Abstract
Consumption of walnuts has slowed breast cancer growth and/or reduced the risk of mammary cancer in mice. The benefit against cancer was associated with altered expression of genes for cancer growth and survival. We hypothesized that walnut consumption would alter gene expression in pathologically confirmed breast cancers of women in a direction that would be expected to decrease breast cancer growth and survival, as was seen in mice. The study was a nonplacebo, 2-arm, clinical trial. Women with breast lumps large enough for research and pathology biopsies were recruited and randomized to walnut consuming or control groups. Immediately after biopsy collection, women in the walnut group began to consume 2 oz of walnuts per day until follow-up surgery. Pathological studies confirmed that lumps were breast cancer in all women who remained in the trial. At surgery, about 2 weeks after biopsy, additional specimens were taken from the breast cancers. Changes in gene expression in the surgical specimen compared to baseline were determined in each individual woman in walnut-consuming (n = 5) and control (n = 5) groups. RNA sequencing expression profiling revealed that expression of 456 identified genes was significantly changed in the tumor due to walnut consumption. Ingenuity Pathway Analysis showed activation of pathways that promote apoptosis and cell adhesion, and inhibition of pathways that promote cell proliferation and migration. These results support the hypothesis that, in humans, walnut consumption could suppress growth and survival of breast cancers.
Keywords: Breast cancer, Human dietary intervention, Walnuts, Prevention, Clinical trial
1. Introduction
Diet is thought to make a large contribution to risk for developing cancer [1,2], with some dietary components increasing and some components decreasing risk. Many research studies involving cancer prevention strategies have used isolated food components to ascertain that the component had cancer preventive properties. Although it is scientifically desirable to use individual components to be able to define the cancer prevention properties of the specific component, people routinely consume whole foods. When combined, the multiple components of whole foods could synergize or antagonize, either increasing or decreasing the effects of the individual components [3]; thus, it is important to test the effects of whole foods on cancer.
Cell culture studies can provide potential mechanisms for slowed tumor growth of individual food components. One cell culture study used extracts from walnuts, and the authors reported that components of walnuts, especially alpha-linolenic acid and beta-sitosterol, are absorbed and either directly or indirectly decreased proliferation of MCF-7 cells [4]. Another cell culture study reported that a potent antioxidant, ellagic acid, present in high amounts in walnuts and many fruits, can inhibit cell proliferation and induce apoptosis [5]. Oxidative stress and the inflammation induced by oxidative stress have been proposed as key promoters of cancer [6]. The abundant and varied forms of antioxidants in walnuts [5,7–14] are likely to be critical to the cancer growth suppression and apoptosis inducing mechanisms of walnuts. However, cell culture studies are not usually designed to test the effects of interactions of components of whole foods.
Preclinical studies can provide tumor development and growth data and might better assess the potential effects of a whole food. We conducted 2 preclinical studies to determine whether consumption of walnuts might affect cancer growth and development. In study 1, we found that addition of a small amount of walnuts (the human equivalent of 2 oz/d, calculated as a fraction of total calories) to the diet of mice significantly slowed the growth of implanted MDA-MB 231 human breast cancers [15]. Because we used the whole walnut meat rather than an isolated component, we could not identify the specific component(s) of the walnut that was (were) effective at slowing cancer growth. Components of walnuts that have individually been shown to slow cancer growth include α-linolenic acid (an 18C omega 3 fatty acid)[16], beta-sitosterol [17], vitamin E [18], and melatonin [19]. We also could not identify a specific mechanism(s) for the slower tumor growth, but proliferation was significantly decreased and apoptosis and antioxidative capacity were slightly increased [15].
In our second study (study 2), we used C(3)1TAg transgenic mice [20] that are genetically programmed to develop mammary gland cancer. We found that, compared to a diet without walnut, consumption of the walnut-containing diet significantly reduced tumor incidence (fraction of mice with at least 1 tumor at 145 days) by about 50%, multiplicity (number of glands with tumor/mouse), and tumor size [21]. Expression analyses of about 80 cancer-associated genes indicated that consumption of the walnut diet altered the expression of multiple genes associated with proliferation and differentiation of mammary epithelial cells and of a few genes associated with cell death. Thus, study 2 results, in a different model, agreed with the results of study 1 that dietary walnut slowed tumor cell proliferation and increased tumor cell death. A comparison with another dietary intervention (canola oil [22]) indicated that the omega 3 content of the walnut alone did not account for the extent of tumor suppression due to the walnut-containing diet, indicating synergism against cancer among the walnut components. The signaling pathways and proteins that were altered by dietary walnut in mammary glands of the mice have been identified as important in the development of human breast cancer; therefore, the mouse study should be relevant to humans. Thus, cell culture and animal studies support the notion that walnut consumption may be beneficial against cancer in humans. However, neither cell culture nor mouse studies can exactly model walnut digestion and metabolism in humans, showing the need for clinical trials to illustrate the potential effects of walnuts on breast cancer.
An important question might be: Can short-term consumption of walnuts change gene expression in humans? Cortes and colleagues reported changes in serum proteins, indicating changes in gene expression, after a single meal containing 40 g (1.4 oz) of walnuts [23]. Given the information that, in humans, a single meal containing walnut can change gene expression and the mouse data that gene expression was changed by walnut consumption, it seems reasonable that consumption of 2 oz of walnuts per day for about 2 weeks could change gene expression in human breast cancer.
We hypothesized that walnut consumption would alter gene expression in the pathologically confirmed breast cancers of women in a direction that would be expected to decrease breast cancer growth and survival, as was seen in mice. If the molecular changes in humans and mice were similar, it seemed reasonable to conclude that the effects on breast cancer in humans should be similar to those seen in mice (ie, slowed cancer cell growth or increased cancer cell death) and that consumption of walnuts could be beneficial to combat occurrence or recurrence of breast cancer in humans.
2. Methods and materials
2.1. Institutional Review Board approval
The Marshall University Office of Research Integrity has an Institutional Review Board, which reviews and monitors all human subject research conducted at Marshall University, St Mary’s Medical Center, Cabell Huntington Hospital, and the Edwards Cancer Center. The research protocol and participant informed consent were approved by the Institutional Review Board (protocol number 339384–3). This study was not listed at ClinicalTrials.gov. Potential study participants were identified from records review by the Research Study Nurse prior to their appointment for a diagnostic biopsy. At the appointment time, the potential participant was interviewed by the study nurse, the study was explained, and informed consent was obtained. The physician obtained 1 or 2 additional biopsies for research use when the biopsy was obtained for pathology studies.
2.1.1. Inclusion criteria
All subjects (1) were female and with a breast mass that, according to standard of care, was to be biopsied for diagnosis and was large enough to obtain the needed biopsies for pathology and research; (2) understood and were willing to sign the informed consent form; (3) had an Eastern Cooperative Oncology Group performance status of 0 or 1 (0: fully active, able to carry on all predisease performance without restriction; 1: restricted in physically strenuous activity but ambulatory and able to carry out work of a light or sedentary nature, eg, light housework, office work); (4) were between 18 and 90 years of age; and (5) were recruited as available without regard to race or ethnicity.
2.1.2. Exclusion criteria
Excluded persons were (1) those who do not like or who were allergic to walnuts or other tree nuts; (2) those with any metabolic disease that could be affected by walnut consumption; (3) those with a life expectancy less than 6 months; and (4) those who were pregnant (to prevent confounding due to pregnancy hormonal factors).
