Abstract
Hereditary hemorrhagic telangiectasia (HHT) is a Mendelian disease characterized by vascular malformations (VMs) including visceral arteriovenous malformations and mucosal telangiectasia. HHT is caused by loss-of-function (LoF) mutations in one of three genes, ENG, ACVRL1, or SMAD4, and is inherited as an autosomal-dominant condition. Intriguingly, the constitutional mutation causing HHT is present throughout the body, yet the multiple VMs in individuals with HHT occur focally, rather than manifesting as a systemic vascular defect. This disconnect between genotype and phenotype suggests that a local event is necessary for the development of VMs. We investigated the hypothesis that local somatic mutations seed the formation HHT-related telangiectasia in a genetic two-hit mechanism. We identified low-frequency somatic mutations in 9/19 telangiectasia through the use of next-generation sequencing. We established phase for seven of nine samples, which confirms that the germline and somatic mutations in all seven samples exist in trans configuration; this is consistent with a genetic two-hit mechanism. These combined data suggest that bi-allelic loss of ENG or ACVRL1 may be a required event in the development of telangiectasia, and that rather than haploinsufficiency, VMs in HHT are caused by a Knudsonian two-hit mechanism.
Keywords: HHT, hereditary hemorrhagic telangiectasia, somatic mutation, mosaicism, two-hit, biallelic, bi-allelic, ENG, ACVRL1
Introduction
Hereditary hemorrhagic telangiectasia (HHT [MIM: 187300, 600376, 175050]) is a Mendelian disease characterized by the development of multiple focal vascular malformations (VMs) consisting of arteriovenous malformations in visceral organs and telangiectasia in mucosal and cutaneous tissue. The genetic etiology of HHT has been established and is caused by mutations in ENG1 (MIM: 131195), ACVRL12 (MIM: 601284), and rarely SMAD43 (MIM: 600993); all of these follow an autosomal-dominant inheritance pattern. Despite our understanding of the genetics of and downstream pathways involved in HHT, the molecular mechanisms that initiate HHT-related VM are poorly understood. Early studies of the functional consequences of HHT causal mutations established that these result in the LoF of the gene product. These findings, in combination with autosomal-dominant inheritance, led to the presumption that VMs result from haploinsufficiency of the mutated gene product.4, 5, 6 However, haploinsufficiency does not explain why HHT-related VMs occur as strictly focal lesions, despite the systemic presence of the causal germline mutation. This disconnect between genotype and phenotype led to an alternative long-standing hypothesis that HHT-related VMs result from a Knudsonian two-hit mechanism in which a local somatic mutation in the wild-type (WT) allele of the affected gene seeds the formation of focal lesions.
The only published study which directly addresses the two-hit hypothesis attempted to determine whether Endoglin was present on the endothelial lining of arteriovenous malformations from an individual with HHT with a causal mutation in the corresponding gene ENG.7 Endoglin immunostaining was visible in the vessel lining, albeit at low levels. The presence of Endoglin in HHT-associated VMs would contradict the hypothesis of a two-hit mechanism; however, complete loss of staining might not be predicted to occur, especially with the heterogeneous—and potentially mosaic—tissue of an arteriovenous malformation that may only contain a minority of cells that harbor the somatic mutation. Previous attempts to address this hypothesis at the DNA level have been hampered by the limitations of past sequencing technology. The advent of next-generation sequencing has drastically increased our sensitivity for detecting low-frequency somatic mutations. Somatic mutations have been identified in a diverse array of VMs8, 9, 10, 11, 12, 13, 14 including recent evidence that sporadic arteriovenous malformations, which are not associated with HHT, harbor somatic activating mutations in KRAS (MIM: 190070) or MAP2K1 (MIM: 176872).15, 16 Notably, a genetic two-hit mechanism is known to contribute to cerebral cavernous malformations (CCM [MIM: 116860])17, 18, 19 and Capillary Malformation-Arteriovenous Malformation Syndrome (CM-AVM [MIM: 608354]);20 like HHT, both diseases are caused by autosomal-dominant LoF mutations. Here we demonstrate that HHT-related telangiectasia contain bi-allelic mutations in ENG or ACVRL1, resulting in homozygous LoF; evidence in support of the long-standing hypothesis that telangiectasia pathogenesis follows a genetic two-hit mechanism.
Material and Methods
Sample Collection
Individuals were enrolled in the study after giving informed consent (approved by either the St. Michael’s Hospital Institutional Review Board [IRB] Committee or the Duke University Health System IRB Committee). Diagnosis of HHT was based on identification of a pathogenic germline mutation or on the patient exhibiting at least three of the four symptoms as per the Curaçao criteria (Table S1).21
Telangiectasia were resected using a 3 mm punch biopsy, after local anesthesia (1% xylocaine with epinephrine), with standard aseptic technique. Sample 6005-1 was immediately formalin fixed (10% formalin) and paraffin embedded (FFPE) and then shipped at room temperature. All other samples were immediately frozen at −80° Celsius and then shipped on dry ice. Saliva samples were obtained using Oragene DNA saliva kits at the time of tissue collection. Blood from individual 6003 was obtained during a subsequent visit, shipped at room temperature, and immediately used for RNA extraction.
DNA and RNA Extraction
DNA from telangiectasia samples was extracted using the DNeasy Blood and Tissue Kit (QIAGEN). DNA from FFPE sample 6005-1 was extracted using QIAamp DNA FFPE Tissue Kit (QIAGEN). Genomic DNA and RNA were extracted from peripheral blood leukocytes from individual 6003 and from a non-HHT control individual through the use of the Gentra PureGene Blood Kit (QIAGEN) and TRIzol Reagent (Invitrogen) extraction protocols, respectively, as per the manufacturers’ directions.
Targeted Sequencing
To enable the detection of somatic mutations in telangiectasia, we used a next-generation sequencing strategy. Somatic mutations involved in the pathogenesis of other VM diseases such as cerebral cavernous malformations often have a low allele frequency due to somatic mosaicism in the malformation. Somatic mosaicism is also present in retinal AVMs from mouse models of HHT; this suggests that low allele frequency may be a confounder when identifying somatic variants in telangiectasia. In addition, the telangiectasia samples collected for this study consisted of bulk biopsied tissue which had not been enriched for any particular cell type. Considering these potential sources of normal (non-mutant) cell contamination, we sequenced telangiectasia to >1000× coverage and incorporated a unique molecular identifier to enable the detection of variants as low as 0.1% allele frequency.
Eighteen fresh-frozen samples and one FFPE sample were sequenced using a custom Agilent SureSelect panel covering 16 genes implicated in various VM disorders: ENG (MIM: 131195), ACVRL1 (MIM: 601284), SMAD4 (MIM: 600993), BRAF (MIM: 164757), CCM2 (MIM: 607929), FLT1 (MIM: 165070), FLT4 (MIM: 136352), GNAQ (MIM: 600998), KDR (MIM: 191306), KRAS (MIM: 190070), KRIT1 (MIM: 604214), MAP2K1 (MIM: 176872), NRAS (MIM: 164790), PDCD10 (MIM: 607118), PIK3CA (MIM: 171834), and PTEN (MIM: 601728). Though GDF2 variants have been identified in an HHT-like phenotype, these cases are extremely rare. Moreover, mutations in GDF2 have not been identified in any individual at the Toronto HHT Centre for Excellence and therefore GDF2 was not included in the panel. To ensure the generation of high-quality sequencing libraries, the Agilent NGS FFPE QC Kit was used to determine the extent of DNA degradation in the FFPE sample. Samples with ΔΔCq values >2 were excluded from the study as per manufacturer recommendation. Sequencing libraries were generated using the Agilent SureSelect XT HS Kit. Samples were then pooled and sequenced on an iSeq 100 (Illumina) with paired-end 150bp reads. Across all samples, target regions were sequenced to a mean depth of 2,803× with 78% of the target region at >1,000× and 96% of the target region at >100×.
Mutation Detection
Sequencing data were processed and analyzed using a custom pipeline based on the Genome Analysis Toolkit (GATK) best practices for somatic short variant discovery. In brief, after analyzing the raw data with fastQC to ensure high-quality data, we trimmed the adaptor sequences from reads through the use of bbduk, we aligned reads to the hg19 human reference genome through the use of bowtie2, we removed duplicates based on UMI sequence through the use of fgbio, we called variants through the use of MuTect2 in tumor-only mode, and we annotated variants through the use of snpSift. The resulting variant call file (VCF) was filtered through several steps to identify somatic mutations. To identify variants that may change the protein sequence or impact splicing, we selected for variants that occur within exons or within 10bp of an exon. From this set, we removed variants that are present in the population at >0.01% frequency by comparing the set to three databases (dbSNP, the 1000 Genomes Project, and the Exome Aggregation Consortium [ExAC]) because these variants are more common than the frequency of HHT (1–5 in 10,000).22 We removed any variants present in <0.02% of sequence reads because this is the reported technical limit of detection for the SureSelect XT technology. We also removed variants in regions with <100× coverage, variants with <5 supporting reads, variants that were strand specific, and variants where <50% of alternative bases had a quality score >30 (>Q30).
