Summary
Arabidopsis FLOWERING LOCUS T (FT) is a pivotal component of florigen, a long-range mobile flowering signal. Here, we determined the 1.0 Å-resolution crystal structure of FT, a significantly higher-resolution crystal structure of FT than previously reported one (2.6 Å). The present crystallographic studies revealed 4 alternative configurations with the precise location of the surrounding water molecules. Using this structural data, computational docking simulation predicted the putative binding sites for phosphatidylcholine (PC), an endogenous ligand that interacts with FT to modulate flowering time. In vitro reconstitution of the lipid–protein interaction showed that mutations at two of the predicted sites significantly compromised the lipid binding ability of FT. In planta, one of the mutant FT proteins significantly affected FT function in flowering, emphasizing the involvement of PC binding in modulating FT function. Our structural, biochemical, and transgenic analyses reveal the molecular mechanism of PC binding in FT-mediated flowering time control.
Subject Areas: Biological Sciences, Plant Biochemistry, Structural Biology, Plant Biology
Graphical Abstract
Highlights
-
•
The crystal structure of FT is determined at 1.0 Å resolution
-
•
A phospholipid-binding site is identified in FT protein
-
•
Phospholipid-binding is demonstrated to modulate FT function
-
•
Molecular basis for FT-mediated flowering time control is proposed
Biological Sciences; Plant Biochemistry; Structural Biology; Plant Biology
Introduction
Control of flowering time is crucial for successful fertilization and propagation of the species in seed plants. Photoperiod is a primary environmental cue that affects flowering time in most plant species. A long-day plant species, Arabidopsis thaliana, perceives day length in leaves and induces transcription and translation of FLOWERING LOCUS T (FT). FT is a component of florigen, a long-range mobile signal expressed in leaf companion cells and transported to the shoot apex to transmit photoperiodic flowering signals (Corbesier et al., 2007, Mathieu et al., 2007, Tamaki et al., 2007). In the shoot apex, FT interacts with a bZIP transcription factor, FD, and induces the expression of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) and APETALA1 (AP1), which triggers flower development (Abe et al., 2005, Wigge et al., 2005, Searle et al., 2006). In rice, the florigen activation complex (FAC) was proposed, in which Hd3a (a rice paralog of FT) interacts with the 14-3-3 protein GF14c and forms a bridge with rice FD (Taoka et al., 2011). FT is homologous with mammalian phosphatidylethanolamine-binding protein (PEBP). We previously demonstrated that FT specifically binds phosphatidylcholine (PC) to promote flowering in vivo (Nakamura et al., 2014). However, phospholipid-binding sites were not defined in the structural model of FAC (Taoka et al., 2013). Although crystal structure of FT at 2.6 Å resolution (Ahn et al., 2006) was a milestone for mechanistic understanding of FT function, even higher-resolution crystal structure of FT is demanded to address the in-depth molecular features of the newly emerging FT functions.
Here, we determined the crystal structure of FT at 1.0 Å resolution, which revealed detailed structures of FT including most of hydrogen atoms along with the precise location of the surrounding water molecules. Using our structural data, we employed a computational modeling approach to predict a putative binding site for PC, an endogenous regulatory ligand of FT (Nakamura et al., 2014). In vitro lipid-binding assay and in vivo functional assay identified a PC-binding site that critically regulates FT function in flowering time control. We propose a structural model of the FT–PC interaction in FAC.
Results
High-Resolution Crystal Structures of FT Protein
We crystallized FT under four different conditions and collected X-ray diffraction data at 1.0–1.5 Å resolution (Table 1). Despite that no electron density was observed for PC in these crystals, the crystal structures at 1.0 Å resolution revealed atomic details of interactions formed within the protein and about 230 water molecules surrounding its molecular surface (Figure 1A, center panel). The overall structure of FT consists of a central anti-parallel β-sheet flanked by short α-helices on one side and a short anti-parallel β-sheet on the other side. A 2Fo-Fc electron density map at 1.0 Å resolution clearly showed that a non-prolyl cis-peptide bond formed between Arg83 and Glu84, where the main-chain carbonyl and amide groups of these two residues and the side-chain carboxyl group of the latter are hydrogen bonded to residues Asn141-Thr144 (Figure 1A, upper right panel). The cis-peptide bond is also observed in other PEBP family proteins (Banfield et al., 1998, Serre et al., 1998, Simister et al., 2002). The 2Fo-Fc map indicated that the Oη of Tyr85, a key residue for flowering (Hanzawa et al., 2005), forms a hydrogen bond with the Oε of Glu109 as part of a hydrogen bond network including five water molecules (Figure 1A, lower right panel). Gln140, located in segment B (residues 128–141), is also involved in this hydrogen bond network, whereas His87 and Arg139 serve to stabilize the configuration of Tyr85 via direct van der Waals contact. Thus, the region around Tyr85 likely maintains a rigid conformation by forming a number of tight interactions with neighboring residues, and hence a Y85H mutation would affect the segment B conformation because of the compromised interactions with Glu109 and Arg139.
