Abstract
Biological age (BA), derived from molecular and physiological measurements, has been proposed to better predict mortality and disease than chronological age (CA). In the present study, a computed estimate of BA was investigated longitudinally in 3,558 individuals using deep phenotyping, which encompassed a broad range of biological processes. The Klemera–Doubal algorithm was applied to longitudinal data consisting of genetic, clinical laboratory, metabolomic, and proteomic assays from individuals undergoing a wellness program. BA was elevated relative to CA in the presence of chronic diseases. We observed a significantly lower rate of change than the expected ~1 year/year (to which the estimation algorithm was constrained) in BA for individuals participating in a wellness program. This observation suggests that BA is modifiable and suggests that a lower BA relative to CA may be a sign of healthy aging. Measures of metabolic health, inflammation, and toxin bioaccumulation were strong predictors of BA. BA estimation from deep phenotyping was seen to change in the direction expected for both positive and negative health conditions. We believe BA represents a general and interpretable “metric for wellness” that may aid in monitoring aging over time.
Keywords: Deep phenotyping, Precision medicine, Healthy aging, Biological age, Chronological age
Age is the most important risk factor for most common diseases. There is considerable interest in mitigating aging-related disease risks through lifestyle, pharmaceutical, and environmental interventions that attenuate biological aging. A hurdle in this quest is the quantification of an individual’s “wellness,” which is not only the absence of disease but also their resilience to future disease, general satisfaction with one’s health and wellbeing, and energy for activities that enrich a person’s life. While a multitude of signals relevant to an individual’s health and wellness can be captured, meaningful clinical relevance remains a challenge. The development of tools and methods for the collation, integration, analysis, and application of these signals is essential to realizing the goals of precision and personalized medicine (1). More sensitive and precise assessments of health status and trajectory, guided by dense longitudinal phenotyping, will enable a transformation in modern health care. Such a paradigm shift can only occur by converting these sophisticated, high-dimensional measures into actionable metrics. Biological age (BA), to the extent it can be estimated, may provide one such personalized and intuitive metric of overall health status that can be communicated effectively to a general population.
Estimation of BA was first proposed in 1969 (2). In 1988, Baker and Sprott proposed that a biomarker of aging is a biological parameter of an organism that either alone or in some multivariate composite will, in the absence of disease, predict physiologically functional capacity at some later stage better than chronological age (CA) (3). More recently, BA has been assessed via epigenetic markers (4), proteomics (5), and Electronic Medical Records (6). The Klemera–Doubal (KD) method has been suggested as a better predictor of all-cause mortality than either CA alone or using multiple linear regression of ten clinical biomarkers (7,8). Studies using small numbers of highly informative clinical variables to develop KD-computed BA measures have demonstrated these measures associated with poor balance, physical weakness, declining cognitive performance, physical appearance, cardiovascular risk, frailty indices, extrinsic epigenetic age, caloric restriction (CR), and gene expression (9–13).
Deep phenotyping offers the opportunity to explore multiple systems that contribute to BA in greater depth, and generate more comprehensive metrics of overall health that change over time, aimed at reflecting an individual’s changing health (14). Herein, we also explore estimated BA by distinct data types: the metabolome, proteome, and clinical labs, as well as a BA calculation that integrates all of these together. BA appears to be modifiable, and thus may be a simple metric that is useful to monitor general health.
In this work, KD was applied to over 900 disparate (principal component analysis, PCA, transformed) biomarkers, including metabolites, proteins, genomics, and clinical measures. This collection of biomarkers is herein termed personal, dense, dynamic data (PD3) clouds (15). Data type (eg, different omics measures)-specific BA estimates were compared to each other and changes in BA over time were examined by data type, and among subgroups that were hypothesized to have different BA trajectories (including stratifications by sex, ethnicity, age group, and baseline BA). Differences between biological and chronological age (BA–CA) were utilized as a metric, noted as ΔAge (more negative indicates scoring younger than CA), and associations between ΔAge and lifetime prevalence of common health conditions were examined.
In this study, we ascertained the effects of conditions and behaviors generally thought of as being healthy or unhealthy upon the introduced BA measure. We found that “healthy” behaviors, such as participation in a scientific wellness program (16), were found to be associated with a decreasing ΔAge over time. Conversely, “unhealthy” conditions, such as self-reported diseases, were found to increase ΔAge in every condition we had data for where there was a significant effect (no significant effects in the opposite direction). The observation indicated that ΔAge was sensitive to changes in the blood associated with common disease states. Association strength and computed BA estimates varied significantly by data type (proteomics, metabolomics, and clinical labs), demonstrating that BA depends on the systems being interrogated. These results support the construction of a BA measure that integrates diverse information across different—omics, biological systems, and disease biomarkers—and/or the use of multiple BA measures to reflect different biological systems—to help assess individual health and for the quantification and exploration of aspects of the aging process in humans.
Methods
Study Population
The sample studied consisted of men and women participating in a consumer data-intensive wellness program (Arivale, now closed) that varied by age and health status (demographics given below). The program involved lifestyle coaching on exercise, nutrition, stress management, and sleep all tailored to the participants’ health goals, specific genetic markers, and clinical metrics as detailed in a prior publication (16). Deidentified data from consenting participants were collected from July 2015 to July 2018. A total of 3,558 participants were observed for an average of 214 days, with an average of 2.1 longitudinal data points with a total of 7,634 observations. In total, 1,354 participants had a single time point, 1,105 had two, 711 had three, and 388 had four or more, with two participants having the maximal (8) number of time points. Average time between observations was 190 days among participants with multiple time points. The study was approved by the Western Institutional Review Board.
Personal, Dense, Dynamic Data Clouds (PD3 Clouds)
We previously developed and published analyses incorporating proteomic, metabolomic, microbiomic, and genetic data (the PD3 cloud) on 108 participants in the context of health and wellness (15). This cohort ultimately expanded to 3,558 individuals at the time data were collected for this study. Participants’ genetic profiles were assessed either by whole genome sequencing (2845) or by single nucleotide polymorphism (SNP) chip (713). Detailed information on the acquisition, storage, generation, and analyte-specific pre-processing of these measures is available in the Supplementary Methods. After pre-processing, the PD3 clouds included genomics plus longitudinal measures from blood, including 54 clinical lab tests from LabCorp or 67 clinical lab tests from Quest, 243 proteins, and 611 metabolites with CA ranging across the adult lifespan (18–89+ years).
Creating the BA Measure
The KD method, with a PCA transformation on the input features, was used to create the BA measure (7). Briefly, KD is a weighted average of independent linear regressions of biomarkers to CA. Ten iterations of 10-fold cross-validation were performed to estimate BA from each data type (clinical labs, metabolites, and proteins). Male and female data were trained separately, as were observations from different laboratory vendors. Training/testing set splits were generated by randomly shuffling participants, partitioning them into ten sets, and iterating over those sets, with one set as the test set and the remaining nine being used for training. Training sets were restricted to baseline measurements, ensuring those participants had minimal wellness coaching, and only one observation of a participant was trained on. All observations of participants in the test set were predicted from the training set. Clinical labs had two vendors, so only the earliest observation among both vendors was included in the training. All samples were z-score normalized using the mean and SD estimated from the training set at each fold.
