Abstract
The presence of ribonucleotides in DNA can lead to genomic instability and cellular lethality. To prevent adventitious rNTP incorporation, the majority of the DNA polymerases (dPols) possess a steric filter. The dPol named MsDpo4 (Mycobacterium smegmatis) naturally lacks this steric filter and hence is capable of rNTP addition. The introduction of the steric filter in MsDpo4 did not result in complete abrogation of the ability of this enzyme to incorporate ribonucleotides. In comparison, DNA polymerase IV (PolIV) from Escherichia coli exhibited stringent selection for deoxyribonucleotides. A comparison of MsDpo4 and PolIV led to the discovery of an additional polar filter responsible for sugar selectivity. Thr43 represents the filter in PolIV and this residue forms interactions with the incoming nucleotide to draw it closer to the enzyme surface. As a result, the 2’-OH in rNTPs will clash with the enzyme surface, and therefore ribonucleotides cannot be accommodated in the active site in a conformation compatible with productive catalysis. The substitution of the equivalent residue in MsDpo4–Cys47, with Thr led to a drastic reduction in the ability of the mycobacterial enzyme to incorporate rNTPs. Overall, our studies evince that the polar filter serves to prevent ribonucleotide incorporation by dPols.
INTRODUCTION
DNA polymerases (dPols) synthesize DNA through template-dependent addition of dNTPs to the growing primer strand. All dPols share a similar core structure with thumb, fingers, and palm domains. Three acidic residues constitute the catalytic center of these enzymes, and these residues are present in the palm domain. The catalytic residues co-ordinate a divalent metal ion, usually Mg2+, that is critical for DNA synthesis activity (1–3).
In the cellular environment, dPols encounter rNTPs much more frequently than dNTPs. The cellular concentrations of rNTPs are up to 10–100-fold higher than those of dNTPs depending on the type of cell, and stage of cell cycle (4–9). Like dNTPs, rNTPs can form Watson-Crick base pairs with DNA nucleotides, as they are sister molecules of dNTPs with an additional 2′-OH on the sugar moiety (4,10,11). Hence it is possible, that the dPols may inadvertently add rNTPs to the growing primer strand. The presence of ribonucleotides in DNA is detrimental to the integrity of the genome as they are less stable due to the reactive nature of the hydroxyl group on the C2′ atom (7–10,12,13). The presence of ribonucleotides in DNA can lead to the spontaneous appearance of double-strand breaks (5,8,13,14). The misincorporation of rNTPs into DNA is known to slow down the rate of DNA replication by Polδ, Polϵ and PolIII (5,6,12,15). Ribonucleotides can alter the conformation of DNA from B to A form, and thus adversely affect target sequence recognition by proteins such as transcription factors (10,16,17). Overall, the presence of ribonucleotides in the genomic DNA is detrimental to cellular physiology and, represents a significant challenge for the basic genomic processes such as replication, repair, and transcription (13,17). It is, therefore, imperative to reduce the frequency of incorporation of rNTPs into the genome during DNA replication.
Most dPols employ a steric filter to minimize rNTP incorporation (4,18–20). The steric filter is represented by an amino acid residue with a bulky side chain which will clash with the 2′-OH group on the ribose ring of the incoming nucleotide (Supplementary Figure S1) (9,19,21). As a result, rNTPs cannot bind stably in the active site of the dPol. The steric gate is represented by Glu in A-family polymerases and Tyr or Phe in members of the B, X, Y and RT families (11,18–20,22–36).
MsDpo4 is a Y-family dPol from Mycobacterium smegmatis (37,38). MsDpo4 does not possess the steric filter and can incorporate ribonucleotides (28,39). In comparison, DNA polymerase IV (PolIV) from Escherichia coli, exhibits stringent sugar selectivity. A rigorous comparison between MsDpo4 and PolIV shows the presence of a strategically located polar residue that aids sugar selectivity during DNA synthesis. The polar residue, therefore, represents a second filter- termed as a polar filter, which acts in tandem with the steric filter to prevent ribonucleotide incorporation by dPols.
MATERIALS AND METHODS
Protein purification
MsDpo4 (C47T, L14Y, C47T+L14Y) and PolIV (F13A and F13A+T43C) mutants were generated using QuikChange Lightning site-directed mutagenesis kit (Agilent). The MsDpo4 and PolIV proteins were expressed and purified as described previously (37,38,40). For phasing, selenomethionine-labeled MsDpo4 was prepared using B834 strain of E. coli. The labeled protein was purified and crystallized using identical protocols as for the native protein.
The PolII gene (E. coli) was amplified from E. coli genomic DNA and cloned into a modified pET-28b vector (pDJN1) (41). The exonuclease deficient mutant of PolII (D335A) (42) was generated using QuikChange Lightning site-directed mutagenesis kit (Agilent). The protein was expressed in the C41(DE3) strain of E. coli. The freshly transformed cells were grown to an OD600 = 0.8, at 37°C. The expression of PolII was induced with 0.4 mM IPTG followed by incubation at 18°C for 16 h. The cells were then harvested by centrifugation, lysed by sonication and the lysate was clarified by centrifugation. The purification was done using pre-packed Ni-NTA column (GE-Healthcare). The binding buffer (Buffer A) composition was 25 mM Tris pH 8.0, 250 mM NaCl, and 5% glycerol. The elution buffer (Buffer B) composition was 25 mM Tris pH 8.0, 250 mM NaCl, 5% glycerol and 1 M Imidazole. The bound protein was eluted using a step gradient of elution Buffer B. The eluted protein was then dialyzed overnight in Buffer A and then, concentrated and stored in −80°C.
Primer extension assay and steady-state kinetic analysis
To prepare the DNA substrate, a 6FAM labeled 15mer primer (5′-6FAM-CGTACTCGTAGGCAT-3′) was annealed to 50mer template (5′- TCCTACCGTGCCTACCTGAACAGCTGGTCACACAXATGCCTACGAGTACG-3′). The nucleosides deoxyadenosine, deoxycytidine, deoxythymidine and deoxyguanosine occupied the underlined position for the templates A, C, T and G respectively. The reaction mixture was composed of enzyme (MsDpo4/PolIV/PolII), dsDNA, dNTPs or rNTPs (Jena Bioscience), 25 mM Tris pH 8.0, 2 mM DTT, 0.1 mM (NH4)2SO4, 2.5 mM MnCl2 and 0.05 mg/ml BSA in a final volume of 20 μl. The reaction was carried out at 37°C for 60 min. The reaction was terminated by adding 10 μl of stop buffer, composed of 80% formamide, 10% bromophenol blue and 10% 0.5 M EDTA (40,43). The reaction products were denatured by heating at 95°C for 10 min and quick cooling in ice for 10 min. The products were then resolved in a 20% polyacrylamide gel with 8 M urea, in 1× TBE buffer and visualized using Typhoon scanner FLA-7000 (GE Healthcare). The band intensities were quantified using ImageQuant software. Relative percentage incorporation (RPI) in each group was calculated with respect to the highest in the group. RPI was calculated using the following equation
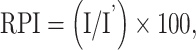 |
where I is the percentage incorporation by the mutant and I’ is the percentage incorporation by the most active version of the protein. For steady-state kinetic analysis, the reaction was carried out for varying time points (30 s to 30 min) at 37°C. The time point at which 20% of the primer had been extended was used for further analysis. Reactions were carried out with varying concentrations of incoming rCTP. After quantification of the resolved products, apparent Km and Vmax values were calculated using a Lineweaver–Burk plot. The measurements were carried out in triplicate, and the standard deviation values were calculated.
Crystallization, structure determination & crystallographic refinement
An 18mer self-complementary oligonucleotide (5′TCTGGGGTCCTAGGACCC 3′) was purified by ion-exchange chromatography using a monoQ column. The purified oligonucleotide was desalted, lyophilized, solubilized in water and annealed. The ternary complexes were reconstituted by mixing MsDpo4 (0.3 mM) with annealed dsDNA in a molar ratio of 1:1.2 followed by addition of 5 mM dCpNHpp or CpCpp (Jena Bioscience) and MgCl2 or MnCl2, respectively. Crystallization trials were carried out by the hanging drop method using commercial screens. After screening and optimization, the best crystals were obtained in 0.2 M Bis–Tris Propane (pH 6.0), 15–25% (w/v) PEG 3350 or PEG 2KMME and 0.1–0.7 M NaCl.
The PolIV-F13A+T43C protein in complex with DNA (template dG) and incoming rCpCpp or dCpNHpp was crystallized using the same conditions as that for wt- protein described previously (40,43). The initial characterization of the crystals was carried out using a Metaljet X-ray generator with a PhotonII detector (Bruker Inc.). The X-ray diffraction data for MsDpo4DNA(dG):dCTP, MsDpo4DNA(dG):rCTP, MsDpo4-L14YDNA(dG):dCTP and MsDpo4-L14YDNA(dG):rCTP were collected at the BM14 beamline of the European Synchrotron Radiation Facility (ESRF) in Grenoble. Data corresponding to MsDpo4-C47TDNA(dG):dCTP, PolIV-F13A+T43CDNA(dG):dCTP and PolIV-F13A+T43CDNA(dG):rCTP were collected at ID30B and ID29 beamline at ESRF, Grenoble, France. The data for MsDpo4-C47T+L14YDNA(dG):dCTP was collected at Elettra Sincrotrone in Trieste.
