Abstract
Human-specific duplications at chromosome 16p11.2 mediate recurrent pathogenic 600 kbp BP4–BP5 copy-number variations, which are among the most common genetic causes of autism. These copy-number polymorphic duplications are under positive selection and include three to eight copies of BOLA2, a gene involved in the maturation of cytosolic iron-sulfur proteins. To investigate the potential advantage provided by the rapid expansion of BOLA2, we assessed hematological traits and anemia prevalence in 379,385 controls and individuals who have lost or gained copies of BOLA2: 89 chromosome 16p11.2 BP4–BP5 deletion carriers and 56 reciprocal duplication carriers in the UK Biobank. We found that the 16p11.2 deletion is associated with anemia (18/89 carriers, 20%, p = 4e−7, OR = 5), particularly iron-deficiency anemia. We observed similar enrichments in two clinical 16p11.2 deletion cohorts, which included 6/63 (10%) and 7/20 (35%) unrelated individuals with anemia, microcytosis, low serum iron, or low blood hemoglobin. Upon stratification by BOLA2 copy number, our data showed an association between low BOLA2 dosage and the above phenotypes (8/15 individuals with three copies, 53%, p = 1e-4). In parallel, we analyzed hematological traits in mice carrying the 16p11.2 orthologous deletion or duplication, as well as Bola2+/− and Bola2−/− animals. The Bola2-deficient mice and the mice carrying the deletion showed early evidence of iron deficiency, including a mild decrease in hemoglobin, lower plasma iron, microcytosis, and an increased red blood cell zinc-protoporphyrin-to-heme ratio. Our results indicate that BOLA2 participates in iron homeostasis in vivo, and its expansion has a potential adaptive role in protecting against iron deficiency.
Keywords: BOLA2, 16p11.2 copy number variants, iron deficiency anemia, human-specific segmental duplications, gene duplication, human evolution
Introduction
The human 16p11.2 chromosomal region is a hotspot of recurrent pathogenic copy-number variants with diverse sizes, breakpoints, and gene content. Most breakpoints map within homologous segmental duplication clusters consistent with non-allelic homologous recombination (NAHR).1 Among these variants, 600 kbp deletions and duplications with breakpoints BP4 and BP5 are among the most frequent genetic causes of neurodevelopmental and psychiatric disorders.2, 3, 4, 5, 6 They are also associated in a dose-dependent manner with head circumference, body mass index, age at menarche, and the size of brain structures associated with reward, language, and social cognition.5, 7, 8, 9, 10, 11, 12 BP4–BP5 rearrangements are mediated by Homo sapiens sapiens-specific duplications. These duplications appeared at the beginning of the modern human lineage, ∼282 thousand years ago, rapidly increased in frequency, and are now nearly fixed in humans, suggesting that a possible evolutionary advantage outweighs the accompanying chromosomal instability.13
The human-specific duplications at BP4 and BP5 contain three genes—BOLA2A/B, SLX1A/B, and SULT1A3/4—that have single-copy orthologs in the mouse genome. Whereas BOLA2A/B and SLX1A/B have copies only in the BP4–BP5 flanking repeats, SULT1A3/4 have paralogs within the neighboring BP2. The BP4–BP5 duplicons also harbor copies of the primate gene family NPIP,14 BOLA2-SMG1 and SLX1-SULT1A fusion genes, and SMG1P and LOC388242 pseudogenes (Figure 1A). These 102-kbp duplications are located at both ends of the single-copy region and are copy-number variants in the genomes of contemporary humans (three to eight diploid copies), but they are single copy in archaic genomes for which DNA sequence data are available.13
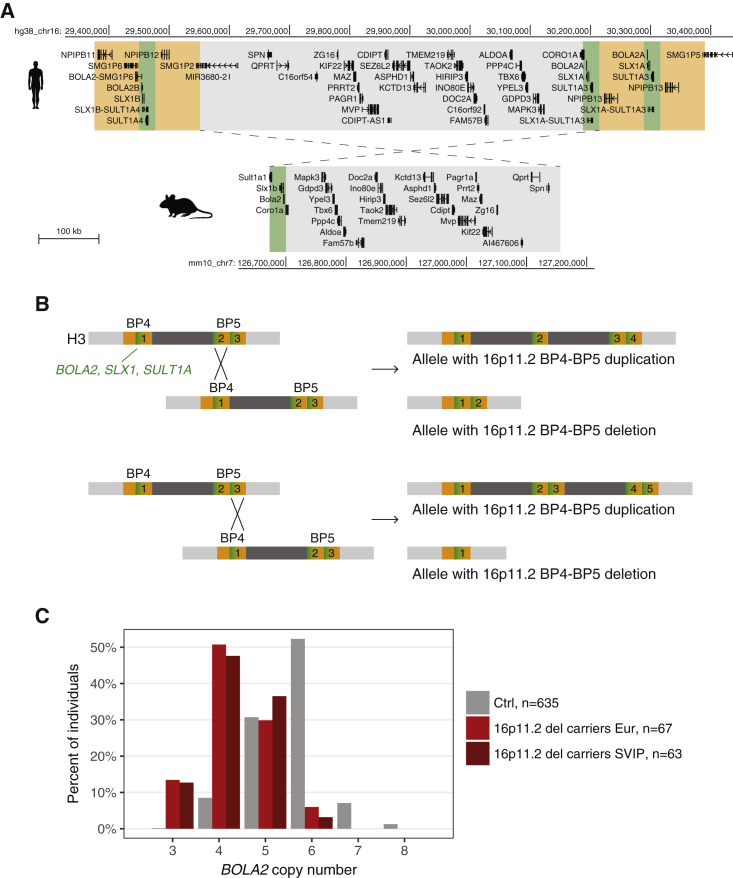
Figure 1.
The 16p11.2 and BOLA2 Locus Architecture
(A) Gene content of the most common human haplotype of the 16p11.2 BP4-BP5 region (top) and comparison with the syntenic single-copy region rearranged in 16p11.2 CNV mouse models30 (bottom). Human single-copy genes are gray shaded; the BOLA2-SLX1-SULT1A human duplicon is green shaded; the NPIP human duplicon is orange shaded. At BP4, the BOLA2B, SLX1B, and SULT1A4 paralogs map; at BP5, two copies of the BOLA2A, SLX1A, and SULT1A3 map. Scale bar 100 kb.
(B) Two examples of outcomes from non-allelic homologous recombination between copies of the BOLA2-SLX1-SULT1A-NPIP segment at BP4 and BP5 are presented. The BP4-BP5 structure corresponds to the most common H3 haplotype. For example, sequence misalignments could occur between one BP4 copy and either the distal (top panel) or proximal (bottom panel) BP5 copy. The resulting alleles with BP4-BP5 duplications and deletions have, respectively, more and fewer copies of BOLA2, SLX1, SULT1A, and NPIP (right panels). Numbers count the number of copies of the BOLA2-SLX1-SULT1A green segment in each haplotype.
(C) BOLA2 copy number in 635 European individuals from the 1000 Genomes Project and Human Genome Diversity Project (Ctrl), 67 16p11.2 BP4-BP5 deletion carriers from the European cohort, and 63 deletion carriers from the SVIP cohort.
In this work, we investigated the potential adaptive role of the human-specific duplications at 16p11.2 BP4 and BP5. We focused our study on the possible benefits associated with an increased dosage of BOLA2, because this is the only gene in the segment with convincing evidence of duplications restricted to Homo sapiens.13 As BOLA2 copy number positively correlates with both RNA expression and protein level in humans, and BOLA2 expression in stem and lymphoblastoid cells is higher in humans than in other large-bodied apes,13, 15 we hypothesize that a possible advantage of BOLA2 expansion derives from increased expression levels in some human cells.
