C. SNPs were genotyped with a 12-plex SNPstream Platform (Beckman Colter Inc.). Genotyping was carried out by the Chinese National Human Genome Center (Shanghai).
Cell culture and stable transfection
TheMCF-7 cell line, 468 cell line, and MDB-MB-231 breast cancer cell line were used in this study. Cells were transfected withLipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer’s instructions. After selection in the presence of 1,000 μg/ml geneticin (G418 sulfate; Invitrogen) for 4 weeks, stably transfected clones were obtained. RT-PCR and western blotting analyzes were performed to measure expression levels.
RT-PCR and relative quantitative real-time PCR
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). First-strand cDNA was synthesized with 1 μg of total RNA, oligo(dT)15 primers and AMV reverse transcriptase (Promega). The primers used in RT-PCR were as follows: TLR3 forward: 5ʹ- TCGAGAGTGCCGTCTATTTGCCACA- 3ʹ, reverse: 5ʹ- CAGGTGGCTGCAGTCAGCAACT-3ʹ; β-actin, forward: 5ʹ-AGCGGGAAATCGTGCGTG-3ʹ, reverse: 5ʹ-CAGGGTACATGGTGGTGCC-3ʹ. Real-time PCR reactions were performed with SYBR Green PCR master mix (Roche). The specificity of the PCR amplification products was checked by performing dissociation melting-curve analysis and by 1% agarose gel electrophoresis. Quantification analysis of TLR3 mRNA was normalized with a housekeeping gene, β-actin, as an internal control. Relative multiples of changes in mRNA expression were determined by calculating 2−ΔΔCT.
Western blot analysis
Proteins were extracted from samples using T-PER Tissue Protein Extraction Reagent (Pierce, Rockford, IL, USA). The protein concentration was measured using the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA). Equal amounts of protein lysate (20–60 μg) were resolved on 10% Tris-HCl polyacrylamide gels and then transferred to a PVDF blotting membrane (Millipore, Billerica, MA, USA). After blocking, each membrane was incubated with antibodies specific for human TLR3 and GAPDH (Abcam). After incubation with peroxidase-conjugated goat anti-mouse secondary antibodies (ZSGB-Bio, Beijing, China), protein bands were detected by chemiluminescence (Pierce).
Electrophoretic mobility shift assay (EMSA)
Nuclear proteins and cytoplasmic proteins were extracted from cells using NE-PER nuclear and cytoplasmic extraction reagents (Pierce Biotechnology, Rockford, IL, USA). The probes and competitors for the −926A allele and the −926T allele were 5ʹ-GCGGaCTAGCTGAAGCTG-3ʹ and 5ʹ-GCGGtCTAGCTGAAGCTG-3ʹ, respectively. Probes were end-labeled with biotin (Invitrogen). Identical, unlabeled oligonucleotides with the same sequences were used as competitors. dsDNA was generated, and EMSAs were performed using the LightShiftChemiluminescent EMSA kit (Pierce, Rockford, IL, USA) according to the manufacturer’s instructions.
Luciferase assays
Human breast cancer cells (MCF-7, MDB-MB-231, and 468 cell lines) were grown in complete medium consisting of DMEM supplemented with 10% heat-inactivated fetal calf serum in a humidified, 5% CO2 incubator at 37°C. Cells were transfected with 500 ng of plasmid DNA and cotransfected with 10 ng of pRL-SV40 as a control for transfection efficiency. Transfections were performed using Lipofectamine2000 (Invitrogen) according to the manufacturer’s protocol. Luciferase activity was measured on a VeritasTMmicroplateluminometer (Turner BioSystems, Sunnyvale, CA, USA) using the Dual-Luciferase Reporter Assay System Kit (Promega, Madison, WI, USA). Each experiment was performed in triplicate at least three times. Luciferase units were calculated using the formula Firefly luciferase units/Renilla luciferase units. A fold increase was reported by defining the activity of the empty pGL3-Basic vector as one.
