Abstract
Introduction:
A critical and as-yet unmet need in Alzheimer’s disease (AD) is the discovery of peripheral small molecule biomarkers. Given that brain pathology precedes clinical symptom onset, we set out to test whether metabolites in blood associated with pathology as indexed by cerebrospinal fluid (CSF) AD biomarkers.
Methods:
This study analyzed 593 plasma samples selected from the European Medical Information Framework for Alzheimer’s Disease Multimodal Biomarker Discovery study, of individuals who were cognitively healthy (n = 242), had mild cognitive impairment (n = 236), or had AD-type dementia (n = 115). Logistic regressions were carried out between plasma metabolites (n = 883) and CSF markers, magnetic resonance imaging, cognition, and clinical diagnosis.
Results:
Eight metabolites were associated with amyloid b and one with t-tau in CSF, these were primary fatty acid amides (PFAMs), lipokines, and amino acids. From these, PFAMs, glutamate, and aspartate also associated with hippocampal volume and memory.
Discussion:
PFAMs have been found increased and associated with amyloid b burden in CSF and clinical measures.
Keywords: EMIF-AD, Alzheimer’s disease, Dementia, Amyloid, Tau, CSF, Brain volume measurements, Cognitive function measurements, Metabolomics, Biomarkers
1. Background
Neurodegenerative dementias are characterized by a progressive decline in cognitive function and memory performance. Alzheimer’s disease (AD) is the most common of the neurodegenerative dementias making it a major source of global morbidity and mortality [1]. The World Alzheimer’s Report has estimated that there are more than 46 million people diagnosed with AD-type dementia and with an aging world population this figure is expected to increase to more than 130 million by 2050 [2]. In addition to a major human cost, AD also poses a significant economic cost estimated to increase to $1 trillion by 2018 [2].
Current clinical diagnosis of AD-type dementia relies on experienced clinicians using a battery of cognitive tests combined with various structural and functional imaging and cerebrospinal fluid (CSF) biomarkers to inform a judgment-based decision, with a definitive AD-type dementia diagnosis only possible at postmortem. Histologic examination of brain tissue during autopsy should contain significant evidence of extracellular amyloid β (Aβ) plaques and intracellular neurofibrillary tangles of hyperphosphorylated tau. The deposition of Aβ plaques has been shown to start up to 20 years before the onset of symptoms [3–5]. There have been numerous drug candidates that have failed clinical trials in symptomatic patients, these have been unsuccessful in producing a reversal of symptoms or a slowing of the progression of the disease [6]. It is thought that one of the reasons for the failure of these candidates is that they were not administered during the preclinical phase of the disease. This introduces the challenge of diagnosing people during the preclinical phase of the disease, when they are cognitively normal. For this to be possible, it is necessary to discover biomarkers that can identify individuals at high risk of developing clinical AD.
Metabolomics is the study of the complete complement of all low molecular weight metabolites (<1500 atomic mass units, Da) [7,8]. In essence, the metabolome represents metabolism in real time, the interaction of both genomic and environmental exchanges. To date there have been numerous AD metabolomic studies performed with different metabolomic platforms (i.e., different metabolome coverage and measurements) that aimed to identify panels of blood biomarkers in AD [9–13]. A handful of studies have included subjects with mild cognition problems who went on to develop AD during follow-up to find early disease biomarkers. These studies have uncovered metabolite panels with potential that are awaiting validation [14,15].
Here, we aimed to identify blood metabolites associating with CSF measures of amyloid and tau (phosphorylated and total). The abundance of metabolites was measured using liquid chromatography-mass spectroscopy to cover ca. 800 metabolites. The selected metabolites were then compared with clinical cognition measures and rate of cognition decline, brain volumes, and diagnosis.
2. Methods
2.1. Subjects
This study used plasma samples from European Medical Information Framework for Alzheimer’s Disease (EMIF-AD) Multimodal Biomarker Discovery study [16]. EMIF-AD Multimodal Biomarker Discovery is a cross-cohort study consisting of collated data from 11 European cohorts that aims to discover novel diagnostic and prognostic markers for AD-type dementia by performing analyses in multiple biomarker modalities. More details on EMIF-AD participants can be found in Section 1 of Supplementary methods and in Bos et al. [16].
2.2. Clinical and cognitive data
In the present study, the 593 plasma samples were from 242 normal cognition (NC), 236 mild cognitive impairment (MCI), and 115 AD participants at sampling (Table 1). Of 236 MCI participants, 83 were later diagnosed with AD-type dementia (defined as AD converting MCI [cMCI]), whereas 78 remained as MCI (defined as stable MCI [sMCI]). The average follow-up length was 2.49 years.
Table 1.
Sample characteristics by clinical diagnosis
| Characteristics | Sample size | NC | MCI | AD | Difference P value |
|---|---|---|---|---|---|
| Size | 593 | 242 | 236 | 115 | NA |
| Gender (M/F) | 281/312 | 50/65 | 116/120 | 115/127 | 6.06 × 10−01* |
| Age | 593 | 65.06 (7.93) | 70.44 (7.86) | 69.55 (8.51) | 5.61 × 10 −04† |
| APOEε4 (+/−) | 306/281 | 103/137 | 131/101 | 72/43 | 6.93 × 10−13* |
| MMSE | 590 | 28.80 (1.13) | 25.60 (2.82) | 21.07 (4.87) | <2.00 × 10−16† |
| Aβ z score | 593 | 0.26 (1.14) | 0.77 (1.04) | 1.19 (0.61) | <2.00 × 10−16† |
| p-tau z score | 538 | 0.10 (0.90) | 0.84 (1.35) | 1.28 (1.67) | <2.00 × 10−16† |
| t-tau z score | 538 | 0.06 (0.83) | 0.90 (1.22) | 1.73 (1.63) | <2.00 × 10−16† |
| MMSE | 590 | 28.80 (1.13) | 25.60 (2.82) | 21.07 (4.87) | <2.00 × 10−16† |
| ROD z score | 405 | 0.86 (0.52) | 0.22 (0.75) | −1.07 (1.07) | <2.00 × 10−16† |
| Attention z score | 543 | 0.24 (1.13) | −0.89 (1.58) | −1.77 (1.96) | <2.00 × 10−16† |
| Executive z score | 343 | 0.20 (1.10) | −0.81 (1.90) | −2.46 (2.07) | <2.00 × 10−16† |
| Language z score | 572 | −0.14 (1.01) | −0.98 (1.26) | −2.13 (1.34) | <2.00 × 10−16† |
| Memory delayed z score | 452 | 0.07 (1.15) | −1.26 (1.18) | −2.29 (1.00) | <2.00 × 10−16† |
| Memory immediate z score | 537 | −0.47 (1.88) | −1.43 (1.29) | −2.24 (1.29) | <2.00 × 10−16† |
| Visuoconstruction z score | 346 | 0.18 (1.20) | 0.13 (1.43) | −1.19 (2.20) | 1.41 × 10−08† |
| Hippocampal left | 387 | 4411.38 (441.14) | 3294.23 (634.68) | 3042.18 (463.92) | <2.00 × 10−16† |
| Hippocampal center | 387 | 3868.75 (429.20) | 3413.52 (628.90) | 3197.99 (496.24) | <2.00 × 10−16† |
| Hippocampal sum | 387 | 7626.65 (837.45) | 6707.78 (1213.79) | 6242.20 (884.14) | <2.00 × 10−16† |
| Cortical thickness in whole brain | 351 | 2.29 (0.12) | 2.30 (0.11) | 2.28 (0.11) | 5.13 × 10−01† |
| Cortical thickness in AD regions | 351 | 2.65 (0.17) | 2.64 (0.16) | 2.57 (0.17) | 7.12 × 10−03† |
| Taking AChEI, yes/no | 210 | 1/40 | 51/66 | 28/24 | 4.90 × 10−07* |
| Taking other AD medications, yes/no | 210 | 0/41 | 16/101 | 7/45 | 4.27 × 10−02* |
NOTE. Results are mean (standard deviation) for continuous variables.
