Abstract
RNA-modifying enzymes targeting mRNA poly(A) tails are universal regulators of post-transcriptional gene expression programs. Current data suggest that an RNA-binding protein (RBP) directed tug-of-war between tail shortening and re-elongating enzymes operates in the cytoplasm to repress or activate specific mRNA targets. While this concept is widely accepted, it was primarily described in the final meiotic stages of frog oogenesis and relies molecularly on a single class of RBPs, i.e. CPEBs, the deadenylase PARN and cytoplasmic poly(A) polymerase GLD-2. Using the spatial and temporal resolution of female gametogenesis in the nematode C. elegans, we determined the distinct roles of known deadenylases throughout germ cell development and discovered that the Ccr4–Not complex is the main antagonist to GLD-2-mediated mRNA regulation. We find that the Ccr4–Not/GLD-2 balance is critical for essentially all steps of oocyte production and reiteratively employed by various classes of RBPs. Interestingly, its two deadenylase subunits appear to affect mRNAs stage specifically: while a Caf1/GLD-2 antagonism regulates mRNA abundance during all stages of oocyte production, a Ccr4/GLD-2 antagonism regulates oogenesis in an mRNA abundance independent manner. Our combined data suggests that the Ccr4–Not complex represents the evolutionarily conserved molecular opponent to GLD-2 providing an antagonistic framework of gene-specific poly(A)-tail regulation.
INTRODUCTION
The ability to regulate gene expression at the mRNA level is crucial for many developmental and physiological processes. In the cytoplasm, gene expression control is achieved by the association of mRNAs with designated mRNA-binding proteins (RBPs) to repress or activate select mRNA targets, forming larger mRNA–protein (mRNP) complexes. Among sequence-specific RBPs also RNA-modifying enzymes that either shorten or elongate the length of mRNA 3′ poly(A) tails were repeatedly identified in previous biochemical purifications of mRNPs, suggesting that endured presence of these enzymes may facilitate efficient mRNA regulation (1,2). In this context, tail shortening is mediated by deadenylases (DeAds) correlating with repression of mRNAs, whereas tail elongation is mediated by cytoplasmic poly(A) polymerases (cytoPAPs) correlating with activation of mRNAs. Hence, opposing activities of DeAds and cytoPAPs might provide an antagonistic frame work for many RBPs to mechanistically regulate mRNAs in the cytoplasm.
In animal development, only one prominent example has been described of how the molecular antagonism between poly(A)-tail modifiers suppresses and reactivates mRNA activities. During frog oocyte maturation, the deadenylase PARN and cytoPAP GLD-2 form an antagonistic pair to regulate the translation of mRNAs whose protein products drive the progression through both meiotic divisions in Xenopus laevis (3). The PARN-GLD-2 antagonism is instructed by the sequence-specific RBP CPEB and serves as a hallmark of how a DeAd/cytoPAP rheostat may be employed in gene-specific mRNA regulation. However, whether the antagonistic PARN-GLD-2 pair has general validity and extends to other stages of germ cell biology or even to other organisms remains unclear. Currently, no studies exist that directly address other potential DeAd–cytoPAP relationships and it remains to be shown how relevant this opposing enzyme pair might be for other RBPs than CPEBs.
In contrast to PARN, which is present in most but not all multi-cellular organisms, two other prominent mRNA deadenylases have been described that are evolutionary conserved from yeast to humans: the Pan2 and the Ccr4–Not complex (1). While the Pan2 complex is composed of two subunits, the single enzyme Pan2 and its regulatory scaffold Pan3, the Ccr4–Not deadenylase is a multi-subunit complex that contains two enzymatic components. Both deadenylases, Caf1 (also known as Pop2) and Ccr4, are attached as a single module to the central scaffolding protein Not1 (4). The Ccr4–Not complex assumes a pivotal role in the general mRNA degradation pathway, and as such provides the major poly(A)-removing activity in all organisms tested to date (5–7). Importantly, Ccr4–Not appears to also participate in gene-specific mRNA repression in many organisms. A number of evolutionary conserved cytoplasmic RBPs, including PUF proteins or Zinc-finger-containing RBPs, utilize this complex to silence and/or degrade mRNA targets (8,9). Whereas the importance of the Ccr4–Not complex for the control of cytoplasmic gene expression regulation is undisputed, a potential antagonistic role in a likely DeAd–cytoPAP relationship and its relevance to other sequence-specific RBPs remains to be determined.
Across species, GLD-2-type poly(A) polymerases provides the major poly(A)-tail elongation activity in the cytoplasm (10,11). These cytoPAPs are terminal nucleotidyl transferases with a preference for efficient A-addition to mRNAs that already end on adenosine (12). The enzymatic activity of GLD-2 is stimulated by interacting proteins and represents the main driver of its molecular and biological functions (13–15). Consistent with its essential roles in various stages of germ cell development, GLD-2-type enzymes are strongly expressed in germ cells, however they are not uniformly abundant throughout gametogenesis (13). Although deadenylases are ubiquitously expressed in most tissues of various organisms, the Ccr4–Not complex is particularly crucial for germ cell biology; loss of its components leads to germ cell defects during female oocyte production in metazoans (5,16,17). However, whether the Ccr4–Not deadenylase complex forms an antagonistic pair with GLD-2-type cytoPAPs in tissue-specific mRNA regulation is currently not known.
In this study, we exploit the simple spatial and temporal resolution of female gametogenesis in the nematode C. elegans to reveal broad-scale antagonistic relationships of poly(A)-tail modifying enzymes. Unlike vertebrates, the gonad of this animal model system facilitates a refined molecular and morphological analysis of all phases of gametogenesis preceding the stage of oocyte maturation. Our work identifies, with a precision that is not possible in vertebrate animals, the Ccr4–Not complex as the major deadenylase that opposes GLD-2 cytoPAP to regulate essentially all phases of early oogenesis, reaching from germ stem cell proliferation to oocyte maturation. Interestingly, in opposition to GLD-2 clear differences exist among the two catalytic subunits of Ccr4–Not. Whereas Caf1, termed CCF-1 in worms, is primarily important for all phases of early female gametogenesis by regulating mRNA abundance, CCR-4 plays a significant role in later phases by promoting gene expression possibly in a translational rather than abundance-dependent manner. This molecular difference coincides with a likely shift of GLD-2 function from promoting mRNA stability to promoting mRNA translatability. Finally, our data suggests that several evolutionary conserved RBPs rely on the antagonistic Ccr4–Not/GLD-2 pair to regulate their target mRNAs. Our combined work reveals that the opposing forces of the deadenylase Ccr4–Not and poly(A) polymerase GLD-2 provide an antagonistic frame work to cytoplasmic gene expression regulation, which is presumably tuned by many diverse RBPs to balance mRNA abundance and translation.
MATERIALS AND METHODS
Nematode strains and transgenesis
Worms were handled according to standard procedures and grown at 20°C (18). The N2 Bristol strain was used as a reference for wild type. Other strains used in this study: Linkage group (LG) I: gld-2(q497), rrf-1(pk1417); II: parn-2(tm1339); III: panl-2(tm1575); IV: ccr-4(tm1312); V: parn-1(tm869). Unless stated otherwise, adult germline phenotypes were scored 24hrs past mid-L4 stage. Transgenic strains EV484 (efIs81[Cbr-unc-119(+) + Pmex-5::rpl-4::FLAG::tbb-2 3′UTR] II) were generated using the Mos1-mediated single copy insertion (MosSCI) protocol (19). Injected constructs were assembled using the multisite Gateway cloning system (Thermo Fisher Scientific). To this end, the entire gene of rpl-4 was amplified from genomic DNA, fused with 2xFLAG-tag encoding sequences via overlap extension PCR, and inserted into the entry vector pDONR221. The size of the proliferative region was analysed by counting nuclei along the distal-proximal axis until a cluster of three to four nuclei with crescent shaped DNA in a circumference was detected.
