Abstract
O-GlcNAcylation is a ubiquitous protein glycosylation playing different roles on variant proteins. O-GlcNAc transferase (OGT) is the unique enzyme responsible for the sugar addition to nucleocytoplasmic proteins. Recently, multiple O-GlcNAc sites have been observed on short-form OGT (sOGT) and nucleocytoplasmic OGT (ncOGT), both of which locate in the nucleus and cytoplasm in cell. Moreover, O-GlcNAcylation of Ser389 in ncOGT (1036 amino acids) affects its nuclear translocation in HeLa cells. To date, the major O-GlcNAcylation sites and their roles in sOGT remain unknown. Here, we performed LC-MS/MS and mutational analyses to seek the major O-GlcNAcylation site on sOGT. We identified six O-GlcNAc sites in the tetratricopeptide repeat domain in sOGT, with Thr12 and Ser56 being two “key” sites. Thr12 is a dominant O-GlcNAcylation site, whereas the modification of Ser56 plays a role in regulating sOGT O-GlcNAcylation, partly through Thr12. In vitro activity and pulldown assays demonstrated that O-GlcNAcylation does not affect sOGT activity but does affect sOGT-interacting proteins. In HEK293T cells, S56A bound to and hence glycosylated more proteins in contrast to T12A and WT sOGT. By proteomic and bioinformatics analyses, we found that T12A and S56A differed in substrate proteins (e.g. HNRNPU and PDCD6IP), which eventually affected cell cycle progression and/or cell proliferation. These findings demonstrate that O-GlcNAcylation modulates sOGT substrate selectivity and affects its role in the cell. The data also highlight the regulatory role of O-GlcNAcylation at Thr12 and Ser56.
Keywords: O-GlcNAcylation, O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT), cell proliferation, cell cycle, enzyme kinetics, G2/M arrest, sOGT, substrate selectivity
Introduction
O-Linked-β-GlcNAc modification (O-GlcNAcylation)4 is a ubiquitous post-translational modification dynamically occurred on thousands of proteins (1, 2). O-GlcNAc transferase (OGT) catalyzes the attachment of GlcNAc to numerous nucleocytoplasmic proteins. In human cells, the unique ogt gene in chromosome X encodes three isoforms of OGT (termed nucleocytoplasmic OGT (ncOGT), mitochondrial OGT (mOGT), and short-form (sOGT)) via post-transcriptional RNA processes. ncOGT and sOGT locate in the nuclear and cytoplasm in cell, whereas mOGT exists in mitochondria. Each isoform has an identical C-terminal catalytic domain but a different N-terminal domain consisting of variant tetratricopeptide repeats (TPRs). Compared with full-length, nucleocytoplasmic ncOGT with 13 N-terminal TPR domains, the N terminus of mOGT starts within the fifth TPR domain, and sOGT starts in the eleventh TPR domain (3).
The TPR domain helps OGT bind to another OGT molecule or variant proteins. In hepatic cells, the TPR domain bridges two ncOGTs and one sOGT to form a heterotrimeric complex, whose role is not yet known (4). N-terminal deletion analyses show that the 2–6 TPRs in ncOGT (5) and the first three TPRs in mOGT (6) help them bind to their substrate proteins. Accordingly, ncOGT and mOGT glycosylate different proteins in vitro (7). TPRs also play a role in recruiting OGT to specific substrate by targeting to a docking protein, such as Track1/Milton (previously known as OIP106) (8). In addition, Sp1 helps ncOGT (but not mOGT and sOGT) glycosylate a mass of Escherichia coli proteins (9), which further expands the role of TPRs.
Once O-GlcNAcylated, a protein may show some new characters. For example, O-GlcNAcylation of the Rpt2 subunit of 26S proteasome represses its ATPase activity (10), whereas the modification on topoisomerase I correlates well with the enzyme activity (11). O-GlcNAcylation of Sp1 disrupts its binding to other proteins and represses Sp1 transcription roles (12–14), whereas O-GlcNAcylation of PGC-1 facilitates its binding to deubiquitinase BAP1 and protects PGC-1 from degradation (15). Moreover, O-GlcNAcylation of the C terminus of co-activator-associated arginine methyltransferase 1 (CARM1) modulates its substrate specificity (16).
OGT is also O-GlcNAcylated (17, 18). Advanced mass technologies have aided in the discovery of multiple O-GlcNAcylation sites on OGT. For example, Seo et al. (9) found the O-GlcNAcylation of ncOGT (1036 aa) at Ser389, which plays a significant role for ncOGT nuclear translocation in HeLa cells. Griffin et al. (19) observed the O-GlcNAcylation of sOGT at Ser10, Thr12, Ser20, Thr38, Ser52, Ser56, and Thr662 in sf9 cells. More recently, Fan et al. (20) observed the O-GlcNAcylation of ncOGT (110-kDa subunit isoform 1, 1046 aa) at Ser437 and the O-GlcNAcylation of sOGT at Ser10, Thr12, Ser18, and Thr38 in huH7 cells. However, the dominant O-GlcNAcylation sites on sOGT and their role in vivo remain largely unknown.
In this work, we report the regulation mechanisms of sOGT substrate selectivity by O-GlcNAcylation. We find six O-GlcNAcylation sites on sOGT via ETD MS and confirm two key sites through mutational analyses. Thr12 is a dominant O-GlcNAcylation site, whereas the modification of Ser56 has a role in regulating sOGT O-GlcNAcylation, partly through Thr12. We also find that O-GlcNAcylation does not affect sOGT activity but alters its substrate selectivity. In HEK293T cells, O-GlcNAcylation of Thr12 or Ser56 modulates sOGT substrate selectivity and hence changes its function in cell cycle progression and/or cell proliferation.
Results
Site mapping of O-GlcNAcylation on sOGT
The gradual increase of sOGT abundance in aging mouse brains suggests the physiological significance of the short-form OGT, which may be involved in age-dependent diseases (21). We have demonstrated that sOGT glycosylates variant proteins in HEK293T cells (22). Moreover, expression of sOGT enables the promotion of cell proliferation (Fig. S1).
sOGT is more easily O-GlcNAcylated than ncOGT (6). Recently, variant O-GlcNAcylation sites have been identified on sOGT via LC-MS/MS analyses (19, 20) (Fig. 1A). To better know the O-GlcNAcylation sites on sOGT, we expressed human sOGT in E. coli BL21 (DE3) cells, where it would be auto-O-GlcNAcylated.
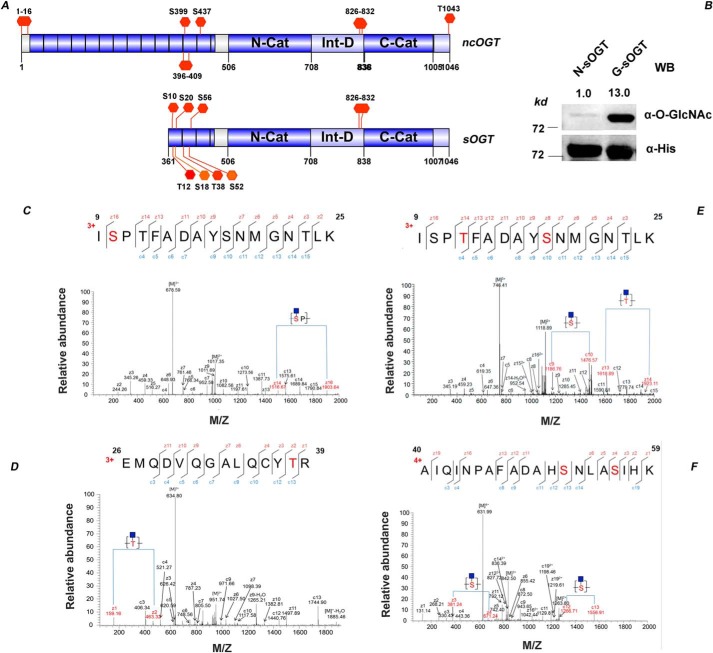
Figure 1.
Site mapping of O-GlcNAcylation on sOGT. A, known O-GlcNAcylation sites on sOGT/ncOGT. B, O-GlcNAc sOGT enriched through WGA affinity chromatography was examined via Western blotting. The blots were quantified with ImageJ, and O-GlcNAcylation of sOGT was normalized to the input (α-His). C–F, O-GlcNAc sOGT was expressed in E. coli cells, pooled with WGA beads, and digested in-gel with trypsin. The resulting peptides were analyzed on an LTQ-Orbitrap Elite mass spectrometer employing ETD fragmentation. ETD spectra revealed 1–2 O-GlcNAc modifications on each of the following peptides: ISPTFADAYSNMGNTLK (C), ISPTFADAYSNMGNTLK (D), EMQDVQGALQCYTR (E), and AIQINPAFADAHSNLASIHK (F) (all of the modification sites shown in underlined boldface letters).
O-GlcNAc sOGT was enriched via the WGA affinity method (Fig. 1B) and applied for O-GlcNAc site mapping. Based on the mass difference of 203 Da in ETD spectra (Fig. 1 (C–F), Sheet S1), we identified six modification sites (Ser10, Thr12, Ser18, Thr38, Ser52, and Ser56). In the Fig. 1C, the mass difference of 387 Da between z14 and z16 (1516.67 versus 1903.64 Da) indicated that O-GlcNAcylation happens at Ser10. In Fig. 1D, the mass difference of 304 Da between z13 and z14 indicated that O-GlcNAcylation happens at Thr12. Similarly, O-GlcNAcylation happens at Ser18 according to c9 and c10. In Fig. 1E, the mass difference of 304 Da between z1 and z2 (159.1 versus 463.32 Da) demonstrated that Thr38 is O-GlcNAcylated. In Fig. 1F, the mass difference of 290 Da between z3 and z4 (361.24 versus 671.24 Da) or c12 and c13 (1266.71 versus 1556.91 Da) demonstrated that Ser52 and Ser56 are O-GlcNAcylated. O-GlcNAcylation of Ser10, Thr12, and Thr38 were also observed on sOGT in different types of cells (19, 20), suggesting that these sites should be easily O-GlcNAcylated.