Abbreviations: Aβ, amyloid β; AChEI, acetylcholine esterase inhibitor; AD, Alzheimer’s disease; APOEε4, apolipoproteinE ε4; MCI, mild cognitive impairment; MMSE, Mini-Mental State Examination; NC, normal cognition; ROD, rate of cognitive decline.
χ2 test.
One-way analysis of variance test.
The Mini-Mental State Examination (MMSE) score and the rate of cognitive decline (ROD) were available for 590 and 405 participants, respectively (n = 405). Neuropsychological tests measuring five different cognitive domains were also available: memory (delayed, n = 452 and immediate, n = 537), language (n = 572), attention (n = 543), executive functioning (n = 434), and visuoconstruction (n = 346). More details on how ROD and neuropsychological test (z score) values were collected can be found in Bos et al. [16] and in Section 2 of Supplementary methods. The clinical design is explained in detail in Bos et al. [16,17]. Details on the amyloid and tau level measurements and magnetic resonance imaging (MRI) and genetic analyses can be found in Section 3 of Supplementary methods.
2.3. Metabolomics data acquisition and treatment
Relative levels of 883 plasma metabolites were measured in fasting blood samples using three different mass spectrometry methods. Details on the analytical method and data treatment can be found in Section 4 of Supplementary methods.
2.4. Statistical analyses
Before statistical analyses, baseline characteristics were compared between the diagnostic group using the %2 test for categorical variables and the analysis of variance for continuous variables (Table 1). To investigate the association of each metabolite with AD clinical variables, regression models were applied, adjusting for age at sampling, gender, and presence of apolipoprotein E (APOE) e4. Adjustment for multiple testing was applied using a Bonferroni correction P value <7.72 × 10−5 (=.05/648), where 648 is the number of metabolites. All associations are reported as the change per one metabolite standard deviation.
A schematic workflow of the primary data analysis used in this study can be found in Supplementary Fig. 1. More details on statistical analyses and the network analysis can be found in Section 5 of Supplementary methods. More details on statistical analyses can be found in Section 5 of Supplementary methods. Finally, a data-driven network analysis was performed post hoc to identify independent associations between all pairs of variables. Missing values were imputed with the k-nearest neighbor algorithm (impute package for R [18]) after standardizing to zero-mean and unit-variance. To infer the partial correlation network between variables, graphical least absolute shrinkage and selection operator algorithm was used [19]. The optimal complexity network was selected with extended Bayesian information criterion (huge package for R [20]) and visualized with qgraph package for R [21]). All statistical analyses were performed using R Statistical Software (version 3.4.1).
3. Results
3.1. Demographic and cognitive comparisons
The current data set comprises 593 participants divided in three diagnostic groups: NC (n = 242), MCI (n = 236), and AD-type dementia (n = 115). Demographic and clinical data are presented in Table 1. There were no differences in gender between the three diagnostic groups. The MCI and AD participants were older when compared with NC participants. AD participants were more frequently APOEε4 carriers and presented higher CSF Aβ, p-tau, and t-tau z score levels (all, P < .01). The z scores for Aβ have inverted positive values, this means lower CSF Aβ42 and CSF Aβ42/40 ratio. All cognitive tests showed values that were lower in AD participants when compared with MCI and NC participants (all, P < .01). Brain volume measurements by MRI analyses showed lower hippocampus volumes (left, right, and sum) and average cortical thickness in AD signature regions in AD participants (all, P < .01). No differences could be observed between the three diagnostic groups for average cortical thickness across the whole brain (P > .05).
3.2. Association of blood metabolites with measures of amyloid and tau in CSF
First, we aimed to identify metabolites that would associate with AD pathologic features, namely Aβ, p-tau and t-tau z scores. For this, we built linear regression models between 648 metabolite levels and (1) Ab levels in participants in all three diagnostic groups (NC + MCI + AD), (2) Aβ in MCI participants, (3) p-tau in participants in all three diagnostic groups (NC + MCI + AD), (4) p-tau in MCI participants, (5) t-tau in participants in all three diagnostic groups (NC + MCI + AD), and (6) t-tau in MCI participants. Associations in MCI participants only were performed to identify metabolites associating with Ab and tau changes in the early stages of AD.
After adjusting for age at sampling, gender, and APOEe4, five metabolites were found to associate with Ab levels across all participants at P <7.72 × 10−5. These five metabolites were palmitamide, oleamide, linoleamide, stearamide, and aspartate. Palmitamide, oleamide, linoleamide, and stearamide are primary fatty acid amides (PFAMs) and they were increased with higher Aβ levels (β = 0.21, β = 0.19, β = 0.18, and β = 0.16) whereas aspartate was decreased with Ab levels (β = −0.18). The P value for each association can be found in Table 2. In addition, a regression model in a subset (N = 467) investigated metabolite associations with CSF Aβ42/40 values (Supplementary Fig. 2). Palmitamide, oleamide, aspartate, linoleamide, stearamide, aspartate, and glutamate associated with CSF Aβ42/40 at P <7.72 × 10−5 level.
Table 2.