RNAi feeding constructs and procedure
The feeding constructs targeting ccf-1, ntl-1 and gld-2 were described previously (5,10). For ccf-1 RNAi treatment in wild type and gld-2(q497), animals were fed from L1 onwards. The same is true for gld-2 RNAi treatments of EV484 animals. For ntl-1 RNAi treatment in wild-type and gld-2(q497), L4 animals were fed until adulthood. For RNAi treatments of rrf-1(pk1417), L4 animals were placed on RNAi plates and F1 progenies were analyzed at the adult stage.
Primary antibodies
Primary antibodies against the following proteins were used: mouse anti-FLAG M2 (Sigma-Aldrich), anti-NPC (Mab414, Covance), anti-tubulin (T5168, Sigma), DAO-5 (20) and anti-GLD-2 (21); guinea pig anti-HIM-3 (22) and anti-OMA-1/2 (5).
Immunocytochemistry
Indirect immunocytochemistry of extruded and 1% paraformaldehyde PFA-fixed gonads was carried out in solution as described (23). Images were taken on a Zeiss Imager M1 equipped with an Axiocam MRm (Zeiss) and processed with AxioVision (Zeiss) and Photoshop CS5 (Adobe). Secondary antibodies coupled to fluorochromes FITC, CY3 and CY5 were purchased from Jackson ImmunoResearch (Dianova).
Western blotting, sucrose gradients and immunoprecipitations
For Western blotting experiments, we collected individual worms by hand and boiled them in Laemmli protein sample loading buffer prior to gel separation. Specific proteins were analyzed by western blotting with ECL detection (GE Healthcare) of HRP-coupled secondary antibodies (Jackson ImmunoResearch). Worm protein extracts for sucrose gradients and ribosome immunoprecipitations were made as described (5). The sucrose gradient centrifugation and fractionation was conducted as previously described (10).
RNA isolation and qPCR
Total RNA was isolated from hand-picked whole worms or immunoprecipitated material using Trizol (Invitrogen). 200 ng of total RNA was reverse transcribed using random hexamer primers and ReverseAid Premium reverse transcriptase (Thermo Fisher Scientific), according to the manufactures protocol. Quantitative PCR (qPCR) was conducted on an iQ™5 (BioRad), using the ABsolute QPCR SYBR Green mix (Thermo Fisher Scientific) and gene-specific primers (sequences available upon request). Immunoprecipitated RNA was also visualized via denaturing agarose gel electrophoresis.
Bulk poly(A)-tail length measurements
One microgram of whole worm total RNA was used to perform bulk poly(A)-tail measurements as previously described (5). Each sample was analyzed from three independent biological replicates; size markers were synthesized RNA oligos of 30 and 45 nucleotides in length, and a loading dye band that corresponds to proximally 65nts. Lane quantifications were performed using Fiji (ImageJ).
Library preparation and next-generation sequencing (NGS)
mRNA was isolated from 1 μg DNAse-treated total RNA using the Ribo-Zero Gold Kit (human, mouse, rat) from Illumina according to the manufacturer's instructions. Final elution was done in 5 μl nuclease free water. Samples were then directly subjected to the workflow for strand-specific RNA-Seq library preparation (Ultra Directional RNA Library Prep II, NEB). For ligation, custom adaptors were used 1: (Adaptor-Oligo 5′-ACA CTC TTT CCC TAC ACG ACG CTC TTC CGA TCT-3′, Adaptor-Oligo 2: 5′-P-GAT CGG AAG AGC ACA CGT CTG AAC TCC AGT CAC-3′). After ligation, adapters were depleted by an XP bead purification (Beckman Coulter) adding beads in a ratio of 1:1. Indexing was done during the following PCR enrichment (15 cycles) using custom amplification primers carrying the index sequence indicated with ‘NNNNNN’. (Primer1: Oligo_Seq AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG ATC T, primer2: GTG ACT GGA GTT CAG ACG TGT GCT CTT CCG ATC T, primer3: CAA GCA GAA GAC GGC ATA CGA GAT NNNNNN GTG ACT GGA GTT. After two more XP beads purifications (1:1) libraries were quantified using Qubit dsDNA HS Assay Kit (Invitrogen). Samples were equimolarly pooled and used for 75bp single read sequencing on a Nextseq 500 (Illumina).
Analysis of NGS data
First, the high quality of the data was ensured using the software fastqc (v. 0.11.5) [FastQC A Quality Control tool for High Throughput Sequence Data http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ by S. Andrews]. The fastq files were than mapped to the Caenorhabditis elegans genome (Ensembl genome version WBcel235) using the STAR algorithm (v. 2.5.2b) (24). Subsequently, the reads were counted per gene using the featureCounts tool from the SubRead package (v. 1.4.6-p4) (25). Reads were counted on a gene level from the bam files based on the ensemble annotations.
Using the DESeq2 package (R version 3.4.0, DESeq2 version 1.18.1) the count data was normalized by the size factor to estimate effective library sizes (26). Following a dispersion calculation across all samples, a pair-wise comparison of various conditions was done resulting in a list of differentially expressed genes for each of the compared groups. Genes with a normalized read count of higher or equal 100, a fold-change higher or equal to 1.5 and with an adjusted P-value lower or equal to 0.05 were defined as differentially expressed (DE).
Immunoprecipiation of ribosomes
Worms expressing rpl4::FLAG were grown on gld-2(RNAi) or control RNAi plates. Extract preparations and immunoprecipitations were conducted as previously described (15) using anti-FLAG M2 affinity agarose (Sigma-Aldrich). RNA was isolated from the matrix material as well as the input material using Trizol (Invitrogen). Equal amounts for each sample of isolated RNA (200 ng) were converted into cDNA and analysed via qPCR as described above. A ribosome association coefficient was calculated for each analysed mRNA (amount of ribosome-bound mRNA/ amount of input mRNA). In order to control for sample to sample variations all measurements were normalized to a negative control (nos-3).
RESULTS
The Ccr4–Not complex is essential for proliferation
The Caenorhabditis elegans adult gonad is a tube-like organ in which female germ cells are organized in a distal-to-proximal gradient of oocyte production, representing all stages of oogenesis. At its most distal end, germline stem cells divide mitotically and occupy the proliferative region together with cells preparing for meiosis. Further proximally, germ cells enter meiosis and begin to differentiate gradually while progressing through all stages of prophase I, before arresting in diakinesis at the proximal end as fully differentiated oocytes (Figure 1A). The terminal oocyte will eventually leave the germline tissue in the process of ovulation and matures to become fertilization competent.
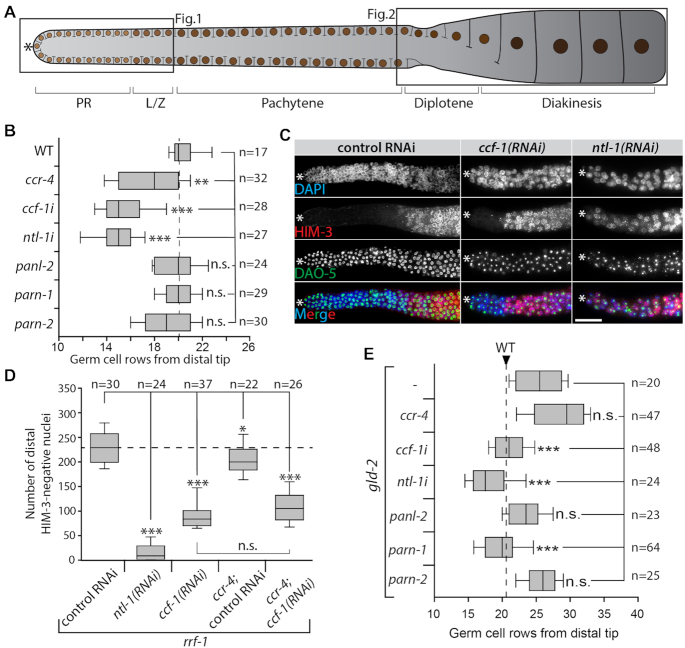
Figure 1.
