Abstract
MicroRNA (miR)-200c functions in antitumorigenesis and mediates inflammation and osteogenic differentiation. In this study, we discovered that miR-200c was upregulated in human bone marrow mesenchymal stromal cells (hBMSCs) during osteogenic differentiation. Inhibition of endogenous miR-200c resulted in downregulated osteogenic differentiation of hBMSCs and reduced bone volume in the maxilla and mandible of a transgenic mouse model. Overexpression of miR-200c by transfection of naked plasmid DNA (pDNA) encoding miR-200c significantly promoted the biomarkers of osteogenic differentiation in hBMSCs, including alkaline phosphatase, Runt-related transcription factor 2, osteocalcin, and mineral deposition. The pDNA encoding miR-200c also significantly enhanced bone formation and regeneration in calvarial defects of rat models. In addition, miR-200c overexpression was shown to downregulate SRY (sex determining region Y)-box 2 (Sox2) and Kruppel-like factor 4 by directly targeting 3′-untranslated regions and upregulate the activity of Wnt signaling inhibited by Sox2. These results strongly indicated that miR-200c may serve as a unique osteoinductive agent applied for bone healing and regeneration.
Keywords: miR-200c, osteogenic, bone, Sox2, Klf4, Wnt
Introduction
Restoring bone tissue through synthetically engineered bone graft substitutes represents a promising alternative approach for clinical treatment of bone defects caused by trauma, dissection of tumors, and congenital deformities.1 The safety and osteoinductive capacity of the substitutes determine the success of bone healing and regeneration in clinical application. Recombinant human bone morphogenetic proteins (rhBMPs), including rhBMP-2 and rhBMP-7, are the only Food and Drug Administration approved synthetic substitutes that have been applied in clinics to induce spinal fusion, heal fractures, and fill bony defects after tumor resection.2–6 However, due to the relatively short half-life of rhBMP-2, a super-physiological dose of rhBMPs is needed to generate sufficient efficacies of new bone formation in many practical circumstances. A growing side effect profile has emerged as the clinical use of rhBMPs has increased, which includes postoperative inflammation and associated adverse effects, ectopic bone formation, osteoclast-mediated bone resorption, and inappropriate adipogenesis.5,7 Severe adverse events have also been reported recently after the use of rhBMPs in oral and maxillofacial procedures and the administration of rhBMP-2.6 Therefore, an alternative osteogenic factor that is safer, more cost-effective, and highly efficient for bone healing and regeneration is needed.
MicroRNAs (miRs), small noncoding RNAs, promote the degradation and/or repress the translation of messenger RNA (mRNA) through sequence-specific interactions with the 3′- or 5′-untranslated regions (UTRs) of specific mRNA targets. miRs play important post-transcriptional regulators in both physiologic and pathophysiological conditions and have emerged as innovative tools for the diagnosis of diseases and treatment.8–10 miR-200c, a member of the miR-200 family, initially was reported to regulate function in epithelium–mesenchyme transition and has been shown to be involved in tumor progression and aggressiveness. Overexpression of miR-200c was revealed to potently inhibit the progression of multiple types of cancers, including breast and bladder cancers.11 miR-200c can also regulate Wnt/β-catenin signaling in cancer development.12,13 Recent studies have revealed that miR-200c is involved in osteogenic differentiation, tooth development, and inflammation.14–18 Specifically, it has been demonstrated that miR-200c mediated the signal pathway of the nuclear factor kappa-light-chain-enhancer of activated B cells in cancer inflammation and potently downregulated interleukin (IL)-8 by targeting the inhibitor of nuclear factor kappa B kinase subunit beta.15 Our recent studies demonstrated that miR-200c effectively downregulated IL-8, IL-6, and chemokine (C–C motif) ligand-5 by targeting their 3′-UTRs, and potently improves osteogenic differentiation markers in primary human bone marrow mesenchymal stromal cells (hBMSCs) and preosteoblasts.16 In addition, miR-200c was revealed to directly target noggin in bone morphogenetic protein (BMP) signaling during tooth development.14 Noggin is an antagonist in the BMP signal pathway, and inhibition of noggin improves osteogenic differentiation and bone formation.19–21 In addition, in cancer research, miR-200c has been found to regulate SRY (sex determining region Y)-box 2 (Sox2), a master transcription factor in affecting the stemness and differentiation of stem cells.22,23 All this evidence strongly indicates that miR-200c might play roles in osteogenic differentiation and craniofacial bone development and can potentially be developed into an osteoinductive agent to promote bone healing and regeneration for clinical application.
In this study, we tested the potential function of miR-200c in osteogenic differentiation and craniofacial bone development and investigated whether miR-200c can effectively improve osteogenesis in vitro and in vivo. We discovered that miR-200c participated in osteogenic differentiation of hBMSCs and played a critical role in craniofacial bone development in an miR-200c-inhibited transgenic mouse models. We also found that naked plasmid DNA (pDNA) encoding miR-200c could be effectively transfected into hBMSCs and significantly improved osteogenic differentiation of hBMSCs. pDNA encoding miR-200c-loaded collagen sponges also exhibited a strong capability for promoting bone formation and regeneration in different sized calvarial defects of rats. We also demonstrated that miR-200c downregulated Sox2 and Kruppel-like factor 4 (Klf4) by directly targeting their 3′-UTRs, and miR-200c upregulated the Wnt activities repressed by Sox2.
Materials and Methods
Construction of plasmids for miR-200c overexpression and inhibition
pSilencer-4.1-CMV puro plasmid (Ambion, Grand Island, NY) was used to generate a vector stably expressing miR-200c according to our previous studies.14,16 In brief, a set of oligonucleotides corresponding to the miR-200c–specific sequence containing BamHI and HindIII restriction site was synthesized with the primers of 3′-UAAUACUGCCGGGUAAUGAUGGA-5′. Oligonucleotides were annealed according to the manufacturer's recommendations (Addgene, Cambridge, MA) and ligated into pSilencer-4.1-CMV puro vector using BamHI and HindIII restriction sites and confirmed by sequencing. Empty pSilencer-4.1-CMV puro vector (EV) was used as a control. The miR-200c inhibitor using plasmid-based microRNA inhibitor system (PMIS) was generously provided by NaturemiRI, (Iowa City, IA) and the construction was established according to previous studies.24 The miR-200c binding sites were annealed and ligated with a central bulge flanked by different sequences into pLL3.7 vector digested with HpaI and XhoI restriction sites.
Transfection of hBMSCs with pDNA encoding miR-200c and PMIS-miR-200c in vitro
The hBMSCs (ScienCell, Carlsbad, CA) were seeded in six-well plates at a concentration of 105 cells per well and cultured with human mesenchymal stem cell growth medium (MSCGM; Lonza, Walkersville, MD). To transfect plasmid encoding miR-200c, the cells were washed twice with phosphate buffered saline (PBS) and then treated with different doses of pDNA encoding miR-200c at 2 and 20 μg/mL in serum-free medium (Opti-MEM; Life Technologies, Camarillo, CA) for 18 h. EV was used as controls for nonspecific effects. PMIS-miR-200c at 10 μg/mL was transfected into hBMSCs to inhibit endogenous miR-200c using the same method. The miR-200c level after transfection with pDNA and PMIS was confirmed using real-time PCR.
