Abstract
Meiotic nondisjunction and resulting aneuploidy can lead to severe health consequences in humans. Aneuploidy rescue can restore euploidy but may result in uniparental disomy (UPD), the inheritance of both homologs of a chromosome from one parent with no representative copy from the other. Current understanding of UPD is limited to ∼3,300 case subjects for which UPD was associated with clinical presentation due to imprinting disorders or recessive diseases. Thus, the prevalence of UPD and its phenotypic consequences in the general population are unknown. We searched for instances of UPD across 4,400,363 consented research participants from the personal genetics company 23andMe, Inc., and 431,094 UK Biobank participants. Using computationally detected DNA segments identical-by-descent (IBD) and runs of homozygosity (ROH), we identified 675 instances of UPD across both databases. We estimate that UPD is twice as common as previously thought, and we present a machine-learning framework to detect UPD using ROH. While we find a nominally significant association between UPD of chromosome 22 and autism risk, we do not find significant associations between UPD and deleterious traits in the 23andMe database.
Keywords: uniparental disomy, runs of homozygosity, identity-by-descent, aneuploidy
Introduction
Meiotic nondisjunction can have severe consequences for human reproduction and health. For example, nondisjunction can lead to aneuploidy, which is the leading cause of both spontaneous miscarriages and severe developmental disabilities.1, 2, 3, 4, 5 Because determining the etiology of aneuploidy is extremely difficult in humans, many studies have instead focused on either studying the consequences of aneuploidy in individual case subjects presenting in the clinic or studying recombination events using population-genomic datasets in order to understand meiotic processes.3, 6, 7 Recombination is an integral part of meiosis, facilitating alignment and then proper segregation of homologous chromosomes. Thus, recombination at each chromosome pair in a human genome is generally regarded as necessary to prevent aneuploidy (with some exceptions3, 6).
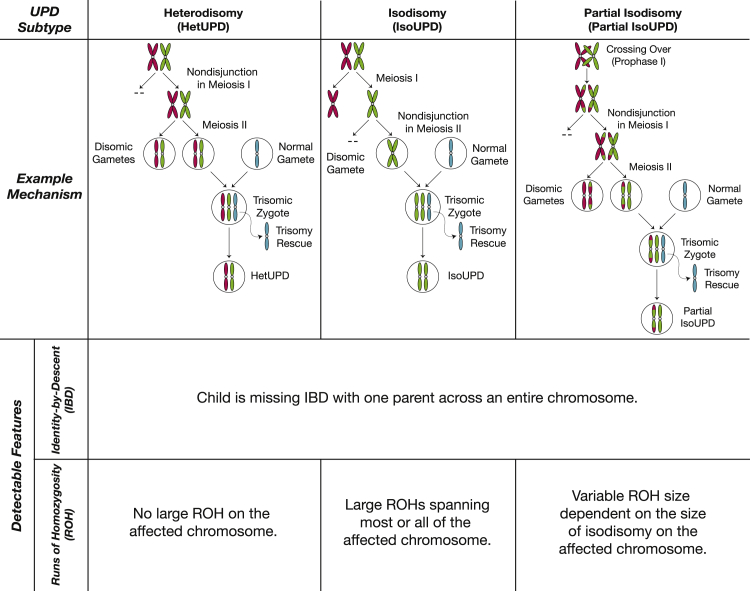
However, viable, euploid humans can result from aneuploid gametes if trisomic rescue, monosomic rescue, or gametic complementation restore normal ploidy during early development.8, 9, 10, 11, 12 These processes can result in uniparental disomy (UPD), which is the inheritance of both homologs of a chromosome from only one parent with no representative copy from the other parent (Figure 1).
Figure 1.
Subtypes of UPD with Example Mechanisms and Detectable Genomic Features for Each UPD Subtype
There are three subtypes of UPD: heterodisomy (hetUPD), isodisomy (isoUPD), and partial isodisomy (partial isoUPD). HetUPD is caused by nondisjunction in meiosis I, and an affected individual will inherit both homologs of a chromosome from the same parent. IsoUPD is caused by nondisjunction in meiosis II, and an affected individual will inherit two identical copies of one homolog from one parent. Partial isoUPD is caused by nondisjunction in either meiosis I or meiosis II after crossing over has happened, resulting in sections of isodisomy and heterodisomy on the UPD chromosome. Given genomic data from a parent-child pair, all UPD subtypes can be detected in the same way based on identity-by-descent (IBD): a parent-child pair will be missing IBD across an entire chromosome. Lastly, isoUPD and some types of partial isoUPD will show large runs of homozygosity (ROHs), which can be detected computationally without the need for parental genotype data.
Since the first report of UPD in 1987,13, 14 ∼3,300 cases of UPD have been described in the scientific literature8 (see Web Resources). To date, UPD of each of the autosomes and the X chromosome has been documented.8, 12, 15 UPD can cause clinical consequences by disrupting genomic imprinting or by unmasking harmful recessive alleles in large blocks of homozygosity on the affected chromosome. Detecting UPD is a useful diagnostic tool for specific imprinting disorders and for rare Mendelian diseases caused by homozygosity.12, 16, 17, 18, 19 UPD has also been implicated in tumorigenesis, particularly in cases of genome-wide UPD, which affects all the chromosomes in the genome.11, 20 Thus, current understanding of UPD is based largely on case reports of individuals in which UPD is detected following suspicion of an imprinting or other clinical disorder, and, in some instances (typically <10 confirmed cases) within larger case-control studies.12, 16, 17, 18, 19, 21
There are three subtypes of UPD resulting from nondisjunction during different stages of meiosis (which we refer to as meiotic-origin UPD): isodisomy (isoUPD), heterodisomy (hetUPD), and partial isodisomy (partial isoUPD), which involves meiotic crossover (Figure 1). UPD can also be classified according to the parent of origin: when the disomic pair originates from the mother, the resulting case is termed maternal UPD (matUPD), and when the disomic pair originates from the father, the case is termed paternal UPD (patUPD). Despite the wealth of clinical UPD cases, prevalence and per-chromosome rates of UPD and its subtypes are not characterized in the general population. Past estimates of UPD prevalence include rates of 1 in 3,500 and 1 in 5,000;8, 22 these estimates were determined by extrapolation from UPD events causing clinical presentation and so do not account for variation in prevalence across chromosomes or for UPD associated with healthy phenotypes.22 Therefore, to obtain an accurate estimate of UPD prevalence, hundreds of thousands of samples from the general population are needed.12 And while chromosome recombination and segregation are regarded as highly constrained processes, population genetic datasets are now reaching large enough sizes to yield insight into normal variability in recombination within and among human genomes.
To address this gap, we detected instances of UPD in consented research participants from the direct to consumer genetics company 23andMe, Inc., whose database consists of single-nucleotide polymorphism (SNP) data from more than 4.4 million individuals, and in 431,094 northern European UK Biobank participants. Here we present estimates of UPD prevalence in the general population, a machine-learning method to identify UPD in individuals without parental genotypes, and previously unrecognized phenotypes associated with UPD. We used both identity-by-descent (IBD) and a supervised classification framework based on runs of homozygosity (ROH) to identify UPD while accounting for parental relatedness and differences in ROH length distributions between ancestral populations. We found that UPD is twice as common in the general population (estimated rate: 1 in 2,000 births) than was previously thought and that, contrary to expectation, many individuals with long isodisomy events (ranging between 12 and 227 Mb) appear to have healthy phenotypes.
Subjects and Methods
Samples
In this study, we analyzed genome-wide SNP genotypes from 4,400,363 research participants from the 23andMe customer base; this research platform has been previously described.23, 24 All research participants included in these analyses provided consent and answered surveys online according to a human subjects protocol approved by Ethical and Independent Review Services, an independent institutional review board (see Web Resources). We also analyzed genotype data from 500,000 participants in the UK Biobank Project25, 26 (see Web Resources). Phenotype data for these individuals were collected through questionnaires, interviews, health records, physical measurements, and imaging carried out at assessment centers across the UK.
