Preface
Decades of research has proven that mutations in the p53 stress response pathway affect the incidence of diverse cancers more than mutations in other pathways. However, most evidence is limited to somatic mutations and rare inherited mutations. Using newly abundant genomic data, we demonstrate that commonly inherited genetic variants in the p53 pathway also affect the incidence of a broad range of cancers more than variants in other pathways. The p53 pathway cancer-associated polymorphisms have strikingly similar genetic characteristics to well-studied p53 pathway mutations. Our results enable insights into p53-mediated tumor suppression in humans and into p53 pathway-based cancer surveillance and treatment strategies.
Introduction
There are substantial differences among individuals in their risk of developing cancer, of progressing to high-grade tumors and in responses to therapies. This heterogeneity is a major obstacle to designing uniformly effective prevention, screening and treatment strategies and motivates the large effort to personalize these strategies using biomarkers1. Commonly inherited genetic variants, such as single nucleotide polymorphisms (SNPs) hold great promise as easily obtainable and measurable biomarkers. More than one thousand SNPs have been shown to associate significantly with human cancer in genome-wide association studies (GWAS) conducted in a broad spectrum of solid and hematological malignancies. However, despite these findings, major challenges remain in translating these associations into clinical applications2,3. For example, discerning the functional consequences of the variant, as well as the genes and molecular pathways connecting the variant to disease, has proven extremely challenging. This limited understanding of the biology behind these very significant associations has clearly constrained our ability to integrate SNP biomarkers into the proper genetic, cellular and clinical context to maximize their effective use in the clinic.
In recent years, the field of cancer genetics has made great strides in generating datasets crucial for revealing the mechanistic relationship between SNPs and tumorigenesis. For example, data generated by the 1000 Genomes Project reveals the genetic diversity in individuals and populations and has been crucial for identifying haplotypes that are linked to diseases studied with GWAS4. Moreover, functional genomic scans of gene regulatory features, such as transcription factor binding or specific histone modifications, by chromatin-immuno-precipitation coupled with sequencing (ChIP-seq) can connect these SNPs with functional biological differences5–8. Additionally, the advent of expression quantitative trait loci (eQTL) mapping approaches that measure gene expression levels for tens of thousands of transcripts in genotyped samples has provided an intermediate biological phenotype useful for interpreting many GWAS associations. In these studies, global gene expression measurements and whole-genome SNP genotypes are correlated in order to connect the abundance of a specific gene transcript with an allelic variant and define eQTL9–11.
Analysis of data sets such as these allows rapid assignment of cancer-associated SNPs to well-studied signaling pathways known to be important in a broad range of cancers and for which somatic genetic variants are currently utilized as biomarkers or drug targets in the laboratory and clinic. Identifying which cancer-associated pathways frequently carry SNPs associated with differential cancer susceptibility could accelerate our biological understanding of the variants’ influence on cancer and the potential clinical utility of SNP biomarkers12,13. One of the most well-studied and important cancer signaling pathways is the p53 tumor suppressor pathway. Decades of intensive research in mice and humans have shown that human genetic variants in the p53 stress response pathway can play key roles in the incidence and survival of many cancers. For example, among individuals with Li-Fraumeni Syndrome (LFS) - who carrying low-frequency, inherited TP53 gene mutations - the penetrance for cancer onset is 100% by the age of 70 years, with the cancers occurring in numerous tissues, such as bone, connective tissues, breast and brain14,15. Indeed, candidate SNP studies have clearly demonstrated that p53 signaling can be affected by functional SNPs that in some cases result in differential cancer susceptibility16–18. Moreover, very similar somatic mutations in the TP53 gene are found in over 50% of all cancer genomes, making it the most frequently mutated gene that is causally implicated in cancer and the gene mutated in the broadest range of cancer types, including epithelial, mesenchymal, and hematological cancers19–21. Importantly, recent network analyses that have taken advantage of advances in high-throughput exome sequencing of cancer genomes, have shown that the p53 pathway represents the largest, most frequently identified network of genes carrying mutations in the broadest spectrum of cancers identified thus far22–27. In fact, common somatic mutations in many genes of the pathway are known to directly affect susceptibility to a broad range of cancer types and are being developed as critical biomarkers in some patient-stratification strategies in the clinic14,16,28–31. Moreover, these somatic mutations and low-frequency inherited mutations have been shown to affect cancer risk, progression and response to therapies for many cancers in humans and many mouse models. Such findings are consistent with the well-defined roles of the p53 stress response pathway in tumor suppression, regulating cell migration and invasion and mediating the cellular response to DNA damage-inducing cancer therapies, primarily mediated through the ability of p53 to regulate transcription32–36.
These observations made in model systems, tumor studies and in a rare familial syndrome (LFS) suggest that genetic variants found in the general human population, such as SNPs, also would be more likely to affect susceptibility to a broader range of cancer types than functional SNPs in other genes and pathways. In this Analysis, we demonstrate that the abundant genomic data generated in the last decade support this prediction. We go on to show that these common SNP variants are similar to well-studied inherited and somatic pathway disease mutations, in that they are frequently found in a high proportion of pathway genes and to associate with multiple types of cancers, but not other diseases. Moreover, they are found almost exclusively in p53 pathway genes that can carry cancer-causing mutations in cancer genomes, thereby suggesting that particular p53 pathway genes are highly sensitive to heritable and somatic genetic variants resulting in altered tumor suppression in many tissue types.
Somatic, causal mutations and inherited, cancer-associated SNPs both occur in a high proportion of p53 pathway genes
Somatic, causal mutations.
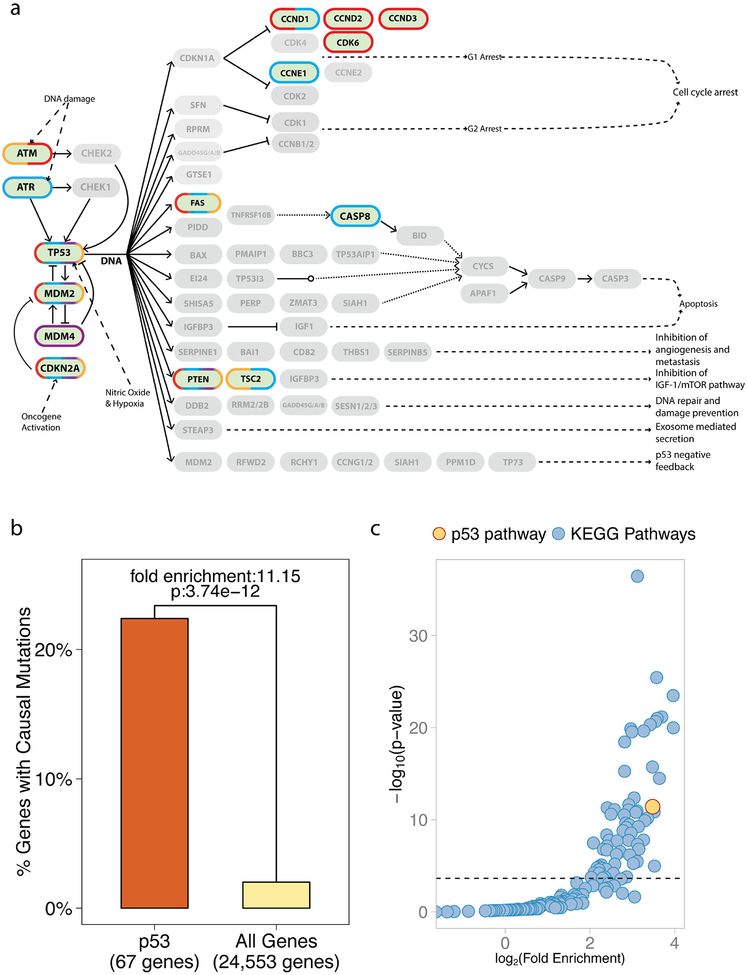
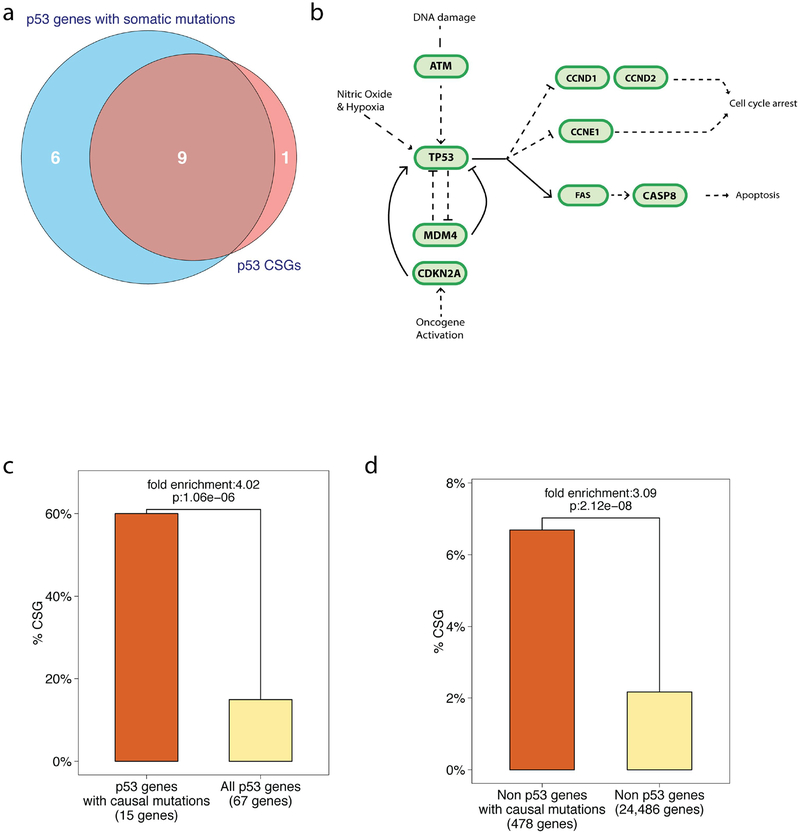
Cancer driver genes, like TP53, when mutated, can promote tumorigenisis and have been identified through studies of inherited cancer predisposition syndromes, cancer genomes, and experimental models of cancer. While estimates regarding the exact number of these genes vary, one well-utilized curated list is the Cancer Gene Census, which utilizes sequencing data to identify genes, whose number and pattern of mutations are highly unlikely to be attributable to chance37. Their current list consists of 493 RefSeq autosomal genes that harbor somatic cancer-promoting mutations (The Cancer Gene Census, The Sanger Institute, http://cancer.sanger.ac.uk/census/, download date 09/11/2015). Out of the 67 autosomal genes attributed to the p53 pathway (Kyoto Encyclopedia of Genes and Genomes, KEGG, http://www.genome.jp/kegg/), 15 have been denoted as harboring somatic, causal mutations in at least one cancer type. This group includes the well-studied oncogenes and tumor suppressor genes: ATM, MDM2, CDKN2A, FAS, and MDM4 (Figure 1A). Thus, 22.38% of p53 pathway genes contain known causal mutations; a significant 11.15-fold enrichment over the 2.01% found in all 24,553 annotated autosomal genes (Figure 1B, RefSeq, p-value: 3.74e-12, adjusted p-value: 8.22e-10).
