Abstract
A way to defeat antimicrobial resistance (AMR) crisis is to supply novel drugs to the pharmaceutical industry. This effort leads to a global call for seeking the beneficial microbes from underexplored habitats. To support this call, we isolated Streptomyces sp. TM32 from the rhizosphere soil of a medicinal plant, turmeric (Curcuma longa L.). TM32 exhibited strong antimicrobial activities against both human and plant pathogens, including an AMR pathogen, Staphylococcus haemolyticus MR‐CoNS. Surprisingly, such antimicrobial results of TM32's autoclaved crude extract remained the same. Based on the genome data analysis, TM32 belongs to the same genomic species with Streptomyces sioyaensis DSM 40032T, supported by the relatively high‐average nucleotide identity values (ANIb: 96.80% and OrthoANIu: 97.14%) and in silico DNA–DNA relatedness value of 75.40%. Importantly, the gene annotation analyses revealed that TM32's genome contains various genes encoding the biosynthesis of either known or unknown antibiotics and some metabolites involved in plant growth‐promoting traits. However, bioactivities and genome data comparison of TM32 and DSM 40032T showed a set of apparent differences, for example, antimicrobial potentials, genome size, number, and occurrence of coding DNA sequences in the chromosomes. These findings suggest that TM32 is a new strain of S. sioyaensis and serves as an emerging source for further discovery of valuable and novel bioactive compounds.
Keywords: antibiotics, antimicrobial resistance, bioactive compounds, genome mining, plant growth promotion, Streptomyces
Streptomyces sp. TM32 can produce thermotolerant antibiotics that inhibit growth of bacteria and fungi. There are many gene clusters in the genome of TM32, encoding unknown secondary metabolites and plant growth‐promoting traits. TM32 is a potential source for novel drug discovery.
1. INTRODUCTION
The inappropriate use of antibiotics in medication and livestock, the anthropogenic release of natural and chemical biocides in agriculture, and the discharge of wastewater effluent containing bioactive residues to the environment, these altogether demonstrate the overuse of antimicrobial substances, which is a significant factor causing the antimicrobial resistance (AMR) crisis (Fiorentino et al., 2019; Rodriguez‐Mozaz et al., 2015; Ventola, 2015). Moreover, existing medicines are less efficient in the treatment of infectious diseases and have limitation for further developments of their modes of action, while the antimicrobial evolution of pathogens occurs more rapidly (Piddock, 2012; Spellberg, Bartlett, & Gilbert, 2013). These critical concerns lead to a global call for encouraging the discovery of novel drugs that possess distinct molecular structures and offer broad‐spectrum functions, as a chance to defeat the AMR problem.
The best‐known natural sources for seeking bioactive compounds (eg, antibiotics, anticancer/antitumor agents, biocatalysts, etc.) are plants and microbes. For centuries, some herbs have served as natural medicines in the treatments of human diseases (Alvin, Miller, & Neilan, 2014; Strobel & Daisy, 2003). These plants synthesize pharmaceutical metabolites that may be stored into their cells and tissues or released in the form of root exudates. Some microbes can live within or in contact with bioactive phytochemicals (Alvin et al., 2014; Nakaew & Sungthong, 2018; Strobel & Daisy, 2003), while some of which also enable to form the same or similar metabolites to those produced by plants (Alvin et al., 2014; Strobel & Daisy, 2003). These plant‐associated niches are yet a challenging source for the discovery of new microbial candidates and their novel secondary metabolites.
With the industrial point of view, antibiotic‐producing microbes require less space for cultivation, shorter time for biosynthesis, and offer higher flexibility in further biotechnological developments. Among the microbial producers, actinobacteria are well‐known cell factories for drug discovery, especially for those belonging to the genus Streptomyces (Bentley et al., 2002; Tiwari & Gupta, 2012). More than two‐thirds of all existing natural antibiotics derive from this actinobacterial genus (Bentley et al., 2002). Streptomyces is a well‐explored genus, which consists of 848 species and 38 subspecies, referring to the list of prokaryotic names with standing in nomenclature (http://www.bacterio.net/streptomyces.html) during the time of writing this article. Regarding these high numbers of published Streptomyces members, the recent trend of drug discovery is more focusing on other microbial resources, such as non‐Streptomyces actinobacteria, so‐called “rare actinomycetes” with the aim to avoid the repetitive finding of the formerly found bioactive metabolites (Tiwari & Gupta, 2012). However, hitherto, it is just a minority of microbes discovered, while the majority of them is hidden in nature and waiting for exploration.
