Abstract
Unlike many cancers, the pattern of tumor evolution in papillary thyroid cancer (PTC) and its potential role in relapse have not been elucidated. In this study, multi-region whole-exome sequencing (WES) was performed on early-stage PTC tumors (n = 257 tumor regions) from 79 individuals, including 17 who had developed relapse, to understand the temporal and spatial framework within which subclonal mutations catalyze tumor evolution and its potential clinical relevance. Paired primary-relapse tumor tissues were also available for a subset of individuals. The resulting catalog of variants was analyzed to explore evolutionary histories, define clonal and subclonal events, and assess the relationship between intra-tumor heterogeneity and relapse-free survival. The multi-region WES approach was key in correctly classifying subclonal mutations, 40% of which would have otherwise been erroneously considered clonal. We observed both linear and branching evolution patterns in our PTC cohort. A higher burden of subclonal mutations was significantly associated with increased risk of relapse. We conclude that relapse in PTC, while generally rare, does not follow a predictable evolutionary path and that subclonal mutation burden may serve as a prognostic factor. Larger studies utilizing multi-region sequencing in relapsed PTC case subjects with matching primary tissues are needed to confirm these observations.
Keywords: papillary thyroid cancer, recurrence, exome sequencing, multi-tissue, evolution, phylogeny, subclonal mutations
Introduction
Papillary thyroid carcinoma (PTC) is the most prevalent endocrine malignancy with a steadily increasing prevalence.1, 2 Although highly curable (∼85% of case subjects) due to its relatively indolent biological nature,3 10%–20% of individuals with PTC tend to develop either local or distant relapse.4, 5 Presence of lymph node metastases, extrathyroidal extension, older age, and BRAF (MIM: 164757) mutations are some of the factors shown to be associated with relapse.6, 7, 8 Nevertheless, the high cure rate overshadows the fact that factors associated with relapse remain poorly understood, especially at the molecular level.
Cancer is a heterogeneous disease, where tumorigenesis is often instigated by a single neoplastic cell propelled by somatic mutation from which it develops into a tumor mass, evolving through a series of sequential events, such as clonal expansions, driven by shifting selective pressures and mutational processes.9, 10, 11 During this intricate biological process, clonal ancestral mutations that are ubiquitous in all clones (a group of tumor cells sharing highly similar mutational profiles) diversify through the Darwinian process of clonal expansions by acquiring additional mutations and forming distinct subclonal populations in the evolutionary lineage, resulting in intratumor heterogeneity.12, 13, 14 Thus, intratumor heterogeneity can provide the evolutionary dynamics of mutations, chronologically accumulated and selected for during the lifetime of a tumor, driven by external pressures and microenvironmental niches such as the immune system, pH changes, chemotherapy, radiation, nutrient deprivation, and anatomical barriers.15, 16, 17, 18, 19, 20 Several recent cancer genomics studies have explored this extensive genetic diversity within tumors, characterizing the genomic landscape of either untreated and treated primary tumors, or comparing the genomic landscapes of primary tumors with that of their corresponding local or distant relapsed tumors.13, 21, 22, 23, 24
Evolutionary processes can be mapped by sequencing tumor samples spatially, through multi-region sampling, and temporally, through multiple time-point sampling, followed by the clustering of somatic mutations and inferring genetic phylogenies according to their cellular prevalence.24, 25 Several competing patterns of tumor evolution exist, such as linear and branching evolution, which can be deduced from phylogenetic trees and cancer cell fractions (CCFs). Linear evolution represents the successive growth of dominant clones by passively accumulating many mutations without expansion before additional fitness is acquired in the form of infrequent driver events conferring a selective advantage to produce clonal expansions.26 Branching evolution, on the other hand, is characterized by the divergence of subclones from a common ancestral clone, leading to the coexistence of subclones sharing no more than partial sets of mutations.27, 28 Some cancers have also exhibited mixed models of tumor evolution.29, 30 Cancer relapses traced evolutionarily demonstrate selective expansion of subpopulations of tumor cells with resistant subclones providing added fitness, leading to the development of relapse.31, 32, 33, 34 Various models of tumor evolution in cancer can have diverse clinical implications for the clinical diagnosis, prognosis, and treatment of the individuals;35 therefore, interpreting intratumor heterogeneity and tumor evolution is of great clinical importance.28
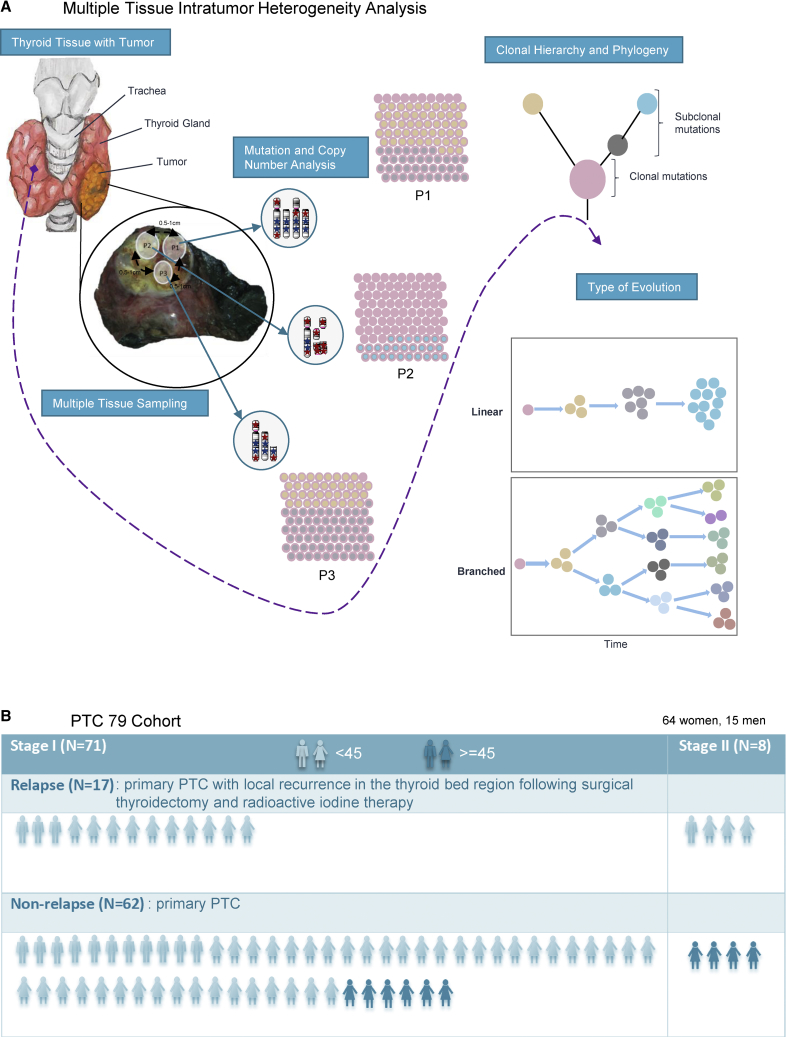
We hypothesized that the evolutionary dynamics of subclonal mutations in PTC might also contribute to relapse. Therefore, we performed relatively large-scale whole-exome sequencing (WES) of 257 PTC tumor tissues from 79 individuals, including 6 individuals sampled at two time points (primary and relapse). This analysis allowed us to study tumor mutations, their clonality and evolution, and their potential correlation with the clinical outcome (Figure 1).
Figure 1.
Overview of the Demographic and Clinical Characteristics of the 79 PTC Cohort
(A) Thyroid gland with tumor. Multiple PTC tumor sections were sequenced, to analyze somatic mutations and copy number alterations, supporting the assessment of intratumor heterogeneity. The type of evolution was determined by phylogenetic reconstruction. Stars indicate mutations, where blue color is clonal mutations and red is subclonal mutations.
(B) Demographic and clinical characteristics of the 79 individuals including tumor stage, age, and relapse status.
Material and Methods
PTC Cohort and Tumor Samples
We collected a total of 257 tumor tissue samples, with a median of 3 (range 2–11) tumor tissues per case subject, along with corresponding normal whole blood, from 79 individuals with early-stage PTC, including 17 with relapse PTC, 6 of whom were sampled at two time points (following primary diagnosis and after incurring local relapse within the thyroid bed), from King Faisal Specialist Hospital and Research Centre and Prince Sultan Military Medical City. All the individuals were treatment naive at the time of surgery for primary tumor. 91% (72/79) of the cohort underwent total thyroidectomy, whereas the remaining 9% (7/79) underwent hemi-thyroidectomy, for removal of primary tumor. Subsequent local relapse was defined as the development of tumor recurrence within the soft tissues of the neck, forming the thyroidectomy bed, following optimized surgery and radioactive iodine therapy.3 To assess intra-tumor heterogeneity, samples of at least two tumor regions, separated by a margin of 0.5 cm to 1.0 cm (depending on the size of the tumor), have been taken for the study. Clinical characteristics of the PTC cohort are provided in Table S1 and Figure 1. Institutional Review Board (IRB) of the King Faisal Specialist Hospital & Research Centre approved the study under the Project RAC # 2110 031 and # 2170 022 on PTC archival clinical samples. Written consent was obtained from all the individuals included in the study.