Figure 1. Somatic, causal mutations occur in a high proportion of p53 pathway genes.
(A) A pathway diagram of the p53 pathway as annotated by KEGG. Genes for which mutations have been causally implicated in cancer appear darker and are outlined (Cancer Gene Census, Sanger). A blue outline indicates causally mutated genes in epithelial cancers, red in leukemia/lymphomas, purple in mesenchymal cancers and orange in other types of cancers. (B) A bar graph of the percent of genes in the p53 pathway with known causal mutations compared to all annotated autosomal genes of the genome. 15 out of 67 genes in the p53 pathway (22.38%) are known to be causally mutated, which represents a significant 11.15-fold enrichment over the rest of the genes in the genome (p-value: 3.74e-12 is also depicted). (C) A scatter plot showing the fold enrichment of causally mutated genes on the x-axis (log-scale), and the p-value on the y-axis (-log10 scale). The horizontal line represents the 5% Family Wise Error Rate threshold (Bonferroni adjusted-p-value: 0.05). The enrichment of causal mutations in p53 pathway genes (in yellow) is the highest and the most significant compared to the other 214 annotated KEGG pathways (in blue). Overall, 30% pathways demonstrated significant enrichment of causally mutated genes.
In order to assess further the importance of the 11.15-fold enrichment, we compared it to potential fold enrichments of causally mutated genes in all well-annotated pathways in the genome. To do this, we determined potential enrichments in all 220 signaling pathways annotated by KEGG in the categories of metabolism, genetic information processing, environmental information processing, cellular processes and organismal systems. We found that 66 of the 220 cellular signaling pathways (30%), including the p53 pathway, demonstrated significant enrichment of causally mutated genes after correction for multiple hypothesis testing (Figure 1C; see Analytical Procedures and Supplementary Table 1 for details). Thus, the enrichment noted in the p53 pathway is amongst the top 8.18% of all well-annotated pathways of the genome. It is important to note that enrichment of causally mutated genes in the p53 pathway was also observed when we used a list of causally mutated genes generated using different criteria by different researchers (Supplementary Figure 1A and B)1.
Cancer-associated SNPs.
Together with the noted high cancer risk among TP53 mutation carriers15, the significant enrichment of causally mutated genes in the p53 pathway suggests that inherited SNPs in p53 pathway could affect cancer susceptibility to a greater extent than SNPs in other annotated signaling pathways. To begin to test this, we utilized the GWAS catalogue (download date: 09/11/2015) and the 10th revision of International Classification of Diseases (ICD10) to identify all cancer susceptibility GWAS studies that have been undertaken to date (Analytical Procedures). Specifically, we first identified all GWAS studies that were designed to study disease susceptibility in European populations (n=756). Subsequently, each of the 756 GWASs was attributed to one of the major ICD10 disease categories, which includes Neoplasms (Supplementary Table 5). We found that 19 different ICD10 disease categories have at least one GWAS study (average: 39.8 studies per disease category). Importantly for this Analysis, Neoplasms had the most studies attributed to it with 165. These 165 studies have been undertaken to assess differential susceptibility for a broad range of cancers (ICD10 Subcategory, Supplementary Table 5) with a median of 11,647.5 individuals with cancer per study. If our hypothesis is correct, we would expect that genes of the p53 pathway would overlap with the cancer susceptibility loci (CSL) identified in these GWAS studies to a greater extent than the genes of other well-defined signaling pathways.
To test this, we first determined which CSLs mapped to the 24,553 autosomal RefSeq genes in the genome. We began by using the 1000 Genomes Phase 3 dataset to identify all known SNPs (MAF ≥ 0.001) from European populations within +/− 10Kb of the gene boundaries of the 24,553 autosomal RefSeq genes and found 7,106,459 SNPs in total. Subsequently, we mined the GWAS catalog, and extracted the 1,034 SNPs (750 unique loci) associated with susceptibility to approximately 17 different types of cancers in European populations, including epithelial, mesenchymal, and hematological cancers (Figure 2). Next, we augmented this dataset with SNPs in linkage disequilibrium (r2=1.0, MAF≥0.001) in European populations using data from the 1000 Genomes Phase 3 dataset. On average, a cancer GWAS tag SNP was in perfect linkage disequilibrium with 3.926 SNPs (range from 1 to 126 SNPs) and the resulting cancer GWAS LD blocks average 6,948.132 bp (range from 1 to 201,267 bp). Our final dataset consists of a total of 3,454 unique cancer GWAS SNPs.
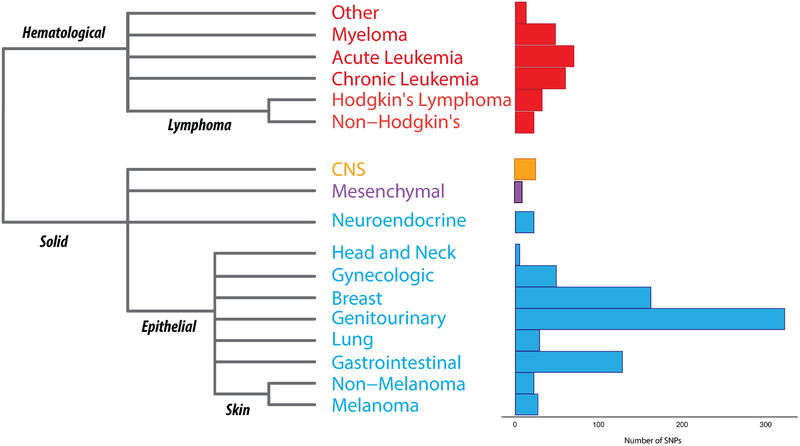
Figure 2. One hundred and sixty five GWAS studies of many cancers types have been performed in European populations.
A histo-pathological classification of all the cancers present in the NHGRI GWAS catalog (download date: 09/11/2015) and a bar graph illustrating the number of tag-SNPs which have been found to significantly associate with differential susceptibility to the particular cancer. Cancers are also classified as epithelial (blue), lymphoma/leukemia (red), mesenchymal (purple) and others (orange).