Cell viability assay and proliferation of hBMSCs with miR-200c overexpression
hBMSCs were transfected with different doses of pDNA miR-200c or with EV and incubated for 3 days in MSCGM medium. Cell viability was evaluated by a LIVE/DEAD® assay according to the manufacturer's protocol (Life Technologies, Carlsbad, CA). A (3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide) (MTT) assay was used to test the effect of miR-200c on hBMSCs proliferation and metabolic activity. After the cells were transfected with different doses of miR-200c or with EV overnight, an MTT assay was performed according to manufacturer's protocol (Sigma-Aldrich, St. Louis, MO).
In vitro osteogenic differentiation of hBMSC with miR-200c overexpression and analysis
Osteogenic differentiation was performed on hBMSCs 24 h after transfection. The cells were seeded in six-well plates at 105 cells per well and cultured with complete Dulbecco's modified Eagle medium (DMEM), supplemented with 1 mM β-glycerophosphate and 0.05 mM l-ascorbic acid 2-phosphate. The medium was changed every 3 days up to 4 weeks. The osteogenic biomarkers including alkaline phosphatase (ALP), Runt-related transcription factor 2 (Runx2), and osteocalcin (OCN) were analyzed after different designed time points. The mRNA expression of osteogenic biomarkers was measured after 4 days using real-time PCR, whereas the enzyme-linked immunosorbent assay (ELISA) measurements to quantify the concentrations of Runx2 and OCN were performed after 2 weeks according to the manufacturer's instruction (MyBiosource, Inc., San Diego, CA). ALP staining was performed after 2 weeks of osteogenic differentiation using a commercially available kit (Millipore Sigma, St. Louis, MO). The intracellular ALP activity was also quantified using the p-Nitrophenyl phosphate (pNPP) phosphatase ELISA according to the manufacturer's instruction (Sigma-Aldrich). Results were normalized to the total proteins content of each well. Mineralization of hBMSCs was analyzed after 4 weeks, and Alizarin red staining (ARS) was used to detect calcium deposition in the extracellular matrix. The red stain was then extracted using 10% cetylpyridinium chloride (Sigma-Aldrich) buffer for 1 h with gentle shaking, and the absorbance at 550 nm was read using a SpectraMax Plus microplate reader to quantitatively measure the mineralization.
Quantitative gene analysis
Total cellular RNA from hBMSCs and from rat explants was extracted using a miRNeasy Mini Kit (Qiagen, Valencia, CA). The concentration and purity of total RNA were quantified using the NanoDrop® ND-2000 spectrophotometer and verified using gel analysis. miR-200c expression was measured using the mirScript II reverse transcription kit and the mirScript SYBR Green PCR Kit (Qiagen) and normalized to RNU6B (U6) by a comparative Ct (ΔΔCt) method. In addition, mRNA expression of osteogenic markers was assessed by quantitative real-time PCR. In total, 2 μg of RNA was reverse transcribed using the PrimeScript™ Reagent Kit. Runx2, ALP, and OCN were expressed on a CFX Connect™ (Bio-Rad, Hercules, CA) using the SYBER® Premix TaqTM II Kit and the following primer sets (Table 1). The Vacuolar protein sorting 29 homolog (VPS29) was used as an internal control for human cells, whereas the ribosomal protein gene (RPS18) was used as housekeeping gene for rat samples.
Table 1.
Sequence for forward and reverse primer sets used for real-time polymerase chain reaction
| Human Primer | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Klf4 | ACGATCGTGGCCCCGGAAAAGGACC | CAACAACCGAAAATGCACCAGCCCCAG |
| Sox2 | GGGAAATGGGAGGGGTGCAAAAGAGG | TTGCGTGAGTGTGGATGGGATTGGTG |
| Runx2 | AACCCACGAATGCACTATCCA | CGGACATACCGAGGGACATG |
| OCN | TAGTGAAGAGACCCAGGCGC | CACAGTCCGGATTGAGCTCA |
| ALP | ACCACCACGAGAGTGAACCA | CGTTGTCTGAGTACCAGTCCC |
ALP, alkaline phosphatase; Klf4, Kruppel-like factor 4; OCN, osteocalcin; Runx2, Runt-related transcription factor 2; Sox2, SRY (sex determining region Y)-box 2.
Generation of PMIS-miR-200c transgenic mice
Transgenic mice with PMIS-miR-200c were created as described in our previous publications.24 In brief, to create mouse lines harboring the PMIS-miR-200c construct, the 900 nucleotide PMIS system was cloned into the transgene B vector using the ASCI and SMAI restriction enzymes. The transgene contains an antirepressor sequence followed by the U6 promoter driving the miR inhibitor construct. After amplification, 10 μg of the vector was linearized and gel-purified for pronuclear injection. The linear DNA was used in the pronuclear injection of fertilized oocytes, which were implanted into pseudopregnant female mice. Several founders were generated using this method and examined for the expression of the PMIS-miR-200c transgene.
Analysis of craniofacial bone development of transgenic PMIS-miR-200c mice
Microcomputed tomography (μCT) was used to assess craniofacial shape and abnormal growth. The skulls of 8-week-old PMIS-miR-200c mice were analyzed using a high-resolution μCT scanner SkyScan model 1272 (Bruker, Belgium) at a voltage of 80 kV, a current of 125 μA, a rotation step of 0.6, and an image pixel size of 21.5 μm. A three-dimensional (3D) virtual image of the skull was reconstructed with the NRecon software version 1.6.10.2. 3D volume rendering and a 3D imaging of the defect area were done using the CTvox software version 3.3. The μCT threshold was first calibrated and then applied to all samples. Skeletal regions including skull, cranial bones, and maxilla/palatine (MP) were segmented and bone mass was calculated by means of a bone volume fraction (bone volume/tissue volume or BV/TV).
Bone formation and regeneration in calvarial defects of rats
Animal experiments were approved by the Office of Animal Resources at the University of Iowa, and performed in accordance with the Institutional Animal Care and Use Committee—policies and guidelines. Under anesthesia using a ketamine/xylazine cocktail, a middle skin incision from the nasofrontal area to the external occipital protuberance was performed on 12-week-old male Sprague Dawley rats (Charles River Laboratories, Wilmington, MA). We used two 5 mm-diameter full-thickness defects created on the parietal bone, on both sides of the sagittal suture of each rat to test the capacity of miR-200c treatment on promoting bone formation,. The defects were randomly filled with a dental collagen plug (Zimmer, Palm Beach Gardens, FL) loaded with different treatments: (1) defect with no treatment; (2) collagen sponge alone; (3–5) with pDNA encoding EV at 1, 10, and 50 μg per defect; (6–8) plasmid miR-200c at 1, 10, and 50 μg per defect, respectively. All operations were performed under sterile conditions and the rats were euthanized after 1 and 4 weeks. The tissues within the original surgical defects, including the collagen sponges, were collected 1 week after surgery and stored at −80°C until processed for total RNA extraction to assess the transfection of pDNA miR-200c in vivo. The bone formation with different treatments at defects was evaluated using μCT and histological analysis after 4 weeks. In addition, critical-sized calvarial defects were used to test the bone regenerative capacities of miR-200c. A full-thickness, 10 mm in diameter, calvarial defect was created in Sprague Dawley rats. The rats were then randomly divided into three groups receiving different treatments: (1) defect with no treatment, (2) collagen sponge loaded with plasmid EV at 10 μg per defect, and (3) collagen sponge loaded with plasmid miR-200c at 10 μg per defect. Six weeks after surgery, animals were sacrificed and the craniums were collected and fixed in 10% formalin for μCT and histological analysis.
