Abstract
Small Angle X-ray Scattering (SAXS) measures comprehensive distance information on a protein’s structure, which can constrain and guide computational structure prediction algorithms. Here we evaluate structure predictions of 11 monomeric and oligomeric proteins for which SAXS data were collected and provided to predictors in the 13th round of the Critical Assessment of protein Structure Prediction (CASP13). The category for SAXS-assisted predictions made gains in certain areas for CASP13 compared to CASP12. Improvements included higher quality data with size exclusion chromatography-SAXS (SEC-SAXS) and better selection of targets and communication of results by CASP organizers. In several cases, we can track improvements in model accuracy with use of SAXS data. For hard multimeric targets where regular folding algorithms were unsuccessful, SAXS data helped predictors to build models better resembling the global shape of the target. For most models however, no significant improvement in model accuracy at the domain level was registered from use of SAXS data, when rigorously comparing SAXS-assisted models to the best regular server predictions. To promote future progress in this category, we identify successes, challenges, and opportunities for improved strategies in prediction, assessment, and communication of SAXS data to predictors. An important observation is that, for many targets, SAXS data were inconsistent with crystal structures, suggesting that these proteins adopt different conformation(s) in solution. This CASP13 result, if representative of PDB structures and future CASP targets, may have substantive implications for the structure training databases used for machine learning, CASP, and use of prediction models for biology.
Keywords: flexibility, unstructured regions, disorder, complexes, structure prediction, SAXS, SAS, experimental restraints, modeling, solution scattering
INTRODUCTION
As assessed in CASP12 and now in CASP131, protein structure prediction algorithms have made major leaps towards improving prediction accuracy. Yet, obstacles remain for novel folds, large proteins, oligomeric complexes, and flexible proteins. To provide additional and realistically achievable constraints on any soluble protein target, CASP12 and CASP13 included an assisted target category where sequence was supplemented with experimental data from cross-linking mass spectrometry, Nuclear Magnetic Resonance (NMR), and Small Angle X-ray Scattering (SAXS). This article focuses on SAXS data. The protein targets chosen for this category were specifically anticipated to be challenging to predictors.
A primary rationale for using SAXS data as experimental input for structure prediction is that collecting SAXS data is high-throughput (HT) and straightforward.2-6 In SAXS, no labeling or crystallization is required. Data collection for basic research is provided for free by all biological SAXS beamlines, with one at every U.S. synchrotron. For fold prediction, samples are ideally stoichiometrically monodisperse, but there are no size limitations, from a few kD to megadaltons. At the SIBYLS beamline and at many other SAXS beamlines, SAXS data can be collected in HT mode with proteins and buffers loaded in 96 well plates or by SEC in-line with SAXS and multi-angle light scattering (MALS). SEC-SAXS with MALS analysis can assure stoichiometric monodispersity for improved confidence in extracted structural information. Importantly, SAXS, as an X-ray scattering technique, provides information on the distances of all electron pairs within the protein in solution6-8 including functional conformational variation.9 This information from SAXS could help constrain and guide computational structure prediction algorithms. This capability and methods for integration were therefore tested in CASP12 and now CASP13.
For the SAXS-assisted category in CASP, analyzed SAXS data in addition to the respective amino acid sequence, were provided to predictors in a report. The SAXS analysis provided predictors with the experimentally validated multimerization state, maximum dimension, radius of gyration, an estimate of flexibility, volume, and radius of cross-section. Furthermore the primary SAXS curve can be converted into the histogram of relative proportion P of electron pairs at distance r, i.e., P(r).7 The P(r) is sensitive to changes as small as 5 Å. The scattering curve, of the atomic model and an approximation of its hydration layer, can be calculated and compared to the SAXS curve (I vs q) or, after Fourier transform, to the P(r), for feedback against experiment. There is enough information within the P(r) function to calculate 3D shapes of ~15 Å resolution.10 The SAXS curve, P(r) curve and shape were provided to predictors.
CASP12 was the first attempt to combine SAXS with CASP.11 Closer analysis of how predictors used SAXS data revealed an underlying assumption within CASP that would be misleading when integrated with SAXS. CASP models are judged based on the crystal structure and even more strictly on domains within the crystal structure. Perhaps as a reflection of this criteria, many CASP12 predictors considered the entire sequence of many protein targets as well-folded and monomeric. However, many CASP12 targets had intrinsically disordered regions and/or were multimeric, as we have found with most proteins that we have studied by SAXS.2 Comparing the sequence of the SAXS sample and what was modeled in the respective crystal structure, the average CASP12 crystal structure was missing 20% of the sequence with an extreme of 44%.12 These were generally terminal ends of the protein and were largely predicted from sequence to be intrinsically disordered. Typically, these regions would not be considered during the assessment – no harm, no foul. However, in the context of SAXS-assisted evaluation, modeling disordered regions as part of the globular fold makes fitting the model to SAXS data misleading. For example, a 5 amino acid disordered terminus can extend the maximum dimension by as much as 12.5 Å.13 To improve awareness of disorder, an intrinsic disorder prediction was attached to SAXS reports in CASP13. Similar discrepancies resulted from CASP12 predictor’s lack of awareness of why modeling the proper multimer to the SAXS data is essential. Over 50% of targets were multimers12 but many predictors fit the data against a monomeric structure. On the data side, there were issues when targets were stoichiometrically heterogeneous, as data were collected by HT SAXS. Although some information could be extracted by varying protein concentration or protein constructs, this was not ideal. Therefore, CASP13 included SEC-SAXS, which can separate out stoichiometrically diverse populations and allow data collection on monodisperse sample. These strategies were suggested following the CASP12 assessment to increase accuracy.12
Below, we describe results and analysis of the SAXS-assisted category for CASP13. Data collection included both HT-SAXS and, if there was enough protein supplied, SEC-SAXS, which increased the reliability of the SAXS data. A target’s multimerization and predicted intrinsically disordered regions were communicated to predictors, and based on model entries, CASP13 predictors generally showed better awareness in treating intrinsically disordered regions and multimerization. There were a few examples where inclusion of SAXS improved the backbone accuracy or domain positioning. However, to rigorously test the potential of SAXS for prediction, many issues still require improvement, and we highlight these issues with exemplary targets to aid future predictions. An unanticipated finding of our analysis is that many crystal structure conformations did not adequately match the respective SAXS data, occurring in 7 out of 11 SAXS-assisted CASP proteins. We discuss those cases when the crystal structure or the crystal structure plus an added unstructured tail, do not match the SAXS data. The discrepancies are not on the scale of small amino acid scale vibrational differences, but rather of domain interactions. If regular or unassisted CASP prediction algorithms are based on training databases with conformations enforced by the crystal lattice or crystallization conditions, they could be biased towards predicting crystal conformations instead of solution conformations. That might reduce biological relevance of prediction results. Our detailed analysis and discussion form a basis to begin considering these and other implications.
METHODS
SAXS Sample Preparation and Data Collection
Proteins were generously provided for SAXS by the crystallographers who had determined the crystal structure. Most of the SAXS data were collected at the SIBYLS beamline (12.3.1) at the Advanced Light Source, part of the Lawrence Berkeley National Laboratory.4 The sample-to-detector distance is 1.5 m, resulting in scattering vectors ranging from 0.01 to 0.5 Å–1. The wavelength of the beam was 1 Å, and the flux was 1013 photons per second. Data were collected by HT-SAXS and/or SEC-SAXS, depending on sample quantity.
Samples generally arrived frozen, which can promote aggregation. For HT-SAXS, just prior to data collection, samples were prepared in 96-well plates, where 20 μL of the consecutive protein concentrations were bracketed with two 20 μL protein-free buffer samples. The protein concentrations used for data collection consisted of the original protein concentration, a 1:2 dilution, and a 1:4 dilution. By collecting data on three protein concentrations, we were able to correct for concentration-dependent behavior. Samples were transferred from a 96-well plate at 10 °C to the sample cuvette, where they are exposed to an X-ray beam for a total of 10 seconds.5 Scattering images are collected by a PILATUS 2M detector every 0.3 seconds, for a total of 33 sample images. For each sample collected, two protein-free buffer samples were also collected to reduce error in subtraction. Each collected image was circularly integrated and normalized for beam intensity to generate a one-dimensional scattering profile by beamline specific software. The one-dimensional scattering profile of each protein sample were buffer-subtracted by each of the two corresponding buffers, producing two sets of buffer subtracted sample profiles. Profiles were examined for radiation damage. Scattering profiles over the ten-second exposure were sequentially averaged together until radiation damage affects were seen to begin changing the scattering curve. Averaging was performed with web-based software (sibyls.als.lbl.gov/ran).
For SEC-SAXS, HPLC SEC was in line with SAXS sample cell and MALS, for simultaneous data collection, to promote the stoichiometrically monodisperse samples with large non-specific aggregation removed. Two second X-ray exposures were collected continuously during an ~25-min elution. The SAXS frames recorded prior to the protein elution peak were used to subtract all other frames. The subtracted frames were investigated by RG and I(0) derived by the Guinier approximation I(q) = I(0) exp(−q2*RG2/3) with the limits q*RG < 1.5. I(0) and RG values were compared for each collected SAXS curve across the entire elution peak. The elution peak was mapped by plotting the scattering intensity at I(0) relative to the recorded frame. Graduate decreasing of RG values across an elution peak was used to indicate transient sample behavior.