Association between metabolites associated with known AD markers
| Aβ z score in all | Aβ z score in MCI group | P-tau z score | P-tau z score in MCI group | T-tau z score | T-tau z score in MCI group | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecule | Beta | P value | Beta | P value | Beta | P value | Beta | P value | Beta | P value | Beta | P value |
| 12,13-DiHOME | −0.049 | 2.09 × 10−01 | −0.279 | 3.11 × 10−05* | −0.009 | 8.76 × 10−01 | −0.148 | 1.31 × 10−01 | 0.017 | 7.66 × 10−01 | −0.191 | 3.04 × 10−02 |
| 9,10-DiHOME | −0.114 | 3.16 × 10−03 | −0.266 | 6.55 × 10−06* | 0.023 | 6.96 × 10−01 | −0.111 | 2.00 × 10−01 | 0.015 | 7.89 × 10 −01 | −0.162 | 3.78 × 10−02 |
| Argininate | −0.083 | 3.52 × 10−02 | −0.005 | 9.44 × 10−01 | −0.179 | 2.38 × 10−01 | −0.038 | 6.95 × 10−01 | −0.225 | 7.65 × 10−05* | −0.068 | 4.32 × 10−01 |
| Aspartate | −0.176 | 1.53 × 10−05* | −0.310 | 3.12 × 10−05* | −0.141 | 2.04 × 10−02 | −0.158 | 1.35 × 10−01 | −0.137 | 1.93 × 10−02 | −0.102 | 2.81 × 10−01 |
| Glutamate | −0.144 | 2.66 × 10−04 | −0.276 | 4.82 × 10−05* | −0.201 | 7.27 × 10−04 | −0.170 | 8.41 × 10−02 | −0.192 | 8.56 × 10−04 | −0.128 | 1.47 × 10−01 |
| Linoleamide (18:2n6) | 0.178 | 1.62 × 10−05* | 0.267 | 5.23 × 10−06* | 0.092 | 1.45 × 10−01 | −0.054 | 5.34 × 10−01 | 0.015 | 8.06 × 10−01 | −0.112 | 1.48 × 10−01 |
| Oleamide | 0.189 | 3.84 × 10−06* | 0.346 | 2.21 × 10−08* | 0.147 | 1.86 × 10−02 | 0.010 | 9.19 × 10−01 | 0.077 | 2.09 × 10−01 | −0.050 | 5.41 × 10−01 |
| Palmitiamide | 0.206 | 3.28 × 10−07* | 0.348 | 2.53 × 10−09* | 0.137 | 2.62 × 10−02 | 0.063 | 4.71 × 10−01 | 0.062 | 3.00 × 10−01 | 0.002 | 9.80 × 10−01 |
| Stearamide | 0.160 | 5.62 × 10−05* | 0.333 | 1.59 × 10−07* | 0.090 | 1.36 × 10−01 | 0.0712 | 4.47 × 10−01 | −0.005 | 9.37 × 10−01 | −0.0123 | 8.82 × 10−01 |
NOTE. Positive beta value denotes that higher metabolite levels associate with lower CSF amyloid and higher cerebrospinal fluid p-tau and t-tau.
Abbreviations: Aβ, amyloid β; AD, Alzheimer’s disease; MCI, mild cognitive impairment.
Passed Bonferroni procedure P < 7.72 × 10−5.
Next, we investigated metabolites associating with Ab levels in MCI participants. Linear regression models were built for all 648 metabolites, eight metabolites were found to associate with Aβ at P <7.72 × 10−5 level. The eight metabolites included the five previous metabolites (palmitamide, oleamide, aspartate, linoleamide, stearamide, and aspartate) and 9,10-DiH0ME, 12,13-DiHOME, and glutamate. Palmitamide (β = 0.35), oleamide (β = 0.35), linoleamide (β = 0.27), and stearamide (β = 0.33) levels were found to be increased, whereas aspartate (β = −0.31), 9,10-DiHOME (β = −0.27), 12,13-DiHOME (β = −0.28), and glutamate (β = −0.28) levels were found to be lower with Aβ levels (P values can be found in Table 2).
We then examined the association of each of the 648 metabolites with the z score of CSF p-tau in all participants (N = 538) and in MCI participants (N = 235). At P <7.72 × 10−5 level, neither model showed metabolites associating with p-tau. Furthermore, we investigated association between 648 metabolites and CSF t-tau levels in all participants (N = 538) and in MCI participants (N = 235). Argininate was found to associate with CSF t-tau levels in all participants at P <7.72 × 10−5, its level was found decreased with higher CSF t-tau levels (β > −0.22). No metabolites associated with CSF t-tau in MCI participants only. Next, we tested if these nine metabolites were associated to any of the drugs detected (n = 2, hydroquinone sulfate and salicylate), and AD medications (n = 2, acetyl-cholinesterase inhibitors or other AD drugs). Regression models showed no associations between the nine metabolites and drugs at P <7.72 × 10−5 (data not shown).
Overall, we investigated associations between 648 metabolites and three well-established AD pathologic markers, Aβ, p-tau, and t-tau in all participants (NC + MCI + AD) and in MCI participants only. From these models, we were able to select nine metabolites, palmitamide, oleamide, lino-leamide, stearamide, 9,10-DiH0ME, 12,13-DiHOME, glutamate, and aspartate, which were found to associate with Ab levels, whereas argininate was found to associate with t-tau levels.
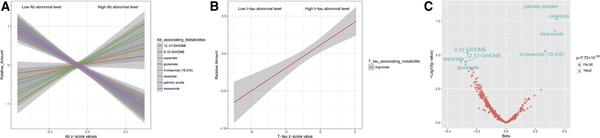
Summary of the regression models between the nine metabolites and clinical Aβ and t-tau levels can be seen in Fig. 1 and Table 2.
Fig. 1.
(A and B) The nine selected metabolite levels vary with AD CSF marker levels in participants in all three diagnostic groups (NC, MCI, and AD). The solid line represents a simple linear regression for the metabolite, whereas the shaded area represents the 95% confidence interval. (C) Volcano plot summarizing results of regression models, which investigated association between 648 plasma metabolites and Aβ levels in MCI participants (N = 236). Blue dots represent metabolites passing Bonferroni correction (P < 7.72 × 10−05).
3.3. Association of blood panel with cognition, ROD, and diagnosis
We next examined if any of these nine metabolites identified in the regression analyses would associate with AD cognitive and diagnosis variables.
First, including all the diagnosis groups we investigated the associations with MMSE scores and the ROD. Linear regression models showed six metabolites associating with MMSE at P <7.72 × 10−5 level. Linoleamide (β = −0.98), oleamide (β = −1.00), and palmitamide (β = −0.94) levels were lower whereas argininate (β = 0.84), aspartate (β = 1.04), and glutamate (β = 0.87) levels were higher with higher MMSE scores. Only glutamate was found to associate with ROD (β = −0.24) at P <7.72 × 10−5 level (P values can be found in Supplementary Table 1).