Genotyping and Quality Control
For the 23andMe dataset, DNA extraction and genotyping were performed on saliva samples by clinical laboratories at Laboratory Corporation of America, which is certified by Clinical Laboratory Improvement Amendments and accredited by College of American Pathologists. Samples were genotyped on one of five Illumina platforms: (1 and 2) two versions of the Illumina HumanHap550+ BeadChip, plus about 25,000 custom SNPs selected by 23andMe (∼560,000 SNPs total), (3) a variation on the Illumina OmniExpress+ BeadChip, with custom SNPs (∼950,000 SNPs total), (4) a fully customized array, including a lower redundancy subset of v2 and v3 SNPs with additional coverage of lower-frequency coding variation (∼570,000 SNPs total), and (5) a customized array based on Illumina’s Global Screening Array (∼640,000 SNPs total), supplemented with ∼50,000 SNPs of custom content. Samples that failed to reach 98.5% call rate were re-analyzed. Individuals whose analyses failed repeatedly were re-contacted by 23andMe customer service to provide additional samples. For all ROH analyses, we limited our analyses to SNPs that are shared between the Illumina platforms 1–4 described above, and then we removed SNPs with a minor allele frequency (MAF) less than 5% and SNPs with genotyping rate less than 99%, resulting in 381,379 SNPs in total. For IBD analyses, we analyzed 579,957 SNPs for all individuals.
For the UK Biobank dataset, quality control was carried out as described in the UK Biobank genotyping quality control document.26 As was done in the 23andMe dataset, we then removed all SNPs with MAF less than 5% and SNPs with genotyping rate less than 99%, resulting in 360,540 SNPs total.
Ancestry Classification
23andMe’s ancestry analysis has been described previously.27 Briefly, the algorithm first partitions phased genomic data into short windows of about 300 SNPs. Within each window, a support vector machine (SVM) was used to classify individual haplotypes into one of 25 reference populations (see Web Resources). The SVM classifications are then fed into a hidden Markov model (HMM) that accounts for switch errors and incorrect assignments and gives probabilities for each reference population in each window. Finally, simulated admixed individuals were used to recalibrate the HMM probabilities so that the reported assignments are consistent with the simulated admixture proportions. The reference population data are derived from public datasets (the Human Genome Diversity Project, HapMap, and 1000 Genomes) as well as 23andMe research participants who have reported having four grandparents from a single country.
Population Structure
For population-specific analyses such as ROH detection in the 23andMe dataset, research participants genotyped on Illumina platforms 1–4 were divided into eight cohorts based on genome-wide ancestry proportions from reference populations, as determined by 23andMe’s Ancestry Composition method: northern Europeans, southern Europeans, African Americans, Ashkenazi Jewish, East Asians, South Asians, Latino/as, and Middle Eastern individuals. The classification criteria have been previously described in Campbell et al.7 Briefly, individuals labeled as northern European met all of the following criteria: greater than 97% European and Middle Eastern/northern African ancestry combined, greater than 90% European ancestry, and greater than 85% northern European ancestry. Southern European individuals satisfied the following requirements: greater than 97% European and Middle Eastern/northern African ancestry combined, greater than 90% European ancestry, and greater than 85% southern European ancestry. Ashkenazi Jewish individuals had greater than 97% European and Middle Eastern/northern African ancestry combined, greater than 90% European ancestry, and greater than 85% Ashkenazi Jewish ancestry. Middle Eastern individuals had greater than 97% European and Middle Eastern/northern African ancestry combined and greater than 70% Middle Eastern/northern African ancestry. East Asian individuals had greater than 97% East Asian and Southeast Asian ancestry combined. South Asians had greater than 97% South Asian ancestry. Individuals were classified as African Americans or Latinos/Latinas if they had greater than 90% European and African and East Asian/Native American and Middle Eastern/northern African ancestry combined as well as greater than 1% African and American ancestry. African Americans and Latino/as were distinguished using a logistic regression classifier trained on self-identified “Black African” and “Hispanic” individuals.7 These classification criteria resulted in 974,511 northern Europeans, 34,508 southern Europeans, 90,349 African Americans, 70,144 Ashkenazi Jewish, 63,683 East Asians, 19,493 South Asians, 208,087 Latino/a individuals, and 16,013 Middle Eastern individuals in the 23andMe dataset. We also identified 87,571 individuals classified as “other” ancestry; we do not analyze this group further.
In the UK Biobank dataset, we focused our analyses on 431,094 individuals of northern European ancestry identified by principal components analysis (PCA) on the genotype data following QC. We arrived at this sample as follows: we first performed PCA using FlashPCA228 (v.2.0) on 2,504 individuals from the 1000 Genomes Project Phase 3 database.29 We pruned the genotype data from the UK Biobank for linkage disequilibrium, resulting in 70,527 SNPs, and we then projected genotype data from 431,102 UK Biobank participants who self-identified as “white British” onto the principal components space derived from PCA of the 1000 Genomes individuals. Lastly, we removed eight individuals who were outliers in PCA with the following thresholds: first principal component value less than 0.03 (PC1 < 0.03) and second principal component value greater than 0.15 (PC2 > 0.15). These filtering steps resulted in 431,094 northern European individuals. We also identified 5,780 South Asian individuals, 3,202 individuals of African ancestry, and 806 East Asian individuals based on self-reported ethnic background in the UK Biobank. We did not analyze these non-northern Europeans individuals from UK Biobank because the sample sizes were smaller than the smallest sample size in the 23andMe dataset (16,013 individuals).
Identification of Parent-Child Duos from Identity-by-Descent Segments
We identified identical-by-descent (IBD) DNA segments for every pair of 4,400,363 individuals in the 23andMe dataset, according to a method that has been previously described by Henn et al.30 Briefly, we compared a pair of individuals’ genotypes at 579,957 SNPs and identified SNPs where the individuals are homozygous for different alleles (also called opposite homozygotes). Long regions (>5 cM) lacking opposite homozygotes were characterized as IBD segments.30
Pairs of individuals that share more than 85% of their genome IBD were classified as parent and child. Theoretically, parent-child pairs should share 100% of their genome IBD on one homologous chromosome, but the threshold is lowered here to 85% to account for the possibility of UPD of chromosome 1, which accounts for ∼10% of the genome, and for the possibility of error or lack of SNP coverage over ∼5% of the genome. Using these criteria, we identified 916,712 parent-child duos in the 23andMe database.
In the UK Biobank, we used the King kinship coefficients provided by the Biobank to identify all third-degree or closer relative pairs (kinship coefficient > 0.044). We determined genome-wide IBD1 and IBD2 values for these pairs using the PLINK31 software (command flag:--genome), and we identified 3,923 parent-child duos using the following thresholds: IBD1 > 0.85 and IBD2 < 0.1. We then calculated IBD1 and IBD2 proportions for each chromosome for each duo using PLINK31 (command flag:--genome).
Identification of Runs of Homozygosity
We calculated runs of homozygosity (ROH) using GARLIC.32, 33 Briefly, GARLIC implements a model-based method for identifying ROH and classifying ROH into length classes. In this method, logarithm of the odds (LOD) scores for autozygosity are calculated in sliding windows of SNPs across the genome; SNP window sizes were chosen automatically by GARLIC based on SNP density. The LOD scores are functions of a user-specified error rate to account for genotyping error and mutation rate, as well as population-specific allele frequencies. The distribution of LOD scores is used to determine a threshold for ROH calling. After ROH are identified, contiguous ROH windows are concatenated. Lastly, we performed Gaussian mixture modeling using the Mclust function from the mclust R package34 (v.5.4) with the same parameters used by Kang et al.35 to classify ROH into three length classes: (1) class A, which are the shortest ROH; (2) class B; and (3) class C, which are the longest (class boundaries shown in Table S1).