All images were integrated using iMOSFLM of CCP4 (44). The structure of the SeMet–MsDpo4DNA(dG):dCTP complex was determined by single-wavelength anomalous dispersion method using the AUTOSOL program in PHENIX (45). The initial model was built using the AUTOBUILD program in PHENIX, and subsequent manual rebuilding was done using COOT (46). The other structures of ternary complexes of wt- or mutant versions of MsDpo4 were determined by Molecular Replacement using PHASER in CCP4 (47,48). Similarly, the structure of PolIV-T43C+F13ADNA(dG):rCTP was determined by PHASER using the structure of PolIVDNA(dG):dCTP (4IRC) as a search model. All the structures were refined using PHENIX until convergence after which TLS refinement was carried out using Refmac in CCP4 (49).
Growth assays
The DNA sequence corresponding to the β-clamp binding peptide from E. coli PolIV (342QMERQLVLGL351) (50) was added to the 3′ end of the MsDpo4 gene, and this gene construct (msdpo4β) was cloned into a modified version of the pET-28b vector (41). The C47T mutation of the msdpo4β gene was generated using QuikChange Lightning site-directed mutagenesis kit (Agilent). The presence of the mutation was confirmed by gene sequencing. The growth assays were conducted using the B834 strain of E. coli. The spot assay was carried out with a serial dilution of culture harvested at the log phase (A600 = 0.6). LB-Ampicillin plates with 0.1 mM IPTG were used to induce the expression of the different constructs and assess their effect on cell survival. LB-Ampicillin plates with 1% glucose were used as the control. The plasmids bearing wt- and mutant msdpo4β gene constructs, and the empty vector were transformed into fresh competent cells of B834, and the transformed cells were grown overnight. Single colonies were picked and used to inoculate a starter culture which was harvested at the log phase (A600 = 0.6). The following serial dilutions were prepared- 10−1, 10−2 and 10−3. For each dilution, 5 μl was spotted on LB ampicillin plates with 0.1 mM IPTG or 1% glucose. The plates were incubated in 37°C for 12 h after which the observations were recorded.
The standard plate assay described by McDonald et al. was performed to quantitate the effect of the C47T mutation on bacterial survival (51). The transformed cells were grown to an OD600 = 0.6, and each culture was induced with 0.1 mM IPTG. Subsequently, the cultures were grown at 37°C for 5 h, following which the cells were plated and incubated overnight. The colonies obtained were counted, and the experiment was repeated three times. The fold change in the number of colony forming units (CFUs) was then calculated with respect to the wild type. The assays were repeated three times, and the standard deviation values were calculated.
RESULTS
Presence of the steric gate does not abrogate the ability of the MsDpo4 to incorporate ribonucleotides
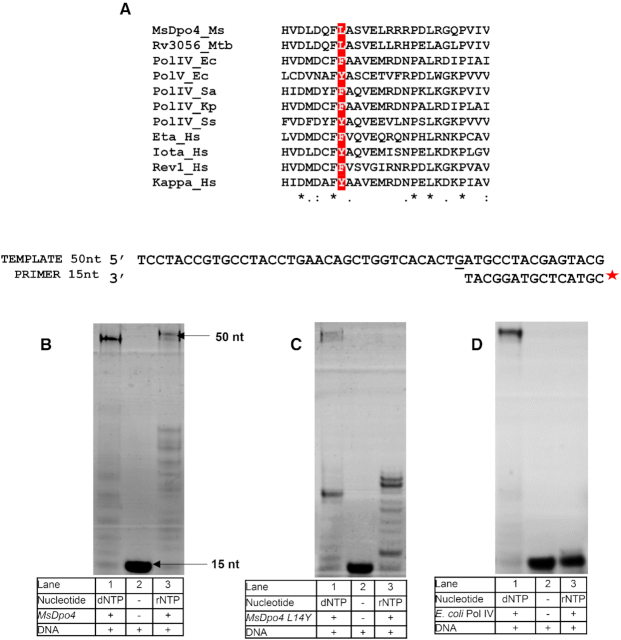
The steric filter is naturally absent in MsDpo4, and it has a Leu at the 14th position, instead of Tyr or Phe (Figure 1A). Primer extension assays show that MsDpo4 can incorporate both dNTPs and rNTPs (Figure 1B). The aromatic steric gate was engineered in MsDpo4, by mutating the Leu14 residue to Tyr. Surprisingly, primer extension assays showed that the MsDpo4-L14Y enzyme still exhibited substantial ability to incorporate rNTP (Figure 1C). In comparison, PolIV-WT did not show significant incorporation of rNTPs and thus exhibited stringent sugar selectivity (Figure 1D). Steady-state kinetic analysis carried out to compare the ability of MsDpo4-WT and MsDpo4-L14Y to incorporate ribonucleotides showed that addition of the steric gate led to only a six-fold reduction in the catalytic efficiency of rCTP incorporation (Table 1).
Figure 1.
Comparison of rNTP incorporation by MsDpo4-WT, MsDpo4-L14Y, and PolIV. (A) Sequence alignment of the N-terminal region of MsDpo4 and homologous proteins from bacteria, archaea, and humans shows that MsDpo4 lacks the steric filter as it has a Leu residue instead of an aromatic residue at the appropriate position (highlighted in red). Primer extension assays show that (B) MsDpo4-WT is capable of rNTP and dNTP incorporation. (C) MsDpo4-L14Y can incorporate rNTP and dNTP and (D) PolIV-WT from E. coli is unable to incorporate rNTP and therefore exhibits stringent sugar selectivity.
Table 1.
Steady-state kinetics of rCTP incorporation by MsDpo4-WT and MsDpo4-L14Y
| Protein | k cat a (min−1) | K m a (μM) | k cat/Kma (μM−1 min−1) |
|---|---|---|---|
| MsDpo4-WT | 0.66 ± 0.07 | 55.3 ± 4.0 | 0.01 ± 0.001 |
| MsDpo4-L14Y | 0.1 ± 0.01 | 59.54 ± 13.4 | 0.0016 ± 0.0003 |
aThe numbers after ± denote standard deviation (n = 3).
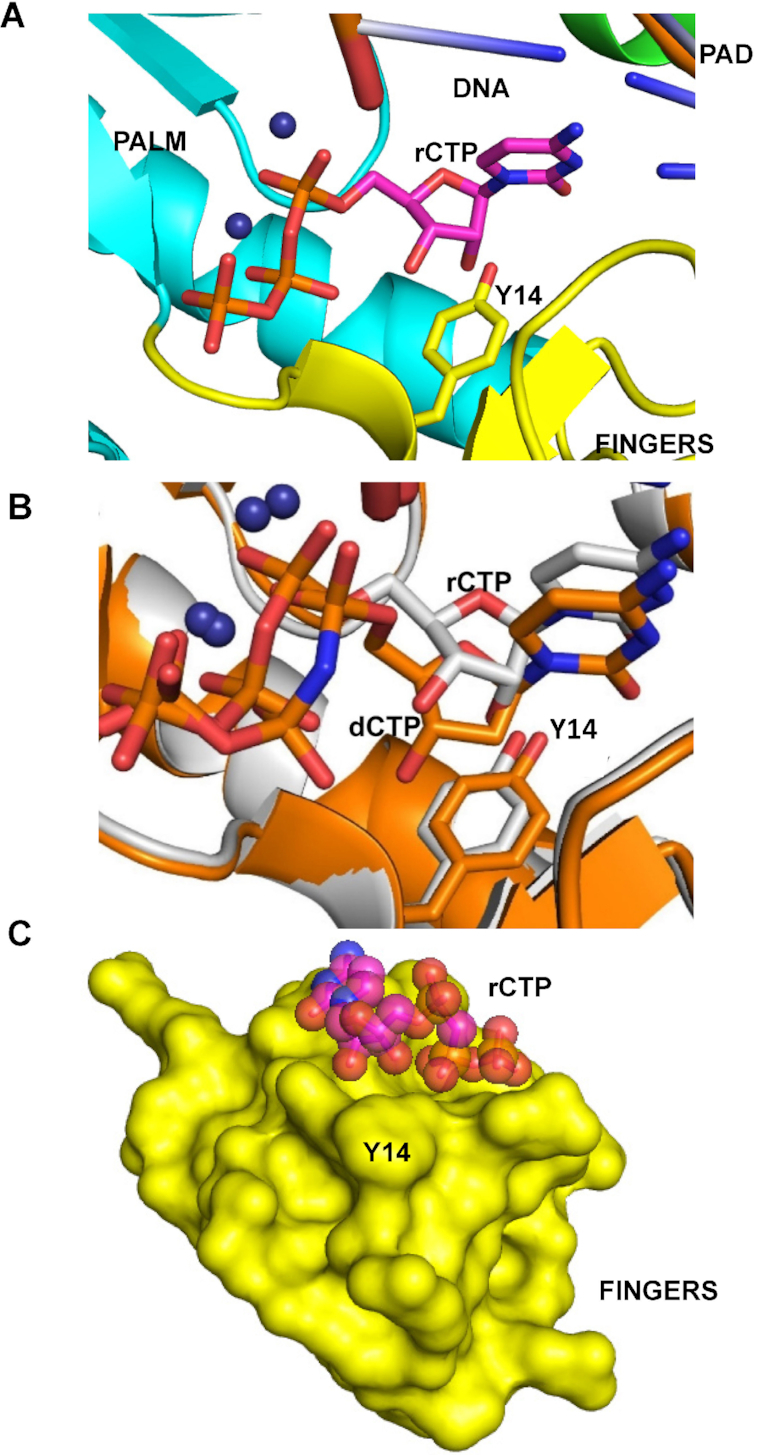
To understand the structural basis of ribonucleotide incorporation by MsDpo4-WT and the L14Y mutant, we first determined the structure of the wt- enzyme in complex with DNA bearing dG at the templating position and rCTP or dCTP as the incoming nucleotide to a resolution of 2.5 and 2.3 Å, respectively (Supplementary Table S2A and B). The structures for MsDpo4-WT showed that both dCTP and rCTP form Watson-Crick base pairs with the template dG, and are present in the location and conformation compatible with productive catalysis (Figure 2A). The two structures superimposed onto each other with an RMSD of 0.36 Å and showed no difference in the position and conformation of incoming nucleotide. These structures, therefore, show that MsDpo4 can bind rCTP and dCTP without any distortion in enzyme and DNA structure (Figure 2B and C). The structure of MsDpo4-L14Y was also determined with DNA bearing dG at the templating position and dCTP or rCTP as the incoming nucleotide to a resolution of 2.16 and 2.06 Å, respectively (Supplementary Table S2C and D). The structure showed that the rCTP was accommodated in the active site of MsDpo4-L14Y without any steric clash between the aromatic residue and 2′- OH (Figure 3A).