Similarly, principal components were estimated using the training, and the transformation was then applied to all samples. Principal components were used to satisfy the biomarker linear independence requirement of the KD algorithm. Slopes, SDs, and intercepts were calculated for each of the strongest components explaining up to 90% of the variance. These variables were then used to calculate BA using KD. The contribution of each analyte to BA was calculated by multiplying the weights learned for each component by the analyte contribution to each component and summing across all components. These representations are equivalent because PCA and KD are linear transformations (see Supplementary Methods). CA was excluded as a biomarker, although KD allows its inclusion. Doing so reduces variance, but adds limited information regarding BA’s relationship to health outcomes (10). For each data type, the 10 predictions were averaged. For each observation of a participant, all available data type predictions were averaged and presented as the overall BA prediction. A total of 2,742 observations had only one data type, 3,634 had two, and 1,258 had all three. See Supplementary Figure 1 and Supplementary Table 2 for details.
Trajectory of the BA Measure Over Time
To determine whether BA changed over time after initiation of health coaching, Generalized Estimating Equations (GEEs) with exchangeable correlation structure were utilized, which accounts for the correlation between multiple observations (time points) per participant (17). Participants with two or more blood draw visits were included in the trajectory models. The primary model assessing linear change in BA included time in the program as the independent variable, starting at time zero (time of first blood draw), and BA as the dependent variable; baseline CA was included as a covariate. Models were stratified by baseline age group, sex, and race (white vs non-white), and interaction terms between study time and sex/race were included to assess for effect modification. Additionally, as factors that may impact BA over time are of interest, models were stratified by CA decade and BA starting point to model BA changes due to differences in initial health status at coaching initiation (model A: participants with an initial BA that was 5 or more years greater than CA; model B: participants with an initial BA that was 5 or more years less than CA. This analysis was repeated with BA ±10 years from CA).
Health History, Behaviors, and Associations With ΔAge
GEE models with exchangeable correlation structure were used to examine associations between ΔAge under combined and independent data modalities and the lifetime prevalence of the 40 most common self-reported health conditions, along with lifetime and/or current smoking. In the minimally adjusted model, ΔAge was modeled as the outcome variable, and self-reported past or current condition was the predictor, with CA at each prediction included as a covariate. Each condition was modeled separately. Since obesity was hypothesized to be strongly associated with ΔAge and many conditions, obesity (0 for body mass index [BMI] < 30, 1 for BMI ≥ 30) was included as a covariate. Association between obesity and BA itself was also calculated. A Bonferroni correction at alpha = .05/(43(conditions)*4 (modalities) = 3E-04 threshold for statistical significance was applied. Many health outcomes are highly correlated with one another, and thus, this correction is highly conservative.
Results
Population Characteristics
Mean age was 47.5 years, with more females than males (58.6% female). Baseline characteristics are presented in Table 1. The percent of obese participants was 27.9%, lower than the Center for Disease Control reported estimate of 37.9% for all U.S. adults. This bias appears to be driven primarily by regional makeup, rather than the self-selection of lower BMI individuals. This cohort is predominantly (~80%) drawn from Washington or California. Given the state/province of residence, the expected percentage of obese individuals is 27.7% (18). Socioeconomic status of participants is presumably higher than the national average, but that information was not captured.
Table 1.
Baseline Self-reported Characteristics of Study Sample
| Characteristic | Alla | Womena | Mena | p Valueb |
|---|---|---|---|---|
| Chronological age, mean years (SD) | 47.6 (12.2) | 48.7 (11.6) | 47.9(11.8) | .1 |
| Non-white, no. (%), n = 3,452 | 784 (22.7) | 416 (20.5) | 368 (25.8) | <.001 |
| BMI, mean (SD), n = 3,227 | 27.7 (6.3) | 27.8 (7.1) | 27.4 (4.9) | .05 |
| Obesec, no. (%), n = 3,227 | 901 (27.9) | 584 (30.7) | 317 (23.9) | <.001 |
| Moderate activity ≥ 3×/wk, no. (%), n = 3,468 | 2,253 (65.0) | 1,278 (62.0) | 975 (69.3) | <.001 |
| Vigorous activity ≥ 3×/wk, no. (%), n = 3,467 | 1,121 (32.3) | 528 (25.6) | 593 (42.1) | <.001 |
| Sitting >8 h/d, no. (%), n = 3,467 | 2,303 (66.4) | 1,392 (67.6) | 911 (64.7) | .09 |
| Ever smoker, no. (%), n = 2,825 | 774 (27.4) | 448 (25.9) | 326 (29.7) | .03 |
| Current smoker, no. (%), n = 3,469 | 174 (5.0) | 73 (3.5) | 101 (7.2) | <.0001 |
| Cholesterol-lowering meds, no. (%), n = 2,817 | 337 (12.0) | 163 (9.5) | 171 (15.7) | <.001 |
| Past and/or current self-report of: | ||||
| High cholesterol, no. (%), n = 3,351 | 788 (23.5) | 408 (20.4) | 380 (28.1) | <.001 |
| Hypertension, no. (%), n = 3,361 | 579 (17.2) | 313 (15.6) | 266 (19.6) | .003 |
| Asthma, no. (%), n = 3,389 | 559 (16.5) | 370 (18.3) | 189 (13.8) | <.001 |
| Type 2 diabetes, no. (%), n = 3,309 | 125 (3.8) | 78 (4.0) | 47 (3.5) | .6 |
| Breast cancer, no. (%), n = 3,235 | 63 (1.9) | 59 (3.0) | 4 (0.3) | <.001 |
| Coronary artery disease, no. (%), n = 3,280 | 50 (1.5) | 19 (1.0) | 31 (2.3) | .003 |
Note: BMI = body mass index.
aTotal N = 3,558; women, N = 2,087; men, N = 1,471. bFor comparisons between men and women, chi-square tests were run for categorical variables and two-sided t-tests for continuous variables. cObese defined as BMI ≥30.
BA Estimation Through PD3 Clouds
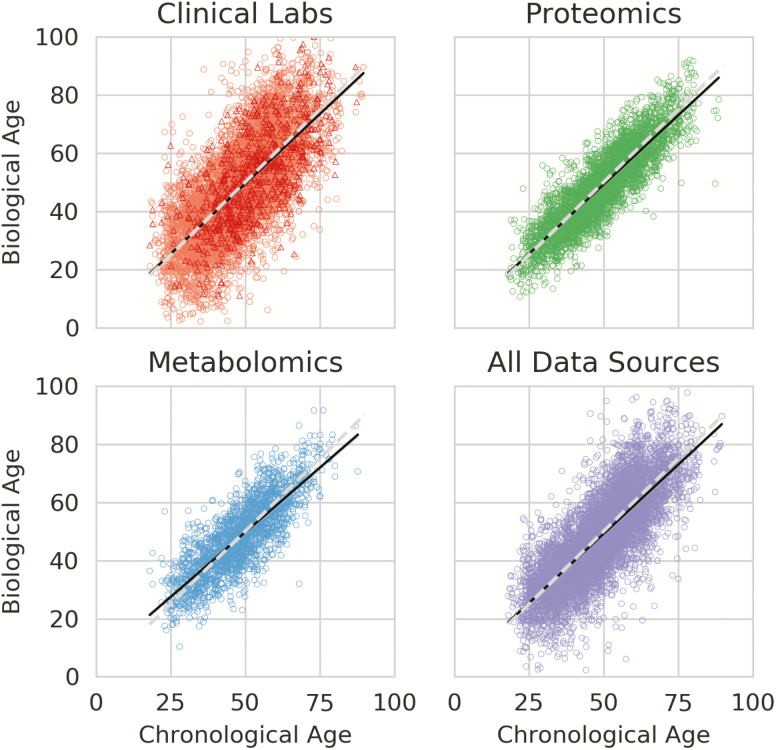
BA estimates using the KD method are shown in Figure 1. The Pearson correlation between BA and CA was .78 overall, .70 for the clinical labs, .81 for the metabolomics, and .88 for the proteomics. The median absolute error, that is, the median absolute difference between BA and CA, of these predictions was 5.54 years overall, 8.04 years for clinical labs, 4.82 years for metabolomics, and 4.39 years for proteomics. Mean (SD) over repeated predictions for the same observation was 3.83 years overall, 1.05 years for clinical labs, 1.52 years for metabolomics, and 1.03 years for proteomics. ΔAge had a mean (SD) of −0.78 (9.28) years overall, −0.43 (12.18) years for clinical labs, −0.11 (7.48) years for metabolomics, and −0.73 (6.57) years for proteomics. ΔAge was largely uncorrelated with CA, at a Pearson r of −.06 overall, −.03 for clinical labs, −.18 for metabolomics, and −.10 for proteomics. See Supplementary Table 2 for summary statistics.
