Abstract
Lactic acid bacteria produce a variety of antimicrobial peptides known as bacteriocins. Most bacteriocins are understood to kill sensitive bacteria through receptor‐mediated disruptions. Here, we report on the identification of the Lactobacillus plantarum plantaricin EF (PlnEF) receptor. Spontaneous PlnEF‐resistant mutants of the PlnEF‐indicator strain L. plantarum NCIMB 700965 (LP965) were isolated and confirmed to maintain cellular ATP levels in the presence of PlnEF. Genome comparisons resulted in the identification of a single mutated gene annotated as the membrane‐bound, magnesium/cobalt efflux protein CorC. All isolates contained a valine (V) at position 334 instead of a glycine (G) in a cysteine‐β‐synthase domain at the C‐terminal region of CorC. In silico template‐based modeling of this domain indicated that the mutation resides in a loop between two β‐strands. The relationship between PlnEF, CorC, and metal homeostasis was supported by the finding that PlnEF‐resistance was lost when PlnEF was applied together with high concentrations of Mg2+, Co2+, Zn2+, or Cu2+. Lastly, PlnEF sensitivity was increased upon heterologous expression of LP965 corC but not the G334V CorC mutant in the PlnEF‐resistant strain Lactobacillus casei BL23. These results show that PlnEF kills sensitive bacteria by targeting CorC.
Keywords: antimicrobial resistance, bacteriocins, lactic acid bacteria, Lactobacillus, probiotics
Small antibacterial peptides, known as bacteriocins, have applications in food preservation and as potential therapeutics for modulating the human microbiome. Here, we identified the bacterial receptor for plantaricin EF (PlnEF), a well‐characterized two peptide bacteriocin produced by strains of Lactobacillus plantarum, as CorC, a putative magnesium/cobalt efflux membrane protein. The sensitivity of L. plantarum strains to PlnEF was magnified when strains were grown in medium containing high concentrations of MgSO4, CoSO4, CuSO4, or ZnSO4.
1. INTRODUCTION
Lactic acid bacteria (LAB) produce a diverse array of bacteriocins. Bacteriocins are ribosomally synthesized peptides with bactericidal activity and are frequently most active against species that are highly related to the producer strains (Chikindas, Weeks, Drider, Chistyakov, & Dicks, 2018). Bacteriocins, and LAB bacteriocins in particular, have received considerable interest for their potential use in food preservation and pathogen inhibition (Cotter, Hill, & Ross, 2005). The most well‐known among these bacteriocins is nisin, a class I bacteriocin produced by Lactococcus lactis which has been approved as a food additive in both Europe and the US since the 1980s (Gharsallaoui, Oulahal, Joly, & Degraeve, 2016). More recently, numerous LAB bacteriocin biosynthetic genes were found in human gastrointestinal tract and vaginal microbiomes, extending the potential functions of bacteriocins beyond foods to human health (Walsh et al., 2015; Zheng, Gänzle, Lin, Ruan, & Sun, 2015). However, the specific host cell targets for many of these peptides have yet to be identified, thereby limiting the expansion of bacteriocin use and accurate predictions of antimicrobial activity.
Although some bacteriocins impair intracellular components of bacteria (Acedo, Chiorean, Vederas, & van Belkum, 2018), LAB bacteriocins typically target the cell surface to cause permeabilization and cell death (Perez, Zendo, & Sonomoto, 2014). Other broad spectrum bacteriocins, such as nisin, are indiscriminate in bacteriocidal activity and bind to cell membrane‐associated components shared across taxa. For example, nisin binds to lipid II, the final precursor in peptidoglycan synthesis (Breukink et al., 1999). Bacteriocins with a narrow inhibitory spectrum are known to affect species‐ or genus‐specific proteins (Kjos et al., 2014). Initial success at identification of such receptors employed comparative genomics of bacteriocin‐sensitive and spontaneous, bacteriocin‐resistant LAB strains (Kjos et al., 2014). In this way, the receptor for the pediocin‐like bacteriocin leucocin A (targeting a mannose‐specific phosphotransferase) (Ramnath, Beukes, Tamura, & Hastings, 2000), the leaderless bacteriocin enterocin K1 (targeting a stress response membrane‐bound Zn‐dependent protease) (Ovchinnikov et al., 2017), the two‐peptide bacteriocins lactococcin G (targeting undecaprenyl pyrophosphate phosphatase) (Kjos et al., 2014), and plantaricin JK (targeting an uncharacterized protein in the amino acid‐polyamine‐organocation APC transporter protein family) were identified (Ekblad, Nissen‐Meyer, & Kristensen, 2017; Oppegård, Kjos, Veening, Nissen‐Meyer, & Kristensen, 2016).
Lactobacillus plantarum is a well‐characterized LAB species and most strains produce several bacteriocins called plantaricins (Nissen‐Meyer, Oppegård, Rogne, Haugen, & Kristiansen, 2010). Plantaricin EF (PlnEF) was one of the first plantaricins to be isolated and characterized for its inhibitory spectrum against LAB (Anderssen, Diep, Nes, Eijsink, & Nissen‐Meyer, 1998). It is a member of the IIb class of bacteriocins, or bacteriocins that require two different peptides in equal quantities for full activity (Nissen‐Meyer et al., 2010). PlnEF consists of the 33 residue PlnE peptide and the 34 residue PlnF peptide. PlnE has a pair of amphiphilic α‐helices at both the N‐ and C‐terminal ends (Fimland, Rogne, Fimland, Nissen‐Meyer, & Kristiansen, 2008). PlnF has a single, central α‐helix that is polar at the N‐terminal end and amphiphilic at the C‐terminus (Fimland et al., 2008). PlnE contains two GXXXG motifs characteristic of class IIb bacteriocins, while PlnF contains a GXXXG‐like motif (SXXXG). Nuclear magnetic resonance analysis of PlnEF suggests these regions provide an interaction point between the peptides (Fimland et al., 2008). Assessment of the molecular orientation that these peptides adopt in artificial, cellular membrane‐mimicking micelles revealed the most likely conformation is an anti‐parallel coupling with the N‐terminus of PlnE pointed into the cellular membrane and the N‐terminus of PlnF closer to the extracellular environment (Kyriakou, Ekblad, Kristiansen, & Kaznessis, 2016).