Figure 2.
Structural mechanism of rCTP and dCTP incorporation by MsDpo4-WT. (A) The structures of MsDpo4-WTDNA(dG):dCTP (colored yellow) and MsDpo4-WTDNA(dG):rCTP (colored violet) superimpose with an RMSD of 0.36 Å. The enzyme structure is shown in cartoon representation, DNA is shown as ribbons, and the incoming nucleotide and the Leu14 residue (in green) are shown in stick representation. A close-up of the region surrounding the incoming nucleotide, is displayed for (B) MsDpo4DNA(dG):rCTP and (C) MsDpo4DNA(dG):dCTP. The comparison shows that the dCTP and rCTP bind the MsDpo4 active site in the same location and conformation with marginal differences in the enzyme structure.
Figure 3.
Mechanism of rCTP incorporation by MsDpo4-L14Y. (A) A close-up of the region surrounding the incoming nucleotide is displayed for MsDpo4-L14YDNA(dG):rCTP structure. (B) The structures of MsDpo4-L14YDNA(dG):dCTP (colored orange) and MsDpo4-L14YDNA(dG):rCTP (colored white), superimpose with an RMSD of 0.37 Å. The comparison shows that the rCTP is repositioned and undergoes a change in the sugar pucker to prevent a steric clash between the 2′-OH and the engineered steric filter. (C) The surface of the fingers domain (in yellow) from the MsDpo4-L14YDNA(dG):rCTP structure is shown along with space filling model of the bound rCTP (colored according to element). The figure shows that there is no steric clashes between the rCTP molecule and the enzyme surface in this structure.
The structures of MsDpo4-L14Y in complex with DNA (template dG) and incoming rCTP or dCTP superimposed onto each other with an RMSD of 0.37 Å (Figure 3B). This comparison showed that, unlike in the case of the wt- enzyme, the incoming rCTP was repositioned. The rCTP is situated slightly upwards such that the C2′ atom of the sugar ring of rCTP is present 1.3 Å above that of the dCTP. Also, the sugar of the rCTP attains a C3′ endo conformation, and due to this, the 3′-OH group is present 2.4 Å above the corresponding location in the case of dCTP (Figure 3B). Due to these structural differences, the 2′-OH of the incoming rCTP is present in the same location as the C2′ atom of dCTP and therefore will not clash with the π-electron cloud over the aromatic side chain of the engineered Tyr14 residue (Figure 3C). The location of the triphosphate moiety is similar in the case of rCTP and dCTP and the changes in the position and conformation of the sugar do not adversely affect the ability of the rCTP to form a productive Watson-Crick base pair with the template dG. Overall, due to the repositioning of the rCTP and change in the sugar pucker, rCTP can bind to the MsDpo4-L14Y enzyme in a conformation compatible with catalysis and without any steric clashes with the enzyme surface (Figure 3C). Thus, the comparison shows that the presence of a steric filter is not adequate to achieve stringent sugar selectivity.
The presence of a polar filter prevents ribonucleotide incorporation
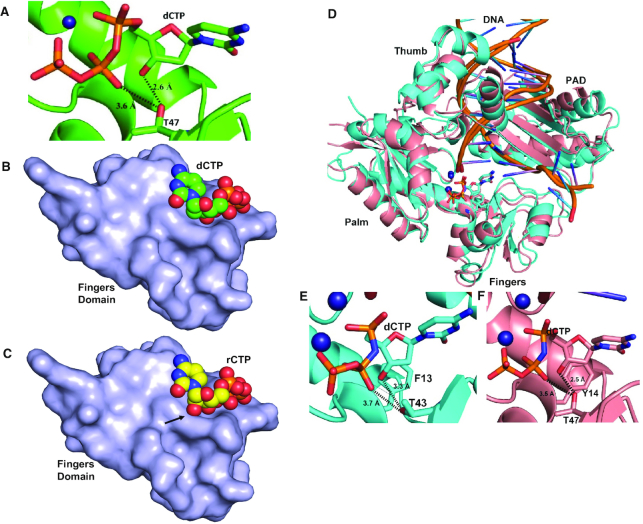
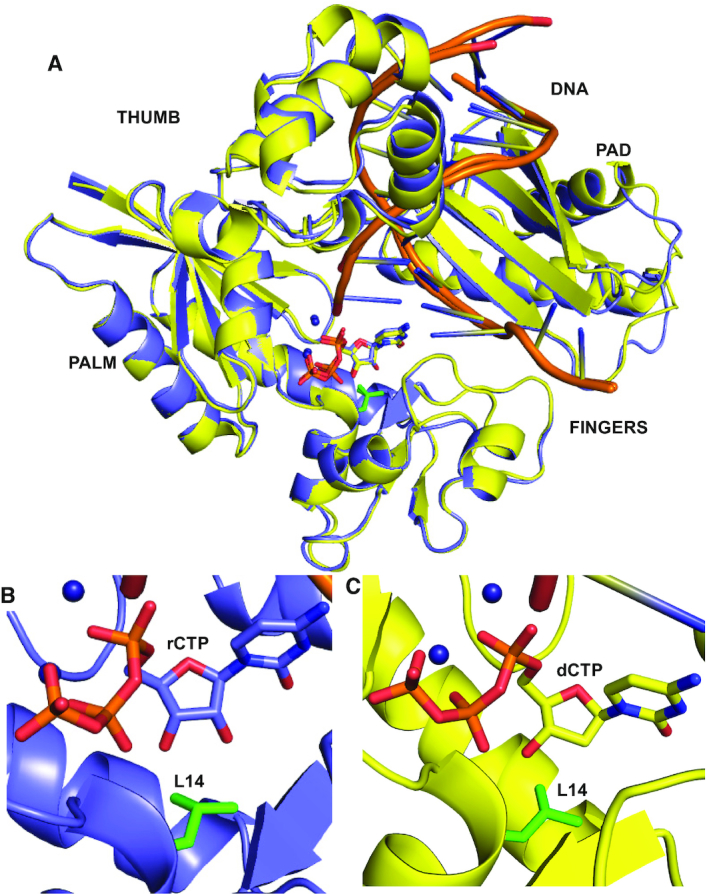
Since PolIV exhibits stringent ribonucleotide exclusion, the structures of ternary complexes of MsDpo4-WT and PolIV-WT (PDB ID 4IRC) (40) were compared to identify additional determinants of sugar selectivity. The structures of MsDpo4-WT and PolIV-WT, in complex with DNA (template dG) and incoming dCTP, were superimposed, and residues located within a distance of 5 Å in the vicinity of dCTP were compared. This comparison showed that at the position equivalent to Cys47 in MsDpo4, there exists a Thr residue in PolIV (Figure 4). This polar residue (Thr43) in PolIV, forms hydrogen bonds with 3′-OH and β-phosphate of the incoming nucleotide (Figure 4B). Since Cys47 in MsDpo4 cannot form equivalent interactions (Figure 4C), it is possible that Thr43 in PolIV may play a role in sugar selectivity.
Figure 4.
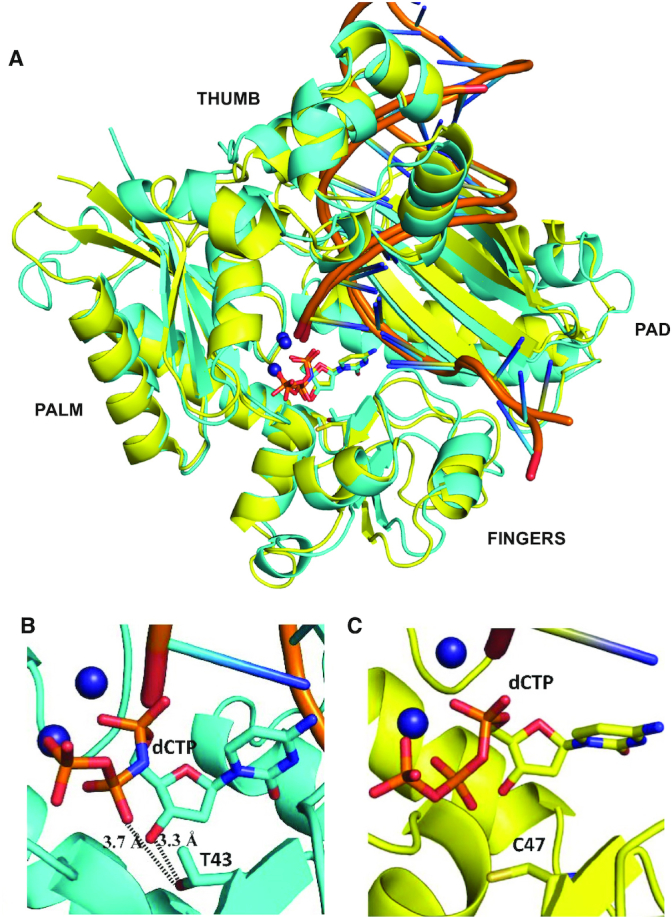
Comparison of the structures of ternary complexes of MsDpo4 and PolIV. (A) The two structures, MsDpo4DNA(dG):dCTP (colored yellow) and PolIVDNA(dG):dCTP (colored cyan) superimpose with an RMSD value of 1.47 Å. The comparison shows that, at the position equivalent to Thr43 residue in PolIV, Cys47 is present in MsDpo4. (B) In the PolIVDNA(dG):dCTP structure, Thr43 residue forms interactions with the 3′-OH and β-phosphate of the incoming dCTP and may play an important role in the ability of PolIV to exclude ribonucleotides. (C) The equivalent residue in the MsDpo4DNA(dG):dCTP structure is Cys47, and the Cys side chain cannot form interactions with the incoming nucleotide.