Candidate somatic variants identified in the targeted sequencing data were then validated by sequencing amplicons generated during a second, independent round of polymerase chain reaction (PCR) amplification. We designed primers for each sample to specifically amplify the position of the somatic mutation and 100–200bp of flanking sequence. When possible, the primers were designed such that they would capture the position of both the germline and somatic mutations within a single amplicon. Each primer was synthesized with the following Illumina flow cell adaptor sequences such that the amplicons could be easily indexed and sequenced: Forward 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-[sequence-specific primer]-3′, Reverse 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-[sequence-specific primer]-3′. Amplicons were prepared from telangiectasia DNA and constitutional DNA if available (Table S1) through the use of two rounds of PCR. The first round of PCR was 25 cycles and served to amplify the target region from genomic DNA. Amplicons from the first round were purified using AMPure XP beads (Beckman Coulter) and used for a second round of PCR using eight cycles to attach a sample index through the use of the Nextera XT Index Kit. Amplicons from the second round were purified, pooled, and sequenced on an iSeq 100 with 150bp paired-end reads to a depth of >10,000×. The frequency of the somatic mutation in telangiectasia and constitutional DNA was determined through the use of custom scripts, excluding bases <Q15.
As these amplicons were sequenced in the same run, it is possible for a low level of index misassignment to cause switching of reads between samples. This could cause some reads from telangiectasia to be assigned as constitutional reads and vice versa. To estimate the rate of index misassignment, we examined pairs of samples that targeted different genomic locations and quantified the proportion of misassigned reads between these samples. Based on this, we estimate that the rate of misassignment is 0.2–0.8% relative to the sample of origin. For example, if one sample had a somatic mutation with a frequency of 1% and was sequenced to 100,000× coverage, then 2–8 reads containing the somatic mutation would be misassigned to each of the other samples in the pool.
Establishing Phase
To establish the phase of the somatic and germline mutations, depending on the distance between the two mutations, we used either short-read sequencing with Illumina chemistry or long-read sequencing with PacBio chemistry. For mutations <500bp apart, during the validation process, we generated amplicons that covered the positions of both the somatic and the germline mutations. These amplicons were sequenced on an iSeq 100 as described above.
For mutations >500bp, we designed primers such that the resulting amplicon would cover the position of both the germline and the somatic mutations for each sample. Each primer was synthesized with the PacBio “universal tag” as follows to enable indexing and sequencing: Forward 5′- /5AmMC6/ GCAGTCGAACATGTAGCTGACTCAGGTCAC-[sequence-specific primer]-3′, Reverse 5′- /5AmMC6/ TGGATCACTTGTGCAAGCATCACATCGTAG-[sequence-specific primer]-3′. We generated amplicons spanning the somatic and the germline mutations through the use of the LongAmp Taq DNA Polymerase Kit as per manufacturer instructions. These amplicons were purified with AMPure XP beads and used for a second round of PCR to attach the sample index. This process used no more than 30 cycles of PCR total. These amplicons were pooled and sequenced across one SMRT cell on a PacBio Sequel System. The sequence reads were aligned to the hg19 human genome through the use of Minimap2 in ava-pb mode.
The single-molecule resolution of these technologies allowed us to determine how the mutant alleles are arranged: if the mutations are in trans, then reads will have either the somatic mutant allele or the germline mutant allele; if the mutations are in cis, then reads will have either no mutant alleles or both mutant alleles. In total, we generated mutation-spanning reads for seven telangiectasia, each with >100 reads which contained the somatic mutation. The p values reported for phase status were calculated using a binomial distribution with the null hypothesis that a random mutation has an equal probability of cis or trans configuration with a nearby variant.
The genomic distance between mutations varied greatly; the closest mutations were in sample 6005-1 with 26 bases between mutations, and the most distant were in 6001-10 with 18.7 kilobases between mutations. Of the nine telangiectasia with identified somatic mutations, the distance between mutations in 6002-2, 6003-1, and 6005-1 was small enough to allow for mutation-spanning reads using illumina chemistry (Table 2). Mutation-spanning reads were generated for 6001-3, 6001-7, 6001-8, and 6002-1 using PacBio chemistry. We were unable to generate amplicons spanning the mutations for 6001-1 and 6001-10. The sequence downstream of the somatic mutation in these telangiectasia contains several repetitive regions which, combined with the genomic distance and limited quantity of input DNA, may have contributed to PCR failure.
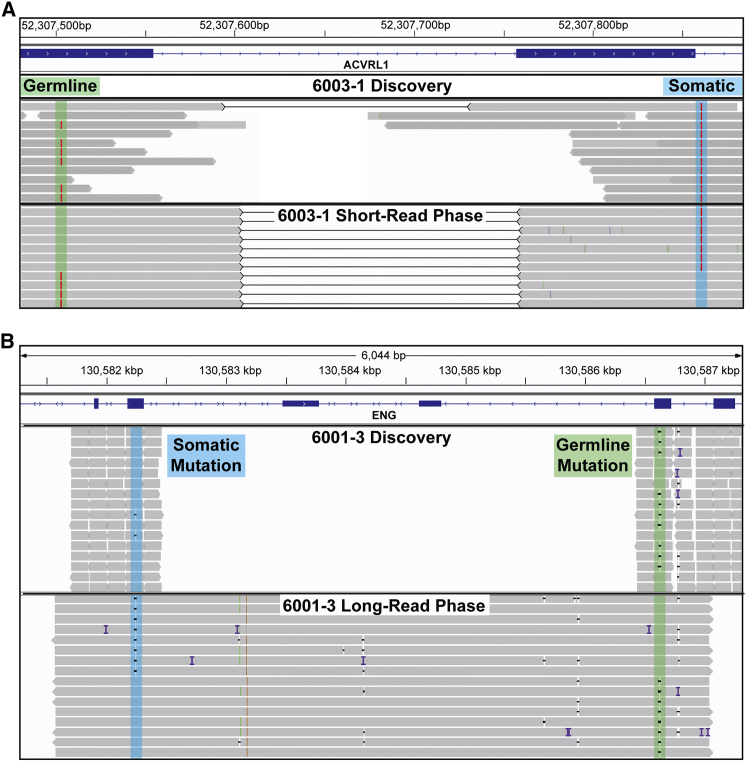
Figure 2.
Somatic and Germline Mutations are Bi-allelic
Integrative Genomics Viewer (IGV) visualization of two samples showing both methods of establishing phase. Each panel shows reads from the initial discovery sequencing and the reads used to establish phase.
(A) Somatic and germline mutations in 6003-1 are both A>T point mutations and highlighted by the blue and green regions respectively. Because the distance between these mutations is relatively small (357bp), phase was established using Illumina short-reads (also used for validation). Black lines between reads denote read pairs, showing that both reads originate from a single molecule of DNA. Each molecule with the somatic mutation contains the wild-type (WT) allele at the germline mutation position; this proves that the mutations are bi-allelic.
(B) Somatic and germline mutations in sample 6001-3, both small deletions. The genomic distance between these mutations is 4377bp. In long reads that span the two mutations, each read with the somatic mutation contains the WT allele at the position of the germline mutation.
One notable confounder in this analysis is the generation of chimeric reads resulting from template switching during PCR. The generation of chimeric reads is known to interfere with amplicon-based haplotype phasing by switching a variant from one strand to another, potentially generating new haplotypes not present in the original sample.23 In practice, chimeric reads randomize the arrangement of the somatic and germline mutations. The frequency of chimeric arrangements is highly dependent on the distance between mutations and the number of PCR cycles used to make the amplicons. To reduce the number of chimeric reads in our libraries, we used no more than 30 cycles for amplification. A previous study reports a chimeric arrangement frequency of 6.5% for 29 cycles of amplification for mutations 9 kbp apart.23 Chimeric reads may account for the very few discordant reads in some of our samples, as shown in Table 2. Nonetheless, these were so minor in comparison to the great majority of the reads that phase could be unequivocally determined.
in vitro Splicing
A 3.8 kb fragment of ACVRL1 genomic DNA spanning from 431 bases upstream of exon 3 to 215 bases downstream of exon 8 was amplified, and this entire insert was ligated into the MCS of pSPL3, a splicing vector.24 Clones were sequenced to ensure that no PCR-induced errors were present in the exons and adjacent intronic regions of the insert. The specific mutation, c.625+4A>T, was introduced using site-directed mutagenesis, and again clones were sequenced to verify that the only sequence difference was at the intended site. Plasmid DNA from empty vector, WT (control) vector, and mutation-containing vector were transfected into HEK293T cells through the use of Lipofectamine 3000 (ThermoFisher Scientific) and incubated for 24 h, and then the RNA was extracted using TRIzol and Direct-zol RNA miniprep kit (Zymo Research).