BOLA2 encodes a protein that physically interacts with GLRX3 (glutaredoxin 3) to form a [2Fe–2S]-bridged heterotrimeric complex.16 This complex participates in the maturation of cytosolic iron-sulfur proteins, transferring [2Fe-2S] clusters to CIAPIN1 (anamorsin), an essential component of the cytosolic iron-sulfur assembly (CIA) system.17, 18, 19 In yeast, the [2Fe-2S]-bridged heterodimeric complex formed by the BolA-like protein Fra2 and the Grx3 and Grx4 glutaredoxins also has essential roles in intracellular iron signaling and regulation.20, 21 GLRX3 and CIAPIN1 support erythropoiesis, as the knockdown of glrx3 in zebrafish impairs heme synthesis and hemoglobin maturation,22 and Ciapin1-deficient mouse embryos are anemic.23
We investigated phenotypes associated with BOLA2 copy-number variation (CNV) in humans and mice and found that reduced BOLA2 dosage associates with mild anemia in both species, suggesting a model wherein increases in BOLA2 dosage might improve systemic iron homeostasis.
Material and Methods
Genotyping BOLA2 Copy Number and Statistical Analysis in Clinical Cohorts
We estimated the BOLA2A and BOLA2B copy number of 16p11.2 BP4–BP5 deletion carriers through the use of a molecular inversion probe (MIP) assay and probes mapping within BOLA2 as described in Nuttle et al.13 We collected iron metabolism-related information (serum iron, ferritin, transferrin, and coefficient of saturation of iron in transferrin), hematological parameters, and/or information about diagnosis of anemia for these individuals. Within the European cohort, a diagnosis of anemia or microcytosis was established if the individual had low blood iron, had low hemoglobin according to WHO guidelines,24 had a mean corpuscular volume (MCV) <80 fl, or had received a diagnosis of anemia. Within the Simons Variation in Individuals Project (SVIP) cohort, a diagnosis of anemia was established if the individual was identified as having anemia per caregiver report or self-report on the standardized medical history interview and was confirmed through medical records review. We used Fisher’s exact test25 to assess the statistical significance of the difference in anemia prevalence between individuals with low and high numbers of BOLA2 copies. The Committee of Human Research Ethics of the Canton of Vaud, Switzerland approved the project. We obtained written informed consent from clinical cohort participants to perform this study.
CNV Calling and Association Statistical Analysis in UK Biobank Population Cohort
The UK Biobank (UKB)26 is a volunteer-based general population biobank of the United Kingdom. Half a million participants aged 40–69 years at the time of recruitment (2006–2010) were enrolled through National Health Service registers. Participants consented to provide personal and health-related information, as well as biological samples, and to have their DNA tested. The UKB governing Research Ethics Committee has given a generic Research Tissue Bank approval to the UKB, which covers the research using this resource.
Single-nucleotide polymorphism genotyping in the UKB was performed using the UK BiLEVE and UKB Affymetrix Axiom platforms. Produced Log R ratio (LRR) and B allele frequency (BAF) values were formatted for CNV calling through the use of hidden Markov model-based software PennCNV v1.0.4.27 We retained samples that passed the post-processing quality-control parameters. To minimize the number of false positive findings, detected CNVs were stratified using a quality score (QS).28 Only validated (QS > |0.8|) carriers were included in downstream analyses. We considered “16p11.2 BP4–BP5 CNV carriers” to be those that had a deletion or duplication starting in the interval 29.4–29.8 Mbp and ending in 30.05–30.4 Mbp (hg19). We restricted the analysis to unrelated participants who declared themselves to be white British.
We searched for evidence of association between gene dosage at 16p11.2 BP4–BP5 (deletion carriers, control individuals, and duplication carriers) and hematological traits relative to red blood cells (RBCs), reticulocytes (immature RBCs), and platelets, using linear models in the statistical package R.25 Using t-tests, we also ran pairwise comparisons of deletion carriers versus controls and duplication carriers versus controls. In linear models, only additive effects for each copy (del = −1, controls = 0, dup = 1) were considered, and age, ageˆ2, sex, the first 40 principal components from the genetic analysis, acquisition route, acquisition time, device ID, and freeze-thaw cycles were included as covariates. Trait measures were normalized by quantile transformation prior to the analyses.
To assess the prevalence of anemia, we analyzed hospital discharge diagnoses coded according to the International Statistical Classification of Diseases and Related Health Problems, 10th revision (ICD-10) together with self-declared illnesses. This information is available under data fields 41202 and 41204 (respectively main and secondary diagnoses) and data field 20002 of self-reported non-cancer medical conditions. The individuals we considered to be anemic were those with ICD-10 codes D50, D51, D52, D53, D55, D58, D59, D60, D61, and D64, and self-reported non-cancer illness codes 1328, 1330, 1331, 1332, and 1446, as in Crawford et al.29 A participant was coded as anemic if he or she had a diagnosis or self-reported the condition. We also used information under data field 6179 (mineral and other dietary supplements) to assess treatment with iron supplementation. We tested the difference in anemia prevalence between 16p11.2 deletion and duplication carriers versus controls using Fisher’s exact test.
Mouse Models
We used the Del(7Sult1a1-Spn)6Yah (Del/+) and Dp(7Sult1a1-Spn)7Yah (Dup/+) mouse models30 and the C57BL/6N-Bola2tm1(KOMP)Wtsi/Nju line (Bola2tm1) produced by the Knockout Mouse Project and International Mouse Phenotyping Consortium.31 Bola2tm1 animals were acquired from the Nanjing Biomedical Research Institute of Nanjing University, China.
For the Del/+, Dup/+, and Bola2tm1 Swiss cohorts, all mice were born and housed in the Animal Facility of the Center for Integrative Genomics. We first removed the agouti allele from the Bola2tm1 line by backcrossing with C57BL/6N mice from Charles River. The animals were maintained on a standard chow diet (Kliba 3436, 250 ppm iron). All procedures were performed in accordance with protocols approved by the veterinary cantonal authority.
For the Bola2tm1 US cohort, heterozygous Bola2tm1 animals with the lacZ and neo cassettes were intercrossed to generate 8- and 15-week-old female animals for analysis. Furthermore, 129S4/SvJaeSor-Gt(ROSA)26Sortm1(FLP1)Dym/J mice were acquired from the Jackson Laboratory and backcrossed (N > 20) to the C57BL/6J background. The lacZ and neo cassettes in the Bola2tm1 animals were excised by breeding to the FLP1 mice and then backcrossed to C57BL/6J for an additional two generations. Resulting heterozygous animals were intercrossed to generate a cohort of 8-week-old female animals for analysis. All genetically modified mice were born and housed in the barrier facility at Boston Children’s Hospital. The animals were maintained on Prolab RMH 3000 diet (380 ppm iron; LabDiet). All procedures were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee. In both animal facilities, mice were provided water and food ad libitum and housed in 12 h long cycles of light and darkness.
SMRT Sequencing of Bola2−/− Mouse Genome
Genomic DNA was isolated from the buffy coat layer of the blood of a male null mouse with the lacZ and neo cassettes excised.32 We prepared one DNA fragment library (40–160 kbp inserts) with no additional shearing. After SMRTbell preparation, the library was size-selected using the BluePippin system (Sage Science) at a minimum fragment length cutoff of 40 kbp. Final library average size was 90 kbp. Single-molecule, real-time (SMRT) sequence data were generated using the PacBio Sequel instrument with Sequel Binding and Internal Ctrl Kit 3.0, Sequel Sequencing Kit 3.0, MagBead cleanup, diffusion loading, and acquisition times of 10 h movies. A total of three SMRT Cell 1M v3 cells were processed, yielding 11.5-fold (ROI/3.2 G) or 11.9-fold (raw/3.2G) whole-genome sequence data. The average subread length was 22.2 kbp with a median subread length of 14.1 kbp and N50 subread length of 35.9 kbp. Raw reads were mapped back to the mm10 mouse reference genome through the use of minimap233 and inspected manually at the Bola2 locus and genome-wide through the use of the PacBio structural variant calling and analysis tool. Local assembly of reads was done with the Canu assembler.34
Blood and Tissue Iron Analysis
For the Del/+, Dup/+, and Bola2tm1 Swiss cohorts, whole blood was collected from the tail vein in ethylenediaminetetraacetic acid (EDTA) or heparin tubes (Sarstedt) for hematological and plasma iron measurements, respectively. Plasma was prepared by centrifugation for 10 min at 2,000 × g, 4 degrees. Measurements were executed by the Centre de PhénoGénomique, Ecole polytechnique fédérale de Lausanne (EPFL). Due to ethical limits imposed by the veterinary office, the low body weight of 7-week-old Del/+ females did not allow for the collection of blood for hematological measurements.
