Abstract
Single-cell patterning technology has revealed significant contributions of single cells to conduct basic and applied biological studies in vitro such as the understanding of basic cell functions, neuronal network formation, and drug screening. Unlike traditional population-based cell patterning approaches, single-cell patterning is an effective technology of fully understanding cell heterogeneity by precisely controlling the positions of individual cells. Therefore, much attention is currently being paid to this technology, leading to the development of various micro-nanofabrication methodologies that have been applied to locate cells at the single-cell level. In recent years, various methods have been continuously improved and innovated on the basis of existing ones, overcoming the deficiencies and promoting the progress in biomedicine. In particular, microfluidics with the advantages of high throughput, small sample volume, and the ability to combine with other technologies has a wide range of applications in single-cell analysis. Here, we present an overview of the recent advances in single-cell patterning technology, with a special focus on current physical and physicochemical methods including stencil patterning, trap- and droplet-based microfluidics, and chemical modification on surfaces via photolithography, microcontact printing, and scanning probe lithography. Meanwhile, the methods applied to biological studies and the development trends of single-cell patterning technology in biological applications are also described.
I. INTRODUCTION
The ability of manipulating and selectively localizing cells into patterns or different microenvironments is critical for the studies of cell behaviors, such as cell migration,1 tissue engineering,2 coculture assay,3 drug screening,4 and cell signaling.5 Conventionally, an experimental result is actually the average of the cell population, which ignores the diversity of phenotypes in the population. In this regard, single-cell patterning technology allows more in-depth studies of cell fundamental characteristics since it has become an ideal tool to research comprehensive heterogeneity from the cellular behavior to molecular expression. Meanwhile, this technology enables the investigation of high-throughput detection. Compared with population-based cell patterning, single-cell patterning is more difficult to be implemented since the cell size is on the micrometer scale. With the development of micro-nanofabrication technology over the last decade, a wide range of methods has been developed in the biological field for achieving efficient single-cell patterning. Considering that many methods for single-cell analysis have been developed in recent years, this review mainly focuses on the developments and applications of single-cell patterning technology.
The fabrication technology of micropatterns for single-cell patterning can be categorized into two types of approaches: physical and physicochemical patterning, each with its own advantages and disadvantages and main applications, as summarized in Table I. Patterning single cells physically can be achieved through physical structures of optimized sizes and shapes that are capable of confining cells, such as the stencil method, or through external forces to manipulate cells, including microrobots, optical and dielectrophoretic traps, acoustic force patterning, and magnetic cell manipulation.6 However, simultaneous implementation of high precision and high throughput is a challenging issue. In general, reaching the accuracy at the single-cell level is difficult for high-throughput methods, while a complex experimental facility is required in high-precision methods. In order to deal with the challenge, single-cell patterning technology has been continuously improved and updated. Over recent years, microfluidic systems are becoming popular in single-cell manipulation. They enable reverting the microenvironment of cell survival in vivo due to the size compatibility between the microchannel and the cell. Additionally, the systems have high accuracy since the working environment is a solution with a volume ranging from picoliters to nanoliters. These advantages make microfluidics a powerful tool for analyzing cellular molecules. Therefore, physical methods, such as the trap- and droplet-based cell patterning, are often combined with microfluidic devices. On the other hand, physicochemical patterning single-cell methods utilize the micro-nanomanufacturing technology that can produce chemical arrays that promote cell adhesion on the substrate and then form the cell patterning according to the corresponding chemical patterns. As one of the commonly used biomolecules, extracellular matrix (ECM) ligands can specifically bind to cell adhesion receptors to fix cells on the surface. Nonbiomolecule polymers are also used to fabricate different substrates, which can indirectly affect cell behaviors through external stimuli, such as heat. Among numerous methods, lithography is common for the fabrication of pattern arrays. It can be divided into two types: mask-based lithography, such as photolithography and soft lithography, and maskless lithography, such as scanning probe lithography. These methods allow high-resolution patterning of arbitrary shapes with feature sizes down to nanometers.
TABLE I.
Comparison of various single-cell patterning methods.
| Methods | Main applications | Advantages | Disadvantages |
|---|---|---|---|
| Stencil patterning (physical) | Single-cell analysis | Simple and multipurpose | Complicated fabrication process |
| Hydrodynamic trap (physical) | Cell culture on chip; cell manipulation; single-cell analysis |
Multifunction in one device | Complicated fabrication process |
| Optical trap (physical) | Cell manipulation; single-cell analysis |
Applicable in many fields of study | Expensive optical system |
| Dielectrophoretic trap (physical) | Cell manipulation; single-cell analysis |
Easy selection of target cells via alternating frequency of AC | Heating problem during long-term manipulation |
| Magnetic trap (physical) | Cell manipulation | Efficient trapping of labeled cells | Require antibodies or primers for magnetic labels |
| Acoustic trap (physical) | Cell manipulation | Good for cell positioning | Limited acoustic-field pattern |
| Droplet-based microfluidics (physical) | Cell manipulation | High-throughput screening for specific single cells | Challenge to encapsulate single cells in each droplet |
| Photolithography (physicochemical) | Single-cell analysis | Precise control of the pattern shape and size | Challenge for applications on nonplanar substrates |
| Microcontact printing (physicochemical) | Single-cell analysis | Applicable to curved surfaces | Complicated fabrication process |
| Dip-pen nanolithography (physicochemical) | Single-cell analysis | Create precise geometries and patterns; direct-write capability |
Substantial time and effort in the optimization process; fragile and expensive pen arrays; labor intensive alignment process |
| Polymer pen lithography (physicochemical) | Single-cell analysis | Large scale and parallel patterns | Nonuniform diffusion of inks |
| Dip-pen nanodisplacement lithography (physicochemical) | Single-cell analysis | High resolution; solution-free and diffusion-limited tool | Complicated fabrication process |
Herein, the physical and physicochemical methods for patterning mammalian cells at the single-cell level are described. In this review, we briefly introduce the basic principles of these methods. In particular, we focus on the recent advances in the single-cell patterning methods and their applications in single cell assays.
II. PHYSICAL AND PHYSICOCHEMICAL PATTERNING METHODS
A. Stencil patterning
Stencil patterning has the advantage over conventional methods for making high-throughput single-cell analysis easy.7 The main strategy of stencil patterning is relying on physical barriers, which reuses stencils and provides prospects for large-scale applications. When using this method, the stencil is fixed to the surface of the substrate before cell seeding. In the through-hole areas, where the diameter of the hole is slightly larger than the cell size or approximately equal, cells can settle through the hole and be patterned onto the accessible regions of the surface at the single-cell level. In other areas that serve as physical barriers, cells are blocked by the stencil film. When the cells adhere to the surface, a high-throughput single cell array is formed after removing the stencil.
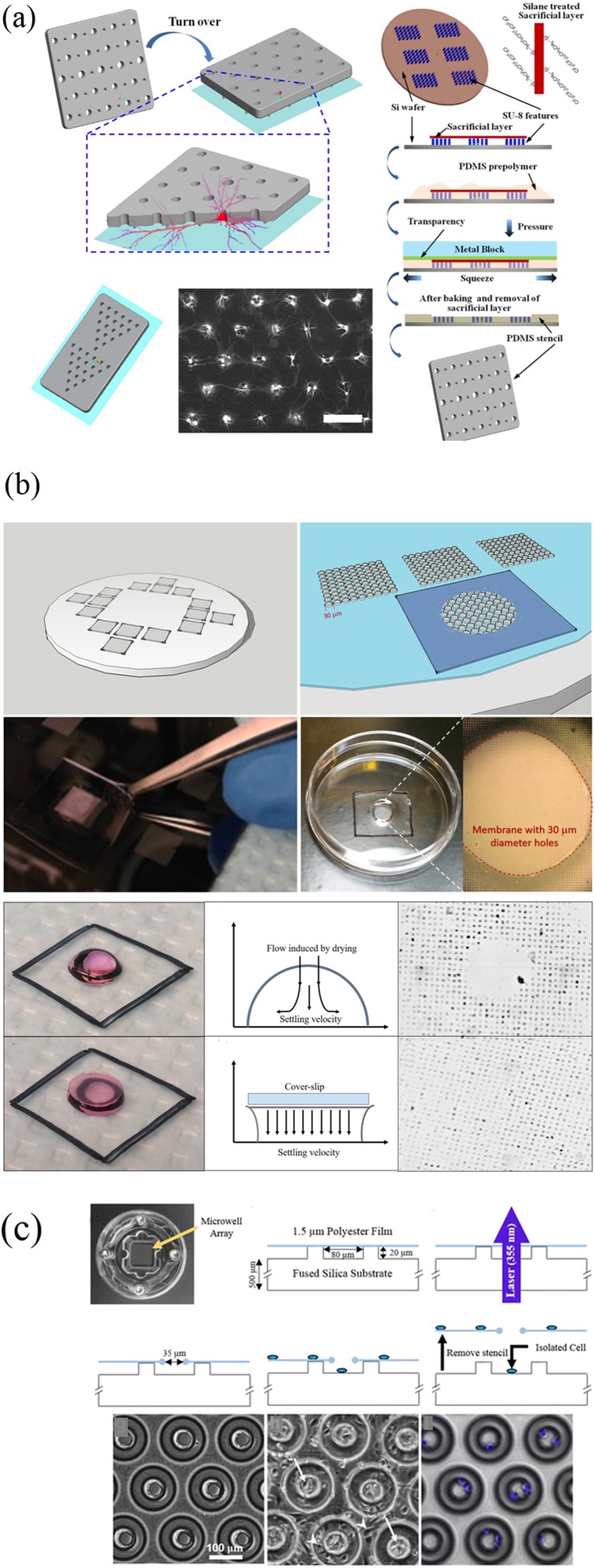
This methodology was first presented by Carter as early as 1967, and the material of the stencil was nickel at that time.8 In the following studies, various materials such as stainless steel,9 Norland Optical Adhesive 73,10 and parylene11,12 have been used. Hard materials such as Si are seldom used because of the complexity in manufacturing processes and the poor surface seal. To address this problem, Wu et al. prepared a silicon stencil by dry etching. A polydimethylsiloxane (PDMS) frame was made to keep the stencil tightly attached to the substrate.13 Up to date, PDMS is the commonly used material for stencil fabrication, which is characterized by soft, cheap, transparent, bendable nature, and fitting for various surfaces, even curved substrates.14 Li et al.15 fabricated a novel PDMS stencil of which a through hole was surrounded by several micropillars of 3-μm height. The stencil can maintain normal morphology of neurons, providing a space between the glass substrate and the stencil membrane for cell axon growth [Fig. 1(a)]. Other methods have been proposed for fabricating PDMS micro through holes, such as soft lithography and laser ablation. The soft lithography method can provide relatively complete arrays of holes. However, difficulty exists in successfully peeling PDMS off from the patterned silicon substrates since the process of cross-linking causes the tendency of membrane breaking. Silanization of silicon substrates is the most widely employed solution. In recent studies, Zaidi and Agrawal16 deposited uncured PDMS around the pattern areas and created a thicker border. The design provided structural robustness to the membrane. Meanwhile, each space can be configured with single, double, triple, or multiple cells as needed by changing the pattern size, cell suspension density, and droplet volume. In addition, a method for suppressing the convection of droplet evaporation is proposed, enhancing the pattern effect [Fig. 1(b)]. Laser ablation directly produces a complete film with holes, eliminating the problems during demolding in soft lithography. Hsieh et al. first used laser sintering to create holes with diameters between 100 μm and 500 μm on PDMS thin films.17 Despite the method is faster compared with the previous methods, flaws are obvious. In the case that the diameter of holes is much larger than the size of single cells, the formation of a single cell array is challenging. To solve this problem, Messner et al.18 produced through holes with 35 μm in diameter using the laser ablation method [Fig. 1(c)]. The polymer film was immobilized and stretched across the silicon microwell array. A comparison of the various stencil patterning methods is listed in Table II.
FIG. 1.
(a) Preparation of NeuroArray and formation of neural networks. Reproduced with permission from W. Li, Z. Xu, J. Huang, X. Lin, R. Luo, C. H. Chen, and P. Shi, Sci. Rep. 4, 4784 (2014). Copyright 2014 Springer Nature. (b) Preparation of microstencils without membrane fragility and a cell pattern formed by droplets in the presence and absence of convection. Reproduced with permission from K. F. Zaidi and N. Agrawal, Biomicrofluidics 12, 064104 (2018). Copyright 2018 AIP Publishing LLC. (c) Device fabrication with a 355 nm UV laser and its application. Reproduced with permission from J. J. Messner, H. L. Glenn, and D. R. Meldrum, BMC Biotechnol. 17, 89 (2017). Copyright 2017 Springer, licensed under a Creative Commons Attribution (CC BY) License.
