Abstract
Colloidal quantum dot (QD) photocatalysts have the electrochemical and optical properties to be highly effective for a range of redox reactions. QDs are proven photo-redox catalysts for a variety of reactions in organic solvent, but are less prominent for aqueous reactions. Aqueous QD photocatalysts require hydrophilic ligand shells that provide long-term colloidal stability but are not so tight-binding as to prevent catalytic substrates from accessing the QD surface. Common thiolate ligands, which also poison many co-catalysts and undergo photo-oxidative desorption, are therefore often not an option. This paper describes a framework for the design of water-solubilizing ligands that are in dynamic exchange on and off the QD surface, but still provide long-term colloidal stability to CdS QDs. The binding affinity and inter-ligand electrostatic interactions of a bifunctional ligand, aminoethyl phosphonic acid (AEP), are tuned with the pH of the dispersion. The key to colloidal stability is electrostatic stabilization of the monolayer. This work demonstrates a means of mimicking the stabilizing power of a thiolate-bound ligand with a zwitterionic tailgroup, but without the thiolate binding group.
Keywords: quantum dots, catalysis, ligand design, nanotechnology, water chemistry
Graphical Abstract
Electrostatic stabilization of CdS QDs in water enabled by alternating charges of the head and tail groups of a sulfur-free dynamically exchanging ligand
Water is the ultimate green solvent, and the choice of medium for many classes of reactions that utilize an emerging methodology for chemical transformations, nanoparticle photocatalysis, that is highly effective in the laboratory and potentially scalable.[1–3] Among the range of nanoparticles that photocatalyze aqueous reactions, semiconductor quantum dots (QDs) stand out for their chemical and electronic tunability and versatility. QDs have demonstrated unprecedented activity in photocatalytic H2 evolution,[4–7] unprecedented sensitization efficiency for CO2 reduction,[8] the ability to serve as triplet exciton donors and scaffolds for stereoselective cycloadditions,[9,10] and the electrochemical potentials to act as both photo-oxidant and photo-reductant in C-C coupling schemes with no sacrificial reagents.[11,12] In water, properly functionalized colloidal QDs form colloidally stable aggregates to mimic the exciton funneling function of photosystem II,[13] and perform chemoselective alcohol oxidations.[14]
The best-quality as-synthesized QDs are capped with hydrophobic ligands, and surface modification is necessary in order to transfer the QDs from nonpolar solvents to aqueous solution. Access of molecular substrates to the inorganic surface of the QD is necessary for applications in photo-redox catalysis, in order to provide sufficient electronic coupling for electron and hole extraction,[15–17] so water-solubilization strategies that involve replacing the hydrophobic shell of the QD with a silica shell or encapsulating the hydrophobic QD in an amphiphilic polymer are not suited for QD-based catalysis.[18,19] Exchange of native hydrophobic ligands for very polar or charged ligands is a versatile strategy for creating water-soluble particles, since not only the chemical structure but also the density of ligands can be controlled. In addition to providing access to the QD surface and, ideally, encouraging catalytic substrates to adsorb in activated geometries, such ligands need to make the particles colloidally stable in water for the lifetime of the illumination (or longer) by inhibiting agglomeration, which decreases catalytically active surface area and eventually leads to precipitation. The most basic requirement for preventing agglomeration is that ligands must not desorb from the QD surface at the catalytically relevant pH, ionic strength, and reagent/product concentrations. Tight-binding thiolates provide long-term stability and have been the headgroups (binding groups) of choice for water-solubilizing ligands for biological applications, but are not a good choice for QD photocatalysts in cases where: (i) the QD photocatalyst is unshelled (for easy charge extraction), and the thiolate traps the excitonic hole and initiates photo-oxidative degradation of the QD and/or desorption of the thiolate to form disulfide,[15,20–23] (ii) there is a inorganic coordination complex present (e.g., as a co-catalyst) that the thiol/thiolate can poison by ligating the metal,[21,22] or (iii) the thiolate ligands bind tightly and densely enough to inhibit the catalytic substrate’s access to the QD surface.[17,24,25]
These considerations prompt the search for a thiolate-free, dynamically exchanging (weak-to-intermediate binding) ligand for aqueous QD-based catalysis. We recently reported the use of phosphonopropionic acid (PPA) to stabilize QDs in aqueous media.[26] This ligand provided enough colloidal stability to carry out very efficient QD-photocatalyzed, air-free oxidation of benzyl alcohol, but, because of its lability, did not consistently effect a high-yield phase transfer of QDs into water, and did not produce dispersions with long-term colloidal stability, especially when the samples were stored in room light and air (which led to ~50% precipitation after 10 h).[14] Aggregation of particles stabilized by thiolate-free ligands has been reported in several aqueous systems.[27,28]
Here, we provide a framework for the design of stable colloidal aqueous dispersions of semiconductor QDs using ligands that are in dynamic exchange on and off the QD surface, and therefore are potential ligands for photocatalytic QDs. Our experimental system is CdS QDs with bifunctional aminoethyl phosphonic acid (AEP) ligands (Figure 1) in water over a range of pH values (6 – 12). Changing the pH changes the protonation state of both the amino and phosphonate groups, which changes their respective charges and affinities for the QD surface. The ligand therefore “flips” (the primary binding group becomes the primary tail group) at certain pH values. Our data indicate that the most important factor in ensuring colloidal stability is electrostatic stabilization of the organic adlayer by minimizing repulsion among binding groups and among tailgroups of the ligands. This strategy mimics the benefits of thiolate-bound ligands with zwitterionic tailgroups,[29,30] without using a tight-binding thiolate.