We applied GARLIC to the eight cohorts in the 23andMe database described earlier: 974,511 northern Europeans, 34,508 southern Europeans, 90,349 African Americans, 70,144 Ashkenazi Jewish, 63,683 East Asians, 19,493 South Asians, 208,087 Latino/as, and 16,013 Middle Eastern individuals as well as 431,094 northern Europeans in the UK Biobank dataset. In the 23andMe dataset, we used a window size of 60 SNPs for autosomes and 30 SNPs for the X chromosome, which were automatically chosen by GARLIC as the best window size given SNP density, and an error rate of 0.001, which was used in previous studies of ROH.32, 33 In the UK Biobank dataset, we used a window size of 60 SNPs for both autosomes and the X chromosome, which were automatically chosen by GARLIC as the best window size for these data. In each dataset, only females were included in analyses of ROH on the X chromosome. In the 23andMe database, population-specific allele frequencies were calculated from individuals who are true negatives for UPD (identified as described in IBD-based UPD Detection); sample sizes of true negatives in the 23andMe cohorts are as follows: 28,338 northern Europeans, 1,018 southern Europeans, 1,500 African Americans, 2,066 Ashkenazi Jewish, 2,031 East Asians, 982 South Asians, 7,639 Latino/as, and 437 Middle Eastern individuals. In the UK Biobank cohort, we calculated allele frequencies for GARLIC from 431,094 individuals of northern European ancestry. All class C ROH were then filtered for deletions as described in Filtering of Deleted Genomic Regions.
IBD-Based UPD Detection
To detect UPD events in children, we looked for parent-child duos who lack IBD segments across an entire chromosome. For the X chromosome, only the following parent-child pairs, which would normally be expected to share IBD on the X chromosome, were considered: mother-daughter pairs, father-daughter pairs, and mother-son pairs. The putative UPD cases were then tested for a deletion spanning the putative UPD chromosome in both the parent and child according to the method described in Filtering of Deleted Genomic Regions below; we refer to children in parent-child pairs without deletion of the putative UPD chromosome as true positives for UPD. IBD segments can also be used to determine true negatives for UPD; we identified children in trios who are completely half identical to both parents and refer to these as true negatives. In order to calculate prevalence of UPD, we focus on trio data since only then can both parent-child pairs be tested for missing IBD.
To distinguish between maternal UPD (matUPD; when the disomic chromosome pair originates from the mother) and paternal UPD (patUPD; when the disomic chromosome pair originates from the father), we labeled the older individual in a parent-child duo as the parent and the younger individual as the child using self-reported age data. If an individual was missing IBD across a chromosome with the father, we labeled the case as maternal UPD (matUPD). If an individual was missing IBD with their mother, we labeled the case as paternal UPD (patUPD).
For UPD case subjects with a mother and father genotyped in the 23andMe database, we were able to use IBD with the parent-of-origin of the disomic chromosome pair to differentiate between the three subtypes of UPD: isodisomy (isoUPD), heterodisomy (hetUPD), and partial isodisomy (partial isoUPD). IsoUPD chromosomes are completely half-identical to the parent of origin, and hetUPD chromosomes are completely identical to the parent of origin. Partial isoUPD chromosomes are some fraction half-identical and some fraction fully identical to the parent of origin. For UPD cases detected in parent-child duos and lacking genotype data for the parent of origin, we use ROH to differentiate between the three subtypes. UPD chromosomes with ROH spanning 100% of the chromosome are labeled isoUPD, UPD chromosomes with 0% class C ROH are labeled hetUPD, and UPD chromosomes with between 0% and 100% class C ROH are labeled partial isoUPD.
Filtering of Deleted Genomic Regions
Large deletions—which can arise from somatic events or be prevalent in low-quality saliva samples—can manifest in genotype data as large regions of homozygosity and missing IBD that confound UPD detection. Thus, we screened all putative UPD case subjects for deletions. We filtered for deletions in one of two ways: by testing for significantly decreased Log R Ratio (LRR) across an ROH, and by using the CNV caller in BCFtools36 (v.1.4.1). LRR, which is a measure of probe intensity, can be used to detect several types of copy number variants.37 Theoretically, LRR is 0 at all loci across the genome and decreases across a deleted region. Therefore, we tested whether the average LRR of a given region (i.e., a run of homozygosity) was significantly lower than the genome-wide average LRR using a two-sample t test. Runs of homozygosity with significantly lower LRR than the genome-wide average (p value < 0.05) were filtered out of all ROH-based analyses.
For a chromosome missing IBD between a parent-child pair, we used the CNV calling function of BCFtools to identify whole-chromosome deletions and trisomies/mosaic trisomies in the parent and child at the putative UPD chromosome.36 The command line option “-l 0.8” was used to upweight LRR within the HMM model relative to BAF; when BAF is given equal weight with LRR, we found that blocks of isodisomy were called as deletions even without a corresponding decrease in LRR across the homozygous region. If more than 40% of a putative chromosome in either parent or child is called with copy number of 1 (deletion), the pair was excluded from further analysis. If more than 50% of the chromosome in either parent or child is called with copy number of 3 (trisomy), the pair was also excluded.
Simulation of Training Data for ROH-Based UPD Classifiers
We generated training data for 23 logistic regression-based classifiers for eight cohorts (described in Population Structure) to detect UPD of each of the autosomes and the X chromosome without parental genotype data, as follows. We simulated 46,000 individuals, consisting of 1,000 UPD case subjects and 1,000 control subjects for each chromosome for each of the eight cohorts, by randomly pairing 46,000 pairs of individuals across all cohorts. We selected individuals without any class C ROH-length deletions and pairs sharing less than 930 cM IBD to be “parents” for 1,000 simulated case subjects and 900 simulated control subjects. To model consanguinity within our training data, we forced 100 pairs of “parents” of simulated control subjects for each chromosome to share between 100 and 930 cM IBD. For our X chromosome classifiers, only female children were simulated since two X chromosomes are required to detect homozygosity on the X chromosome.
We generated 2,000 independent trios with one child each to train each classifier. We randomly sampled recombination breakpoints according to a distribution of crossover probabilities for each locus. The probability of at least one crossover between every pair of adjacent loci was calculated using Equation 1 below, in which we assume that crossovers are Poisson distributed with a rate equal to the difference in genetic distance in cM between two given loci multiplied by 0.01 (since there is approximately one crossover event in 100 cM).
| (Equation 1) |
To calculate genetic distances, we used recombination maps ascertained from the 23andMe research cohort.7 The genetic maps are publicly available online (see Web Resources). We then simulated meiosis by randomly choosing one homolog from each parent to be inherited by the child. For each chromosome’s classifier, we simulated UPD case subjects by deriving both homologs of that chromosome from only one parent. Lastly, we copied genotypes from parental homologs to generate genotypes for the child.
We used GARLIC to detect ROH for each simulated child using the same parameters and population allele frequencies specified in Identification of Runs of Homozygosity. We found that ROH length distributions were not significantly different between the simulated UPD case subjects and true positive UPD case subjects ascertained through IBD-based UPD detection (p value > 0.05) (Figure S1A). However, the distribution of the number of class C ROH differed significantly between the simulated UPD case subjects and real UPD case subjects (p value < 0.05), and thus, we did not use the number of class C ROHs to train the classifiers (Figure S1B). Our simulations produced every subtype of UPD (Figures S2 and S3).
ROH-Based UPD Detection and Performance Assessment
We developed 23 logistic regression classifiers, one for each autosome and the X chromosome, with two independent variables, trained on the simulations described in the previous section. For a given chromosome i, where and n = 23 in females and n = 22 in males, let ci be the total class C ROH length in base pairs. Also, let c(i) be the ith order statistic for n class C ROH lengths across all chromosomes, where c(n) is the maximum ROH length across all chromosomes. The two variables we trained each classifier on are ci for and c(n −1)/c(n). We focused on class C ROH for training the classifiers because, in comparing the distributions of ROH lengths between true positives (UPD case subjects detected through IBD) and true negatives for UPD, we found that only class C ROH length is significantly different (p value < 0.05) between the true positives and true negatives (Figure S4).