TABLE II.
Comparison of stencil methods.
| Cells used | Inlet cell density | Though-hole dimension | Material | Method | Efficiency | Reference |
|---|---|---|---|---|---|---|
| HeLa cells | 1 × 107 cells/ml | 30 μm | Silicon | Dry etching | 60% | 13 |
| Neuronal cells | 30–50 k cells/cm2 | 10 μm, 30 μm, and 50 μm | PDMS | Soft lithography | 80% | 15 |
| HeLa-C3 cells | … | 10–24 μm | PDMS | Soft lithography | ∼90% | 19 |
| HeLa cells | 200 000 cells/ml | 22 μm | PDMS | Soft lithography | >97% | 20 |
| K562, HME1, and MDA-MB-231 | 25 k, 50 k, 100 k, 200 k, 400 k cells/ml | 35 μm | PDMS | Laser ablation | … | 18 |
| CEM cells and HeLa cells | 5 × 105 cells/ml | 10 μm | PDMS | Soft lithography | … | 21 |
| MDA-MB-231 breast cancer cells | 6 × 106 cells/ml | 10 μm | PDMS | Soft lithography | >80% | 16 |
Stencil patterning can be applied in cell apoptosis analysis with single-cell resolution. Apoptosis is a form of programmed cell death resulting from a post-translational pathway driven largely by specific limited proteolysis.22 Cell apoptosis is worth investigating because inappropriate apoptosis causes many human diseases such as autoimmune disorders, neurological diseases, and cancers. A common method for studying apoptosis is based on population. However, due to the enormous heterogeneity within cell populations, the responses of cell groups to drugs, chemicals, or other external stimuli are inaccurate, especially in cancer cells which are known for their high heterogeneity. Hence, a high-throughput single-cell array is a prerequisite for experimentation. Huang et al.19 presented a simple method to monitor the apoptosis of single cells. A truncated cone-shaped microwell array (TCMA) chip was developed by them. After centrifugation, a large number of cells were captured with the capture efficiency of 90%. Stencil patterning is also an attractive approach for the drug metabolism and cell cytotoxicity analyses. Huang et al.20 used the stencil method with a centrifuge to isolate cells and investigate drug-induced phenotypic changes. Hela cells were treated with cisplatin to detect early apoptotic phenotypes in them. With this method, they found the corresponding molecular markers of apoptosis, which was consistent with the results of other methods. Xia et al.21 reported a new toxicity test based on single cell arrays. Under high-throughput conditions, cell responded at the population, single-cell, and subcellular level information could be obtained simultaneously. After exposure to the damaged x-ray radiation, the expression of proteins associated with cell migration, invasion, and production of reactive oxygen species (ROS) was measured by image processing software. The results showed significant molecular heterogeneity of individual cells at the subcellular level.
B. Traps
Single-cell trapping has been used to facilitate cell analysis with efficient capture rates at fixed locations. This trapping strategy is often combined with microfluidics, an effective avenue to assay single cells because of its high throughput and automation. In micron-scale channels, microfluidic systems are capable of transporting and manipulating liquid volumes ranging from microliters to femtoliters, which provide the functions of single-cell capture and release. Several contact and noncontact methods have been commonly used to pattern cells.23 The contact methods such as microrobots and hydrodynamic traps with special structures can capture and enable transport cells through the precise physical contact. However, using a type of method to manipulate cells through the physical contact may cause some problems, such as the difficulty of releasing cells due to the dominant adhesive force at the microscale, and the possibility of cell damage due to the mechanical contact.24 For the noncontact methods, the most common used technologies are optical, dielectrophoretic, magnetic, and acoustic traps. These are all suited for single-cell manipulation applications, offering trapping forces up to a few 100 pN. The noncontact capture and transport of cells are realized by the combination of the fluid medium and the field force. Therefore, the problems existing in the contact process can be solved.
1. Microrobot
Microrobots are promising for a wide range of applications in single-cell manipulation. There are many mechanisms of actuation for microrobots, such as magnetic forces, electrical forces, biohybrid systems, and optothermal forces. Zhang et al. developed a method for precisely manipulating micro-objects in the fluid by the flow of mobile microvortices, which were generated by rotating nickel nanowires with a rotating uniform magnetic field of less than 5 mT as the energy source. The microvortex was precisely moved by propelling the nanowire in a controllable manner.25 Subsequently, due to the relatively small trapping force of the previous method, higher rotation frequencies of the magnetic field were needed to achieve a high trapping efficiency. Then, they proposed a new design for substantially enhanced fluid capture. A dumbbell (DB)-shaped magnetic actuator was developed and assembled by a nickel nanowire and two polystyrene microbeads. Unlike the single-mode of tumbling trapping produced by nickel nanowires, the magnetic dumbbell was able to dynamically capture and transfer micro-objects on demand in three modes: tumbling, wobbling, and rolling.26 Tasoglu et al.27 described a method for encoding complex materials in 2D and 3D utilizing unbound magnetic microrobots under the remote control of magnetic field. Magnetic microrobots encoded cell-encapsulated hydrogels into different geometric shapes as needed without affecting cell viability and proliferation. Lin et al.28 reported a magnetically-actuated peanut-shaped hematite colloid motor. The motor could climb over a steep slope and release cells in a predetermined position and control single-cell patterning in a noncontact manner.
2. Hydrodynamic traps
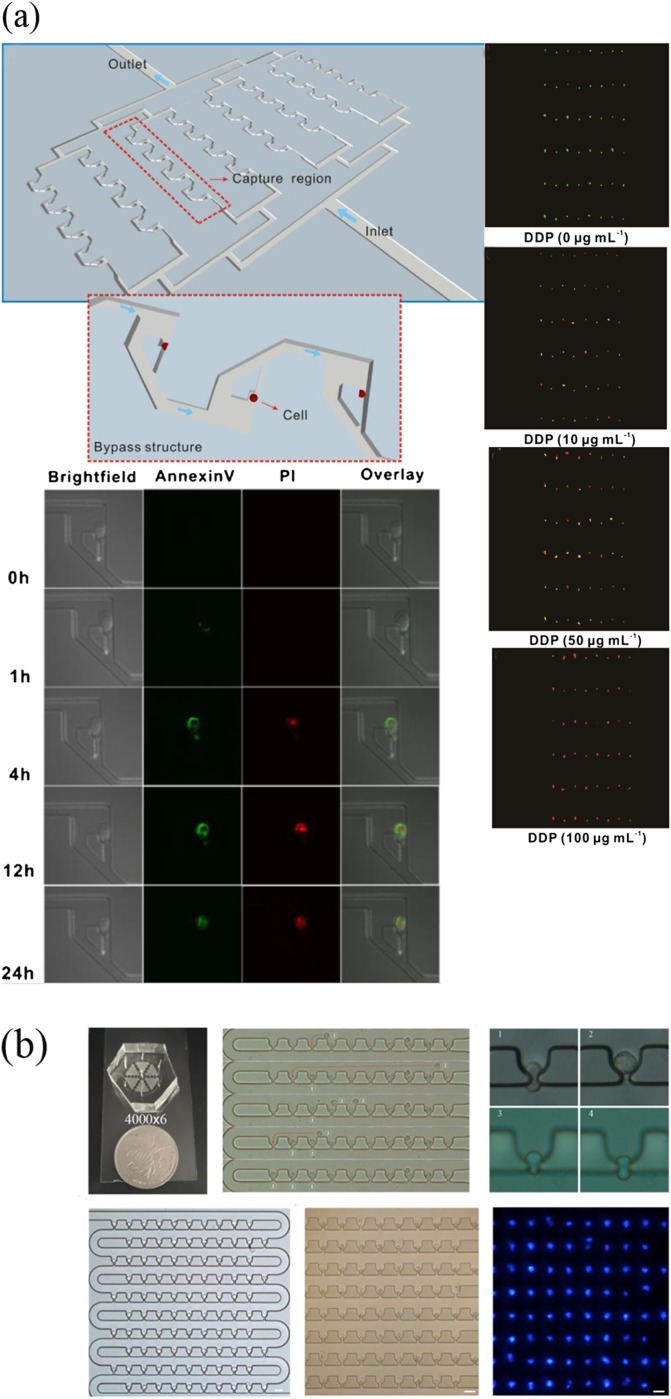
A hydrodynamic flow can be used as a contact approach to confine single cells in a specific microstructure, which is arranged inside the microfluidic channel. This is a common method due to the advantages of easy operation and no auxiliary equipment required.29 Although this hydrodynamic capture mechanism has high throughput compared with previous methods, two aspects that should be concerned and improved are the limited operation of cells after capture and the low capture efficiency with severe cell loss.30,31 Up to date, many types of microstructures have been investigated for optimal single-cell capture. U-shaped microstructures were first reported by Di Carlo et al.32 to trap Hela cells. The traps were molded in PDMS with the size of trapping one or two cells. Then, a serpentine design was proposed to trap cells in sequence.33 Wang et al.34 fabricated a microfluidic device with bypass structures for monitoring cell dynamics [Fig. 2(a)]. To optimize the hydrodynamic traps, three traps were designed by modulating the bypass structures with filleted, vertical, and sharp corners. 90% trapping efficiency was achieved by them, and most trapped cells enabled live for 10 h. The platform was able to monitor cell apoptosis over time and investigated apoptosis caused by chemotherapeutic agents. In recent research, a single-cell trap structure with m-by-n trap units possessing two roundabout channels and one capture channel was proposed by Mi et al.35 The average single-cell capture efficiency was 90% on a large scale (100 × 100 trap sites), but the density of cells was low because many traps lost cells causing a number of inoperative spaces existed. In most designs, a compromise between the trap site density and single-cell capture efficiency on a large scale is significant. Liu et al.36 made an improvement that they fabricated a loop channel trap structure with a high density, high throughput, and high efficiency. The cell capture rate could be up to 100%, while the single-cell efficiency was 95% [Fig. 2(b)]. In addition to the problems mentioned above, the microfluidic traps impose difficulties in delivering single cells to isolated microenvironments for further assay. Hence, the efficiency of releasing captured cells is also a major problem. Many approaches have been made. A multilayer device was developed by Zhou et al.37 It contained a control layer with valve channels, which controlled the release of cells. Chen et al.38 described a novel streamlined structure trap in which the release efficiency was about 99%.
FIG. 2.
(a) Microfluidic trapping system with bypass structures and its application to monitor single cell apoptosis over time. Reproduced with permission from Y. Wang, X. Tang, X. Feng, C. Liu, P. Chen, D. Chen, and B. F. Liu, Anal. Bioanal. Chem. 407, 1139 (2015). Copyright 2014 Springer-Verlag Berlin. (b) Single-cell trapping results of the loop channel trap structures. Reproduced with permission from Y. Liu, D. Ren, X. Ling, W. Liang, J. Li, Z. You, Y. Yalikun, and Y. Tanaka, Sensors 18, 3672 (2018). Copyright 2018 MDPI AG, licensed under a Creative Commons Attribution (CC BY 4.0) License.
Hydrodynamic traps can be applied to various fundamental studies in biology such as cell migration behaviors, which are essential in many biological processes involving immune responses, cancer metastasis, wound healing, inflammation, and embryonic development. In particular, significant attention has been focused on the migration of cancer cells since metastasis is the leading cause of death from cancer.39,40 A single-cell migration platform incorporating a single-cell capture scheme was designed by Chen et al.41 to specifically track and characterize the chemotaxis of each cell over time. Using the above platform, researchers tracked HGF-induced SKOV3 cell chemotaxis, determining the molecular differences between highly chemotactic and nonchemotactic populations of MDA-MB-231 breast cancer cells, and evaluated their effects on tumor cell migration. Ma et al.42 developed a novel microfluidic platform for cancer cell migration assays under periodic mechanical confinement stimulation. When the adherent MHCC-97L cells and suspended OCI-AML cells passed through the microchannel, an array of vertical constrictions produced periodic compressive forces on them. The experimental results revealed that the migration ability of cancer cells was promoted by this mechanical stimulation. Furthermore, Feng et al.43 presented a high-throughput microfluidic chip to study the effect of drug toxicity on cell apoptosis. They conducted drug toxicity studies under five different fluid shear stress (FSS) stimulations on cells. By observing changes in the cell morphology and apoptosis rate, the states of cells stimulated by drugs were observed.