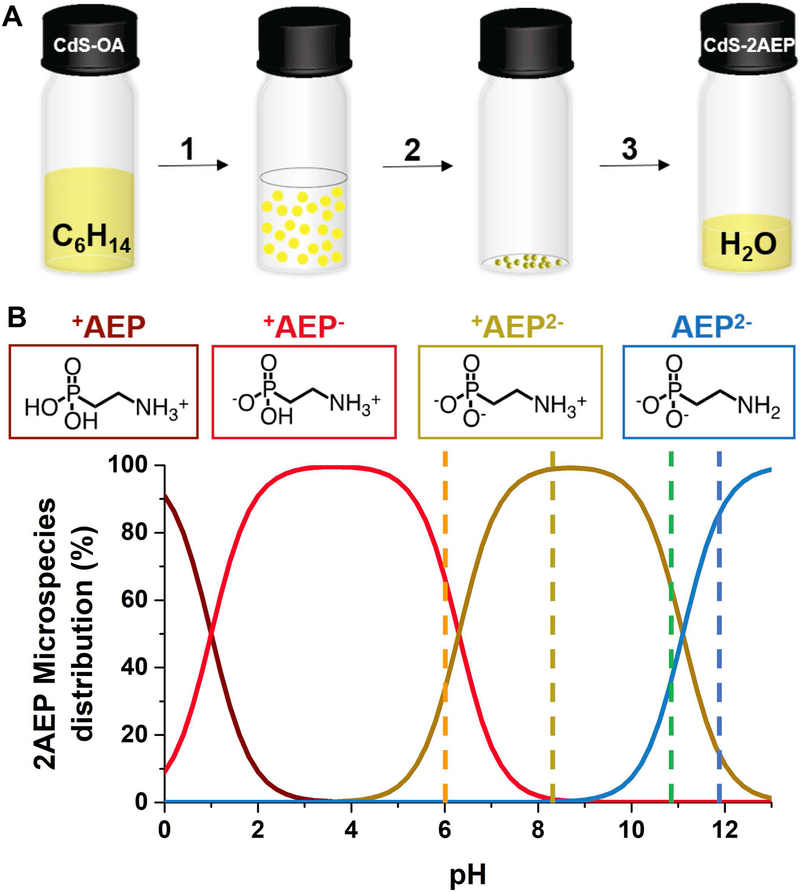
Figure 1.
A) Schematic diagram of the ligand exchange procedure: (1) add 75 μL of a methanolic solution of 0.04 M AEP and 0.1 M KOH (a molar ratio KOH:AEP of 3:1) to 9∙10−7 M CdS QDs in hexanes; (2) mix rapidly, centrifuge at 9500 RPM for 5 minutes and remove the colorless hexanes layer; (3) add 3 mL of (i) DI water to achieve a pH of 10.8, (ii) 2 mM HCl (aq.) to achieve a pH of 6, (iii) 1 mM HCl (aq.) to achieve a pH of 8.2, or (iv) 5.6 mM KOH (aq.) to achieve a pH of 11.8. B) Fractional distribution of each AEP microspecies as a function of pH.
Oleate-capped CdS QDs were synthesized and purified as described elsewhere[26] (see the Supporting Information). Figure 1A shows the three-step phase-transfer of CdS QDs from hexanes to water using AEP. We add 550 equivalents of AEP per QD as a 0.04 M basic solution in MeOH to 6 mL of a 9∙10−7 M suspension of QDs in hexanes to displace the original oleate ligands. The addition of AEP induces the flocculation of the QDs. Rapid mixing of the suspension induces their precipitation, and centrifugation at 9500 rpm for 5 min leaves the hexanes layer completely colorless. We remove the hexanes layer, and redisperse the pellet in 3 mL of water of the desired pH (adjusted with addition of HCl or KOH). The yield of the phase transfer is ~70% at all the pH values (see the Supporting Information).