SAXS Data Analysis and Predictor Data packages
From data collection to analysis, all data were passed to CASP in under 3 weeks. Predictors were provided SAXS curves in reciprocal and real space, a SAXS-based shape prediction, and SAXS scalar values (Table 1). Parameters such as radius of gyration (RG), the Porod Exponent, the Radius of the cross-section (Rxc), and the volume of correlation (Vc) were calculated using Scatter.2,14,15 The P(r), Rg2, and DMax were calculated using PRIMUS and GNOM.10,16 Molecular envelope calculations were performed using GASBOR.17 All data are available at the CASP13 web address (predictioncenter.org) for download in the “Targets” tab under “Assisted structure prediction”. Regions missing in crystal structures were modeled in using Modeller implemented in Chimera.18 Atomic structures were compared to SAXS data using FOXS.19,20 BILBOMD and MultiFOXS were used to create flexible models, with domains defined as rigid bodies.20,21 Models based on crystal structures were modified by nonlinear NOLB normal mode analysis (NMA).16,22
Table 1. SAXS data provided to CASP participants.
Rg, Porod Exponent (PD), Mass calculated from SAXS, Theoretical Mass, Maximum Dimension (Dmax), Radius of Cross-Section (Rxc), and Volume were calculated using SCÅTTER. Rg in real space and Dmax were calculated using PRIMUS and GNOM. Quality of data (Gold, Silver, Bronze) was provided for SAXS data collected at the SIBYLS beamline (12.3.1) at the ALS. S0999 was collected at the Diamond Light Source
| SAXS Target |
RG - Guinier (Å) |
PD | Mass SAXS (kD) |
Mass Theor. (kD) |
Dmax (Å) |
Rxc (Å) |
Volume (Å3) |
Real space RG (Å) |
Sample Quality |
Challenge |
|---|---|---|---|---|---|---|---|---|---|---|
| S0949 | 16.0 | 4.0 | 13 | 16.7 | 53 | 13 | 22158 | 16.5 | Silver | none |
| s0953 | 34.8 | 3.5 | 32 | 25.7,7.3 | 130 | 13 | 63217 | 36.7 | Gold | Elongated 3:1 |
| s0957 | 21.8 | 4.0 | 32 | 18.6,17.7 | 71 | 18.4 | 54545 | 21.58 | Silver | Heteromer |
| s0968 | 25.8 | 3.1 | 36 | 13.9, 13.4 | 83 | 19.2 | 108771 | 26.8 | Bronze | Multimer |
| s0975 | 27.8 | 4.0 | 39 | 38.5 | 89-105 | 17.6 | 82000 | 26.3 | Silver | Fe-S Cluster |
| s0980 | 27.2 | 3.7 | 43 | 13.5, 6.2 | 102 | 18.1 | 83586 | 28.0 | Silver | 2:2 |
| s0981 | 46.6 | 3.7 | 190 | 76 | 176 | 32.7 | 46000 | 47.6 | Silver | Trimer |
| s0985 | 41 | 4.0 | 190 | 98.4 | 136 | 32.6 | 35000 | 40.7 | Gold | Dimer |
| s0987a | 26.8 | 4.0 | 43 | 45.8 | 100 | 18 | 79363 | 27.3 | Gold | Depends on solution |
| S0987b | 24.4 | 4 | 41 | 45.8 | 86.5 | 20 | 73401 | 24.3 | Gold | Depends on solution |
| S0992 | 18.3 | 4.0 | 12 | 13.9 | 65 | 11.3 | 21,000 | 17.5 | Silver | Disorder |
| s0999 | 54.7 | 3.4 | 320 | 170 | 165-170 | 41 | 880,000 | 54.98 | - | Flexible, Dimer |
Correlation between Crystal Structure and Prediction Model Molecular Envelopes
Density correlation score was calculated using programs gmconvert and gmfit.23,24 Number of Gaussian functions was set to 50, number of initial orientations for the global and local searches was set to 50, solutions were sorted by the correlation coefficient, default values were kept for the rest of the parameters.
RESULTS
CASP SAXS data collection
Hard targets were specifically chosen for experimental assistance with an expectation that added experimental information may improve predictor success. These targets were identified using sequence analysis (PSIBLAST, HHsearch). Communication between sample providers and the beamline was minimized to avoid compromising the CASP experiment. Crystallographers generously provided a total of ten protein samples. Marianne Ilbert provided protein for S0949; Petr Leiman, S0953/6F45.PDB25; Karoline Michalska, S0957/6CP8.PDB and S0968/6CP9.PDB; Owen Davies, S0980/6GNX.PDB26, Chi-lin Tsai, S0975; Mark van Raaij, S0981; Jose Henrique Pereira, S0985; Lindsey Spiegelman, S0987, and Andrew Lovering, S0992. An eleventh SAXS data set (S0999) was made available by Marcus Hartmann. All 11 CASP-SAXS targets were based on crystal structures. Seven out of the eleven samples represented multimeric assemblies and were evaluated as such in their entirety. Additionally, the results were evaluated separately for individual peptides or chains. Because four out of the 11 targets were hetero-dimeric, the number of individual peptide targets was 15. Target S0999 was sufficiently large that agreement was judged as five separate domains. All in all, we assessed 19 unique single-sequence targets.
The SAXS-assisted CASP category aimed to test the notion that SAXS may prove useful for experimentally validating structure prediction in general. SAXS would be suitable for this purpose as sample requirements are minimal and can be collected efficiently in HT. This efficiency of data collection was supported in CASP13 since SAXS data were provided for all samples shipped – 100% success rate for data collection and analysis. We collected HT-SAXS and/or SEC-SAXS data at the SIBYLS beamline 12.3.1 in the Advanced Light Source Synchrotron, depending on sample quantity.2-4 HT-SAXS provides the highest signal-to-noise data, while SEC-SAXS was used to purify stoichiometrically monodisperse samples. When sample quantity was low, only HT-SAXS data were collected. When possible, HT-SAXS and SEC-SAXS data were compared. Where SAXS curves overlaid, the higher signal-to-noise HT SAXS data were used and provided. For stoichiometrically polydisperse samples, SEC-SAXS data were provided.
SAXS analysis was coupled with sequence information in reports provided to predictors. Reports included information on whether SEC-SAXS applied, the quality of SAXS data collection, particular challenges relevant to the target, the processed SAXS curves, global parameters extracted from SAXS data, the pair distribution or P(r), 3D shapes and disorder prediction results calculated from DISOPRED. Several factors were considered in determining which value to give an experiment for the three-tier quality scale provided to predictors. The high quality “gold” rating was assigned to experiments where both HT- and SEC-SAXS provided the same scattering curve with low noise. Silver was assigned to curves where SEC-SAXS data were noisy or small discrepancies between anticipated and measured mass were observed. Bronze values were given when only HT-SAXS could be applied or larger inconsistencies were noted. Of the 11 targets, four were rated gold (highest quality), six were silver, and only one was bronze. Target S0968 was ranked bronze, as the molecular mass in solution (36 kD) suggested an ambiguous 1:2 multimeric complex of two similarly sized subunits or protomers (13.9 and 13.4 kDa). A new challenge section highlighted potential stoichiometric heterogeneity, flexibility, and multimerization. When flexibility was indicated by the SAXS signal, a disorder prediction analysis27 was included. In the case of S0975, the protein has a 4Fe-4S group, which was noted in this section.
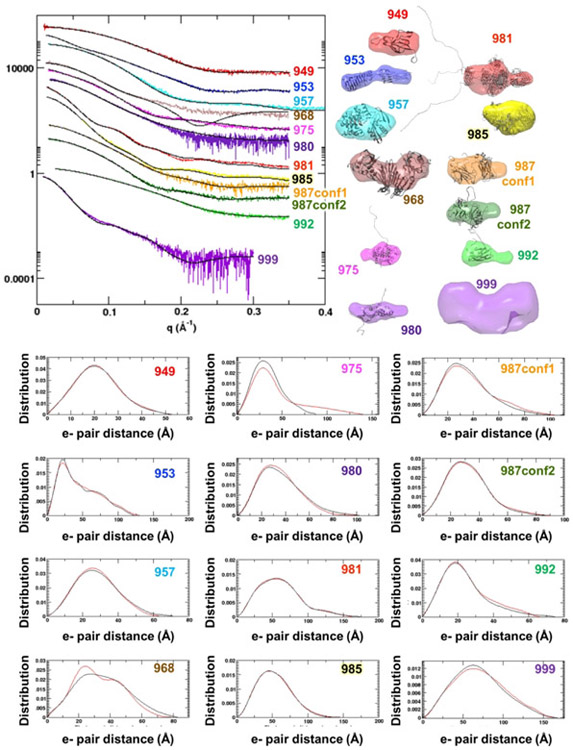
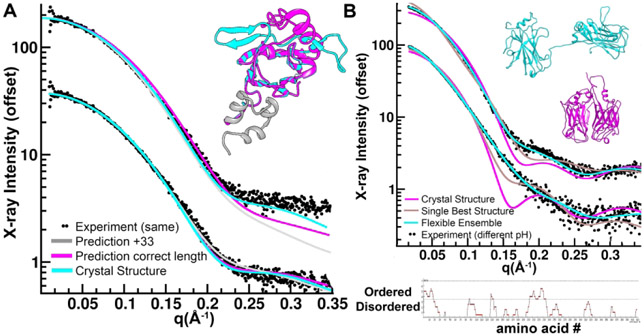
SAXS curves (Reciprocal space I vs q and Real space P(r)) and shapes for the eleven targets show the diversity of targets in CASP13 (Figure 1). In the case of S0987, the SEC-SAXS and HT-SAXS buffers were different yielding significantly different curves describing conformational differences of the monomeric protein. Both curves and analysis were provided to predictors. The global parameters (scalars) (Table 1) reveal information into structure and assembly. The Radius of Gyration (RG) characterization of the first moment of inertia for the samples ranged from 16 to 55 Å. The RG was estimated two ways. First through use of the Guinier region in reciprocal space, and second (Real space RG) through analysis of the P(r) function. All samples had less than 5% difference in these values from both methods, passing this data quality control.
Figure 1.