For the Bola2tm1 US cohorts, whole blood for complete blood counts was collected from the retro-orbital sinus into EDTA-coated microtainer tubes (Becton Dickinson) from animals anesthetized with ketamine/xylazine (100–120 mg/kg ketamine and 10 mg/kg xylazine) in sterile saline. Samples were analyzed on an Avida 120 analyzer (Bayer) in the Boston Children’s Hospital Department of Laboratory Medicine Clinical Core Laboratories. Whole blood for other purposes was collected via retro-orbital bleeding into serum separator tubes (Becton Dickinson), and serum was prepared according to the manufacturer’s instructions. Serum iron values were determined by using the Serum Iron/TIBC kit (Pointe Scientific) according to the manufacturer’s instructions. Liver and spleen tissues were collected, and tissue non-heme iron concentrations were determined as described previously.35 Zinc protoporhyrin (ZPP) values in whole blood were analyzed on a ProtoFluor-Z Hematofluorometer (Helena Laboratories) according to the manufacturer’s instructions.
Statistical analyses (t-test and linear model) were performed using the R software environment.25 Plots were created using the R packages ggplot2 (v.2.2.1),36 gridExtra (v.2.3),37 and reshape2 (v.1.4.2).38
Results
16p11.2 Deletion Carriers Have Fewer Copies of the BOLA2-SLX1-SULT1A Segment
We previously sequenced haplotypes of the 16p11.2 BP4-BP5 interval.13 They differ in the number of copies and positions of a 102 kbp BOLA2-SLX1-SULT1A segment, i.e., its abundance within the BP4 or BP5 low-copy repeats. In particular, H3, the most common haplotype, has one copy of the segment at BP4 (including the paralogs BOLA2B, SLX1B, and SULT1A4) and two copies at BP5 (including the paralogs BOLA2A, SLX1A, and SULT1A3) (Figure 1A). Because alleles with 16p11.2 BP4–BP5 CNVs are generated through NAHR between paralogous copies of this segment,13 we expect that deletion and duplication alleles would lose or gain copies of this segment along with one copy of the intervening unique region containing genes from SPN to CORO1A (Figure 1B). To assess this prediction, we quantified the number of copies of BOLA2 in 16p11.2 BP4–BP5 deletion carriers collected by the 16p11.2 Consortium (European cohort, n = 67) and the SVIP (n = 63).13
Whereas European individuals from the 1000 Genomes Project and Human Genome Diversity Project (n = 635) have a mode of six copies of BOLA2, 16p11.2 deletion carriers showed a left-shifted distribution with a mode of four copies, confirming reduced BOLA2 copy number (Figure 1C). Correspondingly, expression of BOLA2 is positively associated with dosage in lymphoblastoid cells of 16p11.2 CNV carriers.39 We identified nine and eight 16p11.2 deletion carriers with three BOLA2 copies in the European (13.4%) and SVIP (12.7%) cohorts, respectively, compared to 0.2% (one in 635) in control individuals. We never identified individuals with only two copies; one BOLA2 copy per haploid genome is the ancestral copy number state observed both in great apes and archaic hominin genomes.13
Chromosome 16p11.2 Microdeletion Is Associated with Anemia and Mild Hematological Defects
Since 16p11.2 BP4–BP5 CNVs affect the dosage of the copy-number polymorphic BOLA2A/B, SLX1A/B, and SULT1A3/4 genes, it is possible that some associated phenotypes are due to the dosage change of one or a combination of these genes. Because BOLA2 binding partners have been implicated in iron metabolism pathways and erythropoiesis, we evaluated hematological parameters, including RBC indices and anemia, in 16p11.2 BP4–BP5 CNV carriers versus control individuals.
We examined CNVs in 488,366 individuals from the UKB and identified 89 unrelated Europeans (35 females and 54 males) carrying the 16p11.2 BP4–BP5 deletion and 56 (32 females and 24 males) carrying the duplication. We compared 18 hematological parameters to those of 379,385 control individuals through the use of linear models and t-tests combining and separating the genders (Table S1).
Gene dosage at 16p11.2 was negatively correlated with platelet count (PLTc, Beta = −0.685, p = 1e−15) and platelet mass per volume of blood (PLTcrit, Beta = −0.535, p = 4e−10), and positively associated with mean platelet volume (Beta = 0.444, p = 2e−7) (Figure 2A). Deletion carriers showed RBC anisocytosis (increased RBC distribution width [RDW], pfemales = 7e-3 and pmales = 1e-4) and a higher mean reticulocyte volume (MRV) in males (p = 4e−4) (Figure 2A), but all other RBC traits were not different from controls. While female duplication carriers appeared to have lower RBC count, hemoglobin concentration, and hematocrit percentage, these associations did not survive multiple testing correction (Table S1).
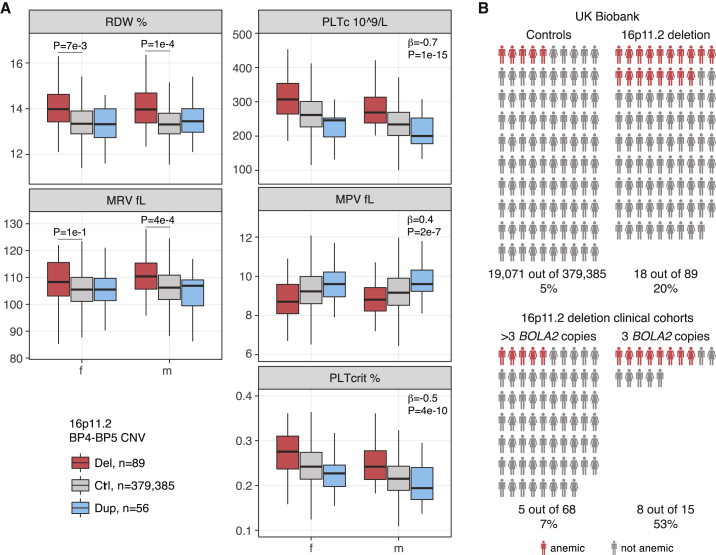
Figure 2.
Hematological Parameters and Anemia Prevalence in 16p11.2 BP4-BP5 CNV Carriers
(A) Red blood cell distribution width (RDW), mean reticulocyte volume (MRV), platelet count (PLTc), mean platelet volume (MPV), and platelet crit (PLTcrit) in 16p11.2 deletion and duplication carriers and control individuals in the UK Biobank (UKB). Beta and p values of linear models and p-values of paired t-tests are shown.
(B) Prevalence of anemia in 379,385 control individuals and 89 16p11.2 deletion carriers in the UKB (top) and 83 16p11.2 deletion carriers in the Simons Variation in Individuals Project (SVIP) and European cohorts stratified by BOLA2 copy number (bottom).
We then used both primary and secondary diagnoses collected for UKB participants to test a possible association between 16p11.2 gene dosage and the prevalence of anemia. We found that the 16p11.2 deletion is associated with a higher prevalence of anemia (Fisher’s exact p = 4e−7, OR = 4.8) with 18 out of 89 carriers referred as anemic (20%) compared to 5% of control individuals (Figure 2B). Of these, four were females and 14 were males, thus there was a trend toward higher prevalence of anemia in males (Fisher’s exact p = 0.1). Out of the 11 cases for which the type of anemia was specified, nine individuals had iron deficiency anemia (six with ICD-10 D50 diagnosis and three who self-reported the condition: three females and six males), one had vitamin B12 deficiency anemia, and one had autoimmune hemolytic anemia. We next assessed the prevalence of iron deficiency anemia (ICD-10 D50) in 16p11.2 deletion carriers versus control individuals and confirmed the higher prevalence in the former group (Fisher’s exact p = 4e−3). In contrast with deletion, 16p11.2 duplication showed no association with anemia (Fisher’s exact p = 0.8).