The pKa of the first acidic proton of PO3H2 within AEP is ∼1;[31] thus it is dissociated throughout the pH range we studied (6–12). In this pH range, AEP has two protonation equilibria, PO3H− ↔ PO32− and NH3+ ↔ NH2. We determined pH-metrically that the acid dissociation constants are 6.3 (pKa1) for the PO3H− group and 11.1 (pKa2) for the NH3+ group (Supporting Information, Figure S1), and we used these values to calculate the microspecies distribution curves for AEP as a function of pH, Figure 1B. We use the notation +AEP− to indicate the molecule in a state with a protonated amine and a mono deprotonated phosphonate (NH3+(CH2)2PO3H−), the notation +AEP2− to indicate the molecule in a state with a protonated amine and a fully deprotonated phosphonate (NH3+(CH2)2PO32−), and the notation AEP2− to indicate the molecule in a state with a neutral amine and a fully deprotonated phosphonate (NH2(CH2)2PO32−). The dashed lines in Figure 1B indicate the four pH values at which we examined the dispersions of AEP-capped CdS QDs, chosen to achieve a distribution of protonation states of AEP. At pH 6, 66% of the ligand is in the +AEP− state and 34% is in the +AEP2− state, so we refer to this mixture as +AEP−/+AEP2−. At pH 8.2, 100% of the ligand is in the +AEP2− state. At pH 10.8, 66% of the ligand is in the +AEP2− state and 34% is in the AEP2− state, so we refer to this mixture as +AEP2−/AEP2−. At pH 11.8, 17% of the ligand is in the +AEP2− state and 83% is in the AEP2− state, so we approximate this state as AEP2−. The values of pKa1 and pKa2 will differ for freely diffusing AEP and AEP bound to the QD surface, but, since the AEP is in dynamic exchange on and off the QD, the distribution in Figure 1B is a sufficient approximation of protonation states.
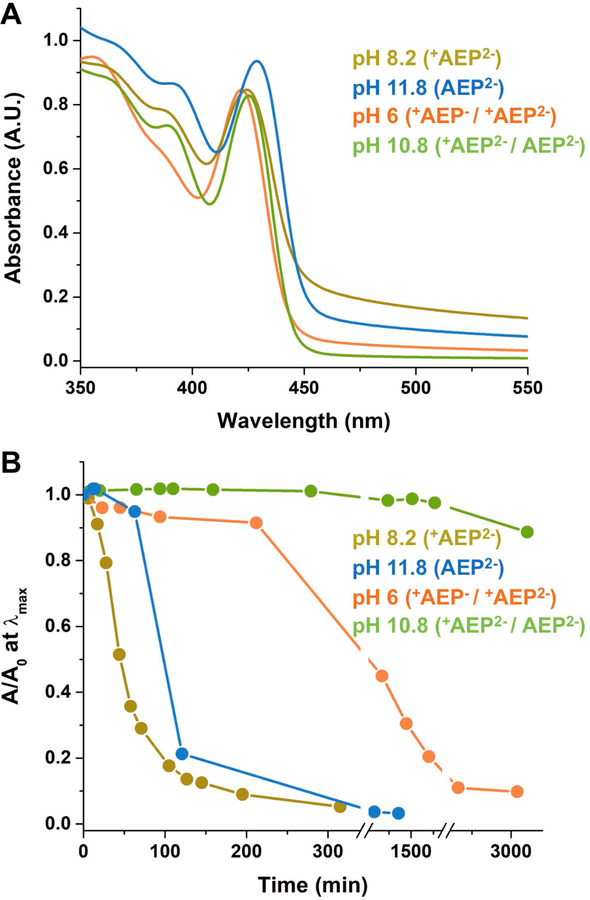
AEP can bind to the surfaces of CdS QDs, which are Cd2+-enriched,[26] through the neutral amine, the mono protonated phosphonate, or the fully deprotonated phosphonate, with different solvent-dependent binding constants. The set of binding modes and orientations of CdS QD-AEP complexes is therefore pH-dependent, as is the colloidal stability of the dispersion. The scattering baselines in the ground state absorption spectra of AEP-capped QDs (aq) (Figure 2A) at the four pH values listed above reflect the degree of aggregation of particles induced by the phase transfer procedure (no scattering baselines are observable in the spectra of the original oleate-capped QDs, Supporting Information, Figure S2). These spectra, measured ≤10 minutes after phase transfer, show that the degree of aggregation increases from pH 10.8 (+AEP2− / AEP2−) to pH 6 (+AEP− / +AEP2−), to pH 11.8 (AEP2−) to pH 8.2 (+AEP2−). The degree of aggregation of the QDs is correlated with the tendency of the particles to precipitate from solution over time, when stored in room light and air, Figure 2B. Figures 2A,B both indicate that the order of the samples from most colloidally stable to least colloidally stable is: pH 10.8 (+AEP2− / AEP2−) > pH 6 (+AEP− / +AEP2−) > pH 11.8 (AEP2−) > pH 8.2 (+AEP2−). At pH 10.8 the system is colloidally stable up to 2 days, at which point we start to observe a decrease in absorbance by 10%. At both pH 8.2 and 11.8 the QDs completely precipitate in a few hours (Supporting Information, Figure S3).
Figure 2.
A) Absorption spectra of AEP-capped CdS QDs at pH 6 (orange), pH 8.2 (dark yellow), pH 10.8 (green) and pH 11.8 (blue). B) Absorbance at the energy of the first excitonic peak as a function of time scaled by its value at time zero (A0) of the same set of samples as in A. In both panels, the legend lists the pH values of the samples in order of increasing colloidal stability, and shows the dominant protonation state(s) of the AEP ligand at that pH.