Sample Processing
Tumor tissue from each region was used for genomic DNA extraction using Gentra DNA isolation kit, following the manufacturer’s recommendations as described previously.36 DNA was quantified by Qubit (Invitrogen).
Whole-Exome Sequencing and Mutation Calling
For each tumor region (n = 257) and matched germline (n = 79), whole-exome sequencing (WES) was performed using SureSelectXT Target Enrichment (Agilent) on Illumina NovaSeq 6000. We sequenced to a median depth of 178×, 257 tumor regions (240 primary and 17 relapse tissues), with a median of 3 regions per tumor (range 2–11), and 79 matched germline samples derived from whole blood. Sequencing reads were aligned to the human reference genome hg19 using Burrows-Wheeler Aligner (BWA) v.0.7.15 algorithm;37 followed by local realignment and PCR duplicate marking via Picard tools (v.1.119). Base-quality recalibration was performed with GATK v.3.8.0.38 All the quality metrics were obtained using GATK and FastQC.
Somatic single-nucleotide variants (SNVs) were identified using MuTect v.1.1.7,39 while somatic small insertions and deletions (indels) were obtained using VarScan v.2.3.9. Different annotations were performed for the identified variants using ANNOVAR.40 SNVs identified by MuTect were filtered with the filter parameter “KEEP” and the indels identified by VarScan with a somatic p value filter of ≤ 0.001. In addition to passing the standard MuTect and VarScan filters, variants presenting with the following features were excluded: common SNPs with a minor allele frequency of >0.01, as found in dbSNP, the National Heart, Lung, and Blood Institute exome-sequencing project, 1000 Genomes, Exome Aggregation Consortium (ExAC), and in our in-house data from exome sequencing of ∼800 normal samples. Variants present in segmental duplication regions were removed from the analyses. All the mutations were also manually analyzed using Integrated Genomics Viewer (IGV) v.2.4.10 to check the relative position and mapping quality of variants in the aligned reads to filter out false positives.
To further reduce false-positive variant calls, additional filtering was performed. A variant was considered a true positive if the VAF was at least 2%. Furthermore, sequencing depth in the variant location region was required to be ≥15 and at least 4 altered sequence reads had to support the variant call. On the other hand, the number of reads supporting the variant in the germline data had to be ≥8 and the VAF < 1%. A variant having more than 2 mapping quality zero reads was considered a false positive and removed from the analysis.
Using SNPs for Sample Mismatch or Swaps
All tumor regions and the corresponding germline sample from a single individual should show a highly similar SNP profile to confirm that the sample is not mixed up or contaminated. The VAF of 24 SNPs identified by Pengelly et al.41 were used and checked in each tumor sample and compared to confirm that no sample swaps had occurred.
Mutation Validation
In order to determine the accuracy of the somatic variant calls, further target capture sequencing using SureSelect DNA Design at a median depth of 3,200× (2,262×–4,355×) was performed to validate the variants (Table S2). The sequencing library was prepared by random fragmentation of the DNA, followed by 5′ and 3′ adaptor ligation. To generate the clusters, the library was loaded into flow cell where fragments were captured on a lawn of surface-bound oligos complementary to the library adapters. Each fragment was then amplified into distinct, clonal clusters through bridge amplification and the templates were ready for sequencing.
Sequencing data were converted into fastq format for downstream analysis as described above. All the filters, as above, were applied with the exception of ≥5 reads supporting the variant call and the tumor VAF of at least 1%. If discrepancies in the start position, end position or length of the indel were seen across multiple tumor regions, the longest predicted indel and the maximum sequence values were taken. Variants absent in any of the tissues of a sample were manually checked to see whether these are present at a very low VAF and uncalled by the algorithm. These were manually added with the VAF filter of at least 1%.
Gene Copy Number Profiling, Cancer Cell Fraction, and Genome Doubling
FACETS v.0.5.1342 was used to determine copy number alterations (CNAs), purity (Figures S1 and S2), and regions of loss of heterozygosity (LOH), while outputs generated by MuTect and VarScan were utilized to calculate mean allelic frequency (MAF). CNA and SNV data were inputted into ABSOLUTE v.1.0.643 to define the integer copy number and cancer cell fractions (CCFs). CNAs were further defined as gains, losses, amplifications, and deletions, in relation to the average ploidy of all tissues from a given individual using the copy number for each segment from facets.
Gene-level amplifications and deletions were called by mean gene copy number ≥2× ploidy + 1 copy or ≤2× ploidy − 1 copy, respectively. To examine intratumor heterogeneity (ITH) of amplified and deleted genes, amplifications and deletions were classified across all tumor regions of each sample. If all the regions of a tumor showed a copy number gain of ploidy +1 copy or a copy number loss of ploidy −1 copy, the gene was classified as clonally amplified or clonally deleted, respectively. The gene was classified as subclonally amplified or deleted if any of the tumor regions in an individual revealed no gain or loss or if clonally amplified and deleted events overlapped with mirrored subclonal allelic imbalance.44
Genome-wide copy number gains and losses were defined by dividing the copy number data of each sample by the sample mean ploidy. ITH for chromosomal arm gains and losses were obtained by requiring at least one region of a tumor to show at least 98% gain or loss. Arm gain or loss was termed clonal if the same chromosomal arm showed at least 75% gain or loss across all remaining tumor regions. For subclonal arm gain or loss, at least one region should show less than 75% chromosomal arm gain or loss or if a chromosomal arm was subjected to mirrored subclonal allelic imbalance.
For clonality, a mutation with a probability of >50% or the lower bound of the 95% confidence interval of its CCF >90% was classified as clonal and subclonal otherwise.45 Genome doubling was calculated based on the mean ploidy of the sample. If the mean ploidy reported by FACETS was greater than 3, the sample was termed to have undergone genome doubling.46
Driver Mutation Classification
Non-silent variants were classified based on a list of potential driver cancer genes (n = 745). The driver gene list includes the genes in the COSMIC cancer gene census (v.87), genes identified in TCGA with q < 0.05 and previous large-scale studies.36, 47, 48, 49, 50, 51 Any variant present in these genes underwent classification based on the following criteria. If the gene was classified as tumor suppressor by COSMIC and the variant was found to be deleterious (stop-gain or predicted deleterious in two of the three computational tools—Sift, PolyPhen, and MutationTaster), the specific variant was classified as a driver. If the gene was classified as tumor oncogene and an exact variant was found ≥3 times in COSMIC, the variant was classed as a driver mutation.44
Detection of Mirrored Subclonal Allelic Imbalance
The samples with copy number segments showing allelic imbalance in at least one region, the B-allele fraction (BAF), was calculated for all the heterozygous SNPs as the ratio of the minor allele to the total allele count. If a given copy number event is shared between different regions of a tumor, the minor allele must always be from the same parental allele across these tumor regions. By analyzing all the heterozygous SNPs across different regions of a tumor, it is possible to determine whether the minor allele is different between regions, demonstrating two independent copy number events. Thus, mirrored subclonal allelic imbalance discovery involves comparing the BAF of the same heterozygous SNPs across multiple tumor regions and check whether the BAF values always follow the same distribution or whether their positions (i.e., high and low frequency) are reversed (mirrored). Mirrored subclonal allelic imbalance can only be detected for the samples having allelic imbalance present in at least two tissues. Mirrored subclonal allelic imbalance arm gain or loss was called when two different parental alleles were affected in at least 75% of a chromosome arm in at least two tumor regions.44
Subclonal Mutations Driven by Copy Number Loss
In the identification of subclonal mutations, driven by copy number loss, the method has been followed as described previously.44 Briefly, mutations have been investigated to identify those whose absence or low CCF values may have driven by copy number loss. Any SNV, residing in genomic segments of copy number heterogeneity across tumor regions, were identified for each tumor, with minor and major aberrations of copy number considered separately. Mutations were grouped into non-contiguous genomic segments with consistent copy number states within tumor regions for each chromosome. In order to restrict analysis to the mutations lost in at least one tumor region, the median CCF value of each SNV group were determined and considered only those SNV groups where the median CCF value was ≤0.25 in at least one tumor region. It has then been evaluated whether copy number loss coincided with lower levels of CCF by utilizing one-sided Wilcoxon test or a one-sided Cochrane Armitage trend test if more than two copy number states were present across tumor regions. Regression analysis was also implemented to ensure that the lower CCF value was driven by copy number and not tumor region by including both copy number and region in the regression model. The entire cluster was classified as copy number driven if more than 85% of mutations within a given cluster were determined to be driven by copy number. In order to avoid overestimating copy number driven losses of mutations, only losses occurring in ≤75% of tumor regions were considered.44
Phylogenetic Trees
Phylogenetic trees were constructed using the algorithm implemented in the tool LICHeE.52 LICHeE reconstructs cancer cell lineages using somatic variants from multiple related tumors of individual cancer cases, estimating heterogeneity in each cancer sample. For a set of high depth somatic variants, it uses the presence patterns of the variants across multiple tumors and their variant allele frequency (VAFs) or cancer cell fractions (CCFs) as lineage markers by relying on the perfect phylogeny model.52 This model describes that mutations do not recur independently in different cells; hence, cells sharing the same mutation must have inherited it from a common ancestral cell. LICHeE divides somatic variants into clones based on their presence in each sample, so that each clone carries the mutations that are present in the same subset of samples. To separate subclone clusters, the mutations of each resulting clone are further clustered based on their CCFs, so that mutations with similar CCFs across multiple samples are clustered together.
