Abstract
Mammalian genomes are pervasively transcribed1,2 to produce thousands of long noncoding RNAs (lncRNAs)3,4. A few of these lncRNAs have been shown to recruit regulatory complexes through RNA-protein interactions to influence the expression of nearby genes5–7, and it has been suggested that many other lncRNAs similarly act as local regulators8,9. Such local functions could explain the observation that lncRNA expression is often correlated with the expression of nearby genes2,10,11. However, such correlations have been challenging to dissect12 and could alternatively result from processes that are not mediated by the lncRNA transcripts themselves. For example, some gene promoters have been proposed to have dual functions as enhancers13–16, and the process of transcription per se has been proposed to contribute to gene regulation by recruiting activating factors or remodeling nucleosomes10,17,18. Here we used genetic manipulations to dissect 12 genomic loci that produce lncRNAs and found that 5 of these loci influence the expression of a neighboring gene in cis. Surprisingly, none of these effects required the specific lncRNA transcripts themselves and instead involved general processes associated with their production, including enhancer-like activity of gene promoters, the process of transcription, and the splicing of the transcript. Importantly, such effects were not limited to lncRNA loci: we found that 4 of 6 protein-coding loci similarly influenced the expression of a neighbor. These results demonstrate that ‘crosstalk’ among neighboring genes is a prevalent phenomenon that can involve multiple mechanisms and cis regulatory signals, including a novel role for RNA splice sites. These mechanisms may explain the function and evolution of some genomic loci that produce lncRNAs and broadly contribute to the regulation of both coding and noncoding genes.
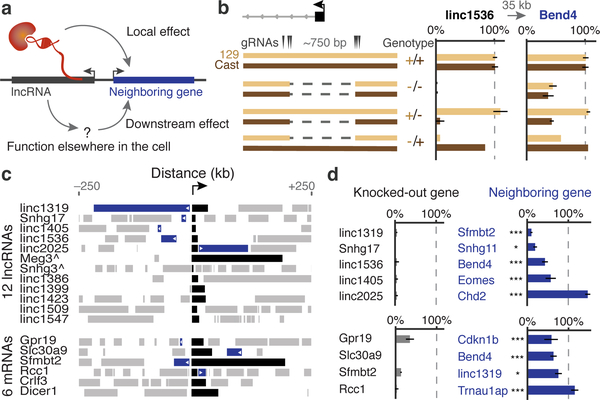
We analyzed 12 lncRNA loci whose RNA transcripts in mouse embryonic stem cells (mESCs) show preferential localization to the nucleus and span a range of abundance levels (Methods, Extended Data Fig. 1). For each locus, we looked for direct regulatory effects on local gene expression by using a genetic approach based on classical cis-trans tests (Fig. 1a, Note S1). Specifically, we generated clonal cell lines carrying heterozygous knockouts of the promoter (~600–1,000 bp deletions) (Fig. 1b) and compared the expression of nearby genes within 1 megabase on the cis and trans alleles (i.e., on the modified and unmodified homologous chromosomes in the same cells) (Note S2). Changes in neighboring gene expression that involve only the cis allele likely result from direct, local functions of the lncRNA locus, while changes that involve both the cis and trans alleles likely result as indirect, downstream consequences of the lncRNA acting elsewhere (Note S1). We performed genetic modifications in 129/Castaneus F1 hybrid mESCs that contain a polymorphic site every ~140 basepairs (bp), enabling us to distinguish the two alleles using RNA sequencing (Extended Data Fig. 2, Note S3).
Fig. 1. Many lncRNA and mRNA loci influence the expression of neighboring genes.
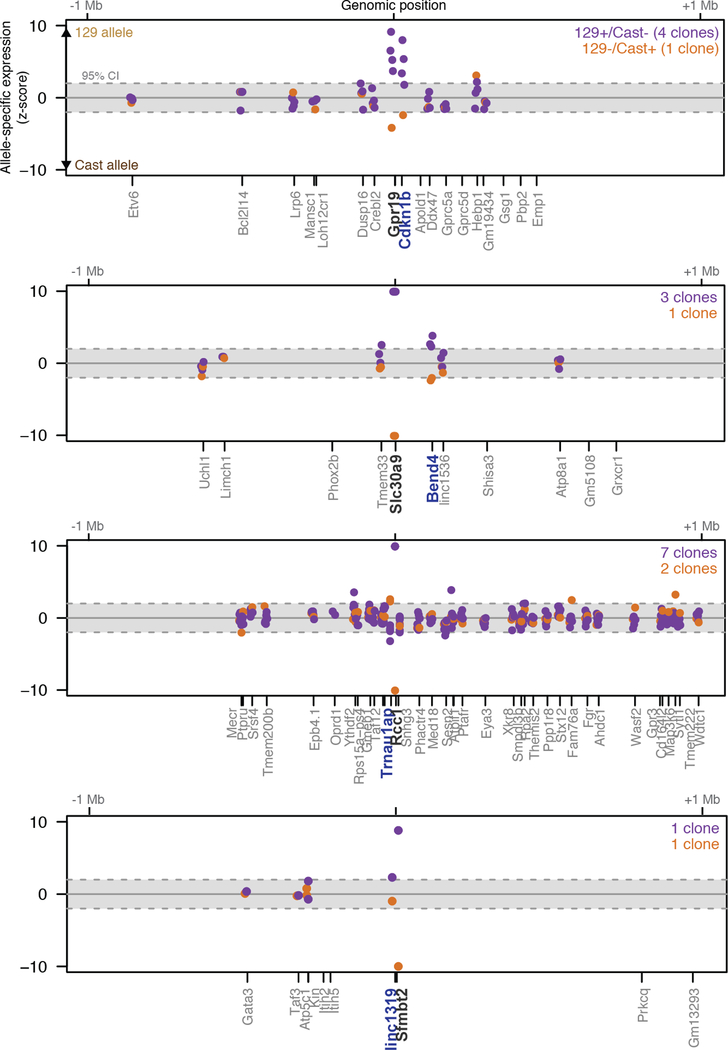
(a) Knocking out a promoter (black) could affect a neighboring gene (blue) directly (local) or indirectly (downstream). (b) Knockout of the linc1536 promoter. Left: genotypes. Right: allele-specific RNA expression for 129 and Castaneus (Cast) alleles normalized to 81 control clones (+/+). Error bars: 95% confidence interval (CI) for the mean (n ≥ 2 clones, see Table S1). (c) Gene neighborhoods oriented so each knocked-out gene (black) is transcribed in the positive direction. Blue neighboring genes show allele-specific changes in expression. ^see Note S3. (d) Average RNA expression on promoter knockout compared to wild-type alleles (n ≥ 2 alleles, see Table S1). *: FDR < 10%. ***: FDR < 0.1%.
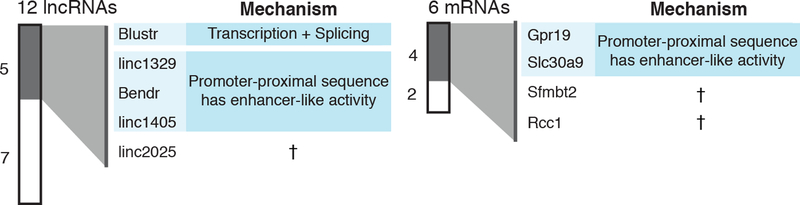
At 5 of these 12 lncRNA loci, promoter knockouts significantly affected the expression of a nearby gene in an allele-specific manner (false discovery rate <10%), including both activating and repressive effects (Fig. 1c,d, Note S4, Extended Data Fig. 3). For each locus, the affected gene was located immediately adjacent to, and within 5–71 kb of, the knocked-out promoter (Fig. 1c, Extended Data Fig. 4). This indicates that a substantial fraction of lncRNA loci influence the expression of a neighboring gene.
To test whether such effects were specific to lncRNA loci, we deleted the promoters of 6 protein-coding genes (Extended Data Fig. 1). Surprisingly, knockouts at 4 of these loci also affected the expression of a neighbor in cis (Fig. 1c,d, Extended Data Fig. 5). Thus, both noncoding and coding loci can directly influence local gene expression. These regulatory connections likely contribute to the observed correlations in the expression of neighboring genes, which have been reported both for lncRNAs and for mRNAs10,11,19,20.
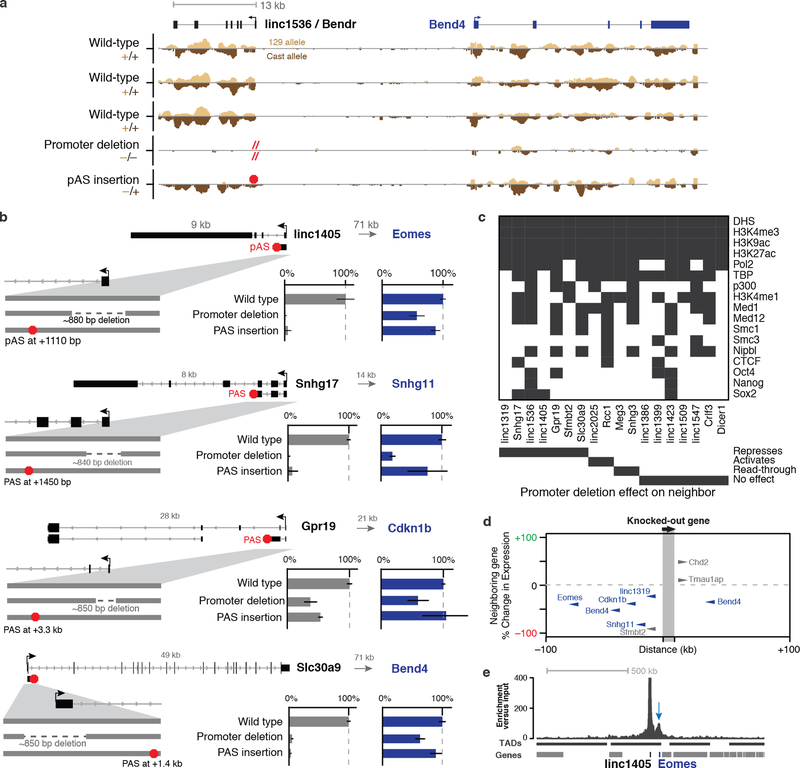
Because in these experiments we deleted gene promoters, the mechanisms underlying such cis effects could in principle involve (i) DNA regulatory elements in gene promoters13–16; (ii) the process of transcription10,17,18; or (iii) the RNA transcripts themselves5–9 (Extended Data Fig. 6a). To begin to distinguish among these possible mechanisms, we inserted early polyadenylation signals (pAS), 0.5–3 kb downstream of each transcription start site (TSS), that eliminated the production of most of the RNA while leaving the promoter sequence intact (Fig. 2, Extended Data Fig. 6b,c, see Methods). We examined 4 lncRNA loci and 2 mRNA loci where promoter deletion affected the expression of a neighboring gene (see Note S5).
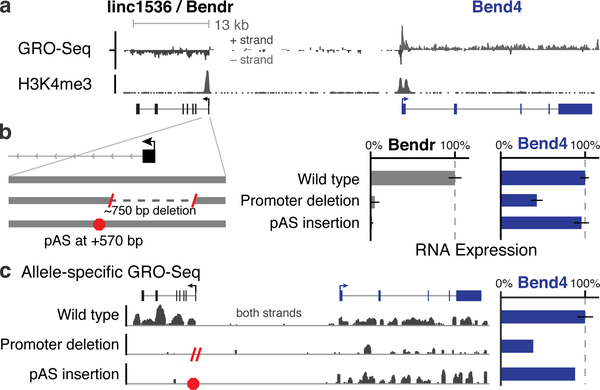
Fig. 2. Enhancer-like function of the Bendr promoter.
(a) Transcriptionally engaged RNA polymerase (GRO-Seq) and H3K4me3 occupancy (ChIP-Seq). (b) p(A)+ RNA expression upon deleting the Bendr promoter or inserting a pAS on modified versus unmodified alleles. Error bars: 95% CI for the mean (n ≥ 2 alleles, see Table S1). (c) Allele-specific GRO-seq signal for clones carrying the indicated modifications. Both clones are modified on the 129 allele, and only reads specifically mapping that allele are shown. Y-axis: normalized read count. Bar plot quantifies signal at Bend4, including 7 additional wild-type controls not shown on left.