In this study, we employed a forward genetics approach using genome comparisons to identify the PlnEF receptor in the sensitive strain L. plantarum NCIMB 700965 (LP965). This strain has been used as an indicator for plantaricin biosynthesis (Kjos, Snipen, Salehian, Nes, & Diep, 2010; Moll et al., 1999) and lacks a functional PlnEF‐immunity protein Plantaricin I. Isolation and characterization of PlnEF‐resistant mutants of LP965 led to the identification of CorC, a putative magnesium/cobalt efflux protein, as a target of PlnEF.
2. RESULTS
2.1. Selection of PlnEF‐resistant LP965 mutants
Plantaricin E (PlnE) and plantaricin F (PlnF) were synthesized and combined in equal molar ratios (PlnEF) prior to measuring for their inhibitory activity against LP965. The MIC50 of PlnEF against LP965 was 12.5 nM, and LP965 did not grow in the presence of PlnEF at concentrations above 50 nM when measured over a 24 hr period (data not shown).
Spontaneous, PlnEF‐resistant mutants of LP965 were enriched by exposing the strain to 15X‐70X the MIC50 (187–875 nM) of PlnEF. A total of five putative PlnEF‐resistant isolates were randomly selected and confirmed for PlnEF resistance using soft agar inhibition (data not shown) and growth assays (Figure 1). All five isolates (designated EF.A to EF.E) exhibited a fourfold increase in PlnEF MIC50 values (MIC50 >50 nM) compared to wild‐type LP965 (WT). Growth rates and maximum optical density (as measured by OD600) of all isolates were increased relative to the WT strain in the presence of 25 nM PlnEF (Figure 1). Conversely, the growth rates of the PlnEF‐resistant mutants did not significantly deviate from WT LP965 when incubated in MRS lacking PlnEF (ANCOVA of growth rates F(3, 148) = 0.06, p = 0.98). The final optical density (as measured by OD600) and growth rates (ANCOVA, F(3, 44) = 1.75, p = 0.17) of all strains was equally reduced in the presence of 1,000 nM of the unrelated bacteriocin plantaricin A (PlnA) (Figure 1) when compared to growth without PlnA, thereby showing that PlnEF resistance was specific and not the result of changes in general, stress‐related responses to antibacterial peptides.
Figure 1.
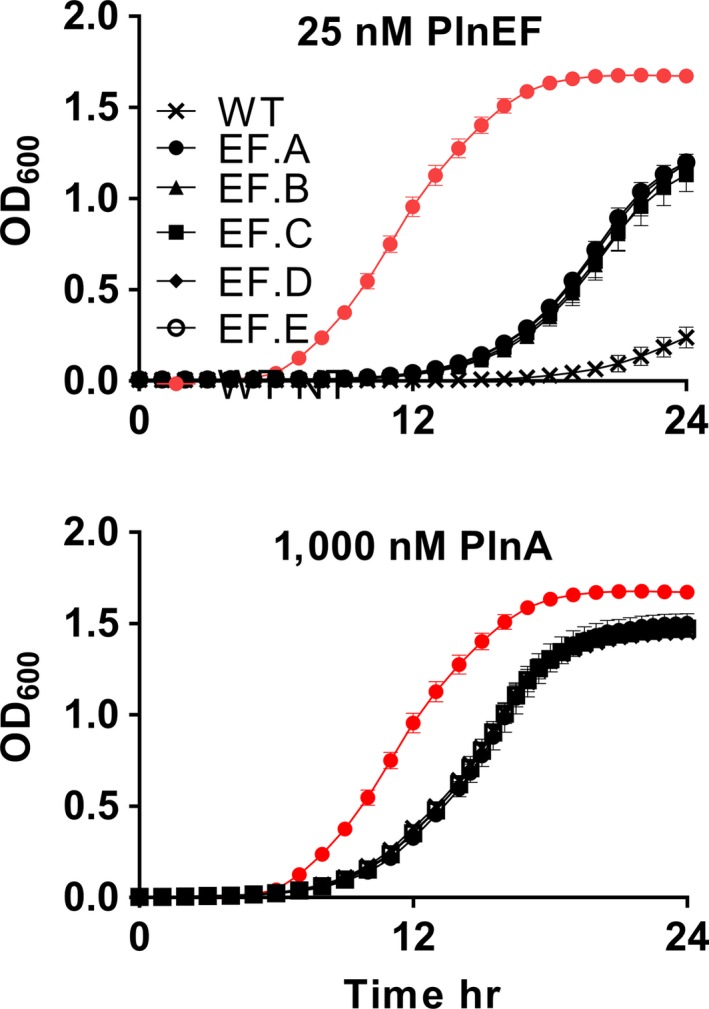
Spontaneous plantaricin EF (PlnEF) resistant mutants of LP965. Lactobacillus plantarum NCIMB 700965 (WT) and PlnEF‐resistant isolates were grown in MRS in the presence of 25 nM PlnEF or 1,000 nM PlnA. Red circles indicate LP965 growth in MRS without PlnEF. The avg ± SD of n = 3 replicates is shown
2.2. Effect of PlnEF on ATP concentrations
Because bacteriocins can disrupt energy metabolism in sensitive cells, intracellular ATP levels were measured to verify PlnEF resistance among the LP965 mutants. In the absence of PlnEF, incubation in 10 mM glucose resulted in increased intracellular ATP levels of all strains by five‐fold within 10 min (Figures 2 and A1). As expected, intracellular ATP concentrations of the WT strain declined upon exposure to 25 nM PlnEF (Figure 2), confirming the negative impact of PlnEF on cell viability. Conversely, intracellular ATP quantities of strains EF.A to EF.E increased until 20 min incubation (Figures 2 and A1), and only slightly declined thereafter. When compared to WT LP965, ATP levels of EF‐resistant strains were approximately 4‐fold higher after 40 min (p < 0.0001, Figures 2 and A1). The effect of PlnEF was specific to intracellular ATP quantities (Figures 2 and A2). There was no change in extracellular ATP concentrations after PlnEF challenge; whereas, all strains rapidly released ATP (approximately 20 nM) when challenged with a growth‐inhibiting concentration (25 µM) of nisin (p < 0.0001, Figures 2 and A2).
Figure 2.
