Abstract
We report the results of residue-residue contact prediction of a new pipeline built purely on the learning of coevolutionary features in the CASP13 experiment. For a query sequence, the pipeline starts with the collection of multiple sequence alignments (MSAs) from multiple genome and metagenome sequence databases using two complementary HMM-based searching tools. Three profile matrices, built on covariance, precision, and pseudolikelihood maximization respectively, are then created from the MSAs, which are used as the input features of a deep residual convolutional neural network architecture for contact-map training and prediction. Two ensembling strategies have been proposed to integrate the matrix features through end-to-end training and stacking, resulting in two complementary programs called TripletRes and ResTriplet, respectively. For the 31 free-modeling (FM) domains that do not have homologous templates in the PDB, TripletRes and ResTriplet generated comparable results with an average accuracy of 0.640 and 0.646, respectively, for the top L/5 long-range predictions, where 71% and 74% of the cases have an accuracy above 0.5. Detailed data analyses showed that the strength of the pipeline is due to the sensitive MSA construction and the advanced strategies for coevolutionary feature ensembling. Domain splitting was also found to help enhance the contact prediction performance. Nevertheless, contact models for tail regions, which often involve a high number of alignment gaps, and for targets with few homologous sequences are still suboptimal. Development of new approaches where the model is specifically trained on these regions and targets might help address these problems.
Keywords: CASP; contact-map prediction; coevolution analysis; deep learning, protein folding
Introduction
For nearly five decades, the success of computational structure prediction has been limited to proteins with homologous templates from solved experimental structures1–3. Significant progress has been recently witnessed on ab initio 3D structure prediction4–7, which is mainly due to the success of sequence-based contact predictions8,9. Due to its significant importance to protein structure prediction, the problem of contact-map prediction has drawn an increasing amount of attention, and as a result, several new approaches have been proposed within the last decade10–21.
Initial studies on protein contact prediction focused on the analysis of the marginal correlation between two positions in the multiple sequence alignment (MSA)22,23, the idea of which is attractive, but the implementations often introduce transitional noise to the predicted contact-map. In other words, if both positions A and B are coupled to position C, the marginal correlation based analysis will report superficial coupling between positions A and B as well. Direct coupling analysis (DCA) methods (e.g., mfDCA10, PSICOV11, CCMpred12, and GREMLIN13), were subsequently proposed to address this problem of transitional noise by excluding effects from other positions. However, for proteins with few sequence homologs, these coevolutionary methods could fail due to the fact that the parameters of the inverse Potts model used in most DCA methods cannot be accurately estimated by limited number of samples. Supervised machine learning based methods, such as MetaPSICOV14,15 and NeBcon16, predict contact-maps by combining features based on the final results of various coevolutionary methods with a variety of one-dimensional sequence properties. These methods outperform the pure DCA methods, especially for proteins with a limited number of sequence homologs. Most recently, deep neural network based contact-map predictors which formulate contactmap prediction as a pixel-level classification problem, have achieved great success17–19. However, there is still room for further improvements, especially in terms of feature representation. It is observed that most machine learning methods use the post-processed scores of coevolutionary analysis methods as features; as a result, there could possibly be information loss in this post-processing step.
Unlike many other machine learning predictors, the recently developed DeepCov20 directly uses the raw sequence covariance matrix as its only feature, followed by convolutional neural networks to predict the contact-map; it achieved comparable results to predictors based on features of postprocessed coevolutionary analysis. Alternatively, ResPRE21 considers the ridge estimation of the inverse of the covariance matrix, which was shown to be capable of wiping out noisy signal from translational interactions; when coupled with a fully residual neural network structure24, the approach demonstrated superiority to the state-of-the-art of other approaches. Here, we further extend this approach during CASP13, where two methods, TripletRes and ResTriplet, are proposed to ensemble a triplet of raw coevolutionary features, including the covariance matrix, the precision matrix, and the parameter matrix of a pseudolikelihood maximized Potts model25,26, by two complementary strategies, based on end-to-end training and stacking.
In this article, we report the results of TripletRes and ResTriplet in the contact prediction section of the CASP13 experiment. Careful analyses will be performed to investigate the strengths and weaknesses of the different components of the pipelines, with particular focuses on the hard free-modeling (FM) targets that lack homologs from the structure and sequence databases. We will also highlight the challenges identified from the CASP experiment to our methods and possible strategies to address these issues.
Materials and methods
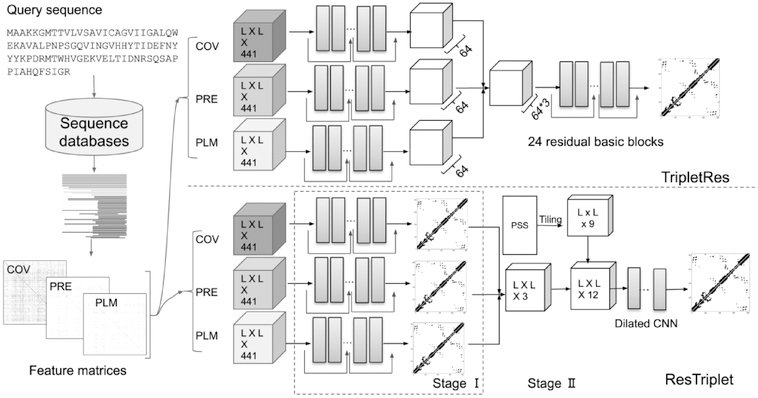
The overall pipelines for TripletRes and ResTriplet are shown in Figure 1. For a given query sequence, a multiple sequence alignment is generated by incrementally searching against multiple sequence databases using DeepMSA27. Three coevolutionary matrix features are then extracted based on the obtained MSA, including the covariance matrix (COV), the precision matrix (PRE), and the coupling parameters of the Potts model by pseudolikelihood maximization (PLM). Two different strategies have been used to integrate coevolutionary features in TripletRes and ResTriplet respectively. In TripletRes, all features are fused directly by neural networks, where all networks are trained end-to-end. In ResTriplet, a two-stage strategy is performed, in which it first learns three individual contact-map predictors from the three feature matrices, and then uses stacking to ensemble the contact-maps from the predictors with secondary structure predictions for the final contact prediction. Here, ResTriplet trains the models in the first stage and the second stage separately. Below we explain the pipelines in more detail.