3. Optical traps
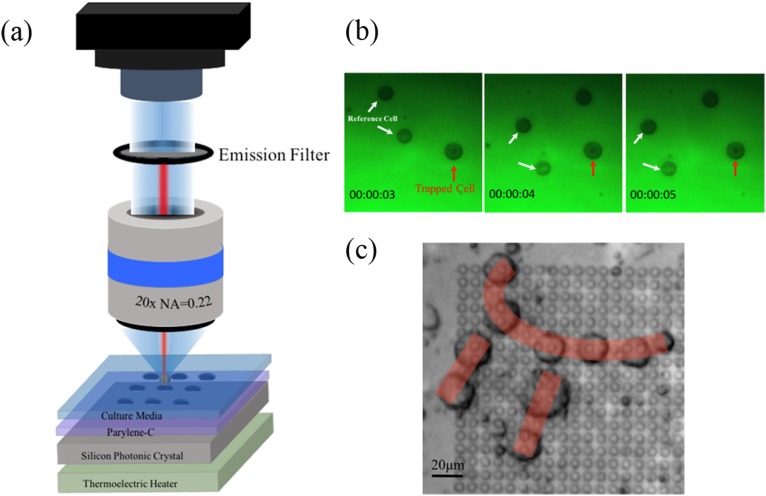
The optical trap (optical tweezers) is a typical example of noncontact single-cell capture. It uses a highly focused laser beam to generate forces for cell trapping. Traditional optical tweezers are based on a single beam, forming a three-dimensional optical well by laser focusing. The movement of the laser beam enables the manipulation of small objects. In general, such an optical trap can spatially capture and position single cells. However, high-intensity laser beams may cause photodamage to cells, especially in the case of prolonged exposure to oxygen.44 In order to minimize the photothermal damage and improve the capture efficiency, Jing et al.45 employed a two-dimensional photonic crystal in optical tweezers, achieving optical capture with high efficiency at low light intensities. For polystyrene beads with the size from 100 nm to 1 μm, the used optical intensities were all lower than that used in typical optical tweezers to trap micrometer-size particles. In addition, the displacement of the 1 μm beads captured at the center of the diffraction pattern in both x and y directions presented a Gaussian distribution. That is, most of the beads were captured. Recently, they also demonstrated a novel type of optical tweezers (Fig. 3), which utilized photonic crystals to manipulate human pluripotent stem cells. The optimal power to manipulate cells was also explored by them.46 Despite the great efforts made for the development of the single optical trap, complex operations of multiple cells are difficult to be achieved by this method. Therefore, multiple optical traps have been proposed to form a single cell array and study the heterogeneity of single cells. This method is called holographic optical tweezers, implemented by a diffractive optical element (DOE) to split a single laser beam.47 Dynamic holographic optical tweezers were reported by Bin et al.48 to rotate single cells. The method generated two separate dynamically controlled optical traps located at the opposite edges of an individual cell.
FIG. 3.
(a) Photonic-crystal optical tweezers system. (b) Manipulation of cells on a hydrophobic parylene-C film with a low-intensity laser beam. (c) Formation of cell “π” pattern with photonic-crystal tweezers. Reprinted with permission from P. Jing, Y. Liu, E. G. Keeler, N. M. Cruz, B. S. Freedman, and L. Y. Lin, Biomed. Opt. Express 9, 771 (2018). Copyright 2018 The Optical Society.
The optical trap can be used for cell manipulation under static environments or combined with a microfluidic chip to manipulate cells under continuous flow conditions. In order to capture single cells, a laser beam has been used as an optical trap to generate force in the microfluidic channel. Liberale et al.49 developed an integrated microfluidic device containing optical traps. Based on fiber bundles, the optical microtweezers used in microfluidics can efficiently capture single cells. The integrated microfluidic device is capable of on-chip manipulation, Raman and fluorescence spectra analysis of single cells. Moreover, a combination of microfluidics and optical traps can be used for quantitative and robust single-cell gene and protein analysis. Magness et al.50 utilized optical tweezers to select living cells and then transferred them to individual analysis chambers. Using this method, they quantitatively evaluated the expression and phosphorylation of the selected tumor suppressor protein p53 at the single cell level, and the results showed a big difference in the variability of protein and phosphorylated quantities between single cancer cells.
4. Dielectrophoresis traps
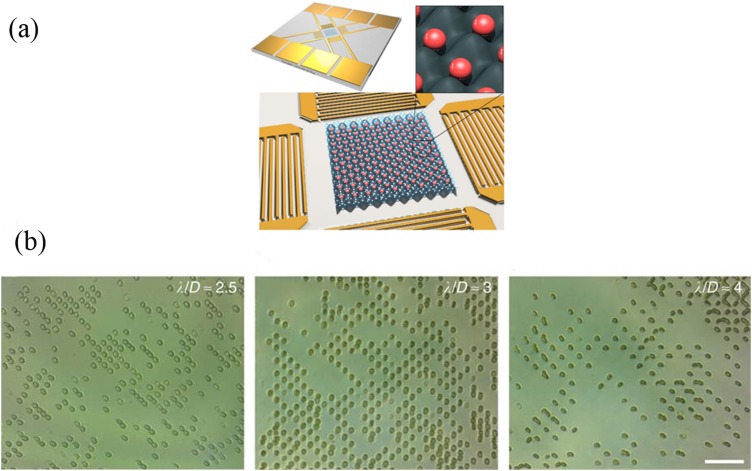
Dielectrophoresis (DEP) trap is a noncontact method for manipulating polarizable objects. It refers to the displacement phenomenon of dielectric particles caused by polarization effects in a nonuniform electric field. This phenomenon is only related to the electrical properties of the particles, the exposed environment, and the applied electric field. Because the particles in the solution are not the same as the fluid around them, their dielectric constants are definitely different. When an electric field is applied, the positive charge inside the particle moves in the direction of the electric field and the negative charge moves in the opposite direction of the electric field. This creates an antielectric field inside the particle, a phenomenon known as polarization. Therefore, the difference between the polarizabilities of object and medium greatly affects the motion of the object. Specifically, when the applied electric field is uniform, the particles are subjected to two forces of equal magnitude and opposite directions, and no displacement occurs. When the applied electric field is not uniform, the induced dipole moment will interact with the electric field to form a dielectrophoretic force, causing the particles to move toward the stronger or weaker side of the electric field. Thus, the DEP can be classified into positive DEP (p-DEP) and negative DEP (n-DEP).51 The p-DEP attracts the particles toward high electric field strength regions, while n-DEP repels the particles from high electric field strength regions. By making microelectrodes, DEP can be combined with microfluidics. Thus, single-cell patterning can be achieved in microfluidics via DEP. The DEP provides a force ranging from 0.1 nN to 1 nN for the manipulation of particles, which is strong enough to trap single cells in a high-speed flow. Iliescu et al.52 reported a dielectrophoretic-hydrodynamic trap located in a microfluidic structure for 2D and 3D cell patterning. The combination of insulator-based DEP and electrode-based DEP would generate a dielectrophoretic force for cell manipulation.
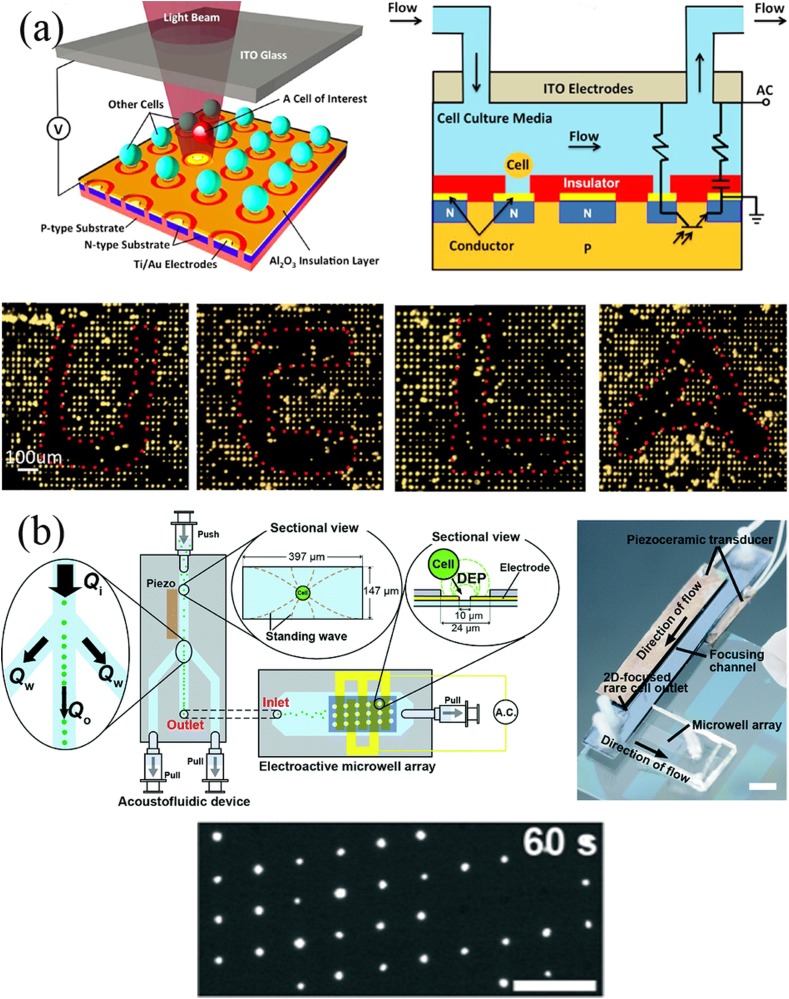
When DEP is used to manipulate cells, incompatibility with high-conductivity media and limited production are two major challenges. Low conductivity solutions are required in most of the cell operating systems based on DEP. However, standard cell culture media are highly conductive. Therefore, cell samples require rigorous pretreatment, which limits the application of dielectrophoresis-based methods. Yang et al.53 developed a novel platform of self-locking optoelectronic tweezers, comprising a periodic phototransistor trap array [Fig. 4(a)]. In the dark state, part of the voltage was reduced by the nanodielectric layer coated on the SLOT surface, enabling large-scale parallel capture of single cells in high-conductivity media. Kim et al.54 integrated an acoustofluidic chip with an electroactive microwell array for cell trapping [Fig. 4(b)]. With a high volumetric flow at the entrance to the microsystem, the velocity of cells is reduced by the acoustofluidic chip. This method results in an increase of 10 times in the sample throughput compared to a single electroactive microwell array chip.
FIG. 4.
(a) Self-locking optoelectronic tweezers (SLOT) platform and the patterning of particles. Reproduced with permission from Y. Yang, Y. Mao, K. S. Shin, C. O. Chui, and P. Y. Chiou, Sci. Rep. 6, 22630 (2016). Copyright 2016 Springer Nature, licensed under a Creative Commons Attribution (CC BY 4.0) License. (b) Integration of the acoustofluidic and EMA chips. Cells are trapped in the EMA chip by DEP. Reproduced with permission from S. H. Kim, M. Antfolk, M. Kobayashi, S. Kaneda, T. Laurell, and T. Fujii, Lab Chip 15, 4356 (2015). Copyright 2015 Royal Society of Chemistry, licensed under a Creative Commons Attribution (CC BY 3.0) License.