Reduced BOLA2 Copy Number More Strongly Associates with Anemia Prevalence in 16p11.2 Deletion Carriers
Candidate genes for anemia in the 16p11.2 region are BOLA2 and ALDOA. ALDOA missense mutations cause an autosomal recessive hemolytic anemia,40 and this is distinctly different from the iron deficiency anemia associated with the 16p11.2 deletion. We thus interrogated the possible involvement of BOLA2 and hypothesized that BOLA2 CNV might contribute to the incomplete penetrance of anemia with the lowest copy number possibly conferring a higher risk. To test this hypothesis, we estimated BOLA2 copy number and collected information regarding the presence of anemia, microcytosis, or iron deficiency in 83 families with chromosome 16p11.2 deletion. This included 63 deletion probands from the SVIP cohort (Table S2) and 25 deletion carriers belonging to 20 families from the European cohort (Table S3).
Six out of 63 probands from the SVIP cohort received a diagnosis of anemia (∼10%). We found a striking and significant difference in the prevalence of anemia between deletion carriers with three copies of BOLA2 (four out of eight, 50%) and those with more than three copies (two out of 55, 4%) (Fisher’s exact p = 1.6e−3). In the European cohort, we collected partial hematological, ferritin, and iron measurements, or information on diagnosed medical conditions. Seven out of 20 (35%) unrelated deletion carriers had low hemoglobin concentration (mild or moderate anemia), low serum iron, and/or microcytosis, or they received a diagnosis of anemia. Of these seven affected individuals, four have three BOLA2 copies and three have four BOLA2 copies. Here, we found a trend of higher anemia prevalence in individuals with three copies (Fisher’s exact p = 0.1) and observed that individuals both with three and with four copies might be more susceptible to anemia, microcytosis, or iron deficiency (Fisher’s exact p = 0.08). We note that the European cohort subgroup for which we have phenotype data is enriched with individuals having three or four BOLA2 copies. This might partly explain the different anemia prevalence in the two clinical cohorts (10% versus 35%). Combining the two cohorts, the occurrence of anemia among deletion carriers with three BOLA2 copies is significantly higher than that among those with more than three copies (Fisher’s exact p = 1e−4, OR = 13.6) (Figure 2B).
Whereas we uncovered an association between 16p11.2 deletion, particularly BOLA2 copy number, and anemia, we cannot rule out the possibility that the genotype of the remaining allele could have an influence. For example, genome-wide association studies for blood cell traits have shown that the variant rs3809627 mapping to the 16p11.2 BP4-BP5 region affects RBC count, MCV, mean corpuscular hemoglobin (MCH), and MRV.41, 42, 43 We assessed the joint effects of the 16p11.2 BP4-BP5 CNV and this SNP on hematological traits and diagnosis of anemia in the UKB (see Supplemental Data and Table S4). We found that the effects of the CNV and the SNP on hematological parameters are independent and the presence or absence of anemia among 16p11.2 deletion carriers is not associated with the rs3809627 genotype on the remaining allele. We next assessed the joint contributions of a rs3809627 proxy (i.e., rs12444415) and BOLA2 copy number on diagnosis of anemia in the SVIP cohort, and we confirmed that the SNP has no significant effect (see Supplemental Data).
Mouse Models of the 16p11.2 Deletion Present with Mild Anemia and Features Indicative of Early Iron Deficiency
To validate our human results, we assessed mouse models that carry a deletion (Del/+) or duplication (Dup/+) of the Sult1a1-Spn region syntenic to the human 16p11.2 BP4-BP5 locus.30 In these models, the rearranged region includes 26 genes that are in single copy in both the human and mouse genomes, together with the mouse single-copy Sult1a1, Slx1b, and Bola2 whose orthologs map to the human-specific duplicated cassette. In contrast to other animal models of the 16p11.2 BP4-BP5 CNV,44, 45 these animals recapitulate the CNV both of the human single-copy genes and the copy-number variant BOLA2, SLX1, and SULT1A genes (Figure 1A). In particular, Del/+ mice allow study of the effects of reduced Bola2 dosage in the context of the whole 16p11.2 BP4-BP5 rearrangement.
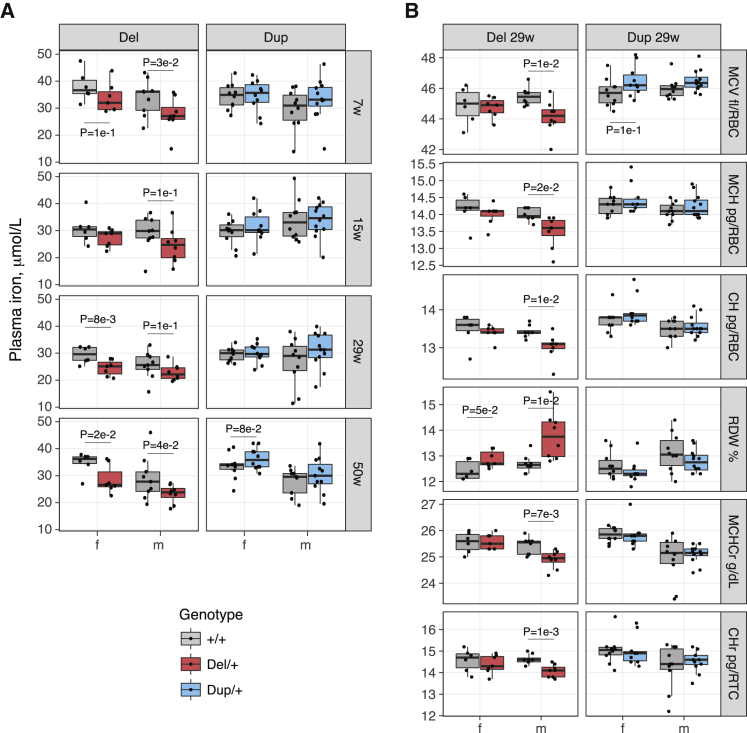
We measured plasma iron and 44 hematological parameters in Del/+ and Dup/+ male and female mice, as well as their respective wild-type (WT) littermates (Del and Dup mouse cohorts), at 7 (hematology only in males), 15, 29, and 50 weeks of age. In parallel, we recorded the body weight from 5 to 53 weeks of age (Figure S1). As reported in Arbogast et al.,30 Del/+ male and female mice had a lower body weight. The weight of Dup/+ males was not different from that of their WT littermates, whereas Dup/+ females showed increased weight at 19 and between 23 to 29 weeks of age (p < 0.05). Del/+ mice showed lower plasma iron levels at 7, 15, 29, and 50 weeks of age and in both genders (Figure 3A). When we included both genotype and weight in the linear model, neither variable was significantly associated with iron level, probably because of the strong effect of the genotype on the weight of Del/+ mice. We observed no significant difference between Dup/+ mice and WT littermates, although we noted a trend toward higher iron level in the former (Figure 3A, Table S5). Some male mice had to be separated because of aggressive behavior. Their husbandry status affected the plasma iron, with male mice housed in single cages showing higher iron levels (p = 0.06 in the Del cohort at 50 weeks of age and p = 0.01 in the Dup cohort at 29 weeks of age) (Table S5).
Figure 3.
Blood Traits of the Del/+ and Dup/+ Mice
(A) Plasma iron level in Del/+ and Dup/+ mice and their wild-type littermates at different ages (f: female, m: male, w: weeks).