Then the selected nine metabolites were examined against neuropsychological tests measuring five different cognitive domains. Lower attention levels were found to associate with higher levels of stearamide (β = −0.38), palmitamide (β = −0.43), oleamide (β = −0.40), and linoleamide (β = −0.40) at P <7.72 × 10−5 level. No metabolite associated with executive function. Lower language levels were found to associate with higher oleamide levels (β = −0.25) and lower levels of glutamate (β = 0.2), aspartate (β = −0.25), and argininate (β = −0.28) levels at P <7.72 X 10−5 level (P values can be found in Supplementary Table 2).
No metabolite was found to associate with delayed memory. Three metabolites associated with immediate memory, linoleamide (β = −0.49), oleamide (β = −0.43), andpalmitamide (β = −0.38). Finally with visuoconstruction scores, no metabolites were found to associate with this function.
We were also interested on how the nine metabolites would associate with clinical diagnosis. Three metabolites associated with AD (vs. NC) at P <7.72 × 10−5 level, lower levels of argininate (odds ratio [OR] = 0.50), aspartate (OR = 0.50), and glutamate (OR = 0.53) were found to associate with AD. Afterward, we combined cMCI and AD participants into one group as cMCI is often defined as early AD. We then measured cMCI and AD against NC as an outcome, six metabolites were found to be associated at P <7.72 × 1025 level. These six metabolites included argininate (OR = 0.49), aspartate (OR = 0.45), glutamate (OR = 0.50), linoleamide (OR = 2.34), oleamide (OR = 2.34), and palmitamide (OR = 2.20). We also looked at AD conversion from MCI participants (cMCI vs. sMCI) and found no association at P <7.72 × 1025 level (Supplementary Table 2). A summary of the regression models can be found in Supplementary Tables 1 and 2.
3.4. Associations of blood panel with brain structural measures
In the present study, MRI data were available for 387 participants (hippocampus volumes left, right, and sum) and 351 participants had cortical thickness measures (average across the whole brain and in AD signature regions).
Of the nine metabolites, four metabolites were found to associate with right hippocampus volume at P <7.72 × 1025 level. These included higher level of glutamate (β = 146.10) and lower levels of linoleamide (β = −154.91), oleamide (β = −145.33), and palmitamide (β = −132.32). When sum of hippocampus volume was examined as an outcome, higher level of glutamate (β = 273.10) and lower levels of linoleamide (β = −274.93), oleamide (β = −268.55), and palmitamide (β = −246.38) were found to associate with higher sum of hippocampus volume (P < 7.72 X 10−5 level). Linear regression analysis with left hippocampus volume or cortical thickness as an outcome showed no metabolite association (Supplementary Table 3).
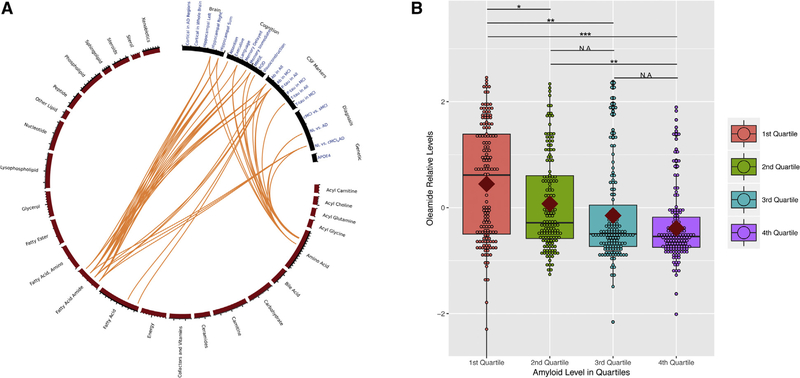
Summary of all the regression models between the nine metabolites and all AD clinical variables can be found in Fig. 2A as a circos plot depicting 25 classes of metabolites and nine metabolites associated with amyloid or tau (Supplementary Fig. 3 as a heat map). Fig. 2B shows relative levels of plasma oleamide versus amyloid levels, Supplementary Fig. 4, relative levels of plasma oleamide versus diagnostic groups.
Fig. 2.
(A) Circos plot to visualize the results of regression models. Orange lines represent association between AD clinical variables and nine selected metabolites (P < 7.72 × 10−5). Here, we examined associations between CSF markers and 648 metabolites in 25 different classes. The circos plot shows nine metabolites in three classes (four fatty acid amides, two fatty acids, and two amino acid metabolites) associating with at least one CSF markers. These nine metabolites associate with other AD clinical variables, brain volumes, cognition, and diagnosis but not APOEε4. (B) Boxplot showing relative levels of plasma oleamide according to the quartiles of amyloid levels. The size of the box represents the 25th and 75th percentiles; the central horizontal line in the boxes represents the median; the red diamond in the boxes represents the mean. Regression models were carried out to test association between oleamide levels between the quartiles after adjusting for age, gender, and APOEε4 status. The asterisks represent the strength of the association after applying multiple test correction. *P < .05;**P < .01;***P < .001.Abbreviations: AD, Alzheimer’s disease; APOE, apolipoprotein E; CSF, cerebrospinal fluid; MCI, mild cognitive impairment; NL, normal; ROD, rate of cognitive decline.
3.5. Predictive models of clinical diagnosis
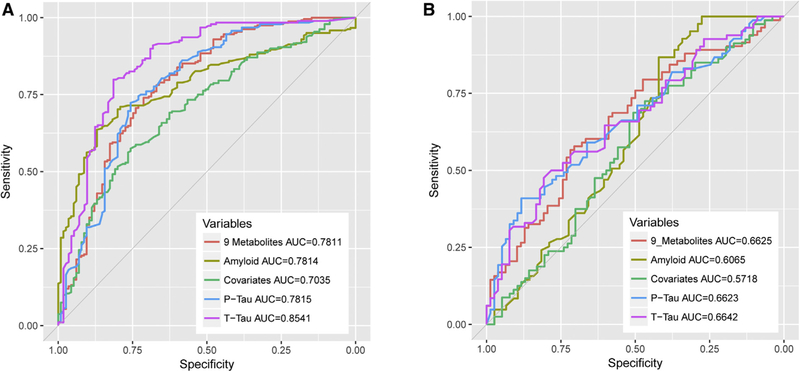
We built receiver operating characteristic (ROC) models for the nine selected metabolites; Fig. 3A depicts how well the panel can discriminate AD-type dementia from control subjects. The resulting area under the curve (AUC) value was 0.7811. We then examined how well other AD clinical variables would discriminate diagnoses groups. Aβ produced AUC value of 0.7814, t-tau produced AUC of 0.7815, p-tau produced AUC of 0.8541, and covariates (known risks of age at sampling and APOEε4 status) produced AUC of 0.7035 (Fig. 3A).
Fig. 3.
Receiver operator curve figures for the selected nine metabolites in (A) AD versus control subjects model and (B) AD cMCI versus non-cMCI (in red). The AUC values for the nine selected metabolites were compared with Aβ (in yellow), p-tau (in green), t-tau (in purple), and covariates consisting of age and APOEε4 status (in green). Abbreviation: AUC, area under the curve.