5. Magnetic traps
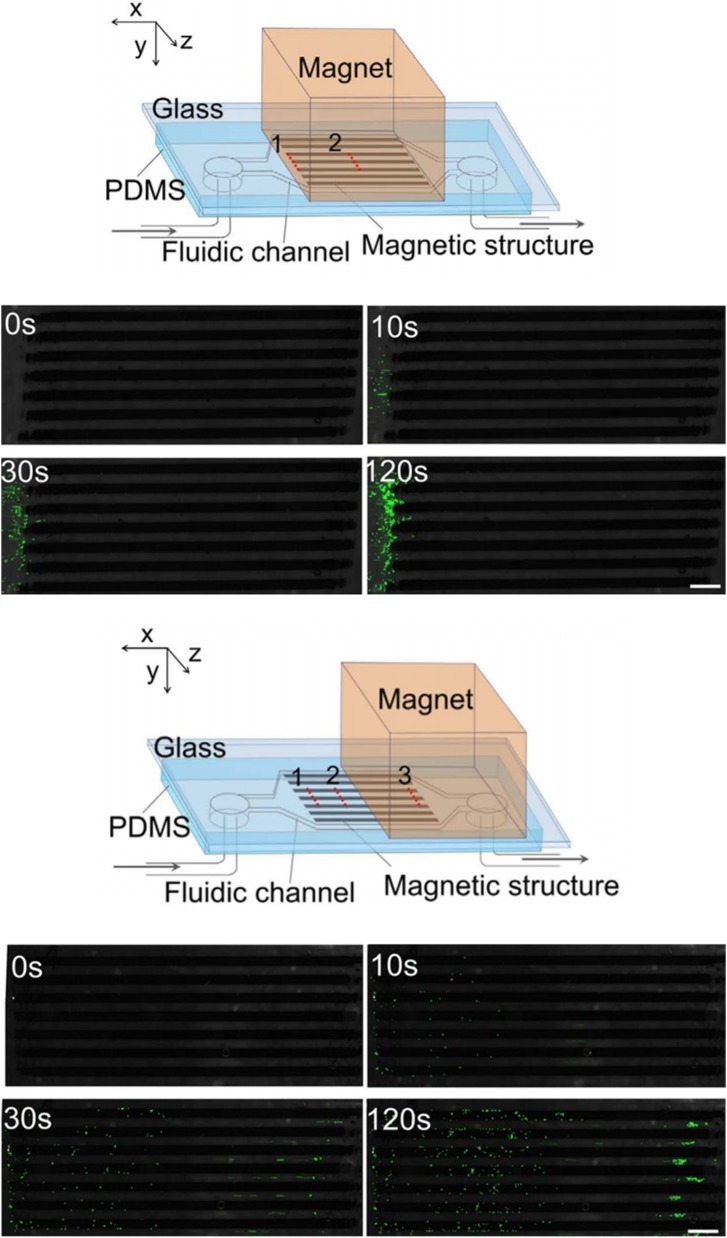
Magnetic manipulation refers to the manipulation of cells using permanent magnets or electromagnets. Since cells are usually lacking in paramagnetic or diamagnetic properties, immunomagnetic beads are used for the surface label of cells. The surface-modified magnetic beads can adhere onto the cell surface by specific binding between the antibody and the antigen. Therefore, this magnetic method is based on labels. A micromagnet array was developed by Pivetal et al.55 for precisely positioning single cells upon large arrays. To manipulate cells, immunomagnetic labels and magnetic in situ hybridization strategies are used by them to magnetically and specifically label cells. A micromagnet can be implanted in a microfluidic channel for future on-chip experimental applications. Shields et al.56 developed a magnetographic device containing micromagnets and microwells to locate labeled single cells into the target spaces. Currently, this labeling method is widely used in scientific research. A combination of magnetic and microfluidic systems has also become a viable method for high throughput and low-cost applications. Placing a large number of permanent magnets near the microfluidic channel is a common strategy. Sun et al.57 developed a simple method in which they embedded paramagnetic structures inside a microfluidic channel to enhance the immunomagnetic separation of cells (Fig. 5). This method increased the capture efficiency up to 4 times compared with the channel without magnetic structures. The layout of the magnetic structures and the position of the external magnet collectively determined the spatial distribution of the captured cells.
FIG. 5.
Microfluidic channels with the magnetic structures and the distributions of cells determined by the location of magnet. Reproduced with permission from C. Sun, H. Hassanisaber, R. Yu, S. Ma, S. S. Verbridge, and C. Lu, Sci. Rep. 6, 29407 (2016). Copyright 2016 Springer Nature, licensed under a Creative Commons Attribution (CC BY 4.0) License.
6. Acoustic traps
Acoustic tweezers manipulate cells through the interaction of sound waves with solids, liquids, and gases, which can be used in almost any medium with the advantages over magnetic tweezers that require magnetic particles. Acoustic traps can be divided into body and surface waves. A surface acoustic wave (SAW) is an elastic sound wave that can only propagate on the substrate surface, and most of the energy is concentrated on the surface depth of several wavelengths. In recent years, SAW chips have been widely used to manipulate cells in microfluidic devices, providing a flexible single-cell localization method. An interdigital transducer (IDT) is commonly used to generate surface acoustic waves, which is an important component of the SAW chip. The resonant frequency of the surface acoustic wave can be controlled by adjusting the interdigital finger pitch of the electrodes, while the distribution of the sound field can be adjusted by changing the shape of the IDT. In microfluidic chips, the fluid gains momentum by absorbing sound waves when the sound waves propagate into a fluid medium. If the size of the particle is much smaller than the wavelength with low density, the particle can move with the fluid. According to this phenomenon, the particles in the fluid can be manipulated. Collins et al.58 utilized high frequency surface acoustic waves (SAWS) to generate two-dimensional (2D) sound fields. Cells were located with one cell per acoustic well (OCPW), which demonstrated that spatially isolating cell patterns are feasible over a relatively narrow particle diameter range (Fig. 6). However, the exposure of cells to ultrasound reduces survival, which is a negative effect of acoustic manipulation.
FIG. 6.
Acoustic single-cell trapping method. (a) The design for OCPW patterning. (b) Red blood cell patterning. Reproduced with permission from D. J. Collins, B. Morahan, J. Garcia-Bustos, C. Doerig, M. Plebanski, and A. Neild, Nat. Commun. 6, 8686 (2015). Copyright 2015 Springer Nature, licensed under a Creative Commons Attribution (CC BY 4.0) License.
Acoustic trapping is a dynamical single-cell patterning method that enables precise control of cell location. In recent research, it has been used to control the morphology of nerve cells, pattern single neurites, and form neural networks. Brugger et al.59 reported a SAW-based microfluidics with a checkerboard-like standing surface acoustic wave pattern on the top of it. Cell growth could be stimulated and manipulated on the chip. This chip enables the active positioning of neurons and guides the growth of neurites, overcoming the limitations of static approaches.
C. Droplets
Similar to analyzing an individual cell in a trap, the cell can be separated by encapsulation in low volume droplets to form microcavities. The risk of cross-contamination is reduced since each cell is stored in its own separate droplet and separated from each other. As a kind of microfluids, droplet-based microfluidics has greatly expanded the potential for single-cell studies since it allows high-throughput single-cell analysis and manipulation by controlling the local environment.60 This method typically requires two immiscible fluids to produce monodisperse water-in-oil (W/O) microdroplets with the size ranging from submicrometers to hundreds of micrometers. Generally, three types of microfluidics for generating droplets are T-joints, flow focusing, and co-flowing. Micronozzles that produce droplets can form a rate from slow dripping to over 10 kHz.61 In order to support the controlled operation of droplets in a high-throughput manner, the droplets can be further manipulated for fusion, mixing, analysis, and sorting.62 Chiu et al.63 first used microfluidics with optical capture and microfluidic T-joints to limit single cells to aqueous droplets, which were surrounded by a continuous oil phase. The optical capture sheared the aqueous phase into individual droplets by applying a pressure pulse. Besides the types of droplets that mentioned above, single cells can be encapsulated by lipid vesicles surrounded by a continuous oil phase/aqueous phase in microfluidic structures,64,65 but the throughput and efficiency of single cell capture are quite low. Recently, a novel hydrogel droplet is presented in which the water phase is substituted by a monomer or polymer solution, maintaining the viability of mammalian cells for several days in them.66,67 This hydrogel can serve as a three-dimensional cell culture matrix which can mimic the extracellular microenvironment for single cells. Further studying in the heterogeneity of single cells is feasible. Due to the excellent selectivity, high reactivity, and strong cell adhesion, microgels can guide osteogenic differentiation and have recently attracted increasing interest. Specifically when combined with droplet-based microfluidics, long-term cell viability can be achieved. Hou et al.68 developed a microfluidic approach based on pH-degradable polyvinyl alcohol microgels, providing a good cell microenvironment for controlling osteogenic differentiation in vitro. Long-term sustained release of BMP-2 from polyvinyl alcohol microgel promoted osteogenic differentiation, which proved that the method can control the osteogenic differentiation of hMSCs for a long term.
Precise control of the number of cells per droplet is challenging due to the random encapsulation nature of cells. That is to say, single-cell encapsulation follows the Poisson distribution, resulting in a great deal of empty droplets or multiple droplets with two or more cells, decreasing the throughput.69 Dilution of the cell suspension can control one cell in a droplet, but the production of a large number of empty droplets leads to reagent waste.70 One way to increase the ratio of cells to empty droplets is to actively sort droplets after encapsulation.71 Electric field,72 dielectrophoresis,73 and localized heating based on the laser74 are common active methods to meet the requirements. Navi et al.75 recently proposed a new active method. They diamagnetically manipulated droplets. An aqueous two-phase system (ATPS) and a conventional oil-in-water droplet were replaced by water-in-water droplets. The purity of 100% single cell encapsulating droplets was achieved. Though the approach is feasible, the active sorting requires a relative complex setup. Passive methods have also been implemented for increasing the single-cell encapsulation efficiency, such as hydrodynamics, which controls the number of cells encapsulated per droplet.76
With the characteristics of fast response and high sensitivity, droplet-based microfluidics has been considered to be potential tools in single cell molecular analysis such as gene and protein. Cellular gene expression levels can be significantly different due to random fluctuations or complex molecular switches that alter the production of mRNA.77 Single-cell RNA-Seq analysis reveals the inherent cellular heterogeneity in cell populations. In recent years, droplet-based microfluidics is an automatic tool, which offers a method for increasing throughput in single-cell RNA-Seq studies. Macosko et al.78 used drop-seq to analyze all cellular RNAs by encapsulating cells in tiny droplets. A molecular barcode strategy was proposed to remember the origin of each mRNA cell. The findings suggested that a large-scale single-cell analysis could deepen our understanding of the complex tissues and cell populations. Klein et al.79 developed a high-throughput and low noise profile droplet-microfluidic approach. Cells were encapsulated by hydrogels with barcoded ssDNA primers. After lysing cells, barcoding mRNAs was achieved by reverse transcription (RT) reaction. Although droplet microfluidics has achieved high-throughput analysis, barcoding errors occurring in cell encapsulation still affected the accuracy of gene expression analysis. To address this, Moon et al.80 designed a new droplet-based microfluidic platform. They encapsulated high-concentration beads containing oligonucleotide barcodes ordered in the droplets, and the cells were also simultaneously encapsulated in droplets.
Compared with genetic studies, protein analysis of single cells is a challenging issue due to the low concentrations. Chen et al.81 fabricated a substrate for multicolor fluorescence resonance energy transfer (FRET), which measures the fluorescence signal produced by protease-substrate reactions. The activities of multiple protease in single cells were determined by encapsulating a multicolor enzymatic substrate and individual cells in droplets. To acquire high throughput, sensitive, and fast profiling methods, Shahi et al.82 presented an Abseq method. Droplet barcoding was similar to barcode DNA molecules and single cell transcriptomes applied before. Thus, this method would allow the simultaneous characterization of genome, transcriptome, and proteome.