Table 1.
Data Collection and Refinement Statistics
| Condition 1 | Condition 2 | Condition 3 | Condition 4 | |
|---|---|---|---|---|
| Data Collection | ||||
| Space group | P212121 | P212121 | P212121 | P321 |
| Cell dimensions | ||||
| a (Å) | 39.62 | 48.48 | 48.89 | 53.5 |
| b (Å) | 48.72 | 51.53 | 60.96 | 53.5 |
| c (Å) | 73.57 | 56.9 | 63.8 | 103.98 |
| Resolution (Å) | 50–1.00 | 50–1.33 | 50–1.01 | 50–1.50 |
| (Outer shell) | (1.02–1.00) | (1.36–1.33) | (1.03–1.01) | (1.53–1.50) |
| Completeness (%) | 97.1 (66.1) | 99.9 (98.0) | 97.4 (94.9) | 99.9 (100) |
| Total reflections | 428,925 | 214,035 | 719,335 | 297,864 |
| Unique reflections | 75,289 | 33,436 | 97,694 | 28,282 |
| Redundancy | 5.7 | 6.4 | 7.4 | 10.5 |
| I/σ | 11.9 (1.2) | 12.9 (1.4) | 9.8 (2.7) | 14.2 (1.5) |
| Rmeas | 9.4 (69.3) | 10.0 (125) | 11.6 (67.3) | 7.3 (171) |
| CC1/2 | 99.8 (65.5) | 99.9 (56.1) | 99.4 (83.3) | 99.9 (70.9) |
| Refinement | ||||
| Rwork/Rfree (%) | 11.6/13.6 | 16.0/18.6 | 11.7/13.9 | 19.0/21.4 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.013 | 0.011 | 0.015 | 0.009 |
| Bond angles (°) | 1.332 | 1.17 | 1.467 | 0.986 |
| Ramachandran plot | ||||
| Favored (%) | 97.6 | 97.5 | 97.6 | 97.6 |
| Allowed (%) | 2.4 | 2.5 | 2.4 | 2.4 |
| Outliers (%) | 0 | 0 | 0 | 0 |
| PDB ID | 6IGG | 6IGI | 6IGH | 6IGJ |
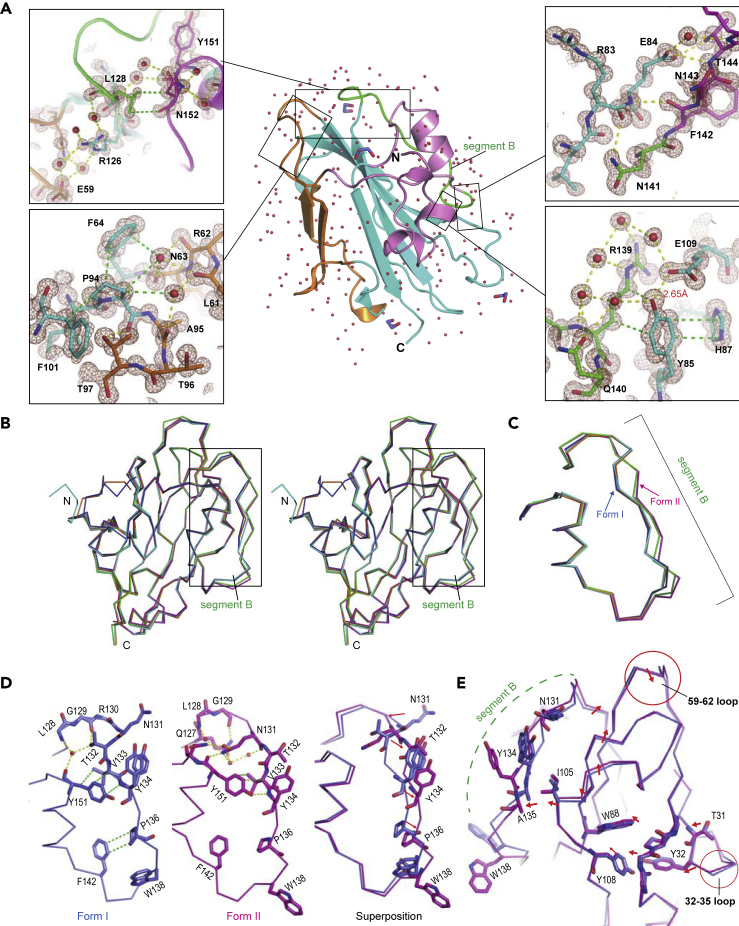
Figure 1.
