Abstract
Radiation-induced gastric injury is a serious concern that may limit the duration and the delivered dose of radiation. However, the genome-wide molecular changes in stomach upon ionizing radiation have not been reported. In this study, mouse stomach was irradiated with 6 or 12 Gy X-ray irradiation and we found that radiation resulted in the atrophy of gastric mucosa and abnormal morphology of chief and parietal cells. Radiation-induced gastric injury was accompanied by an increase in the serum levels of pepsinogen A and pepsinogen C but not gastrin-17. The expression profiles of messenger RNA (mRNA) and long noncoding RNA (lncRNA) in normal and irradiated gastric tissues were measured by microarray analysis. Results revealed 17 upregulated and 10 downregulated mRNAs were consistent in 6 and 12 Gy irradiated gastric tissues, including D site-binding protein (Dbp) and fibrinogen-like protein 1 (Fgl1). Thirteen upregulated and 96 downregulated lncRNAs were commonly changed in 6 and 12 Gy irradiated gastric tissues. The dysregulated mRNAs were implicated in multiple pathways and showed coexpression with lncRNAs. To identify motifs for transcription factors and coactivators in the proximal promoter regions of the dysregulated RNAs, the bioinformatic tool Biopython was used. A variety of common motifs that are associated with transcription factors were identified, including ZNF263, LMX1B, and Dlx1. Our findings illustrate the molecular changes during radiation-induced gastric injury and the potential transcription factors driving this alteration.
Keywords: ionizing radiation, stomach, radiation-induced gastric injury, microarray, lncRNA
Introduction
The stomach is located between the esophagus and the small intestine, which is one of the most sophisticated endocrine organs with unique physiology, biochemistry, and microbiology.1,2 The stomach fulfills important tasks in the mechanic and chemic digestion of food by secretion of digestive enzymes and gastric acids.1,2 Several kinds of cells in the stomach including parietal, chief, gastrin-secreting, enterochromaffin-like, and somatostatin-producing cells participate in the integrity and homeostasis of this organ.3 Radiotherapy is widely used in the treatment of various cancers.4-6 However, radiation-induced gastric injury may occur during radiotherapy of gastric cancer, esophageal cancer, and hepatocellular carcinoma that may limit the duration and the delivered dose of radiation.5,6 Early symptoms include nausea, vomiting, dyspepsia, and abdominal pain, while chronic dyspepsia and abdominal pain due to chronic ulceration have been reported to occur later after radiotherapy. However, the molecular mechanism of radiation-induced gastric injury is still unclear.7 Comprehensive understanding of the response of normal gastric tissue to ionizing radiation and detailed analysis might help to identify biomarkers for the diagnosis of radiation-induced gastric injury or targeted drugs for the prevention of this disease.
The mechanism of radiation-induced tissue damage is complex. Reactive oxygen species and DNA double-strand breaks are well-documented events. Other events including the activation of checkpoint pathway, mitochondrial or ER responses, cell apoptosis, senescence, necrosis, and ferroptosis have been implicated in radiation-induced damage.8-12 Numerous proteins, noncoding RNAs, and pathways are associated with radiation-induced tissue damage, which indicate a tissue-specific molecular alternation in response to ionizing radiation.13,14 Although multiple mechanisms have been proposed for the progression of radiation-induced tissue injury, the molecular signaling events in the stomach in response to radiation remain elusive.
To date, advances in high-throughput profiling methodology have provided a large amount of information regarding gene expression at the transcriptome level, as well as the underlying molecular events in response to irradiation.15,16 To our knowledge, the genome-wide landscape of differentially expressed RNAs of stomach in response to ionizing radiation has not been reported. In this study, we performed microarray analysis of messenger RNA (mRNA) and long noncoding RNA (lncRNA) to investigate irradiation-responsive genes in gastric tissues of mice. We compared the genome-wide expression between normal gastric tissues and irradiated group. Functional categories of differentially expressed mRNAs and differentially expressed lncRNAs were also analyzed. We further characterized key transcription factors involved in mRNA and lncRNA alteration.
Materials and Methods
Animals and Treatments
Male C57 mice (6 weeks old) were purchased from the Shanghai SLAC Laboratory Animal Co, Ltd (Shanghai, China). The animals were housed and maintained in a 12-hour light/dark cycle and had free access to food and water. The mice were anesthetized with an intraperitoneal injection of ketamine (75 mg/kg) and xylazine (10 mg/kg). The mice were immobilized with adhesive tape on a plastic plate to minimize motion during radiation exposure. A 3-cm thick piece of lead was used to shield the mice and localize the radiation field. A single dose of 6 or 12 Gy irradiation was administered to the stomach area at a dose rate of 2 Gy/min using a 6-MeV X-ray irradiation (Clinac 2100EX; Varian Medical Systems, Inc, Palo Alto, California). The control group of mice was sham irradiated. Seven days after irradiation, the gastric tissues were resected for analysis. Protocols for experiments involving animals were approved by the Animal Experimentation Ethics Committee at Soochow University (Suzhou, China).
Hematoxylin and Eosin Staining
Gastric tissues were fixed in 10% neutral-buffered formalin and embedded in paraffin. Three-micrometer paraffin sections were deparaffinized and heat-treated with citrate buffer (pH 6.0) for 7 minutes following an epitope retrieval protocol. The sections of mouse stomach were stained with Hematoxylin and Eosin (H&E). The thickness of gastric mucosa in each treatment group of mice was measured using ImageJ software (National Institutes of Health, Bethesda, Maryland).
