Abstract
Trace amines (TAs) in the mammalian brain have been investigated for four decades. Trace amine‐associated receptors (TAARs) were discovered during the search for receptors activated by TAs. TAARs are considered a second class of vertebrate olfactory receptors and successfully proliferated in conjunction with adaptation to living on the ground to detect carnivore odors. Thus, therian mammals have a high number of TAAR genes due to rapid species‐specific gene duplications. In primate lineages, however, their genomes have significantly smaller numbers of TAAR genes than do other mammals. To elucidate the evolutionary force driving these patterns, exhaustive data mining of TAAR genes was performed for 13 primate genomes (covering all four infraorders) and two nonprimate euarchontan genomes. This study identified a large number of pseudogenes in many of these primate genomes and thus investigated the pseudogenization event process for the TAAR repertoires. The degeneration of TAARs is likely associated with arboreal inhabitants reducing their exposure to carnivores, and this was accelerated by the change in the nose shape of haplorhines after their divergence from strepsirrhines. Arboreal life may have decreased the reliance on the chemosensing of predators, suggestive of leading to the depauperation of TAAR subfamilies. The evolutionary deterioration of TAARs in primates has been reestablished in recently derived primates due to high selection pressure and probably functional diversity.
Keywords: G‐protein‐coupled receptor, olfactory receptor, positive selection, primate evolution
The arboreal lifestyle derived from the Euarchonta ancestor may have reduced reliance on the chemosensing of predators, leading to the depauperation of TAAR subfamilies.
1. INTRODUCTION
Trace amines (TAs) are endogenous biogenic amine compounds that include p‐tyramine, β‐phenylethylamine, tryptamine, para‐octopamine, 3‐iodothyronamine and N,N‐dimethyltryptamine. TAs are present in the brain at very low levels (<10 nM) but are important to the understanding of a number of diseases of the human brain because they are generally considered to be important regulatory elements. For example, elevated TA levels are associated with depression and other neuropsychiatric disorders.1, 2 Recent studies have also suggested that the regulatory role of the TA system affects neurological disorders such as substance abuse, insomnia, depression, attention deficit hyperactivity disorder, bipolar disorder, schizophrenia and other neuropsychiatric diseases.3, 4, 5, 6, 7, 8, 9, 10 The trace amine‐associated receptors (TAARs) were discovered in search of the receptors activated by TAs in the brain.11, 12 The TAAR6 gene (also known as TRAR4 or TA4) is suspected to increase susceptibility to schizophrenia, with evidence of a genetic linkage.3, 4, 13 The rat TAAR1 gene is not only activated by classical TAs but also by synthetic analogues such as 3,4‐methylenedioxymethamphetamine (MDMA, known as ecstasy), d‐lysergic acid diethylamide (LSD) and amphetamine.11 Due to these associations, TAs and TAARs are important in understanding many human psychiatric disorders.
Interestingly, TAAR repertoires are strongly related to danger‐associated behavioral responses in terrestrial mammals and thus play a critical role in sensing predator and prey odors. In particular, Ferrero et al14 showed that TAAR4 is closely associated with odor detection, with rat and mouse TAAR4 shown to be activated by β‐phenylethylamine, a carnivore odor from mountain lions, tigers and jaguars. The removal of a single TAAR gene (TAAR4) in mice eliminated their innate aversion to predator odors such as puma urine volatiles.15
Our previous study found that the size of TAAR gene repertoires varies widely among tetrapods, ranging from 0 in dolphins and 1 in zebra finches to 26 in flying foxes, while terrestrial mammals generally have a higher number of TAARs compared with the platypus, archosaurs and amphibians16; the latter have up to four TAAR subfamilies from among TAAR1‐5, whereas therian mammals have more TAAR subfamilies (TAAR6‐9, E1 and M1‐3; named therian‐specific TAAR subfamilies) with TAAR1‐5.16 These therian‐specific TAAR subfamilies are found to have been subject to rapid species‐specific gene duplications with positive selection and thus adaptation to different ecological niches has been considered.16, 17 For example, the mouse and rat genomes have experienced frequent species‐specific tandem gene duplications, but no functional TAAR gene has been found in the dolphin genome.16, 18, 19 From these results, it is very likely that TAARs proliferated in conjunction with adaptation to living on the ground to detect predator odors. It can be hypothesized that TAAR genes have less critical functions for nonground living mammals, such as arboreal species, due to the lower exposure to ground‐living predators. Arboreality is relatively rare among extant placental mammals, but primates are common arboreal inhabitants of tropical and subtropical forests.20 These primates have special adaptations for that lifestyle, such as their hands and feet being able to grasp twigs and branches rather than grappling tree trunks.20
In contrast to most mammals, some primate species show a conspicuously small number of TAAR gene families. The human genome has six subfamilies (with no gene duplication) and does not have functional TAAR3, TAAR4 and TAAR7 genes.16, 19 Pseudogenization of TAAR3 and TAAR4 occurred before the divergence of humans and orangutans (for TAAR3) or humans and gorillas (for TAAR4).21 Independent pseudogenization has occurred in the marmoset lineage for both TAAR3 and TAAR4,21 leaving the marmoset genome with only two TAAR genes (TAAR1 and TAAR5).16 Therefore, it is of interest whether the reduction in the number of TAAR gene families is specific to a certain group of primates or to all primates as a whole. The pseudogenization events of TAAR3, TAAR4 and TAAR5 happened before the divergence of humans and gorillas,21 but the therian‐specific TAAR repertoires in primates are unknown. In particular, our previous study has found that mammalian TAAR7 genes are subject to positive selection, while this gene has been lost from the human and marmoset genomes.16
Although nearly complete genomes have been released from 13 primates, there have been few in‐depth studies of the chemosensory evolution of these primates, thus they remain poorly understood compared with humans.22, 23, 24, 25, 26, 27, 28, 29 Research into the evolution of the chemosensory system of primates will provide insight into human adaptation. In this study, the complete repertoires of functional TAAR genes and pseudogenes are identified in 13 primate genomes, which include members of all four infraorders (Lorisiformes, Lemuriformes, Tarsiiformes and Simiiformes). The genomes of the Malayan colugo (Sunda flying lemur, Galeopterus variegatus) and the northern treeshrew (Tupaia belangeri) are also examined because these species are crucial to addressing questions regarding chemosensory evolution of primates being members of the most closely related outgroup for primate species.30 The inclusion of 15 euarchontan genomes greatly increases the ability to draw inferences related to the patterns and timing of TAAR evolution. This study is the most comprehensive comparative study to date of TAAR gene family evolution in Grandorder Euarchonta. As such, this study could serve as a critical starting point in the generation of hypotheses related to how different TAARs may have evolved and been depauperated in the adaption to a diverse range of ecological niches.
2. MATERIALS AND METHODS
2.1. Genome sequences and TAAR gene mining
Fifteen euarchontan genomic sequences were obtained from multiple sources (Supporting Information, Table S1). Previously reported TAAR sequences were used as queries.16 A similarity search was performed using tblastn of the Basic Local Alignment Search Tool (BLAST, ver. 2.7.1) programs.31, 32 Putative TAAR genes were searched against genome sequences with an E‐value threshold of 1 × 10−30 (see Eyun et al16 for details). In order to obtain comparable E‐values, a database size of 1.4 × 1010 (using the “‐dbsize” option) was set to be equivalent to the size of the nonredundant (NR) protein database from the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov).33 Based on searches using blastp against the NR database, sequences were considered to be TAAR gene candidates if the top hit from the search was a previously known TAAR. Newly identified primate TAAR candidates were subsequently used as queries against their genomes to find additional candidates. These steps were performed recursively until no other TAAR candidate sequences were detected for each genome. For a more sensitive search, profile hidden Markov models (HMM) were constructed for each TAAR subfamily sequence, including newly identified TAAR subfamilies (TAAR M1‐M3 and E1; see Section 3.1.1). Each genome was searched using the hmmbuild and hmmsearch programs of the HMMER package (ver. 3.0) for building and calibrating HMMs.34 However, no additional sequences were obtained through this search.
The naming of TAAR genes followed the nomenclature in Maguire et al.35 The TAAR genes and their positional information are summarized in Tables 1 and S2. All of the aligned sequences are available in Figure S1.
Table 1.