μCT analysis for bone formation
An μCT analysis was performed to evaluate new bone formation at the defect site. Specimens were analyzed at a voltage of 70 kV, a current of 142 μA, a rotation step of 0.6, and an image pixel size of 10 μm. Reconstruction of 3D virtual models was performed with the NRecon software version 1.6.10.2. Bone mineral density (BMD) and BV/TV were used to calculate new bone formation within a region of interest of 4 and 7 mm from the center of the 5- and 10-mm defects, respectively, using the CTAn software version 1.15.4. 3D volume rendering and 3D imaging of the defect area were done using the CTvox software version 3.3. The μCT threshold was first calibrated and then applied to all samples.
Histological analysis
Fixed calvarial tissues were decalcified with 14% ethylenediaminetetraacetic acid for 14 days, rinsed in PBS, then dehydrated in gradient of alcohol. Samples were cleared with xylene and embedded in paraffin. Coronal sections of 7 μm thick were cut perpendicular to the sagittal suture through the center of the defect. Sections were stained with hematoxylin and eosin staining following standard protocols. Photographs were taken under the microscope to examine bone formation.
Reporter gene analysis of miR-200c on 3′-UTRs of Sox2 and Klf4
Primary hBMSCs cultured in six-well plates were treated overnight with pDNA miR-200c (5 μg/well), or with the same amount of miR-200a or scrambled miRs. After 72 h, RNA was isolated from the cells, and complementary DNA (cDNA) from the RNA was generated using a Biorads PrimePCR cDNA Kit, and the quality of the cDNA was validated by quantitative PCR. The expression of miR-200c, Sox2, and Klf4 was determined using real-time PCR. We performed the reporter gene analysis to test whether miR-200c directly targets 3′-UTR of Sox2 and Klf4 as described in our previous studies.16 In brief, 900 and 850 bp of the 3′-UTRs of Klf4a and Sox2 were cloned into pMIR-reporter and cotransfected into human embryonic palatal mesenchyme (HEPM) cells with overexpression of miR-200c and/or control at a 0.25/1/5 ratio of luciferase reporter, miR, and β-galactosidase reporter, respectively. Cells were lysed and measured for luciferase activity, protein concentration, and β-galactosidase activity at 40 h. Luciferase values were determined, normalized to protein concentration, and β-galactosidase reporter activity.
TOPflash/FOPflash assay
We used TOPflash/FOPflash test to determine the effect of miR-200c and Sox2 on Wnt activity. TOP-Flash (Super 8 × tcf sites) and FOP-Flash (8 × mutant tcf sites) reporters were commercially purchased from Addgene. The TOPflash and FOPflash were added into HEPM cells with overexpression of miR-200c with or without Sox2 at a ratio of 1:5 (reporter to cDNA) for 40 h. Cells were lysed and measured for luciferase activity and protein concentration and β-galactosidase. Values expressed are lactase unit (LU)/μg corrected for β-galactosidase. Lithium chloride (LiCl) was used at a concentration of 8 mM as a positive control for Wnt activation.
Western blot
Sox9 was detected using Western blot to demonstrate the downstream of Wnt/β-Catenin signaling after miR-200c overexpression.25,26 The HEPM cells, HEPM cells with scrambled-miRs, and the HEPM with miR-200c overexpression were grown in DMEM to 80% confluency. The lysates of these cells were prepared using Reporter Lysis Buffer (Promega, Madison, WI) and the protein was detected by standard Western blot technique on 10% nondenaturing acrylamide gels. The primary antibodies of Sox9 (75 kD; Santa Cruz, Dallas, TX) and GAPDH (37 kD; Santa Cruz, Dallas, TX) were used for the detection.
Statistical analyses
Descriptive statistics were conducted. One-way analysis of variance, followed by a post hoc Tukey–Kramer test, was used to detect the difference among the experimental groups. Statistical analyses were performed using the statistical package SAS® System version 9.4 (SAS Institute, Inc., Cary, NC). A significance level of 0.05 was utilized for all the tests.
Results
miR-200c affects osteogenic differentiation in hBMSCs and craniofacial bone development in mice
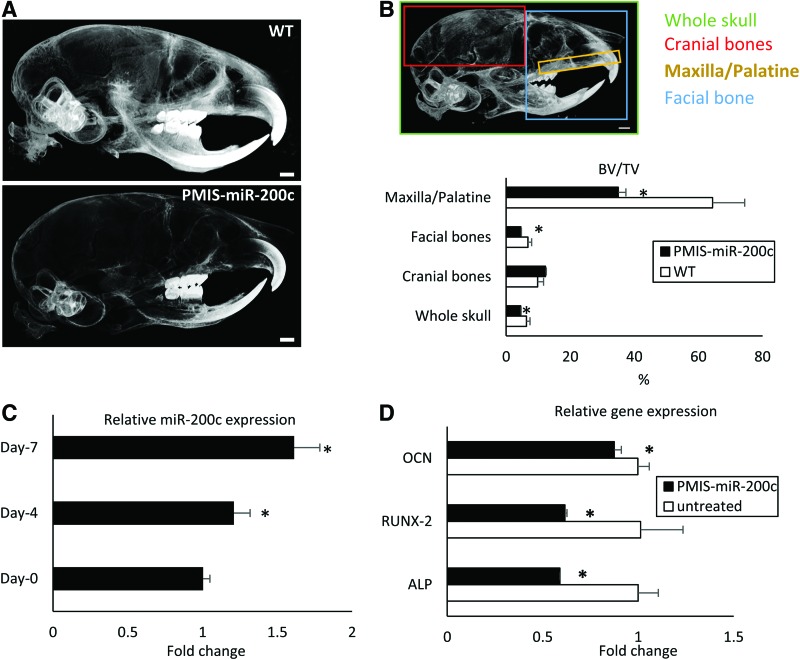
Figure 1A and B summarized the μCT image and quantitative analysis for skulls of the wild-type and transgenic mice with PMIS-miR-200c. The radiopacity of PMIS-miR-200c mouse skulls was lower than that of the wild-type mice, especially at the facial bone and MP region (Fig. 1A). Figure 1B shows how the segment measurement was quantified on mouse craniofacial bone tissues and the quantitative results of BV fractions in the skull, facial bones, and the MP bones. The BV/TV fractions in the location of whole skull, facial, and MP were significantly lower in transgenic PMIS-miR-200c mice than in wild-type mice (Fig. 1B). The roles of miR-200c in osteogenic differentiation were tested in vitro using hBMSCs. After hBMSCs were cultured in osteogenic differentiation medium, the expression of endogenous miR-200c was upregulated by 1.2 and 1.6-fold at days 4 and 7, compared with day 0, respectively (p < 0.05 in each instance) (Fig. 1C). Meanwhile, the transcripts of osteogenic biomarkers, including ALP, Runx2, and OCN, were significantly downregulated in hBMSCs pretreated PMIS-miR-200c to inhibit endogenous miR-200c after 7 days of osteogenic induction (p < 0.05 in each instance) (Fig. 1D).