Streptomyces sp. TM32 was isolated based on the concept of plant–microbe interactions from the rhizosphere soil of turmeric (Curcuma longa L.), a rhizomatous and herbaceous plant, often used in cooking (Nakaew, Rangjaroen, & Sungthong, 2015). A preliminary identification of TM32 using its 16S rRNA gene sequence revealed the closest phylogenetic relation to Streptomyces sioyaensis DSM 40032T, a producer of a peptide thiazole antibiotic, siomycin A, possessing both antibacterial and anticancer functions (Gartel, 2013; Tori et al., 1979). TM32 showed strong antifungal activity and was proven for its plant growth‐promoting (PGP) potentials in the suppression of phytopathogenic fungus, Rigidoporus sp. (Nakaew et al., 2015).
In this study, we aim to report some interesting bioactivities and the draft genomes of the type strain, S. sioyaensis DSM 40032T, and our emerging strain, TM32. We also unveil the biotechnological treasures in both genomes by employing some bioinformatics tools and discuss the novelty of TM32 based on a set of comparative genome data.
2. MATERIALS AND METHODS
The antimicrobial activities of crude extracts derived from DSM 40032T (type strain) or TM32 were tested against either bacteria or fungi and some of which are pathogens of human and plant (Table 1). Each Streptomyces strain was grown in Hickey‐Tresner agar at 30°C for 7 days, which was transferred to grow further in 200 ml of International Streptomyces Project medium II (ISP2) broth under shaking conditions at 150 rpm, 30°C for 14 days (Nakaew et al., 2015). The whole liquid culture was extracted with ethyl acetate at a solute and solvent ratio of 1:2 (v/v) for one night, in which the separated supernatant was collected and evaporated at 45°C using a rotary evaporator (Büchi, Switzerland). The crude extracts obtained were divided into two proportions and one of which was heated by autoclaving at 121°C, 15 psi for 15 min.
Table 1.
Some antimicrobial activities of crude extracts derived from Streptomyces sioyaensis DSM 40032T and Streptomyces sp. TM32
| Test microorganism | DSM 40032T a | TM32a | ||
|---|---|---|---|---|
| Crude extract | Heated‐crude extract | Crude extract | Heated‐crude extract | |
| Gram‐positive bacteria | ||||
| Bacillus subtilis DMST 5871 | + (8.0 ± 0.0) | + (7.5 ± 0.7) | + (8.0 ± 0.0) | + (8.5 ± 0.7) |
| Staphylococcus aureus TISTR 1466 | + (9.5 ± 0.7) | + (10.0 ± 0.0) | + (9.5 ± 0.7) | + (10.0 ± 0.0) |
| Staphylococcus haemolyticus MR‐CoNS 102 | − | − | + (8.0 ± 0.0) | + (8.0 ± 0.0) |
| Gram‐negative bacteria | ||||
| Escherichia coli TISTR 887 | − | − | − | − |
| Phytopathogenic fungi | ||||
| Alternaria alternata TISTR 3435 | + (6.0 ± 0.0) | + (2.0 ± 0.0) | ++++ (37.5 ± 0.7) | +++ (29.0 ± 1.4) |
The antimicrobial activity was determined into five levels (excellent ++++, very good +++, good ++, fair +, and no activity −). The value in parenthesis refers to the average size (ø mm) ± standard deviation of the inhibitory zones derived from the duplicate antagonism assays.
Both nonheated and heated crude extracts were dissolved with dimethyl sulfoxide (DMSO) and tested for their antimicrobial activities by the paper disk‐diffusion method described elsewhere (Nakaew, Sungthong, Ortega‐Calvo, & Lumyong, 2012). The overnight culture of test bacteria in nutrient broth (Himedia, India) and 2‐day‐old culture of test fungi in potato dextrose broth (Himedia, India) were prepared by shaking incubation at 150 rpm and 30°C. The culture of each test microbe was swabbed over its corresponding agar medium mentioned before. Paper disks were pasted on the test agar medium, in which 30 μL of the crude extracts, 2 mg/ml nalidixic acid (positive antibacterial), 1 mg/ml cycloheximide (positive antifungal), or DMSO (negative control) was dropped. The antimicrobial activity was reported with the diameter size of the inhibitory zone appeared on the assayed plates.