(B) Red blood cell traits in Del/+ and Dup/+ mice and their wild-type littermates at the age of 29 weeks. These traits are significantly different in at least two longitudinal measurements (Tables S6 and S7). T-test p ≤ 0.1 are shown (MCV: mean corpuscular volume, MCH: mean corpuscular hemoglobin, CH: red cell hemoglobin content, RDW: red cell distribution width, MCHCr: mean corpuscular hemoglobin concentration reticulocytes, CHr: red cell hemoglobin content reticulocytes).
We describe here differences in hematological parameters between genotypes that differed significantly (p < 0.05) in at least two time points. Del/+ males showed lower MCV, MCH, red cell hemoglobin content (CH), MCH concentration of reticulocytes (MCHCr), and hemoglobin content of reticulocytes (CHr) and higher RDW; these results are consistent with a mild hypochromic microcytosis (Figure 3B, Table S6). Female mice showed a similar trend. No hematological parameter differed significantly between Dup/+ mice and WT littermates in more than one time replicate, in both females and males, except for a lower percentage of basophils in Dup/+ females (Table S7).
Bola2 Haploinsufficient and Deficient Mice Have Lower Blood Iron, Smaller Red Blood Cells, and Higher Zinc Protoporhyrin and Mean Platelet Mass
We assessed the role of Bola2 in organismal iron metabolism and blood cell traits, analyzing mouse models with ablation of Bola2. To control for genetic background and environmental variability, we measured iron-related and hematological parameters in Bola2−/− (homozygous mutants, ko/ko), Bola2+/− (heterozygous mutants, ko/+), and Bola2+/+ mice (WT littermates, +/+). We created three different cohorts: (i) a Bola2tm1 Swiss cohort; (ii) a Bola2tm1 US “neo-in” cohort; and (iii) a Bola2tm1 US cohort in which we excised the lacZ and neomycin cassette to eliminate potential secondary effects of the antibiotic selection gene embedded within the locus.
We first assessed the engineered genotype by sequencing the genome of one neomycin cassette-excised Bola2−/− male. We generated ∼12-fold sequence coverage by using the long-read SMRT sequencing platform, and we mapped and assembled long reads containing the Bola2 locus. All sequence reads (9/9) confirmed the presence of a 563 bp deletion corresponding to exons 2 and 3 (Figure S2), replaced by a 216 bp insertion, which includes one copy of the FRT recognition site. Immunoblotting of liver tissue from Bola2−/−, Bola2+/−, and WT mice (“neo-in” animals) confirmed the absence and reduced level of BOLA2 (Figure S3).
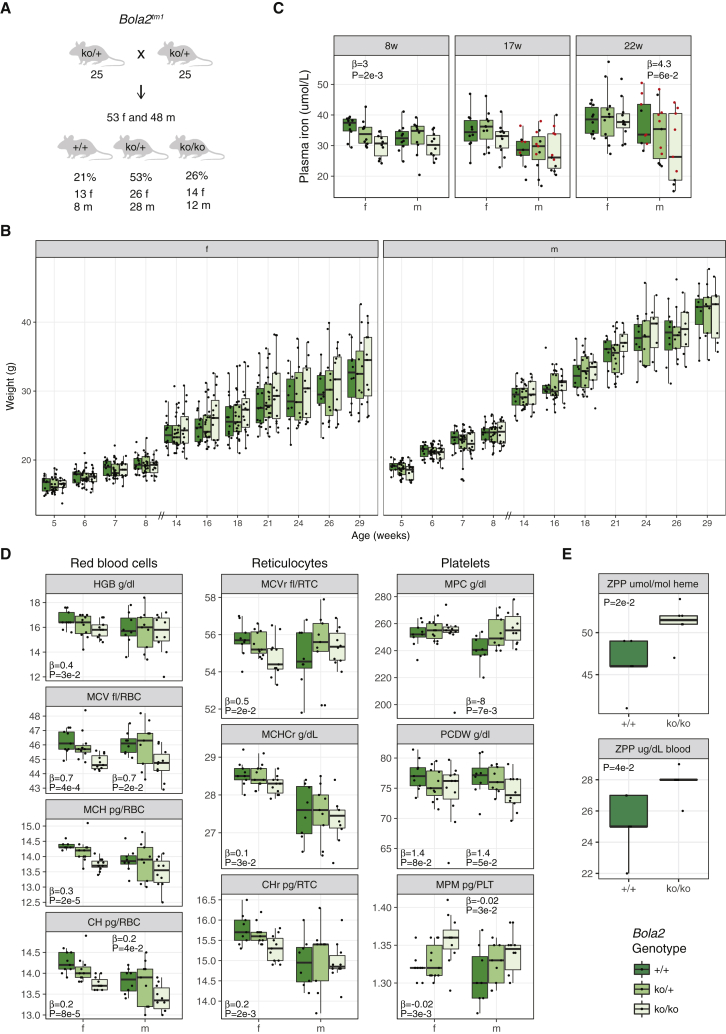
To create the Bola2tm1 Swiss cohort, we set 25 ko/+ × ko/+ pairs that generated 21 +/+, 54 ko/+, and 26 ko/ko mice born within the same 3 day interval from 17 crosses, suggesting no prenatal lethality (Figure 4A). Ko/+ and ko/ko mice showed no weight difference compared to WT littermates between 5 to 29 weeks of age, or in liver weight at 31 weeks (Figure 4B and Figure S4). We tested for a possible association between Bola2 dosage and plasma iron level at three different ages, 8, 17, and 22 weeks, in both genders. We observed a significant positive association in females at 8 weeks (Beta = 3, p = 2e−3) and a trend in males at 22 weeks (Beta = 4.3, p = 6e−2) (Figure 4C, Table S8). When adjusting for weight and/or husbandry condition (males only, single versus shared cage), the association of iron level with gene dosage remained. Of note, animals isolated to a single cage show significantly higher plasma iron levels, as observed in the Del and Dup mouse cohorts. As a result, we constructed two linear models to estimate the effect of the number of Bola2 copies on the iron level, assessing mice in single and shared cages separately. The effect of the genotype on the iron level was stronger and significant only in male mice housed in shared cages. However, the slopes of these two correlations did not differ significantly (p = 0.15).
Figure 4.
Characterization of the Bola2tm1 Knockout Mouse Line
(A) Mouse mating strategy of the Swiss Bola2tm1 line and gender and genotype ratios observed.
(B) Longitudinal body weight profiles. Note: the x axis scale is not continuous.
(C) Plasma iron level in Bola2+/− and Bola2−/− mice and wild-type littermates (f: female, m: male, w: weeks). Red dots represent mice housed in single cages. Beta and p values of linear models (female mice at 8 weeks of age and male mice at 22 weeks of age) are shown.
(D) Red blood cell (HGB: hemoglobin, MCV: mean corpuscular volume, MCH: mean corpuscular hemoglobin, CH: red cell hemoglobin content), reticulocyte (MCVr: mean corpuscular volume reticulocytes, MCHCr: mean corpuscular hemoglobin concentration reticulocytes, CHr: red cell hemoglobin content reticulocytes), and platelet (MPC: mean platelet component concentration, PCDW: platelet component distribution width, MPM: mean platelet mass) parameters of Bola2+/− and Bola2−/− mice and wild-type littermates at 17 weeks of age. Beta and p values of linear models with p ≤ 0.1 are shown.
(E) Blood zinc protoporphyrin (ZPP) levels of Bola2−/− mice and wild-type littermates at 8 weeks of age. P values of t-tests are shown.