FIG. 1.
miR-200c involved in mouse craniofacial development and osteogenic differentiation of hBMSCs. (A) μCT images of 7-week-old mice of transgenic PMIS-miR-200c and WT; bar = 1 mm. (B) Quantitative measurement of BV/TV using μCT at different locations of skull of transgenic mice with PMIS-miR-200c compared with WT mice. *p < 0.05, n = 3. (C) Fold change of miR-200c in hBMSCs after osteogenic differentiation. (D) Fold change of ALP, Runx2, and OCN in hBMSCs with endogenous miR-200c inhibited by PMIS 7 days after osteogenic differentiation. *p < 0.05 versus untreated, performed in triplicate. μCT, microcomputed tomography; ALP, alkaline phosphatase; BV/TV, bone volume/tissue volume; hBMSCs, human bone marrow mesenchymal stromal cells; miR, microRNA; OCN, osteocalcin; PMIS, plasmid-based microRNA inhibitor system; Runx2, Runt-related transcription factor 2; WT, wild type. Color images are available online.
Transfection of naked pDNA encoding miR-200c promotes the miR-200c expression in hBMSCs
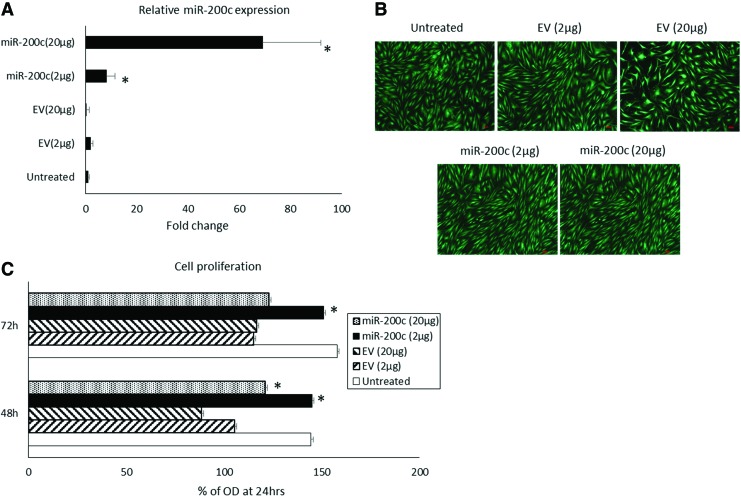
The expression of miR-200c was increased by ∼8- and 69-fold, respectively, 24 h for the different concentrations (2 and 20 μg) of pDNA encoding miR-200c (Fig. 2B). However, EV at the same amount had a limited effect on miR-200c expression. Treatment with pDNA encoding miR-200c and EV showed high viability and less toxicity with no significant difference compared with the untreated group. The cells treated with miR-200c maintained elongated and a spindle-shaped form after transfection (Fig. 2B). All cells with transfection displayed decreased proliferation with a significant and dose-dependent decline after 48 h. However, the cells transfected with miR-200c have significantly higher proliferation rates versus cells transfected with EV at the same concentrations (Fig. 2C).
FIG. 2.
In vitro delivery of miR-200c by naked plasmid transfection. (A) Fold change of miR-200c expression in hBMSCs 3 days after transfected with pDNA encoding miR-200c. (B) LIVE/DEAD® assay of hBMSCs transfected with naked plasmid encoding miR-200c. The scale bar represents 10 μm. (C) Proliferation rates of hBMSCs transfected with miR-200c or EV evaluated by MTT assay. *p < 0.05 versus EVs, performed in triplicate. EV, empty vector; MTT, 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide; pDNA, plasmid DNA. Color images are available online.
pDNA encoding miR-200c promotes hBMSC osteogenic differentiation in vitro
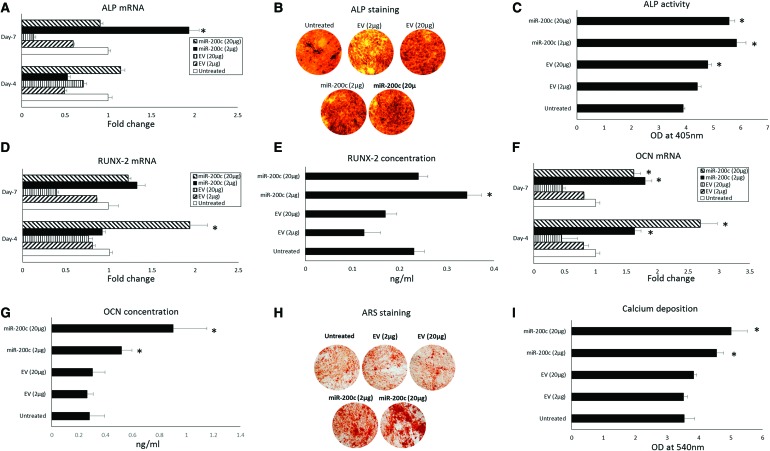
To demonstrate the effect of pDNA encoding miR-200c to enhance osteogenic differentiation, the hBMSCs were transfected with pDNA miR-200c at 2 and 20 μg and subsequently cultured in osteogenic supporting medium up to 28 days. After 4 days from osteogenic differentiation, the mRNA level of the osteogenic markers, including Runx2, ALP, and OCN, in hBMSCs pretransfected with a higher dose of miR-200c pDNA was significantly greater than that of EV-treated hBMSCs (Fig. 3A, D, F). In addition, hBMSCs with miR-200c overexpression displayed denser ALP-positive staining than did the normal and EV-treated hBMSCs after 14 days (Fig. 3B). The extracellular ALP activity was also significantly greater, with an increase of ∼50%, for the hBMSCs pretreated with 2 or 20 μg pDNA encoding miR-200c than it was for the control group (Fig. 3C). miR-200c overexpression also significantly increased Runx2 and OCN secretion in hBMSCs than did untreated controls and the EV after 14 days (Fig. 3E, G). Although the protein level of Runx2 showed no difference when high doses of miR-200c were used (20 μg), OCN protein levels were upregulated after 14 days. ARS at day 28 indicated that the miR-200c overexpression increased bone nodules formation compared with untreated control groups and the cells pretreated with EV (Fig. 3H). Quantitatively, the analysis of calcium content showed that miR-200c overexpression enhanced calcium deposition in a dose-dependent manner (Fig. 3I), whereas there was no significant effect by treatment with EV at 2 and 20 μg.