The biomass of DSM 40032T or TM32 was prepared after cultivation in ISP2 broth under shaking conditions at 150 rpm, 30°C for 5 days. Total genomic DNA of both strains was extracted and used for sequencing with the Illumina HiSeq2500 system, following the services provided by BaseClear BV (Leiden, the Netherlands). The reads generated from the sequencing were assembled using SPAdes version 3.9 (Bankevich et al., 2012) and evaluated by QUAST version 3.2 (Gurevich, Saveliev, Vyahhi, & Tesler, 2013). The gene annotation analysis was carried out based on all contigs of each genome using Prokka (Seemann, 2014). The secondary metabolite‐related genes and gene clusters present in the genomes were predicted using antiSMASH 3.0 (Weber et al., 2015).
Identification of TM32 using its 16S rRNA gene sequence (GenBank accession no. KR534214) was performed using EzBioCloud database (https://www.ezbiocloud.net). The average nucleotide identity between the two genomes by BlastN (ANIb) and the orthologous average nucleotide identity by USEARCH (OrthoANIu) were computed in JSpeciesWS (Richter, Rosselló‐Móra, Oliver Glöckner, & Peplies, 2015) and EzBioCloud (Yoon, Ha, Lim, Kwon, & Chun, 2017), respectively. DNA–DNA relatedness between the two genomes was evaluated by in silico DNA–DNA hybridization (DDH) (Meier‐Kolthoff, Auch, Klenk, & Göker, 2013).
3. RESULTS
The crude extracts provided by ethyl acetate extraction of both Streptomyces strains did not show antimicrobial activity against the test Gram‐negative bacterium, Escherichia coli TISTR 887 (Table 1). Surprisingly, their heated crude extracts by autoclaving at 121°C remained antimicrobial functions. DSM 40032T exhibited less antifungal activity than TM32 and did not inhibit a methicillin‐resistant coagulase negative pathogen, Staphylococcus haemolyticus MR‐CoNS 102 (Seng et al., 2017). The antifungal potential of DSM 40032T's crude extracts was decreased by 67% when the extracts got heated before test, which was much higher compared to that of TM32 (23%). These findings highly prove that TM32 would serve as a promising source for further elucidations of valuable and novel antibiotics.
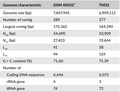
The sequencing generated 10,958,683 reads for DSM 40032T and 6,613,213 reads for TM32. The characteristics of both genomes are listed in Table 2. DSM 40032T has a genome size of 7,847,945 bp in 289 contigs with a G + C content of 71.60%, which is larger than the genome of TM32 (6,909,112 bp in 277 contigs with a G + C content of 71.39%). There are 6,646 coding DNA sequences (CDSs) predicted in the chromosome of DSM 40032T, including 74 tRNA and 4 rRNA genes, while TM32's chromosome contains 6,072 CDSs with 72 tRNA and 3 rRNA genes (Table 2).
Table 2.
Genome characteristics of Streptomyces sioyaensis DSM 40032T and Streptomyces sp. TM32
| Genome characteristic | DSM 40032T | TM32 |
|---|---|---|
| Genome size (bp) | 7,847,945 | 6,909,112 |
| Number of contig | 289 | 277 |
| Largest contig (bp) | 175,362 | 144,590 |
| N 50 (bp) | 54,690 | 33,909 |
| N 75 (bp) | 27,423 | 19,644 |
| L 50 | 41 | 58 |
| L 75 | 94 | 124 |
| G + C content (%) | 71.60 | 71.39 |
| Number of | ||
| Coding DNA sequence | 6,646 | 6,072 |
| rRNA gene | 4 | 3 |
| tRNA gene | 74 | 72 |
Some CDSs of both genomes have relevance in PGP traits, that is, nitrogen assimilation, phosphate solubilization, iron sequestration, and productions of biocatalysts and phytohormones (Table 3). TM32 possesses an extra chitinase gene (chiD), which is absent in the chromosome of DSM 40032T. This finding may result in the stronger antifungal activity of TM32 compared to DSM 40032T (Table 1). However, TM32 lacks the exo‐β‐1,3‐glucanase gene, which is present in DSM 40032T's chromosome. The absence of the glucanase gene was concordant to the phenotype of TM32 previously observed by an in vitro enzymatic assay (Nakaew et al., 2015).
Table 3.
