ABSTRACT
Infection and inflammation are able to induce diet-independent Na+-accumulation without commensurate water retention in afflicted tissues, which favors the pro-inflammatory activation of mouse macrophages and augments their antibacterial and antiparasitic activity. While Na+-boosted host defense against the protozoan parasite Leishmania major is mediated by increased expression of the leishmanicidal NOS2 (nitric oxide synthase 2, inducible), the molecular mechanisms underpinning this enhanced antibacterial defense of mouse macrophages with high Na+ (HS) exposure are unknown. Here, we provide evidence that HS-increased antibacterial activity against E. coli was neither dependent on NOS2 nor on the phagocyte oxidase. In contrast, HS-augmented antibacterial defense hinged on HIF1A (hypoxia inducible factor 1, alpha subunit)-dependent increased autophagy, and NFAT5 (nuclear factor of activated T cells 5)-dependent targeting of intracellular E. coli to acidic autolysosomal compartments. Overall, these findings suggest that the autolysosomal compartment is a novel target of Na+-modulated cell autonomous innate immunity.
Abbreviations: ACT: actins; AKT: AKT serine/threonine kinase 1; ATG2A: autophagy related 2A; ATG4C: autophagy related 4C, cysteine peptidase; ATG7: autophagy related 7; ATG12: autophagy related 12; BECN1: beclin 1; BMDM: bone marrow-derived macrophages; BNIP3: BCL2/adenovirus E1B interacting protein 3; CFU: colony forming units; CM-H2DCFDA: 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester; CTSB: cathepsin B; CYBB: cytochrome b-245 beta chain; DAPI: 4,6-diamidino-2-phenylindole; DMOG: dimethyloxallyl glycine; DPI: diphenyleneiodonium chloride; E. coli: Escherichia coli; FDR: false discovery rate; GFP: green fluorescent protein; GSEA: gene set enrichment analysis; GO: gene ontology; HIF1A: hypoxia inducible factor 1, alpha subunit; HUGO: human genome organization; HS: high salt (+ 40 mM of NaCl to standard cell culture conditions); HSP90: heat shock 90 kDa proteins; LDH: lactate dehydrogenase; LPS: lipopolysaccharide; Lyz2/LysM: lysozyme 2; NFAT5/TonEBP: nuclear factor of activated T cells 5; MΦ: macrophages; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MFI: mean fluorescence intensity; MIC: minimum inhibitory concentration; MOI: multiplicity of infection; MTOR: mechanistic target of rapamycin kinase; NaCl: sodium chloride; NES: normalized enrichment score; n.s.: not significant; NO: nitric oxide; NOS2/iNOS: nitric oxide synthase 2, inducible; NS: normal salt; PCR: polymerase chain reaction; PGK1: phosphoglycerate kinase 1; PHOX: phagocyte oxidase; RFP: red fluorescent protein; RNA: ribonucleic acid; ROS: reactive oxygen species; sCFP3A: super cyan fluorescent protein 3A; SBFI: sodium-binding benzofuran isophthalate; SLC2A1/GLUT1: solute carrier family 2 (facilitated glucose transporter), member 1; SQSTM1/p62: sequestosome 1; ULK1: unc-51 like kinase 1; v-ATPase: vacuolar-type H+-ATPase; WT: wild type
KEYWORDS: Autophagy, cell-autonomous immunity, E. coli, macrophage, salt, sodium
Introduction
Cutaneous infections can induce a diet-independent accumulation of Na+ into the skin without commensurate water retention. This results in an increase in effective osmolyte load approximately 40 mM higher than that in plasma or standard cell culture conditions [1]. Mimicking these conditions, by stimulating macrophages (MΦ) in vitro in the presence of high NaCl concentrations (additional 40 mM NaCl on top of standard cell culture media NaCl concentrations; total [Na+] of approximately 180 mM), resulted in increased inflammatory MФ responses, including NOS2-dependent production of antimicrobial NO (nitric oxide) [1–4]. These changes were dependent on activation of the osmoprotective transcription factor NFAT5/TonEBP. The transcription factor NFAT5 plays a key role in the adaption of various cells to increases in extracellular Na+ levels [5–7]. Moreover, NFAT5 plays also an important role in MΦ immunobiology under standard cell culture conditions [8–11]. High salt conditions increased NFAT5 levels in MΦ, and resulted in improved antiparasitic control of Leishmania major infection both in vitro and in vivo [1]. These findings established a role for local tissue Na+ levels as a protective element to fight infections [12,13]. Of note, enhanced antimicrobial MΦ activity with high Na+ is not restrained to anti-parasitic defenses but is also operative against E. coli [1,14]. Moreover, there is evidence that increased extracellular Na+ helps in antigen degradation [15] and fighting viral infections [16,17]. In addition, elevated Na+ levels prime malignant cells for apoptosis [18,19] and augment the cytotoxic potential of death receptors in cancer cells [20]. However, the mechanism underlying enhanced antibacterial activity in MΦ is unclear.
Recently, Kitada et al demonstrated that, upon a high salt diet, mice generate urea which serves as an alternative osmolyte that facilitates Na+ excretion while avoiding water loss. This metabolic osmoprotective response includes increased autophagic activity in muscles [21]. Autophagy is a cell-intrinsic catabolic mechanism that is essential in the recycling of damaged or aged organelles and proteins, and cellular adaption to starvation and various extracellular stress signals [22]. Moreover, autophagy is involved in rapid, cell-autonomous antimicrobial innate immune response triggered by various pathogens [23,24]. It is known that autophagy is regulated by cell starvation and environmental signals including the class I phosphoinositide 3-kinase (PI3K)-AKT/PKB, MTOR (mechanistic target of rapamycin kinase) and HIF1A (hypoxia inducible factor 1, alpha subunit) [25,26].
Here, we show that elevated extracellular Na+ levels increase the induction of autophagy and target E. coli to acidic autolysosomal compartments, which ultimately favors enhanced antibacterial MΦ activity. We find that the transcription factors HIF1A and NFAT5 govern these activities cooperatively. These results demonstrate that Na+ is a regulator of autophagy and thereby facilitates increased cell-intrinsic antibacterial MΦ function.
Results
Elevated extracellular Na+ boosts antibacterial activity of macrophages
To assess the impact of increased [Na+] on the antibacterial function of MΦ, we infected RAW264.7 MΦ and mouse bone marrow-derived MΦ (BMDM) with E. coli and subjected them to gentamicin protection assays. MΦ were first exposed to E. coli for 2 h in normal/isotonic media (normal salt, NS) to exclude any potential effect of high Na+ on phagocytosis, before switching MΦ cultures to high Na+ conditions (additional 40 mM NaCl on top of standard cell culture NaCl concentrations; high salt, HS). After 24 h of infection, E. coli-recovery was determined. In line with previous observations [1,14] we confirmed that increased Na+ reduced bacterial load both in BMDM and RAW264.7 MΦ (Figure 1(a)). HS neither affected the viability of the infected MΦ (Figure 1(b)) nor impaired the growth of E. coli cultured alone (Figure 1(c)). Moreover, increases in Na+ did not decrease the minimum inhibitory concentration (MIC) of gentamicin directed against the E. coli strain used in our study (Figure 1(d)) and a reference E. coli strain (Fig. S1). This demonstrates that elevated Na+ levels do not increase the susceptibility of E. coli towards gentamicin. Analysis of phagocytic capacity of MΦ using Latex beads revealed that Na+ levels neither affected the adsorption of the beads to MΦ (4°C) nor influenced the internalization of the beads (37°C; Figure 1(e)). Therefore, we tested whether simultaneous MΦ infection and incubation in HS-conditions reduced bacterial recovery. Again, HS reduced intracellular bacterial load (Figure 1(f)). Taken together, these findings demonstrate that HS enhances cell-intrinsic antibacterial activity of MΦ.
Figure 1.
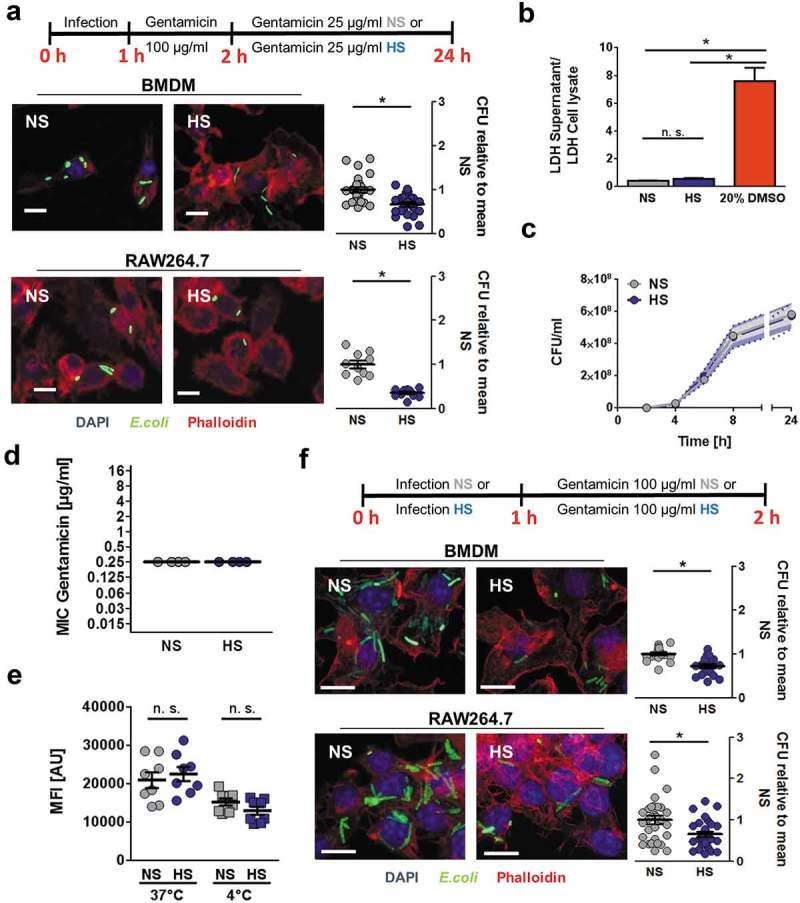
HS boosts antibacterial activity of MΦ against E. coli. (a) BMDM or RAW264.7 MΦ were infected with E. coli (MOI 100) under normal salt conditions (NS) and subjected to gentamicin protection assays (upper panel). Where indicated, cells were exposed to high salt conditions (HS; +40 mM NaCl) 2 h after infection. Confocal images: sfGFP-E. coli, green; Phalloidin, red; DAPI (DNA), blue. Scale bar: 10 µm. Intracellular E. coli load, colony-forming units (CFU) relative to mean CFU under NS conditions (means ± s.e.m; nBMDM = 23–24; nRAW264.7 = 10–11; Student’s t test ± Welch correction, * p < 0.05). (b) Cells treated as in (a) or with 20% DMSO (dimethyl sulfoxide). Relative LDH (Lactate dehydrogenase)-release was quantified (means ± s.e.m; n = 9–12; Kruskal-Wallis test with Dunn’s multiple comparison tests. * p < 0.05). (c) E. coli were grown in the absence of host cells in cell culture media under NS and HS conditions. At the indicated time points, CFU were determined (means ± s.e.m; n = 4–6). (d) Minimal inhibitory concentration of gentamicin against E. coli HB101 under NS and HS conditions (means ± s.e.m; n = 4). (e) BMDM were exposed to Latex beads at 37°C or at 4°C for ½ h under NS or HS and subjected to flow cytometry. Mean fluorescence intensities (MFI) were recorded (means ± s.e.m; n = 8; Student’s t test; * p < 0.05). (f) As in (a), but directly after infection MΦ were incubated under NS or HS for 2 h (upper panel). Confocal images: sfGFP-E. coli, green; Phalloidin, red; DAPI (DNA), blue. Scale bar: 10 µm. Intracellular E. coli load, CFU relative to mean CFU under NS conditions (means ± s.e.m; nBMDM = 18; nRAW264.7 = 29–31; Student’s t test ± Welch correction, * p < 0.05).