Ccr4–Not complex is essential for proliferation. CCF-1 and PARN-1 are opposed by GLD-2 in the regulation of meiotic entry. (A) Scheme of the adult female germline tissue, indicating the proliferative region (PR) and different stages of meiotic prophase I (L/Z: Leptotene and Zygotene). The asterisk indicates its distal tip; fully cellularized oocytes are located at its proximal end. Brown circles represent germ cell nuclei. The two boxed germline regions are analyzed in Figures 1 and 2. (B) Size of the proliferative region shown as germ cell rows until L/Z nuclei are detected in wild-type (WT), mutants, or RNAi-treated adults (indicated with an i, e.g. ccf-1i). Control RNAi is similar to WT and not shown for clarity. (C) DAPI (in blue) and antibody staining of extruded gonads of rrf-1 adults treated with control, ccf-1, or ntl-1 RNAi. HIM-3 (in red) marks nuclei that entered meiosis. Nucleolar DAO-5 (in green) serves as a penetration control; scale bar = 20μm. (D) Quantification of HIM-3–negative nuclei in the distal part of the germ line of indicated RNAi treatments in rrf-1 single or rrf-1;ccr-4 double mutants. (E) Size of the proliferative region in gld-2(q497) mutant animals that were further compromised in deadenylase functions. Data in B, D, and E was tested for statistical significance via a two-tailed Student's t-test: *P < 0.05; **P < 0.01; ***P < 0.001; n.s. = not significant; n = number of analyzed gonads.
In this nematode, homologs have been identified for all major DeAds (5). The two Ccr4–Not associated enzymes, Ccr4 and Caf1, are represented by CCR-4 and CCF-1, respectively. PANL-2 is homologous to the Pan2 deadenylase. PARN-1 and PARN-2 represent two orthologues of worm PARN. Throughout this work the function of ccr-4, panl-2, parn-1 and parn-2 will be addressed using established loss-of-function alleles (5). The function of ccf-1 or ntl-1 (the worm Not1 homolog) will always be assessed upon conditional RNAi-mediated knockdown using feeding RNAi. The founding cytoPAP GLD-2 was initially isolated in C. elegans.
To test which DeAds are functionally important for germ cell proliferation and entry into meiosis, we analyzed the size of the proliferative region of extruded gonads by DAPI staining. In wild-type animals, the proliferative region is between 20 and 21 germ cell rows in size; this did not change in panl-2, parn-1 and parn-2 single mutants (Figure 1B). While the proliferative region was mildly reduced in animals lacking ccr-4, it was strongly reduced in ccf-1 or ntl-1 RNAi-treated animals (Figure 1B). This suggests that Ccr4–Not is the main DeAd complex, important for germ cell proliferation with CCF-1 being the key enzyme in the switch from proliferation to differentiation.
To test how strongly the Ccr4–Not complex is needed in proliferating germ cells, we repeated the analysis in rrf-1 animals, which fail to mount a strong somatic RNAi response (27). This allows to conduct longer periods of RNAi feeding in order to maximize the RNAi effect in germ cells. Strikingly, such ntl-1 or ccf-1 depleted rrf-1 animals contain very small gonads in which it was difficult to distinguish proliferative from meiotic germ cells by DAPI staining alone (Figure 1C). Therefore, to asses cells in meiotic prophase, we visualized the localization of marker protein HIM-3, a synaptonemal complex component, in immunostained extruded gonads (22). In rrf-1 animals treated with control RNAi, we counted ∼230 HIM-3-negative premeiotic cells in the distal end (Figure 1C and D). This number is slightly reduced in rrf-1;ccr-4 and strongly reduced in rrf-1;ccf-1(RNAi) as well as rrf-1;ccr-4; ccf-1(RNAi) (Figure 1C and D). The strongest effect was detected in some rrf-1;ntl-1(RNAi) animals that possessed HIM-3-positive germ cells only (Figure 1C and D), arguing that all of their germ cells had entered meiosis at the expense of mitosis. Together these observations suggest that the Ccr4–Not complex is essential for maintaining germline stem cells in an undifferentiated state.
CCF-1 and PARN-1 are opposed by GLD-2 in the regulation of the switch from proliferation to differentiation
GLD-2 cytoPAP is a known positive regulator of meiotic entry and commitment (15,28,29). To test whether the proliferative defects in Ccr4–Not–compromised germ cells are a consequence of extensive GLD-2-mediated polyadenylation, we removed individual DeAds in gld-2 mutants. In this analysis we additionally included the deadenylases panl-2, parn-1 and parn-2. Compared to wild type, the absence of GLD-2 activity causes germ cells to delay entry into meiosis, which is characterized by a substantial size increase of the proliferative region from 20–21 germ cell rows in wild type to 27–28 in gld-2 mutants (Figure 1E). Interestingly, an additional removal of ccr-4, panl-2, or parn-2 did not change the size of the proliferative region (Figure 1E), arguing that none of these DeAds influence meiotic entry in a gld-2-dependent manner. However, the removal of ccf-1, ntl-1, or parn-1 restored the size of the proliferative region to wild type (Figure 1E). This shows that two different DeAds, CCF-1 and PARN-1, are opposed by GLD-2 in regulating the entry into meiosis. The observation that the removal of PARN-1 in a gld-2 mutant changes the size of the proliferative region is particularly surprising, as the loss of parn-1 by itself had no detectable effect on germ cells in the distal germ line.
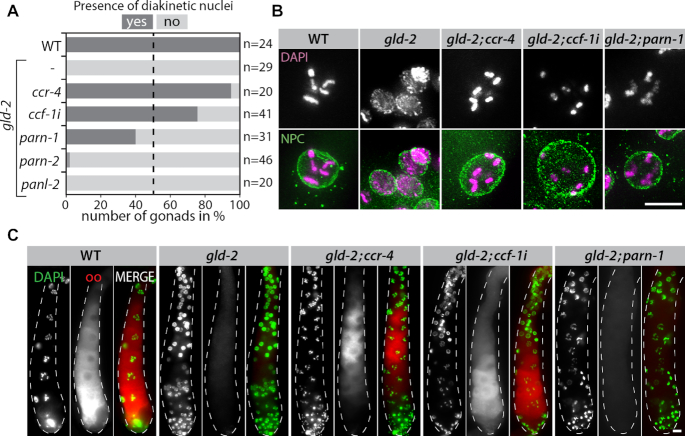
CCR-4 and CCF-1 are opposed by GLD-2 in regulating meiotic progression
To investigate the relationship between the different DeAds and GLD-2 during oocyte production, we analyzed the status of meiotic progression by DAPI staining in the proximal part of the gonad. Loss of GLD-2 activity results in a premature meiotic arrest around the pachytene/diplotene border (28). As a consequence and contrary to wild type, no nuclei with distinct bivalents corresponding to diakinetic nuclei were detected in gld-2 mutants (Figure 2A and B). Whereas the additional removal of parn-2 and panl-2 did not alleviate this meiotic arrest, diakinetic nuclei were detected upon removal of ccr-4, or parn-1, or the reduction of ccf-1 (Figure 2A and B).
Figure 2.
GLD-2 opposes CCR-4 and CCF-1 in the regulation of meiotic progression. (A) Diakinetic nuclei are absent from gonads lacking GLD-2 but form upon the additional removal of certain deadenylase activities. Percentage of germ lines in which diakinetic nuclei can be detected in the proximal part as judged by DAPI staining. (B) Examples of germ cell nuclei in the proximal part of extruded gonads from indicated genetic backgrounds. DAPI staining (purple in merge) marks DNA; NPC = Nuclear pore complex (green in merge). (C) Extruded gonads of indicated genetic backgrounds stained for the oocyte marker proteins OMA-1/2 (oo, red in merge); and DAPI to reveal chromatin morphologies (green in merge). Scale bars = 10 μm.
