Abstract
With the advance of experimental procedures obtaining chemical crosslinking information is becoming a fast and routine practice. Information on crosslinks can greatly enhance the accuracy of protein structure modeling. Here, we review the current state of the art in modeling protein structures with the assistance of experimentally determined chemical crosslinks within the framework of the 13th meeting of Critical Assessment of Structure Prediction approaches. This largest-to-date blind assessment reveals benefits of using data assistance in difficult to model protein structure prediction cases. However, in a broader context it also suggests that with the unprecedented advance in accuracy to predict contacts in recent years, experimental crosslinks will be useful only if their specificity and accuracy further improved and they are better integrated into computational workflows.
Keywords: CASP13, chemical-crosslink-assisted protein structure modeling, chemical crosslinking/mass spectrometry
Introduction
Critical Assessment of Protein Structure Prediction (CASP) is a bi-annual meeting that started in 1994 and uses a blind prediction format to assess the accuracy of various protein structure modeling approaches1. Protein sequences (targets) are released to the public for modeling, while experimental laboratories attempt to solve their structures using X-ray crystallography, NMR spectroscopy or cryo-electron microscopy. The experiments run through the summer months, after which the predicted structures are compared to the experimentally solved ones to identify the approaches that resulted in the most accurate predictions. With the advances in and increased accessibility of high-throughput experimental techniques2-4, data-assisted categories were added to the CASP experiment starting at CASP11 in 2014. Among several data-assisted categories, here we review advances in the chemical crosslinking/mass spectrometry (XL-MS) data-assisted category in CASP13. In this setting, information on chemically-crosslinked residues provides additional restraints that can be incorporated into the modeling of protein structures. Compared to classical structural characterization methods such as X-ray crystallography and NMR spectroscopy, the practical advantages of the XL-MS technique are that it only requires a small amount of sample (nanomoles or less), can be performed on crude, heterogeneous and dilute protein samples and can analyze flexible protein structures. Moreover, crosslinking experiments can be performed in a relatively short timeframe (days). Another possible advantage is that crosslinks are established in solution and therefore can potentially be more informative about the in vivo organization and dynamics of the target protein.
All targets in the XL-assisted modelling category were solved by X-ray crystallography and provided to the XL-MS labs as purified protein samples. CASP organizers asked some of these X-ray crystallography groups to share purified protein samples. The primary focus was on difficult-to-model protein targets, for which there were no trivial templates available in structural databases. The samples were shipped to two research groups specializing in chemical crosslinking and mass spectrometry: Alexander Leitner’s group (Zurich) and Juri Rappsilber’s group (Berlin, Edinburgh). Some proteins were shipped to both groups, while some to only one (A.L.). The two groups used different experimental approaches to generate the crosslinking data. The data were released to modelers after the prediction window for the corresponding regular target (modeling without data assistance) was closed. The predictors were given an opportunity to submit structure models built with the assistance of the crosslinking restraints in a 2-3-week period.
Materials and Methods
Targets
Purified protein samples of 8 regular CASP13 targets - H0953, H0957, H0968, T0975, T0981, T0985, T0987 and T0999 – were provided by Matthew Dunne (ETH Zurich, target H0953), Karolina Michalska (Argonne National Lab, H0957 and H0968), Chi-Lin Tsai (UT MD Anderson Cancer Center, T0975), Mark van Raaij (Centro Nacional de Biotecnologia of Spain, T0981), Jose Henrique Pereira (Lawrence Berkeley Lab, T0985), Lindsey Spiegelman (UCSD, T0987) and Marcus Hartmann (Max Planck Institute, T0999), and shipped to the crosslinking laboratories. Three of these targets were heteromeric complexes (those starting with ‘H’), two homomultimers (T0981 and T0999) and the remaining three – monomers. Alexander Leitner’s group generated crosslinking datasets for all 8 targets, including 3 heterocomplexes (names of the released data-assisted targets start with the uppercase ‘X’, and referred to as ‘BigX’ group in the following), and Juri Rappsilber’s group did so for 4 of the targets, including 2 complexes (targets start with the lowercase ‘x’, and referred to as ‘Smallx’ group). If a protein was a heterocomplex, then the whole complex and its subunits were released as separate crosslink-assisted targets. For instance, a protein corresponding to the regular heterodimeric target H0957 was released for crosslinking-assisted prediction as 6 targets: X0957 and x0957 (whole complex, different datasets), X0957s1 and x0957s1 (first subunit, different datasets) and X0957s2 and x0957s2 (second subunit, different datasets). Overall, 22 crosslinking-assisted targets were released in CASP13, including 5 heteromeric targets (3 different protein complexes) and 17 single-sequence targets (11 different prediction sequences/subunits).
Evaluation units (domains)
As it is customary in CASP, prediction results were evaluated at different levels of protein structural organization, with emphasis on domain-based evaluation. Similarly to regular targets, crosslinking-assisted targets were split into evaluation units5. Eleven different prediction sequences (subunits) were split into 19 distinct-sequence tertiary structure evaluation units (Table 1). Since models were built with the assistance of different crosslinking datasets separately (i.e., ‘x’ and ‘X’ targets), these models were evaluated separately, which brought the total number of evaluation units to 27. The oligomeric targets were evaluated as whole complexes.
Table 1.
Overview of targets in the crosslink assisted modeling category. Upper and lower case X and x refer to different sets of experimental crosslinks provided for the same target. First column lists eight unique targets, sometimes explored by both experimental groups for crosslinks. Second column refers to subunit level dissection of targets while the third column further splits targets into Evaluation Units. The total number of targets were 27 (third column, EUs multiplied by the number of data sets available for each of
| Target /dataset |
Subunits /sequences (#residues) |
Evaluation units /domains (#residues) |
|---|---|---|
| X0953 | X0953s1 (67) | D1 (67) |
| X0953s2 (249) | D1 (46), D2 (127), D3 (77) | |
| X0957, x0957 | {Xx}0957s1 (163) | D1 (108), D2(54) |
| {Xx}0957s2 (155) | D1 (155) | |
| X0968, x0968 | {Xx}0968s1 (119) | D1 (119) |
| {Xx}0968s2 (116) | D1 (116) | |
| X0975, x0975 | D1 (293) | |
| X0981 | D1 (105) | |
| X0985 | D1 (842) | |
| X0987, x0987 | D1 (185), D2(207) | |
| X0999 | D1 (386), D2(453), D3(180), D4(244), D5(288) |
Chemical crosslinking experiments at ETH Zurich (BigX)
Crosslinking reaction and sample processing
For all other targets, except target X0999, the following procedures were followed. Protein stock solutions were provided by CASP contributors and used as received if the buffer was compatible with crosslinking experiments (Supplementary Table 1.). For target X0981, the buffer was exchanged to 20 mM HEPES, 150mM NaCl pH 8.5. Proteins and complexes were crosslinked following previously published procedures 3,6. Conditions were initially optimized using SDS-PAGE as a readout to minimize aggregation or the formation of higher-order oligomers unless it was known that multiple copies of the proteins were present in the target structure. Most final crosslinking experiments were performed with a protein or complex concentration of 1 mg/mL, and samples were crosslinked for 30 minutes at 25 °C at a scale of approximately 50 μg of total protein (Supplementary Table 1).