Figure 1.
The pipeline of TripletRes and ResTriplet for contact-map prediction in CASP13.
Multiple sequence alignment collection
Multiple sequence alignments are critical elements for contact-map prediction based on coevolutionary analysis. DeepMSA is used to generate MSAs from three sequence databases and consists of three steps27. First, HHblits28 is used to search against UniClust3029 for three iterations with a minimal coverage equal to 50%. We will proceed to Step 2 if DeepMSA doesn’t provide enough sequences (i.e., Nf<128) in Step 1. Here, Nf is the normalized number of effective sequences which is calculated by:
| (1) |
where if the sequence identity between m-th sequence and n-th sequence in the MSA is over 0.8, otherwise . In Step 2, Jackhmmer30 is used to search the query sequence against UniRef90 for three iterations with an E-value cutoff of 10. Instead of directly using the sequence alignments obtained by Jackhmmer, the “hhblitdb.pl” script from HH-suite31,32 is used to construct a custom database from the Jackhmmer hits for further HHblits searching. If the Nf of the MSA in Step 2 still lower than 128, Step 3 will be performed, in which HMMbuild from the HMMER package30 is used to search against the Metaclust33 metagenome sequence database with parameters “-E 10 --incE 1e-3”. A custom HHblits database is built from the hits, similar to Step 2. In both Steps 2 and 3, The MSA from the previous steps is used to jump-start an HHblits search against the custom databases. The final MSA obtained thereof can be very large, which would result in long runtimes for the PLM feature calculation. Therefore, an additional filter for low coverage sequences is applied for MSAs with an Nf>128, where we remove sequence homologs with coverage <60% if the resulting MSA retains an Nf>128. We may additionally remove sequence homologs with coverage <75% if the resulting MSA still has an Nf>128.
Three evolutionary matrix features extracted from MSA
There are three coevolutionary features used by our methods. The first one, COV, is the covariance matrix as proposed by DeepCov20. Considering an MSA with N rows and L columns, we can compute a 21∙L by 21∙L sample covariance matrix as follows:
| (2) |
where fi,j(a,b) is the observed relative frequency of residue pair a and b at position i and j and fi(a) is the frequency of occurrence of a residue type a at position i. Each entry of the covariance matrix gives the covariance of residue type a at position i with residue type b at position j. There are in total 21 residue types (20 standard amino acid types plus a gap type).
Since the covariance matrix in Eq. (2) encodes the marginal correlations between variables, we calculate the second feature, the precision matrix (PRE)21, by minimizing the objective function:
| (3) |
where the first two terms can be interpreted as the negative log-likelihood of the inverse covariance matrix, i.e., the precision matrix Θ, under the assumption that the data are under a multivariate Gaussian distribution. Here, tr(SΘ) is the trace of the matrix SΘ and log|Θ| is the log determinant of Θ. The last term in Eq. (3) is the L2 regularization of the precision matrix with ρ being set as e −6. The inverse of the covariance matrix provides direct couplings between pairs of sites conditional on other positions. Thus, the precision matrix has better performance in the prediction of contact-maps than the covariance matrix21.
The negative of the inverse of the covariance matrix can also be interpreted as the Gaussian approximation of the inverse Potts model. Thus, another way to approximate the inverse Potts model through pseudolikelihood maximization (PLM)25,26 is also considered. The starting point for this procedure is approximating the probability of the sequence by the conditional probability of observing one variable conditional on all the other variables. We use CCMpred12 to efficiently calculate the PLM coupling parameters.
The covariance matrix, the precision matrix, and the coupling parameters of the Potts model all assume a form of a 21∙L by 21∙L matrix, representing relationships between the specific residue types of any two positions. In each of the three matrices, the full set of 441 coupling parameters for every position pair is represented as a 21 by 21 sub-matrix. After a reshaping procedure, three input features of size of L by L by 441 are collected for each sequence.
Residual convolutional neural network architectures for contact model training
As shown in Figure 1, we proposed two architectures based on deep residual neural networks (ResNet)24 in CASP13 to investigate the best way of ensembling for contact-map prediction, where the first version of ResNet is used as the basic residual block. Here, each residual block is defined as:
| (4) |
where x and y are the input and output vectors of the residual block considered, f denotes the activation function (ReLU34 is used in this work), and the function F represents the residual mapping to be learned by convolutional operations. Specifically, there are two convolutional layers in a residual block. Thus, the residual function is:
| (5) |
where W1 and W2 are the learnable weights in the first and the second convolutional layers respectively. We also have added instance normalization with default parameters and dropout layers to the basic block to speed up training of the neural networks and to avoid overfitting. The detailed architecture of the basic residual block is shown in Figure S1. Here, the dropout rate was set to 0.2, which means 80% of the input signal before a dropout layer would be randomly masked at each training batch.
The right-upper portion of Figure 1 depicts the architecture of TripletRes, where the three coevolutionary features are ensembled directly by neural networks. Each input feature is fed into a set of 24 residual blocks and transformed into the output feature with 64 channels. The three output features are concatenated along the channel dimension as the input of the last neural networks. The last set of neural networks try to learn patterns from the three transformed features by another 24 residual blocks. All residual blocks have a channel size of 64, and the kernel size of convolutional layers are set to 3 × 3 with padding size equaling to one. Such a padding parameter set-up can keep the spatial information fixed through different layers. Here, we use a convolutional layer of 1 × 1 kernel size to transform each coevolutionary input feature and the concatenated feature into 64 channels. The final contact-map prediction is obtained by a sigmoid activation function over the output of a convolutional layer whose output channel is set to one.