Thr12 and Ser56 are two key O-GlcNAcylation sites on sOGT
To assess key O-GlcNAc sites on sOGT, we performed mutation experiments in E. coli and HEK293T cells. It has been proved that mutation of the active site (H498A) of ncOGT thoroughly abrogates its activity (23, 24). The corresponding mutation (H127A) also abrogated sOGT activity (Fig. 2A). Thus, we made six double-point mutants, including H127A/S10A, H127A/T12A, H127A/S18A, H127A/T38A, H127A/S52A, and H127A/S56A, and co-expressed each of them (with an N-His6 tag) with WT sOGT (without His6 tag) in E. coli cells. Lanes 1 and 2 in Fig. 2B demonstrate the outstanding performance of the glycosylation system, because co-expression with WT sOGT markedly elevated O-GlcNAcylation on H127A. The significant decrease of O-GlcNAcylation on T12A/H127A and S18A/H127A demonstrates that sOGT autoglycosylated mainly at Thr12 and Ser18 in E. coli cells. To our surprise, the modification on S56A/H127A increased.
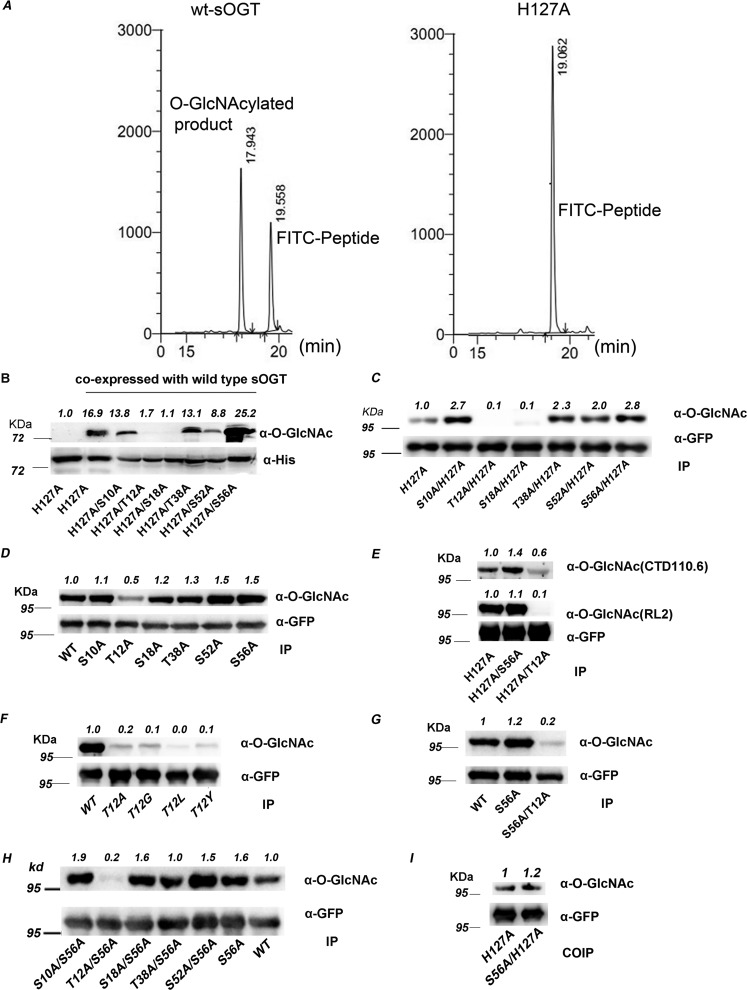
Figure 2.
Thr12 and Ser56 are two key O-GlcNAcylation sites on sOGT. A, H127A mutation abrogated sOGT activity in vitro. B, assessment of the modification sites in E. coli cells. WT sOGT was co-expressed with the indicated sOGT mutants (with an N-terminal His6 tag) in E. coli BL21 (DE3) cells. O-GlcNAcylation of the mutants was examined via Western blotting. C–H, assessment of the modification sites in HEK293T cells. The indicated sOGT mutants were expressed in HEK293T cells. O-GlcNAcylation of the mutants was examined via Western blotting. C, double-point mutants with H127A mutation. D, single-point mutants. E, O-GlcNAcylation was examined with two O-GlcNAc–specific antibodies. F, other Thr12 mutants. G, S56A and T12A/S56A. H, double-point mutants with S56A. I, each indicated mutant (H127A, T12A, or S56A) was co-expressed with WT sOGT (with an N-terminal c-Myc tag) in HEK293T cells. Co-immunoprecipitation/Western blotting was performed to examine the interaction between WT sOGT and each mutant. Western blots were quantified with ImageJ for each blot, and O-GlcNAcylation of target mutants was normalized to the input (α-His in B/α-GFP in others).
To verify these data, we examined these double-point mutants in HEK293T cells. Again, we found that O-GlcNAcylation on T12A/H127A and S18A/H127A sharply declined, but that on other mutants increased, in contrast to H127A (Fig. 2C). An examination of single-point mutants (S10A, T12A, S18A, T38A, S52A, and S56A) revealed the O-GlcNAcylation decline on T12A and the increase on S52A and S56A (Fig. 2D). The data hint that Thr12 and Ser56 might be two key sites for sOGT O-GlcNAcylation: Thr12 should be a major modification site, whereas S56A up-regulates sOGT O-GlcNAcylation via some unknown mechanisms. This was also confirmed by using another O-GlcNAc–specific antibody, CTD 110.6 (Fig. 2E).
To confirm the major O-GlcNAcylation site, we mutated Thr12 to glycine, leucine, or tyrosine, either with a small side chain or a bulkier side chain, with the hope of reducing possible influence from a protein structure change because of the substitution with specific amino acid residues. We found that all of these sOGT mutants (T12G, T12L, or T12Y) sharply reduced sOGT O-GlcNAcylation, besides T12A (Fig. 2F). Thereafter, we concluded that Thr12 is a dominant site for sOGT O-GlcNAcylation.
It is rational to think that S56A affects sOGT O-GlcNAcylation through the dominant modification site. The cutdown O-GlcNAcylation on S56A/T12A and T12A (Fig. 2, C and G) indicates that S56A may increase the modification at Thr12. The rising O-GlcNAcylation on S56A/S10A, S56A/S18A, and S56A/S52A, although not on S56A/T38A, further elucidated that S56A enhanced sOGT O-GlcNAcylation at Thr12 but not at other sites (Fig. 2H). Taking into consideration the O-GlcNAc increase on the majority of double-point mutants with the S56A mutation, we assumed that S56A mutation might increase the binding between two sOGT molecules. To test this point, we performed a co-immunoprecipitation study using WT sOGT and two mutants (H127A and H127A/S56A). We found that H127A/S56A more strongly bound to WT sOGT than H127A (1.2/1) (Fig. 2I), confirming the hypothesis to some extent.
O-GlcNAcylation on sOGT does not impact the enzyme activity
Because O-GlcNAcylation affects the activity of some enzymes (10, 11), we wondered if the O-GlcNAcylation also affects sOGT activity. Therefore, we compared the activity of the naked sOGT and O-GlcNAc sOGT, through in vitro activity assays (Table 1 and Figs. S2–S4). The naked sOGT and O-GlcNAc sOGT showed almost the same product yields with respect to each substrate peptide, demonstrating that O-GlcNAcylation had little influence on sOGT activity. The data were further confirmed by the enzyme kinetics study (Table 1 and Fig. S5), where the naked sOGT and O-GlcNAc sOGT showed a similar Km value against UDP-GlcNAc (175.7 versus 180.5 μm, respectively).
Table 1.
In vitro activity assay of the naked or O-GlcNAc sOGT (Figs. S2–S5)
| Measurements | Naked sOGT | O-GlcNAc sOGT |
|---|---|---|
| Using CK3k peptide | ||
| Yields | 63.6% | 65.4% |
| Using FITC-YAVVPVSK peptide | ||
| Yields | 22.9% | 22.4% |
| Km (μm) | 175.7 ± 18.26 | 180.5 ± 26.17 |
| Vmax (μm/min) | 48.54 ± 18.26 | 27.27 ± 26.17 |
Next, we examined the enzyme kinetics of the mutants S56A and T12A, which differ greatly in O-GlcNAcylation (Table 2 and Fig. S6). The Km values of S56A and T12A (130.2/137.7 μm, respectively) demonstrate that the mutants have similar activity. The data also indicate that O-GlcNAcylation had little influence on sOGT activity.
Table 2.
Enzyme kinetics study of sOGT mutants (Fig. S6)
| Kinetic parameters | WT sOGT | T12A | S56A | Peptide used |
|---|---|---|---|---|
| Km (μm) | 101.9 ± 11.22 | 137.7 ± 7.175 | 130.2 ± 8.957 | YAVVPVSK |
| Vmax (μm/min) | 0.177 ± 0.005555 | 0.1828 ± 0.002985 | 0.223 ± 0.004721 | YAVVPVSK |
O-GlcNAcylation modulates sOGT substrate selectivity
Of note, all of the O-GlcNAcylation sites, including the two key sites, are within the TPR domain (Fig. S7), which is responsible for binding variant proteins. Thus, O-GlcNAcylation may affect sOGT function in glycosylating variant proteins in cell. For this, we expressed the sOGT mutants (S56A or T12A) in HEK293T cells and examined total protein O-GlcNAcylation. We found that expression of S56A significantly increased total protein O-GlcNAcylation in contrast to T12A and the control (Fig. 3A). Because S56A and T12A have a similar enzymatic activity in vitro but differ in O-GlcNAcylation, we reasoned that O-GlcNAcylation possibly affects sOGT substrate selectivity.