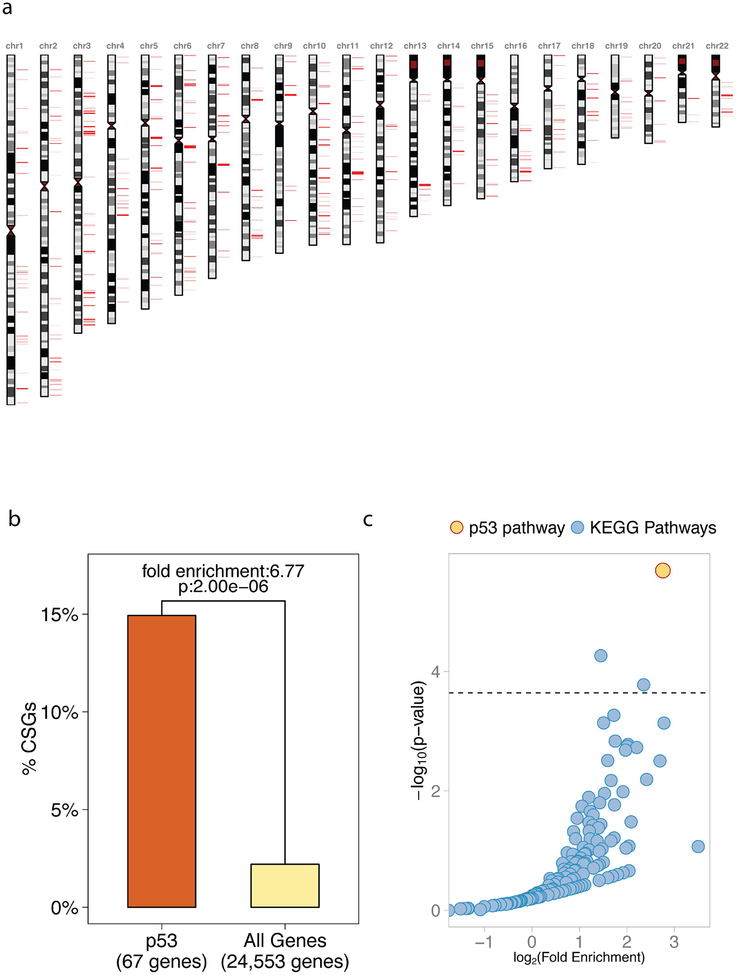
Using our parameters, which require at least one cancer GWAS SNP to reside within +/− 10Kb of an annotated gene body, we determined that 1,904 of 3,454 (55.1%) cancer GWAS SNPs mapped to 541 autosomal genes, which we will refer to as Cancer Susceptibility Genes or CSGs (Figure 3A, Supplementary Table 2). Interestingly, 10 out of the 67 p53 pathway genes (14.93%) are CSGs, namely: ATM, CHEK2, CASP8, CCND1, CCND2, CCNE1, CDKN2A, FAS, MDM4, and TP53. This 14.93% of p53 pathway genes represents a significant 6.77-fold enrichment compared to the 2.2% of 24,553 annotated autosomal genes that are CSGs (p-value: 2.00e-06, adjusted p-value: 4.39e-04, Figure 3B). In order to assess further the importance of the 6.77-fold enrichment of CSGs in the p53 pathway, we compared it to potential enrichment of CSGs in all 220 well-annotated pathways in the genome. Only 3 of these 220 cellular signaling pathways (1.36%), including the p53 pathway, demonstrated significant enrichments of CSGs after correction for multiple hypothesis testing (Figure 3C; see Analytical Procedures and Supplementary Table 3 for details). The 2 other significant pathways are PI3K-AKT and Adherens Junction. Like the p53 pathway, these are also known to be important pan-cancer signaling pathways (KEGG Cancer Signaling Pathways). However, the enrichment of CSGs in the p53 pathway ranks highest for both the level of significance and the fold enrichment (Figure 3C). Specifically, the PI3K-AKT and Adherens Junction pathways are associated with fold enrichments of 2.73 (p-value: 5.42e-05) and 5.11 (p-value: 1.66e-04), respectively, compared to a fold enrichment of 6.77 (p-value: 2.00e-06) for the p53 pathway. Together, the results of these analyses thus far suggest that both p53 pathway somatic, causal mutations and inherited cancer-associated SNPs occur in a high proportion of pathway genes relative to all annotated signaling pathways.
Figure 3. Cancer-associated SNPs occur in a high proportion of p53 pathway genes.
(A) A karyogram of the 541 genes which harbor at least 1 cancer GWAS SNP within 10Kb from their boundaries (Cancer Susceptibility Genes, CSGs). CSGs are noted in red. (B) A bar graph of the percent of CSGs in the p53 pathway compared to all annotated genes of the genome. 10 CSGs are found among the 67 genes of the p53 pathway (14.93%), which represents a significant 6.77-fold enrichment compared to the rest of autosomal genes in the genome (p-value: 2.00e-06 depicted in the figure, adjusted p-value: 4.39e-04). (C) A scatter plot showing the fold enrichment of CSGs on the x-axis (log-scale), and the adjusted p-value on the y-axis (-log10 scale). The horizontal line represents the 5% Family Wise Error Rate threshold (Bonferroni adjusted-p-value: 0.05), which is the pre-fixed significance threshold. The enrichment of CSGs in p53 pathway genes (in yellow) is the highest and the most significant compared to the other 220 annotated KEGG pathways (in blue). Overall, 1.36% pathways demonstrated significant enrichments of CSGs.
Expression quantitative trait loci (eQTL).
The fact that in the above analyses we required that one or more of the cancer GWAS SNPs reside within 10Kb of gene boundaries to define CSGs increases the likelihood that a SNP will be cis-acting and functionally affect its proximal gene. However, in order to gain more certainty that the SNPs can functionally affect the genes in which they reside, we further restricted our analyses to those SNPs, in and around genes, with genotypes that also associate with mRNA levels of their proximal genes in eQTL studies (cis-acting, cis-eSNPs). To do this, we began by curating data from 14 publicly available eQTL studies performed in non-cancerous tissues and cells from European descendants. These studies used 5 different cell types (lymphoblastoid cell lines (LCLs), CD4+ T-cells, primary monocyte samples, B-cells, and peripheral blood cells), as well as cells from 6 primary tissue types (adipose, skin, liver, intestine, heart, and lung). The median number of samples included in these studies is 659, (range from 129 to 1,490)38–52. We selected cis-eQTLs within +/− 10Kb of the gene boundaries of all 24,553 Refseq genes and identified a total of 412,962 unique cis-eSNPs, including 8,891 in B-cells, 71,242 in CD4+ T-cells, 2,130 in cardiac tissue, 4,844 in intestine 33,256 in adipose tissue, 265,671in LCLs, 2,664 in liver, 6,163 in lung, 133,425 in monocytes, 17,345 in peripheral blood cells and 26,417 in skin. Of these, 75.06% were found to be cis-eSNPs in a single tissue, 16.05% in two tissues, 6% in three tissues, and 2.89% in 4 or more tissues.
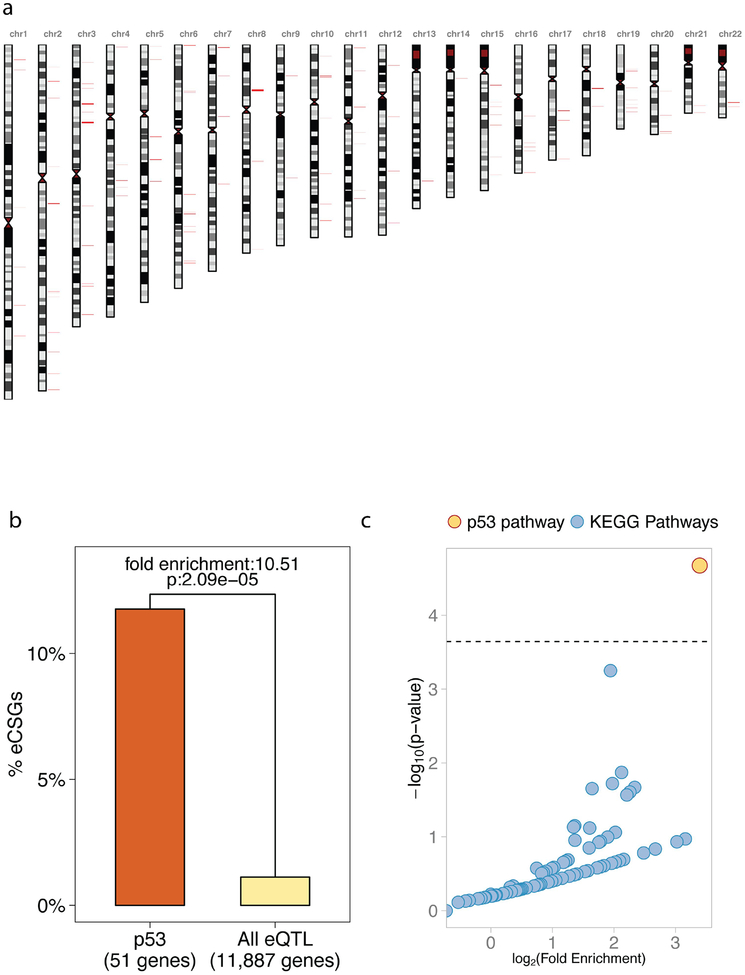
In total, we identified 11,887 genes genome-wide that harbored cis-eSNPs. In 133 of these genes (1.12%), we observed that the eSNPs also overlap (i.e. in complete LD) with cancer GWAS SNPs. Thus, these genes harbor haplotype blocks that associate with both differential cancer susceptibility and proximal gene expression in at least one tissue or cell type analyzed (eCSGs, Figure 4A, Supplementary Table 2). Interestingly, 6 of the eCSGs are among the 51 p53 pathway genes that harbor cis-eSNPs (11.76%): FAS, MDM4, ATM, CCND1, CASP8, and CCNE1. This represents a significant 10.51-fold enrichment compared to the 1.12% found in all 11,887 annotated genes that harbor at least one cis-eSNP (p= 2.09e-05, adjusted p= 4.59e-03, Figure 4B). Importantly, out of the 220 annotated cellular signaling pathways, the p53 pathway is the only pathway that shows a significant enrichment of eCSGs (Figure 4C, Supplementary Table 4); in fact, 61.8% of the remaining pathways have no eCSGs, and 94.1% have no more than 2. These results demonstrate that even when we restrict our analyses to SNPs that can associate with differential gene expression of their proximal genes, cancer-associated SNPs still occur in a high proportion of pathway genes relative to all annotated signaling pathways.
Figure 4. Cancer-associated eQTL occur in a high proportion of p53 pathway genes.