Mannitol-induced osmotic stress does not induce Na+ entry and impairs antibacterial activity
Elevation of extracellular Na+ resulted in rapid influx of Na+ in infected cells (Figure 2(a)) while elevating tonicity with mannitol (a nonionic osmolyte that increases tonicity but does not penetrate the cell membrane [27]) did not result in increased cytosolic Na+ levels (Figure 2(b)). Conforming to earlier studies in proximal tubular cells [28], increases in intracellular Na+ upon HS conditions were paralleled by reduced intracellular Ca2+ contents in MΦ (Figure S2(a)). Activation of MΦ under NS conditions is known to trigger outward K+ currents [29–32]. However, HS conditions did not impact on the gradual decrease of intracellular K+ levels of E. coli-infected MΦ (Figure S2(b)). These findings demonstrate that increased extracellular Na+ impacts on the electrolyte balance of MΦ, while the nonionic osmolyte mannitol does not affect intracellular Na+ levels. We also tested whether mannitol increases MΦ antibacterial activity. In contrast to high salt conditions, mannitol impaired antibacterial MΦ activity (Figure 2(c)).
Figure 2.
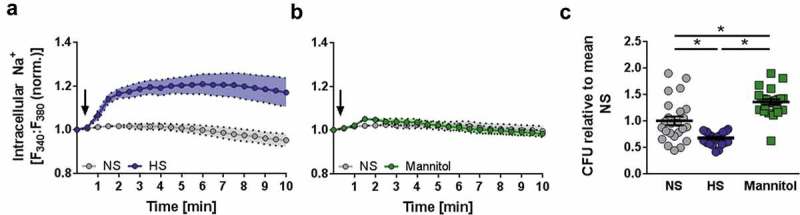
Osmotic stress induced by mannitol does not induce Na+ entry and impairs antibacterial activity. (a) Intracellular Na+ levels were assessed in SBFI-loaded and E. coli-infected RAW264.7 MΦ by ratiometric live cell imaging. The arrow indicates the addition of 40 mM NaCl (corresponding to an increase in osmolality of 80 mOsm/kg) to infected cells (means ± s.e.m; n = 6). (b) As in (a), but instead of NaCl, 80 mM mannitol (corresponding to an increase in osmolality of 80 mOsm/kg) was added where indicated (means ± s.e.m; n = 6). (c) BMDM were infected as in Figure 1(f), but with an additional condition using 80 mM mannitol (means ± s.e.m; n = 24; Kruskal-Wallis test with Dunn’s multiple comparison tests; * p < 0.05).
Increased antibacterial activity of macrophages under high Na+ conditions is not mediated by enhanced NOS2 or PHOX activity
We previously demonstrated that high salt enhanced MФ clearance of the intracellular parasite Leishmania major by increasing the production of leishmanicidal NO [1]. Therefore, we wanted to determine whether the HS-increased antibacterial effects observed here similarly hinge on enhanced NO and/or ROS (reactive oxygen species) production. In line with a recent publication [33], HS treatment decreased ROS production in E. coli-infected MΦ (Figure 3(a)). In MΦ from mice deficient for the Cybb (cytochrome b 245 subunit of phagocyte NADPH oxidase [cybb-/-]), HS treatment similarly increased antibacterial activity (Figure 3(b)). As described earlier [1], HS increased E. coli-triggered NO production at a MOI (multiplicity of infection) of 10 (Figure 3(c)). This was paralleled by enhanced bacterial elimination (Figure 3(d)). However, at higher MOIs of 50 and 100, exposure to high extracellular [Na+] did not further increase NO production (Figure 3(c)), but still facilitated enhanced E. coli elimination (Figure 3(d)). Together, this suggests that the HS-increased antibacterial MΦ activity is not associated to NO production. In line with this, inhibition of NOS2 (and PHOX [phagocyte oxidase]) activity using DPI (diphenyleneiodonium chloride) impaired NO release of infected MΦ (Figure 3(c)), but this did not interfere with HS-mediated antibacterial activity, as HS treatment still enhanced MΦ bacterial elimination (Figure 3(d)). Altogether, these findings indicate that Na+ increased MΦ antibacterial activity does not require PHOX- or NOS2-activity.
Figure 3.
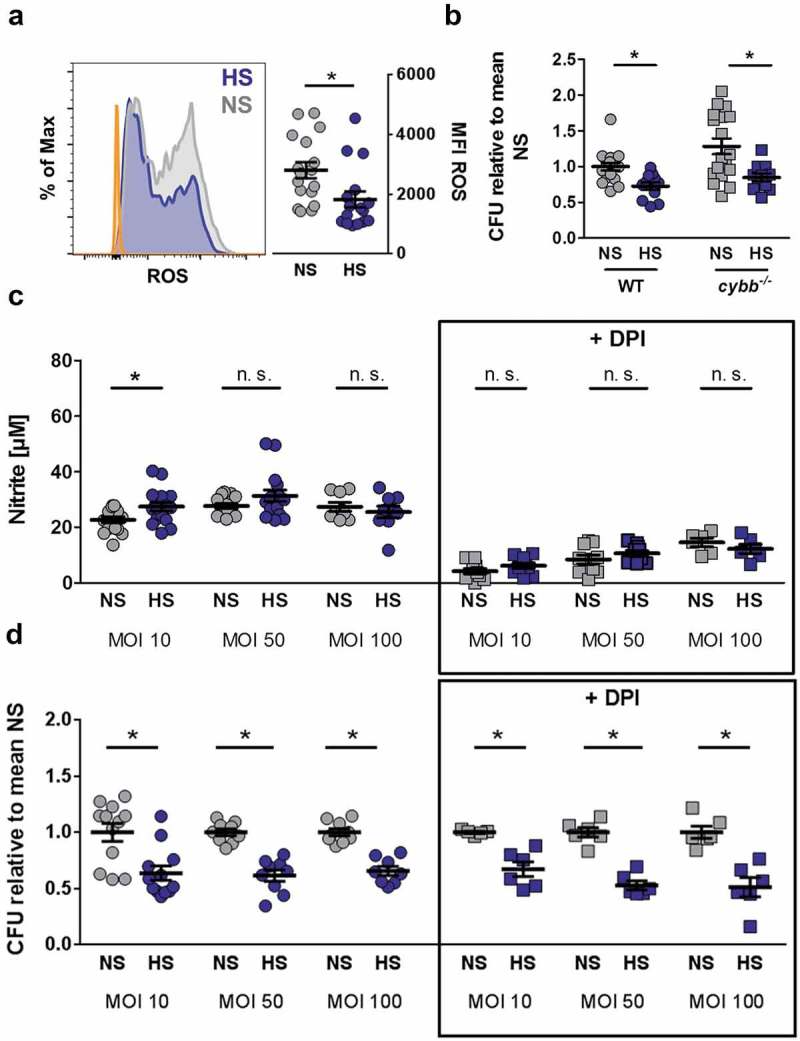
HS-boosted antibacterial activity of MΦ is independent of PHOX and NOS2. (a) As in Figure 1(f), but 6 h after infection, BMDM were stained with CM-H2DCFDA and subjected to flow cytometry. Left section, histogram. Orange area, unstained cells; Gray area, NS; Blue area, HS. Right section, means ± s.e.m; n = 16 −17; Mann Whitney test; * p < 0.05 (b) As Figure 1(f), but cybb-/- and littermate control (WT) BMDM were used. Intracellular E. coli in CFU relative to mean CFU under NS WT conditions (means ± s.e.m; n = 11–18; Student’s t test ± Welch correction; * p < 0.05) (c) BMDM were infected with E. coli at the indicated MOI, subjected to gentamicin protection assays and exposed to NS and HS conditions 2 h after infection ± DPI where indicated. 24 h after infection, nitrite levels were quantified (means ± s.e.m; n = 6–16; Student’s t test and Mann Whitney test; * p < 0.05). (d) As in (c), intracellular E. coli load 24 h after infection in CFU relative to mean CFU under the respective NS conditions (means ± s.e.m; n = 5–12; Student’s t test ± Welch correction; * p < 0.05).
High Na+ conditions increase autophagy of E. coli-infected macrophages
Recently, we demonstrated that high dietary salt intake induces autophagy in muscle cells [21]. Given that autophagy is linked with cell-autonomous elimination of bacteria [23,24], and that increased Na+ induces autophagy in MΦ [34], we hypothesized that HS might mediate increased MΦ antibacterial defense via elevated autophagy. To test this, we performed gene set enrichment analysis (GSEA) with an Illumina Bead chip array dataset of BMDM exposed to high Na+ levels published by Ip et al [2], and our own Agilent microarray dataset obtained from RAW246.7 MΦ stimulated with LPS (lipopolysaccharide) in the absence or presence of HS. By comparing these datasets with the list of genes annotated by the HUGO (Human Genome Organization) as ‘Autophagy related’ or in the ‘Kegg Autophagy pathway’, we found significant positive enrichment for both gene lists in the data from Ip et al (Kegg – NES [normalized enrichment score] = 2.086, FDR [false discovery rate] q value = 0.00480, HUGO – NES = 1.657, FDR q value = 0.0235; HUGO GSEA in Figure 4(a)), and the HUGO gene set in our own microarray (HUGO – NES = 2.323, FDR q value = 0.0085, Kegg – NES – 1.243, FDR q value = 0.1991; Figure 4(b)), indicating an increased expression of genes involved in autophagy in both datasets. Differentially expressed genes from our microarray dataset within the HUGO ‘Autophagy related’ gene set are presented in Figure 4(c). Various transcripts involved autophagy initiation (such as Ulk1 [unc-51 like kinase 1]), vesicle nucleation (such as Becn1 [beclin 1]), vesicle elongation (such as Atg12 [autophagy related 12]) and membrane retrieval (such as Atg2a [autophagy related 2a]) were altered in HS conditions (Figure 4(c)). Several genes were validated by quantitative real-time PCR experiments with E. coli-infected RAW246.7 MΦ, which revealed that the expression of Ulk1, Atg2a and Atg4c (autophagy related 4C, cysteine peptidase) is indeed increased in HS conditions, while the expression of Becn1 and Atg7 (autophagy related 7) was not affected (Figure 4(d)). BMDM were generated from mice that constitutively express GFP (green fluorescent protein)-MAP1LC3/LC3 (microtubule associated protein 1 light chain 3). We detected more GFP-MAP1LC3 puncta upon HS stimulation in E. coli-infected cells (Figure 4(e)), indicating enhanced autophagosome formation. To confirm this finding, we analyzed the abundance of the free cytosolic MAP1LC3B-I/LC3B-I and its lipidated form, MAP1LC3B-II/LC3B-II, which is associated with autophagosome formation. Under HS conditions, E. coli-infected MΦ showed increased MAP1LC3B-II:control ratios (Figure 4(f), quantification and statistical analysis in Figure S3(a)). Given that autophagosomes undergo a maturation process consisting of fusions with endosomes and lysosomes, ultimately forming an autolysosome [35], we assessed the MAP1LC3B-II:control levels in MΦ where the digestive function of the lysosomes was blocked with the vacuolar-type H+-ATPase (v-ATPase) inhibitor, bafilomycin A1. HS exposure resulted in increased MAP1LC3B-II:control levels in the absence or presence of bafilomycin A1-treatment in E. coli-infected MΦ (Figure 4(f)). This suggests that the accumulation of MAP1LC3-II is due to enhanced induction of autophagy, and not an impairment of autolysosomal function. SQSTM1/p62 (sequestosome 1) protein is degraded in autolysosomes and assists in the targeting of cargo to autophagosomes [35]. While HS conditions increased Sqstm1 transcription (Figure 4(g)), HS exposure nevertheless decreased the abundance of SQSTM1 protein in E. coli-infected MΦ (Figure 4(g), quantification and statistical analysis in Figure S3(b)); again, indicating increased autophagy under HS conditions [35]. Of note, the HS-increased formation of MAP1LC3B-II was independent of PHOX, again implying that a HS environment induces canonical autophagy (Figure 4(h), quantification and statistical analysis in Figure S3(c)). In line with this, transmission electron microscopy revealed that infected RAW264.7 MΦ treated with HS contained more double-membraned vacuoles in close proximity to bacteria-containing lysosomes, which were also more often surrounded by double membranes (Figure 4(i)). Collectively, these findings demonstrate that HS promotes autophagy in E. coli-infected MΦ.
Figure 4.