D. Photolithography
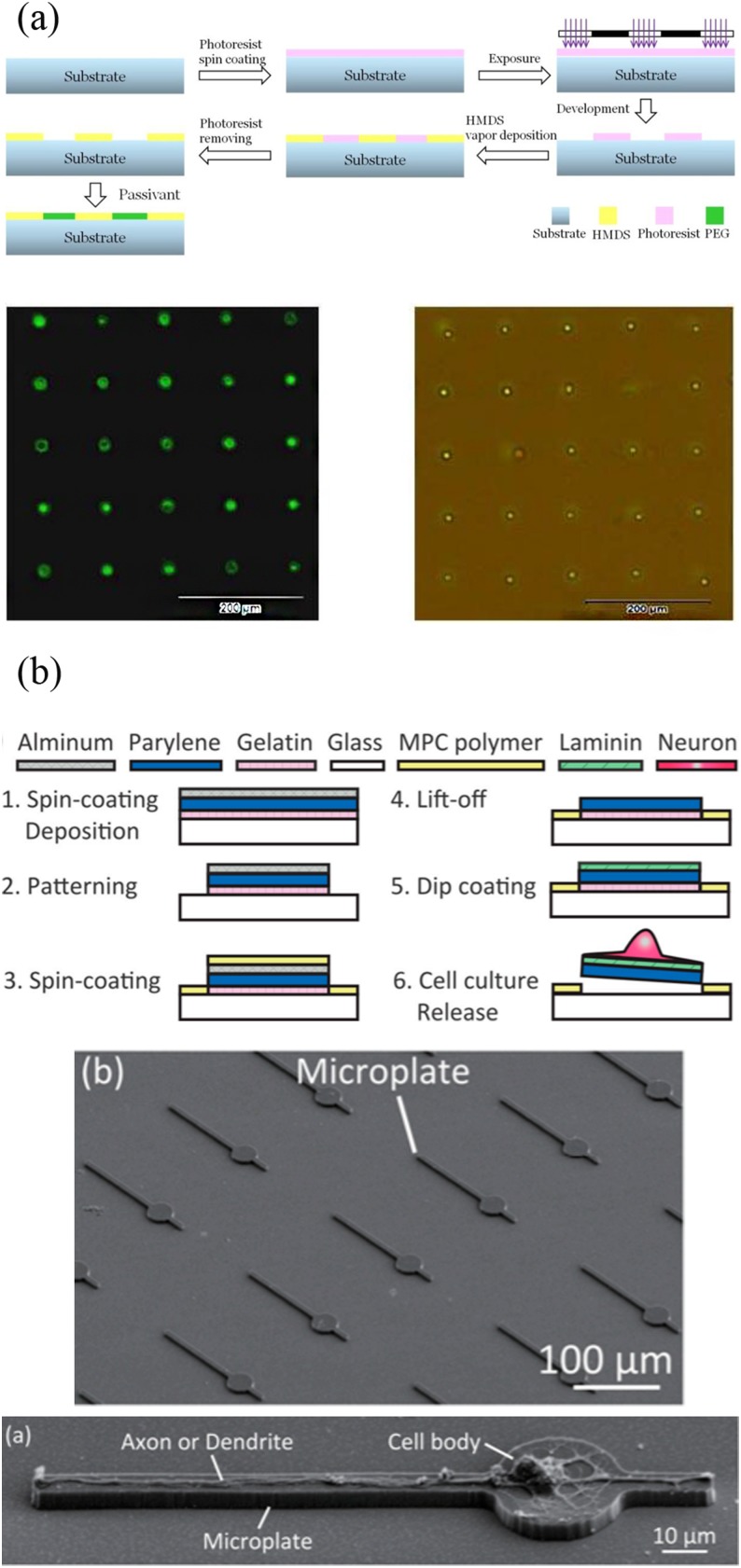
Photolithography is the most popular micropatterning method, which is also the root for most top-down microfabrication approaches. This method is a high-throughput approach enabling large-area surface patterning. Silicon wafers are widely used as the material of the substrate when fabricating semiconductor devices by photolithography. However, in biological applications, transparent wafers are often used because they are suitable for optical microscope studies. The process of photolithography is transferring a geometric pattern from a mask to a substrate by ultraviolet light. In exposed areas, the polymer chain of the photoresist applied to the surface is broken, making it more soluble in a chemical solution called a developer. Then, further processing steps will mainly depend on the type of target chemical patterns. In order to create protein patterns that locate cells, photolithography is often combined with the self-assembly method, which is a bottom-up process. Ayibaike et al.83 fabricated a MEMS-processed substrate in a photolithography-based process [Fig. 7(a)]. Hydrophilic and hydrophobic regions were formed through the chemical vapor deposition (CVD) with hexamethyldisilazane (HMDS) and the passivation treatment with polyethylene glycol (PEG). The bovine serum protein was capable of adhering to the hydrophobic part. Through the self-assembly method, a biotin–Streptavidin (BSA) template layer was fabricated to capture HL 60 cells, and the single cell capture efficiency was 92%. In addition to locating cells at specific locations, cell morphology can be controlled for more in-depth cell behavioral studies, such as the guidance of nerve cell synapses and dendrites. Yoshida et al.84 designed a microplate device which was prepared by photolithography with laminin coated on it. Functional axons and dendrites can be formed and positioned on the device [Fig. 7(b)].
FIG. 7.
(a) The surface modification progress and the patterning of HL-60 cells at the single-cell level. Reproduced with permission from D. Ayibaike, M. Cui, and J. Wei, Appl. Sci. 8, 11 (2018). Copyright 2018 MDPI AG, licensed under a Creative Commons Attribution (CC BY 4.0) License. (b) Preparation process of the microplate device and the SEM images of microplates and neurons. Reproduced with permission from S. Yoshida, M. Kato-Negishi, and S. Takeuchi, Micromachines 9, 235 (2018). Copyright 2018 MDPI AG, licensed under a Creative Commons Attribution (CC BY 4.0) License.
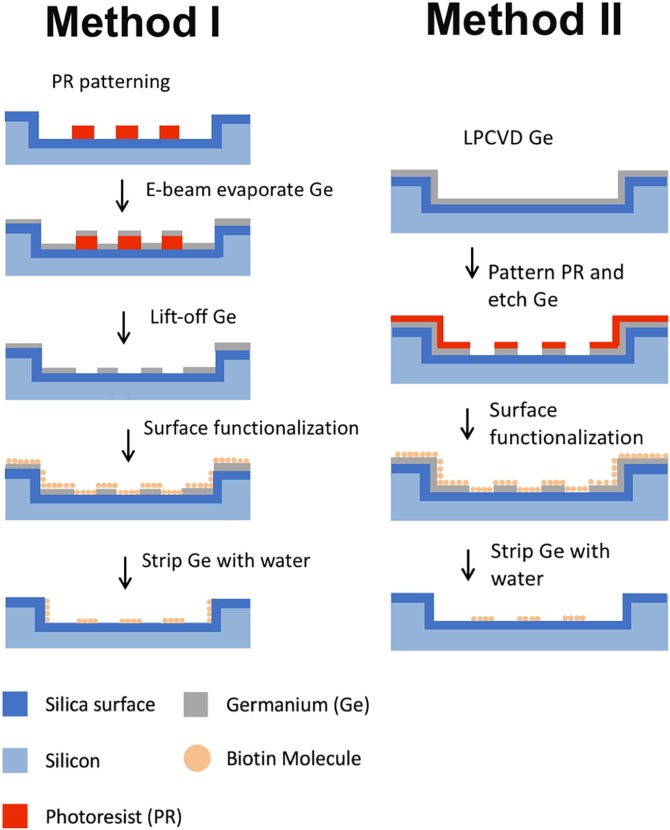
A geometric confinement of cells is provided by this method compared with other patterning approaches. The combination of the features inhibits the deterioration of cell arrays over time. Because of the advantages as well as high precision, photolithography has become one of the main methods for preparing cell patterns on solid surfaces. However, chemicals used in photolithography may denature and inactivate biomolecules. To conquer these, people have made a lot of improvements. For example, Lu and Maharbiz85 utilized germanium (Ge) as a sacrificial layer to protect the substrate during surface functionalization, allowing the formation of protein patterns with submicrometer resolution in common organic solvents using photolithography. Protein activity was not affected after Ge was removed (Fig. 8).
FIG. 8.
The fabrication process of nanoscale protein patterning. Reproduced with permission from B. Lu and M. M. Maharbiz, PLoS One 13, e0195062 (2018). Copyright 2018 PLOS ONE, licensed under a Creative Commons Attribution (CC BY 4.0) License.
E. Microcontact printing
Microcontact printing (μCP) was originally developed by Whiteside and co-workers to form an alkanethiol pattern on gold, which was a simple and effective surface modification method. The desired micropattern was obtained by stamping the desired biomolecule on a modified substrate. This process used a patterned stamp. The earliest stamps were prepared by photolithography, transferring the pattern on the mask to a silicon substrate. However, such a preparation method has a number of disadvantages. For instance, it is not suitable for nonpolar and curved surfaces, and the choice of polymer materials is limited. To overcome this bottleneck, soft lithography was developed by Xia and Whiteside.86 Then, elastomers such as PDMS are commonly used as the material of stamps. However, the initial step of stamp preparation is still implemented by photolithography. Besides, the master plate can no longer be used once blocked when the stamp is removed from the template. Many novel production methods of the stamp have been proposed. Recently, a lithography-free method of making low-cost stamps was proposed by Khadpekar et al.87 Polystyrene beads and PDMS cylinders were used to fabricate circular and stripe-shaped prints. By using the Johnson–Kendall–Roberts (JKR) contact theory, the print size can be controlled.
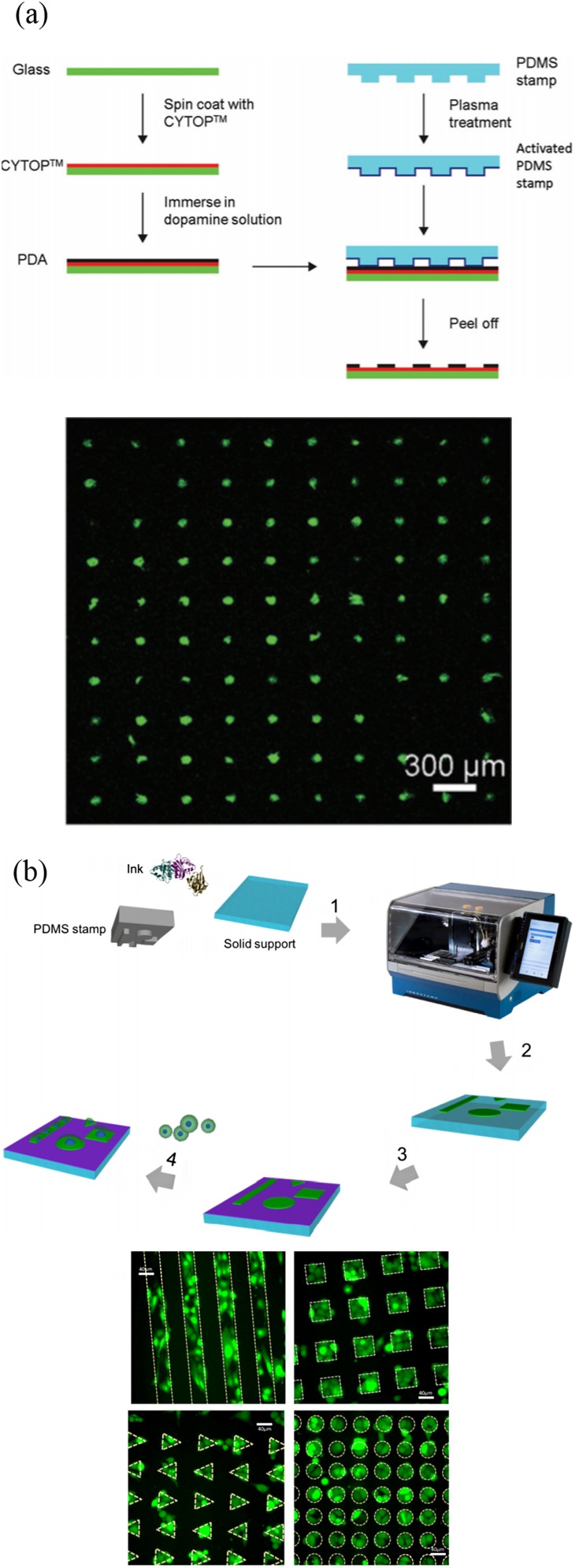
As a powerful and versatile tool, PDMS stamping can print a wide range of biomolecules onto a variety of different background materials. Hence, traditional micropatterning approaches are widely utilized in single-cell patterning. Microcontact printing has become an established single-cell analysis method, which can be used to create a selective extracellular matrix (ECM) environment for cell adhesion and growth. However, this process only reaches medium-throughput. To solve this, many improvements have been made. Wu et al.88 developed a negative microcontact printing method. A hydrophilic PDA pattern was formed on a hydrophobic surface, and its microstructure size was less than 10 μm. The single-cell patterning was achieved with the capture efficiency of about 94% [Fig. 9(a)]. Foncy et al.89 presented an automated microcontact printing method to produce a biomolecular microarray consisting of extracellular matrix proteins. They investigated a microcontact printer, handling the PDMS stamps under the magnetic field. Uniform printing of defined biomolecular patterns was realized under such operation [Fig. 9(b)]. Since the process was effective, fast, and robust, large-scale cell microarrays can be obtained.
FIG. 9.
High-throughput microcontact printing methods and results. (a) Schematic illustration of the fabrication of PDA patterns on the CYTOP-coated glass surface by negative microcontact printing. Single cell array is formed on the PDA-patterned CYTOP surface. Reproduced with permission from H. Wu, L. Wu, X. Zhou, B. Liu, and B. Zheng, Small 14, e1802128 (2018). Copyright 2018 WileyVCH Verlag GmbH & Co. KGaA, Weinheim. (b) Overall microcontact printing of adhesive patterns using InnoStamp. PC3-GFP cell microarrays are formed on fibronectin micropatterns of various shapes. Reproduced with permission from J. Foncy, A. Estève, A. Degache, C. Colin, J. C. Cau, L. Malaquin, C. Vieu, and E. Trévisiol, Methods Mol. Biol. 1771, 83 (2018). Copyright 2018 Springer Science Business Media, LLC, part of Springer Nature.