(A) A karyogram of the 133 genes which harbor an eSNP within 10Kb from their boundaries, which overlaps at least 1 cancer GWAS SNP (cis-eCSG). Cis-eCSGs are shown in red. (B) A bar graph of the percent of e-CSGs in the p53 pathway compared to all annotated genes of the genome. Six cis-eCSG are found among the 67 genes of the p53 pathway, which represents a significant 10.51-fold enrichment compare to the 11,887 genes with at least 1 eSNP (p= 2.09e-05 denoted in the graph, adjusted p= 4.59e-03). (C) A scatter plot showing the fold enrichment of e-CSGs on the x-axis (log-scale), and the adjusted p-value on the y-axis (-log10 scale). The horizontal line represents the 5% Family Wise Error Rate threshold (Bonferroni adjusted-p-value: 0.05), which is the prefixed significance threshold. The enrichment of cis-eCSG in the genes of the p53 pathway (in yellow) is the highest and the most significant compared to all the other KEGG pathways (in blue). Overall, 0.45% pathways demonstrated significant enrichment of cis-eCSG.
p53 Genes are not enriched in SNPs associated with other diseases.
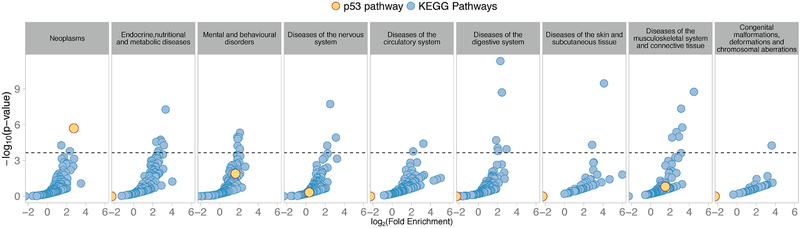
These results clearly lend support to the hypothesis that commonly inherited genetic variation in p53 pathway genes will affect cancer susceptibility to a greater extent than the variation found in genes of other pathways. However, the p53 stress response pathway has also been implicated in the pathogenesis of many other diseases which have been studied in GWAS, including neurological53–57, cardiovascular58,59 and infectious diseases60,61. Therefore, we next explored the potential impact of p53 pathway SNPs on susceptibility to non-cancerous disease. To do this, we took advantage of the 591 GWAS studies that have been carried out to measure the genetic basis of differences in susceptibility to other non-cancer diseases in Europeans. As mentioned above, 18 ICD10 disease categories, other than Neoplasms, had at least one susceptibility GWAS studied attributed to it (Supplementary Table 5). In the same manner described above for our analysis of the 165 cancer GWAS studies (ICD10 Category Neoplasms), we identified a set of genes for each of the other 18 disease categories in which at least one GWAS SNP was found to reside within +/− 10Kb of an annotated gene body (which we term Susceptibility Genes, SGs). All 18 disease categories had at least 4 SGs (median: 88.5, ranging from 4 to 708 SGs). We then explored any potential enrichment of SGs for each disease category in all 220 signaling pathways in the genome, including the p53 pathway. For 8 out of the 18 disease categories, we were able to identify an average of 4.25 signaling pathways that were significantly enriched for non-cancer SGs after correction for multiple hypothesis testing (range from 1 to 8 pathways, Figure 5, Supplementary Table 6). However, in contrast to our findings for cancer (ICD10 Category Neoplasms), the p53 signaling pathway was not significantly enriched in any of these 8 non-cancerous disease categories, which include diseases of the nervous, circulatory, digestive and musculoskeletal systems.
Figure 5. CSGs are significantly enriched in the p53 pathway genes, but not susceptibility genes for other major disease groupings.
Scatter plots showing fold enrichment of susceptibility genes (SGs) in KEGG pathways for the 9 ICD10 disease groups that had at least 1 pathway significantly enriched in SGs out of 19 ICD10 groups. The fold enrichment of SGs is on the x-axis (log-scale), and the adjusted p-value is on the y-axis (-log10 scale). The horizontal line represents the 5% Family Wise Error Rate threshold (Bonferroni adjusted p-value: 0.05), which is the pre-fixed significance threshold. The enrichment of p53 pathway SGs is shown in yellow; a significant enrichment is observed in cancer (Neoplasms), but not in any other disease groups. The y-axis represents the percentage of pathways that demonstrated significant enrichments of SGs for the given disease grouping.
The p53 pathway enrichment is consistent across pathway annotations.
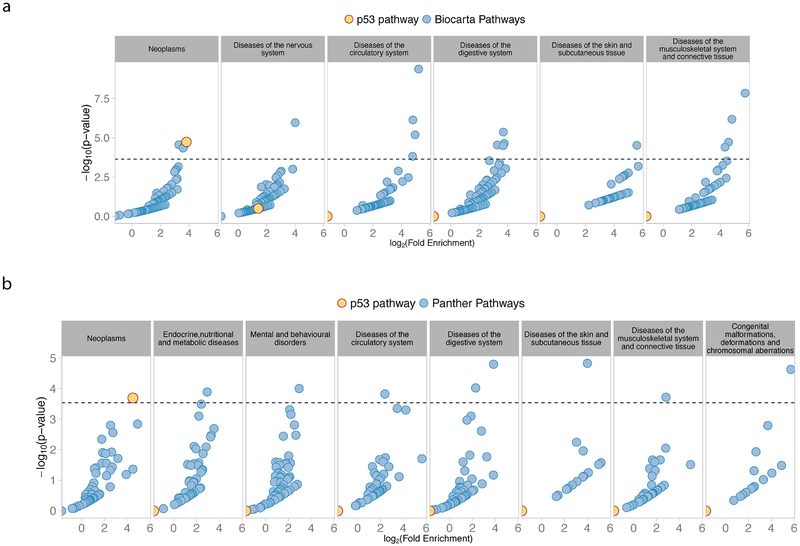
We have demonstrated that the autosomal genes (Figure 3) of the p53 pathway overlap with cancer GWAS loci to a greater extent than the genes of 219 other well-annotated signaling pathways, and that this enrichment is limited to cancer GWASs (Figure 5). Thus far, we have exclusively utilized KEGG pathway annotations for our analyses. In order to explore further the significance of our observations, we extended our analyses to pathway annotations from 2 different well-utilized, curated pathway databases, namely BioCarta (www.biocarta.com) and PANTHER (www.pantherdb.org/pathway/). Similar to the findings utilizing KEGG pathway annotation, the enrichment of CSGs in the p53 pathway ranks highest for the level of significance relative to all other pathways annotated by either BioCarta (Figure 6A, Supplementary Table 7) or PANTHER (Figure 6B, Supplementary Table 8). Also similar to the analyses conducted with KEGG annotation, when we explored the potential enrichment of p53 pathway genes among the susceptibility loci of the other 18 disease groupings defined above, we found no significant enrichment for the p53 signaling pathway annotated by either databases (Figure 6A and 6B, additional panels).
Figure 6. CSG enrichment in p53 pathway genes is not limited to KEGG Pathway Annotation.
Scatter plots showing the fold enrichment of susceptibility genes in Biocarta (A) and Panther (B) annotated pathways for ICD10 disease groups with at least 1 significant pathway. The fold enrichment of SGs is reported on the x-axis (log-scale), and the adjusted p-value on the y-axis (-log10 scale). The horizontal line represents the 5% Family Wise Error Rate threshold (Bonferroni adjusted-p-value: 0.05). The enrichment of SGs in the p53 pathway is observed in cancer (Neoplasms) with both pathway annotations, but never for other diseases. The y-axis represents the percentage of pathways that demonstrated significant enrichments of SGs for the given disease grouping.
Somatic mutations and cancer-associated SNPs occur in a high proportion of p53 pathway genes in multiple cancer types
Causal mutations.
As mentioned above, the ability of p53 to suppress tumor formation in numerous tissues has been demonstrated in numerous mouse models30,31,62. Indeed, the genes of the p53 pathway have been noted to be causally mutated in cancer genomes from all four major annotated tissue type groupings: epithelial, mesenchymal, leukaemia/lymphoma, and other (Cancer Gene Census, Sanger). In Figure 1, we have demonstrated that p53 pathway genes are enriched in genes known to harbor causal mutations in at least one of these four tissue types, whereby the enrichment noted places the p53 pathway amongst the top 8.18% of all annotated pathways of the genome (KEGG). Interestingly, we find similar significant enrichments when we restrict our analyses to causal mutations found in the different cancer types. We find that, in all four major cancer types, p53 pathway genes were enriched in causally mutated genes. (Figure 7A).
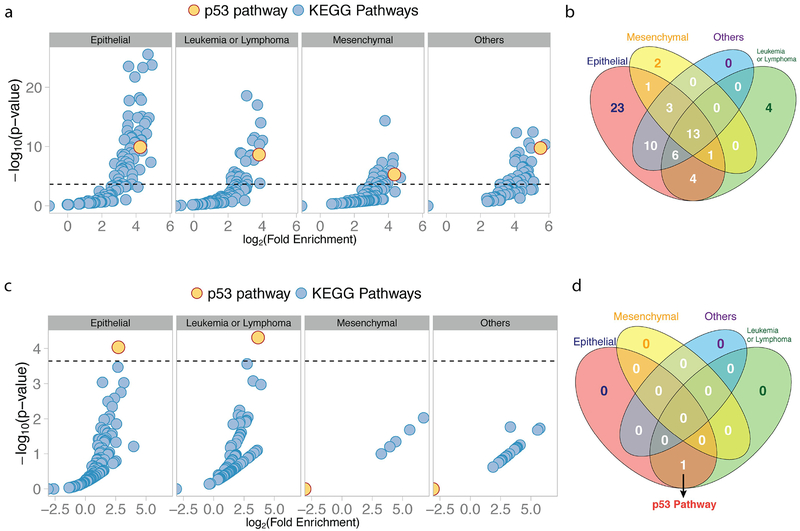
Figure 7. p53 pathway mutations and cancer-associated SNPs both occur in a high proportion of pathway genes in multiple cancer types.