The number of TAAR genes identified in 13 primate and 5 other genomes
| Order (group)/species name | Common name | Total numbera | Number of TAAR subfamily genesb | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | T4 | T5 | T6 | T7 | T8 | T9 | |||
| Primates (Haplorhini) | |||||||||||
| [Simiiformes (Catarrhini)] | |||||||||||
| Homo sapiens | Human | 6 (3) | 1 | 1 | 0 (1) | 0 (1) | 1 | 1 | 0 (1) | 1 | 1 |
| Pan troglodytes | Chimpanzee | 3 (6) | 1 | 0 (1) | 0 (1) | 0 (1) | 1 | 1 | 0 (1) | 0 (1) | 0 (1) |
| Pan paniscus | Bonobo | 2 (7) | 1 | 0 (1) | 0 (1) | 0 (1) | 1 | 0 (1) | 0 (1) | 0 (1) | 0 (1) |
| Gorilla gorilla | Gorilla | 3 (6) | 1 | 1 | 0 (1) | 0 (1) | 1 | 0 (1) | 0 (1) | 0 (1) | 0 (1) |
| Pongo pygmaeus abelii | Sumatran orangutan | 4 (6) | 1 | 0 (1) | 0 (1) | 1 | 1 | 0 (1) | 0 (2) | 0 (1) | 1 |
| Nomascus leucogenys | White‐cheeked gibbon | 1 (3) | 1 | 0 | 0 | 0 (1) | 0 (1) | 0 | 0 | 0 | 0 (1) |
| Macaca mulatta | Rhesus macaque | 6 (3) | 1 | 1 | 1 | 1 | 1 | 1 | 0 (1) | 0 (1) | 0 (1) |
| Papio hamadryas | Hamadryas baboon | 5 (3) | 1 | 1 | 1 | 1 | 0 (1) | 1 | 0 | 0 (1) | 0 (1) |
| Papio anubis | Olive baboon | 6 (2) | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 (1) | 0 (1) |
| [Simiiformes (Platyrrhini)] | |||||||||||
| Callithrix jacchus | Common marmoset | 2 (6) | 1 | 0 (1) | 0 (1) | 0 (1) | 1 | 0 (1) | 0 | 0 (1) | 0 (1) |
| [Tarsiiformes] | |||||||||||
| Tarsius syrichta | Philippine tarsier | 5 (3) | 1 | 1 | 1 | 1 | 0 (1) | 0 (1) | 0 | 0 (1) | 1 |
| Primates (Strepsirrhini) | |||||||||||
| [Lemuriformes] | |||||||||||
| Microcebus murinus | Gray mouse lemur | 7 (1) | 1 | 1 | 1 | 1 | 0 (1) | 1 | 0 | 1 | 1 |
| [Lorisiformes] | |||||||||||
| Otolemur garnettii | Bushbaby | 8 (0) | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| Dermoptera | |||||||||||
| Galeopterus variegatus | Malayan colugo | 8 (2) | 0 (1) | 1 | 1 | 1 | 1 | 1 | 0 | 2 (1) | 1 |
| Scandentia | |||||||||||
| Tupaia belangeri | Northern treeshrew | 10 [2] (5) | 0 (1) | 1 | 1 | 1 | 1 | 2 (2) | 0 | 5 (2) | 1 |
| Rodentia | |||||||||||
| Mus musculus | House mouse | 15 (1)c | 1 | 1 | 1 | 1 | 1 | 1 | 5 (1) | 3 | 1 |
| Rattus norvegicus | Norway rat | 17 (2)c | 1 | 1 | 1 | 1 | 1 | 1 | 7 (2) | 3 | 1 |
| Artiodactyla | |||||||||||
| Bos taurus | Cow | 21 (8)c | 1 | 1 | 1 | 1 | 1 | 5 (2) | 7 (4) | 3 (2) | 1 |
TAAR gene candidates are divided into three categories: intact, incomplete and pseudogenes. The first number shown is that of intact genes, which contain full‐length open reading frames with seven complete transmembrane regions. The numbers of the incomplete genes due to contig ends and pseudogenes due to premature stop codons or frame‐shifting insertions or deletions are given in square brackets and in parentheses, respectively.
T1‐T9: TAAR1 to TAAR9.
These numbers were taken from Eyun et al.16
All TAAR genes are intron‐less and encoded in a single exon. TAAR2 genes (also known as GPR58) are the exception; they have two exons. To determine the exon and intron boundaries for TAAR2, the coding sequences were predicted using GeneWise (ver. 2.2).36 The conserved first exons were found in six primates (humans, chimpanzees, bonobos, gorillas, orangutans and rhesus macaques) (Figure S1B). The average length of the six TAAR2 introns was 6070 bps (6042 bps in orangutans to 6097 in chimpanzees). Chimpanzee TAAR2P (NC_006473.3) from NCBI is a pseudogene due to a nucleotide deletion (nucleotide position 861 in human TAAR2), which is shared by the bonobo TAAR2P (Figure S1B). Bonobo TAAR2P (XM_003827664.1) from NCBI was annotated as an intact gene and had “N” in that position with “low‐quality position.” However, this bonobo gene has a nucleotide deletion in the same position as the chimpanzee TAAR2P, as well as a unique stop codon at N‐terminus (Figure S1B). Therefore, these genes were considered to be pseudogenes and named TAAR2P.
Dong et al37 reported the number of TAARs from five primate genomes (Homo sapiens, Pan troglodytes, Pongo pygmaeus abelii, Macaca mulatta and Callithrix jacchus). However, the number of TAARs in this study differed. For example, the C. jacchus genome has only one TAAR in Dong et al but two in this study. Also, Dong et al identifies five human TAARs (TAAR1, TAAR2, TAAR3, TAAR4 and TAAR5) compared with the six TAARs (TAAR1, TAAR2, TAAR5, TAAR6, TAAR8 and TAAR9) in the present study. This study analyzed three human genome assemblies: the International Human Genome Sequencing Consortium (IHGSC), Celera (an individual human, J. Craig Venter) and Han Chinese (HC) (Table S1).38, 39, 40 TAAR3 was identified as a pseudogene in the three assemblies and TAAR9 was an intact gene in two of the assemblies (For details, see the Sections 3 and 4). Dong et al37 used automatic data‐mining methods, which are likely to be less precise and which have some limitations in terms of correcting frame shift errors.
2.2. Multiple sequence alignments
Multiple alignments of TAAR protein sequences were generated using MAFFT with the L‐INS‐i algorithm (ver. 7.273).41 All of the amino acid positions in this study were numbered based on the human TAAR sequences in the alignment (see Figure S1). The Ballesteros and Weinstein system numbering is presented as a superscript according to the turkey β1‐adrenergic receptor sequence (β1AR, NCBI accession number: P07700.1). All pseudogenes identified in this study were trimmed after removing the codon to have frame‐shifting indels (insertions/deletions) or in‐frame stop codons and were included in the multiple alignments and the phylogenetic analysis.
2.3. Phylogenetic analysis
Phylogenetic relationships were reconstructed using the maximum‐likelihood method with the PROTGAMMAJTT substitution model (JTT matrix with gamma‐distributed rate variation) using RAxML (ver. 8.1.3).42 The neighbor‐joining phylogenetic method was employed using the Phylip package (ver. 3.67).43 Protein distances were estimated using the JTT substitution model with gamma‐distributed rate variation (α = 1.3)44 estimated from the maximum‐likelihood method in RAxML. Bayesian phylogenetic inference was performed using the MrBayes package (ver. 3.2.5)45 with the JTT substitution model with gamma‐distributed rate variation. A Markov chain Monte Carlo search was run for 106 generations, with a sampling frequency of 103, using three heated and one cold chain with a burn‐in of 103 trees. Nonparametric bootstrapping with 1000 pseudo‐replicates46 was used to estimate the confidence of the branching patterns for the maximum‐likelihood and neighbor‐joining methods. The phylogenies were visualized with FigTree (ver. 1.4.3) (http://tree.bio.ed.ac.uk/software/figtree).
To confirm the taxonomic relationships among the 13 primates and 2 outgroup orders, phylogenetic analysis was conducted based on the concatenated TAAR protein supermatrix (2810 amino acids) including the 13 primates, Malayan colugo (G. variegatus), treeshrew (T. belangeri), mouse and rat (all belong to Euarchonta). Cow TAAR sequences were used as an outgroup. The white‐cheeked gibbon (Nomascus leucogenys) was also included despite having only one intact and three pseudogenes and thus its phylogenetic position should be regarded as tentative. The resultant phylogeny shown in Figure S2 is consistent with recent primate phylogenetic studies and the known taxonomical relationship among these species.47, 48
2.4. Transmembrane topology prediction and homology modeling of protein structure
To predict the transmembrane (TM) protein topology, which includes the N‐terminal, TM, intercellular loop (IC), extracellular loop (EC) and C‐terminal regions, HMMTOP (ver. 2.1)49 and Phobius (ver. 1.01)50 were used.