2.2. Clinical protocol
Subjects were consented at their first visit and were randomized into treated (consume walnut) or control (no added walnuts) groups. Routine clinical data were recorded (age, weight, height, family history, etc). A 5-mL blood specimen in ethylenediaminetetraacetic acid was collected for the research laboratory. After the initial biopsy, the subject was asked to continue to consume the usual diet and to not change consumption of any medication or supplements. If she was randomized to the walnut group, the subject was given thirty 1-oz packages of walnuts and was asked to consume 2 packages (2 oz) of walnuts daily and to return remaining packages for counting. If needed, due to extended time for the clinical workup, the subject was given additional packages of walnuts to allow for continued consumption of 2 oz of walnuts per day until surgery (about 2 to 3 weeks). Control group subjects were asked to not intentionally consume walnuts. At the conclusion of the study, each subject was asked to identify whether any changes were made to the usual diet especially in the areas of fruits, vegetables, nuts, or supplement consumption and fats used in cooking and whether walnuts were consumed (walnut group) or not (control group).
At the time that ultrasound-guided core needle biopsies of the breast mass were obtained for diagnosis, 1 or 2 extra cores were taken for research use. In the procedure room, immediately upon removal, the biopsies for research were placed in Qiagen, Germantown, MD, USA All-Protect tissue reagent (Qiagen.com) to preserve RNA, DNA, and protein for up to 7 days at room temperature. Biopsies were delivered to the research laboratory for initial processing within 2 hours.
If the pathology report indicated that the lump was not cancer, no further tissue was collected and the subject was no longer part of the study. If the biopsied tissue was breast cancer and surgery was scheduled without intervening radiation or chemotherapy, another specimen of tumor tissue and blood was collected at surgery. A small section of macroscopically viable tumor, away from the clean margin, was excised then immediately placed in Qiagen All-Protect tissue reagent, as before. Any patient who, according to the clinical care plan, was to receive either chemotherapy or radiation prior to surgery was no longer part of the study so as to not confound analyses.
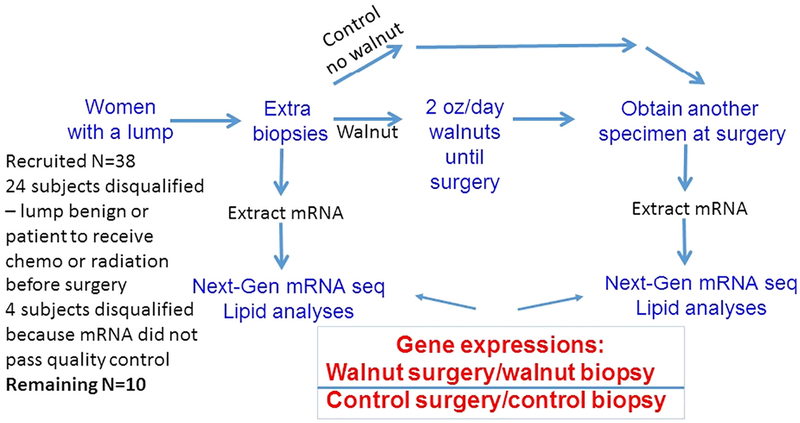
A total of 38 subjects consented to the trial. Of these, 24 were removed because the lump was benign or the cancer was to receive chemotherapy or radiation prior to surgery. The remaining 14 subjects completed the trial. Of these 14, data from four subjects had to be deleted because RNA from at least 1 sample of the pair of specimens did not pass quality control tests. Thus, as shown in Fig. 1, 5 subjects remained in the group that consumed walnuts and 5 subjects remained in the control group.
Fig. 1 –
Experimental design: graphical representation of the experimental design showing the recruitment of women, randomizing to experimental and control groups, reasons for dropout, and the laboratory processing of tumor specimens. A total of 38 subjects consented to the trial. After dropouts, 5 subjects remained in the group that consumed walnuts and 5 subjects remained in the control group.
2.3. Laboratory protocols
Total RNA was extracted using RNeasy, Lipid Tissue kit (Qiagen.com). This microkit is suitable for less than 5 mg of tissue and for extracting up to 45 μg of total mRNA. mRNA was checked for quantity and sent to the Marshall University Genomics Core Facility for further processing. The Genomics Core is a full-service facility and provided RNA quality assessment, RNA sequencing (RNA-Seq) analysis on each specimen, and DESeq2 expression profiling analyses of the data.
2.4. Fatty acid identification by gas chromatography
Gas chromatography was used to determine changes in the lipid composition of red blood cells (RBCs) membranes between the initial biopsy and surgery. RBCs and plasma were separated. About 50 μL of packed RBCs were homogenized in 0.1% BHT buffer to prevent fatty acid oxidation. Lipids were extracted with chloroform/methanol, and the fatty acids were methylated followed by separation and identification using gas chromatography, as previously described [21]. Gas chromatography was done using a Perkin-Elmer Clarus 500 Gas Chromatograph (Shelton, CT, USA) under the following conditions: initial temperature of 150°C, ramp 1 at 175°C for 15 minutes, ramp 2 at 225°C for 50 minutes, ramp 3 at 250°C for 10 minutes, helium carrier gas flow rate of 1.60 mL/min. A GLC Reference Standard Custom Preparation was used for peak identification of 10 fatty acids of interest: sterate (18:0), oleate (18:1n-9), linoleate(18:2n-6), gamma linolenate (18:3n-6), alpha linolenate (18:3n-3), homogamma linolenate (20:3n-6), arachidonate (20:4n-6), eicosapentanoate (20:5n-3), docosapentaenoate (22:5n-3), and docasahexanoate (22:6n-3). The fatty acid methyl esters were reported as the percent of the total methylated fatty acids (area under the curve) of the 10 identified fatty acids.
2.5. RNA sequencing: next-generation sequencing
RNA sample quality was assessed on RNA Pico chips in using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). RNA samples had RNA Integrity Numbers ranging from 2.6 to 9.4. One microgram of total RNA was used to construct RNASeq libraries using a TruSeq stranded total RNA library prep kit with RiboZero(H\M\R) ribosomal RNA reduction (Illumina Inc., San Diego, CA, USA) according to the kit’s instructions. RNA fragmentation times were modified based on RNA samples’ RNA Integrity Number value to generate inserts of equal size across all libraries.
Twenty RNA-Seq libraries were clustered on an Illumina cBot and sequenced on a HiSeq 1500 platform, in a 2 × 50 base paired end design yielding a minimum of 50 million reads per sample. Five matched pairs of samples (initial biopsy and subsequent surgery) were collected from each of the walnut-consuming and control groups.
Reads were trimmed using Trimmomatic v 0.36 [24] to remove low-quality base calls and adapter sequences, and then aligned to the human reference genome GRCh38 using HISAT v2.1.0 [25]. Resulting bam files were sorted and indexed with SamTools v1.3.1 [26], and PCR and optical duplicate reads marked using Picard tools v2.6.0. The numbers of reads mapping to each gene for each sample were counted using the R/Bioconductor package GenomicAlignments, v1.12.2 [27] and the Ensembl gene database for GRCh38, build 84 [28]. Differential gene expression was computed using DESeq2 version 1.10.0 [29], with a statistical model comparing the ratio of expression between surgery and biopsy specimens for the walnut-consuming group to the ratio of expression between surgery and biopsy specimens for the control group, as described in “statistical analyses” below.
2.6. Statistical analyses
Differences between groups (walnut or control) in fractions of individual fatty acids as determined by gas chromatography or in clinical parameters were determined by t test with a significance level of P ≤ .05.
