Abstract
Studies from global loss-of-function mutants suggest that alternative NF-κB downstream of NF-κB inducing kinase (NIK) is a cell-intrinsic negative regulator of osteogenesis. However, the interpretation of the osteoblast and/or osteocyte contribution to the bone phenotype is complicated by simultaneous osteoclast defects in these models. Therefore, we turned to a transgenic mouse model to investigate the direct role of NIK in the osteolineage. Osx-Cre;NT3 animals (NT3-Cre+), which bear a constitutively active NIK allele (NT3) driven by Osx-Cre, were compared to their Cre-negative, Control (Ctrl) littermates. NT3-Cre+ mice had elevated serum P1NP and CTX levels. Despite this high turnover state, microCT showed that constitutive activation of NIK resulted in a net increase in basal bone mass in both cortical and cancellous compartments. Furthermore, NT3-Cre+ mice exhibited a greater anabolic response following mechanical loading compared to controls. We next performed RNA-Seq on non-loaded and loaded tibiae to elucidate possible mechanisms underlying the increased bone anabolism seen in NT3-Cre+ mice. Hierarchical clustering revealed two main transcriptional programs: one loading-responsive and the other NT3 transgene-driven. Gene ontology (GO) analysis indicated a distinct upregulation of receptor, kinase and growth factor activities including Wnts, as well as a calcium-response signature in NT3-Cre+ limbs. The promoters of these GO-term associated genes, including many known to be bone-anabolic, were highly enriched for multiple κB recognition elements (κB-RE) relative to the background frequency in the genome. The loading response in NT3-Cre+ mice substantially overlapped (>90%) with Ctrl. Surprisingly, control animals had 10-fold more differentially expressed genes (DEGs) in response to loading. However, most top DEGs shared between genotypes had a high incidence of multiple κB-RE in their promoters. Therefore, both transcriptional programs (loading-responsive and NT3 transgene-driven) are modulated by NF-κB. Our studies uncover a previously unrecognized role for NF-κB in the promotion of both basal and mechanically-stimulated bone formation.
Introduction
Integration of biophysical stimuli is critical to proper bone formation, both developmentally and in adults, but the mechanisms driving the bone anabolic response are incompletely understood. Bone formation in response to mechanical loading requires the coordinated action of osteocytes, which sense the load, and osteoblasts (OBs), which are directed to form bone by osteocytes. Net accrual of bone mass also requires that the bone-degrading activity of osteoclasts be limited, which is largely accomplished at the level of the osteocyte via expression of osteoprotegerin (OPG) (1).
A long list of developmental, growth factor, and hormonal signaling pathways have been shown to support bone formation during development and/or adult homeostasis, including BMP, Wnt, Hedgehog, and Notch(2). These converge on the master transcriptional regulators of osteoblast differentiation, Runx2 and Osterix (Osx). A variety of other more ubiquitously expressed transcription factors cooperate with Runx2 and Osx and impact osteoblast formation and function (3). Increases in bone formation can be accomplished by increasing generation of OBs from precursors, by stimulating the activity of pre-existing cells, or a combination of the two. Mechanical strain increases OB differentiation, via Wnt/β-catenin signaling, a pathway that also enhances matrix production and mineralization (4). Osteocytes control Wnt activation in OBs via controlled secretion of sclerostin (Sost), a soluble Wnt inhibitor, and mechanical loading downregulates Sost in a strain-dependent manner (1,5–7). However, Sost-independent bone formation after loading has been documented (8). Additionally, the pathway(s) that transduce the mechanical signals to Sost and other bone-stimulating signals are not well-defined, although integrins, PGE2 release, and calcium flux have been implicated (4,9–11). Thus, much remains to be discovered regarding regulation of bone formation in response to mechanical stimuli.
The nuclear factor kappa-B (NF-κB) family of transcription factors has been shown to affect bone formation, primarily in an inhibitory direction, in a number of animal models (12). Classical NF-κB signaling, activated by a wide array of stimuli including the common inflammatory factors TNFα and IL-1β, is largely mediated by the inhibitor of κB kinase complex (IKKα,β,γ) and p65 (13). Its inhibition in the osteoblast lineage through expression of a dominant negative IKKγ increased both bone mass in growing mice and bone formation after ovariectomy, although bone mass normalized in adult mice at baseline (14). Conversely, constitutive activation of classical NF-κB in osteoprogenitors led to abnormal skeletal development and decreased bone formation (15). Alternative (or non-canonical) NF-κB signaling depends on NF-κB inducing kinase (NIK), and IKKα-mediated processing of the precursor p100 to p52 to allow nuclear localization of NF-κB subunit RelB (12). To date, no osteolineage-specific mouse models of alternative NF-κB modulation have been reported, but similar to the classical pathway, available data suggests that it too dampens bone formation. Both aly/aly mice (which globally express an inactive form of NIK) and germline RelB KO mice exhibited an increase in basal bone mass and bone formation rate in vivo, at least transiently (16–20). Furthermore, mesenchymal progenitor cells from RelB KO mice also displayed enhanced bone formation in a tibial cortical defect model (18). The molecular mechanism(s) by which NF-κB inhibits OBs are poorly defined but may include interactions with Runx2, β-catenin, Smads, AP-1 (Fra1), focal adhesion kinase (FAK), JNKs, and ERKs (14,17,18,21–28).
To investigate the role of NF-κB downstream of NIK specifically in the osteolineage, we utilized Osx-Cre (29) to drive a constitutively activated form of NIK. The NIKΔTRAF3 (NT3) mice harbor a mutant allele lacking the negative-regulatory TRAF3 binding domain (30). This allele was knocked into the ROSA26 locus downstream of a loxP/Neo-STOP/loxP cassette allowing for stabilization of NIK protein and continuous activation of alternative NF-κB upon Cre-mediated recombination. Because Osx is upregulated early in osteolineage commitment, activation of the pathway via NT3 transgene expression in our model is targeted to bone-forming osteoblasts as well as osteocytes. In these transgenic mice, we explored both the basal bone phenotype and response to mechanical loading, using RNA-Seq to assess effects on gene expression during bone formation.
Materials and Methods
Reagents and Mice
Mice were housed communally in a temperature-controlled, pathogen-free barrier facility, with 12-hour light/dark cycles, and had ad libitum access to fresh chow and water. Mice were observed daily by Division of Comparative Medicine (DCM) staff and at least once a week by laboratory personnel. Any health concerns were reported promptly to the DCM veterinarian and all treatment or euthanasia recommendations followed. All protocols were approved by Institutional Animal Studies Committee at Washington University School of Medicine (ASC protocol #20170025).
The NT3 transgenic line harbors a mutant allele of human NIK, NIKΔT3 (NT3), which lacks a regulatory TRAF3 binding domain. This construct was placed into the murine ROSA26 locus downstream of a loxP/Neo-STOP/loxP cassette allowing for stabilization of NIK protein and continuous activation of the alternative NF-κB pathway upon Cre-mediated recombination (30). Osx1-GFP::Cre mice express Cre-recombinase under control of a Tet-OFF cassette (29). The NT3 transgenic and Osx1-GFP::Cre (Osx-Cre) mouse lines were maintained separately due to strain differences (C57Bl/6J and mixed C57Bl/6J and CD1, respectively). Homozygous NT3 females were crossed to heterozygous Osx-Cre males to generate Osx-Cre;NT3 (NT3-Cre+) mice and their Cre-negative, Control (Ctrl) littermates. Without any doxycycline in the chow, mating pairs did not yield any NT3-Cre+ pups at time of genotyping (P0-P7). The mechanism for this lethality was not further explored in this study since pups survived to adulthood at the expected Mendelian ratios when fed a 200ppm doxycycline chow until weaning (P21-P22). Experimental mating pairs were maintained on this 200ppm doxycycline chow diet (Purina Test Diet #1816332–203, St. Louis, MO, USA), and pups were switched to standard rodent chow (Purina 5058, St. Louis, MO, USA) at weaning. Age and sex-matched animals were used in all experiments.
Previous reports have shown that the parental Osx-Cre line maintained off doxycycline exhibits a growth delay leading to decreased body weight and reduced cortical bone size and thickness that resolves by 8–12 weeks of age (31–33). Notably, the bone defects in young (6 weeks) Osx-Cre mice were shown to be an indirect effect of their smaller overall size as normalization to body weight accounted for the observed differences (32). To determine whether the Osx-Cre allele had any effects on bone with our doxycycline regimen, we examined both Osx-Cre and WT littermates within our parental Osx1-GFP::Cre strain. At 6 weeks of age, our Osx-Cre mice had slightly reduced body weights (−7.8%) compared to WT (Fig S1A), but this change was much milder than that reported by Davey et al (−22%) without doxycycline administration (32). When normalized to body weight, microCT of our Osx-Cre femurs showed no difference in either cortical or cancellous compartments (Fig S1B-F). Even more importantly, 6-week-old NT3-Cre+ mice had similar body weights to their Ctrl littermates (Fig S1G). Thus, any observed differences in bone mass between NT3-Cre+ and Ctrl mice are driven by the NT3 transgene and not the Cre.
Micro-computed tomography
Scanning settings and thresholding used for analysis were determined empirically to account for known age- and sex-differences in bone microarchitecture. All images were analyzed in a blinded-fashion, and the same threshold was applied for all animals of the same age and sex. Outcome variables are reported in accordance with published guidelines (34). The left femur was scanned ex-vivo (μCT40, Scanco, Brüttisellen, Switzerland) for male mice (10μm, 55kVp, 145μA, 8W, 300ms integration time) and female mice (10μm, 55 kVp, 72μA, 4W, 300ms integration time) at 16 wks. For Osx-Cre mice and their WT littermates, the left femur was scanned ex-vivo at 6 weeks of age (10 μm resolution, 55 kVp, 145μA, 8W, 300ms integration time). For all femur scans, regions of interest were defined at 1 mm proximal to the end of the femoral growth plate or at the mid-shaft (50% femur length) for cancellous and cortical bone, respectively.