Then we built ROC model (Fig. 3B) for the nine selected metabolites for AD cMCI versus non-cMCI groups. The resulting AUC value was 0.6625. This value was compared with other AD clinical variables, Ab (AUC = 0.6065), p-tau (AUC = 0.6623), t-tau (AUC = 0.6642), and covariates (AUC = 0.5718).
3.6. Network analysis
To visualize the interactions in the full data set, including metabolites and all clinical variables, a data-driven network was computed selecting the clinical variables and metabolites that are immediately connected. PFAMs, glutamine, and aspartate were strongly correlated to amyloid. There were two additional metabolites that showed high partial correlation to MMSE and diagnosis, these were tryptophan, betaine, and 2-pyrrolidinone, respectively (see Supplementary Fig. 5).
4. Discussion
At present clinical MCI and AD-type dementia diagnoses are made based on the presentation of symptoms, cognitive assessments, biomarker analyses, and the judgment of the clinician. However, by the time symptoms are present irreversible changes have occurred in the structure of the brain. Hence the ideal plasma biomarker or panel would give information about high amyloid levels and risk of conversion to AD to treat at an earlier stage. In fact very few biomarkers have been approved to this end, the positronemission tomography tracer florbetapir was approved by the Food and Drug Administration in 2012 and it allows for a measurement of amyloid burden in the brain, achieving sensitivity of 82% to 92% and specificity of 95% [22]. CSF measures of amyloid and tau are also widely accepted as biomarkers of AD. A recent meta-analysis of CSF biomarkers (12 cohorts cMCI = 307 and sMCI = 501), using CSF amyloid-42, p-tau, and t-tau, conversion was predicted with average ratios of 0.67, 1.72, and 1.76, respectively [23].
In this study, 883 structurally distinct compounds were measured in 593 plasma samples. We found that metabolites pertaining to three classes of compounds were associated with amyloid or tau measures, namely four PFAMs, two lipid hormones, and three amino acids. The panel predicted AD-type dementia with the same AUC as amyloid (AUC = 0.78 for both) but lower than t-tau for this cohort (AUC = 0.85). For early diagnoses, in the converter group the prediction of the panel was AUC = 0.66, higher than amyloid by itself (AUC = 0.60) but the same as t-tau and p-tau in this cohort.
4.1. PFAMs and AD
In this study, all four PFAMs measured in our analyses were increased with amyloid burden. Oleamide is the most studied and best understood PFAM. Oleamide was first identified in brains of sleep-deprived cats and mice [24] and is an important regulatory lipid in the brain and central nervous system. Oleamide regulates the sleep-wake cycle, memory, locomotion, pain perception, and is anti-inflammatory, anxiolytic, and neuroprotective [25,26]. Administration of oleamide protects against scopolamine-induced cognitive impairment [27] and depression in a rat chronic stress model [28]. Earlier metabolomic studies have shown lower serum concentrations of oleamide in patients with AD and patients with MCI [29,30]. Differences between our results and those reported earlier could arise by using amyloid as the primary outcome.
If oleamide, an endocannabinoid, is neuroprotective and has the ability to induce sleep, it could be synthesized to improve amyloid clearance and restore the sleep-wake cycle disruption that is characteristic of AD [31]. Sleep deprivation and its disorders are suggested to precede and predict dementia [32–34]. During sleep the brain’s interstitial space increases in volume by up to 60% to enable CSF to clear neurotoxic waste into the systemic circulation [35]. It has been now shown that positron-emission tomography amyloid burden increases in the hippocampus with one-night sleep deprivation [36], this is especially interesting because of PFAMs associated with total hippocampal volume in our study. Moreover, it has been shown that early amyloid toxicity can be blocked by the activation of cannabinoid receptors [37].
In addition to oleamide, we find that the plasma levels of three other fatty acid amides, palmitamide, linoleoamide, and stearamide, were also increased. Decreased serum palmitamide in AD was reported earlier [38]. In contrast to oleamide, far less is known about the biological function of palmitamide, linoleamide, and stearamide [26]. Alterations in the blood levels of the PFAMs have been reported for other diseases or disease models. Lower serum oleamide has also been found in a rat model for ischemic stroke [39] and in premutation carriers of the fragile X mental retardation 1 (FMR1) gene [40]. Treatment of rats with qui-nolinic acid increased the serum levels of pentadecanamide, palmitamide, oleamide, and stearamide [41], this is interesting as quinolinic acid is produced by activated microglia [42]. A metabolomic study of the systemic effects of cirrhosis identified increased serum levels of oleamide and stearamide [43].
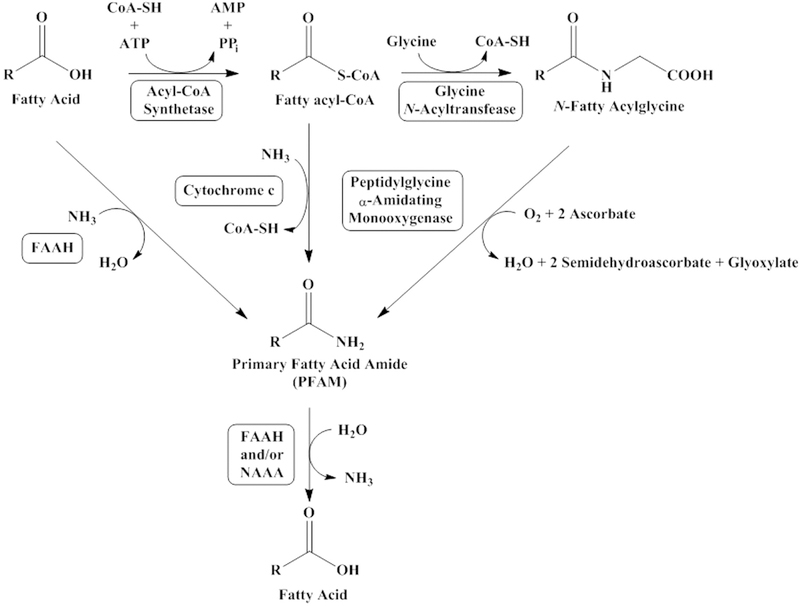
Alterations in plasma PFAM levels in AD are likely resulted from dysfunction in synthesis, degradation, or transport. Little is known about PFAM transport; the major degradative reaction for the PFAMs is their hydrolysis into fatty acid and ammonia, a reaction catalyzed by fatty acid amide hydrolase (FAAH) [44] and/or N-acylethanolamine-hydrolyzing acid amidase [45]. There are conflicting reports regarding FAAH and AD, one report indicating that FAAH is overexpressed [46] and another that FAAH activity is decreased in AD [47]. An AD-related decrease in FAAH activity could lead to increased plasma levels of the PFAMs. In Fig. 4, the three main routes are explained, decreases in peptidylglycine α-amidating monooxygenase expression in cultured neuroblastoma cells yielded decreases in PFAM levels providing evidence in vivo [48]. One report indicated that peptidylglycine α-amidating monooxygenase activity is lower than normal in the brain and CSF of patients with AD [49].