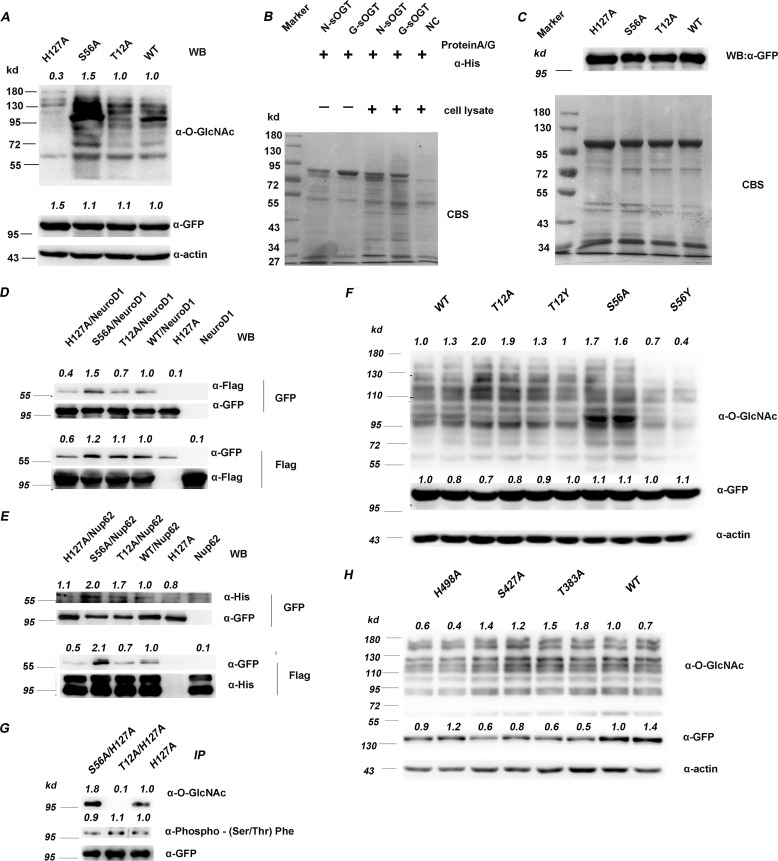
Figure 3.
O-GlcNAcylation modulates sOGT substrate selectivity. A, the indicated sOGT mutants were expressed in HEK293T cells, and total protein O-GlcNAcylation was examined via Western blotting. B, O-GlcNAc sOGT and naked sOGT were applied to pull down proteins from HEK293T cell lysates. The interaction proteins were examined via SDS-PAGE visualized with Coomassie Brilliant Blue staining (CBS). C, the indicated sOGT mutants were expressed in HEK293T cells. Their interaction proteins were visualized by Coomassie Brilliant Blue staining (bottom). sOGT mutants were measured via Western blotting (top). D and E, His-tagged Nup62 (D) and FLAG-tagged NeuroD1 (E) were co-expressed with GFP-tagged sOGT, respectively, and their interaction with sOGT was evaluated via co-immunoprecipitation and Western blotting. F, gain-of-function mutation. Thr12/Ser56 were mutated to tyrosine to mimic O-GlcNAcylation, and their influence was assessed in HEK293T cells via Western blotting. G, examination of sOGT phosphorylation. The indicated sOGT mutants were expressed in HEK293T cells. O-GlcNAcylation and phosphorylation of these mutants were measured. H, influence on ncOGT. The indicated ncOGT mutants (1036 aa) were expressed in HEK293T cells, and O-GlcNAcylation of total proteins was measured via Western blotting. Western blots were quantified with ImageJ for each blot, the O-GlcNAcylation of target mutants or the amount/abundancy of target proteins was normalized to the input (α-actin in A, F, and H; α-FLAG in D and E; α-GFP in D, E, F, and G; α-His in E).
To verify this point, we performed a pulldown study. The naked sOGT or O-GlcNAc sOGT was applied to pulldown proteins from HEK293T cell lysates. Fig. 3B shows that O-GlcNAc sOGT bound to slightly more proteins than naked sOGT, demonstrating that O-GlcNAcylation affects sOGT binding to other proteins. We then expressed sOGT mutants in HEK293T cells and performed co-immunoprecipitation (co-IP) to investigate their interaction proteins. Fig. 3C shows that S56A bound to more proteins. Considering the higher O-GlcNAcylation level on S56A, the data are consistent with the findings in the pulldown study. Unexpectedly, the signals in both of the experiments were too weak, probably due to the weak interaction between OGT and substrate/interacting proteins as well as the limited amounts of some endogenous proteins.
To address this issue, we co-expressed sOGT with two known O-GlcNAc proteins—NeuroD1 (25) or Nup62 (26, 27)—in HEK293T cells and measured their interaction via co-IP. Compared with WT sOGT, S56A strongly bound to NeuroD1 and Nup62, whereas T12A had similar affinity to NeuroD1 but weak affinity to Nup62 (Fig. 3, D and E). The data strongly indicate that S56A bound to more proteins. They also hint that O-GlcNAc sOGT would bind to more proteins.
Next, we performed a gain-of-function study by mutating Thr12 or Ser56 to tyrosine, with the hope of mimicking O-GlcNAcylation on Ser/Thr residues. To determine their function, we examined total protein O-GlcNAcylation upon the expression of each mutant. We found that T12A and T12Y were similar in glycosylating cellular proteins, whereas S56A and S56Y were different. S56A glycosylated more proteins than the WT, whereas S56Y had fewer substrates (Fig. 3F). The contrary influence of S56A and S56Y suggests that the O-GlcNAcylation at Ser56 might affect sOGT function through a steric-hindrance effect. The data verified the role of Ser56 O-GlcNAcylation to some extent (i.e. O-GlcNAcylation of Ser56 may decrease sOGT function in the glycosylation of cellular proteins). As for Thr12, steric hindrance may have little influence at this position, probably due to the protein structure.
OGT is also phosphorylated and extensively cross-talks with O-GlcNAcylation (28, 29). To know the O-GlcNAcylation influence on sOGT phosphorylation, we expressed the three indicated mutants in HEK293T cells and examined their modifications. We found that their phosphorylation slightly increased in a trend: H127A/S56A > H127A > H127A/T12A, contrary to their O-GlcNAcylation changes (Fig. 3G). The data indicate that O-GlcNAcylation had a slight influence on Ser/Thr phosphorylation of sOGT.
We also wondered how a similar mutation would affect ncOGT substrate selectivity, because ncOGT has the same C-terminal amino acid sequence as sOGT, and ncOGT is O-GlcNAcylated at Ser427 (20). To determine the function of S56A in the cell, we examined total protein O-GlcNAcylation upon expression of the ncOGT mutants T383A and S427A (corresponding to T12A and S56A of sOGT). We found that expression of the ncOGT mutants did not affect total protein O-GlcNAcylation (Fig. 3H), although S56A elevated the O-GlcNAcylation significantly. The data suggested that O-GlcNAcylation affecting OGT function might be isoform-dependent. But further studies are required to confirm that.
T12A and S56A glycosylate different proteins/peptides in HEK293T cells
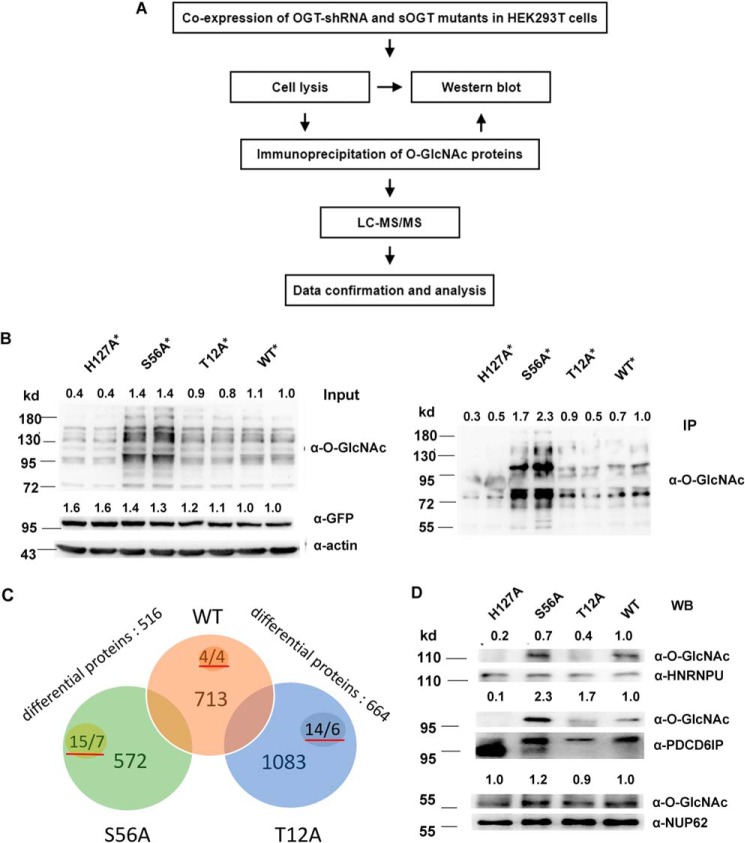
Next, we performed a proteomic study to further investigate the O-GlcNAcylation influence on sOGT substrate selectivity (Fig. 4A). To diminish the influence of endogenous OGT, we transfected cells with a OGT-shRNA, which could efficiently knock down endogenous OGT (Fig. S1). To express sOGT in the presence of the shRNA, we made four shRNA-resistant OGT mutants (H127A*, WT*, T12A*, and S56A*) (Fig. S8). Western blotting shows that the additional mutation did not alter their function in cell (Fig. 4B, Input); the O-GlcNAc patterns of the immunoprecipitated proteins were also consistent with the input (Fig. 4B, IP). The IP proteins were applied for the MS analyses. The mass data gave 713, 1083, and 572 putative O-GlcNAc proteins in WT*, T12A*, and S56A* samples, respectively (Sheet S2, source MS data). Further data analyses yields 516 differential proteins between S56A* and WT* and 664 differential proteins between T12A* and the WT* (Sheets S3 and S4). We found 43 O-GlcNAc peptides with a score of ≥300 in the MS data, which differently distributed in each sample (Sheet S5, supplemental mass spectra). Four O-GlcNAc peptides from four proteins were observed in WT sOGT, 14 peptides from six proteins in T12A, and 15 peptides from seven proteins in S56A. The data definitely demonstrated their different substrate selectivity (Fig. 4C).
