Abstract
Summary
Mislabeling in the process of next generation sequencing is a frequent problem that can cause an entire genomic analysis to fail, and a regular cohort-level checkup is needed to ensure that it has not occurred. We developed a new, automated tool (BAMixChecker) that accurately detects sample mismatches from a given BAM file cohort with minimal user intervention. BAMixChecker uses a flexible, data-specific set of single-nucleotide polymorphisms and detects orphan (unpaired) and swapped (mispaired) samples based on genotype-concordance score and entropy-based file name analysis. BAMixChecker shows ∼100% accuracy in real WES, RNA-Seq and targeted sequencing data cohorts, even for small panels (<50 genes). BAMixChecker provides an HTML-style report that graphically outlines the sample matching status in tables and heatmaps, with which users can quickly inspect any mismatch events.
Availability and implementation
BAMixChecker is available at https://github.com/heinc1010/BAMixChecker
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Increasing use of Next Generation Sequencing (NGS) in clinical practice requires a large number of samples to be processed in a limited time. While improvements in algorithms have provided more accurate means of detecting genomic variants, human errors in sample handling remain a constant concern. Sample mismatch, in particular, is a frequent occurrence detrimental to sequencing analyses (Westra et al., 2011).
In the last few years, several tools have been developed to detect mismatching of samples in NGS datasets. Conpair (Bergmann et al., 2016) detects a mismatched pair of BAM files based on 7387 known polymorphic loci. BAM-matcher (Wang et al., 2016) uses a similar approach, but allows for faster testing as it only uses 1500 exotic single nucleotide polymorphism (SNP) sites. The recently developed NGSCheckMate (Lee et al., 2017) accepts FASTQ, BAM or VCF files as input and provides a list or a tree graph of genotype correlations among the samples. In general, the reported accuracies of these tools are all over 95%.
Despite the good accuracy of these tools, we have found areas for improvement in two major features that would allow for more active use in cohort-level checkup. First, the number and the composition of SNP sites for individual matches need to be optimized. These SNP sites should be applicable to various targeted sequencing panels in order to cope with large-scale clinical genomic tests. Second, the tool should be fast and automated to minimize intervention from users, even with a large number of samples.
Accordingly, we developed BAMixChecker, which facilitates fast and accurate assessment of mismatches in sample-pair assignment from combinations of WGS/WES/RNA-Seq and targeted sequencing panels in NGS cohort. BAMixChecker uses 853 highly informative human polymorphic sites that are optimized for WGS/WES and RNA-Seq data. For targeted sequencing data, BAMixChecker instantly constructs an optimal SNP list specific to the targeted genomic regions; the use of smaller SNP set enabled a reduced running time while maintaining accuracy, even in a small panel. Although the tool was mainly developed for the analysis of human data BAMixChecker provides specific functions for the identification of sets of highly informative polymorphic positions which allow the application of the tool also to non-human species. BAMixChecker categorizes orphan and swapped samples using rules based on genetic distances and file names edit distances. The pipeline is fully automated, allowing users to quickly check abnormal events without the need for further intervention to interpret the result.
2 Materials and methods
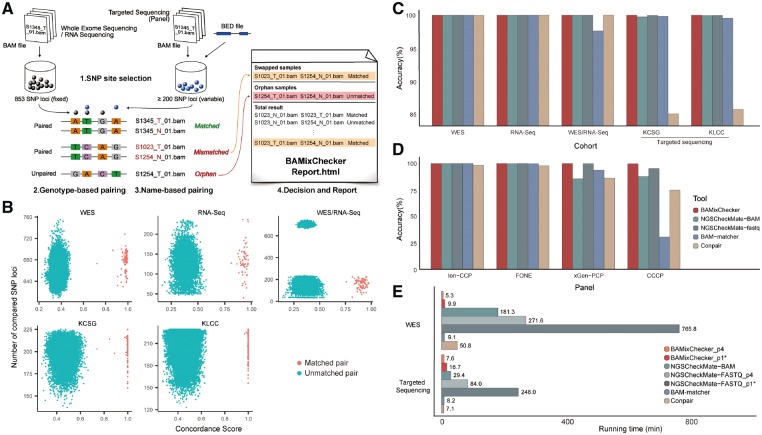
BAMixChecker only takes pairs of BAM/CRAM files as inputs with optional genomic region information (BED file) for targeted sequencing and reports mismatched samples and their types (Fig. 1A). The overall workflow consists of the four major steps described below. Detailed procedures are described in Supplementary Data.
