Bacterial pathogens have evolved to regulate virulence gene expression at critical points in the colonization and infection processes to successfully cause disease. The Shigella species infect the epithelial cells lining the colon to result in millions of cases of diarrhea and a significant global health burden. As antibiotic resistance rates increase, understanding the mechanisms of infection is vital to ensure successful vaccine development. Despite significant gains in our understanding of Shigella infection, it remains unknown how the bacteria initiate contact with the colonic epithelium. Most pathogens harbor multiple adherence factors to facilitate this process, but Shigella was thought to have lost the ability to produce these factors. Interestingly, we have identified conditions that mimic some features of gastrointestinal transit and that enable Shigella to express adherence structural genes. This work highlights aspects of genetic regulation for Shigella adherence factors and may have a significant impact on future vaccine development.
KEYWORDS: Shigella flexneri, adherence factors, long polar fimbriae, type 1 fimbriae, curli, bile salts, glucose, biofilm, in vivo-like conditions, intestinal, epithelial cells
ABSTRACT
The Shigella species are Gram-negative, facultative intracellular pathogens that invade the colonic epithelium and cause significant diarrheal disease. Despite extensive research on the pathogen, a comprehensive understanding of how Shigella initiates contact with epithelial cells remains unknown. Shigella maintains many of the same Escherichia coli adherence gene operons; however, at least one critical gene component in each operon is currently annotated as a pseudogene in reference genomes. These annotations, coupled with a lack of structures upon microscopic analysis following growth in laboratory media, have led the field to hypothesize that Shigella is unable to produce fimbriae or other traditional adherence factors. Nevertheless, our previous analyses have demonstrated that a combination of bile salts and glucose induces both biofilm formation and adherence to colonic epithelial cells. The goal of this study was to perform transcriptomic and genetic analyses to demonstrate that adherence gene operons in Shigella flexneri strain 2457T are functional, despite the gene annotations. Our results demonstrate that at least three structural genes facilitate S. flexneri 2457T adherence for epithelial cell contact and biofilm formation. Furthermore, our results demonstrate that host factors, namely, glucose and bile salts at their physiological concentrations in the small intestine, offer key environmental stimuli required for adherence factor expression in S. flexneri. This research may have a significant impact on Shigella vaccine development and further highlights the importance of utilizing in vivo-like conditions to study bacterial pathogenesis.
IMPORTANCE Bacterial pathogens have evolved to regulate virulence gene expression at critical points in the colonization and infection processes to successfully cause disease. The Shigella species infect the epithelial cells lining the colon to result in millions of cases of diarrhea and a significant global health burden. As antibiotic resistance rates increase, understanding the mechanisms of infection is vital to ensure successful vaccine development. Despite significant gains in our understanding of Shigella infection, it remains unknown how the bacteria initiate contact with the colonic epithelium. Most pathogens harbor multiple adherence factors to facilitate this process, but Shigella was thought to have lost the ability to produce these factors. Interestingly, we have identified conditions that mimic some features of gastrointestinal transit and that enable Shigella to express adherence structural genes. This work highlights aspects of genetic regulation for Shigella adherence factors and may have a significant impact on future vaccine development.
INTRODUCTION
Shigella flexneri is a Gram-negative, facultative anaerobe that infects millions of people each year and that causes watery or bloody diarrhea, cramping, and dehydration. Shigella infection is endemic in developing countries, causing significant mortality and morbidity, particularly in children under the age of 5 years (1). In industrialized nations, infection is episodic and primarily linked to contaminated food or water. Infection in nonimmunocompromised individuals is self-limiting, and most patients recover with oral rehydration therapy and antibiotics (2–4). However, the increasing prevalence of antibiotic resistance (5) highlights the need to pursue effective vaccine strategies in these enteric pathogens that are gaining resistance mechanisms.
The current Shigella infection paradigm is that the bacteria spread through fecal-oral transmission, in which an extremely low infectious dose, with as few as 10 to 100 organisms, initiates infection (2). Once it is ingested, Shigella traverses the digestive tract and localizes to the colon. To invade the colonic epithelium, Shigella transits through M (microfold or membranous) cells, which are specialized antigen-presenting cells of the follicle-associated epithelium (FAE) (6). Transit through M cells allows the bacteria to reach the basolateral pole of the epithelium for invasion (2), and the FAE is considered the major site of entry for Shigella due to the presence of M cells (7). Following basolateral invasion, intracellular replication, and intercellular spread, polymorphonuclear cells are recruited to the site of infection to eliminate the pathogen. The massive tissue destruction that results in the symptoms of bacillary dysentery is due to this intense inflammatory response (2).
While the invasion process and intracellular spread, replication, and survival of Shigella have been thoroughly investigated, much less is known about the virulence dynamics of the bacteria prior to invasion and transcytosis. In fact, there is a critical gap in knowledge regarding how the bacteria target M cells to initiate the invasion process and whether Shigella utilizes adherence factors to adhere to the apical surface of epithelial cells prior to invasion. Due to the mucosal environment encountered on the surface of gastrointestinal epithelial cells, many pathogens, particularly pathogenic Escherichia coli and Salmonella species, often utilize pili, fimbriae, or afimbrial adhesins to efficiently colonize host cells (8–13). Because Shigella and E. coli are closely related (14, 15) and because fimbriae are prevalent among the Enterobacteriaceae (16), it is reasonable to hypothesize that Shigella utilizes fimbriae or other adhesins during colonization. Interestingly, Shigella is thought to have lost the ability to produce traditional E. coli adherence factors as the bacteria adapted to an intracellular lifestyle (2) due to three main reasons. First, Shigella strains grown in standard laboratory media lack visible adhesive structures upon transmission electron microscopy (TEM) (17, 18), unlike some strains of E. coli, in which adherence factors are thought to be constitutively expressed (19, 20). Second, examination of Shigella genomes deposited in GenBank reveals that almost all adherence gene clusters, such as those for type 1 fimbriae (10, 21) and curli (22), contain at least one annotated pseudogene that is crucial for either the adherence factor structure or the assembly process (17, 23, 24). Third, the production of adherence factors is considered counterproductive to the lifestyle of an intracellular pathogen evading immune detection (2, 25, 26).
Despite this null adherence factor hypothesis, a limited number of reports have detected adherence factor expression in S. flexneri (27–29), but, unfortunately, in-depth genetic analyses have not been performed. Furthermore, we have previously demonstrated that tryptic soy broth (TSB) media supplemented with bile salts induce the adherence of S. flexneri 2457T to colonic epithelial cells, which is facilitated at least in part by the type III secretion system effector proteins OspE1 and OspE2 (30). Finally, our recent publication characterizes an adhesive biofilm phenotype following exposure to a combination of bile salts and glucose that represents aspects of the in vivo-like conditions (IVLCs) found in the small intestine (31–36). Given this literature and the fact that deletion of both ospE1 and ospE2 did not completely abrogate adherence (30), we sought to determine if additional adherence genes are expressed by S. flexneri 2457T following exposure to IVLCs. In this study, we performed transcriptomic and genetic analyses to begin to characterize the adherence gene clusters in S. flexneri 2457T. Our results demonstrate that at least three structural genes facilitate adherence for both biofilm formation and colonization of colonic epithelial cells, particularly in the human intestinal organoid-derived epithelial monolayer (HIODEM) model. This work broadens our understanding of S. flexneri 2457T pathogenesis and demonstrates that S. flexneri 2457T likely expresses several traditional adherence factors important for pathogenesis. Insights gained from this work could have an important impact on Shigella-specific therapeutic and vaccine development.
RESULTS
S. flexneri 2457T produces putative adherence structures in IVLCs.
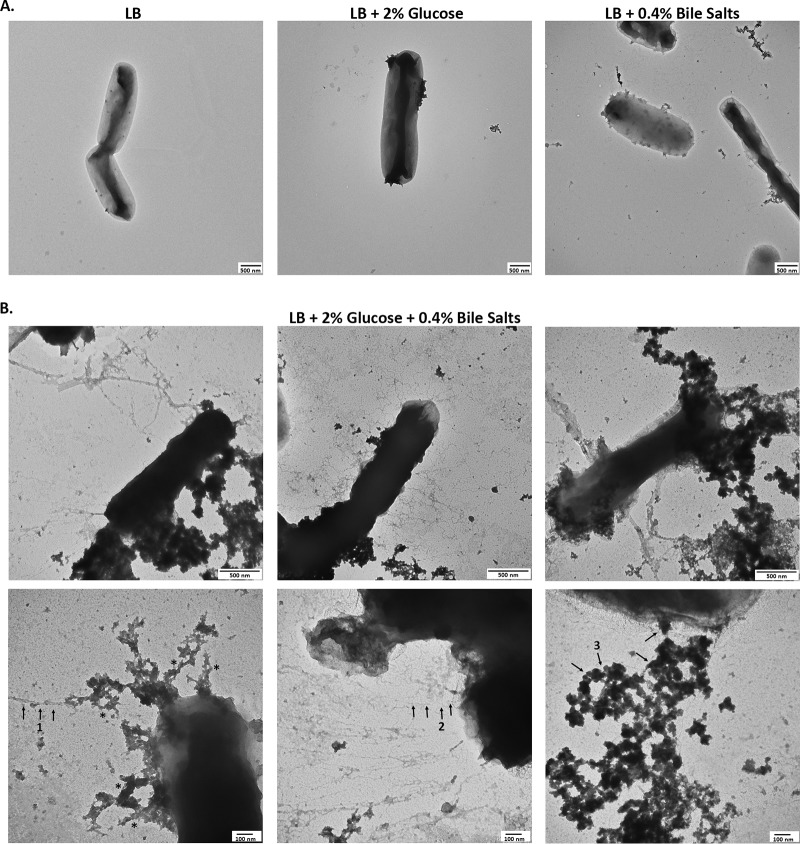
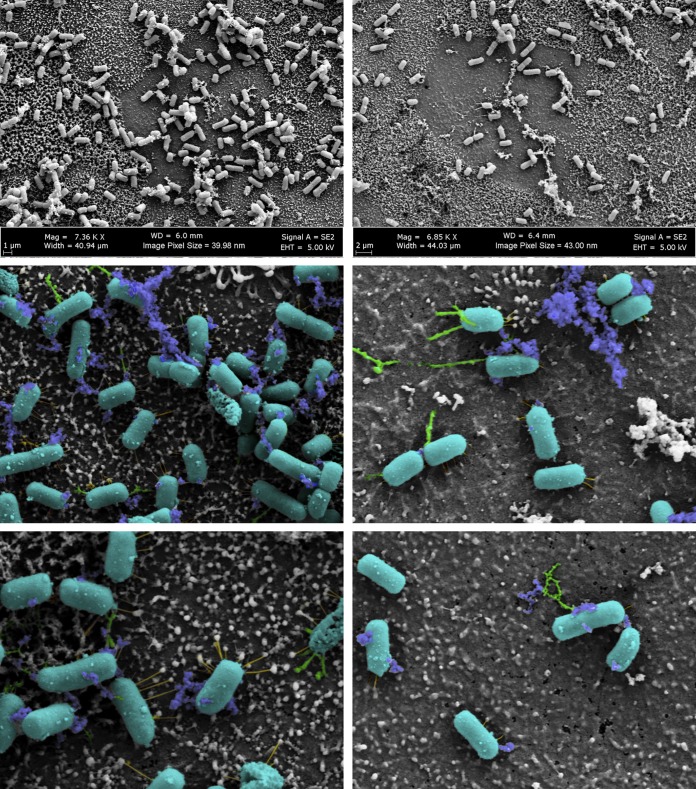
Our previous work demonstrated that S. flexneri 2457T grown in IVLCs produced a biofilm. Furthermore, upon bacterial dispersion from the biofilm, recovered bacteria displayed induced adherence to colonic HT-29 cells. This analysis enabled us to expand the Shigella infection paradigm to incorporate biofilm formation due to exposure to IVLCs during small intestinal passage, biofilm dispersion upon colonic transition following the loss of the bile salts signal, and the subsequently induced infection (31, 35). Since adherence factors are important components of biofilm formation (35, 37), we performed electron microscopy (EM) analysis of bacteria isolated from the IVLC-induced biofilm to visualize possible adherence factors. As shown in Fig. 1 and in Fig. S1 in the supplemental material, bacteria produced thicker and thinner structures of various lengths and electron-dense aggregates in IVLCs. Bacteria grown in Luria broth (LB) and LB supplemented with glucose (2%, wt/vol) lacked structures, while bacteria grown in LB medium supplemented with bile salts (0.4%) produced very minimal structures. The utilization of bile salts in tryptic soy broth (TSB) medium, in which there is additional glucose relative to the amount in LB medium (31), resulted in the appearance of putative adherence structures similar to those seen in LB medium supplemented with both glucose and bile salts (Fig. S1). The data confirmed our observations that glucose and bile salts (IVLCs) are required for S. flexneri 2457T to form an adhesive biofilm (31). To support the biofilm data and our previous induced HT-29 cell adherence observations (30, 31), we performed adherence analysis on a human intestinal organoid-derived epithelial monolayer (HIODEM) model. The model is derived from stem cells isolated from intestinal tissue, propagated as organoids, and subsequently trypsinized and seeded onto transwells to generate a two-dimensional (2-D) polarized, differentiated model of the intestinal epithelium in which enterocytes, mucus-producing goblet cells, and antigen-sampling M cells are present (38–42). With the model derived from the ascending colon, S. flexneri 2457T subcultured in IVLCs displayed putative adherence structures contacting the epithelial cells (Fig. 2). In all, the data suggest that these putative adherence structures are important for both biofilm formation and adherence to colonic epithelial cells.
FIG 1.