(A) Scatter plots show the enrichment of genes with causal, somatic mutations in KEGG pathways grouped by cancer type. Fold enrichment of causally mutated genes is reported on the x-axis (log-scale), and the adjusted p-value on the y-axis (-log10 scale). The horizontal line represents the 5% Family Wise Error Rate threshold (Bonferroni adjusted-p-value: 0.05). The y-axis represents the percentage of pathways that demonstrated significant enrichments of SGs for the given cancer type. (B) A Venn Diagram showing the number of pathways with a significant enrichment of causally mutated genes across the four different types of cancer considered. (C) Analogously, a scatter plot shows the enrichment of CSGs in KEGG pathways grouped by cancer type, and a Venn Diagram (D) showing the number of pathways with a significant enrichment of CSGs grouped by cancer type. For all scatter plots the p53 pathway is in yellow.
In epithelial cancers, 14.93 % of p53 pathway genes can be causally mutated, which represents a 18.98-fold enrichment over the 0.79% causally mutated genes found in all 24,553 annotated autosomal genes (p-value: 1.20e-10, adjusted p-value: 2.64e-08, Figure 7A). 61 of the 220 cellular signaling pathways (27.7%), including the p53 pathway, demonstrated significant enrichments of causally mutated genes after correction for multiple hypothesis testing (Figure 7A; see Analytical Procedures and Supplementary Table 9 for details), thereby placing the enrichment noted in the p53 pathway amongst the top 13.18% of all pathways. Similar significant enrichments were found in leukemia/lymphomas (fold-enrichment: 14.09, p-value: 2.18e-09, adjusted p-value 4.79e-07), mesenchymal cancers (fold-enrichment: 20.36, p-value: 4.77e-06, adjusted p-value: 1.70e-10, adjusted p-value 1.05e-03) and cancers in the Other category (fold-enrichment: 45.80, p-value: 3.75E-8). As seen in Figure 7B, there are only a total of 13 signaling pathways (5.9%), including p53, significantly enriched in causally mutated genes shared by all four major cancer types. These pan-cancer signaling pathways included many other well-studied oncogenic and tumor suppressor pathways, such as the PI3K-AKT, RAS and MAPK signaling pathways (Supplementary Table 10).
Cancer-associated SNPs.
Given the observation that the p53 pathway belongs to the 5.9% of all 220 signaling pathways that are enriched in causally mutated genes in all four major cancer types, we next wanted to explore if similar observations can be found among the cancer-associated SNPs. In Figure 3, we have demonstrated that p53 pathway genes are enriched in genes overlapping CSLs (CSGs), whereby the noted 6.77-fold enrichment places the p53 pathway at the top of all 220 annotated signaling pathways for CSG enrichment (KEGG). However, we also find similar significant enrichments when we restrict our analyses to CSLs found in the individual cancer types.
In epithelial cancers, 10.45% of the 67 p53 pathway genes are CSGs, which represents a 6.59-fold enrichment over the 1.58% CSGs found in all 24,553 annotated autosomal genes (p-value: 9.12e-05, adjusted p-value: 2.01e-02, Figure 7C). Interestingly, only the p53 pathway demonstrated significant enrichments of CSGs after correction for multiple hypothesis testing (Figure 7C; see Analytical Procedures for details). A similar significant enrichment of CSGs in the p53 pathway was found in leukemias/lymphomas (Figure 7C, fold-enrichment: 12.63, p-value: 4.82e-05, adjusted p-value: 1.06e-02). No significant enrichments were noted for any of the 220 pathways in the mesenchymal cancers and cancers in the Other category. However, this is likely due to the relatively fewer studies performed in these cancer type groupings (Figure 2A). Importantly, and as seen in Figure 7D, only the genes of the p53 pathway are significantly enriched in CSGs in more than one cancer type. Together, these results clearly demonstrate that p53 pathway mutations and cancer-associated SNPs both occur in a high proportion of pathway genes in multiple cancer types relative to other cellular signaling pathways.
Somatic mutations and cancer-associated SNPs occur in similar p53 pathway genes
As mentioned above, out of the 67 autosomal genes attributed to the p53 pathway (KEGG), 15 genes have been denoted as harboring somatic, causal mutations in at least one cancer type (Cancer Gene Census, Sanger). In our analysis thus far, we determined that 9 of these genes (60%) are CSGs (Figure 8A): ATM, CASP8, CCND1, CCND2, CCNE1, CDKN2A, FAS, MDM4, and TP53 (Figure 8B, Cancer Gene Census, Sanger)1,31,63–80. This represents a significant 4.02 fold-enrichment compared to the 10 CSG (14.93%) found in all 67 p53 pathway genes (hyper-geometric test p-value: 1.06E-6, Figure 8C). This dramatic enrichment clearly demonstrates that genes in the p53 pathway known to harbor causal somatic mutations in cancer genomes are more likely to also harbor SNPs associated with differential cancer risk as measured in GWAS. However, it is important to note that this is not limited to the genes of the p53 pathway. Specifically, similar enrichments, albeit significantly smaller, can be found among causally mutated genes not in the p53 pathway. For example, when we restrict our analysis to the 478 causally mutated genes not attributed to the p53 pathway (KEGG), we do observe a significant enrichment of CSGs relative to the 24,486 non-p53 pathway genes of the genome, but to a lesser degree (fold enrichment: 3.09, hypergeometric test p-value: 2.12E-08, Figure 8D).
Figure 8. p53 pathway CSGs are frequently causally mutated in cancer.
(A) A Venn diagram depicting the overlap of p53 pathway genes that harbor causal mutations and pathway genes that are CSGs. (B) A pathway diagram of the CSGs annotated to the p53 pathway. (C). A bar graph depicting the percentage of CSGs found among those p53 pathway genes with known causal mutations in cancers and those pathway genes without known mutations. (D) A bar-plot depicting the enrichment of CSGs in genes known harbor causal somatic mutations relative to non-causally mutated genes in non-p53 pathway genes.
Cancer-associated p53 pathway SNPs are enriched in polymorphic regulatory elements for RNA processing
In this study, we have determined that 50 SNPs in 10 p53 pathway genes (KEGG) have been either directly or indirectly found to associate with differential cancer risk in GWAS studies (average of 5 SNPs per gene, ranging from 1 to 16, see Supplementary Table 11–12 for details). In contrast to causal somatic tumor mutations, cancer-associated SNPs are single nucleotide variations that, on average, cannot have negatively affected reproductive success, as they have risen to relatively high frequencies in the human population. This obvious difference between inherited and somatic genetic variation results in the vast majority of SNPs having weaker net effects on protein activity, and ultimately cancer development, relative to somatic mutations. These lower penetrant effects represent a major ongoing challenge in determining the molecular underpinnings of significant SNP associations found in GWASs, along with the fact that the responsible, causal SNP(s) are often linked with many nonfunctional SNPs2. However, for two of the 50 cancer-associated SNPs there is mounting experimental evidence that they reside in RNA processing regulatory elements.
Specifically, one SNP, in the 3’UTR of the TP53 gene (rs78378222, A/C) resides in a canonical polyadenylation signal sequence. This poly(A)-SNP was identified in a GWAS study for basal cell carcinoma, whereby the risk allele (C) is predicted to disrupt the poly(A) signal in the gene by changing AATAAA to AATACA81. Such a disruption of the poly(A) signal could impede cleavage of the nascent mRNA and addition of the poly(A) tail, ultimately resulting in less cellular p53 and potentially less p53 tumor suppression. Two recent studies have provided data supporting that the A to C change does result in aberrant 3’end processing81,82. The other regulatory SNP resides in a 5’splice site (donor) at the exon 4-intron 4 boundary in the CCND1 gene (rs9344, 870G>A). This 5’ splice site-SNP was found to associate with differential risk for t(11;14)(q13;q32) multiple myeloma in a GWAS study, whereby the G-allele, which creates the stronger 5’splice site (CCGgtaagt compared to CCAgtaagt), was found to associate with increased risk83. Alternative splicing of this exon and the potential role of this SNP in affecting the 5’ splice site was first reported over 20 years ago84. Indeed, multiple subsequent studies conducted in various cell types have demonstrated an association of the A-allele with less exon 4 splicing, resulting in the production of the cyclin D1b variant83,85,86. The functional influence of the this variant on cancer risk, relative to the cyclin D1a variant, remains to be further elucidated. However, this heavily studied SNP has consistently been found to associate with differential risk of many cancer types, whereby the A-allele has been found to associate with increased risk87.