Figure 4.
T12A and S56A glycosylate different proteins/peptides in HEK293T cells. A, work flow of the proteomic study. B, *, confirmation of the pooled O-GlcNAc proteins for proteomic analysis. OGT-shRNA2 and indicated shRNA-resistant sOGT mutants were co-expressed in HEK293T cells in duplicate. O-GlcNAcylation of total proteins (Input) or immunoprecipitated proteins (IP) was examined via Western blotting. C, *, raw files from the MS analysis were searched by MaxQuant and processed using an intensity-based absolute quantification (iBAQ) approach. 713, 1083, and 572 proteins were found in WT, T12A, and S56A samples (Sheet S2), respectively. After normalization and blank subtraction, 516 and 664 differential proteins were obtained in two comparison samples (iBAQ ratio ≥ 1.5) (Sheets S3 and S4); the numbers with red underlines indicate confirmed O-GlcNAc peptides/proteins in each sample (Sheet S5). D, the indicated sOGT mutants were expressed in HEK293T cells. Selected proteins were immunoprecipitated from each sample, and their O-GlcNAcylation was measured via Western blotting (WB). *, using OGT-shRNA resistant mutants (Fig. S8). Western blots were quantified with ImageJ for each blot, and the O-GlcNAcylation of target mutants or amount/abundancy of target proteins was normalized to the input.
Among 1476 detected proteins, more than 100 proteins were previously reported as O-GlcNAc proteins (Sheet S6). Most of the other proteins might be unidentified O-GlcNAc proteins and/or proteins interacting with the captured proteins. To verify the MS data, we selected NUP62 (with a score of 323) and two putative O-GlcNAc proteins, HNRNPU and programmed cell death 6–interacting protein (PDCD6IP) (with scores of 313.76 and 138.03) (Sheet S2), and measured their O-GlcNAcylation. We found that these proteins were distinctly O-GlcNAcylated in each sample. HNRNPU was predominantly glycosylated by WT* and S56A* and less likely by T12A*. PDCD6IP and Nup62 were also mainly glycosylated by S56A* but less likely by T12A* (Fig. 4D). The data prove the reliability of the mass data. They also suggest that S56A glycosylates more substrate proteins.
T12A and S56A show different functions in HEK293T cells
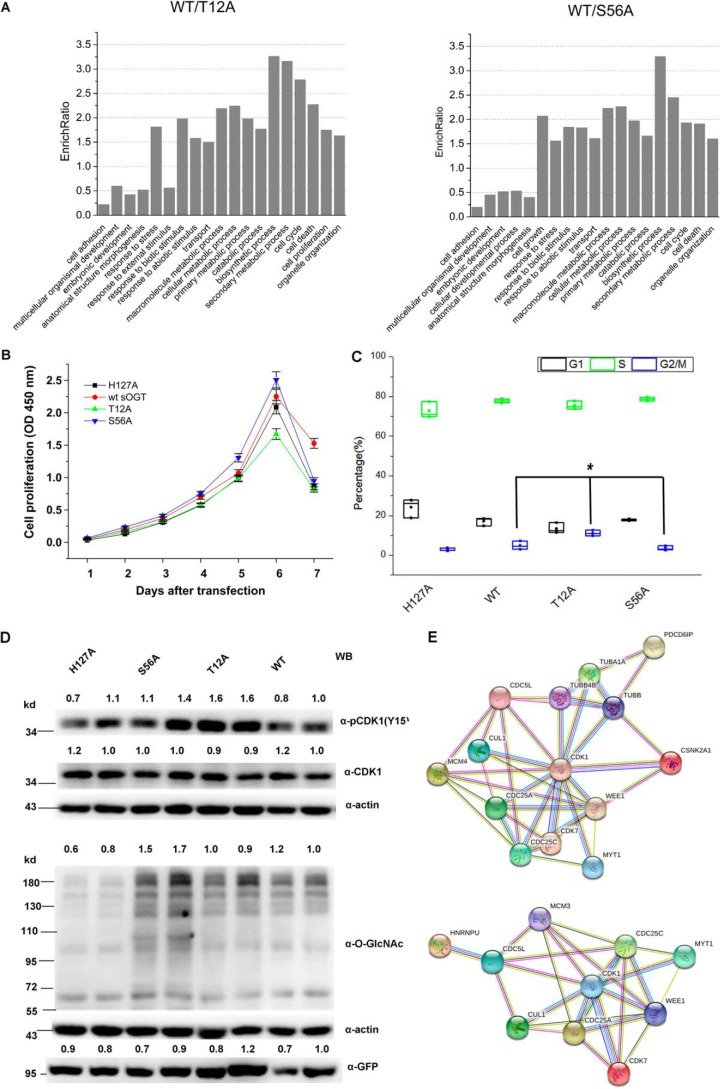
The above data show that T12A and S56A glycosylated different proteins in the cell. Gene ontology (GO) enrichment analyses show that the differential proteins between WT sOGT and the mutants were categorized in various cellular processes, including cell proliferation, cell cycle, and various cellular metabolic process (Fig. 5A). So we assume that expression of T12A or S56A may affect cells in such processes.
Figure 5.
T12A and S56A show different functions in HEK293T cells. A, GO term analysis. Shown is GO enrichment analysis of the differential proteins (Sheets S3 and S4). The histogram shows the output of cellular processes with an EnrichRatio ≥ 1.5. B, measurement of cell proliferation in response to the expression of the indicated sOGT mutants in HEK293T cells. Bars, mean ± S.D. (error bars); p < 0.05. C, measurement of cell cycle progression in response to the expression of the indicated sOGT mutants in HEK293T cells (Fig. S9). Cell percentages in different stages (G1, S, and G2/M) are shown with scatter plots. Bars, mean ± S.D. *, p < 0.05. D, cells were co-transfected with OGT-shRNA and the indicated shRNA-resistant mutants. An equal amount of the cell lysate (30 μg) was applied for Western blotting analyses using antibodies against O-GlcNAc (RL2), CDK1, pCDK1(Tyr15), and β-actin. E, functional PPI analysis was performed against 42 proteins either categorized in cell cycle progression or relevant to G2/M arrest; HNRNPU or PDCD6IP was used as an upstream protein in the networks. Western blots were quantified with ImageJ for each blot, and the O-GlcNAcylation of target mutants or the amount/abundance of target proteins was normalized to the input (α-actin).
To determine the biological relevance, we performed cell proliferation and cell cycle analyses. We found that T12A slightly repressed the cell proliferation, whereas S56A enhanced it. The influence was especially pronounced 3 days after transfection and up to the maximum at day 7 (Fig. 5B). Cell cycle analyses indicated that all groups had a similar cell ratio at S phase, but expression of T12A led to a marked G2/M cell cycle arrest along with a decrease of the cell ratio at G1 phase (Fig. 5C and Fig. S9).
CDK1 (encoded by cdc2) is an important protein in cell cycle progression. Phosphorylation of CDK1 at tyrosine 15 is necessary and sufficient for G2/M cell cycle arrest (30). We found that pCDK1(Tyr15) significantly increased in T12A* (Fig. 5D). The data indicate the G2/M arrest upon T12A expression and confirm the findings in the cell cycle study.
To have a preliminary understanding of the pathways involved in the cell cycle arrest, we performed a functional protein–protein interaction (PPI) analysis based on HNRNPU/PDCD6IP, CDK1, and the differential proteins categorized in cell cycle progression. Finally, we obtained two functional protein association networks, which show possible pathways for T12A-induced G2/M cell cycle arrest (Fig. 5E and Fig. S10).
Discussion
O-GlcNAcylation occurs on vast numbers of nucleocytoplasmic proteins, but there are only three OGT isoforms. Therefore, OGT should be fine-tuned at all sides to meet the requirements and fulfill critical roles in the cells. Here, we report the regulation mechanisms of sOGT by O-GlcNAcylation.
We found six O-GlcNAc sites on sOGT, and five of these sites are consistent with the previous reports (19, 20), suggesting the reliability and frequency of the modification at these sites, especially at Ser10, Thr12, and Thr38. The inconsistencies at some sites (e.g. Thr18/Ser20) are likely because of the application of different expression systems and/or the use of different fragmentation methods (31).