Mutational Signature Analysis
Mutational signatures were predicted using the deconstructSigs package in R.53 Mutational signatures were estimated for both clonal and subclonal mutations separately. Considering the lower mutational burden in PTC, the mutational signature analysis was only applied if at least 10 mutations were present in a sample.54 The signatures were compared to 21 published mutational signatures reported in different cancer types.55 Knowing not all mutations could be classified based on previously identified signatures using deconstructSigs, a small proportion of mutations were classified as “other.”
dN/dS Analysis
Maximum-likelihood method implemented in dNdScv R package was used to estimate dN/dS values for missense (ωmis) and nonsense (ωnon) mutations exome-wide as well as for the cancer genes.56 The method estimates the background mutation rate of each gene by combining local and global information (variation of the mutation rate across genes) and controlling for the sequence composition of the gene and mutational signatures. The substitution models provided in the package were used and maximum-likelihood estimates for ωmis and ωnon were obtained.
Analysis of Pathways
Prospective pathways associated with subclonal mutations were identified using Ingenuity Pathway Analysis by analyzing genes with subclonal mutations.
Tissue Microarray (TMA) Construction
TMA construction was performed from 1,231 formalin-fixed, paraffin-embedded PTC specimens as described earlier.57 Briefly, tissue cylinders with a diameter of 0.6 mm were punched from representative tumor regions of each donor tissue block and brought into recipient paraffin block using a modified semiautomatic robotic precision instrument (Beecher Instruments). Two cores of papillary carcinoma of the thyroid were arrayed from each case.
Immunohistochemistry
Tissue microarray slides were processed and stained manually as described previously.58 Primary antibody against HIF-1α (1:100 dilution, pH 6.0, Novus Biologicals) was used. The localization of HIF-1α was heterogeneous with predominant localization seen in the nucleus; however, low cytoplasmic localization was also noted. HIF-1α scoring was done as described previously.59 Briefly, the proportion of positively stained cells was calculated as a percentage for each core and the scores were averaged across two tissue cores from the same tumor to yield a single percent staining score representing each cancer case. For the purpose of statistical analysis, the scores were dichotomized based on the median proportion level of 20%. Cases showing proportion of more than 20% were classified as increased localization and those with ≤20% as decreased localization.
Statistical Analysis
All statistical analyses were executed on IBM SPSS Statistics (v.21). Mann-Whitney U test, chi-square test, or Fisher’s exact test were utilized to compare continuous and categorical variables, where relevant. Spearman’s rank correlation tests were used to determine associations. The Kaplan-Meier method based on the median fraction of either subclonal mutation or subclonal copy number variant (CNV), was used to perform survival analyses, with p values determined by Mantel-Cox log-rank test. For all the statistical tests performed, p < 0.05 was considered statistically significant.
Results
Intratumor Heterogeneity in PTC
Natural selection and tumor evolution can be propelled by the genetic diversity within tumors. To understand the intratumor heterogeneity, we performed multiregion WES on 257 tumors from 79 PTC-affected individuals. We categorized somatic mutations (single-nucleotide variants [SNVs] and small insertions and deletions [indels]) and copy number variants (CNVs) as clonal (ubiquitous presence in all tumor cells) or subclonal (partially present in all tumor cells) (Figure 1, Tables S3 and S4). Since clonality is based on CCF of the mutation which in turn depends on mutation variant allele frequency (VAF), a good concordance was observed between CCF and VAF (Figures S3 and S4).
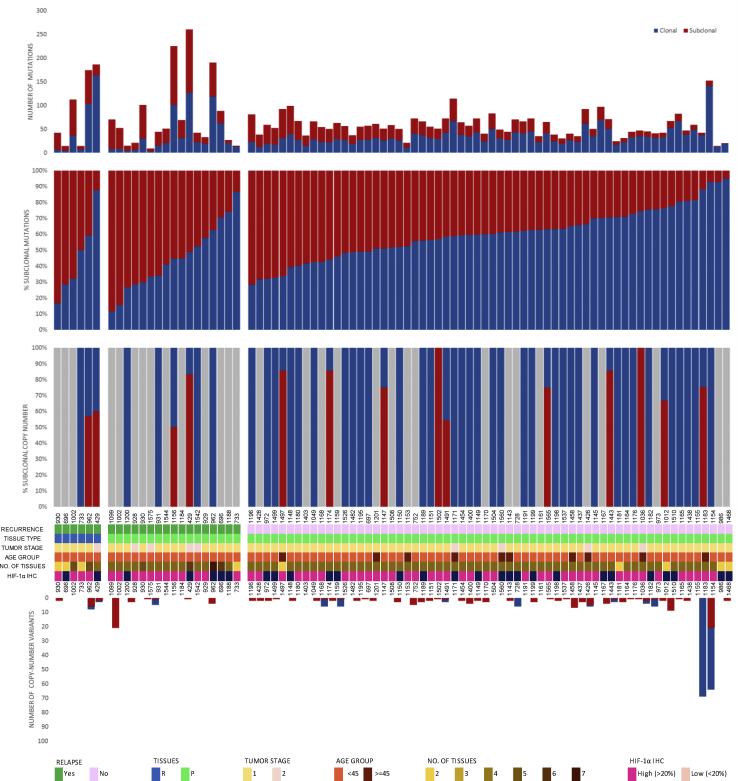
Intratumor heterogeneity was evidenced by a median of 41% (range 5% to 88.6%) and 50% (range 0% to 100%) of mutations and CNVs, respectively, being subclonal (Figure 2). The number of subclonal CNVs ranged from 1 to 133, with the percentage of genome affected ranging from 0% to 8% (Figure 2). Importantly, 40% of the subclonal mutations would have appeared clonal had multiregion WES not been applied, which suggests a degree of selection within individual tumor regions (Figure S5).
Figure 2.
Genetic Heterogeneity of Tumors from PTC-Affected Subjects
The total number of coding and noncoding mutations (top) and copy-number variants (bottom) detected in each tumor region, according to relapse, type of tissue (R, relapsed tissue; p, primary tissue), tumor stage, age, number of tissues, and HIF-1α immunohistochemical staining (IHC), with proportions of clonal and subclonal somatic variants. Grey areas correspond with a lack of mutations/copy number variants.
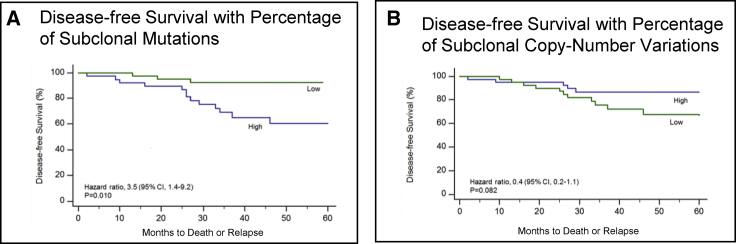
Despite generally low mutation rates in PTC, significantly more mutations were identified through multiregion WES (median 22; range 3 to 114), than using single sample analysis (median 17; range 0 to 57; p = 0.001; Mann-Whitney U test). Subclonal mutations were significantly higher in PTC relapse cases (p = 0.005, Table S6) with 76.5% of PTC relapse cases being categorized as “subclonally high (>41%, the cohort median)” compared to 40.3% of non-relapse cases. Further significant association was also seen between cases with high burden of subclonal mutations (>41%, the cohort median) and clinically aggressive parameters such as distant metastasis (p = 0.014, Table S6). PTC cases with high burden of subclonal mutations also had an increased risk for relapse or death than those with lower proportions (hazard ratio 3.5, 95% confidence interval [CI], 1.4 to 9.2; p = 0.010) (Figure 3A and Table S6). The median time until relapse or death was 26 months. This finding remained significant in multivariate analysis, after adjustment for age, gender, extra-thyroidal extension, and tumor size and stage (hazard ratio 3.62; CI: 1.10 to 11.97, p = 0.035) (Table S5). HIF-1 alpha overexpression showed a trend with the cases harboring a high burden of subclonal mutations (p = 0.076) (Figure 4 and Table S6).
Figure 3.
Disease-free Survival
The percentages of individuals who were disease free over a 60 month period according to whether they had a high proportion (above the median) or low proportion (below the median) of subclonal mutations (A) or of subclonal copy number variations (B). Individuals with a high proportion of subclonal mutations were at a significantly higher risk of relapse than those with a low proportion (p = 0.010), whereas there was no significant association between the proportion of subclonal copy number aberrations and disease-free survival (p = 0.082).