As one example, we describe the linc1536 locus, hereafter called Bendr (Bend4-regulating Effects Not Dependent on the RNA, Fig. 2a). Whereas deleting the Bendr promoter reduced the expression of the adjacent Bend4 gene by 57%, inserting a pAS into the first intron of Bendr (~570 bp downstream of the TSS in this ~13-kb locus) had no effect on Bend4 expression despite eliminating the spliced Bendr RNA (Fig. 2b,c). Furthermore, global run-on sequencing (GRO-seq) did not detect any transcriptionally engaged polymerase upstream of the pAS insertion (Fig. 2c, Extended Data Fig. 7a) — perhaps because the pAS prevents RNA splicing, which may dramatically reduce transcriptional activity in the modified locus21,22. Therefore, cis activation of Bend4 requires neither the mature Bendr RNA transcript nor significant Bendr transcription. Instead, this effect is likely mediated by DNA regulatory elements in the ~750 bp knocked-out promoter-proximal region.
In total, at 5 of the 6 loci examined with pAS insertions (including 3 lncRNAs and 2 mRNAs), DNA regulatory elements in the promoter-proximal sequences appeared to be responsible for activating a neighboring gene (Extended Data Fig. 7b). Although the promoters in these loci would not be classified as “enhancers” based on H3K4me3/H3K4me1 ratios23, they are bound by mESC transcription factors (Extended Data Fig. 7c) and are located in close proximity to their neighboring target genes (Fig. 1c, Extended Data Fig. 7d,e), suggesting that these promoters may affect local gene expression through mechanisms similar or identical to enhancers13,24,25.
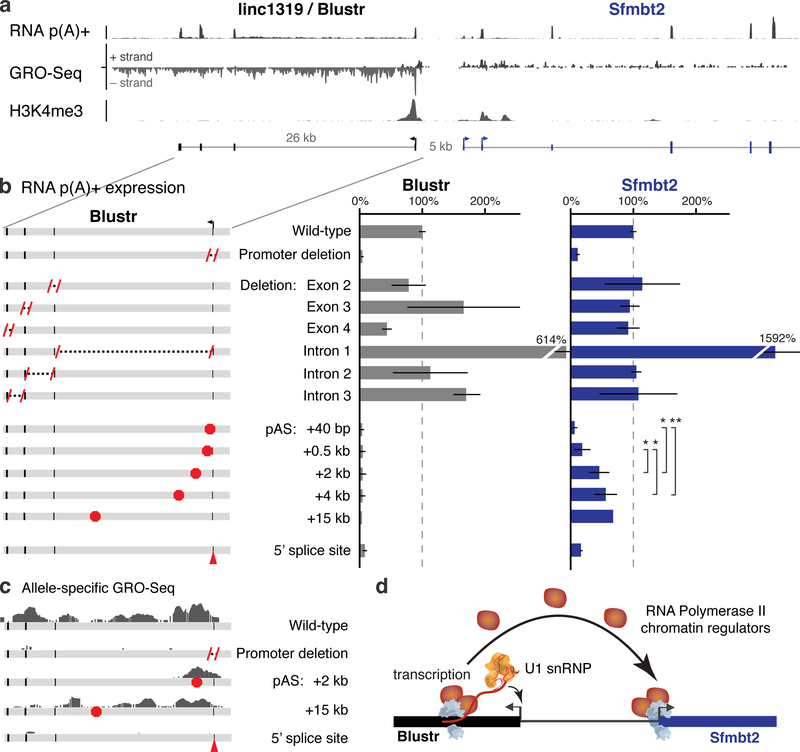
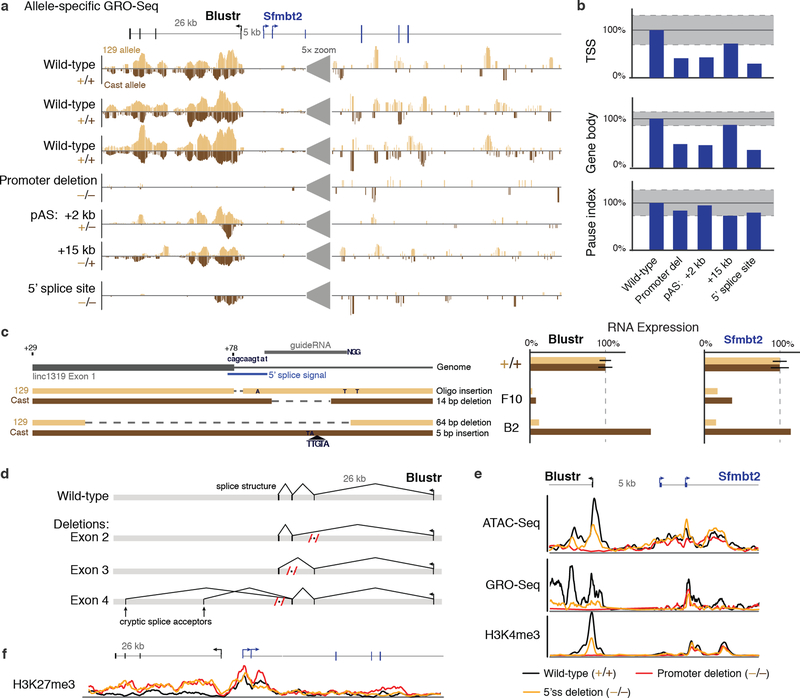
We also identified one locus, linc1319 (renamed Blustr: Bivalent Locus (Sfmbt2) is Up-regulated by the Splicing and Transcription of an RNA), where both promoter deletions and pAS insertions substantially reduced the expression of a neighboring gene, Sfmbt2, located 5 kb upstream (Fig. 3a). To dissect the regulatory mechanism, we tested whether the activation of Sfmbt2 is mediated by (i) a sequence-specific function of the Blustr transcript or (ii) the process of transcription (by which we mean one or more sequence-independent functions associated with transcription, such as changes in chromatin state or recruitment of co-factors). To test the first possibility, we knocked out each of the 3 downstream exons and 3 introns. None of these deletions impaired Sfmbt2 activation (Fig. 3b, Note S6), suggesting that the activation of Sfmbt2 does not require unique sequences or structures in the Blustr transcript itself. To test the second possibility, we engineered pAS insertions at five different locations in the first exon or intron (+40 bp to +15 kb downstream of the TSS) and found that increasing the length of the Blustr transcribed region led to increased activation of Sfmbt2 (Fig. 3b, Extended Data Fig. 8a,b). We note that changing the length of the transcribed region affected the total amount of engaged polymerase in the Blustr locus (Fig. 3c). Thus, Sfmbt2 activation responds to changes in the length/amount of transcriptional activity in the Blustr locus but does not appear to require specific sequence elements in the mature Blustr transcript (Note S7).
Fig. 3. Transcription and splicing of Blustr activates Sfmbt2 expression.
(a) p(A)+ RNA-seq, GRO-seq, and H3K4me3 ChIP-Seq in the Blustr locus. Sfmbt2 has two alternative TSSs. (b) p(A)+ RNA expression on knocked-out alleles compared to controls (arrows). Error bars: 95% CI for the mean (n ≥ 2 alleles, except for pAS +15 kb where n = 1, see Table S1). Sfmbt2 pAS comparisons: two-sided t-test P < 0.05 (*) or < 0.01 (**). (c) Allele-specific GRO-seq signal for clones carrying indicated modifications. Only reads mapping to the modified allele are shown (Cast for pAS +2 kb; 129 for others). (d) Model for how transcription in the Blustr locus activates Sfmbt2.
Because promoter-proximal splice sites and the process of splicing can enhance transcription — in some cases by as much as 100-fold21,22 — we tested whether the splicing of Blustr is involved in Sfmbt2 activation. Upon deleting the 5’ splice site of the first intron of Blustr (Extended Data Fig. 8c), we observed a 94% reduction in Blustr transcription (as assayed by GRO-seq), a 92% reduction in the levels of the mature Blustr transcript, and an 85% reduction in Sfmbt2 expression (Fig. 3b,c, Extended Data Fig. 8a,b), demonstrating that the first 5’ splice site of Blustr has a critical role in activating Blustr and Sfmbt2 transcription. In contrast, downstream splice sites were dispensable: upon deleting downstream Blustr exons, splicing skipped over the removed exon to the next available 3’ splice site (Extended Data Fig. 8d) and Sfmbt2 expression was unaffected (Fig. 3b).
Together, these data demonstrate that the 5’ splice site and the process of transcription in the Blustr locus are important for its ability to regulate Sfmbt2. This indicates that the Blustr RNA is in fact required for Sfmbt2 activation (splicing involves direct interactions between the spliceosome and the nascent transcript), although this mechanism does not appear to depend on the precise sequence of the RNA beyond the presence of initial splice signals. One possibility is that the 5’ splice site promotes transcriptional activity in the Blustr locus, which in turn recruits components of the transcriptional machinery that act on the nearby Sfmbt2 promoter (Fig. 3d, Note S7). Consistent with this model, altering transcription or splicing in the Blustr locus led to changes in chromatin state at the Sfmbt2 promoter (including reductions in H3K4me3 and spreading of H3K27me3) and reduced occupancy of engaged RNA polymerase in the paused position just downstream of the Sfmbt2 TSS (Extended Data Fig. 8b,e,f). Thus, changes in Blustr transcription and splicing may affect Sfmbt2 expression in part by altering chromatin state and RNA polymerase occupancy at the Sfmbt2 promoter (Fig. 3d, Note S7).
In summary, genetic dissection of 12 lncRNA loci and 6 mRNA loci found that 9 loci (50%) regulate the expression of a neighboring gene (Extended Data Fig. 9). In most of these loci, including Bendr, local effects are mediated by enhancer-like functions of DNA elements in promoters. In one locus, Blustr, the processes of transcription and splicing also contribute to cis regulatory functions, perhaps by increasing the local concentration of transcription-associated factors. We did not identify any lncRNA loci in which local effects are mediated by sequence-specific functions of the lncRNA transcript. Because there exist thousands of other loci that fit our selection criteria, we expect that similar mechanisms broadly contribute to gene regulation in many loci (Note S8).
The frequent ‘crosstalk’ between neighboring genes observed in our study indicates that gene loci can encode multiple independent categories of functions. Category I involves functions of the RNA product: mRNAs template protein synthesis, and some noncoding transcripts (e.g., XIST) act as functional lncRNAs. Category II involves the effects of transcription-related processes — including mechanisms mediated by promoters, transcription, and splicing — on the regulation of other nearby genes.
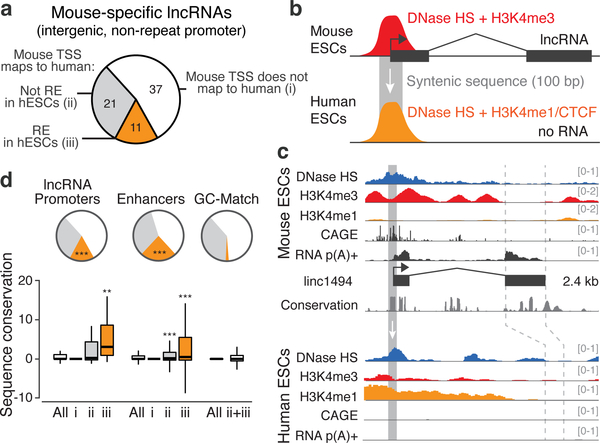
The fact that many lncRNA loci have category II functions does not necessarily mean that they do not also have category I functions, and we note that our experiments do not rule out the possibility that the lncRNAs dissected in this study have RNA-mediated functions other than on local gene regulation. However, the prevalence of category II functions suggests a model for the evolutionary origins of some lncRNAs. In loci where a promoter acts as an enhancer, RNA transcripts may arise as non-functional byproducts16. In loci where co-transcriptional processes have cis regulatory functions, the nascent transcripts might contribute through mechanisms like splicing that require little RNA-sequence specificity. These possibilities are particularly intriguing in light of the patterns of evolutionary conservation of lncRNA loci26–28. For example, although most lncRNA transcripts expressed in mESCs are not conserved (no RNA detected in syntenic loci in other mammals, see Methods), the promoters in some of these loci correspond to conserved DNA sequences that have an enhancer chromatin signature in human ESCs (Fig. 4, Extended Data Fig. 10, Note S9). These sequences may have conserved functional roles as cis regulatory elements, rather than as lncRNA promoters. Thus, mechanisms associated with cis functions by promoters, transcription, and/or RNA processing may contribute to the functions and evolution of an important subset of noncoding loci in mammalian genomes (Extended Data Fig. 10c).