To assess the performance of our classifiers, we generated receiver operating characteristic (ROC) curves by testing each classifier on (1) a simulated set of 11,500 individuals, consisting of 250 case subjects and 250 control subjects for each chromosome, and independent from the training simulations described in the previous section; and (2) the set of true positives and true negatives for UPD ascertained from IBD-based UPD detection. In testing, we found that performance of the classifiers, as measured by area under a ROC curve (auROC), increases with increased proportion of isodisomy on the UPD chromosome (Figure S5). Specifically, detecting hetUPD without parental data is not possible due to the lack of large ROH blocks, and partial isoUPD detection is dependent on the size of the ROH. Thus, we restricted the training set further to comprise only true positives with at least 30% isoUPD on the UPD chromosome and a randomly sampled set of simulated control subjects equal in number to the case subjects for a given chromosome. We then chose initial probability cutoffs for each classifier from its ROC curve; we chose the probability threshold that minimized the false positive rate (FPR) and if there were multiple cutoffs that satisfied these criteria, we then chose the cutoff that also maximized the true positive rate (TPR). We used these cutoffs to classify putative UPD case subjects of each chromosome and remove duplicate cases; this last step removes individuals with large blocks of ROH on multiple chromosomes due to recent consanguinity. We then chose a final probability cutoff for our classifiers of 0.9 to classify ROH-based UPD case subjects.
Phenotypic Association Studies (PheWASs) in the 23andMe Dataset
We regressed 208 phenotypes (Tables S2 and S3) across five categories (cognitive, personality, morphology, obesity, and metabolic traits) onto UPD status, using children with UPD (true positives detected using IBD analysis) as case subjects and true negatives for UPD as control subjects. We tested each subtype of UPD (by chromosome and parent of origin) separately, and we restricted these analyses to individuals of European ancestry. We performed logistic regression for binary traits (Table S2) and linear regression for quantitative traits (Table S3) with the following covariates: age, sex, genotyping platform, and the first five principal components (see Genome-Wide Association Studies (GWAS) in the 23andMe Dataset) to adjust for population substructure. We also tested for differences in parental age between UPD true positives and true negatives.
Genome-Wide Association Studies (GWASs) in the 23andMe Dataset
We conducted GWASs on parents of UPD true positives identified by IBD-based analysis in order to find loci associated with the risk of giving birth to children with UPD. The GWAS was performed on all SNPs that passed quality control by running a logistic regression model correcting for the effects of age, parental age at birth of the child with UPD, first five genetic principal components, and genotype platform, performed separately by sex of the parents (mother or father). For more details on GWASs, imputation, and PCA, please refer to the Supplemental Subjects and Methods.
Results
UPD Prevalence Estimated from Parent-Child Genotypes
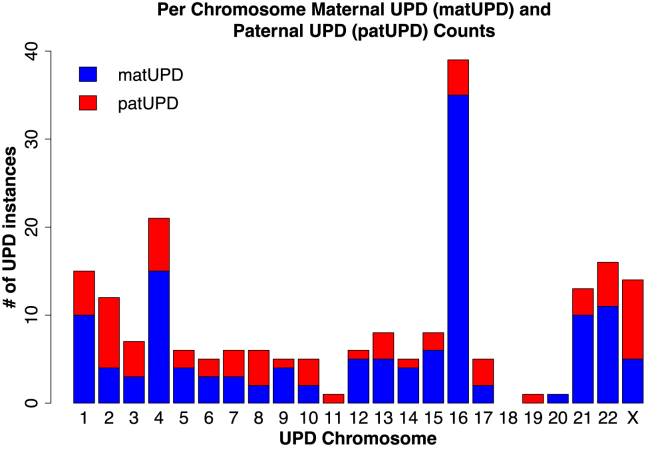
Unlike typical parent-child pairs, individuals with UPD lack IBD segments across an entire chromosome with one parent; thus, we can use parent-child data to identify UPD case subjects. In the 23andMe dataset, we analyzed 916,712 parent-child pairs, which include 214,915 trios, to estimate the prevalence of UPD. We found 199 case subjects of UPD distributed across all 23 chromosomes except chromosome 18 (Figure 2). Within 214,915 trios, we found 105 cases of UPD and estimate that UPD occurs with an overall prevalence rate of roughly 1 in 2,000 births (rate: 0.05%; 99% CI: [0.04%, 0.06%]). Thus, we found that UPD is more common than previously thought (previous estimate based on UPD15 case subjects: 1 in 3,500 births or 0.03%22). Of the 105 true positives observed in 23andMe trios, 26 are patUPD case subjects and 79 are matUPD, suggesting that maternal-origin UPD is three times as prevalent as paternal-origin UPD. Within the 23andMe trios, four were double UPD case subjects, where two chromosomes in the same individual were inherited uniparentally; thus, we estimate that double UPD occurs at a rate of roughly 1 in 50,000 births. We also found that paternal partial isoUPD is the least common UPD subtype, and we observed more hetUPD and partial isoUPD case subjects than isoUPD case subjects (Table 1). We also searched for UPD in 3,923 parent-child pairs in the UK Biobank, and we did not identify any cases of UPD.
Figure 2.
Using IBD-Based UPD Detection, We Found 199 Individuals with 205 Incidences of UPD in 916,712 Parent-Child Duos from the 23andMe Dataset
We identified six individuals with cases of double UPD, when two chromosomes in one individual are inherited uniparentally. This plot shows the per-chromosome distribution of true positives for UPD in the 23andMe dataset; in these true positives, UPD occurs most frequently on chromosomes 1, 4, 16, 21, 22, and X. We also observed three times as many maternal UPD (matUPD) case subjects as paternal UPD (patUPD) case subjects.
Table 1.
All Three Subtypes of UPD
| isoUPD | hetUPD | Partial isoUPD | Total | |
|---|---|---|---|---|
| matUPD | 20 | 45 | 69 | 134 |
| patUPD | 35 | 31 | 5 | 71 |
| Total | 55 | 76 | 74 | 205 |
We identified all three subtypes of UPD in the 205 instances of UPD identified from 916,712 parent-child duos from the 23andMe dataset. We found that paternal partial isoUPD is the least common subtype of UPD. We also observed overall more hetUPD and partial isoUPD case subjects than isoUPD case subjects.
We compared the per-chromosome rates of UPD true positives in the 23andMe database to those from published reports of UPD in the literature (Figure S6). We found that, while UPD true positives in the 23andMe database occur most frequently on chromosomes 1, 4, 16, 21, 22, and X, published UPD cases are most common on chromosomes 6, 7, 11, 14, and 158 (see Web Resources). We failed to reject the null hypothesis of independence between per-chromosome rates of 23andMe UPD true positives and published UPD cases (Fisher’s exact test; p value = 1), and the two per-chromosome distributions are not significantly correlated (Pearson’s correlation; p value = 0.72). The most common UPD chromosomes in the 23andMe duos are significantly depleted for imprinted genes (see Web Resources; Figure S7A), which may cause clinical phenotypes, compared to all other chromosomes (Fisher’s exact test; p value = 1.33 × 10−6). We find that the ratio of per chromosome UPD rates from published cases to the per chromosome UPD rates from 23andMe duos is significantly correlated with the number of imprinted genes on each chromosome (Figure S7B; Pearson’s correlation = 0.70, p value = 0.0003). We also find that the per chromosomes rates of UPD from 23andMe duos are not significantly correlated with the number of imprinted genes (Figure S7C; Pearson’s correlation = −0.32, p value = 0.13). Thus, we conclude that published UPD cases are biased toward chromosomes where UPD causes clinical presentation and do not represent the true distribution of UPD in the general population.
ROH-Based UPD Detection without Parental Genotypes
In many studies, parental genotypes for all probands may be too costly or logistically difficult to generate, and so classification of putative UPD cases may be made based on singleton genotypes only. Several clinical guidelines exist for prioritizing putative UPD cases for further analysis; all of these methods look for a large ROH confined to a single chromosome.38 However, multiple population-genetic studies have shown that relatively large ROH (>1 Mb) are common even in outbred populations;32, 35, 39, 40 we recapitulate this result in eight cohorts in the 23andMe dataset (Figure S8). Thus, an effective ROH-based method for UPD detection must be able to identify UPD chromosomes in the presence of large ROH on non-UPD chromosomes. Further, such a method must be able to distinguish between partial isoUPD and ROH blocks resulting from consanguinity.