The first clue to the nature of AEP binding is that the optical bandgap of the particles shifts to lower energy monotonically by a total ~50 meV with increasing pH, Figure 2A. Based on our previous work with CdS and CdSe QDs dispersed in water with phosphonopropionate ligands,[26,32] this shift is evidence that the surfaces of the QDs are, at least transiently, exposed to negatively charged ions (primarily OH- and AEP itself) that increase the local negative charge with increasing pH. The energy of the dipolar CdS QD decreases with increasing local negative charge. The magnitude of the observed shift is comparable to that we reported for the phosphonopropionate-capped CdS QDs in the same pH range.[33] This shift of bandgap with pH is not observable for the same QDs coated in tightly bound ligands such as MEPA (Supporting Information, Figure S5 and S9) or mercaptopropionate,[26,32] and therefore suggests that the AEP ligands are in fast or intermediate exchange on and off the surface of the QD, such that OH− readily accesses the surface.
While both experiments and simulations have proven useful in determining binding modes of ligands on nanoparticles,[34–37] here we choose to utilize NMR spectroscopy to further specify the binding motifs of AEP at each pH – and thereby determine the binding motifs that optimize colloidal stability. We use 1D and 2D 1H nuclear magnetic resonance (NMR) spectroscopy to study the AEP-capped QD samples at the two pH values at which they are most colloidally stable: pH 10.8 (+AEP2− / AEP2−) and pH 6 (+AEP− / +AEP2−). We compare these spectra to those of reference solutions of free AEP (no QDs) that are prepared following exactly the same procedure as the QD samples. Following our work-up procedure, solutions of free AEP have a pH value that is ~0.3 units higher than dispersions of AEP-capped QDs prepared identically (due primarily to adsorption of AEP to the QD surface); since NMR chemical shifts of AEP are pH-dependent, we use 0.1 M HCl to adjust the pH of the free AEP samples to match those of the corresponding AEP-capped QD samples. We note that resonances corresponding to the original oleate ligands are completely gone in the aqueous samples, and can be observed only in the hexanes supernatant (Supporting Information, Figures S6-S8); this result suggests that the ligand exchange procedure occurred with 100% yield of displacement and residual oleic acid was completely removed during the phase transfer.[38,39]
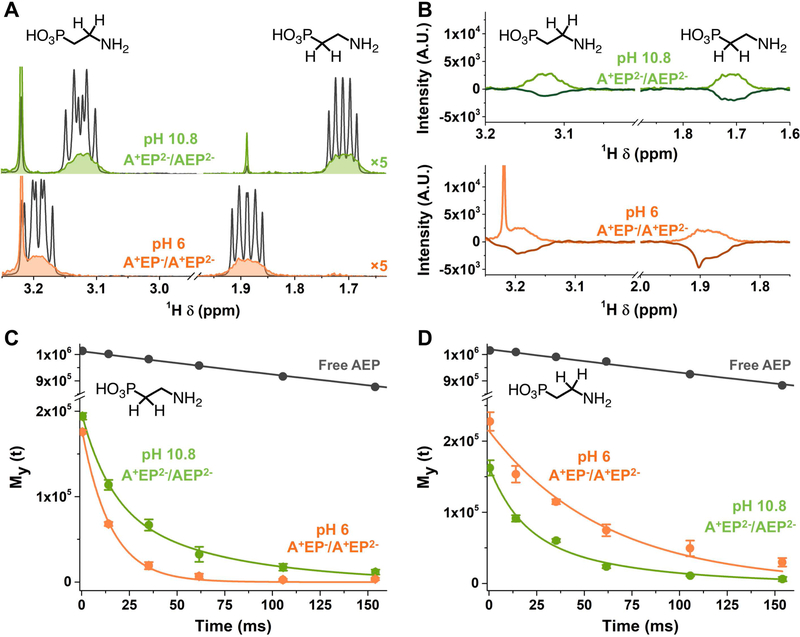
Figure 3A shows the 1H-NMR spectra of AEP-capped QDs at pHs 10.8 (green) and 6 (orange), in the region with resonances for the methylene protons adjacent to the amino group (which we denote CH2-A) and the methylene protons adjacent to the phosphonate group (which we denote CH2-P) of AEP. They both feature spectral lines that are broadened relative to those of free AEP at the same pH (grey), with line widths of ~30 Hz (Supporting Information, Table S1). The degree of broadening and an absence of separate signals for free and bound species confirm that AEP is in intermediate exchange on and off the QD surface (i.e., there is significant interconversion between the free and the bound states during the ~100-ms detection period). The broadening prevents accurate quantification of the AEP in the sample, but only 90 of the added 550 eq AEP per QD were found in the supernatant of the AEP-treated sample after centrifugation, so we assume that 460 eq of AEP per QD are acting as surfactant for the QDs in water. The chemical shifts of these resonances are also slightly different in the samples with and without QDs; however, this observation does not straightforwardly tell us about binding because these shifts are highly sensitive to pH in the regions proximate to pKa1 and pKa2.