Some plant growth‐promoting gene clusters in the chromosomes of Streptomyces sioyaensis DSM 40032T and Streptomyces sp. TM32
| Plant growth‐promoting trait | Relevant gene and product | COGa | DSM 40032T b | TM32b | |
|---|---|---|---|---|---|
| Nitrogen assimilation | nasD | Nitrite reductase | COG1251 | + | + |
| napA‐1 | Nitrate reductase | + | + | ||
| napA‐2 | Nitrate reductase | + | − | ||
| narK‐1 | Nitrate/nitrite transporter | COG2223 | + | + | |
| narK‐2 | Nitrate/nitrite transporter | COG2223 | + | − | |
| Phosphate solubilization | gpm2‐1 | Acid phosphatase | COG0406 | + | + |
| gpm2‐2 | Acid phosphatase | COG0406 | + | + | |
| phoD‐1 | Alkaline phosphatase D | COG3540 | + | + | |
| phoD‐2 | Alkaline phosphatase D | COG3540 | + | − | |
| phoP‐1 | Alkaline phosphatase synthesis transcriptional regulatory protein | COG0745 | + | + | |
| phoP‐2 | Alkaline phosphatase synthesis transcriptional regulatory protein | COG0745 | + | + | |
| phoP‐3 | Alkaline phosphatase synthesis transcriptional regulatory protein | COG0745 | + | − | |
| sphR | Alkaline phosphatase synthesis transcriptional regulatory protein | COG0745 | − | + | |
| ycdX | Phosphatase | + | + | ||
| Putative phosphatase | + | + | |||
| Putative phosphatase | COG0560 | + | − | ||
| Iron sequestration | sbnA | Putative siderophore biosynthesis protein | COG0031 | + | + |
| yusV | Putative siderophore transport system ATP‐binding protein | COG1120 | + | + | |
| yfhA | Putative siderophore transport system permease protein | COG0609 | + | + | |
| yfiY | Putative siderophore‐binding lipoprotein | COG0614 | + | + | |
| Biocatalyst | acdS | 1‐Aminocyclopropane‐1‐carboxylate deaminase | + | + | |
| aml | α‐Amylase | + | + | ||
| bca‐1 | Bromoperoxidase‐catalase | + | + | ||
| bca‐2 | Bromoperoxidase‐catalase | + | + | ||
| katA | Catalase | + | + | ||
| katG | Catalase‐peroxidase | + | + | ||
| cenA | Endoglucanase A (endo‐β‐1,4‐glucanase) | COG5297 | + | + | |
| Exo‐β‐1,3‐glucanase | + | − | |||
| Exochitinase 1 | + | − | |||
| chiA‐1 | Putative bifunctional chitinase/lysozyme | + | + | ||
| chiA‐2 | Putative bifunctional chitinase/lysozyme | − | + | ||
| chiC | Chitinase C | + | + | ||
| chiD | Chitinase D | − | + | ||
| Lipase | + | + | |||
| Lipase 2 | + | + | |||
| lip3‐1 | Lipase 3 | + | + | ||
| lip3‐2 | Lipase 3 | + | + | ||
| Thermostable monoacylglycerol lipase | + | + | |||
| snpA | Extracellular small neutral protease | + | + | ||
| htpX‐1 | Protease | COG0501 | + | + | |
| htpX‐2 | Protease | + | + | ||
| htpX‐3 | Protease | COG0501 | + | + | |
| htpX‐4 | Protease | + | + | ||
| htpX‐5 | Protease | + | − | ||
| prtS | Protease | + | + | ||
| Phytohormone production | trpC | Indole‐3‐glycerol phosphate synthase | COG0134 | + | + |
COG refers to “Clusters of Orthologous Groups” in the Prokka pipeline (Seemann, 2014).
The presence and absence of each gene cluster is determined by + and −, respectively.
DSM 40032T has a higher number (45) of secondary metabolite‐related gene clusters than TM32 (29 gene clusters) (Table 4). Both strains have a siomycin‐encoding gene cluster in their genomes, which is a unique characteristic of S. sioyaensis (Gartel, 2013; Tori et al., 1979). DSM 40032T possesses a lot of micromonolactam gene clusters (8), while TM32 does not have even one. The PGP‐related siderophore gene clusters are present in both genomes. Interestingly, there are many gene clusters of unknown secondary metabolites found in both genomes (indicated with <100% gene cluster similarity to the database), which should be elucidated later for their bioactivities, molecular structures, and biosynthetic pathways.
Table 4.