FIG. 3.
miR-200c promotes osteogenic differentiation of hBMSCs in vitro. (A) Transcripts of Runx2, ALP, and OCN of hBMSCs with different transfection 4 and 7 days after exposure to osteogenic support medium. (B, C) Photographs of ALP staining and the quantitative measurement of ALP activity in hBMSCs pretransfected with pDNA miR-200c. (D, E) Fold change of Runx-2 and its concentration in the supernatant of hBMSCs pretransfected with pDNA miR-200c after osteogenic differentiation. (F, G) Fold change of OCN and its concentration in the supernatant of the hBMSCs after osteogenic differentiation. (H, I) Photographs of ARS and quantitative CPC extraction of ARS staining hBMSCs pretransfected with miR-200c after osteogenic differentiation. *p < 0.05 versus untreated, performed in triplicate. ARS, Alizarin red staining; CPC, cetylpyridinium chloride; mRNA, messenger RNA; OD, optical density. Color images are available online.
pDNA encoding miR-200c promotes bone formation at calvarial defects of rats
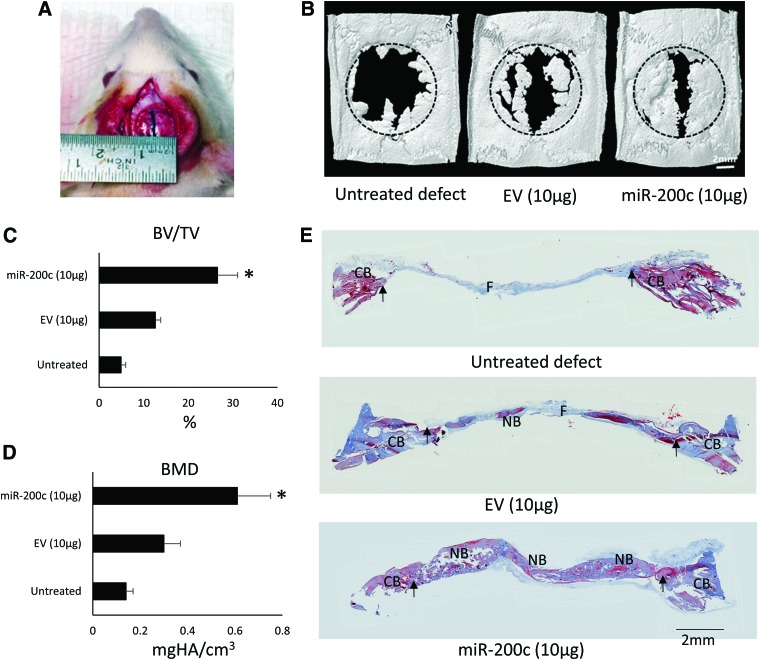
To evaluate the potential of miR-200c to enhance bone formation in vivo, collagen type I sponge trimmed to 5 mm in diameter and 2 mm thickness was used as a scaffold. pDNA encoding miR-200c at 1, 10, and 50 μg was incorporated in collagen sponges and then freeze dried before they were implanted into the 5 mm calvarial bone defects in the rats. After 1 week, the expression of miR-200c in implants loaded with 1, 10, and 50 μg miR-200c was increased by ∼2, 3, and 4.5-fold, respectively, whereas collagen sponge alone or loaded with EV transfection had limited effect on miR-200c overexpression (Fig. 4A). For bone formation enhanced by miR-200c, calvarial bone tissue containing the defects was harvested after 4 weeks. 3D reconstructed images showed complete healing and increased bone tissue for the miR-200c-treated group, whereas the defect in untreated groups and groups treated with collagen alone or EV-loaded collagen was not healed (Fig. 4B). Quantitatively, treatment with collagen and EV-loaded collagen showed a slight, but not statistically significant, increase in bone formation, compared with the empty defect group or without any treatment. However, miR-200c-loaded collagen scaffolds increased the BV formation by ∼205%, 230%, and 245% when 1, 10, and 50 μg pDNA encoding miR-200c were used, respectively (Fig. 4C). Consistently, the BMD of defects treated with miR-200c-loaded collagen sponges was also significantly increased (Fig. 4D). The bone defects treated with 50 μg of miR-200c had the highest BMD and BV/TV. In histological sections, although bone formation was observed inside empty defects because of spontaneous bone healing, miR-200c-loaded collagen sponges induced newly formed bone with greater thickness in defects, compared with treatment with collagen alone and the defects without treatment. The newly formed tissue had woven-like and laminar structures (Fig. 4E).
FIG. 4.
pDNA encoding miR-200c promotes bone formation at rat CB defects. (A) Quantitative measurement of miR-200c expression level in the different implants after 1 week. *p < 0.05; n = 3. (B) μCT top view image of CB defect (5 mm in diameter) 4 weeks after receiving the implantation of collagen sponge loaded with different doses of pDNA miR-200c or controls. (C, D) Quantitative measurement of BV/TV and BMD in the rat CB defects. *p < 0.05 versus untreated; n = 8. (E) Microphotographs of histological cross section of the rat calvarial defects 4 weeks after different treatments. H&E staining; bar = 1 mm; arrows: the edges of defects. BMD, bone mineral density; CB, calvarial bone; H&E, hematoxylin and eosin; NB, new bone. Color images are available online.
miR-200c induces bone regeneration in critical size defects of rat calvaria
We used rat calvarial defects at 10 mm in diameter to test the bone regenerative potential of pDNA encoding miR-200c (Fig. 5A). After 6 weeks of implantation with collagen type I sponge of 10 mm in diameter and 2 mm thickness incorporated 10 μg pDNA miR-200c, limited bone formation was observed in the EV-loaded collagen group as seen by μCT, whereas nearly no bone formation was observed in defects without treatment. However, miR-200c–loaded collagen sponges induced newly formed bone tissue nearly covering the defects (Fig. 5B). Quantitatively, bone defects treated with miR-200c had ∼27% increase in bone formation, whereas the empty defect group with a lower BV/TV only had a ∼4% increase and the EV-loaded collagen groups had a ∼13% increase (Fig. 5C). The BMD of defects treated with miR-200c also significantly increased (Fig. 5D). In histological sections, new bone formation was hardly observed in the defects without treatment. Although the EV-loaded collagen groups presented a connective tissue with minimal bone formation, the defects stayed open and filled with fibrous tissues. Interestingly, collagen sponge loaded with pDNA miR-200c demonstrated a significantly higher amount of newly formed bone in the defects than did those treated with EV and untreated controls (Fig. 5E).
FIG. 5.
pDNA miR-200c promotes bone regeneration in rat calvarial critical-size defect. (A) Surgical photograph of rat calvarial defects at 10 mm in diameter treated with pDNA miR-200c. (B) μCT top view image of critical-sized defect 6 weeks after receiving the implantation of collagen sponge loaded with different doses of pDNA miR-200c. (C, D) Quantitative measurement of BV/TV and BMD in the rat CB defects 6 weeks after receiving different treatments. *p < 0.05; n = 3. (E) Microphotographs of histological cross section of the critical size defects of rat calvaria 6 weeks after different treatments. H&E staining, bar = 1 mm; arrow: the edges of defects. Color images are available online.