Homology‐based structural modeling of two TAAR proteins was performed using the SWISS‐MODEL web server (http://swissmodel.expasy.org).51 The same template, the B‐chain of the turkey (Meleagris gallopavo) β1‐adrenergic receptor (β1AR; 4AMJ), was selected for the human TAAR2 and chimpanzee TAAR6 proteins. The root mean‐squared deviation (RMSD) for the human TAAR2 and chimpanzee TAAR6 was 2.30 and 2.20 Å and the QMEAN score was 0.251 and 0.241, respectively. The graphical representation of TAAR structures was constructed using PyMOL (ver. 1.3; DeLanoScientific, San Carlos, California).
2.5. Tests of selection patterns
Stäubert et al21 pointed out that primate TAAR3, TAAR4 and TAAR5 were subject to purifying selection. This study has expanded to include all primate TAAR subfamilies. Four different approaches were employed using codeml of the Phylogenetic Analysis by Maximum Likelihood (PAML) package (ver. 4.9 g)52: (a) site models, (b) branch models, (c) branch‐site models and (d) Clade models. For site models, the one‐ratio model (M0) for estimating an equal ω (or dN/dS) ratio for all branches in the phylogeny was compared against the discrete model (M3), which allows for heterogeneous ω ratios among sites. This M0/M3 comparison was used to test the heterogeneity of the selective constraints among sites. Branch models were used to determine whether the “foreground” lineage evolved under different selection pressures relative to the rest of the phylogeny (the “background” lineage). Likelihood‐ratio tests (LRTs) were performed between the one‐ratio model (R1; the same ω for all branches) and the two‐ratio model (R2; two independent ω's).53, 54 Branch‐site models were employed to detect positively selected sites along specific branches.54, 55 Positively selected amino acid sites were identified based on Bayes Empirical Bayes posterior probabilities.56 In these models, positive selection was allowed on a specific foreground branch and LRTs were conducted against null models that assumed no positive selection. The branch‐site test of positive selection (“Test 2” in Zhang et al55) has four site classes: 0, 1, 2a and 2b. For site classes 0 and 1, all codons operate under purifying selection (0 < ω0 < 1) and neutral evolution (ω1 = 1), respectively, on all branches. For site classes 2a and 2b, positive selection was allowed on the foreground branches (ω2 ≥ 1) but the other background branches were placed under purifying selection (0 < ω0 < 1) and neutral evolution (ω1 = 1), respectively. For the null model, ω2 was fixed at 1. Clade models take a similar approach to the branch‐site models but also allow among‐site variation using the M2a_rel null model for Clade model C (CmC).55, 57 All PAML analyses were carried out using the F3X4 model of codon frequency.58 The level of significance (P) for the LRTs was estimated using a χ 2 distribution with given degrees of freedom (df). The test statistic was calculated as twice the difference of the log‐likelihood between the models (2ΔlnL = 2[lnL1 − lnL0], where L1 and L0 are the likelihood of the alternative and null models, respectively).
3. RESULTS AND DISCUSSION
3.1. Identification of TAAR genes in Euarchonta genomes
TAAR gene candidates were identified in 13 primate species and two outgroup orders, the Malayan colugo (G. variegatus; Dermoptera) and the northern treeshrew (T. belangeri; Scandentia) (Table S1). From the 13 primate genomes, a total of 107 TAAR genes (including 49 pseudogenes) were identified (Table 1). All of the identified primate TAAR genes can be classified into one of nine TAAR subfamilies (TAAR1‐TAAR9). The two outgroup orders have all TAAR subfamilies except for TAAR1 and TAAR7 (see below; Table 1). A recent study found more TAAR subfamilies (TAAR M1‐M3, and E1) in mammalian genomes.16 However, there are no newly identified TAAR subfamilies (such as TAAR E1 and M1‐M3) in the genomes in the present study (see the next section for details). The number of TAAR genes varies significantly among the primate species, ranging from one in the white‐cheeked gibbon (N. leucogenys) to eight in the bushbaby (Otolemur garnettii) (Table 1). This study found that the primate genomes generally have a smaller number of functional TAAR genes compared with other terrestrial mammalian species. This study also found that many primate TAAR genes have been lost in a lineage‐specific manner and the proportion of TAAR pseudogenes in nonhuman apes (approximately 69%) is significantly larger than that in the mouse and rat genomes (approximately 8%).
For humans, three human genome assemblies (IHGSC, Celera and HC) were analyzed (see Section 2) (Table S1). This study found five intraspecific nucleotide variations: one in TAAR1, three in TAAR5 and one in TAAR9. Of the five variations, four nucleotide sites are synonymous, but one variation (position 181 in human TAAR9) leads the in‐frame stop codon (AAA in IHGSC and HC, but TAA in Celera) (Figure S1H). In the NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/SNP), this position is associated with the allele frequency of rs2842899. The allele frequency of A is set at 0.8057 and that of T is 0.1943. Thus, the majority of human genomes have an intact TAAR9. Note that TAAR9 is a therian‐specific gene family (also known as newer types of mammalian TAARs) and generally consists of single‐copy orthologs (except for the opossum genome).16 TAAR9 is represented by pseudogenes in many mammals16 and all simian primates except for humans and orangutans.
3.2. Phylogenetic analysis of primate TAARs
To clarify the classification and evolutionary relationships among the euarchontan TAAR genes, phylogenetic analyses were conducted (Figure 1). Three sea lamprey TAAR‐like proteins were used as outgroups because they are the most ancestral among all TAAR genes.16, 18 All identified TAAR genes from the 15 euarchontan genomes belong to one of the nine main subfamilies (TAAR1‐TAAR9) but none of TAAR M1‐M3 and E1 (Figure 1). This indicates that TAAR M1‐3 are restricted to metatherian mammals and that TAAR E1 emerged in early eutherians but was lost in many laurasiatherian lineages and the ancestors of euarchontoglireans.16 Seven TAAR7 genes were identified from six primates (Table 1), but these have many premature stop codons and frame‐shifting indels and are thus considered pseudogenes.
Figure 1.