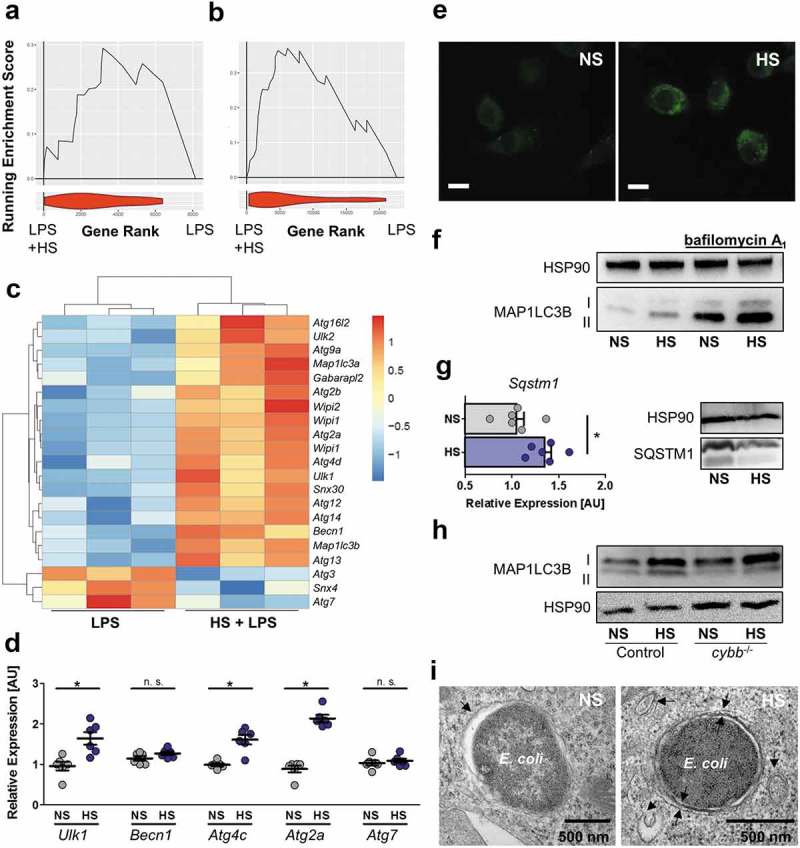
HS increases autophagy in E. coli-infected MΦ. (a and b) GSEA for the HUGO autophagy gene set. Upper section, genes are placed in rank order based on differences in expression on the x-axis and the curve represents the running enrichment score across the dataset. Bottom section, violin plot indicating the frequency distribution of the genes within the HUGO gene set by rank. (a) GSEA using the dataset of Ip et al. [2] (b) GSEA of own Agilent microarray data of RAW264.7 MΦ stimulated with LPS ± 40 mM NaCl for 24 h. (c) Heatmap of gene expression from own Agilent array. Genes plotted are genes within the HUGO autophagy gene set and significantly differentially expressed (Benjamini Hochberg adjusted p value < 0.05) for the addition of Na+ to LPS (10 ng/mL) treated MΦ. (d) As in Figure 1(f), but Ulk1, Becn1, Atg2a, Atg4c and Atg7 mRNA levels were quantified (means ± s.e.m; n = 6; Student’s t test ± Welch correction; * p < 0.05). (e) As Figure 1(f), but GFP-MAP1LC3 expressing BMDM were used. GFP-MAP1LC3 puncta formation was assessed. Representative images of three experiments are displayed. Scale bar: 10 µm. (f) RAW264.7 MΦ were treated as in (d), but for ½ h ± pretreatment with 100 nM bafilomycin A1. A representative immunoblot of MAP1LC3B and HSP90 levels is displayed. (g) As in (d), Sqstm1 was determined on mRNA (means + s.e.m; n = 6; Student’s t test; * p < 0.05) and protein level. (h) As (f), but Cybb-/- and controls BMDM were used. (i) As in (d), but cells were subjected to transmission electron microscopy. Representative E. coli-containing vacuoles of two experiments are displayed.
High Na+ conditions favor autolysosomal-targeting of E. coli
Next, we monitored autophagosome and autolysosome formation in RAW264.7 MΦ expressing a tandem monomeric RFP (red fluorescent protein)-GFP-tagged MAP1LC3 (RAW-Difluo™ mLC3). In these cells, the GFP-signal is more sensitive to quenching than the RFP-signal, which is more stable in the acidic lysosomal environment [36]. HS treatment increased the formation of RFP+ GFP+ puncta (autophagosomes), as well as the amount of RFP+ GFP− vacuoles (autolysosomes; Figure 5(a)). This shows that HS conditions increased the formation of both autophagosomes and autolysosomes. Next, we assessed whether HS favors the targeting of E. coli to RFP-positive vacuoles. HS treatment increased the co-localization of sCFP3A-E. coli with RFP-positive vacuoles from ~33% to ~67% (Figure 5(b)). These findings indicate that HS conditions increase the delivery of E. coli to acidic autolysosomes. Comparably, we found that HS treatment promoted the delivery of E. coli to acidic vacuoles stained with the lysosomotropic dye LysoTracker (Figure 5(c)). Taken together, these findings strongly indicate that HS conditions increased the induction of autophagy and the targeting of E. coli to autolysosomes.
Figure 5.
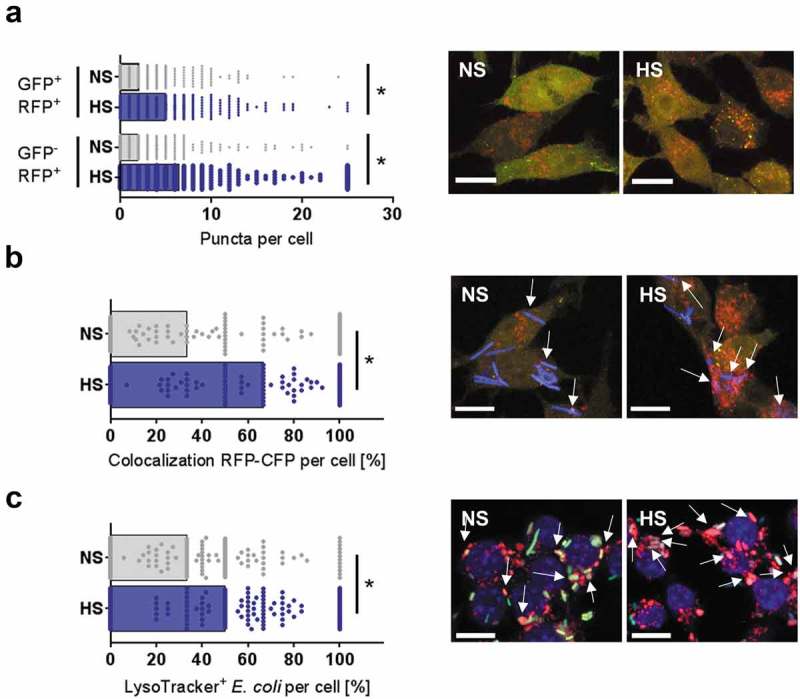
HS facilitates autolysosomal targeting of E. coli. (a) As in Figure 1(f), but RFP-GFP-mLC3 RAW264.7 MΦ were used. RFP+GFP− and RFP+GFP+-puncta were counted per cell. Confocal images: GFP, green; RFP, red. Scale bar: 10 µm. At least 287 MΦ from two independent experiments (bars: median scores; Mann-Whitney test; * p < 0.05). (b) As in (a), but sCFP3A-E. coli were used. Confocal images: GFP, green; RFP, red; sCFP3A-E. coli, blue. Scale bar: 10 µm. Rate of E. coli-sCFP3A/RFP+GFP–colocalization per cell. At least 118 MΦ from two independent experiments (bars: median scores; Mann-Whitney test; *p < 0.05). (c) RAW264.7 MΦ were infected with sfGFP-E. coli as in Figure 1(f), but stained with LysoTracker. Confocal images: sfGFP-E. coli, green; LysoTracker, red; DAPI (DNA), blue. Scale bar: 10 µm. Colocalization of sfGFP-E. coli and LysoTracker per MΦ. At least 166 MΦ from two independent experiments were analyzed (bars: median scores; Mann-Whitney test; * p < 0.05).
Autophagy and acidic compartments are required for high Na+-increased antibacterial activity of macrophages
To determine the importance of autophagy for the increased antibacterial defense of MΦ treated with HS, we generated MΦ from the bone marrow of atg7 cKO (Atg7F/F: Lyz2[lysozyme 2]-Cre) mice, which have severe cellular impairment of autophagy-associated phagosomal clearance [37]. While immunoblotting for ATG7 revealed efficient recombination in Atg7-deficient MΦ, we detected increased ATG7 protein expression upon HS-exposure in controls (Figure 6(a), quantification and statistical analysis in Figure S4(a)), while its mRNA levels remained unaltered (Figure 4(d)), implying that post-transcriptional processes are involved in ATG7 regulation. HS-treatment failed to increase antimicrobial activity of Atg7-deficient MΦ, which was maintained in control MΦ (Figure 6(a)). Next, we determined the contribution of lysosomal acidification on HS-increased MΦ antibacterial function. Pharmacological blockade of the v-ATPase impaired bactericidal activity of MΦ under normal salt conditions, and abolished the HS-increased antibacterial activity of WT MΦ (Figure 6(b)). These data indicate that lysosomal acidification is a key component of the mechanism by which HS increases MΦ antibacterial activity.
Figure 6.
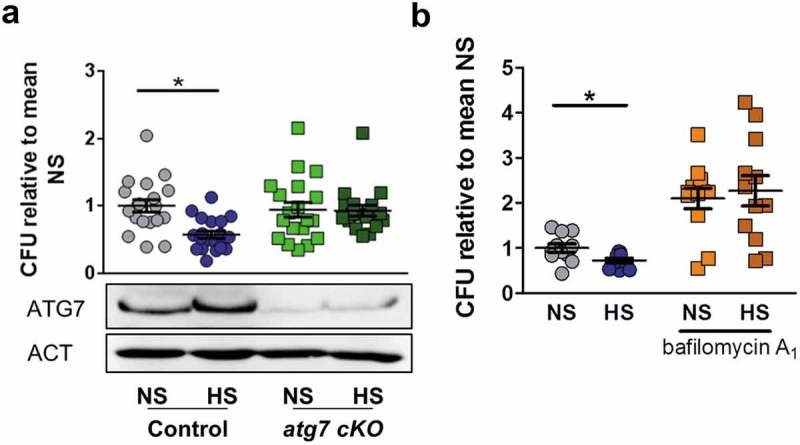
Autophagy and lysosomal acidification are required for high Na+-increased antibacterial activity. (a) As in Figure 1(a), but BMDM from atg7 cKO and littermate controls were used. Upper section, intracellular E. coli load in CFU relative to mean CFU under NS conditions (means ± s.e.m; n = 18–20; Student’s t test + Welch correction or Mann Whitney test, * p < 0.05). Lower section, Immunoblotting of ATG7 and ACT. (b) As in Figure 1(a), but RAW264.7 MΦ were pretreated with 100 nM bafilomycin A1. Intracellular E. coli load in CFU relative to mean CFU under NS conditions (means ± s.e.m; n = 11–12; Student’s t test or Mann Whitney test, * p < 0.05).
Low pH levels are known to inhibit protease activities of several cathepsins such as CTSB (cathepsin B) [38]. Recently Qi et al demonstrated that CTSB impaired MΦ-dependent defenses against Francisella novicida [39]. Therefore, it is conceivable that the observed increase in phagosome maturation and acidification under HS conditions might decrease cathepsin activity and thereby facilitate increased removal of E. coli. We used a cell permeable cysteine protease inhibitor (E-64d) that targets CTSB and other papain-like cysteine proteases [40] in order to address this possible mechanism (Figure S4(b)). CTSB activity was not altered by HS (Figure S4(b)). Moreover, HS still increased the antimicrobial activity of E-64d-treated MΦ (Figure S4(c)). Overall, our data demonstrate that autophagy and lysosomal acidification critically contribute to the increased antibacterial activity under HS conditions, independent of CTSB activity.
NFAT5 is required for the targeting of intracellular E. coli to autolysosomes in high Na+
Given that the transcription factor NFAT5 plays a key role in the adaptation of macrophages to increased extracellular Na+ [5–7], we tested the contribution of this transcription factor to the autophagy-mediated pathogen clearance mechanisms described here. Upon HS treatment of E. coli-infected MΦ, we detected increased Nfat5 mRNA expression (Figure 7(a)) and protein levels (Figure 7(b), quantification and statistical analysis in Figure S5(a)). Silencing of Nfat5 did not affect Atg7 mRNA expression (Figure 7(c)), nor abolish the HS-increased abundance of ATG7 protein (Figure S5(b)). Additionally, the expression of Ulk1, Atg2a or Atg4c mRNA was not influenced by Nfat5-silencing (Figure 7(d)). In line with this, lipidation of MAP1LC3B was not impaired in Nfat5-deficient MΦ. Moreover, while we detected enhanced MAP1LC3B-II:control levels under NS conditions in Nfat5-silenced MΦ, there was still an increase in MAP1LC3B-II:control levels upon HS exposure, indicating that NFAT5 is dispensable for the increased formation of these components of the autophagic machinery (Figure 7(e)).
Figure 7.