Figure 1.
Scatter plots of Biological Age estimates using the Klemera–Doubal algorithm for each data type individually, and in aggregate (All Data Sources). Each point is one observation of an individual. The solid line in each plot is the ordinary least squares regressed line, and the dotted line is biological age = chronological age. The Clinical Lab plot contains estimates from two vendors: Labcorp (circle) and Quest (triangle).
Pearson correlation of ΔAge between multiple observations of the same participant, that is, ρ(ΔAget, ΔAget+1), was .66 overall, .67 for clinical labs, .67 for proteomics, and .64 for metabolomics. These correlations were stronger than the between-data type ΔAge, with clinical labs correlating with metabolomics at an r of .26, clinical labs with proteomics at .25, and metabolomics with proteomics at .27 (Supplementary Figure 2).
BA Changes Over Time
Mean linear trajectory of BA over time, calculated using longitudinal measurements among participants with at least two visits, varied according to whether the predictions were based on clinical labs, metabolites, proteins, or a combination of all three categories (Table 2). In the minimal model adjusted for baseline CA, BA prediction based on all available analytes showed that BA stayed statistically stable over time. On average, BA decreased by 0.16 years for every year of participation in the wellness program (β = −0.16, 95% CI: −0.45, 0.19). This is significantly lower than the expected increase of 1 year/year. BA estimates from all data types had a β coefficient < 1, the natural rate of aging, with all data types except metabolomics being significantly <1.
Table 2.
Regression Coefficients for Changes in Mean Biological Age Over Time for Individuals With Two or More Blood Draws (N = 2,205)
| Data Set Used for BA Predictiona | β Coefficient | SE | 95% CI | Interaction pc |
|---|---|---|---|---|
| All analytes | −0.160 | 0.181 | −0.452, 0.194 | NA |
| Proteins | 0.524 | 0.179 | 0.174, 0.874 | NA |
| Metabolites | 0.274 | 0.471 | −0.648, 1.196 | NA |
| All clinical chemistries | −0.374 | 0.216 | −0.798, 0.049 | NA |
| Stratified Analysesb | ||||
|---|---|---|---|---|
| Sex | ||||
| Males (n = 908) | 0.192 | 0.281 | −0.359, 0.742 | |
| Females (n = 1,297) | −0.484 | 0.229 | −0.932, −0.036 | .043 |
| Self-reported ethnicity | ||||
| White (n = 1,705) | −0.111 | 0.198 | −0.498, 0.276 | |
| Non-white (n = 433) | −0.589 | 0.465 | −1.500, 0.322 | .371 |
| Age at baseline, by decade | ||||
| 18–29 y (n = 130) | −2.720 | 0.744 | −4.180, −1.270 | |
| 30–39 y (n = 388) | −0.055 | 0.419 | −0.877, 0.766 | |
| 40–49 y (n = 658) | −0.356 | 0.332 | −1.010, 0.295 | |
| 50–59 y (n = 638) | 0.530 | 0.356 | −0.168, 1.230 | |
| 60–69 y (n = 322) | −0.285 | 0.393 | −1.050, 0.484 | |
| 70+ y (n = 69) | −0.895 | 0.853 | −2.570, 0.776 | NA |
| Baseline BA prediction | ||||
| BA = 5 y > CA (n = 481) | −0.985 | 0.423 | −1.814, −0.157 | |
| BA = 5 y < CA (n = 540) | 0.024 | 0.377 | −0.715, 0.764 | |
| BA = 10 y > CA (n = 167) | −1.540 | 0.686 | −2.88, −0.194 | |
| BA = 10 y < CA (n = 187) | 1.613 | 0.651 | 0.388, 2.890 | NA |
Note: BA = biological age; CA = chronological age; NA = not applicable.
aGEE model: BA ~ time in wellness program + baseline CA; clustered by client ID, family = Gaussian, with an exchangeable correlation matrix; only individuals with at least two visits were included.
bGEE models, stratified by sex, ethnicity, age group, and baseline BA prediction: ΔAge (BA−CA) ~ time in wellness program + baseline CA; clustered by client ID, family = Gaussian, with an exchangeable correlation matrix; all models use BA predictions based on the “all analyte” data set. cInteraction models: ΔAge (BA−CA) ~ time in wellness program + predictor variable + baseline CA + predictor variable × time in wellness program; clustered by client ID, family = Gaussian, with an exchangeable correlation matrix.
Potential Modifiers of BA Trajectory Over Time
Exploratory analyses to examine several baseline factors (sex, ethnicity, age group, and baseline health status) that were hypothesized a priori might have an impact on BA trajectories were performed. In sex-stratified models based on the “all analyte” BA predictions (clinical labs, metabolites, and proteins combined), BA decreased in women over time (coefficient: −.48, 95% CI: −0.93, −0.04), but stayed constant in men (coefficient: .19, 95% CI: −0.36, 0.74) (Table 2). The sex–time interaction term was weakly significant (p < .05), indicating a difference between men and women in their BA trajectories over time. A similar pattern in BA derived from proteins and clinical labs was observed (Supplementary Table 3), with BA from clinical labs also indicating a weakly significant difference between men and women (interaction p = .02). Since race was unevenly distributed throughout CA, self-reported race (white vs non-white) was also stratified; the interaction term was not significant, and both groups had slowed BA compared to CA. In models stratified by age (in decades), the youngest age group had the slowest rate of biological aging (using BA estimates from the all-analyte data set), though all age groups except 50–59 years indicated slowed aging (upper 95% CI < 1). This effect was not dose dependent (ie, rate of aging did not increase monotonically) and was roughly consistent across data types. This analysis for BA was repeated for each data type and observed similar patterns in BA derived from clinical labs; β coefficients derived from proteins and metabolites were highly variable and had wide CIs, likely due to small N per age group (Supplementary Table 3).