The growth of S. flexneri 2457T in IVLCs produces several putative adherence structures. Cultures of wild-type strain 2457T were grown overnight with static growth in the indicated media. Samples were negatively stained and imaged by electron microscopy. Images are representative of those from at least 3 biological replicates. (A) TEM analysis of 2457T grown in LB, LB supplemented with 2% glucose, or LB supplemented with 0.4% bile salts demonstrated that adherence factors were either not produced or minimally produced under these conditions. Magnification, ×25,000; scale bar, 500 nm. (B) 2457T grown in LB supplemented with both 2% glucose and 0.4% bile salts (IVLCs) revealed three types of putative adherence factors upon TEM analysis. Magnifications, ×50,000 (top row; scale bar, 500 nm) and ×100,000 magnification (bottom row; scale bar, 100 nm). Different structures are highlighted by numbered arrows. Arrow 1 points to thicker structures, arrow 2 points to thinner structures, and arrow 3 points to electron-dense aggregates. The asterisks denote rough, complex structures (refer to Discussion).
FIG 2.
Scanning electron microscopy of S. flexneri 2457T adherence on the human intestinal organoid-derived epithelial monolayer model. S. flexneri 2457T was subcultured in IVLCs, washed, and applied for adherence analysis on the HIODEM model. Following infection, samples were fixed and processed for SEM analysis. The images on the top row display the association of bacteria with the cells of the model. The middle and bottom rows are images from a higher magnification. Magnifications, approximately ×7,000 (top row) and approximately ×20,000 (bottom row). Pseudocoloring was performed to enhance the visualization of the three types of putative adherence factors visualized on the bacteria that interact with the apical surface of the epithelial cells. The bacterial rods are colored teal, thicker structures are colored green, thinner structures are colored yellow, and electron-dense aggregates are colored blue. The images in the left and right columns represent those from two separate biological samples.
Additional images. (A) Additional images of strain 2457T grown in LB with 2% glucose and 0.4% bile salts. Further TEM analyses were performed to capture additional phenotypes of putative adherence structures and provide higher-magnification images to demonstrate that the structures originate from the bacterial cells (white arrows). Asterisks mark areas that displayed possible curli fibrils. The images are from experiments biologically independent of those from which the images provided in Fig. 1 and 7 were obtained. Each image displays a separate bacterial cell unless paired with a matching boxed sample. The magnifications and scale bars are provided for each image or image sets. (B) Wild-type cultures of 2457T were grown overnight by static growth in TSB media supplemented with 0.4% (wt/vol) bile salts. TEM analysis revealed structures similar to those of bacteria grown in LB supplemented with 2% glucose and 0.4% bile salts. Magnification, ×50,000; bar, 500 nm. (C) Two images of control media (LB with 2% glucose and 0.4% bile salts) are on top. Samples were prepared and processed as described in the Materials and Methods section for the biofilm assay and subsequent TEM grid preparation. TEM analysis revealed some precipitates of material from either the media or the uranyl acetate stain; however, the precipitation varied in concentration and did not resemble the structures observed in the presence of bacteria. Magnifications, ×50,000; bars, 500 nm (from separate experiments). (D) An image of an area without bacteria present of a TEM grid prepared from the 2457T biofilm (grown in IVLC medium). The same precipitation seen on the control medium grids tended to be less concentrated in clean areas of the bacterial grids. Magnification, ×50,000; bar, 500 nm. Download FIG S1, PDF file, 1.7 MB (1.7MB, pdf) .
Copyright © 2019 Chanin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
S. flexneri 2457T maintains and transcribes several adherence gene clusters.
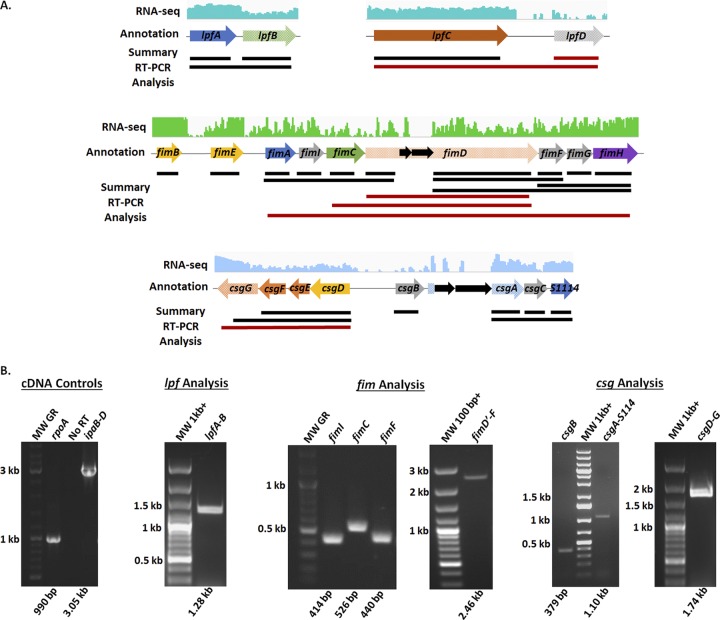
We next examined the transcription of the S. flexneri 2457T adherence genes under various conditions. In silico analyses of the annotated S. flexneri 2457T genome in the NCBI GenBank database identified several adherence gene components (Table 1, Fig. 3, and Fig. S2). These genes are maintained in S. flexneri 2457T, despite examples of full gene and/or operon deletions for some of the adherence gene clusters in other Shigella species (23). As documented in previous studies (17, 23, 24), all S. flexneri 2457T adherence gene clusters contain at least one annotated pseudogene (due to predicted point mutations, truncations, or insertion sequences), supporting hypotheses stating that Shigella cannot produce traditional adherence factors. However, our previous RNA sequencing (RNA-seq) data (31) indicated that despite the gene annotations, most of the adherence genes were transcribed by S. flexneri 2457T (Fig. 3 and Fig. S2). To confirm the RNA-seq results, we performed reverse transcription-PCR (RT-PCR) analysis of the annotated adherence gene clusters (Fig. 3 and Fig. S2). RNA isolated from S. flexneri 2457T broth cultures were positive for transcription of the adherence genes and large segments of the predicted operons. Insertion sequences did not prevent the transcription of large downstream segments. For example, as demonstrated in Fig. 3, we amplified cDNA products from fimD just after the insertion sequences to the end of fimF. Finally, we utilized quantitative RT-PCR (qRT-PCR) analysis to obtain additional data to support the transcription of adherence genes. As described in previously published literature for other pathogens (43–46), glucose induced the expression of the S. flexneri 2457T genes encoding structural subunits (Fig. 4). In all, the data indicate that adherence gene clusters are genomically maintained and transcriptionally regulated in S. flexneri 2457T, despite the pseudogene annotations.
TABLE 1.
Adherence gene clusters identified in the Shigella flexneri 2457T genomea
| Adherence gene | Locus tag | Annotated function/pseudogene |
|---|---|---|
| Long polar fimbriae | ||
| lpfA | S3961 | Major fimbrial subunit |
| lpfB | Chaperone/pseudogene | |
| lpfC | S4048 | Outer membrane usher |
| lpfD | Fimbrial protein/pseudogene | |
| Type 1 fimbriae | ||
| fimB | S4467 | Recombinase |
| fimE | S4466 | Tyrosine recombinase |
| fimA | S4465 | Major fimbrial subunit |
| fimI | S4464 | Pilus biosynthesis protein |
| fimC | S4463 | Chaperone |
| fimD | S4462 | Outer membrane usher/pseudogene |
| fimF | S4458 | Minor subunit |
| fimG | S4457 | Minor subunit |
| fimH | S4456 | Tip adhesin |
| Curli operon | ||
| csgG | S1104 | Assembly protein/pseudogene |
| csgF | S1105 | Assembly protein |
| csgE | S1106 | Assembly protein |
| csgD | S1107 | Transcriptional regulator |
| csgB | S1108 | Minor subunit |
| csgA | S1109 | Major subunit/pseudogene |
| csgC | S1113 | Autoagglutination assembly protein |
| S1114 | S1114 | Hypothetical (fimA homolog) |
| ybg operon | ||
| ybgD | S0591 | Fimbria-like protein (fimA homolog) |
| ybgQ | S0592 | Outer membrane protein (fimD homolog) |
| ybgP | S0593 | Chaperone (fimC homolog) |
| ybgO | S0594 | Putative fimbria-protein/pseudogene |
| ycb operon | ||
| ycbQ | S1003 | Fimbria-like adhesin/pseudogene |
| ycbS | S1006 | Outer membrane usher protein |
| S1007 | S1007 | Fimbrial protein |
| S1008 | S1008 | Fimbria-like protein |
| S1009 | S1009 | Fimbria-like protein (fimA homolog) |
| ycbF | S1010 | Putative pilus assembly chaperone |
| yeh operon | ||
| yehD | S2298 | Fimbria-like protein |
| yehC | S2297 | Chaperone/pseudogene |
| yehB | S2296 | Outer membrane usher protein |
| yehA | S2295 | Putative fimbria-protein/pseudogene |
| yra operon | ||
| yraH | S3396 | Fimbria-like adhesin/pseudogene |
| yraI | S3397 | Chaperone (fimC homolog) |
| yraJ | S3398 | Outer membrane usher/pseudogene |
| yraK | S3403 | Fimbrial protein |
| S4250-S4254 | ||
| S4250 | S4250 | Pilus assembly chaperone (papD_C homolog) |
| S4254 | S4254 | Outer membrane usher/pseudogene |
| S3342-S3341 | ||
| S3342 | S3342 | Hypothetical (fimD homolog) |
| S3341 | S3341 | Hypothetical (pilus biogenesis initiator homolog) |
| sfm operon | ||
| sfmA | S0469 | Fimbria-like protein (fimA homolog) |
| sfmC | S0470 | Chaperone |
| sfmD | Outer membrane usher/pseudogene |
Boldface entries provide information for the genes deleted in this study.
FIG 3.
In silico and transcriptomic analysis of the S. flexneri 2457T adherence gene clusters. (A) Identification of the long polar fimbriae (lpf), type 1 fimbriae (fim), and curli (csg) adherence gene clusters in the S. flexneri 2457T genome was performed using the NCBI GenBank tool. Arrows represent annotated open reading frames (ORF), in which blue arrows represent the annotated major subunits, green arrows represent the annotated chaperones, orange arrows represent the annotated ushers or pores, yellow arrows represent the annotated regulatory subunits, purple arrows represent the annotated tip adhesins, and gray arrows represent additional putative adherence genes involved in assembly or secretion. All arrows with checkered backgrounds are annotated as pseudogenes in NCBI due to predicted insertion sequences, truncations, or frameshifts. Insertion sequences or transposons are represented by solid black arrows. The RNA sequencing trace read data are provided for each gene cluster above the arrows. The best trace read available is presented, with green representing shaking growth conditions (darker green for growth in bile salts) and blue representing static growth conditions (darker blue for growth in bile salts). The solid lines below the arrows represent a summary of the confirmation of gene transcription that resulted from nonquantitative RT-PCR analyses. Single-gene and polycistronic amplifications were performed to obtain the largest products possible for each operon. Red lines represent products that were not obtained. Refer to Fig. S2 in the supplemental material for the results of additional gene cluster analyses. (B) Representative gel electrophoresis images of nonquantitative RT-PCR analyses of the various adherence gene clusters are provided. Biologically independent RNA samples were used in the analysis, in which RNA integrity was verified by amplifying the housekeeping gene rpoA as well as the ipaB to ipaD operon encoded on the virulence plasmid. Control amplifications without reverse transcriptase in the rpoA reaction mixtures ensured that there was no DNA contamination of the RNA samples prior to cDNA synthesis. Each gene is labeled with the expected molecular weight (MW) of the product provided below the gel image. Note that different molecular weight ladders were utilized in the analyses. Refer to the Materials and Methods section for more information.
FIG 4.
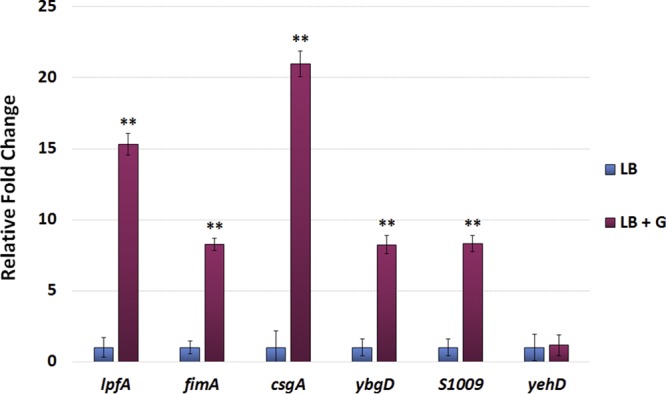
Quantitative RT-PCR analysis of S. flexneri 2457T structural adherence genes. RNA was isolated from S. flexneri 2457T grown in medium (LB) or medium supplemented with 2% (wt/vol) glucose (LB + G). For each primer set, the relative fold change ± the standard error of the ΔCT value is plotted for each gene. Data represent the average for at least three biological independent samples, in which each sample had technical duplicates. yehD (Table 1) expression was not induced in the presence of glucose and therefore served as an internal negative control for the analysis. **, P ≤ 0.01 between LB and LB-glucose conditions.
Additional adherence gene cluster analysis. Analysis was performed as described in Materials and Methods as well as in the Fig. 2 legend. A 10-fold enhanced view of the trace reads are provided for genes S3342 and sfmA to sfmC to demonstrate the presence of RNA-seq reads. Download FIG S2, PDF file, 0.2 MB (226.5KB, pdf) .
Copyright © 2019 Chanin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutational analyses of three S. flexneri 2457T adherence structural genes.