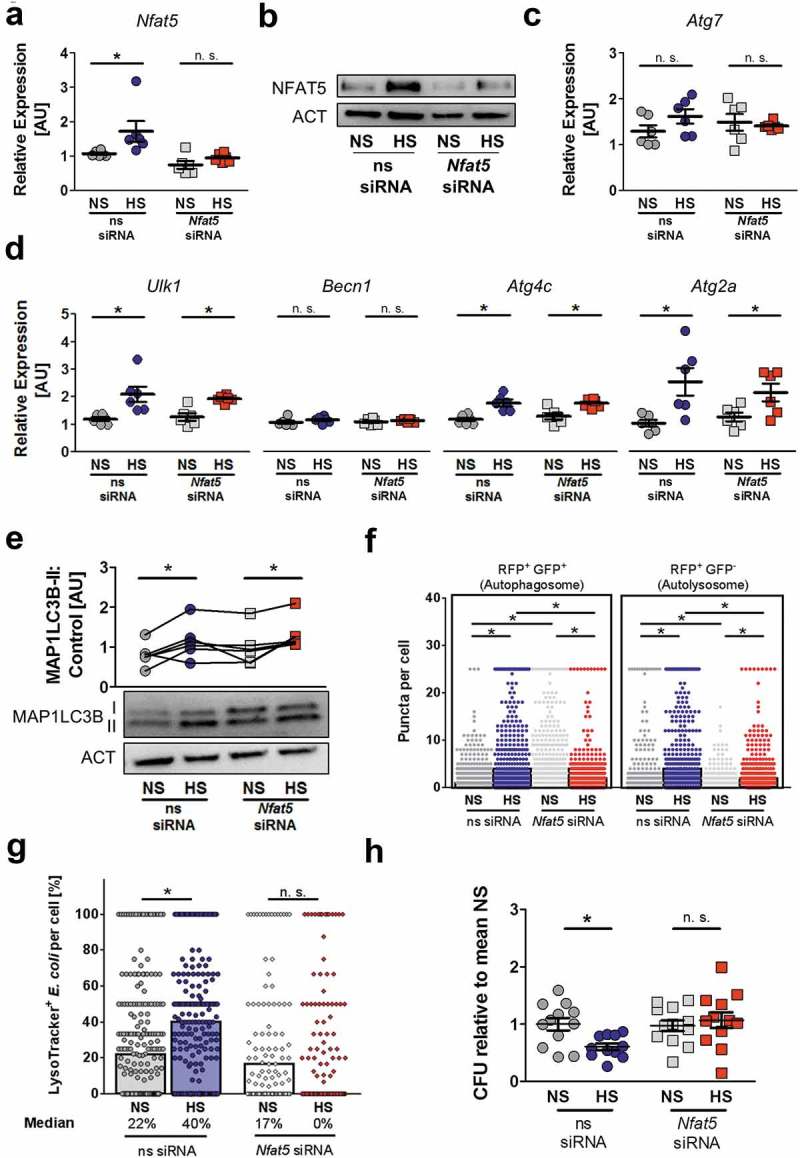
NFAT5 is required for HS-facilitated targeting of intracellular E. coli to autolysosomes. (a to d) As in Figure 1(f), but non-silencing siRNA (ns siRNA) and Nfat5-specific siRNA (Nfat5 siRNA)-treated RAW264.7 MΦ were used. (a) Nfat5 mRNA levels (means ± s.e.m; n = 6; Student’s t test or Mann-Whitney test, * p < 0.05). (b) Immunoblotting of NFAT5 and ACT. (c) As (a), but Atg7 mRNA levels (means ± s.e.m; n = 6; Student’s t test ± Welch correction). (d) As (a), but Ulk1, Becn1, Atg4c and Atg2a mRNA levels (means ± s.e.m; n = 6; Student’s t test ± Welch correction; * p < 0.05). (e) Immunoblotting and densitometry of MAP1LC3B/LC3 and ACT ½ h after infection (n = 6; paired Student’s t test or Wilcoxon signed rank test; *p < 0.05). (f) As in Figure 1(f), but ns siRNA- and Nfat5 siRNA-treated RFP-GFP-mLC3 RAW264.7 MΦ were used. RFP+ and RFP+GFP+-puncta were counted per cell. At least 290 cells from two independent experiments (bars: median scores; Kruskal Wallis test with Dunn’s Multiple Comparison Test, * p < 0.05). (g) As Figure 1(f), but RAW264.7 MΦ were used and stained with LysoTracker. Colocalization of sfGFP-E. coli and LysoTracker per cell. At least 101 cells from at least two independent experiments (bars: median scores; Kruskal Wallis test with Dunn’s Multiple Comparison Test, * p < 0.05). (h) As in Figure 1(a), but ns siRNA- and Nfat5 siRNA-treated RAW264.7 MΦ were used. Intracellular E. coli load in CFU relative to mean CFU under NS conditions (means ± s.e.m; n = 11–12; Student’s t test ± Welch correction, * p < 0.05).
Next, we tested whether NFAT5 is involved in autolysosomal formation. To address this, we determined the impact of NFAT5 on the formation of autophagosomes and autolysosomes in RFP-GFP-mLC3 RAW264.7 MΦ as before. HS treatment increased the formation of RFP+ GFP+ puncta (autophagosomes), as well as the number of RFP+ GFP− vacuoles (autolysosomes) in control cells. By contrast, Nfat5-silencing increased the formation of RFP+ GFP+ vesicles but suppressed the formation RFP+ GFP− puncta under NS conditions (Figure 7(f)), in line with our finding of enhanced MAP1LC3B-II:control levels in Nfat5-deficient MΦ (Figure 7(e)). This suggests that NFAT5 is important for the degradation of MAP1LC3B-II under NS conditions. However, upon HS exposure Nfat5-silencing blunted both the formation of RFP+ GFP+ and RFP+ GFP− vesicles (Figure 7(f)). Overall, this indicates that Nfat5 plays an important role in autophagosome and autolysosome formation, a phenotype which becomes further evident upon exposure to HS. In line with this, we found that the targeting of E. coli to acidic compartments was severely impaired in Nfat5-deficient MΦ upon exposure to HS (Figure 7(g)). Finally, we tested the contribution of Nfat5 in mediating HS-enhanced bacterial elimination. Nfat5-silencing abolished the HS-increased MΦ antibacterial activity, which was preserved in MΦ treated with control ns-siRNA (Figure 7(h)). Altogether, these findings demonstrate that NFAT5 does not mediate the increased expression of components of the autophagy machinery, nor does it play a direct role in increasing MAP1LC3B-II formation upon HS treatment. However, NFAT5 is required for the HS-increased formation of autophagosomes and autolysosomes, and the targeting of E. coli to acidic lysosomal compartments; ultimately, these result in enhanced intracellular bacterial elimination.
Blunted AKT and MTOR activation does not account for HS-increased antibacterial activity
Previously, we demonstrated that HS conditions impair AKT activation in MΦ [27]. Since AKT and MTOR blockade is known to induce autophagy [25], we tested whether pharmacological blockade of cells with the AKT and MTOR inhibitor Torin1 [41] under NS conditions is sufficient to mimic HS-boosted antimicrobial MΦ activity. Administration of Torin1 abolished AKT activation and increased MAP1LC3-II:control protein abundance in NS conditions, i. e. independent of increased extracellular Na+, due to ATP-competitive inhibition of MTORC1/2 [41]. Additionally and confirming earlier findings [27], we found that HS conditions blunted AKT activation in MΦ (Figure 8(a), quantification and statistical analysis in Figure S6(a-b)). In line with an earlier publication [42], Torin1-treatment, did not abrogate the HS-triggered induction of the osmoprotective transcription factor NFAT5 (Figure 8(b), quantification and statistical analysis in Figure S6(c)). Although treatment with Torin1 increased autophagy in MΦ under NS conditions, as assessed with RFP-GFP tagged MAP1LC3 reporter cells (Figure 8(c)), Torin1-treatment did not increase the targeting of intracellular E. coli to autolysosomes under NS conditions (Figure 8(d)). Consequently, MΦ exposed to Torin1 in NS conditions only tended to increase the killing of intracellular E. coli (adjusted p value = 0.08; Figure 8(e)). By contrast, HS increased the targeting of E. coli to autolysosomes (Figure 8(d)) and killing of E. coli (Figure 8(e)), either in the absence or presence of Torin1. Together, these data suggest that increased autolysosome formation alone is not sufficient to improve bacterial elimination, but the targeting of E. coli to autolysosomes is also required for HS-increased antibacterial activity.
Figure 8.
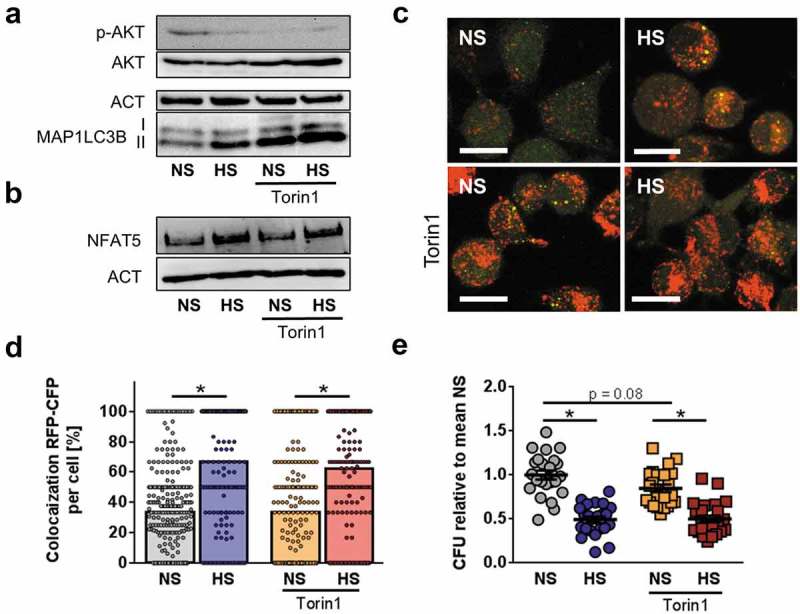
Blunted AKT and MTOR activation does not account for HS-augmented antibacterial activity. (a) As in Figure 1(f), RAW264.7 MΦ were pretreated ± 1 µM Torin1 and infected ± HS. Cells were harvested ½ h post infection. Immunoblots of p-AKT, AKT, MAP1LC3B and ACT levels are displayed. (b) As in (a), but 2 h post infection, NFAT5 and ACT levels were assessed. (c) As in (b), but RFP-GFP-mLC3 RAW264.7 MΦ were used. A representative confocal image out of two experiments is shown. Scale bar = 10 µm. (d) As in (c), but sCFP3A-E. coli were used. Rate of E. coli-sCFP3A/RFP-colocalization per cell. At least 184 MΦ from two independent experiments (bars: median scores; Mann Whitney test; * p < 0.05). (e) As in (b), but intracellular E. coli load is displayed 2 h after infection in CFU relative to mean CFU under NS conditions (means ± s.e.m; n = 23–24; ANOVA with Bonferroni’s test; * p < 0.05).
The presence of HIF1A is necessary for HS-mediated autophagy induction and antibacterial activity
To identify potential mechanisms by which HS mediated its effects on autophagy, we performed a GSEA for positive and negative regulators of autophagy with our microarray data obtained from LPS-stimulated RAW246.7 MΦ in NS and HS as before. We found that HS did not affect the expression of negative regulators of autophagy (NES 1.27, FDR q value 0.191; Figure 9(a)). However, positive regulators of autophagy were increased with HS (NES 2.36, FDR q value 0.000; Figure 9(b)). This included the transcription factor Hif1a and its target Bnip3 (BCL2/adenovirus E1B interacting protein 3; Figure 9(c)) [43,44]. These findings were validated with E. coli-infected MΦ that exhibited similar increased expression of Hif1a mRNA and protein with HS treatment (Figure 9(d), quantification and statistical analysis in Figure S7(a)). In addition to the Hif1a-target Bnip3, HS also increased the expression of the glycolytic Hif1a-targets Slc2a1 (solute carrier family 2 [facilitated glucose transporter], member 1) and Pgk1 (phosphoglycerate kinase 1; Figure S7(b)) [45,46].
Figure 9.