High-Resolution Crystal Structures of FT
(A) Overall structure of FT (middle). A central β-sheet (cyan) is flanked by short α-helices (magenta) on one side and a short anti-parallel β-sheet (orange) on the other side. Segment B (residues 128–141) is shown in green. Water molecules are represented by red spheres. Ethylene glycol molecules (cryoprotectant) are in stick representation. Insets show close-up view of intramolecular interactions in the indicated regions. 2Fo-Fc electron-density maps at 1.5 sigma are in brown. Hydrogen bonds and van der Waals contacts are shown by yellow and green dotted lines, respectively.
(B) Stereo view of Cα backbone superposition of five FT crystal structures under condition 1 (Form I): blue; condition 2 (Form II): magenta; condition 3: cyan; condition 4: orange; and original condition (PDB: 1WKP): green. Note a significant conformational difference in segment B, as indicated by a black box.
(C) The square region in panel (B) is highlighted to demonstrate the different segment B conformations in FT crystal structures under different conditions.
(D) Detailed comparison of the segment B conformations between Form I (blue) and Form II (magenta). Several key residues are shown in stick representation.
(E) Structural correlation between the segment B conformation and 14-3-3 binding loops. 14-3-3 binding loops (32–35 and 59–62 loops) are indicated by red circles. The movements of key residues spatially intervening between these two loops are shown by red arrows.
See also Figures S1 and S2.
The high-resolution crystal structures of FT also provide structural insight into the functional phenotypes of several FT mutants. L128K mutant was reported to have weak activity of TERMINAL FLOWER1 (TFL1; an inhibitor of flowering) (Ho and Weigel, 2014). The side chain of Leu128 makes van der Waals contacts with Asn152 and a water molecule (Figure 1A, upper left panel). Near Leu128, Arg126 forms a hydrogen bond network with four water molecules and Glu59. Replacing Leu128 with positively charged lysine is predicted to cause electrostatic repulsion against Arg126, leading to a disruption of the hydrogen bond network. As a result, the segment B conformation is altered, which has been proposed to discriminate FT and TFL1 activity (Ho and Weigel, 2014). P94L is one of the loss-of-function mutations of FT (Kobayashi et al., 1999). The side chain of Pro94 makes van der Waals contacts with Phe64, Phe101, and two water molecules that form a hydrogen bond network with the carbonyl oxygen of Ala95, Leu61, Arg62, and Asn63 (Figure 1A, lower left panel). The Pro-to-Leu substitution at this position would disrupt this hydrogen bond network and van der Waals contacts, possibly leading to destabilization of the overall structure.
Comparison of the five crystal structures of FT including the previously reported one (Ahn et al., 2006) revealed that segment B, a key regulatory element of FT activity, can adopt two conformations (Forms I and II) (Figures 1B and 1C). Relative to Cα atoms in Form I, those of Try134 and Pro136 in Form II are moved by 3.1 Å and 2.1 Å, respectively (Figure 1D). The Cα atom of Thr132 in Form II is also moved by 3.7 Å from that in Form I, and its side chain is flipped out and exposed to the solvent. By contrast, Asn131 in Form II is flipped in, forming a hydrogen bond with the carbonyl oxygen of Tyr151 and the nearby water molecules. The relative positions of Tyr134 and Trp138, which are proposed to interact with a putative co-activator of FT (Ho and Weigel, 2014), significantly differ between Forms I and II. Superposition of the two forms suggests that the observed alterations in the segment B conformation is caused by the slight but significant displacement of the distant loop composed of residues 32–35 (Figures 1E and S1). The small rotation of the loop is transmitted intramolecularly to segment B via van der Waals contacts, which results in a conformational change in another distant loop of residues 59–62 (Figures 1E and S1). Notably, these two distant loops are involved in the interaction with 14-3-3 protein in the FAC complex (Taoka et al., 2011). Although the local displacement of the loop (residues 32–35) appears to be caused by different crystal packings (Figure S2), the observed structural correlation between segment B and the two 14-3-3 binding loops suggests that the segment B conformation plays a critical role in regulating the FAC complex formation. In this context, the segment B is suggested to be involved in recruitment of a transcriptional coactivator in plants (Taoka et al., 2013). Thus, a third-party protein such as transcription factors may possibly bind the segment B, regulating the formation or dissociation of the FAC complex through conformational changes in the 14-3-3 binding loops.