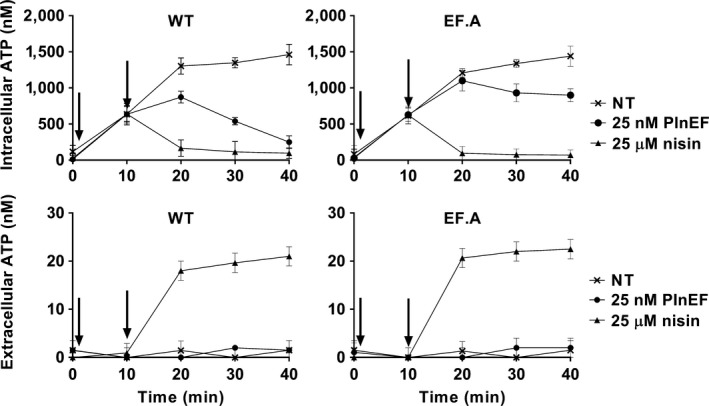
Effects of plantaricin EF (PlnEF) on cellular concentrations of ATP. Cells of LP965 and the PlnEF‐resistant isolate EF.A were initially energized with 10 mM glucose (arrow at 1 min). At 10 min (second arrow), 25 nM of PlnEF, 25 μM nisin, or water (NT) was added to each culture. The avg ± SD of n = 3 replicates is shown
2.3. PlnEF‐resistant isolates contain a mutation in CorC, a membrane‐bound, magnesium/cobalt efflux protein
A high‐confidence reference genome for LP965 was constructed using long (PacBio) and short (Illumina) read sequence data. The DNA sequence of the PlnEF‐resistant strain EF.A was also obtained (PacBio). Alignments between the LP965 and EF.A genomes revealed three chromosomal point mutations and five gaps (a result of putative indels). To rule out sequencing errors, the eight genomic regions with sequence variations were amplified by PCR and subjected to DNA sequencing (Table A1). Only one of the eight putative mutations was verified and this mutation was found to be localized in corC, a gene annotated as a putative membrane‐bound, magnesium/cobalt efflux protein. Compared to WT LP965, a guanine was changed to a thymine in EF.A at position 846,241. This single nucleotide change results in the substitution of a valine (V) instead of a glycine (G) at amino acid residue 334 (G334V) of CorC. The corC gene and 100 bp flanking DNA from the four other PlnEF‐resistant isolates (EF.B–EF.E) was amplified and sequenced. This showed that all five PlnEF‐resistant mutants shared the same corC point mutation (translated to G334V). The expression levels of corC were identical between WT LP965 and the PlnEF‐resistant isolates (p = 0.839), indicating the point mutation was the only factor mediating resistance to PlnEF.
2.4. EF.A CorC G334V mutation resides in a cysteine‐β‐synthase domain
The CorC protein is predicted to be a membrane‐bound protein containing four transmembrane domains (Figure A3). Protein annotation predicts two cysteine‐β‐synthase domains (CBS, also known as Bateman domains (Baykov, Tuominen, & Lahti, 2011)) and two transporter‐associated domains (PFAM03471, Figure A3). The CorC G334V mutation in strains EF.A–EF.E is localized in the second of the two CBS domains.
To elucidate a potential molecular basis for CorC interactions with PlnEF, structural modeling was performed for the WT LP965 CorC protein sequence region from Y216 to G351, encompassing the C‐terminal CBS. This was accomplished using the resolved crystal structure of the CorC protein produced by Oenococcus oeni (PDB ID:3OCO), which shares 41% amino acid identity to CorC in LP965 (Figure A4). Our Y216 to G351 model of LP965 CorC indicated that this region might be involved in a dimerization interface (Figure 3a), a result that is consistent with the model of 3OCO. The template‐based model also revealed that the G334V CorC mutation in PlnEF‐resistant strains of LP965 resides in a loop region between two β‐strands (Figure 3b,c).
Figure 3.
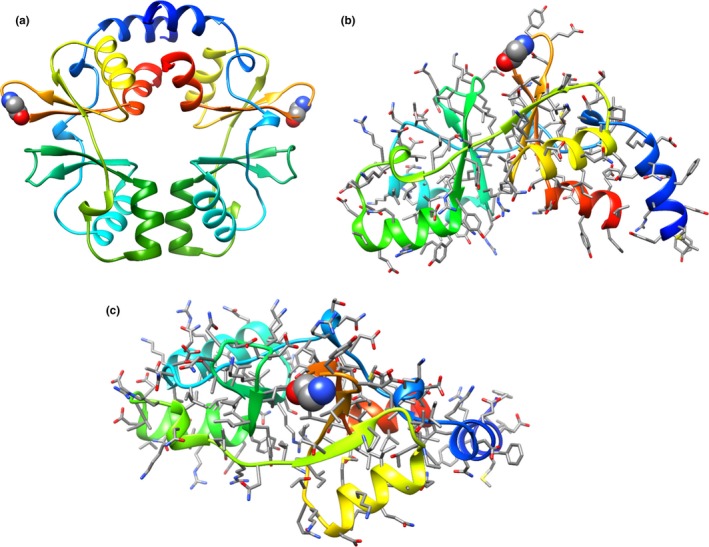
Structural model of the LP965 CorC protein. (a) Top‐down view of proposed homodimer formed by tandem CBS domains. (b) Top‐down and (c) side‐views of (monomer) LP965 CorC protein region from Y216 to G351 shown in ribbon representation and colored by a rainbow scheme from N‐terminus region (blue) to C‐terminus region (red). All sidechains are shown in stick representation, except for G334, which is shown in space‐filling representation. Template‐based modeling was accomplished using PDB: 3OCO
2.5. PlnEF increases the effects of metal stress
Because CorC is annotated as a putative magnesium/cobalt efflux protein, we sought to characterize whether the CorC G334V mutation altered LP965 sensitivity to different divalent metal cations. However, there were no significant differences in the growth rates or final optical densities between LP965 and the CorC mutant when incubated in MRS in the presence of growth‐inhibiting concentrations of MgSO4, CoSO4, C6H8FeNO7, CuSO4, ZnSO4, or MnSO4 (Figure A5).
Next, we investigated whether the CorC mutation altered L. plantarum growth in the presence of both PlnEF and high concentrations of metal cations. Growth of WT LP965 was completely inhibited in MRS containing additional MgSO4, CoSO4, C6H8FeNO7, CuSO4, ZnSO4, or MnSO4 and 25 nM of the PlnEF bacteriocin (Figure 4). Growth of EF.A was not affected by the inclusion of either C6H8FeNO7 or MnSO4 in MRS with 25 nM PlnEF (Figure 4). Surprisingly, however, MRS containing supplemental MgSO4, CoSO4, CuSO4, or ZnSO4, also resulted in significant impairments to the growth of the EF.A mutant when PlnEF was present (Figure 4). A dose dependency was found such that strain EF.A initiated growth by 20 hr in the presence of supplemental 250 mM MgSO4, but not 500 mM MgSO4 (Figure 4). These results show that PlnEF acts synergistically with metals to inhibit the growth of LP965 and that the G334V mutation is not sufficient to prevent cell damage at high concentrations of MgSO4, CoSO4, CuSO4, and ZnSO4.