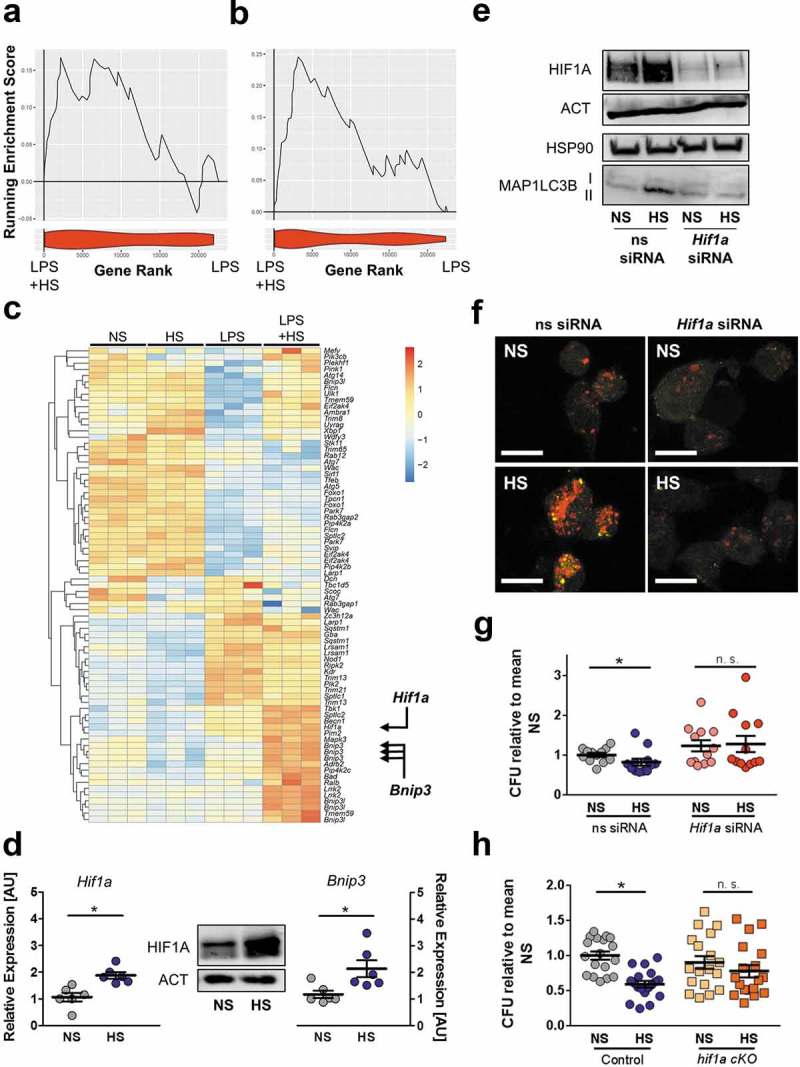
The presence of HIF1A is required for HS-triggered autophagy induction and antibacterial activity. (a and b) GSEA for negative (a) and positive (b) regulators of autophagy. Upper section, genes are placed in rank order based on differential expression between the groups on the x-axis and the curve represents the running enrichment score across the dataset. Bottom section, violin plot indicating the frequency distribution of the genes within the respective GO of positive enrichment of autophagy regulators by rank. (c) Heatmap of gene expression from own Agilent array. Genes plotted are genes within the HUGO autophagy regulatory gene set, which are upregulated and significantly differentially expressed (Benjamini Hochberg adjusted p value < 0.05) with the addition of NaCl to LPS-treated MΦ. (d) Left and right section, as in Figure 1(f), but Hif1a and Bnip3 mRNA levels were quantified (means ± s.e.m; n = 6; Student’s t test or Mann Whitney test; * p < 0.05). Middle section, HIF1A and ACT protein levels. (e to g) As in Figure 1(f), but ns siRNA- and Hif1a-specific siRNA (Hif1a siRNA)-treated RAW264.7 MΦ were used. (e) Upper section: HIF1A and ACT protein levels 4 h after infection. Lower section: MAP1LC3B and HSP90 as Figure 4(f). (f) Instead of RAW264.7 MΦ, RFP-GFP-mLC3 RAW264.7 MΦ were used. Confocal images: GFP, green; RFP, red. Scale bar = 10 µm. Representative images out of two experiments are shown. (g) As in Figure 1(f), intracellular E. coli load, CFU relative to mean CFU under ns siRNA-treated NS conditions (means ± s.e.m; n = 12; Mann Whitney test; * p < 0.05) (h) As in Figure 1(f), but BMDM from hif1a cKO and littermate controls were used. Intracellular E. coli load in CFU relative to mean CFU under NS conditions is displayed (means ± s.e.m; n = 18; Student’s t test or Mann Whitney test; * p < 0.05).
To validate the role of Hif1a in HS-increased autophagy and antibacterial activity, we transfected Hif1a siRNA into RAW264.7 MΦ, which almost completely extinguished Hif1a expression in NS and HS conditions, leading to a suppression of Bnip3 mRNA, and abrogation of the HS-increased Bnip3 expression (Figure S7(c)). In parallel, HS treatment of Hif1a-silenced MΦ diminished MAP1LC3B-II:control levels (Figure 9(e), quantification and statistical analysis in Figure S7(d,e) and impaired the HS-increased formation of RFP+ GFP+ puncta (autophagosomes) and RFP+ GFP− vacuoles (autolysosomes; Figure 9(f)). Together, these data demonstrate that HIF1A is required for HS-increased autophagy. To assess the role of Hif1a in improving bacterial elimination in HS conditions, we next tested the antibacterial activity of Hif1a-silenced RAW264.7 MΦ (Figure 9(g)) and of Hif1a-deficient BMDM (hif1a cKO, Hif1aF/F: Lyz2-Cre; Figure 9(h)). In both the Hif1a knock-down and hif1a cKO situations, the effect of HS in increasing antimicrobial MΦ activity was abolished (Figure 9(g,h)). From these data we conclude that HS-increased induction of autophagy is coordinated by HIF1A, which is critically required for mediating these HS enhanced antibacterial defenses.
Finally, we set out to address the relationship between HIF1A and NFAT5 in mediating the effects of Na+ on MФ activity. We first tested the effect of Hif1a expression on Nfat5 and vice versa. Hif1a-silencing did not affect the expression of Nfat5, nor did it abrogate the HS-increased expression of Nfat5 mRNA (Figure S8(a)) and protein (Figure S8(b)).
By contrast, Nfat5-silencing did not affect Hif1a mRNA and protein expression in normal salt conditions, but it did abolish the HS-increased Hif1a mRNA (Figure S8(c)) expression, and also reduced HIF1A protein levels (Figure S8(d)). Since HS is still able to induce autophagy in Nfat5-silenced cells (Figure 7(d–f)), and Hif1a is required for HS-triggered autophagy induction (Figure 9(e–h)), these findings suggest a separation between the relative expression level of HIF1A and its role in bacterial elimination: while the complete absence of HIF1A abolishes HS-enhanced antibacterial activity, the increased expression of HIF1A induced by HS is dispensable. To corroborate this result, we increased HIF1A accumulation in Nfat5-silenced cells using a chemical stabilizer of HIF1A, DMOG (dimethyloxallyl glycine) [47,48], and tested for its impact on antibacterial activity. These experiments revealed that increasing HIF1A levels in Nfat5-silenced cells is not able to restore the impaired antimicrobial activity under HS conditions (Figure S8(e,f)).
Taken together, we demonstrate here that HS conditions (i) require the presence of HIF1A for increased autophagy induction, and (ii) facilitate targeting of E. coli to acidic autolysosomal compartments via increasing NFAT5 (Figure 10).
Figure 10.
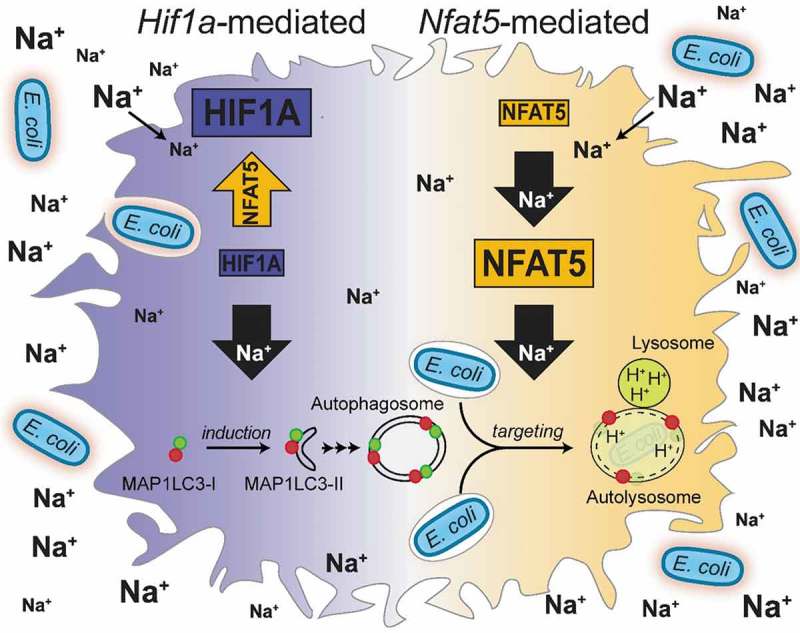
Schematic of mechanisms involved in HS-increased antibacterial activity of MΦ.
Discussion
Components of the autophagy pathway are known to promote phagosome maturation and acidification and thereby e. g. foster degradation of the yeast Saccharomyces cerevisiae [49], Mycobacterium tuberculosis [50] and a virulent adherent-invasive E. coli strain [51]. In this work, we demonstrate that HS induces autophagy, favors autolysosomal formation and targeting of E. coli to acidic lysosomal compartments which, ultimately, results in enhanced intracellular bacterial degradation. Of note, we found in our study that ATG7 was not required for antimicrobial control of E. coli HB101 under normal cell culture conditions (i. e. normal salt conditions). In line with this, Chiu et al reported for instance that under normal cell culture conditions ATG7 was dispensable for control of Salmonella enterica serovar Typhimurium in RAW267.4 MΦ, while an antimicrobial compound increased autophagy in this situation and thereby augmented anti-Salmonella control [52]. Moreover, in line with others [53], our data suggest that compared to an adherent-invasive E. coli [51], the E. coli HB101 strain used in our study is a poor inducer of autophagy under normal salt conditions. Hence autophagy does not participate in removal of E. coli HB101 under normal salt conditions. However, if infected MΦ are incubated under HS conditions, (i) autophagy becomes functional and (ii) ultimately contributes to antimicrobial control by increasing autophagic degradation processes and facilitating the targeting of E. coli to acidic lysosomal compartments. Moreover, our data demonstrate that the autophagic phenotype is largely dependent on the lysosomal acidification. Our data also suggest that modulation of CTSB activity is unlikely to be involved in HS-increased antimicrobial activity. However, it is possible that other hydrolases might be involved in enhanced lysosomal pathogen elimination.
Since autophagy and autophagy-associated pathways play an important role in antigen-processing and presentation in the context of major histocompatibility complex class II and class I molecules [54], our findings might explain enhanced antigen-processing in dendritic cells exposed to high salt concentrations [15]. Several bacteria, viruses and parasites developed strategies to evade autophagy-mediated immune surveillance [24,55–58]. It is conceivable that local increases in extracellular Na+ might help overcome pathogen-triggered strategies to circumvent autophagy-mediated degradation. Moreover, Na+-increased autophagy might facilitate clearing infected and inflamed tissues from dead cells and thereby curtail inflammatory responses upon sterile tissue damage as well [37].
In contrast to LC3-associated phagocytosis [59], we demonstrate that HS-boosted MAP1LC3B-lipidation and autophagy are independent of PHOX. Using our transcriptomic data, we identified HIF1A as a novel regulator of autophagy upon HS stress. In addition to low oxygen tensions, a plethora of other stimuli is able to induce HIF1A accumulation [60–62]. Our finding of HS-increased HIF1A accumulation in infected MΦ conforms to earlier reports that hypertonic conditions are able to induce HIF1A in mouse lung epithelial cells [63]. The mechanisms, however, by which HS conditions boost HIF1A accumulation under conditions of ample oxygen, are unknown. Given that canonical HIF1A accumulation is known to be regulated on a posttranslational manner [60–62], it is possible that HS induced HIF1A stabilization might involve posttranscriptional/posttranslational processes as well.
In MΦ it is largely unexplored how NFAT5- and HIF1A-dependent pathways are intertwined. Hypoxia is reportedly able to induce NFAT5 in synovial MΦ obtained from patients suffering from rheumatoid arthritis [10]. We provide evidence that HIF1A is not important for HS-induced NFAT5 expression, suggesting that mechanisms other than HIF1A may be involved in the hypoxia-induced NFAT5 accumulation. By contrast, while Nfat5-deficient cells had HIF1A present, we find that NFAT5 is essential for the HS-increased HIF1A accumulation. The mechanisms underlying this observation remain elusive and require further studies.