SAXS data for CASP13 targets. (Left upper Panel) Reciprocal space experimental SAXS curves (colored) are overlaid with the predicted scattering (black) from an ensemble of atomic models, found to best match the experimental data. SAXS curves can be scaled without losing information content, so the SAXS curves have been offset for visual clarity. The atomic model(s) are full-length models, based on the crystal structure or when appropriate, multimeric models based on the crystallographic lattice. (Right upper panel) Ab initio shape reconstructions based on the SAXS data and overlaid with a single representative atomic model. (Bottom panel) Real space SAXS curves for different targets (abbreviated CASP target IDs are provided on the graphs).
Only 36% (4 out of 11) of the proteins examined were monomeric: S0949, S0975, S0987, and S0992. The others formed multimeric assemblies. Mass was extracted via two methods. The SAXS curve itself can provide a concentration-independent estimate of mass. The mass of the folded region can be estimated from SAXS (MassSAXS) by defining the Porod-Debye range and calculating the Volume of correlation (Vc).14 SEC-SAXS was coupled to multi-angle light scattering (MALS), which provides an estimate of mass across an elution peak.
The Porod-Debye value (PD, PE or PX) provides objective insights into flexibility.15,28 PD is determined from the rate of decay as a function of q in the mid q range (0.05 < q < 0.2 Å−1) and depends on the volume of the protein. A q−2 dependence indicates largely unfolded structures while a q−4 indicates a globular one. The PD is represented as the negative of the exponent. Seven targets had a PD of 4, indicating a high proportion of folded regions as one would expect for CASP targets that were selected for their crystallizability. S0953, S0980, and S0981 had midrange PDs of 3.6 to 3.7. For S0968 and S0999, the PD scores of 3.1 and 3.4, respectively, indicated significant flexibility. In retrospect, comparisons of the PD scores to the percentage of missing regions in the crystal structure (Table 2), were generally correlated but there were exceptions. S0949, S0975, S0987, and S0992 had PD scores of 4 but had 8-18% of their sequence missing in the respective crystal structures. On the other side, S0953 and S0968 had minimal 2-4% missing, but had flexible PD scores of 3.6 and 3, respectively, suggesting their flexibility comes from domain motions.
Table 2. Crystal structures, Atomic Models and SAXS data.
Stoichiometry (Stoic) and Porod Debye (PD) were calculated from the SAXS data. Flexibility (% Order) was calculated from what the number of amino acids (AA) modeled in the crystal structure (pdb) and what the number of AA present in the SAXS sample. Agreement to the SAXS data (χ2) was determined for the crystal structure, from a single model with missing AA added back (CHIMERA), and with the missing AA and potential flexible domains allowed to move using a version of CHARMM implemented in BILBOMD. The fit of the crystal within the shape was determined by eye.
| Exp SAXS Stoic |
AA- saxs sample |
AA- pdb |
% order |
Predicted Model Sequence vs SAXS |
PD | Crystal, χ2 |
Full- length χ2 |
Fit to SAXS, χ2 |
Fit of crystal within shape |
|
|---|---|---|---|---|---|---|---|---|---|---|
| S0949 | 1 | 151 | 139 | 92% | +32/−10 | 4 | 1.64 | 1.41 | yes | |
| s0953 | 3:1 | 465 | 457 | 98% | ok | 3.6 | 12. | 2.1 | partly | |
| s0957 | 1:1 | 327 | 318 | 97% | ok | 4 | 1.3 | yes | ||
| s0968 | 1:2 or 2:2 | 466 | 484 | 96% | ok | 3.0 | 1.5 | partly | ||
| s0975 | 1 | 343 | 281 | 82% | ok | 4 | 11 | 3.3 | yes | |
| s0980 | 2:2 | 338 | 290 | 86% | −6aa | 3.7 | 15 | 2.4 | partly | |
| s0981 | 3 | 674 | 610 | 90% | −102 aa | 3.7 | 2.84 | 3.24 | partly | |
| s0985 | 2 | 863 | 842 | 98% | 0-42 vary | 4 | 19/1.81 | 16 | partly | |
| s0987 conf1 |
1 | 496 | 381 | 94% | −2-24 vary | 4 | 51 | 52 | 0.94 | yes |
| s0987 conf2 |
4 | 11 | 13 | 1.04 | ||||||
| S0992 | 1 | 126 | 107 | 85% | 0-16 vary | 4 | 114 | 14 | 2.8 | yes |
| s0999 | 2 | 3178 | 3083 | 97% | ok | 3.4 | 7. | 3.9 | partly |
Addition of 5% tetramer for S0985 improved χ2 to 1.8.
The relative ratio of the radius of cross-section (Rxc), the second moment of inertia of the protein to the RG provides information on the overall shape. When Rxc values are comparable to RG, the protein is globular. When Rxc is significantly smaller, the protein is elongated. S0953 had the smallest Rxc to RG (13 to 34.8 Å). Most of the proteins, including S0968, S0975, S0980, S0981, S0985, S0987, S0992, S0999, showed a smaller Rxc to RG, indicating a non-spherical overall organization.
SANS data were also provided to CASP predictors for target S0953 by the Institut Laue-Langevin facility. As the sample was completely hydrogenated, there was no advantage to using SANS data. SAXS has higher signal-to-noise than SANS, and the true advantage of SANS arises when components are differentially hydrogenated/deuterated. If SANS is considered for future CASP, identification of a target complex and a willing collaborator who prepare components under appropriate conditions should be more actively pursued.
After all predictions, assisted and regular, were submitted and finalized the atomic resolution structures were made available and reconciled with SAXS results. For proteins with regions missing in the crystal structure, we made models that included missing regions using Modeller.18 An improvement over CASP12, the targets were missing fewer amino acids: S0949 (8%), S0968 (4%), S0975 (18%), S0980 (14%), S0981 (10%), S0987 (6%), and S0992 (15%). When necessary, we created models with domains set as rigid bodies but with linkers allowed to move and identified ensembles of those models that matched the experimental data.20,21 Based on the χ2 metric < 2, three targets S0949, S0957, and S0968 showed reasonable fit when modeled with missing regions. As described in detail below, the solution state of 9 out of 11 targets (including S0968 – discussed below) differed in varying degrees from the crystallographically determined structures. Flexibility could take the form of disordered tails or that the architecture of the folded regions is adopting multiple conformations in solution. We found that the discrepancy for two of the targets could be explained by addition of unstructured tails, but we believe that the folded regions for 7 of the targets are adopting different conformations in solution. The fits of modified crystallographic structures are shown in Figure 1. It is notable that the P(r) for S0968 SAXS data did not match the crystal structure, despite the χ2 metric < 2.
Assessment of Predictions
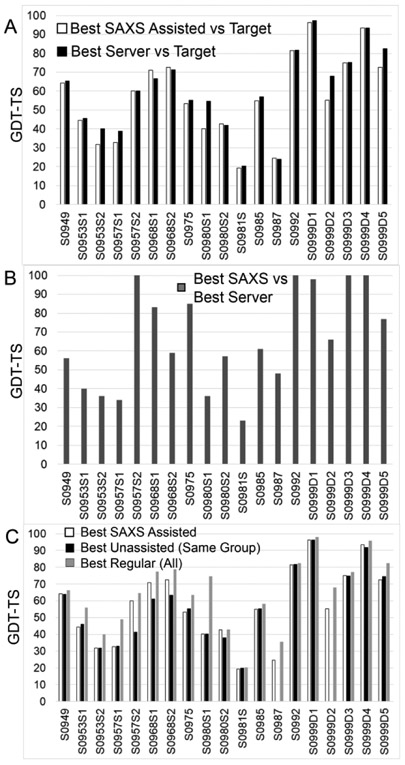
In CASP12 a criterion used to evaluate prediction improvement was the GDT_TS score of assisted predictions vs the regular predictions from the same group. Here we have taken a more stringent approach for domains comparing the best assisted prediction against the best server prediction (Figures 2A and 2B).
Figure 2.
Comparison of assisted predictions compared to regular (unassisted) prediction in CASP13, based on chain or on domain (S0999 only). (A) The GDT_TS scores of the best SAXS-assisted predictions and the best server predictions (regular) against the target crystal structure show that only for S0968 did SAXS-assisted models have higher GDT_TS scores than models from the best servers from the regular prediction. . (B) GDT_TS-based comparison of the SAXS-assisted with the best server prediction suggests the use of server models in the SAXS-assisted category, particularly when the GDT_TS score is 100. (C) The GDT_TS scores of the best SAXS-assisted predictions, the best regular, unassisted from the same group, and the best regular (all groups) against the crystal structure, shows that while SAXS-assisted models sometimes did better than the regular models from the same group, none did better than the best regular from all groups.
We want to note here that predictors had access to server models during both regular and assisted prediction,. However, during the assisted prediction, the best server models for six of the eleven targets were implicitly identified by the CASP committee through releasing these models as starting points in the refinement category. Personal communication with the predictors revealed that some of them used the refinement models. This complicates the analysis of how much SAXS contributed to the assisted models. Removing this uncertainty in future CASPs can help improving clarity of the analysis of results. We want to emphasize here that server models were only available for individual domains, and no server models (including a selected refinement model) were available for multimeric targets. Thus, we compared the SAXS-assisted to regular oligomeric assembly predictions from the same group and to the best regular prediction.
During our assessment, we considered how SAXS can be used to improve prediction models. SAXS can be added to a model accuracy assessment score to select starting models, to alter starting models for improved fit to the solution data, and to rank final models for submission. In the simplest scenario, the predictor can rank server models and submit the top five models. In our analysis, we identified that 20% of the domains submitted were unmodified server models. In eight out of thirteen cases where server models were available, the top GDT_TS-scoring SAXS-assisted model was a resubmitted server model. This is not unexpected as many predictors are testing their model accuracy scoring algorithms or testing their oligomerization or assembly algorithms. These server models could have been the “pre-selected” refinement model or a SAXS-selected server model. For the latter, we consider them a viable entry as SAXS was used for the selection.