Transmission Electron Microscopy
Gastric tissues were fixed for 2 hours with 2.5% glutaraldehyde in 0.05 M sodium cacodylate buffer, pH 7.2 at room temperature, followed by 2 hours in 2% OsO4 in 0.1 M sodium cacodylate buffer and 18 hours in 1% aqueous uranyl acetate solution. After dehydration through an ethanol series, the specimens were embedded in Epon 812 and ultrathin sections were collected on copper grids. After being stained with uranyl acetate and lead citrate, the sections were examined using a Tecnai G2 Spirit BioTwin transmission electron microscope (FEI Company, Hillsboro, Oregon).
LncRNA and MRNA Microarray Expression Profiling
Microarray profiling was performed in the laboratory of OE Biotechnology Company (Shanghai, China). The detailed methods were described in the Supplementary Materials and Methods.
Motif and Transcription Factor Search by Biopython
Two kilobytes upstream promoter sequences of the dysregulated mRNAs and lncRNAs were analyzed to locate known cis-regulatory elements using the sequence analysis tool Biopython,17 with a combined P value of <.01 as the threshold. The matrices generated by Biopython were then compared with the TRANSFAC database18 using the DNA binding motif similarity tool, WebLogo.19
Real-Time PCR Analysis
Total RNA from gastric tissues was reverse transcribed to complementary DNA (cDNA) using an oligo(dT)12 primer and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA). The SYBR green dye (Takara, Japan) was used for amplification of cDNA. Messenger RNA levels as well as that of the internal standard, glyceraldehyde 3-phosphate dehydrogenase (Gapdh), were measured by real-time quantitative PCR in triplicates using a Prism 7900 real-time PCR machine (Applied Biosystems, Foster City, California). The primers used for the real-time PCR analysis are summarized in Supplementary Table 1.
Immunohistochemistry
Gastric tissues of mice were fixed in 10% neutral-buffered formalin and embedded in paraffin. Three-micrometer paraffin sections were deparaffinized and heat treated with citrate buffer (pH 6.0) for 7 minutes following an epitope retrieval protocol. Three-micrometer paraffin sections were incubated with primary antibodies against Lmx1b (Abcam, Cambridge, United Kingdom, ab135734), Dlx1 (Abcam, ab236381), and ZNF263 (Biorbyt Ltd, Cambridgeshire, United Kingdom, orb158791) at 4°C overnight, followed by incubation with biotinylated secondary antibody (Beyotime, Nantong). Immunohistochemistry (IHC) staining was visualized using a substrate solution containing diaminobenzidine and hydrogen peroxide. The counter staining was performed with hematoxylin. All steps were performed at room temperature. Negative controls consisted of tissue sections with similar staining procedures in the absence of primary antibody.
Statistical Analysis
Data were expressed as mean ± standard error of the mean of at least 3 independent experiments. The results were evaluated via 1-way analysis of variance to determine statistical significance. The statistical analyses were performed using SPSS software. The differences were considered significant at P < .05. Other materials and methods are available in the Supplementary Materials and Methods.
Results
Radiation Disrupts the Physiology and Morphology in Gastric Tissues
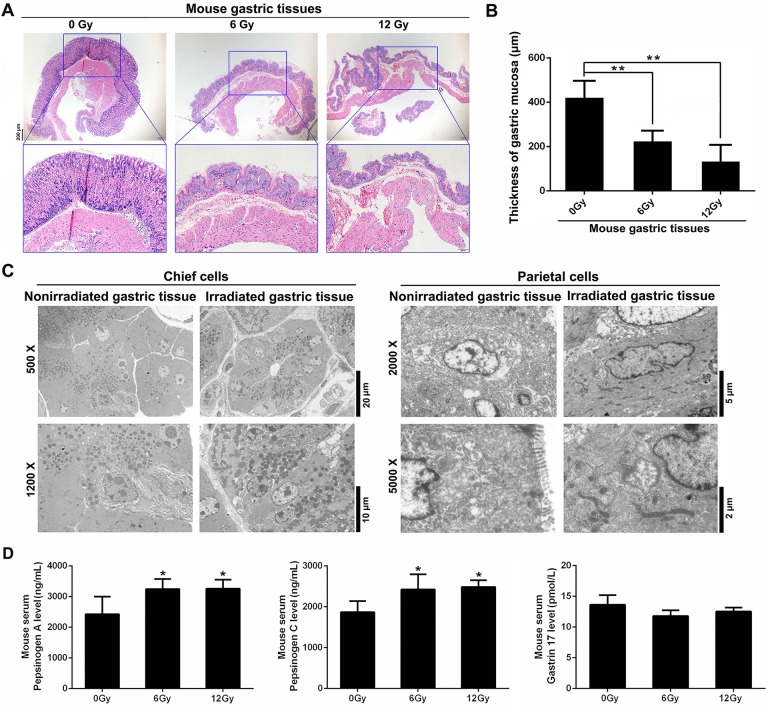
To investigate the effect of radiation on gastric tissues, a mouse model with radiation-induced gastric injury was established by administering 6 and 12 Gy X-ray irradiation. These doses were selected due to their application in larger fractional dose delivery such as stereotactic body radiation therapy (SBRT).20,21 One week after radiation, gastric tissues were collected and observed by H&E staining. Results showed that in irradiated gastric tissues, there was complete atrophy of gastric mucosa, which was replaced by metaplastic squamous epithelium (Figure 1A and B). Under electron microscope, we observed an obvious abnormality in nuclear morphology and aggregation of zymogen granules in 6 Gy X-ray irradiated chief cells (Figure 1C). Abnormal nucleus and mitochondria were observed in the parietal cells of 12 Gy X-ray irradiated gastric tissues (Figure 1C). Next, we investigated whether radiation affected serum biomarkers for gastric mucosa.22 As shown in Figure 1D, radiation increased the serum levels of pepsinogen A and pepsinogen C but not gastrin-17. The above results indicated that normal gastric physiology and morphology may be disrupted by radiation.