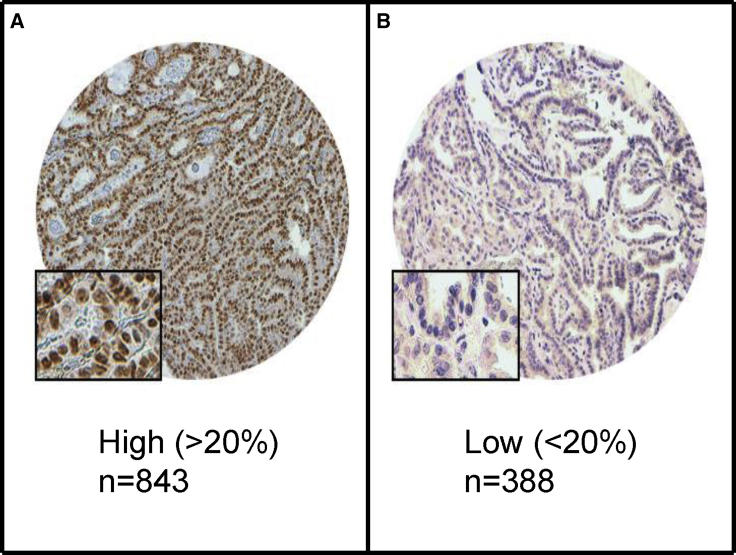
Figure 4.
Immunohistochemical Staining for HIF-1α in PTC
Increased localization (843 case subjects) (A) and decreased localization (388 case subjects) (B). 20×/0.70 objective on an Olympus BX 51 microscope (Olympus America Inc.) with inset showing a 40×/0.85 aperture magnified view of the same.
Clonal and subclonal CNVs were compared to evaluate intratumor heterogeneity in copy number variation. We found more subclonal CNVs compared to clonal, with a median of 100% (0 to 100) found to be subclonal (p < 0.001; Mann-Whitney U test). Furthermore, late-occurring CNVs tended to be deletions, with 100% (17 to 100) of all losses identified as subclonal compared to 50% (9 to 100) of gains (p = 0.053; Mann-Whitney U-test). Clonal CNVs were found to be larger than subclonal CNVs, with a median of 82.4 Mbp (0 to 249) versus 8.1 Mbp (0 to 242; p < 0.001; Mann-Whitney U test). However, the proportion of subclonal CNVs was found to have no prognostic effect (Figure 3B). We also analyzed the burden of subclonal mutations and copy number variations between the histological variants of PTC (classical variant, follicular variant, and tall cell variant), but observed no statistically significant differences.
Evolutionary History and Tumor Clonal Architecture in PTC
Intratumor heterogeneity can reveal the evolutionary history of mutations chronologically accumulated in a tumor.15 Unlike clonal mutations, subclonal mutations are only partially present in all tumor cells and their prevalence in different cancer cells can be used to infer subclonal hierarchy in the tumor phylogeny. Clustering mutations and inferring their phylogenies according to their cellular prevalence, we mapped the evolutionary process of each PTC tumor.52 On the tumor phylogenetic tree, each cluster represents a node, displaying present clones/subclones in the tumor population or clones/subclones having existed over the course of its evolutionary history (Figure S3).
As patterns of tumor evolution can be defined spatially, through multi-region sampling, and temporally, through multiple time-point sampling,24, 25 we deciphered PTC tumor evolution spatially in 79 primary tumors and temporally in 6 primary-relapse tissue pairs.
Our primary PTC evolutionary trees suggested two patterns of evolution. Most cases—54/79 (68%)—followed a linear evolution pattern, where successive clones overgrew or were in the process of overgrowing their ancestral clones, while the remaining cases—25/79 (32%)—followed a branched pattern, with divergence of subclones from a common ancestor clone, leading to the coexistence of subclones sharing only partial sets of mutations.
Comprising a single clone each that was shared in all primary regions, 24 of the 54 tumors showed a linear pattern of evolution with passive accumulation of mutations and driver events seemingly without clonal expansion, and where further subclones may have been too small to discriminate (Figure S3). Therefore, if sampled at a further time point, eventual clonal expansions may lead to further diversification, either continuing a linear path or producing divergent subclones.33 The time at which subclones evolve can vary in different tumor types,34 but this needs to be further studied in PTC.
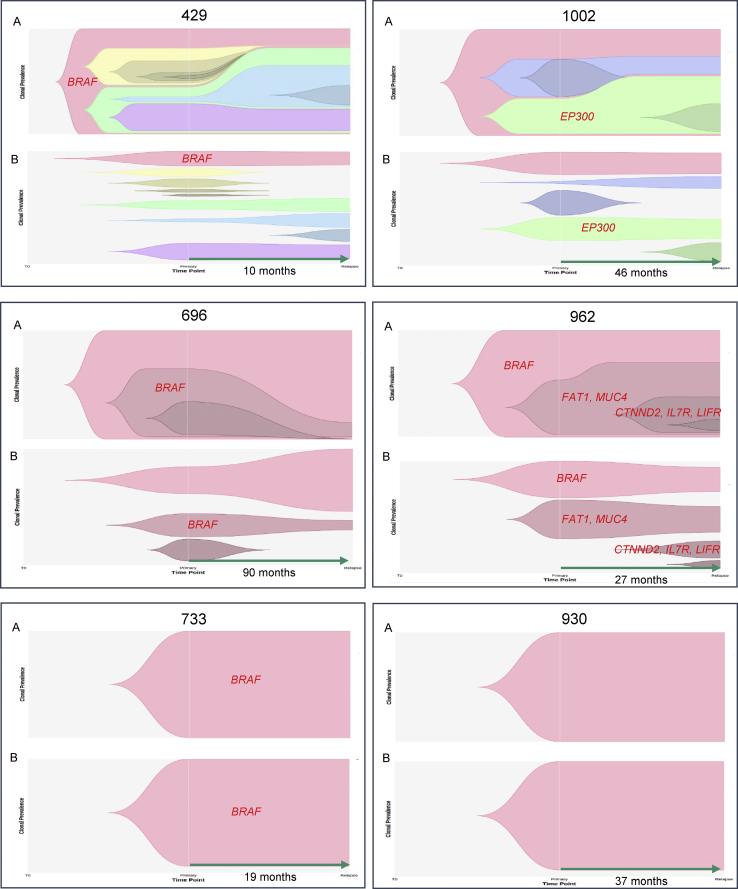
To further consolidate the types of evolutionary patterns observed, we took advantage of the multiple longitudinal relapsed tumor samples acquired from six case subjects by comparing their evolution with their corresponding primary to study the spatial dynamics of their evolution (Figure 5).
Figure 5.
Evolution of Relapsed PTC
Tumor evolution of relapsed PTC based on cell fractions at different time points (time to relapse is in months). Nested view of all clusters together over time (A) and clonal trajectory view (B) depicting the history of individual clusters over time. Different colors represent separate clusters in a sample. Driver genes (in red) are centered in each cluster.
Two case subjects (733 and 930), each with only a single clone shared among all primary and relapse tissue regions, showed no clonal expansion over time. Remarkably, the set of mutations remained unchanged between the two time points, despite ablation and radioiodine therapy between the two time points. We do note, however, that some mutations showed a decline in CCF post-treatment. Both individuals were young females, one 13 years old (733) and the other (930) 33 years old, where both presented with classical PTC variant and stage one tumor. Consistent with young adult and pediatric PTC case subjects,54, 60 case 733 had very few mutations (seven in total, including one BRAF mutation).
Two case subjects (696 and 962) showed linear evolution, where case subject 962 showed successive expansion of clones and acquisition of driver events (MUC4 [MIM: 158372], CTNND2 [MIM: 604275], IL7R [MIM: 146661], and LIFR [MIM: 151443] amplifications) despite treatment, whereas many clones obtained early in tumor evolution remained resistant. In contrast, case subject 696 showed exceptional sensitivity to the treatment given, as most primary subclones had been ablated, with some resistant mutations remaining in the main ancestral clone. More shared mutations were observed between primary and relapse tumors in case subjects displaying linear patterns of evolution.
Two case subjects (429 and 1002) demonstrated a branching clonal evolution pattern, where case subject 1002 initially showed linear evolution in the corresponding primary tissues, but later diverged in parallel, with some relapse tumor regions further expanding from previous primary subclones and others diverging from the main ancestral clone. Lower CCFs in some relapse regions showed some sensitivity to treatment. Case subject 429 had more extensive branching early in the tumor development, only further expanding by a single clone of six mutations post-treatment. Some clones were seen to grow further in the relapse tissues, while the majority that had sufficiently grown in the primary remained resistant in the relapsed tissues. Interestingly, this case had the shortest time to relapse (10 months), higher stage (II), presented a more aggressive tall cell variant, and had metastatic disease.
Newly formed clones in relapse tissues are likely to either contain de novo mutations acquired during the growth of the relapse or were not sampled in the primary tumor blocks. Clusters showing either lower CCFs in the primary tumor but a higher CCF in their relapse counterpart or consistently high CCFs in both the primary tumor and relapse counterpart might contain important radioiodine-resistant mutations. These include mutations in driver genes such as FAT1 (MIM: 600976), in addition to other genes such as RABGEF1 (MIM: 609700), FMN2 (MIM: 606373), SMG5 (MIM: 610962), KCNK17 (MIM: 607370), PDGFRB (MIM: 173410), NFIB (MIM: 600728), and MCM4 (MIM: 602638) (Figure S3). Although branched evolution was observed in our PTC cohort, no evidence of parallel branched evolution, where there is a convergence of somatic events in distinct branches on the same gene, was identified in either primary or relapse tissues.