HIF1A has been implicated in regulating autophagy at various levels in different cells [26,64–68]. While we confirm that HIF1A is essential for inducing autophagy, our data suggest that this is independent of increasing its cellular expression upon HS treatment. It remains unclear, how HIF1A activates autophagy upon exposure to HS conditions. Bellot et al proposed that hypoxia induces HIF1A-dependent expression of Bnip3. This disrupts B-cell lymphoma 2-BECN1 complexes without triggering cell death but ultimately initiates autophagy via free BECN1 [26]. Alternatively, osmotic stress might, for instance, induce subcellular membrane defects [69,70] or activate various ion channels or transporters [71] which result in cellular stress responses including activation of HIF1A.
Confirming earlier findings [27], we showed that elevated extracellular Na+ concentration blunts AKT signaling. In contrast to pharmacological blockade of AKT- and MTOR signaling [72,73], HS-conditions result in reduced AKT activation and simultaneous induction of HIF1A. The molecular mechanisms for this finding require further investigation. In line with others [74], we provide evidence that MTOR-AKT inhibition under NS conditions increases autophagy as well as autophagosome and autolysosome formation. Surprisingly, this is not sufficient to mimic enhanced antimicrobial activity under NS conditions. However, in contrast to HS-treated cells, AKT and MTOR inhibition under NS conditions was not linked with enhanced targeting to autolysosomes. This strongly suggests that increases in autophagy and autolysosome formation are not sufficient but that in addition HS-triggered subcellular targeting of intracellular E. coli to autolysosomes is critically required for HS-enhanced antimicrobial MΦ activity.
In this study, we provide evidence that NFAT5 coordinates targeting of E. coli to autolysosomes under HS-conditions. In contrast to a recent study [75], but in line with findings demonstrating that NFAT5 does not affect expression of genes involved in autophagy such as Becn1 under normal salt conditions [76], our findings suggest that the osmoprotective transcription factor NFAT5 is not directly involved in autophagy induction. Our data demonstrate that NFAT5 is rather involved in regulation of subcellular trafficking, including formation of autolysosomes, and targeting of E. coli to these acidic compartments. In C. elegans, the expression of genes involved in endosomal trafficking and lysosomal biogenesis are required for survival upon exposure to hypertonic conditions [77]. Moreover, hypertonic stress conditions alter intracellular trafficking in the kidney proximal tubule cell line (LLC-PK) and promoted microtubule-dependent cluster formation [34]. Given that microtubular trafficking is implicated in formation of autophagosomes and its fusion with endosomes [78,79], we speculate that NFAT5 might be involved in this state of affair as well. Alternatively, rearrangements of actin cytoskeleton might be involved in this process [14]. However, how NFAT5 governs autophagosome and autolysosome formation and subcellular targeting of E. coli upon exposure to increases in extracellular Na+ requires further investigation.
Materials and methods
Reagents and antibodies
All media (RPMI, DMEM) and PBS buffers were purchased from Gibco (61870–010; 41966–029; 14190–094). NaCl was purchased from Merck. LPS (E. coli O111:B4) and DPI were purchased from Sigma Aldrich (D2926). Torin1 was purchased from Tocris (4247); E-64d was obtained from Hycultec (HY-15282). Ionomycin (407952) was purchased from CalbioChem. ATP (tlrl-atpl) and bafilomycin A1 (tlrl-baf1) were obtained from Invivogen. The prolyl-hydroxylase inhibitor DMOG was purchased from Cayman Chemical (71210). For immunoblotting, the following antibodies were used: rabbit anti-ACT (A2066; Sigma-Aldrich); rabbit-anti-HSP90A/B (Santa Cruz Biotechnology, sc-7947), rabbit anti-NFAT5 (Thermo Scientific, PA1-023), rabbit anti-MAP1LC3B (Novus Biologicals, NB100-2220), rabbit anti-ATG7 (Cell Signaling Technology, D12B11) rabbit anti-p-AKT (Cell Signaling Technology, 4060S), rabbit anti-AKT (Cell Signaling Technology, 9272S), rabbit anti-SQSTM1 (Sigma Aldrich, P0067), rabbit anti-HIF1A (Cayman Chemical, 10006421). As secondary antibody, we used swine anti-rabbit HRP (P0399, Dako).
Cells and bacteria
RAW264.7 MΦ and RFP-GFP-mLC3 RAW264.7 MΦ (RAW-DifluoTM mLC3 cells; purchased from Invivogen and used according to the instructions of the supplier) were used. BMDM from C57BL/6 WT mice and various knock out and transgene mice were generated in Teflon® bags, as described earlier [80]. In addition to C57BL/6 WT mice (Charles River Breeding Laboratories, Sulzfeld, Germany), we used GFP-MAP1LC3 mice [81], cybb-/- mice [82] and their littermate controls, atg7 cKO (Atg7F/F: Lyz2-Cre) mice [83,84] and their littermate controls, and hif1a cKO (Hif1aF/F: Lyz2-Cre) mice [85] and their respective littermate controls as described earlier [86,87]. For infection experiments, we used E. coli HB101 or, for immunofluorescence studies, E. coli harboring pWRG167 [88] and pWRG29 for constitutive expression of superfolder GFP (sfGFP) or super cyan fluorescent protein 3A (sCFP3A), respectively. The gene for sCFP3A [89] was PCR-amplified with primers XbaI-sCFP-for (TCC TCT AGA TTT AAG AAG GAG ATA TAC ATA TGG TGA GCA AGG GCG AGG AG) and sCFP-HindIII-rev (TTA AAG CTT GCA TGC CTG CAG GTC TGG ACA TTT ACT TGT ACA GCT CGT CCA TG). The fragment was digested using XbaI and HindIII and subsequently ligated in the similarly treated vector pFPV25.1 [90] which resulted in pWRG29. For determination of minimal inhibitory concentrations a reference E. coli strain (ATCC 25922) was used in addition to E. coli HB101.
RNA interference in macrophages
RNA interference was performed as described earlier [1,91] and cells were subjected to experimental conditions.
Determination of minimal inhibitory concentrations
Bacteria were grown on blood agar and subjected to serial dilutions of gentamicin in regular cell culture media under NS and HS conditions. Minimal inhibitory concentration was determined after overnight incubation.
Measurement of intracellular ion concentrations
Prior to infection, cells were seeded on FluoroDishTM Plates (World Precision Instruments, FD3510-100) and loaded with fluorescence dyes for Na+ (90 min staining with 0.9 µM SBFI [Thermo Scientific, S1264] at 37°C), K+ (90 min staining with 0.9 µM Potassium-binding benzofuran isophthalate [Thermo Scientific, P1267] at 37°C), or Ca2+ (15 min staining with 1.6 µM Fura-2 [Thermo Scientific, F1221] at room temperature), in Tyrode solution (140 mM NaCl, 4 mM KCl, 1 mM MgCl2, 5 mM HEPES, 1 mM CaCl2, 10 mM Glucose) respectively. After ½ h of infection and additional ½ h of incubation in Tyrode solution containing gentamicin (100 µg/ml), cells were mounted on the stage of an epifluorescence microscope (Motic Model 410E). SBFI, PBFI or Fura-2 was alternatingly excited at 340 and 380 nm. Emitted fluorescence passed through appropriate emission filter sets, was acquired using a photomultiplier for 10 min and analyzed using IonWizard software (IonOptix Cooperation) as described earlier [92]. Fluorescence intensities were background substracted and normalized to F340/F380 at t = 0 min.
Macrophage infection experiments
Intracellular bacterial survival was assessed using a gentamicin protection assay as described earlier [1]. Briefly, RAW264.7 MΦ or BMDM were infected with E. coli HB101 with a MOI of 100 (unless indicated otherwise) for 1 h. Subsequently, extracellular bacteria were removed by washing with PBS. Gentamicin (100 µg/ml) was added to cell culture medium for 1 h. For long long term experiments (24 h) cells were washed with PBS and incubated with 25 µg/ml gentamicin for an additional 22 h. Where indicated, cells were exposed to additional 40 mM NaCl in the medium directly after infection or 2 h after infection. The infection was terminated at 2 h or 24 h post infection by lysis of the cells using 0.1% Triton-X and 0.05% Tween 80 in PBS. The number of remaining intracellular bacteria was determined by generating serial dilutions of the lysate in PBS on Müller-Hinton agar plates. Colony forming units (CFU) were counted after incubation of the plates at 37°C overnight. For immunofluorescence, cells were seeded on cover slips, infected with sfGFP or sCFP3A expressing E. coli HB101 and treated as described above. Instead of lysis, cells were fixed with 3.5% paraformaldehyde (PFA) for 15 min at RT. Subsequently, staining of the actin cytoskeleton using Phalloidin 647 (Invitrogen, A22287) was performed in blocking solution (5% BSA, 2% goat serum in PBS) containing 0.1% saponin for 2h at RT. To visualize DNA and mount the stained cells, we used Prolong Gold antifade reagent containing DAPI (Invitrogen, P36931). LDH release of infected MΦ was performed as described previously [93].
Phagocytosis assays
BMDMs were seeded in low adherence plates (Sigma Aldrich, CLS3473-24EA) and stimulated with FITC-labelled 1 µm Latex beads (Sigma Aldrich, L4655). Cells were kept for ½ h in normal cell culture media ± 40 mM NaCl and kept at 37°C and 4°C to assay internalization and attachment of beads by flow cytometry, respectively. After incubation and washing, cells were assessed by flow cytometry.
Quantification of nitrite and ROS production
As indicator of NO production, accumulation of nitrite in the cell supernatants was quantified by Griess reaction as described earlier [1]. For intracellular ROS measurements, infected cells were kept ± 40 mM NaCl for 6 h in low adherence plates. ½ h before collecting of the cells for flow cytometry, 10 µM of CM-H2DCFDA (Thermo Fisher, C6827) was added and cells were analyzed.
Measurement of CTSB activity
Cells were infected as described. 2 h post infection, CTSB activity was measured using the ‘Cathepsin B Activity Assay Kit’ (abcam, ab65300) according to the manufacturer’s instructions.
Immunoblotting
For immunoblotting, cells were lysed at the indicated time points as described earlier [1,94]. In order to assess the lipidation status of MAP1LC3B, we used Mini-PROTEAN® TGXTM gradient gels from 4% – 20% (Bio-RAD, 456–1094) for sodium dodecyl sulfate polyacrylamide gel electrophoresis and, for blotting, polyvinylidene difluoride membranes (Immobilon®-PSQ, Millipore, ISEQ00005). After staining with appropriate antibodies, signals were visualized by the Chemo Star Imager (Intas Science Imaging Instruments, Göttingen, Germany). Densitometry of western blots was determined via ImageJ measurements (Version 1.50b; Rasband, W.S., ImageJ, U. S. National Institutes of Health, https://imagej.nih.gov/ij/).
Gene expression microarray
RAW 264.7 MΦ were stimulated with or without 10 ng/ml LPS in the absence or presence of additional 40 mM NaCl in the cell culture media for 24 h. Total RNA was extracted with Trifast® (PeqLab) according to the manufacturer’s instruction. Transcriptional profiles were assessed using an Agilent Whole Mouse Genome Oligo Microarrays (8x60K). RNA quality and integrity were determined using the Agilent RNA 6000 Nano Kit on the Agilent 2100 Bioanalyzer (Agilent Technologies). RNA was quantified by measuring A260 on the ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Sample labelling was performed as detailed in the ‘One-Color Microarray-Based Gene Expression Analysis’ protocol (p/n G4140-90040). Briefly, 100 ng of each total RNA samples was used for the amplification and labelling step using the Agilent Low Input Quick Amp Labelling Kit (Agilent Technologies, 5190–2305). Yields of cRNA and the dye-incorporation rate were measured with the ND-1000 Spectrophotometer. The hybridization procedure was performed according to the ‘One-Color Microarray-Based Gene Expression Analysis’ protocol (p/n G4140-90040) using the Agilent Gene Expression Hybridization Kit (Agilent Technologies). Briefly, 0.6 µg Cy3-labeled fragmented cRNA in hybridization buffer was hybridized overnight (17 hours, 65°C) to Agilent Whole Mouse Genome Oligo Microarrays 8x60K (AMADID 028005) using Agilent’s recommended hybridization chamber and oven. Following hybridization, the microarrays were washed once with the Agilent Gene Expression Wash Buffer 1 for 1 min at room temperature followed by a second wash with preheated Agilent Gene Expression Wash Buffer 2 (37°C) for 1 min. Fluorescence signals of the hybridized Agilent Microarrays were detected using Agilent’s Microarray Scanner System G2505C (Agilent Technologies, Santa Clara, CA, USA). The Agilent Feature Extraction Software (FES) 10.7.3.1 was used to read out and process the microarray image files.