Figure 1.
Radiation induces gastric morphological and physiological changes in mouse models. A, Representative H&E staining of normal and irradiated gastric tissues 7 days after radiation. B, Quantification of the thickness of gastric mucosa. C, Electron microscopy analysis of gastric tissues from 6 or 12 Gy X-ray irradiated and nonirradiated mice. Representative images of chief cells and parietal cells are shown. D, Serum levels of pepsinogen A (PGA), pepsinogen C (PGC), and gastrin-17 (G17) were measured by ELISA. *P < .05; **P < .01, compared to the nonirradiated (0 Gy) group. H&E indicates hematoxylin and eosin.
Radiation Modulates the Expression of MRNAs in Gastric Tissues
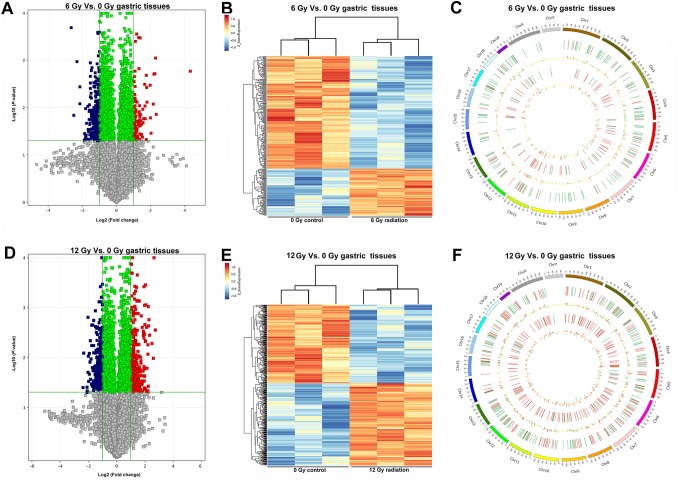
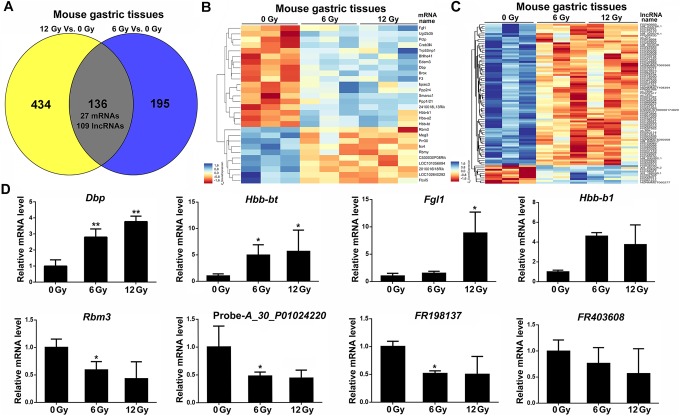
We next investigated the RNA profiles of irradiated and nonirradiated gastric tissues by microarray analysis. The raw array measures are accessible through Gene Expression Omnibus series accession number GSE114246. Messenger RNA profiling detected 92 mRNAs with significant differential expression levels with at least 2-fold change (P < .05) in 6 Gy irradiated gastric tissues compared to nonirradiated normal tissues, with 61 upregulated and 31 downregulated mRNAs (Figure 2A-C). Among the dysregulated mRNA transcripts, hemoglobin, β adult minor chain (Hbb-b2) was the most upregulated with a fold change of 4.99, whereas AI225934 was the most downregulated with a fold change of 6.30. One hundred ninety-four mRNAs (153 upregulated and 41 downregulated) were preferentially expressed in gastric tissues with 12 Gy X-ray radiation. Seventeen upregulated and 10 downregulated mRNAs were consistent in 6 and 12 Gy irradiated gastric tissues (Figure 3A and B). The list of 27 differentially expressed mRNAs in irradiated (both 6 and 12 Gy) gastric tissues is shown in Figure 3B and Table 1. These mRNAs include D site-binding protein (Dbp), fibrinogen-like protein 1 (Fgl1), proline rich 30 (Prr30), and RNA binding motif protein 3 (Rbm3).
Figure 2.
mRNA and lncRNA expression profiling of gastric tissue samples subjected to 6 and 12 Gy X-ray irradiation. A, The volcano plot of mRNA and lncRNA profiling between 6 Gy irradiation and 0 Gy control gastric tissues of mice. B, The heatmap of the significant mRNA and lncRNA expression patterns in the control (0 Gy) and 6 Gy group of gastric tissue samples. C, Differential expressed gene distribution in mouse chromosomes following 6 Gy irradiation. Red and blue bars represent upregulated and downregulated differential expressed genes, respectively. D, The volcano plot of expressed mRNAs and lncRNAs between control and 12 Gy irradiated gastric tissues. E, The heatmap of the significant mRNA and lncRNA expression patterns in the control and 12 Gy group of gastric tissue samples. F, Differential expressed gene distribution in mouse chromosomes following 12 Gy irradiation. lncRNA indicates long noncoding RNA; mRNA, messenger RNA.