Lastly, baseline ΔAge was stratified, with the idea that participants with higher ΔAge at study entry would be less healthy (under the assumption that ΔAge is an adequate summary metric for health and wellness), and therefore experience greater benefit from health coaching. Participants with BA 5 or more years higher than their CA at baseline experienced approximately 1-year decline in BA for every 1 year in the program (coefficient: −.99, 95% CI: −1.81, −0.16), while participants who entered the program with BA at least 5 years less than CA maintained their youthful BA over time based on the all-analyte BA estimates (coefficient: .02, 95% CI: −0.72, 0.76). These effects were more pronounced in individual data modalities, though many stratified analyses had small N, which likely inflated estimates. Regression-to-the-mean effects could not be ruled out in the absence of a control group, particularly when N was small or when baseline deviations were extreme (ie, the >10 years plus or minus for ΔAge) (19).
ΔAge Is Associated With Health and Behavior and Especially With Type 2 Diabetes
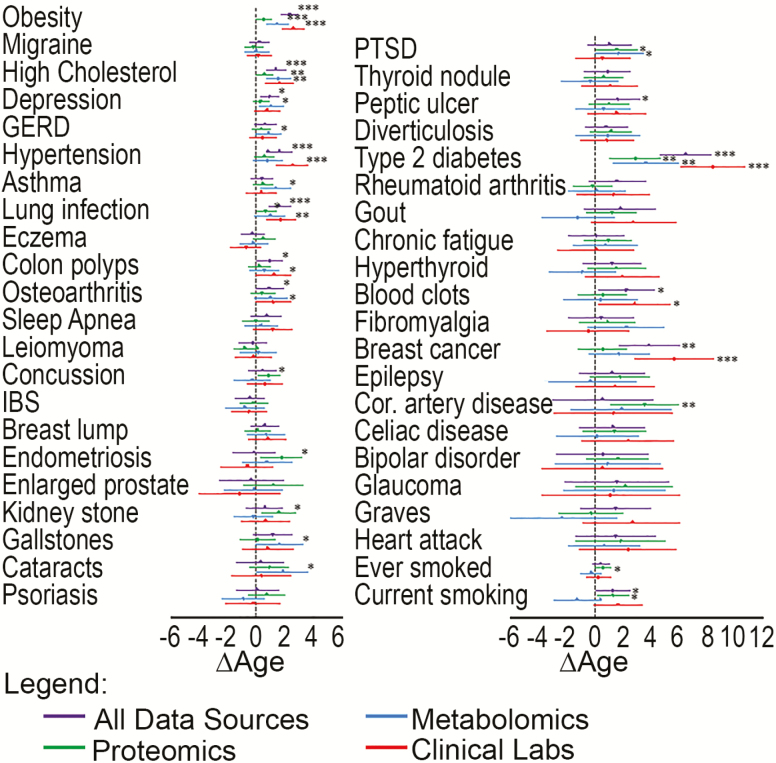
Among the top 43 most common health conditions and behaviors in our cohort, after correcting for multiple comparisons, obesity, hypertension, high cholesterol, lung infection, type 2 diabetes (T2D), and breast cancer were associated with increased ΔAge in models adjusted for CA and obesity (Figure 2). T2D had the highest increase in ΔAge in combined models, such that these participants had a BA that was higher than their CA by an average of 6.4 years (95% CI: 4.6, 8.2). This effect was consistent among data types for T2D. However, effects varied slightly among the different data types. For instance, the combined data type–derived BA provided highest statistical significance for increased ΔAge among participants with high cholesterol, the estimates based on metabolomics and clinical labs were also associated with increased ΔAge, though below the threshold for multiple corrections. Estimates derived from proteomics did not show as pronounced an effect, with the coefficient having a p-value >.05. Since the all-data modality and the clinical labs had the largest N, these associations were well powered and most likely to show significant associations. However, several nonsignificant trends of interest were observed indicating potential disease-specific differences in sensitivity among different data modalities, with some health conditions having a trending association (p < .05) with only one of the four modalities (such as concussion, endometriosis, kidney stone, gallstones, cataracts, and coronary artery disease). While these trends were not strong enough to be significant individually after a conservative Bonferroni correction for multiple hypothesis testing, collectively, every one is in the direction of increasing ΔAge with none in the opposite direction, adding confidence in their likely validity.
Figure 2.
Forest plot of ΔAge estimates and 95% confidence intervals associated with the 40 most common health conditions, plus ever smoking, current smoking, and obesity. Each condition or behavior was modeled individually, with ΔAge as the dependent variable, the health condition/behavior as the independent variable, and further adjustment for chronological age (CA) and obesity (body mass index > 30) in Generalized Estimating Equation models clustered by client ID with an exchangeable correlation matrix to account for multiple observations from individual clients. The obesity outcome was adjusted for CA only. Biological age (BA) estimates for each data type are shown. The blue dotted line at 0 indicates no difference between BA and CA; point estimates to the right of the blue line indicate higher BA than CA associated with the health condition/behavior (eg, based on the all-data-type BA estimate, individuals with type 2 diabetes have BAs that are, on average, 6.4 years greater [95% CI: 4.6, 8.2] than their CAs, after adjustment for CA and obesity). ***p < .0003 (Bonferroni threshold); **p < .005; *p < .05. GERD = gastroesophageal reflux disease; IBS = irritable bowel syndrome; PTSD = Post-traumatic stress disorder.