Based on this “best server” criterion, SAXS assisted predictors generally had equivalent best predictions as the best regular servers (Figures 2A and 2B). The best regular server models are a high bar as several server models on these targets also scored best in CASP13 overall. Only one assisted prediction from the SBROD method run by the Grudinin group, the first subunit of the S0968 heteromer (S0968S1), showed modest 4 point improvement in GDT_TS score. This model was ten GDT_TS points better than the best regular model from the same group. In a comparison of the best SAXS-assisted models on all targets, five were closely similar to the best regular server models, suggesting that predictors used these server models without significantly altering them (Figure 2B). Three of these (T0957S2, T0992, and T0999S3) were released as refinement models and could simply be refinement models resubmitted into the SAXS category. The other two domain targets were not released as refinement models, and the high degree of GDT_TS similarity could have been from the SAXS data-based selection from among the server models.
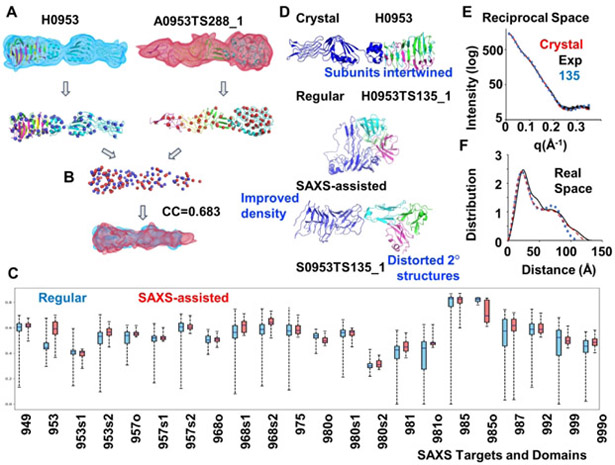
For difficult targets, a global density correlation method provides alternative perspective (Figure 3). This score captures global shape similarity of prediction model to the crystal structure, while placement of the local elements of structure, such as secondary and even tertiary structure, have little effect.23,24 The mean density correlation improved for ten targets, was worse in four and had no change in the remaining targets (Figure 3). Using this criterion, several predictions had a better score than any of the regular predictions. The improvement in the global density correlation reflects the ability to predict the protein envelope from SAXS data, the most recognized attribute of SAXS, and provides indirect evidence that SAXS data is being applied by the predictors. Getting the shape correct does not help if the topology is grossly incorrect, as discussed below for S0953. If the topology is correct, we suggest that it could help to shift secondary structure elements or promote conversion from compact helices to longer helices. Indeed, we identified individual examples (S0957, S0968, S0985, S0999) where the predictors had a roughly correct topology in their models and their SAXS-assisted model was better than the same group’s regular or all regular. We discuss them in the individual sections.
Figure 3.
Density correlation score. (A.) Atomic models of the target (left) and a prediction (right) are converted into low-resolution density maps and fitted with Gaussian mixture models (GMM) - shown in blue and red respectively. Centers (mean values) of the Gaussians are shown as spheres. (B) Two GMMs are superimposed so that overlap of the two distributions is maximized. Density correlation score is the correlation of the corresponding superposition of the simulated densities from A. (C) Box plots for the density correlation scores for all regular (by any group) and SAXS-assisted targets. (D) Target H0953 atomic models of the crystal structure and of an example of regular and the corresponding SAXS-assisted model from the Grudinin group. The SAXS-assisted model has a similar elongated shape, but secondary structure elements are clearly disrupted. This example also highlights how the subunits are entwined, and the predicted models appear to have been folded independently and placed together.(E and F) Corresponding to D, overlay of H0953 experimental data with SAXS curves predicted for crystal structure and for SAXS-assisted models in reciprocal and real space.
None of the SAXS assisted predictions at the monomeric or domain level were as good as the best regular predictions from the entire CASP13 predictor pool using the GDT_TS metric. The best assisted GDT_TS scores were plotted against the best regular scores (from the same group or from all groups) in Figure 2C.
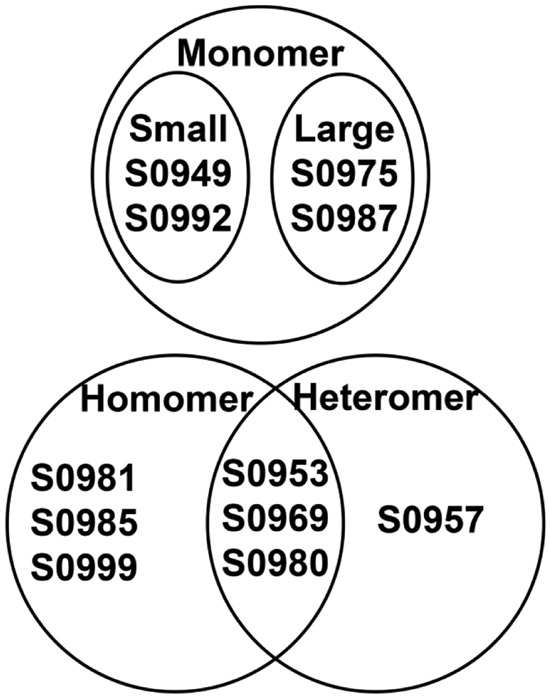
To highlight successes and challenges in the SAXS-assisted prediction, we perform case studies below for each of the targets. The assessment is separated into five categories based on the assembly of the protein and difficulty as indicated by best server GDT_TS scores: small monomers, large monomers, 1:1 heteromer, homo-oligomer and multimers of heteromers (Figure 4). Each type will require a unique adjustment to the prediction algorithm. The results below also detail modest improvements in prediction in the SAXS-assisted multimeric category of CASP13.
Figure 4.
Venn diagram highlighting oligomerization state of CASP13 targets in the SAXS-assisted category.
Small Monomeric Proteins (S0949 & S0992)
Only two targets, S0949 and S0992, were small monomers. SAXS data were consistent with crystallographic results for both targets, providing accurate guidance.
T0992 server predictions had GDT_TS scores in the 80s. The SAXS data reflected that of a small protein with a flexible tail, consistent with the 18 residues presumably too disordered to be modeled in the crystal structure. Small proteins are more sensitive to positioning of flexible termini in target crystal structure. Therefore, predictions for S0992 were arguably already highly accurate before SAXS information was added, and no improvement could be tracked with GDT_TS.
For S0949, the best SAXS-assisted prediction had a GDT_TS score of 64, nearly equivalent to the best regular server score of 65. . In comparing SAXS-assisted versus the regular predictions from the same group, all predictors did equivalent (less than 2 GDT_TS improvement in score) or worse than their best regular prediction. We suspect that the use of a sequence not consistent with what was in the SAXS sample is the reason. The sequence provided in the SAXS report conflicted with that listed at the Prediction Center. The discrepancy originated from the truncation the target provider made to the construct between the time of agreeing to send sample for SAXS analysis and data collection. All assisted predictors used either a 20% longer or 7% shorter sequence than the actual sequence in the SAXS sample, which likely had a significant negative impact given the small size (Table 2). The S0949 SAXS sample had only 151 amino acids in total, compared to 183 listed at the Prediction Center.
Despite the sequence disparity, first models for S0949 (i.e., top models as ranked by predictors) improved by average 4 GDT_TS points over the same group’s top-ranked regular models. Thus, SAXS data may have helped predictors in ranking models.
To effectively use SAXS data to improve predictions of small proteins where regular predictions are reasonably accurate (e.g. GDT_TS > 50), several factors should be carefully considered including the sequence correspondence between the measured and predicted construct (Table 2 and Figure 5). The top scoring prediction from MULTICOM was the only prediction using a sequence that was 11 amino acids shorter than the SAXS sample and therefore suffered the least from having an incorrect sequence. Yet, this prediction did not match the SAXS data to within error of the experiment and perhaps higher weighting of the fit to SAXS would have led to a better model.
Figure 5.
Improvements in sequence accuracy, hydration layer and flexibility are required for SAXS-assisted predictions. (A) Most predictors used a sequence that was 33 amino acids longer (grey model) than the sequence of the SAXS sample for S0949. Most predictions placed a helix (magenta and grey) in place of a sheet structure (cyan) on an otherwise correctly predicted model. These discrepancies are marginally discernable using SAXS calculators that adjust the hydration layer (bottom curves) but the correct model is a better fit when hydration layer is fixed (top curves). (B) Predictors did not include flexibility in fitting SAXS data. S0987 crystal structure is compact (magenta model). This crystal structure does not fit the either SAXS data set collected at two different pHs (top and bottom curves). Allowing the model to flex at positions where disorder is predicted (bottom DISOPRED result) and create an ensemble of models resembling the cyan model fits both data sets well varying in the relative proportion of compact configurations. The best single conformation generated by BILBOMD (grey) cannot fit either curve. Fitting the SAXS data with a rigid model can only be done by severely compromising the prediction of the domains.
The largest deviation from the target for all top scoring predictors was a 40 amino acid stretch where predictors had a helix in place of a two-stranded beta sheet structure. The volumes occupied by both helix and sheet topologies are similar. Using the FOXS SAXS calculator in default mode, both topologies fit the SAXS data nearly equivalently and therefore provide no discrimination. To achieve discrimination, assuming sequences are correct, a consistent treatment of the hydration layer, turning off the default option, is required. FOXS, and most other calculators will adjust the hydration layer to fit the data.20 However, at this level of resolution, allowing hydration layer parameters to drift compromises discrimination. Not allowing the FOXS hydration parameters to vary would have been sufficient to provide guidance to the crystal structure (Figure 5).