Fig. 1.
(A) Overall workflow of BAMixChecker. (B) Score distribution of BAMixChecker in five datasets. Each dot reflects a comparison result between two samples. Red dots indicate unmatched pairs; blue dots are matched pairs. (C) Accuracies of the four tools in five cohorts. NGSCheckMate contains two different modes (BAM and FASTQ input). WES/RNA-Seq represents a WES-RNA-Seq pair. (D) Accuracy of the four tools in downsampled cohorts. (E) Running times of the four tools. The running times of BAMixChecker and NGSCheckMate were measured in two different modes (p1: single-thread, p4: multi-thread with four processors).*: default
1. SNP site selection: To select only highly informative SNP loci and to reduce ambiguous calls, we considered two criteria: (i) mappability and (ii) population allele-frequency. From gnomAD v2.0.2 (Lek et al., 2016), we collected 57 582 candidate exonic SNPs that passed filters, including variant quality, mapping quality and genomic mappability depending on position like not in a low complex region, segment duplicated region and sample repeat region. Out of the 57 582 candidates, 853 SNPs with a global minor allele frequency (gMAF) between 0.45 and 0.55, and also population-specific MAF between 0.35 and 0.65 for eight populations (Supplementary Methods) were selected to build a fixed list for WGS/WES and RNA-seq data. For targeted sequencing, BAMixChecker automatically adjusts MAF condition of SNPs from higher global MAF and MAF in each population by downing the values, ranging 0.45–0.1, until at least 200 SNPs overlap given targeted genomic region (Supplementary Methods).
2. Genotype-based pairing: For selected SNP sites, BAMixChecker calls genotypes of samples using GATK HaplotypeCaller with further filtering (Supplementary Methods). Genotype concordance scores are then calculated between all pairs in the cohort. Sample pairs with a concordance score of >0.7 are considered matched. The use of the fixed cut-off value is supported by a large margin in the observed concordance scores between matched and unmatched samples from large-scale databases (Fig. 1B). Although a perfect concordance (1.0) is expected between matched samples in general, we assumed that many confounding factors including contamination, copy number variation, allele-specific expression and poor sample quality allowed the lenient cut-off. Unpaired samples in this step are considered orphans.
3. Name-based paring: Assuming that file names are rule-based within a cohort, sample relationships can be inferred from the names, just as a human would do. BAMixChecker emulates this using entropy-based file matching (Supplementary Methods and Supplementary Fig. S1). Briefly, the uncertainty of values in the same position of a delimited file name is measured. Positions with high uncertainty tend to represent sample- or individual-specific information (e.g. sample id), while low uncertainty reflects global information (e.g. cohort id). File-name similarity is calculated by adding or subtracting positional entropy for each matched or mismatched value: file names of matched samples only differ in low entropy positions (e.g. T versus N) and gain a high score in high entropy positions (e.g. sample id), thereby being considered as the best match in the cohort. We have confirmed that this approach perfectly identifies true matches for 463 sample pairs in four different cohorts (Supplementary Table S2). The file-name based matching algorithm searches matched paired sample with the best similarity score. Otherwise, a user can directly offer matched samples information with a list if the matched samples are not a pair (Supplementary Methods).
4. Decision and report: After genotype-based and file-name-based pairing, BAMixChecker categorizes all samples into three classes: matched (match for genotype and file-name pairing), swapped (genotype match that is not file-name matched, or vice versa) and orphan (no genotype match found). BAMixChecker outputs the final judgment in an HTML file, with an additional Heatmap that describes the overall sample concordance (Supplementary Fig. S2).