Figure 4.
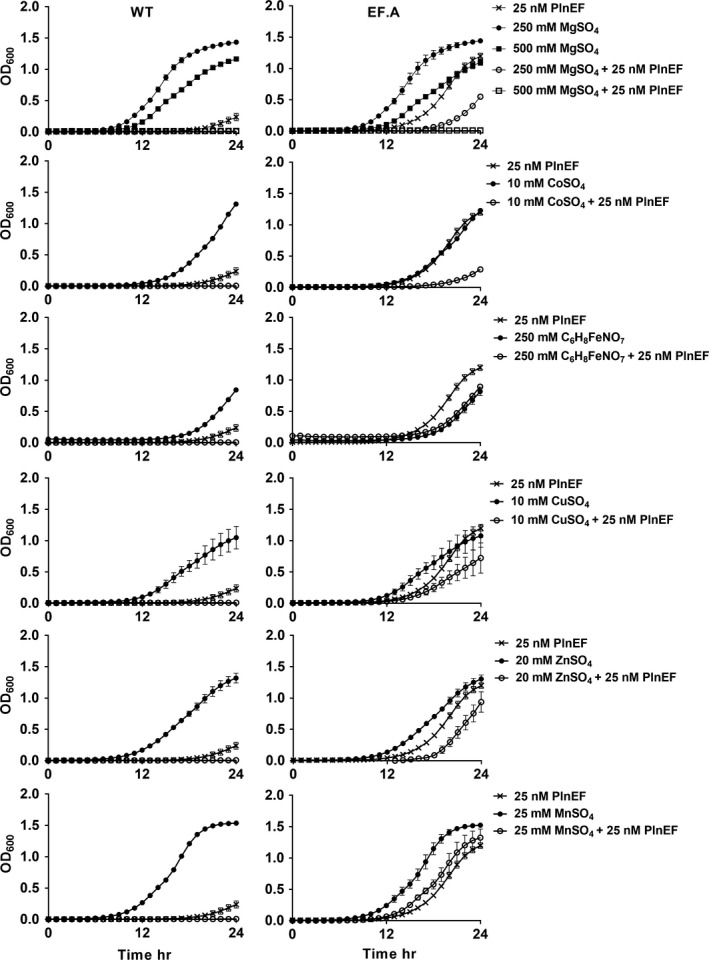
Additive effects of plantaricin EF (PlnEF) and metals on Lactobacillus plantarum growth. WT LP965 and the EF‐resistant strain EF.A were incubated in MRS supplemented with 25 nM PlnEF or MRS with 25 nM PlnEF and the indicated metal salt. The avg ± SD of n = 3 replicates is shown
2.6. Heterologous expression of wild‐type LP965 corC increases L. casei BL23 sensitivity to PlnEF
Numerous attempts to truncate or delete LP965 corC were unsuccessful using methods commonly applied for genetic modification of L. plantarum (Aukrust, Brurberg, & Nes, 1995). Efforts to delete or truncate corC in L. plantarum NCIMB 8,826 (LP8826) were also unsuccessful. Because LP8826 is amenable to genetic manipulation (Yin et al., 2017), we concluded that CorC is likely an essential protein for L. plantarum.
Therefore, to confirm that the G334V CorC mutation is required for PlnEF resistance, we introduced corC from WT LP965 and the EF.A mutant into L. casei strain BL23. Strain BL23 contains a homolog to CorC (59% amino acid identity), but is at least twice as resistant to PlnEF as LP965 (MIC50 = 25 nM). Expression of wild‐type LP965 corC in L. casei BL23 increased the sensitivity of that organism to PlnEF from 25 nM to 12.5 nM (Figure 5). Conversely, no increase in PlnEF sensitivity was found when the EF.A CorC G334V mutant was expressed (MIC50 = 25 nM) (Figure 5). The growth rates (ANCOVA, F(2, 134) = 0.03, p = 0.90) and final OD600 of BL23 or BL23 harboring either corC variant were equivalent in MRS lacking PlnEF. The expression levels of LP965 and EF.A corC in L. casei were also equivalent (p = 0.745). These results confirm that wild‐type LP965 corC increases sensitivity to PlnEF and that the single G334V amino acid substitution is sufficient to confer resistance.
Figure 5.
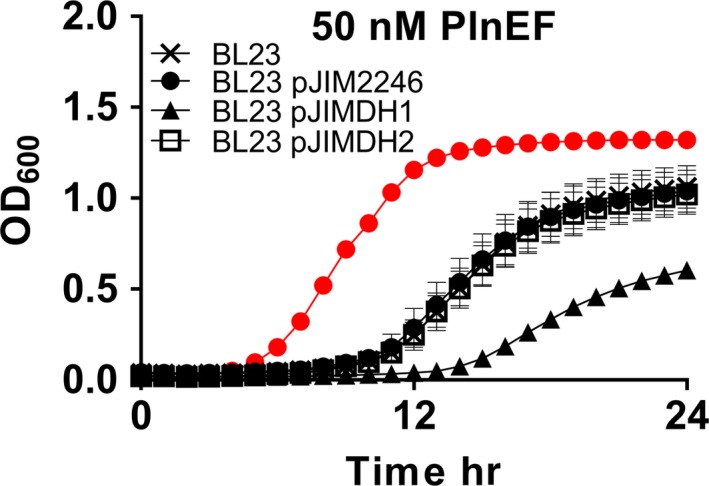
Heterologous expression of LP956 and EF.A CorC in L. casei BL23. L. casei was grown in MRS with 50 nM of plantaricin EF (PlnEF). pJIMDH1 contains corC from WT and pJIMDH2 contains CorC from strain EF.A. Red circles indicate BL23 growth in MRS without PlnEF. The avg ± SD of n = 3 replicates is shown
3. DISCUSSION
We identified CorC, a putative magnesium/cobalt exporter, as the receptor for the L. plantarum bacteriocin PlnEF. These findings are in agreement with previous studies on PlnEF showing that this bacteriocin causes cation efflux (Moll et al., 1999). We hypothesize that PlnEF anchors to CorC and either inserts into the lipid bilayer directly or, alternatively, PlnEF inserts through CorC to cause disrupted metal homeostasis. In either case, the activity of PlnEF is distinct from the L. plantarum bacteriocin Plantaricin JK which is known to induce anion efflux and for which the cell surface receptor was identified as a protein in the APC transporter family (Oppegård et al., 2016). The results are similarly consistent with prior work showing the bacterial receptor for PlnEF is different from lactococcin A and other class II pediocin‐like bacteriocins known to bind to proteins in the mannose phosphotransferase system (Diep, Skaugen, Salehian, Holo, & Nes, 2007). Our findings also conform with the expectation that there is a specific protein receptor for PlnEF which causes targeted damage to the cell, as opposed to receptors for broad‐spectrum bacteriocins like nisin which cause more pervasive effects on the cell membrane and the release of intracellular ATP (Breukink et al., 1999).