Figure 3.
Common dysregulated mRNAs and lncRNAs in irradiated gastric tissues. A, Venn diagram significant differently expressed genes (lncRNA and mRNA) in 0, 6, and 12 Gy irradiated gastric tissue samples. B, The heatmap of the common significant mRNA expression patterns in control and irradiated groups. C, The heatmap of the common significant lncRNA expression patterns in control and irradiated groups. D, Real-time PCR analysis of relative mRNAs and lncRNAs levels. Relative RNA level was normalized to that of GAPDH. *P < .05; **P < .01, compared with the nonirradiated (0 Gy) group. lncRNA indicates long noncoding RNA; mRNA, messenger RNA.
Table 1.
Differentially Expressed MRNAs in Irradiated Gastric Tissues of Mice.
| No. | Gene Symbol | Fold Change (12 vs 0 Gy) | P Value (12 vs 0 Gy) | Fold Change (6 vs 0 Gy) | P Value (6 vs 0 Gy) | Regulation |
|---|---|---|---|---|---|---|
| 1 | Dbp | 4.736 | .002 | 3.368 | .004 | Upregulation |
| 2 | Hbb-bt | 3.786 | .015 | 3.890 | .003 | Upregulation |
| 3 | Fgl1 | 3.759 | .017 | 2.330 | .048 | Upregulation |
| 4 | Hbb-b1 | 3.486 | .033 | 4.851 | .001 | Upregulation |
| 5 | Trp53inp1 | 2.912 | .035 | 2.130 | .002 | Upregulation |
| 6 | Bhlhe41 | 2.718 | .028 | 2.270 | .020 | Upregulation |
| 7 | F3 | 2.644 | .032 | 2.023 | .042 | Upregulation |
| 8 | Ugt2b35 | 2.542 | .024 | 2.996 | .017 | Upregulation |
| 9 | Edem3 | 2.538 | .004 | 2.474 | .002 | Upregulation |
| 10 | Hba-a2 | 2.518 | .045 | 2.543 | .009 | Upregulation |
| 11 | Ppp1r21 | 2.512 | .001 | 2.083 | .022 | Upregulation |
| 12 | 2410018L3Rik | 2.502 | .019 | 3.185 | .009 | Upregulation |
| 13 | Creb3l4 | 2.350 | .031 | 2.884 | .011 | Upregulation |
| 14 | Brox | 2.277 | .004 | 2.040 | .014 | Upregulation |
| 15 | Smarcc1 | 2.202 | .022 | 2.007 | .045 | Upregulation |
| 16 | Pctp | 2.201 | .041 | 2.194 | .049 | Upregulation |
| 17 | Ppp2r4 | 2.190 | .015 | 2.552 | .020 | Upregulation |
| 18 | LOC101056094 | 2.089 | .018 | 2.085 | .003 | Downregulation |
| 19 | Prr30 | 3.666 | .005 | 2.537 | .029 | Downregulation |
| 20 | Rbm3 | 3.012 | .018 | 2.020 | .001 | Downregulation |
| 21 | Rbmy | 2.880 | .016 | 2.606 | .028 | Downregulation |
| 22 | LOC102640292 | 2.851 | .029 | 2.695 | .039 | Downregulation |
| 23 | Meg3 | 2.639 | .007 | 2.466 | .022 | Downregulation |
| 24 | Fbxl5 | 2.493 | .038 | 2.527 | .019 | Downregulation |
| 25 | Irx4 | 2.306 | .014 | 2.222 | .020 | Downregulation |
| 26 | 2010016I18Rik | 2.266 | .005 | 2.180 | .030 | Downregulation |
| 27 | C530030P08Rik | 2.208 | .007 | 2.028 | .003 | Downregulation |
Long Noncoding RNAs Are Dysregulated in Irradiated Gastric Tissues
Using the same criteria as the mRNAs, we found 239 lncRNAs (36 upregulated and 203 downregulated) with significant differential expression levels between 6 Gy irradiated and mock irradiated gastric tissues (P < .05). Besides, we found 376 differentially expressed lncRNA, and there are 138 upregulated and 238 downregulated lncRNAs in gastric tissues with 12 Gy X-ray radiation. Thirteen upregulated and 96 downregulated lncRNAs were commonly changed in 6 and 12 Gy irradiated gastric tissues (Figures 1A-1F, 3A and C). The list of the common differentially expressed lncRNAs is shown in Supplementary Table 2. These dysregulated lncRNAs included FR112177, FR198137, XR_002334.2, and FR403608.
Validation of Dysregulated RNAs by Real-Time PCR Analysis
We next validated the expression of 8 dysregulated genes using real-time PCR. The results showed that Dbp, Hbb-bt, Fgl1, and Hbb-b1 transcripts were significantly higher in irradiated gastric tissues, whereas the expression levels of Rbm3, FR198137, and probe A_30_P01024220-related gene were significantly reduced (Figure 3D). Excluding the expression levels of FR403608 and probe A_30_P01024220-related gene, the results from real-time PCR analysis were consistent with that from microarray analysis.
