Abstract
Summary
Due to the rapidly increasing scale and diversity of epigenomic data, modular and scalable analysis workflows are of wide interest. Here we present snakePipes, a workflow package for processing and downstream analysis of data from common epigenomic assays: ChIP-seq, RNA-seq, Bisulfite-seq, ATAC-seq, Hi-C and single-cell RNA-seq. snakePipes enables users to assemble variants of each workflow and to easily install and upgrade the underlying tools, via its simple command-line wrappers and yaml files.
Availability and implementation
snakePipes can be installed via conda: `conda install -c mpi-ie -c bioconda -c conda-forge snakePipes’. Source code (https://github.com/maxplanck-ie/snakepipes) and documentation (https://snakepipes.readthedocs.io/en/latest/) are available online.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The decreasing price of sequencing and increasing multiplexing ability has allowed researcher to easily produce large datasets. To understand genetic and epigenetic regulation, researchers routinely perform multiple assays, such as RNA-seq and Bisulfite-seq in the same project, necessitating scalable data processing workflows. Since exploratory studies demand more flexibility in data processing, and standards evolve rapidly, conventional rigid pipelines become quickly outdated. Computational frameworks, such as Galaxy (Goecks et al., 2010), Nextflow (Di Tommaso et al., 2017) and snakemake (Köster and Rahmann, 2012) address this issue to some extent by allowing users to create their own workflows, or adopt workflows from public repositories. However, these frameworks are still challenging for novice users, as they require training in their specific programing language or syntax and assembling workflows themselves. This leads to a conundrum, how can we offer the flexibility of assembling and upgrading analyses workflows to the novice users, while still keeping them scalable and reproducible?
We developed snakePipes to address this issue. snakePipes provides a set of best-practices workflows for processing, quality control and downstream analysis of data from the most common assays used in epigenomic studies: ChIP-seq, RNA-seq, whole-genome bisulfite-seq (WGBS), ATAC-seq, Hi-C and single-cell RNA-seq (Supplementary Fig. S1a; Supplementary Methods). However, unlike conventional pipelines, workflows in snakePipes are based on a repository of modular rules, such that multiple variations of each workflow can be assembled on-the-fly by changing the parameters on their command-line wrappers. This novel approach allows novice users to perform exploratory analysis in a reproducible way without manually assembling workflows.
2 Implementation
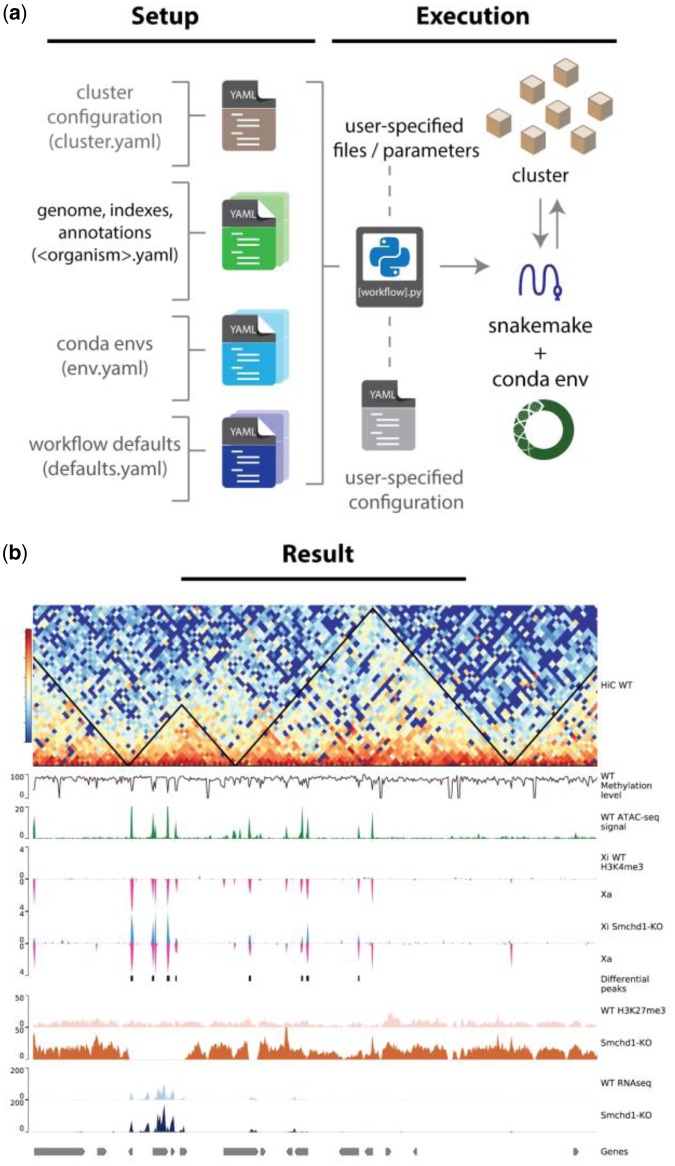
snakePipes employs snakemake (Köster and Rahmann, 2012) as its core workflow language, which benefits from easy readability of the code, widespread adoption and scalability to most clusters and cloud platforms. snakePipes also makes use of conda environments and the bioconda platform (Grüning et al., 2018), which allows hassle-free installation and upgrade of known-compatible and known-functional tools (Fig. 1a;Supplementary Methods). Conda environments alleviate the need to manually manage tools or have administrator permissions.