Figure 3.
A) 1H-NMR spectra of AEP-capped CdS at pH 6 (orange) and pH 10.8 (green) and of AEP when no QDs are in solution (grey). B) 1H-NMR and 1D-NOESY spectra of AEP-capped CdS at pH 6 (light and dark orange) and pH 10.8 (light and dark green). C) T2 exponential decay and associated curve fits of resonances adjacent to the phosphonate group at pH 6 (orange, monoexponential fit) and pH 10.8 (green, biexponential fit). D) T2 exponential decay and associated curve fits of resonances adjacent to the amino group at pH 6 (orange, monoexponential fit) and pH 10.8 (green, biexponential fit). All the peak integrals used to plot these decays are averaged among 3 experiments.
Figure 3B shows that, at both pH values studied by NMR (6 and 10.8), the selective excitation of CH2-P resonances generates a negative NOE signal relative that for the CH2-A resonances. The negative NOE signal indicates that the rotational mobility of the ligand is dominated by the rotational diffusion of the QDs; thus AEP spends a significant fraction of its time adsorbed to the QD surface, consistent with the intermediate exchange regime.[40–42]
Comparison of the T2 relaxation times of the CH2-A and CH2-P resonances tells us which functional group (A or P) is the binding group and which functional group is the terminal, solubilizing group, because T2 relaxation is affected by local molecular rotations, which are additionally constrained at and near the binding group.[43–45] At pH 6 (+AEP− / +AEP2−), the amine is protonated, so the ligand can only bind to the QDs through the phosphonate group. At pH 10.8 (+AEP2− / AEP2−), 34% of ligands in solution have a neutral amino group, so the ligand can bind through either group, or through both simultaneously (in a chelating geometry). We measured the T2 relaxation times using the Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence.[46,47] In this method, an initial 90° pulse is applied, followed by a series of 180° spin-echo pulses separated by a relaxation delay period (τ) that is varied. The T2 is obtained by fitting the transverse magnetization My (Gaussian−Lorentzian peak area) to the exponential function My = M0e−τ/T2, where M0 is the equilibrium magnetization that is proportional to the population of the corresponding species.
Figures 3C and D shows the T2 decay curves for the CH2-P and CH2-A protons within (i) free AEP, (ii) AEP-capped QDs at pH 6 (+AEP− / +AEP2−), and (iii) AEP-capped QDs and pH 10.8 (+AEP2− / AEP2−). The decays obtained from the QD samples at pH 6, where AEP can only bind through the phosphonate, are satisfactorily fit with a monoexponential function, which indicates the presence of a single time-averaged population of ligands with relaxation times of T2 = 14.9 ± 0.6 ms for CH2-P protons and 60.9 ± 5.9 ms for CH2-A protons. The reduction of these T2 values from that of the free AEP (~1000 ms, Supporting Information, Figure S10 and Table S2) is consistent with molecules in intermediate exchange on and off the QD surface. The faster T2 for the CH2-P protons than for the CH2-A protons is consistent with binding of AEP through the phosphonate, as expected. Since there is only one binding configuration here, we conclude that, for the QD-AEP system at any pH, T2 = ~15 ms is characteristic of CH2-P protons when the phosphonate serves as the binding group (head group) (T2-HEAD), and T2 = ~61 ms is characteristic of CH2-A protons when the amine serves as the tail group (T2-TAIL).[48]
At pH 10.8 (+AEP2− / AEP2−), where AEP can bind through either the amino or phosphonate group, monoexponential fits to the decay curves for the QD samples (Supporting Information, Figure S10) yield T2 = 33.0 ± 2.5 ms for CH2-P resonances and 32.3 ± 2.6 ms for CH2-A resonances. The similarity of these values and the fact that they are both approximately the average of T2-HEAD and T2-TAIL measured at pH 6 suggest that, at pH 10.8, the phosphonate and amino groups both experience an average of head group and tail group configurations - that is, a fraction of the AEP is bound through the phosphonate and a fraction is bound through the neutral amine. In order to estimate these fractions, we fit the T2 decay curves for each type of proton (adjacent to phosphonate or adjacent to amine) at pH 10.8 with a biexponential function: M0,HEADe−τ/T2-HEAD + M0,TAILe−τ/T2-TAIL, Figure 3C,D. The residuals for biexponential fitting are smaller than those for monoexponential fitting (Supporting Information, Figure S10). In the biexponential fit of the decay for CH2-P protons, we fix the value of T2-HEAD to 14.9 ms (the value we measured at pH 6, where we know the phosphonate is the head group), and allow T2-TAIL and the two corresponding M0 values to float. In the biexponential fit of the decay for CH2-A protons, we fix T2-TAIL to 60.9 ms (the value we measured at pH 6, where we know the amine is the tail group), and allow T2-HEAD and the two corresponding M0 values to float.