Reverse-Transcription PCR
RNA extracted from peripheral blood leukocytes and from transfected cells was used as template for cDNA synthesis using the Maxima H Minus First Strand cDNA kit (ThermoFisher Scientific). A reverse transcription (RT) primer in ACVRL1 exon 8 was used in the RNA from blood, and a vector-specific RT primer was used for the RNA from transfected cells to ensure that only RNA from the transfected vectors was being used as template for cDNA synthesis. cDNA from peripheral blood leukocytes was PCR amplified using primers in exons 3 and 6, and cDNA from the transfected cells was PCR amplified using primers in exons 3 and 8. PCR reactions were run on 1% agarose gels, and cDNA extracted from excised bands was Sanger sequenced.
Results
To determine whether a genetic two-hit mechanism underlies HHT pathogenesis, we tested three underlying expectations of the two-hit mechanism: (1) telangiectasia contain a somatic mutation in the same gene as a germline mutation which causes HHT, (2) the somatic and germline mutations are bi-allelic, and (3) both mutations result in LoF.
Telangiectasia Harbor a Somatic Mutation in ENG or ACVRL1
We used capture-based library preparations to sequence 19 telangiectasia for the three genes mutated in HHT (ENG, ACVRL1, and SMAD4) and 13 other VM-related genes (see Material and Methods for the identities of genes). These 13 additional genes were chosen in part due to the possibility that they also might harbor somatic mutations, but primarily to serve as control genes because the two-hit mechanism requires a mutation in the corresponding HHT-associated gene harboring the causal germline mutation. Somatic mutations in these other genes may or may not contribute to HHT pathogenesis, but absence of a somatic mutation in the HHT-associated gene would violate the first expectation of the genetic two-hit mechanism.
In each telangiectasia, we identified a pathogenic germline mutation in either ENG or ACVRL1. Although in most cases the individuals’ germline mutation was already known from clinical diagnostic sequencing, we intentionally remained blinded to this information until after our own sequence analysis of the tissue samples. In 6003-1, the individual harbors a silent germline mutation which was found by the clinical lab and noted as a variant of unknown significance (VUS). Below, we show that this variant is indeed the pathogenic germline variant in this individual.
We used the MuTect2 variant caller to detect variants present in the sequence data. To identify candidate somatic mutations, we removed variants on the basis of several stringent filtering criteria including (in brief): intronic or intergenic variants; population frequency >0.01%; <0.1% reads with alternate allele; <5 total supporting reads; low coverage; strand specificity; and low base quality scores.
To validate or refute the authenticity of each candidate somatic mutation, we performed an independent round of amplification using primers flanking each putative variant position for each sample, and we sequenced the resulting amplicons to >10,000× coverage. In each tissue sample, the identical somatic variant was re-identified. Thus, these variants were bona fide somatic mutations existing in the telangiectatic tissue. In total, we identified somatic variants in nine of 19 telangiectasia: five in ENG (RefSeq accession number NM_001114753.1 [ENG_v001]) and four in ACVRL1 (RefSeq NM_000020.2 [ACVRL1_v001]) (Table 1) (Figure 1) (see Material and Methods). In each case, the somatic mutation was found in the same gene as the pathogenic germline mutation. Somatic mutations were not found in any of the other 15 genes sequenced, not even in any of the other HHT-associated genes. Importantly, no telangiectasia harbored more than a single somatic mutation. The lack of mutational noise suggests these mutations are pathobiologically significant. Importantly, all are consistent with LoF; five of the variants are small indels that result in a frameshift, three are in-frame indels, and one is a point mutation four bases after an exon-intron boundary that is predicted to impact RNA splicing.
Table 1.
Summary of Somatic Mutation Discovery and Validation Sequencing
| Sample ID | Germline Mutation | Somatic Mutation | Discovery Readsa | Validation Readsa | Constitutional Readsa |
|---|---|---|---|---|---|
| 6001-1 |
ENG c.1080_1083delGACA p.Thr361Serfs∗7 |
ENG c.293_304del p.Val98_Asn102delinsAsp |
33/1318 (2.5%) | 1067/100268 (1.1%) | 0/26462 (0%)b |
| 6001-3 | same as above |
ENG c.1195_1196del p.Arg399Glyfs∗2 |
5/1080 (0.46%) | 723/115963 (0.62%) | 0/24357 (0%)b |
| 6001-7 | same as above |
ENG c.1237_1238insCA p.Gly413Alafs∗9 |
27/5127 (0.53%) | 341/115570 (0.30%) | 0/23066 (0%)b |
| 6001-8 | same as above |
ENG c.578delinsTGCG p.Thr193delinsMetArg |
111/4845 (2.3%) | 1142/142572 (0.80%) | 0/21315 (0%)b |
| 6001-10 | same as above |
ENG c.205del p.Leu69Trpfs∗12 |
33/3389 (1.0%) | 3575/326894 (1.1%) | 0/22098 (0%)b |
| 6001-2,4,5,6,9,11,12,13 | same as above | NF | - | - | - |
| 6002-1 | ACVRL1 c.1451G>A p.Arg484Gln |
ACVRL1 c.349delinsTT p.Gly117Leufs∗52 |
20/2217 (0.90%) | 309/24018 (1.3%) | 0/65818 (0%) |
| 6002-2 | same as above |
ACVRL1 c.[1378-3del; 1381_1398del; 1402A>C]c p.[Leu461_Gln466del; Met468Leu]c |
26/1649 (1.6%) | 3189/202550 (1.6%) | 6/155855 (0.0038%) |
| 6003-1 |
ACVRL1 c.474A>T p.Gly158 = |
ACVRL1 c.625+4A>T | 101/3392 (3.0%) | 372/16303 (2.3%) | 2/38924 (0.0051%) |
| 6004-1 |
ACVRL1 c.1232G>A p.Arg411Gln |
NF | - | - | - |
| 6004-2 | same as above | NF | - | - | - |
| 6005-1 |
ACVRL1 c.1232G>A p.Arg411Gln |
ACVRL1 c.1206del p.Leu403Trpfs∗12 |
133/1664 (8.0%) | 2671/189690 (1.4%) | N/A |
ENG: RefSeq accession number NM_001114753.1 (ENG_v001)
ACVRL1: RefSeq NM_000020.2 (ACVRL1_v001)
For multiple telangiectasia collected from one individual, the sample ID is listed as (Individual#)-(Telangiectasia#)
NF = None Found
Only bases >Q15 included
Allele frequency in other telangiectasia from 6001
Components of a single complex mutation
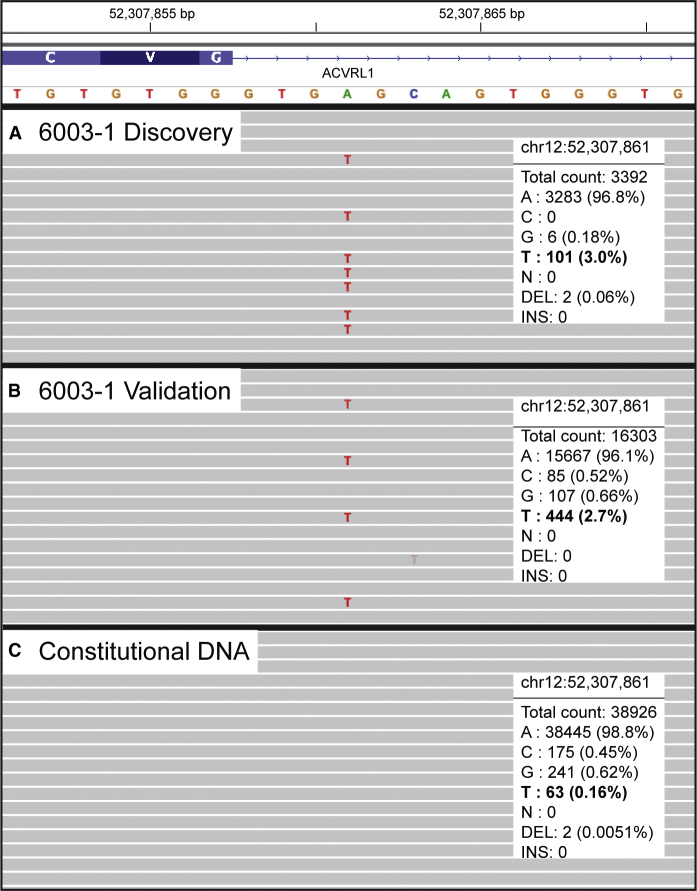
Figure 1.