Fig. 4.
PFAM biosynthesis. PFAM biosynthesis route is not clearly defined with possible routes including the direct conjugation of ammonia to a fatty acid as catalyzed by FAAH [50], a pathway that might be unfavorable in vivo [51]. The cytochrome c catalyzed reaction of ammonia with oleoyl-Coenzyme A to produce oleamide and coenzyme A was reported as one possible in vivo route for oleamide [52]. Peptidylglycine α-amidating monooxgenase-catalyzed cleavage of N-fatty acid glycines to the PFAMs has been proven in vivo [53]. Abbreviations: FAAH, fatty acid amide hydrolase; NAAA, N-acylethanolamine-hydrolyzing acid amidase.
We also found two lipokines that were decreased in MCI participants with higher amyloid burden in the brain. Although there are no brain studies in which lipokines were detected, in blood, these were negatively correlated with both body mass index and insulin resistance in the obese, and in another study increased after exercise [54,55]. These findings would point out toward a metabolic explanation that could be investigated in future studies.
The amount of glutamate and aspartate was decreased in plasma and associated with amyloid whereas arginine correlated with t-tau. We and others have found in brain that excitatory neurotransmitters glutamate and aspartate were decreased with increasing amyloid and tau burden [56], whereas plasma arginine was not found to correlate with CSF p-tau [57], but it has been consistently found to be decreased in the brain of patients with AD [58,59].
A limitation of this study is that although it is a relatively large study for metabolomics, the number of participants is still small. In addition, the amounts of PFAMs alone predicted AD-type dementia with an ROC AUC of 0.63 (data not shown), which is much lower than that predicted by the panel together with amino acids and lipokines.
In sum, our data points toward a panel of metabolites including PFAMs, amino acids, and lipokines that could help understand AD pathways and achieve prediction in blood equivalent to that achieved by amyloid measures in CSF. PFAMs are endocannabinoids that could potentially have an anti-inflammatory and sleep-inducing role as amyloid burden increases toxicity in the brain.
Supplementary Material
RESEARCH IN CONTEXT.
Systematic review: The authors reviewed the literature using PubMed and reported key publications. Most studies were small and relatively heterogeneous in the metabolites identified. Most blood me- tabolomic studies have highlighted the role of lipid metabolites as important in Alzheimer’s disease (AD). However, we still need to identify peripheral early stage AD biomarkers that reflect pathology and/or that inform on its biochemistry and potential targets. For this, we used a comprehensive range me- tabolomic approach to identify small molecules in blood associating with pathology as indexed by cerebrospinal fluid AD biomarkers.
Interpretation: The results show that primary fatty acid amides (PFAMs) associated with AD pathology and phenotype. There are two important implications to this finding: first, the lipids in question are thought to be synthesized in the brain; and second, they are also thought to be natural endocannabinoids. With the exception of oleamide, the biology behind endogenous PFAMs is largely unknown; it is thought that they are synthesized to induce sedation. Neurotransmitters were also flagged and these could be depleted because of compromised synaptic signaling.
Future directions: Results of this study should be integrated with proteomics and genetics to find more about mechanisms involved in blood. Future studies should address whether PFAMs are causally related to AD, and if it is brain or other organs with the brain that are involved in the synthesis of PFAMs.
Acknowledgments
The authors thank the individuals and families who took part in this research. The authors would also like to thank all people involved in data and sample collection and/or logistics across the different centers, and in particular Marije Benedictus, Wiesje van de Flier, Charlotte Teunissen, Ellen De Roeck, Naomi De Roeck, Ellis Niemantsverdriet, Charisse Somers, Babette Reijs, Andrea Izagirre Otaegi, Mirian, Ecay Torres, Sindre Rolstad, Eva Bringman, Domiie Tautvydaite, Barbara Moullet, Charlotte Evenepoel, Isabelle Cleynen, Bea Bosch, Daniel Alcolea Rodriguez, Moira Marizzoni, Alberto Redolfi, and Paolo Bosco.
Funding: The present study was conducted as part of the EMIF-AD project, which has received support from the Innovative Medicines Initiative Joint Undertaking under EMIF grant agreement no. 115372, resources of which are composed of financial contribution from the European Union’s Seventh Framework Program (FP7/2007-2013) and EFPIA companies’ in-kind contribution. The DESCRIPA study was funded by the European Commission within the fifth framework program (QLRT-2001-2455). The EDAR study was funded by the European Commission within the fifth framework program (contract no. 37670). The San Sebastian GAP study is partially funded by the Department of Health of the Basque Government (allocation 17.0.1.08.12.0000.2.454.01.41142.001.H).
D.J.M was partially supported by a grant from the National Institutes of Health (R15-GM107864).
S.E. reports research funding from Janssen Pharmaceutica N.V. and ADx Neurosciences (paid to institution). A.N.-H. holds a funding award from Ono Pharmaceutical Co Ltd. L.F. has participated in advisory boards for Allergan, Eli Lilly, Avraham Pharmaceuticals, Axon Neuroscience, Axovant, Biogen, Boehringer Ingelheim, Eisai, Functional Neuromodulation, Lundbeck, MerckSharpe & Dohme, Novartis, Pfizer, Pharnext, Roche, Schwabe and has received research grants from Novartis. H.Z. has served at advisory boards for Eli Lilly, Roche Diagnostics, Wave, Samumed and CogRx, has received travel support from Teva, and is a cofounder of Brain Biomarker Solutions in Gothenburg AB, a GU Ventures-based platform company at the University of Gothenburg (all unrelated to this study). F.B. has served at advisory boards for Bayer-Schering Pharma, Biogen, Teva, Merck-Serono, Novartis, Roche, Jansen Research, Genzyme-Sanofi, IXICO, GeNeuro, Apitope.
Footnotes
Supplementary data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.jalz.2019.03.004.
Conflict of interest: All other authors report no biomedical financial interests or potential conflicts of interest.