Identification of FT-PC-Binding Sites by Computational Modeling
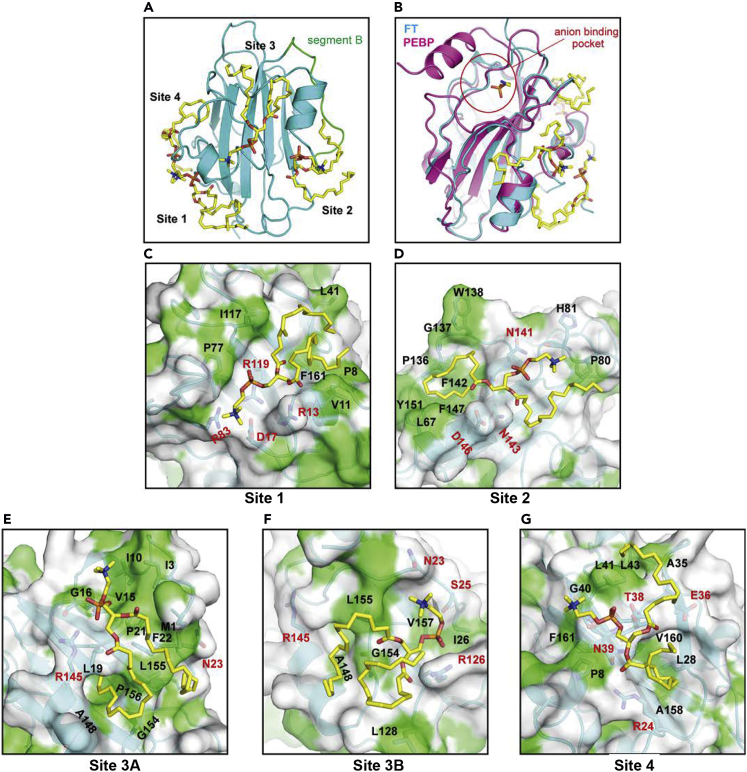
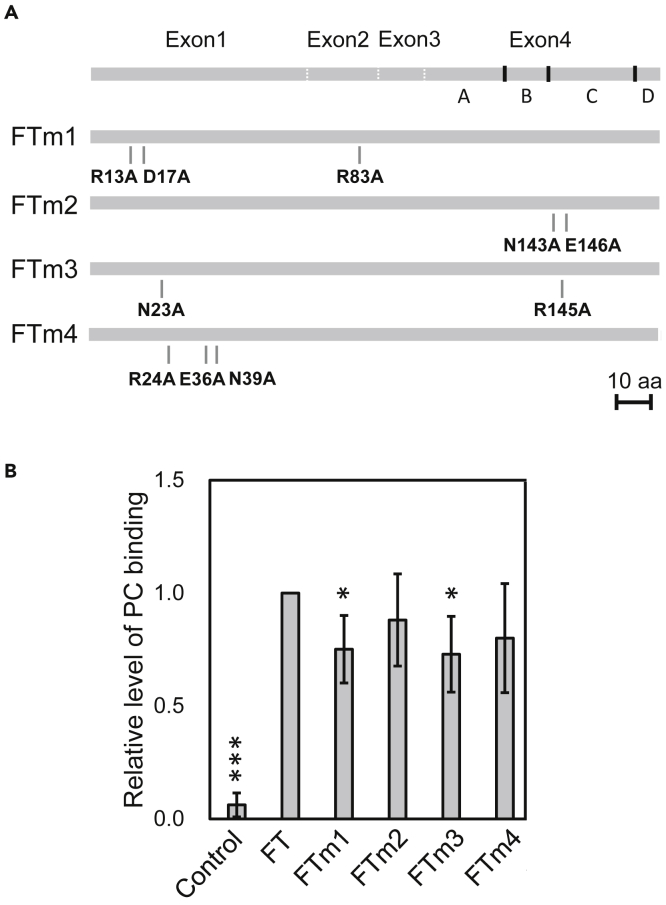
The anion-binding pocket of FT was originally proposed on the basis of the crystal structure of the PEBP in complex with phosphorylethanolamine (Figure S3) (Serre et al., 1998, Ahn et al., 2006). However, our recent study revealed that FT actually binds PC, not phosphatidylethanolamine (PE), to accelerate flowering. Notably, the larger head group of PC seems unlikely to be accommodated in the proposed anion-binding pocket of FT. In line with this, our docking simulation predicted other potential PC-binding sites (Figures 2A and 2B). In all docking models, the glycerophosphoric acid moiety of PC interacts with charged residues of FT, whereas the acyl groups interact with hydrophobic patches or clefts of FT (Figures 2C–2G). The choline moiety in site 1 interacts with charged residues, whereas those at the other three sites weakly interact with FT through van der Waals contacts with the nearby residues. To verify whether the predicted amino acid residues are important in PC binding in vitro, we performed lipid binding assay with four different versions of mutant FT (FTm1 to FTm4). Figure 3A illustrates amino acid substitutions by Ala in each FT mutant. We expressed recombinant FT proteins fused N-terminally to maltose-binding protein (MBP) and purified them for liposome association assay with 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, liposome size: 400 nm) (Cabrera et al., 2010). In this assay, PC liposomes were incubated with purified MBP-tagged FT protein, then precipitated by centrifugation. Because proteins that bind the liposome can be co-precipitated and thus recovered from the precipitates after centrifugation, the FT-PC binding can be analyzed quantitatively. A control experiment with MBP precipitated a negligible amount of PC (Figure 3B), indicating that MBP had no PC binding under this assay condition. As compared with wild-type FT, PC binding was significantly compromised for FTm1 and FTm3 but not FTm2 and FTm4, which suggests that amino acid residues mutated in FTm1 or FTm3 are involved in PC binding in vitro. Collectively, we identified two sites (Site 1 and Site 3) important for PC binding in vitro.