Some gene clusters encoding secondary metabolite biosynthesis in the chromosomes of Streptomyces sioyaensis DSM 40032T and Streptomyces sp. TM32
| Type of secondary metabolite | Most similar known cluster | MIBiG BGC‐IDa | DSM 40032T b | TM32b |
|---|---|---|---|---|
| Bacteriocin | + (3 clusters) | + (3 clusters) | ||
| Butyrolactone | Blasticidin | BGC0000874 | + (7%) | + (7%) |
| Coelimycin | BGC0000038 | + (12%) | − | |
| Unknown | + (1 cluster) | + (1 cluster) | ||
| Butyrolactone‐Phenazine‐other PKS | Esmeraldin | BGC0000935 | + (64%) | − |
| Ectoine | Ectoine | BGC0000853 | + (100%) | + (100%) |
| Ladderane | Colabomycin | BGC0000213 | + (4%) | + (4%) |
| Lanthipeptide | Kanamycin | BGC0000703 | − | + (1%) |
| Unknown | − | + (1 cluster) | ||
| Lassopeptide | Chaxapeptin | BGC0001307 | + (42%) | + (42%) |
| FD‐594 | BGC0000222 | − | + (4%) | |
| Linaridin | Legonaridin | BGC0001188 | + (55%) | + (55%) |
| Unknown | + (1 cluster) | − | ||
| Melanin | A‐500359s | BGC0000949 | + (5%) | − |
| Non‐ribosomal peptide synthase (NRPS) | A‐503083 | BGC0000288 | + (7%) | − |
| Lipstatin | BGC0000382 | + (28%) | + (28%) | |
| Neocarzilin | BGC0000111 | − | + (14%) | |
| Unknown | − | + (1 cluster) | ||
| Polyketide synthase (PKS) | ||||
| Type 1 PKS | Hedamycin | BGC0000233 | + (6%) | − |
| Maklamicin | BGC0001288 | + (13%) | − | |
| Micromonolactam | BGC0000095 | + (100%, 8 clusters) | − | |
| Phoslactomycin B | BGC0000123 | + (100%) | − | |
| Pladienolide | BGC0000126 | + (50%) | − | |
| PM100117/PM100118 | BGC0001359 | + (26%, 40%, 47%) | − | |
| Stambomycin | + (36%) | − | ||
| Unknown | BGC0000151 | + (1 cluster) | + (1 cluster) | |
| Vicenistatin | BGC0000167 | + (25%, 60%) | − | |
| Type 2 PKS | Lysolipin | BGC0000242 | − | + (50%) |
| Spore pigment | BGC0000271 | + (75%) | + (75%) | |
| Type 3 PKS | Naringenin | BGC0001310 | + (100%) | + (100%) |
| Betalactone‐NRPS‐Type 1 PKS | Lomaiviticin | BGC0000240 | − | + (3%) |
| Other PKS‐Type 1 PKS | Roseoflavin | BGC0000927 | + (100%) | − |
| Sanglifehrin A | BGC0001042 | − | + (4%) | |
| Unknown | + (1 cluster) | + (1 cluster) | ||
| Trans‐AT PKS | Dorrigocin/Migrastatin | BGC0000177 | + (100%) | − |
| Siderophore | Desferrioxamine B | BGC0000941 | + (80%) | + (80%) |
| Unknown | + (2 clusters) | + (2 clusters) | ||
| Terpene | Hopene | BGC0000663 | + (61%) | + (69%) |
| Isorenieratene | BGC0000664 | + (71%) | − | |
| Salinomycin | BGC0000144 | − | + (6%) | |
| Unknown | − | + (1 cluster) | ||
| Thiopeptide‐Terpene | Siomycin | BGC0000611 | + (100%) | + (100%) |
| Total | 45 clusters | 29 clusters | ||
MIBiG refers to “Minimum Information about a Biosynthetic Gene cluster” in the antiSMASH 3.0 (Weber et al., 2015).
The presence (+) or absence (−) of each gene cluster is determined, while the value indicated in the parenthesis is the similarity percentage to the known cluster or the number of cluster found.