The right-lower part shows a two-stage ensemble model, ResTriplet, using stacking strategy. In Stage I, three individual base models are trained separately based on the three different sets of coevolutionary features, PRE, PLM and COV, as described above. The base models have the same training data and the same neural network structure consisting of 22 residual basic blocks. In Stage II, we use a shallow neural network structure to combine the predictions of base models from Stage I. Thus, the predicted contact-maps of base models are considered as the input features in stage II. The predicted secondary structures, denoted as PSS in Figure 1, by PSIPRED35 are also adopted as an extra feature for the neural network model in Stage II. For shallow convolutional neural networks, the size of receptive fields is usually limited. Hence, a collection of 5 dilated convolutional neural network layers36 with dilation value set to 2 and channel size set to 16 is employed in order to enlarge the size of receptive fields. Different from TripletRes, ResTriplet employs a dilated convolutional neural network layer with dilation value set to 2 and output channel set to 1 as the last layer. The final output of ResTriplet is then obtained by applying a sigmoid function over the last convolutional layer.
It should be noted that we did not apply any kind of pre-normalization operation to the input features; instead, an instance normalization layer is added after each convolutional layer except the last convolutional layer in both TripletRes and ResTriplet. Both TripletRes and ResTriplet share the same training set as described in Text S1. For TripletRes, a total of 10 models were trained. The training set was divided into 10 subsets. Each subset was considered as a validation set, and the remaining subsets were considered as the training set of each model. Finally, the output is the average of all 10 models. In Stage I of ResTriplet, to reduce the risk of over-fitting, predicted contact-maps produced by each base model are also generated by 10-fold cross validation. In other words, we build 10 models for each coevolutionary feature using the same data splitting strategy as that in TripletRes. For each specific coevolutionary feature type, the predicted contact-maps of the validation set of each model are considered as the features of Stage II. However, in Stage II, ResTriplet does not perform cross-validation because of limited time before the CASP experiment. The neural networks in both TripletRes and ResTriplet are implemented in Pytorch37 and trained by Adam optimizer38 with a default initial learning rate, i.e., 1e-3, for 50 epochs. The training of TripletRes requires 4 GPUs running concurrently, while the training procedures of ResTriplet can be handled with only one GPU. A dynamic batch size strategy during training is considered due to the limited of GPU resources. A batch size of 1 is used for sequences with length L > 300, 2 for in 200–300, and 4 for LL < 200. The choice of the hyperparameters of the two models, especially the number of layers, is a compromise between memory usage and performance. In particular, while deeper CNN models can, in theory, yield better performance, only a limited number of layers can fit into the GPU memory for efficient training.
Domain splitting and domain-based contact prediction
Protein domains are subunits that can fold and evolve independently. Due to the independent evolution of domains, constructing MSAs and predicting contacts for individual domains can often lead to improved accurate intra-domain contact prediction compared to that of the full-length sequence. Since the domain boundary of CASP targets are not known a priori, for a given CASP full-length target, ThreaDom39 is used before contact prediction in order to identify domain boundary locations. The core methodology of ThreaDom is threading the query sequence through the PDB library using LOMETS40 to construct a template structure-based multiple sequence alignment. Following this alignment, a domain conservation score is calculated, where a target-specific scoring cut-off strategy is then used to assign the domain boundaries. The final contact-map prediction is derived from both the full-length sequence and each of its domains. The inter-domain contacts are the results from the prediction of the full-length sequence, and the intra-domain contacts are replaced by the prediction of individual domains.
Results
Overall performance
CASP13 had a total of 90 full-length protein targets, where 82 have had their final structure released, which have been split into 122 domains by the assessors. In Table SI in the Supporting Information, we give a list of Nf values of MSAs and the top L, L/2 and L/5 contact prediction accuracy by ResTriplet and TripletRes for all the 122 domains in three ranges (short, medium and long), where L is the length of the query sequence. Among them, the average results of FM domains are summarized in Table I, following the official assessment of the CASP accessors, for three categories of short, medium and long-range contacts. Here, a contact is defined as two residues (i and j) whose Cβ atoms are less than 8Å apart. A short-range contact is where 6 ≤ |i − i| ≤ 11, a medium-range contact is where 12 ≤ |i − j| ≤ 23, and a long-range contact is where |i − i| ≥ 24. It is shown that the two proposed methods have comparable performance, although the TripletRes has a slightly higher accuracy than the ResTriplet program, by 1.2% and 0.9% for the top L and L/5 long-range contacts, respectively. Nevertheless, the corresponding p-values of the difference are 0.82 and 0.65, showing that the difference is statistically insignificant. If we count the number of targets with a top L long-range contact accuracy >0.5, TripletRes met this criterion in 23 out of the 31 domains, which is also slightly higher than ResTriplet (22). This difference is probably due to the fact that the TripletRes pipeline is trained end-to-end and therefore does not suffer from the shortcomings of the stacking strategy used by ResTriplet, such as the fact that since the feature ensemble in ResTriplet is optimized separately, the predicted contact-map and the secondary structure features could be less reliable in some extreme cases where sequence homologs are barely obtained. Therefore, this strategy suffers a slightly higher loss of contact accuracy for the FM targets compared to TripletRes.
Table I.