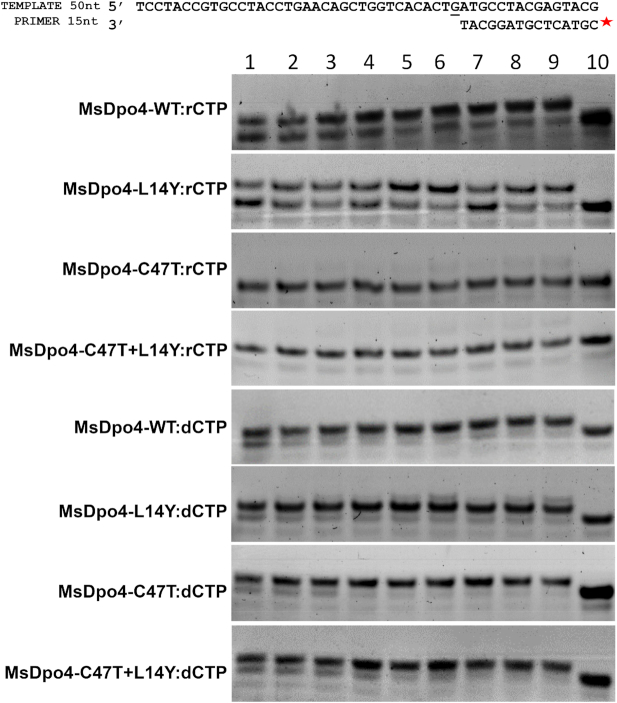
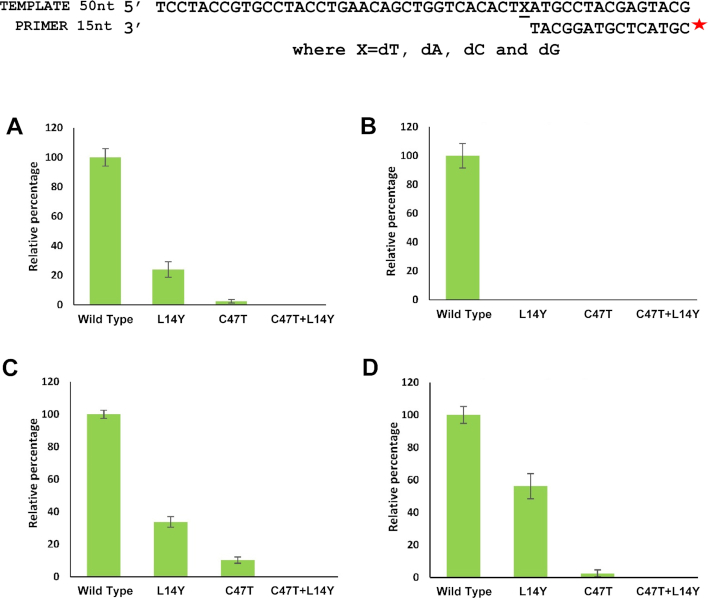
To test this hypothesis, two additional mutant versions of the MsDpo4 enzyme were prepared- one wherein the Cys47 residue was mutated to Thr, and another with the double mutation C47T+L14Y. It was observed that the MsDpo4-C47T enzyme exhibited substantially higher sugar selectivity than the wt- enzyme. The double mutant of MsDpo4 with both the steric filter and the polar residue showed almost no ribonucleotide incorporation activity at different concentrations of enzyme and nucleotide (Figure 5). The ability of the mutant versions of MsDpo4 to prevent ribonucleotide incorporation was true for all four rNTPs (Figure 6). There was no difference in the ability of the wt- and mutant enzymes to incorporate dNTPs (Supplementary Figure S2).
Figure 5.
The polar residue reduces rCTP incorporation by MsDpo4. The results of primer extension assays conducted using different concentrations of wt- MsDpo4 and variants (MsDpo4-L14Y, MsDpo4-C47T & MsDpo4-C47T+L14Y) at different concentrations of rCTP or dCTP are shown. The concentrations of enzyme used were 5 nM (Lanes 1, 2 and 3), 10 nM (Lanes 4, 5 and 6) and 15 nM (lanes 7, 8 and 9). The concentrations of incoming nucleotide (rCTP or dCTP) used were 1 μM (lanes 1, 4 and 7), 2 μM (lanes 2, 5, and 8) and 4 μM (lanes 3, 6 and 9). The assays show that mutation of Cys47 to Thr enhances the ability of MsDpo4 to exclude ribonucleotides. The MsDpo4-C47T and MsDpo4-C47T+L14Y proteins show higher selectivity for dCTP than MsDpo4-WT or MsDpo4-L14Y. These experiments shows that engineered residue (Thr47) enhances sugar selectivity.
Figure 6.
The presence of the polar residue results in drastic reduction in incorporation of all rNTPs. The level of rNTP incorporation by MsDpo4-WT and variants (MsDpo4-C47T, MsDpo4-L14Y & MsDpo4-L14Y+C47T) are displayed. (A) The level of incorporation of rATP opposite template dT by MsDpo4-WT and variants is displayed. (B) The figure exhibits the level of incorporation of UTP opposite template dA by MsDpo4-WT and variants. (C) The level of incorporation of rGTP opposite template dC by MsDpo4-WT and variants is displayed. (D) The ability of MsDpo4 and variants to incorporate rCTP opposite dG is displayed. For all graphs, the error bars indicate standard deviation (n = 3). For MsDpo4, the presence of the polar filter (C47T) results in a drastic reduction in rNTP incorporation and the presence of both filters in MsDpo4 gives rise to stringent sugar selectivity.
To ascertain if the engineered polar residue interacts with the incoming dNTP, the structure of MsDpo4-C47T in complex with DNA (with dG in the templating position) and dCTP was determined to a resolution of 1.8 Å (Supplementary Table S2F). The structure showed that, as in the case of PolIV, the Thr47 residue forms polar interactions with 3′-OH and the β-phosphate of the incoming dCTP (Figure 7A).
Figure 7.
A polar filter aids sugar selectivity. (A) The crystal structure of the MsDpo4-C47TDNA(dG):dCTP complex shows that the 3′-OH and the β-phosphate of the incoming nucleotide form interactions with the engineered T47 residue. (B) The surface of the fingers domain from the structure of the MsDpo4-C47TDNA(dG):dCTP complex along with the space-filling representation of the incoming dCTP is displayed. The figure shows that the dCTP molecule is accommodated in the catalytic site cavity without any steric clashes. (C) The surface of the fingers domain from the structure of the MsDpo4-C47TDNA(dG):dCTP complex along with the space-filling representation of incoming rCTP modeled in the catalytic site is displayed. The 2′-OH of the modeled rCTP forms steric clashes with the surface of the fingers domain (highlighted by an arrow). The engineered residue (Thr47) therefore represents a polar filter that aids sugar selectivity. (D) The PolIVDNA(dG):dCTP (Cyan) and MsDpo4-C47T+L14YDNA(dG):dCTP (peach) structures superimpose with an RMSD of 1.44 Å. The protein structure is shown in cartoon representation, DNA in ribbon form, and ions in the form of spheres. The incoming nucleotide and the steric plus polar filters are shown in stick representation. The dCTP molecules in the two structures are present in identical locations. (E) Thr43 forms interactions with the 3′–OH and β-phosphate of incoming dCTP in the PolIVDNA(dG):dCTP structure. (F) The engineered T47 residue in the MsDpo4-C47T+L14YDNA(dG):dCTP structure also forms interactions with the 3′-OH and β-phosphate of the incoming dCTP. Overall, the presence of the steric and polar filters in MsDpo4 lead to similar interactions as that seen in PolIV.
The structures of the ternary complexes of PolIV and MsDpo4-C47T show that the interaction with the polar residue holds the incoming dCTP closer to the polymerase surface (Figure 7B). As a result, it is not possible to reposition the incoming nucleotide, such that the 2′-OH of rNTP will not clash with the enzyme surface as was seen in the ternary complex of MsDpo4-L14Y with DNA and rCTP. Thus, the residue represented by Thr43 in PolIV constitutes a polar filter that mediates a closer association of the enzyme with the incoming nucleotide and thus prevents the repositioning required to accommodate rNTP in the active site (Figure 7C).
Further, the structure of MsDpo4-L14Y+C47T in complex with DNA (template dG) and incoming dCTP was determined to a resolution of 2.5 Å (Supplementary Table S2E) and compared with that of the similar complex of PolIV (PDB ID: 4IRC (40)). This comparison showed that the orientation and conformation of incoming nucleotide in the two structures were nearly identical (Figure 7D–F). The presence of the steric and polar filters in MsDpo4, therefore, led to a similar interactions as that for PolIV-WT and complete loss of ribonucleotide incorporation activity.