Although the 16S rRNA gene sequence similarity of TM32 and DSM 40032T was 99.23%, this percentage is insufficient to determine the difference in genomic species. Therefore, the average nucleotide identity values (ANIb and OrthoANIu) between the genomes of TM32 and DSM 40032T were computed, whereas ANIb and OrthoANIu were 96.80% and 97.14%, respectively. Besides, DNA–DNA relatedness value between the two genomes evaluated by in silico DDH was 75.40%. Following the recent standards for using the genome data in the taxonomy of prokaryotes proposed by Chun et al. (2018), TM32 belongs to the same species with DSM 40032T. However, the apparent differences in bioactivities and genotypes of both strains would be sufficient to propose TM32 as a new subspecies of S. sioyaensis, in which a set of polyphasic approaches should be performed further to complete the taxonomy of TM32.
We believe that our proposing new strain of Streptomyces TM32 with its potentials to form thermotolerant antibiotics and genome insights could be a challenging cell factory that advances various biotechnological applications, for instance, novel drug discovery to defeat AMR crisis and green technology to optimize organic agriculture.
CONFLICT OF INTERESTS
The authors declare not to have any conflict of interest.
AUTHOR CONTRIBUTIONS
NN provided the bacteria, performed bioactivity tests, and prepared the manuscript. SL and WTS gave advices regarding the work. RS supervised the work, wrote and edited the manuscript.
ETHICS STATEMENT
None required.
ACKNOWLEDGMENTS
The authors acknowledge the research grants from the National Research Council of Thailand (NRCT Grant: R2560B070) and the Center of Excellence on Biodiversity – Office of Higher Education Commission (BDC‐PG2‐159010). We also thank Dr. Worarat Kruasuwan (BIOTEC – National Science and Technology Development Agency, Pathum Thani, Thailand) and Mr. Thanarat Salahong (Chiang Mai University) for their assistance in the gene annotation analysis. WTS and RS have financial support from the Engineering and Physical Sciences Research Council of the UK (EPSRC Grant: EP/K038885/1).
Nakaew N, Lumyong S, Sloan WT, Sungthong R. Bioactivities and genome insights of a thermotolerant antibiotics‐producing Streptomyces sp. TM32 reveal its potentials for novel drug discovery. MicrobiologyOpen. 2019;8:e842 10.1002/mbo3.842
DATA ACCESSIBILITY
These whole genome shotgun projects have been deposited at DDBJ/ENA/GenBank under the accessions SDIF00000000 (BioProject: PRJNA516206) and SDIG00000000 (BioProject: PRJNA516236) for Streptomyces sioyaensis DSM 40032T and Streptomyces sp. TM32, respectively. The versions described in this paper are SDIF01000000 and SDIG01000000, accordingly.
REFERENCES
- Alvin, A. , Miller, K. I. , & Neilan, B. A. (2014). Exploring the potential of endophytes from medicinal plants as sources of antimycobacterial compounds. Microbiological Research, 169, 483–495. 10.1016/j.micres.2013.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich, A. , Nurk, S. , Antipov, D. , Gurevich, A. A. , Dvorkin, M. , Kulikov, A. S. , … Pevzner, P. A. (2012). SPAdes: A new genome assembly algorithm and its applications to single‐cell sequencing. Journal of Computational Biology, 19, 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, S. D. , Chater, K. F. , Cerdeño‐Tárraga, A.‐M. , Challis, G. L. , Thomson, N. R. , James, K. D. , … Hopwood, D. A. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature, 417, 141–147. 10.1038/417141a [DOI] [PubMed] [Google Scholar]
- Chun, J. , Oren, A. , Ventosa, A. , Christensen, H. , Arahal, D. R. , da Costa, M. S. , … Trujillo, M. E. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. International Journal of Systematic and Evolutionary Microbiology, 68, 461–466. 10.1099/ijsem.0.002516 [DOI] [PubMed] [Google Scholar]
- Fiorentino, A. , Di Cesare, A. , Eckert, E. M. , Rizzo, L. , Fontaneto, D. , Yang, Y. , & Corno, G. (2019). Impact of industrial wastewater on the dynamics of antibiotic resistance genes in a full‐scale urban wastewater treatment plant. Science of the Total Environment, 646, 1204–1210. 10.1016/j.scitotenv.2018.07.370 [DOI] [PubMed] [Google Scholar]