Bola2 dosage affects RBCs, reticulocytes, and platelets (Figure 4D, Table S9). Specifically, the MCV (Beta = 0.7, pmales = 2e−2, pfemales = 4e−4 at 17 weeks), MCH (Beta = 0.3, pmales = 2e−4 at 22 weeks, pfemales = 2e−5 at 17 weeks), and CH (Beta = 0.2, pmales = 4e−2, pfemales = 8e−5 at 17 weeks) decrease in Bola2+/− and Bola2−/− mice, in both genders. This shows that Bola2 haploinsufficiency and deficiency are associated with a mild hypochromic microcytosis. Reticulocytes showed the same features, i.e., their size and hemoglobin content are positively associated with Bola2 copy number. We observe that some platelet traits show differences: the platelet component distribution width (PCDW, Beta = 1.4, pmales = 5e−2, pfemales = 8e−2) is positively associated with Bola2 copy number, while the mean platelet component concentration (MPC, Betamales = −8, pmales = 7e−3) and mean platelet mass (MPM, Beta = −0.02, pmales = 3e−2, pfemales = 3e−3) are negatively associated.
To generate the US cohorts, heterozygous Bola2tm1 mice on a pure C57BL/6N background were intercrossed. Bola2tm1 animals with the lacZ and neo cassettes excised with the Gt(ROSA)26Sortm1(FLP1)Dym/J FLP transgenic line on the C57BL/6J background were backcrossed to C57BL/6J for two more generations. Resulting heterozygous animals were intercrossed to yield (B6N)B6J N3F1 animals. We assessed complete blood counts and performed additional tests, including serum and tissue iron levels (Document S2. Tables S1–S15, Document S1). We replicated the mild microcytosis in the “neo-in” cohort (p = 1e−2) and observed no alteration in iron levels or spleen-to-body weight ratio which would suggest abnormal iron storage or an anemia compensated by erythroid hyperplasia. There were no morphological differences on peripheral blood smears. However, there were elevated ZPP normalized to heme levels in the blood of Bola2−/− mice (p = 2e−2 in the “neo-in” and p = 1e−3 in the “neo-excised” cohort, Figure 4E), suggesting functional iron deficiency in maturing erythroid cells that could account for the mild microcytosis.
These data show that Bola2 haploinsufficiency and deficiency cause mild hypochromic, microcytic, iron-deficiency anemia and suggest that a decrease in BOLA2 dosage might be causative for the iron-deficiency anemia seen in 16p11.2 deletion carriers.
Discussion
This work sheds light on some possible benefits of increased BOLA2 dosage in humans, with six copies (the most common copy number), on average, per genome, compared to two copies in other hominins, primates, and mammals.13 Duplications containing BOLA2 map to the 16p11.2 locus and generate chromosomal instability associated with neurodevelopmental and psychiatric disorders. BOLA2 duplications are thus another example of a genetic trade-off because their potential gain comes with the cost of predisposition to pathogenic rearrangements,2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 13 similar to the positively selected 17q21.31 inversion, which is associated with increased fecundity but predisposing to a deletion syndrome.46, 47
We assessed blood-related phenotypes associated with BOLA2 copy number and found that, as recently reported,29 the 16p11.2 BP4-BP5 deletion is associated with anemia (18/89, ∼20% of carriers), particularly iron deficiency anemia (9/11 cases with specified type of anemia), in a large adult population cohort, and men seemed to be more affected than women. In 16p11.2 deletion clinical cohorts, ∼50% of carriers with three BOLA2 copies are anemic, suggesting a contribution of lower BOLA2 dosage. We validated these results in mouse models of the 16p11.2 deletion (These models encompass the BOLA2 ortholog.) and in mouse models of Bola2-specific haploinsufficiency and deficiency. All models presented iron deficiency and a mild form of microcytic and hypochromic anemia. Recalling the higher susceptibility of men, among Del/+ mice, only males had microcytosis, lower corpuscular hemoglobin, and anisocytosis. Transcriptome profiling of the brain cortex shows that Del/+ females, but not males, have increased expression of Glrx3 and Bola1, thus possibly compensating for the decreased Bola2 expression through increased expression of BOLA2 binding partner (GLRX3) or mitochondrial counterpart (BOLA1) (De Nittis, et al., unpublished data). Finally, Bola2 knockout mice showed mild microcytosis, higher ZPP level, and lower plasma iron, although the last phenotype was not observed in all longitudinal measurements; this may be due to compensatory mechanisms. Our data suggest that BOLA2 is a driver of iron deficiency anemia; however, it is still possible that other genes in the 16p11.2 single-copy region or its flanking human-specific duplicon contribute to this phenotype, as the number of BOLA2 copies coincides with the number of copies of the entire 102 kbp segment. The effects of BOLA2 copy-number polymorphism on the phenotypic spectrum and incomplete penetrance of phenotypes of the 16p11.2 deletion is a striking example of the contribution of genes in the flanking low-copy repeats to traits associated with a genomic disorder, pointing out the evolutionary and medical relevance of this work.
We found discrepancies in hematological parameters between human and mouse. Both Del/+ male and Bola2−/− and Bola2+/− mice had significantly lower MCV and MCH compared to WT mice. We did not find the same features in RBCs of 16p11.2 deletion carriers; this result may be due to an unreported iron supplementation therapy for anemia. In fact, we found no difference in dietary iron supplementation between 16p11.2 deletion carriers and controls from the UKB (data field 6179, comparing iron versus “none of the above,” Fisher’s exact p = 0.6). We note that this information might be partial because it was collected by a touchscreen questionnaire. RBC traits and/or iron levels were not modified in 16p11.2 duplication carriers and duplication mouse models. It is possible that cells are sensitive to low BOLA2 levels, but there is no effect when these are above a certain threshold. Alternatively, opposite balancing effects and/or negative epistasis might be at play between BOLA2 copy numbers and the duplication of another gene or other genes of the 16p11.2 region. Platelet traits were modified in human and mouse; however, humans showed negative effects of gene dosage on PLTc and PLTcrit and positive effects on platelet volume, whereas Del/+ male, Bola2−/−, and Bola2+/− mice showed higher platelet mass. Because BOLA2 dosage modifies two hematopoietic lineages, the RBCs and the platelets, it is possible that this variant acts in the megakaryocyte-erythroid progenitor. Alternatively, these platelet alterations could be secondary to the iron deficiency, as serum iron is inversely related to PLTc and PLTcrit in iron deficiency anemia.48, 49, 50 Human versus mouse discrepancies further confirm the fundamental divergence in gene expression between human and mouse hematopoiesis and the limitations of mouse systems to model human hematopoiesis.51, 52
Overall, our data suggest that increased BOLA2 dosage in humans might protect against iron deficiency. Iron deficiency is still the most common micronutrient deficiency in the world and the most common cause of anemia;53 this suggests that it is difficult to precisely tune iron metabolism in humans. Iron is an essential micronutrient and serves as a cofactor for numerous enzymes, including those involved in energy metabolism, synthesis of DNA and proteins, and a range of other biochemical functions in cells. It is important in brain development, kidney function, immune responses, and growth, and as a component of hemoglobin and myoglobin, it is essential for oxygen transport and delivery.54
Veterinary hematological data for chimpanzee, bonobo, gorilla, and orangutan55 (Table S14) show that hemoglobin and iron levels in the blood do not differ between humans and great apes; this suggests that humans need higher BOLA2 levels to guarantee the same level of iron supply to the organism. Menstrual blood loss is much larger in humans than in great apes. Different hypotheses have been proposed to explain this difference: elimination of pathogens from the reproductive tract, economy of energy because it is more costly to maintain the endometrium than to grow a new one, elimination of defective embryos, and adaptation to increased amounts of easily absorbable iron from meat.56 It is possible that the increased copy number of BOLA2 is necessary to cope with the increased menstrual iron loss, although males seemed to be more affected by anemia among 16p11.2 deletion carriers.
Higher BOLA2 levels might have been important in the past in order for humans to thrive in nutrient-poor environments or during the transition from a red meat hunter-gatherer Paleolithic diet to the iron-reduced cereal grain Neolithic diet, although BOLA2 expansion is dated much earlier.13 These shifts likely resulted in an increased risk of iron deficiency anemia, especially in women of reproductive age. The rapid expansion and selection of BOLA2 copy number at the root of the Homo lineage may have evolved to deal with this pressure as our species successfully expanded its ecological range at the cost of increased predisposition to rearrangements associated with autism.