Because metal concentrations vary significantly throughout microenvironments, bacteria contain dedicated proteins for the import of essential metals and export of excess/toxic metals (Barwinska‐Sendra & Waldron, 2017). The L. plantarum reference strain WCFS1 contains 42 annotated metal cation transporters (Kleerebezem et al., 2003). L. plantarum strains have long been known to have a relatively high requirement for manganese (MacLeod & Snell, 1947), and the best understood metal transport systems in this species are those with affinity for Mn2+ (Groot et al., 2005). Presently, transport systems for magnesium and the trace metals Zn2+, Co2+, Cu2+ and Fe2+ have not been well characterized in L. plantarum. In other bacteria, magnesium import, in particular, has been extensively studied and linked to four classes of transporters (Groisman et al., 2013; Moomaw & Maguire, 2008; Shin et al., 2014). CorA, the most widely distributed of these transporters, has been shown to maintain both Mg2+ and Co2+ homeostasis in L. lactis (Mills et al., 2005). Some bacteria also carry systems for Mg2+ export, although this function is not as well understood. Putative Mg2+ efflux proteins have been identified as CorB, CorC, and CorD in Salmonella (Gibson, Bagga, Miller, & Maguire, 1991) and Shigella (Zhang, Ren, Zhu, Li, & Wang, 2010) and YhdP in Bacillus (Akanuma et al., 2014). These proteins might also export other metals such as Zn2+, as indicated for CorB and CorC in Pseudomonas stutzeri (Vaccaro et al., 2016).
CBS domains are frequently found in two to four tandem copies in both cytosolic‐ and membrane‐associated proteins and are present in proteins from all domains of life (Baykov et al., 2011). In eukaryotes, the cyclin M (CNNM) family of proteins mediate Mg2+ transport and share some structural similarities to bacterial CorC and CorB including the C‐terminal CBS pairs (Hirata, Funato, Takano, & Miki, 2014). The CBS domains of magnesium transporters are reported to be important for magnesium and ATP binding (Armitano, Redder, Guimarães, & Linder, 2016; Baykov et al., 2011). Recently, a Staphylococcus aureus protein with a domain structure similar to Salmonella CorB (magnesium protection factor A (MpfA); SA00657) was found to be essential for S. aureus growth in magnesium concentrations as low as 10 mM (Armitano et al., 2016). Point mutations in a conserved glycine residue at position 326 in an intracellular CBS domain of MpfA resulted in increased Mg2+ sensitivity. The LP965 CorC G334V mutation is also in a CBS domain, however, because the growth rates of the CorC mutant EF.A and WT LP965 were equivalent in MRS with high concentrations of MgSO4, our data suggest that the glycine residue at position 334 does not participate in metal binding. Instead of binding metals, the G334V mutation could result in steric inhibition altering the target site of PlnEF. Therefore it was notable the EF.A CorC mutation was not sufficient to sustain resistance to PlnEF in the presence of high levels of either MgSO4 or the trace metals CoSO4, CuSO4, and ZnSO4. This increase in PlnEF sensitivity could be due to changes in CorC conformation induced by external metal concentrations which result in increased bacteriocin binding. Such a possibility is supported by experiments visualizing CorC from Shigella flexneri altering conformations in response to ATP binding (Zhang et al., 2010). The magnesium import protein CorA has also been shown to change conformational states in response to magnesium binding (Matthies et al., 2016), thereby indicating a dynamic process.
PlnEF‐resistant mutants were only incrementally (4X) more resistant to the bacteriocin than WT LP965. By comparison, lactococcin G‐resistant mutants of L. lactis had MIC50 values at least 1,000 to 10,000 times greater than the wild‐type strain (Kjos et al., 2014). This difference could be due to the fact that those mutations were predicted to encode truncated forms of the Upp receptor protein (Kjos et al., 2014); whereas only a single amino acid substitution in CorC was found among the five LP965 PlnEF‐resistant mutants examined here. Heterologous expression of the LP965 and EF.A CorC proteins in L. casei also confirmed the importance of the single G334V point mutation in conferring PlnEF resistance. Because we did not find other mutants and because we were unable to construct a CorC deletion mutant in either LP965 or LP8826, our results indicate that similar to S. aureus MpfA (Armitano et al., 2016), CorC is essential for maintaining magnesium metal homeostasis in L. plantarum.
Determining how bacteriocins exert antimicrobial activity is important for elucidating microbe‐microbe interactions and application potential in food and intestinal ecosystems. Identification of bacteriocin cellular receptors will advance our understanding of the inhibitory spectra, functional significance, and resistance mechanisms associated with the myriad of bacteriocins currently known. We recently demonstrated that the PlnEF system is important for L. plantarum mediated protection against diet induced obesity in mice (Heeney et al., 2018). The gut microbiota and colonic metabolomes were not altered with L. plantarum consumption. Instead, the L. plantarum plantaricin system was correlated with increased production of the tight junction protein ZO‐1 in the intestinal epithelium. Application of the purified peptides on differentiated Caco‐2 monolayers also prevented disruptions to epithelial barrier integrity caused by proinflammatory cytokines (Heeney et al., 2018), thereby indicating direct interactions of the bacteriocin with epithelial cells. Identification of the PlnEF‐binding sites should clarify the exact mechanisms for which the bacteriocin alters metal homeostasis and supports the identification of other molecular targets for these peptides in the mammalian intestine.