Low-Frequency Somatic Mutations Detected in Telangiectasia
Integrative Genomics Viewer (IGV) visualization of next-generation sequencing data for one representative sample with a somatic mutation in ACVRL1. The somatic mutation in 6003-1, an A>T point mutation, is present at low frequency in DNA from telangiectatic tissue used for (A) discovery (capture-based sequencing) and (B) validation (amplicon-based sequencing). The mutation is below the level of sequencing noise in (C) constitutional DNA (amplicon-based sequencing); this confirms that the mutation is somatic.
Although these variants fall well below the 50% allele frequency expected for germline variants, it is formally possible that these variants exist constitutionally as very rare, somatic mosaic variation in the individual. We next investigated whether these somatic mutations were present in constitutional DNA from the individual. A source of constitutional DNA (saliva) was available for three of the nine mutation-positive samples, and in each, we found that the somatic mutation was completely absent or present at a level no higher than technical sequencing noise in that sample (see Material and Methods). Saliva was not available for individual 6001, but we obtained and sequenced DNA from multiple telangiectasia collected from this same individual. This enabled us to determine whether any of the five somatic mutations that we identified in individual samples was present in tissue of near identical pathobiology from the same individual; compared with saliva as a control, this is a more powerful test for somatic mosaicism. We found that the somatic mutations identified in five of the telangiectasia for which we identified a mutation were entirely absent in all other telangiectasia from this same individual. Finally, sample 6005-1 is a single archived FFPE telangiectasia, and no source of constitutional DNA is available. In total, we found that nine of 19 telangiectasia harbor a somatic mutation specifically in the same gene as a pathogenic germline mutation and that these mutations are not present constitutionally. The presence of somatic mutations in telangiectasia fulfills the first expectation of the genetic two-hit mechanism.
Somatic and Germline Mutations are Bi-allelic
The second expectation of the genetic two-hit mechanism is that the somatic and germline mutations are bi-allelic; the somatic mutation occurs on the WT allele of the affected gene in trans with the germline mutation. To determine if the mutations are bi-allelic, we examined whether they were arranged in a cis or trans configuration by sequencing amplicons that cover the nucleotide positions of both somatic and germline mutations in a single molecule. The amplicons were sequenced with either short-read (Illumina) or long-read (PacBio) chemistry, depending on the amplicon size, in order to generate reads that would span the two mutations. In contrast to traditional Sanger sequencing, which measures the population average at each position, both Illumina and PacBio chemistries output sequences of single DNA molecules. In total, we generated mutation-spanning reads for seven telangiectasia, each with more than 100 reads that contained the somatic mutation. From these mutation-spanning reads, we established that >95% of reads with the somatic mutation possessed the WT allele at the position of the germline mutation, showing that all seven mutation pairs are in trans configuration (Table 2) (Figure 2A–2B). Any two variants in a chromosome must be arranged in cis or trans with an equal probability of either arrangement. Considering this, our observation that 7/7 mutation pairs are arranged in trans corresponds to a p value of 0.008, which demonstrates significant bias towards a trans configuration. These data show that the somatic and germline mutations are bi-allelic, fulfilling the second expectation of the genetic two-hit mechanism.
Table 2.
Summary of Phase Status for Pairs of Somatic and Germline Mutations
| Sample ID | Reads with Somatic Mutation | trans Reads | cis Reads | p Value |
|---|---|---|---|---|
| 6001-1 | N/A | N/A | N/A | N/A |
| 6001-3 | 112 | 112 (100%)a | 0 (0%) | 1.9e-34 |
| 6001-7 | 155 | 153 (98.7%)a | 2 (1.3%) | 2.6e-43 |
| 6001-8 | 593 | 590 (99.5%)a | 3 (0.5%) | 1.0e-171 |
| 6001-10 | N/A | N/A | N/A | N/A |
| 6002-1 | 125 | 120 (96.0%)a | 5 (4.0%) | 5.5e-30 |
| 6002-2 | 3189 | 3160 (99.0%)b,c | 29 (1.0%) | 4.2e-890 |
| 6003-1 | 372 | 364 (97.8%)b,c | 6 (1.4%) | 1.4e-99 |
| 6005-1 | 2671 | 2653 (99.3%)b,c | 18 (0.7%) | 6.3e-759 |
Reads generated with PacBio long-read chemistry
Reads generated with Illumina short-read chemistry
These reads were also used for validation shown in Table 1
Mutations are Consistent with Homozygous LoF
The third expectation of the genetic two-hit mechanism is that the bi-allelic somatic and germline variants both result in LoF. Due to the functional studies and extensive allelic series of mutations in each of the HHT-associated genes, HHT is known to be caused by LoF mutations. The germline mutations in four of the five individuals in this study have been identified previously in individuals with HHT and are reported in ClinVar (6001: VCV000213214.2, 6002: VCV000212796.2, and 6004 and 6005: VCV000008243.2). There are also several publications confirming the pathogenicity of these mutations.25, 26, 27, 28, 29 These are all therefore bona fide LoF mutations. The germline mutation in 6003-1, a silent mutation in ACVRL1 exon 4, has been identified before in an individual with HHT, however, it was classified as a VUS. We used the in silico tool Human Splicing Finder 3.130 to analyze this variant and found that it was predicted to both disrupt an exonic splice enhancer and create an internal splice donor site, potentially activating a cryptic splice site. Based on this prediction, we extracted RNA from peripheral blood leukocytes of 6003 and a control individual and performed RT-PCR to examine the splicing of ACVRL1 transcripts. RNA from 6003 shows a new splice variant that is not present in control WT RNA (Figure 3B). As predicted by the Splice Finder program, the aberrant transcript is spliced precisely at the internal splice donor created by the mutation. The resulting transcript is missing the portion of exon 4 downstream of the germline mutation, and it skips exon 5, resulting in the in-frame deletion of 52 amino acids (Figure 3D–3E). This deleted region contains several codons with known pathogenic missense mutations, suggesting that the 52-amino-acid deletion would likely also result in LoF. It is possible that the skipping of exon 5 is due to alternative splicing observed only in peripheral blood leukocytes, rather than a result of the mutation. If exon 5 is retained, the mutation would then generate a protein that would lack 17 amino acid residues from exon 4 but then be frameshifted for the remainder of the transcript. With this evidence, all of the identified germline mutations meet the American College of Medical Genetics (ACMG) criteria for pathogenic mutations,31 fulfilling the first half of the third expectation of the genetic two-hit mechanism.
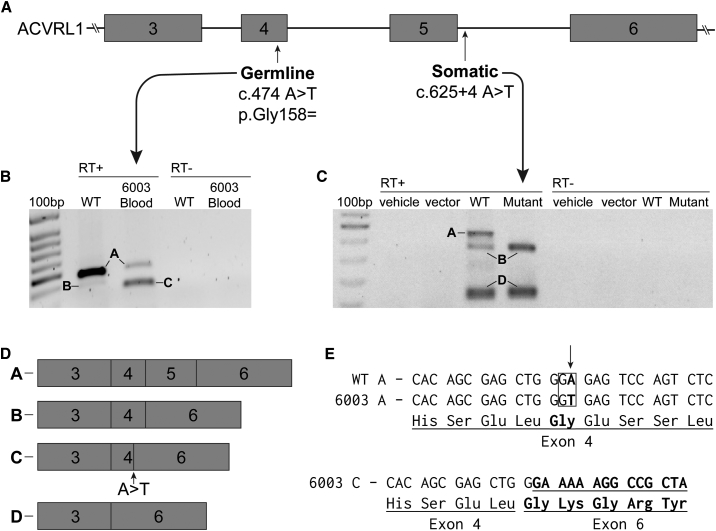
Figure 3.
Germline and Somatic Mutations in 6003-1 Both Cause Aberrant Splicing of ACVRL1 RNA
(A) The gene structure of ACVRL1 exons 3-6 marked with the location of the germline and somatic mutations found in 6003-1.
(B) RT-PCR showing ACVRL1 transcripts from peripheral blood leukocytes taken from wild-type (WT) and peripheral blood leukocytes containing the germline mutation. The labeled bands were excised and sequenced. Full-length transcript (Band A) is present in both the control and leukocytes from 6003 although the level in leukocytes from 6003 is greatly reduced. Band B in the control contains complete exons 3, 4, and 6. This splice variant has been seen previously and differs from band C in 6003, which splices from the newly created splice donor site within exon 4 directly to exon 6.
(C) As the somatic mutation is only present in 3.0% of reads, it would be challenging to detect misspliced RNA from the biopsied tissue. Therefore, we inserted WT and mutant sequences of ACVRL1 into an in vitro splicing vector, pSPL3-ACVRL1, and used RT-PCR to visualize the impact of the mutation on splicing. Only WT band A shows the full-length transcript containing exon 5; there is no corresponding full-length transcript from the plasmid containing the somatic mutation.