For CH2-P protons, these fits yield T2-TAIL = 64.1 ± 13.5 ms (M0,TAIL = 92 ± 16) and T2-HEAD = 14.9 ms (fixed) (M0,HEAD of 106 ± 19). For CH2-A protons, these fits yield T2-HEAD = 15.7 ± 4.8 ms (M0,HEAD = 89 ± 16) and T2-TAIL = 60.9 ms (fixed) (M0,TAIL = 75 ± 16). The extracted M0 values tell us that the 53% ± 20% of the CH2-P protons and the 54% ± 20% of the CH2-A protons are adjacent to headgroups. In other words, ~50% of the AEP molecules are bound through the amine and ~50% are bound through the phosphonate (Supporting Information, Table S2).
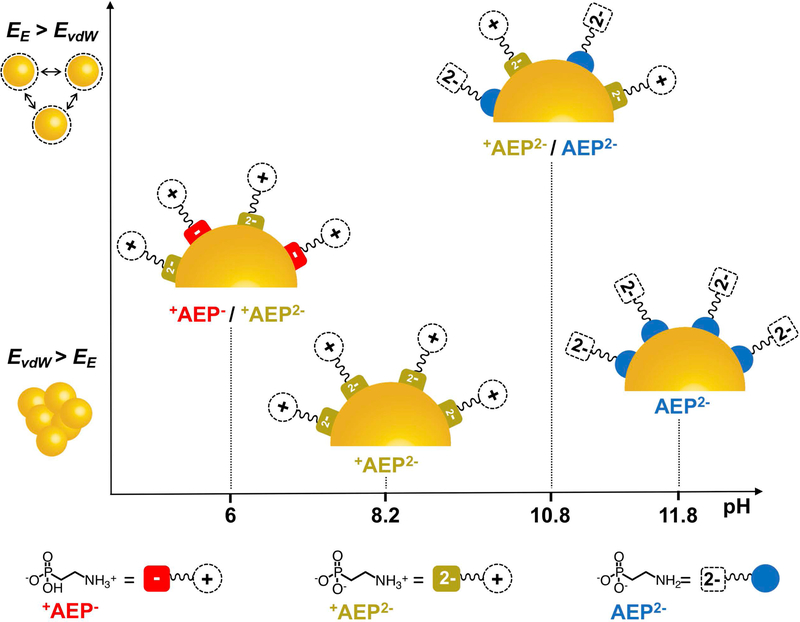
Figure 4 summarizes the binding modes of QD-AEP complexes at the four pH values we examined, as determined by NMR. Recall the order of colloidal stability is (from most stable to least stable): pH 10.8 (+AEP2− / AEP2−) > pH 6 (+AEP− / +AEP2−) > pH 11.8 (AEP2−) > pH 8.2 (+AEP2−). The two most stable configurations occur at pH values very close to the pKa’s of the hydrogen phosphonate (pKa2 = 11.1) and ammonium (pKa1 = 6.3), such that there are significant populations of two forms of the ligand (both of which are able to bind to the QD) in the dispersion simultaneously. The greater stability of the pH 10.8 and pH 6 samples over the pH 11.8 sample indicates that the amine is a too labile head group to stabilize the particles in the presence of a high density of repulsive dianionic tail groups. This result is reasonable given that the amine forms a dative “L-type” bond with Cd2+ that has no additional contribution from the electrostatic binding present in “X-type” interactions like PO32−-Cd2+. The very poor stability of the pH 8.2 sample indicates that even the X-type binding of the PO32− group cannot overcome repulsive interactions of both the tail groups and the more densely packed head groups. The greater stability of the pH 10.8 sample over the pH 6 sample is due to the favorable interactions of alternating positive and negative charges of the tail groups combined with alternating charged and uncharged headgroups. The stability of this configuration coupled with the binding of the amine (a σ-donor that passivates Cd2+ electron trapping sites) results in the only detectably photoluminescent sample of those we studied, with a quantum yield of 0.04% in water (Supporting Information, Figure S4).
Figure 4.
Schematic of the ligand binding motifs for AEP-capped CdS QDs in water for the four pH values we studied: 6, 8.2, 10.8, and 11.8, derived from optical and NMR data. Figure 2B shows the timescales of precipitation for the particles at various pH values; here the particles are ordered from top (most stable, pH 10.8) to bottom (least stable, pH 8.2) EE ≡ electrostatic force; EvdW ≡ vdW attractive energy between the particles.
In summary, one can achieve colloidal stability of QDs in water, at any given pH, with a ligand that binds more weakly than does a thiolate and is in exchange on and off the QD surface, if the ligand monolayer is itself electrostatically stabilized through alternating charges, or at least not destabilized by repulsive interactions of the tail groups or head groups. Such an electrostatically stabilized monolayer could be achieved with a zwitterionic tail group (where ligands arrange such that the negative charge on one ligand is proximate to the positive charge on a neighboring ligand), but it is probable that even such a monolayer would not be stable without a charged headgroup to promote an X-type interaction with the surface of the particle. Charged headgroups however mutually repel, decreasing the stability of the monolayer. We therefore believe that the ideal arrangement of ligands is that shown in Figure 4 at pH 10.8, where the portion of the monolayer is adsorbed through an L-type interaction and the entire monolayer is reinforced by the electrostatic stabilization offered by alternating charges of the tail groups. These considerations present a blueprint for designing water-soluble QDs with sulfur-free, weak-binding ligands, which will open new design pathways for improved photocatalytic systems. None of these considerations are necessary of course if a tightly binding head group like a thiolate is used, but thiolates are generally not an option if the application requires a large degree of access to the QD surface by a catalytic substrate, or involves a co-catalyst that is poisoned by thiolates.