References
- [1].Reitz C, Brayne C, Mayeux R. Epidemiology of Alzheimer disease. Nat Rev Neurol 2011;7:137–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Prince MJ. World Alzheimer Report 2015: The global Impact of Dementia: An Analysis of Prevalence, Incidence, Cost and Trends 2015. United Kingdom: Alzheimer’s Disease International; 2015. [Google Scholar]
- [3].Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 2011;7:280–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jack CR, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol 2010;9:119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Visser PJ, Tijms B. Brain amyloid pathology and cognitive function: Alzheimer disease without dementia? JAMA 2017;317:2285–7. [DOI] [PubMed] [Google Scholar]
- [6].Cummings JL, Morstorf T, Zhong K. Alzheimer’s disease drug-development pipeline: few candidates, frequent failures. Alzheimers Res Ther 2014;6:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol Biol 2002;48:155–71. [PubMed] [Google Scholar]
- [8].Whiley L, Legido-Quigley C. Current strategies in the discovery of small-molecule biomarkers for Alzheimer’s disease. Bioanalysis 2011;3:1121–42. [DOI] [PubMed] [Google Scholar]
- [9].Han X, Rozen S, Boyle SH, Hellegers C, Cheng H, Burke JR, et al. Metabolomics in early Alzheimer’s disease: identification of altered plasma sphingolipidome using shotgun lipidomics. PLoS One 2011;6:e21643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Trushina E, Nemutlu E, Zhang S, Christensen T, Camp J, Mesa J, et al. Defects in mitochondrial dynamics and metabolomic signatures of evolving energetic stress in mouse models of familial Alzheimer’s disease. PLoS One 2012;7:e32737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Orešič M, Hyötyläinen T, Herukka S, Sysi-Aho M, Mattila I, Seppanan-Laakso T, et al. Metabolome in progression to Alzheimer’s disease. Transl Psychiatry 2011;1:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kim M, Legido-Quigley C. Small Molecule Biomarkers in Alzheimer’s Disease 2018. France: OCL; 2018. [Google Scholar]
- [13].de Leeuw FA, Peeters CFW, Kester MI, Harms AC, Struys EA, Hankemeier T, et al. Blood-based metabolic signatures in Alzheimer’s disease. Alzheimers Dement (Amst) 2017;8:196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mapstone M, Cheema AK, Fiandaca MS, Zhong X, Mhyre TR, MacArthur LH, et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med 2014;20:415–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Casanova R, Varma S, Simpson B, Kim M, An Y, Saldana S, et al. Blood metabolite markers of preclinical Alzheimer’s disease in two longitudinally followed cohorts of older individuals. Alzheimers Dement 2016;12:815–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bos I, Vos S, Vandenberghe R, Scheltens P, Engelborghs S, Frisoni G, et al. The EMIF-AD multimodal biomarker discovery study: design, methods and cohort characteristics. Alzheimers Res Ther 2018;10:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jack CR Jr, Wiste HJ, Weigand SD, Therneau TM, Lowe VJ, Knopman DS, et al. Defining imaging biomarker cut points for brain aging and Alzheimer’s disease. Alzheimers Dement 2017;13:205–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].TrHastie T, Tibshirani R, Narasimhan B, Chu G. Impute: imputation for microarray data. R package version 1501; 2017. [Google Scholar]
- [19].Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics 2008;9:432–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhao T, Liu H, Roeder K, Lafferty J, Wasserman L. The huge package for high-dimensional undirected graph estimation in R. J Mach Learn Res 2012;13:1059–62. [PMC free article] [PubMed] [Google Scholar]
- [21].Epskamp S, Cramer AOJ, Waldorp LJ, Schmittmann VD, Borsboom D. qgraph: Network visualizations of relationships in psychometric data. J Stat Softw 2012;48:18. [Google Scholar]
- [22].Souslova T, Marple TC, Spiekerman AM, Mohammad AA. Personalized medicine in Alzheimer’s disease and depression. Contemp Clin Trials 2013;36:616–23. [DOI] [PubMed] [Google Scholar]
- [23].Herukka S-K, Simonsen AH, Andreasen N, Baldeiras I, Bjerke M, Blennow K, et al. Recommendations for cerebrospinal fluid Alzheimer’s disease biomarkers in the diagnostic evaluation of mild cognitive impairment. Alzheimers Dement 2017;13:285–95. [DOI] [PubMed] [Google Scholar]
- [24].Cravatt B, Proserogarcia O, Siuzdak G, Gilula N, Henriksen S, Boger D. Chemical characterization of a family of brain lipids that induce sleep. Science 1995;268:1506–9. [DOI] [PubMed] [Google Scholar]
- [25].Farrell EK, Merkler DJ. Biosynthesis, degradation and pharmacological importance of the fatty acid amides. Drug Discov Today 2008; 13:558–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Waluk DP, Battistini MR, Dempsey DR, Farrell EK, Jeffries KA, Mitchell P, et al. Chapter 9—Mammalian fatty acid amides of the brain and CNS A2—Watson, Ronald Ross In: Meester FD, ed. Omega-3 fatty acids in brain and neurological health. Boston: Academic Press; 2014. p. 87–107. [Google Scholar]
- [27].Heo HJ, Park YJ, Suh YM, Choi SJ, Kim MJ, Cho HY, et al. Effects of oleamide on choline acetyltransferase and cognitive activities. Biosci Biotechnol Biochem 2003;67:1284–91. [DOI] [PubMed] [Google Scholar]
- [28].Ge L, Zhu MM, Yang JY, Wang F, Zhang R, Zhang JH, et al. Differential proteomic analysis of the anti-depressive effects of oleamide in a rat chronic mild stress model of depression. Pharmacol Biochem Behav 2015;131:77–86. [DOI] [PubMed] [Google Scholar]
- [29].Gonzalez-Dominguez R, Garcia-Barrera T, Gomez-Ariza JL. Metabolomic study of lipids in serum for biomarker discovery in Alzheimer’s disease using direct infusion mass spectrometry. J Pharm Biomed Anal 2014;98:321–6. [DOI] [PubMed] [Google Scholar]
- [30].Gonzalez-Dominguez R, Ruperez FJ, Garcia-Barrera T, Barbas C, Gomez-Ariza JL. Metabolomic-driven elucidation of serum disturbances associated with Alzheimer’s disease and mild cognitive impairment. Curr Alzheimer Res 2016;13:641–53. [DOI] [PubMed] [Google Scholar]
- [31].Widner B, Ledochowski M, Fuchs D. Sleep disturbances and tryptophan in patients with Alzheimer’s disease. Lancet 2000;355:755–6. [DOI] [PubMed] [Google Scholar]
- [32].Villa C, Ferini-Strambi L, Combi R. The synergistic relationship between Alzheimer’s disease and sleep disorders: an update. J Alzheimers Dis 2015;46:571–80. [DOI] [PubMed] [Google Scholar]