Functional Annotation of Differentially Expressed mRNAs
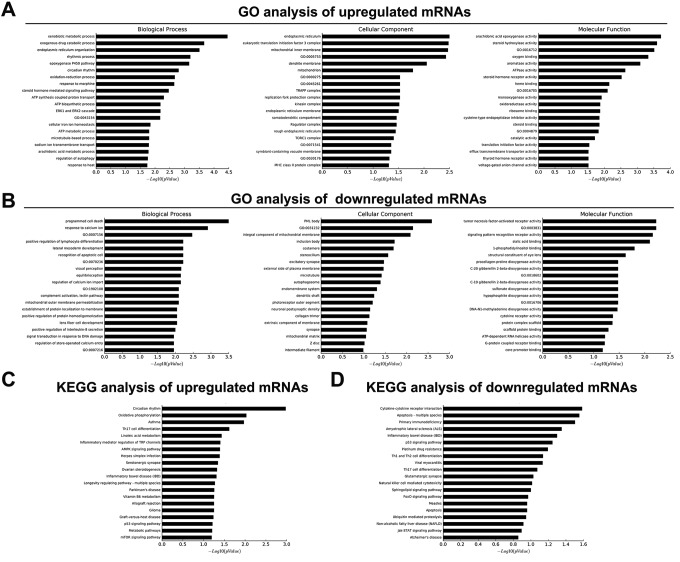
Upregulated and downregulated mRNAs were assigned into functional groups according to the Gene Ontology (GO) analysis, respectively. For upregulated mRNAs, the most dominant groups were “xenobiotic metabolic process,” “exogenous drug catabolic process,” and “endoplasmic reticulum organization” in the biological process ontology. While, in the cellular component ontology, the dominant groups were “endoplasmic reticulum.” The molecular function ontology analysis also convinced us that differentially expressed proteins mainly function in “arachidonic acid epoxygenase activity,” “steroid hydroxylase activity,” and “oxidoreductase activity” (Figure 4A).
Figure 4.
Gene Ontology (GO) category and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of the differentially expressed mRNA from the mouse gastric tissues in response to radiation. A, GO category enrichment of the upregulated expressed mRNAs from mouse gastric tissues in response to radiation. B, GO category enrichment of the downregulated expressed mRNAs from mouse gastric tissues in response to radiation. C, KEGG pathway enrichment of the upregulated mRNAs from mouse gastric tissues in response to radiation. D, KEGG pathway enrichment of the downregulated mRNAs from mouse gastric tissues in response to radiation. mRNA indicates messenger RNA.
For downregulated mRNAs, the most dominant groups were “programmed cell death” and “response to calcium ion.” While, in the cellular component ontology, the dominant groups were “PML body” and “extrinsic component of external side of plasma membrane.” In the molecular function ontology, the dominant groups were “tumor necrosis factor-activated receptor activity” and “signaling pattern recognition receptor activity” (Figure 4B).
Then, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed to investigate pathways that possibly involved in radiation-induced gastric injury. The enriched pathways in upregulated mRNAs included circadian rhythm and oxidative phosphorylation, Th17 cell differentiation, and linoleic acid metabolism (Figure 4C). The pathways associated with downregulated mRNAs included cytokine–cytokine receptor interaction, apoptosis, and primary immunodeficiency (Figure 4D).
Construction of Coexpression Network
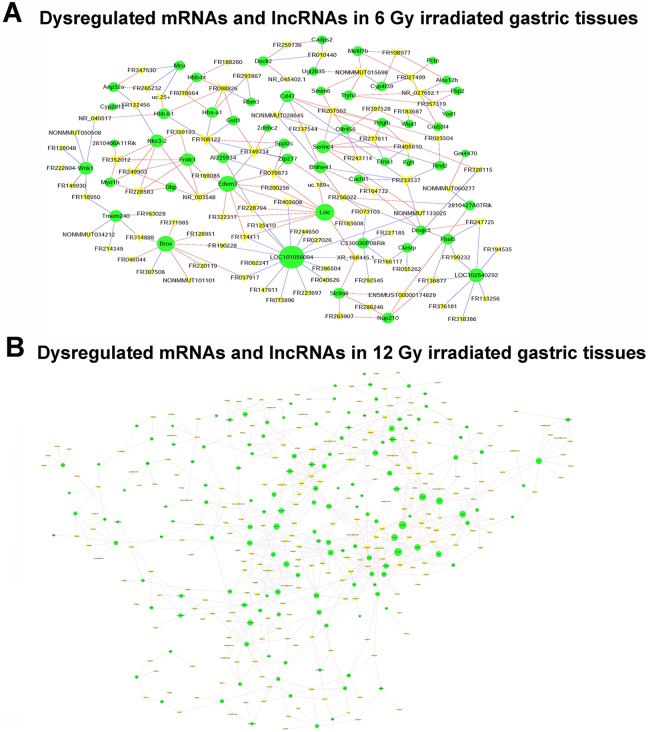
To identify those lncRNAs and mRNAs that might play a critical role in radiation-induced gastric injury progression, we constructed a coexpression network based on the correlation analysis between the differentially expressed lncRNAs and mRNAs. The coexpression networks are shown in Figure 5A and B. The coexpression network indicated that a single mRNA or lncRNA might correlate with 1 to 16 lncRNAs suggesting that the interregulation of lncRNAs and mRNAs is involved in radiation-induced gastric injury. There was a difference between 6 and 12 Gy irradiated coexpression network, indicating different response mechanism following radiation.
Figure 5.
The coexpression network between dysregulated lncRNAs and mRNAs. A, Coexpression network between dysregulated lncRNAs and mRNAs in 6 Gy irradiated gastric tissues. B, Coexpression network between dysregulated lncRNAs and mRNAs in 12 Gy irradiated gastric tissues. The green circles indicate mRNAs and yellow arrow indicates lncRNAs. lncRNA indicates long noncoding RNA; mRNA, messenger RNA.