Figure 2.
Docking Model of the FT–PC Complex
(A) Four potential PC-binding sites in FT. PC molecules are represented by stick models.
(B) Superposition of putative PC (yellow stick)-bound FT (cyan ribbon) onto the phosphorylethanolamine-bound PEBP (magenta ribbon, PDB ID: 1B7A).
(C–G) Detailed structure of each potential PC-binding site. FT is shown in molecular surface representation, with hydrophobic residues in green (except main chain amide and oxygen). Residues that are predicted to interact with PC are represented by stick models.
See also Figure S3.
Figure 3.
Mutation of Putative PC-binding Domains Compromised FT–PC Binding In Vitro
(A) A schematic illustration of amino acid residues substituted by Ala in four different versions of mutated FT protein (FTm1 to FTm4). Scale bar = 10 amino acid residues.
(B) Quantitative analysis of FT-PC binding by protein–liposome association assay. The recombinant FT–maltose-binding protein (FT–MBP) fusion protein including different mutations (shown in panel A) was examined for binding to liposome consisting of dioleoyl (18:1)-PC. Binding with MBP (without FT) was a control. Data are relative to binding with wild-type FT (set as 1), with mean ± SD from three biologically independent experiments. ∗p < 0.05, ∗∗∗p < 0.001 by Student's t-test.
PC Binding at Site 1 Is Required for FT Function In Vivo
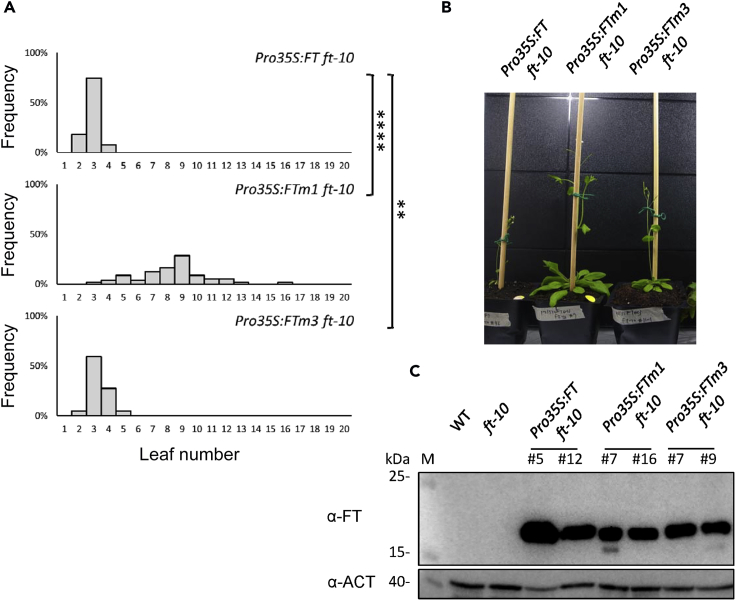
To examine whether the FT-PC binding is required for FT function in flowering time control in vivo, we used Pro35S:FT ft-10 transgenic line for the following reasons. First, overexpression of wild-type FT leads to very early flowering with narrow distribution of flowering time (Kobayashi et al., 1999) and thus is useful to map functionally important amino acid residues (Ho and Weigel., 2014). Second, endogenous FT protein is barely detectable in wild type due to its extremely low expression level but is clearly detected in Pro35S:FT plants by immunoblotting (Kim et al., 2016). Expression level of a mutant protein can thus be analyzed by using Pro35S:FT ft-10 transgenic line. We constructed Pro35S:FT ft-10, Pro35S:FTm1 ft-10, and Pro35S:FTm3 ft-10 transgenic lines.
For each transgenic plant, we observed the flowering time phenotype in at least 60 independent T1 transgenic lines for Pro35S:FT ft-10 and Pro35S:FTm1 ft-10 and 26 lines for Pro35S:FTm3 ft-10 to obtain average flowering time. Under long-day condition, as compared with Pro35S:FT ft-10, Pro35S:FTm1 ft-10, but not Pro35S:FTm3 ft-10, plants showed significantly delayed flowering time (Figures 4A and 4B). To assess the levels of the FT mutants, we performed immunoblot assay with a commercially available antibody against FT (AS06 198, Agrisera). We examined two representative transgenic lines for each overexpressing plant line (Pro35S:FT ft-10, Pro35S:FTm1 ft-10, and Pro35S:FTm3 ft-10) and found no marked reduction in FT protein level (Figure 4C). Thus, altered flowering time observed in the plants overexpressing FT mutants was not likely due to a considerable reduction in FT protein level.