We next performed mutational analyses of the genes encoding major structural subunits to demonstrate functional roles in epithelial cell adherence and biofilm formation. We concentrated our analyses on genes that encode long polar fimbriae, type 1 fimbriae, and curli, based on our in silico analyses, the combined appearance of the structures in Fig. 1 and 2, and the known functional roles of these structures in initial biofilm formation and epithelial cell adherence in other pathogens (8, 28, 47–49). Thus, we constructed ΔlpfA, ΔfimA, and ΔcsgA mutants. We also constructed a double ΔcsgAB mutant due to the additional role of the csgB minor subunit in adherence (50).
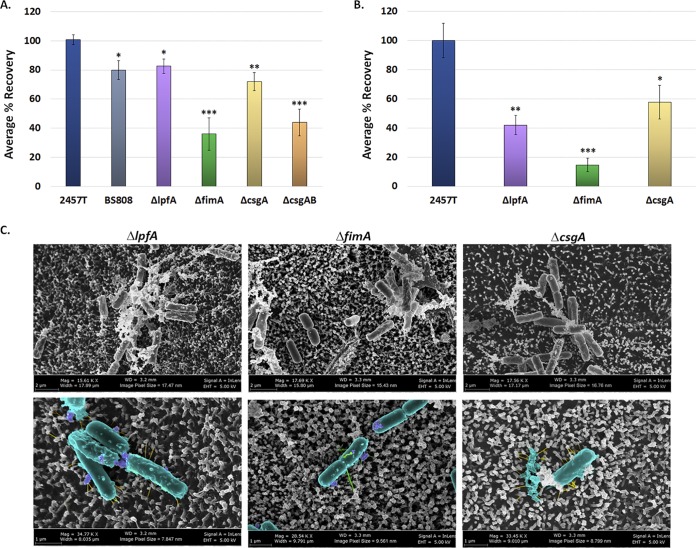
Functional analyses of the mutants were performed to evaluate the role of each factor in adhesion to epithelial cells and biofilm formation. First, we hypothesized that the adherence factors expressed in the IVLCs would facilitate epithelial cell contact. This hypothesis was supported by our previous observations of induced S. flexneri 2457T adherence to HT-29 cells following biofilm dispersion under conditions that mimicked the loss of the bile salts signal during the terminal ileum-to-colon transition (31). All mutants had significant reductions in adherence relative to wild-type bacteria, with the ΔfimA and ΔcsgAB mutants having the greatest reductions (Fig. 5A). The double ΔospE1 ospE2 mutant (strain BS808) served as an adherence mutant control, given our previous analysis of the role of OspE1 and OspE2 in bile salt-mediated adherence (30). To ensure that the mutations did not affect the overall invasive ability of each strain, invasion assays were performed using conventional methods of centrifugation to initiate host cell contact (51). All mutants retained wild-type levels of invasion following centrifugation of the bacteria onto the HT-29 cells (data not shown), which confirmed that the mutations did not affect the basic invasion phenotype of the strains. Finally, to confirm the HT-29 cell adherence data, we evaluated the ΔlpfA, ΔfimA, and ΔcsgA mutants in the HIODEM model and found that each mutant had significantly reduced adherence relative to wild-type bacteria (Fig. 5B). EM analysis of infected samples enabled the visualization of mutants with a smoother surface and less visible structures than wild-type bacteria (Fig. 2 and 5C). Combined, the data demonstrate that the S. flexneri 2457T lpfA, fimA, and csgAB gene products have functional roles in adherence to colonic epithelial cells.
FIG 5.
Analysis of epithelial cell adherence for each S. flexneri 2457T adherence mutant. (A) Wild-type S. flexneri strain 2457T, the control strain BS808 (ΔospE1 ΔospE2), and the adherence mutants (the ΔlpfA, ΔfimA, ΔcsgA, and ΔcsgAB mutants) were grown overnight as described in the text for the biofilm assay. On the next day, bacteria were collected, washed with 1× PBS, and analyzed for adherence to HT-29 cells. Each mutant had a significant reduction in adherence relative to wild-type strain 2457T, with the most significant reductions being seen with the ΔfimA and ΔcsgAB mutants. (B) To verify the data obtained with HT-29 cells, the ΔlpfA, ΔfimA, and ΔcsgA mutants were evaluated for adherence in the HIODEM model. Like with the HT-29 cells, each mutant displayed a significant reduction in adherence relative to wild-type strain 2457T, with the ΔfimA mutant having the greatest reduction. For both panels A and B, the average percent recovery of adherent bacteria ± standard error relative to that of wild-type strain 2457T is plotted. The results for HT-29 cell infections represent those from three independent experiments in which each experiment had technical triplicates. The results for HIODEM infections represent those from two independent experiments in which each experiment had at least two technical replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Scanning electron microscopy analysis of the ΔlpfA, ΔfimA, and ΔcsgA mutants on the HIODEM model enabled visualization of the adherence structures for each mutant. Magnifications, ×15,600 to ×17,700 (top row) and ×28,500 to ×34,800 (bottom row). To facilitate the visualization of adherence structures, pseudocoloring was performed for the images in the bottom row, as described in the legend to Fig. 2. The ΔlpfA mutant lacked the thicker structures, the ΔfimA mutant lacked the thinner structures, and the ΔcsgA mutant lacked the electron-dense aggregates.
Second, given the importance of adherence in the initiation of biofilms (37, 52), we analyzed the mutants in the biofilm assay (31, 35). All mutants exhibited reduced biofilm formation at 3 h (Fig. 6), a time point used to examine the role of adherence factors in early biofilm formation (35). A quadruple mutant (the ΔcsgAB ΔlpfA ΔfimA mutant) was constructed to confirm the results of the biofilm analyses and displayed the greatest reduction in biofilm formation relative to wild-type bacteria. Thus, we concluded that the lpfA, fimA, and csgAB gene products also have roles in the adhesion process for IVLC-induced biofilm formation in S. flexneri 2457T.
FIG 6.
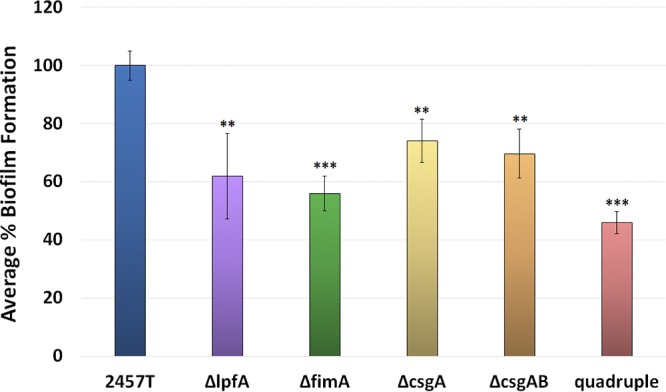
Analysis of biofilm formation for each S. flexneri 2457T adherence mutant. Wild-type S. flexneri strain 2457T and the ΔlpfA, ΔfimA, ΔcsgA, ΔcsgAB, and quadruple mutants were analyzed for biofilm formation at 3 h postinoculation to examine the adherence phase of biofilm formation in IVLCs. Each mutant had a significant reduction in biofilm formation. The average percent biofilm formation ± standard error relative to that for 2457T is plotted for each mutant. All data represent those from three biological independent experiments in which each experiment had at least three technical replicates. **, P < 0.01; ***, P < 0.001.
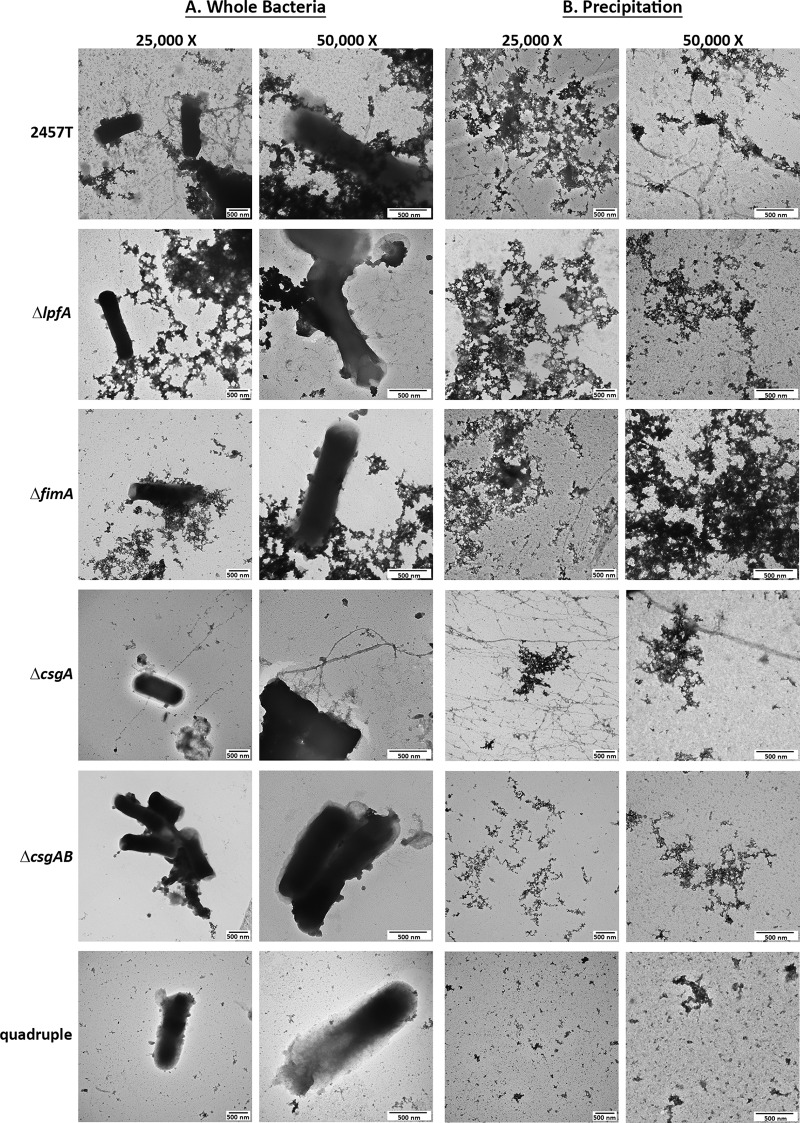
Finally, the mutants were evaluated by EM for the loss of surface structures. For the single mutants, each mutation resulted in the loss of a structure, while it also facilitated visualization of two other predominant structures. No apparent structures were visualized in the quadruple mutant (Fig. 7A). Furthermore, ammonium sulfate precipitation for the isolation of adherence factors (53) was performed to verify our results (Fig. 7B). Finally, to provide additional data for the presence of a csgA gene product despite the disorganized appearance, the Congo red (CR) binding assay was performed, given the ability of Congo red dye to bind the amyloid structures of curli and produce a birefringence signal under polarized light (54–56). A positive birefringence signal was detected for both wild-type strain 2457T and the ΔlpfA mutant, which was used as a mutation control for this assay. However, the ΔcsgA mutant produced significantly less signal (Fig. S3). In all, these analyses suggest that we identified the genes encoding the structural subunits of the putative adherence structures expressed by S. flexneri 2457T.
FIG 7.
TEM analysis for each S. flexneri 2457T adherence mutant. (A) The ΔlpfA, ΔfimA, ΔcsgA, ΔcsgAB, and quadruple mutants were grown statically overnight in IVLCs, subsequently processed for TEM analysis, and analyzed with wild-type strain 2457T as a control. Each mutation resulted in the loss of either the thicker structures (ΔlpfA mutant), the thinner structures (ΔfimA mutant), or the electron-dense aggregates (ΔcsgA and ΔcsgAB mutants). The quadruple mutant did not display visible structures. Images for wild-type strain 2457T are from experiments separate and biologically independent of those used to obtain the images provided in Fig. 1. (B) To verify the results, ammonium sulfate precipitation was performed to isolate and visualize structures from wild-type S. flexneri strain 2457T and each of the five mutants. The three types of factors can be visualized in wild-type bacteria; however, only two of the three structures were present for the single mutants. Each mutation resulted in the expected loss of structure, and no structures were visualized in the quadruple mutant. The data verify that the correct structural subunit was deleted for each mutant. All images are representative of those from at least two biological independent experiments. Different fields are presented for the images with ×25,000 and ×50,000 magnifications for all images in panels A and B, in which both sets of images display the 500-nm scale bar.
Congo red binding assay. The assay was performed to test for the presence of the CsgA protein due to the ability of the Congo red dye to bind amyloid fibers and produce a birefringence signal under polarized light. The apple green color indicates the Congo red fluorescence that occurs when amyloid fibers are present. Wild-type 2457T produced a positive signal, while a significant reduction in the signal was detected for the ΔcsgA mutant. The ΔlpfA mutant was analyzed as a mutation control, and as seen, the mutation did not affect the birefringence signal. Images are representative of those from at least two biological independent experiments. Download FIG S3, PDF file, 0.2 MB (222.1KB, pdf) .
Copyright © 2019 Chanin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MS analysis to evaluate secretion of adherence structural proteins in IVLCs.