The crosslinked samples were further processed using standard procedures. Steps included unfolding by urea (6 M), reduction of disulfide bonds with TCEP (2.5 mM), alkylation of free cysteine thiol groups with iodoacetamide (5 mM) in the dark, and a two-step digestion with endoproteinase Lys-C (Wako, 1:100, w/w) and trypsin (Promega, 1:50, w/w). The digested protein samples were purified with solid-phase extraction (Waters tC18 cartridges) and directly analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) without further enrichment or fractionation.
Target X0999 was crosslinked in a collaboration with the group of Marcus Hartmann (MPI Tübingen, Germany) prior to the start of CASP13 (more details will be published elsewhere).
MS data acquisition
LC-MS/MS analysis was performed on a Thermo Easy nLC 1000 LC system coupled to a Thermo Orbitrap Elite mass spectrometer equipped with a nano-electrospray source. The instrument was operated in data dependent acquisition mode (DDA). MS data were acquired in the Orbitrap at resolution 120,000, followed by fragmentation of the 10 highest intensity ions by CID, before mass analysis in the ion trap. The samples were analyzed in three technical replicates, where a single run included ions with a charge state ≥ +2, while the rest only included ions with a charge state ≥ +3.
Data analysis
Thermo raw files were converted into the mzMXL format using msconvert (ProteoWizard version 3.0.7494). MS/MS spectra were searched using xQuest 7 (version 2.1.4), against the target protein sequence(s) as provided and including contaminants identified from a search with Mascot (v. 2.1.5, MatrixScience) against the SwissProt database. xQuest search settings were as follows: Enzyme: trypsin, maximum number of missed cleavages: 2, MS mass tolerance: 5 ppm, MS/MS mass tolerance: 0.2 Da for “common”-type fragment ions and 0.3 Da for “xlink”-type fragment ions. All putative identifications were manually assessed.
Data deposition in PRIDE
All mass spectrometry data have been deposited in the PRIDE Archive 8 with the following dataset identifiers and are accessible al https://www.ebi.ac.Uk/pride/archive/projects/PXD######, the targets and corresponding web links are as follows: X0953: PXD010094; X0957: PXD010003; X0968: PXD010004; X0975: PXD010385; X0981: PXD010384; X0985: PXD010483; X0987: PXD010410; X0999: PXD010479.
Chemical crosslinking experiments at Berlin (Smallx)
Crosslinking reaction and sample processing
T0975 and T0987 had been forwarded to the Rappsilber Laboratory as previously thawed-frozen samples by Esben Trabjerg from the Leitner Laboratory at ETH Zurich.
Crosslinking was carried out according to previously described procedures 9-11. Briefly, target proteins were crosslinked separately using sulfosuccinimidyl 4,4’-azipentanoate (sulfo-SDA) (Thermo Scientific Pierce, Rockford IL) in a two-stage reaction (using eight different crosslinker-to-protein ratios: 0.13:1, 0.19:1, 0.25:1, 0.38:1, 0.5:1, 0.75:1, 1:1 and 1.5:1 (w/w), a protein concentration of 0.5 mg/mL and using 20 μg protein aliquots), with reaction of the NHS-ester firstly, subsequently followed by UV photoactivation at 365 nm, from a UVP CL-1000 UV Crosslinker (UVP Inc.).
Following crosslinking, reaction conditions were mixed and resulting crosslinked proteins separated by electrophoresis using NuPAGE 4-12% Bis-Tris gels, with MES SDS running buffer and staining using InstantBlue™ (Expedeon). Protein gel bands were digested using trypsin via standard protocols 12. Resulting peptides were desalted using StageTips 13,14.
MS data acquisition
Samples were analyzed using an HPLC (UltiMate 3500RS Nano LC system, Thermo Fisher Scientific, San Jose, CA) coupled to a tribrid mass spectrometer (Orbitrap Fusion Lumos Tribrid Mass Spectrometer, fitted with an EASY-Spray Source, Thermo Fisher Scientific, San Jose, CA). Peptides were loaded onto a 500 mm C18 EASY-Spray LC column (Thermo Fisher Scientific, San Jose, CA), operating at 45 °C. Mobile phase A consisted of water and 0.1% formic acid, mobile phase B of 80% acetonitrile, 0.1% formic acid and 19.9% water. Peptides were loaded and eluted at a flow-rate of 0.3 μL/min, using a linear gradient starting at 2% mobile phase B and increasing over 109 min to 40%, followed by a linear increase over 11 min, from 40% to 95% mobile phase B.
MS data were acquired in the Orbitrap at resolution 120,000, using the top-speed data-dependent mode. Selected precursor ions were fragmented using higher-energy collisional dissociation (HCD), using a normalized collision energy of 30%. Fragmentation spectra were then recorded in the Orbitrap at resolution 30,000, AGC target set to 5 × 104 and maximum injection time of 70 ms.
Data processing
Raw files were processed into mgf files using ProteoWizard msconvert (3.0.9576), with the inclusion of a MS2 peak filter for the 20 most intense peaks in a 100 m/z window15. The resulting peak lists were searched against FASTA sequence files using Xi16 (https://github.com/Rappsilber-Laboratory/XiSearch) version 1.6.731, using the following settings: MS accuracy, 3 ppm; MS/MS accuracy, 15 ppm; missing mono-isotopic peaks, 2; enzyme, trypsin; maximum allowed missed cleavages, 4; crosslinker, SDA; fixed modifications, none; variable modifications, carbamidomethylation on cysteine, oxidation on methionine, SDA-loop (SDA crosslink within a peptide that is also crosslinked to a separate peptide, mass modification: 82.041865). The linkage specificity for sulfo-SDA was assumed to be at lysine, serine, threonine, tyrosine and protein N-termini at one end, with the other end having specificity for any amino acid residue. False discovery rates (FDR) 5%, 10%, 20% (corresponding to reported confidence scores provided to modelers: 0.95, 0.9, 0.8) were estimated using xiFDR 17 (a target-decoy approach to false discovery rate error estimation), version 1.1.26.58.
Data deposition in PRIDE
Mass spectrometry data was deposited to the ProteomeXchange Consortium via the PRIDE partner repository 8 with the dataset identifier PXD010884 (accessible at https://www.ebi.ac.uk/pride/archive/) (Reviewer account details: Username: reviewer91980@ebi.ac.uk, Password: Ow22Vk9d).
Participants and predictions
In CASP13, 14 prediction groups submitted 576 crosslinking-assisted models on 17 tertiary structure prediction targets. In addition, 41 quaternary structure predictions were submitted on 2 homo-oligomeric targets and 157 predictions on 5 heteromeric targets. The number of groups that provided models both with and without crosslinks ranged from 3 to 6 per target.
The number of attempted targets and predictions varies significantly by group. Six prediction groups were evaluated on 20 or more domains, while the remaining eight – on twelve or fewer domains.
Evaluation measures
To assess accuracy of crosslinking-assisted models and their improvement over the corresponding non-assisted predictions, we employed the GDT_TS measure 18,19 for monomeric predictions, and the LDDT measure 20 for multimeric ones. Comparative analysis of these measures is provided in a recently published paper 21.
To rank groups, we initially transformed per-target raw scores into Z-scores considering only the first ranked models. However, the number of predicted targets per group varied widely, from 3 to 27: this could heavily influence any Z-score-based ranks, averages, or cumulative scores. Therefore, we employed a pairwise comparison among all groups, where a one-tailed Wilcoxon statistical test was used at a significance cutoff of 0.05 to assess the significance of differences in performance between two groups on the common set of targets shared between them. This test was not possible to perform if less than two common targets were shared between any two groups.