Statistical analysis
Statistical analyzes were performed with Student’s t test. Values in the figures are expressed as the means±SE. Values of P < .05 were considered statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary materials
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Fan L, Strasser-Weippl K, Li -J-J, St Louis J, Finkelstein DM, Yu K-D, Chen W-Q, Shao Z-M, Goss PE.. Breast cancer in China. Lancet Oncol. 2014;15:1–8. doi: 10.1016/S1470-2045(13)70567-9. [DOI] [PubMed] [Google Scholar]
- 2.Chen DN, Song CG, Yu KD, Jiang YZ, Ye FG, Shao ZM.. A prospective evaluation of the association between a single nucleotide polymorphism rs3775291 in toll-like receptor 3 and breast cancer relapse. PLoS One. 2015;10:e0133184. doi: 10.1371/journal.pone.0133184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Standish LJ, Sweet ES, Novack J, Wenner CA, Bridge C, Nelson A, Martzen M, Torkelson C. Breast cancer and the immune system. J Soc Integr Oncol. 2008;6:158–168. [PMC free article] [PubMed] [Google Scholar]
- 4.Hammerich L, Marron TU, Upadhyay R, Svensson-Arvelund J, Dhainaut M, Hussein S, Zhan Y, Ostrowski D, Yellin M, Marsh H, et al. Systemic clinical tumor regressions and potentiation of PD1 blockade with in situ vaccination. Nat Med. 2019;25:814–824. doi: 10.1038/s41591-019-0410-x. [DOI] [PubMed] [Google Scholar]
- 5.Gosu V, Son S, Shin D, Song KD. Insights into the dynamic nature of the dsRNA-bound TLR3 complex. Sci Rep. 2019;9:3652. doi: 10.1038/s41598-019-39984-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 7.Salaun B, Coste I, Rissoan MC, Lebecque SJ, Renno T. TLR3 can directly trigger apoptosis in human cancer cells. Journal of Immunol. 2006;176:4894–4901. doi: 10.4049/jimmunol.176.8.4894. [DOI] [PubMed] [Google Scholar]
- 8.Patchett AL, Tovar C, Corcoran LM, Lyons AB, Woods GM. The toll-like receptor ligands Hiltonol((R)) (polyICLC) and imiquimod effectively activate antigen-specific immune responses in Tasmanian devils (Sarcophilus harrisii). Dev Comp Immunol. 2017;76:352–360. doi: 10.1016/j.dci.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Fan L, Zhou P, Hong Q, Chen AX, Liu GY, Yu KD, Shao Z-M. Toll-like receptor 3 acts as a suppressor gene in breast cancer initiation and progression: a two-stage association study and functional investigation. Oncoimmunology. 2019;8:e1593801. doi: 10.1080/2162402X.2019.1593801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia D, Wang L. The other face of TLR3: A driving force of breast cancer stem cells. Mol Cell Oncol. 2015;2:e981443. doi: 10.4161/23723556.2014.981443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Souza PPC, Lerner UH. Finding a toll on the route: the fate of osteoclast progenitors after toll-like receptor activation. Front Immunol. 2019;10:1663. doi: 10.3389/fimmu.2019.01663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olmos-Ortiz A, Flores-Espinosa P, Mancilla-Herrera I, Vega-Sanchez R, Diaz L, Zaga-Clavellina V. Innate immune cells and toll-like receptor-dependent responses at the maternal-fetal interface. Int J Mol Sci. 2019;20:e3654.doi: 10.3390/ijms20153654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korppi M, Tormanen S. Toll-like receptor 1 and 10 variations increase asthma risk and review highlights further research directions. Acta Paediatr. 2019;108:1406–1410. [DOI] [PubMed] [Google Scholar]
- 14.Schurz H, Daya M, Moller M, Hoal EG, Salie M. TLR1, 2, 4, 6 and 9 variants associated with tuberculosis susceptibility: a systematic review and meta-analysis. PLoS One. 2015;10:e0139711. doi: 10.1371/journal.pone.0139711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanki M, Seppanen H, Mustonen H, Hagstrom J, Haglund C. Toll-like receptor 1 predicts favorable prognosis in pancreatic cancer. PLoS One. 2019;14:e0219245. doi: 10.1371/journal.pone.0219245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patidar A, Selvaraj S, Sarode A, Chauhan P, Chattopadhyay D, Saha B. DAMP-TLR-cytokine axis dictates the fate of tumor. Cytokine. 2018;104:114–123. doi: 10.1016/j.cyto.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Chi H, Li C, Zhao FS, Zhang L, Ng TB, Jin G, Sha O. Anti-tumor activity of toll-like receptor 7 agonists. Front Pharmacol. 2017;8:304. doi: 10.3389/fphar.2017.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seya T, Takeda Y, Matsumoto M. A Toll-like receptor 3 (TLR3) agonist ARNAX for therapeutic immunotherapy. Advanced drug delivery reviews. Adv Drug Deliv Rev. 2019. doi: 10.1016/j.addr.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Ultimo A, Giménez C, Bartovsky P, Aznar E, Sancenón F, Marcos MD, Amorós P, Bernardo AR, Martínez-Máñez R, Jiménez-Lara AM, et al. Targeting Innate Immunity with dsRNA-conjugated mesoporous silica nanoparticles promotes antitumor effects on breast cancer cells. Chemistry. 2016;22:1582–1586. doi: 10.1002/chem.201504629. [DOI] [PubMed] [Google Scholar]
- 20.Salaun B, Zitvogel L, Asselin-Paturel C, Morel Y, Chemin K, Dubois C, Massacrier C, Conforti R, Chenard MP, Sabourin J-C, et al. TLR3 as a biomarker for the therapeutic efficacy of double-stranded RNA in breast cancer. Cancer Res. 2011;71:1607–1614. doi: 10.1158/0008-5472.CAN-10-3490. [DOI] [PubMed] [Google Scholar]
- 21.Di S, Zhou M, Pan Z, Sun R, Chen M, Jiang H, Shi B, Luo H, Li Z. Combined adjuvant of poly I:C improves antitumor effects of CAR-T cells. Front Oncol. 2019;9:241. doi: 10.3389/fonc.2019.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Etokebe GE, Knezevic J, Petricevic B, Pavelic J, Vrbanec D, Dembic Z. Single-nucleotide polymorphisms in genes encoding toll-like receptor −2, −3, −4, and −9 in case-control study with breast cancer. Genet Test Mol Biomarkers. 2009;13:729–734. doi: 10.1089/gtmb.2009.0045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