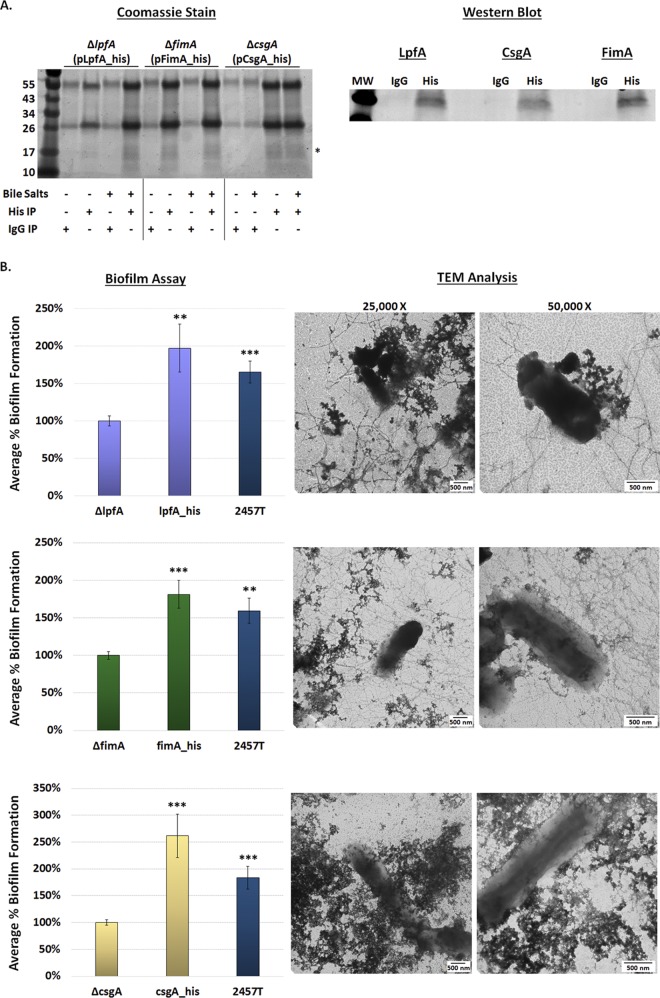
As a method to detect the presence and secretion of the LpfA, FimA, and CsgAB proteins, proteomic analyses were performed on culture supernatants from the biofilm assay. Both intact mass spectrometry (MS) analysis and peptide fingerprinting MS/MS analysis of trypsin-digested samples confirmed the presence of LpfA, FimA, CsgA, and CsgB, with each protein having high levels of sequence coverage upon the fingerprinting MS/MS analysis (Table 2), indicating that the proteins were secreted in IVLCs. Due to the complexity of the samples for MS analysis, especially those from the extracellular polymeric substance (EPS) matrix produced by the IVLC-induced biofilm (31), a higher than expected mass error was observed. Therefore, we cloned the lpfA, fimA, and csgA genes from S. flexneri 2457T, added a histidine tag to the genes, and transformed each respective mutant to perform immunoprecipitation and complementation analyses. As shown in Fig. 8, the tagged LpfA, FimA, and CsgA proteins were expressed in the respective mutants, secreted, and purified from IVLC-induced biofilm culture supernatants, which confirmed the MS data. Biofilm assay analyses and TEM visualization of the overexpressed structures not only verified that these tagged constructs were functional (Fig. 8) but also provided additional data to confirm the findings of the mutational and EM analyses and thus demonstrate that the lpfA, fimA, and csgAB genes produce functional adherence proteins in S. flexneri 2457T.
TABLE 2.
Mass spectrometry analysis of the bile salt-induced biofilm supernatants
| Protein | Intact avg mass (Da) |
Mass error (ppm) |
Response (total ion count) |
Digested peptide coverage (%) |
|
|---|---|---|---|---|---|
| Observed | Expected | ||||
| LpfA | 19,644.38 | 19,642.80 | 86.8 | 583,652,552 | 100 |
| FimA | 18,348.20 | 18,347.28 | 49.8 | 221,292,058 | 82 |
| CsgA | 12,389.58 | 12,388.92 | 53.4 | 203,325,206 | 84 |
| CsgB | 15,870.81 | 15,869.62 | 74.9 | 369,128,136 | 91 |
FIG 8.
Immunoprecipitation and functional complementation of the histidine-tagged proteins. (A) To confirm the results of mass spectrometry analysis, bacteria expressing His-tagged proteins were cultured in the overnight biofilm assay and the culture supernatants were immunoprecipitated. (Left) IgG negative-control and anti-His immunoprecipitation (IP) experiments were performed in media with or without IVLCs with analysis by SDS-PAGE with Coomassie blue staining. Proteins in the 17-kDa range (*) were immunoprecipitated with the anti-His resin and in the presence of bile salts. (Right) Western blot analysis of proteins in the 17-kDa range confirmed that the His-tagged LpfA, FimA, and CsgA were secreted into the supernatant of the biofilm culture. No proteins were detected in the negative-control samples in which IgG was used in the immunoprecipitation. (B) Biofilm and TEM analyses were used to verify that the His-tagged proteins were functional and could complement the respective mutants. The complemented strains (strains complemented with lpfA [top], fimA [middle], and csgA [bottom]) produced significantly higher biofilms than the mutants to indicate the restoration of function, while TEM analysis enabled visualization of the overexpressed structures. The results of the biofilm assays were analyzed at 4.5 h. The data represent the average from two independent experiments, each with six technical replicates, ± standard error. **, P < 0.01; ***, P < 0.001. The level for each complemented strain averaged the same or above the level for wild-type strain 2457T (the differences were not significant). Note the differences in the y axis range between the analysis with the csgA mutant and the analyses with the lpfA and fimA mutants.
DISCUSSION
Characterization of the three structural genes in this study provides evidence that S. flexneri 2457T utilizes traditional adherence factors to initiate biofilm formation and to facilitate contact with colonic epithelial cells. Several observations influenced the investigation, including the lack of an adherence-null mutant in OspE1 and OspE2 analysis (30), the subsequent biofilm formation and induced adherence observed following IVLC exposure (31, 35), as well as the presence of the various adherence gene clusters in the S. flexneri 2457T genome. The literature on traditional Shigella adherence factors is contradictory. Numerous studies have suggested that the various gene clusters have been lost during evolution as a pathoadaptive response to the host. Notably, the laboratory growth methods consistently used to demonstrate fimbrial production in strains of E. coli (19, 20) were not successful for either lab strains or clinical isolates of Shigella (17, 18). Our analyses with control medium, in which the combination of glucose and bile salts was absent, confirmed many of these previous findings on the phenotypic level. The visualization of putative adherence structures required the addition of both glucose and bile salts to the media, signals that are present in the small intestine during host transit (31–36). Interestingly, glucose induced the transcription of the structural subunits (Fig. 4), yet adherence factors were not visible in LB medium-glucose treatment, while minimal adherence factors were visualized in the LB medium-bile salts treatment (Fig. 1). Thus, based on the data presented in Fig. 1 and 4, we hypothesize that glucose induces structural gene transcription, while bile salts serve as a secretion signal. The amount of glucose required for signaling can vary, as is evident by the different percentages of glucose in TSB and the glucose-supplemented LB medium, a finding which is consistent with our previous observations (31). Nevertheless, this work highlights the importance of using physiological conditions to study bacterial pathogenesis, especially for human-adapted pathogens like Shigella.
The combined RNA-seq and RT-PCR analyses of the adherence gene clusters demonstrate that some of the gene annotations are accurate, while other annotations require refinement. For example, the csgG gene is annotated as a pseudogene due to a point mutation that creates an in-frame stop codon. The RT-PCR analysis confirmed this annotation, since a partial csgG product was detected prior to the stop codon; however, no product was detected with a reverse primer that annealed downstream of this mutation. As another example, there was significant transcription of the ycbQ gene, despite the truncated pseudogene annotation. Finally, while the full ybgO gene could not be amplified under the conditions examined, inspection of the primary genomic sequence (GenBank accession number AE014073.1) combined with the RNA-seq read mapping indicates that two separate open reading frames or small RNAs may be transcribed in this region. The effects of transcription of these partial gene fragments on S. flexneri 2457T gene regulation or adherence factor expression will require additional analyses.
The mutational and complementation analyses demonstrated functional adherence roles for the products of the lpfA, fimA, and csgAB structural genes. While additional direct evidence of each structure is required and such comprehensive studies are under way, the EM, mutational, and complementation analyses provided significant support for the appearance and function of each structure. Long polar fimbriae have been shown to be important for pathogenic E. coli and Salmonella interactions with M cells during intestinal colonization and can facilitate biofilm formation in pathogenic E. coli strains. The lpfA genes have also been demonstrated to be induced by bile salts (13, 57–61). As seen in Fig. 2, thicker structures are bound to the surface of cells lacking microvilli, which is a hallmark of M cells (62). Additionally, the ΔlpfA mutation had a greater effect on adherence in the HIODEM model, in which M cells are present (38–41), than on adherence to HT-29 cells (Fig. 5). For type 1 fimbriae, previous studies support our observations of both fimA gene transcription and soluble FimA expression. First, clinical isolates of Shigella produced fimbria-like adhesins after periods of prolonged static growth; however, the genes encoding the factors were not identified (28). Second, another RNA-seq study detected significant induction of the type 1 fim operon in a ΔicgR mutant of S. flexneri 2457T during the intracellular phase of the Shigella lifestyle (63). Finally, soluble S. flexneri FimA protects mitochondrial integrity and epithelial cell survival during infection (64). It is worth noting that the predicted type 1 fimbria-like structures visualized from the biofilm assays (Fig. 1 and 7) appeared to be thinner than the fimbria-like structures visualized during infection (Fig. 2 and 5). We hypothesize that the structures may appear atypical relative to observed E. coli structures, especially since a truncated or substituted FimD (see below) could affect assembly. While the ΔfimA mutant analyses resulted in fewer visualized fimbria-like structures (Fig. 5 and 7), we currently cannot rule out the possibility of the contribution of or compensation by the additional S. flexneri 2457T fimA homologs or other genes (Table 1), particularly under bile salt conditions that induce such a strong biofilm response (31, 65). For example, it was recently demonstrated that the IcsA autotransporter protein facilitates S. flexneri cell-cell interactions in bile salt-induced biofilms (66). Nevertheless, as was the case for strains complemented with lpfA and csgA, analysis of the histidine-tagged fimA-complemented strain provided further data on the appearance of the structures while verifying their function (Fig. 8). Thus, there is strong evidence that the type 1-like fimbriae visualized in our analyses are due to expression from the fimA structural gene.
The proteins generated by the csgAB genes in S. flexneri 2457T appear to be disorganized and to have a limited organization compared to the conventional curli fiber structures detected in other pathogens (47, 54). This lack of complete assembly could be due to a truncated CsgA protein or due to the incomplete production of CsgG, the outer membrane lipoprotein involved in the stability of the curlin proteins during assembly (22, 47, 67). Furthermore, a truncated CsgG may prevent appropriate interaction with CsgF, thereby affecting curli assembly (47, 68). Our analyses indicate that a soluble portion of CsgA is produced in S. flexneri 2457T and that it is sufficient to provide function in adherence, particularly in the establishment of the IVLC-induced biofilm. This soluble portion of the CsgA protein is likely facilitated by a functional CsgB minor subunit protein, given the further reduction in phenotypes of the double ΔcsgAB mutant, the visualization of electron-dense aggregates in the ΔcsgA mutant (Fig. 5 and 7), and the demonstration that CsgB has a role in adherence (50). Interestingly, our EM images suggest that csgAB products may exploit other adherence structures as a scaffold for a more appropriate organization (e.g., see the rough, complex structures marked by asterisks in Fig. 1B). Moreover, the additional electron-dense material visualized in the mutants, particularly with the ΔcsgAB mutant and the quadruple mutants in Fig. 7, is likely from the cellulose component of the EPS matrix, which is also controlled by the transcriptional regulator CsgD (69). Treatment of the S. flexneri 2457T IVLC-induced biofilm with cellulase, which hydrolyzes β-1,4 glycosidic linkages (70), resulted in a significant reduction in the IVLC-induced biofilm (see Fig. S4 in the supplemental material). Further characterization of the electron-dense material is required, but the combined EM analyses (Fig. 5, 7, and 8) demonstrate that much of this material is due to the presence of the csgA and csgB genes. Thus, regardless of the appearance, our data demonstrate that the csgAB products are produced in S. flexneri 2457T and have functional roles in adherence.
Treatment of the IVLC-induced biofilm with cellulase. The biofilm formation analysis was performed with or without cellulase to analyze the contribution of cellulose. A significant reduction in biofilm formation was detected in the presence of cellulase. All data represent those for three average OD540 readings from three biological independent experiments in which each experiment had technical triplicates. ***, P < 0.001. Download FIG S4, PDF file, 0.04 MB (40.3KB, pdf) .