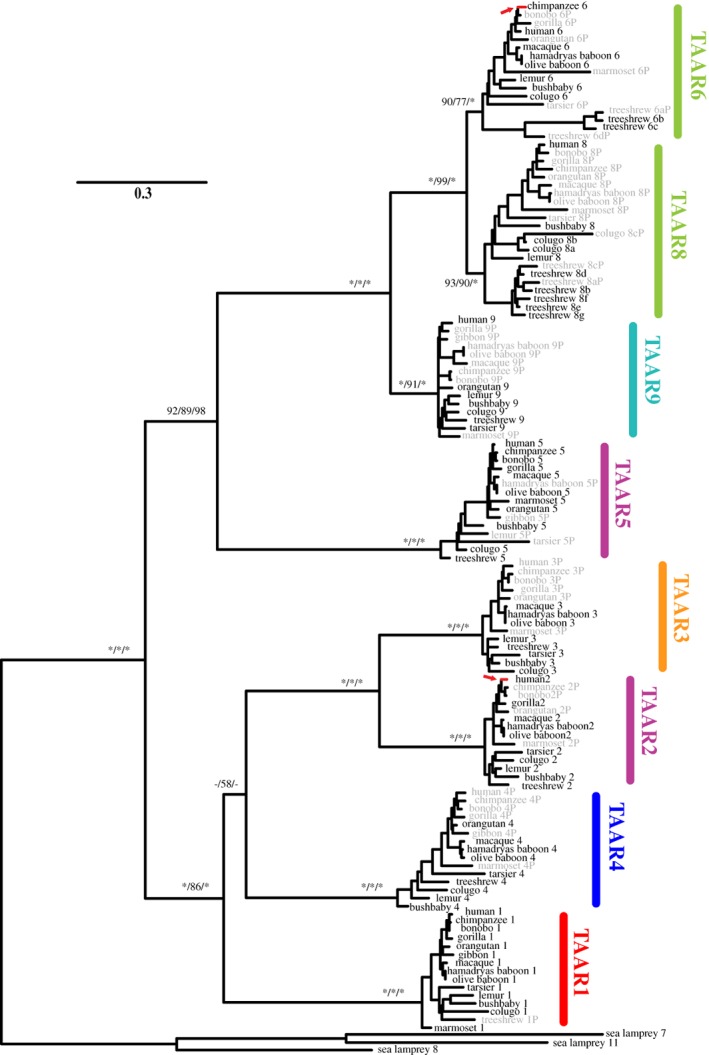
Evolutionary relationship among the TAAR subfamilies from 13 primates, the Malayan colugo, and the northern treeshrew. The phylogenetic tree is reconstructed using the maximum‐likelihood method. All TAAR7 pseudogenes were excluded from the analysis. Three sea lamprey TAAR‐like proteins were used as outgroups. The numbers for the internal branches show the bootstrap support values (%) for the neighbor‐joining and maximum‐likelihood phylogenies and the posterior probability (%) for the Bayesian phylogeny in this order, with asterisks indicating scores of 100%. Supporting values are shown only for the major internal branches. Gray names indicate pseudogenes. The two red‐colored branches and arrows indicate those that were identified to be subject to positive selection by PAML branch‐site models (see Table S5). The TAAR subfamily names are shown in different colors based on their taxonomic distribution as follows: TAAR1 found in jawed vertebrates in red, tetrapod‐specific TAAR4 in dark blue, amniote‐specific TAAR2 and TAAR5 in purple, mammalian‐specific TAAR3 in orange, therian‐specific TAAR9 in cyan and eutherian‐specific TAAR6 and TAAR8 in light green
In Figure 1, each cluster of the eight main subfamilies (TAAR1‐TAAR9) is strongly supported by high bootstrap values (>77% in the maximum likelihood phylogeny, >90% in the neighbor‐joining phylogeny and 100% posterior probability in the Bayesian phylogeny). Based on functional assays, TAARs can be classified into two groups (TAAR1‐4 vs TAAR5‐9) based on whether they preferentially detect primary or tertiary amines.59 Phylogenetic analysis also supports this two‐group classification (>86% by the maximum‐likelihood, neighbor‐joining and Bayesian inference) (Figure 1). Tertiary‐amine detecting TAARs except for TAAR5 are the recently diverged TAAR subfamilies, emerging after the divergence between prototherian and therian mammals (230‐166 million years ago; MYA).16 They have been subjected to rapid species‐specific tandem gene duplication in most mammalian species. For example, the cow (Bos taurus) genome possesses 16 therian‐specific TAARs (5 TAAR6s, 7 TAAR7s, 3 TAAR8s and 1 TAAR9). In euarchontoglirean species, two Rodentia genomes (the mouse and rat) also have all four therian‐specific TAARs and a high number of TAAR7 (5‐7 genes) and TAAR8 genes (3 genes) (Table 1). This pattern is consistently observed in the Malayan colugo and northern treeshrew genomes, which have colugo‐specific and treeshrew‐specific duplicated copies of TAAR6 and TAAR8 (Figure 1). In the primate lineage, however, no extra copy (gene gain or duplication) of any TAAR gene was found. A possible gene duplication event was identified only for two orangutan TAAR7Ps. However, this event appears to have occurred very recently after pseudogenization because they have 100% identical nucleotide sequences but different genomic locations. All functional primate TAAR genes have apparently remained as single‐copy genes. This is obviously different from what has been typically observed in mammalian TAAR evolution.
3.3. Pseudogenization of TAAR subfamilies
All primate TAARs have only experienced gene losses with no gene gains. Pseudogenization events were identified in all 15 euarchontan genomes, whereas gene duplications have only occurred in Scandentia and Dermoptera. In Figure 2, all pseudogenization (or gene loss) events that have occurred during primate evolution are summarized. Frequent gene losses have occurred particularly in haplorhine primates, and a higher proportion of pseudogenization can be observed in Hominoidea after the divergence from Cercopithecoidea, reaching approximately 69% in nonhuman apes. These results are consistent with Stäubert et al,21 who reported that pseudogenization events in TAAR3‐5 are more frequent in primates than in other mammals.
Figure 2.
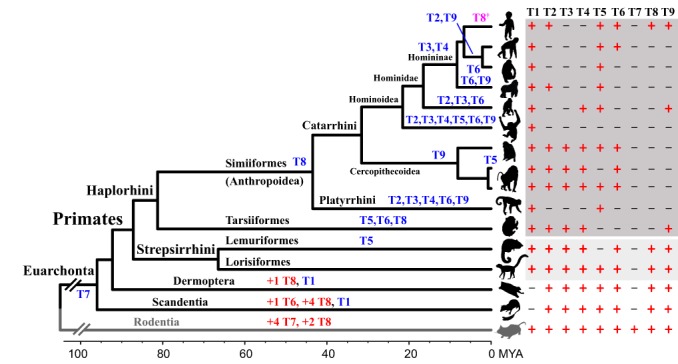
TAAR gene gains and losses in euarchontan genomes. TAAR gene gain (red) and gene loss (including pseudogenization; blue) events are shown along the branches. T1 to T9 indicate TAAR1 to TAAR9. The current consensus for primate phylogenies with their approximate divergence times (MYA) was obtained from Perelman et al.47 All species belong to Euarchonta except for the mouse (gray, Mus musculus), which is used as an outgroup. The species are listed in the same order as shown in Table 1 (from the top, except for the rat and the cow)
TAAR1 is highly conserved in sequence and represents the oldest TAAR subfamily, as its origin can be traced back to jawed vertebrates.16 This study shows that all 13 primate TAAR1s are intact, but the TAAR1s are pseudogenes in the Malayan colugo (G. variegatus) and the northern treeshrew (T. belangeri) due to the presence of four different indel nucleotides and a premature stop codon, respectively. In tetrapods, only a few lineages (the bottlenosed dolphin, Tursiops truncatus; the dog, Canis familiaris; and the tammar wallaby, Macropus eugenii) have been reported as TAAR1 pseudogenes.16, 60 The dog TAAR1 is thought to be a case of independent pseudogenization in a recent event (~54.2 MYA) because the TAAR1s are all pseudogenes in the dog, the wild gray wolf, and four other caniforms, but they remain intact in cats.60
The results of the present study show the TAAR7 subfamily to be completely absent from all 15 euarchontan genomes. In mammals, TAAR7 genes have the greatest variation in gene number (1 in the horse and armadillo to 16 in the flying fox). In Euarchontoglires, the mouse and rat genomes have multiple TAAR7 genes (5‐7 genes) (Table 1). This subfamily has also been shown to evolve under the influence of positive selection.16 In contrast, none of the 13 primate genomes examined in this study had an intact TAAR7. The bushbaby (O. garnettii) belongs to the suborder Strepsirrhini and, with lemurs, diverged from other primates earlier. The bushbaby genome possesses eight TAAR subfamilies, but no identifiable TAAR7 gene nor pseudogene exists in the bushbaby genome. The order Dermoptera, which includes the Malayan colugo (G. variegatus), is the closest outgroup to the primates.30 The Malayan colugo genome possesses all intact TAAR gene subfamilies except for TAAR1 and TAAR7, as does the northern treeshrew (T. belangeri; Scandentia) genome. Therefore, TAAR7 genes emerged in a common ancestor of eutherians, was maintained until the euarchontoglirean lineages, but was then subsequently lost in the common ancestor of the Euarchonta (Primates, Dermoptera and Scandentia) (Figure 2).
The white‐cheeked gibbon (N. leucogenys) possesses only one intact TAAR1 gene and three pseudogenes (Table 1). Hylobatidae is known to have experienced extremely rapid chromosome evolution.61, 62 All tetrapod TAAR genes are located in a region of a single chromosome.16, 18 The syntenic relationships of all TAARs and the adjacent genes are highly conserved as a single gene cluster.16 All great ape TAAR genes are located on chromosome 6 (Table S2). Human chromosome 6 corresponds to six chromosomes (NLE1, NLE3, NLE8, NLE17, NLE18 and NLE22) in the white‐cheeked gibbon genome,61 suggesting an association with TAAR gene losses. Although all gibbon TAAR genes are located on a single chromosome (chromosome 3, NC_019818.1) (Table S2), a higher rate of segmental rearrangement may have led to the relaxation of negative selection and acted as a driving force in TAAR gene loss in the white‐cheeked gibbon.