There is an experimental rule that protein O-GlcNAcylation mostly occurs in a random-coil region with low-molecular-weight amino acids around the modification site (24). With regard to its location and nearby amino acid sequence, Thr12 should be a major O-GlcNAcylation site on sOGT. This was confirmed by extensive mutation analyses in this work. Because all of the mutants related to Thr12, including T12A, T12G, T12L, T12Y, H127A/T12A, and S56A/T12A, significantly decreased their O-GlcNAcylation, Thr12 is undoubtedly a dominant O-GlcNAcylation site on sOGT.
At first sight, we were very surprised by the increased O-GlcNAcylation on S56A in E. coli cells. Then when we found that S56A mutants also increased O-GlcNAcylation in HEK293T cells, either the double-point mutant or the single-point mutant, we began to believe the fact. Simultaneously, another question was raised. How did the S56A mutation increase sOGT O-GlcNAcylation? The following co-IP study showed that relative to H127A, H127A/S56A enhanced binding to WT sOGT. This explains well why the S56A mutants were heavily O-GlcNAcylated.
Actually, S56A mutants not only increase binding to WT sOGT; they also strongly bind to other substrate proteins, such as Nup62 and NeuroD1. Because O-GlcNAc sOGT binds to more proteins than naked sOGT, and S56A is heavily O-GlcNAcylated, we think that the enhanced binding capability of S56A may be attributable to its heavy O-GlcNAcylation, at least in part.
Because Thr12 is the dominant O-GlcNAcylation site on sOGT and S56A mutation increases the modification of sOGT, we hypothesize that there would be an O-GlcNAcylation increase at Thr12 in S56A-related mutants. However, we could not directly check the increase via Western blotting, due to the lack of specific antibodies. The rising O-GlcNAcylation of double-point mutants with S56A mutation, except T12A/S56A, help to confirm that S56A up-regulates sOGT O-GlcNAcylation at T12A.
All of the O-GlcNAcylation sites detected here are within the TPR domain but not the catalytic domain. This explains why O-GlcNAcylation did not affect sOGT activity in the in vitro activity assay but impacted sOGT substrate selectivity in the pulldown study.
Increasing evidence indicates that phosphorylation plays significant roles in regulating OGT activity or substrate selectivity. For instance, insulin treatment of 3T3-L1 adipocytes activates OGT and enhances O-GlcNAcylation on various proteins. Insulin receptor, which shows a tyrosine kinase activity, accounts for the activation of OGT (32). CaMKIV also phosphorylates OGT and enhances its activity in neuroblastoma NG-108-15 cells (33). More detailed studies indicate that GSK3β phosphorylates ncOGT at S3/S4 sites and enhances its activity (28), whereas AMPK phosphorylates ncOGT at Thr-444 and alters its substrate selectivity in several cell lines (29). We find that O-GlcNAcylation slightly affects sOGT phosphorylation at Ser/Thr residues. Therefore, O-GlcNAcylation may also influence sOGT substrate selectivity through phosphorylation.
We have applied WGA, cpOGA-Y298L or RL-2 to enrich O-GlcNAc proteins for subsequent proteomic studies. In WGA enrichment, we pooled more than 1000 proteins for each sample, but <1% of these proteins were previously identified as O-GlcNAc proteins (data not shown). We think that this is likely due to the nonspecific affinity of the WGA beads. Later, we applied cpOGA-Y298L for the enrichment of O-GlcNAc proteins (34), but only a few proteins were detected in MS analysis, probably due to the weak affinity of the cpOGA mutant to O-GlcNAc proteins (data not shown). Last, we performed the enrichment with RL-2. This time, we found more than 100 O-GlcNAc proteins previously reported, suggesting that the IP method with RL-2 antibody is relatively suitable for O-GlcNAc protein enrichment.
The verification of the O-GlcNAcylation of two randomly selected proteins (HNRNPU and PDCD6IP) demonstrates the dependability of the MS data to some extent. It also predicts that sOGT mutants would glycosylate different proteins in HEK293T cells.
It is reported that sOGT could not glycosylate protein substrates in vitro. Here, we found that sOGT could glycosylate various proteins, including Nup62 and NeuroD1. The discrepancy is probably due to the different reaction system. In HEK293T cells, sOGT function might be regulated by specific interacting proteins, such as SP1 (35). In addition, distinct modification of sOGT may also modulate its substrate selectivity.
Both Western blotting analyses and MS data show that WT sOGT, T12A, and S56A glycosylated different proteins in HEK293T cells. GO term analyses predicted that expression of these proteins may affect multiple cellular events, including cell cycle and cell proliferation. This was verified by the following cell proliferation and cell cycle studies. Notably, T12A causes a significant G2/M cell cycle arrest, which is confirmed by the increasing level of pCDK1(Tyr15).
To delineate the pathway in T12A-induced G2/M cell cycle arrest, we performed the PPI analysis. We select PDCD6IP and HNRNPU as the initiation factors, because they are less O-GlcNAcylated in the T12A sample. Moreover, PDCD6IP is a protein relevant to cell apoptosis and G2/M arrest (36–38). HNRNPU also plays some roles in mitosis (39). Differential proteins categorized in cell cycle progression are selected as junction proteins, which may link the initiation factors to the termination factor, PDK1, whose phosphorylation at Tyr15 is a biomarker in G2/M arrest. Because there are no known connections between PDCD6IP and HNRNPU, we obtained two possible functional protein association networks, both with two nexuses: CDC5L and CUL1.
CDC5L is a protein highly expressed in various cancers. Overexpression of CDC5L favors cell cycle progress in HCC cells (40), whereas silence of this protein significantly reduces cell proliferation and leads to a G2/M arrest in osteosarcoma cells (41). We find that CDC5L was slightly decreased in T12A-expressing cells (Fig. S11), which might play a role in G2/M cell cycle arrest and cell proliferative reduction.
CUL1 is a critical part of the Skp1-Cul1-Fbox (SCF) E3 ligase complex, which controls the amount of p27 and p21 through ubiquitination and proteasomal degradation (42). p21 and p27 are two members of the family of CDK inhibitors (43), which can inhibit the function of cyclin-CDK complex in G1 phase and lead to G1 phase cell cycle arrest (44, 45). The MS data (Sheet S2) showed a much higher intensity-based absolute quantification (iBAQ) value of CUL1 (29,140:0) in a T12A sample relative to WT sOGT. With regard to the protective role of O-GlcNAcylation against protein (e.g. c-Myc) degradation (46), there would be more CUL1 protein in the T12A sample, which may degrade more p27 and/or p21 (42) and eventually promote the cell transition from G1 to S phase. This may explain the decreased cell ratio at G1 phase in T12A-expressing cells. But further studies are still required to elucidate the role of the junction proteins and characterize the pathway.
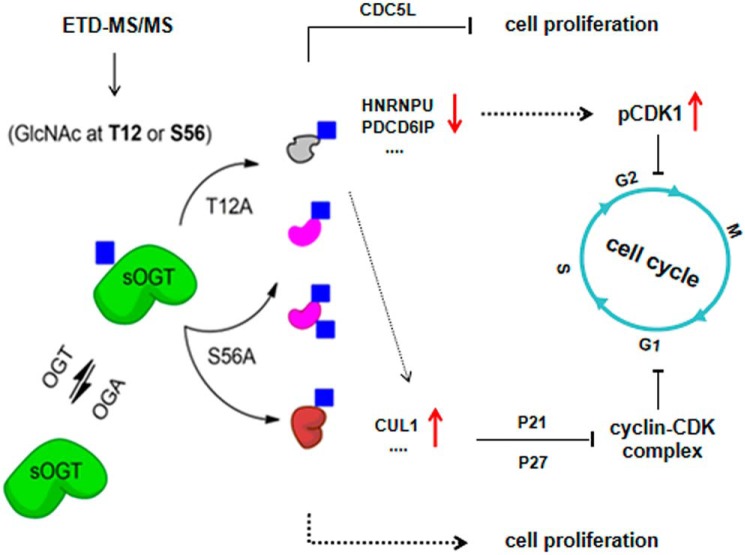
In conclusion, we have found two important O-GlcNAc sites on sOGT: Thr12 and Ser56. Thr12 is a dominant O-GlcNAcylation site on sOGT, whereas the modification of Ser56 plays a role in regulating sOGT O-GlcNAcylation, partly through Thr12. O-GlcNAcylation of sOGT modulates its substrate selectivity, which ultimately affects cell cycle process and/or cell proliferation. Expression of T12A would cause O-GlcNAc decline on specific proteins, such as HNRNPU and PDCD6IP, and change the protein level of some nexus proteins (e.g. CDC5L or CUL1), subsequently leading to elevated pCDK1(Tyr15), G2/M cell cycle arrest, and cell proliferative reduction (Fig. 6). We also find that O-GlcNAcylation of sOGT slightly influences its phosphorylation. Our results provide new insights into the regulation mechanisms of sOGT in the cell. O-GlcNAcylation and phosphorylation may cooperate to elaborately manipulate OGT function and control various cellular events.
Figure 6.
Hypothesis of functional sOGT regulation by O-GlcNAcylation. sOGT is O-GlcNAcylated at Thr12 and/or Ser56, which affects sOGT substrate selectivity. Expression of T12A results in less O-GlcNAcylation of HNRNPU and PDCD6IP as well as low abundance of CDC5L and, hence, reduces cell proliferation and causes G2/M cell cycle arrest. The O-GlcNAcylation loss would also elevate CUL1 abundance, hence promoting G1-to-S-phase transition. Expression of S56A enhances O-GlcNAcylation of HNRNPU and PDCD6IP, which may cause cell proliferation through unknown mechanisms.