Transcriptional Analysis of Dysregulated mRNAs and lncRNAs by Radiation
Since the differentially expressed mRNAs and lncRNAs are possibly driven by transcription factors or coactivators, which are mediated by the recruitment of transcription factors or coactivators to cis-regulatory elements,23,24 we therefore analyzed the proximal promoter regions of the dysregulated RNAs by Biopython to identify motifs for transcription factors and coactivators. Evaluation of 2.0 kb upstream promoter region (−1 to −2000) of the promoters of the dysregulated mRNAs and lncRNAs using Biopython resulted in the identification of several common transcription factors and corresponding cis-regulatory elements. Multiple motifs with binding sites for putative transcription factors and coactivators were found. The putative transcription factors and corresponding motifs are shown in Figure 6 and Supplementary Table 3, respectively. For example, ZNF263, LMX1B, THAP1, GSX2, and GSX1 showed over 500 binding sites in the promoter region of the commonly upregulated mRNAs and lncRNAs. A variety of transcription factors including GSX2, GSX1, and Barhl1 were possibly associated with downregulated RNAs. Interestingly, multiple transcription factors including LMX1B, GSX1/2, Dlx1, and Barhl1 were associated with both upregulated and downregulated mRNAs and lncRNAs. These transcriptional factors are likely to be activated by radiation and function along with their coactivators/cosuppressors.
Figure 6.
Transcriptional analysis of mRNA and lncRNA alteration of radiation-induced gastric injury. The differentially expressed mRNAs and lncRNAs were analyzed by Biopython for the –1 bp to –2.0 kb (relative to the transcription start site) sequences of the promoters. Shown were the top 20 putative transcriptional factors and corresponding motifs for the upregulated (A) or the downregulated RNAs (B). lncRNA indicates long noncoding RNA; mRNA, messenger RNA.
The Involvement of ZNF263, LMX1B, and Dlx1 in Radiation-Induced Gastric Injury
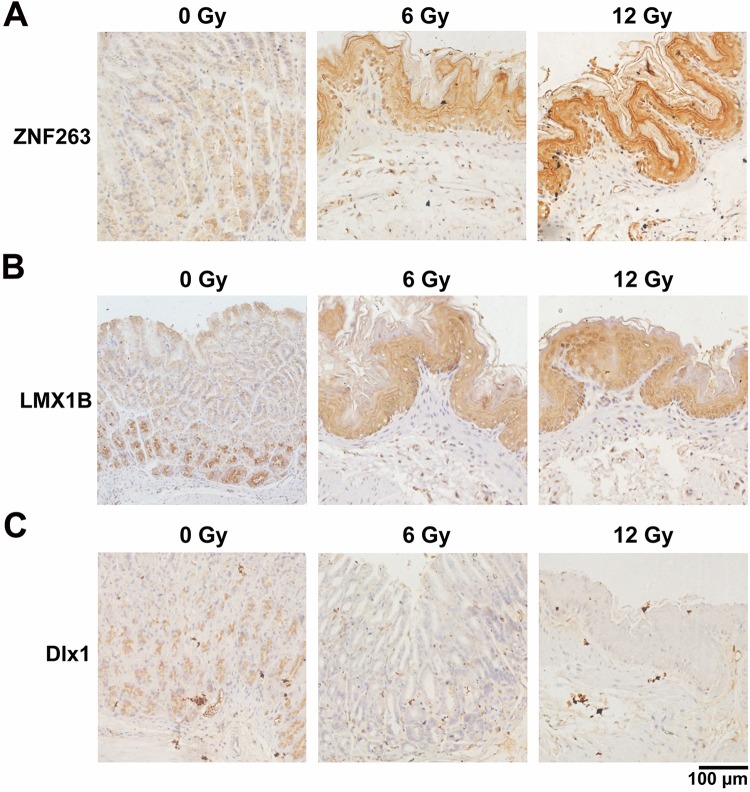
To confirm our predicted bioinformatics result by Biopython, IHC was performed to investigate the expression levels of ZNF263, LMX1B, and Dlx1 in gastric tissues with or without radiation. Results showed that 6 and 12 Gy irradiation caused a marked increase in the expression levels of ZNF263 and LMX1B in gastric mucosa of mice, especially in the nucleus (Figure 7A and B). In contrast, the expression of Dlx1 was decreased in irradiated gastric tissues (Figure 7C). However, the expression levels of ZNF263, LMX1B, and Dlx1 did not show significant change, suggesting a protein-level regulation of these transcriptional factors upradiation. These results indicated the potential involvement of these transcription factors in radiation-induced gastric injury.
Figure 7.
Involvement of ZNF263, LMX1B, and Dlx1 in radiation-induced gastric injury. IHC staining of (A) ZNF263, (B) LMX1B, and (C) Dlx1 in normal and irradiated gastric tissues of mice. Representative images are shown. Scale bar represents 100 µm (magnification, ×200). IHC indicates immunohistochemistry.