It was expected that there would be large interpatient heterogeneity; thus, the baseline mRNA expression of individual genes would be highly variable between patients. The analyses of biopsy and surgical specimens allowed each patient to serve as her own control. Gene expressions for the sample collected at initial biopsy and the sample collected at surgery were determined, and the ratios of these expressions were calculated for each patient. Then, the means of the ratio of expressions were compared between the walnut-consuming group and the control group. The comparison was performed using DESeq2, which models the read count per gene using a negative binomial distribution and moderates the estimated expression changes to account for the dependence on overall read count [29]. Each patient was assigned a unique ID within their treatment (walnut or control) group, and the statistical model Eq. (1):
| (1) |
was passed to DESeq2, with extraction taking values “biopsy” or “surgery” and treatment taking values “walnut” or “control.” Genes that were significant for the extraction: treatment interaction parameter at a Benjamini-Hochberg (B-H) controlled false discovery rate of 10% were considered to be differentially expressed. The corresponding moderated fold change computed by DESeq2 is an estimate of this parameter and can thus be considered to be an estimate of the quantities:
| (2) |
In Eq. (2), ‘g’ represents the expression level of gene ‘g’, the subscripts ‘w’ and ‘c’ represent samples in the walnut and control groups, respectively, and the subscripts ‘s’ and ‘b’ represent samples from surgery and biopsy, respectively. Thus, the ratio of expression of gene ‘g’ from surgical specimens verses biopsy specimens of walnut patients was divided by the ratio of expression of gene ‘g’ from surgical specimens verses biopsy specimens of control patients. These analyses determined whether, across the group, there were significant and consistent changes in the mRNA expression of specific genes due to walnut consumption and provide the input for subsequent Ingenuity Pathway Analyses (IPAs).
2.7. Ingenuity Pathway Analyses
The complex data resulting from RNA seq expression profiling requires complex analyses. Data were further analyzed by use of IPA [30]. Final downstream phenotypic effects are due to the balance of positive and negative influences on expression of genes in a pathway. The goal of the IPA Downstream Effects Analysis is to identify genes and the resulting functions that are expected to increase or decrease given the observed gene expression changes in the experimental dataset [30]. Downstream Effects Analysis is based on expected causal effects between genes and functions; the expected causal effects are derived from the literature compiled in the Ingenuity Knowledge Base [30]. The analysis examines genes in the dataset that are known to affect functions, compares the genes’ direction of change to expectations derived from the literature, and then issues a prediction for each function based on the direction of changes in the dataset [30]. IPA uses a z-score algorithm to make predictions which is designed to reduce the chance that random data will generate significant predictions [30]. A publication further describing Downstream Effects Analyses can be found at [31].
The P values for networks were calculated using a Fisher exact test with B-H multiple testing corrections. The networks were generated through the use of IPA [30].
3. Results
3.1. Subject profiles
Data presented in Tables 1 and 2 show that there were no significant differences between the walnut and control groups in mean ages, initial body weight, body weight at surgery, tumor size, or menopausal status. All subjects were post-menopausal. No subjects had received either chemo- or radiation therapy for their tumors prior to entry into the study. The molecular characteristics of each group were also similar. There were 1 estrogen receptor (ER)–negative subject in the walnut group and 1 in the control group. Progesterone receptor (PR)+ and human epidermal growth factor receptor-2 (HER2)/Neu+ ratios were similar in each group.
Table 1 –
Patient tumor type, molecular characteristics, diameter at surgery, and pathological staging
| ID | Groupa | Age | Body weight at biopsy | Body weight at surgery | Tumor type | Molecular characteristics | Tumor diameter at surgery | Menopausal status | Pathological stagingb | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | W | 46 | 87.7 | 86.7 | IDC, grade2 spindle cell differentiation& focal squamous differentiation |
ER | + | 19 | Postmenopausal | pT1cpN0 |
| PR | + | |||||||||
| Her2neu | −,1+ | |||||||||
| 2 | W | 64 | 98.7 | 102.7 | IDC, grade 3 | ER | + | 25 | Postmenopausal | pT2pN0 |
| PR | + | |||||||||
| Her2neu | −, 1+ | |||||||||
| 3 | W | 56 | 110.4 | 108.9 | IDC, grade 3 | ER | + | 30 | Postmenopausal | pT2pN1a |
| PR | + | |||||||||
| Her2neu | −, 1+ | |||||||||
| 4 | W | 56 | 113.4 | 113.4 | IDC | ER | + | 29 | Postmenopausal | T2N0M0 |
| PR | − | |||||||||
| Her2neu | + | |||||||||
| 5 | W | 52 | 88 | 87 | Metaplastic carcinoma | ER | − | 25 | Postmenopausal | T2N0M0 |
| PR | − | |||||||||
| Her2neu | + | |||||||||
| 6 | C | 51 | 113.0 | z | IDC, grade3 | ER | + | 59 | Postmenopausal | pT3pN0 |
| PR | + | |||||||||
| Her2neu | −, 1+ | |||||||||
| 7 | C | 45 | 44.4 | 44.5 | ILC, grade1 | ER | + | 22 | Premenopausal | pT2pN0 |
| PR | + | |||||||||
| Her2neu | −, 0 | |||||||||
| 8 | C | 51 | 86.3 | 85.8 | IDC, moderately differentiated | ER | + | 23 | Postmenopausal | pT2pN1a |
| PR | + | |||||||||
| Her2neu | 1+ | |||||||||
| 9 | C | 67 | 105.6 | 103.1 | IMC, grade 3 | ER | − | 32 | Postmenopausal | pT2pN0 |
| PR | − | |||||||||
| Her2neu | −, 1+ | |||||||||
| 10 | C | 56 | 92.9 | 94.8 | IDC | ER | + | 25 | Postmenopausal | pT2pN1M0 |
| PR | + | |||||||||
| Her2neu | − | |||||||||
The age (years), initial body weight (kg), body weight at surgery (kg), tumor and molecular characteristics, tumor size (mm), menopausal status, and pathological staging of each subject in the walnut-consuming and control groups. w = Walnut consuming group; c = Control group. Pathological staging: tumor/node/metastasis scale. pT (size) pT1 < 20 mm, pT2 > 20 mm, pT3 > 50 mm; pN0 or pN1: no lymph node metastasis or 1–2 lymph nodes metastasis; pM0: no distant metastasis.
Table 2 –
Patient age, body weight, and tumor aspects
| Walnut group | Control group | P value | |
|---|---|---|---|
| Age (y) | 54.8 ± 6.6 | 54.0 ± 8.2 | .87 |
| Initial body weight (kg) | 99.6 ± 12.1 | 88.4 ± 26.7 | .41 |
| Body weight at surgery (kg) | 99.7 ± 12.4 | 88.5 ± 26.7 | .41 |
| Body weight change (surgery − initial, kg) | 0.1 ± 2.2 | +0.06 ± 1.7 | .97 |
| Tumor diameter (mm) | 25.6 ± 0.4 | 32.2 ± 1.5 | .38 |
| Postmenopausal | 5 of 5 | 5 of 5 | |
| Tumor molecular characteristics | |||
| ER+, PR+, HER2+ | 3 of 5 | 2 of 5 | |
| ER+. PR+, HER− | 0 of 5 | 2 of 5 | |
| ER+, PR−, HER2+ | 1 of 5 | 0 of 5 | |
| ER−, PR−. HER2+ | 1 of 5 | 1 of 5 |
Values are means ± SD. P value by t test walnut vs control group. There were no significant differences between groups for the tested characteristics.