Together, these data clearly demonstrate that 2 out of the 50 p53 pathway cancer GWAS SNPs reside in regulatory elements that affect differential RNA processing. If there is a causal relationship between the RNA processing SNPs and the noted differential cancer risk, we could expect similar RNA processing SNPs among the 16,890 p53 pathway SNPs to be significantly enriched in cancer GWAS SNPs. To test this, we determined the occurrence of similar poly(A) and 5’ splice site SNPs among all 16,890 SNP in and around all 67 p53 pathway genes (KEGG, Analytical Procedures). We identified only 2 additional SNPs that, like CCND1 rs9344, also reside in the exonic region of the 9-mer 5’splice site sequences and are predicted to demonstrate similar allelic differences in splice site recognition. The additional 2 SNPs reside in the CCNB2 and THBS1 pathway genes (Supplementary Table 13). Thus, of the 3 SNPs in the 5’splice sites of p53 pathway genes, 1 (33.3%) is a cancer GWAS SNPs (the abovementioned CCND1 rs9344, 870G>A). This represents a significant enrichment compared to both the 50 (0.29%) cancer GWAS SNPs found among the total 16,890 p53 pathway SNPs (fold-enrichment:112.6, hypergeometric test p-value: 0.0088) and the 3 (1.03%) cancer GWAS SNPs found among the 290 SNPs in coding exons (fold-enrichment: 32.22, hypergeometric test p-value: 0.030). We identified 4 SNPs in AAUAAA poly(A) sites in four different pathway genes (TP53, PPM1D, CCNG2, RRM2B, Supplementary Table 13). Of these 4 SNPs, only the one in TP53 (25%, rs78378222) is a cancer GWAS SNP; this represent a significant enrichment compared to both the 50 (0.29%) cancer GWAS SNPs found among the total 16,890 p53 pathway SNPs (fold-enrichment: 84.45, p-value: 0.011) and the 3 (0.54%) cancer GWAS SNPs found among the 555 SNPs found in 3’UTRs (fold-enrichment: 46.25, p-value: 0.021). It is important to note that no such significant enrichment was found among the missense coding SNPs, whereby out of the 143 missense SNPs identified amongst the p53 pathway SNPs, 2 are cancer GWAS SNPs (1.4%, hypergeometric test p-value: 0.07 when compared to all pathway SNPs). Together, these results support a causal relationship between these classes of RNA processing SNPs in p53 pathway genes and the noted differential cancer risk.
Discussion
Decades of research have clearly shown that genetic manipulation of p53 signaling can dramatically affect susceptibility to a broad range of cancers in mice and humans, and the topic is well-reviewed14,15,21,29,31,88. However, most evidence has been restricted to the characterization of rare inherited mutations found in families with LFS and common somatic mutations found in cancer genomes (Figure 1). In this analysis, we aimed to explore the possibility that commonly inherited genetic variants in the pathway also have a significant role in cancer susceptibility to a broad range of cancers. We took advantage of the wealth of data from studies mapping genome-wide susceptibility loci for a broad range of cancer types (Figure 2). Specifically, we took an integrated bioinformatics approach that linked SNPs and LD blocks from the 1000 Genomes Project to cancer GWAS SNPs and eQTLs in genes expressed in many different tissues. Our results demonstrate that p53 pathway genes are the more significantly enriched in cancer susceptibility loci compared to other signaling pathways, regardless of the annotation database (Figures 3 and 6). Indeed when we restricted our analyses to SNPs that reside in known cis-eQTLs, only the enrichment of the p53 pathway genes remained significant after multiple hypothesis correction (Figure 4). Moreover, we found that only the p53 pathway genes were significantly enriched in cancer susceptibility loci for different cancer types (Figure 7). Thus, the convergence of multiple lines of evidence, both genetic and functional, strongly suggests that the p53 signaling pathway is highly sensitive to inherited genetic variation, whether it is rare, highly penetrant mutations (LFS)15 or common, lower-penetrant variants (SNPs) studied in this Analysis, and that this sensitivity can contribute to the observed heterogeneity of cancer risk in the broader population. It is intriguing to speculate that the identified cancer GWAS SNPs in p53 pathway genes could help to define the heterogeneity of age-dependent and organ-dependent cancer risk seen among families with LFS, which remains a major hurdle in designing effective screening programs to reduce the substantial morbidity and mortality associated with a genetically weakened p53 pathway14. Moreover, in order to maximize the clinical utility of SNPs in the broader population, the known interactions of the p53 pathway members (Figure 8A) can serve as starting point to explore possible interactions among cancer GWAS SNPs, as well as possible interactions of SNPs with the somatic mutations frequently found in the identical genes.
The p53 stress response pathway has been implicated in the pathogenesis of many different diseases, including neurological53–57, cardiovascular58,59 and infectious diseases60,61. Indeed in our Analysis, we have found SNPs in p53 pathway genes that have overlapped susceptibility loci for other, non-cancerous diseases (Supplementary Table 14). However, we did not find p53 pathway genes to be significantly enriched in susceptibility loci for any other major disease groupings, other than cancer, for which sufficient GWAS studies were available to interrogate. These included diseases of the nervous, circulatory, digestive and musculoskeletal systems (Figures 5 and 6). Similar observations have been made in LFS families and mice carrying highly penetrant TP53 mutations, whereby carriers have a dramatically high risk of developing a broad range of cancers, but not other diseases14,89. A clearer picture will emerge with the accrual of more data on the genetic basis of human disease susceptibility. Indeed, a challenge of the GWAS design is the necessity for large patient cohorts to compensate for the much needed multiple hypothesis testing correction. Thus, rarer diseases and syndromes for which p53 (mis-)activity has been attributed to, have yet to be thoroughly examined90–93. However, the data generated in GWAS thus far suggest that genetic differences in the p53 pathway primarily affect susceptibility to cancer, rather than other major diseases such as Alzheimer’s disease, multiple sclerosis, coronary heart disease, type 2 diabetes and schizophrenia. As agents that modulate the levels of p53 signaling are entering the clinic29, such information could prove useful to help to predict and monitor potential side effects.
One of the most striking findings of our analyses is the strong similarities between the causal, somatic mutations and the inherited, cancer-associated SNPs of the p53 pathway. We have found that both classes of genetic variants occur in a high proportion of p53 pathway genes (Figures 1 and 3) in multiple cancer types (Figure 7), and in similar genes (Figure 8). Together, these observations suggest that certain genes in p53 signaling are highly sensitive to both heritable and somatic genetic variants resulting in differential tumor suppression in multiple tissue types. Indeed, for all of these genes their importance in cancer has been demonstrated in mouse models94–113. This group of genes encode important known regulators and effectors of p53-dependent stress signaling71,114–127. Indeed, it has been conclusively demonstrated that moderate alterations in expression levels of these genes through genome engineering of mice can lead to differences in cancer risk and progression97,128–133. Our results provide added human genetic evidence of their central roles in regulating or affecting p53 signaling and provide further evidence that targeting these genes and their protein products could prove an efficient method to modulate p53 signaling in a clinical setting. Our observations also suggest that the high frequency inherited genetic variants of these genes should be considered when designing and testing patient stratification strategies, which are currently under development and are aimed at predicting responsiveness to conventional and targeted cancer therapies29,31. The added information about the inherent differences in p53 signaling gained by these easily accessible and measurable biomarkers, could contribute to improving the efficacy of p53 pathway-based surveillance and treatment strategies, which has been lacking in the vast majority of cases29.
Supplementary Material
Acknowledgements
This work was funded in part by the Ludwig Institute for Cancer Research, the Nuffield Department of Medicine, the Development Fund-Oxford Cancer Research Centre-University of Oxford, and by the Intramural Research Program of National Institute of Environmental Health Sciences, National Institutes of Health (projects: Z01ES100475 and Z01ES46008). We would like to thank Joel S. Bader and Mary Muers for their critical reading of the manuscript.