Results
Types of crosslinks
Crosslinking experiments were carried out using complementary strategies (Fig. 1.). The group at ETH Zurich (a.k.a. BigX group) performed reactions with residue-specific linkers: disuccinimidyl suberate (DSS), which predominantly crosslinks primary amines on Lys residues and the N-terminus of proteins, and a combination of pimelic acid dihydrazide (PDH) and the coupling reagent DMTMM, resulting in crosslinks between residues with carboxyl groups (Asp, Glu, and the C-terminus) and “zero-length” links between Asp or Glu and Lys. These crosslinking strategies are typically applied to multi-subunit assemblies and may not be the optimal choice for small proteins or complexes of small proteins, where there may be too few crosslinkable residues.
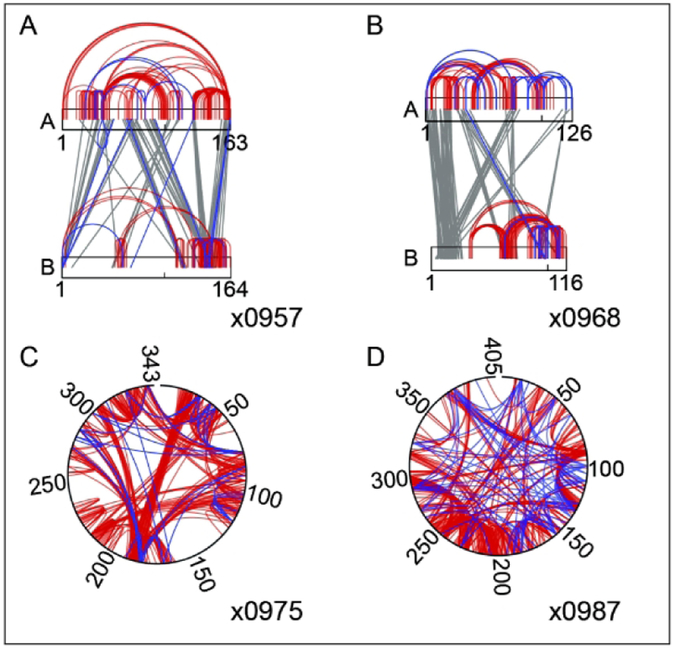
Figure 1.
Crosslinking mass spectrometry data provided by the two contributing labs on the four targets that were processed by both groups: Non-specific crosslinks from Rappsilber lab (SmallX, red and grey) and residue-specific crosslinks from the Leitner lab (BigX, blue). Targets x0957 and x0968 are heteromeric complexes, while x0975 and x0987 are single chain proteins, as indicated in the figure.
Reaction conditions were optimized using SDS-PAGE to minimize the formation of homo-oligomers or non-native stoichiometries of complexes, although the “true” oligomeric state was not known in all cases. A single crosslinking experiment with the best conditions was performed per target (for DSS and PDH in combination with DMTMM, respectively).
Data analysis was performed using the in-house software xQuest7 and results were provided to the CASP participants with an expected error (false discovery) rate of <5%, although accurate FDR estimation is difficult if only very few crosslinks are identified. The final reports were published on the CASP website and listed the crosslinked residues along with the xQuest identification score (the higher, the better), so that participating groups could adjust their stringency thresholds, if desired. The main score of xQuest is a weighted composite score from several sub-scores that reflect the similarity of the experimentally observed and the predicted MS/MS spectrum (e.g., cross-correlation of fragment ions, percentage of cumulative intensity that in the spectrum that is assigned to fragment ions), much like score of conventional proteomics search engines. Therefore, it is important to note that the xQuest identification score is only a measure of the confidence of the mass spectrum identification and is not related to any structural/distance property. In addition, the group in Zurich pointed out regions in the protein sequences that were not adequately covered by trypsin (for example, even complete trypsin digestion of target X0953 would result in some very long peptides that are unlikely to be identified by mass spectrometric analysis under the conditions used for this study). Furthermore, the group in Zurich also provided a list of residues that were found to be modified by the crosslinking reagents, but for which only one side of the linker reacted (“dead-end” products, “mono-links”). These residues may be considered solvent accessible/exposed, a fact that could also be exploited during modeling22.
In contrast, the other source for crosslinking experiments, the Rappsilber group (a.k.a. Smallx), used heterobifunctional, photoactivatable crosslinking chemistry, where the reaction occurs firstly on (predominantly) lysine residue side chains (but also the side chains of serine, tyrosine and threonine), and following photoactivation, completes crosslinking by inserting non-specifically into vicinal bonds. This semi-specificity has been shown to allow greater data density, which can be beneficial for protein structure prediction 9. This approach provided the first experimental data in CASP history, in CASP11, in the form of high-density XL-MS (HD-XL-MS) data 4,10,23 and has been subsequently re-used for targets in CASP1211 and in the present study for CASP13.
Structure based evaluation of crosslinking information
Once the experimental protein structures became available, we explored the general question of whether the crosslinks provided had the potential to benefit the modeling. Two issues were explored: first, if a crosslink is ”valid”, and second, if it is ”informative”. A crosslink was assumed to be valid if it connected residues in the structure within 30 Å of each other, once measured along the shortest path on the surface of the protein 24. This general and generous cutoff was selected based on earlier observation about the crosslinkable positions in proteins3. Arguably a variable definition could be used for different types of crosslinks, for instance a shorter cutoff distance could be applied to zero length crosslinks, but only about by 5 Å, according to earlier studies3. Using a shorter cutoff would increase the fraction of invalid crosslinks at the price of incorrectly assigning some. As we show later, there is no trivial drop in the distribution of observed crosslinked distances and the definition we use here is intentionally inclusive and renders crosslinks invalid only if these bridge really long distances. The informativeness of crosslinks is a more subjective definition. Arguably, information on all crosslinks are informative, for instance, to gain insight about surface accessibility 22. However, for the current purpose, to model protein structures where even just identifying the general topology of the fold is challenging, we assumed that crosslinks that formed between more distant positions, preferably beyond a supersecondary structure motif, were more informative than the ones that connected residues within the same short motif or within a well-defined secondary structure. We subjectively required a minimum sequential separation of 50 residues to define informative crosslinks. This excludes the possibility that crosslinks between two adjacent helices of a typical length (4-6 turns each, plus a connecting loop between them) are considered.