Fig. 4. Evolutionary conservation of mESC lncRNAs and their promoters.
(a) Classification of a subset of lncRNAs expressed in mESCs (see Note S9, Methods). (b) 11 have promoters whose syntenic sequence corresponds to putative DNA regulatory elements (REs) marked by DNase I hypersensitivity (HS) in human ESCs. (c) Example: linc1494. (d) Enhancers and lncRNA promoters are significantly enriched for corresponding to human REs (pie chart, ***: P < 10−10, Chi-squared test versus GC-matched random regions) and show elevated sequence conservation compared to GC-matched regions (bar plot, **: P < 0.01, ***: P < 0.001, Mann-Whitney test versus ii+iii).
Beyond the implications for lncRNAs, these cis regulatory connections between neighboring genes occur in both protein-coding and noncoding loci and thus appear to represent a fundamental property of mammalian gene regulatory networks. The properties of these cis regulatory connections — including mechanisms for specificity and the potential for cooperative dynamics of gene activation — represent key areas for future investigation.
Methods
Cell lines and cell culture.
F1 hybrid 129/Castaneus female mouse embryonic stem cells (gift from Kathrin Plath) were cultured in serum-free N2B27-based medium (250 ml Neurobasal media (Gibco), 250 ml DMEM/F12 (Gibco), 5 ml 100× N2 supplement (Gibco), 5 ml 50× B27 supplement (Gibco), 5 ml 200 mM L-Glutamine (Gibco), 3.6 μl 2-mercaptoethanol, 50 μg human leukemia initiation factor (5 × 105 units, EMD Millipore), 7.4 μg Progesterone, 10 mg Bovine Insulin (Sigma), 350 μl 7.5% BSA Fraction V (Gibco), supplemented with MEK inhibitor PD0325901 (50 μl 10 mM, SelleckChem), and GSK3b inhibitor CHIR99021 (150 μl 10 mM, SelleckChem)). Prior to plating cells, tissue culture dishes were pretreated with PBS + 0.2% gelatin (Sigma) and 1.75 μg/ml laminin (Sigma) for 2–10 hours at 37°C. At each passage, cells were trypsinized for 3–5 minutes in TVP Solution (0.025% trypsin, 1% Chicken Serum (Sigma), and 1 mM EDTA in PBS pH 7.4) at room temperature. Cells tested negative for mycoplasma contamination and were authenticated by comparing polymorphisms to 129S1 and Castaneus genomes.
Cellular fractionation.
To estimate the relative abundance of lncRNAs in different cellular compartments and to characterize transcriptional activity in Blustr knockouts, we performed cellular fractionation to isolate chromatin-associated, soluble nuclear, and cytoplasmic fractions essentially as described29. Briefly, we first lysed 5 million cells in 200 μl cold cell lysis buffer (10 mM Tris-HCl pH 7.5, 0.05% IGEPAL CA-630, 150 mM NaCl), incubating on ice for 5 minutes. We layered the cell lysate over 2.5 volumes of chilled sucrose cushion (24% sucrose in cell lysis buffer) and centrifuged at 15,000 × g for 10 minutes. The supernatant from this spin became the cytoplasmic fraction. After washing the pellet of nuclei with PBS (pH 7.5) + 1 mM EDTA, we resuspended the pellet in 100 μl of cold glycerol buffer (20 mM Tris-HCl pH 7.5, 75 mM NaCl, 0.5 mM EDTA, 0.85 mM DTT, 0.125 mM PMSF, 50% glycerol) by gently flicking the tube. We added 100 μl of cold nuclei lysis buffer (10 mM HEPES pH 7.5, 1 mM DTT, 7.5 MgCl2, 0.2 mM EDTA, 0.3 M NaCl, 1 M urea, 1% IGEPAL CA-630), then vortexed for four seconds. After 2 minutes on ice, we spun the nuclear lysate at 15,000 × g for 2 minutes. This supernatant was collected as the soluble nuclear (nucleoplasm) fraction. We rinsed the remaining pellet (chromatin fraction) in PBS + 1 mM EDTA, then resuspended the chromatin in 300 μl chromatin DNase buffer (20 mM Tris-HCl pH 7.5, 50 mM KCl, 4 mM MgCl2, 0.5 mM CaCl2, 2 mM TCEP, 0.5 mM PMSF, 0.4% sodium deoxycholate, 1% IGEPAL CA-630, 0.1% N-lauroylsarcosine) plus 15 μl murine RNase inhibitor (NEB) and 30 μl TURBO DNase (Ambion). The DNase digestion proceeded for 20 minutes at 37°C and was halted by adding 10 mM EDTA and 5 mM EGTA. Protein was digested with proteinase K for 1 hour at 37°C. RNA was isolated using Zymo RNA Concentrator-25 columns (two columns for the cytoplasmic fraction). With this method, nuclear-associated endoplasmic reticulum is known to fractionate with the nucleoplasm 29, and we observed that nucleolar RNAs fractionated with chromatin (data not shown). From each cellular fraction, we sequenced total RNA and polyadenylated RNA (selected using oligo d(T)25 magnetic beads, NEB) using a strand-specific RNA-sequencing protocol for Illumina instruments described previously30.
Selection criteria for knocked-out lncRNAs.
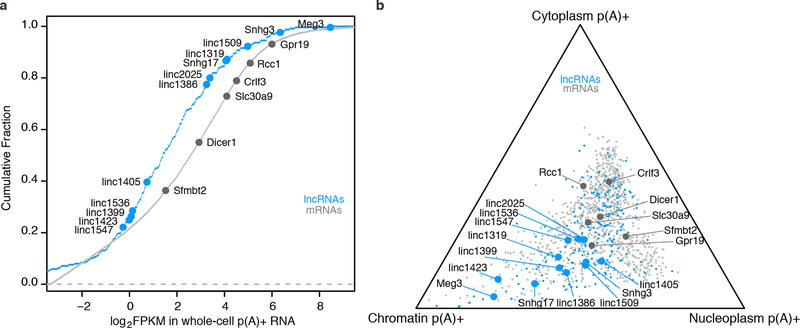
We selected lncRNA loci initially identified and defined by a chromatin signature of H3K4me3 at promoters and H3K36me3 through gene bodies3. We further required that lncRNAs selected for knockout analysis have TSSs, as defined by capped analysis of gene expression (CAGE), located >5 kb from other genes (for epigenomic annotation of each locus, see http://pubs.broadinstitute.org/neighboring-genes/). To prioritize intergenic lncRNA loci that may regulate local gene expression, we focused on lncRNAs that have subcellular localization biased toward the nucleus versus the cytoplasm (Extended Data Fig. 1). We performed cellular fractionation experiments in V6.5 male mESCs as described above and sequenced RNA from chromatin-associated, soluble nuclear, and cytoplasmic fractions (GEO Accession GSE80262). We calculated a relative nuclear-to-cytoplasmic ratio (chromatin RPKM + soluble nuclear RPKM divided by cytoplasmic RPKM) and focused on lncRNAs with ratios above the median (1.5): these lncRNAs are preferentially localized to the nucleus compared to other lncRNAs and mRNAs. We selected nuclear-biased lncRNAs that span a range of abundance levels (Extended Data Fig. 1). We also included some lncRNAs that are conserved across mammalian evolution (Snhg3, Snhg17, Meg3, and linc2025).
Selection criteria for knocked out mRNAs.
We selected 6 mRNAs for promoter knockouts based on the following criteria. We knocked out 2 mRNAs that are moderately expressed and are not expected to be essential for mESC growth (Dicer1 and Crlf3). We knocked out 2 mRNAs that are located adjacent to knocked-out lncRNAs (Sfmbt2 and Rcc1), in order to look for reciprocal regulatory effects between the lncRNA and the affected mRNA. We knocked out 2 mRNAs that are located adjacent to a gene that is itself adjacent to a lncRNA (Gpr19 and Slc30a9), in order to determine whether affected genes are specifically responsive to lncRNA promoters or are generally responsive to other promoters in the locus. Similar to the lncRNAs selected, the TSSs of these selected mRNAs are located >5 kb from other genes.
CRISPR sgRNA design.
To design single-guide RNAs (sgRNAs), we built custom software to calculate a specificity score (based on potential off-target sites using the algorithm described at crispr.mit.edu31) and an efficacy score (based on a sequence model for sgRNA efficiency as previously described32) for each 20-nt targeting sequence. We removed guides with specificity scores <20 or efficacy scores >0.7. To avoid T-rich sequences that result in premature termination of Pol III-mediated sgRNA transcription, we removed guides with more than 1 “T” in the 4 bases closest to the seed region, guides with more than 3 consecutive T’s, and guides with more than 8 T’s total. We removed guides with homopolymer stretches of 5 or more bases and guides with GC content <20% or >90%. We removed guides that overlapped a known 129/Castaneus SNP33. Within a given region, we typtically chose the three remaining guides with the highest specificity scores. The sequences of all sgRNAs used in this study are listed in Table S2.
Promoter deletion guide placement.
To knock out a lncRNA or mRNA promoter, we chose 2–3 sgRNAs located in windows 300–500 bp upstream and downstream of the TSS, leading to deletions of approximately 600–1000 bp surrounding the TSS. We adjusted the precise deletion boundaries outward if we could not successfully design guides in these regions (e.g., because they were located in repetitive sequences). We note that we often found that the “wild-type” alleles in heterozygous knockouts were affected by scars from repair of sgRNA double-stranded breaks. Accordingly, we adjusted the bounds if necessary to cut outside of the exons of the mRNA or lncRNA and thus avoid damaging the exonic sequences on the “wild-type” alleles in heterozygous knockouts. We note that the presence of these scars (and their lack of allele-specific effects on the expression of neighboring genes) indicate that the cis effects observed upon deleting promoters are not merely a result of CRISPR-mediated cutting and subsequent DNA repair.
Genetic deletions with CRISPR/Cas9.
To delete specific sequences, we co-transfected 100 ng of Cas9-expressing plasmids (“PX330-NoGuide”), 300 ng of a pool of sgRNA-expressing plasmids (“pZB-Sg3”), and 100 ng of a plasmid expressing EGFP and a puromycin selectable marker from a CAG promoter (pS-pp7-GFPiP). To create PX330-NoGuide, we modified PX330 (gift from Feng Zhang, Addgene plasmid #4423034) to remove the sgRNA expression cassette. To generate pZB-Sg3, we cloned a human U6 promoter and optimized sgRNA scaffold sequence35 into a minimal vector with an ampicillin-selectable marker and a ColE1 replication origin. We transfected batches of 250,000 mouse embryonic stem cells using the Neon Transfection System (Invitrogen), using 1 pulse of 40 milliseconds at 1200 V and plated two batches of cells (500,000 total) into a 96-well plate in 200 μl media. As an internal control for each set of transfections, we performed a transfection using 4 guides with no predicted target sites in the mouse genome.
We verified efficient transfection by examining GFP expression after 24 hours. To select for transfected cells, we replaced the media 24 hours after transfection with 200 μl 2i + 1 μg/ml puromycin. One day later, we split the cells into a 10-cm plate with 8 ml of 0.5 μg/ml puromycin. One day later, we replaced the media with 10 ml of 2i with no puromycin. We allowed cells to grow for 7–8 days, replacing the media every 2–3 days. We hand-picked 88 individual colonies and 8 control colonies for each transfection in 5 μl media, added 20 μl of TVP for ~10–20 minutes at 37°C to dissociate the colonies, and then split the colonies into two identical plates. We grew the cells in these plates for 4–5 days. We harvested one of the plates for DNA and RNA extraction by removing most of the media and adding 3.5× volume Buffer RLT (Qiagen) and froze the other plate for later recovery in Freezing Media (2i media + 10% fetal bovine serum + 10% DMSO).
Genotyping by PCR and sequencing.