Analytes That Are Most Predictive of a High or Low BA Measure
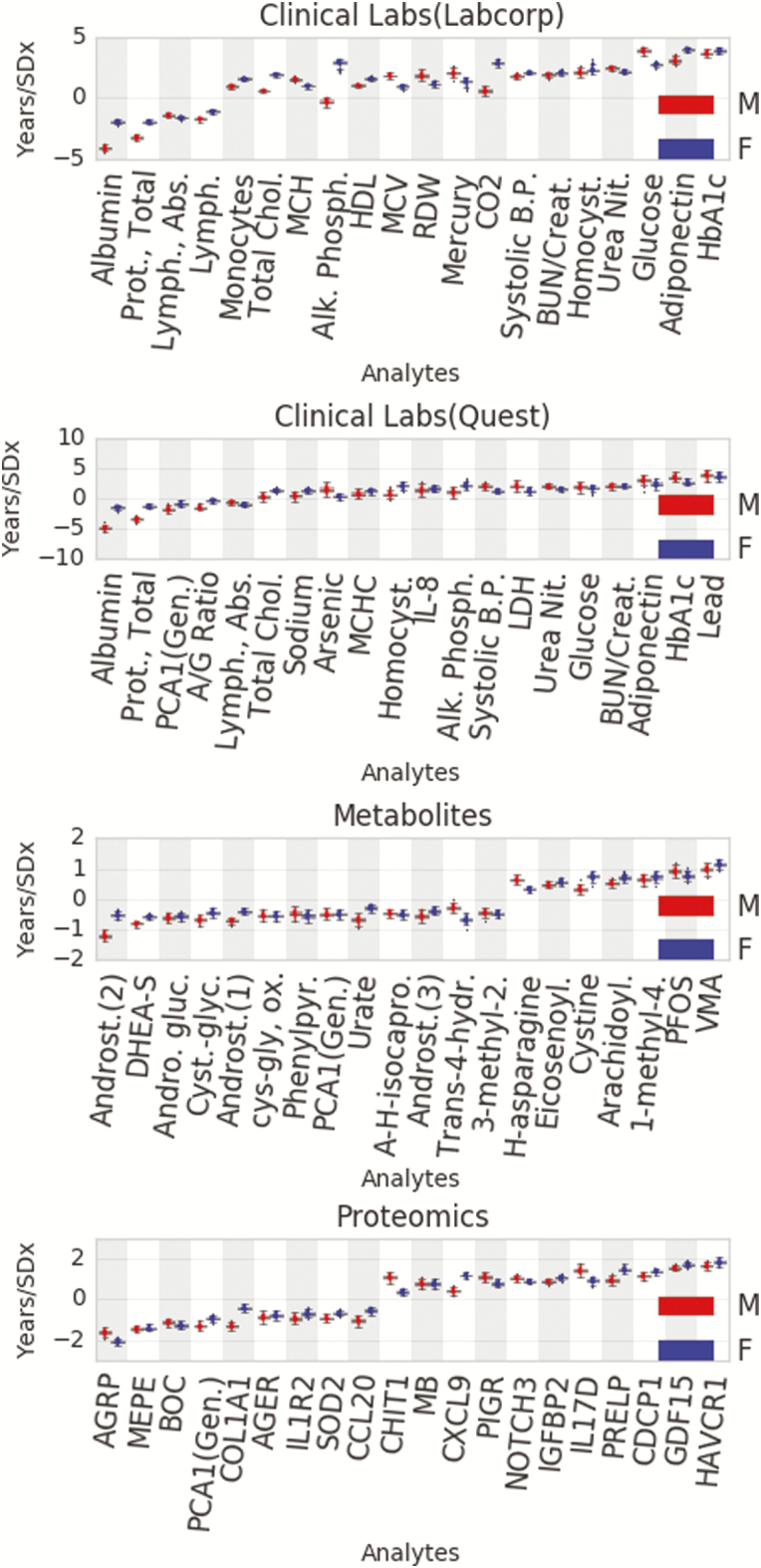
The top mean model coefficients, representing the importance of individual analytes in the model, are shown in Figure 3. The value of each analyte coefficient corresponds to the contribution of that analyte to the computed BA. For instance, a coefficient of +1 indicates an increase in BA of 1 year per SD higher than the mean, while a coefficient of −1 indicates a corresponding decrease in BA of 1 year per SD above the mean. Most markers that were strongly predictive of BA were dominated by three axes of aging: metabolic health, inflammation, and bioaccumulation of toxins.
Figure 3.
Shown are the strongest 20 analytes per data type by average effect for males (red) and females (blue). The y-axis demonstrates the effect in years per SD of the analyte, that is, 1 SD greater than the mean value for HbA1c in the Labcorp clinical labs would result in a roughly 4-year increase in biological age, all other values held constant.
In clinical labs, glycated hemoglobin (HbA1c) was the strongest positive predictor of BA independent of sex, with other (highly correlated) metabolic health markers demonstrating similar effects, that is, adiponectin and glucose. Metabolic health was also reflected in proteomics, where agouti-related peptide (AgRP) was the strongest negative predictor of BA for both men and women. AgRP is involved in energy balance through regulation of appetite and energy expenditure (20). Similarly, analytes reflective of redox balance, an integral component of metabolic homeostasis, were strongly predictive of BA. The metabolite subfamily of glutathione was one of the strongest predictors of BA for both men and women (Supplementary Figure 3).
Multiple proteomic markers of inflammation were associated with BA, including chemokine C-X-C motif ligand 9 (CXCL9), interleukin 17D (IL17D), and growth/differentiation factor 15 (GDF15); additionally, the lymphocyte produced lymphotoxin alpha (LTA) was a negative predictor of BA in men, but not in women. Inflammation plays a crucial role in BA prediction in the clinical labs as well, where monocyte count was a strong positive predictor of BA and lymphocytes were a strong negative predictor.
Several environmental pollutants, including the heavy metals lead and mercury, were identified as strong predictors of BA. Within the metabolomics, the bioaccumulated toxin perfluorooctanesulfonic acid (PFOS) emerged as the second strongest positive predictor. The related metabolite, perfluorooctanoic acid (PFOA) was a strong positive predictor in men but not in women (Figure 3).
Sex steroid hormones dominated the calculation of BA in metabolites for men and women. Dehydroepiandrosterone (DHEA-S) and its direct metabolite androstenediol monosulfate were strong negative predictors with the stress-related hormone vanillylmandelic acid (VMA) being a strong positive predictor. Similar to PFOA and LTA, several other analytes demonstrated sex-specific differences (Supplementary Table 4). Alkaline phosphatase (ALP) proved to be a strong positive predictor in women but not in men (Figure 3). In contrast to ALP, creatine metabolites emerged as an important subfamily for calculation of BA in men, but not women (Supplementary Figure 3). Within the creatine metabolite subfamily, creatinine was one of the stronger negative predictors in men. Several elements of the immune system also showed sex-specific differences in calculating BA, including Spondin 2 (SPON2), which was a negative predictor in women, but not in men. Macrophage receptor with collagenous structure (MARCO) was a positive predictor in women, but a negative predictor in men. IL-16 was a positive predictor in women, but a negative predictor in men. The intestinal mucosa secreted protein trefoil factor 3 (TFF3) was a positive predictor in men, but a negative predictor in women.
Discussion
The BA measure was generated by integrating and comparing diverse data types, including clinical labs, proteomics, metabolomics, and genetics. The key findings of this paper are as follows (1). Higher ΔAge was shown to be associated with lifetime prevalence of common disease conditions, and BA was seen to decrease over time after joining a wellness program, supporting the hypothesis that BA is reflective of increasing or decreasing health as commonly understood (2). The degree of plasticity in BA is dependent on several factors such as sex, current health status, and CA (3). Blood factors corresponding to metabolic health, inflammation, and bioaccumulation of toxins were found to be the most strongly related to BA across data types (4). Men and women showed distinct differences in the features most relevant to the determination of BA, especially those related to aspects of sex-specific physiology, such as bone density, muscle mass, immune system function, and sex-related metabolism of environmental pollutants (5). BA was affected by the data type used in their determination, and different BAs can thus be derived from different data sources.
The complexity and variability of the aging process justify the development of system-level predictive and analytical models to describe it, with the ultimate goal of maintaining healthy aging and improving the extent and quality of healthspan through actionable lifestyle, environmental, and pharmaceutical interventions. Following an unscreened sample enabled the observation of health-related changes across the spectrum of commonly observed health conditions. The highest ΔAges, perhaps indicating poor wellness relative to CA, are in the T2D subpopulation (+6 years) which is consistent with studies observing 5–9 years shortened life expectancy with T2D (21). Average ΔAge was higher among participants self-reporting multiple types of current or past health conditions. This finding does not suggest that BA is a useful diagnostic for any specific disease, but instead that ΔAge maps consistently to the concept of general wellness, where every disease condition in which a statistically significant association was discovered (or even a lower-threshold trend observed) was in the direction of increasing ΔAge. Stationary or negative BA trajectories over time, after initiation of a wellness program, was consistent with the potential utility of BA as an aggregate marker (metric) of increasing wellness. In general, the all-analyte predictions of BA did not increase or decrease over time in this sample on average, despite increasing CA. Further study is required to determine the persistence of these effects or efficacy relative to other interventions.