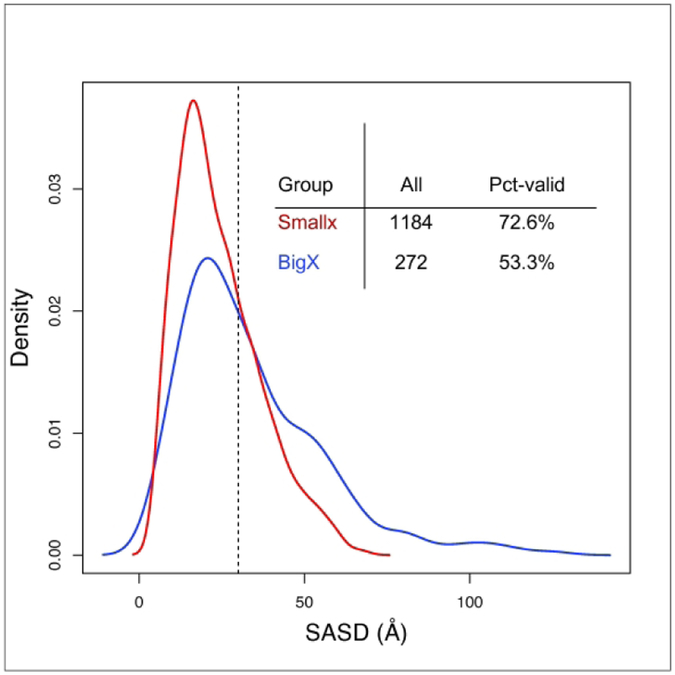
The distribution of crosslinks shows that a substantial fraction (27-47%) were formed between residues more than 30 Å away as measured by the shortest Solvent Accessible Surface Distance 24 (Fig. 2). This large fraction of inconsistent crosslinks made it challenging for modelers to simultaneously satisfy as many crosslinks as they could. In this assessment we are evaluating crosslinks on the experimental crystal structure, which cannot reflect various levels of flexibility and dynamic movements of the protein. Crosslinks are established in solution therefore a substantial fraction of crosslinks that we deemed invalid in this assessment actually may reflect the real dynamic nature of some of the target protein structures. When exploring the fraction of informative crosslinks, which were formed between residues 50 positions or more apart, we found that about 40-60% of all crosslinks satisfy this condition (Fig. 3). If one combines these two requirements it appears that about 23% and 27% (Smallx: 277/1184 and BigX: 73/272) of crosslinks fall into this combined category, respectively. However, from a practical point of view, the informativeness of crosslinks is known to all users, because the sequence separation is easy to check; therefore, a more practical measure is the fraction of valid and informative crosslinks over all of the known-to-be-informative ones, which results in 58% and 44% of crosslinks for the Smallx and BigX data sources, respectively.
Figure 2.
Distribution of crosslinks from the two experimental sources, Smallx (red) and BigX (blue), as a function of the solvent accessible surface distance (SASD) in angstroms. The table inset shows the number of all crosslinks determined and the percent fraction that fall within 0-30 Å (vertical dashed line on plot).
Figure 3.
Distribution of crosslinks from the two experimental sources, Smallx (red) and BigX (blue), as a function of the sequential separation between crosslinked residues. The table inset shows the number of all crosslinks determined and the percent fraction that connects residues >50 positions apart.
Assessing the usefulness of confidence scores of crosslinks
We also explored how much the provided confidence scores can help to filter and enrich the set for valid crosslinks. Different types of confidence scores were provided by the two experimental labs. The BigX group gave scores between 15 and 50 where the larger numbers indicate higher confidence of the mass spectrum identification. When we count the enrichment of valid crosslinks as a function of increasing confidence cutoff, we see a notable improvement once we require a score of at least 35. At this point the fraction of valid crosslinks increases from 45 to 64%. (Table 2). However, this comes at a price of keeping only 26% of the original set of informative crosslinks, meaning that a large fraction of valuable data is discarded. In case of the Smallx group, three different confidence levels were provided, 80, 90 and 95 (Table 3). Here, a slightly more informative selection can be made based on the confidence values. The enrichment of valid crosslinks among the informative ones starts already at a higher value of ~59%, and at a 95% confidence level cutoff value it increases to 71%. This latter set still contains most of the original information (60% of total); hence, the information loss is not as significant as in the case of filtering the BigX input.
Table 2.
Relationship between confidence levels (first column) and the number of crosslinks above each cutoff value within the set of informative crosslinks for data from BigX group.
| Confidence cutoff |
# Xlinks left |
% of total |
% valid |
|---|---|---|---|
| 15 | 163 | 100% | 45% |
| 20 | 156 | 96% | 44% |
| 25 | 129 | 79% | 44% |
| 30 | 73 | 45% | 51% |
| 35 | 42 | 26% | 64% |
| 40 | 14 | 9% | 79% |
| 45 | 6 | 4% | 100% |
| 50 | 0 | 0% | 0% |
Table 3.
Relationship between confidence levels (first column) and the number of crosslinks above each cutoff value within the set of informative crosslinks for data from Smallx group.
| Confidence cutoff | # Xlinks left | % of total | % valid |
|---|---|---|---|
| All (80% and up) | 471 | 100% | 58.8% |
| 90% | 336 | 71% | 67.0% |
| 95% | 282 | 60% | 70.6% |
Overall group performance at CASP13
Following our analysis on the valid and informative crosslinks, we decided to focus only on those targets where at least a single valid and informative crosslink was provided. We did this in order to remove from the group performance comparison the effect that comes from targets where information on crosslinks does not play any role and all differences are due to the quality of initial models generated by the groups. Out of the 27 evaluation units and 5 complex targets there were twelve for which there was not a single valid and informative crosslink (Table 4).
Table 4.
List of targets and the corresponding number of valid and informative crosslinks available.
| Target | All | Valid | Informative | Valid-Inf | valid- inf/inform |
|---|---|---|---|---|---|
| X0953S1D1 | 0 | 0 | 0 | 0 | 0.00% |
| X0953S2 | 5 | 3 | 0 | 0 | 0.00% |
| X0953S2D1 | 0 | 0 | 0 | 0 | 0.00% |
| X0953S2D2 | 2 | 2 | 0 | 0 | 0.00% |
| X0953S2D3 | 1 | 1 | 0 | 0 | 0.00% |
| x0957S1D2 | 12 | 9 | 0 | 0 | 0.00% |
| X0957S1D2 | 2 | 2 | 0 | 0 | 0.00% |
| x0957S2D1 | 83 | 68 | 13 | 0 | 0.00% |
| X0957S2D1 | 0 | 0 | 0 | 0 | 0.00% |
| X0968S2D1 | 5 | 5 | 0 | 0 | 0.00% |
| X0981D1 | 0 | 0 | 0 | 0 | 0.00% |
| X0999D5 | 0 | 0 | 0 | 0 | 0.00% |
| X0968S1D1 | 9 | 8 | 1 | 1 | 100.00% |
| X0957S1 | 7 | 7 | 2 | 2 | 100.00% |
| x0957S1D1 | 73 | 66 | 6 | 2 | 33.30% |
| X0957S1D1 | 2 | 2 | 2 | 2 | 100.00% |
| X0999D3 | 8 | 3 | 5 | 2 | 40.00% |
| X0999D4 | 5 | 4 | 2 | 2 | 100.00% |
| X0987D1 | 15 | 9 | 4 | 3 | 75.00% |
| X0999D1 | 12 | 10 | 5 | 3 | 60.00% |
| X0999D2 | 10 | 5 | 7 | 3 | 42.90% |
| x0968S2D1 | 76 | 69 | 5 | 5 | 100.00% |
| X0987D2 | 20 | 12 | 6 | 6 | 100.00% |
| X0975D1 | 19 | 14 | 10 | 7 | 70.00% |
| X0985D1 | 37 | 21 | 19 | 9 | 47.40% |
| x0968S1D1 | 68 | 50 | 20 | 16 | 80.00% |
| X0987 | 66 | 28 | 37 | 16 | 43.20% |
| x0987D1 | 147 | 108 | 29 | 20 | 69.00% |
| x0957S1 | 144 | 116 | 41 | 26 | 63.40% |
| x0987D2 | 246 | 193 | 96 | 77 | 80.20% |
| x0975D1 | 272 | 192 | 144 | 90 | 62.50% |
| x0987 | 539 | 362 | 248 | 140 | 56.50% |
If we compare the targets in this subset (with at least one valid and informative crosslink) that were modelled with and without crosslink information we see a strong shift to higher quality models (90% of the time) (Fig. 4). Even when considering all targets with an without crosslink information we observe a considerable shift towards higher quality models (76% of the time). This suggest that crosslinks connecting shorter sequential distances were also beneficial (76% of the time) but when more informative crosslinks were provided it really tilted the balance towards systematic improvement (90% of the time). The corresponding average GDT_TS changes are 4.71 and 5.23, respectively, but the actual range goes up to nearly 20 GDT_TS scores (Fig. 4).