Microcontact printing can be applied to cell proliferation studies. Yao et al.90 investigated the proliferation of single cells on cell-adhesive microislands using the micropatterning method. Two critical spreading areas related to cell proliferation was first reported by them, which were the critical diffusion regions of cells from almost no proliferation to limited proliferation, and the cells proliferated from the restricted to almost free proliferation. In addition, microcontact printing is more suitable than photolithography for precisely controlling the direction of signal propagation with single-cell resolution. Yamamoto et al.91 investigated a micropatterned surface to induce the functional synapse of rat hippocampal neurons. Microcontact printing was utilized by them to produce a cell permissive protein pattern. The micropattern consisted of a circular island for somatic cell growth, and a long and a short path for axon and dendritic generation, respectively.
F. Scanning probe lithography (SPL)
1. Dip-pen nanolithography (DPN)
Scanning probe lithography (SPL) can create patterns with high resolution to control cell behavior effectively. Among the methods of lithography, Dip-pen nanolithography (DPN) is widely applied. DPN holds multiple advantages over other traditional nanofabrication approaches. Neither has the complicated steps of nanocontact printing,92–95 nor requires a vacuum environment for electron-beam lithography and FIB,96,97 the DPN method uses an atomic force microscope (AFM) tip with molecules on it and directly transfers the molecules in a humid environment. Overall, its implementation is the same as writing on the paper using a pen with ink but more complicated. The water meniscus is critical, which is formed when an ink-coated AFM tip contacts with the surface under suitable ambient humidity. Then, the biomolecular ink transfers to the surface via the water bridge.
DPN was originally introduced by Mirkin's group as an attractive method for nanopatterning small alkanethiol molecules on Au surfaces.98 Subsequently, different molecular inks such as collagens, DNA molecules, and proteins are used to achieve the desired function. DPN can produce regions that promote/prevent cell adhesion on the surface of a substrate by producing protein or polymer patterns. However, the denaturation of protein always exists due to the interaction with the bare solid AFM tip or substrate. To overcome it, the indirect and direct methods have been tried to print protein structures with nanoresolution while retaining their bioactivity. The indirect DPN approach provides a perfect way to prevent the denaturation of biomolecules. However, it consumes more time to passivate the background without patterns. To keep functional immobilization, the direct method has been improved. Multimaterial substrates have been utilized for protein nanopatterning. In general, there are about two main types: gold and silicon surfaces.99 Ti/Au coating or chemical modification on the tip surface has also been studied to maintain the bioactivity of protein. Simultaneously, restrictions such as slow transportation from the AFM tip to the surface and difficulties of short operation time due to the limited amount of ink can also be solved by a chemically modified tip. In addition, adding a carrier to biomolecule ink is a common method. Glycerol is routinely added to biomolecule solutions to promote the transport process of ink molecules and slow down the evaporation of water. Therefore, achieving the transfer of glycerol in a relatively low humidity environment becomes easy. Recently, hydrogels and phospholipids have also been demonstrated as good biomolecular ink carriers to reduce the evaporation of highly volatile ink molecules.
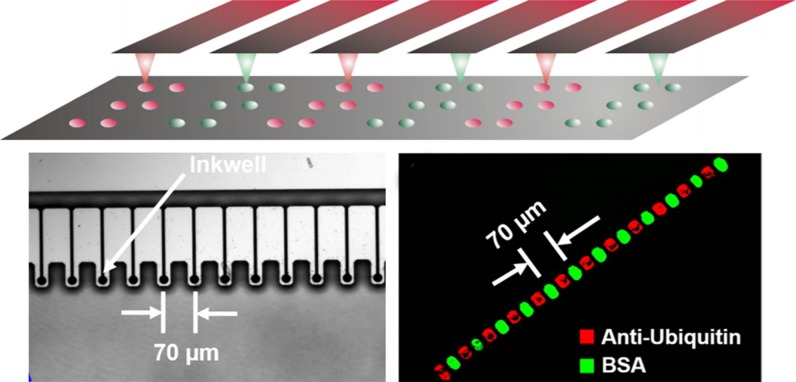
Apart from the restrictions mentioned above, there are still other limitations. The capability of large-area parallelization is needed for widespread applications like single cell assays. However, drawing a square centimeter area with a single probe takes long time. To address this, high-throughput DPN has been reported. 1D and 2D cantilever arrays have been developed to transit the technology from the serial writing process to the parallel writing process.100 Mirkin's group also developed a novel DPN holding 55000 multiple probes. This DPN can generate massively two-dimensional protein pattern arrays with a resolution better than 100 nm within a square centimeter area.101 Recently, a special inkwell array has been designed combining with a parallel AFM tip array.102 The dimensional design of ink well matched the AFM tip array, and tips were coated with a specific ink when immersed in the well (Fig. 10). Generally, these multiplexed DPN approaches enable rapid fabrication of large-area array patterns, greatly simplifying the experimental process and revealing great potential in adhesion studies and single-cell drug screening. In biological applications, DPN can control various parameters efficiently at the molecular level, which allows for studying a single focal adhesion at a finer scale than previous approaches. Various functional group nanopatterns prepared by DPN can control the differentiation of human mesenchymal stem cells (hMSCs). The formed nanodot pattern has an optimum fixed diameter of 70 nm with the spacing of 140–1000 nm.103 Laing et al.104 reported a novel thermosensitive hydrogel array made from DPN. The polymer has different thermal response behaviors as the temperature of aqueous solution changes. Cells are able to adhere and interact with the dynamic characteristics, exhibiting behavioral changes.
FIG. 10.
Multiple AFM tips with a designed inkwell array. Reproduced with permission from H. Ma, Z. Jiang, X. Xie, L. Huang, and W. Huang, ACS Appl. Mater. Interfaces 10, 25121 (2018). Copyright 2018 American Chemical Society.
2. Polymer pen lithography (PPL)
Considering the deficiency of the throughput and the high cost that DPN holds, a new patterning method of polymer pen lithography (PPL) is proposed. PPL is a massively parallel direct writing approach, developed by Huo et al.105 In the method, a free cantilever and an elastomeric pyramidal tip array are utilized instead of using an AFM cantilever. The feature size of the pattern generated by PPL is changed by adjusting the force applied to the pen array or tip-to-substrate contact time.106–108 Since the mechanical compliance of the probe is achieved without relying on a fragile and expensive cantilever array, producing large-area protein patterns on the substrate becomes simple and low cost. Protein patterns with different characteristic densities or feature sizes are important for studying the molecular processes of cells adhering to the patterns. Kumar et al. utilized lipids as a biomimetic cell interface and prepared large-area gradient patterns by PPL, increasing the throughput. There are two strategies for printing phospholipid patterns, which can produce patterns with different morphological features by controlling the printing parameters.109
PPL is widely used in cell-ECM interaction studies. To make the screening of mesenchymal stem cell (MSC) adhesion rapidly and systematically, Giam et al.110 used PPL to prepare nano- and microscale fibronectin patterns on the substrate and investigated the influence of fibronectin feature size on the differentiation of MSCs. By measuring the expression level of osteogenic markers and relative content of FAK phosphorylation, the result showed that fibronectin promoted cell differentiation while the effect of nanopatterns was more obvious.
3. Dip-pen nanodisplacement lithography (DNL)
Despite the above two methods have met most of the requirements, nonuniform diffusion of inks is still a common deficiency. To overcome it, Zheng et al.111 developed a lithography method, namely, Dip-pen nanodisplacement lithography (DNL). DNL combines the attributes of DPN and nanoshaving to produce the controlled structure, topography, size, and shape of patterns in an air environment. Grafting density, chain configuration, hierarchical structure, and multiplexing of the polymer brushes can also be easily realized. DNL can prepare 3D nanostructures that mimic the natural environment of cells in vivo, which can more accurately control the interaction between cells and extracellular matrix. However, the studies of cell adhesion require a large amount of extracellular matrices to collect statistical results. In order to replicate a large number of three-dimensional polymer structures in macroscopic regions, developing a parallel dip-pen nanodisplacement lithography with continuity and high resolution is necessary. Zheng's group improved DNL on the basis of the previous work, and a parallel dip-pen nanodisplacement lithography (p-DNL) was reported by them with multicantilevers. Despite p-DNL achieves high-throughput patterning in a large area, the uniformity and resolution are still not satisfactory for many applications.112 Zheng et al.113 continued to improve p-DNL and conducted detailed research on the DNL molecular displacement process to obtain good uniformity and high reliability. To acquire a large area for cell activity studies, a 2D 55 000-tip cantilever array was developed instead of the previous parallel DNL with an 8-tip cantilever array in 2016. This binary system allows the nanopatterning over 1 × 1 cm2 within 60 min.114 In addition to achieving cell localization, DNL has an advantage over other methods in which it can produce multiscale surfaces for the studying of cell behaviors. Chen et al.114 reported a biomimicking nano-microbinary polymer brush system. To obtain the improved biomimetic ECM, they used a poly(N-isopropylacrylamide) (PNIPAm) brush to fabricate gelatin-poly(glycidyl methacrylate) (PGMA) nanowire arrays. Gelatin-PGMA can direct cells, and the temperature response behavior of the PNIPAm brush regulates cell adhesion.
III. CONCLUSIONS AND PERSPECTIVES
Over the past few decades, many single-cell patterning methods have been developed and utilized extensively for a wide range of biological applications. Compared with the traditional methods based on cell populations, cell patterning at the single-cell level can show the individual differences of cells. In this review, various physical and physicochemical single-cell patterning methods have been summarized, including physical methods such as stencils, traps, and droplets patterning single cells on the surface or space with high throughput. However, the physical methods may damage cells. Identifying the drawbacks of each method and improving them can preserve cell viability and functions. The study of the effects of cell-surface interactions on cell behaviors is primarily based on physicochemical methods. Physicochemical methods prepare chemical patterns by physical means and mainly use biomolecules to specifically recognize chemical bonds between the cellular receptors and different functional groups, attaching individual cells to the surface. The methods create patterns in a continuous manner. However, the main drawback of the methods is the limited throughput due to the slow serial processing nature, making them inappropriate for mass production. This review aims to outline some novel methodologies to improve the methods that are currently used. The physical and physicochemical methods are widely used in current biological research. In particular, the single-cell microarrays made by them perfectly solve the problem of cellular heterogeneity in traditional analysis.
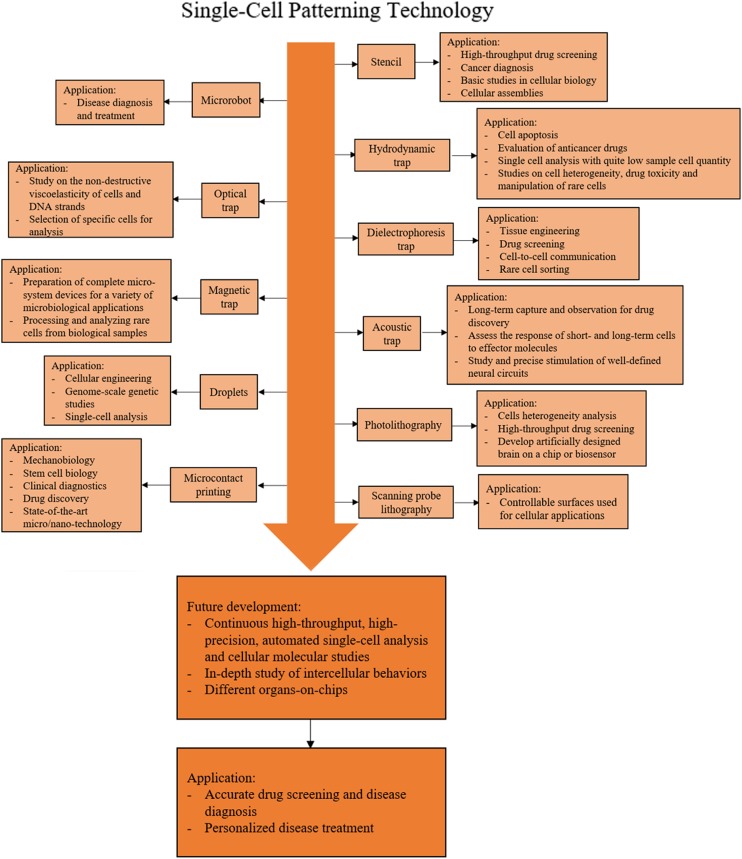
A roadmap for single-cell patterning technology and applications is shown in Fig. 11. The figure outlines the single-cell patterning methods developed in recent years, their current applications, and future developments. In the coming years, the advances in high-throughput methods will offer opportunities to investigate more fundamental mechanisms of cells and cellular molecules. Future technology will enable automated high-precision, high-throughput single-cell analysis. In order to conduct more in-depth research at the molecular level, there are still great challenges to be dealt with such as the systematic understanding of DNAs and proteins. Microfluidics has the potential to solve some key problems in the future. However, more improvements are needed to turn it into a more robust and easy-to-use tool. This will ultimately promote a more accurate and comprehensive understanding of the function and regulatory mechanisms of intercellular molecules in biological processes. At the same time, the precise positioning of single cells in a nondestructive manner is the key to study intercellular behaviors and develop organs-on-chips. Through continuous research on the topics, accurate drug screening, disease diagnosis, and personalized disease treatment can be achieved.