A total of 188 mutation clusters with a median of 2 per tumor (range 1 to 9) were identified in our PTC cohort, with more than half (52%) of tumor regions found to carry subclones from only a single branch of the phylogenetic tree. Furthermore, 89% of branched subclone clusters could have inaccurately appeared as clonal, without the use of spatially distinct multi-sampling approaches. This revealed the limitations of single diagnostic biopsy samples for the accurate assessment of intratumor heterogeneity, particularly in tumors with branched evolutionary patterns. Although linear evolution patterns generally imply limited intratumor heterogeneity, where single biopsy samples can represent the entire tumor, simplifying diagnostic assays, incomplete clonal expansions in linear evolution also produce intratumor heterogeneity, indicating the need for multi-sampling approaches in diagnosis and for informing the appropriate treatment options.35
Differential Selection during Evolution
Exploring the ongoing selection during PTC evolution can help in identifying evolutionary constraints, eventually dictating the evolutionary routes of this tumor. Due to the lack of parallel evolution, we sought to estimate positive selection at the mutation level via a dN/dS ratio, which parallels substitution rates at nonsynonymous sites to those at synonymous sites, thus accounting for the trinucleotide context of each mutation and determining the enrichment of protein-altering mutations compared to the background mutation rate.61 Positive selection (dN/dS,>1) was observed, when considering all exonic missense mutations (Figure S6 and Table S7). However, upon temporal dissection, significant positive selection was observed only in clonal mutations. Without temporal dissection, non-relapse case subjects also showed positive selection in both exonic missense and nonsense clonal mutations with depletion of missense and nonsense subclonal mutations, suggesting mutations may be shaped by positive selection in early but not late stages of PTC, with particularly strong positive selection seen early in relapse PTC.
Causes of Intratumor Heterogeneity in PTC Mutational Process
QA Alternating mutational stresses over tumor history result in varying subclones and patterns of trinucleotide signatures.62 It is particularly important to understand the effect of these mutational processes that shape PTC tumor evolution, given the association we found between subclonal mutation burden and shorter relapse-free survival (Spearman’s rank correlation rho = 0.369, p = 0.013), potentially informing clinical strategies to limit tumor adaptation.
Published mutational signatures55 were used to analyze clonal and subclonal mutations, to determine the mutational processes contributing to intratumor heterogeneity. However, due to limited number of mutations (< 10), only 57% (45/79) and 61% (48/79) primary PTC cases were assigned mutational signatures for clonal and subclonal mutations, respectively. In addition, only 28% (33/79) of primary tumors and additional two (cases 429 and 962) matching relapse tissues, harboring sufficient mutations for clonal and subclonal pairs, could be assessed for mutational signature enrichment in clonal and/or subclonal mutations by allocating the contribution of individual mutational signatures over molecular time to each tumor (Figures S7–S9).
We identified several signatures within the tested tumors, of which the most prominent were signature 1A (21/33, 64%), involving the endogenous spontaneous deamination of methylated cytosines correlating with the individual’s age at the time of cancer diagnosis; signature 3 (16/33, 48%), associated with failure of DNA double-strand break-repair by homologous recombination; signature 11 (16/33, 48%), associated with DNA alkylating agent temozolomide treatment; and signature 15 (17/33, 52%), which is one of the four signatures related to defective mismatch repair (MMR).
The age-related signatures 1A and/or 1B, dominated by C to T mutations at CpG loci, were consistently seen in most cases (27/33, 82%). Unlike increased clonal mutation burden, which was significantly correlated with the number of mutations assigned to signature 1B (Spearman’s rank correlation, rho = 0.338, p = 0.023), subclonal mutation burden was significantly correlated with signature 1A (Spearman’s rank correlation, rho = 0.381, p = 0.008).
Interestingly, we noticed a relatively consistent contribution of MMR deficiency signatures (6, 15, 20 and 21) across clonal and subclonal mutations (25/33, 76%), four of which were primary cases with relapse showing continued dominant contribution over time. One particular relapse case (429) showed further increased subclonal contribution, 35% to 42%, from primary to relapse (Figure S8). Furthermore, only signature 20 was significantly more enriched in clonal, compared to subclonal mutations (Wilcoxon’s paired sign-rank test, p = 0.043). However, unlike previous reports, tumors showing MMR deficiency signatures completely lacked loss-of-function mutations in MMR genes, MLH1 (MIM: 120436), MSH2 (MIM: 609309) and MSH6 (MIM: 600678), as seen in more aggressive forms of thyroid cancer.3 However these tumors showed a trend toward more small insertions and deletions (<3 bp) compared to the tumors without MMR deficiency signatures (p = 0.070; Mann-Whitney U test) which is the proposed etiology of these signatures. Similarly, APOBEC (a family of cytidine deaminase enzymes involved in messenger RNA editing) signatures (2 and 13) also showed relatively consistent contribution (14/33, 42%) – where a single case with relapse (1156) showed prominent increase in subclonal mutation contribution, from 18% in clonal to 81% in subclonal (Figure S8). Remarkably, signature 13 was significantly correlated with increased clonal mutational burden (Spearman’s rank correlation, rho = 0.369, p = 0.013), albeit in a small number of samples (3/45, 7%; Figure S9). Collectively, to investigate whether these observations are significant for relapse cases, a larger study is required.
There were also significant inverse correlations between the subclonal mutation burden and the number of mutations assigned to signature 5 (Spearman’s rank correlation, rho = −0.317, p = 0.028) and 14 (Spearman’s rank correlation, rho = −0.322, p = 0.026), although the etiologies of these signatures have not been elucidated (Figure S9).
Furthermore, samples carrying subclonal driver mutations were enriched for signature 4, related to tobacco smoking (p = 0.022).
Interpreting these temporal differences should be considered with caution, owing to the low number of mutations in some of these tumors. Nonetheless, the overall data indicates various mutational processes might have important roles in subclonal diversification during PTC evolution.
Clonal and Subclonal Driver Alterations and Timing of Genomic Events
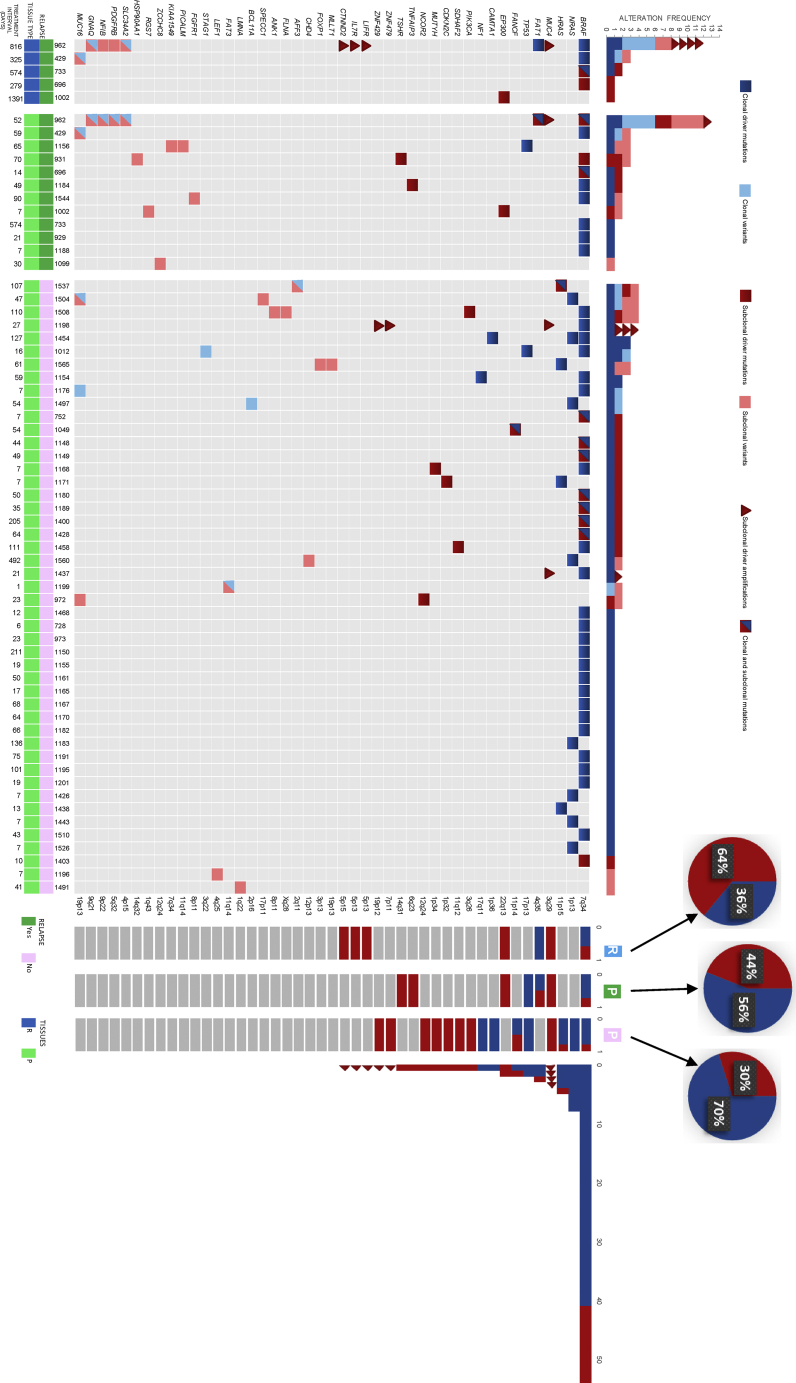
Only a small portion of somatic mutations are cancer drivers that confer clonal fitness and are positively selected over the course of tumor evolution.63, 64 Determining whether cancer driver events occur early or late can suggest whether they have a role in tumor initiation or tumor maintenance. Also, determining clonality of these driver events might help in identifying potential therapeutic strategies. We identified 94 driver events (range in primary PTC, 1 to 5; range in relapsed PTC, 1 to 6). Of these events 34 were found to be subclonal (range in primary PTC 1 to 3; range in relapse 1 to 4) and 60 to be clonal (range in Primary PTC, 1 to 3; range in relapse PTC, 1 to 2) (Figure 6). Again, significantly more driver alterations were identified with the use of multiple region WES than with single sample analysis (p = 0.027).