Microarray analysis
Microarray data was analyzed using the R package Limma within the R statistical environment using the standard work flow recommended for Agilent arrays. Briefly the data was read in, background corrected and quantile normalized between arrays. Probes of low expression were removed from the dataset. Low expressed probes were defined as probes which did not have an expression value greater than 110% of the 95th quantile of the negative control probes on each chip for at least 3 chips in the dataset. To identify differential expression of genes an appropriate liner model was fitted.
Gene set enrichment analyzes
GSEA was carried out using the java based graphical user interface [95] using a pre-ranked list with the classic enrichment statistic. For the data from Ip et al as there was no replicates, a ranked list was produced based on log2 fold change between conditions. For our dataset, genes were ranked based on the inverse of the unadjusted p value with the sign of the log2-fold change. GSEA was ran against 2 gene lists. The HUGO gene list consisted of all the genes annotated as ‘Autophagy associated’ by HUGO and the Kegg gene list is the ‘Kegg pathway’ for autophagy. For Autophagy regulation the gene ontology lists GO:1903599 ‘positive regulation of Autophagy’ and GO:0010507 ‘negative regulation of autophagy’ were used.
RNA isolation, reverse transcriptions, real-time PCR, and relative quantification
RNA extraction and reverse transcription were performed as described earlier [94]. We used the following TaqMan assays for quantitative real-time PCR (Applied Biosystems): Hprt1 (Mm00446968_m1), Atg2a (Mm01212087_m1), Atg4c (Mm01259886_m1), Ulk1 (Mm00437238_m1), Becn1 (Mm01265461_m1), Nfat5 (Mm00467257_m1), Atg7 (Mm00512209_m1), Hif1a (Mm01283760_m1), Bnip3 (Mm01275600_g1), Pgk1 (Mm01225301_m1), Slc2a1 (Mm00441480_m1). Quantitative real-time PCR was carried out with an ABI Prism 7900 sequence detector (Applied Biosystems, Darmstadt, Germany). Subsequently, data were processed and analyzed using the ΔΔCT method. We set normalized ratio of target mRNA:control Hprt1 in infected cells kept under normal salt conditions to 1.
Analysis of autophagosome and autolysosome formation as well as subcellular localization of E. coli
To monitor autophagosome and autolysosome formation as well as autolysosomal targeting of E. coli, RFP-GFP-mLC3 RAW264.7 MΦ were seeded on coverslips and infected with E. coli HB101 or sCFP3A-E. coli HB101 and subjected to gentamicin-protection assays as described above. 2 h post infection, cells were fixed with 3.5% PFA and mounted with Prolong Gold containing DAPI (Invitrogen). To assess targeting of E. coli to acidic vacuoles, RAW264.7 MΦ were seeded on coverslips and infected with sfGFP-E. coli HB101 and treated as described above. 1.5 h after infection, cells were stained with 0.5 µM LysoTracker (LysoTracker Red® DND-99, Thermo Fisher, L7528). 2 h post infection, cells were fixed with 3.5% PFA and mounted with Prolong Gold containing DAPI. Images were collected with a Leica TCS SP5 confocal laser microscope. Data were acquired and processed using Leica Application Suite (v2.7.3.9723), Power Point (v14.0.7192.5000) and Photo Shop CS6.
For autophagosome and autolysosome formation GFP+ RFP+ and GFP− RFP+ puncta per cell were counted. RFP+ GFP+ and RFP+ GFP− puncta per cell were manually counted in RFP-GFP-mLC3 RAW264.7 MΦ. The maximum number of puncta per cell counted was 25. To assess targeting of E. coli to autolysosomes, colocalization of RFP+ puncta (autolysosomes) with intracellular sCFP3A-E. coli was determined as per cent of total intracellular bacteria per cell. To assess targeting of E. coli to acidic vacuoles, colocalization of LysoTracker-positive vacuoles with intracellular sfGFP-E. coli was determined as per cent of total intracellular bacteria per cell.
Transmission electron microscopy
RAW264.7 MΦ were infected as described above. Two hours after infection, cells were washed twice with PBS and harvested in PBS containing 2% gelatin (Sigma Aldrich, G9391-100 g). Afterwards, the infected cells were routinely fixed in 0.1 M cacodylate-buffered Karnovsky solution (2.5% glutaraldehyde [Electron Microscopy Sciences, 16,400] and 2% paraformaldehyde [Electron Microscopy Sciences, 19,200]) overnight at room temperature, followed by postfixation in 1% osmium tetroxide (Electron Microscopy Sciences, 19,110) for 2 h. Samples were then dehydrated in graded ethanols (Sigma Aldrich, 32221-1l-M), and embedded in the EMbed-812 epoxy resin (Electron Microscopy Sciences, 14,900, 13,710, 19,000, 13,600]). After 48 h heat polymerization at 60°C, semithin (0.8 µm) sections were cut, stained with a toluidine blue (AgarScientific, R1727) and basic fuchsine (Polysciences, 635) solution, and after selection of appropriate areas of interest the epon block was trimmed for ultrathin sectioning. Ultrathin (80 nm) sections were cut with a diamond knife on a Reichert Ultracut-S ultramicrotome (Leica, Wetzlar, Germany) and double contrasted with aqueous 2% uranyl acetate (Fluka, 94,260) and lead citrate solutions (Leica, 169,707,235) for 10 min each. The sections were examined in a LEO912AB transmission electron microscope (Zeiss, Oberkochen, Germany) operating at 100 kV.
Statistical analysis
Results are expressed as mean ± s.e.m. (unless indicated otherwise). Statistical significance was tested with Prism v5.0 or v6.0 software (GraphPad). We tested normality distribution with the Kolmogorov-Smirnov test. When comparing normal distributed datasets, two groups were compared using the unpaired two-tailed Student t test if equal variances were assumed. If unequal variances were detected, unpaired two-tailed Student t test with Welch correction was used. Mann-Whitney U test was applied whenever the data points were not normally distributed. When comparing more than two groups of non-normally distributed or normally distributed datasets the Kruskal-Wallis test followed by the Dunn multiple-comparison test or ANOVA with Bonferroni’s multiple-comparisons test was used, respectively. Densitometry of immunoblots was analyzed by paired two-tailed Student t test or Wilcoxon signed rank tests for normally distributed and non-normally distributed data sets, respectively. p values < 0.05 were deemed statistically significant.
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft [WA 2539/4-1, 5-1, 7-1]; Deutsche Forschungsgemeinschaft (DE) [JA 1993/4-1]; Universitätsklinikum Regensburg [Reform C]; NIHR Cambridge Blood and Transplant Research Unit Organ Donation.
Acknowledgments
This work was supported by grants from Deutsche Forschungsgemeinschaft to JJ (JA1993/4-1) and SW (WA 2539/4-1, 5-1 and 7-1), faculty grants from the University of Regensburg to JJ, KH, SW (ReForM C program), and NIHR Cambridge Blood and Transplant Research Unit Organ Donation to JRF. We are grateful for the excellent technical assistance provided by Monika Nowottny and Heiko Siegmund.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Jantsch J, Schatz V, Friedrich D, et al. Cutaneous Na+ storage strengthens the antimicrobial barrier function of the skin and boosts macrophage-driven host defense. Cell Metab. 2015;21:493–501. PMID:25738463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ip WK, Medzhitov R.. Macrophages monitor tissue osmolarity and induce inflammatory response through NLRP3 and NLRC4 inflammasome activation. Nat Commun. 2015;6:6931 PMID:25959047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hucke S, Eschborn M, Liebmann M, et al. Sodium chloride promotes pro-inflammatory macrophage polarization thereby aggravating CNS autoimmunity. J Autoimmun. 2016;67:90–101. PMID:26584738. [DOI] [PubMed] [Google Scholar]
- [4].Zhang WC, Zheng XJ, Du LJ, et al. High salt primes a specific activation state of macrophages, M(Na). Cell Res. 2015;25:893–910. PMID:26206316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Miyakawa H, Woo SK, Dahl SC, et al. Tonicity-responsive enhancer binding protein, a Rel-like protein that stimulates transcription in response to hypertonicity. Proc Natl Acad Sci U S A. 1999;96:2538–2542. PMID:ISI:000078956600131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lopez-Rodriguez C, Aramburu J, Rakeman AS, et al. NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc Natl Acad Sci U S A. 1999;96:7214–7219. PMID:10377394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ko BC, Turck CW, Lee KW, et al. Purification, identification, and characterization of an osmotic response element binding protein. Biochem Biophys Res Commun. 2000;270:52–61. PMID:10733904. [DOI] [PubMed] [Google Scholar]
- [8].Buxade M, Huerga Encabo H, Riera-Borrull M, et al. Macrophage-specific MHCII expression is regulated by a remote Ciita enhancer controlled by NFAT5. J Exp Med. 2018;215:2901–2918. PMID:30327417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Buxade M, Lunazzi G, Minguillon J, et al. Gene expression induced by Toll-like receptors in macrophages requires the transcription factor NFAT5. J Exp Med. 2012;209:379–393. PMID:22312110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Choi S, You S, Kim D, et al. Transcription factor NFAT5 promotes macrophage survival in rheumatoid arthritis. J Clin Invest. 2017;127:954–969. PMID:28192374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tellechea M, Buxade M, Tejedor S, et al. NFAT5-regulated macrophage polarization supports the proinflammatory function of macrophages and T lymphocytes. J Immunol. 2018;200:305–315. PMID:29150563. [DOI] [PubMed] [Google Scholar]
- [12].Schatz V, Neubert P, Schroder A, et al. Elementary immunology: Na+ as a regulator of immunity. Pediatr Nephrol. 2017;32:201–210. PMID:26921211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Muller DN, Wilck N, Haase S, et al. Sodium in the microenvironment regulates immune responses and tissue homeostasis. Nat Rev Immunol. 2019. PMID:30644452 DOI: 10.1038/s41577-018-0113-4 [DOI] [PubMed] [Google Scholar]
- [14].Berry MR, Mathews RJ, Ferdinand JR, et al. Renal sodium gradient orchestrates a dynamic antibacterial defense zone. Cell. 2017;170:860–74 e19. PMID:28803730. [DOI] [PubMed] [Google Scholar]
- [15].Popovic ZV, Embgenbroich M, Chessa F, et al. Hyperosmolarity impedes the cross-priming competence of dendritic cells in a TRIF-dependent manner. Sci Rep. 2017;7:311 PMID:28331179 DOI: 10.1038/s41598-017-00434-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang WC, Du LJ, Zheng XJ, et al. Elevated sodium chloride drives type I interferon signaling in macrophages and increases antiviral resistance. J Biol Chem. 2018;293:1030–1039. PMID:29203528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ramalingam S, Cai B, Wong J, et al. Antiviral innate immune response in non-myeloid cells is augmented by chloride ions via an increase in intracellular hypochlorous acid levels. Sci Rep. 2018;8:13630 PMID:30206371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Calance DN, Steixner C, Gross S, et al. Hypertonicity primes malignant melanoma cells for apoptosis. Apoptosis. 2018;23:201–209. PMID:29435687. [DOI] [PubMed] [Google Scholar]
- [19].Heimer S, Knoll G, Steixner C, et al. Hypertonicity-imposed BCL-XL addiction primes colorectal cancer cells for death. Cancer Lett. 2018;435:23–31. PMID:30075205. [DOI] [PubMed] [Google Scholar]
- [20].Sirtl S, Knoll G, Trinh DT, et al. Hypertonicity-enforced BCL-2 addiction unleashes the cytotoxic potential of death receptors. Oncogene. 2018;37:4122–4136. PMID:29706657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kitada K, Daub S, Zhang Y, et al. High salt intake reprioritizes osmolyte and energy metabolism for body fluid conservation. J Clin Invest. 2017;127:1944–1959. PMID:28414295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Galluzzi L, Baehrecke EH, Ballabio A, et al. Molecular definitions of autophagy and related processes. Embo J. 2017;36:1811–1836. PMID:28596378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cadwell K. Crosstalk between autophagy and inflammatory signalling pathways: balancing defence and homeostasis. Nat Rev Immunol. 2016;16:661–675. PMID:27694913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Deretic V. Autophagy in leukocytes and other cells: mechanisms, subsystem organization, selectivity, and links to innate immunity. J Leukoc Biol. 2016;100:969–978. PMID:27493243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. PMID:20034776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bellot G, Garcia-Medina R, Gounon P, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–2581. PMID:19273585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Binger KJ, Gebhardt M, Heinig M, et al. High salt reduces the activation of IL-4- and IL-13-stimulated macrophages. J Clin Invest. 2015;125:4223–4238. PMID:26485286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Raat NJ, van Os CH, Bindels RJ. Effects of osmotic perturbation on [Ca2+]i and pHi in rabbit proximal tubular cells in primary culture. Am J Physiol. 1995;269:F205–F211. PMID:7653594. [DOI] [PubMed] [Google Scholar]