To genotype each promoter knockout, we extracted genomic DNA and performed PCR using primers spanning the deleted sequence. We genotyped each clone by running the PCR products on agarose gels and comparing PCR amplicon sizes to predicted wild-type and deletion band sizes. We confirmed the sequences of wild-type and deletion bands by Sanger sequencing or high-throughput sequencing through barcoded amplicon sequencing on an Illumina MiSeq (see Table S2). Where possible, we used known polymorphic sites from 129S1 and Castaneus genomes33 to determine the haplotype-resolved genotype of each clone. Based on the genotyping data, we nominated clones for RNA sequencing. We eliminated clones showing evidence of (i) polyclonal or subclonal mutations or (ii) complex mutations such as inversion or duplication of the genomic sequence between the sgRNAs. The sequences of all genotyping primers are listed in Table S2.
RNA sequencing libraries.
We generated RNA sequencing libraries as previously described30,36, with some modifications for high sample throughput. We isolated RNA from harvested mESCs using RNeasy 96 columns. We enriched for poly(A)+ RNA using oligo d(T)25 magnetic beads (NEB) and eluted in 18 μl H2O. We fragmented RNA to an average of ~150-nt by adding 2 μl Ambion Fragmentation Buffer and incubating at 70°C for exactly 2.5 minutes. After transferring quickly to ice, we added 40 μl of a master mix containing 12 μl 5× FNK Buffer (50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 0.6 mM CaCl2, 50 mM KCl, 10 mM DTT, 0.01% Triton X-100), 1 μL Murine RNase Inhibitor (NEB), 3 μL FastAP Thermosensitive Alkaline Phosphatase (Thermo Scientific), 3 μL T4 Polynucleotide Kinase (NEB), and 1 μL TURBO DNase (Life Technologies). We incubated this reaction for 37°C for 30 minutes, then cleaned the reaction with MyOne SILANE magnetic beads37 and eluted in 6 μl of H2O.
We proceeded with the library preparation as previously described30, with one additional modification. To simplify the library preparation for many samples, we added unique sample barcodes (8 nt) during the first adapter ligation36. We used 12 pools each with 4 barcodes in order to mitigate differences in the efficiency of ligation for different adapter sequences. Following the first adapter ligation, we pooled 12 samples together, including up to 9 clones corresponding to a single target gene as well as 3 control clones, during the first 70% ethanol wash of the SILANE-bead purification. We performed an extra SILANE purification using the same beads to remove excess adapter and then proceeded with reverse transcription.
Hybrid selection of RNA sequencing libraries.
To measure allele-specific expression for hundreds of genes in a cost-effective manner, we developed a hybrid selection strategy to enrich for allele-informative reads at target genes (Extended Data Fig. 2). We designed oligo pools to capture allele-informative sequences in the ~1600 RNAs located in the genome within 1 Mb of one of the knockout targets. These target RNAs were divided into two independent pools: #140820 and #141203. We used RefSeq RNA annotations for mRNAs and our custom annotations for most lncRNAs. We identified SNPs that would distinguish the 129S1 and Castaneus genomes33. We designed 120-bp capture oligos in the vicinity of each 129/Castaneus polymorphic site, tiling every 15 bp across either 600 bp (pool #140820) or 240 bp (pool #141203) centered on the SNP. We included probes targeting both alleles to minimize differences in capture efficiency between the two alleles. We filtered capture probe sequences as previously described37. We included up to 10 oligos per targeted RNA, duplicating probes where necessary to include the sequences corresponding to each allele. Empirically, this probe design strategy in combination with the protocol described below enabled assessing allele-specific expression for 84% (611 of 731) of the targeted expressed genes in mESCs (RPKM ≥ 2) at a sequencing depth of <5 million reads per sample. Target genes and oligos sequences for these pools are listed in Table S3.
We synthesized pools of 12,000 capture oligos using CustomArray technology. Oligos in each pool were flanked by unique primers (Left primer sequence: CTTCCTACGAGCAGTTTGCC; Right primer sequence: AGTTTACGCATTACGGGCAC). After one round of PCR to add a T7 promoter (GGATTCTAATACGACTCACTATAGGG), we generated biotinylated RNA probes as described previously38, adding in 20% Biotin-16-UTP (Roche) and 20% Biotin-14-CTP (Life Technologies) to the in vitro transcription reactions. We generated RNA probes targeting both strands by incorporating the T7 promoter into either side of the PCR product and performing two separate in vitro transcription reactions per oligo pool.
To capture the allele-informative regions, we pooled the final, barcoded RNA sequencing libraries from all samples in the batch and performed a modified version of solution hybrid selection39. We first combined 500 ng dsDNA library pool with 1 nmol of Illumina P5 and P7 primer mix in 21 μl total. We denatured this mix at 94°C for 10 minutes and transferred immediately to ice. We added 7.5 μl 20× SSPE, 0.5 μl Murine RNase Inhibitor (NEB), and 1 μl of 500 ng/μl biotinylated RNA probe, for a total volume of 30 μl. We set up at least two reactions per 10 libraries, including at least one reaction with each strand of probes. We incubated the hybridization reaction at 65°C for 24–48 hours. For each capture sample, we washed 30 μl Streptavidin C1 MyOne magnetic beads (Invitrogen) in 5× SSPE and aliquoted them into PCR tubes. After removing the wash from the beads, we added the hybridization reaction and mixed to resuspend the beads. We captured the biotinylated probes by shaking at 65°C for 20 minutes. We washed the beads twice in 150 μl Low Stringency Wash Buffer (1× SSPE, 0.1% SDS, 1% NP-40, 4 M urea) at 62°C for 3–4 minutes, and twice in 150 μl High Stringency Wash Buffer (0.1× SSPE, 0.1% SDS, 1% NP-40, 4 M urea). To elute, we removed the final wash and resuspended beads in 10 μl 100 mM NaOH and heated to 70°C for 10 minutes. To complete the elution, we added 1 μl 1 M acetic acid and 14 μl NLS Elution Buffer (20 mM Tris-HCl pH 7.5, 10 mM EDTA, 2% N-lauroylsarcosine, 2.5 mM TCEP) and heated to 94°C for 4 minutes. While hot, we placed samples on magnet, removed eluate, and then placed the eluate on ice for at least 30 seconds. We cleaned the eluates with 20 μl MyOne SILANE magnetic beads as described37, using 75 μl RLT and 61 μl 100% ethanol for the initial precipitation. We eluted in 23 μl H2O, and used this as input for a 50 μl NEBNext High Fidelity PCR reaction using 500 pmol each P5 and P7 Illumina primers (98°C for 30 s; 13 cycles of 98°C for 15 s, 68°C for 30 s, 72°C for 30s; 72°C for 2 minutes, 4°C hold). We cleaned the PCR reaction twice with 1× volume Agencourt Ampure XP magnetic beads and eluted in 20 μl H2O.
Allele-specific gene expression measurements from RNA sequencing.
We sequenced RNA libraries on an Illumina HiSeq 2500 (Read 1: 38 cycles; Read 2: 30 cycles; Index: 8 cycles). The first read includes the 8-nt barcode added during the first adapter ligation (see above). Following processing to separate samples based on the inline barcodes, we filtered out sequencing reads that aligned to highly abundant RNA transcripts, including ribosomal RNAs, snRNAs, and repetitive elements, as defined by RefSeq and RepeatMasker. A FASTA file containing these sequences is available at the Gene Expression Omnibus (GSE55914).
We developed a computational pipeline to estimate allele-specific expression from RNA-sequencing data. We created two separate reference files for the 129S1 and Castaneus haplotypes, starting with the mm9 genome build and layering on SNPs based on whole-genome sequencing of each of the two mouse strains33. We aligned RNA-sequencing data separately to each of the two haplotypes using Tophat (version 2.0.8). We combined the results of the two alignments using PySuspenders40, which identifies reads that map specifically to one or the other allele and splits them into separate BAM files. We discarded duplicate reads and reads with MAPQ < 30. After generating separate BAM files containing the reads mapping to each allele, we counted reads that mapped to each RefSeq transcript (including both spliced and unspliced isoforms) using Scripture41 and calculated “allelic expression ratios” for each gene (counts from 129 allele divided by total counts from both 129 and Castaneus alleles). The distribution of allelic expression ratios for all active genes in mESCs was centered on 0.5, indicating that on average each gene is expressed equally from the 129 and Castaneus alleles (Extended Data Fig. 2b). This indicates that there is not systematic bias in our mapping procedure toward one allele or the other.
RNA-seq data analysis.
We processed RNA-sequencing datasets in batches corresponding to sets of libraries made on the same day with the same hybrid selection probe pool. We removed samples with fewer than 100,000 non-repetitive, unique, allele-informative reads. For within-batch quality control, we performed hierarchical clustering on all samples by their allelic expression ratios and removed the 2–5% of outlier samples, which were largely comprised of clones that showed monoallelic expression from the X chromosome.
Assessment of gene knockout by expression analysis.
The PCR genotyping procedure described above provided putative genotypes for the cell clones. We confirmed the genotype of cells by analyzing the allele-specific expression of the knocked out gene in each clone. We required that clones show >80% reduction of expression of the knocked out gene on the appropriate allele in order to include the clone in downstream analysis. Incomplete reduction of expression in some cases appeared to result from use of alternative TSSs that were not included in the deleted sequence. In other cases, incomplete reduction of expression appeared to result from subclonal genetic mosaicism within the cell line, which likely resulted from deletions that occurred after several cell divisions, leading to genetic differences between individual cells in a colony. For further analysis, we focused on gene loci where we obtained at least 2 heterozygous knockout clones.
Identifying significant changes in allele-specific expression.
In developing a statistical approach to identify local, cis effects of these genetic manipulations, we sought to distinguish local effects of the genetic deletion from downstream effects that result as a consequence of either lncRNA/mRNA functions elsewhere in the cell, off-target effects, or biological/technical variation between clonal cell lines (Note S1). Our power to detect these effects varies between different measured genes (due to their level of expression and availability of SNPs) and between different knockout targets (due to differences in the numbers of knockout clones analyzed).
To account for these two variables, we developed a statistical approach to empirically estimate the false discovery rate of allele-specific changes in the expression neighboring genes using hundreds of genes on other chromosomes as controls. For each gene in the neighborhood of one of our promoter deletions, we calculated three statistics: (i) a T-test statistic comparing the average change in expression for each of the knockout alleles (including both heterozygous and homozygous knockout clones), normalized to the expression of the gene on the wild-type allele of the heterozygous clones; (ii) a z-score statistic comparing the expression of the knockout allele in heterozygous clones to the expression of the wild-type allele in the same clone; and (iii) a T-test statistic comparing the heterozygotes to the wild-type control clones using the allelic expression ratio after applying a variance-stabilizing transformation (arcsin of the square root of the allelic expression ratio). For a given gene, only samples with at least 20 allele-informative reads were considered, in order to enable accurate estimates of allele-specific expression. These three tests differ in whether they incorporate information from homozygous clones and how they normalize between knockout and wild-type alleles. We required that a gene perform significantly in each of the three tests in order to regard the gene as significant, as described below. We note that each underlying measure was approximately normally distributed, with some apparent outliers across hundreds of control clones; we conservatively included these outliers in calculating each test statistic. We examined differences in variation between knockout and control alleles with Levene’s test. For estimates of the variance of distributions presented in figures, see Table S1.
Because the distributions are only approximately normal, we assessed the significance of each of these gene-level statistics by permutation, sampling other cell lines from the same experimental batch and randomly assigning them as heterozygous or homozygous knockout clones to match the distribution of genotypes of the real samples. We calculated an empirical false discovery rate for the sum of these permutation ranks, testing each of the neighboring genes and using all of the genes on other chromosomes as the background model. Neighboring genes with FDR < 10%, a transformed allelic expression ratio >0.03, and an effect size of >10% in heterozygotes were considered significant.
Transcriptional read-through for Meg3 and Snhg3.