To test whether compromising either Ccr4–Not or PARN-1 activity in a gld-2 mutant would also restore the oocyte differentiation program, we analyzed the respective gonads for the presence of the two paralogous oocyte marker proteins, OMA-1 and OMA-2, jointly referred to as OMA-1/2 (30). In wild type but not in gld-2 single mutants, OMA-1/2 expression is detected in proximally differentiating oocytes (Figure 2C). Interestingly, OMA-1/2 expression recovered by the additional removal of ccr-4 or ccf-1, but not parn-1 (Figure 2C). Overall this suggests that the entire Ccr4–Not deadenylase module is the primary opposing poly(A)-removing activity to GLD-2, guiding meiotic oocyte progression and differentiation.
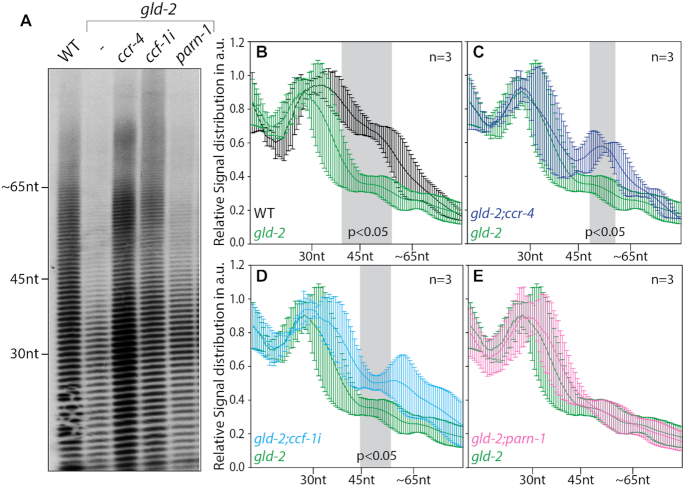
The Ccr4–Not complex counteracts gld-2-mediated polyadenylation at the global level
GLD-2 is important for the polyadenylation of many mRNAs in the germ line (10). To test a likely influence of CCR-4, CCF-1 or PARN-1 on the polyadenylation status of mRNAs at the global level, we conducted from whole worms bulk poly(A)-tail length measurements upon their removal. Consistent with our previous findings (10), bulk poly(A) tails were significantly shorter in the absence of GLD-2 (Figure 3A and B), also illustrating that that global changes in tail length can be detected specifically form germ cells using this method. Compared to gld-2 single mutants, no significant difference in tail length distribution was detected in gld-2;parn-1 double mutants (Figure 3A and E), whereas longer tails were present in gld-2;ccr-4 and gld-2;ccf-1(RNAi) animals (Figure 3A, C and D). This suggests that both deadenylases of the Ccr4–Not complex, CCR-4 and CCF-1, oppose GLD-2-mediated polyadenylation of target mRNAs. Contrary to this, PARN-1 has no significant global impact on the polyadenylation status of GLD-2-dependent target mRNAs, suggesting that either a small subset or no mRNAs are targeted by PARN-1.
Figure 3.
Reduced activities of the Ccr4–Not complex but not PARN rescue bulk polyadenylation defects in gld-2 mutants. (A) The global status of poly(A)-tail lengths were analyzed in the indicated genetic backgrounds using bulk poly(A) measurements. A representative gel image is shown. (B–E) Line scans of bulk poly(A) measurements of three independent biological samples (n) showing the relative signal distribution in the lane in arbitrary units (a.u.). Values of statistical difference, calculated with a two-tailed Student's t-test, are highlighted in gray. nt = nucleotides.
The balance between gld-2 and ccf-1 regulates mRNA levels
In a previous study, we found that a large number of germline mRNAs depends on GLD-2 for efficient expression (10). To identify which DeAd is responsible for the degradation of these putative gld-2-target mRNAs, we assessed the abundance of mRNAs by Next-Generation Sequencing in the following genetic backgrounds: wild type, gld-2, gld-2;ccr-4, gld-2;ccf-1(RNAi) (henceforth called gld-2;ccf-1) and gld-2;parn-1. For each genetic background, three independent biological samples were prepared and mRNAs from hand-picked whole worms were analyzed.
First, we compared wild type to gld-2 single mutants focusing our analysis on germline-expressed mRNAs (31). The mRNA abundance changes detected in the mutant are similar to the ones detected in our previous RNAi experiments (10), with one notable difference. Spermatogenic mRNAs are more strongly upregulated in the mutant compared to the RNAi treatment (Supplementary Figure S1A). This is consistent with the observation that an RNAi knockdown of gld-2 has a less severe impact on germ cell development than a protein null mutant (10,15,32), paralleling its spermatogenic defect (28,32). Regardless of the extent in expression changes the correlation is high between the two data sets (r = 0.81; Supplementary Figure S1A), arguing that the gene expression changes are comparable between gld-2 mutants and gld-2(RNAi) animals.
To assess which genes are significantly changed in the gld-2 mutant, we chose the following parameter for our analysis: minimal normalized read count ≥100; fold change ≥1.5 and P-adjusted < 0.05, again focusing only on germ line expressed genes. As a result, we found that 406 mRNAs are significantly down and 1056 mRNAs are significantly up-regulated (Figure 4A, Supplementary FileS1). Consistent with described phenotypic changes (28), the up-regulated mRNAs are to a large extent from spermatogenic genes, whereas down-regulated mRNAs are primarily from sex-neutral and oogenic genes (Figure 4B). To gauge the behavior of direct GLD-2 target mRNAs in our data set, we analyzed the expression changes of mRNAs that have been described to physically be associated with GLD-2 protein in an RNP complex (32). We find that the abundance of direct GLD-2 targets is significantly reduced in gld-2 null mutants (Figure 4C). This observation suggests that the up-regulated mRNAs are most-likely indirect GLD-2 targets and their expression changes are a consequence of altered germ cell physiology. Overall these data support the previously suggested function of GLD-2 in promoting mRNA stability (10), and argues that the 406 down-regulated mRNAs are significantly enriched in direct GLD-2 targets. For this study, we define these less abundant mRNAs as GLD-2-stability targets.
Figure 4.