FIG. 11.
A roadmap for single-cell patterning technology and applications.
ACKNOWLEDGMENTS
This work was supported by the National Key R&D Program of China (No. 2017YFE0112100), the EU H2020 Program (MNR4SCell No. 734174), the Jilin Provincial Science and Technology Program (Nos. 20160623002TC, 20180414002GH, 20180414081GH, 20180520203JH, and 20190702002GH), and the “111” Project of China (No. D17017).
REFERENCES
- 1.Gorelik R. and Gautreau A., Nat. Protoc. 9, 1931 (2014). 10.1038/nprot.2014.131 [DOI] [PubMed] [Google Scholar]
- 2.Frisman I., Seliktar D., and Bianco-Peled H., Acta Biomater. 8, 51 (2012). 10.1016/j.actbio.2011.07.030 [DOI] [PubMed] [Google Scholar]
- 3.Snyder-Talkington B. N., Schwegler-Berry D., Castranova V., Qian Y., and Guo N. L., Part. Fibre Toxicol. 10, 35 (2013). 10.1186/1743-8977-10-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egawa N., Kitaoka S., Tsukita K., Naitoh M., Takahashi K., Yamamoto T., Adachi F., Kondo T., Okita K., and Asaka I., Sci. Transl. Med. 4, 145ra104 (2012). 10.1126/scitranslmed.3004052 [DOI] [PubMed] [Google Scholar]
- 5.Lee M. J., Ye A. S., Gardino A. K., Heijink A. M., Sorger P. K., MacBeath G., and Yaffe M. B., Cell 149, 780 (2012). 10.1016/j.cell.2012.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez-Rivas A., Gonzalez-Quijano G. K., Proa-Coronado S., Severac C., and Dague E., Micromachines 8, 347 (2017). 10.3390/mi8120347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mooney R., Haeger S., Lawal R., Mason M., Shrestha N., Laperle A., Bjugstad K., and Mahoney M., Tissue Eng. Part A 17, 2805 (2011). 10.1089/ten.tea.2010.0654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter S. B., Exp. Cell Res. 48, 189 (1967). 10.1016/0014-4827(67)90298-4 [DOI] [PubMed] [Google Scholar]
- 9.Jimbo Y., Robinson H. P., and Kawana A., IEEE Trans. Biomed. Eng. 40, 804 (1993). 10.1109/10.238465 [DOI] [PubMed] [Google Scholar]
- 10.Zheng Y., Dai W., Ryan D., and Wu H., Biomicrofluidics 4, 36504 (2010). 10.1063/1.3491474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright D., Rajalingam B., Selvarasah S., Dokmeci M. R., and Khademhosseini A., Lab Chip 7, 1272 (2007). 10.1039/b706081e [DOI] [PubMed] [Google Scholar]
- 12.Selvarasah S., Chao S. H., Chen C. L., Sridhar S., Busnaina A., Khademhosseini A., and Dokmeci M. R., Sens. Actuators A 145–146, 306 (2008). 10.1016/j.sna.2007.10.053 [DOI] [Google Scholar]
- 13.Wu J., Zhang M., Chen L., Yu V., Tin-Yum Wong J., Zhang X., Qin J., and Wen W., RSC Adv. 1, 746 (2011). 10.1039/c1ra00520k [DOI] [Google Scholar]
- 14.Liu Y., Du K., Wathuthanthri I., Xu W. and Choi C.-H., in 25th IEEE International Conference on Micro Electro Mechanical Systems (MEMS) (IEEE, 2012), p. 192. [Google Scholar]
- 15.Li W., Xu Z., Huang J., Lin X., Luo R., Chen C. H., and Shi P., Sci. Rep. 4, 4784 (2014). 10.1038/srep04784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaidi K. F. and Agrawal N., Biomicrofluidics 12, 064104 (2018). 10.1063/1.5061922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsieh C. H., Huang C. J., and Huang Y. Y., Biomed. Microdevices 12, 897 (2010). 10.1007/s10544-010-9444-z [DOI] [PubMed] [Google Scholar]
- 18.Messner J. J., Glenn H. L., and Meldrum D. R., BMC Biotechnol. 17, 89 (2017). 10.1186/s12896-017-0408-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang L., Chen Y., Chen Y., and Wu H., Anal. Chem. 87, 12169 (2015). 10.1021/acs.analchem.5b03031 [DOI] [PubMed] [Google Scholar]
- 20.Huang L., Chen Y., Weng L. T., Leung M., Xing X., Fan Z., and Wu H., Anal. Chem. 88, 12196 (2016). 10.1021/acs.analchem.6b03170 [DOI] [PubMed] [Google Scholar]
- 21.Xia J., Qiu Y., Xun X., Ma L., Guan J., and Su M., Anal. Chim. Acta 1007, 26 (2018). 10.1016/j.aca.2017.11.044 [DOI] [PubMed] [Google Scholar]
- 22.Fuentes-Prior P. and Salvesen G. S., Biochem. J. 384, 201 (2004). 10.1042/BJ20041142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nilsson J., Evander M., Hammarstrom B., and Laurell T., Anal. Chim. Acta 649, 141 (2009). 10.1016/j.aca.2009.07.017 [DOI] [PubMed] [Google Scholar]
- 24.Liu X., Shi Q., Lin Y., Kojima M., Mae Y., Huang Q., Fukuda T., and Arai T., Sensors 18, 2002 (2018). 10.3390/s18072002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petit T., Zhang L., Peyer K. E., Kratochvil B. E., and Nelson B. J., Nano Lett. 12, 156 (2011). 10.1021/nl2032487 [DOI] [PubMed] [Google Scholar]
- 26.Zhou Q., Petit T., Choi H., Nelson B. J., and Zhang L., Adv. Funct. Mater. 27, 1604571 (2017). 10.1002/adfm.201604571 [DOI] [Google Scholar]
- 27.Tasoglu S., Diller E., Guven S., Sitti M., and Demirci U., Nat. Commun. 5, 3124 (2014). 10.1038/ncomms4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Z., Fan X., Sun M., Gao C., He Q., and Xie H., ASC Nano 12, 2539 (2018). 10.1021/acsnano.7b08344 [DOI] [PubMed] [Google Scholar]
- 29.Liu W., Li L., Wang J. C., Tu Q., Ren L., Wang Y., and Wang J., Lab Chip 12, 1702 (2012). 10.1039/c2lc00034b [DOI] [PubMed] [Google Scholar]
- 30.Rettig J. R. and Folch A., Anal. Chem. 77, 5628 (2005). 10.1021/ac0505977 [DOI] [PubMed] [Google Scholar]
- 31.Delincé M. J., Bureau J.-B., López-Jiménez A. T., Cosson P., Soldati T., and McKinney J. D., Lab Chip 16, 3276 (2016). 10.1039/C6LC00649C [DOI] [PubMed] [Google Scholar]
- 32.Di Carlo D., Wu L. Y., and Lee L. P., Lab Chip 6, 1445 (2006). 10.1039/b605937f [DOI] [PubMed] [Google Scholar]
- 33.Tan W.-H. and Takeuchi S., Proc. Natl. Acad. Sci. U.S.A. 104, 1146 (2007). 10.1073/pnas.0606625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Tang X., Feng X., Liu C., Chen P., Chen D., and Liu B. F., Anal. Bioanal. Chem. 407, 1139 (2015). 10.1007/s00216-014-8325-3 [DOI] [PubMed] [Google Scholar]
- 35.Mi L., Huang L., Li J., Xu G., Wu Q., and Wang W., Lab Chip 16, 4507 (2016). 10.1039/C6LC01120A [DOI] [PubMed] [Google Scholar]
- 36.Liu Y., Ren D., Ling X., Liang W., Li J., You Z., Yalikun Y., and Tanaka Y., Sensors 18, 3672 (2018). 10.3390/s18113672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Y., Basu S., Wohlfahrt K. J., Lee S. F., Klenerman D., Laue E. D., and Seshia A. A., Sens. Actuators B 232, 680 (2016). 10.1016/j.snb.2016.03.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y., Austin R. H., and Sturm J. C., Biomicrofluidics 11, 054107 (2017). 10.1063/1.4985771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanahan D. and Weinberg R. A., Cell 144, 646 (2011). 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 40.Steeg P. S., Nat. Med. 12, 895 (2006). 10.1038/nm1469 [DOI] [PubMed] [Google Scholar]
- 41.Chen Y. C., Allen S. G., Ingram P. N., Buckanovich R., Merajver S. D., and Yoon E., Sci. Rep. 5, 9980 (2015). 10.1038/srep09980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma D., Wang R., Chen S., Luo T., Chow Y. T., and Sun D., Biomicrofluidics 12, 024118 (2018). 10.1063/1.5030135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng S., Mao S., Zhang Q., Li W., and Lin J. M., ACS Sens. 4, 521 (2019). 10.1021/acssensors.8b01696 [DOI] [PubMed] [Google Scholar]
- 44.Neuman K. C., Chadd E. H., Liou G. F., Bergman K., and Block S. M., Biophys. J. 77, 2856 (1999). 10.1016/S0006-3495(99)77117-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jing P., Wu J., and Lin L. Y., ACS Photonics 1, 398 (2014). 10.1021/ph500041m [DOI] [Google Scholar]
- 46.Jing P., Liu Y., Keeler E. G., Cruz N. M., Freedman B. S., and Lin L. Y., Biomed. Opt. Express 9, 771 (2018). 10.1364/BOE.9.000771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jordan P., Leach J., Padgett M., Blackburn P., Isaacs N., Goksor M., Hanstorp D., Wright A., Girkin J., and Cooper J., Lab Chip 5, 1224 (2005). 10.1039/b509218c [DOI] [PubMed] [Google Scholar]
- 48.Bin C., Kelbauskas L., Chan S., Shetty R. M., Smith D., and Meldrum D. R., Opt. Lasers Eng. 92, 70 (2017). 10.1016/j.optlaseng.2016.12.019 [DOI] [Google Scholar]
- 49.Liberale C., Cojoc G., Bragheri F., Minzioni P., Perozziello G., La Rocca R., Ferrara L., Rajamanickam V., Di Fabrizio E., and Cristiani I., Sci. Rep. 3, 1258 (2013). 10.1038/srep01258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magness A. J., Squires J. A., Griffiths B., Khan K., Swain A., Willison K. R., Cunningham D., Gerlinger M., and Klug D. R., Converg. Sci. Phys. Oncol. 3, 024003 (2017). 10.1088/2057-1739/aa6aae [DOI] [Google Scholar]
- 51.Gossett D. R., Weaver W. M., Mach A. J., Hur S. C., Tse H. T., Lee W., Amini H., and Di Carlo D., Anal. Bioanal. Chem. 397, 3249 (2010). 10.1007/s00216-010-3721-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iliescu C., Xu G., Tong W. H., Yu F., Bălan C. M., Tresset G., and Yu H., Microfluidics Nanofluidics 19, 363 (2015). 10.1007/s10404-015-1568-2 [DOI] [Google Scholar]
- 53.Yang Y., Mao Y., Shin K. S., Chui C. O., and Chiou P. Y., Sci. Rep. 6, 22630 (2016). 10.1038/srep22630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim S. H., Antfolk M., Kobayashi M., Kaneda S., Laurell T., and Fujii T., Lab Chip 15, 4356 (2015). 10.1039/C5LC01065A [DOI] [PubMed] [Google Scholar]
- 55.Pivetal J., Royet D., Ciuta G., Frenea-Robin M., Haddour N., Dempsey N. M., Dumas-Bouchiat F., and Simonet P., J. Magn. Magn. Mater. 380, 72 (2015). 10.1016/j.jmmm.2014.09.068 [DOI] [Google Scholar]