Overall performance of TripletRes and ResTriplet on CASP13 FM targets
| Method | Short range | Medium range | Long-range | ||||||
|---|---|---|---|---|---|---|---|---|---|
| L | L/2 | L/5 | L | L/2 | L/5 | L | L/2 | L/5 | |
| ResTriplet | 0.278 | 0.449 | 0.691 | 0.358 | 0.533 | 0.759 | 0.404 | 0.529 | 0.640 |
| TripletRes | 0.280 | 0.453 | 0.671 | 0.360 | 0.541 | 0.744 | 0.409 | 0.534 | 0.646 |
Since the FM targets have on average a lower number of homologous sequences than the template-based modeling (TBM) targets, the contact prediction accuracy for FM is expected to be lower. To examine this, we also listed the results of contact-map prediction for all domains in Table SI. In our case, the average Nf is 57.4 and 390.8 for the FM targets and all targets, respectively. Accordingly, the average accuracy of the top L long-range contacts of ResTriplet and TripletRes predictions for all targets is 29% and 25% higher compared to that of FM targets. Interestingly, the increase in ResTriplet is slightly larger, which results in a slightly higher accuracy by ResTriplet than by TripletRes in all targets, contrary to the trend present in the FM targets only. The larger discrepancy in accuracy is likely because of the consideration of secondary structure information in ResTriplet; the prediction of secondary structure is relatively reliable when more homologous sequences are found.
Impact of DeepMSA on contact prediction accuracy
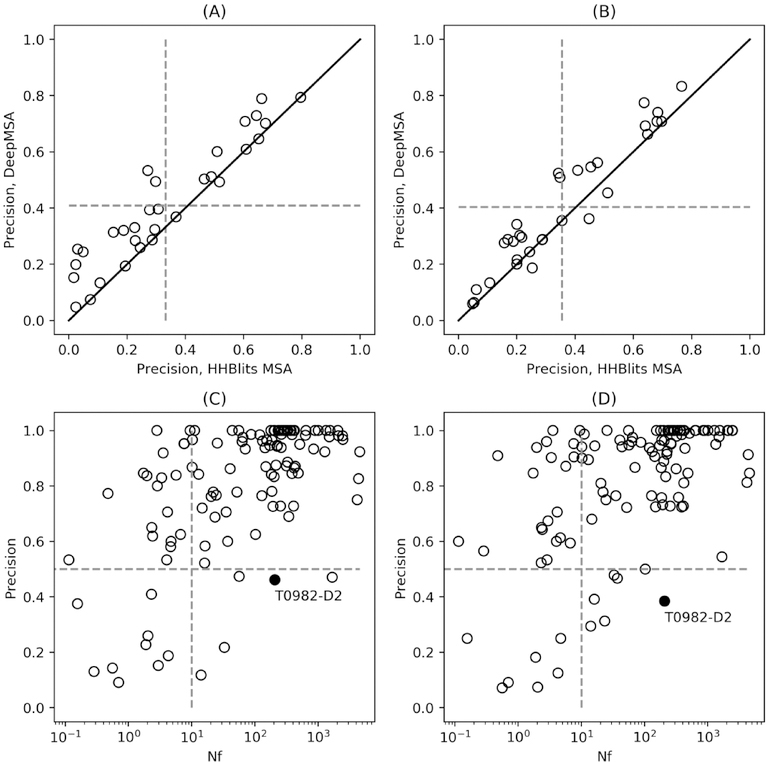
The quality of the MSA is highly correlated with the predictive accuracy of a protein contact-map, especially for TripletRes and ResTriplet due to their dependence on coevolutionary features derived directly from the MSA. In CASP13, we utilized DeepMSA to construct MSAs by searching across multiple databases using complementary sequence searching engines27. To examine the impact of such pipeline on the final contact predictions, Figure 2A and Figure 2B show a head-to-head comparison of the top L long-range predictions on the FM targets by TripletRes and ResTriplet using two different pipelines of MSA collection, one with DeepMSA and another with a routine approach of HHBlits searching through UniClust30 sequence database. The result shows that 27 (28) out of the 31 FM domains have improved contact predictions using DeepMSA relative to the HHBlits pipeline for TripletRes (ResTriplet). On average, DeepMSA improved the precision of TripletRes/ResTriplet from 33.2%/35.4% to 40.9%/40.4%. The p-values from a Student’s t-test are 2.9e-06/1.1e-04, suggesting that the improvement is statistically significant. Since the only factor that was changed between these two pipelines was the method of MSA collection, the difference can be attributed solely to this factor. In this regard, the average Nf by DeepMSA is 57.4 for the 31 FM domains, compared to 11.6 from HHBlits, indicating that DeepMSA indeed generates more diverse sequences with a deeper alignment.
Figure 2.
Illustration of the effect of MSAs on the performance of TripletRes and ResTriplet. (A) and (B) Comparison of top L long-range contact prediction results using MSAs by DeepMSA versus those by the routine HHblits search for TripletRes and ResTriplet, respectively. (C) and (D) Precision of long-range top L/5 contact prediction versus Nf of MSAs for TripletRes and ResTriplet, respectively.