Declaration of Interests
Evan E. Eichler is on the scientific advisory board (SAB) of DNAnexus. The other authors declare no competing interests.
Consortia
16p11.2 Consortium members: Giuliana Giannuzzi, Katrin Männik, Catia Attanasio, Pasquelena De Nittis, Sandra Martin, Sébastien Jacquemont, Armand Bottani, Marion Gérard, Sacha Weber, Aurélia Jacquette, Fabien Lesne, Bertrand Isidor, Cédric Le Caignec, Mathilde Nizon, Catherine Vincent-Delorme, Brigitte Gilbert-Dussardier, Aurora Currò, Alessandra Renieri, Daniela Giachino, Alfredo Brusco, Alexandre Reymond. The affiliations and email addresses of the members of the 16p11.2 Consortium are listed in Table S15.
Acknowledgments
We thank families participating in the Simons Variation in Individuals Project (SVIP) and the 16p11.2 Consortium cohorts. G.G. is recipient of a Pro-Women Scholarship from the Faculty of Biology and Medicine, University of Lausanne. This work was supported by grants from the Swiss National Science Foundation (31003A_160203 and 31003A_182632 to A.R.), the National Institutes of Health (R01HG002385 to E.E.E.), Horizon2020 Twinning Project ePerMed (692145 to A.R.), Simons Foundation Autism Research Initiative (SFARI #303241 to E.E.E., #198677 to R.A.B., and #274424 to A.R.), INSERM, CNRS, University of Strasbourg (ANR-10-LABX-0030-INRT and ANR-10-INBS-07-03 PHENOMIN to Y.H.), and framework program Investissements d’Avenir (ANR-10-IDEX-0002-02 to Y.H.). This research has been conducted using the UK Biobank resource (#16389). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. E.E.E. is an investigator of the Howard Hughes Medical Institute. We thank Jacques S. Beckmann for helpful discussions.
Published: October 24, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.09.023.
Supplemental Data
Figures S1–S4, List of Supplemental Tables, Supplemental Methods, and Supplemental References
References
- 1.Loviglio M.N., Arbogast T., Jonch A.E., Collins S.C., Popadin K., Bonnet C.S., Giannuzzi G., Maillard A.M., Jacquemont S., 16p11.2 Consortium The immune signaling adaptor LAT contributes to the neuroanatomical phenotype of 16p11.2 BP2-BP3 CNVs. Am. J. Hum. Genet. 2017;101:564–577. doi: 10.1016/j.ajhg.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zufferey F., Sherr E.H., Beckmann N.D., Hanson E., Maillard A.M., Hippolyte L., Macé A., Ferrari C., Kutalik Z., Andrieux J., Simons VIP Consortium. 16p11.2 European Consortium A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J. Med. Genet. 2012;49:660–668. doi: 10.1136/jmedgenet-2012-101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCarthy S.E., Makarov V., Kirov G., Addington A.M., McClellan J., Yoon S., Perkins D.O., Dickel D.E., Kusenda M., Krastoshevsky O., Wellcome Trust Case Control Consortium Microduplications of 16p11.2 are associated with schizophrenia. Nat. Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss L.A., Shen Y., Korn J.M., Arking D.E., Miller D.T., Fossdal R., Saemundsen E., Stefansson H., Ferreira M.A., Green T., Autism Consortium Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 5.D’Angelo D., Lebon S., Chen Q., Martin-Brevet S., Snyder L.G., Hippolyte L., Hanson E., Maillard A.M., Faucett W.A., Macé A., Cardiff University Experiences of Children With Copy-number variants (ECHO) Study. 16p11.2 European Consortium. Simons Variation in Individuals Project (VIP) Consortium Defining the effect of the 16p11.2 duplication on cognition, behavior, and medical comorbidities. JAMA Psychiatry. 2016;73:20–30. doi: 10.1001/jamapsychiatry.2015.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall C.R., Howrigan D.P., Merico D., Thiruvahindrapuram B., Wu W., Greer D.S., Antaki D., Shetty A., Holmans P.A., Pinto D., Psychosis Endophenotypes International Consortium. CNV and Schizophrenia Working Groups of the Psychiatric Genomics Consortium Contribution of copy-number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 2017;49:27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacquemont S., Reymond A., Zufferey F., Harewood L., Walters R.G., Kutalik Z., Martinet D., Shen Y., Valsesia A., Beckmann N.D. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maillard A.M., Ruef A., Pizzagalli F., Migliavacca E., Hippolyte L., Adaszewski S., Dukart J., Ferrari C., Conus P., Männik K., 16p11.2 European Consortium The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Mol. Psychiatry. 2015;20:140–147. doi: 10.1038/mp.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shinawi M., Liu P., Kang S.H., Shen J., Belmont J.W., Scott D.A., Probst F.J., Craigen W.J., Graham B.H., Pursley A. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J. Med. Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walters R.G., Jacquemont S., Valsesia A., de Smith A.J., Martinet D., Andersson J., Falchi M., Chen F., Andrieux J., Lobbens S. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin-Brevet S., Rodríguez-Herreros B., Nielsen J.A., Moreau C., Modenato C., Maillard A.M., Pain A., Richetin S., Jønch A.E., Qureshi A.Y., 16p11.2 European Consortium. Simons Variation in Individuals Project (VIP) Consortium Quantifying the effects of 16p11.2 copy-number variants on brain structure: A multisite genetic-first study. Biol. Psychiatry. 2018;84:253–264. doi: 10.1016/j.biopsych.2018.02.1176. [DOI] [PubMed] [Google Scholar]
- 12.Männik K., Arbogast T., Lepamets M., Lepik K., Pellaz A., Ademi H., Kupchinsky Z.A., Ellegood J., Attanasio C., Messina A. Leveraging biobank-scale rare and common variant analyses to identify ASPHD1 as the main driver of reproductive traits in the 16p11.2 locus. bioRxiv. 2019 [Google Scholar]
- 13.Nuttle X., Giannuzzi G., Duyzend M.H., Schraiber J.G., Narvaiza I., Sudmant P.H., Penn O., Chiatante G., Malig M., Huddleston J. Emergence of a Homo sapiens-specific gene family and chromosome 16p11.2 CNV susceptibility. Nature. 2016;536:205–209. doi: 10.1038/nature19075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson M.E., Viggiano L., Bailey J.A., Abdul-Rauf M., Goodwin G., Rocchi M., Eichler E.E. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 15.Marchetto M.C.N., Narvaiza I., Denli A.M., Benner C., Lazzarini T.A., Nathanson J.L., Paquola A.C.M., Desai K.N., Herai R.H., Weitzman M.D. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 2013;503:525–529. doi: 10.1038/nature12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Mapolelo D.T., Randeniya S., Johnson M.K., Outten C.E. Human glutaredoxin 3 forms [2Fe-2S]-bridged complexes with human BolA2. Biochemistry. 2012;51:1687–1696. doi: 10.1021/bi2019089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banci L., Camponeschi F., Ciofi-Baffoni S., Muzzioli R. Elucidating the molecular function of human BOLA2 in GRX3-dependent anamorsin maturation pathway. J. Am. Chem. Soc. 2015;137:16133–16143. doi: 10.1021/jacs.5b10592. [DOI] [PubMed] [Google Scholar]