Figure 6.
Heterogeneity of Driver Mutations in PTC
Driver alterations detected for each tumor. All genes containing driver alterations across the cohort are included. The color represents the clonal status (blue, clonal; red, subclonal). Number of driver alterations identified across tumors (top) and the barplots to the right are proportion of variants found as clonal/subclonal in relapsed tissue (R), primary relapse (P; green), and primary non-relapse (P; pink) tissues for each gene, along with total count of driver alterations. Pie charts represent the overall proportion of subclonal driver events per category. Treatment intervals are in days, from diagnosis to surgical intervention.
Mutations in MAP kinase genes (BRAF, NRAS [MIM: 164790], HRAS [MIM: 190020]) and TP53 (MIM: 191170) were predominantly clonal in both primary and relapse PTC tumors, which suggests their involvement in tumor initiation rather than maintenance or progression. On the other hand, we have identified genes that were enriched for subclonal alterations such as CDKN2C (MIM: 603369), FANCF (MIM: 613897), MUTYH (MIM: 604933), EP300 (MIM: 602700), PIK3CA (MIM: 171834), along with ZNF and MUC4 amplifications (Figure 6). Relevant to our finding of significant association between subclonal mutations and the disease-free survival, we found enrichment for subclonal driver events in relapse PTC (64% versus 30%, p = 0.030) (Figure 6).
Although we found no evidence of convergence when analyzing subclonal mutations at the individual gene level, we searched for potential convergence at the pathway level, to investigate whether the subclonal genes were associated with specific biological pathways. Pathway analysis of the mutations that appeared to be subclonal revealed enrichment for genes associated with the HIF-1α signaling pathway (p = 0.001) using ingenuity pathway analysis (IPA) (Table S8). To further explore the prognostic value of HIF-1α in PTC, we screened the protein localization of HIF-1α in > 1000 PTC (inclusive of our 79 cases) using tissue microarray. Interestingly, PTC tumors that showed increased localization of HIF-1α protein tended to be clustered in older age individuals (p < 0.001) and were significantly associated with higher stage (p < 0.001), as well as extra-thyroidal extension (p = 0.027). Most importantly, localization of HIF-1α was strongly correlated with adverse prognosis (p = 0.037) (Table S9). However, on multivariate analysis, the disease-free survival was not significant after adjusting for age, gender, extra-thyroidal extension and stage of tumor (p = 0.104, HR = 1.29, 95% CI = 0.95–1.81). We have further analyzed the pathways involved in different histological variants of PTC, (classical variant, follicular variant and tall cell variant), via their subclonal mutations. The HIF1α Signaling pathway was significantly seen in tall cell variant. Conversely, no related significant pathways were observed for the other two variants of PTC.
Chromosomal Instability and Genomic Doubling
Although we did not observe any association between subclonal CNVs and clinical parameters in the tumors analyzed, we explored the relation of copy number intratumor heterogeneity and the underlying forces of chromosomal aberrations in tumor regions. By identifying germline heterozygous single nucleotide polymorphisms (SNPs) in tumors, we can conclude whether the gain or loss of parental alleles in distinct parallel subclones has ensued mirrored subclonal allelic imbalance. However, no such event was observed in our PTC cohort.
Furthermore, we explored the effects of chromosomal instability, by the loss of genomic segments harboring clonal mutations, on mutational heterogeneity. Interestingly, none of the subclonal mutations were found subclonal as a result of copy number loss events, suggesting there is no role of chromosomal instability on mutational heterogeneity.
In addition, whole-genome doubling events were observed in two case subjects (1036 and 1183), where events were shared in 2/3 tumor regions in both cases. All clonal mutations were present at ploidy ≥ 2 in both cases, suggesting mutational burden occurred before genome doubling. In one case subclonal mutations were also present at ploidy ≥ 2, which indicates two independent genome doubling events. The fact that we had little evidence of WGD in PTC, is consistent with previous reports21 and is expected, given their oncogene-driven but otherwise quiet genome. In summary, chromosomal instability seems to play no role in driving PTC genome evolution.
Discussion
Although most PTC driver mutations are clonal, a portion occurs subclonally. This pattern of PTC clonality has clear clinical utility, since we found that individuals with high subclonal mutations have a significantly higher rate of relapse. Our findings, therefore, have potential implications on individualizing prognosis for individuals with PTC.
Key aspects of tumor evolution have been revealed using multiregion sequencing in different solid tumors.13, 23, 25, 29, 30 We pursued this approach in a large cohort of PTC case subjects with relapse to understand spatial and temporal aspects of tumor evolution and the population characteristics of their subclonal evolution. Our study provides an overview of PTC evolution by demonstrating that intratumor heterogeneity is relatively common. Consistent with previous expectations,18 neutral evolution was not observed in PTC; instead, Darwinian-based linear and branched evolution was seen across the cohort. Specifically, we have identified intratumor heterogeneity of somatic mutations in multiple regions (n = 257) from 79 PTC tumors and demonstrated that 23.1% of the somatic mutations were not identified in all regions of the tumor. Our data also confirm the previously known role of MAP Kinase genes in PTC tumorigenesis especially in tumor initiation since most of these driver genes, including BRAF, NRAS, HRAS, as well as TP53 were almost exclusively clonal (Figure 6).
Although TCGA study on PTC sampled a single site from each tumor, we compared the driver genes to see their presence in TCGA study.49 A good concordance was observed in the frequency of genes in our study verses TCGA (NRAS [10.12% versus 8.47%], HRAS [5.01% versus 3.49%], TP53 [2.53% versus 0.74%], CDKN2C [1.26% versus 0.24%], FANCF [1.26% versus 0.24%], NF1 [MIM: 613113] [1.26% versus 0.49%], PIK3CA [1.26% versus 0.49%], TSHR [MIM: 603372] [1.26% versus 0.49%]) except BRAF which was higher in TCGA (50.63% versus 59.35%). The slight higher range in our cohort might be due to the chance of getting more mutations in multi region sequencing as described in this study. In our previous landscaping study on PTC, a concordance to TCGA was observed for BRAF mutation (59.41% versus 59.35%).35 No mutations in EIF1AX (MIM: 300186), RBM10 (MIM: 300080), PPM1D (MIM: 605100), CHEK2 (MIM: 604373), and a low frequency of TERT (MIM: 187270) promoter mutations (6.3%) were observed in this study. The absence of these mutations might be due to the ethnicity of the cohort population as reported previously65 and low frequency of TERT due to the selected cohort of early stage as we have previously shown TERT promoter mutations to be associated with advanced tumors.66 A high frequency of RECQL5 (MIM: 603781) splice site recurrent mutation (16.4% case subjects) and three frameshift mutations in ZNF717 (MIM: 618405) (30.4% case subjects) were observed in the cohort, but these genes were not associated with any clinico-pathological characteristics. The RECQL5 splice site mutation (c.1586−1>GT) is an insertion of GT nucleotides but there are two GT repeats after the insertion keeping the sequence same up to c.1586−5, so it might not play any role in splicing. The ZNF717 mutations have been previously reported by Molenaar et al. and others67 with no clinical associations reported.
In our cohort, 29% of PTC tumors carried subclonal driver alterations, with a portion of subclonal driver mutations seemingly clonal in a single region but absent or subclonal in other regions. These data reiterate the limitations of single biopsy sampling, while underlining the importance of multi-region WES to address these limitations in the detection of clonal and subclonal events for potential drug targeting and improved clinical characterization of PTC tumors. Furthermore, as per our association between elevated subclonal mutations and shorter relapse-free survival, individuals with early-stage PTC and a high subclonal burden may signify a high-risk group that could benefit from close observation and early treatment. Our data have also shown further significant association between increased clinical aggressiveness, due to distant metastasis and relapse, and genetic evolution in PTC and may eventually help in the selection of therapeutic strategies to prevent the expansion of these subclones.
Advances in cancer biology have highlighted the HIF-1α pathway as a critical survival pathway, which can be regulated at various levels, such as translation, transcriptional activation, and degradation. Studies have shown an altered HIF-1α pathway can lead to the downstream upregulation of proteins promoting angiogenesis and anerobic metabolism among other survival pathways (Figure S10). We extended our knowledge of the survival effect of HIF-1α pathway by identifying a significant correlation with increased subclonal mutation burden. The apparent involvement of this important pathway as a result of increased subclonal mutational burden potentially makes it an attractive target for therapeutic intervention and prevention of tumor relapse, especially since there are many drugs that can inhibit this pathway.