Serum markers of bone turnover
Osteoblast and osteoclast activities were measured in the serum of animals using Rat/Mouse P1NP EIA ((Immunodiagnostic Systems, Gaithersburg, MD, USA; AC-33F1) or RatLaps (CTX-I) EIA (Immunodiagnostic Systems, Gaithersburg, MD, USA; AC-06F1) according to manufacturer’s instructions. Mice were fasted overnight of food, but not water, before blood collection to prevent the release of post-prandial hormones, which affect collagen degradation (35). Samples were collected either by mandibular bleed or under isoflurane/oxygen anesthesia by cardiac puncture prior to euthanasia.
Bone histomorphometry for basal studies
At 6 weeks, male mice were injected intraperitoneally (IP) with 10 mg/kg calcein (C0875; Sigma, USA) and 30 mg/kg alizarin red (A3882; Sigma, USA) for a 5-day labeling period. The left tibiae were fixed in 10% neutral buffered formalin for 16–24h, washed briefly with water, then stored in 70% ethanol. For tissue processing, the left tibiae were submitted to the Washington University Musculoskeletal Histology and Morphometry Core. Tibia were embedded in methyl methacrylate and sagittal sections either imaged unstained or after TRAP staining. A NanoZoomer 2.0 HT whole slide scanner (Hamamatsu Photonics, Japan) was used to acquire images for basal phenotyping using standard brightfield settings or FITC (level 1)/TRITC (level 8) at 20x magnification. Bioquant Osteo software (v18.2.6; Bioquant Image Analysis Corp., Nashville, TN, USA) was used for image analysis by a blinded user. Dynamic indices of bone formation were measured at the anterior endocortical surface in a 2 mm region of interest defined 5mm from the end of the growth plate. Static indices were measured in a 2 mm region of interest defined 100 μm from the end of the primary spongiosa. All measurements are reported in accordance with published standards (36).
Quantitative real-time PCR
Tibiae of 12-week-old male mice were flushed of marrow and the distal epiphyses removed. Total RNA was isolated as outlined in RNA-Seq methods. cDNA synthesis was performed using 300 ng total RNA with a double-primed cDNA kit (mix of random hexamer and oligo(dT)18 primers) per the manufacturer’s instructions (639549, Takara Bio, USA). qRT-PCR was performed using iTaq universal SYBR green supermix (172–5121, BioRad, USA) and a QuantStudio 3 Real-time PCR system (Applied Biosystems, USA) using standard settings followed by melt-curve analysis. Biological replicates (n=4–7 per genotype) were run in technical duplicates. Relative expression of target genes was calculated using the 2-ΔΔCT method with Ipo8 as the reference gene. Primer sequences: Tnfsf11/Rankl (Forward: ACT TTC GAG CGC AGA TGG AT; Reverse: CCA GAG TCG AGT CCT GCA AA), Tnfrsf11b/Opg (Forward: GTT TCC CGA GGA CCA CAA T; Reverse: CCA TTC AAT GAT GTC CAG GAG), Ipo8 (Forward: GACCATGTGGAGTTCCCTGT; Reverse: CTGGCCAGTATTGTGTCACC).
Strain gauging, in vivo loading and dynamic histomorphometry
Unilateral axial tibial compression was performed on the right tibiae of 16-week-old, male animals. The left tibia served as a non-loaded control. One cohort of mice (n=5) was used for strain gauging to empirically determine peak periosteal strain values as previously described (37). A force of 8.7N or 10.0N was used to generate a −2000 microstrain deformation in Ctrl and NT3-Cre+ animals, respectively. This level of microstrain has been shown to be anabolic for cortical bone in similar aged mice without causing a damage/woven bone response (38). Mice were subjected to 1 loading bout per day (1200 cycles, 4 Hz triangle waveform with 0.1 s rest-insertion after each cycle) for a total of 9 days (Tuesday-Friday, week 1; Monday-Friday, week 2) using an Electropulse 1000 materials testing apparatus (Instron, Norwood, MA, USA). Mice were anesthetized with isoflurane/oxygen before the procedure and their level of sedation monitored by hind paw reflex. After loading, mice received either buprenorphine (0.1 mg/kg, SQ) or buprenorphine SR (1.0 mg/kg, SQ) for pain control and were monitored until they could ambulate normally.
Calcein and alizarin labels were given IP on study days 4 and 11, respectively (7-day labeling period). Mice were euthanized for tissue harvest on study day 14. Both non-loaded and loaded tibiae were sectioned at a distance 5 mm proximal to the distal TFJ to assess the area of peak strain (37,39). FITC and TRITC images were captured at 20x and overlaid using an Olympus IX51 fluorescent microscope (Olympus, Center Valley, PA) and Leica EC3 camera/software (Leica Microsystems Inc., Buffalo Grove, IL). Resulting images were stitched together using an image composite editor (Microsoft corp., USA). Bioquant Osteo software (v18.2.6; Bioquant Image Analysis Corp., Nashville, TN, USA) was used for image analysis by a blinded user to determine dynamic indices of bone formation. In samples where the MAR was zero, an imputed value of 0.1 was used to allow for statistical comparisons. All measurements are reported in accordance with published standards (36).
RNA-Seq
A separate cohort of Ctrl and NT3-Cre+ (n=4) were subjected to the same mechanical loading protocol (described above) for 5 consecutive days. Four hours after the last loading bout, right and left tibiae were stripped of all muscle tissue and flushed by centrifugation (10,000g x 2 min) at room temperature to remove bone marrow. The flushed tibiae were then cut 2 mm from the articular surface and again at the distal TFJ to isolate the area of peak strain. Tissue samples were flash-frozen in liquid nitrogen and stored at −80C°. Frozen tibiae were pulverized in liquid nitrogen using a Mikro Dismembrator (Mikro-Dismembrator S; B. Braun Biotech International, Melsungen, Germany), resuspended in TRIzol (15596026; Invitrogen, USA), and stored at −80C°. Total RNA was isolated using a Direct-zol kit per manufacturer instructions (R2072; Zymo Research, USA). RNA concentration was determined using a Nanodrop (ND-2000; ThermoFisher, USA). RIN values were used to determine RNA quality (Bioanalyzer 2100; Agilent Technologies, USA), and the average RIN score was 5.4.
RNA sequencing was performed on the Total RNAble platform (Cofactor Genomics, http://cofactorgenomics.com, St. Louis, MO, USA). Briefly, species-specific rRNA-probes were hybridized to total RNA to remove nuclear-encoded and mitochondrial contaminating ribosomal RNA from the sample. The resulting ribo-depleted RNA was then fragmented. First-strand cDNA synthesis was performed using reverse transcriptase and random primers in the presence of Actinomycin D, followed by second-strand cDNA synthesis with DNA polymerase I and RNase H. Double-stranded cDNA was end-repaired and A-tailed for subsequent adaptor ligation. Indexed adaptors were ligated to the A-tailed c-DNA. Enrichment by PCR was performed to generate the final cDNA sequencing library. Libraries were sequenced as single-end 75 base pair reads on an Illumina NextSeq500 following the manufacturer’s protocols. Raw sequence data was analyzed by the Bioinformatics Research Core at the Washington University Center of Regenerative Medicine. RNA-Seq reads were aligned to the mouse genome (assembly mm10) with STAR version 2.4.2a (40). Gene counts were derived from the number of uniquely aligned unambiguous reads by Subread:featureCount (41), with GENCODE gene annotation (V14) (42). Only non-redundant uniquely aligned reads were used to estimate the expression level of genes. All gene-level transcript counts were then imported into the R/Bioconductor package DESeq2 (43) and normalized to adjust for differences in library size. Genes not expressed in any sample were excluded from further analysis. Differentially expressed genes were then filtered for those having fold-changes (FC) > 1.5 together with a BH adjusted p-value less than or equal to 0.05. A negative log2FC value indicates higher gene expression in the second group vs the first group for all DEG lists and data presented except for Figures 6–7, where the FC was flipped for visualization purposes. The genotype of samples was verified using Cre expression data normalized per 10 million raw reads (NT3-Cre+: 18–66; Ctrl: 0). To generate the top 20 DEG lists, we further refined our analysis to only those genes with an adjusted p-value less than 0.01. RNA-Seq data have been deposited in NCBI’s Gene Expression Omnibus under the GEO Series accession number GSE133212 (https://www.ncbi.nlm.nih.gov/geo/).
Figure 6: The transcriptional response to mechanical loading is augmented in NT3-Cre+ mice.
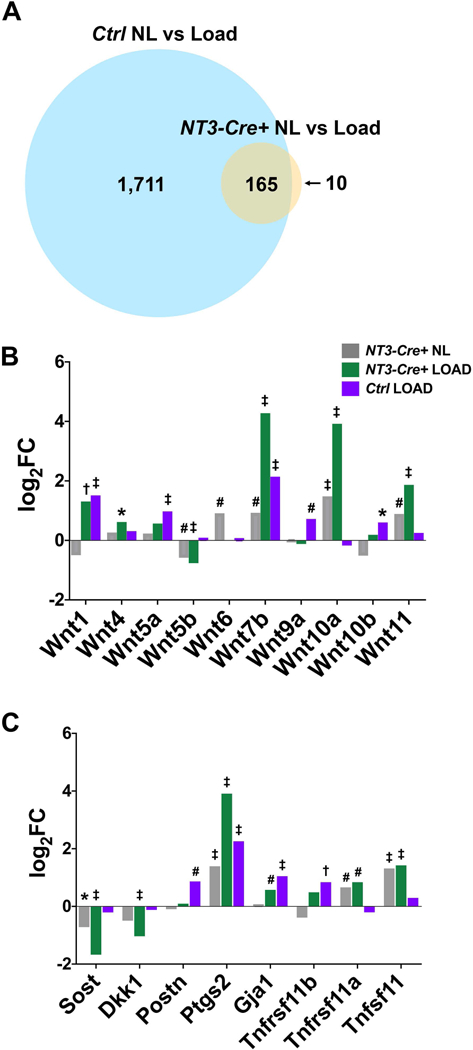
(A) Venn diagram depicting the extensive overlap in DEG lists between Ctrl vs NT3-Cre+ in response to loading (adjusted p-value <0.01). Fold change (expressed as Log2FC) for each genotype and/or loading condition compared to Ctrl NL samples for (B) Wnt genes and (C) other loading-related genes of interest. Significance (adjusted p-value): *p<0.05, #p<0.01, †p<0.001, ‡p<0.0001. Tnfrsf11b (OPG); Tnfrsf11a (RANK); Tnfsf11 (RANKL).