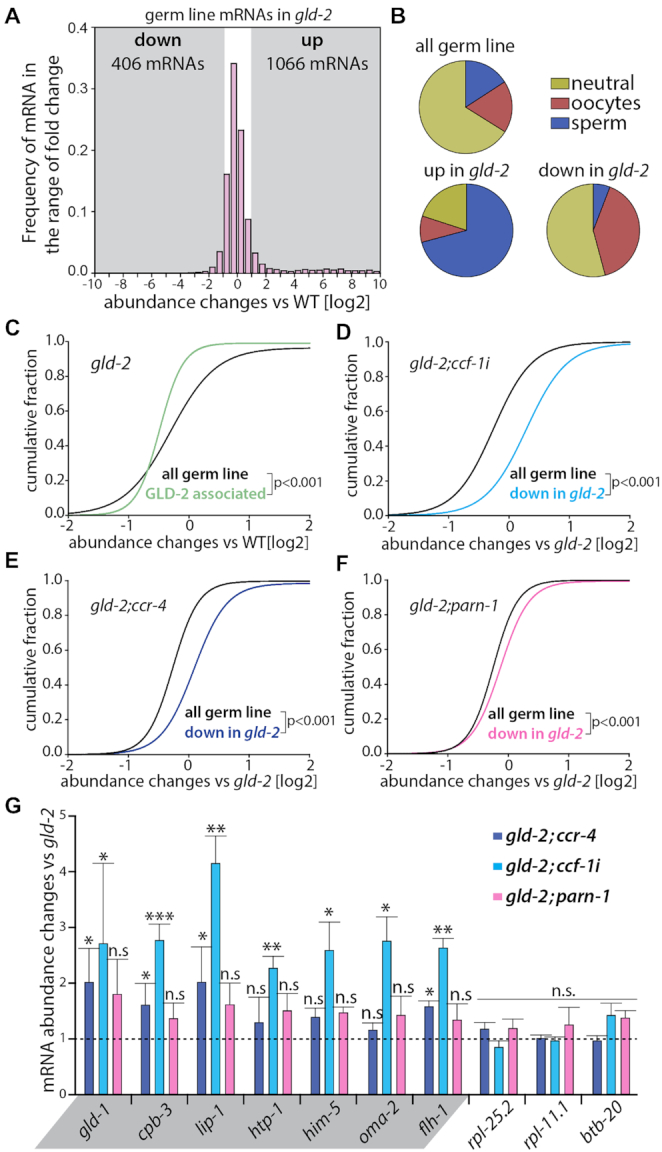
GLD-2-dependent mRNA targets are stabilized by the additional knockdown of CCF-1 deadenylase. (A) Histogram of germline mRNA abundance changes in gld-2 animals compared to wild type. All detected 8286 germline mRNAs are shown. Gray areas mark regions of at least a twofold change. The numbers of significantly up- or down-regulated mRNAs fulfil the following parameters: read number ≥ 100, fold change ≥1.5×, P-adj ≤ 0.05. (B) Distribution of sex-neutral, sperm-, or oocyte-enriched mRNAs in the indicated data sets, according to the classification by Oritz at al. 2014. (C–F) Cumulative fractions of mRNA abundance changes comparing GLD-2-associated mRNAs to all germline mRNAs in the gld-2 single mutant (C), and gld-2-down-regulated mRNAs to all germline mRNAs in gld-2;ccf-1(RNAi) (D), gld-2;ccr-4 (E), or gld-2;parn-1 (F) animals. Given statistically significant differences were calculated with a Mann–Whitney Rank Sum Test. (G) Abundance measurements of putative GLD-2-dependent target mRNAs (highlighted in gray) or nontarget mRNAs. A selection of candidates is shown; for all tested mRNAs see Supplementary Figure S1C. All quantitative real-time PCR measurements (n = 3) were normalized to the expression values of pab-1. Statistically significant differences were calculated via the Student's t-test. *P < 0.05; **P < 0.01; ***P < 0.001; n.s. = not significant.
To see which DeAd might be responsible for the down regulation of mRNAs in gld-2, we investigated the expression changes of GLD-2-stabilty targets in various gld-2;DeAd animals by comparing them to the gld-2 single mutant. In comparison to the changes of all germline mRNAs, the additional removal of ccf-1, ccr-4 or parn-1 led to a statistically significant increase in the levels of GLD-2-stability targets in gld-2 mutants (Figure 4D-F). However, quantitative differences are detected among the three analyzed DeAds: a minor increase in abundance was detected by removal of parn-1; a moderate increase by removal of ccr-4; and the largest increase upon reduction of ccf-1 (Supplementary Figure S1B). In addition, the RNA-sequencing data were validated by analyzing the expression changes of twelve GLD-2-stability targets by RT-qPCR. These measurements showed that the mRNA levels were significantly increased for 12/12 and 5/12 mRNAs in gld-2;ccf-1 and gld-2;ccr-4 (Figure 4G; Supplementary Figure S1C), respectively. No significant increase was detected in gld-2;parn-1 animals for the tested candidate mRNAs (Figure 4G; Supplementary Figure S1C). This suggests that PARN-1-mediated expression changes might be very subtle, and argues that PARN-1 most-likely plays no major role in the degradation of GLD-2-stability targets. Furthermore, this expression analysis supports the view that GLD-2 overall protects its target mRNAs from Ccr4–Not-mediated degradation, which is primary due to the activity of CCF-1.
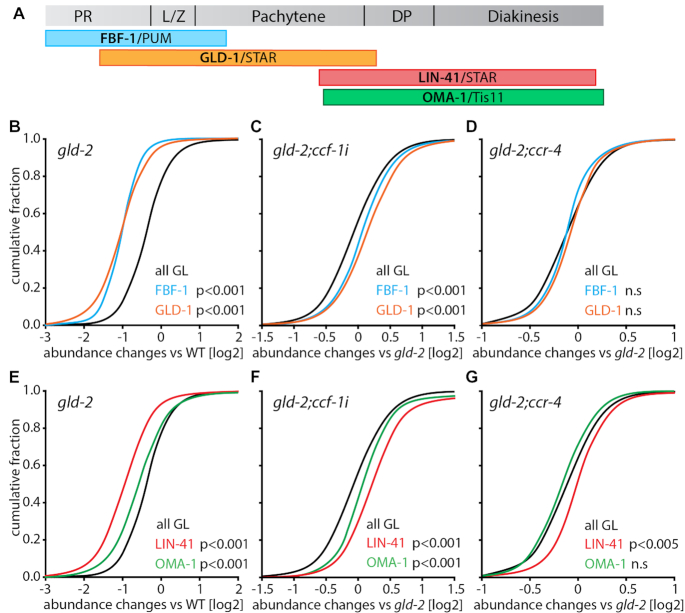
GLD-2 and the Ccr4–Not complex regulate RNA stability during all stages of meiotic prophase I
Next, we investigated whether Ccr4–Not and GLD-2 regulate mRNAs during all stages of oocyte production. To this end, we analyzed abundance changes of mRNAs in our data set that are presumed targets of well-defined sequence-specific RBPs. We used data from representatives of four evolutionary conserved RBPs: the PUF protein FBF-1 (33), the STAR protein GLD-1 (34), the TRIM-NHL protein LIN-41 and the Tis11-type Zinc-finger protein OMA-1 (35). All of these proteins are expressed in a stage-specific manner during oocyte production (Figure 5A). FBF-1 and GLD-1 are mRNA regulators during early stages of oocyte production and are mainly expressed in proliferating or pachytene germ cells, respectively (36,37). LIN-41 and OMA-1 are mRNA regulators during late stages of oocyte production and are primarily expressed in diplotene and diakinetic germ cells (30,38). Large data sets of putative target mRNAs are available for all four RBPs that were previously co-purified with each respective RBP (33–35).
Figure 5.
The Ccr4–Not complex and GLD-2 influence mRNA abundance during all meiotic stages of oocyte production. (A) Scheme of adult germline tissue with indicated protein expression patterns of four evolutionary conserved RNA-binding proteins that guide oogenesis. (B–G) Cumulative fractions of the abundance changes of all germline mRNAs (all GL) in the indicated genetic backgrounds and the top 100 mRNAs bound by either FBF-1/PUF, GLD-1/STAR, OMA-1/Tis11, or LIN-41/TRIM-NHL. Only mRNAs classified as sex-neutral or oocyte-enriched were considered, following the classification by Oritz et al. 2014. Statistically significant differences between the RBP-bound and all germline mRNAs are indicated and were calculated via a Mann–Whitney Rank Sum test.