4. EXPERIMENTAL PROCEDURES
4.1. Bacterial strains and growth conditions
Strains and plasmids used in this study are listed in Table 1. Lactobacillus strains were grown in Lactobacilli MRS (MRS, BD Biosystems) broth. LP965 strains were grown without aeration at 30°C. L. casei BL23 and L. plantarum NCIMB 8,826 (LP8826) were grown without aeration at 37°C. Escherichia coli DH5α was grown in Lennox lysogeny broth (Teknova) with aeration (250 rpm) at 37°C. When appropriate, 5 or 100 µg/ml chloramphenicol (CmR) and 5 or 300 µg/ml erythromycin (EryR) (Sigma‐Aldrich) was included in the media for Lactobacillus and E. coli, respectively. To test the effects of plantaricin A (PlnA) on L. plantarum growth, PlnA was added to MRS broth at a final concentration of 1,000 nM. This concentration was selected because it resulted in some inhibition, reducing the final optical density of LP965 compared to when grown in MRS without bacteriocin. To measure sensitivity to metal cations, sterile solutions of anhydrous MgSO4, CoSO4.7H2O, ZnSO4.7H2O, CuSO4.5H2O, MnSO4.H2O, or ferric ammonium citrate (C6H8FeNO7) were added to MRS to reach final supplemental concentrations of 500 mM or 1 M MgSO4, 10 mM or 20 mM CoSO4.7H2O, 20 mM ZnSO4.7H2O, 10 mM CuSO4.5H2O, 25 mM MnSO4.H2O, and 500 mM or 1 M C6H8FeNO7. The concentrations were selected to test the range of LP965 sensitivity to those metal salts.
Table 1.
Strains and plasmids used in this study
| Description | Reference | |
|---|---|---|
| Strains | ||
| L. plantarum NCIMB 700,965 | PlnEF‐sensitive strain | (Sherwood, 1939) |
| L. plantarum NCIMB 8,826 | PlnEF‐producing strain, parent of WCFS1 | (Hayward & Davis, 1956) |
| L. casei BL23 | PlnEF‐resistant reference strain | (Maze et al., 2010) |
| E. coli DH5α | fhuA2 lac(del)U169 phoA glnV44 Φ80' lacZ(del)M15 gyrA96 recA1 relA1 endA1 thi−1 hsdR17, amplification of cloning vector | (Taylor, Walker, & McInnes, 1993) |
| Plasmids | ||
| pJIM2246 | CmR, low‐copy cloning vector | (Renault et al., 1996) |
| pJIMDH1 | pJIM2246 derivative containing strain 965 corC | This work |
| pJIMDH2 | pJIM2246 derivative containing strain EF.A corC | This work |
| pRV300 | EryR AmpR, E. coli Ori pMB1, integrative vector | (Leloup, Ehrlich, Zagorec, & Morel‐Deville, 1997) |
| pRVDH1 | pRV300 derivative used for deletion of corC | This work |
4.2. Bacteriocin peptide synthesis
The full‐length peptide sequences of plantaricins taken from the published genome of L. plantarum WCFS1 (Refseq: NC_004567.2) were downloaded from NCBI and trimmed to delete export‐signal peptide sequences. Leaderless forms of plantaricin E (FNRGGYNFGKSVRHVVDAIGSVAGIRGILKSIR), and plantaricin A (PlnA) (KSSAYSLQMGATAIKQVKKLFKKWGW) were chemically synthesized by Genscript. Plantaricin F (VFHAYSARGVRNNYKSAVGPADWVISAVRGFIHG) was synthesized by Thermo‐Fisher. Peptides were 98%–99% pure and diluted in ultra‐pure, molecular grade water (Ambion) prior to being stored at −20°C until use.
4.3. Bacteriocin activity assays
The antimicrobial activity of PlnEF against target cells was tested as previously described (Kjos et al., 2014) using a spectrophotometer to monitor optical density (Synergy 2, Biotek instruments). The minimum inhibitory concentration (MIC50) was defined as the peptide concentration (the sum of both peptides [in a 1:1 ratio]) that inhibited growth by 50% after 6 hr incubation.
4.4. Isolation of PlnEF‐resistant mutants
LP965 PlnEF‐resistant strains were selected as previously described (Kjos et al., 2014). Five wild‐type (WT) LP965 colonies were inoculated into separate 10 ml volumes of MRS broth containing 70X the MIC50 of PlnEF (875 nM) and incubated at 30°C for 72 hr. Aliquots of 100 µl from each culture were then plated onto MRS agar plates containing 15X the MIC50 of PlnEF (187 nM) and incubated for 48 hr. One putative resistant colony from each plate (out of a total of approximately 30 colonies across all plates) was then incubated in MRS broth for 24 hr. Overnight cultures were diluted in phosphate buffered saline (Corning) (PBS) and then 100 µl of each culture was plated onto MRS agar containing 15X PlnEF at an estimated density of 1 × 106 CFU ml−1 and incubated for 24 hr. Out of a total of 420 putative resistant mutants, one PlnEF resistant colony was picked from each PlnEF‐containing MRS plate and grown in MRS broth without the bacteriocin for approximately 36 generations. PlnEF resistance was confirmed for each of the isolates prior to preparation of glycerol stocks and storage at −80°C.
4.5. Quantification of corC expression levels
RNA was extracted from three separate exponential‐phase cultures of WT LP965, EF.A, or L. casei BL23 strains grown in MRS as previously described (Tachon, Lee, & Marco, 2014). cDNA was synthesized with the RETROscript kit (Ambion) according to the manufacturer's instructions. Expression of corC was quantified for each strain by quantitative PCR (qPCR). Reactions were performed on an Applied Biosystems 7500 Real‐time thermocycler using 4 ng of RNA, 0.2 µM of each primer (Table A2), and Fast Sybr Green Master Mix (Thermo) under the following conditions: 10 min at 95°C and then 40 cycles of 10 s 95°C and 30 s 60°C followed by melt curve analysis. Expression was quantified by the 2−∆∆Ct method using rpoB as an internal control and WT expression levels as the reference condition.
4.6. Soft‐agar growth inhibition assay
Single colonies of naïve and PlnEF‐resistant LP965 strains were used as indicator strains in soft agar assays as previously described (Kjos et al., 2014) with strain L. plantarum NCIMB 8826 used as the producer. A resistant LP965 phenotype was detected by a reduced zone of inhibition compared to the naïve strain after overnight incubation at 37°C.