Large Monomeric Proteins (S0975 & S0987)
The two large monomeric proteins (S0975 and S0987: 343 and 408 amino acids respectively) had disordered sequence sections, based on residues not modeled in the crystal structure but present in the protein used in the crystallization; complicating predictors’ task. The SAXS data for both targets were of high quality as both HT- and SEC-SAXS were applied. In S0975, 18% of the protein was missing in the crystallographic structure: 35 residues at the N-terminus, 13 in the middle and 14 at the C-terminus. For S0987: 12 residues at the N-terminus, 10 in the middle and 3 at the C-terminus were missing. Sequence-based prediction indicated the missing termini were disordered. Predictors generally used folded and rigid models to represent the missing regions falling into a common trap where fold prediction algorithms will create folds even when a protein is intrinsically disordered. However, upon deeper investigation, this was not the only type of flexibility required to match the data. For peptides longer than 200 amino acids that are not allosterically and symmetrically stabilized, flexibility may be a factor for matching crystallographic targets.
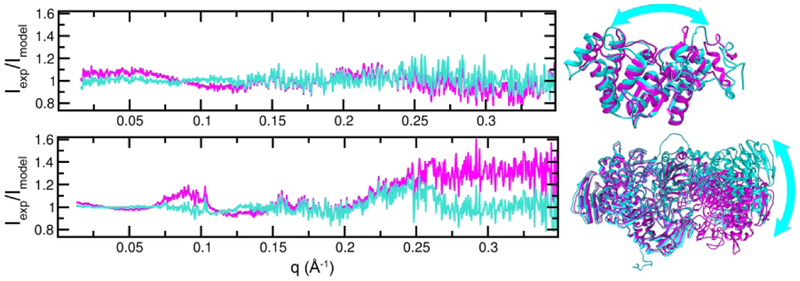
For S0975, SAXS-assisted models matched the SAXS data better than the reference crystal structure. The crystallographically-determined structure of S0975 is elongated (Fig. 6) and did not fit the SAXS data within the statistical error (χ2 > 2). Assisted models were more elongated conformations with mostly correct secondary structure elements. A model generated from a nonlinear NOLB normal mode analysis (NMA) of the crystal structure and consistent with the SAXS data (χ2 = 1) had this flatter shape. Comparing the NMA model to the crystal yielded a GDT_TS score of 73 relative to the crystal structure. If predictors are generating conformations based on the SAXS data, then ~73 is potentially the limit to the GDT_TS score they can achieve when scored against the crystal structure for this case. Crystal contacts or other factors likely compressed the structure.
Figure 6.
Conformations consistent with the SAXS data differ from those found in the target crystal structures. (Top) A flattened conformation (cyan) of S0975 fits the SAXS better than the crystal structure (magenta) as shown by a plot of the ratio between experiment and the two models, which is more sensitive than the simple overlay of curves in reciprocal space. An identical fit produces a ratio of 1.0 for all values of q. The models are GDT_TS = 72 apart. (Bottom) An asymmetric conformation (cyan) of dimeric S0985 fits the experimental SAXS data better than the symmetric form found in the crystal (magenta). The models are GDT_TS = 53 apart.
For S0987 and looking at all assisted predictors as a group, the mean GDT_TS improved with SAXS data for domain 1 of S0987D1 but not for the complete structure. Group 3Dbio, led by Dina Schneidman, scored best for domain one S0987D1 (GDT_TS = 50), compared to all the other groups participating in the assisted category. This model was slightly better than the best server model (GDT_TS=48), and 3Dbio models were significantly different from all server models. 3Dbio did not submit a model for the regular category. Looking at domain two (S0987D2) and the target as a whole, assisted predictions were same or worse than the respective group’s regular. A negative observation for both domains was that some predictors with GDT_TS scores over 50 for their regular had SAXS-assisted scores that dropped by as much as 30 points. Comparing the prediction models for the entire monomer (2 domains), these SAXS-assisted models were expanded while maintaining globularity, causing the internal fold to distort.
This expansion, not observed in the crystal lattice, could be explained by the solution data. SAXS experiments showed interdomain flexibility (Figure 5). S0987 was collected in two buffer conditions varying pH from 6 to 8. The SAXS profiles were markedly different changing the maximum dimension from 100 to 87 Å retaining the same molecular weight. This data indicates flexibly linked domains that shift relative to each other in differing conditions. Moreover, disorder predictions show a disordered region midway through the structure. The crystal structure indeed shows two large domains separated by a linker. In the crystal, the domains are in direct contact and the proteins maximum dimension is ~60 Å. Reconciling the crystal structure with the SAXS data suggests an ensemble of structures rather than a single structure should be used to measure prediction accuracy (Figure 5). To fit the SAXS data assuming flexible sections are rigid rather than flexible would require adjustments in protein parts that are deleterious relative to the regular predictions. No prediction group used an ensemble to fit SAXS data. Attempting to fit a single model to the SAXS data might have caused the observed distortion as the model tries to fit both longer and shorter distances.
For long peptides (> 150 amino acids) that are not allosterically stabilized through symmetric contacts, flexibility may be a consistent feature. Above 150 residues, the proteins often have multiple domains. Therefore, the best predictors can do with a static structure and fit SAXS data is to produce the average conformation. However, when the goal is to match a crystal structure, the most compact member of an ensemble may be the better choice.
One-to-One Heteromeric Complex S0957
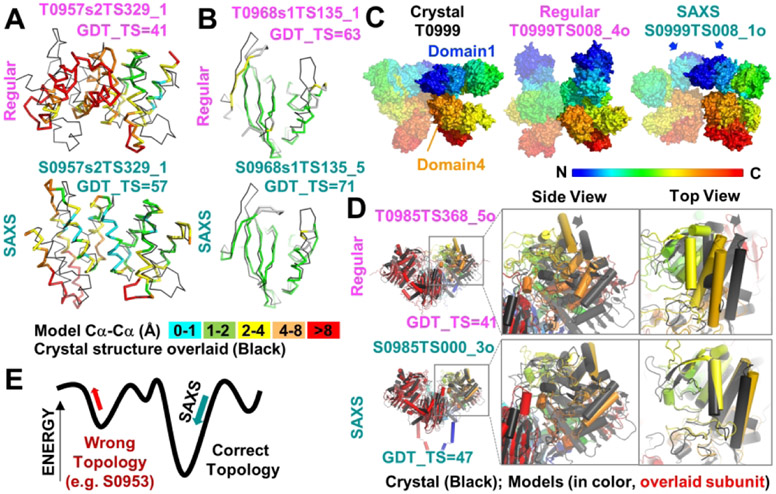
For this only 1:1 heteromeric complex with three domains, the top scoring SAXS-assisted models were worse or equivalent in GDT_TS to the top scoring server models. Discussion with some predictors revealed use of the refinement models released for these domains. Sergei Grudinin, one of our coauthors, used the same starting server models for target T0957S2 in the regular and the SAXS-assisted category and inclusion of SAXS data enabled him to identify a different server model, (Figure 7A). This is an example of where SAXS-assisted assessment of model accuracy was used to identify a better server model. For all predictors, target S0957 showed an overall improvement in density correlation (Figure 3). The elongated shape characteristic of the complex was captured by the SAXS-assisted predictors whereas regular were universally more globular. S0957 was also one of two targets where the crystal structure matched the SAXS data without additional modifications.
Figure 7.
Examples where SAXS-assisted models were better than best regular model from all groups or from the same group. (A and B) SAXS-assisted models for targets S0957s2 and S0968s1, respectively, had higher GDT_TS scores than regular models from the same group. Ribbon diagrams of domain models (colored by Cα-Cα deviation from the crystal) are overlaid onto the respective crystal structure (black). (C) The Pierce-group SAXS assisted assembly model for target S0999 was visually better than the best regular model from the same group. Surface models are colored by rainbow from the N to C terminus. One subunit of homodimer is partially transparent so that chains can be distinguished. Arrows highlight the domain 1 dimer interface that is predicted in the SAXS-assisted model. (D) The 3Dbio SAXS-assisted homodimer model for target S0985 had a better GDT_TS score for the entire ensemble than the best regular model from any group and better overlaid on the crystal structure. Arrows on regular model highlight rotation needed for proper overlay. Cartoon depiction with cylindrical helices of models when the left subunit (red) is overlaid onto crystal structure. The right subunit is colored by rainbow as in C and is the focus of the zoom views. (E) A model for how inclusion of SAXS data would have opposing effects on the fold energy term, depending on the starting model topology. If the starting model has the wrong topology, SAXS data would distort the wrong topology into the right shape. If the starting model has the right topology, SAXS data would lead to an improved fold with no deterioration of the folding elements.
Three Homomeric Complexes (S0999, S0981 & S0985)
The CASP13 pure homomeric proteins in the assisted category were all composed of large chains (target/monomer weight: S0999/170kDa, S0981/76kDa and S0985/98kDa). The large size made prediction and assessment challenging.
As with other large multidomain proteins, the T0999 crystal structure did not fit the SAXS data well, with a χ2 of 7. We were only able to improve the fit to χ2 of 4 by creating conformations derived from the crystal structure and defining flexible linker regions based on global B-factors and the Translation-Libration-Screw-rotation (TLS) from the crystallographic refinement. More advanced molecular dynamics analysis is required to obtain a better model for the T0999 homodimer. Based on the SAXS envelope prediction, these movements although significant are small and would likely not have negatively impacted the predictions, at their current precision level.