Discussion
Radiation-induced gastric injury is a dose-limiting factor for radiotherapy or chemoradiotherapy.5-7 Till now, only a few studies reported countermeasures against this disease, including extract of Aegle marmelos25 and stem cell therapy.26 Therefore, it is necessary to explore novel mechanisms, biomarkers, and intervention targets to understand and identify strategies against radiation-induced gastric injury.27 Comparing the transcriptome profiles in response to radiation, we identified 136 differentially expressed genes (27 mRNAs and 109 lncRNAs) between the control and irradiated gastric tissues including 29 upregulated and 107 downregulated RNAs. To our knowledge, this study is the first to show RNA profiles of gastric tissues in response to ionizing radiation. Also, we investigated the expression landscape across whole chromosomes. We found that the genes showing radiation-altered expression were evenly located on every chromosome except the sex chromosomes X and Y. This is the first report to describe the changes in mRNA and lncRNA expression in response to ionizing radiation in gastric tissues, providing insights into the molecular pathogenesis of radiation-induced gastric injury. Moreover, these dysregulated mRNAs and lncRNAs might play an important role in the occurrence of radiation-induced gastric injury and emerge as hallmarks of radiation-induced gastric injury. The application of molecular biomarkers to assist with the early detection of radiation-induced gastric injury has the potential to substantially improve our ability to select patients for radiation-induced gastric injury screening. As expected, pathway analysis revealed that radiation affected multiple pathways in the mouse stomach. For example, oxidative phosphorylation has been shown to be modulated by ionizing radiation.28,29 Circadian rhythm has recently been shown to affect the efficacy of radiotherapy,30 and it has also been suggested as a radioprotective approach.31
Cis-regulatory elements play a central role in regulating gene expression by integrating signals at the DNA level, upstream of a target gene. Our study presents an in silico analysis of upstream promoter sequences that regulate radiation-affected mRNAs and lncRNAs, which contributes to the identification of radiation-responsive transcriptional factors. Using this analysis, we identified multiple putative transcription factors for the dysregulated genes. The differentially expressed mRNAs and lncRNAs are possibly regulated as a result of these trans-acting factors. For example, LMX1B strongly promotes migration of cancer cells in culture and promotes xenograft growth in nude mice.32 DLX1, a binding protein of β-catenin, promoted the growth and migration of prostate cancer cells.33 THAP1 regulates embryonic stem cell potential, survival, and proliferation.34 However, the majority of these transcription factors have rarely been reported to be associated with radiation response, suggesting a set of potential radiation-responsive transcription factors in the stomach. The mechanism of radiation-induced activation of these transcription factors and identification of their coactivators/cosuppressors merit further investigation.
In conclusion, we have characterized pathological alterations in radiation-induced gastric injury and found that a substantial number of specific mRNAs and lncRNAs are involved in this process. A variety of common motifs that are associated with transcription factors were identified, including ZNF263, LMX1B, and Dlx1. Taken together, our findings illustrate the pathological and molecular changes during radiation-induced gastric injury and the potential transcription factors driving this alteration.
Supplemental Material
Supplementary_Materials for mRNA and lncRNA Expression Profiling of Radiation-Induced Gastric Injury Reveals Potential Radiation-Responsive Transcription Factors by Guangxia Chen, Yang Feng, Zhiqiang Sun, Yiying Gao, Chuannan Wu, Haihan Zhang, Jinming Cao, Zhuo Chen, Jianping Cao, Yaqun Zhu and Shuyu Zhang in Dose-Response
Acknowledgments
This work is supported by the National Natural Science Foundation of China (81602101, 81502038, 81773227, and 31770911), Key Scientific Development Program of China (2016YFC0904702), Social Development Program of Jiangsu Province (BE2017634 and BE2017652), and Post graduate Research & Practice Innovation Program of Jiangsu Province (No KYCX19_1969).
Authors’ Note: Microarray data Gene Expression Omnibus accession number: GSE114246.
Author Contribution: Guangxia Chen and Yang Feng are contributed equally to this work.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Shuyu Zhang  https://orcid.org/0000-0003-1419-3635
https://orcid.org/0000-0003-1419-3635
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Hunt RH, Camilleri M, Crowe SE, et al. The stomach in health and disease. Gut. 2015;64(10):1650–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arin RM, Gorostidi A, Navarro-Imaz H, Rueda Y, Fresnedo O, Ochoa B. Adenosine: direct and indirect actions on gastric acid secretion. Front Physiol. 2017;8:737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Choi E, Roland JT, Barlow BJ, et al. Cell lineage distribution atlas of the human stomach reveals heterogeneous gland populations in the gastric antrum. Gut. 2014;63(11):1711–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brizel DM. Pharmacologic approaches to radiation protection. J Clin Oncol. 2007;25(26):4084–4089. [DOI] [PubMed] [Google Scholar]