3 Results
We evaluated the accuracy of BAMixChecker in comparison to previously reported tools (NGSCheckMate, BAM-matcher and Conpair) in five real NGS cohorts with tumor-normal pairs: (1) TCGA WES pair cohort (n = 202), (2) TCGA RNA-Seq pair cohort (n = 130), (3) TCGA WES/RNA-Seq pair cohort (n = 168), (4) Korean Cancer Study Group (KCSG) panel sequencing pair cohort (n = 192) (Lim et al., 2019) and (5) Korean Lung Cancer Consortium (KLCC) panel sequencing pair cohort (n = 402) (Supplementary Table S1).
For TCGA WES and RNA-Seq, all tools exhibited good accuracy, except for a few miscalls by BAM-matcher and Conpair (Fig. 1C and Supplementary Table S3). However, there was a noticeable drop in accuracy for the targeted sequencing cohorts (KCSG and KLCC in Fig. 1C) with Conpair. For all cohorts, only BAMixChecker showed perfect accuracy.
For evaluation of smaller panels, TCGA WES data were downsampled to gene lists of four popular commercial panels: Ion AmpliSeq Comprehensive Cancer Panel (Ion-CCP, 409 genes), Foundation One (FONE, 315 genes), xGen Pan-Cancer Panel (xGen-PCP, 127 genes) and Comprehensive Common Cancer Panel (CCCP, 46 genes). We found BAMixChecker showed almost perfect accuracy in all panels (>99.8%), while the other tools showed lower accuracy in smaller panels (Fig. 1D). BAMixChecker showed robust performance even with a family dataset (Supplementary Methods and Supplementary Fig. S3).
For additional validation, we generated artificial mismatches by intentionally changing file names (Supplementary Methods). For each NGS cohort, 10% of files were randomly selected and simulated to be swapped (by switching file names) or orphan (by assigning a file name to a wrong sample), which were repeated 100 times with different randomization. Testing by BAMixChecker confirmed that all mismatches were perfectly reported (100% accuracy), regardless of the mismatch type or used NGS cohort (Supplementary Methods and Supplementary Fig. S4).
Finally, running times were assessed for all tools (Fig. 1E and Supplementary Methods). BAMixChecker and NGSCheckMate can be run in a multiprocessing mode and were tested with two different CPU numbers (single and 4-CPUs). BAMixChecker exhibited comparable or faster speed than BAM-matcher and Conpair, and was remarkably faster (∼18×) than NGSCheckMate. Considering the reduced need for intervention from users, we expect that the practical hands-on time would be much shorter with BAMixChecker.
Supplementary Material
Acknowledgements
The authors thank the KCSG TRIUMPH for providing sequencing data.
Funding
This research was supported by the grant of the Korea Health Technology R&D Project [HI14C1324] through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea, and the Bio & Medical Technology Development Program [NRF-2016M3A9B6903439] through the National Research Foundation of Korea (NRF), funded by the Ministry of Science and ICT.
Conflict of Interest: none declared.
References
- Bergmann E.A. et al. (2016) Conpair: concordance and contamination estimator for matched tumor-normal pairs. Bioinformatics, 32, 3196–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. et al. (2017) NGSCheckMate: software for validating sample identity in next-generation sequencing studies within and across data types. Nucleic Acids Res., 45, e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M. et al. (2016) Analysis of protein-coding genetic variation in 60 706 humans. Nature, 536, 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.M. et al. (2019) Investigating the feasibility of targeted next-generation sequencing to guide the treatment of head and neck squamous cell carcinoma. Cancer Res Treat, 51, 300–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.P. et al. (2016) BAM-matcher: a tool for rapid NGS sample matching. Bioinformatics, 32, 2699–2701. [DOI] [PubMed] [Google Scholar]
- Westra H.J. et al. (2011) MixupMapper: correcting sample mix-ups in genome-wide datasets increases power to detect small genetic effects. Bioinformatics, 27, 2104–2111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.