Supplementary Material
Acknowledgements
We kindly acknowledge the National Institutes of Health (R21GM127919) for funding. This work made use of the IMSERC at Northwestern University, which has received support from the NIH (1S10OD012016-01/1S10RR019071-01A1); the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF ECCS-1542205); the State of Illinois and the International Institute for Nanotechnology (IIN).
References
- [1].Pinaud BA, Benck JD, Seitz LC, Forman AJ, Chen Z, Deutsch TG, James BD, Baum KN, Baum GN, Ardo S, et al. , Energy Environ. Sci 2013, 6, 1983–2002. [Google Scholar]
- [2].Fabian DM, Hu S, Singh N, Houle FA, Hisatomi T, Domen K, Osterloh FE, Ardo S, Energy Environ. Sci 2015, 8, 2825–2850. [Google Scholar]
- [3].Wang Q, Hisatomi T, Jia Q, Tokudome H, Zhong M, Wang C, Pan Z, Takata T, Nakabayashi M, Shibata N, et al. , Nat. Mater 2016, 15, 611–615. [DOI] [PubMed] [Google Scholar]
- [4].Li XB, Tung CH, Wu LZ, Nat. Rev. Chem 2018, 2, 160–173. [Google Scholar]
- [5].Chen X, Shen S, Guo L, Mao SS, Chem. Rev 2010, 110, 6503–6570. [DOI] [PubMed] [Google Scholar]
- [6].Li X-B, Tung C-H, Wu L-Z, Angew. Chem. Int. Ed 2019, 2–10.
- [7].Wang F, Wang WG, Wang XJ, Wang HY, Tung CH, Wu LZ, Angew. Chem. Int. Ed 2011, 50, 3193–3197; Angew. Chem. 2011, 123, 3251–3255. [DOI] [PubMed] [Google Scholar]
- [8].Lian S, Kodaimati MS, Dolzhnikov DS, Calzada R, Weiss EA, J. Am. Chem. Soc 2017, 139, 8931–8938. [DOI] [PubMed] [Google Scholar]
- [9].Jiang Y, Wang C, Rogers C, Kodaimati MS, Weiss E, Jiang C, Mohamad S, ChemRxiv 2019, 10.26434/chemrxiv.7713632.v1. [DOI]
- [10].Mongin C, Garakyaraghi S, Razgoniaeva N, Zamkov M, Castellano FN, Science 2016, 351, 369–372. [DOI] [PubMed] [Google Scholar]
- [11].Zhang Z, Edme K, Lian S, Weiss EA, J. Am. Chem. Soc 2017, 139, 4246–4249. [DOI] [PubMed] [Google Scholar]
- [12].Caputo JA, Frenette LC, Zhao N, Sowers KL, Krauss TD, Weix DJ, J. Am. Chem. Soc 2017, 139, 4250–4253. [DOI] [PubMed] [Google Scholar]
- [13].Kodaimati MS, Lian S, Schatz GC, Weiss EA, Proc. Natl. Acad. Sci 2018, 115, 8290–8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].McClelland KP, Weiss EA, ACS Appl. Energy Mater 2019, 2, 92–96. [Google Scholar]
- [15].Kodaimati MS, McClelland KP, He C, Lian S, Jiang Y, Zhang Z, Weiss EA, Inorg. Chem 2018, 57, 3659–3670. [DOI] [PubMed] [Google Scholar]
- [16].Weiss EA, ACS Energy Lett 2017, 2, 1005–1013. [Google Scholar]
- [17].Chang CM, Orchard KL, Martindale BCM, Reisner E, J. Mater. Chem. A 2016, 4, 2856–2862. [Google Scholar]
- [18].Smith JG, Jain PK, J. Am. Chem. Soc 2016, 138, 6765–6773. [DOI] [PubMed] [Google Scholar]
- [19].Zhu H, Song N, Lian T, J. Am. Chem. Soc 2010, 132, 15038–15045. [DOI] [PubMed] [Google Scholar]
- [20].Reisner E, Powell DJ, Cavazza C, Fontecilla-Camps JC, Armstrong FA, J. Am. Chem. Soc 2009, 131, 18457–18466. [DOI] [PubMed] [Google Scholar]
- [21].Han Z, Qiu F, Eisenberg R, Holland PL, Krauss TD, Science 2012, 338, 1321–1324. [DOI] [PubMed] [Google Scholar]
- [22].Das A, Han Z, Haghighi MG, Eisenberg R, Proc. Natl. Acad. Sci 2013, 110, 16716–16723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Brown KA, Wilker MB, Boehm M, Dukovic G, King PW, J. Am. Chem. Soc 2012, 134, 5627–5636. [DOI] [PubMed] [Google Scholar]