- Gartel, A. L. (2013). Thiazole antibiotics siomycin A and thiostrepton inhibit the transcriptional activity of FOXM1. Frontiers in Oncology, 3 10.3389/fonc.2013.00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich, A. , Saveliev, V. , Vyahhi, N. , & Tesler, G. (2013). QUAST: Quality assessment tool for genome assemblies. Bioinformatics, 29, 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier‐Kolthoff, J. P. , Auch, A. F. , Klenk, H.‐P. , & Göker, M. (2013). Genome sequence‐based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics, 14, 60 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaew, N. , Rangjaroen, C. , & Sungthong, R. (2015). Utilization of rhizospheric Streptomyces for biological control of Rigidoporus sp. causing white root disease in rubber tree. European Journal of Plant Pathology, 142, 93–105. 10.1007/s10658-015-0592-0 [DOI] [Google Scholar]
- Nakaew, N. , & Sungthong, R. (2018). Seed phytochemicals shape the community structures of cultivable actinobacteria‐inhabiting plant interiors of Thai pigmented rice. MicrobiologyOpen, 7(4), e00591 10.1002/mbo3.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaew, N. , Sungthong, R. , Ortega‐Calvo, J. J. , & Lumyong, S. (2012). Antibiotic activity and anticancer potential of a new Nonomuraea sp. strain PT708 originated from Thai cave soil In Mendez‐Vilas A. (Ed.), Microbes in applied research: Current advances and challenges (pp. 474–780). Singapore: World Scientific Publishing; 10.1142/9789814405041_0096 [DOI] [Google Scholar]
- Piddock, L. J. (2012). The crisis of no new antibiotics—What is the way forward? The Lancet Infectious Diseases, 12, 249–253. 10.1016/S1473-3099(11)70316-4 [DOI] [PubMed] [Google Scholar]
- Richter, M. , Rosselló‐Móra, R. , Oliver Glöckner, F. , & Peplies, J. (2015). JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics, 32, 929–931. 10.1093/bioinformatics/btv681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Mozaz, S. , Chamorro, S. , Marti, E. , Huerta, B. , Gros, M. , Sànchez‐Melsió, A. , … Balcázar, J. L. (2015). Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Research, 69, 234–242. 10.1016/j.watres.2014.11.021 [DOI] [PubMed] [Google Scholar]
- Seemann, T. (2014). Prokka: Rapid prokaryotic genome annotation. Bioinformatics, 30, 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Seng, R. , Kitti, T. , Thummeepak, R. , Kongthai, P. , Leungtongkam, U. , Wannalerdsakun, S. , & Sitthisak, S. (2017). Biofilm formation of methicillin‐resistant coagulase negative staphylococci (MR‐CoNS) isolated from community and hospital environments. PLoS ONE, 12, e0184172 10.1371/journal.pone.0184172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellberg, B. , Bartlett, J. G. , & Gilbert, D. N. (2013). The future of antibiotics and resistance. New England Journal of Medicine, 368, 299–302. 10.1056/NEJMp1215093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel, G. , & Daisy, B. (2003). Bioprospecting for microbial endophytes and their natural products. Microbiology and Molecular Biology Reviews, 67, 491–502. 10.1128/MMBR.67.4.491-502.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, K. , & Gupta, R. K. (2012). Rare actinomycetes: A potential storehouse for novel antibiotics. Critical Reviews in Biotechnology, 32, 108–132. 10.3109/07388551.2011.562482 [DOI] [PubMed] [Google Scholar]
- Tori, K. , Tokura, K. , Yoshimura, Y. , Okabe, K. , Otsuka, H. , Inagaki, F. , & Miyazawa, T. (1979). 1H NMR spectral evidence for the structure and conformation of peptide antibiotic siomycin‐A. The Journal of Antibiotics, 32, 1072–1077. 10.7164/antibiotics.32.1072 [DOI] [PubMed] [Google Scholar]
- Ventola, C. L. (2015). The antibiotic resistance crisis: Part 1: Causes and threats. Pharmacy and Therapeutics, 40, 277. [PMC free article] [PubMed] [Google Scholar]
- Weber, T. , Blin, K. , Duddela, S. , Krug, D. , Kim, H. U. , Bruccoleri, R. , … Medema, M. H. (2015). antiSMASH 3.0–A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Research, 43, W237–W243. 10.1093/nar/gkv437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, S. H. , Ha, S. M. , Lim, J. M. , Kwon, S. J. , & Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek, 110, 1281–1286. 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
These whole genome shotgun projects have been deposited at DDBJ/ENA/GenBank under the accessions SDIF00000000 (BioProject: PRJNA516206) and SDIG00000000 (BioProject: PRJNA516236) for Streptomyces sioyaensis DSM 40032T and Streptomyces sp. TM32, respectively. The versions described in this paper are SDIF01000000 and SDIG01000000, accordingly.