For our analysis, we focused on the top 100 FBF-1-, GLD-1-, LIN-41- or OMA-1-associated mRNAs. These target mRNAs were always compared to the behaviour of all germline genes, and only mRNAs that have previously been classified as sex-neutral or oogenic were considered in our analysis (31). We found that target mRNAs of FBF-1/PUF and GLD-1/STAR behaved similarly: they were significantly decreased in the gld-2 single mutant, increased in gld-2;ccf-1, and unchanged in gld-2;ccr-4 (Figure 5B–D). However, target mRNAs of LIN-41/TRIM-NHL and OMA-1/Tis11 behaved differently: although both were decreased in the gld-2 single mutant and increased in gld-2;ccf-1 (Figure 5E, F), OMA-1 targets were unchanged in gld-2;ccr-4 as compared to gld-2, and a small but significant increase was detected for LIN-41 targets (Figure 5G). Taken together, this suggests that GLD-2 and CCF-1 form a functionally antagonistic pair to regulate the abundance of mRNAs during all stages of oocyte production, with a small contribution of CCR-4 during late stages. Overall, this data shows that balancing Ccr4–Not and GLD-2 activities mediate mRNA levels essentially during all different stages of early oogenesis.
GLD-2 promotes ribosome association of mRNA targets during late prophase I
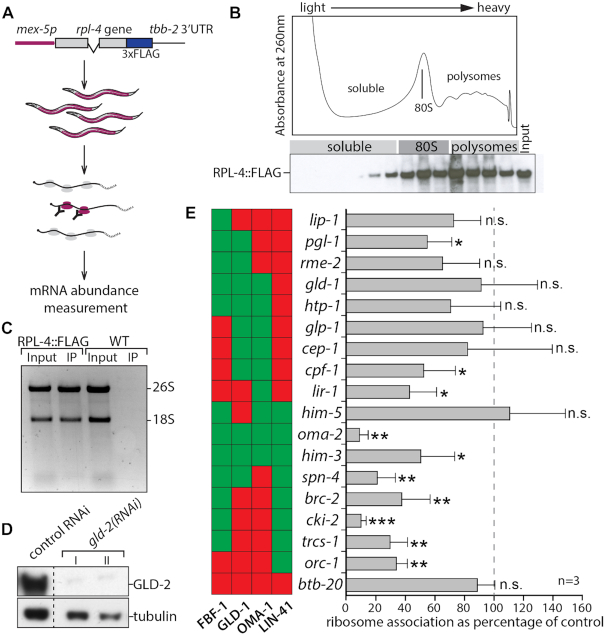
Finally, we investigated whether GLD-2 cytoPAP also influences the translation of mRNAs during any stage of oocyte production. To allow tissue-specific isolation of ribosomes from multi-cellular organisms, we adopted the Translating Ribosome Affinity Purification (TRAP) method to C. elegans germ cell analysis (39), and expressed from a germ cell-specific promoter the FLAG-tagged RPL-4 large ribosomal subunit protein (Figure 6A). Western blot analysis of polysome gradient fractions showed that the epitope tagged ribosomal protein RPL-4::FLAG is enriched in heavy polysomal fractions (Figure 6B), indicating its functional incorporation into translating ribosomes. This was further confirmed by immunoprecipitating ribosomes from RPL::FLAG-expressing but not wild-type worm extracts using anti-FLAG antibodies. Subsequent analysis of associated RNA material revealed that both ribosomal RNAs (18S and 26S) were abundantly detected in purifications from transgenic but not in control animals (Figure 6C). As FLAG-tagged RPL-4 worms have no apparent fecundity defects [number of offspring N2: 320±47(N = 23), rpl4::FLAG: 308±61 (N = 21)], we concluded that RPL-4::FLAG expressing worms are suitable for TRAP experiments.
Figure 6.
GLD-2 promotes the translation of LIN-41 targets. (A) Scheme of germ cell-specific ribosome immunopurification, for details see Materials and Methods. (B) Absorbance profile at 260nm of a typical polysome gradient from RPL-4::FLAG-expressing adult worms (top). Western blot of gradient fractions tested for the presence of tagged RPL-4 (bottom). The sedimentation positions of major ribonucleoprotein complexes are indicated. (C) A denaturing agarose gel of RNA isolated from anti-FLAG immunoprecipitation (IP) experiments from crude extracts (Input) of either RPL-4::FLAG-expressing or non-FLAG-expressing wild-type (WT) animals. Bands corresponding to the large (26S) and small (18S) ribosomal RNAs are indicated. (D) Western blot analysis of the indicated proteins of RPL-4::FLAG expressing animals treated with control or gld-2 RNAi. Extracts of two out of three independent gld-2(RNAi) treatments (I and II) are shown for comparison. The control sample corresponds to RNAi-treatment I. Additional lanes were removed for clarity (dashed line). (E) A selection of FBF-1, GLD-1, OMA-1 or LIN-41 target mRNAs analyzed for changes in ribosome association, comparing gld-2 to control RNAi-treated animals. The abundance of 18 mRNA was measured by RT-qPCR. Statistically significant differences are indicated and were calculated via a two-tailed Student's t-test. *P < 0.05; **P < 0.01; ***P < 0.001; n.s. = not significant; n = 3 biologically independent experiments. The color-coded columns to the left of each gene name indicates the target status of the corresponding mRNAs towards the respective RBPs (red = non-target; green = target). The complete data set of 30 mRNAs is shown in Supplementary Figure S2A.
To test whether GLD-2 influences the translation efficiency of mRNAs during any stage of oocyte production, we examined the changes in ribosome association of FBF-1/PUF, GLD-1/STAR, LIN-41/TRIM-NHL or OMA-1/Tis11 mRNA targets upon GLD-2 knockdown by feeding RNAi. Comparative western blot analysis of control RNAi to gld-2(RNAi) protein extracts showed that GLD-2 protein could be efficiently reduced in RPL-4::FLAG expressing worms (Figure 6D). Following immunoprecipitations of RPL-4::FLAG from three independent RNAi treatments, co-purified mRNAs and the corresponding input materials were analyzed by quantitative real-time PCR. To account for mRNA translation changes driven by variations in general mRNA abundance, a ribosome association coefficient was calculated for each individual mRNA by normalizing the amounts of ribosome-associated mRNA to the signal from the input. Thirty candidates were chosen that represent prominent mRNA targets of the four different RBPs. While two third of the tested mRNAs (20/30) remained unchanged, one third (10/30) were significantly less associated with ribosomes in gld-2(RNAi) (Figure 6E and Supplementary Figure S2A). Interestingly, the majority of the translation-reduced mRNAs are targets of LIN-41 (7/9) compared to FBF-1 (7/19), GLD-1 (4/17) and OMA-1 (4/16) (Figure 6E and Supplementary Figure S2). This suggests that especially LIN-41-associated mRNAs require GLD-2 for efficient translation, further arguing that in addition to mRNA stability, GLD-2 promotes also the translation of mRNAs prior to oocyte maturation stages.
DISCUSSION
The regulation of gametogenesis in many organisms relies heavily on post-transcriptional mechanisms involving the deadenylases of the Ccr4–Not complex and the cytoPAP GLD-2. Removal of one of the two opposing activities has devastating consequences for germ cell development. We show that the removal of both poly(A) modifying forces restores aspects of germ cell development back to wild type arguing that an intricate balance between these opposing enzymes is a key to regulate mRNAs in this tissue. Interestingly, these two opposing activities represent the major polyadenylation and deadenylation forces in female germ cells, and are likely recruited by a number of different translational repressor RBPs to stabilize and translationally regulate the oogenic mRNAs. Due to the evolutionary conservation of the poly(A) modifiers, as well as many of the RBPs investigated in this study, it is likely that Ccr4–Not and GLD-2 provide the regulatory frame work for poly(A)-mediated mRNA regulations also in other organisms.