Figure 7: κB-RE are enriched in response to the NT3 transgene and with loading.
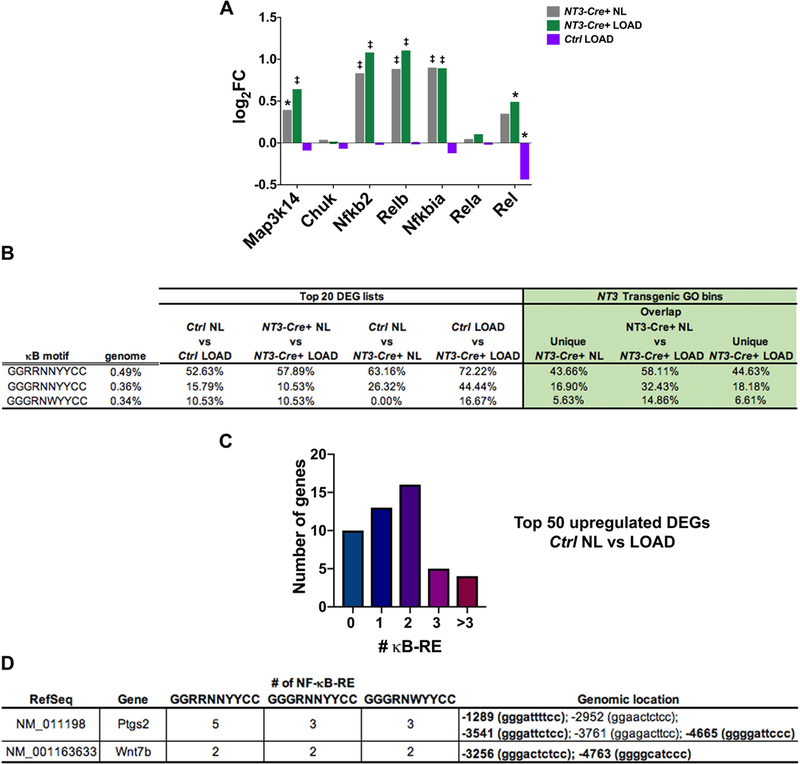
(A) Log FC for each genotype and/or loading condition compared to Ctrl NL samples for NF-κB signaling pathway components. Significance (adjusted p-value): *p<0.05, ‡p<0.0001. (B) (left) Incidence of genes with ≥2 κB recognition elements (κB-RE) in the top 20 upregulated DEG lists and (right) NT3 transgenic GO bins is much higher than the background frequency in the genome in both loading and NT3 transgene settings. Genome background frequency is defined as the probability of at least 1 out of 121 randomly selected genes (size of largest NT3 transgenic GO bin) containing ≥2 κB-RE in 10 million iterations. (C) Number of genes with the indicated number of κB-RE in the top 50 Ctrl NL vs LOAD upregulated DEG list, indicating an enrichment of κB-RE in the normal loading response. (D) Both Ptgs2 and Wnt7b contain multiple κB-RE. Genomic coordinates are listed relative to the transcription start site (position zero). Bold genomic location indicates sequence is found across all 3 motifs tested. R = A/G; N = A/G/C/T; Y= C/T; W = A/T
Gene ontology (GO) analysis was performed across all four groups with optimal sequential multi-way partitioning using a probabilistic method script written in Python (44). Significant GO terms were determined using a cut-off value of p<1e-3. GO-term associated genes and the top DEG lists were queried for NF-κB binding motifs (45–47) within 5kb of their transcriptional start site (UCSC Golden Path database for the mouse genome). For technical reasons, microRNAs, GM predicted genes, genes with complex promoters, or genes not fully mapped to the genome (Mir6950, Gm8276, Gm15677, Gm17140, Gm20056, Gm20465, Gm22615, Gm28530, Gm37226, Gm42679, Gm45745, Gm46572, Ccdc170, Cnr1, Emid1, Nrg2, Sdk2, Trdc) were not included in the analysis for κB recognition elements (κB-RE).
Statistical Analysis
Except for RNA-Seq analyses, all statistics were computed using GraphPad Prism v8 software (GraphPad Software, Inc., La Jolla, CA, USA). Values of p<0.05 were considered significant, and data are presented as mean ± SD. For pairwise comparisons, either a student’s, unpaired, two-tailed t-test or student’s paired, two-tailed t-test was used. A Welch’s correction was applied to the t-test if the F-test to compare variances was significantly different. For microCT and body weight data, either a Grubbs’ (alpha 0.05) or ROUT outlier test (Q2%) was employed to remove a maximum of 1 outlier per group, if indicated, only in sample sizes n≥12. For multiple group comparisons, a 2-way ANOVA followed by Tukey’s multiple comparison test was performed. Sample sizes are indicated in the respective figure legends.
Results
Basal bone mass is enhanced in NT3-Cre+ mice
To investigate the role of alternative NF-κB specifically in the osteolineage, we mated the NT3 line (NT3f/f) (30) to the Tet-OFF Osx-Cre (Cre+/−) line (29). The resulting litters, maintained on doxycycline until weaning, contained Control (Ctrl) mice with no NT3 transgene expression and NT3-Cre+ mice with activation of alternative NF-κB signaling after removal of doxycycline. Recombination of the NT3 transgenic allele was detected in crushed long bones (flushed of marrow) by PCR in NT3-Cre+ bones but not in Ctrl (Fig S2).
We utilized ex-vivo microCT to assess both male and female mice at 16 weeks. Male NT3-Cre+ femurs displayed increased cortical thickness and total area (Fig 1A). Additionally, these bones had significantly greater cancellous bone volume fraction (BV/TV) and volumetric bone mineral density (vBMD) compared to Ctrl (Fig 1B-C). Likewise, cortical thickness, total area, and cancellous parameters (BV/TV, vBMD) were also increased in NT3-Cre+ female mice (Fig 1D-F). In addition, NT3-Cre+ bones of both sexes had higher cortical bone area and polar moment of inertia (Fig S3), which is positively correlated to bone strength (48). Thus, Osx-Cre driven alternative NF-κB signaling in the osteolineage enhances bone accrual in both adult male and female mice.
Figure 1: Basal bone mass is enhanced in both male and female NT3-Cre+ mice.
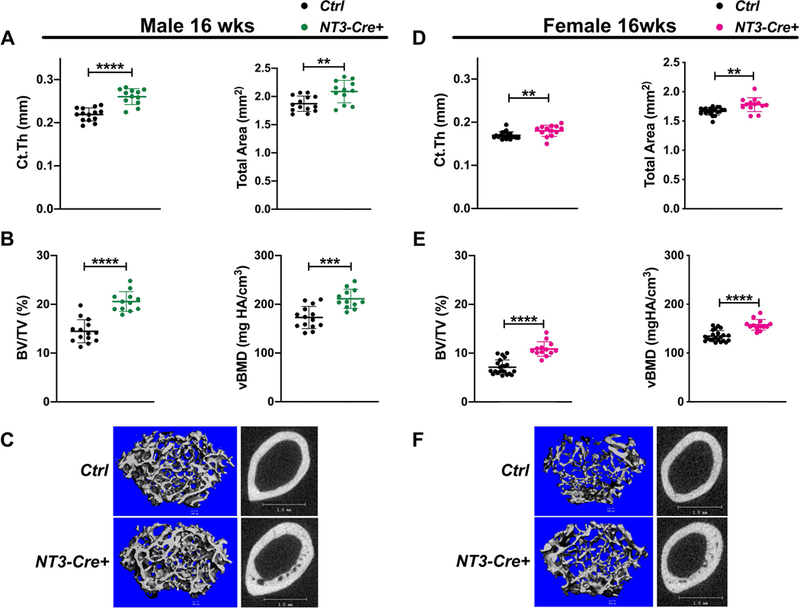
(A) MicroCT analysis (ex-vivo) of the femur in male mice at 16 wks showing increased cortical thickness and cortical total area in NT3 transgenic mice. n=12–14 per group; black circles = male Ctrl and green circles = male NT3-Cre+. (B) Cancellous BV/TV and vBMD are higher in male NT3-Cre+ animals. (C) Representative images for cancellous (left) and cortical (right) femoral bone indicating heightened bone mass in male NT3-Cre+ mice. (D) MicroCT analysis (ex-vivo) of the femur in female mice at 16 wks also shows increased cortical thickness and cortical total area in NT3 transgenic mice. n=13–22 per group; black circles = female Ctrl and pink circles = female NT3-Cre+. (E) Cancellous BV/TV and vBMD are also elevated in female NT3-Cre+ animals. (F) Representative images for cancellous (left) and cortical (right) femoral bone illustrating greater bone mass in female NT3-Cre+ mice. Data are represented as mean ± SD. Unpaired, student’s two-tail t-test: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Ct.Th = cortical thickness; BV/TV = bone volume/total volume; vBMD = volumetric bone mineral density
To monitor the development of the bone phenotype over time, in vivo microCT was used to assess mice at 6, 12, and 16 weeks encompassing both rapid growth and peak bone mass stages. The tibiae of 6-week-old, NT3-Cre+ animals had similar cortical thickness compared to Ctrl, but were larger in total area (Fig S4A). By 12 weeks of age, both cortical thickness and total area were increased at the mid-diaphysis (Fig S4B). At the time of peak bone mass (16 weeks), cortical thickness and total area remained increased in NT3-Cre+ mice relative to controls (Fig S4C). In the cancellous compartment, BV/TV was similar in young NT3 transgenic animals (Fig S4D). BV/TV and vBMD, while trending upwards, did not reach statistical significance in these 12- or 16-week-old cohorts (Fig S4E-F). However, both tibial BV and TV were greater at these ages (Fig S4G). Considering the entirety of the microCT data, we conclude that constitutive activation of alternative NF-κB in the osteolineage leads to an increase in basal bone mass.