Figure 4.
Head to head comparison of changes in model accuracy (ΔGDT_TS) for each group and each model. The total set of targets is colored red, while the subset of targets with at least one valid and informative crosslink is colored green.
If we focus on specific group performances we need to address the issue that groups submitted significantly different numbers of targets (in the range of 3-27). This prevents general Z-score averaging or summing approaches from being informative as the results will depend on how many and which targets certain groups decided to submit. In order to address significance, we performed a pairwise comparison among all groups and assessed whether the performance of one group was significantly better than that of the other group, using a one-tailed Wilcoxon test at a significance level of 0.0525,26. This comparison could not be performed between pairs of groups that shared less than 2 common targets (Fig. 5). A relatively clear split appears between groups that systematically over- and underperformed in this exercise (groups with many blue vs red squares). From this ranking we provide more detailed description from the top two performers, groups 208 and 196, in the coming sections. Along with the performance of group 208, we also discuss that of groups 288 and 492, which used a similar methodology and, although did not perform as well regarding the accuracy of models, they achieved much greater relative model quality improvement upon introducing the crosslink information.
Figure 5.
Comparison and ranking of Group performance in the subset of data assisted targets where at least one valid and informative crosslink was provided. One-tailed Wilcoxon tests were performed at a 0.05 significance cutoff between all pairs of groups. Vertical axis lists groups, ranked by performance from top to bottom. Blue: vertical performed better than horizontal; Red: vertical not significantly better than horizontal; White: not enough shared targets between groups; Gray: vertical and horizontal are the same group.
Modeling with crosslinks by group 208 (KIAS-Gdansk) and two related groups
The data-assisted-prediction protocol developed in the laboratory of the KIAS-Gdansk group and described in 27 was used. The main step of this protocol is extensive conformational search by using the multiplexed replica-exchange molecular dynamics (MREMD) method28,29 with the coarse-grained UNRES force field30-32. MD33,34 and MREMD35 were implemented in UNRES in our earlier work. A total of 48 replicas at 12 temperatures were run for each target using 20,000,000 4.89 fs MD time steps, which correspond to about 0.1 ms of real time per trajectory because of time-scale extension in UNRES 33. The conformational space of the simulations was restrained by the crosslinks provided for the data-assisted targets. Use of the coarse-grained approach makes the conformational search more efficient as the time-scale is extended by at least 3 orders of magnitude due to averaging out most of the degrees of freedom 33. The conformational ensembles thus obtained are clustered into 5 families, from which conformations closest to cluster centers are selected and converted to all-atom representations to give the final models.
We used both the non-specific 36 and specific 3 restraints, corresponding to the Smallx and BigX type targets, respectively. Non-specific crosslink restraints were used together with specific crosslink restraints for most of the targets. For non-specific restraints provided by the Rappsilber lab10,36 a bounded flat-bottom function was used 37,38 (eq. 1).
| (1) |
where d is the distance between the Cα atoms of the two crosslinked residues in the computed structure and dl, and du are the lower and upper contact-distance boundaries, respectively (we set dl=2.5 Å, du=25 Å), σ(set at 1 Å) is the width of the transition region between zero and the maximum restraint height, and A is the height of the restraint well, which we assume to be equal to the confidence of a contact, which was taken from the XLMS-information files deposited at the CASP13 web page. This function generates no gradient if a restraint is grossly not satisfied, which naturally eliminates the incompatible XLMS restraints from consideration.
The specific restraints provided by the Leitner lab 3, were incorporated in a form of statistical potentials derived based on the data in Figure 3 of 3. The functional form is given by eq. 2.
| (2) |
where d is the distance between the UNRES side-chain centers of the two crosslinked residues, X denotes the type of crosslink (ZL, PDH or DSS) 3, and αx, βx, δx and σx are the parameters obtained by least-squares fitting of the statistical potentials of mean force derived from the distributions in Figure 3 of ref 3., and A is the confidence of a crosslink restraint. The parameters of the expression of eq. 2 were obtained by nonlinear least squares fitting V(x) to the logs of the distributions from Figure 3 of ref. 3, as given by eq. (3)
| (3) |
where Px;k is the distribution value for the cross link of type X at the kth bin, dk is the distance at the center of that bin, and β=1/RT, R being the universal gas constant and T the absolute temperature set at T = 298 K. The experimental and fitted Px are plotted in Fig. 6.
Figure 6:
Comparison of the experimental (ref. 3; bars) and fitted by using eq. 3 (lines) distributions of Cα-Cα distances in model proteins for 4 different types of crosslinks: zero-length crosslinks (ZL; orange), adipic acid dihydrazide (ADH; green; not used in CASP13), pimelic acid dihydrazide (PDH; purple), and disuccinimidyl suberate (DSS; blue).
The XLMS restraints were applied together with the SAXS or SANS restraints, which were available for all crosslink-assisted targets. Data of both kinds were used because the objective of CASP exercises is to produce the best predictions possible and, consequently, the organizers encouraged the predictors to use all available data while processing the data-assisted targets. The starting structures were the final models obtained in the non-data-assisted mode by the respective group.
The SAXS/SANS restraints were incorporated in the form of a maximum-likelihood function introduced in ref. 27, which is given by eq. 4.
| (4) |
where r is the distance, rk is the distance at the center of the kth bin of the histogram of the distance distribution from SAXS measurements, M is the number of bins, PSAXS(r) is the value of the probability distribution determined by SAXS at r, Pcalc(r) is the value of the probability distribution calculated from simulations at r, dmax is the maximum distance in the molecule, and Δr is the bin size taken as 1 Å. The SAXS-derived values of the probability distribution, PSAXS(r), were only normalized and no quality check was performed. and Pcalc is defined by eq. 5
| (5) |
with
| (6) |
| (7) |
where rij is the distance between the Cα atoms of residues i and j in the calculated conformation, σij is the standard deviation of the respective Gaussian, σi, and σj being the Stokes’ radii of residues i and j, respectively; in this work we use the values as in Langevin-dynamics simulations with UNRES33, s is the radius scaling factor set at s = 5, and A is the factor normalizing the calculated probability to 1.
We submitted predictions for 11 out of 12 crosslink-assisted targets (all except for X0981) from three UNRES-related groups: UNRES (group 288; no knowledge-based information except for secondary-structure prediction), KIAS-Gdansk (group 208; homology-assisted modeling with UNRES), and wf-BAKER-UNRES (group 492; contact-assisted modeling with UNRES). The GDT_TS improvement between un-assisted and crosslink assisted models is moderate (Fig. 7), with many models being deteriorated for the KIAS-Gdansk models, but significant for the UNRES and wf-BAKER-UNRES models, which can be explained by better quality of the crosslink-unassisted KIAS-Gdansk models due to the introduction of homology-based restraints. It can also be seen that the improvement is more significant for predictions with only specific crosslinks than for those with non-specific and specific crosslinks. The reason for this difference in model quality is that many restraints from non-specific crosslinks are invalid or ambiguous.