Interestingly, DeepMSA has a slightly greater improvement with TripletRes than ResTriplet. Even though TripletRes has lower accuracy for long-range top L contacts compared to ResTriplet based on HHblits MSAs, the final accuracy of TripletRes is higher than that of ResTriplet after both employing deep MSAs. To provide a more quantitative analysis about the impact of MSAs on the performance of the proposed methods, we present the precision of long-range top L/5 contact prediction by TripletRes and ResTriplet versus the Nf of MSAs for all targets in Figure 2C and Figure 2D. Note that 8 targets (i.e., T0952-D1, T0953s1-D1, T0960-D1, T0960-D4, T0963-D1, T0963-D4, T0979-D1, and T0980s2-D1) with no long-range contacts in the experimental structures are excluded from the figure. The Pearson correlation coefficients between precision and the logarithm of Nf are 0.584 and 0.551 for TripletRes and ResTriplet, respectively, for all 122 domains, indicating that the correlations are both modest. The reason is mainly because of the occurrence of targets with low Nf values but high precision. 19/21 out of 28 domains that have an Nf lower than 10 predicted by TripletRes/ResTriplet achieve precisions over 0.5, as shown in the left-upper blocks in Figure 2C and Figure 2D. To further investigate this phenomenon, we performed direct coupling analysis by the CCMpred program on domains with an Nf less than 10; the precision of long-range L/5 contact prediction was found to be 0.149, 74.8%/75.5% lower than 0.591/0.608 achieved by TripletRes and ResTriplet. The large gap between the performance of the pure DCA method and deep-learning based methods demonstrates the effectiveness of supervised deep neural network training. However, some targets with highly populated MSAs still have lower precision. Taking T0982-D2 as an example, the long-range top L/5 precision of contact-maps predicted by TripletRes and ResTriplet are 0.462 and 0.385 respectively, despite the Nf value of 207.15. According to the top template (PDB ID: 3tfzB) provided by CASP, it is highly possible that the domain interacts with a specific ligand in vivo. Therefore, the structure determination of this target should be conditional not only on the sequence information, but also information of the binding ligand. The negligence of ligand information in our pipeline can lead to bias in the contact map prediction. Another possible reason for the low precision values is the inherent roughness of using contacts as the ground truth during the training process. The binary contact-map representation may cause the loss of detailed distance information and cannot faithfully represent the elasticity of the protein. The mean ground truth distance of false positives predicted by ResTriplet in long-range top L/5 predicted contacts are 10.86, which means the false positives are still close enough to be in contact. Such phenomenon can be caused by the fact that the models trained by binary contact-maps may not be able to distinguish residue pairs whose distance is near the contact threshold in this case. The studies of training with distance information are under progress.
While DeepMSA improves contact precision on average, it occasionally has negative effects on targets where MSAs are too aggressively collected. In particular, for the domain T0982-D2, for example, DeepMSA went through all 3 steps to obtain the final multiple sequence alignment, and the Nf values of MSAs of three steps are 39.7, 91.6, and 288.9 respectively. TripletRes and ResTriplet achieved long-range top L/5 precisions of 0.962 and 0.923 respectively based on the MSA generated by Step 1. When the MSA generated from Step 2 is used, the precision of ResTripet slightly improves to 0.962 while that of TripletRes drops to 0.615. The precision of TripletRes and ResTiplet both drops to 0.577 and 0.423 based on the MSA of Step 3 and become 0.462 and 0.385 at last. The downward trend of the precision for the two predictors in response to the deeper searching of the DeepMSA pipeline indicates that deeper MSAs do not necessarily lead to better contact prediction, partly because there could be the alignment noise introduced with a deeper MSA. How to quantitatively measure and reduce the alignment noise of homologous sequences contained within an MSA is still an important issue worthy of further study.
When only FM targets are counted, we found that the correlations between precision and Nf value were 0.559 and 0.659 for TripletRes and ResTriplet, respectively. In other words, the performance of TripletRes is less dependent on the quality of MSAs for FM targets. Such an observation further proves the robustness of TripletRes, in particular for FM targets. We also observed that the correlation between the precision of ResTriplet and Nf is much higher than that of TripletRes on FM targets, which confirms that the performance of ResTriplet is more sensitive to the content of the MSAs than TripletRes. As pointed out previously, this is probably because ResTriplet uses extra predicted one-dimensional features based on MSAs. The quality of the input MSA thus has more impact on the performance of the final prediction for ResTriplet. Nevertheless, 11/12 out of the 18 FM targets that have a very low Nf (<10) for TripletRes/ResTriplet achieve a reasonable contact accuracy >0.5. These data show that the neural networks fed with coevolutionary features still have the ability to learn the underlying contact patterns even from a very limited number of sequence homologs, which is important for the modeling of FM targets that lack homologous sequences.
Comparison of ensemble methods with their components
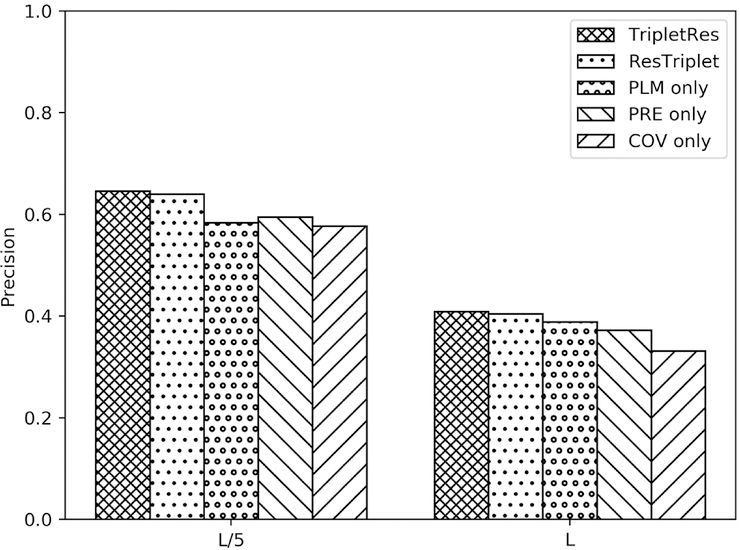
Both TripletRes and ResTriplet are hybrid approaches integrating information from three different components. To examine the effect of the information ensembling method, we present a comparison of the results of the hybrid methods against three predictors using the individual component input features on CASP13 FM targets in Figure 3. The prediction of each component predictor is the average of 10 models for the corresponding coevolutionary feature type. It is observed that the impact of the ensemble is quite significant for FM targets. For example, the mean precision of long-range top L/5 contacts was 64.6% for TripletRes, which is 10.6%, 8.6%, and 12.0% higher than those of predictors based on PLM, PRE, and COV, respectively. Similarly, the stacked ensemble of ResTriplet also brought a mean precision of 64.0%, which is 9.6%, 7.6%, and 10.9% higher than the mean precisions of each component. The same pattern was also observed in the mean precisions of long-range top L contacts. When we consider all targets (Figure S2), the results showed that the ensemble methods (TripletRes and ResTriplet) only slightly outperformed PLM, PRE, and COV based component predictors. For example, the top L/5 long-range average precision of TripletRes and ResTriplet for all targets are 75.4% and 76.2%, which were only marginally higher than 75.2%, 74.6%, and 71.0% achieved by predictors based on PLM, PRE, and COV features, respectively. The same is true when considering the top L long-range predictions. Such an observation indicates that the ensemble is particularly necessary for FM targets. This is partly because the FM targets usually have a lower number of homologous sequences, where complementary information from different components can help enhance the overall accuracy of the final contact models. For TBM targets, however, the MSA is usually deep enough for each of the component predictors to generate satisfactory contacts and the effectiveness of ensembling is therefore less pronounced.