Both osteoblast and osteoclast activity are upregulated in NT3-Cre+ mice
Consistent with unchanged bone mass, serum P1NP, a marker of bone formation, was indistinguishable at 6 weeks (Fig S5A), although dynamic histomorphometry already revealed an increase in bone formation, with 2-fold greater BFR/BS than that seen in Ctrl sections at the endocortical surface (Fig 2A-B). MS/BS and MAR were 1.5-fold and 1.3-fold higher, respectively (Fig 2C-D). Neither Ob.N/BS nor Ob.S/BS were changed in the cancellous compartment (Fig S5B-C; cancellous BFR could not be analyzed due to extensive calcified cartilage and a lack of distinct double-labels at this young age. At 12 weeks, serum P1NP was markedly increased (+38%) in NT3-Cre+ compared to Ctrl littermates (Fig 2E). Serum P1NP and cancellous BFR/BS were similar between genotypes at 16 weeks (Fig S6A-B); however, sLS, dLS, MS, and BS were all elevated in NT3-Cre+ mice (Fig S6C). Thus, although not all indices of bone formation are significantly increased in NT3-Cre+ cohorts, all at least trend in this direction.
Figure 2: Both osteoblast and osteoclast activity are upregulated in NT3-Cre+ mice.
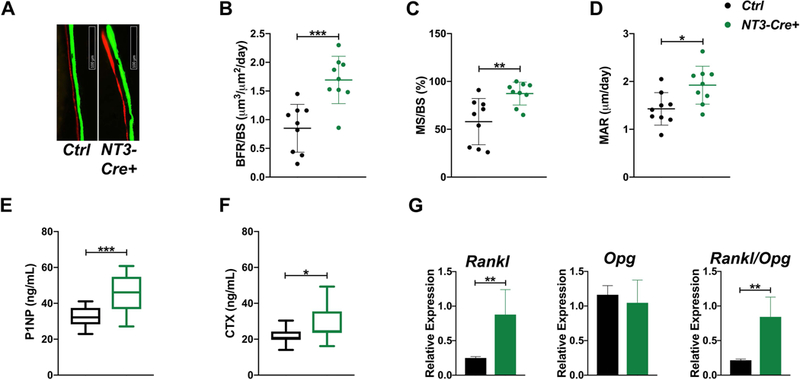
(A) Representative images of calcein and alizarin 5-day double-labeling for each genotype at 6 wks. (B) Dynamic histomorphometry at the tibial endocortical surface shows increased BFR/BS, (C) MS/BS, and (D) MAR in 6-wk-old NT3 transgenic animals (n=9–10 per group). Serum P1NP (E) and CTX (F) levels are markedly elevated in 12-wk-old NT3-Cre+ mice (n=12–15 per group). (G) qPCR showing increases in Rankl expression and the Rankl/Opg ratio in flushed, crushed bone of 12-wk-old NT3-Cre+ mice (n=4–7 per group). Results are presented as mean ± SD. black = male Ctrl and green = male NT3-Cre+. Unpaired, student’s two-tail t-test: *p<0.05, **p<0.01, ***p<0.001. BFR/BS = bone formation rate per bone surface; MS/BS = mineralizing surface per bone surface; MAR = mineral apposition rate; OPG (Tnfrsf11b); RANK (Tnfrsf11a); RANKL (Tnfsf11).
In order to determine if decreases in bone resorption also contributed to the observed increases in bone mass, we measured serum CTX. In NT3-Cre+ mice, CTX was similar to controls at 6 weeks (Fig S5D), and histomorphometry showed no change in OC indices (Fig S5E-F). By 12 weeks of age, NT3 transgenic animals displayed elevated serum CTX (Fig 2F) as well as greater than 2-fold increases in both Rankl expression and the Rankl/Opg ratio in crushed bone (Fig 2G). Heightened circulating CTX levels were also seen in 16-week-old NT3-Cre+ mice (Fig S6D). Therefore, activation of NIK in the osteolineage leads to increased bone formation and high bone mass, in the face of significant activation of osteoclastic bone resorption.
NT3-Cre+ mice have an enhanced anabolic response to mechanical loading
To determine the role of NT3 in bone formation in a condition that is not normally coupled to OC activation, we employed an adaptive mechanical loading model which stimulates periosteal bone formation in the mid-diaphysis without clear signs of resorption (49). The right tibiae of 16 week Ctrl and NT3-Cre+ mice were subjected to the loading protocol outlined in Fig 3A. The left tibiae served as non-loaded (NL) contralateral controls. Coincident with the increase in cortical bone size and thickness seen by microCT, a higher absolute force was needed to achieve the same resultant microstrain in NT3-Cre+ tibiae compared to Ctrl (10.0N vs 8.7N). Mechanical loading induced robust double-labeling in both genotypes at the peak compressive and tensile surfaces (Fig 3B). Consistent with the observed increase in basal bone mass, NT3-Cre+ mice showed 2-fold greater increases in MS/BS and Ps.BFR/BS following loading compared to Ctrl mice (Fig 3C, E). The change in Ps.MAR trended upwards, but did not reach statistical significance (Fig 3D). These results indicate that constitutive activation of alternative NF-κB using Osx-Cre promotes bone formation in response to loading, either via direct effects on OBs or by enhancing mechanosensitivity of osteocytes, or a combination of both mechanisms.
Figure 3: NT3-Cre+ mice have an enhanced anabolic response to mechanical loading.
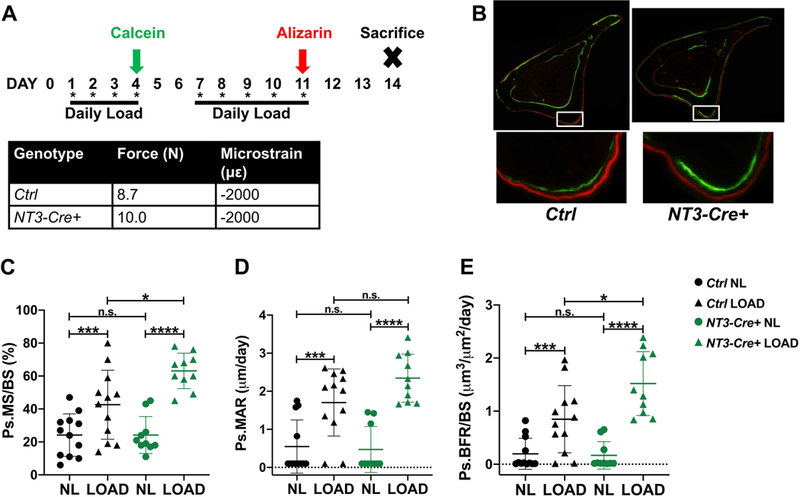
(A) Study design and adaptive mechanical loading protocol for 16-week-old Ctrl and NT3-Cre+ mice. One bout of cyclic loading per day was performed a total of 9 times. Calcein green and alizarin red fluorochromes were given on study days 4 and 11, respectively. Strain gauging was used to determine the force required to achieve an equivalent microstrain in each genotype, as shown. (B) (top) Representative cross-section of the loaded tibia for each genotype and (bottom) inset view of the area of peak compressive strain highlighting the enhanced bone formation seen in NT3-Cre+ mice. Quantification of fluorochrome labeling shown as (C) Ps.MS/BS, (D) Ps.MAR, and (E) Ps.BFR/BS (n=10–12 per group). Results are presented as mean ± SD. black = male Ctrl and green = male NT3-Cre+. Right tibiae were loaded (LOAD, triangles) and left tibiae served as non-loaded (NL, circles) controls. Student’s, paired, two-tailed t-test (LOAD vs NL within each genotype) or 2-way ANOVA followed by Tukey’s multiple comparison test (cross-genotype comparisons): n.s. - not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Ps.BFR/BS = periosteal bone formation rate per bone surface; Ps.MS/BS = periosteal mineralizing surface per bone surface; Ps.MAR = periosteal mineral apposition rate
RNAseq analysis reveals two main transcriptional programs: one loading-responsive and the other driven by the NT3 transgene
To capture the gene program responsible for initiation and production of new bone in response to mechanical loading seen with constitutive activation of NIK, we harvested RNA from a separate cohort of animals loaded for 5 days(50). Four hours after the last loading bout, tibiae were flushed, and cortical bone in the area of peak compressive strain was isolated and processed for RNA-Seq analysis (Fig 4A). The same region in the contralateral limb served as the NL control in all analyses. A heatmap showing changes in gene expression across all samples is shown in Fig 4B. Hierarchical clustering reveals a unique relationship dependent on genotype as Ctrl and NT3-Cre+ samples segregate regardless of loading. Furthermore, there are 4 distinct patterns of interest, (numbered blocks 1–4 in Fig 4B). Block 1 genes are vastly upregulated with loading in Ctrl, but more modestly upregulated and with more sample variability in NT3-Cre+ mice. Genes in blocks 2a and 2b are potently downregulated with loading in Ctrl, and have minimal changes in transgenic animals. The expression patterns in blocks 3 and 4 are mostly unchanged with loading in both genotypes. However, in NT3-Cre+ samples, block 3 genes have higher expression and block 4 genes have lower expression compared to control mice. Thus, two main transcriptional programs predominate, one loading-responsive (blocks 1 and 2) and the other NT3 transgene- driven (blocks 3 and 4).
Figure 4: RNAseq analysis reveals two main transcriptional programs: one loading-responsive and the other driven by the NT3 transgene.