- [29].Nelson DJ, Jow B, Jow F. Lipopolysaccharide induction of outward potassium current expression in human monocyte-derived macrophages: lack of correlation with secretion. J Membr Biol. 1992;125:207–218. PMID:1556735. [DOI] [PubMed] [Google Scholar]
- [30].Haslberger A, Romanin C, Koerber R. Membrane potential modulates release of tumor necrosis factor in lipopolysaccharide-stimulated mouse macrophages. Mol Biol Cell. 1992;3:451–460. PMID:1498365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gerth A, Grosche J, Nieber K, et al. Intracellular LPS inhibits the activity of potassium channels and fails to activate NFkappaB in human macrophages. J Cell Physiol. 2005;202:442–452. PMID:15389581. [DOI] [PubMed] [Google Scholar]
- [32].Vicente R, Escalada A, Coma M, et al. Differential voltage-dependent K+ channel responses during proliferation and activation in macrophages. J Biol Chem. 2003;278:46307–46320. PMID:12923194. [DOI] [PubMed] [Google Scholar]
- [33].Nadesalingam A, Chen JHK, Farahvash A, et al. Hypertonic saline suppresses NADPH oxidase-dependent neutrophil extracellular trap formation and promotes apoptosis. Front Immunol. 2018;9:359 PMID:29593709 DOI: 10.3389/fimmu.2018.00359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nunes P, Ernandez T, Roth I, et al. Hypertonic stress promotes autophagy and microtubule-dependent autophagosomal clusters. Autophagy. 2013;9:550–567. PMID:23380587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1–222. PMID:26799652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. PMID:17534139. [DOI] [PubMed] [Google Scholar]
- [37].Martinez J, Almendinger J, Oberst A, et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A. 2011;108:17396–17401. PMID:21969579. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [38].Turk B, Turk D, Turk V. Lysosomal cysteine proteases: more than scavengers. Biochim Biophys Acta. 2000;1477:98–111. PMID:10708852. [DOI] [PubMed] [Google Scholar]
- [39].Qi X, Man SM, Malireddi RK, et al. Cathepsin B modulates lysosomal biogenesis and host defense against Francisella novicida infection. J Exp Med. 2016;213:2081–2097. PMID:27551156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hook G, Jacobsen JS, Grabstein K, et al. Cathepsin B is a new drug target for traumatic brain injury therapeutics: evidence for E64d as a promising lead drug candidate. Front Neurol. 2015;6:178 PMID:26388830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Thoreen CC, Kang SA, Chang JW, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. PMID:19150980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ortells MC, Morancho B, Drews-Elger K, et al. Transcriptional regulation of gene expression during osmotic stress responses by the mammalian target of rapamycin. Nucleic Acids Res. 2012;40:4368–4384. PMID:22287635: gks038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bruick RK. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci U S A. 2000;97:9082–9087. PMID:10922063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sowter HM, Ratcliffe PJ, Watson P, et al. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001;61:6669–6673. PMID:11559532. [PubMed] [Google Scholar]
- [45].Iyer NV, Kotch LE, Agani F, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. PMID:9436976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. Embo J. 1998;17:3005–3015. PMID:9606183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. PMID:11292862. [DOI] [PubMed] [Google Scholar]
- [48].Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. PMID:11292861. [DOI] [PubMed] [Google Scholar]
- [49].Sanjuan MA, Dillon CP, Tait SW, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. PMID:18097414. [DOI] [PubMed] [Google Scholar]
- [50].Singh SB, Davis AS, Taylor GA, et al. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. PMID:16888103. [DOI] [PubMed] [Google Scholar]
- [51].Lapaquette P, Bringer MA, Darfeuille-Michaud A. Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cell Microbiol. 2012;14:791–807. PMID:22309232. [DOI] [PubMed] [Google Scholar]
- [52].Chiu HC, Kulp SK, Soni S, et al. Eradication of intracellular Salmonella enterica serovar Typhimurium with a small-molecule, host cell-directed agent. Antimicrob Agents Chemother. 2009;53:5236–5244. PMID:19805568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Deuretzbacher A, Czymmeck N, Reimer R, et al. Beta1 integrin-dependent engulfment of Yersinia enterocolitica by macrophages is coupled to the activation of autophagy and suppressed by type III protein secretion. J Immunol. 2009;183:5847–5860. PMID:19812190. [DOI] [PubMed] [Google Scholar]
- [54].Randow F, Munz C. Autophagy in the regulation of pathogen replication and adaptive immunity. Trends Immunol. 2012;33:475–487. PMID:22796170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Casanova JE. Bacterial autophagy: offense and defense at the host-pathogen interface. Cell Mol Gastroenterol Hepatol. 2017;4:237–243. PMID:28660242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yordy B, Tal MC, Hayashi K, et al. Autophagy and selective deployment of Atg proteins in antiviral defense. Int Immunol. 2013;25:1–10. PMID:23042773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Matsuzawa-Ishimoto Y, Hwang S, Cadwell K. Autophagy and Inflammation. Annu Rev Immunol. 2018;36:73–101. PMID:29144836. [DOI] [PubMed] [Google Scholar]
- [58].Kimmey JM, Stallings CL. Bacterial pathogens versus autophagy: implications for therapeutic interventions. Trends Mol Med. 2016;22:1060–1076. PMID:27866924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Huang J, Canadien V, Lam GY, et al. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci U S A. 2009;106:6226–6231. PMID:19339495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Greer SN, Metcalf JL, Wang Y, et al. The updated biology of hypoxia-inducible factor. Embo J. 2012;31:2448–2460. PMID:22562152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kaelin WG Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. PMID:18498744. [DOI] [PubMed] [Google Scholar]
- [62].Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. PMID:22304911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhou B, Ann DK, Li X, et al. Hypertonic induction of aquaporin-5: novel role of hypoxia-inducible factor-1alpha. Am J Physiol. 2007;292:C1280–C1290. PMID:17108010. [DOI] [PubMed] [Google Scholar]
- [64].Mimouna S, Bazin M, Mograbi B, et al. HIF1A regulates xenophagic degradation of adherent and invasive Escherichia coli (AIEC). Autophagy. 2014;10:2333–2345. PMID:25484075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhang H, Bosch-Marce M, Shimoda LA, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. PMID:18281291. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [66].Wu HM, Jiang ZF, Ding PS, et al. Hypoxia-induced autophagy mediates cisplatin resistance in lung cancer cells. Sci Rep. 2015;5:12291 PMID:26201611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Jeong JK, Gurunathan S, Kang MH, et al. Hypoxia-mediated autophagic flux inhibits silver nanoparticle-triggered apoptosis in human lung cancer cells. Sci Rep. 2016;6:21688 PMID:26867977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Wang X, Ribeiro M, Iracheta-Vellve A, et al. Macrophage-specific HIF-1alpha contributes to impaired autophagic flux in non-alcoholic steatohepatitis. Hepatology. 2018. PMID:30102772 DOI: 10.1002/hep.30215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Florey O, Gammoh N, Kim SE, et al. V-ATPase and osmotic imbalances activate endolysosomal LC3 lipidation. Autophagy. 2015;11:88–99. PMID:25484071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kreibich S, Emmenlauer M, Fredlund J, et al. Autophagy proteins promote repair of endosomal membranes damaged by the Salmonella type three secretion system 1. Cell Host Microbe. 2015;18:527–537. PMID:26567507. [DOI] [PubMed] [Google Scholar]
- [71].Kondratskyi A, Kondratska K, Skryma R, et al. Ion channels in the regulation of autophagy. Autophagy. 2018;14:3–21. PMID:28980859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Knaup KX, Jozefowski K, Schmidt R, et al. Mutual regulation of hypoxia-inducible factor and mammalian target of rapamycin as a function of oxygen availability. Mol Cancer Res. 2009;7:88–98. PMID:19147540. [DOI] [PubMed] [Google Scholar]
- [73].Hudson CC, Liu M, Chiang GG, et al. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol Cell Biol. 2002;22:7004–7014. PMID:12242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–448. PMID:15928708. [DOI] [PubMed] [Google Scholar]
- [75].Zhu H, Cao W, Zhao P, et al. Hyperosmotic stress stimulates autophagy via the NFAT5/mTOR pathway in cardiomyocytes. Int J Mol Med. 2018;42:3459–3466. PMID:30221680. [DOI] [PubMed] [Google Scholar]
- [76].Liu C, Choi H, Johnson ZI, et al. Lack of evidence for involvement of TonEBP and hyperosmotic stimulus in induction of autophagy in the nucleus pulposus. Sci Rep. 2017;7:4543 PMID:28674405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Choe KP, Strange K. Genome-wide RNAi screen and in vivo protein aggregation reporters identify degradation of damaged proteins as an essential hypertonic stress response. Am J Physiol. 2008;295:C1488–C1498. PMID:18829898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kochl R, Hu XW, Chan EY, et al. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 2006;7:129–145. PMID:16420522. [DOI] [PubMed] [Google Scholar]
- [79].Di Bartolomeo S, Corazzari M, Nazio F, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–168. PMID:20921139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Schleicher U, Bogdan C. Generation, culture and flow-cytometric characterization of primary mouse macrophages. Methods Mol Biol. 2009;531:203–224. PMID:19347320. [DOI] [PubMed] [Google Scholar]
- [81].Mizushima N, Yamamoto A, Matsui M, et al. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. PMID:14699058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Pollock JD, Williams DA, Gifford MA, et al. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat Genet. 1995;9:202–209. PMID:7719350. [DOI] [PubMed] [Google Scholar]
- [83].Clausen BE, Burkhardt C, Reith W, et al. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. PMID:10621974. [DOI] [PubMed] [Google Scholar]
- [84].Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. PMID:15866887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Ryan HE, Poloni M, McNulty W, et al. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 2000;60:4010–4015. PMID:10945599. [PubMed] [Google Scholar]
- [86].Weigert A, Weichand B, Sekar D, et al. HIF-1alpha is a negative regulator of plasmacytoid DC development in vitro and in vivo. Blood. 2012;120:3001–3006. PMID:22936665. [DOI] [PubMed] [Google Scholar]
- [87].Roda JM, Sumner LA, Evans R, et al. Hypoxia-inducible factor-2alpha regulates GM-CSF-derived soluble vascular endothelial growth factor receptor 1 production from macrophages and inhibits tumor growth and angiogenesis. J Immunol. 2011;187:1970–1976. PMID:21765015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Jennewein J, Matuszak J, Walter S, et al. Low-oxygen tensions found in Salmonella-infected gut tissue boost Salmonella replication in macrophages by impairing antimicrobial activity and augmenting Salmonella virulence. Cell Microbiol. 2015;17:1833–1847. PMID:26104016. [DOI] [PubMed] [Google Scholar]
- [89].Kremers GJ, Goedhart J, van Munster EB, et al. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Forster radius. Biochemistry. 2006;45:6570–6580. PMID:16716067. [DOI] [PubMed] [Google Scholar]
- [90].Valdivia RH, Falkow S. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol. 1996;22:367–378. PMID:8930920. [DOI] [PubMed] [Google Scholar]
- [91].Siegert I, Schatz V, Prechtel AT, et al. Electroporation of siRNA into mouse bone marrow-derived macrophages and dendritic cells. Methods Mol Biol. 2014;1121:111–119. PMID:24510816. [DOI] [PubMed] [Google Scholar]
- [92].Mustroph J, Wagemann O, Lucht CM, et al. Empagliflozin reduces Ca/calmodulin-dependent kinase II activity in isolated ventricular cardiomyocytes. ESC Heart Fail. 2018;5:642–648. PMID:30117720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Wiese M, Gerlach RG, Popp I, et al. Hypoxia-mediated impairment of the mitochondrial respiratory chain inhibits the bactericidal activity of macrophages. Infect Immun. 2012;80:1455–1466. PMID:22252868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Siegert I, Schodel J, Nairz M, et al. Ferritin-mediated iron sequestration stabilizes hypoxia-inducible factor-1alpha upon LPS activation in the presence of ample oxygen. Cell Rep. 2015;13:2048–2055. PMID:26628374. [DOI] [PubMed] [Google Scholar]
- [95].Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. PMID:12808457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