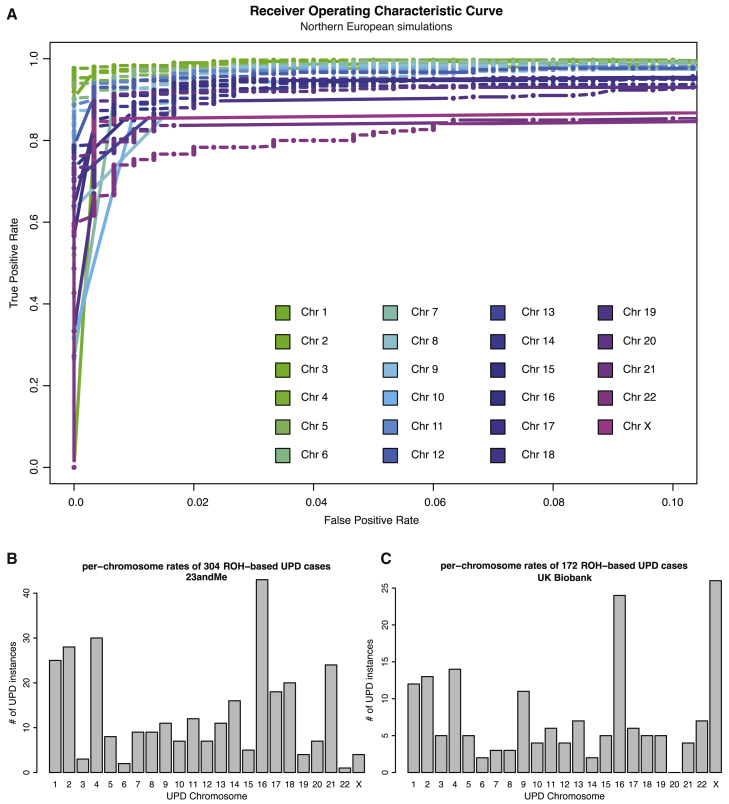
To address these challenges, here we introduce a supervised logistic regression classification framework that accounts for ROH length distributions within ancestral populations and is able to identify partial isoUPD and isoUPD on all autosomes and the X chromosome. In simulations across five cohorts in the 23andMe dataset (northern European, southern European, Latino, African American, and East Asian individuals), we demonstrated that our classifiers achieve high power while minimizing the false positive rate (auROC > 0.9 for all classifiers; Figures 3A and S9). We also found that classifiers for larger chromosomes perform better than those for smaller chromosomes (Figures 3A and S9). In three cohorts (Ashkenazi Jewish, Middle Eastern, and South Asians), we found that our classifiers performed poorly on simulated genotype data (auROC < 0.9; Figure S9); when analyzing individual genotype data from these three cohorts in the 23andMe dataset, we classified thousands of putative cases which appeared to be false positives. Therefore, we ignored these three cohorts for further ROH-based UPD detection. As another form of validation, we applied our classifiers to northern European true positives with ROH spanning at least 20% of the UPD chromosome and northern European true negatives from IBD-based UPD detection. We find that our classifiers also achieved high power while minimizing false positive rate (auROC > 0.95 for 5 chromosomes tested; Figure S10) when applied to true positives and true negatives from IBD analysis. Using our chosen probability cutoff of 0.9, we identified 85% of the northern European true positives (TPR) and we did not classify any of the northern European true negatives as putative UPD cases (FPR). In 1,371,138 singletons from five cohorts in the 23andMe dataset (northern European, southern European, Latino, African American, and East Asian individuals), we classified 304 putative ROH-based UPD cases using our ROH-based method, 297 of which were newly discovered using ROH analysis (Figure 3B). The chromosome distribution of the ROH-based cases (Figure 3B) recapitulates the chromosome distribution of true positives identified through IBD analysis (Figure 2; Pearson’s correlation = 0.67; p value = 0.0005).
Figure 3.
Using ROH-Based UPD Detection, We Identified 304 UPD Case Subjects across Five Cohorts in the 23andMe Dataset and 172 UPD Case Subjects in the UK Biobank
We developed a per-chromosome simulation-based classification framework to search for putative UPD case subjects using ROH across five cohorts (northern European, southern European, Latino, African American, and East Asian individuals) in the 23andMe and in northern Europeans in the UK Biobank.
(A) Receiver operating characteristic (ROC) curves show the performance of our per-chromosome UPD classifiers on simulated testing data, based on genotype data from northern Europeans in the 23andMe dataset. Our classifiers identified UPD with high accuracy (area under the ROC curve [auROC] > 0.9; TPR between 0.75 and 0.98 when FPR is fixed at 0.01). At fixed FPR, power is inversely related to chromosome length.
(B) The chromosome distribution of the ROH-based cases found in the 23andMe dataset recapitulates features of the chromosome distribution of true positives for UPD, which are identified through IBD analysis (Figure 2; Pearson’s correlation = 0.67; p value = 0.0005).
(C) The chromosome distribution of the ROH-based cases found in the UK Biobank also recapitulates features of the chromosome distribution of true positives for UPD identified through IBD analysis (Figure 2; Pearson’s correlation = 0.74; p value = 4.79 × 10−5). We note that parent-of-origin cannot be identified for ROH-based cases.
We also applied these 23 classifiers to data from 431,094 northern European individuals from the UK Biobank Project and identified 172 ROH-based UPD case subjects, observing cases of each chromosome except chromosome 20 (Figure 3C). The chromosome distribution of ROH-based cases (Figure 3C) in the UK Biobank also recapitulates the chromosome distribution of UPD cases identified through IBD analysis in the 23andMe database (Figure 2; Pearson’s correlation = 0.74; p value = 4.785 × 10−5). Karyograms of ROH in the 172 putative UPD cases from the UK Biobank show large blocks of homozygosity ranging from 12.7 Mb to 231 Mb (Figure S11; Data S1). The classifier models and code developed in this study are publicly available online (see Web Resources).
Phenotypic Consequences of UPD
UPD can cause phenotypic consequences in multiple ways, including (1) disrupting imprinting and (2) uncovering recessive alleles in blocks of isodisomy. We tested for phenotypic associations between UPD of each of the 23 chromosomes in true positives in the 23andMe dataset and 208 phenotypes (Tables S2 and S3) across five categories (cognitive, personality, morphology, obesity, and metabolic traits) obtained from self-reported survey answers. We found 23 nominally significant (p value < 0.01) phenotype associations with UPD of chromosomes 1, 3, 6, 7, 8, 15, 16, 21, and 22 (Table S4). While some of these 23 associations were driven by a single UPD case, three associations had multiple cases (or multiple measurements, in the case of quantitative traits), representing a more robust signal: we found that UPD6 is associated with lower weight (p value = 0.0038) and shorter height (p value = 0.0055) and that UPD22 is associated with a higher risk for autism (p value = 2.557 × 10−5) (Table 2). We note that none of these associations remain significant after Bonferroni correction for the number of phenotypes (208) and chromosomes (22) tested.
Table 2.
PheWAS in the 23andMe Dataset Identified Phenotypes Significantly Associated with UPD of Chromosomes 6 and 22
| UPD Type | Phenotype | Effect Size (95% CI) | p Values (Uncorrected) |
|---|---|---|---|
| UPD6 | weight | −2.02 (−3.38 −0.65) | 0.0038 |
| UPD6 | height | −1.99 (−3.40 −0.59) | 0.0055 |
| UPD22 | autism spectrum | 3.61 (1.93 5.30) | 2.557 × 10−5 |
Only traits with at least two case subjects (or two measurements for quantitative traits) are shown; the full list of all associations is shown in Table S4. Effect sizes shown are odd ratios. We tested for association between UPD on each of the autosomes and 208 self-reported phenotypes (Tables S2 and S3) across five categories (cognitive, personality, morphology, obesity, and metabolic traits). We uncover associations between UPD of chromosome 6 and weight and height, and UPD of chromosome 22 and autism spectrum (p value < 0.01); we note that none of these associations remain significant after Bonferroni correction for the number of phenotypes (208) and chromosomes (22) tested.