Figure 4.
Functional Assay of Mutated FT with Reduced PC Binding In Vivo
(A) Distribution of flowering time in T1 transgenic plants of Pro35S:FT ft-10, Pro35:FTm1 ft-10, and Pro35:FTm3 ft-10. At least 26 independent T1 transgenic lines were tested for each. Statistical significance was examined by Student's t-test (∗∗p < 0.01; ∗∗∗∗p < 0.0001).
(B) A representative image of each transgenic plant analyzed in (A) at the onset of flowering.
(C) Detection of endogenous FT protein in 8-day-old seedlings of Pro35S:FT ft-10, Pro35:FTm1 ft-10, and Pro35:FTm3 ft-10. Two independent lines were examined for each transgenic line. Image is representative of three biological replicates.
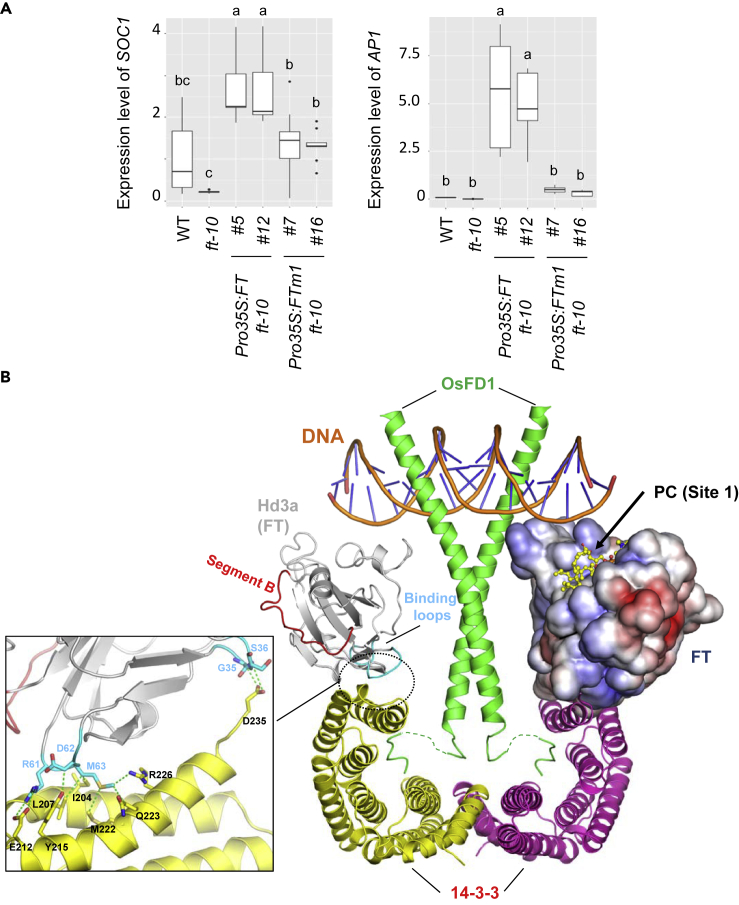
Next, to examine whether the altered flowering time was associated with FT function, we analyzed the expression of 2 effector genes of FT, AP1 and SOC1, in shoot apices of 7- and 14-day-old seedlings. Overexpression of FT (Pro35S:FT ft-10) upregulated SOC1 and AP1 as compared with the wild type (Figure 5A). Conversely, the expression levels of SOC1 and AP1 were reduced in Pro35S:FTm1 ft-10, indicating that the ability of FT to promote flowering was compromised in the transgenic plants expressing the FT Site 1 mutant. Taken together, our data suggest a PC-binding site in FT protein that is required for the flowering time control in vivo.
Figure 5.
FT–PC Interaction Affects Regulation of Effector Genes
(A) The expression of SOC1 and AP1 in shoot apices of 7- and 14-day-old seedlings, respectively. Data are mean ± SD of three biological replicates. Different letters indicate significant difference at p < 0.05 determined by one-way ANOVA with Tukey's post-test.