Figure 7:
Scatter plot of the differences in GDT_TS values of the best models of the assisted and regular predictions as a function of the highest GDT_TS corresponding to the regular prediction of the respective group for the specific crosslink-assisted (X; filled symbols) and non-specific only or non-specific plus specific crosslink-assisted (x; open symbols) prediction of the UNRES (group 288, red circles), wf-BAKER-UNRES(group 492, green triangles) and KIAS-Gdansk(group 208, blue squares) groups, respectively.
The most significant qualitative improvement of the models was obtained by the UNRES group for targets T0968s1 and T0968s2 following the introduction of specific crosslink restraints (Fig 8). It should be noted that prediction simulations were run for the whole tetramer (dimer of dimers) and subunit coordinates were extracted from the final models. It can be seen that, for X0968s1, specific-crosslink information resulted in reorientation of the α-helical section of the subunit with respect to the β-sheet, resulting in native-like orientation of these sections. Likewise, unassisted UNRES simulations resulted in orthogonal packing of two β-sheets forming the structure of T0968s2, while introducing specific crosslinks reduced the angle between the β-sheet sections, as also observed in the experimental structure.
Figure 8:
Cartoon drawings of the best UNRES (left, blue) and best specific crosslink-assisted UNRES (right, dark orange) models of the first (T0968s1; A) and second (T0968s2; B) subunit of target T0968 superposed on the respective portions of the experimental structure of CASP13 target H0968 (gray).
Modeling with crosslinks by group 196 (Grudinin) and related group 135
In our approach we integrated information from crosslink experiments to a combination of a physics-based and a knowledge-based model. Let us first consider two residues, represented by the corresponding Cα, for which the XL experiment has detected a putative contact. First, we estimated the probability of the presence of one Cα atom with respect to the distance to the second Cα atom. We approximated this probability with a Gaussian distribution, with the center and the standard deviation specific to each type of XL experiment 3. Fig. 9 shows these distributions fitted to the data provided in Leitner et al 3. We could not fit data from the zero-length (ZL) experiments with a single Gaussian, and thus used a sum of two Gaussians. We then made a Boltzmann-like hypothesis and considered that there is pseudo-potential associated with each of the XL constraints, whose value is given by the logarithm of the probability of a certain Cα-Cα distance. Since we made the hypothesis of a Gaussian distribution of one alpha carbon with respect to the other, this pseudo-potential is a harmonic, with the exception of ZL potentials that we did not have in the experimental CASP13 data. We collected initial models from CASP13 stage-2 server submissions and ranked them using the SBROD orientation-dependent backbone-only scoring function39. We picked the top five models and refined them iteratively using a gradient-based optimization. When moving the model atoms along the raw gradient of the XL pseudo-potential fXL, we observed that the bonds may break, unrealistic local topology may occur, and as a result, the initial secondary structure can get severely distorted. To preserve the local model topology, we added an energy term from the Gaussian network model, represented by the Hessian matrix H, whose equilibrium is always at the current structure. As a result, we were iteratively solving the following problem with respect to atomic displacements Δx,
| (8) |
which can be transformed to a linear system of equations. The coefficient λ determines the relative importance of XL restraints with respect to the Gaussian network model. Its value was adjusted such that the final structure had a meaningful overall RMSD difference compared to the initial one (on average of several Å). The Gaussian network model was computed by the NOLB library 40 and is often used in the normal mode analysis. It allows large-amplitude realistic motions, with marginal modification of the local topology. However, the accumulation of small perturbations of the local topology over the course of several iterations may still produce unrealistic final structures. To tackle this problem, we added to our iterative process an additional minimization of a simple force-field containing bond length, bond angle, and van der Waals interaction terms. We continued the refinement until the convergence of the total energy.
Figure 9:
Distribution of Cα-Cα distances in model proteins for 4 different types of crosslink experiments. Data points for pimelic acid dihydrazide (PDH), disuccinimidyl suberate (DSS), adipic acid dihydrazide (ADH, not used in CASP13), as crosslinking reagents, and zero-length crosslinks (ZL) are shown. Solid lines represent Gaussian fits to the experimental data points. The ZL fit is described with a sum of two Gaussians. A logarithm of the presented fits is used as a pseudo-potential. The bin size of 3 Å to calculate the probabilities was adapted from Leitner et al3.
We did not use additional SAXS or SANS restraints in our protocol, even though these were available for most of crosslink-assisted targets.
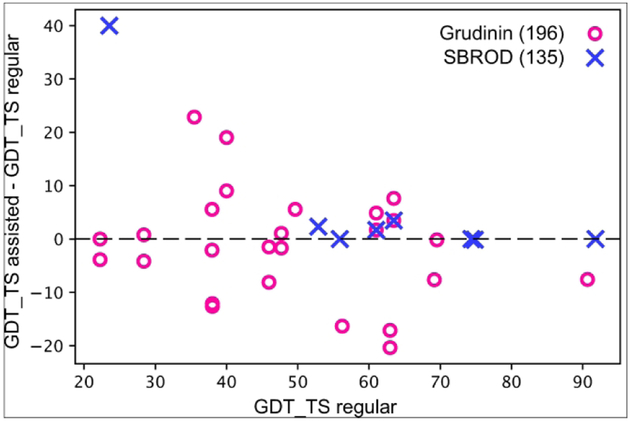
Similarly to the UNRES groups, we also submitted predictions for 11 out of 12 crosslink-assisted targets (except for X0953). We used two slightly different protocols. The first one submitted by the Grudinin group (196) ranked final models by the XL energy restraints. We applied it to 11 out of 12 targets. The second one, submitted by the SBROD group (135), rescored the final predictions with the SBROD score. This one was applied to only 4 targets. Fig. 10 presents the GDT_TS differences between regular and XL-assisted predictions for the two groups. We can draw several conclusions from this plot. First, rescoring of the final models with the knowledge-based SBROD potential seems to help select models with slightly better quality. On the other hand, trying to satisfy the XL restraints as much as possible may improve the model quality more significantly, but very often results in models of lower quality compared to the starting templates. This is likely caused by the ambiguity of the XL restraints that to some extent might reflect the in-solution dynamics of the investigated protein targets.
Figure 10.
Scatter plot of the differences in GDT_TS values between the first ranked models of the assisted and regular predictions as a function of the GDT_TS value corresponding to the first model of the regular prediction. Results of two groups are shown, Grudinin with red circles (group 196), and SBROD with blue crosses (group 135).
Assessing complexes
There will be a separate article devoted to assessing the modeling of complexes with data assistance in this issue of the journal. We just briefly summarize here the narrow category of chemical-crosslinks-assisted complex modeling. In general we followed the same evaluation as before for single chain targets, but in adjusting to the presence of multiple chains we do not define informative crosslinks. Also, distances were measured directly in Euclidean space as opposed to considering the accessible protein surface as before. The number of targets was very limited, at 7. We compared the improvement to models in terms of LDDT measure 20 with and without crosslinking assistance within the subcategory of assisted modeling and also against the entire CASP general category (Fig. 11). Out of the 7 targets, two had no valid crosslinks i.e. 0% on the figure, (interchain crosslinks were connecting distances longer than 30 Å), and one had no interchain crosslinks determined, i.e. No, on the figure. Out of the remaining 4 targets, 3 improved upon adding crosslink information (Fig. 11). The few available examples prevent us from making statistically strong statements, but overall the general trend on these few cases is that modeling complexes benefits from crosslink information, even when compared to the general modeling category of CASP (blue marks on figure) where 99 groups submitted models without assistance of experimental data.