(D) Exon structure of ACVRL1 transcripts determined by sequencing the excised bands.
(E) Sequence of DNA showing the nature of the germline mutation. In ACVRL1 transcripts containing the germline mutation, exon 4 is shortened due to the activation of a cryptic splice site.
In contrast to the germline mutations, many of which have been previously identified, the somatic mutations we identified have not been reported. The ACMG guidelines for establishing pathogenicity are not applicable to somatic variants, however, several lines of evidence support our hypothesis that each somatic mutation results in LoF. Five of the nine somatic mutations result in a frameshift; all of those frameshifts result in premature termination codons which would generate transcripts susceptible to nonsense-mediated decay. Frameshift mutations in ENG or ACVRL1 are the most common mechanisms for LoF leading to HHT. Based on this, the five somatic frameshift mutations likely result in LoF. Other than frameshift mutations, the other four somatic mutations we identified consisted of three in-frame deletions and one intronic mutation predicted to impact splicing. These four mutations are not present in the genome aggregation database (gnomAD), and this shows that the population allele frequency of these variants is extremely low or zero. For the somatic in-frame deletion mutation found in 6001-8, there are two reports of different in-frame deletions which have overlap at this position and are known to cause HHT; this suggests that the somatic deletion in 6001-8 is likely to result in LoF. The somatic mutations in 6001-1 and 6002-2 also result in in-frame deletions; these deletions delete four and seven amino acids respectively. Comparing the crystal structures of ENG and ACVRL1, we determined that the somatic mutations in 6001-1 and 6002-2 delete portions of a beta strand and helix respectively, potentially impacting protein folding (Table 3). The in silico tool Protein Variation Effect Analyzer (PROVEAN) was used to predict how the protein would tolerate these deletions. −2.5 or lower (more negative) is considered to be the threshold for a deleterious change. The scores for 6001-8, 6001-1, and 6002-2 were −6.106, −14.903, and −25.903 respectively, strongly suggesting that all three are deleterious. The remaining somatic mutation is intronic, and it occurs four nucleotides from the exon-intron boundary. We used Human Splicing Finder 3.1 to predict the effect of this variant on splicing, and we found that it is likely to disrupt the donor site. This prediction was confirmed by RT-PCR using an in vitro splicing construct which revealed that the somatic mutation prevents the formation of full-length ACVRL1 transcripts (Figure 3C). In summary, we present evidence supporting our hypothesis that the bi-allelic germline and somatic mutations all likely result in LoF, fulfilling the third expectation of the genetic two-hit mechanism.
Table 3.
Evidence that Identified Germline and Somatic Mutations Result in LoF
| Sample ID | Germline Mutation | Somatic Mutation |
|---|---|---|
| 6001-1 | frameshift: ACMG PVS1 6 supporting publications |
in-frame deletion (−4 residues) PROVEAN prediction: deleterious (−14.903) deletes region in β-sheet gnomAD AF: 0 |
| 6001-3 | same as above | frameshift common ENG LOF mechanism, expect NMD |
| 6001-7 | same as above | frameshift common ENG LOF mechanism, expect NMD |
| 6001-8 | same as above | in-frame delins (−1 +2 residues) 2 reported pathogenic in-frame indels overlapping this codon54, 55 PROVEAN prediction: deleterious (−6.106) gnomAD AF: 0 |
| 6001-10 | same as above | frameshift common ENG LOF mechanism, expect NMD |
| 6002-1 | missense: ACMG PS1 6 supporting publications |
frameshift common ACVRL1 LOF mechanism, expect NMD |
| 6002-2 | same as above | in-frame delins (−7 +1 residues) PROVEAN prediction: deleterious (−25.903) deletes region in α-helix gnomAD AF: 0 |
| 6003-1 | cryptic splice site: ACMG PS3 in silico predicted to activate cryptic site in vitro evidence (Fig. 4) |
Splice Site in silico predicted to disrupt donor site gnomAD AF: 0 |
| 6005-1 | missense: ACMG PS1 16 supporting publications |
Frameshift common ACVRL1 LOF mechanism, expect NMD |
AF = Allele Frequency; NMD = Nonsense Mediated Decay
Germline variant classification according to ACMG guidelines, not applicable to somatic variants31
PVS1 = Very strong evidence for pathogenicity; PS1-4 = Strong evidence
PROVEAN scores below −2.5 are predicted deleterious
Telangiectasia from the Same Individual Harbor Unique Somatic Mutations
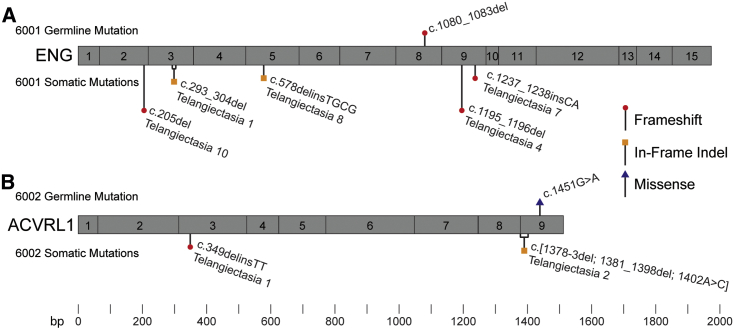
We next sought to determine whether mutant cells in different telangiectasia derive from a somatic mutation in a common ancestor cell, or whether the mutant cell population in each telangiectasia derives from an independent somatic mutation event. To test this, we examined the somatic mutations present in multiple telangiectasia from single individuals. In 6001, for whom we had obtained 13 different telangiectasia, we identified a somatic mutation in five telangiectasia tissue samples. In each case, the somatic mutation in each telangiectasia was unique. Likewise, for 6002, for whom we had two telangiectasia, we identified a unique somatic mutation in each (Figure 4). These results are consistent with independent mutation events rather than the somatic mutation occurring in a progenitor cell or clonality due to a metastasis from a single initial lesion.
Figure 4.
Multiple Telangiectasia from Single Individuals Contain Unique Somatic Mutations
Schematic representation of exons in ENG and ACVRL1 with germline and somatic mutations identified in (A) fie telangiectasia collected from 6001 and (B) two telangiectasia collected from 6002. In each panel, the common germline mutation is listed above the gene and somatic mutations in each telangiectasia are listed below the gene. Gene structure and mutation position are drawn to scale.
Discussion
In this study, we present strong evidence that VMs associated with HHT, specifically cutaneous telangiectasia, follow a genetic two-hit mechanism of pathogenesis. HHT is also associated with arteriovenous malformations in the lungs, liver, brain, and gastrointestinal tract, but these tissues are not available for prospective collection. We postulate that the visceral, deeper VMs that occur in HHT also follow this two-hit mechanism.
The two-hit hypothesis for HHT pathogenesis has persisted for decades without evidence, but these low-frequency somatic mutations are challenging to identify using traditional sequencing methods. The only published study to address this topic employed immunohistochemical staining in an attempt to identify endothelial cells lacking staining in the lining of HHT-related arteriovenous malformations.32 However, the absence of staining as a proxy for the gain of a mutation could be difficult to discover, especially if only a fraction of a cells would exhibit this lack of signal.
Using next-generation sequencing with unique molecular identifiers, we successfully identified somatic mutations in multiple telangiectasia from different individuals with HHT. The somatic mutations we identified were present at frequencies ranging between 0.46% and 8.0% in the tissue, with an average frequency of 2.3%. The low allele frequency is likely a result of two main contributing factors: the presence of normal tissue in the skin biopsy, and somatic mosaicism within the telangiectasia. The telangiectasias in this study were sampled as skin punch biopsies, and although some of the surrounding skin tissue was removed before DNA extraction, an undetermined amount of normal tissue invariably remained. However, after we removed the surrounding tissue, the enrichment for the vascular component of the tissue was subjectively greater than the often low somatic mutation allele frequency might suggest. We posit that a second explanation for the low mutant allele frequency is that telangiectasia are mosaic for the somatic mutation. This agrees with existing data from mouse models of HHT, which show that induced retinal AVMs are mosaic: consisting of both heterozygous and homozygous null cells.33 It is also consistent with the heterogeneity seen in cerebral cavernous malformations34, 35 and the low mutant allele frequency in somatic mutations of other VM disorders.8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 VMs, similar to tumors in cancer, appear to be seeded by somatic mutations; however, unlike tumors, VMs do not appear to consist of pure populations of clonally expanded mutant cells, but instead contain a substantial percentage of unmutated cells.