We find that the previously characterized GLD-2 antagonist PARN plays no major role in regulating mRNAs during oocyte production. This observation is in strong contrast to the model proposed for oocyte maturation in frogs (40). The difference of the oogenic developmental stages could be one possible explanation for this finding. In general, PARN is assumed to be predominantly nuclear and by analogy to Xenopus oocytes is only present in the cytoplasm upon nuclear envelop breakdown during oocyte maturation (41). Hence, PARN might not be able to participate in cytoplasmic mRNA regulation during the oocyte production phase. Two PARN proteins exist in C. elegans, PARN-1 and PARN-2, and the subcellular localization of either endogenous protein has not been clarified yet. Our own attempts to generate specific antibodies failed to date. While our experimental setup precluded us from detecting potential oocyte maturation functions of PARN, neither deadenylase appears to be a major factor for mRNA regulation during early female gametogenesis. In animals lacking GLD-2, only the removal of PARN-1 rescues some aspects of early oocyte development. This observation is likely indirect and might point at poly(A)-length control mechanisms in the nucleus that precede further length control in the cytoplasm. Alternatively, the rescue might be explained by other PARN-1 functions that influence germ cell development, such as piRNA biogenesis (42). Although, no data exist addressing the functionality of PARN during early female germ cell development in other organisms, a growing number of studies from tissue culture systems supports the view that PARN proteins primarily target non-coding RNAs rather than mRNAs (43–47).
The catalytic module of the Ccr4–Not complex is of central importance for animal gametogenesis. It has previously been suggested that the deadenylase CCF-1/Caf1 is important for promoting germ cell proliferation in nematodes (48). Here, we confirm this observation and extend it to other components of the Ccr4–Not complex. First, downregulation of either the structural component NTL-1/Not1 or the catalytic component CCF-1/Caf1 elicits in all tested aspects of germ cell development similar defects, arguing that both components together comprise the active nuclease core module of the Ccr4–Not complex. Second, for regulating the transition from proliferation to differentiation the activity of CCF-1 is more important than the activity of CCR-4. Third, next to promoting germ cell proliferation, Ccr4–Not is also essential for maintaining undifferentiated germ cells. Similar observations have been made in D. melanogaster, where CAF1-driven CCR4-Not activity is employed to promote germline stem cell self-renewal (49). In mice, Ccr4–Not–mediated activity is required for spermatogenic stem regulation, however it is currently unknown which catalytic subunit is important for this function (50). Therefore, the evolutionary conserved Ccr4–Not complex represents the main deadenylase activity in germline stem and differentiation cells to regulate gene expression programs in metazoans.
Many RBPs that negatively regulate specific sets of target mRNAs form mRNPs that recruit the Ccr4–Not complex to facilitate translational repression (1). However, the majority of such mRNPs contain many additional RNA-associated regulators, including poly(A)-tail length modifiers. All four RBPs investigated in this work, FBF-1/PUF, GLD-1/STAR, LIN-41/TRIM-NHL, and OMA-1/Tis11 belong to distinct RBP families and were initially described to act as translational repressors (38,51–53). Although existing biochemical data on FBF-1-, LIN-41- and OMA-1-containing RNPs suggest that Ccr4–Not components and GLD-2 cytoPAP are part of each respective mRNP (35,48,54), it was unknown whether the presence of the tail modifiers has any functional relevance for the target mRNAs. Moreover no biochemical data on GLD-1/STAR-containing RNP exists currently that links their repressive function to enzymes that edit poly(A)-tail lengths. Our data predicts that all four RBPs, including GLD-1/STAR, utilize the antagonism of poly(A)-tail modifiers to regulate their mRNA targets, reminiscent of CPEB-target mRNAs during frog oocyte maturation (40). Our findings on members of four other RNA-binding protein classes suggest that a fine balance of opposing tail-length modifiers is reiteratively employed during all stages of female germ cell development to achieve differential gene expression regulation at the mRNA level. Furthermore, this concept argues that translational repressor RBPs employing deadenylases may in addition have to recruit cytoPAPs to ensure mRNA stability, either during the period of active repression or immediately after release from repression. It is conceivable that this antagonistic frame work of Ccr4–Not/GLD-2 could be a widespread phenomenon in poly(A)-mediated mRNA regulation and its use via RBPs extends beyond germ cells.
During the different stages of oocyte development, poly(A) function might shift from promoting mRNA stability towards promoting translatability. In Drosophila, strong GLD-2-dependent polyadenylation of mRNAs is detected in maturing oocytes (11,55,56). Furthermore, during this developmental stage the translational efficiency of mRNAs is mainly driven by its poly(A)-tail length (55,56), arguing that regulation of translation is the primary mechanisms of GLD-2-mediated gene expression control during oocyte maturation. Our current work in C. elegans provides evidence that GLD-2 cytoPAP promotes translation of mRNAs already prior to oocyte maturation, indicating that aspects of tail-mediated translational regulation may start to appear already during late stages of oocyte production in preparation for embryogenesis. This may reflect a change in general RNA degradation competence of female germ cells, a potentially conserved feature of animal gametogenesis.
DATA AVAILABILITY
NGS data was deposited in the GEO data base (accession number: GSE130685).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Elmar Wahle and Tosin Oyewale for critical reading of the manuscript, Andreas Dahl (Dresden Genome Centre) for NGS sequencing and Johannes Müller for technical support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation [EC369/2-3, EC369/3-1 and EC369/6-1 to C.R.E.; NO1402/1-1 to M.N.]. Funding for open access charge: German Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Goldstrohm A.C., Wickens M.. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008; 9:337–344. [DOI] [PubMed] [Google Scholar]
- 2. Minasaki R., Eckmann C.R.. Subcellular specialization of multifaceted 3′end modifying nucleotidyltransferases. Curr. Opin. Cell Biol. 2012; 24:314–322. [DOI] [PubMed] [Google Scholar]
- 3. Richter J.D., Lasko P.. Translational control in oocyte development. Cold Spring Harb. Perspect. Biol. 2011; 3:a002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wahle E., Winkler G.S.. RNA decay machines: deadenylation by the Ccr4–Not and Pan2-Pan3 complexes. Biochim. Biophys. Acta. 2013; 1829:561–570. [DOI] [PubMed] [Google Scholar]
- 5. Nousch M., Techritz N., Hampel D., Millonigg S., Eckmann C.R.. The Ccr4–Not deadenylase complex constitutes the main poly(A) removal activity in C. elegans. J. Cell Sci. 2013; 126:4274–4285. [DOI] [PubMed] [Google Scholar]
- 6. Temme C., Zhang L., Kremmer E., Ihling C., Chartier A., Sinz A., Simonelig M., Wahle E.. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA. 2010; 16:1356–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yamashita A., Chang T.C., Yamashita Y., Zhu W., Zhong Z., Chen C.Y., Shyu A.B.. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005; 12:1054–1063. [DOI] [PubMed] [Google Scholar]
- 8. Goldstrohm A.C., Hook B.A., Seay D.J., Wickens M.. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006; 13:533–539. [DOI] [PubMed] [Google Scholar]
- 9. Lykke-Andersen J., Wagner E.. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005; 19:351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nousch M., Yeroslaviz A., Habermann B., Eckmann C.R.. The cytoplasmic poly(A) polymerases GLD-2 and GLD-4 promote general gene expression via distinct mechanisms. Nucleic Acids Res. 2014; 42:11622–11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cui J., Sartain C.V., Pleiss J.A., Wolfner M.F.. Cytoplasmic polyadenylation is a major mRNA regulator during oogenesis and egg activation in Drosophila. Dev Biol. 2013; 383:121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakel K., Bonneau F., Eckmann C.R., Conti E.. Structural basis for the activation of the C. elegans noncanonical cytoplasmic poly(A)-polymerase GLD-2 by GLD-3. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:8614–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang L., Eckmann C.R., Kadyk L.C., Wickens M., Kimble J.. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002; 419:312–316. [DOI] [PubMed] [Google Scholar]