References
- 1.Vogelstein B et al. Cancer genome landscapes. Science 339, 1546–1558, doi: 10.1126/science.1235122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edwards SL, Beesley J, French JD & Dunning AM Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet 93, 779–797, doi: 10.1016/j.ajhg.2013.10.012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manolio TA Bringing genome-wide association findings into clinical use. Nat Rev Genet 14, 549–558, doi: 10.1038/nrg3523 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Genomes Project C et al. A global reference for human genetic variation. Nature 526, 68–74, doi: 10.1038/nature15393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunham I et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74, doi: 10.1038/nature11247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schodel J et al. Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nat Genet 44, 420–425, S421–422, doi: 10.1038/ng.2204 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sur IK et al. Mice lacking a Myc enhancer that includes human SNP rs6983267 are resistant to intestinal tumors. Science 338, 1360–1363, doi: 10.1126/science.1228606 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Zeron-Medina J et al. A polymorphic p53 response element in KIT ligand influences cancer risk and has undergone natural selection. Cell 155, 410–422, doi: 10.1016/j.cell.2013.09.017 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cookson W, Liang L, Abecasis G, Moffatt M & Lathrop M Mapping complex disease traits with global gene expression. Nat Rev Genet 10, 184–194, doi: 10.1038/nrg2537 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stranger BE et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet 8, e1002639, doi: 10.1371/journal.pgen.1002639 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veyrieras JB et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet 4, e1000214, doi: 10.1371/journal.pgen.1000214 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nica AC et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet 6, e1000895, doi: 10.1371/journal.pgen.1000895 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicolae DL et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet 6, e1000888, doi: 10.1371/journal.pgen.1000888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McBride KA et al. Li-Fraumeni syndrome: cancer risk assessment and clinical management. Nat Rev Clin Oncol 11, 260–271, doi: 10.1038/nrclinonc.2014.41 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Merino D & Malkin D p53 and hereditary cancer. Subcell Biochem 85, 1–16, doi: 10.1007/978-94-017-9211-0_1 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Vazquez A, Bond EE, Levine AJ & Bond GL The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov 7, 979–987, doi: 10.1038/nrd2656 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Grochola LF, Zeron-Medina J, Meriaux S & Bond GL Single-nucleotide polymorphisms in the p53 signaling pathway. Cold Spring Harb Perspect Biol 2, a001032, doi:cshperspect.a001032 [pii] 10.1101/cshperspect.a001032 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whibley C, Pharoah PD & Hollstein M p53 polymorphisms: cancer implications. Nat Rev Cancer 9, 95–107 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Kandoth C et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339, doi: 10.1038/nature12634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leiserson MD et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet, doi: 10.1038/ng.3168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leroy B et al. The TP53 website: an integrative resource centre for the TP53 mutation database and TP53 mutant analysis. Nucleic acids research 41, D962–969, doi: 10.1093/nar/gks1033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068, doi: 10.1038/nature07385 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70, doi: 10.1038/nature11412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49, doi: 10.1038/nature12222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525, doi: 10.1038/nature11404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kandoth C et al. Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73, doi: 10.1038/nature12113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leiserson MD et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet 47, 106–114, doi: 10.1038/ng.3168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grochola LF, Zeron-Medina J, Meriaux S & Bond GL Single-nucleotide polymorphisms in the p53 signaling pathway. Cold Spring Harb Perspect Biol 2, a001032, doi: 10.1101/cshperspect.a001032 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khoo KH, Verma CS & Lane DP Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov 13, 217–236, doi: 10.1038/nrd4236 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Muller PA & Vousden KH Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25, 304–317, doi: 10.1016/j.ccr.2014.01.021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wade M, Li YC & Wahl GM MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 13, 83–96, doi: 10.1038/nrc3430 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ventura A et al. Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Xue W et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riley T, Sontag E, Chen P & Levine A Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9, 402–412 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Muller PA, Vousden KH & Norman JC p53 and its mutants in tumor cell migration and invasion. The Journal of cell biology 192, 209–218, doi: 10.1083/jcb.201009059 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller PA & Vousden KH p53 mutations in cancer. Nat Cell Biol 15, 2–8, doi: 10.1038/ncb2641 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Futreal PA et al. A census of human cancer genes. Nat Rev Cancer 4, 177–183, doi: 10.1038/nrc1299 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown CD, Mangravite LM & Engelhardt BE Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs. PLoS Genet 9, e1003649, doi: 10.1371/journal.pgen.1003649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fairfax BP et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet 44, 502–510, doi: 10.1038/ng.2205 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fehrmann RS et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet 7, e1002197, doi: 10.1371/journal.pgen.1002197 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaffney DJ et al. Dissecting the regulatory architecture of gene expression QTLs. Genome Biol 13, R7, doi: 10.1186/gb-2012-13-1-r7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grundberg E et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet 44, 1084–1089, doi: 10.1038/ng.2394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hao K et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet 8, e1003029, doi: 10.1371/journal.pgen.1003029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Innocenti F et al. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet 7, e1002078, doi: 10.1371/journal.pgen.1002078 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang L et al. A cross-platform analysis of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res 23, 716–726, doi: 10.1101/gr.142521.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schadt EE et al. Mapping the genetic architecture of gene expression in human liver. PLoS biology 6, e107, doi: 10.1371/journal.pbio.0060107 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeller T et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One 5, e10693, doi: 10.1371/journal.pone.0010693 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bryois J et al. Cis and trans effects of human genomic variants on gene expression. PLoS Genet 10, e1004461, doi: 10.1371/journal.pgen.1004461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fairfax BP et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343, 1246949, doi: 10.1126/science.1246949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kabakchiev B & Silverberg MS Expression quantitative trait loci analysis identifies associations between genotype and gene expression in human intestine. Gastroenterology 144, 1488–1496, 1496 e1481–1483, doi: 10.1053/j.gastro.2013.03.001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koopmann TT et al. Genome-wide identification of expression quantitative trait loci (eQTLs) in human heart. PLoS One 9, e97380, doi: 10.1371/journal.pone.0097380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raj T et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344, 519–523, doi: 10.1126/science.1249547 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alves da Costa C et al. Presenilin-dependent gamma-secretase-mediated control of p53-associated cell death in Alzheimer’s disease. J Neurosci 26, 6377–6385, doi: 10.1523/JNEUR0SCI.0651-06.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fogarty MP et al. A role for p53 in the beta-amyloid-mediated regulation of the lysosomal system. Neurobiol Aging 31, 1774–1786, doi: 10.1016/j.neurobiolaging.2008.09.018 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Perier C et al. Two molecular pathways initiate mitochondria-dependent dopaminergic neurodegeneration in experimental Parkinson’s disease. Proc Natl Acad Sci U S A 104, 8161–8166, doi: 10.1073/pnas.0609874104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.da Costa CA et al. Transcriptional repression of p53 by parkin and impairment by mutations associated with autosomal recessive juvenile Parkinson’s disease. Nat Cell Biol 11, 1370–1375, doi: 10.1038/ncb1981 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng Z et al. p53 tumor suppressor protein regulates the levels of huntingtin gene expression. Oncogene 25, 1–7 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Matsumoto S et al. Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ Res 113, 322–326, doi: 10.1161/CIRCRESAHA.113.301209 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Sano M et al. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature 446, 444–448, doi: 10.1038/nature05602 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Munoz-Fontela C et al. p53 serves as a host antiviral factor that enhances innate and adaptive immune responses to influenza A virus. J Immunol 187, 6428–6436, doi: 10.4049/jimmunol.1101459 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takaoka A et al. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 424, 516–523, doi: 10.1038/nature01850 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Garcia PB & Attardi LD Illuminating p53 function in cancer with genetically engineered mouse models. Semin Cell Dev Biol 27, 74–85, doi: 10.1016/j.semcdb.2013.12.014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guarini A et al. ATM gene alterations in chronic lymphocytic leukemia patients induce a distinct gene expression profile and predict disease progression. Haematologica 97, 47–55, doi: 10.3324/haematol.2011.049270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Renwick A et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet 38, 873–875, doi: 10.1038/ng1837 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Kim HS et al. Inactivating mutations of caspase-8 gene in colorectal carcinomas. Gastroenterology 125, 708–715 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Soung YH et al. Caspase-8 gene is frequently inactivated by the frameshift somatic mutation 1225_1226delTG in hepatocellular carcinomas. Oncogene 24, 141–147, doi: 10.1038/sj.onc.1208244 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Wiestner A et al. Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood 109, 4599–4606, doi: 10.1182/blood-2006-08-039859 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao YB et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet 46, 1097–1102, doi: 10.1038/ng.3076 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068, doi: 10.1038/nature07385 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sieuwerts AM et al. Which cyclin E prevails as prognostic marker for breast cancer? Results from a retrospective study involving 635 lymph node-negative breast cancer patients. Clinical Cancer Research 12, 3319–3328, doi: 10.1158/1078-0432.CCR-06-0225 (2006). [DOI] [PubMed] [Google Scholar]