Presence of polar filter reduces cellular lethality due to ectopic expression of MsDpo4
It has been observed previously that, incorporation of rNTPs into genomic DNA, leads to cellular lethality in E. coli and higher organisms (7,10,13,33,51,52). To assess the importance of the polar filter in ribonucleotide incorporation, growth assays were performed using E. coli as a surrogate host. To enable the recruitment of the MsDpo4 protein to the replication fork in E. coli, the β-clamp binding motif of PolIV (50) (342QMERQLVLGL351) was added to C-terminus of MsDpo4. Wt- and mutant versions (C47T) of this construct were prepared.
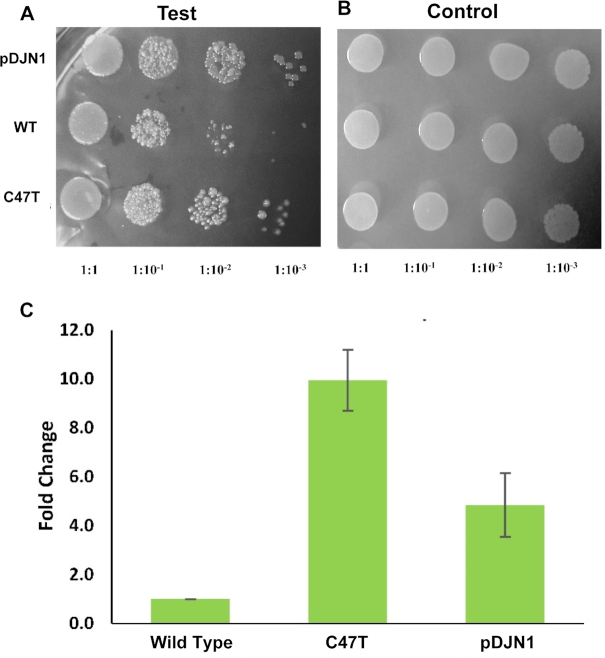
It was seen that when the expression of these constructs was induced in E. coli, fewer colonies were obtained in case of wt- protein as compared to the construct bearing the C47T mutation or empty vector (Figure 8A-B). In the quantitative plate assay, cells expressing the C47T mutant exhibited 10-fold higher survival with respect to wild type (Figure 8C). The only difference between the wt- and mutant constructs is the presence of the polar filter in the latter. Hence, the low level of survival observed in the case of wt- enzyme may be due to the higher incorporation of rNTP into the genome.
Figure 8.
Presence of the polar filter enhances bacterial survival. Spot Assay was conducted to assess the effect of MsDpo4-WT and MsDpo4-C47T on bacterial survival. The pDJN1 (vector alone), MsDpo4-WT and MsDpo4-C47T are shown in rows from top to bottom respectively. (A) Serial dilutions of cells expressing MsDpo4-WT and variants were spotted on LB ampicillin plates with 0.1mM IPTG and it was seen that bacteria expressing MsDpo4-C47T exhibit better survival than those expressing MsDpo4-WT.(B) Serial dilutions of cells expressing MsDpo4-WT and MsDpo4-C47T were spotted on LB ampicillin plates with 1% glucose and the displayed picture shows that equal amount of cells were spotted for all the samples tested. (C) Quantitative growth assay was done to assess the contribution of the polar filter towards bacterial survival. The effect of expression of wt- and mutant versions of MsDpo4 on the survival of the E. coli were measured and compared. The cells expressing MsDpo4-C47T exhibited 10-fold higher number of Colony Forming Units than cells expressing MsDpo4-WT. The error bars indicate standard deviation (n = 3).
PolIV lacking polar and steric filter can incorporate ribonucleotides
The variant of MsDpo4 with the polar filter showed substantially enhanced sugar selectivity. To observe the effect of removal of the polar filter in PolIV on ribonucleotide incorporation, the polar residue Thr43 was replaced with Cys (corresponding to Cys47 in MsDpo4) and Phe13 (steric filter) was replaced by Ala. The ability of the mutant proteins (PolIV-F13A and PolIV-F13A+T43C) to incorporate rCTP and dCTP was compared to that of PolIV-WT using primer extension assays. The level of rCTP incorporation by PolIV-F13A+T43C was 50-fold higher than the wt- enzyme; while that of the F13A mutant was ∼35-fold higher (Figure 9A). However, the mutations did not show a significant difference in the ability of PolIV to incorporate dCTP (Figure 9B). The structure of the complex of PolIV-F13A+T43C with DNA (template G) and incoming rCTP or dCTP was determined to a resolution of 2.77 and 2.44 Å, respectively (Supplementary Table S2G and H). The comparison of the two structures shows that the dCTP and rCTP superimpose well on each other (Figure 9C). These structures show that the loss of polar residue helps accommodation of rCTP in a catalytically competent conformation without any steric clash between the 2′–OH and the enzyme surface. Thus, the mutant version of PolIV without the polar and steric filters exhibits a considerably heightened ability to incorporate ribonucleotides as seen in the case of MsDpo4-WT. These experiments also show that the polar and aromatic filters act in tandem to achieve stringent dNTP selectivity in PolIV-WT.
Figure 9.
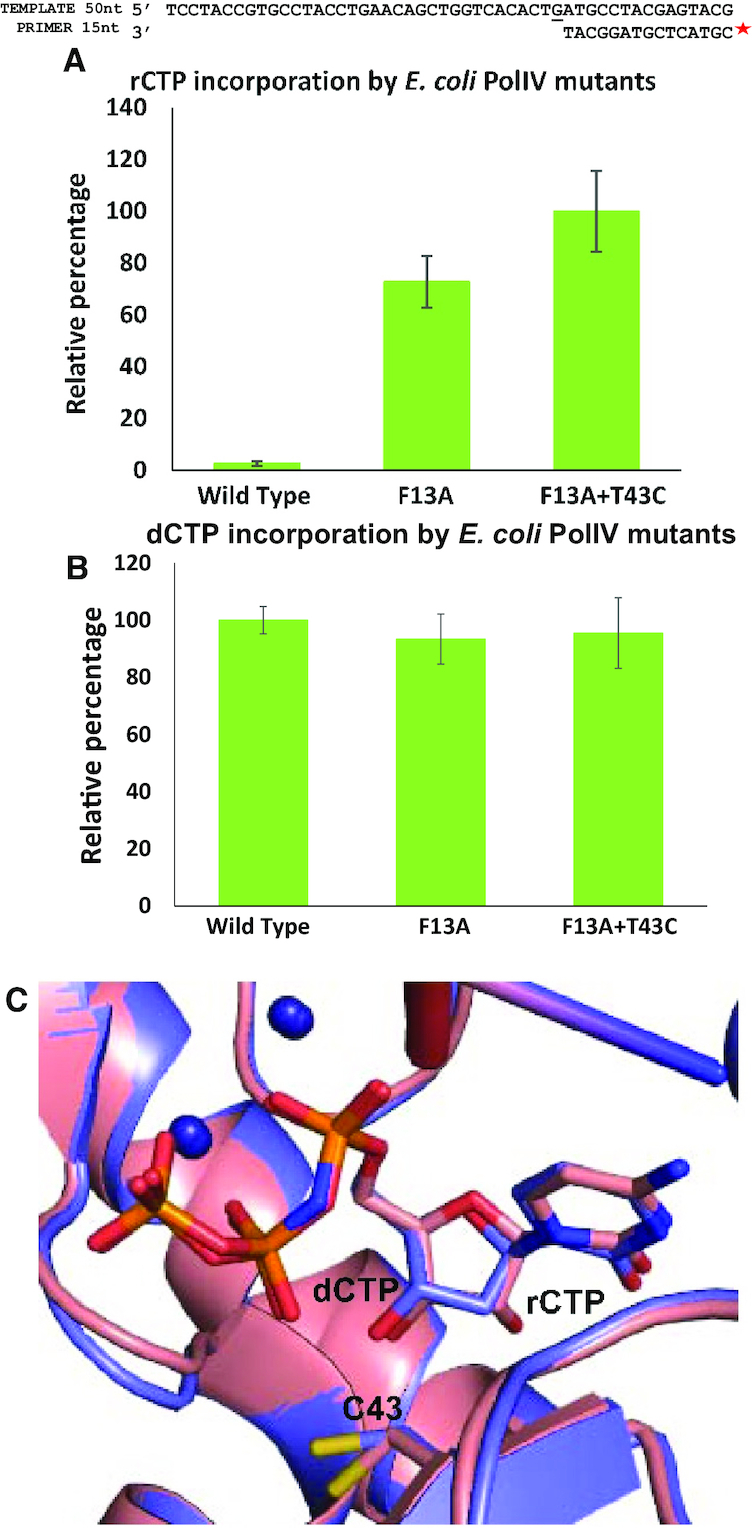
The absence of polar and aromatic steric filter in PolIV results in a drastic reduction in sugar selectivity. (A) Primer extension assays showed that the mutant version of PolIV without the steric and polar filter exhibits a 50-fold enhancement in rCTP incorporation. The error bars indicate standard deviation (n = 3). (B) Primer extension assays showed that the absence of the steric and polar filters did not affect the ability of PolIV to incorporate dCTP. The error bars indicate standard deviation (n = 3). (C) The PolIV-F13A+T43CDNA(dG):rCTP (peach) structure superimposed with PolIV-F13A+T43CDNA(dG):dCTP (violet) structure with an RMSD of 0.26 Å. The position and conformation of the dCTP and rCTP in the two structures are identical.
The polar filter is conserved in B and Y family of DNA Polymerases
The structures of ternary complexes of different dPols (enzyme:DNA:dNTP) available in PDB were analyzed to ascertain if the presence of the polar filter is a general feature of dPols. It was observed that the polar and aromatic residues were conserved in the majority of the dPols belonging to Y- and B- families.