miR-200c regulates Sox2, Klf4, and associated Wnt activity
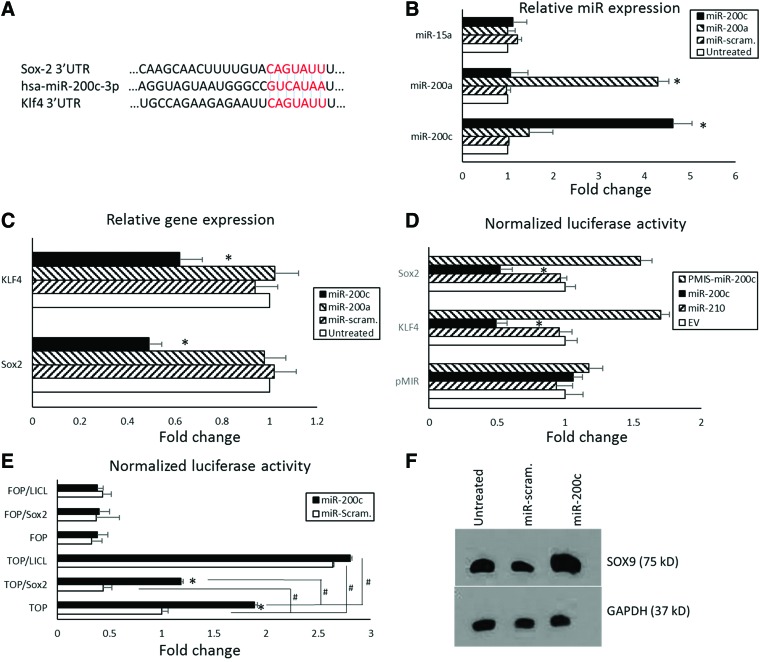
miR target predication indicates that miR-200c directly targets the 3′-UTR of Sox2 and Klf4 (Fig. 6A). To further confirm it, primary hBMSCs cultured in six-well plates were treated overnight with pDNA encoding miR-200c (5 μg/well), or the same amount of miR-200a or scrambled miRs. After 72 h, expression of miR-200c was approximately fivefold greater for hBMSCs treated with miR-200c than for the cells treated with pDNA encoding miR-200a and those with scrambled miRs (Fig. 6B). The transcripts of Sox2 and Klf4 were significantly reduced in the hBMSCs treated with miR-200c compared with the cells treated with miR-200a and those treated with scrambled miRs (Fig. 6C). We cloned the 3′-UTR sequence of Sox2 and Klf4 after the luciferase gene and measured luciferase activity with and without miR-200c. After treatment with miR-200c, the normalized luciferase activity of the luciferase reporter with 3′-UTR of Sox2 and Klf4 was significantly lower, whereas EV and the miR-210 (a negative control) have no effects on 3′-UTR of Sox2 and Klf4 (Fig. 6D). Inhibiting miR-200c using PMIS-miR-200c effectively inhibited endogenous miR-200c and increased the luciferase activity of Sox2 and Klf4 reporter constructs. There is no effectiveness of miR-200c treatment in luciferase activity when using the parental vector with no 3′-UTR after the luciferase gene. In addition, we used the TOP/FOPflash analysis to investigate the effect of miR-200c and Sox2 on Wnt activity. Under the stimulation of LiCl as a positive control (a known inhibitor of GSK3), TOP (Wnt signaling) was activated in both HEPM with miR-200c and scrambled miRs. However, LiCl had no function on FOPflash, a negative control. Cells with the overexpression of miR-200c had significantly higher TOPflash activation than did the cells with scrambled miRs. Although overexpression of Sox2 reduced TOPflash activation in both cells with miR-200c overexpression and those with scrambled miRs (Fig. 6E), the TOP activation in HEPM with miR-200c overexpression was higher than that of HEPM with scrambled miRs. This evidence strongly indicates that miR-200c potently upregulates Wnt activity repressed by Sox2. Figure 6F shows the protein level of Sox9 in HEPM cells with overexpression of miR-200c compared with controls. The Western blot band intensity of Sox9 in the HEPM cells with miR-200c overexpression was higher than in control cells, which further demonstrated that the Wnt/β-Catenin signaling was upregulated after miR-200c overexpression in the HEPM cells. Figure 7 shows the potential signal pathways in miR-200c regulation on osteogenic differentiation and bone formation.
FIG. 6.
miR-200c targets Sox2 and Klf4 and upregulates Wnt activity. (A) The sequence and miR-200c binding region located in the 3′-UTR of Sox2 and Klf4. (B) Fold change of expression of miR-200c, 200a, and 15a in hBMSCs treated with pDNA miR-200c, miR-200a, or the same amount of scrambled miRs. (C) Fold change of Sox2 and Klf4 in hBMSCs treated with pDNA miR-200c, miR-200a, or the same amount of scrambled miRs. (D) Normalized luciferase activities of the 3′-UTR of Sox2 and Klf4-luciferase reporters treated with EV, miR-210, miR-200c, and PMIS-miR-200c. *p < 0.05 versus untreated; performed in triplicate. (E) TOP/FOPflash luciferase activity of HEPM cells with overexpression of miR-200c and scrambled miRs with or without Sox2. *p < 0.05 versus scrambled miR; #p < 0.05; performed in triplicate. (F) A Western blot analysis of Sox9 in untreated HEPM cells and the cells with miR-200c overexpression and scrambled miRs. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HEPM, human embryonic palatal mesenchyme; Klf4, Kruppel-like factor 4; Sox2, SRY (sex determining region Y)-box 2; UTR, untranslated region. Color images are available online.
FIG. 7.
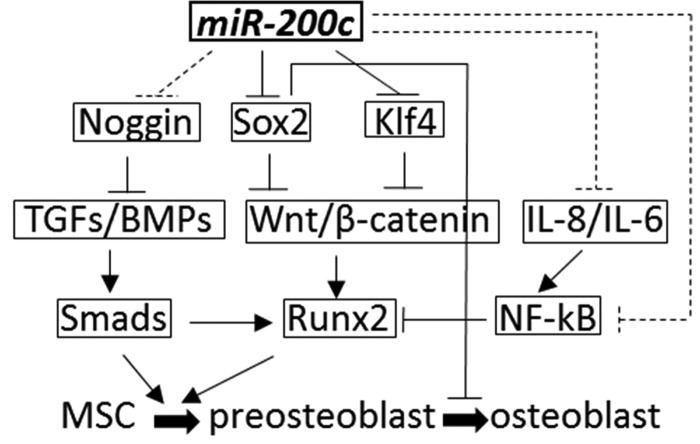
A graphic summary of the multiple roles of miR-200c in bone formation by mediating Wnt signaling through targeting Sox2 and Klf4. BMPs, bone morphogenetic proteins; IL, interleukin; MSC, mesenchymal stromal cell; NF-κB, nuclear factor-kappa B; TGF, transforming growth factor.
Discussion
Our previous studies have reported that miR-200c enhanced osteogenic differentiation and mediated BMP signaling by targeting noggin.14,16 In this study, we extended that line of research and revealed that pDNA encoding miR-200c can be transfected into hBMSCs in vitro and calvarial defects in a rat model, and doing so effectively improved osteogenic differentiation of hBMSCs and enhanced bone formation in vivo. The pDNA encoding miR-200c loaded in collagen sponges effectively promoted bone regeneration in the critical-sized defects of the rats. We have also demonstrated that miR-200c downregulated mRNAs of Klf4 and Sox2 by directly targeting their 3′-UTRs. miR-200c potently upregulated the activity of Wnt signaling inhibited by Sox2. miR-200c effectively promoted Sox9, a readout of Wnt signal pathway. These data strongly indicate that miR-200c may potentially serve as an osteoinductive factor to promote bone healing and regeneration for clinical therapeutic purposes.