3.2. Compliance with walnut consumption and diet
Candidate subjects who did not like or who were allergic to tree nuts, including walnuts, were screened out prior to study consent. Of subjects who consented to the trial and received walnuts, none reported difficulty with consuming 2 oz of walnuts per day. Bag counts and patient reports indicated that 2 oz of walnuts were consumed daily. No patient reported changes to the usual diet composition during the time of walnut consumption, except for the walnuts. No subject in the control group reported intentional consumption of walnuts during the 2 weeks of the trial.
3.3. RBC fatty acid composition
Gas chromatography results (data not shown) revealed no significant differences in the lipid composition of RBC membranes due to walnut consumption for about 2 weeks. However, there was a not-significant trend toward increased total omega 3 lipids in the red blood cells of walnut-consuming subjects, as would be expected in subjects who consumed walnuts.
3.4. RNA Seq Expression Profiling
Differential (between biopsy and surgery of walnut and control groups) expression analysis of RNA-Seq results showed that the expression of 456 mapped genes was significantly and consistently altered by incorporation of walnuts in the diet. The list of all 456 mapped gene identifications, the expression log ratio, and the false discovery rate is included in the related Data in Brief article, Table 1 [32]. A list of 281 unmapped identifications which were also changed is included in the related Data in Brief article, Table 2 [32]. Files containing the raw reads, along with a shell script describing the complete data analysis pipeline, were deposited to the Gene Expression Omnibus at the National Center for Biotechnology Information and can be obtained via accession number GSE111073 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111073).
3.5. Ingenuity Pathway Analyses
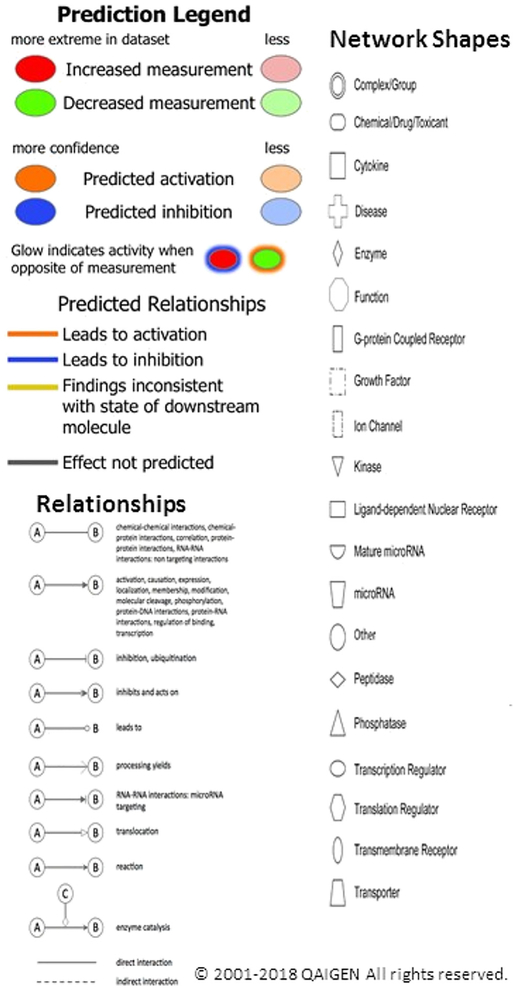
IPAs were critical for understanding the meaning of the 456 gene expression changes in this experiment. Through IPAs, 25 gene networks were identified that were influenced by walnut consumption. The Pathway Legend for Figs. 3, 4, and 5 is shown in Fig. 2. Note that the reds and greens, which represent the measured change in mRNA expression, tend to be at the outside of each network diagram. The blues and oranges, which represent the predicted changes in activity, are downstream of the measured changes and tend to be toward the center of the network. The 3 most significant networks, shown in Figs. 3, 4, and 5, with network scores greater than 30, will be discussed below. Genes of all 25 networks are included in the related Data in Brief article Table 3, [33]. The network score is based on the hypergeometric distribution and is calculated with the right-tailed Fisher exact test with B-H multiple testing corrections. For example, for a network with a P value of 1 × 10−30, the network’s score = [−log(Fisher exact test result)] =30. Thus, a score of 30 can be interpreted as meaning that if there were no associations between walnut consumption and the gene expression changes seen in the network, an overlap between the network and the differentially expressed gene set would only occur 1 in 1030 times in similar experiments. The phenotypic changes expected due to significant changes in expression of genes at the nodes are described in the discussion.
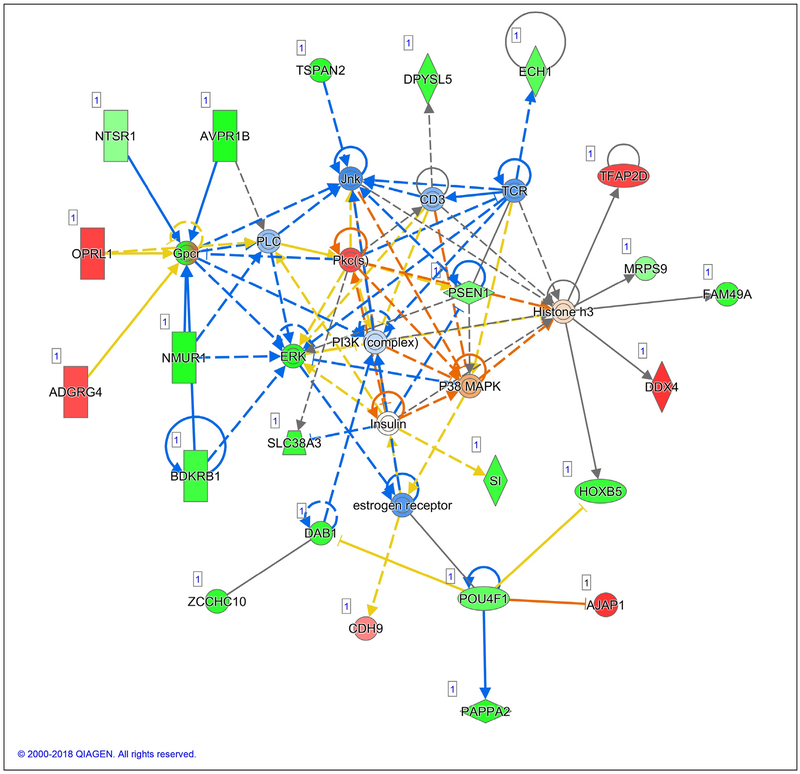
Fig. 3 –
Network 1 with a score of 41 shows genes involved in reproductive system development and function, carbohydrate metabolism, and lipid metabolism. Involved in breast cancer development are the expected downregulation of the estrogen receptor; decreased ERK; and increased P38/MAPK [35,45], JNK [45,57,58], and the PI3K complex.