Variants Associated with UPD Incidence
Although heritability of UPD, or chromosomal aneuploidy, has not been reported, there may be genetic variants that predispose individuals to produce aneuploid germ cells, thus increasing the likelihood of giving birth to an offspring with UPD. We tested this hypothesis by performing a genome-wide association study (GWAS) comparing 221 parents of UPD case subjects to 205,141 parents of UPD true negatives from IBD analysis. We performed the analysis in all parents, adjusted for age, and stratified by parental sex. No association reached genome-wide significance (p value = 5 × 10−8), and the heritability estimated by LD score regression41 was non-significant across all three analyses (Figure S12; p value > 0.05). Given the small sample size, this likely reflects our lack of power to detect genetic associations even for common variants associated with UPD.
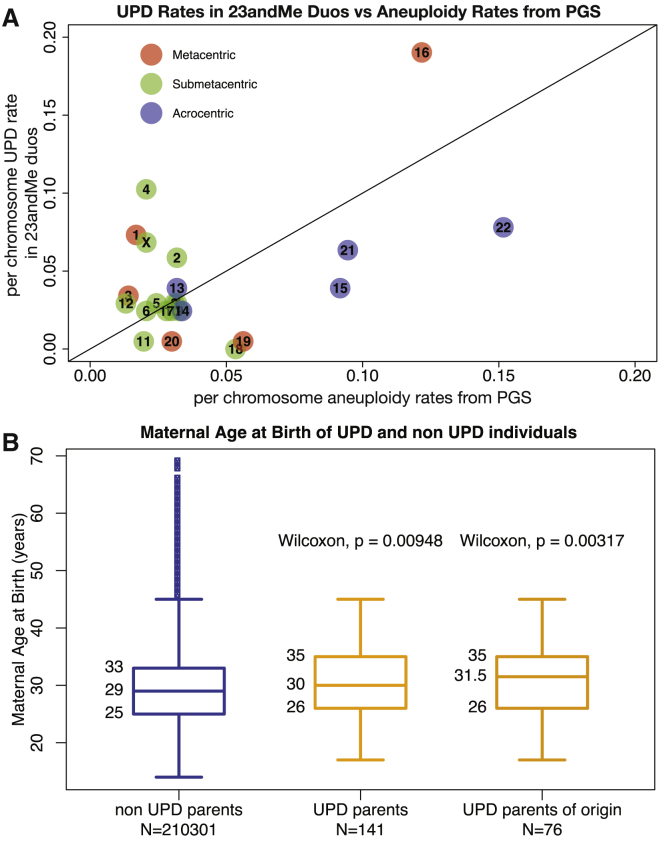
In order to further investigate the etiology of UPD, we assess the relationship between per-chromosome UPD rates and per-chromosomes rates of aneuploidy in pre-implantation embryos (PGS). We found that UPD rates in the 23andMe database are significantly correlated with published aneuploidy rates from PGS4 (Figure 4A; Pearson’s correlation = 0.49; p value = 0.02). We also found that mothers who are parents of origin of UPD true positives in the 23andMe dataset are significantly older than those of UPD true negatives (Figure 4B; Wilcoxon p value = 0.00317), whereas paternal age does not show a robust association with UPD (Figure S13; Wilcoxon p value = 0.286).
Figure 4.
Aneuploidy Rates and Maternal Age Are Correlated with UPD
(A) The correlation between per-chromosome UPD rate in true positives from the 23andMe database and per chromosome aneuploidy rate in published pre-implantation genetic screening data;4 chromosomes are colored by centromeric type: metacentric chromosomes are shown in red, submetacentric chromosomes in green, and acrocentric chromosomes in blue. These two rates are significantly correlated (Pearson’s correlation = 0.49; p value = 0.02) and this correlation remains significant after correction for chromosome length and centromeric type (Pearson’s correlation = 0.73; p value = 0.006), suggesting that meiotic nondisjunction occurs more frequently on some chromosomes (such as 15, 16, 21, and 22) than others, resulting in more instances of both UPD and aneuploidy on these chromosomes. We also note that the acrocentric chromosomes have among the highest per-chromosome rates of both UPD and aneuploidy.
(B) The age distribution of mothers of UPD true negatives (blue) and that of mothers who are parents of origin of UPD true positives (matUPD cases, yellow) in the 23andMe dataset. We find that mothers of UPD case subjects are significantly older than mothers of UPD true negatives (Wilcoxon p value = 0.00948) and that this associations holds when restricted to cases of matUPD, where mothers are the parents of origin of the UPD case subjects (Wilcoxon p value = 0.00317).
Discussion
The recombination process has long been studied by evolutionary, medical, molecular, and population geneticists, in part to gain insight into meiotic nondisjunction. It is difficult to directly study meiotic nondisjunction in humans because errors in meiosis often lead to fetal loss or serious health consequences. However, UPD is a detectable genomic signature of meiotic nondisjunction and aneuploidy in euploid, liveborn individuals. In this study we show that, given large genomic datasets, detecting UPD offers new insight into recombination and meiosis in humans.
First, using 214,915 trios in the 23andMe customer database, we obtained an estimate of UPD prevalence in the general population: 1 in 2,000 births, 1.75 times higher than the current clinical estimate of 1 in 3,500 births. The current estimate of UPD prevalence is derived from UPD15 prevalence in clinical cohorts, which might not be representative of the general population and also does not account for differences in UPD prevalence between chromosomes.22 The 23andMe customer base comprises, for the most part, healthy individuals from the general population and so our estimate is more representative of overall UPD prevalence. We also found that the per-chromosome prevalence rate of UPD is significantly correlated with per-chromosome aneuploidy rates calculated from published PGS data (Figure 4A; Pearson’s correlation = 0.49; p value = 0.02) whereas per-chromosome rates from clinical UPD cases are not (Figure S14; Pearson’s correlation = 0.2; p value = 0.34). Since a liveborn individual with UPD results from the restoration of euploidy in an aneuploid zygote, we expect the true per-chromosome rates of UPD to be correlated with those of aneuploidy, providing further evidence that our estimated rates are closer to the true prevalence and per chromosome distribution of UPD than existing clinical rates. We note that participation in 23andMe may be cost prohibitive for many and also that the customer base may be biased toward geographic regions or other covariates. Furthermore, individuals with severe health problems may be unlikely or unable to participate in 23andMe, and so the UPD cases in this study may be depleted for UPD causing serious health consequences.
Second, we have introduced a machine-learning method to find UPD cases using genomic data from singleton data only, without requiring parental genotypes. Existing guidelines for classification of putative UPD cases without parental genotypes consist of a hard ROH length threshold for all chromosomes.38 However, ROH length distributions vary (1) by the demographic history of an individual’s ancestral population(s), (2) by the history of consanguinity in the individual’s recent ancestors, and (3) by chromosome. Our method learns the distributions of ROH lengths on each chromosome from simulated data based on each of eight global population cohorts in the 23andMe dataset while also modeling recent consanguinity and is able to classify UPD with high accuracy. Using our method, we were able to find 297 additional UPD case subjects in 1,371,138 individuals in the 23andMe cohort. Our classifiers can also be readily applied to other genomic datasets such as the UK Biobank,25, 26 in which we identified 172 additional ROH-based UPD case subjects (Figures 3C and S11, Data S1). This underscores that an effective ROH-based method for UPD detection offers crucial insight into UPD when combined with large-scale genomic datasets; these putative cases can then be further investigated by genotyping parents, cytogenetic techniques, or DNA methylation studies. One limitation of our ROH-based detection method is that we can only identify isoUPD and partial isoUPD cases that contain large blocks of homozygosity (spanning greater than 30% of the chromosome). In that respect, UPD per-chromosome rates estimated using our ROH-based method are conservative. In order to minimize the false positive rate, we did not try to refine classification of small partial isoUPDs (Figure S5). Also, we were unable to identify UPD in populations that are historically known to practice endogamy and thus have higher than average levels of homozygosity (Ashkenazi Jewish, Middle Eastern, and South Asian individuals; Figure S9).