Materials and methods
Reagents and antibodies
All of the peptides in Tables 1 and 2 (purity ≥95%) were synthesized by GL Biochem (Shanghai, China). UDP-GlcNAc (U4375) was obtained from Sigma. Thiamet-G (1009816-48-1) was purchased from MedChemExpress. Protein A/G PLUS-agarose (sc-2003) was purchased from Santa Cruz Biotechnology, Inc. GFP-Trap (gta-20) was purchased from ChromoTek. All antibodies used here were purchased from Abcam, Cell Signaling Technology, or Proteintech Group (Table S1).
Preparation of naked sOGT/O-GlcNAc sOGT
Human sOGT was expressed in E. coli BL21 (DE3) cells and purified by the one-step nickel chelating affinity method (47). sOGT protein was then separated into two parts through WGA chromatography (vector, AL-1023) (48). O-GlcNAc sOGT was eluted with 300 mm GlcNAc. The naked sOGT (flow-through) and O-GlcNAc sOGT were applied for buffer change using a 30-kDa cutoff filter (Millipore) and hence kept for the next step.
In vitro activity assays
In vitro activity assays were performed as described previously (24, 47), using variant peptide substrates: FITC-YAVVPVSK, YAVVPVSK, or CK3k (KKKYPGGSTPVSSANMM). The reaction system contained a mixture of 40 μg of sOGT, 2 mm peptide, 500 μm or variant concentrations (for the enzyme kinetics study) of UDP-GlcNAc in 100 μl of reaction buffer. The reaction was performed at 37 °C for 45 min. To identify the O-GlcNAc peptides, the product purified through HPLC was lyophilized and determined on an FTICR-MS instrument (Varian 7.0T FTMS). The O-GlcNAc modification was confirmed by the mass increase of 203 Da compared with substrate peptides. The yields of O-GlcNAc peptides was calculated based on the integrated areas of product and substrate peptides on HPLC spectra. Apparent kinetic parameters were obtained by fitting the data into the Michaelis–Menten equation using GraphPad Prism version 5.
Cell proliferation and cell cycle assays
For cell proliferation assays, ∼2000 HEK293T cells were seeded in 96-well plates with six parallel wells for each group. All of the cells were transfected with the indicated plasmids on day zero, and the cell viability was measured every 24 h using a cell-counting kit (CCK-8) (Sigma-Aldrich, catalog no. 96992) from day 1 up to day 7.
For cell cycle assays, HEK293T cells were seeded in 6-well plates in triplicate and transfected with the indicated plasmids. 72 h after the transfection, cells were harvested and fixed in chilled 70% ethanol overnight at −20 °C. The cells were stained with propidium iodide (100 μg/ml) in the dark for 30 min at room temperature and analyzed with FACS using a Guava apparatus from Millipore (Molsheim, France).
Pulldown assay
An equal amount (20 μg) of naked sOGT/O-GlcNAc sOGT was added to 500 μg of HEK293T cell lysates supplemented with 5 μg of anti-His antibody. The mixture was mixed overnight on a rotator at 4 °C. 50 μl of slurry of PierceTM Protein A/G Magnetic Beads was then added into the mixture and mixed in a rotator for 1 h at room temperature. After two washes with PBS, the enriched proteins were boiled in 30 μl of 2× SDS-PAGE loading buffer for 5 min, and the supernatant was used for SDS-PAGE analysis.
IP and Co-IP
HEK293T cells expressing the indicated proteins were normally cultured for 48 h and were lysed using a mammalian cell lysis kit (Beyotime, Nanjing, China). After a centrifugation at 15,000 × g for 30 min at 4 °C, the supernatant protein was quantified with a BCA protein assay kit (Beyotime) and subsequently applied for IP or co-IP analysis.
IP was performed to pool target proteins. 3 μg of antibody was used to enrich target proteins from 500 μg of cell lysates, and the solution was mixed overnight on a rotator at 4 °C. 50 μl of slurry of PierceTM Protein A/G Magnetic Beads was added into the mixture and mixed in a rotator for 1 h at room temperature. For GFP-fusion sOGT immunoprecipitation, 30 μl of slurry of GFP-trap beads was used to bind sOGT in 500 μg of cell lysates. After two washes with PBST to remove nonspecific binding proteins, the enriched protein was boiled in 60 μl of 2× SDS-PAGE loading buffer for 5 min, and the supernatant was subjected to Western blot analysis.
Co-IP was performed to test protein interaction in vivo. Unlike in the IP process, the removal of nonspecific binding proteins in co-IP was achieved by two washes with PBS.
SDS-PAGE analysis and Western blotting
SDS-PAGE analysis was performed on 12% SDS-polyacrylamide gels with a constant running for 1 h at 300 mA. The gel was visualized by Coomassie Blue staining.
For Western blotting, protein samples were resolved on 12% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes. The membranes were blocked with blocking buffer (TBS + 0.05% Tween + 5% nonfat milk powder) and incubated overnight with primary antibodies at 4 °C. After three washes with washing buffer (TBS + 0.05% Tween), the membranes were incubated with their respective secondary antibodies at room temperature for 1 h. After another three washes, the blots were visualized using an enhanced chemiluminescence detection system (Millipore) on a ChemiDocXRS (Bio-Rad).
Site-mapping of O-GlcNAcylation on sOGT
O-GlcNAc sOGT was purified with SDS-PAGE and digested in-gel with trypsin. The resultant peptides were fractionated with an EASY-Spray source and Nano-LC UltiMate 3000 HPLC system equipped with EASY-Spray PepMap C18 columns (15 cm; particle size, 3 μm; pore size, 100 Å; Thermo Fisher Scientific) and analyzed on an LTQ-Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). HPLC separation was achieved with a linear gradient from 3 to 40% buffer B for 30 min at a flow rate of 300 nl/min (mobile phase A: 1.95% acetonitrile (ACN), 97.95% H2O, 0.1% formic acid (FA); mobile phase B: 79.95% ACN, 19.95% H2O, 0.1% FA). The mass spectrometer was operated in data-dependent mode. A full-scan survey MS experiment (m/z range from 375 to 1600; automatic gain control target, 1,000,000 ions; resolution at 400 m/z, 60,000; maximum ion accumulation time, 50 ms) was acquired with an Orbitrap mass spectrometer, and the 10 most intense precursor ions were each fragmented by CID and ETD. The CID fragmented ion spectra were acquired using an ion trap analyzer (automatic gain control target, 10,000 ions; maximum ion accumulation time, 100 ms). The ETD fragmentation ion spectra were also acquired using the ion trap analyzer, and the activation time of ETD was set to 100 ms. The MS2 scanning model was set to the centroid mode. The other conditions included a capillary temperature of 200 °C and an S-lens RF level of ∼60%.
The raw file was identified using the pFind 2.1 software to search the database and its reversed database. The protein sequence database was downloaded from Uniprot_swissprot plus Uniprot_TrEMBL (release 2012-04, 65,493 entries), and the following modifications were defined: static modification of carbamidomethyl (Cys); dynamic modification of GlcNAc 203.079 Da (Ser and Thr), deamination (Asn), oxidation (Met), and acetylation (Lys). Trypsin was selected as the enzyme, and two missed cleavages were allowed. The mass tolerance of the precursor ion was set to 20 ppm, and the fragmentation ions were set to 0.5 Da. A false discovery rate of 1% was estimated and applied to all data sets at the total peptide level (49).
Proteomic analysis of O-GlcNAc proteins
OGT-shRNA2 (50) and shRNA-resistant sOGT mutants were co-expressed in HEK293T cells for 48 h in duplicate. The cells were treated with 2 μm TMG (OGT inhibitor) for 12 h before the harvest. After cell disruption, 1 mg of cell lysates was pretreated with 50 μl of protein A/G beads. O-GlcNAc proteins were pooled via immunoprecipitation using RL2 antibody, followed with Western blot analysis to confirm the consistency of the duplicate samples. O-GlcNAc proteins in duplicate samples were combined and further purified by running SDS-PAGE.
O-GlcNAc proteins were digested in-gel with trypsin, the resultant peptides were fractionated using an online nanoflow LC and analyzed with an electrospray ionization MS system (EASY-nLC Orbitrap Fusion Lumos). The sample was separated by a 15-cm-long, 150-μm inner diameter analytical column packed with reversed-phase 1.9-μm C18 material. The peptide separation was achieved over a 78-min gradient (buffer A: 0.1% FA in water; buffer B: 0.1% FA in ACN) at a flow rate of 700 nl/min (0–8 min, 5–8% B; 8–58 min, 8–22% B; 58–70 min, 22–32% B; 70–71 min, 32–90% B; and 71–78 min, 90% B). The MS ion source was operated at 2.1 kV. For full MS survey scans, the AGC target was 5e5, and the scan range was from m/z 300 to 2000 with a resolution of 120,000. The instrument was run in the top-speed mode with a cycle time of 3 s. The HCD fragmentation was performed at a normalized collision energy of 32%. The MS2 AGC target was set to 2e5 with a maximum injection time of 50 ms, resolution of 30,000, and 12-s dynamic exclusion.
The raw data were searched against the human UniProt database (release 2014-09, 20,193 entries) using MaxQuant software (version 1.5.3.8) as described previously (51). To explore the O-HexNAc–modified glycopeptides, the MS data files were searched against the human UniProt database (release 2014-09, 20,193 entries) using Byonic (version 2.10.21, Protein Metrics, Inc.). For glycopeptides, a Byonic score of 300 was considered a good, which reflects the absolute quality of the peptide-spectrum match. To rank the relative abundance of different proteins, we used an iBAQ approach (52).