Conclusion
Overall, our study highlights the importance of using multi-region WES of primary and relapsed PTC tumors to discern the patterns of PTC progression and relapse. More importantly, our study underscores the importance of evolutionary development as the engine driving cancer relapse. This knowledge will pave the road to develop therapeutic strategies that can target not only specific gene alterations but also the trajectory landscape of these alterations.
Declaration of Interests
The authors declare that they have no competing interests.
Acknowledgments
We thank the participants of this study. The authors would also like to acknowledge Maha Al Rasheed, Khadija Al Obaisi, Mark Ranier Diaz, Ingrid Garcia Victoria, Dionne Rae Rala, Allianah Benito, Maria Angelita Sabido, Padmanaban Annaiyappanaidu, Valorie Balde, Nabil Siraj, and Mary Beleno Galvez for their technical assistance. This project was conducted by a generous fund from the Saudi Government to the Thyroid Cancer Genome Project as part of the International Cancer Genome Consortium - Accelerate Research in Genomic Oncology (ICGC-ARGO) under the Project RAC # 2110 031 and # 2170 022.
Published: October 24, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.09.026.
Contributor Information
Fowzan S. Alkuraya, Email: falkuraya@kfshrc.edu.sa.
Khawla S. Al-Kuraya, Email: kkuraya@kfshrc.edu.sa.
Accession Numbers
All the raw data are submitted to NCBI Sequence Read Archive (SRA) with accession number PRJNA550230.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
ExAC Browser, http://exac.broadinstitute.org/
FastQC, http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Ingenuity Variant Analysis, http://www.ingenuity.com/products/variant-analysis
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, https://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Sequence Read Archive (SRA), http://www.ncbi.nlm.nih.gov/sra
TCGA Portal, https://cancergenome.nih.gov/
VarScan, http://varscan.sourceforge.net/
Supplemental Data
Figures S1–S10 and Tables S1, S5, S6, and S9
Tables S2–S4, S7, and S8
References
- 1.Wiltshire J.J., Drake T.M., Uttley L., Balasubramanian S.P. Systematic review of trends in the incidence rates of thyroid cancer. Thyroid. 2016;26:1541–1552. doi: 10.1089/thy.2016.0100. [DOI] [PubMed] [Google Scholar]
- 2.Alzahrani A.S., Alomar H., Alzahrani N. Thyroid cancer in Saudi Arabia: a histopathological and outcome study. Int. J. Endocrinol. 2017;2017:8423147. doi: 10.1155/2017/8423147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grant C.S. Recurrence of papillary thyroid cancer after optimized surgery. Gland Surg. 2015;4:52–62. doi: 10.3978/j.issn.2227-684X.2014.12.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howell G.M., Carty S.E., Armstrong M.J., Lebeau S.O., Hodak S.P., Coyne C., Stang M.T., McCoy K.L., Nikiforova M.N., Nikiforov Y.E., Yip L. Both BRAF V600E mutation and older age (≥ 65 years) are associated with recurrent papillary thyroid cancer. Ann. Surg. Oncol. 2011;18:3566–3571. doi: 10.1245/s10434-011-1781-5. [DOI] [PubMed] [Google Scholar]
- 5.Alzahrani A.S., Xing M. Impact of lymph node metastases identified on central neck dissection (CND) on the recurrence of papillary thyroid cancer: potential role of BRAFV600E mutation in defining CND. Endocr. Relat. Cancer. 2013;20:13–22. doi: 10.1530/ERC-12-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czarniecka A., Kowal M., Rusinek D., Krajewska J., Jarzab M., Stobiecka E., Chmielik E., Zembala-Nozynska E., Poltorak S., Sacher A. The risk of relapse in papillary thyroid cancer (PTC) in the context of BRAFV600E mutation status and other prognostic factors. PLoS ONE. 2015;10:e0132821. doi: 10.1371/journal.pone.0132821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing M., Westra W.H., Tufano R.P., Cohen Y., Rosenbaum E., Rhoden K.J., Carson K.A., Vasko V., Larin A., Tallini G. BRAF mutation predicts a poorer clinical prognosis for papillary thyroid cancer. J. Clin. Endocrinol. Metab. 2005;90:6373–6379. doi: 10.1210/jc.2005-0987. [DOI] [PubMed] [Google Scholar]
- 8.Vini L., Hyer S.L., Marshall J., A’Hern R., Harmer C. Long-term results in elderly patients with differentiated thyroid carcinoma. Cancer. 2003;97:2736–2742. doi: 10.1002/cncr.11410. [DOI] [PubMed] [Google Scholar]
- 9.Vogelstein B., Kinzler K.W. The multistep nature of cancer. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 10.Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L.A., Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allison K.H., Sledge G.W. Heterogeneity and cancer. Oncology (Williston Park) 2014;28:772–778. [PubMed] [Google Scholar]
- 12.Alizadeh A.A., Aranda V., Bardelli A., Blanpain C., Bock C., Borowski C., Caldas C., Califano A., Doherty M., Elsner M. Toward understanding and exploiting tumor heterogeneity. Nat. Med. 2015;21:846–853. doi: 10.1038/nm.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerlinger M., Rowan A.J., Horswell S., Math M., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis A., Gao R., Navin N. Tumor evolution: Linear, branching, neutral or punctuated? Biochimic. Biophys. Acta. Rev. Can. 2017;1867:151–161. doi: 10.1016/j.bbcan.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Navin N.E., Hicks J. Tracing the tumor lineage. Mol. Oncol. 2010;4:267–283. doi: 10.1016/j.molonc.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amirouchene-Angelozzi N., Swanton C., Bardelli A. Tumor evolution as a therapeutic target. Cancer Discov. 2017;7:805–817. doi: 10.1158/2159-8290.CD-17-0343. [DOI] [PubMed] [Google Scholar]
- 17.McGranahan N., Swanton C. Cancer evolution constrained by the immune microenvironment. Cell. 2017;170:825–827. doi: 10.1016/j.cell.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Williams M.J., Werner B., Barnes C.P., Graham T.A., Sottoriva A. Identification of neutral tumor evolution across cancer types. Nat. Genet. 2016;48:238–244. doi: 10.1038/ng.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quail D.F., Joyce J.A. The microenvironmental landscape of brain tumors. Cancer Cell. 2017;31:326–341. doi: 10.1016/j.ccell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiménez-Sánchez A., Memon D., Pourpe S., Veeraraghavan H., Li Y., Vargas H.A., Gill M.B., Park K.J., Zivanovic O., Konner J. Heterogeneous tumor-immune microenvironments among differentially growing metastases in an ovarian cancer patient. Cell. 2017;170:927–938.e20. doi: 10.1016/j.cell.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah S.P., Roth A., Goya R., Oloumi A., Ha G., Zhao Y., Turashvili G., Ding J., Tse K., Haffari G. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Findlay J.M., Castro-Giner F., Makino S., Rayner E., Kartsonaki C., Cross W., Kovac M., Ulahannan D., Palles C., Gillies R.S. Differential clonal evolution in oesophageal cancers in response to neo-adjuvant chemotherapy. Nat. Commun. 2016;7:11111. doi: 10.1038/ncomms11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhai W., Lim T.K.-H., Zhang T., Phang S.-T., Tiang Z., Guan P., Ng M.-H., Lim J.Q., Yao F., Li Z. The spatial organization of intra-tumour heterogeneity and evolutionary trajectories of metastases in hepatocellular carcinoma. Nat. Commun. 2017;8:4565. doi: 10.1038/ncomms14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caswell-Jin J.L., McNamara K., Reiter J.G., Sun R., Hu Z., Ma Z., Ding J., Suarez C.J., Tilk S., Raghavendra A. Clonal replacement and heterogeneity in breast tumors treated with neoadjuvant HER2-targeted therapy. Nat. Commun. 2019;10:657. doi: 10.1038/s41467-019-08593-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J., Cazzato E., Ladewig E., Frattini V., Rosenbloom D.I., Zairis S., Abate F., Liu Z., Elliott O., Shin Y.-J. Clonal evolution of glioblastoma under therapy. Nat. Genet. 2016;48:768–776. doi: 10.1038/ng.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowell P.C. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 27.Grove C.S., Vassiliou G.S. Acute myeloid leukaemia: a paradigm for the clonal evolution of cancer? Dis. Model. Mech. 2014;7:941–951. doi: 10.1242/dmm.015974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGranahan N., Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Saito T., Niida A., Uchi R., Hirata H., Komatsu H., Sakimura S., Hayashi S., Nambara S., Kuroda Y., Ito S. A temporal shift of the evolutionary principle shaping intratumor heterogeneity in colorectal cancer. Nat. Commun. 2018;9:2884. doi: 10.1038/s41467-018-05226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang D., Niu X., Wang Z., Song C.-L., Huang Z., Chen K.-N., Duan J., Bai H., Xu J., Zhao J. Multiregion sequencing reveals the genetic heterogeneity and evolutionary history of osteosarcoma and matched pulmonary metastases. Cancer Res. 2019;79:7–20. doi: 10.1158/0008-5472.CAN-18-1086. [DOI] [PubMed] [Google Scholar]