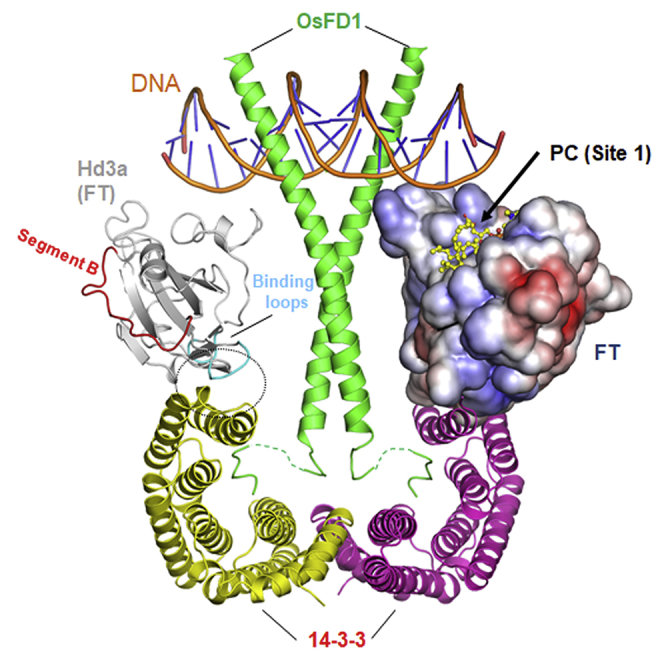
(B) A proposed structural model of florigen activation complex (FAC) comprising two Hd3a (FT homologue) molecules, a 14-3-3 dimer (yellow and magenta), bZIP domains of OsFD1 (green), and DNA (orange). The proposed model is based on the crystal structure of Hd3a in complex with the 14-3-3 protein and the C-terminal region of OsFD1 peptide (PDB: 3AXY). A present structure of FT is superposed onto the right-side Hd3a, as shown in surface representation colored by electrostatic surface potential. A complex model of the bZIP domain of OsFD1 and DNA was built based on the complex structure of the mouse CREB bZIP region (residues 285–339) and C-box DNA (Taoka et al., 2011; PDB: 1DH3). The predicted bound PC molecule is shown in a ball-and-stick model on the right-side FT molecule. Segment B and the 34–37 and 61–64 binding loops of Hd3a (the 32–35 and 59–62 loops in FT) are shown in red and cyan, respectively, on the left-side Hd3a molecule. The interaction between the binding loops of Hd3a and 14-3-3 protein is highlighted in the inset. Van der Waals contacts between them are shown by green dashed lines. Other interactions at the interface between Hd3a and 14-3-3 are omitted for clarity.
Discussion
The present crystal structures of FT reveal atomic details about the protein. Docking models and systematic mutation analyses suggest that Site 1 in FT is involved in PC binding. Compromised PC binding of FTm1 and FTm3 in vitro is consistent with computational modeling. In vivo, the ability of FT to promote flowering was compromised in the transgenic plants expressing the FTm1 but not the FTm3. This observation is not likely due to the altered stability of the mutant protein because a comparable amount of FT was detected in transgenic plants that expressed different versions of mutant FT (Figure 4C). Although the remaining PC-binding property of FTm1 suggests the presence of yet unidentified PC-binding site(s) in FT, our in vivo and in vitro data demonstrate that FT-PC interaction at Site 1 is involved in flowering time control.
PEBP proteins have a putative anion-binding pocket, and the phosphate-containing polar head group of phospholipids was considered to be a possible ligand for this pocket (Taoka et al., 2013). However, the amino acid residues we demonstrated to be involved in PC binding indicate that this putative anion-binding pocket is not likely a primary site that accommodates the polar head group. This notion is supported by the fact that phosphocholine, a polar head group of PC that binds to FT, is not anionic and also by the observation that no anionic phospholipid (e.g., phosphatidylglycerol) was capable of binding to FT (Nakamura et al., 2014). Moreover, FT–PC binding involves specificity toward the hydrophobic acyl chain of PC, which can hardly be explained by only the anion-binding pocket (Nakamura et al., 2014). Although the current crystallographic study and in silico modeling cannot fully address the differential interaction of diurnally changing acyl species of PC (Nakamura et al., 2014), our proposed model of PC binding at the peripheral region of FT agrees with the finding that the phospholipid acyl chains as well as the polar head group confer binding specificity in the FT–PC interaction (Nakamura et al., 2014).
It remains an important open question how the FT-PC interaction triggers FT function in flowering time control. The previously proposed model in rice (Taoka et al., 2013) suggests that the bZIP domain of OsFD1 tightly interacts with DNA, and FT appears to serve as an additional DNA binding protein that further stabilizes the FAC complex. In this regard, the positively charged surface of the FT Site 1 likely plays an auxiliary role in DNA binding in the complete FAC complex (Figure 5B). Of note, PC is predicted to bind to the positively charged surface (Site 1) situated close to DNA. Indeed, FTm1, with compromised PC binding, significantly affected the upregulation of effector gene expression (Figure 5A). Although several alternative mechanisms can be inferred, these observations suggest a possibility that PC binding may facilitate the interaction of FT with DNA, likely serving for the gene regulation in flowering time control. This notion awaits further experimental verification by cell biological approaches including an imaging of cellular FT movement under altered PC content. In conclusion, we revealed higher-resolution crystal structures of FT than previously reported one and used the data to elucidate the molecular mode of FT-PC interaction in flowering time control.