Figure 11.
Accuracy of protein complex modeling with and without XL-MS data. Accuracy (LDDT) of best XL-MS assisted model (vertical axis) vs the best TS model (without XL-MS information) from the corresponding assisted group (grey) or all structure modeling groups (blue). Grey data was selected from a subset of assisted modeling groups that submitted models both with and without crosslink assistance. Information about Xlinks are added to blue points: % valid or NO suitable crosslinks available.
Discussion
Comparing crosslinks
On the subset of targets where crosslinks were provided by both experimental groups we compared the accuracy of models for the same targets by focusing on the single best model produced by any group given the same set of crosslink information (Fig. 12). There seems to be a clear tendency that more accurate models were generated using BigX group generated crosslinks. Reasons for this can be speculated upon, however solid conclusions from the comparison of different crosslink datasets are difficult for two main reasons: 1. Different sample history. Protein samples were analyzed first by the BigX group and the remnants were forwarded to the Smallx group for a subsequent analysis. 2. Biased data release. Data from both groups were made available at different time points (BigX generated crosslinks released weeks-months before Smallx) and for different time durations (for example, for X0975 it was 21 days, compared to 14 days for x0975).
Figure 12.
Accuracy of structure modeling with XL-MS data utilizing different sources. Head-to-head comparison of accuracies (GDT_TS) of best models from assisted modeling groups using data from BigX group (x-axis) vs. Smallx group (y-axis).
Comparing the best crosslink-assisted models vs the best models
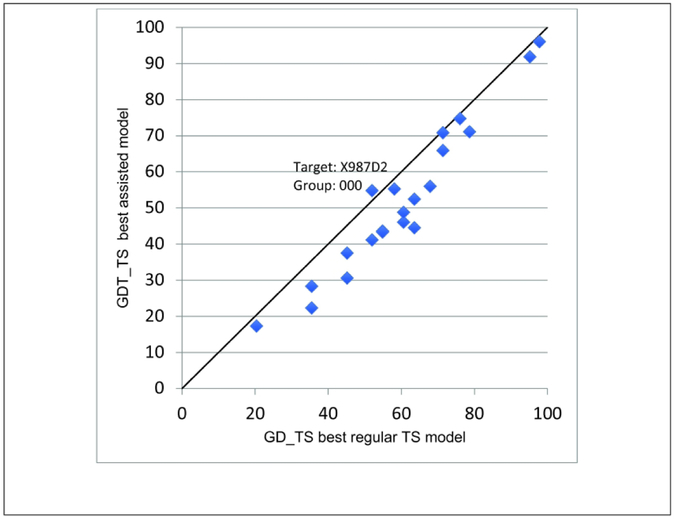
In our analysis so far (except analysis on complex modeling), we made comparisons among the 14 groups that submitted crosslink-assisted models and we drew conclusions about the relative improvements within this group. While systematic improvements were observed, the performance of these groups is primarily limited by their ability to sample correct conformations for the target proteins. The results are less impressive if we compare the accuracy of crosslink-assisted models to those in the general competition where 99 groups submitted predictions (Fig. 13). Clearly, the general category decidedly outperforms the 14 groups even though they were not using crosslinking data. This contrast can be explained in a broader context if we consider that the last three CASP meetings have witnessed a renaissance of predicting and incorporating predicted contacts in structure modeling, which culminated (so far) in CASP13 with never-before-seen contact prediction accuracies and correspondingly highly accurate models even in the free modeling category. Obviously, the purpose of using predicted contacts and experimental crosslinks is very similar, but one could argue that contact prediction, if accurate, provides a higher resolution information due to the shorter spatial distances of direct residue interactions and without the experimental limitation of the residues that can be considered.
Figure 13.
Head-to-head comparison of model accuracies between XL-MS assisted and regular modeling groups. Horizontal axis, accuracy of models in the general modeling category, vertical axis, accuracy of same models in data-assisted category. The plot displays targets with at least one valid and informative crosslink. The single best first models generated by any group are compared. The text marks the only superior performance, when an XL-MS assisted model (X987D2 by group 000) outperformed every single regular model.
Besides a general comparison between the 99 groups that submitted targets in the general category and the 14 that submitted in the data-assisted category, it is difficult to assess the possibility of additional synergy. The 14 groups in general were not among the top performers in the general modeling category, and therefore it is unclear how much they could have improved by using a more accurate starting conformation. While overall, the general modeling category models outperformed the XLMS models, there were anecdotal bright spots, where the best models were tied in accuracy and at least in one case (X987D2) when the data-assisted model was better than any model from the general category (Fig. 13). While statistically significant results cannot be reported for XLMS assisted complex modeling due to the small number of cases, the majority of complexes were more accurate than any of the general category results. A further refinement of the experimental procedures to generate crosslinks and of the algorithms that make use of experimentally derived distance information should increase the relevance of the method even for single proteins. The results from the data-assisted modeling category from CASP13 should help to direct such efforts.
A more thorough review of the general impact of data-assisted CASP experiments is both necessary and opportune but is beyond the scope of this article focusing solely on CASP13. It will therefore be the subject of a dedicated article to be published elsewhere.
Supplementary Material
Acknowledgments
We than Dr. I. Anishchanka and Dr. S. Ovchinnikov, University of Washington, for assistance in preparing contact-distance restraints for group 492. This work was supported by the following grants and agencies: National Institutes of Health (NIH) grant GM118709, GM100482 and AI141816; L'Agence Nationale de la Recherche (grant number ANR-15-CE11-0029-03); National Science Center of Poland (Narodowe Centrum Nauki) (NCN), grants UMO-2017/25/B/ST4/01026, UMO-2017/26/M/ST4/00044 and UMO-2017/27/B/ST4/00926; and the Benzon Foundation. Computational resources were provided by (a) the Interdisciplinary Centre of Mathematical and Computer Modeling (ICM) at the University of Warsaw (b) the Centre of Informatics - Tricity Academic Supercomputer & networK (CI TASK) (c) the Polish Grid Infrastructure (PL-GRID), and (d) our Beowulf cluster at the Faculty of Chemistry, University of Gdańsk. A.L. and E.T. would like to thank Ruedi Aebersold (ETH Zurich) for access to the Orbitrap Elite mass spectrometer and laboratory infrastructure. The CASP organizers and the Leitner and Rappsilber labs would also like to thank all groups that shared proteins and protein complexes for crosslinking experiments.
Footnotes
The authors declare no conflict of interests.