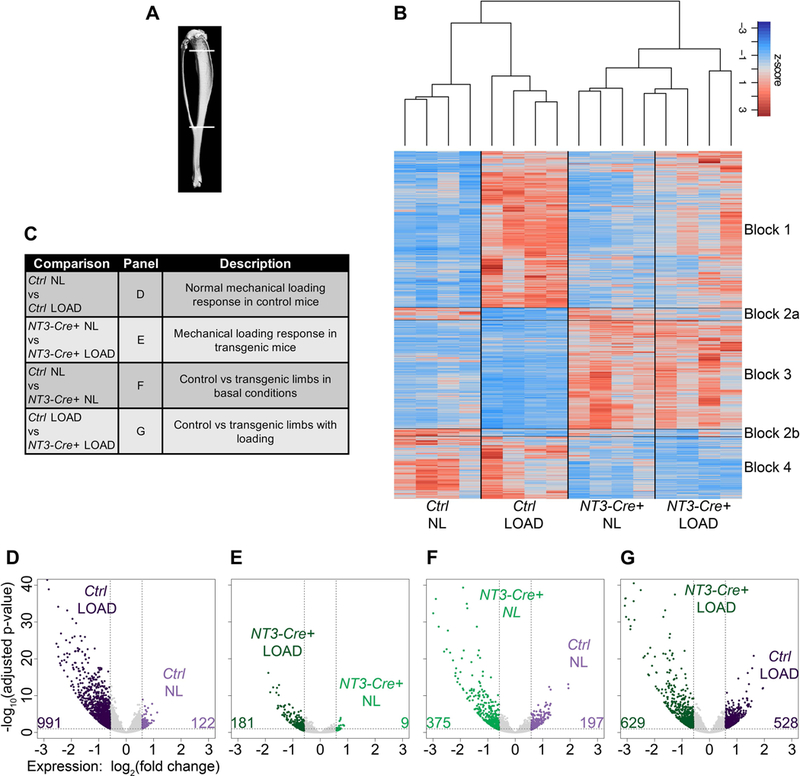
(A) Schematic representing the tibial region of interest harvested for RNA-Seq analysis (top line: 2 mm distal to the articular surface and bottom line: distal tibia-fibula junction). (B) Heatmap displaying z-scores across all samples highlighting two main transcriptional programs: one induced by loading (blocks 1–2) and the other by the NT3 transgene (blocks 3–4). (C) Table listing RNAseq analysis comparisons and description of resulting differentially expressed genes (DEGs). (D-G) Volcano plots for each of the comparisons in (C) showing number of upregulated DEGs in each pairwise comparison. DEGs were defined as having a fold-change > 1.5 together with an adjusted p-value ≤ 0.05. n=4 per genotype/loading condition. light purple = Ctrl NL; dark purple = Ctrl LOAD; light green = NT3-Cre+ NL; dark green = NT3-Cre+ LOAD
Four main comparisons were made to parse out differentially expressed genes (DEGs) associated with each experimental perturbation (Fig 4C). A select set of low and high expressing genes (per RPKM) was validated by qPCR and showed the expected relationships between groups (Fig S7). Volcano plots for each comparison are shown in Fig 4D-G. Mechanical stimulus elicits an activating genomic program more than an inhibitory one in both Ctrl and NT3-Cre+ mice. There are far fewer DEGs in the NT3-Cre+ genotype with loading (Fig 4E). In addition, both the NL and loaded limbs of NT3-Cre+ animals show a greater number of upregulated genes that are more statistically significant compared to Ctrl NL and loaded sides, respectively (Fig 4F-G). Based on the global expression patterns depicted in the heat map and volcano plots, we conclude that constitutive activation of alternative NF-κB in NT3-Cre+ mice drives a primarily activating differential gene program to enhance bone anabolism both at baseline and in response to mechanical loading.
The NT3 transgene modulates expression of known bone anabolic factors
To characterize the transcriptional effect of the NT3 transgene in an unbiased manner, gene ontology (GO) analysis for molecular function was performed across all 4 experimental groups. For each condition, instead of analyzing transcript levels, we used a multi-way partitioning algorithm (44) to identify patterns of GO molecular-function terms specific to genotype, loading and their combinations. The number of unique GO IDs for each comparison is depicted in Fig 5A. We examined the top GO IDs (p-value <10−3) in the NL, NL vs LOAD overlap, and LOAD bins for NT3-Cre+ samples to determine which molecular functions were driven by the NT3 transgene (Fig 5B-D). GO terms involved at baseline (NL limbs) and regardless of loading (NL vs LOAD overlap) pointed to an enrichment of receptor, kinase, and growth factor activities. GO terms unique to both the NT3 transgene and loading were enriched for a calcium signature, likely reflecting enhanced mechanosensation, which is known to rely on calcium flux (4).
Figure 5: Gene ontology analysis reveals a bone anabolic signature in NT3-Cre+ mice.
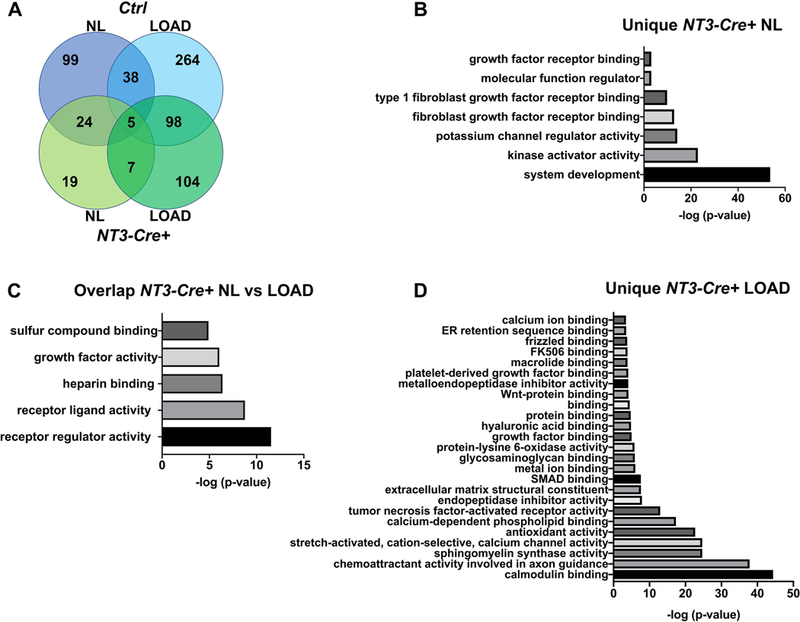
(A) Venn diagram for the gene ontology (GO) analysis of molecular function across all 4 experimental groups using a multi-way partitioning algorithm. The number of unique GO terms is listed for each bin. The top GO terms (p<10−3) in NT3-Cre+ mice unique to the (B) NL side or (C) shared between loading conditions is enriched for receptor, kinase, and growth factor activities. (D) Top GO terms unique to NT3-Cre+ loaded tibiae indicate involvement of calcium signaling. NL = non-loaded tibia; LOAD = loaded tibia
To further compare the transcriptional response to loading across genotypes, we evaluated the DEG lists from NL vs loaded tibiae in Ctrl and NT3-Cre+ mice and generated a Venn diagram to depict their relationships (Fig 6A). Although ~10-fold fewer genes were differentially regulated by loading in NT3 transgenic mice, 94% of DEGs were shared with the controls. We next, focused on the top 20 up- and downregulated genes for each comparison (Tables S1–4). Hapln4, Ptgs2 (encoding Cox2), and Wnt7b were upregulated in both genotypes in response to loading, consistent with previous reports for in vivo mechanical loading in wild type mice (9,51–54). Cxcl13 is the top upregulated gene in NT3-Cre+ bones regardless of loading. Previous work has documented Cxcl13 in the early response to mechanical loading (4h after a single loading session), and this chemokine can induce OB activity in vitro (50,55). Furthermore, expression of Cxcl13 is dependent on alternative NF-κB, requiring signaling through both NIK and IKKα (56,57). The top downregulated genes in response to loading in both genotypes include Fgf23 and the TGF-β inhibitor, Grem1, both of which have been previously shown to be reduced during the matrix formation phase after loading (50). The extensive overlap between genotypes in response to loading and preponderance of DEGs with known roles in bone formation suggest that NT3-Cre+ mice modulate the normal bone anabolic program to a greater degree than controls.
Because other studies have identified a strong Wnt signature with loading, we investigated the expression levels of Wnt ligands and inhibitors (54,58) in all groups relative to Con:NT3 NL limbs (Fig 6B). NT3 transgene-responsive Wnt ligands include Wnt5b (downregulated) and Wnts 7b, 10a and 11 (upregulated). Wnt 1 and Wnt7b were induced both with loading and by the NT3 transgene. Sost was downregulated in both the NL and loaded NT3-Cre+ limbs compared to Ctrl NL tibiae, while Dkk1 was significantly downregulated only with loading. Although others have described suppression of Sost with loading in wild type mice (1,5–7), its expression trended downwards in Ctrl loaded samples, although it did not reach statistical significance at the timepoint we evaluated. Taking all these factors together, the Wnt-mediated response to loading appears to be enhanced in NT3-Cre+ limbs.
We also examined other candidate genes previously described as loading-responsive (Fig 6C). Ptgs2 (encoding Cox2), which is required for a maximal anabolic loading response (9,51), was increased by both the NT3 transgene and loading. Postn (59) was upregulated only in loaded controls, while Gja1 (60) increased with loading in both genotypes, suggesting that neither is regulated by the NT3 transgene. As expected based on previous studies (61), Tnfrsf11b (OPG) was potently upregulated in Ctrl in response to loading, but it did not reach statistical significance in the NT3 transgenic setting. Confirming that our adaptive loading protocol was not coupled to resorption, Ctrl limbs did not show changes in Tnfrsf11a (RANK) or Tnfsf11 (RANKL). However, NT3-Cre+ tibiae showed increased expression of both Tnfrsf11a (RANK) and Tnfsf11 (RANKL) regardless of loading, consistent with basal phenotyping data.