In addition to the data presented here, the two-hit hypothesis of HHT-related VMs is consistent with observations from mouse models of the disease. Whereas constitutional loss of both copies of Eng or Acvrl1 in mice is embryonic lethal, mice heterozygous for constitutional deletion of either gene show extremely mild phenotypes with relatively few, if any, detectable VMs.36, 37 Robust mouse models of HHT that recapitulate VM phenotypes require the use of Cre-Lox technology to delete both copies of ENG in a temporally controlled (postnatal), cell-type specific (endothelial cells) manner.
In these mouse models of HHT, it is also required that bi-allelic knockout (KO) of the HHT gene occur in endothelial cells. Several groups have experimented with expressing Cre recombinase in different vascular-related cell types including pericytes (NG2-Cre), vascular smooth muscle cells (Myh11-Cre), and endothelial cells (Scl-Cre & Pdgfb-Cre); however, mice only develop VMs when Cre is expressed in endothelial cells.38, 39, 40, 41 We have used laser capture microdissection in an attempt to confirm that the somatic mutations we identified in human lesions occur in the endothelium, however, these efforts have been hampered by the small quantity of tissue in telangiectasia biopsies and the difficulty of isolating a single layer of cells by microdissection. This question may be more easily addressed in larger arteriovenous malformations; however, these samples have been thus far inaccessible due to the rarity of their removal in individuals with HHT.
Interestingly, in addition to the local requirement for loss of both alleles, VMs in this model only develop after injury, such as an ear punch, or by vascular endothelial growth factor injection.42 This requirement of an angiogenic stimulus is consistent in mouse models for all HHT genotypes: Eng, Acvrl1, and Smad4.43 The requirement for KO of both copies of the gene supports the genetic two-hit mechanism we describe here. In addition, the necessity for an angiogenic stimulus suggests that loss of both copies of the relevant HHT gene is necessary, but not sufficient, for the development of the VM.
We might have expected to find somatic mutations in every telangiectasia, however, we only found somatic mutations in nine of the 19 we sequenced. The next-generation sequencing strategy we employ for discovering somatic mutations is extremely sensitive for the detection of point mutations and small indels. However, there are several other types of genetic alterations that would result in bi-allelic LoF due to loss of heterozygosity (LOH). LOH is a common occurrence in many tumors in cancer, and it is a predominant mechanism of somatic loss and/or mutation. LOH can occur due to a variety of genetic mechanisms: large deletions, chromosome loss, and mitotic recombination. Given the apparent capacity for even a low fraction of somatically mutant cells to initiate the VM, it follows that the same would be true for LOH-associated mutational events; therefore, these mutations would appear instead as allelic imbalance rather than outright LOH. But if the level of allelic imbalance is as low as the frequency of somatic mutations we have observed in this study, we might expect linked marker haplotype ratios in the range of 48% to 52% at nearby markers; this slight and even trivial imbalance would be difficult if not impossible to detect and validate. It is also possible that non-genetic mechanisms such as loss of expression due to epigenetic silencing account for bi-allelic LoF. This process, like the LOH associated events, would not be detected by our sequencing strategy; a problem that is only exacerbated by low allele frequency. Thus, it may not be surprising that we identified a somatic mutation in only nine of the VMs that were sequenced. We postulate that it is highly likely that all of the 10 telangiectasia with no identified somatic mutations have bi-allelic loss by one of these other mechanisms.
One consequence of the genetic two-hit mechanism that might appear to be improbable is that, if true, a new somatic mutation must occur in every one of the numerous VMs in HHT. For example, some affected individuals have dozens or more visible telangiectasia on the skin and mucocutaneous surfaces alone.44, 45, 46 An attractive hypothesis to reconcile this conundrum would be that a somatic mutation first occurs in a circulating progenitor cell which then proliferates and seeds the formation of multiple telangiectasia. There is precedence for this mechanism in another VM syndrome, Blue Rubber Bleb Nevus syndrome. Individuals with this syndrome display multiple small vascular lesions which harbor an identical somatic double-mutation in TEK (MIM: 600221). These vascular lesions appear to be anatomically dispersed clones arising from an original, dominant, large lesion.9 In HHT-related VMs, we report evidence that contradicts this hypothesis: different telangiectasia collected from the same individual harbor different, unique somatic mutations. This observation does not exclude the possibility that circulating cells may in some cases spread telangiectasia, however, the data thus far suggest that the primary mechanism is independent somatic mutation events.
The dilemma of the requirement for numerous independent somatic mutation events in a single gene can be resolved with a probabilistic argument. Considering the size of the human genome (3.23 × 109 bp), the probability that a random somatic mutation would occur in the coding sequence of ENG (3201 bp) in trans (50% likelihood) with a pathogenic ENG germline mutation is ∼0.00005%. Compounding this value with empirical evidence that single cells have anywhere from 100–1,500 somatic mutations per cell,47, 48, 49 and an estimate that 5.66% of exonic somatic mutations result in LOF;47 we calculate a conservative estimate that 0.00028% of cells have bi-allelic LOF ENG mutations. An adult human has at least 6 × 1011 endothelial cells,50 therefore we estimate that an individual with HHT and a germline mutation in ENG has bi-allelic LOF ENG mutations in ∼1.5 million endothelial cells. It is clear that not every cell with ENG bi-allelic LOF results in VM, as individuals with HHT have at most hundreds, not millions, of telangiectasia. This disparity is consistent with the idea that telangiectasia only develop under very specific conditions: likely that the somatic mutation must occur in a specific type of vascular bed, in an endothelial cell, and must be followed by local angiogenic stimulus.
Our samples consisted of telangiectasia from individuals with HHT from a single HHT Centre of Excellence. This cohort had germline mutations in either ENG or ACVRL1, and somatic mutations were identified in telangiectasia from both genotypes. Mutations in SMAD4 cause the combined syndrome HHT and Juvenile Polyposis (JP-HHT), however, individuals with SMAD4 mutations only account for ∼2% of HHT cases.3 Unfortunately, no individuals with JP-HHT were present in our cohort, but we believe it is likely that JP-HHT-related telangiectasia follow an identical genetic two-hit mechanism resulting from somatic mutations in SMAD4.
As HHT is caused by germline mutations in any one of three genes, an interesting question is whether the compound effect of a LOF mutations in two different HHT genes could drive pathogenesis (e.g., a germline mutation in ENG and a somatic mutation in ACVRL1). However, thus far, all of the somatic mutations we identified in HHT-related telangiectasia occur in the same gene as that harboring the germline mutation. This human genetic data is consistent with the observation that mice with combined deficiency for one allele each of Acvrl1 and Eng are fertile, viable, and not teeming with VMs51 (Srinivasan and Marchuk, unpublished), as might be expected if trans-heterozygosity of mutations in these two HHT-genes could initiate VM development.
A genetic two-hit mechanism for HHT pathogenesis has therapeutic implications. Certain efforts to develop therapies for HHT assume a model of haploinsufficiency of the relevant HHT gene. These strategies attempt to increase the amount of the affected gene product by increasing the level of transcription of the WT allele.52 We show here that some fraction of cells within the malformation do not possess a WT allele. Even if expression in surrounding heterozygous cells could be increased, the null cells would remain devoid of protein, suggesting that this avenue of therapy may be ineffective. In contrast, a more effective strategy may be gene replacement, reintroducing a fully WT allele into the mutated cells,53 as this would simultaneously provide an extra copy of the gene to both the heterozygous and null cells. This strategy is particularly attractive as it might inhibit new VM formation by adding back a second WT copy of the mutated gene.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank all participants and their families who donated samples for this study. We thank Dr. Nicolas Devos and the Duke Sequencing and Genomic Technologies Core for assistance in designing experiments and sequencing. This study was supported by U.S. Department of Defense grant W81XWH-17-1-0429 and a Fondation Leducq Transatlantic Network of Excellence Grant in Neurovascular Disease (17 CVD 03). M.E.F. also received financial support from the Nelson Arthur Hyland Foundation.
Published: October 17, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.09.010.