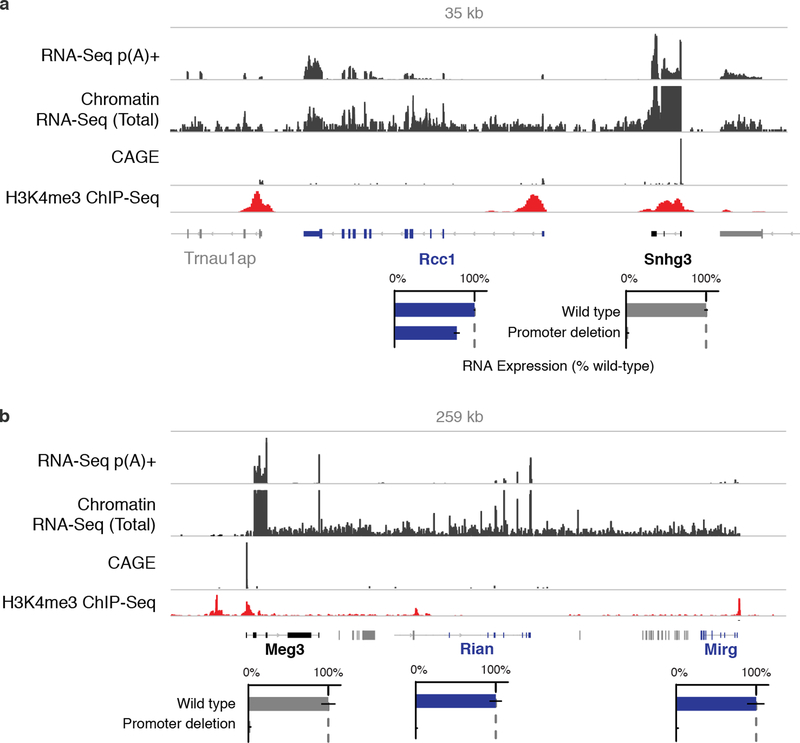
Promoter knockouts of Meg3 and Snhg3 led to reductions in one or more downstream genes oriented in the same direction as the knockout target gene. We attributed these changes to transcriptional read-through based on the following evidence (Note S4, Extended Data Fig. 3). For both Meg3 and Snhg3, we observed evidence for transcription continuing past the annotated 3’ end of the knockout target, through intergenic regions, and into the downstream gene (as assayed by RNA sequencing of chromatin-associated RNA). For the Meg3 locus, we did not observe H3K4me3 or CAGE reads at the 5’ ends of Rian and Mirg (downstream of Meg3), indicating that they are not expressed from their own promoters. In the Snhg3 locus, the downstream affected gene (Rcc1) is in fact expressed from its own promoter, but we found evidence for reads splicing from just downstream of Snhg3 into the first splice acceptor of Rcc1, indicating that at least some fraction of Rcc1 transcripts begin at the Snhg3 promoter.
Insertion of polyadenylation signals.
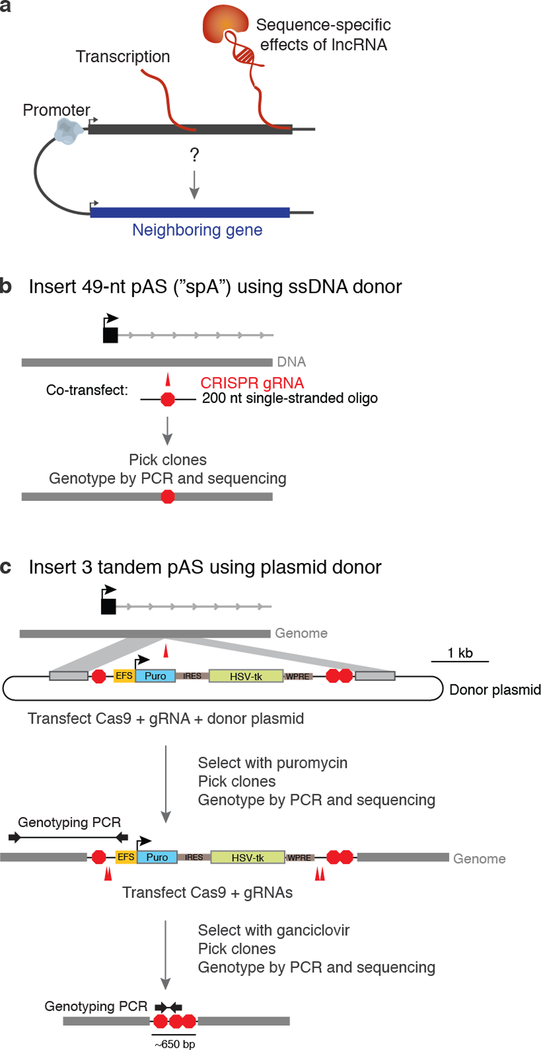
To halt transcription, we initially attempted to use a short 49-bp synthetic polyadenylation signal (spA) sequence42 to minimize the amount of genomic sequence added (Extended Data Fig. 6b). For a given gene, we designed a guide 0.5–3 kb downstream of the transcription start site. We designed 200-nt ssDNA oligos including the spA sequence flanked by 75- and 76-bp homologous arms, centered on the sgRNA cut site (~4 bp upstream of the PAM sequence), and ordered these as ultramers from Integrated DNA Technologies (Table S2). To knock in polyadenylation signals, we transfected 100 ng PX330-NoGuide, 100 ng pZB, 100 ng pS-pp7-GFPiP, and 100–200 ng of donor ssDNA oligo and followed the selection procedure described for the promoter knockouts. To genotype these insertions, we used a combination of PCR and high-throughput amplicon sequencing as described above. We identified clones that had heterozygous insertions of the full 49-bp spA sequence on one allele; we typically observed that the other allele had a short insertion or deletion, consistent with non-homologous end joining (NHEJ)-mediated repair. This short pAS sequence (spA) succeeded in halting the transcription of three RNAs: Blustr (pAS at +40bp and +0.5 kb in Fig. 3), Gpr19, and Bendr. However, for other genes, transcription was unaffected despite pAS knock-in, consistent with the location-dependent efficiency previously observed for this pAS sequence42.
Accordingly, we built a larger construct containing three polyadenylation signals (p3PA, Extended Data Fig. 6c). The structure of this construct upon insertion into the genome through homologous recombination is as follows: spA – EFS promoter – Puromycin resistance gene IRES thymidine kinase – WPRE – SV40 pAS – PGK pAS (“p3PA-Puro-iTk”). We co-transfected 300 ng of this construct with 100 ng of pZB and 100 ng of PX330-NoGuide, waited three days, and then selected for cells with integrations with 1 μg/mL puromycin for one week. We picked individual colonies and used PCR to genotype clones, using primers spanning the insertion junctions. We sequenced these PCR products to determine the allele of insertion. Following genotyping, we expanded clonal cell lines and transfected with PX330 and a pool four sgRNAs to delete the selection cassette, leaving behind three tandem pASs. Following selection with 2 μg/mL ganciclovir, we again picked individual colonies, used PCR to confirm loss of the cassette, and sequenced RNA from multiple clones. PCR primer sequences for cloning homology arms and genotyping p3PA insertions are listed in Table S2.
Knockouts of Blustr exons and introns.
To delete each exon and intron of Blustr, we transfected cells with pools of guides as described for the promoter deletions, using 2 guides on each side. We assessed the genotype of clonal cell lines as described above for promoter deletions. To confirm exon knockout from RNA sequencing data, we examined SNPs in each of the exons. Upon knockout of exon 2, for example, we observed loss of RNA sequencing reads mapping to exon 2, while reads mapping to other exons were still present. We also identified reads spanning a new splice junction between exon 1 and exon 3, further confirming that exon 2 was removed from the mature transcript. For barplots in Fig. 3 measuring Blustr expression, the values represent the normalized read counts of the remaining exons that were not deleted in that experiment. To confirm intron knockout, we used PCR primers spanning the deletion junction and sequenced the resulting PCR products. We note that the intron knockouts, by design, do not affect the sequence of the spliced Blustr RNA.
5’ splice site knockout.
To knock out the 5’ splice site of Blustr, we co-transfected mESCs as described above, using a single sgRNA pZB plasmid and 200 ng of ssDNA oligonucleotide donor for homologous recombination (Extended Data Fig. 8c). The oligo was ordered as an ultramer from Integrated DNA Technologies (Table S2). We genotyped these insertions through amplicon sequencing using an Illumina MiSeq (primers in Table S2).
Transcriptional activity with GRO-Seq.
We used precision run-on sequencing (PRO-seq)43, a variant of global run-on sequencing44, to map transcriptionally engaged RNA polymerase for a subset of clones. Clones for PRO-seq (as well as ChIP-Seq and ATAC-Seq) were chosen from among the recoverable knockout cell lines with a preference for clones with homozygous knockouts or knockouts on the 129 allele only. We performed PRO-seq as previously described45, with modifications. We harvested 10 million mESCs by scraping, washing in cold PBS, and spinning at 330 × g for 3 minutes. The cell pellet was resuspended in 1 ml cold Douncing Buffer (10 mM Tris-HCl pH 7.4, 300 mM Sucrose, 3 mM CaCl2, 2 mM MgCl2, 0.1% (v/v) Triton X-100, and 0.5 mM DTT) per 1 million cells. The cells were incubated on ice in the cold room for 5 minutes and dounced 25 times. The nuclei were pelleted at 500 × g for 2 minutes, washed twice in 5 ml Douncing Buffer, and centrifuged at 500 × g for 2 minutes. The nuclei were then gently resuspended in 100 μl of cold Storage Buffer (10 mM Tris-HCl, pH 8.0, 25% (v/v) glycerol, 5 mM MgAc2, 0.1 mM EDTA, and 0.5 mM DTT), immediately flash frozen, and stored at −80°C until use.
A 28 μl 2× Nuclear Run-On (NRO) mix was prepared as follows: 1 M Tris-HCl, pH 8.0, 1M MgCl2, 2M KCl, and 0.1 M DTT. 5 μl of 1 mM Biotin-11-CTP (Perkin Elmer), 1 μl of 0.05 mM CTP, 2.5 μl of 2 mM ATP, 2.5 μl of 2 mM GTP, 2.5 μl of 2 mM UTP (Sigma Aldrich), 6.5 μl of nuclease free water, and 2 μl of SUPERaseIn (Ambion) were added to the 2× NRO mix and mixed well prior to the addition of 50 μl of 2% NLS. The NRO reaction mix was mixed well and preheated to 37°C. 100 μl of NRO mix was added to 100 μl of nuclei in Storage Buffer. The reaction was mixed gently by pipetting and incubated at 37°C for 3 minutes, mixing halfway through. To halt the reaction 500 μl of Trizol LS (Thermo Fisher) was added, mixed well, and incubated at room temperature for 5 minutes. RNA was isolated through a chloroform extraction and ethanol precipitation, and resuspended in 20 μl of H2O. The RNA was heat denatured at 65°C for 40 seconds and fragmented on ice for 10 minutes with 5 μl of 1N NaOH. To stop the reaction, 5 μl of 1 M Acetic Acid and 20 μl of 1 M Tris-HCl, pH 7.4 were added. To remove unincorporated biotinylated nucleotides, the sample was passed through a P-30 exchange column (BioRad). 1 μl of RNase inhibitor was added to the ~50 μl of RNA and the first biotin enrichment was then performed.
Each biotin enrichment was performed as follows. To prepare the Streptavidin M280 Beads (Invitrogen) for biotin enrichment, 100 μl of beads were taken per sample and washed once in 0.1 N NaOH with 50 mM NaCl and twice in 100 mM NaCl. Beads were resuspended in 160 μl of Binding Buffer (10 mM Tris-HCl, pH 7.4, 300 mM NaCl, and 0.1% (v/v) Triton X-100). To each sample an equal volume of Streptavidin M280 beads was added, mixed, and incubated on a rotator for 20 minutes at room temperature. The beads were magnetically separated and washed twice in 500 μl of ice cold High Salt Wash Buffer (50 mM Tris-HCl, pH 7.4, 2 M NaCl, and 0.5% (v/v) Triton X-100), twice in 500 μl of Binding Buffer, and once in 500 μl of Low Salt Wash Buffer (50 mM Tris-HCl, pH 7.4 and 0.1% (v/v) Triton X-100). To harvest the RNA, 300 μl of Trizol (Thermo Fisher) was added to the beads, vortexed for 20 seconds, and incubated at room temperature for 3 minutes. 60 μl of chloroform was added and mixture was incubated at room temperature for 3 minutes. The samples were centrifuged at 14,000 × g for 5 minutes at 4°C. The aqueous phase was collected and transferred to a new tube; the remaining organic phase was removed from the beads. The Trizol extraction was then repeated as above and the two aqueous phases were combined. RNA was purified with a chloroform extraction and ethanol precipitation, and resuspended in nuclease free water. RNA sequencing libraries were then prepared as described above, except that SILANE clean-ups were replaced with Streptavidin-biotin capture enrichments until after reverse transcription (a total of 3 enrichments).