Alternative NF-κB orchestrates a bone anabolic program
Map3k14 (NIK), Nfkb2 (p100), and RelB transcripts were all significantly upregulated in NT3-Cre+ compared to Ctrl NL limbs, regardless of loading, indicating robust expression of the NT3 transgene and downstream alternative NF-κB pathway components. Expression levels of core NF-κB pathway factors, except for c-Rel, were not changed in response to loading in control mice (Fig 7A).
To predict if NT3 transgene-responsive target genes were directly NF-κB regulated, we searched for NF-κB consensus motifs within 5 kb upstream of each transcription start site since previous studies have shown that the majority of κB binding sites are found in this region (62). The NF-κB consensus binding motif is degenerate, so we used 3 variants with different stringencies (45–47). First, to determine the background frequency in the genome of each 10-mer sequence, we queried sets of 121 randomly selected genes for 10 million iterations to find how often at least one gene had 2 or more κB recognition elements (κB-RE) (Fig 7B). As expected, only a very small percentage of these sets, which are the size of the largest group analyzed below, have even one such gene. The first motif [GGRRNNYYCC] is the most permissive and is found more frequently, while the second [GGGRNNYYCC] and third [GGGRNWYYCC] motifs are increasingly more stringent and somewhat less prevalent in the genome.
Given that the NT3 transgene activates NF-κB signaling, we expected our analysis would show enrichment in the number of κB-RE in NT3-Cre+ samples. In pairwise comparisons between Ctrl and NT3-Cre+ limbs, either with or without loading, the top 20 DEGs show a strong enrichment for genes containing at least 2 κB-RE, compared to the background genome frequencies for each motif (Fig 7B). Likewise, examination of the genes in the top GO terms for the NT3-Cre+ bins (Unique NT3-Cre+ NL, Overlap NT3-Cre+ NL vs LOAD, and Unique NT3-Cre+ LOAD, Fig 5B-D) also yields a very high prevalence of targets with multiple κB-RE. Of particular interest are the 10 genes contained within the top GO term in the overlap between NT3-Cre+ NL and LOAD bins (“receptor regulator activity”) since these target genes may represent a common mechanism by which activation of NF-κB increases both basal and stimulated bone anabolism. Over half of these genes (Bmp2, Cxcl13, Gal, Inhba, Ngf, Npy, Ptn, Timp1, Wnt1, Wnt7b) contain multiple κB-RE and have been implicated in skeletal biology, mostly with bone anabolic functions (52,54,63,64).
Whether NF-κB contributes to the normal loading response has not previously been investigated. Consistent with the high degree of overlap in the entire loading response (Fig 6A), we found that over half the genes in the top 20 DEGs in response to loading were enriched for κB-RE, not only in NT3-Cre+, but also in Ctrl animals (Fig 7B). Further, 75% of these DEGs were shared between genotypes, and 8 out of 15 had ≥ 2 κB-RE (GGRRNNYYCC). The 5 DEGs with ≥2 kB-RE that were not found in common in the top 20 lists were nevertheless upregulated by loading in the other genotype (by at least 2-fold). These results suggest that the normal loading response may include NF-κB activation. Indeed, extension of our analysis to the top 50 DEGs in Ctrl mice (+/− loading) also revealed a very high incidence of κB-RE (Fig 7C). Both Ptgs2 and Wnt7b, which have been identified as upregulated genes in other mechanical loading studies (9,51–54), have multiple κB-RE across all motifs tested (Fig 7D). The prevalence of κB-RE in genes regulated by loading, including those known to be anabolic, provides evidence that NF-κB is activated by biophysical stimuli to promote bone formation.
Discussion
To date, previous reports have positioned both classical and alternative NF-κB signaling pathways as solely anti-bone anabolic in vivo. Conditional inhibition of classical NF-κB in the osteolineage using a IKKγDN allele (14) or global deficiency in alternative NF-κB (aly/aly or RelB) (16,18,20) led to transient increases in bone formation in young mice. Basal bone mass returned to baseline over time with inhibition of classical NF-κB, but the elevation persisted up to 14 weeks in RelB KO mice, although the phenotype was constrained to diaphyseal bone. Both classical and alternative NF-κB loss of function mutants were shown to negatively regulate bone anabolism following ovariectomy (14), ectopic bone formation (17), or a cortical defect model (18). Notably, overexpression of c-Rel or Relb was not found to inhibit OB differentiation in vitro (14). In a complementary study, constitutive activation of classical NF-κB using a IKK2ca allele resulted in a low bone mass phenotype with reduced bone formation rates at 4 weeks (15). In contrast to these previous reports, we find that constitutive activation of alternative NF-κB in the osteolineage using Osx-Cre can have positive effects on bone. In both male and female NT3-Cre+ mice, indices of cortical and cancellous bone mass are increased. Although not all metrics of bone formation are increased in all cases, the totality of the data suggests that the NT3 transgene drives bone anabolism similarly in both sexes.
The increased bone mass in NT3 transgenic mice is accompanied by upregulation of bone resorption as well. Serum CTX and the Rankl:Opg ratio from crushed bone are both higher in NT3-Cre+ animals compared to Ctrl littermates. This is analogous to our previous observation that conditional activation of NIK in the osteoclast lineage leads to osteoclast activation and low bone mass, despite increased bone formation rates (65). Thus, whether expressed exclusively in OB or OC lineages, the NT3 transgene seems to enhance, or at least maintain, coupling between the two cell types. Inhibition of classical NF-κB using either Bglap2-Cre or Col1α1-Cre drivers did not alter the OC phenotype at 2– 8 weeks of age or following ovariectomy (14), but constitutive activation of classical NF-κB using Col2-Cre increased the number of OCs (15). Under systemic inflammatory conditions with NF-κB activation, bone formation is typically suppressed in the face of enhanced resorption, while in other contexts such as osteophyte formation in osteoarthritis (66) or formation of a sequestrum in osteomyelitis (67), new bone is formed despite local inflammation. Further work is required to determine if NF-κB, and perhaps particularly its alternative arm, plays a role in these settings.
To examine bone formation independent of coupling to bone resorption, we turned to an adaptive mechanical loading model and found that NT3-Cre+ animals formed about twice as much periosteal bone as Ctrl littermates. RNA-Seq on both NL and loaded tibiae revealed two major genomic programs, one that was loading-responsive and the other driven by the NT3 transgene. Interestingly, despite the greater accrual of bone in NT3 transgenic mice, the controls had almost 10-fold more DEGs. Nevertheless, the DEGs in the NT3 transgenic loading response were highly overlapping with those in controls. Indeed, the top 20 DEG lists had 75% identity between genotypes. GO analysis of molecular function in NT3-Cre+ NL and loaded limbs revealed a distinct upregulation of receptor, kinase, and growth factor activities, as well as a calcium-response signature. Taken together, our gene expression and pathway analyses support the conclusion that activation of NIK has bone anabolic effects at baseline and with loading.
To ascertain if the bone anabolic effects due to the NT3 transgene and/or in response to loading were part of a direct κB-responsive program, we next searched both the DEG lists and GO bins for multiple κB consensus motifs. As expected, in both basal and loading conditions, DEGs from NT3-Cre+ limbs displayed a high prevalence of κB-RE. Quite surprisingly, we also observed a similar incidence of κB-RE in the top 20 DEGs in Ctrl mice in response to loading. Further analysis encompassing the top 50 DEGs in Ctrl limbs still indicated a strong enrichment of κB-RE. Although functional κB binding sites are not restricted to the 3 motifs used in this study and can also be found more remotely (further upstream, within introns, etc), comparison of RNA-Seq and ChIP-Seq data suggests that most NF-κB-regulated genes have multiple κB sites in this classical promoter region (5kB upstream of the transcription start site) {Wong:2011gi, (62). Thus, our data strongly suggests a previously unrecognized physiologic contribution of NF-κB to the anabolic loading response.
The Wnt signaling pathway is a major driver of new bone formation (68), and we find evidence of Wnt enhancements in our dataset, driven by both the NT3 transgene and loading. Wnt7b is upregulated by loading as well as by the NT3 transgene, and has multiple κB-RE, while Wnt1 is upregulated by loading in both genotypes and has a single κB-RE. Expression of Sost, an inhibitor of Wnt signaling, is well-documented to decrease in response to mechanical loading (1,5,6). In our study, Sost expression was trending downward but did not reach statistical significance in control animals. However, Sost does contain one κB-RE, and we observed strong downregulation of Sost in NT3-Cre+ limbs, further supporting Wnt activation by the NT3 transgene. Regulation of transcription by NF-κB is complex, and DNA binding by NF-κB dimers is but one of many factors, including binding of other transcription factors and epigenetic modifications, that determine activation of specific targets (69). Nevertheless, the presence of κB-RE in genes known to play a role in bone formation is strongly suggestive of a role for NF-κB in this process.
One limitation of our study is that we have not used any approaches such as ChIP-Seq to demonstrate NF-κB binding to specific genes in the bone anabolic response. Unfortunately, there are very significant technical hurdles to performing ChIP-Seq in whole bone samples as we did for the RNA-Seq , simply due to the challenges of working with a matrix-rich mineralized tissue. Additionally, we have not established which NF-κB subunits are involved, or what timepoints after a loading bout(s) would be most informative. Mouse models lacking specific NF-κB components could be examined with mechanical loading in future work, although redundancy between subunits could complicate this approach. In vitro models of fluid shear stress have been used in an attempt to recapitulate the in vivo mechanosensing environment (70,71). Unfortunately, due to cellular differences, nature of fluid flow, and nuances in perfusion chamber mechanics, results have been conflicting on whether fluid shear stress activates or inhibits NF-κB signaling (23,72–74). Therefore, it is unlikely that an in vitro approach could be used to demonstrate a definitive role for NF-κB in mechanical loading.