Copyright © 2019 Chanin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
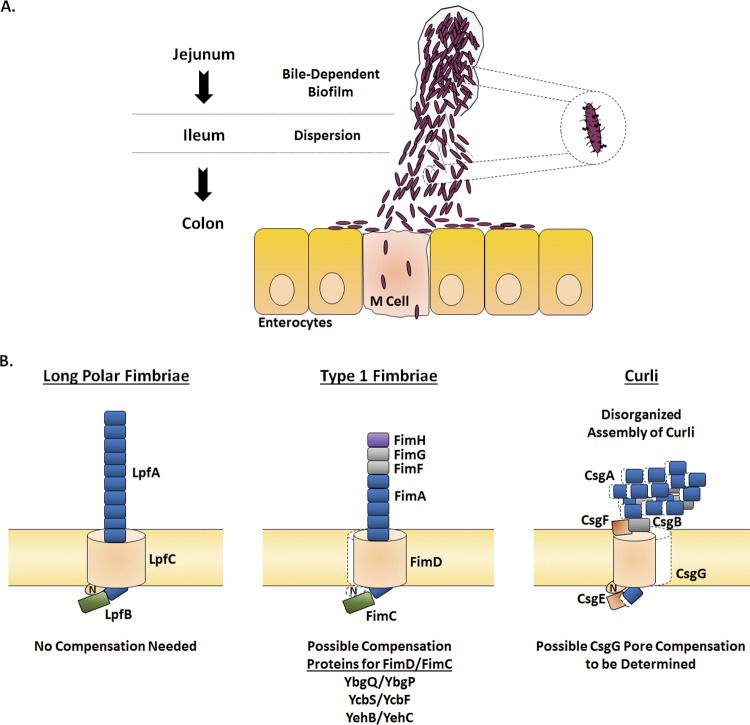
The pseudogene annotations, particularly for the genes encoding the pores or chaperone-usher components required for assembly of the major structural subunits, warrant future investigations into determining how S. flexneri 2457T assembles adherence structures (Fig. 9). If fimD is nonfunctional, we hypothesize that homologous genes located in other genomic locations may compensate for a pseudogene in an operon, if needed. For example, the ybgQ, ycbS, or yehB ushers and accompanying chaperone genes may compensate for the truncated expression of fimD in the fim operon to enable FimA secretion and assembly. This hypothesis is supported by the demonstration of fimbrial promiscuity in biogenesis in E. coli, in which heterologous structural subunits or secretion systems from different operons are utilized to generate and assemble intact structures (71, 72). Fimbrial promiscuity has also been suggested for Proteus mirabilis, since soluble Fim14A was detected by MS in the extracellular environment, despite an incomplete operon in which the chaperone is absent and the usher is annotated as a pseudogene. Proteus mirabilis encodes 17 chaperone-usher fimbrial operons, and therefore, compensation by one of the other operons is hypothesized to enable Fim14A secretion (73). Thus, functional products are likely produced by the other S. flexneri 2457T operons, especially for the ushers, given the transcriptomic analyses performed and the identification of at least three fimD homologs throughout the genome, as denoted by the color coding in Fig. 3 and Fig. S2.
FIG 9.
Hypothesized model of Shigella flexneri adherence factor expression during infection. (A) Based on our combined analyses performed previously (31, 35) and here, we propose the following infection model. Shigella enters the small intestine and resists the initial bile salts exposure that occurs in the duodenum. As food is digested in the duodenum and jejunum, free glucose becomes available. The combined bile salts and glucose signals result in adherence factor expression and biofilm formation. Following transit through the majority of the small intestine, biofilm dispersion is triggered by loss of the bile salts signal in the terminal ileum and colon. Subsequently, Shigella adheres to enterocytes and M cells to initiate the infection process. The figure was adapted from our previous publication (31). (B) The hypothesized protein organization for each of the three adherence factors based on the analyses performed in this study is provided. First, the major components for the long polar fimbriae are likely intact and functional. Second, we hypothesize that the major and minor structural subunits for type 1 fimbriae are intact to form a full structure. Due to the insertion sequence in fimD, we predict either a truncated protein in which the N terminus is missing (highlighted by the white shadow relative to E. coli) or compensation by one of the other FimD homologues listed to enable secretion of the structural subunits. Depending on the truncation, FimC may still be able to interact with FimD or the homologues to serve the appropriate chaperone function. Otherwise, the corresponding FimC homologue would be utilized. Third, due to the N-terminal CsgA truncation and/or the C-terminal CsgG truncation, we hypothesize secretion but not the complete assembly of CsgA and CsgB for curli. Curli assembly may also be affected by a possible lack of CsgF interaction with a truncated CsgG. Colors for the proteins correspond to the color scheme described in the legend to Fig. 3.
In conclusion, we have demonstrated that at least three S. flexneri 2457T adherence structural genes generate functional products for IVLC-induced biofilm formation and adherence to colonic epithelial cells, despite the presence of any mutations that would normally inhibit expression. Future investigations, including in-depth analyses defining the mechanism of adherence factor production and secretion in IVLCs, the effects of an anerobic environment as the bacteria transition to the colon, and studies with other Shigella species, will enhance our understanding of the evolution of this pathogen. Analysis of two clinical S. flexneri isolates thus far demonstrated conserved phenotypes (Fig. S5). We hypothesize that the pathoadaptive changes that Shigella sustained were not the loss of adhesion expression but, rather, a precise control of gene expression to enable the production of adhesins only when necessary and in instances that are most beneficial to the pathogen. We agree that constitutive expression of these adherence factors would possibly interfere with the pathogenic lifestyle of Shigella and impair critical immune evasion tactics. A similar regulation of adhesion genes has been described for other bacterial pathogens, such as enterotoxigenic, enterohemorrhagic, and uropathogenic E. coli strains (74–78). Clearly, human-adapted pathogens have efficiently evolved to regulate virulence gene expression for efficient colonization and infection tactics in the human host. Our work provides an example of this concept and highlights the importance of utilizing IVLCs to study bacterial pathogens. Finally, this work has profound effects on the development of therapeutics against Shigella. The adherence factors provide innovative targets and promise for novel therapies and new strategies to ultimately control and prevent Shigella infection.
Analysis of clinical isolates of Shigella flexneri. Both TEM (top) and biofilm formation (bottom) analyses were performed with clinical isolates of S. flexneri serotype 3a (strain AF11) and S. flexneri serotype 2a (strain AF16) to confirm the results of the study. Magnification for TEM, ×50,000 (on the left for strain AF11) and ×25,000 (on the right for strain AF16). For the biofilm assay, the average OD540 for the strains grown in either medium (light blue bars) or medium supplemented with IVLCs (dark blue bars) is plotted. All data represent those from three biological independent experiments in which each experiment had technical triplicates. ***, P < 0.001 for TSB versus TSB-bile salts for each strain. Download FIG S5, PDF file, 0.5 MB (541.4KB, pdf) .
Copyright © 2019 Chanin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MATERIALS AND METHODS
Ethics statement.
Human sample collection was approved by Institutional Review Board (IRB) protocol 2015P001908 of the Massachusetts General Hospital, Boston, MA. Donor tissue was obtained from consenting patients undergoing medically required surgical resections, as determined by a licensed physician. All subjects provided written informed consent.
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are presented in Table 3. Bacteria were routinely cultured at 37°C in either Luria broth (LB; Lennox) or tryptic soy broth (TSB; which contains an additional 2.5 g/liter glucose relative to LB) with aeration or in tissue culture-treated plates to represent static growth conditions (31, 35). Plating for determination of the number of CFU was performed using tryptic soy broth plates with 1.5% agar and 0.025% Congo red (CR; catalog number C6277; Sigma-Aldrich). Bile salts (catalog number B8756; Sigma-Aldrich) were used at a concentration of 0.4% (wt/vol). All media were filter sterilized with a 0.22-μm-pore-size filter following the addition of bile salts and/or glucose. Chloramphenicol was used at 5 μg/ml, kanamycin was used at 50 μg/ml, and ampicillin was used at 100 μg/ml, where indicated.
TABLE 3.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Shigella strains | ||
| 2457T | S. flexneri serotype 2a | 92 |
| BS808 | 2457T ospE1::aphA-3 ospE2::cat | 30 |
| BS766 | 2457T transformed with pKM208 | 89 |
| ΔlpfA | 2457T lpfA::cat, chloramphenicol resistance | This study |
| ΔfimA | 2457T fimA::aph-3, kanamycin resistance | This study |
| ΔcsgA | 2457T csgA::cat, chloramphenicol resistance | This study |
| ΔcsgAB | 2457T csgB csgA::aph-3, kanamycin resistance | This study |
| ΔcsgAB ΔlpfA ΔfimA | 2457T ΔcsgAB lpfA::cat fimA::aph-3, chloramphenicol and kanamycin resistance |
This study |
| ΔlpfA(pLpfA_his) | 2457T lpfA::cat transformed with pLpfA_his | This study |
| ΔfimA(pFimA_his) | 2457T fimA::aph-3 transformed with pFimA_his | This study |
| ΔcsgA(pCsgA_his) | 2457T csgA::cat transformed with pCsgA_his | This study |
| AF11 | S. flexneri serotype 3a (recent clinical isolate) | AFRIMSa |
| AF16 | S. flexneri serotype 2a (recent clinical isolate) | AFRIMS |
| Plasmids | ||
| pKD3 | oriR6K bla cat | 93 |
| pKD4 | oriR6K bla aph-3 | 93 |
| pKM208 | Temperature-sensitive red-, gam-, and lacI- expressing plasmid driven by Ptac promoter, bla |
93 |
| pCP20 | FLP+ λ cI857+ λ ρR RepTs bla cat | 90 |
| pGEMT | PCR cloning vector, bla, high copy number | Promega |
| pLpfA_his | pGEMT::lpfA lpfA with C-terminal His tag | This study |
| pFimA_his | pGEMT::fimA fimA with C-terminal His tag | This study |
| pCsgA_his | pGEMT::csgA csgA with C-terminal His tag | This study |
AFRIMS, Department of Enteric Diseases, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand.
Biofilm assays.
The biofilm assay was performed as previously described (31, 35). Single colonies of each bacterial strain were inoculated into IVLC medium in a single well of a 96-well plate. The plates were incubated at 37°C without shaking. At the time points indicated below, the wells were gently washed twice with 1× phosphate-buffered saline (PBS) and either fixed for electron microscopy (see below) or stained with 0.5% crystal violet for 5 min. Afterwards, the wells were gently washed five times with sterile distilled H2O and then set to air dry. Biofilm formation was quantified by adding 95% ethanol to the wells to solubilize the crystal violet stain. After 30 min of incubation at room temperature on an orbital shaker at 70 rpm, the absorbance reading at an optical density of 540 nm (OD540) was measured with a plate reader (79). Absorbance readings at OD600 were taken to ensure that there were no significant differences in growth prior to the washing steps. For experiments in which cellulase (catalog number C1184; Sigma-Aldrich) was used, 60 units/ml of enzyme was added to the wells at the start of the biofilm assay. For complementation analysis, the assays were performed at 4.5 h to enable appropriate expression of the genes from the pGEMT plasmid. Cellulase and complementation biofilms were subsequently processed as described above. Statistical significance was determined by comparing the findings for wild-type S. flexneri 2457T to those for each mutant using Student's t test, and a P value of ≤0.05 was considered significant.
Adherence assays.
The isolation and preparation of human intestinal epithelial cells were performed as previously described (38–41, 80, 81). The excess healthy margins of the ascending colon, as verified by a pathologist, were used to obtain the intestinal crypts. The tissue was washed once in cold 1× PBS (Thermo Fisher Scientific, MA), and then tissue strips were cut and placed into a dissociation buffer consisting of 1× PBS, penicillin-streptomycin (pen-strep; Thermo Fisher Scientific), 1 mM dithiothreitol (DTT; Sigma-Aldrich, MO), and 0.5 mM EDTA (Sigma-Aldrich). Intestinal strips were incubated at 4°C for 30 min and then vigorously shaken to promote epithelium dissociation from the basal membrane. This procedure was repeated five times to collect multiple fractions. Subsequently, the fractions containing the intestinal crypts were further processed and plated in Matrigel matrix (Corning, NY) as previously described (38, 80). Intestinal crypt-derived organoids were incubated at 37°C with 5% CO2 in medium that consisted of a 1:1 ratio of stem cell medium and L-WRN (ATCC CRL-3276)-derived conditioned medium, in which both medium types were prepared as previously described (38, 82). The culture medium was replenished every other day, and the organoids were passaged every 7 to 9 days using standard trypsin-based techniques. Approximately 2.0 × 106 cells/ml were replated in Matrigel matrix to ensure robust propagation of the organoids (38).
Organoid-derived cell monolayers were generated as previously described (38–41). Single-cell suspensions derived from the organoids were plated on polyethylene terephthalate (PET) membrane transwell inserts with a 0.4-μm pore size (Corning) at 1.0 × 106 cells/ml and incubated in the 1:1 stem cell medium–L-WRN medium at 37°C with 5% CO2. The culture medium was changed every other day until the cultures reached confluence, as determined by transepithelial electrical resistance (TEER) monitoring and microscopic observation. At 48 h prior to each experiment, the media in the apical chamber were replaced with complete Dulbecco modified Eagle medium (DMEM)–Ham’s F-12 medium plus 5 μM γ-secretase inhibitor IX (DAPT; Calbiochem), while the basolateral media were replenished with the 1:1 stem cell medium–L-WRN medium with 10 μM Y-27632 (Calbiochem) and 100 to 500 ng/ml of the receptor activator of the NF-κB ligand (RANKL; Peprotech). This process was utilized to promote cell differentiation (38, 40), especially for M cells (83). On the day of each experiment, the monolayers were washed with 1× PBS, both the apical and basolateral media were replaced with DMEM without phenol red, and the monolayers were incubated for at least 2 h before the initiation of the experiment. S. flexneri 2457T or the various mutants were subcultured in TSB-bile salts and then were washed in 1× PBS, resuspended in DMEM without phenol red, applied to the apical surface of the monolayers without centrifugation, and incubated for 3 h as previously described for polarized T84 cells (30). Afterwards, infected cells were processed for adherence quantification (30) or fixed for electron microscopy (see below). The average percent recovery was calculated from two independent experiments, each of which was performed with at least two technical duplicates, as (recovered bacterial titer/infecting titer) × 100% and was plotted relative to the results for wild-type strain 2457T. Statistical significance was determined by comparing the result for wild-type S. flexneri 2457T to that for each mutant using Student's t test, and a P value of ≤0.05 was considered significant.