- [24].Kuehnel MF, Wakerley DW, Orchard KL, Reisner E, Angew. Chem. Int. Ed 2015, 54, 9627–9631; Angew. Chem. 2015, 127, 9763–9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kuehnel MF, Creissen CE, Sahm CD, Wielend D, Schlosser A, Orchard KL, Reisner E, Angew. Chem. Int. Ed 2019, 58, 5059–5063; Angew. Chem. 2019, 131, 5113–5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Calzada R, Thompson CM, Westmoreland DE, Edme K, Weiss EA, Chem. Mater 2016, 28, 6716–6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Varga K, Tannir S, Haynie BE, Leonard BM, Dzyuba SV, Kubelka J, Balaz M, ACS Nano 2017, 11, 9846–9853. [DOI] [PubMed] [Google Scholar]
- [28].Puri M, Ferry VE, ACS Nano 2017, 11, 12240–12246. [DOI] [PubMed] [Google Scholar]
- [29].Zhan N, Palui G, Mattoussi H, Nat. Protoc 2015, 10, 859–874. [DOI] [PubMed] [Google Scholar]
- [30].Huo S, Jiang Y, Gupta A, Jiang Z, Landis RF, Hou S, Liang XJ, Rotello VM, ACS Nano 2016, 10, 8732–8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Freedman LD, Doak GO, Chem. Rev 1957, 57, 479–523. [Google Scholar]
- [32].Westmoreland DE, Nap RJ, Arcudi F, Szleifer I, Weiss EA, Chem. Commun 2019, 55, 5435–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Thompson CM, Kodaimati M, Westmoreland D, Calzada R, Weiss EA, J. Phys. Chem. Lett 2016, 7, 3954–3960. [DOI] [PubMed] [Google Scholar]
- [34].Li Y, Cheng J, Li J, Zhu X, He T, Chen R, Tang Z, Angew. Chem. Int. Ed 2018, 57, 10236–10240; Angew. Chem. 2018, 130, 10393-10397. [DOI] [PubMed] [Google Scholar]
- [35].Zhou Y, Yang M, Sun K, Tang Z, Kotov NA, J. Am. Chem. Soc 2010, 132, 6006–6013. [DOI] [PubMed] [Google Scholar]
- [36].Cheng J, Hao J, Liu H, Li J, Li J, Zhu X, Lin X, Wang K, He T, ACS Nano 2018, 12, 5341–5350. [DOI] [PubMed] [Google Scholar]
- [37].Choi JK, Haynie BE, Tohgha U, Pap L, Elliott KW, Leonard BM, Dzyuba SV, Varga K, Kubelka J, Balaz M, ACS Nano 2016, 10, 3809–3815. [DOI] [PubMed] [Google Scholar]
- [38].Knauf RR, Lennox JC, Dempsey JL, Chem. Mater 2016, 28, 4762–4770. [Google Scholar]
- [39].Kessler ML, Starr HE, Knauf RR, Rountree KJ, Dempsey JL, Phys. Chem. Chem. Phys 2018, 20, 23649–23655. [DOI] [PubMed] [Google Scholar]
- [40].Moreels I, Justo Y, De Geyter B, Haustraete K, Martins JC, Hens Z, ACS Nano 2011, 5, 2004–2012. [DOI] [PubMed] [Google Scholar]
- [41].Fritzinger B, Moreels I, Lommens P, Koole R, Hens Z, J. Am. Chem. Soc 2009, 3024–3032. [DOI] [PubMed]
- [42].Malicki M, Knowles KE, Weiss EA, Chem. Commun 2013, 49, 4400–4402. [DOI] [PubMed] [Google Scholar]
- [43].De Roo J, Yazdani N, Drijvers E, Lauria A, Maes J, Owen JS, Van Driessche I, Niederberger M, Wood V, Martins JC, et al. , Chem. Mater 2018, 30, 5485–5492. [Google Scholar]
- [44].Shen L, Soong R, Wang M, Lee A, Wu C, Scholes GD, Macdonald PM, Winnik MA, J. Phys. Chem. B 2008, 112, 1626–1633. [DOI] [PubMed] [Google Scholar]
- [45].Morris-Cohen AJ, Malicki M, Peterson MD, Slavin JWJ, Weiss EA, Chem. Mater 2013, 25, 1155–1165. [Google Scholar]
- [46].Carr HY, Purcell EM, Phys. Rev 1954, 94, 630–638. [Google Scholar]
- [47].Meiboom S, Gill D, Rev. Sci. Instrum 1958, 29, 688–691. [Google Scholar]
- [48].Zhang Y, Fry CG, Pedersen JA, Hamers RJ, Anal. Chem 2017, 89, 12399–12407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.