Figure 3.
Mean precisions of long-range top L and top L/5 contacts of TripletRes and ResTriplet on FM targets, compared to the predictors trained on the component features from the covariance matrix feature (COV), the precision matrix feature (PRE) and the coupling matrix of the inverse Potts model feature (PLM).
In addition, it was observed that predictors based on PLM and PRE features performed better than the COV based feature in all the comparisons. This is because the PLM and the PRE features are both built on direct coupling analysis, while the COV feature is based on marginal correlation analysis, which suffers more from indirect transitional noise.
Impact of domain splitting on contact predictions
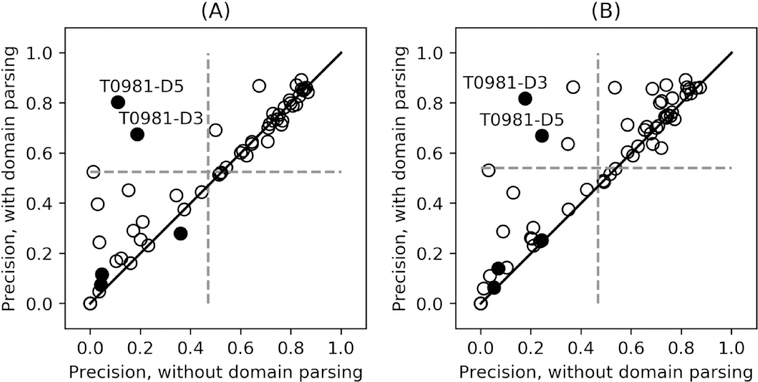
Both TripletRes and ResTriplet use DeepMSA to create MSAs from the sequences of individual domains as parsed by ThreaDom. To examine the effect of the domain splitting on contact prediction, Figure 4 compares the long-range top L precision before and after the domain splitting procedure on 26 whole-length proteins that were assigned as multi-domain sequences by ThreaDom. Out of these 26 proteins, 59 domains have their structure released, where Figure 4 lists the contact prediction results of these 59 domains following the official domain definitions from CASP13.
Figure 4.
Comparison of precisions of long-range top L contact predictions with domain partitioning versus those without using domain partitioning. (A) TripletRes; (B) ResTriplet.
The ThreaDom-based domain partitions generated an obviously positive impact on the contact predictions. There are overall 23 (or 36) out of the 59 domains that have an increased precision after the domain splitting procedure for TripletRes (or ResTriplet) relative to the whole-chain based predictions, while the opposite occurs only in 12 (or 7) cases. On average, the top L long-range precision of TripletRes and ResTriplet (52.6% and 54.1%) are also higher than that of the whole-chain prediction (46.9% and 46.6%), which corresponds to a p-value of 1.8e-03 and 9.9e-05, respectively, showing that the difference is statistically significant.
Among the 59 domains in Figure 4, two domains, T0981-D3 and T0981-D5, stand out with a very large difference between using and not using the domain splitting. Both of the domains are from T0981, which contains five domains, but ThreaDom split the target sequence into four domains (see Figure S3). A closer look showed that the normalized effective number of sequences in the MSA (Nf) is 0.2, with only 9 homologous sequences being present in the MSA when the whole-chain sequence is used. The Nf value increased to 229.6 and 2.8, respectively, for T0981-D3 and T0981-D5 after the domain splitting by ThreaDom, which eventually increased the contact prediction accuracy of TripletRes/ResTriplet from 0.187/0.177 to 0.675/0.818 for T0981-D3, and 0.110/0.244 to 0.803/0.669 for T0981-D5. These data suggest that the benefit of domain partitioning mainly manifests as improvement of the MSA construction, since domain splitting helps DeepMSA to detect more homologous sequences for each individual domain.
However, theoretically, domain splitting may also result in bias in the estimation of DCA models. Before domain splitting, the probability of a certain amino acid at a certain position in DCA models is conditional on all other positions of the full-length sequence. However, after domain splitting, this probability is only conditional on other positions of the domain. Moreover, alignment quality of positions close to the domain boundary could also be negatively affected after domain splitting. These two factors could be the reason that a small number of domains have lower precision even with more sequence homologs being present in the MSA.
What went right?
We found that using raw coevolutionary features can provide high-precision contact-map predictions when coupled with deep convolutional neural networks. Among the coevolutionary features, the DCA-based features, i.e., the precision matrix and the coupling parameters of the pseudolikelihood maximized Potts model, outperform the marginal correlation feature, i.e., the covariance matrix. Considering that many methods in literature have used the covariance matrix feature, this conclusion may help push the boundaries of contact-map prediction. Moreover, although a single raw coevolutionary feature can provide relatively accurate contact-maps, multiple feature fusion/ensembling with deep convolutional neural networks is still needed to achieve even better performance.
According to our self-test benchmark and the CASP13 results, a combination of diverse multiple sequence alignment generation protocols (search algorithms and sequence databases) can significantly improve contact prediction, especially for FM targets. Another important aspect of our prediction procedure is domain splitting. Even when the predicted domain boundary is not exact, domain splitting still improves the precision by providing more diverse and deeper MSAs.
What went wrong?