Errors in recombination typically, with few exceptions,3, 6 lead to aneuploidy and severe health consequences, and so are largely viewed as deleterious. However, the majority of UPD types, including the most common UPD (UPD16), did not show significant, plausible associations with deleterious traits in the 23andMe database (Table 2). Our work challenges the typical view that errors in recombination are strongly deleterious, showing that even in extreme cases where individuals are homozygous for an entire chromosome, those individuals can be, to the best of our knowledge, phenotypically normal and healthy (Table 2). We note that phenotype data in the 23andMe database is self-reported and so depends on customers answering surveys about their health and traits. We also note that there has yet to be a prospective study of the long-term consequences of UPD since most current studies focus on special syndromes and recessive disorders that are apparent in childhood; future studies could extend our work in this direction.
To interrogate the role of genetics in UPD etiology, we performed a GWAS of UPD. Though our results are mostly suggestive, with increased sample sizes or deep sequencing, future studies may find plausible, significant loci underlying UPD incidence.
Lastly, we expect the etiology of meiotic nondisjunction and UPD to be similar since UPD is caused by rescue of aneuploid zygotes. Here, we found that UPD rates in the 23andMe database are significantly correlated with aneuploidy rates from PGS (Figure 4A; Pearson’s correlation = 0.49; p value = 0.02). Also, similarly to aneuploidy, we found that mothers who are parents of origin of UPD true positives in the 23andMe dataset are significantly older than those of UPD true negatives (Figure 4B; Wilcoxon p value = 0.00317). Previous studies have shown elevated escape from crossover interference on certain chromosomes (8, 9, and 16) and especially in older mothers; future studies could test whether crossover interference rates vary between UPD case subjects and UPD true negatives.7 And though we focused in this study on meiotic-origin UPD, future studies could also extend our work to characterize the prevalence and chromosomal distribution of segmental (or mitotic-origin) UPD case subjects in the general population; segmental UPD is also currently studied only in clinical settings.9
Consortia
Members of the 23andMe Research Team: Michelle Agee, Adam Auton, Robert K. Bell, Katarzyna Bryc, Sarah L. Elson, Pierre Fontanillas, Nicholas A. Furlotte, Barry Hicks, David A. Hinds, Ethan M. Jewett, Yunxuan Jiang, Keng-Han Lin, Jennifer C. McCreight, Karen E. Huber, Aaron Kleinman, Nadia K. Litterman, Matthew H. McIntyre, Elizabeth S. Noblin, Carrie A.M. Northover, Steven J. Pitts, G. David Poznik, Janie F. Shelton, Suyash Shringarpure, Chao Tian, Joyce Y. Tung, Vladimir Vacic, and Xin Wang.
Declaration of Interests
J.F.S., K.F.M., J.L.M., and members of 23andMe Research Team are employees of 23andMe, Inc., and are share or option holders of the company. P.N. was an employee of 23andMe, Inc., while conducting the research. All other authors declare no competing interests.
Acknowledgments
We thank the 23andMe research participants who made this work possible. We also thank the employees of 23andMe who developed the infrastructure that made this research possible. This research has been conducted using the UK Biobank Resource under Application Number 44606. We gratefully acknowledge Uta Francke, Shai Carmi, Aaron Carrel, Kirk Lohmueller, Priya Moorjani, John Novembre, Ben Raphael, Suyash Shringarpure, Janie Shelton, and the Ramachandran Lab for helpful conversations. This research was supported in part by US National Institutes of Health (NIH) grant R01GM118652, NIH COBRE award P20GM109035, and National Science Foundation (NSF) CAREER award DBI-1452622 to S.R. Support was also provided by the NIH National Child Health and Development Institute training grant K12HD052896 to A.H.O.-L.
Published: October 10, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.09.016.
Contributor Information
Priyanka Nakka, Email: pnakka@wellesley.edu.
J. Fah Sathirapongsasuti, Email: fsathirapongsasuti@23andme.com.
23andMe Research Team:
Michelle Agee, Adam Auton, Robert K. Bell, Katarzyna Bryc, Sarah L. Elson, Pierre Fontanillas, Nicholas A. Furlotte, Barry Hicks, David A. Hinds, Ethan M. Jewett, Yunxuan Jiang, Keng-Han Lin, Jennifer C. McCreight, Karen E. Huber, Aaron Kleinman, Nadia K. Litterman, Matthew H. McIntyre, Elizabeth S. Noblin, Carrie A.M. Northover, Steven J. Pitts, G. David Poznik, Janie F. Shelton, Suyash Shringarpure, Chao Tian, Joyce Y. Tung, Vladimir Vacic, and Xin Wang
Web Resources
23andMe’s Ancestry Composition, https://www.23andme.com/ancestry-composition-guide/
23andMe Genetic Maps, https://github.com/auton1/Campbell_et_al
Ethical and Independent Review Services, http://www.eandireview.com
Geneimprint, http://www.geneimprint.com/site/genes-by-species.Homo+sapiens.any
UK Biobank, https://www.ukbiobank.ac.uk
UPD cases, http://upd-tl.com/upd.html
UPD Detector, https://github.com/ramachandran-lab/UPD_Detector
Supplemental Data
We tested for association between UPD of each of the chromosomes with at least one UPD case and 172 binary phenotypes across five categories (cognitive, personality, morphology, obesity and metabolic traits). Where possible, we also tested for association between matUPD and patUPD of each of the chromosomes separately.
References
- 1.Koehler K.E., Hassold T.J. Human aneuploidy: lessons from achiasmate segregation in Drosophila melanogaster. Ann. Hum. Genet. 1998;62:467–479. doi: 10.1046/j.1469-1809.1998.6260467.x. [DOI] [PubMed] [Google Scholar]
- 2.Hassold T., Chen N., Funkhouser J., Jooss T., Manuel B., Matsuura J., Matsuyama A., Wilson C., Yamane J.A., Jacobs P.A. A cytogenetic study of 1000 spontaneous abortions. Ann. Hum. Genet. 1980;44:151–178. doi: 10.1111/j.1469-1809.1980.tb00955.x. [DOI] [PubMed] [Google Scholar]
- 3.Fledel-Alon A., Wilson D.J., Broman K., Wen X., Ober C., Coop G., Przeworski M. Broad-scale recombination patterns underlying proper disjunction in humans. PLoS Genet. 2009;5:e1000658. doi: 10.1371/journal.pgen.1000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez-Purata J., Lee J., Whitehouse M., Moschini R.M., Knopman J., Duke M., Sandler B., Copperman A. Embryo selection versus natural selection: how do outcomes of comprehensive chromosome screening of blastocysts compare with the analysis of products of conception from early pregnancy loss (dilation and curettage) among an assisted reproductive technology population? Fertil. Steril. 2015;104 doi: 10.1016/j.fertnstert.2015.08.007. 1460–66.e1, 12. [DOI] [PubMed] [Google Scholar]
- 5.Popadin K., Peischl S., Garieri M., Sailani M.R., Letourneau A., Santoni F., Lukowski S.W., Bazykin G.A., Nikolaev S., Meyer D. Slightly deleterious genomic variants and transcriptome perturbations in Down syndrome embryonic selection. Genome Res. 2018;28:1–10. doi: 10.1101/gr.228411.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coop G., Wen X., Ober C., Pritchard J.K., Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 7.Campbell C.L., Furlotte N.A., Eriksson N., Hinds D., Auton A. Escape from crossover interference increases with maternal age. Nat. Commun. 2015;6:6260. doi: 10.1038/ncomms7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liehr T. Cytogenetic contribution to uniparental disomy (UPD) Mol. Cytogenet. 2010;3:8. doi: 10.1186/1755-8166-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotzot D. Complex and segmental uniparental disomy updated. J. Med. Genet. 2008;45:545–556. doi: 10.1136/jmg.2008.058016. [DOI] [PubMed] [Google Scholar]