The HT-29 cell adherence assay was performed as previously described (31). HT-29 cells (ATCC HTB-38) were seeded in DMEM to establish a semiconfluent monolayer of approximately 75%. For bacterial cultures, single colonies of S. flexneri 2457T or the various mutants were inoculated into media with or without IVLCs in tissue culture plates and grown statically at 37°C. Following overnight growth, the bacteria were collected, washed with 1× PBS, standardized to an OD600 of 0.35, resuspended in DMEM, and applied to the HT-29 cell monolayers without centrifugation. The cells were incubated at 37°C with 5% CO2 for 3 h. Afterwards, the monolayers were washed five times with 1× PBS and lysed with 1% Triton X-100. Serial dilutions were made to determine the number of cell-associated bacteria. The average percent recovery was calculated from three independent experiments as (recovered bacterial titer/infecting titer) × 100% and was plotted relative to the results for wild-type 2457T. Statistical significance was determined by comparing the result for wild-type S. flexneri 2457T to the result for each mutant using Student's t test, and a P value of ≤0.05 was considered significant.
Electron microscopy analyses.
For the biofilm culture analysis, single colonies of S. flexneri 2457T or the various mutants were added to tissue culture-treated plates containing medium with or without IVLCs. The cultures were grown statically overnight at 37°C. On the following day, samples were collected and prepared for transmission electron microscopy (TEM) imaging by fixing in 2.5% glutaraldehyde and staining with uranyl acetate (84). Samples were mounted on Formvar/carbon 100-mesh grids (Electron Microscopy Services) and imaged with a JEOL transmission electron microscope. For scanning electron microscopy (SEM) analysis of the HIODEM adherence assay, samples were fixed in 0.5× Karnovsky fixative and subsequently stored in 1× PBS. All sample processing occurred at the Massachusetts Eye and Ear Infirmary core facility. All SEM imaging was performed at the Harvard University Center for Nanoscale Systems (CNS) using a FESEM Supra55VP microscope. The SEM images were pseudocolored according to protocols listed at http://www.nuance.northwestern.edu/docs/epic-pdf/Basic_Photoshop_for_Electron_Microscopy_06-2015.pdf.
For TEM analysis of isolated adherence structures, the wild-type and mutant strains were cultured statically in IVLC medium and an ammonium sulfate precipitation was performed (53). Briefly, samples were collected and pelleted by centrifugation at 4,000 rpm for 10 min. The bacterial pellet was resuspended in 1× PBS, and the mixture was heated at 65°C for 30 min and subsequently centrifuged at 4,000 rpm for 10 min. The supernatants were transferred to a new tube and precipitated by mixing the samples with 40% ammonium sulfate on an end-over-end mixer for 10 min at room temperature. Afterwards, the samples were dialyzed in 1× PBS using 3.5-molecular-weight-cutoff dialysis cassettes for 1 h at room temperature on a rotating shaker at 50 rpm. The 1× PBS was then changed and the cassettes were transferred to 4°C for overnight dialysis. The dialyzed fraction was collected and stored at −20°C. A fraction of each sample was fixed and processed for TEM analysis.
RNA isolation.
RNA was isolated from the bacterial cultures as previously described (85) with Qiagen RNeasy kits. DNA was digested with Turbo DNase (Invitrogen), and the concentrations of total RNA were determined using a NanoDrop ND-1000 spectrophotometer. The cDNA was synthesized from total RNA using a SuperScript III first-strand synthesis kit (Invitrogen) or a RevertAid cDNA first-strand synthesis kit (Thermo Fisher Scientific) according to the manufacturers’ protocols. All RNA was first confirmed to be free of DNA contamination by performing separate cDNA synthesis reactions with and without reverse transcriptase in the reaction mixture, followed by PCR amplification of the housekeeping gene rpoA as described previously (31).
RNA-seq analysis.
The data for the RNA sequencing (RNA-seq) trace reads were obtained from our previous study (31). Duplicate cultures were grown either by static or shaking growth in TSB or TSB supplemented with 0.4% bile salts as previously described (31). The RNA-seq trace read data were generated using Integrative Genomics Viewer (IGV) software (version 2.3.67) (86, 87). Each RNA-seq data set was loaded into IGV software, and the traces were normalized to the trace for the S. flexneri 2457T rpoA gene on the autoscale setting. The zoomed-in traces for two genes provided in Fig. S1 in the supplemental material represents a 10-fold magnification in the scale setting. The genes of interest were searched for using the publicly available S. flexneri 2457T genome (GenBank accession number AE014073.1) and S. flexneri serotype 2a strain 301 virulence plasmid annotations (GenBank accession number AF386526.1).
RT-PCR analysis.
For nonquantitative reverse transcription-PCR (RT-PCR) analysis, cDNA was synthesized from total RNA isolated from broth cultures using the RevertAid cDNA first-strand synthesis kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. All RNA was first confirmed to be free of DNA contamination as described above. The various PCRs were performed using the 2× Taq-Pro complete PCR mix (Denville Scientific). All primer sets were validated and tested for proper DNA amplification prior to the experiment (data not shown). The annealing temperatures were adjusted accordingly for each primer set, and the extension time was adjusted for the size of each product. The products of the reactions were visualized by gel electrophoresis on 1% agarose gels stained with ethidium bromide on a Syngene GelDoc system. The molecular weight markers used in the analysis included GeneRuler, 1 kb Plus, and 100 bp Plus (Thermo Fisher Scientific). For quantitative RT-PCR (qRT-PCR) analysis, biologically independent RNA samples were isolated, and it was ensured that they were DNA free as described above. Analysis by qRT-PCR was performed as previously described (85), and all data were normalized to the levels of rpoA and analyzed using the comparative cycle threshold (ΔCT) method (88). The expression levels of the target genes under the various conditions were compared using the relative quantification method (88). Real-time data are expressed as the relative changes in expression levels compared with the levels in the media without glucose and/or bile salts. Statistical significance was determined using Student's t test to compare the expression of each gene in control versus treatment media, and a P value of ≤0.05 was considered significant. Due to the significant number of primers used in this analysis, primer sequences are not presented here but are available upon request.
Mutant construction.
The single gene deletion mutants used in this study were constructed using the bacteriophage λ Red linear recombination method as previously described (89). Briefly, PCR was used to amplify a chloramphenicol resistance cassette gene (cat) from pKD3 or the kanamycin resistance gene cassette (aph-3) from pKD4 (Table 3) with 5′ and 3′ overhangs identical to the 5′ and 3′ regions of each gene of interest. Antibiotic-resistant recombinants were identified and selected on chloramphenicol or kanamycin plates and subsequently screened via PCR using confirmation primers that annealed to unique regions up- and downstream of each gene to detect the size difference due to the insertion of the antibiotic resistance cassette. The sequences of the primers used for mutant construction and confirmation are also available upon request. For the quadruple mutant, the kanamycin resistance cassette was removed from the ΔcsgAB mutant by transforming the strain with pCP20 (Table 3) and incubating positive transformants at 42°C as previously described (90). The strain was retransformed with pKM208, and a ΔlpfA deletion was constructed by inserting the chloramphenicol resistance cassette with the bacteriophage λ Red linear recombination method. Finally, this ΔcsgAB ΔlpfA strain was used as the recipient strain in a bacteriophage P1L4 transduction in which the ΔfimA mutant was used as the donor strain and kanamycin was used to select for positive transductants. The resulting strain has a quadruple mutation in the lpfA, fimA, csgA, and csgB structural genes harboring both chloramphenicol and kanamycin resistance cassettes (Table 3). At each step of the construction, the same confirmation primers used for the single gene deletions were again used to confirm the removal or addition of the antibiotic resistance cassettes as previously described (89).
Plasmid construction.
The plasmids harboring the sequences encoding histidine-tagged LpfA, FimA, and CsgA were constructed as previously described (30). Briefly, each gene and respective native promoter region were amplified by PCR with high-fidelity Taq polymerase (Invitrogen) from wild-type strain 2457T. For FimA, a 6× His tag was added to the C terminus followed by a stop codon. For LpfA and CsgA, a glycine linker sequence was added upstream of the 6× His tag. The PCR products were ligated into pGEMT, and the plasmids were subsequently transformed into the appropriate adherence mutant. Selection for positive transformants occurred on tryptic soy broth plates containing 1.5% agar, 0.025% Congo red, and 100 μg/ml ampicillin. Sequencing was performed to ensure that no mutations were introduced during the cloning process. All primers used for the plasmid constructions and sequencing verification will also be made available upon request.
Congo red binding assay for curli detection.
Samples for the Congo red binding assay were collected by gentle scraping of samples from the biofilm, and the samples were processed for ammonium sulfate precipitation as detailed above and placed on a clean, dry glass slide. The specimens were air dried, subsequently stained with alkaline Congo red solution (catalog number HT603; Sigma-Aldrich), and incubated at room temperature for approximately 10 min. The smears were rinsed in water until excess stain was drained, and the slides were observed under polarized light for apple green birefringence (55, 56). Samples were imaged with a Nikon Ci-E microscope with an attached camera.
Mass spectrometry analysis.
Shigella flexneri 2457T was cultured in IVLC media as described above for the biofilm assay. Following overnight incubation, culture supernatants were collected and concentrated by trichloroacetic acid (TCA) precipitation. The protein pellet was stored at −20°C until analyzed. For mass spectrometry (MS) analysis, first, intact mass analysis was performed by reconstituting the lyophilized sample in 0.1% trifluoroacetic acid. Ultra-high-pressure liquid chromatography–quantitative time of flight (Q-TOF) MS was performed to detect the masses of intact molecules present in the mixture. The samples were analyzed using reversed-phase liquid chromatography (RPLC) and a Xevo G2-S Q-TOF system (Waters Corp., Milford, MA). Liquid chromatography was performed at 0.200 ml/min using an H-Class Acquity UPLC system (Waters Corp., Milford, MA) on a BEH300-C4 column (2.1 mm by 150 mm; pore size, 1.7 μm; Waters Corp., Milford, MA). Buffer A consisted of 0.1% (vol/vol) formic acid in UPLC-grade water, and buffer B consisted of 0.1% (vol/vol) formic acid in 100% UPLC-grade acetonitrile. In all analyses, a gradient separation was performed as follows: 0 min 90% buffer A, 5 min 90% buffer A, 80 min 10% buffer A, 90 min 10% buffer A, 91 min 90% buffer A, and 100 min 90% buffer A. After RPLC, samples were introduced via an electrospray ion source in-line with the Xevo G2-S Q-TOF system. External calibration of the m/z scale was performed using sodium cesium iodide. The Q-TOF parameters were run in sensitivity mode with m/z scanning from 400 to 4,000, a 3.00-kV capillary voltage, a 40-V cone voltage, a 150°C source temperature, a 350°C desolvation temperature, and a desolvation gas flow rate of 800 liters/h. MS data were collected at a scan speed of 1.0 s. Liquid chromatography (LC) solvents were UPLC grade, and all other chemicals were of analytical grade. Intact masses were calculated using the Waters UNIFI software package and deconvolved using the MaxEnt algorithm.
For peptide analysis, samples were digested with trypsin at 37°C for 1.5 h, and the resulting peptides were subsequently extracted for analysis. UPLC–Q-TOF MS/MS was performed to detect the masses of the digested peptides and the respective fragments. Samples were analyzed using RPLC as described above on a BEH300-C18 column (2.1 mm by 150 mm; pore size, 1.7 μm; Waters Corp., Milford, MA) using buffer A and buffer B with the same compositions described above. In all analyses, a gradient separation was performed as follows: 0 min 95% buffer A, 2 min 95% buffer A, 55 min 40% buffer A, 64 min 10% buffer A, 74 min 10% buffer A, 75 min 95% buffer A, and 90 min 95% buffer A. After RPLC, samples were introduced via an electrospray ion source in-line with the Xevo G2-S Q-TOF system. External calibration of the m/z scale was performed using sodium cesium iodide. The Q-TOF parameters were run in resolution mode with m/z scanning from 50 to 2,000, a 3.00-kV capillary voltage, a 30-V cone voltage, a 130°C source temperature, a 250°C desolvation temperature, and a desolvation gas flow rate of 800 liters/h. MS/MS data were collected at a scan speed of 0.1 s. LC solvents were UPLC grade, and all other chemicals were of analytical grade. Peptide fingerprinting was completed by use of the Waters UNIFI software package. Parameters were set to restrict matches only to those peptide fragments where the primary ion exhibited a >+1 charge and at least 1 daughter ion was detected, confirming the presence of each particular peptide. Any peptide maps with less than 10% coverage were excluded from the analysis.
Immunoprecipitation analysis.
Each strain harboring the His-tagged constructs (Table 3) was grown in static overnight biofilm cultures as described above. For plasmid maintenance, ampicillin was added. Culture supernatants were subsequently collected, filtered sterilized, and TCA precipitated. The total protein pellets were resuspended in 1 ml NP-40 with protease inhibitor cocktail (Roche Diagnostics GmbH). Samples were precleared using protein A/G plus agarose beads (Pierce), followed by immunoprecipitation with a mouse anti-His affinity resin (GenScript) or a negative-control mouse IgG antibody (Santa Cruz). Samples were incubated overnight at 4°C with rotation. On the following day, protein A/G plus agarose beads were added to the negative-control IgG samples, and the mixture was incubated for 1 h at 4°C with rotation. Afterwards, the beads or resin samples were pelleted, washed six times, and boiled in acidified Laemmli lysis buffer as previously described for adherence proteins (91). After boiling, the samples were neutralized with basic Laemmli lysis buffer. Samples were run on a 4 to 20% SDS-PAGE gel (Bio-Rad), and Western blot analysis was performed as previously described (30) using a primary anti-His antibody (Qiagen) and a secondary Alexa Fluor 700 goat anti-mouse immunoglobulin antibody. The Western blots were scanned using an Odyssey infrared detection system (Li-Cor).
ACKNOWLEDGMENTS
We thank Stefania Senger, Alessio Fasano, and Laura Ingano of the Mucosal Immunology and Biology Research Center for the use of and assistance with the human intestinal organoid-derived epithelial monolayer model. We also thank Brett Swierczewski, Walter Reed Army Institute of Research, for the clinical isolates of S. flexneri, and Marcia Goldberg, Massachusetts General Hospital, for the P1L4 phage used in this study. We thank Erin Jones for technical support with the TEM core.