Fig. 1.
Setup, execution and results from snakePipes. (a) All configurable parameters for snakepipes are defined as YAML files during setup. However, most parameters can be overwritten during execution by providing another YAML file, adding flexibility to the analysis. (b) Output of HiC (track 1), WGBS (track 2), ATAC-seq (track 3), allele-specific ChIP-seq (tracks 3–7) and RNA-seq (tracks 8–9) workflows, plotted using pyGenomeTracks (Ramírez et al., 2018)
snakePipes’ modular architecture allows various tools and resources to be shared between workflows, simplifying data integration since data from multiple assays are processed using identical tool versions. Genome annotations and indices are shared by all workflows, and can also be generated directly via snakePipes, facilitating easy setup as well as integrative analysis. Finally, all workflows in snakePipes calculate extensive quality control metrics and produce reports using multiQC (Ewels et al., 2016) and R, that inform the user of processing and analysis results.
Apart from conventional processing steps such as mapping and peak calling, workflows in snakePipes also include various downstream analyses. All workflows (except scRNA-seq workflow) optionally accept a sample information (tab-separated) file that can be used to define groups of sample. This allows comparative analysis, such as differential expression (RNA-seq), differential peak calling (ChIP-Seq), differential accessibility (ATAC-seq) and differential methylation (WGBS). Complex design formulas are supported using additional columns of the sample sheet. The HiC workflow uses sample information to merge groups and can perform TAD calling with parameters adapted to the resolution of the produced matrix [using HiCExplorer (Ramírez et al., 2018)]. Most workflows also allow allele-specific processing of data via SNPSplit (Krueger and Andrews, 2016) where a single or dual-hybrid genome can be created on-the-fly using the ‘allelic-mapping’ mode and a Variant Call Format file (Danecek et al., 2011). Further downstream analysis, such as allele-specific differential expression can be performed automatically. This preliminary analysis, combined with visualization-ready BED and bigWig files, allows users to quickly interpret their data (Fig. 1b). Our comparison with other recently released workflows and pipelines suggests that snakePipes offers the most extensive processing and analysis options under a single package. Further, it compares equally well to the other available alternatives in terms of installation, ease of use and scalability (Supplementary Table S1).
3 Application
To demonstrate how snakePipes can simplify analysis of data from multiple epigenomic assays, we processed data from a study of the mammalian X-chromosome (Wang et al., 2018). The knock-out of Smchd1 in mouse neural progenitor cells affects the X-chromosome organization and leads to a loss of H3K27me3 domains, gain of H3K4me3, along with de-repression of genes on the inactive X-chromosome. These changes are apparent directly from the snakePipes output (Fig. 1b;Supplementary Fig. S1b and c). We further combined these results with those obtained from online ATAC-seq (Giorgetti et al., 2016) and WGBS data (GSE101090) processed via snakePipes, and find that these de-repressed genes have a higher open chromatin signature compared to the downregulated or unchanged genes (Supplementary Fig. S1d). These genes also show a methylation status similar to the downregulated but lower than unchanged genes (Supplementary Fig. S1e), corroborating previous (Schübeler, 2015) and recent (Lea et al., 2018) links between promoter CpG methylation and gene repression.
4 Conclusion
In summary, snakePipes simplifies the analysis of large-scale epigenomic studies by allowing fast and reproducible processing of data from several assays. While further downstream analysis would still be required to integrate the results depending upon biological questions, snakePipes’ outputs allow biologists to quickly interpret and understand their results, facilitating integrative analysis.
Supplementary Material
Acknowledgements
We thank Gina Renschler and Jana Böhm for testing workflows and Chen-Yu Wang for useful comments on our preprint.
Funding
L.R. and T.M. acknowledge funding from the German Science Foundation [CRC992 ‘Medical Epigenetics’].
Conflict of Interest: none declared.
References
- Danecek P. et al. (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P. et al. (2017) Nextflow enables reproducible computational workflows. Nat. Biotechnol., 35, 316–319. [DOI] [PubMed] [Google Scholar]
- Ewels P. et al. (2016) MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics, 32, 3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L. et al. (2016) Structural organization of the inactive X chromosome in the mouse. Nature, 535, 575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J. et al. (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol., 11, R86.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüning B. et al. (2018) Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat. Methods, 15, 475–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster J., Rahmann S. (2012) Snakemake–a scalable bioinformatics workflow engine. Bioinformatics, 28, 2520–2522. [DOI] [PubMed] [Google Scholar]
- Krueger F., Andrews S.R. (2016) SNPsplit: allele-specific splitting of alignments between genomes with known SNP genotypes. F1000Res., 5, 1479.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea A.J. et al. (2018) Genome-wide quantification of the effects of DNA methylation on human gene regulation. Elife, 7, e37513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F. et al. (2018) High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun., 9, 189.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D. (2015) Function and information content of DNA methylation. Nature, 517, 321–326. [DOI] [PubMed] [Google Scholar]
- Wang C.-Y. et al. (2018) SMCHD1 merges chromosome compartments and assists formation of super-structures on the inactive X. Cell, 174, 406–421.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.