- 31.Turke A.B., Zejnullahu K., Wu Y.-L., Song Y., Dias-Santagata D., Lifshits E., Toschi L., Rogers A., Mok T., Sequist L. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Misale S., Yaeger R., Hobor S., Scala E., Janakiraman M., Liska D., Valtorta E., Schiavo R., Buscarino M., Siravegna G. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding L., Ley T.J., Larson D.E., Miller C.A., Koboldt D.C., Welch J.S., Ritchey J.K., Young M.A., Lamprecht T., McLellan M.D. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson B.E., Mazor T., Hong C., Barnes M., Aihara K., McLean C.Y., Fouse S.D., Yamamoto S., Ueda H., Tatsuno K. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S., Han Y., Kim S.I., Kim H.-S., Kim S.J., Song Y.S. Tumor evolution and chemoresistance in ovarian cancer. NPJ Precis Oncol. 2018;2:20. doi: 10.1038/s41698-018-0063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siraj A.K., Masoodi T., Bu R., Beg S., Al-Sobhi S.S., Al-Dayel F., Al-Dawish M., Alkuraya F.S., Al-Kuraya K.S. Genomic profiling of thyroid cancer reveals a role for thyroglobulin in metastasis. Am. J. Hum. Genet. 2016;98:1170–1180. doi: 10.1016/j.ajhg.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pengelly R.J., Gibson J., Andreoletti G., Collins A., Mattocks C.J., Ennis S. Erratum to: a SNP profiling panel for sample tracking in whole-exome sequencing studies. Genome Med. 2015;7:44. doi: 10.1186/s13073-015-0163-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen R., Seshan V.E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carter S.L., Cibulskis K., Helman E., McKenna A., Shen H., Zack T., Laird P.W., Onofrio R.C., Winckler W., Weir B.A. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jamal-Hanjani M., Wilson G.A., McGranahan N., Birkbak N.J., Watkins T.B.K., Veeriah S., Shafi S., Johnson D.H., Mitter R., Rosenthal R., TRACERx Consortium Tracking the Evolution of Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 45.Ng C.K.Y., Bidard F.C., Piscuoglio S., Geyer F.C., Lim R.S., de Bruijn I., Shen R., Pareja F., Berman S.H., Wang L. Genetic Heterogeneity in Therapy-Naïve Synchronous Primary Breast Cancers and Their Metastases. Clin. Cancer Res. 2017;23:4402–4415. doi: 10.1158/1078-0432.CCR-16-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez P., Mallo D., Paulson T.G., Li X., Sanchez C.A., Reid B.J., Graham T.A., Kuhner M.K., Maley C.C. Evolution of Barrett’s esophagus through space and time at single-crypt and whole-biopsy levels. Nat. Commun. 2018;9:794. doi: 10.1038/s41467-017-02621-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan W., Zhou L., Ge M., Zhang B., Yang X., Xiong X., Fu G., Zhang J., Nie X., Li H. Whole exome sequencing identifies lncRNA GAS8-AS1 and LPAR4 as novel papillary thyroid carcinoma driver alternations. Hum. Mol. Genet. 2016;25:1875–1884. doi: 10.1093/hmg/ddw056. [DOI] [PubMed] [Google Scholar]
- 48.Fang Y., Ma X., Zeng J., Jin Y., Hu Y., Wang J., Liu R., Cao C. The Profile of Genetic Mutations in Papillary Thyroid Cancer Detected by Whole Exome Sequencing. Cell. Physiol. Biochem. 2018;50:169–178. doi: 10.1159/000493966. [DOI] [PubMed] [Google Scholar]
- 49.Cancer Genome Atlas Research Network Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Capdevila J., Mayor R., Mancuso F.M., Iglesias C., Caratù G., Matos I., Zafón C., Hernando J., Petit A., Nuciforo P. Early evolutionary divergence between papillary and anaplastic thyroid cancers. Ann. Oncol. 2018;29:1454–1460. doi: 10.1093/annonc/mdy123. [DOI] [PubMed] [Google Scholar]
- 51.Dong W., Nicolson N.G., Choi J., Barbieri A.L., Kunstman J.W., Abou Azar S., Knight J., Bilguvar K., Mane S.M., Lifton R.P. Clonal evolution analysis of paired anaplastic and well-differentiated thyroid carcinomas reveals shared common ancestor. Genes Chromosomes Cancer. 2018;57:645–652. doi: 10.1002/gcc.22678. [DOI] [PubMed] [Google Scholar]
- 52.Popic V., Salari R., Hajirasouliha I., Kashef-Haghighi D., West R.B., Batzoglou S. Fast and scalable inference of multi-sample cancer lineages. Genome Biol. 2015;16:91. doi: 10.1186/s13059-015-0647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenthal R., McGranahan N., Herrero J., Taylor B.S., Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pozdeyev N., Gay L.M., Sokol E.S., Hartmaier R., Deaver K.E., Davis S., French J.D., Borre P.V., LaBarbera D.V., Tan A.-C. Genetic analysis of 779 advanced differentiated and anaplastic thyroid cancers. Clin. Cancer Res. 2018;24:3059–3068. doi: 10.1158/1078-0432.CCR-18-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., Australian Pancreatic Cancer Genome Initiative. ICGC Breast Cancer Consortium. ICGC MMML-Seq Consortium. ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martincorena I., Raine K.M., Gerstung M., Dawson K.J., Haase K., Van Loo P., Davies H., Stratton M.R., Campbell P.J. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2017;171:1029–1041.e21. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siraj A.K., Bavi P., Abubaker J., Jehan Z., Sultana M., Al-Dayel F., Al-Nuaim A., Alzahrani A., Ahmed M., Al-Sanea O. Genome-wide expression analysis of Middle Eastern papillary thyroid cancer reveals c-MET as a novel target for cancer therapy. J. Pathol. 2007;213:190–199. doi: 10.1002/path.2215. [DOI] [PubMed] [Google Scholar]
- 58.Uddin S., Ahmed M., Bavi P., El-Sayed R., Al-Sanea N., AbdulJabbar A., Ashari L.H., Alhomoud S., Al-Dayel F., Hussain A.R., Al-Kuraya K.S. Bortezomib (Velcade) induces p27Kip1 expression through S-phase kinase protein 2 degradation in colorectal cancer. Cancer Res. 2008;68:3379–3388. doi: 10.1158/0008-5472.CAN-07-6109. [DOI] [PubMed] [Google Scholar]
- 59.Qin R., Smyrk T.C., Reed N.R., Schmidt R.L., Schnelldorfer T., Chari S.T., Petersen G.M., Tang A.H. Combining clinicopathological predictors and molecular biomarkers in the oncogenic K-RAS/Ki67/HIF-1α pathway to predict survival in resectable pancreatic cancer. Br. J. Cancer. 2015;112:514–522. doi: 10.1038/bjc.2014.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanden Borre P., Schrock A.B., Anderson P.M., Morris J.C., 3rd, Heilmann A.M., Holmes O., Wang K., Johnson A., Waguespack S.G., Ou S.I. Pediatric, adolescent, and young adult thyroid carcinoma harbors frequent and diverse targetable genomic alterations, including kinase fusions. Oncologist. 2017;22:255–263. doi: 10.1634/theoncologist.2016-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martincorena I., Roshan A., Gerstung M., Ellis P., Van Loo P., McLaren S., Wedge D.C., Fullam A., Alexandrov L.B., Tubio J.M. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Espiritu S.M.G., Liu L.Y., Rubanova Y., Bhandari V., Holgersen E.M., Szyca L.M., Fox N.S., Chua M.L.K., Yamaguchi T.N., Heisler L.E. The evolutionary landscape of localized prostate cancers drives clinical aggression. Cell. 2018;173:1003–1013.e15. doi: 10.1016/j.cell.2018.03.029. [DOI] [PubMed] [Google Scholar]
- 63.Stratton M.R. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 64.Stratton M.R., Campbell P.J., Futreal P.A. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alzahrani A.S., Murugan A.K., Qasem E., Alswailem M.M., AlGhamdi B., Moria Y., Al-Hindi H. Absence of EIF1AX, PPM1D, and CHEK2 mutations reported in Thyroid Cancer Genome Atlas (TCGA) in a large series of thyroid cancer. Endocrine. 2019;63:94–100. doi: 10.1007/s12020-018-1762-6. [DOI] [PubMed] [Google Scholar]
- 66.Bu R., Siraj A.K., Divya S.P., Kong Y., Parvathareddy S.K., Al-Rasheed M., Al-Obaisi K.A.S., Victoria I.G., Al-Sobhi S.S., Al-Dawish M. Telomerase reverse transcriptase mutations are independent predictor of disease-free survival in Middle Eastern papillary thyroid cancer. Int. J. Cancer. 2018;142:2028–2039. doi: 10.1002/ijc.31225. [DOI] [PubMed] [Google Scholar]
- 67.Molenaar J.J., Koster J., Zwijnenburg D.A., van Sluis P., Valentijn L.J., van der Ploeg I., Hamdi M., van Nes J., Westerman B.A., van Arkel J. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S10 and Tables S1, S5, S6, and S9
Tables S2–S4, S7, and S8