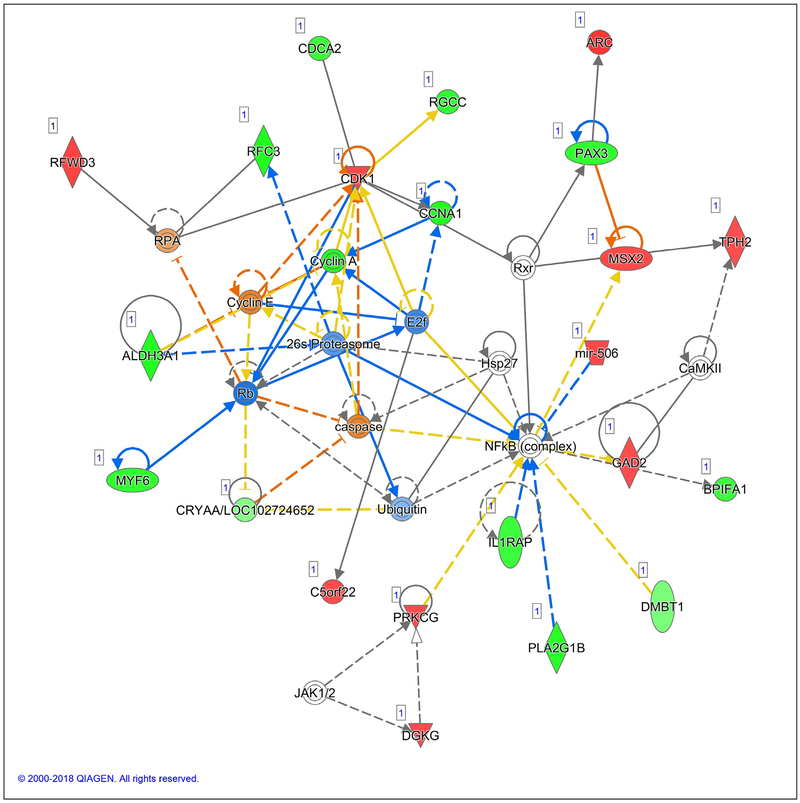
Fig. 4 –
Network 2 with a score of 39 includes genes important in cell-to-cell signaling and interaction, cellular growth and proliferation, and cellular movement. Involved in breast cancinogenesis are caspase, cyclin A and E, and E2f.
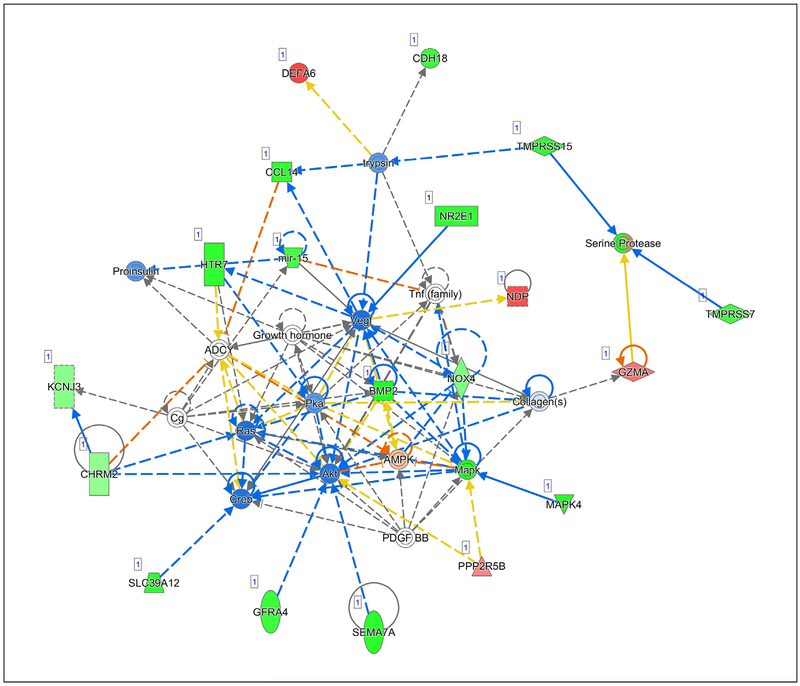
Fig. 5 –
Network 3 with a score of 32 contains genes important in embryonic development, nervous system development and function, and organ development. Involved in breast cancinogenesis are Vegf, Akt, Ras, Pka, and Creb.
Fig. 2 –
Legend for IPA analyses (from the IPA Web site: www.ingenuity.com): Molecules are coded by measured expression (reds for increased expression and greens for decreased expression) or predicted activation (oranges for predicted gene activation and blues for predicted gene inhibition). The intensity of green and red molecule colors indicates the degree of down or upregulation, respectively. The intensity of the orange or blue colors indicates the strength of confidence in activation or inhibition, respectively. The relationships between molecules are indicated by the solid or broken lines. The shapes indicate the type of the molecule.
4. Discussion
RNA-Seq Expression Profiling analyses from a pilot clinical trial in which women with breast cancer consumed 2 oz of walnuts per day for 2 to 3 weeks between the diagnostic biopsy and subsequent surgery showed that gene expression in the tumor was modified in ways expected to slow proliferation, reduce inflammation, reduce metastasis, and increase cancer cell death. It seems likely that if gene expression was modified in the tumor due to consumption of walnuts, then gene expression of any metastatic cells should also be modified, thus decreasing risk for cancer recurrence.
Gene expression modifications resulting from walnut consumption that influence breast cancer suppression can be identified in the 3 illustrated gene networks with the 3 highest network scores. Some of these expression changes will be highlighted below. Additional modified pathways can be found in Data in Brief Table 3 [33].
In Network 1, suppression of the ER stands out as an action that is important for breast cancer. Excess activity of ER driven by binding to estrogen is an important driver of breast cancer initiation and progression [33] and promotes proliferation of breast epithelial cells [33]. Importantly, basal ER activity, in the absence of estrogen binding, promotes proliferation and must be blocked to prevent cell cycle progression [34]. Modulation of ER activity or degradation of ER is recognized as important therapeutic strategies [35].
It has been reported that, in breast cancer, a change in the ratio of extracellular signal regulated–kinase (ERK) to P38 (a mitogen-activated protein kinase) determines whether these changes will promote tumor growth or tumor arrest. A higher p38/ERK ratio induces tumor growth arrest compared to the lower ratio [36]. In Network 1, p38 activity was predicted to increase and ERK activity to decrease; thus, the higher p38/ERK ratio should lead to arrest of growth of breast cancer cells.
Network 1 and Network 3 both indicate that phosphatidylinositol 3-kinase/serine/threonine kinase 1 (PI3K/Akt) is significantly suppressed by walnut consumption. PI3K/Akt has been reported to promote breast cancer cell survival and resistance to chemotherapy [37]. Because PI3K and Akt were suppressed in the cancers of patients who consumed walnuts, it would be expected that apoptosis in residual tumor cells might increase and that these patients might also respond better to chemotherapy following surgery.
Bradykinin, which can be generated by inflammation, is active in migration and cell proliferation and activates ERK [38]. Network 1 shows that the measured expression of bradykinin receptor B1 (BDKRB1) mRNA was decreased by walnut. This decrease in BDKRB1 may be one of the factors leading to the decrease in ERK expression. It has been suggested that antagonism of BDKRB1 may be a useful treatment for breast cancer by disrupting the BDKRB1/epidermal growth factor receptor cross talk that maintains tumor growth [38].
The Wnt/planar cell polarity pathway is a highly conserved proliferative signaling cascade. It is upregulated in breast cancer patients with shorter survival and can be inactivated in patient-derived xenograft cells by suppression of c-Jun N-terminal kinase (JNK). Interruption of this pathway by suppression of JNK has been proposed as a therapeutic strategy [39]. Walnut consumption effectively suppressed JNK mRNA.
Network 2 shows a predicted significant increase in caspase, a marker for active apoptosis in cancer cells. Caspase has been shown to be induced in breast cancer cells by beta-sitosterol [17], the highest phytosterol in walnuts at 87 mg/100 g [40]. An increase in cell death would slow tumor growth, and the increase in factors required for cell death might allow chemotherapy to be more effective.