- 18.Frey A.G., Palenchar D.J., Wildemann J.D., Philpott C.C. A Glutaredoxin·BolA Complex Serves as an Iron-Sulfur Cluster Chaperone for the Cytosolic Cluster Assembly Machinery. J. Biol. Chem. 2016;291:22344–22356. doi: 10.1074/jbc.M116.744946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., Lyver E.R., Nakamaru-Ogiso E., Yoon H., Amutha B., Lee D.W., Bi E., Ohnishi T., Daldal F., Pain D., Dancis A. Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol. Cell. Biol. 2008;28:5569–5582. doi: 10.1128/MCB.00642-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H., Outten C.E. Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe-2S] binding partners in iron homeostasis. Biochemistry. 2012;51:4377–4389. doi: 10.1021/bi300393z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumánovics A., Chen O.S., Li L., Bagley D., Adkins E.M., Lin H., Dingra N.N., Outten C.E., Keller G., Winge D. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J. Biol. Chem. 2008;283:10276–10286. doi: 10.1074/jbc.M801160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haunhorst P., Hanschmann E.M., Bräutigam L., Stehling O., Hoffmann B., Mühlenhoff U., Lill R., Berndt C., Lillig C.H. Crucial function of vertebrate glutaredoxin 3 (PICOT) in iron homeostasis and hemoglobin maturation. Mol. Biol. Cell. 2013;24:1895–1903. doi: 10.1091/mbc.E12-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibayama H., Takai E., Matsumura I., Kouno M., Morii E., Kitamura Y., Takeda J., Kanakura Y. Identification of a cytokine-induced antiapoptotic molecule anamorsin essential for definitive hematopoiesis. J. Exp. Med. 2004;199:581–592. doi: 10.1084/jem.20031858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.World Health Organization . 2011. Haemoglobin concentrations for the diagnosis of anaemia and assessment of severity.https://www.who.int/vmnis/indicators/haemoglobin.pdf [Google Scholar]
- 25.R Core Team . 2017. R: A language and environment for statistical computing.https://www.r-project.org/ [Google Scholar]
- 26.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F., Hakonarson H., Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macé A., Tuke M.A., Beckmann J.S., Lin L., Jacquemont S., Weedon M.N., Reymond A., Kutalik Z. New quality measure for SNP array based CNV detection. Bioinformatics. 2016;32:3298–3305. doi: 10.1093/bioinformatics/btw477. [DOI] [PubMed] [Google Scholar]
- 29.Crawford K., Bracher-Smith M., Owen D., Kendall K.M., Rees E., Pardiñas A.F., Einon M., Escott-Price V., Walters J.T.R., O’Donovan M.C. Medical consequences of pathogenic CNVs in adults: analysis of the UK Biobank. J. Med. Genet. 2019;56:131–138. doi: 10.1136/jmedgenet-2018-105477. [DOI] [PubMed] [Google Scholar]
- 30.Arbogast T., Ouagazzal A.M., Chevalier C., Kopanitsa M., Afinowi N., Migliavacca E., Cowling B.S., Birling M.C., Champy M.F., Reymond A., Herault Y. Reciprocal Effects on Neurocognitive and Metabolic Phenotypes in Mouse Models of 16p11.2 Deletion and Duplication Syndromes. PLoS Genet. 2016;12:e1005709. doi: 10.1371/journal.pgen.1005709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skarnes W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kronenberg Z.N., Fiddes I.T., Gordon D., Murali S., Cantsilieris S., Meyerson O.S., Underwood J.G., Nelson B.J., Chaisson M.J.P., Dougherty M.L. High-resolution comparative analysis of great ape genomes. Science. 2018;360:eaar6343. doi: 10.1126/science.aar6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torrance J.D., Bothwell T.H. Tissue iron stores. In: Cook J.D., editor. Methods in Hematology. Churchill Livingstone Press; New York: 1980. pp. 104–109. [Google Scholar]
- 36.Wickham H. Springer-Verlag; New York: 2009. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 37.Auguie B. 2017. gridExtra: Miscellaneous Functions for “Grid” Graphics. [Google Scholar]
- 38.Wickham H. Reshaping data with the reshape package. J. Stat. Softw. 2007;21:1–20. [Google Scholar]
- 39.Migliavacca E., Golzio C., Männik K., Blumenthal I., Oh E.C., Harewood L., Kosmicki J.A., Loviglio M.N., Giannuzzi G., Hippolyte L., 16p11.2 European Consortium A potential contributory role for ciliary dysfunction in the 16p11.2 600 kb BP4-BP5 Pathology. Am. J. Hum. Genet. 2015;96:784–796. doi: 10.1016/j.ajhg.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beutler E., Scott S., Bishop A., Margolis N., Matsumoto F., Kuhl W. Red cell aldolase deficiency and hemolytic anemia: a new syndrome. Trans. Assoc. Am. Physicians. 1973;86:154–166. [PubMed] [Google Scholar]
- 41.Ulirsch J.C., Lareau C.A., Bao E.L., Ludwig L.S., Guo M.H., Benner C., Satpathy A.T., Kartha V.K., Salem R.M., Hirschhorn J.N. Interrogation of human hematopoiesis at single-cell and single-variant resolution. Nat. Genet. 2019;51:683–693. doi: 10.1038/s41588-019-0362-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Astle W.J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A.L., Mead D., Bouman H., Riveros-Mckay F., Kostadima M.A. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429 e1419. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanai M., Akiyama M., Takahashi A., Matoba N., Momozawa Y., Ikeda M., Iwata N., Ikegawa S., Hirata M., Matsuda K. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018;50:390–400. doi: 10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
- 44.Horev G., Ellegood J., Lerch J.P., Son Y.E., Muthuswamy L., Vogel H., Krieger A.M., Buja A., Henkelman R.M., Wigler M., Mills A.A. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc. Natl. Acad. Sci. USA. 2011;108:17076–17081. doi: 10.1073/pnas.1114042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Portmann T., Yang M., Mao R., Panagiotakos G., Ellegood J., Dolen G., Bader P.L., Grueter B.A., Goold C., Fisher E. Behavioral abnormalities and circuit defects in the basal ganglia of a mouse model of 16p11.2 deletion syndrome. Cell Rep. 2014;7:1077–1092. doi: 10.1016/j.celrep.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stefansson H., Helgason A., Thorleifsson G., Steinthorsdottir V., Masson G., Barnard J., Baker A., Jonasdottir A., Ingason A., Gudnadottir V.G. A common inversion under selection in Europeans. Nat. Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 47.Koolen D.A., Vissers L.E., Pfundt R., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat. Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 48.Park M.J., Park P.W., Seo Y.H., Kim K.H., Park S.H., Jeong J.H., Ahn J.Y. The relationship between iron parameters and platelet parameters in women with iron deficiency anemia and thrombocytosis. Platelets. 2013;24:348–351. doi: 10.3109/09537104.2012.699641. [DOI] [PubMed] [Google Scholar]
- 49.Dan K. Thrombocytosis in iron deficiency anemia. Intern. Med. 2005;44:1025–1026. doi: 10.2169/internalmedicine.44.1025. [DOI] [PubMed] [Google Scholar]
- 50.Kadikoylu G., Yavasoglu I., Bolaman Z., Senturk T. Platelet parameters in women with iron deficiency anemia. J. Natl. Med. Assoc. 2006;98:398–402. [PMC free article] [PubMed] [Google Scholar]
- 51.Pishesha N., Thiru P., Shi J., Eng J.C., Sankaran V.G., Lodish H.F. Transcriptional divergence and conservation of human and mouse erythropoiesis. Proc. Natl. Acad. Sci. USA. 2014;111:4103–4108. doi: 10.1073/pnas.1401598111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beer P.A., Eaves C.J. Modeling normal and disordered human hematopoiesis. Trends Cancer. 2015;1:199–210. doi: 10.1016/j.trecan.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Kassebaum N.J., GBD 2013 Anemia Collaborators The global burden of anemia. Hematol. Oncol. Clin. North Am. 2016;30:247–308. doi: 10.1016/j.hoc.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 54.Srai S.K., Sharp P. Humana Press; 2012. Iron Physiology and Pathophysiology in Humans. vii–xi. [Google Scholar]
- 55.Miller R.E., Fowler M.E. Vol. 8. Elsevier; 2015. Fowler’s Zoo and Wild Animal Medicine. [Google Scholar]
- 56.Denic S., Agarwal M.M. Nutritional iron deficiency: an evolutionary perspective. Nutrition. 2007;23:603–614. doi: 10.1016/j.nut.2007.05.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S4, List of Supplemental Tables, Supplemental Methods, and Supplemental References