Stratified analyses highlight differences between groups in response to their engagement in a wellness program. Both men and women experienced slowed BA on average, but the effect was stronger in women with their BA decreasing over time, while men maintained their BA. On average, the youngest participants (18–29) tended to show more ability to reduce their BA, while older participants decreased their ΔAge but maintained their initial BA. Importantly, a dose-dependent response was not observed, that is, participants over 29 have roughly similar trends. Participants tended to maintain a high degree of consistency in ΔAge over time for all data types.
Of interest is baseline health status. Participants with high ΔAge experienced a greater decline in BA over time in the program, which may be expected, given that less healthy participants had more actionable “wellness targets” to work on. Diminishing returns were also observed as those with extremely low baseline BA relative to CA had a slope of approximately the expected standard rate of BA (though confidence intervals were wide). While this seems expected from a biological perspective, the direction and magnitude of these trends are consistent with regression-to-the-mean effects, especially at the most extreme strata (ie, > 10 years |ΔAge|). An independent control group, not undergoing wellness coaching, would be required to differentiate these two effects regression to the mean and improvement from the wellness program.
Metabolic health, inflammation, and bioaccumulation of toxins represent dominant themes under our BA models across data types. The importance of metabolic health is well supported in aging literature, and a major concern in the developed world with nearly 40% of Americans expected to develop T2D in their lifetime and diagnosed diabetes patients accounting for one in four health care dollars in the United States in 2017 (22,23). The substantial effect of HbA1c, where 1 SD increase corresponded to a roughly 4-year increase in BA, partially explained the considerable effect on BA observed in participants that self-reported T2D. Adiponectin and AgRP are involved in the regulation of appetite and energy balance, with their levels in the blood rising in response to fasting and CR (24,25). Interestingly, adiponectin was a positive predictor of BA in our models, despite its aforementioned beneficial role in metabolic regulation. This is consistent with the proposed “adiponectin paradox,” where despite its beneficial role throughout the life span, increased circulating adiponectin levels in elderly populations are associated with a higher risk of mortality (26). The purported health benefits of CR are, in part, attributed to its ability to slow down metabolic decline and decrease oxidative stress. Consistently, strong beneficial effects from the anti-oxidant glutathione subfamily observed in the metabolites are consistent with these inter-relationships. Chronic inflammation is a common risk factor in many age-associated diseases, including heart disease, depression, cancer, osteoarthritis, and diabetes (27,28). Concordantly, changes in immune activity as people age were reflected in BA (5,29). CXCL9, a strong positive predictor of BA, is involved in the chemo-attraction of T cells and NK cells and has been demonstrated as a biomarker for the development of heart failure (30,31). CXCL9 and GDF15 were shown to explain significant variability in arterial stiffness and myocardial relaxation (32). The negative association of LTA with BA is aligned with its broad anti-tumor active, via multiple pathways, including the recruitment of NK cells (33). Bioaccumulation of toxins is known to be detrimental to human health, especially in Alzheimer’s disease, and a growing concern as people age (34–37). Several environmental pollutants, including the heavy metals lead and mercury, were identified as strong positive predictors of BA.
While most of the strongest predictors of BA were shared, sex-specific analyte contributions illuminate some differences in the biological aging process. For example, ALP was a strong positive predictor of BA in women, but not in men (Figure 3). Circulating ALP levels are commonly used as a marker for liver or bone disease, as total ALP consists mainly of bone and liver-derived isoforms. Particularly relevant to bone, increase in total and bone-specific ALP levels has been associated with increased rates of bone turnover (38,39). Given postmenopausal women experience higher bone turnover rate and accelerated bone mineral density loss with age compared to men, the difference in the effect of ALP on BA between men and women may result from sex-specific differences in bone physiology across the life span (40).
In contrast to ALP, creatine metabolites emerged as a notable subfamily for BA estimation in men, but not in women (Supplementary Figure 3). Within the creatine metabolite subfamily, creatinine was one of the stronger negative predictors for men. While creatinine build-up can be an indicator of reduced kidney function, it is also commonly used as a surrogate marker for muscle mass (41,42). This difference may reflect age-related muscle loss (sarcopenia) that is generally more pronounced in males than females (43). PFOA was also a strong positive predictor of BA in men only. Kinetic studies suggest sex differences in the excretion of PFO metabolites, which may in part explain the observed effects (36,44). Additionally, animal studies have shown that higher testosterone levels increase the rate of elimination of PFOA (45). Decreasing testosterone levels as men age or with obesity may partially explain the predictive capacity of PFOA levels in men but not in women.
Particularly intriguing is the fact that different data types illuminate different facets of wellness (Supplementary Figure 2), even though each data type was independently effective at estimating CA (Figure 1 and Supplementary Table 2). While each data type provides rich information about an individual’s biological state, the view into that state is inextricably affected by the modality of those measures. It has previously been demonstrated that different omics profiles of the same individuals do not cluster together (46). Data type–dependent differences among associations between ΔAge and some health conditions were observed (Table 2). For instance, ΔAge estimates derived from proteomics were associated with coronary artery disease, while estimates from the other data types had CIs showing little effect. While this association was not significant after FDR correction (unadjusted p = .004), the protein panels used were heavily focused on inflammation and cardiovascular disease, and so this result is not surprising. Determining which data types are most appropriate for certain diseases may help create condition-specific calculations of BA, and lead to greater precision based on an individual’s specific health concerns and history. This study argues that a fuller picture of an individual’s health emerges by incorporating multiple views of aging systems. As costs decrease over the next 10–15 years, expanding the protein, clinical chemistry, and metabolites panels to the largest extent reasonable will enable each of the different analyte classes to reflect in the broadest possible manner the “integrated” aging process.
This study confirms previously identified biomarkers that also estimated BA. Eight of the 10 biomarkers identified in Levine (2012) are measured in the clinical labs, with 6 being top predictors in our clinical lab models (Figure 3 and Supplementary Table 4) (8). Creatinine was not directly a top predictor in the clinical labs, but the blood urea nitrogen and creatinine ratio was, and creatinine is one of the strongest predictors of BA in men in the metabolomics. Presence of a large number of inflammatory markers may explain why C-reactive protein does not emerge as a particularly strong predictor. Another study demonstrated GDF15 as a potent predictor of BA (5). These verifications reinforce the generalizability and relevance of these biomarkers to BA.
Strengths of this study include deep phenotyping, large cohort size, a broad age distribution (18–89+), and longitudinal measurement of participants actively improving their health through lifestyle changes. Limitations of this study include the lack of many aging-specific covariates (such as grip strength, balance, and cognition), the short duration of observation relative to earlier epidemiological studies, and the lack of uniformity of measures across all people and observations. As mentioned, since a suitable control population (individuals not enrolled in a wellness program) was not available, regression-to-the-mean effects in analyses stratified by baseline BA subgroups could not be ruled out, particularly those with the largest deviation (outside of ±10) of ΔAge away from zero. The lack of a control group additionally raises issues for interpretation of these results. Neither causality nor the relative efficacy of this program compared to other interventions can be determined. Data type–specific stratified analyses were often underpowered, yielding large CIs and inconsistent estimates. Nevertheless, these exploratory analyses demonstrate intriguing trends for future studies. This study focuses on the applicability of BA to the whole adult life span as a general measure of wellness by assessing through hundreds of blood analytes literally 100s of biological networks. The lack of uniformity of measured variables over time presents challenges in integration and analysis, which are inevitable in the process of utilizing real-world data. Interest in repurposing incidental measures, electronic medical records, patient-contributed data, and mining of public databases is high. Thus, developing flexible methods that robustly integrate existing data is a superior strategy to ignoring essential features of human health due to partial missingness.