- 71.Nakayama N et al. Gene amplification CCNE1 is related to poor survival and potential therapeutic target in ovarian cancer. Cancer 116, 2621–2634, doi: 10.1002/cncr.24987 (2010). [DOI] [PubMed] [Google Scholar]
- 72.Gronbaek K et al. Concurrent disruption of p16INK4a and the ARF-p53 pathway predicts poor prognosis in aggressive non-Hodgkin’s lymphoma. Leukemia 14, 1727–1735 (2000). [DOI] [PubMed] [Google Scholar]
- 73.Holzelova E et al. Autoimmune lymphoproliferative syndrome with somatic Fas mutations. N Engl J Med 351, 1409–1418, doi: 10.1056/NEJMoa040036 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Dowdell KC et al. Somatic FAS mutations are common in patients with genetically undefined autoimmune lymphoproliferative syndrome. Blood 115, 5164–5169, doi: 10.1182/blood-2010-01-263145 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park WS et al. Somatic mutations in the death domain of the Fas (Apo-1/CD95) gene in gastric cancer. J Pathol 193, 162–168 (2001). [DOI] [PubMed] [Google Scholar]
- 76.Gembarska A et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med 18, 1239–1247, doi: 10.1038/nm.2863 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Forbes SA et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic acids research 39, D945–950, doi: 10.1093/nar/gkq929 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dotsch V, Bernassola F, Coutandin D, Candi E & Melino G p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol 2, a004887, doi: 10.1101/cshperspect.a004887 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schuetz JM et al. BCL2 mutations in diffuse large B-cell lymphoma. Leukemia 26, 1383–1390, doi: 10.1038/leu.2011.378 (2012). [DOI] [PubMed] [Google Scholar]
- 80.Marchini S et al. DeltaNp63 expression is associated with poor survival in ovarian cancer. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO 19, 501–507, doi: 10.1093/annonc/mdm519 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Stacey SN et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet 43, 1098–1103, doi: 10.1038/ng.926 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Z et al. Further Confirmation of Germline Glioma Risk Variant rs78378222 in TP53 and Its Implication in Tumor Tissues via Integrative Analysis of TCGA Data. Hum Mutat 36, 684–688, doi: 10.1002/humu.22799 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weinhold N et al. The CCND1 c.870G>A polymorphism is a risk factor for t(11;14)(q13;q32) multiple myeloma. Nat Genet 45, 522–525, doi: 10.1038/ng.2583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Betticher DC et al. Alternate splicing produces a novel cyclin D1 transcript. Oncogene 11, 1005–1011 (1995). [PubMed] [Google Scholar]
- 85.Comstock CE et al. Cyclin D1 splice variants: polymorphism, risk, and isoform-specific regulation in prostate cancer. Clin Cancer Res 15, 5338–5349, doi: 10.1158/1078-0432.CCR-08-2865 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Olshavsky NA et al. Identification of ASF/SF2 as a critical, allele-specific effector of the cyclin D1b oncogene. Cancer Res 70, 3975–3984, doi: 10.1158/0008-5472.CAN-09-3468 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Knudsen KE, Diehl JA, Haiman CA & Knudsen ES Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene 25, 1620–1628, doi: 10.1038/sj.onc.1209371 (2006). [DOI] [PubMed] [Google Scholar]
- 88.Kruse JP & Gu W Modes of p53 regulation. Cell 137, 609–622, doi: 10.1016/j.cell.2009.04.050 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bieging KT & Attardi LD Deconstructing p53 transcriptional networks in tumor suppression. Trends in cell biology 22, 97–106, doi: 10.1016/j.tcb.2011.10.006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fumagalli S & Thomas G The role of p53 in ribosomopathies. Semin Hematol 48, 97–105, doi: 10.1053/j.seminhematol.2011.02.004 (2011). [DOI] [PubMed] [Google Scholar]
- 91.McGowan KA & Mason PJ Animal models of Diamond Blackfan anemia. Semin Hematol 48, 106–116, doi: 10.1053/j.seminhematol.2011.02.001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Van Nostrand JL & Attardi LD Guilty as CHARGED: p53’s expanding role in disease. Cell Cycle 13, 3798–3807, doi: 10.4161/15384101.2014.987627 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Van Nostrand JL et al. Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature 514, 228–232, doi: 10.1038/nature13585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Adachi M et al. Targeted mutation in the Fas gene causes hyperplasia in peripheral lymphoid organs and liver. Nature genetics 11, 294–300, doi: 10.1038/ng1195-294 (1995). [DOI] [PubMed] [Google Scholar]
- 95.Adachi M et al. Enhanced and accelerated lymphoproliferation in Fas-null mice. Proc Natl Acad Sci U S A 93, 2131–2136 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barlow C et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell 86, 159–171 (1996). [DOI] [PubMed] [Google Scholar]
- 97.Bieging KT, Mello SS & Attardi LD Unravelling mechanisms of p53-mediated tumour suppression. Nature reviews. Cancer 14, 359–370, doi: 10.1038/nrc3711 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen L et al. CD95 promotes tumour growth. Nature 465, 492–496, doi: 10.1038/nature09075 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Deane NG et al. Hepatocellular carcinoma results from chronic cyclin D1 overexpression in transgenic mice. Cancer Res 61, 5389–5395 (2001). [PubMed] [Google Scholar]
- 100.Geng Y et al. Kinase-independent function of cyclin E. Molecular cell 25, 127–139, doi: 10.1016/j.molcel.2006.11.029 (2007). [DOI] [PubMed] [Google Scholar]
- 101.Geng Y et al. Cyclin E ablation in the mouse. Cell 114, 431–443 (2003). [DOI] [PubMed] [Google Scholar]
- 102.Hakem A et al. Caspase-8 is essential for maintaining chromosomal stability and suppressing B-cell lymphomagenesis. Blood 119, 3495–3502, doi: 10.1182/blood-2011-07-367532 (2012). [DOI] [PubMed] [Google Scholar]
- 103.Jiang H et al. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev 23, 1895–1909, doi: 10.1101/gad.1815309 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kamijo T et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91, 649–659 (1997). [DOI] [PubMed] [Google Scholar]
- 105.Kwong LN, Weiss KR, Haigis KM & Dove WF Atm is a negative regulator of intestinal neoplasia. Oncogene 27, 1013–1018, doi: 10.1038/sj.onc.1210708 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu SC et al. Overexpression of cyclin D2 is associated with increased in vivo invasiveness of human squamous carcinoma cells. Mol Carcinog 34, 131–139, doi: 10.1002/mc.10057 (2002). [DOI] [PubMed] [Google Scholar]
- 107.Salmena L et al. Essential role for caspase 8 in T-cell homeostasis and T-cell-mediated immunity. Genes & development 17, 883–895, doi: 10.1101/gad.1063703 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Varfolomeev EE et al. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 9, 267–276 (1998). [DOI] [PubMed] [Google Scholar]
- 109.Xiong S et al. Spontaneous tumorigenesis in mice overexpressing the p53-negative regulator Mdm4. Cancer Res 70, 7148–7154, doi: 10.1158/0008-5472.CAN-10-1457 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bahassi el M et al. Mice with the CHEK2*1100delC SNP are predisposed to cancer with a strong gender bias. Proc Natl Acad Sci U S A 106, 17111–17116, doi: 10.1073/pnas.0909237106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hirao A et al. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol Cell Biol 22, 6521–6532 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kwak EL et al. Mammary tumorigenesis following transgenic expression of a dominant negative CHK2 mutant. Cancer Res 66, 1923–1928, doi: 10.1158/0008-5472.CAN-05-1237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Takai H et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. Embo J 21, 5195–5205 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Balint EE & Vousden KH Activation and activities of the p53 tumour suppressor protein. Br J Cancer 85, 1813–1823, doi: 10.1054/bjoc.2001.2128 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Feng Z et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res 67, 3043–3053, doi: 10.1158/0008-5472.CAN-06-4149 (2007). [DOI] [PubMed] [Google Scholar]
- 116.Harris SL & Levine AJ The p53 pathway: positive and negative feedback loops. Oncogene 24, 2899–2908, doi: 10.1038/sj.onc.1208615 (2005). [DOI] [PubMed] [Google Scholar]
- 117.Hermeking H & Benzinger A 14-3-3 proteins in cell cycle regulation. Semin Cancer Biol 16, 183–192, doi: 10.1016/j.semcancer.2006.03.002 (2006). [DOI] [PubMed] [Google Scholar]
- 118.Hofseth LJ, Hussain SP & Harris CC p53: 25 years after its discovery. Trends Pharmacol Sci 25, 177–181, doi: 10.1016/j.tips.2004.02.009 (2004). [DOI] [PubMed] [Google Scholar]
- 119.Levine AJ, Hu W & Feng Z The P53 pathway: what questions remain to be explored? Cell Death Differ 13, 1027–1036, doi: 10.1038/sj.cdd.4401910 (2006). [DOI] [PubMed] [Google Scholar]
- 120.Pietenpol JA & Stewart ZA Cell cycle checkpoint signaling: cell cycle arrest versus apoptosis. Toxicology 181–182, 475–481 (2002). [DOI] [PubMed] [Google Scholar]
- 121.Sherr CJ Divorcing ARF and p53: an unsettled case. Nat Rev Cancer 6, 663–673, doi: 10.1038/nrc1954 (2006). [DOI] [PubMed] [Google Scholar]
- 122.Taylor WR & Stark GR Regulation of the G2/M transition by p53. Oncogene 20, 1803–1815, doi: 10.1038/sj.onc.1204252 (2001). [DOI] [PubMed] [Google Scholar]
- 123.Tokino T & Nakamura Y The role of p53-target genes in human cancer. Crit Rev Oncol Hematol 33, 1–6 (2000). [DOI] [PubMed] [Google Scholar]
- 124.Budhram-Mahadeo V et al. p53 suppresses the activation of the Bcl-2 promoter by the Brn-3a POU family transcription factor. J Biol Chem 274, 15237–15244 (1999). [DOI] [PubMed] [Google Scholar]
- 125.Gaiddon C, Lokshin M, Ahn J, Zhang T & Prives C A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol 21, 1874–1887, doi: 10.1128/MCB.21.5.1874-1887.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mihara M et al. p53 has a direct apoptogenic role at the mitochondria. Molecular cell 11, 577–590 (2003). [DOI] [PubMed] [Google Scholar]
- 127.Hirao A et al. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287, 1824–1827 (2000). [DOI] [PubMed] [Google Scholar]
- 128.Donehower LA & Lozano G 20 years studying p53 functions in genetically engineered mice. Nat Rev Cancer 9, 831–841 (2009). [DOI] [PubMed] [Google Scholar]
- 129.Kamijo T, Bodner S, van de Kamp E, Randle DH & Sherr CJ Tumor spectrum in ARF-deficient mice. Cancer Res 59, 2217–2222 (1999). [PubMed] [Google Scholar]
- 130.Spring K et al. Mice heterozygous for mutation in Atm, the gene involved in ataxia-telangiectasia, have heightened susceptibility to cancer. Nature genetics 32, 185–190, doi: 10.1038/ng958 (2002). [DOI] [PubMed] [Google Scholar]
- 131.Spring K et al. Atm knock-in mice harboring an in-frame deletion corresponding to the human ATM 7636del9 common mutation exhibit a variant phenotype. Cancer Res 61, 4561–4568 (2001). [PubMed] [Google Scholar]
- 132.Wang YV, Leblanc M, Wade M, Jochemsen AG & Wahl GM Increased radioresistance and accelerated B cell lymphomas in mice with Mdmx mutations that prevent modifications by DNA-damage-activated kinases. Cancer cell 16, 33–43, doi: 10.1016/j.ccr.2009.05.008 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yamamoto K et al. Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice. The Journal of cell biology 198, 305–313, doi: 10.1083/jcb.201204098 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.