Another limitation is that our study utilized the doxycycline repressible Osx-Cre line to drive activation of the NT3 transgene (29). On its own, this particular Cre allele is known to proportionally decrease body size and bone mass in young mice, resolving by 8–12 weeks (31–33). To reduce these effects, we fed mice a 200ppm doxycycline chow until weaning, observing milder decreases in body weight and bone mass in 6 week Osx-Cre mice, compared to previous reports without doxycycline feeding. Importantly, our NT3-Cre+ mice had similar body weights to their Cre-negative littermate controls at this age. Since the Cre-driven phenotype is low bone mass proportional to body weight prior to 8–12 weeks of age, it is highly unlikely that the increased bone mass in NT3 transgenic mice at 12 and 16 weeks is caused by the Cre.
In conclusion, our results provide compelling evidence that activation of NF-κB in the osteolineage can modulate both basal and mechanically-stimulated bone formation by fine-tuning the expression of a broad array of bone anabolic factors. However, because we expressed the NT3 transgene early in osteoblast differentiation using Osx-Cre, we have not yet determined if this pathway drives changes in OBs themselves, enhances mechanosensitivity of osteocytes, or both. Future studies using an osteocyte-specific Cre such as Sost-Cre (75) may help elucidate cell autonomous effects of NF-κB in these cell types.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants R01 AR052705 and R01 AR070030 (to DJV), F31 AR068853 (to JLD), and R01AR064821 and R01AR068438 (to RA); the Molecular Oncology Training Program at Washington University (T32CA113275) and by the Shriners Hospitals for Children grant 85117 (to DJV). Microcomputed tomography and histomorphometry were supported by the Musculoskeletal Research Center (P30 AR057235) and image analysis by the Alafi Neuroimaging Center (S10 RR027552) at Washington University. Partial support for bioinformatics was provided by the Washington University Center of Regenerative Medicine. We thank the Civitelli, Faccio, Long, and Silva labs for technical assistance, Crystal Idleburg and Samantha Coleman for histology, Daniel Leib and Gary London for imaging support, as well as Dr. Gabriel Mbalaviele and Anna Ballard for critical reading of the manuscript.
Footnotes
Disclosures
All authors state that they have no conflicts of interest.
References
- 1.Galea GL, Lanyon LE, Price JS. Sclerostin“s role in bone”s adaptive response to mechanical loading. Bone. The Authors; 2017. March 1;96(C):38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long F Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2012. January;13(1):27–38. [DOI] [PubMed] [Google Scholar]
- 3.Bradley EW, Westendorf JJ, van Wijnen AJ, Dudakovic A. 4 Osteoblasts: Function, Development, and Regulation. ASBMR primer. Ninth. 2018. September 19;:31–7. [Google Scholar]
- 4.Robling AG, Turner CH. Mechanical signaling for bone modeling and remodeling. Crit Rev Eukaryot Gene Expr. 2009;19(4):319–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robling AG, Niziolek PJ, Baldridge LA, Condon KW, Allen MR, Alam I, et al. Mechanical stimulation of bone in vivo reduces osteocyte expression of Sost/sclerostin. J Biol Chem. American Society for Biochemistry and Molecular Biology; 2008. February 29;283(9):5866–75. [DOI] [PubMed] [Google Scholar]
- 6.Tu X, Rhee Y, Condon KW, Bivi N, Allen MR, Dwyer D, et al. Sost downregulation and local Wnt signaling are required for the osteogenic response to mechanical loading. Bone. Elsevier B.V; 2012. January 1;50(1):209–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robling AG, Kang KS, Bullock WA, Foster WH, Murugesh D, Loots GG, et al. Sost, independent of the non-coding enhancer ECR5, is required for bone mechanoadaptation. Bone. Elsevier Inc; 2016. November 1;92(C):180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morse A, McDonald MM, Kelly NH, Melville KM, Schindeler A, Kramer I, et al. Mechanical Load Increases in Bone Formation via a Sclerostin-Independent Pathway. Journal of Bone and Mineral Research. Wiley-Blackwell; 2014. October 20;29(11):2456–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forwood MR. Inducible cyclo-oxygenase (COX-2) mediates the induction of bone formation by mechanical loading in vivo. J Bone Miner Res. Wiley-Blackwell; 1996. November;11(11):1688–93. [DOI] [PubMed] [Google Scholar]
- 10.Thompson WR, Rubin CT, Rubin J. Mechanical regulation of signaling pathways in bone. Gene. Elsevier B.V; 2012. July 25;503(2):179–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sebastian A, Loots GG. Transcriptional control of Sost in bone. Bone. The Authors; 2017. March 1;96(C):76–84. [DOI] [PubMed] [Google Scholar]
- 12.Novack DV. Role of NF-κB in the skeleton. Cell Res. Nature Publishing Group; 2010. November 16;21(1):169–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Begalli Federica, Bennett Jason, Capece Daria, Verzella Daniela, Daniel D’Andrea Laura Tornatore, et al. Unlocking the NF-κB Conundrum: Embracing Complexity to Achieve Specificity. Biomedicines. 2017. December;5(4):50–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang J, Wang Z, Tang E, Fan Z, McCauley L, Franceschi R, et al. Inhibition of osteoblastic bone formation by nuclear factor-κB. Nature Medicine. 2009. May 17;15(6):682–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swarnkar G, Zhang K, Mbalaviele G, Long F, Abu-Amer Y. Constitutive Activation of IKK2/NF-κB Impairs Osteogenesis and Skeletal Development. Awad HA, editor. PLoS ONE. 2014. March 11;9(3):e91421–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maruyama T, Fukushima H, Nakao K, Shin M, Yasuda H, Weih F, et al. Processing of the NF-kappa B2 precursor p100 to p52 is critical for RANKL-induced osteoclast differentiation. Journal of Bone and Mineral Research. 2010. May;25(5):1058–67. [DOI] [PubMed] [Google Scholar]
- 17.Seo Y, Fukushima H, Maruyama T, Kuroishi KN, Osawa K, Nagano K, et al. Accumulation of p100, a precursor of NF-κB2, enhances osteoblastic differentiation in vitro and bone formation in vivo in aly/aly mice. Mol Endocrinol. 2012. March;26(3):414–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao Z, Li Y, Yin X, Dong Y, Xing L, Boyce BF. NF-κB RelB Negatively Regulates Osteoblast Differentiation and Bone Formation. Journal of Bone and Mineral Research. Wiley-Blackwell; 2014. March 19;29(4):866–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soysa NS, Alles N, Weih D, Lovas A, Mian AH, Shimokawa H, et al. The pivotal role of the alternative NF-kappaB pathway in maintenance of basal bone homeostasis and osteoclastogenesis. Journal of Bone and Mineral Research. 2010. April;25(4):809–18. [DOI] [PubMed] [Google Scholar]
- 20.Zarei A, Yang C, Gibbs J, Davis JL, Ballard A, Zeng R, et al. Manipulation of the Alternative NF-κB Pathway in Mice Has Sexually Dimorphic Effects on Bone. JBMR Plus. John Wiley & Sons, Ltd; 2018. August 23;3(1):14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamazaki M, Fukushima H, Shin M, Katagiri T, Doi T, Takahashi T, et al. Tumor Necrosis Factor Represses Bone Morphogenetic Protein (BMP) Signaling by Interfering with the DNA Binding of Smads through the Activation of NF- B. Journal of Biological Chemistry. 2009. December 11;284(51):35987–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirata-Tsuchiya S, Fukushima H, Katagiri T, Ohte S, Shin M, Nagano K, et al. Inhibition of BMP2-Induced Bone Formation by the p65 Subunit of NF-κB via an Interaction With Smad4. Mol Endocrinol. 2014. September;28(9):1460–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young SRL, Gerard-O’Riley R, Harrington M, Pavalko FM. Activation of NF-κB by fluid shear stress, but not TNF-α, requires focal adhesion kinase in osteoblasts. Bone. Elsevier Inc; 2010. July 1;47(1):74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan Y-X, Gong Y- W, Guo Y, Lv Q, Guo C, Zhuang Y, et al. Mechanical Strain Regulates Osteoblast Proliferation through Integrin-Mediated ERK Activation. Pintus G, editor. PLoS ONE. 2012. Apr 23;7(4):e35709–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang J, Zhao L-G, Teng Y- J, Chen S- L, An L- P, Ma J- L, et al. ERK5 signalling pathway is essential for fluid shear stress-induced COX-2 gene expression in MC3T3-E1 osteoblast. Molecular and Cellular Biochemistry. Springer US; 2015. May 14;406(1):237–43. [DOI] [PubMed] [Google Scholar]
- 26.Chang J, Liu F, Lee M, Wu B, Ting K, Zara JN, et al. NF-κB inhibits osteogenic differentiation of mesenchymal stem cells by promoting β-catenin degradation. Proc Natl Acad Sci USA. 2013. June 4;110(23):9469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarapore RS, Lim J, Tian C, Pacios S, Xiao W, Reid D, et al. NF-κB Has a Direct Role in Inhibiting Bmp- and Wnt-Induced Matrix Protein Expression. Journal of Bone and Mineral Research. 2015. August 6;:n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sui Y, Liu Z, Park S-H, Thatcher SE, Zhu B, Fernandez JP, et al. IKKβ is a β-catenin kinase that regulates mesenchymal stem cell differentiation. JCI Insight. 2018. January 25;3(2):341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodda SJ. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006. August 15;133(16):3231–44. [DOI] [PubMed] [Google Scholar]
- 30.Sasaki Y, Calado DP, Derudder E, Zhang B, Shimizu Y, Mackay F, et al. NIK overexpression amplifies, whereas ablation of its TRAF3-binding domain replaces BAFF:BAFF-R-mediated survival signals in B cells. Proc Natl Acad Sci USA. National Acad Sciences; 2008. August 5;105(31):10883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berman SD, Calo E, Landman AS, Danielian PS, Miller ES, West JC, et al. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci USA. National Academy of Sciences; 2008. August 19;105(33):11851–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davey RA, Clarke MV, Sastra S, Skinner JP, Chiang C, Anderson PH, et al. Decreased body weight in young Osterix-Cre transgenic mice results in delayed cortical bone expansion and accrual. Transgenic Res. Springer Netherlands; 2011. December 13;21(4):885–93. [DOI] [PubMed] [Google Scholar]