Based on shared indel positions, the timing of pseudogenization events can be estimated using the previous phylogenetic analyses. For the TAAR2 gene, one shared nucleotide deletion (nucleotide position 861 in human TAAR2) can be observed in the genus Pan (chimpanzees and bonobos) (Figure S1B). Thus, the pseudogenization of the TAAR2 gene in the genus Pan may have happened very recently after the divergence of humans and the genus Pan but before the divergence of chimpanzees and bonobos (4.5‐1.0 MYA)63 (see Section 2 for details regarding pseudogene assignment). Species of the subfamily Homininae (African apes) share the same positions of the indel events in the TAAR3 and TAAR4 genes, which is consistent with results from Stäubert et al.21 Two nucleotide deletions (position between 136 and 137 in human TAAR3P), and one or two nucleotide insertions (position 749 in human TAAR4P) can be observed in the African apes (Figure S1C,D). Thus, the indels associated with pseudogenization in TAAR3 and in TAAR4 seem to have occurred in the lineage leading to the subfamily Homininae. Independent pseudogenization events for TAAR4 can be observed in the white‐cheeked gibbon (N. leucogenys) and common marmoset (C. jacchus). This indicates that the pseudogenization of TAAR4 is not shared ancestral event but rather the result of multiple lineage‐specific independent events.
3.4. TAAR8 pseudogenes in Haplorhini except for humans
Except for the white‐cheeked gibbon (N. leucogenys), 12 TAAR8 sequences were identified from the 13 primate genomes (Table 1). All Haplorhini TAAR8s are characterized by multiple premature stop codons and frame‐shifting indels except for human TAAR8, although the four TAAR8s in Strepsirrhini and the two outgroup orders are intact (Figure S1G). Seven additional TAAR8 sequences were obtained from the NCBI database; these sequences belong to Haplorhini (Figure 3). All TAAR8s identified in haplorhines are pseudogenes except for those in humans. These pseudogenization events were mapped to the consensus primate phylogeny (Figure 3). Of the multiple pseudogenization events in the TAAR8 sequences, two nucleotide deletions at positions 748 and 749 are shared in the lineage leading to the infraorder Simiiformes, except for human TAAR8 (Figures 3 and S1G). Note that the three human genome assemblies have 100% of the TAAR8 nucleotides without variation (Figure S1G). Two positions are associated with rs537864861 (NCBI dbSNP database) and ss1376213897 (1000 genomes), but the allele frequency of the deletion is less than 0.002. Furthermore, the Neanderthal genome has not experienced a deletion in this position (http://www.eva.mpg.de/neandertal).64 This suggests two possibilities. First, two nucleotide deletions occurred independently at the same position in each simian species. Second, these two deletions occurred in a common ancestor of simians and were subsequently resurrected in humans by regaining the two missing nucleotides. The second explanation is more likely because it is more parsimonious than multiple independent events. In addition, based on the number of TAAR8 pseudogenization events, the functional constraint was likely relaxed earlier than other TAAR subfamilies because the primate TAAR8 subfamily has had the most frequent indel events among the TAAR pseudogenes (Figure 3). For example, once a functional gene became a pseudogene, the gene usually accumulates deleterious mutations over time.65 All simian primates except humans and bonobos have experienced other pseudogenization events in addition to the shared two nucleotide deletion event. This suggests that TAAR8 pseudogene events may have a long history in the haplorrhine lineage. Although it is a very rare case, similar resurrection events have been reported in cows and humans.66, 67, 68 The ruminant pancreatic seminal ribonuclease gene became a pseudogene before the divergence of ruminants (~39 MYA),66, 67 thus gaining deleterious mutations or not being expressed, but it was revived recently in cows.66, 67 Also, a resurrection event has been posited for the IRGM gene, a member of immunity‐related GTPases (IRGs), in some primates.68 The single‐copy IRGM gene became pseudogenized in the common ancestor of Simiiformes but was restored in the common ancestor of humans and great apes, possibly due to the insertion of a retroviral element.68 These results indicate that some pseudogenes are likely to have been restored.
Figure 3.
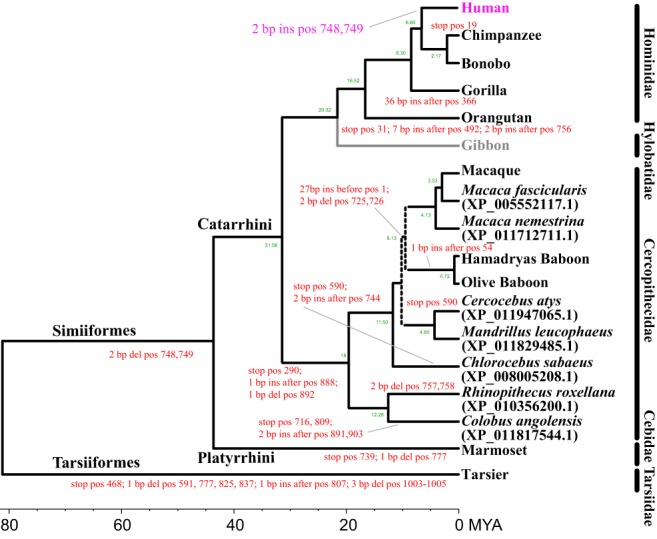
TAAR8 pseudogenization events for 17 Haplorhini species. The position numbers (in red) for the ORF disruptions such as stop codons and indel events are based on the human TAAR nucleotide sequence. All species belong to the Haplorhini. The TAAR8 gene was not identified in the white‐cheeked gibbon (Nomascus leucogenys) genome (gray). It is likely that TAAR8 was pseudogenized in Simiiformes but subsequently resurrected in humans (magenta). The tree is obtained from recent primate phylogenetic studies47, 48, 84 and their approximate divergence times (MYA) are obtained from Perelman et al47
3.5. The degeneration of TAAR gene repertoires in Euarchonta
The evolutionary deterioration of TAARs appears to be a major trend in Euarchonta (Figure 2). The present study found that the initial breakdown of TAARs occurred in basal euarchontan species and has happened more often in primates than in other mammals. Mammalian TAARs are known to have a lower proportion of pseudogenes compared with odorant receptor (OR) genes.18 For instance, mouse and rat genomes have 15 and 17 intact TAARs but only 1 and 2 pseudogenes, respectively, while their genomes have more than 1000 OR genes, more than 23.5% of which are pseudogenes.69 The degeneration of TAAR gene repertoires likely began in the common ancestor of Euarchonta and is highly likely to be related to arboreal living. The two nonprimate euarchontan genomes have a smaller number of TAARs compared with Rodentia (the mouse and rat) and Artiodactyla (the cow) (Table 1). They have already lost TAAR1 and TAAR7. The euarchontan ancestor probably arose from arboreal animals, and primates became more arboreal than this euarchontan ancestor. Certain characteristics of the primate ancestor remain as adaptations to this lifestyle, such as a shortened rostrum with stereoscopic vision, an opposable hallux and pollex, and a highly mobile radius and ulna in the forelimb.20, 70, 71
Living in trees significantly reduces exposure to predators and facilitates escape from ground‐living predators.72 Thus, it is conceivable that arboreal life may have decreased the reliance on the chemosensing of predators, leading to the nonfunctionalization of primate TAARs. For instance, TAAR4 is stimulated by β‐phenylethylamine, which is a carnivore odor that evokes physiological and behavioral responses in two prey species (the mouse and rat).14 The genetic deletion of TAAR4 in mice specifically eliminates high‐sensitivity responses to β‐phenylethylamine and puma urine volatiles.15 Interestingly, all African apes (Homininae) have lost TAAR4. Two independent TAAR4 pseudogenization events in two arboreal primates, the white‐cheeked gibbon and the marmoset, were also observed. In contrast, three cercopithecid species, including the macaque and two baboon species have functional TAAR4 genes and a higher number of TAARs (approximately five genes) compared with other haplorhines. They still face a wide array of predators such as leopards, tigers and cheetahs, thus they display a variety of behaviors in response to the threat of these predators.73, 74 Therefore, reduced pressure from ground‐dwelling predators for primate TAARs has altered evolutionary selection processes. Whether the low number of TAARs is specific to euarchontan species or whether it is shared with other arboreal species requires further testing.
3.6. Sensory trade‐offs for primate chemosensory receptors
This study has found that pseudogenization events were more frequent in Haplorhini (including Simiiformes and Tarsiiformes) after the divergence from Strepsirrhini (including Lemuriformes and Lorisiformes) (87 MYA). More than half of TAARs have been lost due to multiple independent pseudogenization events, particularly in Hominoidea genomes outside of humans (Table 1). The proportion of TAAR pseudogenes in nonhuman apes (approximately 69%) is significantly larger than in other primates (approximately 31%) and significantly larger than in the mouse and rat (approximately 8%). Although primates are traditionally divided into two suborders, Simiiformes (Anthropoidea) and Prosimii (Tarsiiformes, Lemuriformes and Lorisiformes), Haplorhini and Strepsirrhini are divided on the basis of the shape of the nose.26, 75 The name Strepsirrhini derives from the “curly” nostrils of the rhinarium (the moist area of the nasal tip in mammals or wet nose, an ancestral condition), while Haplorhini means “simple nose” in that it lacks a rhinarium.26 Mammals with a rhinarium are known to have a very sensitive and more acute olfaction capacity. In addition to the loss of the rhinarium, the size of the main olfactory epithelium (MOE, the back of the nose into which air flows) is reduced in haplorhines compared with strepsirrhines.76 In the mammalian MOE, the sensory neurons have two types of chemosensory receptor, ORs and TAARs.77 Thus, the loss of the rhinarium and the smaller MOE in haplorhines is very likely associated with a reduction in the reliance on olfaction sensitivity.26, 78 Haplorhini OR gene repertoires have also exhibited consistent deterioration compared with other mammals.24, 25, 26 Therefore, the frequent pseudogenization of TAARs and ORs in the Haplorhini lineage can be considered a result of relaxed selection due to the lower reliance on olfaction.