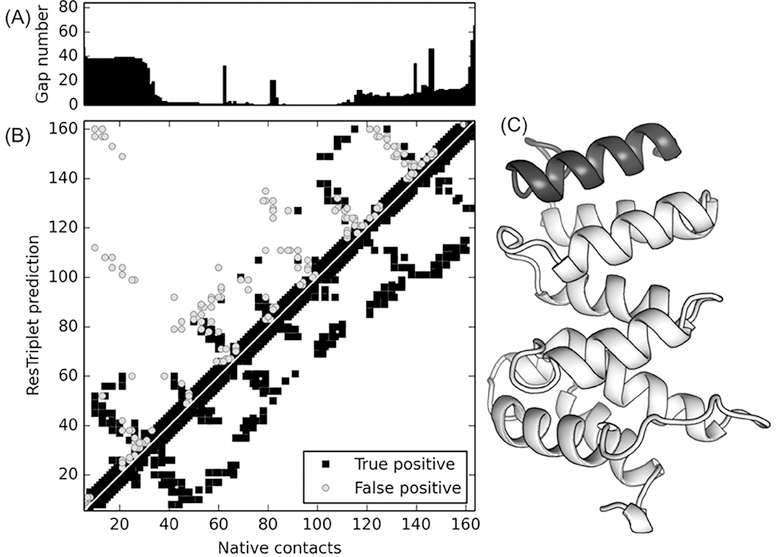
Since all evolutionary coupling are inferred from the MSA, strong noise can be introduced in terminal regions where long stretches of gaps in the MSA leads to a false positive coupling signal. This is because gaps are treated as an additional amino acid type in constructing coevolutionary features. This issue has been amplified by T0957s2-D1, where the top L long-range accuracies are 39.4% and 34.2% for TripletRes and ResTriplet respectively. As shown in Figure 5A, there is a significantly high number of alignment gaps in the first 30 residues. Accordingly, the majority of the false positive predictions (marked as grey circles in the upper-left corner of the contact map in Fig. 5B) are from the contacts involving these N-terminal residues. Figure 5C presents the 3D structure of the domain, which shows that the N-terminal is well-packed with the rest of the domain and has the same secondary structure as other residues, indicating that the gaps are not due to the irregular local structure or motif disordering. In this regard, how to appropriately consider large gaps in MSAs is still an important problem.
Figure 5.
An illustrative example of CASP13 domain T0957s-D1 showing false positive contact prediction in the N-terminal tail region due to the higher number of gaps in the alignment. (A) Bar plot of the number of gaps along the query sequence. (B) Contacts from the native structure (lower-right triangle section) versus predicted contacts by ResTriplet (upper-left section) where gray circles and black squares denote false and true positive predictions respectively. (C) 3D experimental structure of the T0957s-D1 with the N-terminal tail marked in black.
Conclusion
We have introduced two hybrid contact prediction methods, TripletRes and ResTriplet, which have been tested in CASP13. Unlike other methods which use post-processed coevolutionary analysis coupling potentials, these two methods use the raw coevolutionary matrices as the only input features, which can result in advanced contact-map predictions when coupled with deep residual neural networks. Part of the success of these methods is due to the feature ensembling strategies: feature fusion by neural networks, denoted as TripletRes, and multiple predictor stacking, denoted as ResTriplet. Meanwhile, the iterative MSA construction procedure DeepMSA, which combines multiple sources of sequence databases, and domain specific MSA collection also contributed to the improvement of the final contact-map prediction performance. The effects and usefulness of these approaches are particularly pronounced for FM targets, which typically involve a smaller number of homologous sequences compared to easier TBM targets.
Nevertheless, there are still issues in contact-map prediction particularly in the tail regions of sequences, which often have a higher number of alignment gaps that result in a lower contact accuracy than other regions. Meanwhile, contact prediction for hard targets with a lower number of homologous sequences is still far from satisfactory. Development of new pipelines with models specifically trained on these tail regions and hard targets might help address these shortcomings.
Supplementary Material
Acknowledgements
We thank Dr. Wei Zheng for insightful discussion. TripletRes and ResTriplet were trained using the Extreme Science and Engineering Discovery Environment (XSEDE)41, which is supported by National Science Foundation (ACI-1548562).
Funding
This work was supported in part by the National Natural Science Foundation of China (61772273, 61373062, and 31628003), the Fundamental Research Funds for the Central Universities (30918011104), National Institute of General Medical Sciences (GM083107 and GM116960), and National Science Foundation (DBI1564756).