- 10.Conlin L.K., Thiel B.D., Bonnemann C.G., Medne L., Ernst L.M., Zackai E.H., Deardorff M.A., Krantz I.D., Hakonarson H., Spinner N.B. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum. Mol. Genet. 2010;19:1263–1275. doi: 10.1093/hmg/ddq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalish J.M., Conlin L.K., Bhatti T.R., Dubbs H.A., Harris M.C., Izumi K., Mostoufi-Moab S., Mulchandani S., Saitta S., States L.J. Clinical features of three girls with mosaic genome-wide paternal uniparental isodisomy. Am. J. Med. Genet. A. 2013;161A:1929–1939. doi: 10.1002/ajmg.a.36045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King D.A., Fitzgerald T.W., Miller R., Canham N., Clayton-Smith J., Johnson D., Mansour S., Stewart F., Vasudevan P., Hurles M.E., DDD Study A novel method for detecting uniparental disomy from trio genotypes identifies a significant excess in children with developmental disorders. Genome Res. 2014;24:673–687. doi: 10.1101/gr.160465.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Créau-Goldberg N., Gegonne A., Delabar J., Cochet C., Cabanis M.O., Stehelin D., Turleau C., de Grouchy J. Maternal origin of a de novo balanced t(21q21q) identified by ets-2 polymorphism. Hum. Genet. 1987;76:396–398. doi: 10.1007/BF00272452. [DOI] [PubMed] [Google Scholar]
- 14.Engel E. A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am. J. Med. Genet. 1980;6:137–143. doi: 10.1002/ajmg.1320060207. [DOI] [PubMed] [Google Scholar]
- 15.Yeung K.S., Ho M.S.P., Lee S.L., Kan A.S.Y., Chan K.Y.K., Tang M.H.Y., Mak C.C.Y., Leung G.K.C., So P.L., Pfundt R. Paternal uniparental disomy of chromosome 19 in a pair of monochorionic diamniotic twins with dysmorphic features and developmental delay. J. Med. Genet. 2018;55:847–852. doi: 10.1136/jmedgenet-2018-105328. [DOI] [PubMed] [Google Scholar]
- 16.Bruno D.L., White S.M., Ganesamoorthy D., Burgess T., Butler K., Corrie S., Francis D., Hills L., Prabhakara K., Ngo C. Pathogenic aberrations revealed exclusively by single nucleotide polymorphism (SNP) genotyping data in 5000 samples tested by molecular karyotyping. J. Med. Genet. 2011;48:831–839. doi: 10.1136/jmedgenet-2011-100372. [DOI] [PubMed] [Google Scholar]
- 17.Carmichael H., Shen Y., Nguyen T.T., Hirschhorn J.N., Dauber A. Whole exome sequencing in a patient with uniparental disomy of chromosome 2 and a complex phenotype. Clin. Genet. 2013;84:213–222. doi: 10.1111/cge.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiszniewska J., Bi W., Shaw C., Stankiewicz P., Kang S.-H.L., Pursley A.N., Lalani S., Hixson P., Gambin T., Tsai C.H. Combined array CGH plus SNP genome analyses in a single assay for optimized clinical testing. Eur. J. Hum. Genet. 2014;22:79–87. doi: 10.1038/ejhg.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brun B.N., Willer T., Darbro B.W., Gonorazky H.D., Naumenko S., Dowling J.J., Campbell K.P., Moore S.A., Mathews K.D. Uniparental disomy unveils a novel recessive mutation in POMT2. Neuromuscul. Disord. 2018;28:592–596. doi: 10.1016/j.nmd.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borgulová I., Soldatova I., Putzová M., Malíková M., Neupauerová J., Marková S.P., Trková M., Seeman P. Genome-wide uniparental diploidy of all paternal chromosomes in an 11-year-old girl with deafness and without malignancy. J. Hum. Genet. 2018;63:803–810. doi: 10.1038/s10038-018-0444-9. [DOI] [PubMed] [Google Scholar]
- 21.Wilfert A.B., Chao K.R., Kaushal M., Jain S., Zöllner S., Adams D.R., Conrad D.F. Genome-wide significance testing of variation from single case exomes. Nat. Genet. 2016;48:1455–1461. doi: 10.1038/ng.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson W.P. Mechanisms leading to uniparental disomy and their clinical consequences. BioEssays. 2000;22:452–459. doi: 10.1002/(SICI)1521-1878(200005)22:5<452::AID-BIES7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 23.Eriksson N., Macpherson J.M., Tung J.Y., Hon L.S., Naughton B., Saxonov S., Avey L., Wojcicki A., Pe’er I., Mountain J. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010;6:e1000993. doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tung J.Y., Do C.B., Hinds D.A., Kiefer A.K., Macpherson J.M., Chowdry A.B., Francke U., Naughton B.T., Mountain J.L., Wojcicki A., Eriksson N. Efficient replication of over 180 genetic associations with self-reported medical data. PLoS ONE. 2011;6:e23473. doi: 10.1371/journal.pone.0023473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durand E.Y., Do C.B., Mountain J.L., Macpherson J.M. Ancestry Composition: A Novel, Efficient Pipeline for Ancestry Deconvolution. bioRxiv. 2014 [Google Scholar]
- 28.Abraham G., Qiu Y., Inouye M. FlashPCA2: principal component analysis of Biobank-scale genotype datasets. Bioinformatics. 2017;33:2776–2778. doi: 10.1093/bioinformatics/btx299. [DOI] [PubMed] [Google Scholar]
- 29.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H., 1000 Genomes Project Consortium An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henn B.M., Hon L., Macpherson J.M., Eriksson N., Saxonov S., Pe’er I., Mountain J.L. Cryptic distant relatives are common in both isolated and cosmopolitan genetic samples. PLoS ONE. 2012;7:e34267. doi: 10.1371/journal.pone.0034267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pemberton T.J., Absher D., Feldman M.W., Myers R.M., Rosenberg N.A., Li J.Z. Genomic patterns of homozygosity in worldwide human populations. Am. J. Hum. Genet. 2012;91:275–292. doi: 10.1016/j.ajhg.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szpiech Z.A., Blant A., Pemberton T.J. GARLIC: Genomic Autozygosity Regions Likelihood-based Inference and Classification. Bioinformatics. 2017;33:2059–2062. doi: 10.1093/bioinformatics/btx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scrucca L., Fop M., Murphy T.B., Raftery A.E. mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. R J. 2016;8:289–317. [PMC free article] [PubMed] [Google Scholar]
- 35.Kang J.T.L., Goldberg A., Edge M.D., Behar D.M., Rosenberg N.A. Consanguinity Rates Predict Long Runs of Homozygosity in Jewish Populations. Hum. Hered. 2016;82:87–102. doi: 10.1159/000478897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Danecek P., McCarthy S.A., Durbin R., HipSci Consortium A Method for Checking Genomic Integrity in Cultured Cell Lines from SNP Genotyping Data. PLoS ONE. 2016;11:e0155014. doi: 10.1371/journal.pone.0155014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peiffer D.A., Le J.M., Steemers F.J., Chang W., Jenniges T., Garcia F., Haden K., Li J., Shaw C.A., Belmont J. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoppman N., Rumilla K., Lauer E., Kearney H., Thorland E. Patterns of homozygosity in patients with uniparental disomy: detection rate and suggested reporting thresholds for SNP microarrays. Genet. Med. 2018;20:1522–1527. doi: 10.1038/gim.2018.24. [DOI] [PubMed] [Google Scholar]
- 39.McQuillan R., Leutenegger A.-L., Abdel-Rahman R., Franklin C.S., Pericic M., Barac-Lauc L., Smolej-Narancic N., Janicijevic B., Polasek O., Tenesa A. Runs of homozygosity in European populations. Am. J. Hum. Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirin M., McQuillan R., Franklin C.S., Campbell H., McKeigue P.M., Wilson J.F. Genomic runs of homozygosity record population history and consanguinity. PLoS ONE. 2010;5:e13996. doi: 10.1371/journal.pone.0013996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bulik-Sullivan B.K., Loh P.-R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
We tested for association between UPD of each of the chromosomes with at least one UPD case and 172 binary phenotypes across five categories (cognitive, personality, morphology, obesity and metabolic traits). Where possible, we also tested for association between matUPD and patUPD of each of the chromosomes separately.