We sequenced PRO-seq libraries to a depth of ~10 million 30-bp paired-end reads. To analyze the data, we mapped and processed the RNA sequencing data as described above, including aligning individually to the 129 and Castaneus genomes. Figures showing “Allele-specific GRO-seq” depict coverage for reads that uniquely map to the specific allele indicated in the figure. To assess the relative read density in the promoter-proximal region and gene body of Sfmbt2, we counted reads in the 2 kb region downstream of the first Sfmbt2 TSS and in the remainder of the gene body46. We calculated the pause index as the ratio of these two quantities, normalized to total read count. We noticed that different PRO-seq libraries had subtle biases in the relative fraction of reads aligning to the TSS versus the gene body, leading to slightly offset distributions of pause indices across all genes, and so we corrected for these biases in each library by normalizing TSS and gene body RPKMs to the median of the ~5,000 genes with coverage across all samples.
Chromatin accessibility with ATAC-Seq.
Libraries were generated as previously described47 using 50,000 mESCs. We generated duplicate ATAC-Seq libraries for each clonal cell line examined and sequenced each to a depth of ~40 million 30-bp paired end reads. We aligned paired-end DNA sequencing reads using bowtie248 to each of the 129 and Castaneus genomes with the following parameters: “--met-stderr --maxins 1000”, removed duplicate reads using Picard (http://picard.sourceforge.net), and filtered to uniquely aligning reads using samtools (MAPQ < 30, https://github.com/samtools/samtools). For plotting normalized read coverage at the Blustr and Sfmbt2 promoters, we combined data from the two biological replicates (two independent measures of the same cell line) and connected paired-end reads to generate fragments. Fragment coverage was normalized by the total number of uniquely mapping reads.
Chromatin immunoprecipitation.
ChIP-seq for H3K4me3 and H3K27me3 was performed using monoclonal antibodies as previously described49. Sequencing data was analyzed as for ATAC-Seq described above.
Validation of allele-specific RNA expression with ddPCR.
To validate our RNA-seq based measurements of allele specific expression, we used a quantitative allele-specific PCR assay to verify measurements for Blustr and Sfmbt2. We isolated RNA from harvested mESCs using RNeasy 96 columns and performed a DNase treatment followed by reverse transcription of 500 ng of RNA (total reaction volume 20 μl). We performed droplet digital PCR (ddPCR) using Bio-Rad Custom ddPCR Assays that involve qPCR primers flanking a polymorphic site and two allele-specific fluorescent probes. For Blustr: Left primer sequence: GACAAATACTCCCTTCAACA; Right primer sequence: GAACAGTTTGTCCTGCC; Probe sequence: TAAGTGAGGTGAACTCCAAG (129 allele, FAM) or AGTGAGGCGAACTTCAAG (Castaneus, HEX). For Sfmbt2: Left primer sequence: TGTAAGTTTGCCTGATACTC; Right primer sequence: TCTAATGTACCTCAGCCC; Probe sequence: TTTCCTATGAGCAGTTCAAC (129 allele, FAM) or TCCTATGAACCGTTCAGC (Castaneus, HEX). ddPCR was done with 2.2 μl of cDNA, 11 μl of Supermix (BioRad), 1.1 μl of each probe, and 7.7 μl of water per reaction followed by droplet generation. PCR was performed as follows: 95°C for 10 minutes; and cycling at 94°C for 30 s and 55°C for 1 minute for a total of 40 cycles; and 98°C for 10 minutes. Readout was done using the QX200 Droplet Reader and Quantasoft Software (BioRad) to determine the total number of droplets containing each allele. We calculated allelic expression ratios from these values and compared it to values generated through RNA-sequencing and hybrid selection of the same RNA samples (Extended Data Fig. 2d,e).
External ChIP-Seq, RNA-Seq, and DNase HS data.
We utilized the following data from ENCODE50: H3K4me3, H3K4me1, H3K27ac, and CTCF ChIP-Seq in mESCs (ES-Bruce4); DNase hypersensitivity sequencing in mESCs (E14); H3K4me3, H3K4me1, and CTCF ChIP-Seq and DNase HS data in H1-hESCs; and RNA-sequencing data in H1-hESCs (nuclear p(A)+, nuclear total). To assess transcription factor binding to mRNA and lncRNA promoters (Extended Data Fig. 7c), we examined mESC ChIP-seq peaks available from Kagey et al. at the Gene Expression Omnibus (GSE22562)51.
DNA purification for examining proximity contacts.
To examine the proximity contacts of the linc1405 locus, we used the RAP-DNA protocol, which we initially developed in order to map RNA localization to chromatin, to capture linc1405 DNA37. Briefly, we crosslinked live cells to fix endogenous chromatin complexes, then purified a target DNA region using a pool of oligonucleotides targeting the linc1405 locus (Table S3). Here, we used probes that are the same strand as the linc1405 RNA – in this way, we specifically capture the linc1405 DNA and do not directly capture the linc1405 RNA itself. We mapped the 3-D proximity contacts of the linc1405 locus through high-throughput sequencing of co-purified DNA and calculated the normalized enrichment to an input DNA library in 1-kb windows (Extended Data Fig. 7e). Annotations for topologically associated domains (TADs) were downloaded from the Ren Lab (http://chromosome.sdsc.edu/mouse/hi-c/download.html)52.
LncRNA transcript annotations.
For evolutionary conservation analysis, we used lncRNA annotations and isoforms previously defined based on RNA sequencing in mouse embryonic stem cells, combining annotations generated with multiple methods (Scripture41 and slncky28). We filtered the combined list using slncky28 to eliminate transcripts predicted to encode proteins or micropeptides by UCSC, transcripts that partially align to protein-coding genes (e.g., pseudogenes or incomplete reconstructions), and species-specific coding gene duplications. Subsequently we performed several manual curation steps. We examined each isoform using a combination of long-read RNA-sequencing data, total chromatin-associated RNA sequencing data, capped analysis of gene expression (CAGE) data, and poly(A+) 3’-end sequencing data from mESCs28,30,41,53. We eliminated transcripts that appeared to result from an extended 3’UTR of an upstream protein-coding transcript. Because the precise 5’ ends of transcripts are imprecisely assigned by based on RNA-sequencing data alone, we re-assigned 5’ ends (TSSs) using a sliding-window approach to find the 10-bp window with the highest number of same-strand CAGE reads within 300-bp of the initial calculated TSS. We additionally manually curated the TSS of each lncRNA, some of which were incorrectly assigned by more than 300 bp, based on CAGE and H3K4me3 ChIP-Seq data, and eliminated any where we could not identify the TSS (e.g., due to unmappable sequence or very low abundance).
Analysis of lncRNA and promoter conservation.
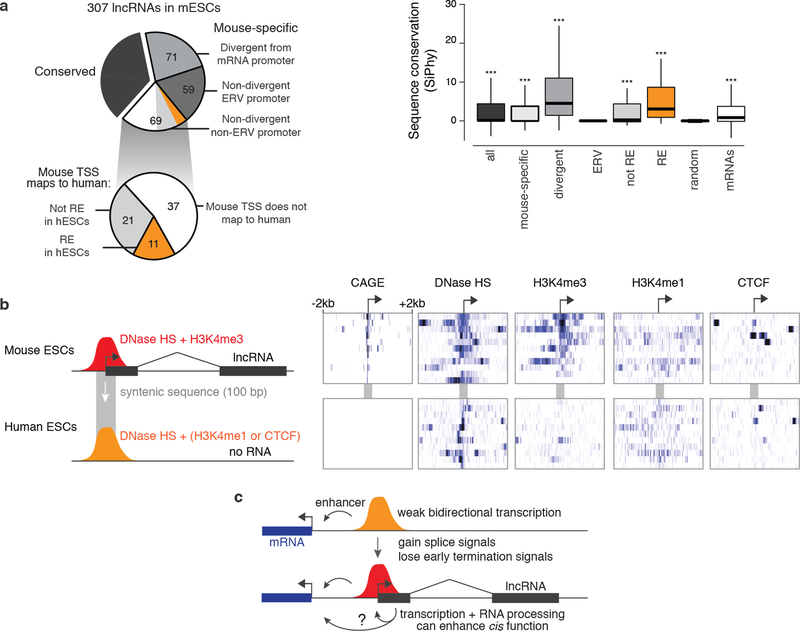
To categorize lncRNAs by their conservation properties and promoter locations, we examined a set of 307 lncRNAs expressed in mESCs as described above. We assessed the conservation of each lncRNA through a two-step approach. We first used slncky to look in syntenic locations for evidence of lncRNA transcripts in deep p(A)+ RNA-seq of rat, chimp, and human induced pluripotency stem cells (iPSCs)28. LncRNAs called “conserved” by this first filter have substantial evidence based on RNA-seq that allows for independent reconstruction of the transcript in one or more of these other organisms. We categorized the remaining lncRNAs by the location of their TSS: 71 lncRNAs originate within 500-bp of an mRNA TSS on the opposite strand (“divergent”); 59 lncRNAs originate within the long-terminal repeats (LTRs) of endogenous retroelements; and 79 lncRNAs have their promoters in intergenic regions that do not overlap with LTRs and do not emerge from a bidirectional mRNA promoter (henceforth, “intergenic”).
Because some conserved lncRNAs might be too lowly expressed to assemble a transcript de novo in a given species, we examined more closely the 79 intergenic lncRNAs that were called “mouse-specific” in the initial slncky analysis. We applied a second, more stringent threshold to remove lncRNAs misclassified as mouse-specific due to low abundance. For each intergenic lncRNA locus, we used liftOver54 to map the 10 bp surrounding the mouse TSS (mm9) to the human genome (hg19) (minMatch=0.1, UCSC chain). 37 of these transcripts did not lift over at this step, and thus were considered mouse-specific. For the 42 that did lift over, we examined the syntenic region for evidence of p(A)+ RNA-seq data from human iPSCs28 or p(A)+ nuclear-fraction RNA-seq from hESCs (–100 to +900 bp relative to the TSS), or for evidence of p(A)+ nuclear-fraction or whole-cell CAGE from hESCs (–250 to +250 bp relative to the TSS), and removed from consideration any lncRNAs that showed evidence for RNA-seq or CAGE above a certain threshold. We chose this threshold based on a set of random intergenic regions, which were matched to the set of intergenic mouse-specific lncRNAs based on GC content. We eliminated from consideration the 10 lncRNAs that showed RNA-seq or CAGE signal greater the 90th percentile of random regions, corresponding to approximately 2 CAGE or RNA-seq reads in the windows described above. These 10 lncRNAs were added to the “conserved” section of the pie chart in Fig. 4a. Several of these 10 lncRNAs correspond to substantially shortened, single-exon p(A)+ transcripts that show minimal overlap with the syntenic exons in mouse; although a majority of the exonic sequence of these transcripts are not in fact conserved between human and mouse, we excluded these from consideration as putative mouse-specific lncRNAs.
For the purposes of examining the conservation properties of these intergenic mouse-specific lncRNAs, we defined a matched set of “enhancer” elements. We first generated a list of regulatory elements in mESCs using the DNase hotspots called by ENCODE-UW in ES-E14 cells. As an estimate of the activity of each element, we calculated the density of H3K27ac reads in the region. From the set of intergenic elements that did not overlap a promoter, lncRNA promoter, or LTR, we selected a random subset matched to the intergenic lncRNA promoters for H3K27ac density (binned by 10 reads / bp) and distance to the TSS of the closest active gene (binned by 5 kb). We call these elements “enhancers” because they are marked by DNase hypersensitivity and H3K27ac but do not overlap a known gene promoter.