- 56.Shields C. W., Livingston C. E., Yellen B. B., Lopez G. P., and Murdoch D. M., Biomicrofluidics 8, 041101 (2014). 10.1063/1.4885840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun C., Hassanisaber H., Yu R., Ma S., Verbridge S. S., and Lu C., Sci. Rep. 6, 29407 (2016). 10.1038/srep29407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Collins D. J., Morahan B., Garcia-Bustos J., Doerig C., Plebanski M., and Neild A., Nat. Commun. 6, 8686 (2015). 10.1038/ncomms9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brugger M. S., Grundeen S., Doyle A., Theogarajan L., Wixforth A., and Westerhausen C., Phys. Rev. E 98, 012411 (2018). 10.1103/PhysRevE.98.012411 [DOI] [PubMed] [Google Scholar]
- 60.Zhu Z. and Yang C. J., Acc. Chem. Res. 50, 22 (2017). 10.1021/acs.accounts.6b00370 [DOI] [PubMed] [Google Scholar]
- 61.Lagus T. P. and Edd J. F., J. Phys. D: Appl. Phys. 46, 114005 (2013). 10.1088/0022-3727/46/11/114005 [DOI] [Google Scholar]
- 62.Seemann R., Brinkmann M., Pfohl T., and Herminghaus S., Rep. Prog. Phys. 75, 016601 (2012). 10.1088/0034-4885/75/1/016601 [DOI] [PubMed] [Google Scholar]
- 63.He M., Edgar J. S., Jeffries G. D., Lorenz R. M., Shelby J. P., and Chiu D. T., Anal. Chem. 77, 1539 (2005). 10.1021/ac0480850 [DOI] [PubMed] [Google Scholar]
- 64.Tan Y. C., Hettiarachchi K., Siu M., Pan Y.-R., and Lee A. P., J. Am. Chem. Soc. 128, 5656 (2006). 10.1021/ja056641h [DOI] [PubMed] [Google Scholar]
- 65.Luo D., Pullela S. R., Marquez M., and Cheng Z., Biomicrofluidics 1, 034102 (2007). 10.1063/1.2757156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsu M. N., Wei S. C., Guo S., Phan D. T., Zhang Y., and Chen C. H., Small 14, e1802918 (2018). 10.1002/smll.201802918 [DOI] [PubMed] [Google Scholar]
- 67.Zimny P., Juncker D., and Reisner W., Biomicrofluidics 12, 024107 (2018). 10.1063/1.5020571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hou Y., Xie W., Achazi K., Cuellar-Camacho J. L., Melzig M. F., Chen W., and Haag R., Acta Biomater. 77, 28 (2018). 10.1016/j.actbio.2018.07.003 [DOI] [PubMed] [Google Scholar]
- 69.Thompson A. M., Paguirigan A. L., Kreutz J. E., Radich J. P., and Chiu D. T., Lab Chip 14, 3135 (2014). 10.1039/C4LC00175C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Collins D. J., Neild A., deMello A., Liu A. Q., and Ai Y., Lab Chip 15, 3439 (2015). 10.1039/C5LC00614G [DOI] [PubMed] [Google Scholar]
- 71.Baret J. C., Miller O. J., Taly V., Ryckelynck M., El-Harrak A., Frenz L., Rick C., Samuels M. L., Hutchison J. B., Agresti J. J., Link D. R., Weitz D. A., and Griffiths A. D., Lab Chip 9, 1850 (2009). 10.1039/b902504a [DOI] [PubMed] [Google Scholar]
- 72.Link D. R., Grasland-Mongrain E., Duri A., Sarrazin F., Cheng Z., Cristobal G., Marquez M., and Weitz D. A., Angew. Chem. Int. Ed. Engl. 45, 2556 (2006). 10.1002/anie.200503540 [DOI] [PubMed] [Google Scholar]
- 73.Ahn K., Kerbage C., Hunt T. P., Westervelt R. M., Link D. R., and Weitz D. A., Appl. Phys. Lett. 88, 024104 (2006). 10.1063/1.2164911 [DOI] [Google Scholar]
- 74.Baroud C. N., Delville J.-P., Gallaire F., and Wunenburger R., Phys. Rev. E 75, 046302 (2007). 10.1103/PhysRevE.75.046302 [DOI] [PubMed] [Google Scholar]
- 75.Navi M., Abbasi N., Jeyhani M., Gnyawali V., and Tsai S. S. H., Lab Chip 18, 3361 (2018). 10.1039/C8LC00867A [DOI] [PubMed] [Google Scholar]
- 76.Rakszewska A., Tel J., Chokkalingam V., and Huck W. T. S., NPG Asia Mater. 6, e133 (2014). 10.1038/am.2014.86 [DOI] [Google Scholar]
- 77.Luo T., Fan L., Zhu R., and Sun D., Micromachines 10, 104 (2019). 10.3390/mi10020104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Macosko E. Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A. R., Kamitaki N., Martersteck E. M., Trombetta J. J., Weitz D. A., Sanes J. R., Shalek A. K., Regev A., and McCarroll S. A., Cell 161, 1202 (2015). 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klein A. M., Mazutis L., Akartuna L., Tallapragada N., Veres A., Li V., Peshkin L., Weitz D. A., and Kirschner M. W., Cell 161, 1187 (2015). 10.1016/j.cell.2015.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moon H. S., Je K., Min J. W., Park D., Han K. Y., Shin S. H., Park W. Y., Yoo C. E., and Kim S. H., Lab Chip 18, 775 (2018). 10.1039/C7LC01284E [DOI] [PubMed] [Google Scholar]
- 81.Ng E. X., Miller M. A., Jing T., and Chen C. H., Biosens. Bioelectron. 81, 408 (2016). 10.1016/j.bios.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 82.Shahi P., Kim S. C., Haliburton J. R., Gartner Z. J., and Abate A. R., Sci. Rep. 7, 44447 (2017). 10.1038/srep44447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ayibaike D., Cui M., and Wei J., Appl. Sci. 8, 2152 (2018). 10.3390/app8112152 [DOI] [Google Scholar]
- 84.Yoshida S., Kato-Negishi M., and Takeuchi S., Micromachines 9, 235 (2018). 10.3390/mi9050235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu B. and Maharbiz M. M., PLoS One 13, e0195062 (2018). 10.1371/journal.pone.0195062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xia Y. and Whitesides G. M., Annu. Rev. Mater. Sci. 28, 153 (1998). 10.1146/annurev.matsci.28.1.153 [DOI] [Google Scholar]
- 87.Khadpekar A. J., Khan M., Sose A., and Majumder A., Sci. Rep. 9, 1024 (2019). 10.1038/s41598-018-36521-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu H., Wu L., Zhou X., Liu B., and Zheng B., Small 14, e1802128 (2018). 10.1002/smll.201802128 [DOI] [PubMed] [Google Scholar]
- 89.Foncy J., Estève A., Degache A., Colin C., Cau J. C., Malaquin L., Vieu C., and Trévisiol E., Methods Mol. Biol. 1771, 83 (2018). 10.1007/978-1-4939-7792-5_7 [DOI] [PubMed] [Google Scholar]
- 90.Yao X., Liu R., Liang X., and Ding J., ACS Appl. Mater. Interfaces 11, 15366 (2019). 10.1021/acsami.9b03780 [DOI] [PubMed] [Google Scholar]
- 91.Yamamoto H., Matsumura R., Takaoki H., Katsurabayashi S., Hirano-Iwata A., and Niwano M., Appl. Phys. Lett. 109, 043703 (2016). 10.1063/1.4959836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li H.-W., Muir B. V., Fichet G., and Huck W. T., Langmuir 19, 1963 (2003). 10.1021/la0269098 [DOI] [Google Scholar]
- 93.Rozkiewicz D. I., Brugman W., Kerkhoven R. M., Ravoo B. J., and Reinhoudt D. N., J. Am. Chem. Soc. 129, 11593 (2007). 10.1021/ja073574d [DOI] [PubMed] [Google Scholar]
- 94.Rozkiewicz D. I., Gierlich J., Burley G. A., Gutsmiedl K., Carell T., Ravoo B. J., and Reinhoudt D. N., ChemBioChem 8, 1997 (2007). 10.1002/cbic.200700402 [DOI] [PubMed] [Google Scholar]
- 95.Mrksich M., Dike L. E., Tien J., Ingber D. E., and Whitesides G. M., Exp. Cell Res. 235, 305 (1997). 10.1006/excr.1997.3668 [DOI] [PubMed] [Google Scholar]
- 96.Escalante M., Zhao Y., Ludden M. J., Vermeij R., Olsen J. D., Berenschot E., Hunter C. N., Huskens J., Subramaniam V., and Otto C., J. Am. Chem. Soc. 130, 8892 (2008). 10.1021/ja802843m [DOI] [PubMed] [Google Scholar]
- 97.Escalante M., Maury P., Bruinink C. M., van der Werf K., Olsen J. D., Timney J. A., Huskens J., Hunter C. N., Subramaniam V., and Otto C., Nanotechnology 19, 025101 (2007). 10.1088/0957-4484/19/02/025101 [DOI] [PubMed] [Google Scholar]
- 98.Piner R. D., Zhu J., Xu F., Hong S., and Mirkin C. A., Science 283, 661 (1999). 10.1126/science.283.5402.661 [DOI] [PubMed] [Google Scholar]
- 99.Senesi A. J., Rozkiewicz D. I., Reinhoudt D. N., and Mirkin C. A., ACS Nano 3, 2394 (2009). 10.1021/nn9005945 [DOI] [PubMed] [Google Scholar]
- 100.Salaita K., Lee S. W., Wang X., Huang L., Dellinger T. M., Liu C., and Mirkin C. A., Small 1, 940 (2005). 10.1002/smll.200500202 [DOI] [PubMed] [Google Scholar]
- 101.Salaita K., Wang Y., Fragala J., Vega R. A., Liu C., and Mirkin C. A., Angew. Chem. Int. Ed. 45, 7099 (2006). 10.1002/anie.200690148 [DOI] [PubMed] [Google Scholar]
- 102.Ma H., Jiang Z., Xie X., Huang L., and Huang W., ACS Appl. Mater. Interfaces 10, 25121 (2018). 10.1021/acsami.8b07369 [DOI] [PubMed] [Google Scholar]
- 103.Curran J. M., Stokes R., Irvine E., Graham D., Amro N. A., Sanedrin R. G., Jamil H., and Hunt J. A., Lab Chip 10, 1662 (2010). 10.1039/C004149A [DOI] [PubMed] [Google Scholar]
- 104.Laing S., Suriano R., Lamprou D. A., Smith C. A., Dalby M. J., Mabbott S., Faulds K., and Graham D., ACS Appl. Mater. Interfaces 8, 24844 (2016). 10.1021/acsami.6b03860 [DOI] [PubMed] [Google Scholar]
- 105.Huo F., Zheng Z., Zheng G., Giam L. R., Zhang H., and Mirkin C. A., Science 321, 1658 (2008). 10.1126/science.1162193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Giam L. R. and Mirkin C. A., Angew. Chem. Int. Ed. Engl. 50, 7482 (2011). 10.1002/anie.201100839 [DOI] [PubMed] [Google Scholar]
- 107.Zheng Z., Daniel W. L., Giam L. R., Huo F., Senesi A. J., Zheng G., and Mirkin C. A., Angew. Chem. Int. Ed. Engl. 48, 7626 (2009). 10.1002/anie.200902649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liao X., Braunschweig A. B., Zheng Z., and Mirkin C. A., Small 6, 1082 (2010). 10.1002/smll.200901538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kumar R., Urtizberea A., Ghosh S., Bog U., Rainer Q., Lenhert S., Fuchs H., and Hirtz M., Langmuir 33, 8739 (2017). 10.1021/acs.langmuir.7b01368 [DOI] [PubMed] [Google Scholar]
- 110.Giam L. R., Massich M. D., Hao L., Shin Wong L., Mader C. C., and Mirkin C. A., Proc. Natl. Acad. Sci. U.S.A. 109, 4377 (2012). 10.1073/pnas.1201086109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu X., Li Y., and Zheng Z., Nanoscale 2, 2614 (2010). 10.1039/c0nr00565g [DOI] [PubMed] [Google Scholar]
- 112.Zhou X., Liu Z., Xie Z., Liu X., and Zheng Z., Small 8, 3568 (2012). 10.1002/smll.201201544 [DOI] [PubMed] [Google Scholar]
- 113.Chen C., Zhou X., Xie Z., Gao T., and Zheng Z., Small 11, 613 (2015). 10.1002/smll.201400642 [DOI] [PubMed] [Google Scholar]
- 114.Chen L., Xie Z., Gan T., Wang Y., Zhang G., Mirkin C. A., and Zheng Z., Small 12, 3400 (2016). 10.1002/smll.201600634 [DOI] [PubMed] [Google Scholar]