Target T0999 was a 340 kd homodimer with 5 domains in each subunit. At the domain level, the top scoring server scores were already exceptionally high. Domains 1-5 had GDT_TS scores of 97, 66, 75, 93, and 80 respectively. No SAXS-assisted model scored better than the server models. The high accuracy of the prediction models enabled a test of whether SAXS could aid in the assembly of relatively well-predicted domains. However, quantitative comparison of the scores gave a conflicting message. For the highest scoring models from the Pierce group, the Jaccard coefficient went from 0.15 for the regular model to 0.62 for the SAXS-assisted model and the QS globular from 0.20 to 0.70. These were both interface scores. Yet, the lDDT oligomer barely changed from .70 to .69, respectively and the GDT_TS score went from an inconclusive 16 to 23, respectively. Nonetheless, visual examination of the models revealed a significant improvement for one set of models (Figure 7C). In the regular category, the Pierce group correctly predicted the domain 4 interface but mispredicted that domain 1 was not interacting. The shape of the Pierce regular prediction was mistakenly tall. However, in the SAXS-assisted Pierce model, domains 1 and 4 were correctly placed at the dimer interface. During the SAXS-assisted prediction window, the team identified a homodimer template for Domain 1 (Brian Pierce, pers. comm). Domain 2 appears to be flipped although otherwise positioned correctly. Domains 3 and 5 were incorrectly shifted and were better in the Pierce regular. Although one can argue that the homodimer template helped at the later timepoint of the SAXS prediction window, the Pierce group included their regular models with their new models, used an interdomain hinge program, and ranked the entire set against multiple information from the SAXS data (RG, χ2, and SAXS envelope). Pertinent to the potential of SAXS to act in model accuracy assessment, their top ranked model was indeed the closest in quaternary orientation to the crystal structure.
SAXS data for S0981 were of high quality. With 10% of the structure added back in a compact conformation to the crystal structure the fit of the data is excellent. The residues missing in the crystal structure are likely causing the PD of 3.7.
A challenge for the predictors is that the subunits of the S0981 trimer are interwoven with one another. Thus, taking a hierarchical approach of predicting the subunit structure as independently folded and assembling the trimer thereafter is problematic. Many prediction algorithms aim to first predict the fold of each domain within a polypeptide chain, followed by assembly of domains together completing each unique polypeptide chain, followed by assembling the polypeptide chains together to form a multimer and finally assembling the multimers into heteromers. This approach fails when folding of multimers relies on interweaving of the components. The configurations of the domains within the subunit depend on the trimeric structure. GDT_TS scores of the subunit and the full trimeric structure were all below 20 and therefore an atomistic comparison of prediction to model is not informative. Based on a density correlation approach, SAXS-assisted predictions were better than regular predictions (Figure 3). The range of scores were narrower, indicating the SAXS data provided guidance to predictors, and the mean density correlation showed better matching of the shape. Given the excellent match of crystal to SAXS results, reviewing the strategies for using SAXS in predicting this structure should be informative.
In the case of the homomeric assembly of S0985, the 3Dbio group led by Dina Schneidman had a standout prediction using SAXS, outscoring all regular and assisted CASP13 participants (GDT_TS = 47 vs 41 for best regular, both calculated for the entire assembly) (Figure 7D). As found with S0999, a visual confirmation is more accessible as some scoring methods improved with SAXS (GDT_TS and lDDT-oligomer) while others got worse (interface scores, RMSD-glob). When one subunit of the homodimer is overlaid (colored red), the best regular is shifted relative to the crystal structure (see arrows). The 3Dbio model overlays better onto the crystal structure than the best regular. Unlike S0999, the SAXS-assisted model got significantly worse when comparing QS globular (0.30 SAXS vs 0.41 regular) and Jaccard scores (0.29 SAXS vs 0.37 regular). At the subunit level, the top five predictors scored equivalently assisted vs regular with GDT_TS scores in the 50s.
A difference between the solution and crystal conformation played a role, as SAXS data did not match the crystal structure. SEC-SAXS data quality was excellent and the single elution peak had a MALS mass measurement in agreement with a dimeric structure. The subunit to subunit interface is large and SAXS data suggests alternate rotations of the subunits relative to one another (Fig. 6). A comparison of a best fitting SAXS conformation (applying normal modes analysis) to the MX structure yielded a GDT_TS score of 47 - comparable to 3Dbios result.
Despite the monomer to monomer orientational differences in the crystal structure and the solution state, the interface was consistent. In a post-CASP analysis, we tested if we could obtain the correct interface with the SAXS data based on a prediction model. Using the best monomeric predictions with GDT_TS > 50, exhaustive and blind docking of monomers using C2 symmetry generated 600 symmetric dimer models. Ranking models by a χ2 comparison of calculated and experimental SAXS data alone provided excellent guidance on the correct interface and is exemplary of how SAXS might benefit predictors in monomeric structures even with conformational variation.
Heteromeric Complexes that form larger multimers (S0953, S0968, & S0980)
SAXS benefited predictors on two of three targets (S0953 and S0968) that formed multimers of heteromers. SAXS-assisted models showed a modest 2-4 point improvement in GDT_TS scores on predictions of the individual subunits of S0968 (a 2:2 heteromer) compared to best server model from the regular category. Improvements in S0953 (a 3:1 heteromer) were best measured using a density correlation approach, as all CASP13 fold predictions were significantly far from the target (Figure 3). For S0980 (a 2:2 heteromer), there were no obvious improvements from the SAXS data.
For domain 1 (S0968S1), the top scoring group’s best SAXS-assisted model for SBROD (GDT_TS = 71) is the only SAXS-assisted model that was better than the best server model (by 4 points). It outperformed SBROD’s best regular prediction by 10 points (Figure 7A) and showed 83% similarity to the refinement model. The outer beta strands were better placed in the SAXS-assisted model. This SAXS-assisted SBROD model was not better than the best regular by A7D (GDT_TS=78). In considering what can go wrong, MULTICOM, while scoring well regular, did not score well assisted. Discussion with the MULTICOM team revealed that the SAXS data were fit assuming that the heteromer did not further multimerize. Fitting SAXS data with a 1:1 model for a sample that is 2:2 will confound the algorithm as only an over expanded 1:1 can fit the volume of a compact 2:2 complex. This was particularly apparent in the second subunit of S0968S2 where MULTICOM scored well regular (GDT_TS = 71) but poorly assisted (GDT_TS = 43).
For domain 2 (S0968S2), the top scoring SBROD model in the SAXS-assisted category (GDT_TS=73) was nearly equivalent in score to the top server (GDT_TS=71). We view the 2-point improvement as equivalent.
Despite improving model accuracy for the individual subunits of S0968, SAXS data did not benefit predictors for the total complex. This is possibly due to the SAXS data fitting to a different 2:2 assembly in solution. Cross-linking contacts agreed with the crystallographic orientation of parts of the assembly. In depth analysis will be required to ascertain which complex is occurring in solution. Different buffer conditions could induce transitions in multimeric assembly though further experiments are required to rule out possible systematic errors. Regardless of the assembly, SAXS data informed on a flat compact object, which constrained predictions to tighter, more compact structures than were provided in the regular category.
SAXS data had a positive impact on predictions for S0953, though not from the GDT_TS perspective. S0953 was a difficult free modeling target forming a 3:1 heteromeric multimer. SAXS data indicate that the extended beta sheet region is bent relative to the heterotetramer interface region, compared to the more linear configuration observed in the crystal lattice (Figure 3).
As found for S0981, folding approaches where domains are individually folded before assembly were confounded by the trimeric intertwined beta structure. The best GDT_TS score for the full complex from all CASP predictors came from the assisted Grudinin algorithm. However, the score was very low (<18) and was only marginally better than its un-assisted score. Low scores of this kind indicate that predictions were not accurate. However, when viewed from a density correlation perspective (Figure 3), predictors benefited from SAXS data. Examination of the H0953 prediction models reveals that the regular atomic models were often globular, and all the SAXS-assisted models were elongated (one example in Figure 3). However, some of the secondary structures were distorted, as if the atomic model was being squashed into the envelope. Notably the topology of the regular model was wrong, and conversion to the correct topology would have required unfolding and overcoming large energy barriers (Fig. 7E). This example suggests how SAXS can be misleading when the topology is incorrect and furthermore, that these false positives may be detected by examining the effect of SAXS data on model accuracy parameters (fit to optimal secondary structure parameters, nearest neighbor, evolutionary covariance, etc.). When we examine the similarity of the experimental data to the predicted data from the model in reciprocal space, it shows how well the Grudinin group fit the curve in reciprocal space. However, comparison of the model to the experimental data in real space revealed significant differences in the curve, suggesting real space as an alternative strategy for fitting the SAXS data. This is another notable example where the crystal structure did not closely fit the SAXS experimental data, indicating that the target had a different conformation in solution. Yet the crystal was closer to the solution data than the incorrect prediction model, indicating room for computational improvements.
Target H0980 was a 2:2 heteromer. The top scoring SAXS-assisted models for S0980s1 scored below or similar to the top scoring server models. Visual examination of the structure shows that one chain folds into a globular fold with a central beta sheet that forms the major dimer interface on itself and that the other chain has minimal secondary structure, packing along the surface of the first chain. All predictors folded the second chain in isolation from the first chain and thus could not predict the extended chain properly. Using models based on the crystal structure with the missing residues replaced, we were unable to conclusively distinguish between different oligomerization states. The best fit that we could obtain had a χ2 of 2.4. The Porod Debye number was 3.7, suggesting some flexibility. Thus, the protein in solution was adopting multiple conformations masking a definitive identification of the assembly state or there was an error in the data collection (e.g. buffer subtraction error). Further analysis is needed to distinguish the possibilities.
DISCUSSION
SAXS-assisted prediction showed some bright spots during CASP13 and identified areas for further improvement. In one case, the predictor used SAXS for model accuracy assessment, thereby experimentally validating one server model over another. In another case, the edges of the protein were improved. For the most difficult targets like S0981 and S0953, where all predictors were challenged at the fold level, assisted predictors generated models with higher density correlation to the target (Figure 3). Density correlation is not beneficial when the starting topology is wrong. However, for predictions with the right gross topology, the ability to fit models within the envelope could twist folds into the correct structure, correct the secondary structure at the edges, or reorient domains within an assembly (S0985 and S0999 examples). Thus, SAXS has potential value to prediction algorithms in defining interdomain and intersubunit orientations and/or conformational plasticity, which are critically important, unsolved areas of protein structure prediction. Improvements in assisted algorithms, experimental data quality and in how SAXS results were communicated to predictors by CASP organizers all contributed to this success. However, for fold accuracy of the domains based on GDT_TS, no SAXS-assisted model was better than best regular model. Below we discuss factors that could be addressed for further improvement and the importance of continuing assisted prediction in CASP14.