- 14. Kim K.W., Nykamp K., Suh N., Bachorik J.L., Wang L., Kimble J.. Antagonism between GLD-2 binding partners controls gamete sex. Dev. Cell. 2009; 16:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nousch M., Minasaki R., Eckmann C.R.. Polyadenylation is the key aspect of GLD-2 function in C. elegans. RNA. 2017; 23:1180–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morris J.Z., Hong A., Lilly M.A., Lehmann R.. twin, a CCR4 homolog, regulates cyclin poly(A) tail length to permit Drosophila oogenesis. Development. 2005; 132:1165–1174. [DOI] [PubMed] [Google Scholar]
- 17. Sha Q.Q., Yu J.L., Guo J.X., Dai X.X., Jiang J.C., Zhang Y.L., Yu C., Ji S.Y., Jiang Y., Zhang S.Y. et al.. CNOT6L couples the selective degradation of maternal transcripts to meiotic cell cycle progression in mouse oocyte. EMBO J. 2018; 37:e99333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974; 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frokjaer-Jensen C., Davis M.W., Ailion M., Jorgensen E.M.. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods. 2012; 9:117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hadwiger G., Dour S., Arur S., Fox P., Nonet M.L.. A monoclonal antibody toolkit for C. elegans. PLoS One. 2010; 5:e10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Millonigg S., Minasaki R., Nousch M., Novak J., Eckmann C.R.. GLD-4-mediated translational activation regulates the size of the proliferative germ cell pool in the adult C. elegans germ line. PLoS Genet. 2014; 10:e1004647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zetka M.C., Kawasaki I., Strome S., Muller F.. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 1999; 13:2258–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rybarska A., Harterink M., Jedamzik B., Kupinski A.P., Schmid M., Eckmann C.R.. GLS-1, a novel P granule component, modulates a network of conserved RNA regulators to influence germ cell fate decisions. PLoS Genet. 2009; 5:e1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liao Y., Smyth G.K., Shi W.. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013; 41:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sijen T., Fleenor J., Simmer F., Thijssen K.L., Parrish S., Timmons L., Plasterk R.H., Fire A.. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001; 107:465–476. [DOI] [PubMed] [Google Scholar]
- 28. Kadyk L.C., Kimble J.. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development. 1998; 125:1803–1813. [DOI] [PubMed] [Google Scholar]
- 29. Eckmann C.R., Crittenden S.L., Suh N., Kimble J.. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics. 2004; 168:147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Detwiler M.R., Reuben M., Li X., Rogers E., Lin R.. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell. 2001; 1:187–199. [DOI] [PubMed] [Google Scholar]
- 31. Ortiz M.A., Noble D., Sorokin E.P., Kimble J.. A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3. 2014; 4:1765–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim K.W., Wilson T.L., Kimble J.. GLD-2/RNP-8 cytoplasmic poly(A) polymerase is a broad-spectrum regulator of the oogenesis program. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:17445–17450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Porter D.F., Prasad A., Carrick B.H., Kroll-Connor P., Wickens M., Kimble J.. Toward Identifying Subnetworks from FBF Binding Landscapes in Caenorhabditis Spermatogenic or Oogenic Germlines. G3. 2019; 9:153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wright J.E., Gaidatzis D., Senften M., Farley B.M., Westhof E., Ryder S.P., Ciosk R.. A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. EMBO J. 2011; 30:533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tsukamoto T., Gearhart M.D., Spike C.A., Huelgas-Morales G., Mews M., Boag P.R., Beilharz T.H., Greenstein D.. LIN-41 and OMA ribonucleoprotein complexes mediate a translational repression-to-activation switch controlling oocyte meiotic maturation and the oocyte-to-embryo transition in caenorhabditis elegans. Genetics. 2017; 206:2007–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Crittenden S.L., Bernstein D.S., Bachorik J.L., Thompson B.E., Gallegos M., Petcherski A.G., Moulder G., Barstead R., Wickens M., Kimble J.. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002; 417:660–663. [DOI] [PubMed] [Google Scholar]
- 37. Jones A.R., Francis R., Schedl T.. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol. 1996; 180:165–183. [DOI] [PubMed] [Google Scholar]
- 38. Spike C.A., Coetzee D., Eichten C., Wang X., Hansen D., Greenstein D.. The TRIM-NHL protein LIN-41 and the OMA RNA-binding proteins antagonistically control the prophase-to-metaphase transition and growth of Caenorhabditis elegans oocytes. Genetics. 2014; 198:1535–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heiman M., Kulicke R., Fenster R.J., Greengard P., Heintz N.. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat. Protoc. 2014; 9:1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim J.H., Richter J.D.. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006; 24:173–183. [DOI] [PubMed] [Google Scholar]
- 41. Korner C.G., Wormington M., Muckenthaler M., Schneider S., Dehlin E., Wahle E.. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 1998; 17:5427–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tang W., Tu S., Lee H.C., Weng Z., Mello C.C.. The RNase PARN-1 trims pirna 3′ ends to promote transcriptome surveillance in C. elegans. Cell. 2016; 164:974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berndt H., Harnisch C., Rammelt C., Stohr N., Zirkel A., Dohm J.C., Himmelbauer H., Tavanez J.P., Huttelmaier S., Wahle E.. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA. 2012; 18:958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yi H., Park J., Ha M., Lim J., Chang H., Kim V.N.. PABP cooperates with the CCR4-NOT complex to promote mRNA deadenylation and block precocious decay. Mol. Cell. 2018; 70:1081–1088. [DOI] [PubMed] [Google Scholar]
- 45. Shukla S., Parker R.. PARN modulates Y RNA stability and its 3′-end formation. Mol. Cell Biol. 2017; 37:e00264-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Montellese C., Montel-Lehry N., Henras A.K., Kutay U., Gleizes P.E., O’Donohue M.F.. Poly(A)-specific ribonuclease is a nuclear ribosome biogenesis factor involved in human 18S rRNA maturation. Nucleic Acids Res. 2017; 45:6822–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moon D.H., Segal M., Boyraz B., Guinan E., Hofmann I., Cahan P., Tai A.K., Agarwal S.. Poly(A)-specific ribonuclease (PARN) mediates 3′-end maturation of the telomerase RNA component. Nat. Genet. 2015; 47:1482–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suh N., Crittenden S.L., Goldstrohm A., Hook B., Thompson B., Wickens M., Kimble J.. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics. 2009; 181:1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Joly W., Chartier A., Rojas-Rios P., Busseau I., Simonelig M.. The CCR4 deadenylase acts with Nanos and Pumilio in the fine-tuning of Mei-P26 expression to promote germline stem cell self-renewal. Stem Cell Rep. 2013; 1:411–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yamaji M., Jishage M., Meyer C., Suryawanshi H., Der E., Yamaji M., Garzia A., Morozov P., Manickavel S., McFarland H.L. et al.. DND1 maintains germline stem cells via recruitment of the CCR4-NOT complex to target mRNAs. Nature. 2017; 543:568–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang B., Gallegos M., Puoti A., Durkin E., Fields S., Kimble J., Wickens M.P.. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997; 390:477–484. [DOI] [PubMed] [Google Scholar]
- 52. Jan E., Motzny C.K., Graves L.E., Goodwin E.B.. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 1999; 18:258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Guven-Ozkan T., Robertson S.M., Nishi Y., Lin R.. zif-1 translational repression defines a second, mutually exclusive OMA function in germline transcriptional repression. Development. 2010; 137:3373–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Spike C.A., Coetzee D., Nishi Y., Guven-Ozkan T., Oldenbroek M., Yamamoto I., Lin R., Greenstein D.. Translational control of the oogenic program by components of OMA ribonucleoprotein particles in Caenorhabditis elegans. Genetics. 2014; 198:1513–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lim J., Lee M., Son A., Chang H., Kim V.N.. mTAIL-seq reveals dynamic poly(A) tail regulation in oocyte-to-embryo development. Genes Dev. 2016; 30:1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Eichhorn S.W., Subtelny A.O., Kronja I., Kwasnieski J.C., Orr-Weaver T.L., Bartel D.P.. mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. eLife. 2016; 5:e16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGS data was deposited in the GEO data base (accession number: GSE130685).