For the Y- family, it was seen that the polar filter is represented by a Ser or Thr residue, and a Tyr or Phe residue is present as the steric filter (Supplementary Figure S3A) (53–56) with both filters present in the fingers domain. In the B-family, Asn (fingers domain) represents the polar filter, and Tyr (palm domain) is present as the steric filter (Supplementary Figure S3B) (29,42,57–60).
To validate the role of the polar filter in ribonucleotide exclusion by B-family dPols, DNA polymerase II (PolII) from E. coli was utilized as a model enzyme. In this enzyme, Tyr424 is present as the steric filter, and Asn497 is predicted to represent the polar filter (Supplementary Figure S3B3). Primer extension assays showed that, compared to PolII-WT, the mutant proteins PolII-Y424L and PolII-Y424L+N497A exhibit considerably enhanced ability to incorporate ribonucleotides (Figure 10A). The levels of rCTP incorporation by the single and double mutants were 3 and 4.5 times higher than the wild type. In comparison, the ability of the mutant proteins to incorporate dNTPs was unaffected (Figure 10B). The results show that polar residue Asn497 contributes significantly towards ribonucleotide exclusion by PolII. In summary, the studies on MsDpo4, PolIV, and PolII described here point to the presence of a polar filter in dPols that acts in tandem with the steric filter to exclude rNTPs during DNA synthesis.
Figure 10.
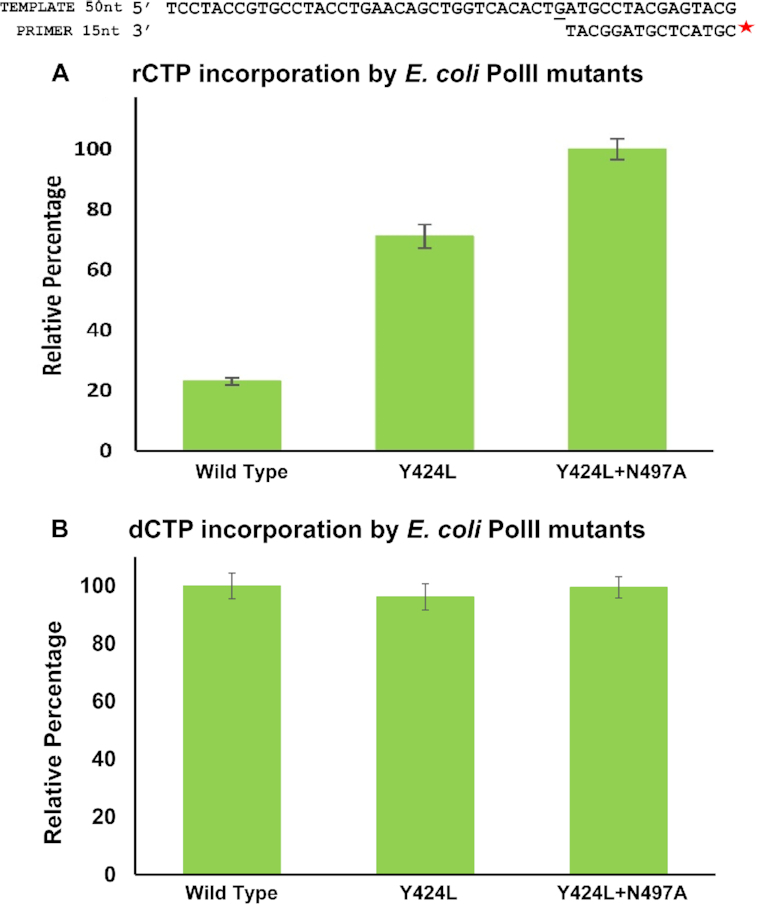
The contribution of polar and steric filters towards sugar selectivity in B-family DNA polymerases. Y424 and N497 represent the steric and polar filters in PolII (E. coli), a representative member of the B-family of dPols. (A) Primer extension assays were carried out to compare the ability of PolII-WT, PolII-Y424L and PolII-Y424L+N497A to incorporate rCTPs. These assays show that the polar filter contributes substantially towards ribonucleotide exclusion in this family. (B) Further, primer extension assays showed that the mutations Y424L and Y424L+N497A did not have a significant effect on the ability of PolII to incorporate dCTPs. For both the graphs, the error bars indicate standard deviation (n = 3).
DISCUSSION
Our results evince that, to achieve stringent sugar selectivity, dPols possess two filters. The first one, known as the steric filter, is composed of a bulky aliphatic or aromatic side chain that is present underneath the ribose sugar and will clash with the 2′-OH of the rNTP molecule to reduce the chances of it occupying the active site in a conformation compatible with productive catalysis. The present study shows the existence of a second polar filter, which interacts with the 3′-OH and triphosphate moiety of the incoming nucleotide and orients it closer to the surface of the fingers domain. As a result, the presence of the polar filter ensures that the 2′-OH of an rNTP molecule will clash with the surface of the fingers domain and consequently there is a limited possibility of the rNTP binding in a catalytically competent conformation. The steric filter is present below the sugar, and the polar filter is present close to the 3′-OH and the triphosphate moiety of the incoming nucleotide. Hence, the filters fall on nearly perpendicular planes to each other, and synergize to ensure high sugar selectivity.
The presence of the steric and polar filters that adopt chemically distinct strategies to prevent ribonucleotide incorporation appears to be a conserved feature of the Y- and B-family of dPols. Polη is a Y-family dPol and exhibits a heightened ability to incorporate ribonucleotides (61). It is believed that this is because it has an extraordinarily spacious active site (30). However, the ability of the enzyme to incorporate ribonucleotides may also be due to the fact that it does not possess a polar filter. An alignment of hPolη with PolIV (RMSD = 2.36 Å) shows that an Ala residue (Ala49) is present at the position equivalent to the Thr43 residue (Supplementary Figure S4). The alignment of Polη incorporating rCTP and dCTP (RMSD = 0.19 Å) shows that the rCTP is repositioned away from the aromatic filter (Supplementary Figure S4D and E) (30). The presence of a polar filter that interacts with the rNTP will prevent such a repositioning. It has been reported before Polη has a heightened ability to incorporate ribonucleotides, and these observations are in line with the present study (61).
The strategy utilized by Y- and B- family dPols to exclude ribonucleotides is distinct from that observed in the case of A, X and C- family members. In A-family, it is suggested that dual steric filters exist in the form of Glu residue below the sugar of the incoming nucleotide and an aromatic residue lining the fingers domain (19,62–64). The two steric filters are positioned such that the 2′-OH of an rNTP molecule will clash with these residues, thus preventing its incorporation (65,66). Based on the structures of DNA polymerase III (PolIII) from E. coli and Geobacillus kaustophilus, C-family dPols may utilize a histidine residue to perform the function of the steric filter. It was seen that the side chains of His760 and His 1275 in PolIII from E. coli and G. kaustophilus, respectively (67,68) stack against the ribose sugar of the incoming nucleotide. A number of X-family members exhibit lower selectivity for dNTPs and even though, some members show the presence of an aromatic residue at the position equivalent to the steric filter, the structures show that it does not stack with the ribose sugar (18,69). Instead, it appears that the polypeptide backbone in the region which is present close to the C2′ atom of the sugar acts as a steric filter to provide some level of sugar selectivity (18). For the PrimPol family, it is suggested that the main chain of residues 289–291 in human PrimPol may act as a steric filter to prevent ribonucleotide incorporation (32). Also, Y100 present in the fingers domain, which does not stack with the sugar of the incoming nucleotide is known to play an important role in nucleotide selectivity (70,71). The members of the RT family of dPols do possess the steric filter to reduce ribonucleotide incorporation (72–74). The D-family of dPols represents the least studied family of dPols and strategy utilized by these enzymes to prevent ribonucleotide incorporation is unclear (75).
B-family dPols are highly accurate and responsible for DNA synthesis during replication and repair, especially in eukaryotes (76–80). In humans, members of the B-family include Polα, Polδ, Polϵ, and Polζ. A thorough examination of different databases such as NIH-NCI GDC data portal, NCBI ClinVar and BIH-DB SNP Short Genetic Variation, revealed a correlation between mutations at the polar filter site and disease in human dPols δ and ζ (Supplementary Table S1). Thermostable members of B-family dPols have been used as enzymes for PCR-based applications (81,82). The results presented in the present study may be useful in the development of engineered enzymes that can directly amplify RNA without the need for cDNA formation. Such engineered dPols may be a valuable tool for a number of applications in biotechnology (83).