Although we previously revealed that miR-200c enhanced osteogenic differentiation of hBMSCs in vitro and miR-200c targeted noggin to effect BMP signaling, its roles in osteogenesis and potentially underlying mechanism(s) were not completely known. Also the potential of miR-200c to regenerate bone tissue in vivo had not been tested. In this study, we first extended our previous studies to investigate the potential roles of miR-200c in osteogenic differentiation of hBMSCs and craniofacial development of mice. We observed that miR-200c is upregulated at the early stage of osteogenic differentiation in hBMSCs, and repression of miR-200c using miR-200c inhibitor resulted in the downregulation of osteogenic biomarkers in hBMSCs. Furthermore, in a mouse model with miR-200c inhibition, we observed that it had an underdeveloped craniofacial bone with lower BV and BMD than did wild-type mice. These data strongly indicate that miR-200c plays important roles in osteogenic differentiation and craniofacial bone development. We further detected that miR-200c downregulated the expression of Klf4 and Sox2 in hBMSCs and our reporter gene analysis revealed that miR-200c effectively targeted 3′-UTR of Klf4 and Sox2, which is consistent with the predicted targets using the miR target predication software (Targetscan 7.0; miRbase). Sox2 and Klf4 are transcription factors that regulate stem cell proliferation and differentiation; however, both of them are involved the maintenance of the stemness in these stem cells and work to inhibit differentiation. Although they are required for the multiple differentiation capacities of stem cells, inhibition of Sox2 and Klf4 has been demonstrated to improve differentiation capabilities.27–32 Besides targeting noggin in BMP signaling, these findings also identify additional underlying mechanism(s) of the function of miR-200c in osteogenic differentiation and craniofacial bone development. In addition, our Wnt activity studies demonstrated that Sox2 effectively downregulated Wnt activity, which is consistent to previous studies.27 It has been well documented that Wnt signaling plays a critical role in osteogenesis and bone formation.29,30,33 Inhibition of Wnt activity effectively repressed osteogenic differentiation and bone formation, whereas the activation of Wnt signaling pathways can accelerate bone regeneration. Many miRs have been reported to regulate Wnt signaling in osteogenic differentiation.34–37 In this study, we demonstrated that Sox2 inhibited Wnt activity and miR-200c could effectively increase the Wnt activity inhibited by Sox2. Therefore, the inhibition of Sox2 and Klf4 targeted by miR-200c can potentially improve osteogenic differentiation and bone development. In addition, noggin is an antagonist in BMP signaling and plays inhibitory roles in Wnt signaling.38 miR-200c may also upregulate Wnt signaling indirectly by targeting noggin. Taken together, by directly targeting noggin, Sox2, Klf4, and a number of proinflammatory cytokines, miR-200c effectively improves the activities of BMP and Wnt signaling to promote osteogenic differentiation and bone regeneration (Fig. 7).
Our previous studies have demonstrated that overexpression of miR-200c induced by pDNA encoding miR-200c delivered using polyethylenimine (PEI) nanoparticles promoted osteogenic differentiation of hBMSCs. In this study, we further demonstrated that naked pDNA encoding miR-200c also exhibited strong capabilities to enhance osteogenic differentiation of hBMSCs. The transfection efficiency of miR-200c was dose dependent and we determined that limited or no toxicity of pDNA transfection was observed even at a higher concentration of pDNA. However, because the plasmids do not contain the green fluorescent protein, we could not visually measure the transfection efficiency of the plasmid encoding miR-200c in this study. Although the transfection of pDNA reduced the proliferation of hBMSCs, the proliferation rate in the cells with miR-200c overexpression was higher than in the cells with EV. miR-200c was reported to reduce the proliferation in many types of cancer cells. Although the mechanism(s) that miR-200c uses to increase hBMSC proliferation is not clear, it may be due to the initial differentiation of hBMSCs induced by miR-200c. Although the transfection efficiency induced by naked pDNA miR-200c is lower than that delivered using PEI in our previous studies,16 miR-200c significantly increased the biomarkers of osteogenic differentiation in hBMSCs in vitro and promoted bone formation in rat calvarial defects in vivo. In addition, no dose-dependent enhancement of osteogenic differentiation and bone formation was observed in the study, which indicated that the osteogenic potential induced by pDNA encoding miR-200c was sufficient to significantly improve bone formation. Also, the newly formed bone structure was similar to the natural bone, and no overgrowth of bone formation was observed. In the critical-sized defects, we also observed that naked pDNA encoding miR-200c significantly improved bone regeneration. These data indicated that miR-200c at a relatively low transfection efficiency has a strong capability to promote bone regeneration, and naked pDNA encoding miR-200c may be safely applied in the clinical applications of bone healing and regeneration. To achieve this long-term goal, the underlying mechanism(s) of miR-200c and its potential off-target effects need to be further investigated to ensure the safety of miR-200c applications. Also, a safe and efficient delivery system is needed to reduce the dose and postpone the release of pDNA encoding miR-200c, which may further promote the efficiency of bone regeneration induced by miR-200c.
Conclusion
Our findings in this study strongly indicated that miR-200c has an important role in osteogenic differentiation and craniofacial bone development. Overexpression of miR-200c using naked pDNA as vectors can efficiently promote osteogenic differentiation and bone regeneration in vitro and in vivo, which indicates that pDNA encoding miR-200c may serve as an effective gene therapy tool for human clinical application of bone healing and regeneration. In addition, the mechanism(s) of osteogenesis induced by miR-200c is targeting Sox2 and Klf4 and upregulating Wnt signal.
Acknowledgments
The authors thank Dr. J. Michael Tilley for proofreading and editing the article. This study was supported by the National Institute of Dental and Craniofacial Research (Grant Nos. R21 DE024799, R21 DE025328, and R01 DE026433) of the National Institutes of Health (NIH). M.R.-B. would like to acknowledge the support received from the NIH under the R90 DE024296-03 grant to pursue a career in dental science.
Authors' Contributions
B.A.A. and L.H. designed the research; A.A., S.E., M.E.S., M.R.B., M.Z., and L.H. performed the research; A.A., F.Q., and L.H. analyzed and interpreted the data; and A.A. and L.H. wrote the article.
Author Disclosure
L.H. and B.A.A. have filed the U.S. patent for miR-200c-based bone regeneration. B.A.A. is the founder of the NaturemiRI Company.