Most haplorhines except tarsiers are diurnal and have well‐developed color vision systems, while most strepsirrhine species are nocturnal.26, 79, 80 This observation suggests a relationship between the acquisition of well‐developed trichromatic vision and reduced chemosensory function, such as olfaction.25, 26 This has been referred to as a sensory trade‐off hypothesis, in which lower olfaction sensitivity can be compensated for by other sensory mechanisms such as better vision. All apes, three Old World monkeys, and the common marmoset (depending upon gender) possess full trichromatic vision, but the proportion of TAAR pseudogenes in these species has a large variation (25% in the olive baboon to 78% in the bonobo). Thus, the sensory trade‐off hypothesis may not be a factor in haplorhine TAAR evolution. In addition, the gradual degeneration of OR gene repertoires in primates has been observed in every lineage and thus cannot be directly linked to full trichromatic vision.26 Furthermore, a relatively smaller proportion (37%‐46%) of OR pseudogenes are found in macaques and marmosets,26 but TAAR pseudogenes exhibit a large variation (33% in the macaque to 75% in the marmoset). Therefore, the relationships among the three chemosensory receptors (opsins, ORs and TAARs) in primates cannot be simply explained by the trade‐off hypothesis.
3.7. Selection patterns and selective forces operating on TAAR subfamilies
In order to examine selective constraint patterns, the ratio of nonsynonymous to synonymous distances (ω or dN/dS) was estimated for each TAAR subfamily. Overall, the average ω for the TAAR subfamilies was ~1.5 times higher in primates than in nonprimate mammalian orthologs (0.2232 for primates and 0.1523 for nonprimate mammals) (Table S3). Furthermore, the overall average ω (0.3813) was significantly higher when estimated using only haplorhines than when using nonprimate mammals (P = 0.0367 with the Mann‐Whitney U test) (Table S3). This indicates that primate TAARs have been subject to relaxed purifying selection, a process which has been accelerated in haplorhines.
In order to confirm the hypothesis that there are different selective pressures within primate TAAR subfamilies, PAML tests based on the branch and Clade models were conducted (Table S4).52, 55, 57 The branch model tests an alternative hypothesis where two ω's are allowed within specific branches against the null hypothesis that there is a single ω for all branches. Using this test, the patterns of selective pressure are compared between Haplorhini TAARs and Strepsirrhini TAARs. The alternative hypothesis with two ω's was found to be significantly stronger than the null hypothesis for most of the TAAR subfamilies (P < 0.01 for TAAR2, TAAR3, TAAR4, TAAR6 and TAAR9; Table S4). The estimated ω's were about two or three times higher in Haplorhini (ω1 in R2) compared with that of the Strepsirrhini lineages (ω0 in R2) (Table S4) indicating further relaxed selective constraints in Haplorhini TAAR subfamilies after the divergence from Strepsirrhini TAARs.
Statistically significant differences also appeared in the results for the Clade models. Large increases in the ω's of haplorhine primates from Clade model analysis were observed (Table S4). The divergent ω ratio estimates for haplorhine TAARs (TAAR4, TAAR5, TAAR6 and TAAR9) were significantly greater than ω = 1 (P < 0.01), indicating a small proportion of relaxed constraints or positive diversifying selection (Table S4). Taken together, these results indicate that Haplorhini TAARs have been subject to significantly relaxed selection constraints and more derived primate TAARs have evolved under more relaxed selection. This is most likely associated with the major morphological transition from Strepsirrhini to Haplorhini described above. Relaxed selection pressure on TAAR subfamilies in haplorhines has led to the accumulation of multiple independent mutations, resulting in the nonfunctionalization of numerous TAAR genes.
3.8. Positive‐selection sites located in potential ligand‐binding sites
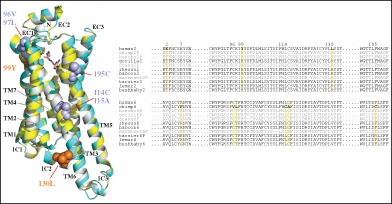
Amino acid sites exhibiting positive selection signatures were identified using PAML branch‐site models with Bayes Empirical Bayes inference.56 PAML tests with branch‐site models can detect a short episode of positive selection in a small fraction of amino acids.55 The tests were conducted both with and without pseudogenes. The models that allowed ω > 1 exhibited a significant fit for the data for chimpanzee TAAR6 (P < 0.0001) and a marginally significant fit for human TAAR2 (P < 0.05) (Table S5). In analyses including all TAAR sequences and excluding pseudogenes, six sites were identified in chimpanzee TAAR6 (positions 7, 963.25, 973.26, 1143.43, 1153.44 and 1955.43) and three sites in human TAAR2 (positions 2, 993.28 and 1303.59) (Table S5). Four of the six sites identified in chimpanzee TAAR6 (positions 963.25, 973.26, 1143.43 and 1153.44) had posterior probabilities higher than 95%, indicating strong positive selection. Three positively selected sites in chimpanzee TAAR6 (positions 963.25 and 1955.43) and human TAAR2 (position 993.28) corresponded to residues identified to be directly involved with ligand‐binding on β‐adrenergic receptors 1 and 2 (Figure S3).81, 82, 83 To determine the spatial distribution of these nine positive‐selection sites, homology modeling of the TAAR protein structure was conducted (Figure 4). Six of the nine amino acids identified as being under positive selection are located in or near the extracellular regions of the receptors (including two in the N‐terminal region) (Figure 4). Thus, the locations of positive selected sites are found in the area around ligand‐binding site pockets in human TAAR2 and chimpanzee TAAR6. These substitutions may have affected ligand‐binding activity and the specificity of these TAARs. The TAARs in more recently derived primates have likely been rebuilt with possible positive selection.
Figure 4.
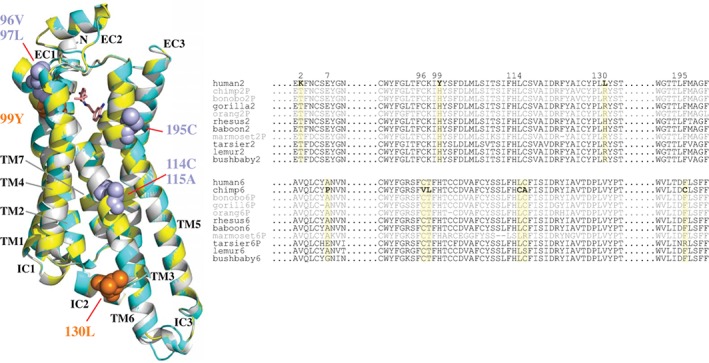
Three‐dimensional‐structural model and partial sequence alignments for TAAR2 and TAAR6 proteins. (A) A 3D‐structural model of the human TAAR2 (yellow) and chimpanzee TAAR6 protein (cyan) superimposed with the Turkey β1‐adrenergic receptor (β1AR, gray). The ligand β1AR (dobutamine) is shown as a stick model. Positively selected sites are indicated in orange (human TAAR2) and dark cyan (chimpanzee TAAR6). Two positive selection sites (positions 2 and 7) are not shown due to the lack of a 3D protein model. (B) The partial sequence alignment of primate TAAR2 and TAAR6. The nine residues predicted to be under positive selection are shown in boldface (indicated by the yellow boxes). The pseudogenes are in gray
4. CONCLUSION
This study identified the complete TAAR gene repertoires in 15 euarchontan genomes and showed that gradual to rapid degeneration of TAARs in primates has occurred without gene duplication. The arboreal lifestyle derived from the Euarchonta ancestor may have reduced reliance on the chemosensing of predators, leading to the depauperation of TAAR subfamilies. This was likely to have been accelerated after the change in nose shape in Haplorhini species. Relaxed selection in primate TAARs has resulted in multiple independent mutations and thus smaller numbers of TAAR genes compared with other mammalians. In recently derived primates, human TAAR2 and chimpanzee TAAR6 experienced positive selection. Human TAAR8 is likely to have been restored in an unknown resurrection event. Although TAARs are likely to be associated with adaptation to ground living, some primate TAAR genes have been reestablished under high selection pressure, probably due to functional divergence.