4.7. Isolation of genomic DNA and whole genome sequencing
DNA was isolated from WT and EF.A stationary phase cultures grown in MRS by phenol‐chloroform extractions, followed by ethanol precipitation (Sambrook & Russell, 2006). DNA Sequencing was performed using a PacBio RSII instrument with a desired insert size of 10 kb and P6C4 chemistry according to manufacturer's instructions at the University of California, Davis, DNA Technologies Core (http://dnatech.genomecenter.ucdavis.edu).
Sequence SMRTcell files were downloaded from the UC Davis Sequencing core and imported to the PacBio SMRT portal graphical interface unit (http://www.pacb.com). The genome of LP965 was assembled first de novo using the hierarchical genome assembly protocol 2 (RS_HGAP_assembly.2) with default parameters. The final assembly resulted in an average 310X coverage (chromosome of 3,015,426 bp, and plasmids with the following sizes, plasmid 1:66,439 bp, plasmid 2:52,109 bp, plasmid 3:41,818 bp, plasmid 4:23,484 bp, and plasmid 5:16,940 bp). To ensure a high‐quality reference genome, short read archive data generated from LP965 (accession number: SRR1553345) was downloaded from NCBI (https://www.ncbi.nlm.nih.gov/sra). A total of 4.7 million 75 bp (349.3 Mb) single end Illumina reads were assembled into 307 contigs (Cellera assembler, default parameters, https://sourceforge.net/projects/wgs-assembler/). These contigs were then aligned to the PacBio assembly data (RS_AHA_Scaffolding.1) to generate a single high confidence FASTA file with one chromosome and five plasmids (LP965). The LP965 genome sequence was then uploaded to RAST (http://rast.nmpdr.org/) (Aziz et al., 2008) for annotation and further analyzed with Seqbuilder (DNASTAR).
The LP965.EF.A strain genome was assembled using the high‐quality, wildtype genome of LP965 as a reference and the program (RS_Resequencing.1) with default parameters except for the following adjustments: minimum subread length of 8 kb, minimum read quality 75, minimum polymerase read length of 12 kb. This resulted in one chromosome and five plasmids with an average coverage of 193X. To identify variants between the wild type and EF.A strain, each genome was compiled into single FASTA files and aligned with MAUVE 2.4.0 (Darling, Mau, & Perna, 2010). This resulted in five indels and three single nucleotide polymorphisms found on the chromosome.
Each region with conflicting base calls was used to design PCR primers to amplify a 250 bp segment of DNA (Table A1). PCR was conducted with Takara ex‐Taq according to manufacturer's instructions. The PCR products were purified with the Wizard SV gel and PCR Clean‐Up system (Promega) before being submitted for bi‐directional DNA sequencing at the University of California Davis Sequencing core (http://dnaseq.ucdavis.edu/).
4.8. Cellular ATP quantification
Intracellular and extracellular ATP levels were determined in triplicate cell cultures. Overnight cultures were washed in PBS and suspended in PBS with 10 mM glucose and either water (sham), 25 nM PlnEF or 25 µM nisin (Sigma). Nisin was added at concentrations that limited growth of LP965 to a similar extent as 25 nM PlnEF. At indicated time points, 120 µl samples were collected and centrifuged for 2 min at 20,000X g. The supernatant (extracellular ATP) was aspirated and mixed at a ratio of 1:1 with dimethyl sulfoxide (DMSO, Sigma). The cell pellet (intracellular ATP) was then mixed with 100 µl DMSO. ATP concentrations were determined with the Invitrogen ATP Quantification kit according to manufacturer's instructions and luminescence was quantified according to a standard curve on a Biotek Synergy 2 spectrophotometer.
4.9. CorC mutagenesis
LP965 was not amenable to genetic manipulation with techniques described to transform this strain previously (Aukrust et al., 1995) or by adjusting parameters (glycine concentrations, recovery times, voltage) described therein. Repeated attempts to delete the corC open reading frame (ORF) in LP8826 (WCFS1 lp_2671) using methods previously established for clean deletions in this strain (Yin et al., 2017) or variations of those methods (glycine concentrations, recovery times, voltage, incubation temperature) were equally unsuccessful. Primers used for these attempts are listed in Table A3.
4.10. Heterologous expression of L. plantarum corC in L. casei BL23
The corC genes from LP965 and EF.A were amplified with primers A and D using genomic DNA as a template (Table A3). The product was digested with EcoR1‐HF and SacI‐HF (New England Biolabs) and ligated into the multiple cloning site of pJIM2246 (Renault, Corthier, Goupil, Delorme, & Ehrlich, 1996) resulting in pJIMDH1 (WT LP965 CorC) and pJIMDH2 (EF.A CorC). E. coli DH5α was transformed and selected for as previously described (Yin et al., 2017). Plasmids were isolated from E. coli and amplified by PCR using corC primers (Table A1) and DNA sequencing for verification. L. casei BL23 electrocompetent cells were prepared and transformed as previously described (Welker, Hughes, Steele, & Broadbent, 2015). Briefly, freshly prepared BL23 cells were electroporated with 400 ohms, 2 kV, and 25 μF and then immediately transferred to MRS supplemented with 0.5 M sucrose. The cells were then incubated for 4 hr and transformants were selected by plating serial dilutions on selective medium. PCR and DNA sequencing confirmed the presence of the plasmids in L. casei BL23.
4.11. Structural modeling of CorC protein
Co‐evolutionary constraints were determined using the GREMLIN server (http://gremlin.bakerlab.org/) (Kamisetty, Ovchinnikov, & Baker, 2013). A 3D structure model of the CorC protein from LP965 was performed using relax application (Conway, Tyka, DiMaio, Konerding, & Baker, 2014; Khatib et al., 2011; Nivón, Moretti, & Baker, 2013; Tyka et al., 2011) in Rosetta structural modeling software (Alford et al., 2017; Bender et al., 2016; Rohl, Strauss, Misura, & Baker, 2004) and the x‐ray structure of a CorC‐like protein from of Oenococcus oeni PSU (PDB ID: 3OCO) as a template. Sequences were aligned with MUSCLE (Edgar, 2004) and % identity between the CorC protein sequence and 3OCO was 41% (Figure A4). All structural modeling figures were generated using the UCSF Chimera package (Pettersen et al., 2004). The highest ranked Rosetta model of CorC protein was used to investigate the mutational hotspot region. Further domain annotation was accomplished by submitting the LP965 CorC protein sequence to the TMHMM server (Krogh, Larsson, Heijne, & Sonnhammer, 2001) and the InterPro online server http://www.ebi.ac.uk/interpro/(Finn et al., 2017).