Reference
- 1.Browne WJ, North AC, Phillips DC, Brew K, Vanaman TC, Hill RL. A possible three-dimensional structure of bovine alpha-lactalbumin based on that of hen’s egg-white lysozyme. J Mol Biol 1969;42(1):65–86. [DOI] [PubMed] [Google Scholar]
- 2.Levitt M, Warshel A. Computer-Simulation of Protein Folding. Nature 1975;253(5494):694–698. [DOI] [PubMed] [Google Scholar]
- 3.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 1993;234(3):779–815. [DOI] [PubMed] [Google Scholar]
- 4.Wu S, Szilagyi A, Zhang Y. Improving protein structure prediction using multiple sequence-based contact predictions. Structure 2011;19(8):1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ovchinnikov S, Kim DE, Wang RY, Liu Y, DiMaio F, Baker D. Improved de novo structure prediction in CASP11 by incorporating coevolution information into Rosetta. Proteins 2016;84 Suppl 1:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ovchinnikov S, Park H, Varghese N, Huang PS, Pavlopoulos GA, Kim DE, Kamisetty H, Kyrpides NC, Baker D. Protein structure determination using metagenome sequence data. Science 2017;355(6322):294–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C, Mortuza SM, He B, Wang Y, Zhang Y. Template-based and free modeling of ITASSER and QUARK pipelines using predicted contact maps in CASP12. Proteins 2018;86 Suppl 1:136–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinch LN, Li W, Monastyrskyy B, Kryshtafovych A, Grishin NV. Evaluation of free modeling targets in CASP11 and ROLL. Proteins 2016;84 Suppl 1:51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abriata LA, Tamo GE, Monastyrskyy B, Kryshtafovych A, Dal Peraro M. Assessment of hard target modeling in CASP12 reveals an emerging role of alignment-based contact prediction methods. Proteins 2018;86 Suppl 1:97–112. [DOI] [PubMed] [Google Scholar]
- 10.Morcos F, Pagnani A, Lunt B, Bertolino A, Marks DS, Sander C, Zecchina R, Onuchic JN, Hwa T, Weigt M. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proceedings of the National Academy of Sciences 2011;108(49):E1293–E1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones DT, Buchan DW, Cozzetto D, Pontil M. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics 2011;28(2):184–190. [DOI] [PubMed] [Google Scholar]
- 12.Seemayer S, Gruber M, Söding J. CCMpred—fast and precise prediction of protein residue–residue contacts from correlated mutations. Bioinformatics 2014;30(21):3128–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue–residue contact predictions in a sequence-and structure-rich era. Proceedings of the National Academy of Sciences 2013;110(39):15674–15679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones DT, Singh T, Kosciolek T, Tetchner S. MetaPSICOV: combining coevolution methods for accurate prediction of contacts and long range hydrogen bonding in proteins. Bioinformatics 2014;31(7):999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchan DW, Jones DT. Improved protein contact predictions with the MetaPSICOV2 server in CASP12. Proteins: Structure, Function, and Bioinformatics 2018;86:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He B, Mortuza S, Wang Y, Shen H-B, Zhang Y. NeBcon: protein contact map prediction using neural network training coupled with naïve Bayes classifiers. Bioinformatics 2017;33(15):2296–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Sun S, Li Z, Zhang R, Xu J. Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS computational biology 2017;13(1):e1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adhikari B, Hou J, Cheng J. DNCON2: Improved protein contact prediction using two-level deep convolutional neural networks. Bioinformatics 2017;34(9):1466–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Palmedo P, Ye Q, Berger B, Peng J. Enhancing evolutionary couplings with deep convolutional neural networks. Cell systems 2018;6(1):65–74. e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones DT, Kandathil SM. High precision in protein contact prediction using fully convolutional neural networks and minimal sequence features. Bioinformatics 2018;34(19):3308–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Hu J, Zhang C, Yu D-J, Zhang Y. ResPRE: high-accuracy protein contact prediction by coupling precision matrix with deep residual neural networks. Bioinformatics 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Göbel U, Sander C, Schneider R, Valencia A. Correlated mutations and residue contacts in proteins. Proteins: Structure, Function, and Bioinformatics 1994;18(4):309–317. [DOI] [PubMed] [Google Scholar]
- 23.Shindyalov I, Kolchanov N, Sander C. Can three-dimensional contacts in protein structures be predicted by analysis of correlated mutations? Protein Engineering, Design and Selection 1994;7(3):349–358. [DOI] [PubMed] [Google Scholar]
- 24.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. 2016. p 770–778. [Google Scholar]
- 25.Ekeberg M, Lövkvist C, Lan Y, Weigt M, Aurell E. Improved contact prediction in proteins: using pseudolikelihoods to infer Potts models. Physical Review E 2013;87(1):012707. [DOI] [PubMed] [Google Scholar]
- 26.Ekeberg M, Hartonen T, Aurell E. Fast pseudolikelihood maximization for direct-coupling analysis of protein structure from many homologous amino-acid sequences. Journal of Computational Physics 2014;276:341–356. [Google Scholar]
- 27.Zhang C, Zheng W, Mortuza S, Li Y, Zhang Y. DeepMSA: Constructing deep multiple sequence alignment to improve contact prediction and fold-recognition for distant-homology proteins. 2019:In preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remmert M, Biegert A, Hauser A, Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nature methods 2012;9(2):173. [DOI] [PubMed] [Google Scholar]
- 29.Mirdita M, von den Driesch L, Galiez C, Martin MJ, Söding J, Steinegger M. Uniclust databases of clustered and deeply annotated protein sequences and alignments. Nucleic acids research 2016;45(D1):D170–D176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eddy SR. Accelerated profile HMM searches. PLoS computational biology 2011;7(10):e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinegger M, Meier M, Mirdita M, Voehringer H, Haunsberger SJ, Soeding J. HH-suite3 for fast remote homology detection and deep protein annotation. bioRxiv 2019:560029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Remmert M, Biegert A, Hauser A, Soding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods 2011;9(2):173–175. [DOI] [PubMed] [Google Scholar]
- 33.Steinegger M, Soding J. Clustering huge protein sequence sets in linear time. Nature communications 2018;9(1):2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nair V, Hinton GE. Rectified linear units improve restricted boltzmann machines. 2010. p 807–814. [Google Scholar]
- 35.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics 2000;16(4):404–405. [DOI] [PubMed] [Google Scholar]
- 36.Yu F, Koltun V. Multi-scale context aggregation by dilated convolutions. arXiv preprint arXiv:151107122 2015. [Google Scholar]
- 37.Paszke A, Gross S, Chintala S, Chanan G, Yang E, DeVito Z, Lin Z, Desmaison A, Antiga L, Lerer A. Automatic differentiation in pytorch. 2017. [Google Scholar]
- 38.Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980 2014. [Google Scholar]
- 39.Xue Z, Xu D, Wang Y, Zhang Y. ThreaDom: extracting protein domain boundary information from multiple threading alignments. Bioinformatics 2013;29(13):i247–i256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu S, Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic acids research 2007;35(10):3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Towns J, Cockerill T, Dahan M, Foster I, Gaither K, Grimshaw A, Hazlewood V, Lathrop S, Lifka D, Peterson GD. XSEDE: accelerating scientific discovery. Computing in Science & Engineering 2014;16(5):62–74. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.