We compared the sequence conservation and functional conservation of three classes of elements: intergenic mouse-specific lncRNAs, matched intergenic enhancer elements, and GC-matched random intergenic elements. First, we computed the rate at which each set maps to human sequence. We centered each element and used liftOver (--minMatch=0.1) to identify the syntenic region in the human genome. Elements that did not lift over at this step correspond to the white segment of the pie charts in Fig. 4 (iii – “did not map”). For elements that did lift over to human, we next defined the subset that map to putative regulatory elements in human. We examined a 500-bp window centered on the lifted over region and counted reads in hESC DNase-seq data from ENCODE. We defined regions showing DNase HS scores higher than 95% of the mappable random intergenic regions as putative DNA regulatory elements. We note that these random intergenic regions include some enhancers – they are matched to lncRNA promoters for GC content, and thus frequently correspond to regulatory elements (which are GC-rich) that happen to be active in hESCs. For both intergenic mouse-specific lncRNAs and enhancers, ~33% of elements corresponded to putative DNA regulatory elements in human (Fig. 4d), representing a ~6.6-fold enrichment versus the random intergenic controls. To compare sequence conservation of these classes of elements, we calculated the average SiPhy score55 across each 500-bp region surrounding the mouse TSS or the center of the enhancer element, using the 29 mammals alignment from the mouse perspective56. We used a two-sided Mann-Whitney U-test to look for changes in the distributions of SiPhy scores to the set of mappable random intergenic regions (Fig. 4d – random ii+iii).
Impact of expression level on conservation analysis.
Although the set of intergenic mESC lncRNAs examined above does not show any significant evidence for p(A)+ RNA in the syntenic locus in human, some of these transcripts may not be detected in human and yet still be truly conserved. These transcripts might be misclassified as “mouse-specific” lncRNAs for several reasons, including: (i) low expression level in hESCs and iPSCs such that the lncRNA, by chance, is not detected based on the depth of sequencing data available; or (ii) the lncRNA is not expressed in hESCs or iPSCs, but is expressed in a different human cell type and thus may have a conserved function.
To estimate the false positives resulting from these and other scenarios, we examined the properties of a set of 853 conserved mRNAs matched to the intergenic “mouse-specific” lncRNAs based on expression in mESCs. We counted the frequency at which these mRNAs would be called “not conserved” by the same procedures described above: we applied the nuclear p(A)+ CAGE and RNA-seq filters to eliminate transcripts that show detectable transcription in the 1-kb region near the TSS. While 87% of the intergenic lncRNAs described above passed these filters (and thus appeared to be mouse-specific), only 22% of the expression-matched mRNAs passed; this indicates that the set of 69 mouse-specific intergenic lncRNAs are approximately 3.9-fold enriched for human elements that are not transcribed in hESCs. Thus, the mouse-specific lncRNAs defined above appear to consist largely of transcripts that are not conserved.
We performed the following additional analyses to ensure the robustness of our conclusions regarding the existence of lncRNAs that evolved from ancestral regulatory elements. First, we examined the conservation of the first 5’ splice sites of this set of lncRNAs. In 7 of these 11 loci, the “GT” dinucleotide in the first 5’ splice site is not conserved, suggesting that a similar spliced transcript cannot be produced from this locus. Second, we re-performed the entire conservation analysis focusing on the 50% of mESC intergenic lncRNAs with the highest expression levels – these lncRNAs are less likely to be missed in hESCs due to low abundance. We also adjusted our p(A)+ RNA and CAGE filters to require a complete absence of reads in the corresponding regions in hESCs and iPSCs. Using these filters, 79% of the intergenic lncRNAs are not detectably expressed in human cells, representing a ~12-fold enrichment over mRNAs matched for expression level. Therefore we are confident that most of these lncRNAs are correctly classified as mouse-specific. Of the 30 intergenic lncRNAs called mouse-specific by this more conservative analysis, 5 do indeed correspond to putative DNA regulatory elements, including linc1494 (Fig. 4c), representing a >8-fold enrichment versus GC-matched random sequences (Chi-squared P < 10−10). Thus, our conclusions that some lncRNAs appear to evolve from ancestral regulatory elements are robust even with stringent thresholds.
Software for data analysis and graphical plots.
We used the following software for data analysis and graphical plots: R Bioconductor (version 3.0)57, Gviz (version 1.10.11), gplots (version 2.17.0), GenomicRanges (version 1.18.4)58, rtracklayer (version 1.26.3)59, BEDTools60, Integrative Genomics Viewer (version 2.3.26)61, and vcftools (version 0.1.12)62.
Extended Data
Extended Data Fig. 1. Expression and subcellular localization of knocked-out lncRNAs and mRNAs.
(a) Expression of lncRNAs and mRNAs in F1 129/Castaneus female mESCs, reported in fragments per kilobase per million (FPKM) in whole-cell p(A)+ RNA-seq. Cumulative fraction is plotted for all mRNAs expressed in mESCs. Large dots represent transcripts whose promoters we deleted in this study. LncRNAs and mRNAs span a >20-fold range of abundance levels. (b) Relative subcellular localization of lncRNAs and mRNAs. We sequenced p(A)+ RNA from chromatin, soluble nuclear, and cytoplasmic fractions (see Methods) and plotted the relative abundance of mature transcripts in each fraction. We selected lncRNAs that showed localization biased toward the nuclear fractions relative to most mRNAs. For comparison, we plotted 1,000 randomly selected mRNAs (light gray).
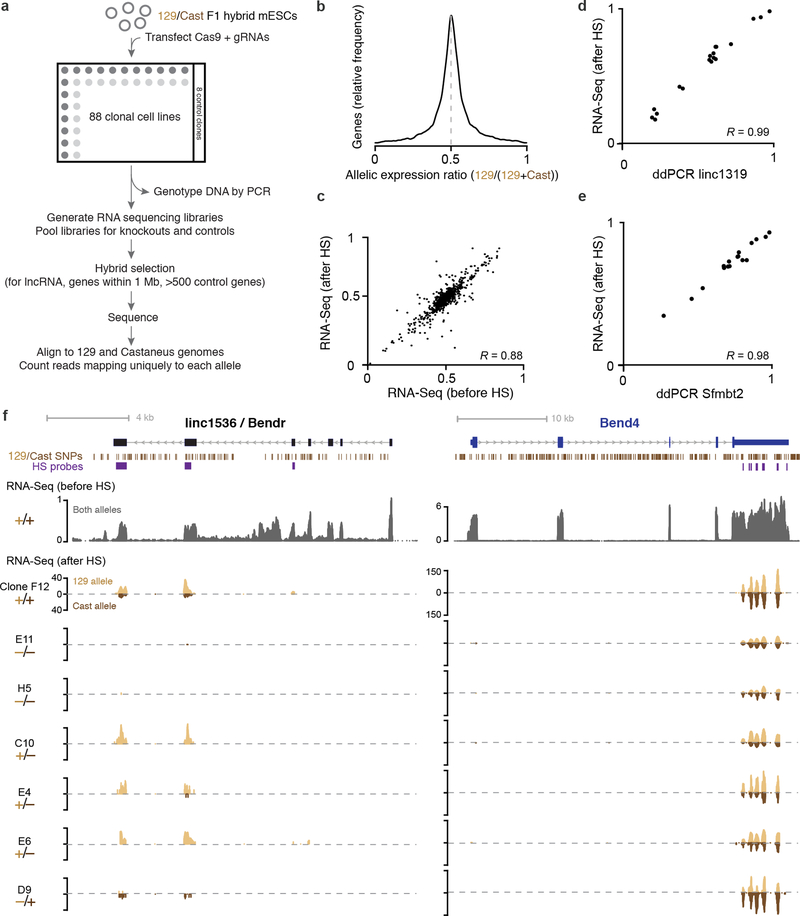
Extended Data Fig. 2. Generation of knockout clones and measurement of allele-specific RNA expression.
(a) Overview of knockout and measurement protocol. (b) Distribution of allelic expression ratios (number of informative reads mapping to 129S1 allele divided by the number mapping to either the 129S1 or the Castaneus allele) across active genes in mESCs. (c) Scatterplot of allelic expression ratios for genes with RPKM ≥ 2 that have more than 100 allele-informative reads across all libraries. Allelic expression ratios are consistent in RNA sequencing data before and after hybrid selection (HS). (d) Allelic expression ratios as measured by two independent methods for Blustr and (e) Sfmbt2 expression in 15 clonal cell lines containing genetic modifications in the Blustr locus. (f) Example locus showing hybrid selection strategy and RNA-seq coverage for cell lines with the indicated genotype for deletion of the Bendr promoter. Y-axis scales represent normalized read counts and are the same for all hybrid selection tracks. The absolute level of expression for any given gene varies among clonal cell lines; throughout this work, we instead consider the relative level of expression between the two alleles in heterozygous knockout cells. For similar plots of each gene studied, see http://pubs.broadinstitute.org/neighboring-genes/.
Extended Data Fig. 3. Read-through transcription at Meg3 and Snhg3 loci.
(a) Snhg3 promoter knockout reduces the levels of Rcc1 mRNA by 23%. However, sequencing of chromatin-associated RNA shows that transcription continues past the annotated 3’ end of Snhg3 into the downstream Rcc1 gene (see Methods). This read-through transcription creates a fusion transcript containing exons of both Snhg3 and Rcc1, as well as intergenic RNA. We note that this fusion transcript is also annotated in the syntenic human locus as an alternative isoform of RCC1. Bars: relative p(A)+ RNA expression on modified versus unmodified alleles. Error bars: 95% CI for the mean (n ≥ 2 alleles, see Table S1). (b) Meg3 promoter knockout eliminates the expression not only of Meg3 but also of two additional lncRNAs encoded downstream in a tandem orientation (Rian and Mirg). Although these three lncRNAs are annotated as separate genes, they appear to be derived from a single transcript driven by the Meg3 promoter. This is consistent with the presence of continuous chromatin-associated RNA throughout the locus and a lack of CAGE reads at the 5’ ends of Rian and Mirg3.
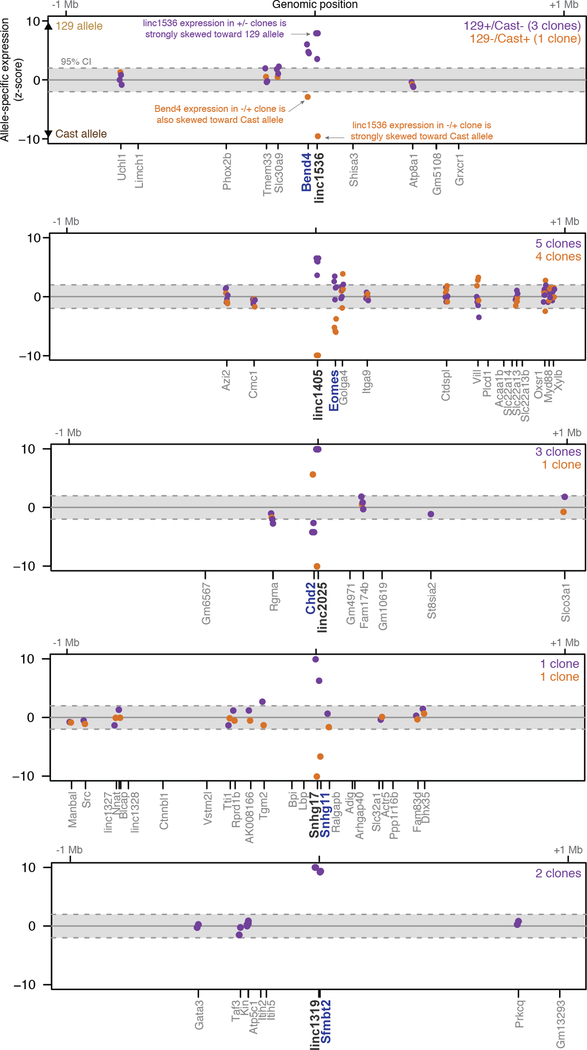
Extended Data Fig. 4. Promoter knockouts for 5 intergenic lncRNAs affect the expression of a neighboring gene.
Significance (z-score) of allele-specific expression ratios at all genes within 1 Mb of each of 5 lncRNA loci. Each dot represents a different heterozygous promoter knockout clone for a given gene. Dots are shown only for genes that are sufficiently highly expressed to assess allele-specific expression (see Methods). The y-axis is capped at –10 to +10 standard deviations from the mean. Black: knocked-out lncRNA. Blue: Gene with significant allele-specific change in gene expression (FDR < 10%). Independent clones are not expected to yield the same significance value (z-score), in part because read depth differs between samples.
Extended Data Fig. 5. Promoter knockouts for 4 mRNAs affect the expression of a neighboring gene.
Significance (z-score) of allele-specific expression ratios at all genes within 1 Mb of each of 4 mRNA loci. Each dot represents a different heterozygous promoter knockout clone for a given gene. Dots are shown only for genes that are sufficiently highly expressed to assess allele-specific expression (see Methods). The y-axis is capped at –10 to +10 standard deviations from the mean. Black: knocked-out lncRNA. Blue: Gene with significant allele-specific change in gene expression (FDR < 10%). Independent clones are not expected to yield the same significance value (z-score), in part because read depth differs between samples.