One question raised by application of deep phenotyping to calculate BA is whether measuring these large sets of variables is justified. They are at the level of discovery—that is, you want to survey the largest possible set of analytes to discover those which have the dominant effects on BA. Once these are discovered, far more limited feature tests can likely be assembled to calculate BA. Notably, a perfect predictor of CA would be useless as a wellness marker, giving no more information than the individual’s birthday (47). The main point is whether deviations in prediction represent deviations from wellness states and the extent to which this measure is modifiable. Longitudinal, deep phenotyping of individuals allow us to fully realize the broad dimensionality of a given population—and they allow us to stratify the population based on personal data clouds of the individuals and not on averaged data from populations. Additionally, if BA or ΔAge were used as a summary metric for wellness, a drop in BA over time may encourage participants to persist with healthful behaviors in order to maintain their “healthy” progress and allow one to carry out individual N = 1 studies on interesting compounds to facilitate healthy aging with lower BA as a target measure. Thus, it is proposed that ΔAge can be a useful metric to facilitate healthy aging. While the population insights herein are robust, reducing the high variance in the metric, however, is clearly an important factor in how such a measure might be used in the future on an individual basis.
This study estimates BA measures from PD3 clouds as gross, aggregate measures of health and wellness, which are useful because they constitute the averaging of many different biological systems. Importantly, BA has the potential to serve as a metric that can be used to track progress towards healthy aging. The factors affecting BA represent acute and cumulative damage that occurs over an individual’s lifetime and are mostly actionable through lifestyle, environmental, and pharmaceutical intervention. BA measures may be positive or negative wellness markers that can be used in instances where an individual lacks any specific disease conditions but is still interested in increasing wellness and preventing disease. Additionally, as CA is used to determine risk categories for many prophylactic tests such as colonoscopies, prostate exams, and mammograms, so too might BA provide personalized guidance on the relevance of those tests. As health care moves its focus from treatment to prevention, this actionable, holistic, and easily interpretable metric of wellness can be a valuable tool.
Funding
This paper was published as part of a supplement sponsored and funded by AARP. The statements and opinions expressed herein by the authors are for information, debate, and discussion, and do not necessarily represent official policies of AARP.
Supplementary Material
Acknowledgments
The authors acknowledge important contributions to this project made by Scott Lundberg, James Yurkovich, and Matt Conomos. The authors also gratefully acknowledge the participants that consented to share their deidentified data for this study. J.C.E., N.D.P., and L. Hood conceived and directed the study. J.C.E., N.R., L. Heath, and T.W. performed the analyses and generated figures and tables. All authors drafted and revised the manuscript and aided in the interpretation of the results.
Conflict of Interest
At the time some of this research was conducted, L. Hood and N.D.P. were co-founders of Arivale (where these data come from) and held stock in the company. N.D.P. was on the board of directors; L. Hood was chair of, and G.S.O. a member of, Arivale’s scientific advisory board. A.T.M. and J.L. were employees and had stock options in the company, as did G.S.O. and J.C.E. Arivale is now closed and the authors declare no ongoing competing financial interests.
References
- 1. National Research Council. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, DC: National Academies Press; 2011. [PubMed] [Google Scholar]
- 2. Comfort A. Test-battery to measure ageing-rate in man. Lancet. 1969;2:1411–1414. doi: 10.1016/s0140-6736(69)90950-7 [DOI] [PubMed] [Google Scholar]
- 3. Baker GT III, Sprott RL. Biomarkers of aging. Exp Gerontol. 1988;23:223–239. doi: 10.1016/0531-5565(88)90025-3 [DOI] [PubMed] [Google Scholar]
- 4. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tanaka T, Biancotto A, Moaddel R, et al. ; CHI Consortium. Plasma proteomic signature of age in healthy humans. Aging Cell. 2018;17:e12799. doi: 10.1111/acel.12799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Z, Li L, Glicksberg BS, Israel A, Dudley JT, Ma’ayan A. Predicting age by mining electronic medical records with deep learning characterizes differences between chronological and physiological age. J Biomed Inform. 2017;76:59–68. doi: 10.1016/j.jbi.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klemera P, Doubal S. A new approach to the concept and computation of biological age. Mech Ageing Dev. 2006;127:240–248. doi: 10.1016/j.mad.2005.10.004 [DOI] [PubMed] [Google Scholar]
- 8. Levine ME. Modeling the rate of senescence: can estimated biological age predict mortality more accurately than chronological age? J Gerontol A Biol Sci Med Sci. 2013;68:667–674. doi: 10.1093/gerona/gls233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Belsky DW, Caspi A, Houts R, et al. . Quantification of biological aging in young adults. Proc Natl Acad Sci USA. 2015;112:E4104–E4110. doi: 10.1073/pnas.1506264112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mitnitski A, Howlett SE, Rockwood K. Heterogeneity of human aging and its assessment. J Gerontol A Biol Sci Med Sci. 2017;72:877–884. doi: 10.1093/gerona/glw089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murabito JM, Zhao Q, Larson MG, et al. . Measures of biologic age in a community sample predict mortality and age-related disease: the Framingham Offspring Study. J Gerontol A Biol Sci Med Sci. 2018;73:757–762. doi: 10.1093/gerona/glx144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin H, Lunetta KL, Zhao Q, et al. . Whole blood gene expression associated with clinical biological age. J Gerontol A Biol Sci Med Sci. 2019;74(1):81–88. doi:10.1093/gerona/gly164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Belsky DW, Huffman KM, Pieper CF, Shalev I, Kraus WE. Change in the rate of biological aging in response to caloric restriction: CALERIE Biobank analysis. J Gerontol A Biol Sci Med Sci. 2017;73:4–10. doi: 10.1093/gerona/glx096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen R, Mias GI, Li-Pook-Than J, et al. . Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Price ND, Magis AT, Earls JC, et al. . A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat Biotechnol. 2017;35:747–756. doi: 10.1038/nbt.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zubair N, Conomos MP, Hood L, et al. . Genetic predisposition impacts clinical changes in a lifestyle coaching program. Sci Rep. 2019;9:6805. doi: 10.1038/s41598-019-43058-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liang K-Y, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. doi:10.1093/biomet/73.1.13 [Google Scholar]