Supporting information
Figure S1. The nucleotide sequence alignment of primate TAAR1 (A) to TAAR9 (H)
Figure S2. The maximum‐likelihood phylogeny inferred from the supermatrix dataset
Figure S3. Multiple alignment of the three TAARs with turkey β1‐adrenergic receptor proteins
Table S1. Taxonomic classification and the genomes used in this study
Table S3. The results of PAML site model analysis and likelihood ratio statistics within primate TAAR subfamilies
Table S4. PAML branch‐model and Clade model tests between Haplorhini TAARs and Strepsirrhini TAARs
Table S5. The results of PAML branch‐site model analysis
Table S2. TAAR sequence information
ACKNOWLEDGMENTS
The author sincerely thanks Dr. Etsuko Moriyama (University of Nebraska‐Lincoln, USA) and two anonymous reviewers whose comments greatly improved an earlier draft of this manuscript. This work was supported by National Research Foundation of Korea grants (2018R1C1B3001650 and 2018R1A5A1025077) (to S.E.).
Eyun S. Accelerated pseudogenization of trace amine‐associated receptor genes in primates. Genes, Brain and Behavior. 2019;18:e12543 10.1111/gbb.12543
Funding information National Research Foundation of Korea, Grant/Award Number: 2018R1C1B3001650, 2018R1A5A1025077
REFERENCES
- 1. Berry MD. Mammalian central nervous system trace amines. Pharmacologic amphetamines, physiologic neuromodulators. J Neurochem. 2004;90:257‐271. [DOI] [PubMed] [Google Scholar]
- 2. Miller GM. The emerging role of trace amine‐associated receptor 1 in the functional regulation of monoamine transporters and dopaminergic activity. J Neurochem. 2011;116:164‐176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Duan J, Martinez M, Sanders AR, et al. Polymorphisms in the trace amine receptor 4 (TRAR4) gene on chromosome 6q23.2 are associated with susceptibility to schizophrenia. Am J Hum Genet. 2004;75:624‐638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Serretti A, Pae C‐U, Chiesa A, Mandelli L, De Ronchi D. Influence of TAAR6 polymorphisms on response to aripiprazole. Prog Neuro‐Psychopharmacol Biol Psychiatry. 2009;33:822‐826. [DOI] [PubMed] [Google Scholar]
- 5. Pae C‐U, Drago A, Kim J‐J, et al. TAAR6 variations possibly associated with antidepressant response and suicidal behavior. Psychiatry Res. 2010;180:20‐24. [DOI] [PubMed] [Google Scholar]
- 6. Premont RT, Gainetdinov RR, Caron MG. Following the trace of elusive amines. Proc Natl Acad Sci USA. 2001;98:9474‐9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wolinsky TD, Swanson CJ, Smith KE, et al. The trace amine 1 receptor knockout mouse: an animal model with relevance to schizophrenia. Genes Brain Behav. 2007;6:628‐639. [DOI] [PubMed] [Google Scholar]
- 8. Simmler LD, Buchy D, Chaboz S, Hoener MC, Liechti ME. In vitro characterization of psychoactive substances at rat, mouse, and human trace amine‐associated receptor 1. J Pharmacol Exp Ther. 2016;357:134‐144. [DOI] [PubMed] [Google Scholar]
- 9. Gainetdinov RR, Hoener MC, Berry MD. Trace amines and their receptors. Pharmacol Rev. 2018;70:549‐620. [DOI] [PubMed] [Google Scholar]
- 10. Christian SL, Berry MD. Trace amine‐associated receptors as novel therapeutic targets for immunomodulatory disorders. Front Pharmacol. 2018;9:680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bunzow JR, Sonders MS, Arttamangkul S, et al. Amphetamine, 3,4‐methylenedioxymethamphetamine, lysergic acid diethylamide, and metabolites of the catecholamine neurotransmitters are agonists of a rat trace amine receptor. Mol Pharmacol. 2001;60:1181‐1188. [DOI] [PubMed] [Google Scholar]
- 12. Borowsky B, Adham N, Jones KA, et al. Trace amines: identification of a family of mammalian G protein‐coupled receptors. Proc Natl Acad Sci USA. 2001;98:8966‐8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vladimirov V, Thiselton DL, Kuo PH, et al. A region of 35 kb containing the trace amine associate receptor 6 (TAAR6) gene is associated with schizophrenia in the Irish study of high‐density schizophrenia families. Mol Psychiatry. 2007;12:842‐853. [DOI] [PubMed] [Google Scholar]
- 14. Ferrero DM, Lemon JK, Fluegge D, et al. Detection and avoidance of a carnivore odor by prey. Proc Natl Acad Sci USA. 2011;108:11235‐11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dewan A, Pacifico R, Zhan R, Rinberg D, Bozza T. Non‐redundant coding of aversive odours in the main olfactory pathway. Nature. 2013;497:486‐489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eyun S, Moriyama H, Hoffmann FG, Moriyama EN. Molecular evolution and functional divergence of trace amine‐associated receptors. PLoS One. 2016;11:e0151023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eyun S, Moriyama EN. Species‐specific duplications of trace amine‐associated receptors in vertebrate genomes Proceedings of Biotechnology and Bioinformatics Symposium 2009 (BIOT‐2009), Lincoln, NE; 2009:117. [Google Scholar]
- 18. Hashiguchi Y, Nishida M. Evolution of trace amine‐associated receptor (TAAR) gene family in vertebrates: lineage‐specific expansions and degradations of a second class of vertebrate chemosensory receptors expressed in the olfactory epithelium. Mol Biol Evol. 2007;24:2099‐2107. [DOI] [PubMed] [Google Scholar]
- 19. Lindemann L, Ebeling M, Kratochwil NA, Bunzow JR, Grandy DK, Hoener MC. Trace amine‐associated receptors form structurally and functionally distinct subfamilies of novel G protein‐coupled receptors. Genomics. 2005;85:372‐385. [DOI] [PubMed] [Google Scholar]
- 20. Martin RD. Primates. Curr Biol. 2012;22:R785‐R790. [DOI] [PubMed] [Google Scholar]
- 21. Stäubert C, Böselt I, Bohnekamp J, Römpler H, Enard W, Schöneberg T. Structural and functional evolution of the trace amine‐associated receptors TAAR3, TAAR4 and TAAR5 in primates. PLoS One. 2010;5:e11133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang J, Webb DM. Evolutionary deterioration of the vomeronasal pheromone transduction pathway in catarrhine primates. Proc Natl Acad Sci USA. 2003;100:8337‐8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Melin AD, Wells K, Moritz GL, et al. Euarchontan opsin variation brings new focus to primate origins. Mol Biol Evol. 2016;33:1029‐1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dong D, He G, Zhang S, Zhang Z. Evolution of olfactory receptor genes in primates dominated by birth‐and‐death process. Genome Biol Evol. 2009;1:258‐264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gilad Y, Wiebe V, Przeworski M, Lancet D, Pääbo S. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2004;2:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsui A, Go Y, Niimura Y. Degeneration of olfactory receptor gene repertories in primates: no direct link to full trichromatic vision. Mol Biol Evol. 2010;27:1192‐1200. [DOI] [PubMed] [Google Scholar]
- 27. Hayakawa T, Suzuki‐Hashido N, Matsui A, Go Y. Frequent expansions of the bitter taste receptor gene repertoire during evolution of mammals in the euarchontoglires clade. Mol Biol Evol. 2014;31:2018‐2031. [DOI] [PubMed] [Google Scholar]
- 28. Go Y, Niimura Y. Similar numbers but different repertoires of olfactory receptor genes in humans and chimpanzees. Mol Biol Evol. 2008;25:1897‐1907. [DOI] [PubMed] [Google Scholar]
- 29. Niimura Y, Matsui A, Touhara K. Acceleration of olfactory receptor gene loss in primate evolution: possible link to anatomical change in sensory systems and dietary transition. Mol Biol Evol. 2018;35:1437‐1450. [DOI] [PubMed] [Google Scholar]
- 30. Janečka JE, Miller W, Pringle TH, et al. Molecular and genomic data identify the closest living relative of primates. Science. 2007;318:792‐794. [DOI] [PubMed] [Google Scholar]
- 31. Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389‐3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Biochem. 2009;10:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eyun S, Soh HY, Posavi M, et al. Evolutionary history of chemosensory‐related gene families across the Arthropoda. Mol Biol Evol. 2017;34:1838‐1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maguire JJ, Parker WAE, Foord SM, Bonner TI, Neubig RR, Davenport AP. International union of pharmacology. LXXII. Recommendations for trace amine receptor nomenclature. Pharmacol Rev. 2009;61:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Birney E, Clamp M, Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988‐995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dong D, Jin K, Wu X, Zhong Y. CRDB: database of chemosensory receptor gene families in vertebrate. PLoS One. 2012;7:e31540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. International Human Genome Sequencing Consortium . Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931‐945. [DOI] [PubMed] [Google Scholar]
- 39. Li Y, Zhu J, Tian G, et al. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 2010;8:e1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Levy S, Sutton G, Ng PC, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772‐780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics. 2014;30:1312‐1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. PHYLIP (Phylogeny Inference Package) version 3.6. [computer program]. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle. 2005.