Network 2 also indicates that cyclin A is decreased, which would slow proliferation. Cyclin E is expected to be activated. Cyclin E does promote proliferation, which would not seem to be beneficial against breast cancer. However, increased activity of cyclin E also suppresses invasion [41] by blocking a transcription factor (E2F). E2F is required for induction of hypoxia inducible factor 1A (an initiator for angiogenesis) by cyclin E [42]. Because in Network 2, E2F is predicted to be downregulated by high cyclin E, hypoxia inducible factor 1A would not be available to induce angiogenesis and invasion. Tumors must induce angiogenesis to grow and suppression of invasion is important because it is usually the cancer metastasis that kills, not a primary tumor. Sengupta et al further report that dysregulation of cyclin E induces cellular senescence [42], thereby slowing proliferation.
In Network 3, Vascular ENDOTHELIAL growth factor (Vegf), Akt, rat sarcoma virus (Ras), protein kinase a (Pka), and cAMP responsive element binding protein (Creb) are especially important for cancer initiation and progression. Vegf is critical for the formation of blood vessels in a growing tumor and for cell migration and metastasis of breast cancer [43–45]. Increased AKT increases cell motility and expression of matrix metalloprotease 9 to increase metastasis [46] and development of epithelial mesenchymal transition [47], and promotes resistance to chemotherapy [37]. Hyperactivation of Ras is found in only about 5% of breast cancers, yet there is much experimental evidence that activated Ras can promote breast cancer growth and development [48]. Activation of Pka (protein kinase a) is required for the growth factor stimulation of migration of cancer cells [49] and may lead to multiple drug resistance via the PI3K/AKT pathway [50]. Downregulation of Creb and Pka has been associated with increased apoptosis in breast cancer cell lines [51]. Consumption of walnut was predicted to significantly suppress Vegf, AKT, Ras, Pka, and Creb for multiple benefits against breast cancer.
Multiple in vitro studies have shown that walnuts may have benefit against cancer and have described potential mechanisms, including suppression of proliferation, induction of apoptosis, and decreasing oxidative damage [4–6]. Walnuts are rich in the bioactive molecules that have been shown to be active against cancer, as reviewed in [52]. Our animal studies demonstrated that the human equivalent of 2 oz of walnut in the diet (1) slowed growth of implanted human breast cancers [15] and (2) prevented cancer development in a transgenic model [21]. The identified mechanisms in [15] included slowed proliferation, increased apoptosis, and increased antioxidative capacity, in agreement with the in vitro studies. In study 2 [21], the list of genes that were significantly altered by walnut consumption included genes related to proliferation, inflammation, metastasis, and cell death. Another study, from a different laboratory, shows that suppressing angiogenesis was a mechanism by which walnut inhibited colon cancer [53]. These mechanisms are expected to be altered in human breast cancer as indicated by the mRNA expression changes seen in this pilot clinical trial. Additionally, the IPAs showed the alteration of several genes by walnut consumption that are expected to decrease the development of chemoresistance in breast cancer tumors. Thus, the results of multiple studies in multiple models indicate that consumption of walnut should provide benefit against human breast cancer, strongly supporting the results of the current study. The IPAs of the current study indicate alteration of expression of multiple genes by walnut consumption, thus providing multiple and overlapping targets for the effects of walnut against cancer.
There is concern that the high fat content of walnuts would tend to increase the body weight of persons who consumed them as part of the routine diet. In this study, persons in the walnut-consuming group did not gain weight in the short term. That walnuts do not cause weight gain is supported by studies in diabetic patients [54] and in over-weight men [55]. It was shown that in a longer term, the walnuts displaced other foods in the diet [55]. The current study did not show significant changes in RBC lipids due to walnut in the diet for about 2 weeks. Other studies including our mouse studies [15,21] and longer-term (up to 6 months) human studies [56] have shown a change in RBC lipids due to walnut consumption.
A recent clinical trial indicates that walnuts, indeed, may modulate breast cancer growth or development [57]. Secondary analyses of the PREDIMED study showed that the hazard ratio for breast cancer for the Mediterranean diet with nuts group vs the control group (Mediterranean diet only) was a not quite significant 0.53 (95% confidence interval, 0.23–1.26) decrease [57]. Reasons for the “not quite significant” results in the PREDIMED study could be because (1) there were very few incident cases of breast cancer in the PREDIMED study and (2) the background diet was a Mediterranean diet, which in itself was likely to suppress cancer development [58]. The current study had a “Western diet in West Virginia” as a background diet; thus, benefit and gene expression changes due to the addition of walnuts to the diet were likely to be more pronounced than if the background diet had been a Mediterranean diet.
A recent meta-analysis [59] concluded that nut consumption, including peanuts, was associated with reduced risk of cancer and reduced all-cause mortality, in agreement with the results of this study. Many of the genes seen to be modified in the current study are promotional to all types of cancers, not just breast cancer, indicating that there may be benefit against many cancer types.
Strengths of this study include mechanistic agreement with the previous cell culture and animal studies on the effects of walnut against cancer. The in vitro studies have identified specific molecular mechanisms for reduced cancer cell growth and increased cancer cell death for various components of the walnut. The in vivo, animal studies have provided phenotypic mechanisms demonstrating that consumption of walnut does slow cancer cell proliferation, increase cancer cell death, and decrease oxidative stress in animals. Furthermore, the animal studies provide measurable endpoints of decreased tumor size and numbers due to walnut consumption that could not be determined in humans. Another strength is that the human (and animal) subjects were able to consume the walnut without detrimental side effects.
Limitations of this study include the small numbers of subjects in the clinical trial. More human subjects would be desirable to increase confidence in the results. However, due to the power of RNA-seq, even with the small number of individuals, large significant differences in expression of multiple genes were shown. All the subjects were postmenopausal; it is not known if the results would be the same for premenopausal women. No Western blots could be done due to very small sample sizes. Assessment of differences in protein would increase confidence in the meaning of the gene expression changes. A person who is allergic to walnut would have detrimental side effects and could not benefit from walnut consumption. Consumption of a different background diet might result is a different degree of benefit because the usual diet itself could have positive or negative effects on breast cancer.
A definitive clinical trial should be conducted to verify that these gene expression changes actually translate to reduced tumor recurrence or slowed tumor growth. Due to good agreement between the animal studies (with identified mechanisms and a tumor endpoint showing suppression of tumor growth and/or development), the cell culture studies for mechanistic data, the verified changes in RNA expression, and the mechanisms identified in this pilot clinical trial, the results of this study support our hypothesis that walnut consumption would alter gene expression in breast cancers of women in a direction that would be expected to decrease breast cancer growth and survival. Because many of the beneficial components of walnuts are also found in fruits and vegetables, incorporation of walnuts in a healthy diet should provide additional and likely synergistic benefits by increasing the total consumption of antioxidants, phytosterols, dietary fibers, and polyunsaturated fats to reduce morbidity and mortality from breast cancer.
Acknowledgment
With profound appreciation to the women who agreed to participate in this clinical trial at a very stressful time of life. This trial could not have been conducted without the professional assistance of the Clinical Research Nurses at the Edwards Cancer Center and at the St Mary’s Cancer Center who coordinated the patient identification, consenting, and sample collection for the study. RNA sequencing was performed at the Marshall University Genomics Core Facility, and data analysis was performed in part by the WV-INBRE Bioinformatics Core Facility. These facilities, and access to Ingenuity Pathway Analysis, are supported by the West Virginia IDeA Networks of Biomedical Research Excellence (WV-INBRE) NIH P20 GM103434. Primary funding for this work was from the California Walnut Commission to WEH (no grant number). This work was supported in part by NIH/NIGMS P20GM103434 which funds the IDeA WV-INBRE program and partially supports the Marshall University Genomics Core Facility (DAP and JD). WEH, DAP, and JD also receive support from the Marshall University COBRE, NIH/NIGMS P20GM121299. Neither the California Walnut Commission nor the NIH had input in the study design; in the collection, analysis, or interpretation of data; in the writing of the report; or in the decision to publish these research results.