Web Resources
Human Splice Finder, https://umd.be/HSF/
Online Mendelian Inheritance in Man, https://omim.org
SNPsift, https://github.com/pcingola/SnpSift
Supplemental Data
References
- 1.McAllister K.A., Grogg K.M., Johnson D.W., Gallione C.J., Baldwin M.A., Jackson C.E., Helmbold E.A., Markel D.S., McKinnon W.C., Murrell J. Endoglin, a TGF-beta binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat. Genet. 1994;8:345–351. doi: 10.1038/ng1294-345. [DOI] [PubMed] [Google Scholar]
- 2.Johnson D.W., Berg J.N., Baldwin M.A., Gallione C.J., Marondel I., Yoon S.J., Stenzel T.T., Speer M., Pericak-Vance M.A., Diamond A. Mutations in the activin receptor-like kinase 1 gene in hereditary haemorrhagic telangiectasia type 2. Nat. Genet. 1996;13:189–195. doi: 10.1038/ng0696-189. [DOI] [PubMed] [Google Scholar]
- 3.Gallione C.J., Repetto G.M., Legius E., Rustgi A.K., Schelley S.L., Tejpar S., Mitchell G., Drouin E., Westermann C.J., Marchuk D.A. A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4) Lancet. 2004;363:852–859. doi: 10.1016/S0140-6736(04)15732-2. [DOI] [PubMed] [Google Scholar]
- 4.Pece N., Vera S., Cymerman U., White R.I., Jr., Wrana J.L., Letarte M. Mutant endoglin in hereditary hemorrhagic telangiectasia type 1 is transiently expressed intracellularly and is not a dominant negative. J. Clin. Invest. 1997;100:2568–2579. doi: 10.1172/JCI119800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdalla S.A., Pece-Barbara N., Vera S., Tapia E., Paez E., Bernabeu C., Letarte M. Analysis of ALK-1 and endoglin in newborns from families with hereditary hemorrhagic telangiectasia type 2. Hum. Mol. Genet. 2000;9:1227–1237. doi: 10.1093/hmg/9.8.1227. [DOI] [PubMed] [Google Scholar]
- 6.Ola R., Künzel S.H., Zhang F., Genet G., Chakraborty R., Pibouin-Fragner L., Martin K., Sessa W., Dubrac A., Eichmann A. SMAD4 Prevents Flow Induced Arteriovenous Malformations by Inhibiting Casein Kinase 2. Circulation. 2018;138:2379–2394. doi: 10.1161/CIRCULATIONAHA.118.033842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pece-Barbara N., Cymerman U., Vera S., Marchuk D.A., Letarte M. Expression analysis of four endoglin missense mutations suggests that haploinsufficiency is the predominant mechanism for hereditary hemorrhagic telangiectasia type 1. Hum. Mol. Genet. 1999;8:2171–2181. doi: 10.1093/hmg/8.12.2171. [DOI] [PubMed] [Google Scholar]
- 8.Al-Olabi L., Polubothu S., Dowsett K., Andrews K.A., Stadnik P., Joseph A.P., Knox R., Pittman A., Clark G., Baird W. Mosaic RAS/MAPK variants cause sporadic vascular malformations which respond to targeted therapy. J. Clin. Invest. 2018;128:1496–1508. doi: 10.1172/JCI98589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soblet J., Kangas J., Nätynki M., Mendola A., Helaers R., Uebelhoer M., Kaakinen M., Cordisco M., Dompmartin A., Enjolras O. Blue Rubber Bleb Nevus (BRBN) syndrome is caused by somatic TEK (TIE2) mutations. J. Invest. Dermatol. 2017;137:207–216. doi: 10.1016/j.jid.2016.07.034. [DOI] [PubMed] [Google Scholar]
- 10.Limaye N., Kangas J., Mendola A., Godfraind C., Schlögel M.J., Helaers R., Eklund L., Boon L.M., Vikkula M. Somatic activating PIK3CA mutations cause venous malformation. Am. J. Hum. Genet. 2015;97:914–921. doi: 10.1016/j.ajhg.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Limaye N., Wouters V., Uebelhoer M., Tuominen M., Wirkkala R., Mulliken J.B., Eklund L., Boon L.M., Vikkula M. Somatic mutations in angiopoietin receptor gene TEK cause solitary and multiple sporadic venous malformations. Nat. Genet. 2009;41:118–124. doi: 10.1038/ng.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirley M.D., Tang H., Gallione C.J., Baugher J.D., Frelin L.P., Cohen B., North P.E., Marchuk D.A., Comi A.M., Pevsner J. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N. Engl. J. Med. 2013;368:1971–1979. doi: 10.1056/NEJMoa1213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couto J.A., Vivero M.P., Kozakewich H.P., Taghinia A.H., Mulliken J.B., Warman M.L., Greene A.K. A somatic MAP3K3 mutation is associated with verrucous venous malformation. Am. J. Hum. Genet. 2015;96:480–486. doi: 10.1016/j.ajhg.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luks V.L., Kamitaki N., Vivero M.P., Uller W., Rab R., Bovee J.V., Rialon K.L., Guevara C.J., Alomari A.I., Greene A.K. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J. Pediatr. 2015;166:1048–1054. doi: 10.1016/j.jpeds.2014.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikolaev S.I., Vetiska S., Bonilla X., Boudreau E., Jauhiainen S., Rezai Jahromi B., Khyzha N., DiStefano P.V., Suutarinen S., Kiehl T.R. Somatic activating KRAS mutations in arteriovenous malformations of the brain. N. Engl. J. Med. 2018;378:250–261. doi: 10.1056/NEJMoa1709449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Couto J.A., Huang A.Y., Konczyk D.J., Goss J.A., Fishman S.J., Mulliken J.B., Warman M.L., Greene A.K. Somatic MAP2K1 mutations are associated with extracranial arteriovenous malformation. Am. J. Hum. Genet. 2017;100:546–554. doi: 10.1016/j.ajhg.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akers A.L., Johnson E., Steinberg G.K., Zabramski J.M., Marchuk D.A. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum. Mol. Genet. 2009;18:919–930. doi: 10.1093/hmg/ddn430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald D.A., Shi C., Shenkar R., Gallione C.J., Akers A.L., Li S., De Castro N., Berg M.J., Corcoran D.L., Awad I.A., Marchuk D.A. Lesions from patients with sporadic cerebral cavernous malformations harbor somatic mutations in the CCM genes: evidence for a common biochemical pathway for CCM pathogenesis. Hum. Mol. Genet. 2014;23:4357–4370. doi: 10.1093/hmg/ddu153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gault J., Awad I.A., Recksiek P., Shenkar R., Breeze R., Handler M., Kleinschmidt-DeMasters B.K. Cerebral cavernous malformations: somatic mutations in vascular endothelial cells. Neurosurgery. 2009;65:138–144, discussion 144–145. doi: 10.1227/01.NEU.0000348049.81121.C1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macmurdo C.F., Wooderchak-Donahue W., Bayrak-Toydemir P., Le J., Wallenstein M.B., Milla C., Teng J.M., Bernstein J.A., Stevenson D.A. RASA1 somatic mutation and variable expressivity in capillary malformation/arteriovenous malformation (CM/AVM) syndrome. Am. J. Med. Genet. A. 2016;170:1450–1454. doi: 10.1002/ajmg.a.37613. [DOI] [PubMed] [Google Scholar]
- 21.Shovlin C.L., Guttmacher A.E., Buscarini E., Faughnan M.E., Hyland R.H., Westermann C.J., Kjeldsen A.D., Plauchu H. Diagnostic criteria for hereditary hemorrhagic telangiectasia (Rendu-Osler-Weber syndrome) Am. J. Med. Genet. 2000;91:66–67. doi: 10.1002/(sici)1096-8628(20000306)91:1<66::aid-ajmg12>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 22.Grosse S.D., Boulet S.L., Grant A.M., Hulihan M.M., Faughnan M.E. The use of US health insurance data for surveillance of rare disorders: hereditary hemorrhagic telangiectasia. Genet. Med. 2014;16:33–39. doi: 10.1038/gim.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laver T.W., Caswell R.C., Moore K.A., Poschmann J., Johnson M.B., Owens M.M., Ellard S., Paszkiewicz K.H., Weedon M.N. Pitfalls of haplotype phasing from amplicon-based long-read sequencing. Sci. Rep. 2016;6:21746. doi: 10.1038/srep21746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Church D.M., Stotler C.J., Rutter J.L., Murrell J.R., Trofatter J.A., Buckler A.J. Isolation of genes from complex sources of mammalian genomic DNA using exon amplification. Nat. Genet. 1994;6:98–105. doi: 10.1038/ng0194-98. [DOI] [PubMed] [Google Scholar]
- 25.Johnson D.W., Berg J.N., Gallione C.J., McAllister K.A., Warner J.P., Helmbold E.A., Markel D.S., Jackson C.E., Porteous M.E., Marchuk D.A. A second locus for hereditary hemorrhagic telangiectasia maps to chromosome 12. Genome Res. 1995;5:21–28. doi: 10.1101/gr.5.1.21. [DOI] [PubMed] [Google Scholar]
- 26.Bossler A.D., Richards J., George C., Godmilow L., Ganguly A. Novel mutations in ENG and ACVRL1 identified in a series of 200 individuals undergoing clinical genetic testing for hereditary hemorrhagic telangiectasia (HHT): correlation of genotype with phenotype. Hum. Mutat. 2006;27:667–675. doi: 10.1002/humu.20342. [DOI] [PubMed] [Google Scholar]