- [33].Ng KP, Pascoal TA, Mathotaarachchi S, Chung C-O, Benedet AL, Shin M, et al. Neuropsychiatric symptoms predict hypometabolism in preclinical Alzheimer disease. Neurology 2017;88:1814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hita-Yañez E, Atienza M, Cantero JL. Polysomnographic and subjective sleep markers of mild cognitive impairment. Sleep 2013; 36:1327–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, et al. Sleep drives metabolite clearance from the adult brain. Science 2013; 342:373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shokri-Kojori E, Wang GJ, Wiers CE, Demiral SB, Guo M, Kim SW, et al. beta-Amyloid accumulation in the human brain after one night of sleep deprivation. Proc Natl Acad Sci U S A 2018;115:4483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Currais A, Quehenberger O, Armando AM, Daugherty D, Maher P. Schubert D. Amyloid proteotoxicity initiates an inflammatory response blocked by cannabinoids. NPJ Aging Mech Dis 2016;2:16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chu H, Zhang A, Han Y, Lu S, Kong L, Han J, et al. Metabolomics approach to explore the effects ofKai-Xin-San on Alzheimer’s disease using UPLC/ESI-Q-TOF mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 2016;1015–1016:50–61. [DOI] [PubMed] [Google Scholar]
- [39].Su L, Zhao H, Zhang X, Lou Z, Dong X. UHPLC-Q-TOF-MS based serum metabonomics revealed the metabolic perturbations of ischemic stroke and the protective effect of RKIP in rat models. Mol Biosyst 2016;12:1831–41. [DOI] [PubMed] [Google Scholar]
- [40].Giulivi C, Napoli E, Tassone F, Halmai J, Hagerman R. Plasma biomarkers for monitoring brain pathophysiology in FMR1 premutation carriers. Front Mol Neurosci 2016;9:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kalaska B, Ciborowski M, Domaniewski T, Czyzewska U, Godzien J, Miltyk W, et al. Serum metabolic fingerprinting after exposure of rats to quinolinic acid. J Pharm Biomed Anal 2016;131:175–82. [DOI] [PubMed] [Google Scholar]
- [42].Guillemin GJ, Smith DG, Smythe GA, Armati PJ, Brew BJ. Expression of the kynurenine pathway enzymes in human microglia and macrophages. Adv Exp Med Biol 2003;527:105–12. [DOI] [PubMed] [Google Scholar]
- [43].Lian JS, Liu W, Hao SR, Guo YZ, Huang HJ, Chen DY, et al. A serum metabonomic study on the difference between alcohol-and HBV-induced liver cirrhosis by ultraperformance liquid chromatography coupled to mass spectrometry plus quadrupole time-of-flight mass spectrometry. Chin Med J (Engl) 2011; 124:1367–73. [PubMed] [Google Scholar]
- [44].McKinney MK, Cravatt BF. Structure and function of fatty acid amide hydrolase. Annu Rev Biochem 2005;74:411–32. [DOI] [PubMed] [Google Scholar]
- [45].Tsuboi K, Takezaki N, Ueda N. The N-acylethanolamine-hydrolyzing acid amidase (NAAA). Chem Biodivers 2007;4:1914–25. [DOI] [PubMed] [Google Scholar]
- [46].Benito C, Nunez E, Tolon RM, Carrier EJ, Rabano A, Hillard CJ, et al. Cannabinoid CB2 receptors and fatty acid amide hydrolase are selectively overexpressed in neuritic plaque-associated glia in Alzheimer’s disease brains. J Neurosci 2003;23:11136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pascual AC, Martin-Moreno AM, Giusto NM, de Ceballos ML, Pasquare SJ. Normal aging in rats and pathological aging in human Alzheimer’s disease decrease FAAH activity: modulation by cannabinoid agonists. Exp Gerontol 2014;60:92–9. [DOI] [PubMed] [Google Scholar]
- [48].Jeffries KA, Dempsey DR, Farrell EK, Anderson RL, Garbade GJ, Gurina TS, et al. Glycine N-acyltransferase-like 3 is responsible for long-chain N-acylglycine formation in N18TG2 cells. J Lipid Res 2016;57:781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wand GS, May C, May V, Whitehouse PJ, Rapoport SI, Eipper BA. Alzheimer’s disease: low levels of peptide alpha-amidation activity in brain and CSF. Neurology 1987;37:1057–61. [DOI] [PubMed] [Google Scholar]
- [50].Arreaza G, Devane WA, Omeir RL, Sajnani G, Kunz J, Cravatt BF, et al. The cloned rat hydrolytic enzyme responsible for the breakdown of anandamide also catalyzes its formation via the condensation of arachidonic acid and ethanolamine. Neurosci Lett 1997;234:59–62. [DOI] [PubMed] [Google Scholar]
- [51].Bisogno T, Sepe N, De Petrocellis L, Mechoulam R, Di Marzo V. The sleep inducing factor oleamide is produced by mouse neuroblastoma cells. Biochem Biophys Res Commun 1997;239:473–9. [DOI] [PubMed] [Google Scholar]
- [52].Driscoll WJ, Chaturvedi S, Mueller GP. Oleamide synthesizing activity from rat kidney: identification as cytochrome c. J Biol Chem 2007; 282:22353–63. [DOI] [PubMed] [Google Scholar]
- [53].Merkler DJ, Merkler KA, Stern W, Fleming FF. Fatty acid amide biosynthesis: a possible new role for peptidylglycine alpha-amidating enzyme and acyl-coenzyme A: glycine N-acyltransferase. Arch Biochem Biophys 1996;330:430–4. [DOI] [PubMed] [Google Scholar]
- [54].Lynes MD, Leiria LO, Lundh M, Bartelt A, Shamsi F, Huang TL, et al. The cold-induced lipokine 12,13-diHOME promotes fatty acid transport into brown adipose tissue. Nat Med 2017;23:631–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Stanford KI, Lynes MD, Takahashi H,Baer LA, Arts PJ, May FJ, et al. 12,13-diHOME: an exercise-induced lipokine that increases skeletal muscle fatty acid uptake. Cell Metab 2018;27:1111–1120.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Snowden SG, Ebshiana AA, Hye A, An Y, Pletnikova O, O’Brien R, et al. Association between fatty acid metabolism in the brain and Alzheimer disease neuropathology and cognitive performance: a nontargeted metabolomic study. PLoS Med 2017;14:e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Arlt S, Schwedhelm E, Kolsch H, Jahn H, Linnebank M, Smulders Y, et al. Dimethylarginines, homocysteine metabolism, and cerebrospinal fluid markers for Alzheimer’s disease. J Alzheimers Dis 2012; 31:751–8. [DOI] [PubMed] [Google Scholar]
- [58].Xu J, Begley P, Church SJ, Patassini S, Hollywood KA, Jullig M, et al. Graded perturbations of metabolism in multiple regions of human brain in Alzheimer’s disease: snapshot of a pervasive metabolic disorder. Biochim Biophys Acta 2016;1862:1084–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Gueli MC, Taibi G. Alzheimer’s disease: amino acid levels and brain metabolic status. Neurol Sci 2013;34:1575–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.