- 44. Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39:306‐314. [DOI] [PubMed] [Google Scholar]
- 45. Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754‐755. [DOI] [PubMed] [Google Scholar]
- 46. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783‐791. [DOI] [PubMed] [Google Scholar]
- 47. Perelman P, Johnson WE, Roos C, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fabre PH, Rodrigues A, Douzery EJP. Patterns of macroevolution among primates inferred from a supermatrix of mitochondrial and nuclear DNA. Mol Phylogenet Evol. 2009;53:808‐825. [DOI] [PubMed] [Google Scholar]
- 49. Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849‐850. [DOI] [PubMed] [Google Scholar]
- 50. Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction‐the Phobius web server. Nucleic Acids Res. 2007;35:W429‐W432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS‐MODEL workspace: a web‐based environment for protein structure homology modelling. Bioinformatics. 2006;22:195‐201. [DOI] [PubMed] [Google Scholar]
- 52. Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586‐1591. [DOI] [PubMed] [Google Scholar]
- 53. Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568‐573. [DOI] [PubMed] [Google Scholar]
- 54. Yang Z, Nielsen R. Codon‐substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908‐917. [DOI] [PubMed] [Google Scholar]
- 55. Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch‐site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472‐2479. [DOI] [PubMed] [Google Scholar]
- 56. Yang Z, Wong WSW, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107‐1118. [DOI] [PubMed] [Google Scholar]
- 57. Weadick CJ, Chang BSW. An improved likelihood ratio test for detecting site‐specific functional divergence among clades of protein‐coding genes. Mol Biol Evol. 2012;29:1297‐1300. [DOI] [PubMed] [Google Scholar]
- 58. Goldman N, Yang Z. A codon‐based model of nucleotide substitution for protein‐coding DNA sequences. Mol Biol Evol. 1994;11:725‐736. [DOI] [PubMed] [Google Scholar]
- 59. Ferrero DM, Wacker D, Roque MA, Baldwin MW, Stevens RC, Liberles SD. Agonists for 13 trace amine‐associated receptors provide insight into the molecular basis of odor selectivity. ACS Chem Biol. 2012;7:1184‐1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vallender E, Xie Z, Westmoreland S, Miller G. Functional evolution of the trace amine associated receptors in mammals and the loss of TAAR1 in dogs. BMC Evol Biol. 2010;10:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Roberto R, Capozzi O, Wilson RK, et al. Molecular refinement of gibbon genome rearrangements. Genome Res. 2007;17:249‐257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Misceo D, Capozzi O, Roberto R, et al. Tracking the complex flow of chromosome rearrangements from the Hominoidea ancestor to extant Hylobates and Nomascus gibbons by high‐resolution synteny mapping. Genome Res. 2008;18:1530‐1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Prüfer K, Munch K, Hellmann I, et al. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486:527‐531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Green RE, Krause J, Briggs AW, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710‐722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292‐298. [Google Scholar]
- 66. Kleineidam RG, Jekel PA, Beintema JJ, Situmorang P. Seminal‐type ribonuclease genes in ruminants, sequence conservation without protein expression? Gene. 1999;231:147‐153. [DOI] [PubMed] [Google Scholar]
- 67. Breukelman HJ, van der Munnik N, Kleineidam RG, Furia A, Beintema JJ. Secretory ribonuclease genes and pseudogenes in true ruminants. Gene. 1998;212:259‐268. [DOI] [PubMed] [Google Scholar]
- 68. Bekpen C, Marques‐Bonet T, Alkan C, et al. Death and resurrection of the human IRGM gene. PLoS Genet. 2009;5:e1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9:951‐963. [DOI] [PubMed] [Google Scholar]
- 70. Cartmill M, Smith F. The Human Lineage. Hoboken, NJ: Wiley‐Blackwell; 2009. [Google Scholar]
- 71. Chadwell BA, Young JW. Angular momentum and arboreal stability in common marmosets (Callithrix jacchus). Am J Phys Anthropol. 2015;156:565‐576. [DOI] [PubMed] [Google Scholar]
- 72. Hart D. Predation on primates: a biogeographical analysis In: Gursky S, Nekaris KAI, eds. Primate Anti‐Predator Strategies. New York, NY: Springer; 2007:27‐59. [Google Scholar]
- 73. Enstam K. Effects of habitat structure on perceived risk of predation and anti‐predator behavior of vervet (Cercopithecus aethiops) and patas (Erythrocebus patas) monkeys In: Gursky S, KAI N, eds. Primate Anti‐Predator Strategies. New York, NY: Springer; 2007:308‐338. [Google Scholar]
- 74. Cords M. The behavior, ecology, and social evolution of cercopithecine monkeys In: Mitani JC, Call J, Kappeler PM, Palombit RA, Silk JB, eds. The Evolution of Primate Societies. Chicago, IL: University of Chicago Press; 1987:91‐112. [Google Scholar]
- 75. Schmitz J, Ohme M, Zischler H. SINE insertions in cladistic analyses and the phylogenetic affiliations of Tarsius bancanus to other primates. Genetics. 2001;157:777‐784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Barton RA. Olfactory evolution and behavioral ecology in primates. Am J Primatol. 2006;68:545‐558. [DOI] [PubMed] [Google Scholar]
- 77. Kaupp UB. Olfactory signalling in vertebrates and insects: differences and commonalities. Nat Rev Neurosci. 2010;11:188‐200. [DOI] [PubMed] [Google Scholar]
- 78. Smith T, Rossie J. Primate olfaction: anatomy and evolution In: Brewer W, Castle D, Pantelis C, eds. Olfaction and the Brain. New York, NY: Cambridge University Press; 2006:135‐166. [Google Scholar]
- 79. Regan BC, Julliot C, Simmen B, Vienot F, Charles‐Dominique P, Mollon JD. Fruits, foliage and the evolution of primate colour vision. Philos Trans R Soc Lond Ser B Biol Sci. 2001;356:229‐283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Perry GH, Martin RD, Verrelli BC. Signatures of functional constraint at aye‐aye opsin genes: the potential of adaptive color vision in a nocturnal primate. Mol Biol Evol. 2007;24:1963‐1970. [DOI] [PubMed] [Google Scholar]
- 81. Warne T, Moukhametzianov R, Baker JG, et al. The structural basis for agonist and partial agonist action on a β1‐adrenergic receptor. Nature. 2011;469:241‐244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Warne T, Edwards PC, Leslie AG, Tate CG. Crystal structures of a stabilized β1‐adrenoceptor bound to the biased agonists bucindolol and carvedilol. Structure. 2012;20:841‐849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kleinau G, Pratzka J, Nurnberg D, et al. Differential modulation of beta‐adrenergic receptor signaling by trace amine‐associated receptor 1 agonists. PLoS One. 2011;6:e27073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Liedigk R, Roos C, Brameier M, Zinner D. Mitogenomics of the old world monkey tribe Papionini. BMC Evol Biol. 2014;14:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The nucleotide sequence alignment of primate TAAR1 (A) to TAAR9 (H)
Figure S2. The maximum‐likelihood phylogeny inferred from the supermatrix dataset
Figure S3. Multiple alignment of the three TAARs with turkey β1‐adrenergic receptor proteins
Table S1. Taxonomic classification and the genomes used in this study
Table S3. The results of PAML site model analysis and likelihood ratio statistics within primate TAAR subfamilies
Table S4. PAML branch‐model and Clade model tests between Haplorhini TAARs and Strepsirrhini TAARs
Table S5. The results of PAML branch‐site model analysis
Table S2. TAAR sequence information