- 18. Centers for Disease Control and Prevention. Centers for Disease Control and Prevention Behavioral Risk Factor Surveillance System. Washington, DC: Center for Disease Control and Prevention; 2010. https://data.cdc.gov/api/views/hn4x-zwk7 [Google Scholar]
- 19. Barnett AG, van der Pols JC, Dobson AJ. Regression to the mean: what it is and how to deal with it. Int J Epidemiol. 2005;34:215–220. doi: 10.1093/ije/dyh299 [DOI] [PubMed] [Google Scholar]
- 20. Yu JH, Kim MS. Molecular mechanisms of appetite regulation. Diabetes Metab J. 2012;36:391–398. doi: 10.4093/dmj.2012.36.6.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leal J, Gray AM, Clarke PM. Development of life-expectancy tables for people with type 2 diabetes. Eur Heart J. 2009;30:834–839. doi: 10.1093/eurheartj/ehn567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. American Diabetes Association. Economic costs of diabetes in the U.S. in 2017. Diabetes Care. 2018;41:917–928. doi:10.2337/dci18-0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gregg EW, Zhuo X, Cheng YJ, Albright AL, Narayan KM, Thompson TJ. Trends in lifetime risk and years of life lost due to diabetes in the USA, 1985-2011: a modelling study. Lancet Diabetes Endocrinol. 2014;2:867–874. doi: 10.1016/S2213-8587(14)70161-5 [DOI] [PubMed] [Google Scholar]
- 24. Shimokawa I, Higami Y. Leptin signaling and aging: insight from caloric restriction. Mech Ageing Dev. 2001;122:1511–1519. doi: 10.1016/s0047-6374(01)00284-6 [DOI] [PubMed] [Google Scholar]
- 25. Shen CP, Wu KK, Shearman LP, et al. . Plasma agouti-related protein level: a possible correlation with fasted and fed states in humans and rats. J Neuroendocrinol. 2002;14:607–610. doi: 10.1046/j.1365-2826.2002.00825.x [DOI] [PubMed] [Google Scholar]
- 26. Poehls J, Wassel CL, Harris TB, et al. ; Health ABC Study. Association of adiponectin with mortality in older adults: the Health, Aging, and Body Composition Study. Diabetologia. 2009;52:591–595. doi: 10.1007/s00125-009-1261-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dregan A, Charlton J, Chowienczyk P, Gulliford MC. Chronic inflammatory disorders and risk of type 2 diabetes mellitus, coronary heart disease, and stroke: a population-based cohort study. Circulation. 2014;130:837–844. doi: 10.1161/CIRCULATIONAHA.114.009990 [DOI] [PubMed] [Google Scholar]
- 28. Kim YK, Na KS, Myint AM, Leonard BE. The role of pro-inflammatory cytokines in neuroinflammation, neurogenesis and the neuroendocrine system in major depression. Prog Neuropsychopharmacol Biol Psychiatry. 2016;64:277–284. doi: 10.1016/j.pnpbp.2015.06.008 [DOI] [PubMed] [Google Scholar]
- 29. Weiskopf D, Weinberger B, Grubeck-Loebenstein B. The aging of the immune system. Transpl Int. 2009;22:1041–1050. doi: 10.1111/j.1432-2277.2009.00927.x [DOI] [PubMed] [Google Scholar]
- 30. Altara R, Manca M, Hessel MH, et al. . CXCL10 is a circulating inflammatory marker in patients with advanced heart failure: a pilot study. J Cardiovasc Transl Res. 2016;9:302–314. doi: 10.1007/s12265-016-9703-3 [DOI] [PubMed] [Google Scholar]
- 31. Altara R, Gu YM, Struijker-Boudier HA, Thijs L, Staessen JA, Blankesteijn WM. Left ventricular dysfunction and CXCR3 ligands in hypertension: from animal experiments to a Population-Based Pilot Study. PLoS One. 2015;10:e0141394. doi: 10.1371/journal.pone.0141394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bouajila S, Moneghetti K, Kobayashi Y, et al. . Immune profile of healthy cardiovascular aging: insights from a population-based study and network modeling. J Am Coll Cardiol. 2018;71:A1657. doi:10.1016/S0735-1097(18)32198-3 [Google Scholar]
- 33. Takei K, Ikeda S, Arai T, Tanaka N, Muramatsu M, Sawabe M. Lymphotoxin-alpha polymorphisms and presence of cancer in 1,536 consecutive autopsy cases. BMC Cancer. 2008;8:235. doi: 10.1186/1471-2407-8-235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Papanikolaou NC, Hatzidaki EG, Belivanis S, Tzanakakis GN, Tsatsakis AM. Lead toxicity update. a brief review. Med Sci Monit. 2005;11:RA329–RA336. PMID: 16192916. [PubMed] [Google Scholar]
- 35. Houston MC. Role of mercury toxicity in hypertension, cardiovascular disease, and stroke. J Clin Hypertens (Greenwich). 2011;13:621–627. doi: 10.1111/j.1751-7176.2011.00489.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Donohue JMaD TM, Wambaugh J.. Health Effects Support Document for Perfluorooctanoic Acid. Washington, DC: Environmental Protection Agency; 2016. EPA Document ID: 822-R-16-003. [Google Scholar]
- 37. Genuis SJ, Kelln KL. Toxicant exposure and bioaccumulation: a common and potentially reversible cause of cognitive dysfunction and dementia. Behav Neurol. 2015;2015:620143. doi: 10.1155/2015/620143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mukaiyama K, Kamimura M, Uchiyama S, Ikegami S, Nakamura Y, Kato H. Elevation of serum alkaline phosphatase (ALP) level in postmenopausal women is caused by high bone turnover. Aging Clin Exp Res. 2015;27:413–418. doi: 10.1007/s40520-014-0296-x [DOI] [PubMed] [Google Scholar]
- 39. Ureña P, Hruby M, Ferreira A, Ang KS, de Vernejoul MC. Plasma total versus bone alkaline phosphatase as markers of bone turnover in hemodialysis patients. J Am Soc Nephrol. 1996;7:506–512. PMID: 8704118. [DOI] [PubMed] [Google Scholar]
- 40. Riggs BL, Melton Iii LJ III, Robb RA, et al. . Population-based study of age and sex differences in bone volumetric density, size, geometry, and structure at different skeletal sites. J Bone Miner Res. 2004;19:1945–1954. doi: 10.1359/JBMR.040916 [DOI] [PubMed] [Google Scholar]
- 41. Patel SS, Molnar MZ, Tayek JA, et al. . Serum creatinine as a marker of muscle mass in chronic kidney disease: results of a cross-sectional study and review of literature. J Cachexia Sarcopenia Muscle. 2013;4:19–29. doi: 10.1007/s13539-012-0079-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schutte JE, Longhurst JC, Gaffney FA, Bastian BC, Blomqvist CG. Total plasma creatinine: an accurate measure of total striated muscle mass. J Appl Physiol Respir Environ Exerc Physiol. 1981;51:762–766. doi: 10.1152/jappl.1981.51.3.762 [DOI] [PubMed] [Google Scholar]
- 43. Iannuzzi-Sucich M, Prestwood KM, Kenny AM. Prevalence of sarcopenia and predictors of skeletal muscle mass in healthy, older men and women. J Gerontol A Biol Sci Med Sci. 2002;57:M772–M777. doi: 10.1093/gerona/57.12.m772 [DOI] [PubMed] [Google Scholar]
- 44. Lee JJ, Schultz IR. Sex differences in the uptake and disposition of perfluorooctanoic acid in fathead minnows after oral dosing. Environ Sci Technol. 2010;44:491–496. doi: 10.1021/es901838y [DOI] [PubMed] [Google Scholar]
- 45. Kudo N, Kawashima Y. Toxicity and toxicokinetics of perfluorooctanoic acid in humans and animals. J Toxicol Sci. 2003;28:49–57. doi: 10.2131/jts.28.49 [DOI] [PubMed] [Google Scholar]
- 46. Gao LL, Bien J, Witten D. Are clusterings of multiple data views independent? Biostatistics; Oxford, England. 2019. doi:10.1093/biostatistics/kxz001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hertel J, Frenzel S, König J, et al. . The informative error: a framework for the construction of individualized phenotypes. Stat Methods Med Res. 2019;28:1427–1438. doi: 10.1177/0962280218759138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.