- 5. Hartgrink HH, Jansen EP, Van Grieken NC, Van de Velde CJ. Gastric cancer. Lancet. 2009;374(9688):477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buergy D, Lohr F, Baack T, et al. Radiotherapy for tumors of the stomach and gastroesophageal junction—a review of its role in multimodal therapy. Radiat Oncol. 2012;7:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shadad AK, Sullivan FJ, Martin JD, Egan LJ. Gastrointestinal radiation injury: symptoms, risk factors and mechanisms. World J Gastroenterol. 2013;19(2):185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Panganiban RA, Snow AL, Day RM. Mechanisms of radiation toxicity in transformed and non-transformed cells. Int J Mol Sci. 2013;14(8):15931–15958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ravanat JL, Breton J, Douki T, et al. Radiation-mediated formation of complex damage to DNA: a chemical aspect overview. Br J Radiol. 2014;87(1035):20130715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Q, Xia X, Wu S, et al. Apoptosis, necrosis, and autophagy in mouse intestinal damage after 15-Gy whole body irradiation. Cell Biochem Funct. 2014;32(8):647–656. [DOI] [PubMed] [Google Scholar]
- 11. Nakajima T. Signaling cascades in radiation-induced apoptosis: roles of protein kinase C in the apoptosis regulation. Med Sci Monit. 2006;12(10):RA220–RA224. [PubMed] [Google Scholar]
- 12. Xu Y, Rong X, Hu W, et al. Bevacizumab monotherapy reduces radiation-induced brain necrosis in nasopharyngeal carcinoma patients: a randomized controlled trial. Int J Radiat Oncol Biol Phys. 2018;101(5):1087–1095. [DOI] [PubMed] [Google Scholar]
- 13. Datta K, Suman S, Fornace AJ., Jr Radiation persistently promoted oxidative stress, activated mTOR via PI3K/Akt, and downregulated autophagy pathway in mouse intestine. Int J Biochem Cell Biol. 2014;57:167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang S, Wang W, Gu Q, et al. Protein and miRNA profiling of radiation-induced skin injury in rats: the protective role of peroxiredoxin-6 against ionizing radiation. Free Radic Biol Med. 2014;69:96–107. [DOI] [PubMed] [Google Scholar]
- 15. Belling KC, Tanaka M, Dalgaard MD, et al. Transcriptome profiling of mice testes following low dose irradiation. Reprod Biol Endocrinol. 2013;11:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xie L, Zhou J, Zhang S, et al. Integrating microRNA and mRNA expression profiles in response to radiation-induced injury in rat lung. Radiat Oncol. 2014;9:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cock PJ, Antao T, Chang JT, et al. Biopython: freely available python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25(11):1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khan A, Fornes O, Stigliani A, et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46(D1):D260–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Amini A, Yeh N, Gaspar LE, Kavanagh B, Karam SD. Stereotactic body radiation therapy (SBRT) for lung cancer patients previously treated with conventional radiotherapy: a review. Radiat Oncol. 2014;9:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Milano MT, Constine LS, Okunieff P. Normal tissue toxicity after small field hypofractionated stereotactic body radiation. Radiat Oncol. 2008;3:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Broutet N, Plebani M, Sakarovitch C, Sipponen P, Mégraud F, Eurohepygast Study Group. Pepsinogen A, pepsinogen C, and gastrin as markers of atrophic chronic gastritis in European dyspeptics. Br J Cancer. 2003;88(8):1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu Z, Liu X, Liu L, et al. Regulation of lncRNA expression. Cell Mol Biol Lett. 2014;19(4):561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoon JH, Kim J, Gorospe M. Long noncoding RNA turnover. Biochimie. 2015;117:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jagetia GC, Venkatesh P, Baliga MS. Fruit extract of Aegle marmelos protects mice against radiation-induced lethality. Integr Cancer Ther. 2004;3(4):323–332. [DOI] [PubMed] [Google Scholar]
- 26. Sémont A, François S, Mouiseddine M, et al. Mesenchymal stem cells increase self-renewal of small intestinal epithelium and accelerate structural recovery after radiation injury. Adv Exp Med Biol. 2006;585:19–30. [DOI] [PubMed] [Google Scholar]
- 27. Carrington R, Staffurth J, Warren S, et al. The effect of dose escalation on gastric toxicity when treating lower oesophageal tumours: a radiobiological investigation. Radiat Oncol. 2015;10:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nosov AV, Ivnitsky YY, Malakhovsky VN. Metabolic correction of cerebral radiation syndrome. Radiat Res. 1999;152(5):523–529. [PubMed] [Google Scholar]
- 29. Scaife JF, Hill B. The uncoupling of oxidative phosphorylation by ionizing radiation. Can J Biochem Physiol. 1962;40:1025–1042. [PubMed] [Google Scholar]
- 30. Chan S, Zhang L, Rowbottom L, et al. Effects of circadian rhythms and treatment times on the response of radiotherapy for painful bone metastases. Ann Palliat Med. 2017;6(1):14–25. [DOI] [PubMed] [Google Scholar]
- 31. Gupta D, Shukla P, Munshi A, Aggarwal JP. Cardioprotective radiotherapy: the circadian way. Med Hypotheses. 2012;78(3):353–355. [DOI] [PubMed] [Google Scholar]
- 32. He L, Guo L, Vathipadiekal V, et al. Identification of LMX1B as a novel oncogene in human ovarian cancer. Oncogene. 2014;33(33):4226–4235. [DOI] [PubMed] [Google Scholar]
- 33. Liang M, Sun Y, Yang HL, Zhang B, Wen J, Shi BK. DLX1, a binding protein of beta-catenin, promoted the growth and migration of prostate cancer cells. Exp Cell Res. 2018;363(1):26–32. [DOI] [PubMed] [Google Scholar]
- 34. Aguilo F, Zakirova Z, Nolan K, et al. THAP1: Role in mouse embryonic stem cell survival and differentiation. Stem Cell Rep. 2017;9(1):92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_Materials for mRNA and lncRNA Expression Profiling of Radiation-Induced Gastric Injury Reveals Potential Radiation-Responsive Transcription Factors by Guangxia Chen, Yang Feng, Zhiqiang Sun, Yiying Gao, Chuannan Wu, Haihan Zhang, Jinming Cao, Zhuo Chen, Jianping Cao, Yaqun Zhu and Shuyu Zhang in Dose-Response