References
- 1. Damien CJ, Parsons JR. Bone graft and bone graft substitutes: a review of current technology and applications. J Appl Biomater 1991;2:187–208 [DOI] [PubMed] [Google Scholar]
- 2. Bhatt RA, Rozental TD. Bone graft substitutes. Hand Clin 2012;28 457–468 [DOI] [PubMed] [Google Scholar]
- 3. Agarwal R, Williams K, Umscheid CA, et al. . Osteoinductive bone graft substitutes for lumbar fusion: a systematic review. J Neurosurg Spine 2009;11:729–740 [DOI] [PubMed] [Google Scholar]
- 4. Boden SD. Biology of lumbar spine fusion and use of bone graft substitutes: present, future, and next generation. Tissue Eng 2000;6:383–399 [DOI] [PubMed] [Google Scholar]
- 5. Woo EJ. Recombinant human bone morphogenetic protein-2: adverse events reported to the Manufacturer and User Facility Device Experience database. Spine J 2012;12:894–899 [DOI] [PubMed] [Google Scholar]
- 6. Boyne PJ. Application of bone morphogenetic proteins in the treatment of clinical oral and maxillofacial osseous defects. J Bone Joint Surg Am 2001;83-A(Suppl. 1, Pt. 2):S146–S150 [PubMed] [Google Scholar]
- 7. James AW, LaChaud G, Shen J, et al. . A review of the clinical side effects of bone morphogenetic protein-2. Tissue Eng Part B Rev 2016;22:284–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development 2005;132:4653–4662 [DOI] [PubMed] [Google Scholar]
- 9. Hassan MQ, Tye CE, Stein GS, et al. . Non-coding RNAs: epigenetic regulators of bone development and homeostasis. Bone 2015;81:746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Song JL, Nigam P, Tektas SS, et al. . microRNA regulation of Wnt signaling pathways in development and disease. Cell Signal 2015;27:1380–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar S, Nag A, Mandal CC. A comprehensive review on miR-200c, a promising cancer biomarker with therapeutic potential. Curr Drug Targets 2015;16:1381–1403 [DOI] [PubMed] [Google Scholar]
- 12. Dermani FK, Amini R, Saidijam M, et al. . Zerumbone inhibits epithelial-mesenchymal transition and cancer stem cells properties by inhibiting the beta-catenin pathway through miR-200c. J Cell Physiol 2018;233:9538–9547 [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y, Wang J, Wu D, et al. . IL-21-secreting hUCMSCs combined with miR-200c inhibit tumor growth and metastasis via repression of Wnt/beta-catenin signaling and epithelial-mesenchymal transition in epithelial ovarian cancer. Onco Targets Ther 2018;11:2037–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cao HJ, Jheon A, Li X, et al. . The Pitx2:miR-200c/141: noggin pathway regulates BMP signaling and ameloblast differentiation. Development 2013;140:3348–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chuang TD, Khorram O. miR-200c regulates IL8 expression by targeting IKBKB: a potential mediator of inflammation in leiomyoma pathogenesis. PLoS One 2014;9:e95370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hong L, Sharp T, Khorsand B, et al. . MicroRNA-200c represses IL-6, IL-8, and CCL-5 expression and enhances osteogenic differentiation. PLoS One 2016;11:e0160915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Howe EN, Cochrane DR, Cittelly DM, et al. . miR-200c targets a NF-kappa B up-regulated TrkB/NTF3 autocrine signaling loop to enhance Anoikis sensitivity in triple negative breast cancer. PLoS One 2012;7:e49987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wendlandt EB, Graff JW, Gioannini TL, et al. . The role of MicroRNAs miR-200b and miR-200c in TLR4 signaling and NF-kappa B activation. Innate Immun 2012;18:846–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tsialogiannis E, Polyzois I, Oak Tang Q, et al. . Targeting bone morphogenetic protein antagonists: in vitro and in vivo evidence of their role in bone metabolism. Expert Opin Ther Targets 2009;13:123–137 [DOI] [PubMed] [Google Scholar]
- 20. Rosen V. BMP and BMP inhibitors in bone. Ann N Y Acad Sci 2006;1068:19–25 [DOI] [PubMed] [Google Scholar]
- 21. Reddi AH. Interplay between bone morphogenetic proteins and cognate binding proteins in bone and cartilage development: noggin, chordin and DAN. Arthritis Res 2001;3:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu YX, Yuan L, Xue XL, et al. . Regulation of colorectal carcinoma stemness, growth, and metastasis by an miR-200c-Sox2-negative feedback loop mechanism. Clin Cancer Res 2014;20:2631–2642 [DOI] [PubMed] [Google Scholar]
- 23. Ma K, Song G, An X, et al. . miRNAs promote generation of porcine-induced pluripotent stem cells. Mol Cell Biochem 2014;389:209–218 [DOI] [PubMed] [Google Scholar]
- 24. Cao H, Yu W, Li X, et al. . A new plasmid-based microRNA inhibitor system that inhibits microRNA families in transgenic mice and cells: a potential new therapeutic reagent. Gene Ther 2016;23:634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blache P, van de Wetering M, Duluc I, et al. . SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J Cell Biol 2004;166:37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu JA, Wu MH, Yan CH, et al. . Phosphorylation of Sox9 is required for neural crest delamination and is regulated downstream of BMP and canonical Wnt signaling. Proc Natl Acad Sci U S A 2013;110:2882–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mansukhani A, Ambrosetti D, Holmes G, et al. . Sox2 induction by FGF and FGFR2 activating mutations inhibits Wnt signaling and osteoblast differentiation. J Cell Biol 2005;168:1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schonitzer V, Wirtz R, Ulrich V, et al. . Sox2 is a potent inhibitor of osteogenic and adipogenic differentiation in human mesenchymal stem cells. Cell Reprogram 2014;16:355–365 [DOI] [PubMed] [Google Scholar]
- 29. Seo E, Basu-Roy U, Gunaratne PH, et al. . SOX2 regulates YAP1 to maintain stemness and determine cell fate in the osteo-adipo lineage. Cell Reports 2013;3:2075–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li J, Dong J, Zhang ZH, et al. . miR-10a restores human mesenchymal stem cell differentiation by repressing KLF4. J Cell Physiol 2013;228:2324–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barba M, Pirozzi F, Saulnier N, et al. . Lim mineralization protein 3 induces the osteogenic differentiation of human amniotic fluid stromal cells through Kruppel-like factor-4 downregulation and further bone-specific gene expression. J Biomed Biotechnol 2012;2012:813894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu L, Zheng L, Wang Z, et al. . TNF-alpha-induced SOX5 upregulation is involved in the osteogenic differentiation of human bone marrow mesenchymal stem cells through KLF4 signal pathway. Mol Cells 2018;41:575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chang J, Liu F, Lee M, et al. . NF-kappaB inhibits osteogenic differentiation of mesenchymal stem cells by promoting beta-catenin degradation. Proc Natl Acad Sci U S A 2013;110:9469–9474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li S, Hu C, Li J, et al. . Effect of miR-26a-5p on the Wnt/Ca(2+) pathway and osteogenic differentiation of mouse adipose-derived mesenchymal stem cells. Calcif Tissue Int 2016;99:174–186 [DOI] [PubMed] [Google Scholar]
- 35. Zhang J, Tu Q, Bonewald LF, et al. . Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J Bone Miner Res 2011;26:1953–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duan L, Zhao H, Xiong Y, et al. . miR-16-2* interferes with WNT5A to regulate osteogenesis of mesenchymal stem cells. Cell Physiol Biochem 2018;51:1087–1102 [DOI] [PubMed] [Google Scholar]
- 37. Wang T, Xu Z. miR-27 promotes osteoblast differentiation by modulating Wnt signaling. Biochem Biophys Res Commun 2010;402:186–189 [DOI] [PubMed] [Google Scholar]
- 38. Gkotzamanidou M, Dimopoulos MA, Kastritis E, et al. . Sclerostin: a possible target for the management of cancer-induced bone disease. Expert Opin Ther Targets 2012;16:761–769 [DOI] [PubMed] [Google Scholar]