REFERENCES
- 1.Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A. Critical assessment of methods of protein structure prediction (CASP)-Round XII. Proteins. 2018;86 Suppl 1:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan FN, Che FY, Nieves E, Weiss LM, Angeletti RH, Fiser A. Photo-assisted peptide enrichment in protein complex cross-linking analysis of a model homodimeric protein using mass spectrometry. Proteomics. 2011;11(20):4109–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leitner A, Joachimiak LA, Unverdorben P, et al. Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. P Natl Acad Sci USA. 2014;111(26):9455–9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belsom A, Schneider M, Brock O, Rappsilber J. Blind Evaluation of Hybrid Protein Structure Analysis Methods based on Cross-Linking. Trends in Biochemical Sciences. 2016;41(7):564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinch L, Monastyrskyy B, Kryshtafovych A. CASP13 domain definition and classification. Proteins. 2019;This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leitner A, Walzthoeni T, Aebersold R. Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nature Protocols. 2014;9(1):120–137. [DOI] [PubMed] [Google Scholar]
- 7.Walzthoeni T, Claassen M, Leitner A, et al. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat Methods. 2012;9(9):901–903. [DOI] [PubMed] [Google Scholar]
- 8.Vizcaino JA, Csordas A, Del-Toro N, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(22):11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belsom A, Schneider M, Fischer L, Brock O, Rappsilber J. Serum Albumin Domain Structures in Human Blood Serum by Mass Spectrometry and Computational Biology. Mol Cell Proteomics. 2016;15(3):1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belsom A, Schneider M, Fischer L, et al. Blind testing cross-linking/mass spectrometry under the auspices of the 11(th) critical assessment of methods of protein structure prediction (CASP11). Wellcome Open Res. 2016;1:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogorzalek TL, Hura GL, Belsom A, et al. Small angle X-ray scattering and cross-linking for data assisted protein structure prediction in CASP 12 with prospects for improved accuracy. Proteins. 2018;86Suppl 1:202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiolica A, Cittaro D, Borsotti D, et al. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol Cell Proteomics. 2007;6(12):2200–2211. [DOI] [PubMed] [Google Scholar]
- 13.Rappsilber J, Friesen WJ, Paushkin S, Dreyfuss G, Mann M. Detection of arginine dimethylated peptides by parallel precursor ion scanning mass spectrometry in positive ion mode. Anal Chem. 2003;75(13):3107–3114. [DOI] [PubMed] [Google Scholar]
- 14.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature Protocols. 2007;2(8):1896–1906. [DOI] [PubMed] [Google Scholar]
- 15.Lenz S, Giese SH, Fischer L, Rappsilber J. In-Search Assignment of Monoisotopic Peaks Improves the Identification of Cross-Linked Peptides. J Proteome Res. 2018;17(11):3923–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giese SH, Fischer L, Rappsilber J. A Study into the Collision-induced Dissociation (CID) Behavior of Cross-Linked Peptides. Molecular & Cellular Proteomics. 2016;15(3):1094–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer L, Rappsilber J. Quirks of Error Estimation in Cross-Linking/Mass Spectrometry. Anal Chem. 2017;89(7):3829–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zemla A LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31(13):3370–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zemla A, Venclovas, Moult J, Fidelis K. Processing and evaluation of predictions in CASP4. Proteins. 2001;Suppl 5:13–21. [DOI] [PubMed] [Google Scholar]
- 20.Mariani V, Biasini M, Barbato A, Schwede T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics. 2013;29(21):2722–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olechnovic K, Monastyrskyy B, Kryshtafovych A, Venclovas C. Comparative analysis of methods for evaluation of protein models against native structures. Bioinformatics. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bullock JMA, Sen N, Thalassinos K, Topf M. Modeling Protein Complexes Using Restraints from Crosslinking Mass Spectrometry. Structure. 2018;26(7):1015-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider M, Belsom A, Rappsilber J, Brock O. Blind testing of cross-linking/mass spectrometry hybrid methods in CASP11. Proteins-Structure Function and Bioinformatics. 2016;84:152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bullock JMA, Schwab J, Thalassinos K, Topf M. The Importance of Non-accessible Crosslinks and Solvent Accessible Surface Distance in Modeling Proteins with Restraints From Crosslinking Mass Spectrometry. Molecular & Cellular Proteomics. 2016;15(7):2491–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marti-Renom MA, Madhusudhan MS, Fiser A, Rost B, Sali A. Reliability of assessment of protein structure prediction methods. Structure. 2002;10(3):435–440. [DOI] [PubMed] [Google Scholar]
- 26.Rykunov D, Fiser A. New statistical potential for quality assessment of protein models and a survey of energy functions. BMC Bioinformatics. 2010;11(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karczynska A, Mozolewska MA, Krupa P, et al. Use of the UNRES force field in template-assisted prediction of protein structures and the refinement of server models: Test with CASP12 targets. J Mol Graph Model. 2018;83:92–99. [DOI] [PubMed] [Google Scholar]
- 28.Hansmann UHE. Parallel tempering algorithm for conformational studies of biological molecules. Chem Phys Lett. 1997;281(1–3):140–150. [Google Scholar]
- 29.Rhee YM, Pande VS. Multiplexed-replica exchange molecular dynamics method for protein folding simulation. Biophysical Journal. 2003;84(2):775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liwo A, Baranowski M, Czaplewski C, et al. A unified coarse-grained model of biological macromolecules based on mean-field multipole-multipole interactions. J Mol Model. 2014;20(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krupa P, Halabis A, Zmudzinska W, Oldziej S, Scheraga HA, Liwo A. Maximum Likelihood Calibration of the UNRES Force Field for Simulation of Protein Structure and Dynamics. J Chem Inf Model. 2017;57(9):2364–2377. [DOI] [PubMed] [Google Scholar]
- 32.Sieradzan AK, Makowski M, Augustynowicz A, Liwo A. A general method for the derivation of the functional forms of the effective energy terms in coarse-grained energy functions of polymers. I. Backbone potentials of coarse-grained polypeptide chains. J Chem Phys. 2017;146(12). [DOI] [PubMed] [Google Scholar]
- 33.Khalili M, Liwo A, Jagielska A, Scheraga HA. Molecular dynamics with the united-residue model of polypeptide chains. II. Langevin and Berendsen-Bath dynamics and tests on model alpha-helical systems. Journal of Physical Chemistry B. 2005;109(28):13798–13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rakowski F, Grochowski P, Lesyng B, Liwo A, Scheraga HA. Implementation of a symplectic multiple-time-step molecular dynamics algorithm, based on the united-residue mesoscopic potential energy function. J Chem Phys. 2006;125(20). [DOI] [PubMed] [Google Scholar]
- 35.Czaplewski C, Kalinowski S, Liwo A, Scheraga HA. Application of Multiplexed Replica Exchange Molecular Dynamics to the UNRES Force Field: Tests with alpha and alpha plus beta Proteins. J Chem Theory Comput. 2009;5(3):627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rappsilber J, Siniossoglou S, Hurt EC, Mann M. A generic strategy to analyze the spatial organization of multi-protein complexes by cross-linking and mass spectrometry. Analytical Chemistry. 2000;72(2):267–275. [DOI] [PubMed] [Google Scholar]
- 37.Sieradzan AK, Jakubowski R. Introduction of Steered Molecular Dynamics into UNRES Coarse-Grained Simulations Package. Journal of Computational Chemistry. 2017;38(8):553–562. [DOI] [PubMed] [Google Scholar]
- 38.Lubecka EA, Liwo A. Introduction of a bounded penalty function in contact-assisted simulations of protein structures to omit false restraints. J Comput Chem. 2019;40(25):2164–2178. [DOI] [PubMed] [Google Scholar]
- 39.Karasikov M, Pages G, Grudinin S. Smooth orientation-dependent scoring function for coarse-grained protein quality assessment. Bioinformatics. 2018. [DOI] [PubMed] [Google Scholar]
- 40.Hoffmann A, Grudinin S. NOLB: Nonlinear Rigid Block Normal-Mode Analysis Method. J Chem Theory Comput. 2017;13(5):2123–2134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.