Quantitative pathway analysis on the enriched proteins
Each iBAQ value was normalized, followed with the blank (H127A) subtraction. The subtraction values (>0) were used to calculate differential proteins in two comparison samples (T12A compared with WT or S56A compared with WT). For each comparison, the protein with a -fold change > 1.5 was considered to be a differential protein. The differential proteins were used for GO enrichment analyses online. GO associations were made based on NCBI gene2go and the GO consortium's OBO, and the enrichment of specific GO terms was identified using a hypergeometry test. Pathway assignment was based on the KEGG data set (release 53.0).
PPI network generation and module analysis
Functional PPI analysis is critical to explain the molecular mechanisms of key cellular activities. In this study, we searched the interacting gene database (STRING) to obtain PPIs for 42 genes. The target hub genes used here had to meet the following two criteria: (i) they were genes relevant to cell cycle progression, or (ii) they were found in differential proteins in the proteomic analysis.
Statistics
The statistical significance of differences between groups was assessed using the OriginPro (version 9.0) or GraphPad Prism version 5 software. A two-sample t test was used to compare parameters between groups in cell proliferation and cell cycle studies. The level of significance was set at p < 0.05.
Other procedures
Detailed methods for plasmid construction, protein preparation, knockdown of OGT, and in-gel digestion of proteins are described in the supporting information.
Author contributions
L. Liu, L. Li, and C. M performed all experiments except as otherwise indicated. Y. Z., F. T., and W. Y. performed proteomic analyses. C. L. performed the functional protein–protein interaction (PPI) analysis, Y. S. and Z. X. participated in plasmid construction and the in vitro activity assay. Y. G. and J. Z. participated in cell cycle analyses. P. G. W. participated in the design of the work. L. Z. designed the work and prepared the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Dong Li (Beijing Institute of Radiation Medicine) for help with bioinformatics analyses and Dr. Wen Yi (Zhejiang University) for the supply of OGT-shRNA.
This work was supported by National Natural Science Foundation of China Grant 31470795, Tianjin Municipal Science and Technology Commission Grant 15JCYBJC24100, and “Fundamental Research Funds for the Central Universities,” Nankai University, Grant 63191148. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Sheets S1–S6, Table S1, and Figs. S1–S11.
Mass spectrometry proteomics data were deposited into the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD009731.
- O-GlcNAcylation
- O-linked-β-GlcNAc modification
- OGT
- O-GlcNAc transferase
- mOGT
- mitochondrial OGT
- sOGT
- short-form OGT
- ncOGT
- nucleocytoplasmic OGT
- TPR
- tetratricopeptide repeat
- HCD
- higher-energy collisional dissociation
- aa
- amino acids
- WGA
- wheat germ agglutinin
- ETD
- electron-transfer dissociation
- IP
- immunoprecipitation
- GO
- gene ontology
- PPI
- protein–protein interaction
- PDCD6IP
- programmed cell death 6–interacting protein
- CDK
- cyclin-dependent kinase
- ACN
- acetonitrile
- FA
- formic acid
- CID
- collision-induced dissociation
- iBAQ
- intensity-based absolute quantification
- AGC
- automatic gain control.
References
- 1. Hart G. W., Housley M. P., and Slawson C. (2007) Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446, 1017–1022 10.1038/nature05815 [DOI] [PubMed] [Google Scholar]
- 2. Yang W. H., Park S. Y., Nam H. W., Kim D. H., Kang J. G., Kang E. S., Kim Y. S., Lee H. C., Kim K. S., and Cho J. W. (2008) NFκB activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc. Natl. Acad. Sci. U.S.A. 105, 17345–17350 10.1073/pnas.0806198105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hanover J. A., Yu S., Lubas W. B., Shin S. H., Ragano-Caracciola M., Kochran J., and Love D. C. (2003) Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch. Biochem. Biophys. 409, 287–297 10.1016/S0003-9861(02)00578-7 [DOI] [PubMed] [Google Scholar]
- 4. Kreppel L. K., and Hart G. W. (1999) Regulation of a cytosolic and nuclear O-GlcNAc transferase: role of the tetratricopeptide repeats. J. Biol. Chem. 274, 32015–32022 10.1074/jbc.274.45.32015 [DOI] [PubMed] [Google Scholar]
- 5. Iyer S. P., and Hart G. W. (2003) Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J. Biol. Chem. 278, 24608–24616 10.1074/jbc.M300036200 [DOI] [PubMed] [Google Scholar]
- 6. Lubas W. A., and Hanover J. A. (2000) Functional expression of O-linked GlcNAc transferase: domain structure and substrate specificity. J. Biol. Chem. 275, 10983–10988 10.1074/jbc.275.15.10983 [DOI] [PubMed] [Google Scholar]
- 7. Lazarus B. D., Love D. C., and Hanover J. A. (2006) Recombinant O-GlcNAc transferase isoforms: identification of O-GlcNAcase, yes tyrosine kinase, and tau as isoform-specific substrates. Glycobiology 16, 415–421 10.1093/glycob/cwj078 [DOI] [PubMed] [Google Scholar]
- 8. Iyer S. P. N., Akimoto Y., and Hart G. W. (2003) Identification and cloning of a novel family of coiled-coil domain proteins that interact with O-GlcNAc transferase. J. Biol. Chem. 278, 5399–5409 10.1074/jbc.M209384200 [DOI] [PubMed] [Google Scholar]
- 9. Seo H. G., Kim H. B., Kang M. J., Ryum J. H., Yi E. C., and Cho J. W. (2016) Identification of the nuclear localisation signal of O-GlcNAc transferase and its nuclear import regulation. Sci. Rep. 6, 34614 10.1038/srep34614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang F., Su K., Yang X., Bowe D. B., Paterson A. J., and Kudlow J. E. (2003) O-GIcNAc modification is an endogenous inhibitor of the proteasome. Cell 115, 715–725 10.1016/S0092-8674(03)00974-7 [DOI] [PubMed] [Google Scholar]
- 11. Noach N., Segev Y., Levi I., Segal S., and Priel E. (2007) Modification of topoisomerase I activity by glucose and by O-GlcNAcylation of the enzyme protein. Glycobiology 17, 1357–1364 10.1093/glycob/cwm105 [DOI] [PubMed] [Google Scholar]
- 12. Roos M. D., Su K., Baker J. R., and Kudlow J. E. (1997) O-Glycosylation of an Sp1-derived peptide blocks known Sp1 protein interactions. Mol. Cell Biol. 17, 6472–6480 10.1128/MCB.17.11.6472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang X., Su K., Roos M. D., Chang Q., Paterson A. J., and Kudlow J. E. (2001) O-Linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc. Natl. Acad. Sci. U.S.A. 98, 6611–6616 10.1073/pnas.111099998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lim K., and Chang H. I. (2010) O-GlcNAc inhibits interaction between Sp1 and sterol regulatory element binding protein 2. Biochem. Biophys. Res. Commun. 393, 314–318 10.1016/j.bbrc.2010.01.128 [DOI] [PubMed] [Google Scholar]
- 15. Ruan H. B., Han X., Li M. D., Singh J. P., Qian K., Azarhoush S., Zhao L., Bennett A. M., Samuel V. T., Wu J., Yates J. R. 3rd, Yang X. (2012) O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability. Cell Metab. 16, 226–237 10.1016/j.cmet.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Charoensuksai P., Kuhn P., Wang L., Sherer N., and Xu W. (2015) O-GlcNAcylation of co-activator-associated arginine methyltransferase 1 regulates its protein substrate specificity. Biochem. J. 466, 587–599 10.1042/BJ20141072 [DOI] [PubMed] [Google Scholar]
- 17. Kreppel L. K., Blomberg M. A., and Hart G. W. (1997) Dynamic glycosylation of nuclear and cytosolic proteins: cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315 10.1074/jbc.272.14.9308 [DOI] [PubMed] [Google Scholar]
- 18. Tai H. C., Khidekel N., Ficarro S. B., Peters E. C., and Hsieh-Wilson L. C. (2004) Parallel identification of O-GlcNAc-modified proteins from cell lysates. J. Am. Chem. Soc. 126, 10500–10501 10.1021/ja047872b [DOI] [PubMed] [Google Scholar]
- 19. Griffin M. E., Jensen E. H., Mason D. E., Jenkins C. L., Stone S. E., Peters E. C., and Hsieh-Wilson L. C. (2016) Comprehensive mapping of O-GlcNAc modification sites using a chemically cleavable tag. Mol. Biosyst. 12, 1756–1759 10.1039/C6MB00138F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fan Q., Moen A., Anonsen J. H., Bindesbøll C., Sæther T., Carlson C. R., and Grønning-Wang L. M. (2018) O-GlcNAc site-mapping of liver X receptor-α and O-GlcNAc transferase. Biochem. Biophys. Res. Commun. 499, 354–360 10.1016/j.bbrc.2018.03.164 [DOI] [PubMed] [Google Scholar]