DATA AVAILABILITY
The structure factors and the refined co-ordinates have been deposited in the PDB with the following codes: 6JUL MsDpo4-WTDNA(dG):dCTP, 6JUM MsDpo4-C47TDNA(dG):dCTP, 6JUN MsDpo4-C47T+L14YDNA(dG):dCTP, 6JUO MsDpo4-L14YDNA(dG):dCTP, 6JUP PolIV-F13A+T43CDNA(dG):dCTP, 6JUQ PolIV-F13A+T43CDNA(dG):rCTP, 6JUR MsDpo4-L14YDNA(dG):rCTP and 6JUS MsDpo4-WTDNA(dG):rCTP.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the X-ray diffraction facilities located at the Regional Centre for Biotechnology, Faridabad and NII, New Delhi. We thank Dr Hassan Belrhali and Dr Babu Manjashetty (BM14 beamline, ESRF), Dr Nicolas Foos (ID 30B Beamline, ESRF) and Dr Danielle de Sanctis (ID29 beamline, ESRF) for help with X-ray diffraction data collection. We thank Dr Babu Manjashetty and Dr Annie Heroux at for help with data collection at the XRD2 beamline of the Elettra Sincrotrone in Trieste, Italy. We thank Prof. S. Ramaswamy (inStem, Banglore) for critically reading the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Science & Engineering Research Board of the Department of Science & Technology, Government of India [EMR/2016/000986]. Data collection at the BM14 beamline of ESRF (Grenoble, France) was supported by the BM14 project—a collaboration between DBT, EMBL and ESRF; Data collection at ID29 and ID30B was facilitated by the ESRF Access Program of RCB which is supported by Department of Biotechnology, Government of India [BT/INF/22/SP22660/2017]. Funding for open access charge: Intramural Funding from Regional Centre for Biotechnology. Access to the XRD2 beamline at Elettra was made possible through grant-in-aid from the Department of Science and Technology, India [DSTO-1668].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jain R., Aggarwal A.K., Rechkoblit O.. Eukaryotic DNA polymerases. Curr. Opin. Struct. Biol. 2018; 53:77–87. [DOI] [PubMed] [Google Scholar]
- 2. Steitz T.A. A mechanism for all polymerases. Nature. 1998; 391:231–232. [DOI] [PubMed] [Google Scholar]
- 3. Kottur J., Nair D.T.. Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction. Nucleic Acids Res. 2018; 46:5875–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yao N.Y., Schroeder J.W., Yurieva O., Simmons L.A., O'Donnell M.E.. Cost of rNTP/dNTP pool imbalance at the replication fork. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:12942–12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nick McElhinny S.A., Watts B.E., Kumar D., Watt D.L., Lundstrom E.-B., Burgers P.M.J., Johansson E., Chabes A., Kunkel T.A.. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schroeder J.W., Randall J.R., Hirst W.G., O’Donnell M.E., Simmons L.A.. Mutagenic cost of ribonucleotides in bacterial DNA. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:11733–11738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reijns M.A., Rabe B., Rigby R.E., Mill P., Astell K.R., Lettice L.A., Boyle S., Letich A., Keighren M., Kilanowski F. et al.. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012; 149:1008–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mcelhinny S.A.N., Kumar D., Clark A.B., Watt D.L., Watts B.E., Lundström E., Johansson E., Chabes A., Kunkel T.A.. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biolology. 2010; 6:774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Joyce C.M. Choosing the right sugar: How polymerases select a nucleotide substrate. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caldecott K.W. Ribose-An internal threat to DNA. Science. 2014; 343:260–261. [DOI] [PubMed] [Google Scholar]
- 11. Dahl J.M., Wang H., Lazaro J.M., Salas M., Lieberman K.R.. Kinetic mechanisms governing stable ribonucleotide incorporation in individual DNA polymerase complexes. Biochemistry. 2014; 53:8061–8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kennedy E.M., Amie S.M., Bambara R.A., Kim B.. Frequent incorporation of ribonucleotides during HIV-1 reverse transcription and their attenuated repair in macrophages. J. Biol. Chem. 2012; 287:14280–14288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Potenski C.J., Klein H.L.. How the misincorporation of ribonucleotides into genomic DNA can be both harmful and helpful to cells. Nucleic Acids Res. 2014; 42:10226–10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Williams J.S., Lujan S.A., Kunkel T.A.. Processing ribonucleotides incorporated during eukaryotic DNA replication. Nat. Rev. Mol. cell Biol. 2016; 17:351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Watt D.L., Johansson E., Burgers P.M., Kunkel T.A.. Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair (Amst). 2011; 10:897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Egli M., Usman N., Rich A.. Conformational influence of the ribose 2 ′ -Hydroxyl group: Crystal structures of DNA-RNA chimeric duplexes. Biochemistry. 1993; 32:3221–3237. [PubMed] [Google Scholar]
- 17. Mcginness K.E., Joyce G.F.. Substitution of ribonucleotides in the T7 RNA polymerase promoter element *. J. Biol. Chem. 2002; 277:2987–2991. [DOI] [PubMed] [Google Scholar]
- 18. Brown J.A., Fiala K.A., Fowler J.D., Sherrer S.M., Newmister S.A., Duym W.W., Suo Z.. A novel mechanism of sugar selection utilized by a human X-family DNA polymerase. J. Mol. Biol. 2010; 395:282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Astatke M., Ng K., Grindley N.D.F., Joyce C.M.. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:3402–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vaisman A., Woodgate R.. Ribonucleotide discrimination by translesion synthesis DNA polymerases. Crit. Rev. Biochem. Mol. Biol. 2018; 53:382–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brown J.A., Suo Z.. Unlocking the sugar ‘steric gate’ of DNA polymerases. Biochemistry. 2011; 50:1135–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cavanaugh N.A., Beard W.A., Wilson S.H.. DNA polymerase B ribonucleotide discrimination insertion, misinsertion, extension, and coding. J. Biol. Chem. 2010; 285:24457–24465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang G., Franklin M., Li J., Lin T., Konigsberg W.. A Conserved tyr residue is required for sugar selectivity in a pol alpha DNA polymerase. Biochemistry. 2002; 41:10256–10261. [DOI] [PubMed] [Google Scholar]
- 24. Sherrer S.M., Beyer D.C., Xia C.X., Fowler J.D., Suo Z.. Kinetic basis of sugar selection by a Y-Family DNA polymerase from sulfolobus solfataricus P2 †. Biochemistry. 2010; 49:10179–10186. [DOI] [PubMed] [Google Scholar]
- 25. Ogawa M., Tosaka A., Ito Y., Yoshida S., Suzuki M.. Enhanced ribonucleotide incorporation by an O-helix mutant of Thermus aquaticus DNA polymerase I. DNA Repair (Amst). 2000; 485:197–207. [DOI] [PubMed] [Google Scholar]
- 26. Randrianjatovo-gbalou I., Rosario S., Sismeiro O., Varet H., Legendre R., Coppée J., Huteau V., Pochet S., Delarue M.. Enzymatic synthesis of random sequences of RNA and RNA analogues by DNA polymerase theta mutants for the generation of aptamer libraries. Nucleic Acids Res. 2018; 46:6271–6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen T., Hongdilokkul N., Liu Z., Adhikary R., Tsuen S.S., Romesberg F.E.. Evolution of thermophilic DNA polymerases for the recognition and amplification of C2′-modified DNA. Nat. Chem. 2016; 8:556–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ordonez H., Uson M.L., Shuman S.. Characterization of three mycobacterial DinB (DNA polymerase IV) paralogs highlights DinB2 as naturally adept at ribonucleotide incorporation. Nucleic Acids Res. 2014; 42:11056–11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Swan M.K., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K.. Structural basis of high fidelity DNA synthesis by yeast DNA polymerase delta. Nat. Struct. Mol. Biol. 2009; 16:979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Su Y., Egli M., Guengerich F.P.. Mechanism of ribonucleotide incorporation by human DNA polymerase η. J. Biol. Chem. 2016; 291:3747–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nevin P., Engen J.R., Beuning P.J.. Steric gate residues of Y-family DNA polymerases DinB and pol kappa are crucial for dNTP-induced conformational change. DNA Repair (Amst). 2015; 344:1173–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rechkoblit O., Gupta Y.K., Malik R., Rajashankar K.R., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K.. Structure and mechanism of human PrimPol, a DNA polymerase with primase activity. Sci. Adv. 2016; 2:e1601317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crespan E., Furrer A., Rösinger M., Bertoletti F., Mentegari E., Chiapparini G., Imhof R., Ziegler N., Sturla S.J., Hübscher U. et al.. Impact of ribonucleotide incorporation by DNA polymerases β and lambda on oxidative base excision repair. Nat. Commun. 2016; 7:10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bonnin A., Lázaro J.M., Blanco L., Salas M.. A single tyrosine prevents insertion of ribonucleotides in the eukaryotic-type φ29 DNA polymerase. J. Mol. Biol. 1999; 290:241–251. [DOI] [PubMed] [Google Scholar]
- 35. Moon A.F., Pryor J.M., Ramsden D.A., Kunkel T.A., Bebenek K., Pedersen L.C.. Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu. Nucleic Acids Res. 2017; 45:9138–9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Delucia A.M., Grindley N.D.F., Joyce C.M.. An error-prone family Y DNA polymerase (DinB homolog from Sulfolobus solfataricus) uses a ‘steric gate' residue for discrimination against ribonucleotides. Nucleic Acids Res. 2003; 31:4129–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharma A., Subramanian V., Nair D.T.. The PAD region in the mycobacterial DinB homologue MsPolIV exhibits positional heterogeneity. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012; 68:960–967. [DOI] [PubMed] [Google Scholar]
- 38. Sharma A., Nair D.T.. Cloning, expression, purification, crystallization and preliminary crystallographic analysis of MsDpo4: a Y-family DNA polymerase from Mycobacterium smegmatis. Acta Crystallogr. Sect. F. 2011; 67:812–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ordonez H., Shuman S.. Mycobacterium smegmatis DinB2 misincorporates deoxyribonucleotides and ribonucleotides during templated synthesis and lesion bypass. Nucleic Acids Res. 2014; 42:12722–12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sharma A., Kottur J., Narayanan N., Nair D.T.. A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli. Nucleic Acids Res. 2013; 41:5104–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jain D., Nair D.T.. Spacing between core recognition motifs determines relative orientation of AraR monomers on bipartite operators. Nucleic Acids Res. 2013; 41:639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang F., Yang W.. Structural insight into translesion synthesis By DNA Pol II. Cell. 2009; 139:1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kottur J., Sharma A., Gore K.R., Narayanan N., Samanta B., Pradeepkumar P.I., Nair D.T.. Unique structural features in DNA polymerase IV enable efficient bypass of the N2 adduct induced by the Nitrofurazone Antibiotic. Structure. 2015; 23:56–67. [DOI] [PubMed] [Google Scholar]