Extended Data Fig. 6. Dissecting mechanisms for how gene loci regulate a neighbor.
(a) Three categories of possible mechanisms by which a gene locus might regulate the expression of a neighbor. (b) We used two strategies to insert pAS downstream of gene promoters. In the first strategy, we inserted a 49-bp synthetic pAS (“spA”) using a single-stranded DNA oligo with 75-bp homology arms (see Methods). (c) In the second pAS insertion strategy, we cloned a donor plasmid containing a selection cassette and three different pAS sequences (see Methods). Homology arms of 300–800 bp were used to integrate the cassette. After isolating clones with successful insertions, we used a second round of transfections to remove the selection cassette, leaving behind three tandem pASs. EFS = elongation factor 1 promoter. Puro = puromycin resistance gene (pac). HSV-tk = herpes simplex virus thymidine kinase.
Extended Data Fig. 7. Promoters of lncRNAs and mRNAs have enhancer-like functions.
(a) Allele-specific GRO-seq signal for clones with the indicated modifications at the Bendr locus. Only reads specifically mapping to one of the two alleles are shown. Y-axis scale represents normalized read count and is the same for all tracks. (b) Allele-specific p(A)+ RNA expression for genetic modifications at the linc1405, Snhg17, Gpr19, and Slc30a9 loci. Bars: Average RNA expression on modified compared to unmodified (wild-type) alleles. Error bars: 95% CI for the mean (n ≥ 2 alleles, see Table S1). Gray arrows indicates distance from the targeted locus promoter to the affected neighboring gene. We note that, based on their location, the Snhg17 and Gpr19 pAS insertions likely allow more substantial splicing and transcription; for these loci, it is clear that the majority of the transcript is dispensable but it is possible that transcription close to the promoter may be involved in the cis regulatory function. (c) Presence (gray) or absence (white) of various chromatin marks and transcription factors in mESCs in a 1.5-kb window centered on the TSS of each targeted gene. (d) Distance from each knocked-out gene to its neighboring target gene (x-axis) versus the magnitude of the effect on the expression of the neighboring gene (% compared to wild-type, y-axis). Blue genes represent those discussed in main text; gray genes are discussed in Note S5. (e) Proximity-based contacts between the linc1405 and Eomes loci (the pair of loci separated by the greatest linear distance). The y-axis shows enrichment in a sequencing-based proximity assay in which we used antisense oligos to capture linc1405 DNA and any interacting, crosslinked proximal DNA (see Methods). TAD annotations are derived from Hi-C experiments in mESCs (see Methods). Blue arrow: focal contact between the linc1405 and Eomes loci.
Extended Data Fig. 8. Characterization of genetic modifications in the Blustr locus.
(a) Allele-specific GRO-seq signal for clones with the indicated modifications at the Blustr locus. Only reads specifically mapping to one of the two alleles are shown. Y-axis scale represents normalized read count and is the same for all tracks, and is magnified 5 times at the indicated location to better visualize the reads in the Sfmbt2 locus. (b) Quantification of allele-specific GRO-seq signal in the Sfmbt2 locus on alleles modified as indicated. TSS: region including the two alternative TSSs of Sfmbt2 and 2 kb downstream. Gene body: region containing the remainder of the Sfmbt2 gene locus. Pause index: ratio of TSS to gene body. Dashed gray lines indicate the 95% CI for the mean of 8 wild-type clones. (c) Schematic of the 5’ end of the Blustr locus and genotypes of two knockout clones. The 5’ splice site is located 78 bp downstream of the Blustr transcription start site (in this panel, Blustr is transcribed from left to right). One of the alleles from the two clones contains insertion of the oligo mediated by homologous recombination; the remaining three alleles contain insertions or deletions resulting from non-homologous end joining repair of sgRNA-mediated double-strand breaks, some of which also disrupt the 5’ splice site. Barplots show allele-specific RNA expression for knockout clones and control clones (+/+). (d) Schematic of the observed splice structures of Blustr RNA transcripts in p(A)+ RNA sequencing of the exon deletion clones. Each deletion removes a region including ~50–200 bp on either side of the exon, thereby removing both the exon and its splice sites. The Exon 4 deletion removes the endogenous pAS, leading to new isoforms of the lncRNA transcript that splice into two cryptic splice acceptors downstream. (e) GRO-Seq, H3K4me3 ChIP-Seq, and chromatin accessibility (ATAC-Seq FPKM) at the Blustr and Sfmbt2 promoters in cell lines with the indicated genotypes. Deletion of the first 5’ splice site leads to a significant reduction in H3K4me3, RNA polymerase occupancy, and chromatin accessibility at the Blustr promoter, as well as H3K4me3 and RNA polymerase occupancy (but not accessibility) at the Sfmbt2 promoter. (f) H3K27me3 ChIP-seq at the Blustr and Sfmbt2 loci in cell lines with the indicated genotypes. Deletion of the Blustr promoter or 5’ splice site leads to spreading of the repression-associated H3K27me3 modification across a ~30 kb region.
Extended Data Fig. 9. Mechanisms for crosstalk between neighboring lncRNAs and mRNAs.
Proposed mechanisms based on pAS insertion experiments and other genetic manipulations (see text). †For proposed mechanisms, see Note S5.
Extended Data Fig. 10. Classification of lncRNAs based on conservation and promoter location.
(a) Classification of 307 lncRNAs expressed in mESCs. “Conserved” transcripts are those that show significant evidence of capped analysis of gene expression (CAGE) data and/or p(A)+ RNA in syntenic loci (see Methods). Divergent: initiating within 500 bp of an mRNA TSS, on the opposite strand. ERV: endogenous retroviral repetitive element (see Note S9). Boxplot shows sequence-level conservation of the promoters of subsets of lncRNAs expressed in mESCs. Random intergenic regions are matched to lncRNA promoters by GC content. Positive SiPhy score indicates evolutionary constraint on functional sequences. Orange category corresponds to mouse-specific lncRNAs that appear to have evolved from ancestral regulatory elements (REs) and correspond to sequences that show evidence for DNase I hypersensitivity in human embryonic stem cells. Significance is calculated compared to random intergenic regions using a Mann-Whitney U-test. ***: P < 0.001. Whiskers represent data within 1.5× the interquartile range of the box. (b) Chromatin and RNA data for 11 mouse-specific lncRNAs that appear to have evolved from ancestral regulatory elements. In mouse, these elements show evidence for CAGE, H3K4me3, and DNase I hypersensitivity, consistent with their roles as promoters. The syntenic sequences in human do not show evidence for CAGE but nonetheless are DNase I hypersensitive and are frequently marked by H3K4me1 and/or CTCF. (c) Model for evolution of lncRNAs from pre-existing enhancers, which often initiate weak bidirectional transcription to produce eRNA. Spliced transcripts may neutrally appear through the appearance of splice signals and loss of polyadenylation signals. In some cases, transcription, splicing, or other RNA processing mechanisms may feed back and contribute to the cis regulatory function of the promoter, producing a lncRNA as a byproduct.
Supplementary Material
Acknowledgements:
We thank S. Grossman, J. Rinn, M. Yassour, P. Sharp, L. Boyer, M. Ray, C. Fulco, M. Munschauer, T. Wang, and N. Friedman for discussions; A. Goren and Broad Technology Labs for ChIP; J. Lis, D. Mahat, and A. Shishkin for technical advice and reagents; and J. Flannick for computational tools. J.M.E. is supported by the Fannie and John Hertz Foundation and the National Defense Science and Engineering Graduate Fellowship. M.G. is supported the NIH Director’s Early Independence Award (DP5OD012190), the Edward Mallinckrodt Foundation, the Sontag Foundation, and the Searle Scholars Program. Work in the Lander Lab is supported by the Broad Institute.
Footnotes
Conflict of interest statement: The Broad Institute holds patents and has filed patent applications on technologies related to other aspects of CRISPR.
Data availability. Sequencing data for this study is available at the Gene Expression Omnibus (GSE80262 and GSE85798), and additional visualizations of the data are available at http://pubs.broadinstitute.org/neighboring-genes/.
References:
- 1.Okazaki Y. et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420, 563–573 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Kapranov P. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Guttman M. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carninci P. et al. The transcriptional landscape of the mammalian genome. Science 309, 1559–1563 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Lee JT Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev 23, 1831–1842 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagano T. et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Wang KC et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ørom UA et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 143, 46–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guil S. & Esteller M. Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol 19, 1068–1075 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Ebisuya M, Yamamoto T, Nakajima M. & Nishida E. Ripples from neighbouring transcription. Nat. Cell Biol. 10, 1106–1113 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Cabili MN et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25, 1915–1927 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassett AR et al. Considerations when investigating lncRNA function in vivo. Elife 3, e03058 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li G. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajagopal N. et al. High-throughput mapping of regulatory DNA. Nat Biotechnol 34, 167–174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin Y. et al. Opposing Roles for the lncRNA Haunt and Its Genomic Locus in Regulating HOXA Gene Activation during Embryonic Stem Cell Differentiation. Cell Stem Cell 16, 504–516 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Paralkar VR et al. Unlinking an lncRNA from Its Associated cis Element. Mol Cell 62, 104–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martens JA, Laprade L. & Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429, 571–574 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Shearwin KE, Callen BP & Egan JB Transcriptional interference--a crash course. Trends Genet 21, 339–345 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purmann A. et al. Genomic organization of transcriptomes in mammals: Coregulation and cofunctionality. Genomics 89, 580–587 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Kosak ST et al. Coordinate gene regulation during hematopoiesis is related to genomic organization. PLoS Biol 5, e309 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brinster RL, Allen JM, Behringer RR, Gelinas RE & Palmiter RD Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci U S A 85, 836–840 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fong YW & Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414, 929–933 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Calo E. & Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell 49, 825–837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersson R, Sandelin A. & Danko CG A unified architecture of transcriptional regulatory elements. Trends Genet 31, 426–433 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Kim T-K & Shiekhattar R. Architectural and Functional Commonalities between Enhancers and Promoters. Cell 162, 948–959 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Necsulea A. et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Hezroni H. et al. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep 11, 1110–1122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J. et al. Evolutionary analysis across mammals reveals distinct classes of long non-coding RNAs. Genome Biol 17, 19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional references:
- 29.Bhatt DM et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150, 279–290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engreitz JM et al. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell 159, 188–199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu PD et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31, 827–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T, Wei JJ, Sabatini DM & Lander ES Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keane TM et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cong L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen B. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shishkin AA et al. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat Methods 12, 323–325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engreitz J, Lander ES & Guttman M. RNA antisense purification (RAP) for mapping RNA interactions with chromatin. Methods Mol Biol 1262, 183–197 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Engreitz JM et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gnirke A. et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 27, 182–189 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang S, Holt J, Kao C-Y, McMillan L. & Wang W. A novel multi-alignment pipeline for high-throughput sequencing data. Database (Oxford) 2014, bau057–bau057 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guttman M. et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28, 503–510 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levitt N, Briggs D, Gil A. & Proudfoot NJ Definition of an efficient synthetic poly(A) site. Genes Dev 3, 1019–1025 (1989). [DOI] [PubMed] [Google Scholar]
- 43.Kwak H, Fuda NJ, Core LJ & Lis JT Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339, 950–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Core LJ, Waterfall JJ & Lis JT Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahat DB et al. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat Protoc 11, 1455–1476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adelman K. & Lis JT Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13, 720–731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B. & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Busby M. et al. Systematic comparison of monoclonal versus polyclonal antibodies for mapping histone modifications by ChIP-seq. (2016). doi: 10.1101/054387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mouse ENCODE Consortium et al. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 13, 418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kagey MH et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dixon JR et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fort A. et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet 46, 558–566 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Kent WJ et al. The human genome browser at UCSC. Genome Res 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garber M. et al. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics 25, i54–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindblad-Toh K. et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gentleman RC et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5, R80 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lawrence M. et al. Software for computing and annotating genomic ranges. PLoS Comput Biol 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lawrence M, Gentleman R. & Carey V. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25, 1841–1842 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson JT et al. Integrative genomics viewer. Nat Biotechnol 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Danecek P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.