This work was supported by National Institute of Allergy and Infectious Diseases grants K22-AI104755 (to C.S.F.), 5T32-AI095190-04 (to J.R.S.), and U19-AI110820 (to D.A.R.). Funding for B.J.D.K., J.D., K.L., and the Mucosal Immunology and Biology Center’s Summer Internship Program is provided by National Institute of Diabetes and Digestive and Kidney Diseases grant R25 DK103579. Funding for the human intestinal organoid-derived epithelial monolayer model is supported by National Institute of Allergy and Infectious Diseases grant U19-AI082655. The TEM core is supported by National Institute of Neurological Disorders and Stroke grant P30-NS045776. Support for the Philly Dake Electron Microscope Facility was provided by National Institutes of Health grant 1S10RR023594S10 and by funds from the Dake Family Foundation.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
REFERENCES
- 1.Kotloff KL, Riddle MS, Platts-Mills JA, Pavlinac P, Zaidi AKM. 2017. Shigellosis. Lancet 391:801–812. doi: 10.1016/S0140-6736(17)33296-8. [DOI] [PubMed] [Google Scholar]
- 2.Schroeder GN, Hilbi H. 2008. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev 21:134–156. doi: 10.1128/CMR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 4.Livio S, Strockbine NA, Panchalingam S, Tennant SM, Barry EM, Marohn ME, Antonio M, Hossain A, Mandomando I, Ochieng JB, Oundo JO, Qureshi S, Ramamurthy T, Tamboura B, Adegbola RA, Hossain MJ, Saha D, Sen S, Faruque ASG, Alonso PL, Breiman RF, Zaidi AKM, Sur D, Sow SO, Berkeley LY, O'Reilly CE, Mintz ED, Biswas K, Cohen D, Farag TH, Nasrin D, Wu Y, Blackwelder WC, Kotloff KL, Nataro JP, Levine MM. 2014. Shigella isolates from the global enteric multicenter study inform vaccine development. Clin Infect Dis 59:933–941. doi: 10.1093/cid/ciu468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya D, Bhattacharya H, Sayi DS, Bharadwaj AP, Singhania M, Sugunan AP, Roy S. 2015. Changing patterns and widening of antibiotic resistance in Shigella spp. over a decade (2000–2011), Andaman Islands, India. Epidemiol Infect 143:470–477. doi: 10.1017/S0950268814000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baranov V, Hammarstrom S. 2004. Carcinoembryonic antigen (CEA) and CEA-related cell adhesion molecule 1 (CEACAM1), apically expressed on human colonic M cells, are potential receptors for microbial adhesion. Histochem Cell Biol 121:83–89. doi: 10.1007/s00418-003-0613-5. [DOI] [PubMed] [Google Scholar]
- 7.Sansonetti PJ, Arondel J, Cantey JR, Prevost MC, Huerre M. 1996. Infection of rabbit Peyer’s patches by Shigella flexneri: effect of adhesive or invasive bacterial phenotypes on follicle-associated epithelium. Infect Immun 64:2752–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kline KA, Dodson KW, Caparon MG, Hultgren SJ. 2010. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol 18:224–232. doi: 10.1016/j.tim.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soto GE, Hultgren SJ. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J Bacteriol 181:1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torres AG, Zhou X, Kaper JB. 2005. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect Immun 73:18–29. doi: 10.1128/IAI.73.1.18-29.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Townsend SM, Kramer NE, Edwards R, Baker S, Hamlin N, Simmonds M, Stevens K, Maloy S, Parkhill J, Dougan G, Baumler AJ. 2001. Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect Immun 69:2894–2901. doi: 10.1128/IAI.69.5.2894-2901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chessa D, Winter MG, Jakomin M, Baumler AJ. 2009. Salmonella enterica serotype Typhimurium Std fimbriae bind terminal alpha(1,2)fucose residues in the cecal mucosa. Mol Microbiol 71:864–875. doi: 10.1111/j.1365-2958.2008.06566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner C, Hensel M. 2011. Adhesive mechanisms of Salmonella enterica. Adv Exp Med Biol 715:17–34. doi: 10.1007/978-94-007-0940-9_2. [DOI] [PubMed] [Google Scholar]
- 14.Pupo GM, Lan R, Reeves PR. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci U S A 97:10567–10572. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahl JW, Morris CR, Emberger J, Fraser CM, Ochieng JB, Juma J, Fields B, Breiman RF, Gilmour M, Nataro JP, Rasko DA. 2015. Defining the phylogenomics of Shigella species: a pathway to diagnostics. J Clin Microbiol 53:951–960. doi: 10.1128/JCM.03527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bloch CA, Stocker BA, Orndorff PE. 1992. A key role for type 1 pili in enterobacterial communicability. Mol Microbiol 6:697–701. doi: 10.1111/j.1365-2958.1992.tb01518.x. [DOI] [PubMed] [Google Scholar]
- 17.Bravo V, Puhar A, Sansonetti P, Parsot C, Toro CS. 2015. Distinct mutations led to inactivation of type 1 fimbriae expression in Shigella spp. PLoS One 10:e0121785. doi: 10.1371/journal.pone.0121785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gbarah A, Mirelman D, Sansonetti PJ, Verdon R, Bernhard W, Sharon N. 1993. Shigella flexneri transformants expressing type 1 (mannose-specific) fimbriae bind to, activate, and are killed by phagocytic cells. Infect Immun 61:1687–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenstein BI, Clements JR, Dodd DC. 1983. Isolation and characterization of a monoclonal antibody directed against type 1 fimbriae organelles from Escherichia coli. Infect Immun 42:333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knutton S, Lloyd DR, Candy DC, McNeish AS. 1984. In vitro adhesion of enterotoxigenic Escherichia coli to human intestinal epithelial cells from mucosal biopsies. Infect Immun 44:514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pizarro-Cerdá J, Cossart P. 2006. Bacterial adhesion and entry into host cells. Cell 124:715–727. doi: 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang F, Yang J, Zhang X, Chen L, Jiang Y, Yan Y, Tang X, Wang J, Xiong Z, Dong J, Xue Y, Zhu Y, Xu X, Sun L, Chen S, Nie H, Peng J, Xu J, Wang Y, Yuan Z, Wen Y, Yao Z, Shen Y, Qiang B, Hou Y, Yu J, Jin Q. 2005. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res 33:6445–6458. doi: 10.1093/nar/gki954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakellaris H, Hannink NK, Rajakumar K, Bulach D, Hunt M, Sasakawa C, Adler B. 2000. Curli loci of Shigella spp. Infect Immun 68:3780–3783. doi: 10.1128/iai.68.6.3780-3783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergsten G, Wullt B, Svanborg C. 2005. Escherichia coli, fimbriae, bacterial persistence and host response induction in the human urinary tract. Int J Med Microbiol 295:487–502. doi: 10.1016/j.ijmm.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Ashkar AA, Mossman KL, Coombes BK, Gyles CL, Mackenzie R. 2008. FimH adhesin of type 1 fimbriae is a potent inducer of innate antimicrobial responses which requires TLR4 and type 1 interferon signalling. PLoS Pathog 4:e1000233. doi: 10.1371/journal.ppat.1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Utsunomiya A, Naito T, Ehara M, Ichinose Y, Hamamoto A. 1992. Studies on novel pili from Shigella flexneri. I. Detection of pili and hemagglutination activity. Microbiol Immunol 36:803–813. doi: 10.1111/j.1348-0421.1992.tb02082.x. [DOI] [PubMed] [Google Scholar]
- 28.Snellings NJ, Tall BD, Venkatesan MM. 1997. Characterization of Shigella type 1 fimbriae: expression, FimA sequence, and phase variation. Infect Immun 65:2462–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gillies RR, Duguid JP. 1958. The fimbrial antigens of Shigella flexneri. J Hyg (Lond) 56:303–318. doi: 10.1017/s0022172400037803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faherty CS, Redman JC, Rasko DA, Barry EM, Nataro JP. 2012. Shigella flexneri effectors OspE1 and OspE2 mediate induced adherence to the colonic epithelium following bile salts exposure. Mol Microbiol 85:107–121. doi: 10.1111/j.1365-2958.2012.08092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nickerson KP, Chanin RB, Sistrunk JR, Rasko DA, Fink PJ, Barry EM, Nataro JP, Faherty CS. 2017. Analysis of Shigella flexneri resistance, biofilm formation, and transcriptional profile in response to bile salts. Infect Immun 85:e01067-16. doi: 10.1128/IAI.01067-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ridlon JM, Kang DJ, Hylemon PB. 2006. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Kamada N, Chen GY, Inohara N, Nunez G. 2013. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomson AB, Keelan M, Thiesen A, Clandinin MT, Ropeleski M, Wild GE. 2001. Small bowel review: normal physiology part 1. Dig Dis Sci 46:2567–2587. doi: 10.1023/a:1012794505897. [DOI] [PubMed] [Google Scholar]
- 35.Nickerson KP, Faherty CS. 6 May 2018. Bile salt-induced biofilm formation in enteric pathogens: techniques for identification and quantification. J Vis Exp doi: 10.3791/57322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borgstrom B, Dahlqvist A, Lundh G, Sjovall J. 1957. Studies of intestinal digestion and absorption in the human. J Clin Invest 36:1521–1536. doi: 10.1172/JCI103549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta P, Sarkar S, Das B, Bhattacharjee S, Tribedi P. 2015. Biofilm, pathogenesis and prevention—a journey to break the wall: a review. Arch Microbiol 198:1–15. doi: 10.1007/s00203-015-1148-6. [DOI] [PubMed] [Google Scholar]
- 38.Senger S, Ingano L, Freire R, Anselmo A, Zhu W, Sadreyev R, Walker WA, Fasano A. 2018. Human fetal-derived enterospheres provide insights on intestinal development and a novel model to study necrotizing enterocolitis (NEC). Cell Mol Gastroenterol Hepatol 5:549–568. doi: 10.1016/j.jcmgh.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nickerson KP, Senger S, Zhang Y, Lima R, Patel S, Ingano L, Flavahan WA, Kumar DKV, Fraser CM, Faherty CS, Sztein MB, Fiorentino M, Fasano A. 2018. Salmonella Typhi colonization provokes extensive transcriptional changes aimed at evading host mucosal immune defense during early infection of human intestinal tissue. EBioMedicine 31:92–109. doi: 10.1016/j.ebiom.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.VanDussen KL, Marinshaw JM, Shaikh N, Miyoshi H, Moon C, Tarr PI, Ciorba MA, Stappenbeck TS. 2015. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 64:911–920. doi: 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Llanos-Chea A, Citorik RJ, Nickerson KP, Ingano L, Serena G, Senger S, Lu TK, Fasano A, Faherty CS. 2019. Bacteriophage therapy testing against Shigella flexneri in a novel human intestinal organoid-derived infection model. J Pediatr Gastroenterol Nutr 68:509–516. doi: 10.1097/MPG.0000000000002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szabady RL, Louissaint C, Lubben A, Xie B, Reeksting S, Tuohy C, Demma Z, Foley SE, Faherty CS, Llanos-Chea A, Olive AJ, Mrsny RJ, McCormick BA. 2018. Intestinal P-glycoprotein exports endocannabinoids to prevent inflammation and maintain homeostasis. J Clin Invest 128:4044–4056. doi: 10.1172/JCI96817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller CM, Aberg A, Straseviciene J, Emody L, Uhlin BE, Balsalobre C. 2009. Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor CRP-cAMP. PLoS Pathog 5:e1000303. doi: 10.1371/journal.ppat.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin CT, Lin TH, Wu CC, Wan L, Huang CF, Peng HL. 2016. CRP-cyclic AMP regulates the expression of type 3 fimbriae via cyclic di-GMP in Klebsiella pneumoniae. PLoS One 11:e0162884. doi: 10.1371/journal.pone.0162884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De la Cruz MA, Ruiz-Tagle A, Ares MA, Pacheco S, Yáñez JA, Cedillo L, Torres J, Girón JA. 2017. The expression of Longus type 4 pilus of enterotoxigenic Escherichia coli is regulated by LngR and LngS and by H-NS, CpxR and CRP global regulators. Environ Microbiol 19:1761–1775. doi: 10.1111/1462-2920.13644. [DOI] [PubMed] [Google Scholar]
- 46.Romao FT, Hernandes RT, Yamamoto D, Osugui L, Popi AF, Gomes TA. 2014. Influence of environmental factors in the adherence of an atypical enteropathogenic Escherichia coli strain to epithelial cells. BMC Microbiol 14:299. doi: 10.1186/s12866-014-0299-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu Rev Microbiol 60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baumler AJ, Heffron F. 1995. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J Bacteriol 177:2087–2097. doi: 10.1128/jb.177.8.2087-2097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lloyd SJ, Ritchie JM, Torres AG. 2012. Fimbriation and curliation in Escherichia coli O157:H7: a paradigm of intestinal and environmental colonization. Gut Microbes 3:272–276. doi: 10.4161/gmic.20661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bian Z, Normark S. 1997. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J 16:5827–5836. doi: 10.1093/emboj/16.19.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hale TL, Formal SB. 1981. Protein synthesis in HeLa or Henle 407 cells infected with Shigella dysenteriae 1, Shigella flexneri 2a, or Salmonella typhimurium W118. Infect Immun 32:137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 53.Chattopadhyay S, Tchesnokova V, McVeigh A, Kisiela DI, Dori K, Navarro A, Sokurenko EV, Savarino SJ. 2012. Adaptive evolution of class 5 fimbrial genes in enterotoxigenic Escherichia coli and its functional consequences. J Biol Chem 287:6150–6158. doi: 10.1074/jbc.M111.303735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Gerven N, Klein RD, Hultgren SJ, Remaut H. 2015. Bacterial amyloid formation: structural insights into curli biogenesis. Trends Microbiol 23:693–706. doi: 10.1016/j.tim.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puchtler H, Waldrop FS, Meloan SN. 1985. A review of light, polarization and fluorescence microscopic methods for amyloid. Appl Pathol 3:5–17. [PubMed] [Google Scholar]