Limitations of the Study
Although our structural, biochemical, and transgenic analyses reveal the molecular mechanism of PC binding in FT-mediated flowering time control, the proposed model (Figure 5B) requires thorough experimental verification by testing the interaction between protein and DNA in the presence or absence of PC and observing the actual movement of the FT-PC complex into the nucleus. Also, the model needs to be developed so that it can account for the differential interaction of diurnally changing acyl species of PC (Nakamura et al., 2014).
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This research was supported by Academia Sinica (grant ID: AS-CDA-107-L02 to YN) and Ministry of Science and Technology, Taiwan (grant ID: 105-2628-B-001-006-MY3 to YN) and performed under the Cooperative Research Program of the “Network Joint Research Center for Materials and Devices” awarded to YN & KI.
Author Contributions
Y.N, Y-C. L, S.W, K.K, and K.I designed the experiments. K.I supervised structural experiments (Figures 1, 2, and S1–S3, and Table 1) and Y.N supervised biochemical (Figure 3) and in planta (Figures 4 and 5A and Table S1) experiments. Y.N, Y-C. L, S.W, Y-c. L, K. K, and K.K performed experiments. Y.N, Y-C. L, S.W, K.K, and K.I wrote the paper.
Declaration of Interests
The authors declare no competing interests.
Published: November 22, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.10.045.
Contributor Information
Yuki Nakamura, Email: nakamura@gate.sinica.edu.tw.
Kenji Inaba, Email: kenji.inaba.a1@tohoku.ac.jp.
Data and Code Availability
Coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 6IGG (condition 1), 6IGI (condition 2), 6IGH (condition 3), and 6IGJ (condition 4), respectively.
Supplemental Information
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- Ahn J.H., Miller D., Winter V.J., Banfield M.J., Lee J.H., Yoo S.Y., Henz S.R., Brady R.L., Weigel D. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 2006;25:605–614. doi: 10.1038/sj.emboj.7600950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield M.J., Barker J.J., Perry A.C., Brady R.L. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure. 1998;6:1245–1254. doi: 10.1016/s0969-2126(98)00125-7. [DOI] [PubMed] [Google Scholar]
- Cabrera M., Langemeyer L., Mari M., Rethmeier R., Orban I., Perz A., Bröcker C., Griffith J., Klose D., Steinhoff H.-J. Phosphorylation of a membrane curvature–sensing motif switches function of the HOPS subunit Vps41 in membrane tethering. J. Cell Biol. 2010;191:845–859. doi: 10.1083/jcb.201004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Hanzawa Y., Money T., Bradley D. A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. U S A. 2005;102:7748–7753. doi: 10.1073/pnas.0500932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho W.W., Weigel D. Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell. 2014;26:552–564. doi: 10.1105/tpc.113.115220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.J., Hong S.M., Yoo S.J., Moon S., Jung H.S., Ahn J.H. Post-translational regulation of FLOWERING LOCUS T protein in Arabidopsis. Mol. Plant. 2016;9:308–311. doi: 10.1016/j.molp.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Kuttner F., Schmid M. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 2007;17:1055–1060. doi: 10.1016/j.cub.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Andres F., Kanehara K., Liu Y.C., Dormann P., Coupland G. Arabidopsis florigen FT binds to diurnally oscillating phospholipids that accelerate flowering. Nat. Commun. 2014;5:3553. doi: 10.1038/ncomms4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Krober S., Amasino R.A., Coupland G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre L., Vallee B., Bureaud N., Schoentgen F., Zelwer C. Crystal structure of the phosphatidylethanolamine-binding protein from bovine brain: a novel structural class of phospholipid-binding proteins. Structure. 1998;6:1255–1265. doi: 10.1016/s0969-2126(98)00126-9. [DOI] [PubMed] [Google Scholar]
- Simister P.C., Banfield M.J., Brady R.L. The crystal structure of PEBP-2, a homologue of the PEBP/RKIP family. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1077–1080. doi: 10.1107/s090744490200522x. [DOI] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Taoka K., Ohki I., Tsuji H., Furuita K., Hayashi K., Yanase T., Yamaguchi M., Nakashima C., Purwestri Y.A., Tamaki S. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Taoka K., Ohki I., Tsuji H., Kojima C., Shimamoto K. Structure and function of florigen and the receptor complex. Trends Plant Sci. 2013;18:287–294. doi: 10.1016/j.tplants.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 6IGG (condition 1), 6IGI (condition 2), 6IGH (condition 3), and 6IGJ (condition 4), respectively.