Abbreviations:
- Akt
serine/threonine kinase 1
- B-H
Benjamini-Hochberg
- BDKRB1
bradykinin receptor
- Creb
cAMP responsive element binding protein
- E2F
a transcription factor
- ER
estrogen receptor
- ERK/P38
extracellular signal–regulated kinase/P38
- HER2
human epidermal growth factor receptor-2
- IPA
Ingenuity Pathway Analysis
- JNK
c-Jun N-terminal kinase
- P38
a member of the mitogen activated protein kinase family
- PI3K
phosphatidylinositol 3-kinase
- Pka
protein kinase a
- PR
progesterone receptor
- Ras
rat sarcoma virus
- RBC
red blood cell
- RNA-Seq
RNA sequencing
- Vegf
Vascular endothelial growth factor
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- [1].Abdulla M, Gruber P. Role of diet modification in cancer prevention. Biofactors 2000;12(1–4):45–51. [DOI] [PubMed] [Google Scholar]
- [2].Harris HR, Bergkvist L, Wolk A. Adherence to the World Cancer Research Fund/American Institute for Cancer Research recommendations and breast cancer risk. Int J Cancer 2016;138(11):2657–64. [DOI] [PubMed] [Google Scholar]
- [3].Jacobs DR Jr, Gross MD, Tapsell LC. Food synergy: an operational concept for understanding nutrition. Am J Clin Nutr 2009;89(5):1543S–8S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Vanden Heuvel JP, Belda BJ, Hannon DB, Kris-Etherton PM, Grieger JA, Zhang J, et al. Mechanistic examination of walnuts in prevention of breast cancer. Nutr Cancer 2012;64 (7):1078–86. [DOI] [PubMed] [Google Scholar]
- [5].Chen HS, Bai MH, Zhang T, Li GD, Liu M. Ellagic acid induces cell cycle arrest and apoptosis through TGF-beta/Smad3 signaling pathway in human breast cancer MCF-7 cells. Int J Oncol 2015;46(4):1730–8. [DOI] [PubMed] [Google Scholar]
- [6].Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic Biol Med 2010;49(11):1603–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ahmad H, Khan I, Wahid A. Antiglycation and antioxidation properties of Juglans regia and Calendula officinalis: possible role in reducing diabetic complications and slowing down aging. J Tradit Chin Med 2012;32(3):411–4. [DOI] [PubMed] [Google Scholar]
- [8].Anderson KC, Teuber SS. Ellagic acid and polyphenolics present in walnut kernels inhibit in vitro human peripheral blood mononuclear cell proliferation and alter cytokine production. Ann N Y Acad Sci 2010;1190:86–96. [DOI] [PubMed] [Google Scholar]
- [9].Anderson KJ, Teuber SS, Gobeille A, Cremin P, Waterhouse AL, Steinberg FM. Walnut polyphenolics inhibit in vitro human plasma and LDL oxidation. J Nutr 2001;131(11): 2837–42. [DOI] [PubMed] [Google Scholar]
- [10].Bati B, Celik I, Dogan A. Determination of hepatoprotective and antioxidant role of walnuts against ethanol-induced oxidative stress in rats. Cell Biochem Biophys 2015;71(2): 1191–8. [DOI] [PubMed] [Google Scholar]
- [11].Blomhoff R, Carlsen MH, Andersen LF, Jacobs DR Jr. Health benefits of nuts: potential role of antioxidants. Br J Nutr 2006; 96:S52–60. [DOI] [PubMed] [Google Scholar]
- [12].Carvalho M, Ferreira PJ, Mendes VS, Silva R, Pereira JA, Jeronimo C, et al. Human cancer cell antiproliferative and antioxidant activities of Juglans regia L. Food Chem Toxicol 2010;48(1):441–7. [DOI] [PubMed] [Google Scholar]
- [13].Chen N, Yang H, Sun Y, Niu J, Liu S. Purification and identification of antioxidant peptides from walnut (Juglans regia L.) protein hydrolysates. Peptides 2012;38(2):344–9. [DOI] [PubMed] [Google Scholar]
- [14].Derosa G, Maffioli P, Sahebkar A. Ellagic acid and its role in chronic diseases. Adv Exp Med Biol 2016;928:473–9. [DOI] [PubMed] [Google Scholar]
- [15].Hardman WE, Ion G. Suppression of implanted MDA-MB 231 human breast cancer growth in nude mice by dietary walnut. Nutr Cancer 2008;60(5):666–74. [DOI] [PubMed] [Google Scholar]
- [16].Hardman WE. Dietary canola oil suppressed growth of implanted MDA-MB 231 human breast tumors in nude mice. Nutr Cancer 2007;57(2):177–83. [DOI] [PubMed] [Google Scholar]
- [17].Awad AB, Roy R, Fink CS. Beta-sitosterol, a plant sterol, induces apoptosis and activates key caspases in MDA-MB-231 human breast cancer cells. Oncol Rep 2003;10(2):497–500. [PubMed] [Google Scholar]
- [18].Kline K, Yu W, Sanders BG. Vitamin E: mechanisms of action as tumor cell growth inhibitors. J Nutr 2001;131:161S–3S. [DOI] [PubMed] [Google Scholar]
- [19].Blask DE, Sauer LA, Dauchy RT, Holowachuk EW, Ruhoff MS, Kopff HS. Melatonin inhibition of cancer growth in vivo involves suppression of tumor fatty acid metabolism via melatonin receptor-mediated signal transduction events. Cancer Res 1999;59:4693–701. [PubMed] [Google Scholar]
- [20].Green JE, Shibata MA, Yoshidome K, Liu ML, Jorcyk C, Anver MR, et al. The C3(1)/SV40 T-antigen transgenic mouse model of mammary cancer: ductal epithelial cell targeting with multistage progression to carcinoma. Oncogene 2000;19(8): 1020–7. [DOI] [PubMed] [Google Scholar]
- [21].Hardman WE, Ion G, Akinsete JA, Witte TR. Dietary walnut suppressed mammary gland tumorigenesis in the C(3)1 TAg mouse. Nutr Cancer 2011;63(6):960–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ion G, Akinsete JA, Hardman WE. Maternal consumption of canola oil suppressed mammary gland tumorigenesis in C3 (1) TAg mice offspring. BMC Cancer 2010;10:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cortes B, Nunez I, Cofan M, Gilabert R, Perez-Heras A, Casals E, et al. Acute effects of high-fat meals enriched with walnuts or olive oil on postprandial endothelial function. J Am Coll Cardiol 2006;48(8):1666–71. [DOI] [PubMed] [Google Scholar]
- [24].Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30 (15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12(4): 357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol 2013;9(8):e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Aken BL, Achuthan P, Akanni W, Amode MR, Bernsdorff F, Bhai J, et al. Ensembl 2017. Nucleic Acids Res 2017;45(D1):D635–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550–8. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Qiagen. Assessing and using downstream effects analyses. http://qiagen.force.com/KnowledgeBase/KnowledgeIPAPage?id=kA1D0000000PIoeKAG; 2017, Accessed date: 20 February 2018.