Solution structure vs Crystal Structure
Of particular relevance to future CASPs is that SAXS-based models of CASP targets, many of which are selected by the prerequisite of having been crystallized, are not usually monomeric and rigid. This discrepancy between solution and crystal structures has precedent but have been limited to anecdotal examples in the case of SAXS.29-34 In light of the game changing accuracy gains CASP predictors have made in the free modelling regular categories in CASP12 and 13 and their use of crystallographic databases, a surprising new realization is how few proteins are in their crystallographic conformation in solution, based on agreement with SAXS data. In CASP13, over half of the proteins (S0953, S0968, S0975, S0980, S0985, S0987, and S0999) were found to be in a different architectural conformation than that found in the crystal, a number consistent with a database study on differences between NMR and crystal structures.29 These cases are considered different when a full-length model, based on the crystal structure and with missing regions replaced, does not match the SAXS data. Importantly, these were not conformational differences of disordered regions but rather differences in the relative position of one domain or sub-domain to another. Although we cannot exclude the possibility that the disagreement is from inaccuracies in modeling the disordered region, it is our experience that it is more often the other way around - that the disordered region modeling can mask domain movements. Thus, we view our assessments that certain targets are in a different conformation in solution as fairly reliable but not conclusive. Additional experimental analysis, such as NMR, would be required for a conclusive assessment. For these proteins, models based on crystal structures adjusted through domain reorientation or normal modes analysis better fit the SAXS data. Models that fit the SAXS data of these proteins therefore cannot match the crystal structure exactly (GDT_TS of 100). Based on CASP13 target S0985, the solution conformation may differ from that of its crystal by as much as GDT_TS of 50, which is on par with prediction accuracy on many targets. In other words, a prediction may accurately represent the conformation in solution but would not score well against the crystal structure. More emphasis on non-rigid evaluation scores, such as lDDT, CAD, SphereGrinder, or RPF may in part address these structural discrepancies.35-37
Including SAXS data is thus a double-edged sword. CASP often uses not-yet-released crystal structures as a source for their targets and, for those targets, aims for a perfect fit to the precisely determined crystal structure. Given the conformational differences between solution and crystallographic conditions, predictors cannot reach a GDT_TS of 100 by accurately fitting SAXS data. However, SAXS data provides information on the structure adopted in arguably more physiologically and functionally relevant conditions. For example, recent comparisons of SEC-SAXS data taken across the peak unveils functional DNA repair complex conformations in solution can sample the compact crystal structure conformations, but these interconvert with more extended conformations that enable the functional release of contacts.38
In the short term, moving away from crystallography as the gold standard, which has formed the backbone of CASP, is likely unwise. Small targets are less likely to have these challenges, and models fitting SAXS data may hope to achieve GDT_TS > 80. However, for large targets where conformational flexibility is more likely, reconciling a solution-guided prediction with a crystallographic target may only be possible by adjusting the SAXS conformation. Predictors may need to compact or make commensurate adjustments that consider crystallographic lattice packing. A normal modes analysis of each prediction may be helpful to produce the most compact configuration.
In the longer term, conformationally flexible structures as indicated by the SAXS data are likely to be an increasingly important consideration. This is particularly true as machine learning becomes a central tool for prediction. Machine learning is particularly prone to learning inherent flaws in training data sets and will only reinforce what is likely to be a view of proteins that is systematically misrepresented. Perhaps inclusion of SAXS data to training databases could improve algorithms to model solution conformations.
Fitting SAXS data with ensembles for flexible systems
S0987 was an example where an ensemble was required to fit the SAXS data rather than one rigid structure (Figure 5). However, the same issue will occur for protein disordered regions and those undergoing conformational changes. For disordered regions, several predictors continue to fold these regions despite clear indications provided to the contrary by disorder prediction algorithms. As the accuracy of predictions becomes better, the inherently flexible nature of proteins will require more consideration. Some conformational modes are indistinguishable by SAXS, others like those discussed in the preceding section have observable impacts. Fitting a SAXS curve from a flexible or disordered system with a single rigid structure will impact other parts of the model. If the protein is flexible and the experimental structure is a crystal structure, the CASP community may have to decide between keeping the crystal structure as the reference structure for assessment or generating reference model(s) based on the crystal structure but modified to fit the SAXS data. If the former, then predictors may need to compact their models before submitting. If the latter, development of methods to generate realistic SAXS-based models with proper geometry in silico is needed. These methods should be capable of identifying regions of the protein that artifactually pack in the crystal lattice.33
Algorithms for multimeric structures
When we first introduced high-throughput SAXS analysis, we were surprised by the number of oligomers.2 At least half of the proteins we interrogated formed multimers. In this round of CASP this was further accentuated as 63% were multimers: homomers or heteromers. In SAXS, information on the monomeric target is convoluted with information on higher order assembly. Predictors must become more aware of the oligomerization state and assemble models accordingly.
Predictors were aware of heteromeric structure designation and appropriate steps were taken to fit the SAXS data as heteromeric. However consideration of homomeric structures was less uniform among predictors. Several predictors fit monomers into SAXS data from a homomer with detrimental consequences on their model. Monomeric proteins are likely to become the exception in CASP as most new folds may come from multimeric assemblies.
One reason we expect many new folds will come from multimeric structures is that multimerization enables intertwined polypeptides or domains; opening up new folding possibilities. With the hard targets for the assisted category in CASP13, many folds were obligate homo- or hetero-oligomers, meaning that the subunits likely fold cooperatively. In contrast, predictor models of these targets were assemblies of independent folds that were rigidly assembled. In predicting these structures, the commonly used hierarchical approach of first folding domains independently, then assembling domains, and finally bringing subunits together will typically fail. On the other hand, SAXS can provide insight into whether straightforward independent folding of each subunit has generated an accurate topology or if a more sophisticated approach is required.
Distinguishing incorrect vs correct starting model topology with SAXS data
While many arrangements of the same number of atoms can fit a SAXS profile, most are energetically impossible. Scoring functions provide constraints on allowable configurations. If protein topology is distorted to an energetically unfavorable configuration to fit SAXS data, this distortion signals that the starting model may have the wrong topology. So, new starting models with different topologies should be considered.
For example, many predictors utilized starting models from regular prediction approaches. When these starting models did not fit the SAXS data, movements of secondary structures were made to improve fit. Few truly topological changes were made between assisted and regular. These CASP results show that SAXS may drive a topologically-correct model towards a better energy score with better features of a folded protein, but can also drive a topologically-incorrect model towards a worse energy score (Figure 6). Topologically diverse starting models would increase the chances that there is a topologically correct model that the SAXS data can identify and improve. Using a similar paradigm could aid in assessing whether multimeric models generated by hierarchical methods need to be re-evaluated with an obligate multimer fold topology.
Many assisted algorithms explicitly included a SAXS comparison term between model and experiment in their scoring function. SAXS comparison can be done in reciprocal or in real space. The two are related through a Fourier transform. A challenge for using reciprocal space is the exponential decay as a function of the scattering angle q which is characteristic of the scattering contrast between solvent and protein and can be affected by hydration layer considerations or buffer subtraction errors rather than fold. Many reciprocal comparison methods allow this feature to dominate the outcome. Using a comparison in real space removes this strong bias (Figure 3), and features related to fold become more strongly weighted. In addition, the real space function has a relationship to contact distances used in many prediction approaches providing interesting options for score function construction.
If reciprocal space is used, accuracy of prediction has reached the stage where the hydration layer impacts the ability of SAXS to discriminate between close models. Many SAXS calculators allow the hydration layer to adjust in both how ordered the structure is and how much scattering contrast relative to bulk water it has. In the case of S0949 (Figure 5), fixing the hydration layer to default values for all models provided the necessary discrimination between the target and another fold of equivalent volume. Prediction of the hydration layer in SAXS, crystallography and EM is an active area of research and will benefit the structure prediction community.
Sequences, Assessment and Experimental Considerations
Variation in the sequence of predicted models and the SAXS protein construct was much smaller than in CASP12 (Table 2).12 However the margin of improvement that predictors were looking for with SAXS data in CASP13 also became more constricted, particularly for small monomeric systems. For 5 of 11 targets, the model entries matched the SAXS sample (Table 2). For 4 targets (S0949, S0980, S0981, S0987), the model sequence entries all were different from the SAXS samples and varied from each other. Predictors are allowed to submit incomplete models, but using an incorrect sequence does not make sense for fitting to SAXS data. For S0985 and S0992, some prediction entries had the correct sequence and some did not. Completely correct sequences are particularly important for small proteins and proteins with disordered regions. For these systems a disordered and extended 5 amino acid terminus can increase the maximum dimension by 12.5Å, for example.13
Assessment of predictor success remains somewhat complex. Target size and difficulty require more than one scoring criterion. Herein we utilized GDT_TS and density correlation however alternate metrics may have improved assessment. In addition, models for the refinement category were released at the same time as SAXS data. These models potentially provide additional information beyond what the regular predictors used as input complicating assessment in some cases. Delaying the release of these models during prediction would remove this uncertainty in assessment.
On the experimental side, several improvements can be made in SAXS data collection to better aid structure prediction. Buffer subtraction from sample scattering can lead to systematic errors. In CASP13 measurements, buffer subtraction differences between HT-SAXS and SEC-SAXS were observed. Significant and recent improvements have been made at the SIBYLS beamline that have reduced these errors.