- 44. Battye T.G.G., Johnson O., Powell H.R., Leslie A.G.W.. iMOSFLM: a new graphical interface for diffraction- image processing with MOSFLM. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011; D67:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Adams P.D., Pavel V., Chen V.B., Ian W., Echols N., Moriarty N.W., Read R.J., Richardson D.C., Jane S., Thomas C.. PHENIX: a comprehensive Python-based system for macromolecular structure. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010; D66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004; D60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 47. Mccoy A.J., Grosse-kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Winn M.D., Charles C., Cowtan K.D., Dodson E.J., Leslie A.G.W., Mccoy A., Stuart J., Garib N., Powell H.R., Randy J.. Overview of the CCP 4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011; 4449:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Murshudov G.N., Nicholls R.A.. REFMAC 5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011; D67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bunting K.A., Roe S.M., Pearl L.H.. Structural basis for recruitment of translesion DNA polymerase Pol IV / DinB to the b -clamp. EMBO J. 2003; 22:5883–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McDonald J.P., Vaisman A., Kuban W., Goodman M.F., Woodgate R.. Mechanisms employed by escherichia coli to prevent ribonucleotide incorporation into genomic DNA by Pol V. PLoS Genet. 2012; 8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moss C.F., Rosa I.D., Hunt L.E., Yasukawa T., Young R., Jones A.W.E., Reddy K., Desai R., Virtue S., Elgar G. et al.. Aberrant ribonucleotide incorporation and multiple deletions in mitochondrial DNA of the murine MPV17 disease model. Nucleic Acids Res. 2017; 45:12808–12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lone S., Townson S.A., Uljon S.N., Johnson R.E., Brahma A., Nair D.T., Prakash S., Prakash L., Aggarwal A.K.. Human DNA polymerase κ encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell. 2007; 25:601–614. [DOI] [PubMed] [Google Scholar]
- 54. Nair D.T., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K.. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005; 309:2219–2222. [DOI] [PubMed] [Google Scholar]
- 55. Nair D.T., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K.. Hoogsteen base pair formation promotes synthesis opposite the 1, N6 -ethenodeoxyadenosine lesion by human DNA polymerase iota. Nat. Struct. Mol. Biol. 2006; 13:619–625. [DOI] [PubMed] [Google Scholar]
- 56. Gaur V., Vyas R., Fowler J.D., Efthimiopoulos G., Feng J.Y., Suo Z.. Structural and kinetic insights into binding and incorporation of L-nucleotide analogs by a Y-family DNA polymerase. Nucleic Acids Res. 2014; 42:9984–9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Berman A.J., Kamtekar S., Goodman J.L., Lázaro J.M., De Vega M., Blanco L., Salas M., Steitz T.A.. Structures of phi29 DNA polymerase complexed with substrate: The mechanism of translocation in B-family polymerases. EMBO J. 2007; 26:3494–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xia S., Wang J., Konigsberg W.H.. DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase. J. Am. Chem. Soc. 2013; 135:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hogg M., Osterman P., Bylund G.O., Ganai R.A., Lundström E.B., Sauer-Eriksson A.E., Johansson E.. Structural basis for processive DNA synthesis by yeast DNA polymerase ϵ. Nat. Struct. Mol. Biol. 2014; 21:49–55. [DOI] [PubMed] [Google Scholar]
- 60. Baranovskiy A.G., Duong V.N., Babayeva N.D., Zhang Y., Pavlov Y.I., Anderson K.S., Tahirov T.H.. Activity and fidelity of human DNA polymerase α depend on primer structure. J. Biol. Chem. 2018; 293:6824–6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mentegari E., Crespan E., Bavagnoli L., Kissova M., Bertoletti F., Sabbioneda S., Imhof R., Sturla S.J., Nilforoushan A., Hübscher U. et al.. Ribonucleotide incorporation by human DNA polymerase eta impacts translesion synthesis and RNase H2 activity. Nucleic Acids Res. 2017; 45:2600–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Joyce C.M., Potapova O., Delucia A.M., Huang X., Basu V.P., Grindley N.D.F.. Fingers-Closing and other rapid conformational changes in DNA Polymerase I (Klenow Fragment) and their role in nucleotide selectivity †. Biochemistry. 2008; 47:6103–6116. [DOI] [PubMed] [Google Scholar]
- 63. Astatke M., Grindley N.D.F., Joyce C.M.. How E. coli DNA polymerase I (Klenow fragment) distinguishes between Deoxy- and dideoxynucleotides. J. Mol. Biol. 1998; 278:147–165. [DOI] [PubMed] [Google Scholar]
- 64. Patel P.H., Suzuki M., Adman E., Shinkai A., Loeb L.A.. Prokaryotic DNA polymerase I: Evolution, structure, and “base flipping” mechanism for nucleotide selection. J. Mol. Biol. 2001; 308:823–837. [DOI] [PubMed] [Google Scholar]
- 65. Kropp H.M., Diederichs K., Marx A.. Structural biology the structure of an archaeal B-Family DNA polymerase in complex with a chemically modified nucleotide. Angew. Chem. 2019; 58:5457–5461. [DOI] [PubMed] [Google Scholar]
- 66. Zahn K.E., Averill A.M., Aller P., Wood R.D., Doublié S.. Human DNA polymerase θ grasps the primer terminus to mediate DNA repair. Nat. Struct. Mol. Biol. 2015; 22:304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fernandez-leiro R., Conrad J., Scheres S.H.W., Lamers M.H.. cryo-EM structures of the E. coli replicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease and tau. Elife. 2015; 4:e11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Evans R.J., Davies D.R., Bullard J.M., Christensen J., Green L.S., Guiles J.W., Pata J.D., Ribble W.K., Janjic N., Jarvis T.C.. Structure of PolC reveals unique DNA binding and fidelity determinants. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20695–20700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gosavi R.A., Moon A.F., Kunkel T.A., Pedersen L.C., Bebenek K.. The catalytic cycle for ribonucleotide incorporation by human DNA Pol Lambda. Nucleic Acids Res. 2012; 40:7518–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Díaz-talavera A., Calvo P.A., González-acosta D., Díaz M., Sastre-moreno G., Blanco-franco L., Guerra S., Martínez-jiménez M.I., Méndez J., Blanco L.. A cancer-associated point mutation disables the steric gate of human PrimPol. Sci. Rep. 2019; 9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bianchi J., Rudd S.G., Jozwiakowski S.K., Bailey L.J., Soura V., Taylor E., Stevanovic I., Green A.J., Stracker T.H., Lindsay H.D. et al.. PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol. Cell. 2013; 52:566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gao G., Orlova M., Georgadis M.M., Hendrickson W.A., Goff S.P.. Conferring RNA polymerase Activity to a DNA polymerase: A single residue in reverse transcriptase controls substrate selection. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cases-gonza C.E., Mene L.. Coupling ribose selection to fidelity of DNA synthesis. J. Biol. Chem. 2000; 275:19759–19767. [DOI] [PubMed] [Google Scholar]
- 74. Beck J., Vogel M., Nassal M.. dNTP versus NTP discrimination by phenylalanine 451 in duck hepatitis B virus P protein indicates a common structure of the dNTP-binding pocket with other reverse transcriptases. Nucleic Acids Res. 2002; 30:1679–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sauguet L., Raia P., Henneke G., Delarue M.. Shared active site architecture between archaeal PolD and multi-subunit RNA polymerases revealed by X-ray crystallography. Nat. Commun. 2016; 7:12227–12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gómez-Llorente Y., Malik R., Jain R., Choudhury J.R., Johnson R.E., Prakash L., Prakash S., Ubarretxena-belandia I., Aggarwal A.K.. The architecture of yeast DNA polymerase ζ. Cell Rep. 2013; 5:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Coloma J., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K.. Human DNA polymerase α in binary complex with a DNA: DNA template - primer. Sci. Rep.-Nat. 2016; 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Johnson R.E., Klassen R., Prakash L., Prakash S.. A Major Role of DNA polymerase Delta in replicaton of Both the laeading and Lagging DNA strands. Mol. Cell. 2015; 59:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jain R., Rajashankar K.R., Buku A., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K.. Crystal structure of yeast DNA polymerase epsilon catalytic domain. PLoS One. 2014; 9:e94835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jozwiakowski S.K., Keith B.J., Gilroy L., Doherty A.J., Connolly B.A.. An archaeal family-B DNA polymerase variant able to replicate past DNA damage: occurrence of replicative and translesion synthesis polymerases within the B family. Nucleic Acids Res. 2014; 42:17–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Biles B.D., Connolly B.A.. Low-fidelity Pyrococcus furiosus DNA polymerase mutants useful in error-prone PCR. Nucleic Acids Res. 2004; 32:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pavlov A.R., Pavlova N.V., Kozyavkin S.A., Slesarev A.I.. Recent developments in the optimization of thermostable DNA polymerases for efficient applications. Trends Biotechnol. 2004; 22:253–260. [DOI] [PubMed] [Google Scholar]
- 83. Blatter N., Bergen K., Nolte O., Welte W., Diederichs K., Mayer J., Wieland M., Marx A.. Structure and function of an RNA-reading thermostable dna polymerase. Angew. Chem. 2013; 52:11935–11939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure factors and the refined co-ordinates have been deposited in the PDB with the following codes: 6JUL MsDpo4-WTDNA(dG):dCTP, 6JUM MsDpo4-C47TDNA(dG):dCTP, 6JUN MsDpo4-C47T+L14YDNA(dG):dCTP, 6JUO MsDpo4-L14YDNA(dG):dCTP, 6JUP PolIV-F13A+T43CDNA(dG):dCTP, 6JUQ PolIV-F13A+T43CDNA(dG):rCTP, 6JUR MsDpo4-L14YDNA(dG):rCTP and 6JUS MsDpo4-WTDNA(dG):rCTP.