- 27.Gallione C.J., Klaus D.J., Yeh E.Y., Stenzel T.T., Xue Y., Anthony K.B., McAllister K.A., Baldwin M.A., Berg J.N., Lux A. Mutation and expression analysis of the endoglin gene in hereditary hemorrhagic telangiectasia reveals null alleles. Hum. Mutat. 1998;11:286–294. doi: 10.1002/(SICI)1098-1004(1998)11:4<286::AID-HUMU6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 28.Ricard N., Bidart M., Mallet C., Lesca G., Giraud S., Prudent R., Feige J.J., Bailly S. Functional analysis of the BMP9 response of ALK1 mutants from HHT2 patients: a diagnostic tool for novel ACVRL1 mutations. Blood. 2010;116:1604–1612. doi: 10.1182/blood-2010-03-276881. [DOI] [PubMed] [Google Scholar]
- 29.Olivieri C., Pagella F., Semino L., Lanzarini L., Valacca C., Pilotto A., Corno S., Scappaticci S., Manfredi G., Buscarini E., Danesino C. Analysis of ENG and ACVRL1 genes in 137 HHT Italian families identifies 76 different mutations (24 novel). Comparison with other European studies. J. Hum. Genet. 2007;52:820–829. doi: 10.1007/s10038-007-0187-5. [DOI] [PubMed] [Google Scholar]
- 30.Desmet F.O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bourdeau A., Cymerman U., Paquet M.E., Meschino W., McKinnon W.C., Guttmacher A.E., Becker L., Letarte M. Endoglin expression is reduced in normal vessels but still detectable in arteriovenous malformations of patients with hereditary hemorrhagic telangiectasia type 1. Am. J. Pathol. 2000;156:911–923. doi: 10.1016/S0002-9440(10)64960-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Y., Muhl L., Burmakin M., Wang Y., Duchez A.C., Betsholtz C., Arthur H.M., Jakobsson L. Endoglin prevents vascular malformation by regulating flow-induced cell migration and specification through VEGFR2 signalling. Nat. Cell Biol. 2017;19:639–652. doi: 10.1038/ncb3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Detter M.R., Snellings D.A., Marchuk D.A. Cerebral cavernous malformations develop through clonal expansion of mutant endothelial cells. Circ. Res. 2018;123:1143–1151. doi: 10.1161/CIRCRESAHA.118.313970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malinverno M., Maderna C., Abu Taha A., Corada M., Orsenigo F., Valentino M., Pisati F., Fusco C., Graziano P., Giannotta M. Endothelial cell clonal expansion in the development of cerebral cavernous malformations. Nat. Commun. 2019;10:2761. doi: 10.1038/s41467-019-10707-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bourdeau A., Dumont D.J., Letarte M. A murine model of hereditary hemorrhagic telangiectasia. J. Clin. Invest. 1999;104:1343–1351. doi: 10.1172/JCI8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srinivasan S., Hanes M.A., Dickens T., Porteous M.E., Oh S.P., Hale L.P., Marchuk D.A. A mouse model for hereditary hemorrhagic telangiectasia (HHT) type 2. Hum. Mol. Genet. 2003;12:473–482. doi: 10.1093/hmg/ddg050. [DOI] [PubMed] [Google Scholar]
- 38.Tual-Chalot S., Oh S.P., Arthur H.M. Mouse models of hereditary hemorrhagic telangiectasia: recent advances and future challenges. Front. Genet. 2015;6:25. doi: 10.3389/fgene.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi E.J., Chen W., Jun K., Arthur H.M., Young W.L., Su H. Novel brain arteriovenous malformation mouse models for type 1 hereditary hemorrhagic telangiectasia. PLoS ONE. 2014;9:e88511. doi: 10.1371/journal.pone.0088511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garrido-Martin E.M., Nguyen H.L., Cunningham T.A., Choe S.W., Jiang Z., Arthur H.M., Lee Y.J., Oh S.P. Common and distinctive pathogenetic features of arteriovenous malformations in hereditary hemorrhagic telangiectasia 1 and hereditary hemorrhagic telangiectasia 2 animal models--brief report. Arterioscler. Thromb. Vasc. Biol. 2014;34:2232–2236. doi: 10.1161/ATVBAHA.114.303984. [DOI] [PubMed] [Google Scholar]
- 41.Mahmoud M., Allinson K.R., Zhai Z., Oakenfull R., Ghandi P., Adams R.H., Fruttiger M., Arthur H.M. Pathogenesis of arteriovenous malformations in the absence of endoglin. Circ. Res. 2010;106:1425–1433. doi: 10.1161/CIRCRESAHA.109.211037. [DOI] [PubMed] [Google Scholar]
- 42.Choi E.J., Walker E.J., Shen F., Oh S.P., Arthur H.M., Young W.L., Su H. Minimal homozygous endothelial deletion of Eng with VEGF stimulation is sufficient to cause cerebrovascular dysplasia in the adult mouse. Cerebrovasc. Dis. 2012;33:540–547. doi: 10.1159/000337762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim Y.H., Choe S.W., Chae M.Y., Hong S., Oh S.P. SMAD4 deficiency leads to development of arteriovenous malformations in neonatal and adult mice. J. Am. Heart Assoc. 2018;7:e009514. doi: 10.1161/JAHA.118.009514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gonzalez C.D., Cipriano S.D., Topham C.A., Stevenson D.A., Whitehead K.J., Vanderhooft S., Presson A.P., McDonald J. Localization and age distribution of telangiectases in children and adolescents with hereditary hemorrhagic telangiectasia: A retrospective cohort study. J. Am. Acad. Dermatol. 2019;81:950–955. doi: 10.1016/j.jaad.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 45.Letteboer T.G., Mager H.J., Snijder R.J., Lindhout D., Ploos van Amstel H.K., Zanen P., Westermann K.J. Genotype-phenotype relationship for localization and age distribution of telangiectases in hereditary hemorrhagic telangiectasia. Am. J. Med. Genet. A. 2008;146A:2733–2739. doi: 10.1002/ajmg.a.32243. [DOI] [PubMed] [Google Scholar]
- 46.Plauchu H., de Chadarévian J.P., Bideau A., Robert J.M. Age-related clinical profile of hereditary hemorrhagic telangiectasia in an epidemiologically recruited population. Am. J. Med. Genet. 1989;32:291–297. doi: 10.1002/ajmg.1320320302. [DOI] [PubMed] [Google Scholar]
- 47.Milholland B., Dong X., Zhang L., Hao X., Suh Y., Vijg J. Differences between germline and somatic mutation rates in humans and mice. Nat. Commun. 2017;8:15183. doi: 10.1038/ncomms15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lodato M.A., Woodworth M.B., Lee S., Evrony G.D., Mehta B.K., Karger A., Lee S., Chittenden T.W., D’Gama A.M., Cai X. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015;350:94–98. doi: 10.1126/science.aab1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lo Sardo V., Ferguson W., Erikson G.A., Topol E.J., Baldwin K.K., Torkamani A. Influence of donor age on induced pluripotent stem cells. Nat. Biotechnol. 2017;35:69–74. doi: 10.1038/nbt.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sender R., Fuchs S., Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eleftheriou N.M., Sjölund J., Bocci M., Cortez E., Lee S.J., Cunha S.I., Pietras K. Compound genetically engineered mouse models of cancer reveal dual targeting of ALK1 and endoglin as a synergistic opportunity to impinge on angiogenic TGF-β signaling. Oncotarget. 2016;7:84314–84325. doi: 10.18632/oncotarget.12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruiz-Llorente L., Gallardo-Vara E., Rossi E., Smadja D.M., Botella L.M., Bernabeu C. Endoglin and alk1 as therapeutic targets for hereditary hemorrhagic telangiectasia. Expert Opin. Ther. Targets. 2017;21:933–947. doi: 10.1080/14728222.2017.1365839. [DOI] [PubMed] [Google Scholar]
- 53.Seki T., Yun J., Oh S.P. Arterial endothelium-specific activin receptor-like kinase 1 expression suggests its role in arterialization and vascular remodeling. Circ. Res. 2003;93:682–689. doi: 10.1161/01.RES.0000095246.40391.3B. [DOI] [PubMed] [Google Scholar]
- 54.Shovlin C.L., Hughes J.M., Scott J., Seidman C.E., Seidman J.G. Characterization of endoglin and identification of novel mutations in hereditary hemorrhagic telangiectasia. Am. J. Hum. Genet. 1997;61:68–79. doi: 10.1086/513906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Argyriou L., Twelkemeyer S., Panchulidze I., Wehner L.E., Teske U., Engel W., Nayernia K. Novel mutations in the ENG and ACVRL1 genes causing hereditary hemorrhagic teleangiectasia. Int. J. Mol. Med. 2006;17:655–659. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.