Data will be provided to higher angle. CASP13 measurements were generally stopped at q < 0.35 Å−1 as few SAXS calculators showed accuracy beyond this range. However, as the CASP community has become more sophisticated with SAXS analysis and the increased need for resolution of prediction, treatment of wider angles may provide some additional discrimination.
Based on this CASP13 analysis, we recommend more attention paid to eight points: 1) the correct sequence corresponding to the SAXS sample, 2) the solution oligomerization state, 3) intrinsic disorder predictions, 4) ensembles of conformations when necessary, 5) cooperative folding possible for obligate homo or hetero-oligomers, 6) a topologically diverse set of starting models; 7) the effect of SAXS data on model accuracy, and 8) post-SAXS compaction to mimic crystallographic conditions or a change in how CASP scores model accuracy. As providers of SAXS data, we will work in parallel with predictors to create tools and improve SAXS data quality for CASP scientists.
Conclusion
The experimentally assisted category seeks to supplement sequence information with realistically attainable experimental data for prediction of any soluble protein target. We identified clear examples where SAXS aided predictors in model accuracy assessment of their models at the domain fold level and for assembly. For some easier folds, we found that CASP13 prediction has reached an accuracy approaching the differences between solution conformation and crystallographic conformation. This will limit the impact of SAXS in assisting prediction algorithms in cases where the reference structure is a crystal structure and the crystal structure is not consistent with the SAXS data. Predictors may be able to take steps that modify their SAXS-assisted prediction into a more crystallographic one, or what the models are scored against may be changed in future CASPs. Prediction algorithms need and will continue to benefit from the precision of crystallography for accurate residue interactions, but defining the solution conformation by SAXS and/or NMR is likely important for biological relevance. Biology occurs in the active sites and interfaces on the protein surface, indicating that the ultimate bar for predicted models is not only the right fold but also the correct surface. These functional surfaces are impacted by oligomerization orientation and disordered regions - the features on which solution data can be informative. Structure prediction has the powerful potential to provide biologically relevant models with atomic accuracy that encodes the inherent conformations of proteins in solution.
Acknowledgements
The authors declare no conflict of interest. For financial support, we thank NIH (PO1CA092584 to J.A.T., G.L.H, M.H. and S.E.T. R35CA22043 to J.A.T, R01GM110387 to S.T., and R01GM100482 to K.F.). J.A.T. acknowledges support by a Robert A. Welch Chemistry Chair, the Cancer Prevention and Research Institute of Texas, the University of Texas System Science and Technology Acquisition and Retention. DG and JD were supported by the RCSB PDB, jointly funded by the NSF, the NIH and the DOE (NSF-DBI 1338415; Principal Investigator Stephen K. Burley). This research used resources of the Advanced Light Source, which are DOE Office of Science User Facilities under contract no. DE-AC02-05CH11231. The SIBYLS beamline 12.3.1 and our CASP efforts are supported by the DOE-BER IDAT program, the NIH supported ALS-ENABLE (P30 GM124169) and a CRG3 from KAUST. Molecular graphics and analyses performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.
REFERENCES
- 1.Kryshtafovych A, Schwede T, Topf M, Fidelis K, Moult J. Critical Assessment of Methods of Protein Structure Prediction (CASP) – Round XIII. Proteins. This issue 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL 2nd, Tsutakawa SE, Jenney FE Jr., Classen S, Frankel KA, Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MW, Tainer JA. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nature methods 2009;6(8):606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Classen S, Rodic I, Holton J, Hura GL, Hammel M, Tainer JA. Software for the high-throughput collection of SAXS data using an enhanced Blu-Ice/DCS control system. Journal of synchrotron radiation 2010;17(6):774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Classen S, Hura GL, Holton JM, Rambo RP, Rodic I, McGuire PJ, Dyer K, Hammel M, Meigs G, Frankel KA, Tainer JA. Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. Journal of applied crystallography 2013;46(Pt 1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyer KN, Hammel M, Rambo RP, Tsutakawa SE, Rodic I, Classen S, Tainer JA, Hura GL. High-throughput SAXS for the characterization of biomolecules in solution: a practical approach. Methods in molecular biology 2014;1091:245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosey CA, Tainer JA. Evolving SAXS versatility: solution X-ray scattering for macromolecular architecture, functional landscapes, and integrative structural biology. Current opinion in structural biology 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Quarterly reviews of biophysics 2007;40(3):191–285. [DOI] [PubMed] [Google Scholar]
- 8.Rambo RP, Tainer JA. Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annual review of biophysics 2013;42:415–441. [DOI] [PubMed] [Google Scholar]
- 9.Rambo RP, Tainer JA. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Current opinion in structural biology 2010;20(1):128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svergun DI. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. Journal of applied crystallography 1992;25:495–503. [Google Scholar]
- 11.Tamo GE, Abriata LA, Fonti G, Dal Peraro M. Assessment of data-assisted prediction by inclusion of crosslinking/mass-spectrometry and small angle X-ray scattering data in the 12(th) Critical Assessment of protein Structure Prediction experiment. Proteins 2018;86 Suppl 1:215–227. [DOI] [PubMed] [Google Scholar]
- 12.Ogorzalek TL, Hura GL, Belsom A, Burnett KH, Kryshtafovych A, Tainer JA, Rappsilber J, Tsutakawa SE, Fidelis K. Small angle X-ray scattering and cross-linking for data assisted protein structure prediction in CASP 12 with prospects for improved accuracy. Proteins 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soranno A, Longhi R, Bellini T, Buscaglia M. Kinetics of contact formation and end-to-end distance distributions of swollen disordered peptides. Biophysical journal 2009;96(4):1515–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature 2013;496(7446):477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reyes FE, Schwartz CR, Tainer JA, Rambo RP. Methods for using new conceptual tools and parameters to assess RNA structure by small-angle X-ray scattering. Methods in enzymology 2014;549:235–263. [DOI] [PubMed] [Google Scholar]
- 16.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of applied crystallography 2003;36:1277–1282. [Google Scholar]
- 17.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophysical journal 2001;80(6):2946–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, Goddard TD, Meng EC, Sali A, Ferrin TE. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. Journal of structural biology 2012;179(3):269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic acids research 2010;38(Web Server issue):W540–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic acids research 2016;44(W1):W424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. General physiology and biophysics 2009;28(2):174–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann A, Gradinin S. NOLB: Nonlinear Rigid Block Normal-Mode Analysis Method. J Chem Theory Comput 2017;13(5):2123–2134. [DOI] [PubMed] [Google Scholar]
- 23.Kawabata T Multiple subunit fitting into a low-resolution density map of a macromolecular complex using a Gaussian mixture model. Biophysical journal 2008;95(10):4643–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawabata T Gaussian-input Gaussian mixture model for representing density maps and atomic models. Journal of structural biology 2018;203(1):1–16. [DOI] [PubMed] [Google Scholar]
- 25.Dunne M, Denyes JM, Arndt H, Loessner MJ, Leiman PG, Klumpp J. Salmonella Phage S16 Tail Fiber Adhesin Features a Rare Polyglycine Rich Domain for Host Recognition. Structure 2018;26(12):1573–1582 e1574. [DOI] [PubMed] [Google Scholar]
- 26.Dunce JM, Milburn AE, Gurusaran M, da Cruz I, Sen LT, Benavente R, Davies OR. Structural basis of meiotic telomere attachment to the nuclear envelope by MAJIN-TERB2-TERB1. Nat Commun 2018;9(1):5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones DT, Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 2015;31(6):857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 2011;95(8):559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrec M, Snyder DA, Zhou Z, Young J, Montelione GT, Levy RM. A large data set comparison of protein structures determined by crystallography and NMR: statistical test for structural differences and the effect of crystal packing. Proteins 2007;69(3):449–465. [DOI] [PubMed] [Google Scholar]
- 30.Faraggi E, Dunker AK, Sussman JL, Kloczkowski A. Comparing NMR and X-ray protein structure: Lindemann-like parameters and NMR disorder. J Biomol Struct Dyn 2018;36(9):2331–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai YT, Reading E, Hura GL, Tsai KL, Laganowsky A, Asturias FJ, Tainer JA, Robinson CV, Yeates TO. Structure of a designed protein cage that self-assembles into a highly porous cube. Nat Chem 2014;6(12):1065–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsutakawa SE, Van Wynsberghe AW, Freudenthal BD, Weinacht CP, Gakhar L, Washington MT, Zhuang Z, Tainer JA, Ivanov I. Solution X-ray scattering combined with computational modeling reveals multiple conformations of covalently bound ubiquitin on PCNA. Proceedings of the National Academy of Sciences of the United States of America 2011;108(43):17672–17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsutakawa SE, Yan C, Xu X, Weinacht CP, Freudenthal BD, Yang K, Zhuang Z, Washington MT, Tainer JA, Ivanov I. Structurally distinct ubiquitin- and sumo-modified PCNA: implications for their distinct roles in the DNA damage response. Structure 2015;23(4):724–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vestergaard B, Sanyal S, Roessle M, Mora L, Buckingham RH, Kastrup JS, Gajhede M, Svergun DI, Ehrenberg M. The SAXS solution structure of RF1 differs from its crystal structure and is similar to its ribosome bound cryo-EM structure. Molecular cell 2005;20(6):929–938. [DOI] [PubMed] [Google Scholar]
- 35.Huang YJ, Mao B, Aramini JM, Montelione GT. Assessment of template-based protein structure predictions in CASP10. Proteins 2014;82 Suppl 2:43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mariani V, Biasini M, Barbato A, Schwede T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 2013;29(21):2722–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olechnovic K, Monastyrskyy B, Kryshtafovych A, Venclovas C. Comparative analysis of methods for evaluation of protein models against native structures. Bioinformatics 2019;35(6):937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y, Millott R, Kim HJ, Peng S, Edwards RA, Skene-Arnold T, Hammel M, Lees-Miller SP, Tainer JA, Holmes CFB, Glover JNM. Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis. Structure 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]