- 56.Puchtler H, Sweat F, Levine M. 1962. On the binding of Congo red by amyloid. J Histochem Cytochem 10:355–364. doi: 10.1177/10.3.355. [DOI] [Google Scholar]
- 57.Gonzales AM, Wilde S, Roland KL. 2017. New insights into the roles of long polar fimbriae and Stg fimbriae in Salmonella interactions with enterocytes and M cells. Infect Immun 85:e00172-17. doi: 10.1128/IAI.00172-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chassaing B, Etienne-Mesmin L, Bonnet R, Darfeuille-Michaud A. 2013. Bile salts induce long polar fimbriae expression favouring Crohn’s disease-associated adherent-invasive Escherichia coli interaction with Peyer’s patches. Environ Microbiol 15:355–371. doi: 10.1111/j.1462-2920.2012.02824.x. [DOI] [PubMed] [Google Scholar]
- 59.Arenas-Hernández MM, Rojas-López M, Medrano-López A, Nuñez-Reza KJ, Puente JL, Martínez-Laguna Y, Torres AG. 2014. Environmental regulation of the long polar fimbriae 2 of enterohemorrhagic Escherichia coli O157:H7. FEMS Microbiol Lett 357:105–114. doi: 10.1111/1574-6968.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cordonnier C, Etienne-Mesmin L, Thevenot J, Rougeron A, Renier S, Chassaing B, Darfeuille-Michaud A, Barnich N, Blanquet-Diot S, Livrelli V. 2017. Enterohemorrhagic Escherichia coli pathogenesis: role of Long polar fimbriae in Peyer’s patches interactions. Sci Rep 7:44655. doi: 10.1038/srep44655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ross BN, Rojas-Lopez M, Cieza RJ, McWilliams BD, Torres AG. 2015. The role of Long polar fimbriae in Escherichia coli O104:H4 adhesion and colonization. PLoS One 10:e0141845. doi: 10.1371/journal.pone.0141845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Araujo F, Pereira C, Costa J, Barrias C, Granja PL, Sarmento B. 2016. In vitro M-like cells genesis through a tissue-engineered triple-culture intestinal model. J Biomed Mater Res B Appl Biomater 104:782–788. doi: 10.1002/jbm.b.33508. [DOI] [PubMed] [Google Scholar]
- 63.Morris CR, Grassel CL, Redman JC, Sahl JW, Barry EM, Rasko DA. 2013. Characterization of intracellular growth regulator icgR by utilizing transcriptomics to identify mediators of pathogenesis in Shigella flexneri. Infect Immun 81:3068–3076. doi: 10.1128/IAI.00537-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sukumaran SK, Fu NY, Tin CB, Wan KF, Lee SS, Yu VC. 2010. A soluble form of the pilus protein FimA targets the VDAC-hexokinase complex at mitochondria to suppress host cell apoptosis. Mol Cell 37:768–783. doi: 10.1016/j.molcel.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 65.Sistrunk JR, Nickerson KP, Chanin RB, Rasko DA, Faherty CS. 2016. Survival of the fittest: how bacterial pathogens utilize bile to enhance infection. Clin Microbiol Rev 29:819–836. doi: 10.1128/CMR.00031-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koseoglu VK, Hall CP, Rodriguez-Lopez EM, Agaisse H. 2019. The autotransporter IcsA promotes Shigella flexneri biofilm formation in the presence of bile salts. Infect Immun 87:e00861-18. doi: 10.1128/IAI.00861-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goyal P, Krasteva PV, Van Gerven N, Gubellini F, Van den Broeck I, Troupiotis-Tsailaki A, Jonckheere W, Pehau-Arnaudet G, Pinkner JS, Chapman MR, Hultgren SJ, Howorka S, Fronzes R, Remaut H. 2014. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature 516:250–253. doi: 10.1038/nature13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nenninger AA, Robinson LS, Hultgren SJ. 2009. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc Natl Acad Sci U S A 106:900–905. doi: 10.1073/pnas.0812143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tursi SA, Tukel C. 2018. Curli-containing enteric biofilms inside and out: matrix composition, immune recognition, and disease implications. Microbiol Mol Biol Rev 82:e00028-18. doi: 10.1128/MMBR.00028-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fleming D, Chahin L, Rumbaugh K. 2017. Glycoside hydrolases degrade polymicrobial bacterial biofilms in wounds. Antimicrob Agents Chemother 61:e01998-16. doi: 10.1128/AAC.01998-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klemm P, Christiansen G, Kreft B, Marre R, Bergmans H. 1994. Reciprocal exchange of minor components of type 1 and F1C fimbriae results in hybrid organelles with changed receptor specificities. J Bacteriol 176:2227–2234. doi: 10.1128/jb.176.8.2227-2234.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klemm P, Jorgensen BJ, Kreft B, Christiansen G. 1995. The export systems of type 1 and F1C fimbriae are interchangeable but work in parental pairs. J Bacteriol 177:621–627. doi: 10.1128/jb.177.3.621-627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuan L, Schaffer JN, Zouzias CD, Pearson MM. 2014. Characterization of 17 chaperone-usher fimbriae encoded by Proteus mirabilis reveals strong conservation. J Med Microbiol 63:911–922. doi: 10.1099/jmm.0.069971-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Njoroge JW, Nguyen Y, Curtis MM, Moreira CG, Sperandio V. 2012. Virulence meets metabolism: Cra and KdpE gene regulation in enterohemorrhagic Escherichia coli. mBio 3:e00280-12. doi: 10.1128/mBio.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spurbeck RR, Stapleton AE, Johnson JR, Walk ST, Hooton TM, Mobley HL. 2011. Fimbrial profiles predict virulence of uropathogenic Escherichia coli strains: contribution of ygi and yad fimbriae. Infect Immun 79:4753–4763. doi: 10.1128/IAI.05621-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saldaña Z, De la Cruz MA, Carrillo-Casas EM, Durán L, Zhang Y, Hernández-Castro R, Puente JL, Daaka Y, Girón JA. 2014. Production of the Escherichia coli common pilus by uropathogenic E. coli is associated with adherence to HeLa and HTB-4 cells and invasion of mouse bladder urothelium. PLoS One 9:e101200. doi: 10.1371/journal.pone.0101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wurpel DJ, Totsika M, Allsopp LP, Webb RI, Moriel DG, Schembri MA. 2016. Comparative proteomics of uropathogenic Escherichia coli during growth in human urine identify UCA-like (UCL) fimbriae as an adherence factor involved in biofilm formation and binding to uroepithelial cells. J Proteomics 131:177–189. doi: 10.1016/j.jprot.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 78.Nicklasson M, Sjoling A, von Mentzer A, Qadri F, Svennerholm AM. 2012. Expression of colonization factor CS5 of enterotoxigenic Escherichia coli (ETEC) is enhanced in vivo and by the bile component Na glycocholate hydrate. PLoS One 7:e35827. doi: 10.1371/journal.pone.0035827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nickerson KP, McDonald C. 2012. Crohn’s disease-associated adherent-invasive Escherichia coli adhesion is enhanced by exposure to the ubiquitous dietary polysaccharide maltodextrin. PLoS One 7:e52132. doi: 10.1371/journal.pone.0052132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 81.Jung P, Sato T, Merlos-Suarez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA, Sancho E, Clevers H, Batlle E. 2011. Isolation and in vitro expansion of human colonic stem cells. Nat Med 17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 82.Miyoshi H, Stappenbeck TS. 2013. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc 8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wood MB, Rios D, Williams IR. 2016. TNF-alpha augments RANKL-dependent intestinal M cell differentiation in enteroid cultures. Am J Physiol Cell Physiol 311:C498–C507. doi: 10.1152/ajpcell.00108.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumar DK, Choi SH, Washicosky KJ, Eimer WA, Tucker S, Ghofrani J, Lefkowitz A, McColl G, Goldstein LE, Tanzi RE, Moir RD. 2016. Amyloid-beta peptide protects against microbial infection in mouse and worm models of Alzheimer’s disease. Sci Transl Med 8:340ra72. doi: 10.1126/scitranslmed.aaf1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Faherty CS, Wu T, Morris CR, Grassel CL, Rasko DA, Harper JM, Shea-Donohue T, Fasano A, Barry EM. 2016. The synthesis of OspD3 (ShET2) in Shigella flexneri is independent of OspC1. Gut Microbes 7:486–502. doi: 10.1080/19490976.2016.1239682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kendall MM, Rasko DA, Sperandio V. 2010. The LysR-type regulator QseA regulates both characterized and putative virulence genes in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol 76:1306–1321. doi: 10.1111/j.1365-2958.2010.07174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Faherty CS, Maurelli AT. 2009. SpaXV of Shigella flexneri is secreted through the type III secretion system and prevents staurosporine-induced apoptosis. Infect Immun 77:5281–5290. doi: 10.1128/IAI.00800-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 91.Floyd KA, Mitchell CA, Eberly AR, Colling SJ, Zhang EW, DePas W, Chapman MR, Conover M, Rogers BR, Hultgren SJ, Hadjifrangiskou M. 2016. The UbiI (VisC) aerobic ubiquinone synthase is required for expression of type 1 pili, biofilm formation, and pathogenesis in uropathogenic Escherichia coli. J Bacteriol 198:2662–2672. doi: 10.1128/JB.00030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Formal SB, Dammin GJ, Labrec EH, Schneider H. 1958. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J Bacteriol 75:604–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional images. (A) Additional images of strain 2457T grown in LB with 2% glucose and 0.4% bile salts. Further TEM analyses were performed to capture additional phenotypes of putative adherence structures and provide higher-magnification images to demonstrate that the structures originate from the bacterial cells (white arrows). Asterisks mark areas that displayed possible curli fibrils. The images are from experiments biologically independent of those from which the images provided in Fig. 1 and 7 were obtained. Each image displays a separate bacterial cell unless paired with a matching boxed sample. The magnifications and scale bars are provided for each image or image sets. (B) Wild-type cultures of 2457T were grown overnight by static growth in TSB media supplemented with 0.4% (wt/vol) bile salts. TEM analysis revealed structures similar to those of bacteria grown in LB supplemented with 2% glucose and 0.4% bile salts. Magnification, ×50,000; bar, 500 nm. (C) Two images of control media (LB with 2% glucose and 0.4% bile salts) are on top. Samples were prepared and processed as described in the Materials and Methods section for the biofilm assay and subsequent TEM grid preparation. TEM analysis revealed some precipitates of material from either the media or the uranyl acetate stain; however, the precipitation varied in concentration and did not resemble the structures observed in the presence of bacteria. Magnifications, ×50,000; bars, 500 nm (from separate experiments). (D) An image of an area without bacteria present of a TEM grid prepared from the 2457T biofilm (grown in IVLC medium). The same precipitation seen on the control medium grids tended to be less concentrated in clean areas of the bacterial grids. Magnification, ×50,000; bar, 500 nm. Download FIG S1, PDF file, 1.7 MB (1.7MB, pdf) .
Copyright © 2019 Chanin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Additional adherence gene cluster analysis. Analysis was performed as described in Materials and Methods as well as in the Fig. 2 legend. A 10-fold enhanced view of the trace reads are provided for genes S3342 and sfmA to sfmC to demonstrate the presence of RNA-seq reads. Download FIG S2, PDF file, 0.2 MB (226.5KB, pdf) .
Copyright © 2019 Chanin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Congo red binding assay. The assay was performed to test for the presence of the CsgA protein due to the ability of the Congo red dye to bind amyloid fibers and produce a birefringence signal under polarized light. The apple green color indicates the Congo red fluorescence that occurs when amyloid fibers are present. Wild-type 2457T produced a positive signal, while a significant reduction in the signal was detected for the ΔcsgA mutant. The ΔlpfA mutant was analyzed as a mutation control, and as seen, the mutation did not affect the birefringence signal. Images are representative of those from at least two biological independent experiments. Download FIG S3, PDF file, 0.2 MB (222.1KB, pdf) .
Copyright © 2019 Chanin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Treatment of the IVLC-induced biofilm with cellulase. The biofilm formation analysis was performed with or without cellulase to analyze the contribution of cellulose. A significant reduction in biofilm formation was detected in the presence of cellulase. All data represent those for three average OD540 readings from three biological independent experiments in which each experiment had technical triplicates. ***, P < 0.001. Download FIG S4, PDF file, 0.04 MB (40.3KB, pdf) .
Copyright © 2019 Chanin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of clinical isolates of Shigella flexneri. Both TEM (top) and biofilm formation (bottom) analyses were performed with clinical isolates of S. flexneri serotype 3a (strain AF11) and S. flexneri serotype 2a (strain AF16) to confirm the results of the study. Magnification for TEM, ×50,000 (on the left for strain AF11) and ×25,000 (on the right for strain AF16). For the biofilm assay, the average OD540 for the strains grown in either medium (light blue bars) or medium supplemented with IVLCs (dark blue bars) is plotted. All data represent those from three biological independent experiments in which each experiment had technical triplicates. ***, P < 0.001 for TSB versus TSB-bile salts for each strain. Download FIG S5, PDF file, 0.5 MB (541.4KB, pdf) .
Copyright © 2019 Chanin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.