- 33.Huang W, Olsen BR. Skeletal defects in Osterix-Cre transgenic mice. Transgenic Res. 2014. August 20;24(1):167–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Müller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. Journal of Bone and Mineral Research. 2010. June 7;25(7):1468–86. [DOI] [PubMed] [Google Scholar]
- 35.Scott JPR, Sale C, Greeves JP, Casey A, Dutton J, Fraser WD. Effect of fasting versus feeding on the bone metabolic response to running. Bone. Elsevier Inc; 2012. December 1;51(6):990–9. [DOI] [PubMed] [Google Scholar]
- 36.Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H, et al. Standardized nomenclature, symbols, and units for bone histomorphometry: A 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. Journal of Bone and Mineral Research. 2012. December 18;28(1):2–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel TK, Brodt MD, Silva MJ. Experimental and finite element analysis of strains induced by axial tibial compression in young-adult and old female C57Bl/6 mice. Journal of Biomechanics. Elsevier; 2014. January 22;47(2):451–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holguin N, Brodt MD, Sanchez ME, Silva MJ. Aging diminishes lamellar and woven bone formation induced by tibial compression in adult C57BL/6. Bone. Elsevier Inc; 2014. August 1;65(C):83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stadelmann VA, Hocké J, Verhelle J, Forster V, Merlini F, Terrier A, et al. 3D strain map of axially loaded mouse tibia: a numerical analysis validated by experimental measurements. Computer Methods in Biomechanics and Biomedical Engineering. 2009. February;12(1):95–100. [DOI] [PubMed] [Google Scholar]
- 40.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012. October 25;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013. April 3;41(10):e108–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Research. Cold Spring Harbor Lab; 2012. September;22(9):1760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. BioMed Central; 2014. December 5;15(12):31–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korf RE, Schreiber EL, ISAIM MM, 2014. Optimal Sequential Multi-Way Number Partitioning. International Symposium on Artificial Intelligence and Mathematics. 2014. ed. 2013. [Google Scholar]
- 45.Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kB bound to DNA. Nature. Nature Publishing Group; 1998. January 22;391(6665):410–3. [DOI] [PubMed] [Google Scholar]
- 46.Wang VY-F, Huang W, Asagiri M, Spann N, Hoffmann A, Glass C, et al. The Transcriptional Specificity of NF-kB Dimers Is Coded within the kB DNA Response Elements. CellReports. The Authors; 2012. October 25;2(4):824–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Q, Lenardo MJ, Baltimore D. 30 Years of NF-κB: A Blossoming of Relevance to Human Pathobiology. Cell. Elsevier; 2017. January 12;168(1–2):37–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jepsen KJ, Silva MJ, Vashishth D, Guo XE, van der Meulen MC. Establishing Biomechanical Mechanisms in Mouse Models: Practical Guidelines for Systematically Evaluating Phenotypic Changes in the Diaphyses of Long Bones. Journal of Bone and Mineral Research. Wiley-Blackwell; 2015. May 18;30(6):951–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McBride SH, Silva MJ. Adaptive and injury response of bone to mechanical loading. BoneKEy Reports. Nature Publishing Group; 2012. October 10;1:192–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mantila Roosa SM, Liu Y, Turner CH. Gene expression patterns in bone following mechanical loading. J Bone Miner Res. 2011. January;26(1):100–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lara-Castillo N, Kim-Weroha NA, Kamel MA, Javaheri B, Ellies DL, Krumlauf RE, et al. In vivo mechanical loading rapidly activates β-catenin signaling in osteocytes through a prostaglandin mediated mechanism. Bone. Elsevier Inc; 2015. July 1;76(C):58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelly NH, Schimenti JC, Ross FP, van der Meulen MCH. Transcriptional profiling of cortical versus cancellous bone from mechanically-loaded murine tibiae reveals differential gene expression. Bone. Elsevier Inc; 2016. May 1;86(C):22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holguin N, Brodt MD, Silva MJ. Activation of Wnt Signaling By Mechanical Loading Is Impaired in the Bone of Old Mice. Journal of Bone and Mineral Research. 2016. June. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sebastian A, Loots GG. Chapter 5 - Genomic Profiling in Bone. Second Edition. Genetics of Bone Biology and Skeletal Disease. Elsevier Inc; 2018. 21 p. [Google Scholar]
- 55.Lisignoli G, Toneguzzi S, Piacentini A, Cattini L, Lenti A, Tschon M, et al. Human osteoblasts express functional CXC chemokine receptors 3 and 5: Activation by their ligands, CXCL10 and CXCL13, significantly induces alkaline phosphatase and ?-N-acetylhexosaminidase release. J Cell Physiol. John Wiley & Sons, Ltd; 2002. November 22;194(1):71–9. [DOI] [PubMed] [Google Scholar]
- 56.Dejardin E, Droin NM, Delhase M, Haas E, Cao Y, Makris C, et al. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity. 2002. October;17(4):525–35. [DOI] [PubMed] [Google Scholar]
- 57.Yang J, Zhang S, Zhang L, Xie X, Wang H, Jie Z, et al. Lymphatic endothelial cells regulate B-cell homing to lymph nodes via a NIK-dependent mechanism. Nature Publishing Group. Nature Publishing Group; 2018. March 5;:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kang KS, Robling AG. New Insights into Wnt-Lrp5/6-β-Catenin Signaling in Mechanotransduction. Front Endocrinol (Lausanne). Frontiers; 2014;5(4):246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonnet N, Garnero P, Ferrari S. Periostin action in bone. Molecular and Cellular Endocrinology. 2016. September 5;432:75–82. [DOI] [PubMed] [Google Scholar]
- 60.Stains JP, Civitelli R. Seminars in Cell & Developmental Biology. Seminars in Cell and Developmental Biology. Elsevier Ltd; 2016. February 1;50:31–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kramer I, Halleux C, Keller H, Pegurri M, Gooi JH, Weber PB, et al. Osteocyte Wnt/beta-catenin signaling is required for normal bone homeostasis. Mol Cell Biol. American Society for Microbiology Journals; 2010. June;30(12):3071–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xing Y, Yang Y, Zhou F, Wang J. Characterization of genome-wide binding of NF-κB in TNFα-stimulated HeLa cells. Gene. Elsevier B.V; 2013. September 10;526(2):142–9. [DOI] [PubMed] [Google Scholar]
- 63.Wasserman E, Webster D, Kuhn G, Attar-Namdar M, Müller R, Bab I. Differential load-regulated global gene expression in mouse trabecular osteocytes. Bone. Elsevier Inc; 2013. March 1;53(1):14–23. [DOI] [PubMed] [Google Scholar]
- 64.Tu X, Joeng KS, Nakayama KI, Nakayama K, Rajagopal J, Carroll TJ, et al. Noncanonical Wnt Signaling through G Protein-Linked PKCδ Activation Promotes Bone Formation. Dev Cell. 2007. January;12(1):113–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang C, McCoy K, Davis JL, Schmidt-Supprian M, Sasaki Y, Faccio R, et al. NIK stabilization in osteoclasts results in osteoporosis and enhanced inflammatory osteolysis. PLoS ONE. 2010;5(11):e15383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schett G, Zwerina J, David J-P. The role of Wnt proteins in arthritis. Nat Rev Rheumatol. Nature Publishing Group; 2008. September 1;4(9):473–80. [DOI] [PubMed] [Google Scholar]
- 67.Lew DP, Waldvogel FA. Osteomyelitis. The Lancet. 2004. July;364(9431):369–79. [DOI] [PubMed] [Google Scholar]
- 68.Maupin KA, Droscha CJ, Williams BO. A Comprehensive Overview of Skeletal Phenotypes Associated with Alterations in Wnt/β-catenin Signaling in Humans and Mice. Nature Publishing Group. Sichuan University; 2013. April 14;1(1):27–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cieślik M Genome-wide predictors of NF-κB recruitment and transcriptional activity. BioData Mining. BioData Mining; 2015. November 25;:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fritton SP, Weinbaum S. Fluid and Solute Transport in Bone: Flow-Induced Mechanotransduction. Annu Rev Fluid Mech. 2009. January;41(1):347–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wittkowske C, Reilly GC, Lacroix D, Perrault CM. In Vitro Bone Cell Models: Impact of Fluid Shear Stress on Bone Formation. Front Bioeng Biotechnol. 3rd ed. Frontiers; 2016. November 15;4(Pt 7):87–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kurokouchi K Oscillating Fluid Flow Inhibits TNF-alpha -induced NF-kB Activation via an IkB Kinase Pathway in Osteoblast-like UMR106 Cells. Journal of Biological Chemistry. 2000. November 28;276(16):13499–504. [DOI] [PubMed] [Google Scholar]
- 73.Agarwal S, Long P, Seyedain A, Piesco N, Shree A, Gassner R. A central role for the nuclear factor-kB pathway in anti-inflammatory and proinflammatory actions of mechanical strain. FASEB J. 2003. May;17(8):899–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen NX, Geist DJ, Genetos DC, Pavalko FM, Duncan RL. Fluid shear-induced NFκB translocation in osteoblasts is mediated by intracellular calcium release. Bone. 2003. September;33(3):399–410. [DOI] [PubMed] [Google Scholar]
- 75.Xiong J, Piemontese M, Onal M, Campbell J, Goellner JJ, Dusevich V, et al. Osteocytes, not Osteoblasts or Lining Cells, are the Main Source of the RANKL Required for Osteoclast Formation in Remodeling Bone. Heymann D, editor. PLoS ONE. Public Library of Science; 2015. September 22;10(9):e0138189–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