- [31].Kramer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30(4):523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hardman WE, Primerano DA, Legenza MA, Morgan J, Fan J, Denvir J. Gene expression in breast cancers before and after consumption of walnut by women. Data in Brief; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Velloso FJ, Bianco AF, Farias JO, Torres NE, Ferruzo PY, Anschau V, et al. The crossroads of breast cancer progression: insights into the modulation of major signaling pathways. Onco Targets Ther 2017;10:5491–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gaben AM, Sabbah M, Redeuilh G, Bedin M, Mester J. Ligand-free estrogen receptor activity complements IGF1R to induce the proliferation of the MCF-7 breast cancer cells. BMC Cancer 2012;12:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Patel HK, Bihani T. Selective estrogen receptor modulators (SERMs) and selective estrogen receptor degraders (SERDs) in cancer. Pharmacol Ther 2017;17:10. [DOI] [PubMed] [Google Scholar]
- [36].Aguirre-Ghiso JA, Estrada Y, Liu D, Ossowski L. ERK(MAPK) activity as a determinant of tumor growth and dormancy; regulation by p38(SAPK). Cancer Res 2003;63(7):1684–95. [PubMed] [Google Scholar]
- [37].Clark AS, West K, Streicher S, Dennis PA. Constitutive and inducible Akt activity promotes resistance to chemotherapy, trastuzumab, or tamoxifen in breast cancer cells. Mol Cancer Ther 2002;1(9):707–17. [PubMed] [Google Scholar]
- [38].Molina L, Matus CE, Astroza A, Pavicic F, Tapia E, Toledo C, et al. Stimulation of the bradykinin B(1) receptor induces the proliferation of estrogen-sensitive breast cancer cells and activates the ERK1/2 signaling pathway. Breast Cancer Res Treat 2009;118(3):499–510. [DOI] [PubMed] [Google Scholar]
- [39].Puvirajesinghe TM, Bertucci F, Jain A, Scerbo P, Belotti E, Audebert S, et al. Identification of p62/SQSTM1 as a component of non-canonical Wnt VANGL2-JNK signaling in breast cancer. Nat Commun 2016;7:10318 10.1038/ncomms10318.:10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].United States Department of Agriculture Agricultural Research Service. National nutrient database for standard reference release 28. https://ndb.nal.usda.gov/ndb/foods; 2018, Accessed date: 9 January 2018.
- [41].Qian X, Hulit J, Suyama K, Eugenin EA, Belbin TJ, Loudig O, et al. p21CIP1 mediates reciprocal switching between proliferation and invasion during metastasis. Oncogene 2013;32 (18):2292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sengupta T, Abraham G, Xu Y, Clurman BE, Minella AC. Hypoxia-inducible factor 1 is activated by dysregulated cyclin E during mammary epithelial morphogenesis. Mol Cell Biol 2011;31(18):3885–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chen X, Zheng Z, Chen L, Zheng H. MAPK, NFkappaB, and VEGF signaling pathways regulate breast cancer liver metastasis. Oncotarget 2017;8(60):101452–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gampenrieder SP, Westphal T, Greil R. Antiangiogenic therapy in breast cancer. Memo 2017;10(4):194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Weddell JC, Chen S, Imoukhuede PI. VEGFR1 promotes cell migration and proliferation through PLCgamma and PI3K pathways. NPJ Syst Biol Appl 2017;4:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Byun HJ, Hong IK, Kim E, Jin YJ, Jeoung DI, Hahn JH, et al. A splice variant of CD99 increases motility and MMP-9 expression of human breast cancer cells through the AKT-, ERK-, and JNK-dependent AP-1 activation signaling pathways. J Biol Chem 2006;281(46):34833–47. [DOI] [PubMed] [Google Scholar]
- [47].Chou CC, Lee KH, Lai IL, Wang D, Mo X, Kulp SK, et al. AMPK reverses the mesenchymal phenotype of cancer cells by targeting the Akt-MDM2-Foxo3a signaling axis. Cancer Res 2014;74(17):4783–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Eckert LB, Repasky GA, Ulku AS, McFall A, Zhou H, Sartor CI, et al. Involvement of Ras activation in human breast cancer cell signaling, invasion, and anoikis. Cancer Res 2004;64(13):4585–92. [DOI] [PubMed] [Google Scholar]
- [49].O’Connor KL, Mercurio AM. Protein kinase A regulates Rac and is required for the growth factor-stimulated migration of carcinoma cells. J Biol Chem 2001;276(51):47895–900. [DOI] [PubMed] [Google Scholar]
- [50].Yu T, Yang G, Hou Y, Tang X, Wu C, Wu XA, et al. Cytoplasmic GPER translocation in cancer-associated fibroblasts mediates cAMP/PKA/CREB/glycolytic axis to confer tumor cells with multidrug resistance. Oncogene 2017;36(15):2131–45. [DOI] [PubMed] [Google Scholar]
- [51].Sheikholeslami A, Nabiuni M, Arefian E. Suppressing the molecular signaling pathways involved in inflammation and cancer in breast cancer cell lines MDA-MB-231 and MCF-7 by miR-590. Tumor Biol 2017;39(4). 10.1177/1010428317697570. [DOI] [PubMed] [Google Scholar]
- [52].Catanzaro E, Greco G, Potenza L, Calcabrini C, Fimognari C. Natural products to fight cancer: a focus on Juglans regia. Toxins (Basel) 2018;10(11) [toxins10110469]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nagel JM, Brinkoetter M, Magkos F, Liu X, Chamberland JP, Shah S, et al. Dietary walnuts inhibit colorectal cancer growth in mice by suppressing angiogenesis. Nutrition 2012;28(1):67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tapsell LC, Batterham MJ, Teuss G, Tan SY, Dalton S, Quick CJ, et al. Long-term effects of increased dietary polyunsaturated fat from walnuts on metabolic parameters in type II diabetes. Eur J Clin Nutr 2009;63(8):1008–15. [DOI] [PubMed] [Google Scholar]
- [55].Kranz S, Hill AM, Fleming JA, Hartman TJ, West SG, Kris-Etherton PM. Nutrient displacement associated with walnut supplementation in men. J Hum Nutr Diet 2014;27(S2):247–54. [DOI] [PubMed] [Google Scholar]
- [56].Tapsell LC, Gillen LJ, Patch CS, Batterham M, Owen A, Bare M, et al. Including walnuts in a low-fat/modified-fat diet improves HDL cholesterol-to-total cholesterol ratios in patients with type 2 diabetes. Diabetes Care 2004;27(12): 2777–83. [DOI] [PubMed] [Google Scholar]
- [57].Toledo E, Salas-Salvado J, Donat-Vargas C, Buil-Cosiales P, Estruch R, Ros E, et al. Mediterranean diet and invasive breast cancer risk among women at high cardiovascular risk in the PREDIMED trial: a randomized clinical trial. JAMA Intern Med 2015;175(11):1752–60. [DOI] [PubMed] [Google Scholar]
- [58].Schwingshackl L, Schwedhelm C, Galbete C, Hoffmann G. Adherence to Mediterranean diet and risk of cancer: an updated systematic review and meta-analysis. Nutrients 2017;9(10) [nu9101063]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Aune D, Keum N, Giovannucci E, Fadnes LT, Boffetta P, Greenwood DC, et al. Nut consumption and risk of cardiovascular disease, total cancer, all-cause and cause-specific mortality: a systematic review and dose–response meta-analysis of prospective studies. BMC Med 2016;14(1):207–0730. [DOI] [PMC free article] [PubMed] [Google Scholar]