- 21. Liu Y., Li X. J., Yu Y., Shi J., Liang Z., Run X., Li Y., Dai C. L., Grundke-Iqbal I., Iqbal K., Liu F., and Gong C. X. (2012) Developmental regulation of protein O-GlcNAcylation, O-GlcNAc transferase, and O-GlcNAcase in mammalian brain. PLoS One 7, e43724 10.1371/journal.pone.0043724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feng Z., Hui Y., Ling L., Xiaoyan L., Yuqiu W., Peng W., and Lianwen Z. (2013) FBXW10 is negatively regulated in transcription and expression level by protein O-GlcNAcylation. Biochem. Biophys. Res. Commun. 438, 427–432 10.1016/j.bbrc.2013.07.091 [DOI] [PubMed] [Google Scholar]
- 23. Lazarus M. B., Nam Y., Jiang J., Sliz P., and Walker S. (2011) Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469, 564–567 10.1038/nature09638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu X., Li L., Wang Y., Yan H., Ma X., Wang P. G., and Zhang L. (2014) A peptide panel investigation reveals the acceptor specificity of O-GlcNAc transferase. FASEB J. 28, 3362–3372 10.1096/fj.13-246850 [DOI] [PubMed] [Google Scholar]
- 25. Andrali S. S., Qian Q., and Ozcan S. (2007) Glucose mediates the translocation of NeuroD1 by O-linked glycosylation. J. Biol. Chem. 282, 15589–15596 10.1074/jbc.M701762200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miller M. W., Caracciolo M. R., Berlin W. K., and Hanover J. A. (1999) Phosphorylation and glycosylation of nucleoporins. Arch. Biochem. Biophys. 367, 51–60 10.1006/abbi.1999.1237 [DOI] [PubMed] [Google Scholar]
- 27. Mizuguchi-Hata C., Ogawa Y., Oka M., and Yoneda Y. (2013) Quantitative regulation of nuclear pore complex proteins by O-GlcNAcylation. Biochim. Biophys. Acta 1833, 2682–2689 10.1016/j.bbamcr.2013.06.008 [DOI] [PubMed] [Google Scholar]
- 28. Kaasik K., Kivimäe S., Allen J. J., Chalkley R. J., Huang Y., Baer K., Kissel H., Burlingame A. L., Shokat K. M., Ptáček L. J., and Fu Y. H. (2013) Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 17, 291–302 10.1016/j.cmet.2012.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bullen J. W., Balsbaugh J. L., Chanda D., Shabanowitz J., Hunt D. F., Neumann D., and Hart G. W. (2014) Cross-talk between two essential nutrient-sensitive enzymes: O-GlcNAc transferase (OGT) and AMP-activated protein kinase (AMPK). J. Biol. Chem. 289, 10592–10606 10.1074/jbc.M113.523068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ayeni J. O., and Campbell S. D. (2014) “Ready, Set, Go”: checkpoint regulation by Cdk1 inhibitory phosphorylation. Fly 8, 140–147 10.4161/19336934.2014.969147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chalkley R. J., Thalhammer A., Schoepfer R., and Burlingame A. L. (2009) Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc. Natl. Acad. Sci. U.S.A. 106, 8894–8899 10.1073/pnas.0900288106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Whelan S. A., Lane M. D., and Hart G. W. (2008) Regulation of the O-linked β-N-acetylglucosamine transferase by insulin signaling. J. Biol. Chem. 283, 21411–21417 10.1074/jbc.M800677200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Song M., Kim H. S., Park J. M., Kim S. H., Kim I. H., Ryu S. H., and Suh P. G. (2008) O-GlcNAc transferase is activated by CaMKIV-dependent phosphorylation under potassium chloride-induced depolarization in NG-108-15 cells. Cell. Signal. 20, 94–104 10.1016/j.cellsig.2007.09.002 [DOI] [PubMed] [Google Scholar]
- 34. Selvan N., Williamson R., Mariappa D., Campbell D. G., Gourlay R., Ferenbach A. T., Aristotelous T., Hopkins-Navratilova I., Trost M., and van Aalten D. M. F. (2017) A mutant O-GlcNAcase enriches Drosophila developmental regulators. Nat. Chem. Biol. 13, 882–887 10.1038/nchembio.2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Riu I. H., Shin I. S., and Do S. I. (2008) Sp1 modulates ncOGT activity to alter target recognition and enhanced thermotolerance in E. coli. Biochem. Biophys. Res. Commun. 372, 203–209 10.1016/j.bbrc.2008.05.034 [DOI] [PubMed] [Google Scholar]
- 36. Vito P., Pellegrini L., Guiet C., and D'Adamio L. (1999) Cloning of AIP1, a novel protein that associates with the apoptosis-linked gene ALG-2 in a Ca2+-dependent reaction. J. Biol. Chem. 274, 1533–1540 10.1074/jbc.274.3.1533 [DOI] [PubMed] [Google Scholar]
- 37. Yu Q., Zhou C., Wang J., Chen L., Zheng S., and Zhang J. (2013) A functional insertion/deletion polymorphism in the promoter of PDCD6IP is associated with the susceptibility of hepatocellular carcinoma in a Chinese population. DNA Cell Biol. 32, 451–457 10.1089/dna.2013.2061 [DOI] [PubMed] [Google Scholar]
- 38. Zhan Q., Tsai S., Lu Y., Wang C., Kwan Y., and Ngai S. (2013) RuvBL2 is involved in histone deacetylase inhibitor PCI-24781-induced cell death in SK-N-DZ neuroblastoma cells. PLoS One 8, e71663 10.1371/journal.pone.0071663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang L., Song D., Zhu B., and Wang X. (2019) The role of nuclear matrix protein HNRNPU in maintaining the architecture of 3D genome. Semin. Cell Dev. Biol. 90, 161–167 10.1016/j.semcdb.2018.07.006 [DOI] [PubMed] [Google Scholar]
- 40. Qiu H., Zhang X., Ni W., Shi W., Fan H., Xu J., Chen Y., Ni R., and Tao T. (2016) Expression and clinical role of Cdc5L as a novel cell cycle protein in hepatocellular carcinoma. Digest. Dis. Sci. 61, 795–805 10.1007/s10620-015-3937-9 [DOI] [PubMed] [Google Scholar]
- 41. Wang Y., Chang H., Gao D., Wang L., Jiang N., and Yu B. (2016) CDC5L contributes to malignant cell proliferation in human osteosarcoma via cell cycle regulation. Int. J. Clin. Exp. Pathol. 9, 10451–10457 [Google Scholar]
- 42. Kulinski M., Achkar I. W., Haris M., Dermime S., Mohammad R. M., and Uddin S. (2018) Dysregulated expression of SKP2 and its role in hematological malignancies. Leukemia Lymphoma 59, 1051–1063 10.1080/10428194.2017.1359740 [DOI] [PubMed] [Google Scholar]
- 43. Garcia-Gutierrez L., Delgado M. D., and Leon J. (2019) MYC oncogene contributions to release of cell cycle brakes. Genes (Basel) 10, 244 10.3390/genes10030244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Abbastabar M., Kheyrollah M., Azizian K., Bagherlou N., Tehrani S. S., Maniati M., and Karimian A. (2018) Multiple functions of p27 in cell cycle, apoptosis, epigenetic modification and transcriptional regulation for the control of cell growth: a double-edged sword protein. DNA Repair (Amst.) 69, 63–72 10.1016/j.dnarep.2018.07.008 [DOI] [PubMed] [Google Scholar]
- 45. Srivastava R. K., Chen Q., Siddiqui I., Sarva K., and Shankar S. (2007) Linkage of curcumin-induced cell cycle arrest and apoptosis by cyclin-dependent kinase inhibitor p21(/WAF1/CIP1). Cell Cycle 6, 2953–2961 10.4161/cc.6.23.4951 [DOI] [PubMed] [Google Scholar]
- 46. Luanpitpong S., Angsutararux P., Samart P., Chanthra N., Chanvorachote P., and Issaragrisil S. (2017) Hyper-O-nGlcNAcylation induces cisplatin resistance via regulation of p53 and c-Myc in human lung carcinoma. Sci. Rep. 7, 10607 10.1038/s41598-017-10886-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ma X., Liu P., Yan H., Sun H., Liu X., Zhou F., Li L., Chen Y., Muthana M. M., Chen X., Wang P. G., and Zhang L. (2013) Substrate specificity provides insights into the sugar donor recognition mechanism of O-GlcNAc transferase (OGT). PLoS One 8, e63452 10.1371/journal.pone.0063452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gao Y., Miyazaki J., and Hart G. W. (2003) The transcription factor PDX-1 is post-translationally modified by O-linked N-acetylglucosamine and this modification is correlated with its DNA binding activity and insulin secretion in min6 beta-cells. Arch. Biochem. Biophys. 415, 155–163 10.1016/S0003-9861(03)00234-0 [DOI] [PubMed] [Google Scholar]
- 49. Wang L. H., Li D. Q., Fu Y., Wang H. P., Zhang J. F., Yuan Z. F., Sun R. X., Zeng R., He S. M., and Gao W. (2007) pFind 2.0: a software package for peptide and protein identification via tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21, 2985–2991 10.1002/rcm.3173 [DOI] [PubMed] [Google Scholar]
- 50. Caldwell S. A., Jackson S. R., Shahriari K. S., Lynch T. P., Sethi G., Walker S., Vosseller K., and Reginato M. J. (2010) Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene 29, 2831–2842 10.1038/onc.2010.41 [DOI] [PubMed] [Google Scholar]
- 51. Zhang Y., Xie X., Zhao X., Tian F., Lv J., Ying W., and Qian X. (2018) Systems analysis of singly and multiply O-glycosylated peptides in the human serum glycoproteome via EThcD and HCD mass spectrometry. J. Proteomics 170, 14–27 10.1016/j.jprot.2017.09.014 [DOI] [PubMed] [Google Scholar]
- 52. Smits A. H., Jansen P. W. T. C., Poser I., Hyman A. A., and Vermeulen M. (2013) Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 41, e28 10.1093/nar/gks941 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.