4.12. Statistics
Growth assays were conducted with technical replicates in triplicate and data are representative of at least two independent experiments. Data are presented as average values ± SD. Analysis of covariance (ANCOVA) was used to determine whether the growth rates of cultures in logarithmic phase were statistically different from one‐another. Student's t test with Welch's correction was used to determine significant differences between final optical densities of cultures and levels of corC expression.
CONFLICT OF INTERESTS
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AUTHORS CONTRIBUTION
DDH wrote the manuscript and designed and conducted experiments. VYY produced 3D models and edited. MLM designed experiments and edited the manuscript.
ETHICS STATEMENT
None required.
ACKNOWLEDGEMENTS
This manuscript is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. (1148897), Henry A. Jastro graduate research award (DDH), and The Agriculture and Food Research Initiative Grant no. CA‐D‐FST‐2281‐CG from the USDA National Institute of Food and Agriculture (MLM).
APPENDIX 1.
Table A1.
Primers used for DNA sequencing
| Primers | DNA Sequence | Description of gene | Putative mutation location in CP023490 | Reference |
|---|---|---|---|---|
| lipPFor | CAAAAATACCGGAGGCGTAA | FIG00745599, lipoprotein precursor | SNP: G1,960,045A | This study |
| lipPRev | ATTGCGACGCCTCATAATTC | |||
| corCFor | CCCGCTGACTACGATAGACC | putative magnesium efflux protein | SNP: G846,241T | This study |
| corCRev | TGCCATTTTAAGTAGTGCGTGT | |||
| topoFor | CCGGCAATACTTTGTTCGAT | DNA topoisomerase I | SNP: G119,959A | This study |
| topoRev | GAAATTGCGGACCAGGTCTA | |||
| oppAFor | CAACATAGCCGGTCCAGTTT | Oligopeptide ABC transporter | Gap: A del 1,596,580 | This study |
| oppARev | CGTCGGCCTTATTAGGATCA | |||
| abcTFor | TGGTTCAAATGGTGCTGGTA | ABC transporter ATP‐binding protein | Gap: in G 2,679,932 | This study |
| abcTRev | TTGGGCTTTTTCCTCATCAC | |||
| mutLFor | CCTTGAATGGCAACCATCTT | DNA mismatch repair protein | Gap: del G 550,025 | This study |
| mutLRev | TTGATGACGCTGAAGTCCTG | |||
| carBFor | CCGGCAATACTTTGTTCGAT | Carbamoyl‐phosphate synthase large chain | Gap: del C 880,235 | This study |
| carBRev | GAAATTGCGGACCAGGTCTA | |||
| ptsPFor | TCGTGTCCTTCGCCTTAACT | Promoter region of phosphotransferase system | Gap: del T 2,558,123 | This study |
| ptsPRev | AACTGGTGCCCAGATTTGAC |
Table A2.
Primers used for qPCR
| Primers | DNA sequence | Description of gene | Reference |
|---|---|---|---|
| corCFor | GCCAAGGTGAAGTTGACGAT | putative magnesium efflux protein | This study |
| corCRev | CCACTGGTCCCACCATATTC | ||
| Lplant_rpoBFor | GGCAGAACAGATCAAGGAAGG | RNA‐directed polymerase subunit B | (Marco, Bongers, Vos, & Kleerebezem, 2007) |
| Lplant_rpoBRev | TATCCACTTCGGCACGCTTA | ||
| Lcasei_rpoBFor | GACCTTGCGTTTAACCAACC | RNA‐directed polymerase subunit B | This study |
| Lcasei_rpoBRev | AACGGACCAACTGAGAAACG |
Table A3.
Primers used for cloning
| Primer | Sequencea | Description of gene region | Plasmid construction | Reference |
|---|---|---|---|---|
| EcoR1‐corC‐For (A) | CCGGAATTCAGCACCATTCATTGGAAACG | Upstream of corC. Restriction site | pJIMDH1, pJIMDH2, pRVDH1 | This study |
| CorC_up_rev_linker (B) | GAAGCAGCTCCAGCCTACACCGGTTGAGACGGATAGGTTG | Upstream of corC. linker sequence | pRVDH1 | This work |
| CorC_down_for_linker (C) | GTGTAGGCTGGAGCTGCTTCCCGATGTTGTCACTTTGACC | Downstream of corC. linker sequence | pRVDH1 | This study |
| SacI‐corC‐Rev (D) | GGGGAGCTCTCCCTTAAATCAACCGCATC | Downstream of corC. Restriction site | pJIMDH1, pJIMDH2, pRVDH1 | This study |
Restriction enzyme sites are underlined; linker DNA sequences are italicized.
Figure A1.
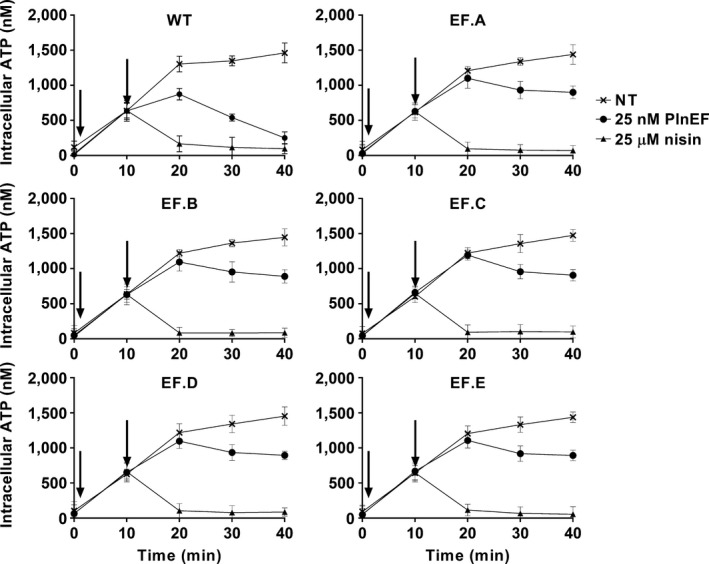
Effects of plantaricin EF (PlnEF) on intracellular concentrations of ATP. Cells of WT and PlnEF‐resistant isolates were initially energized with 10 mM glucose (arrow at 1 min). At 10 min (second arrow), 25 nM of PlnEF, 25 µM nisin, or water (NT) was added to each culture. The avg ± SD of n = 3 replicates is shown
Figure A2.
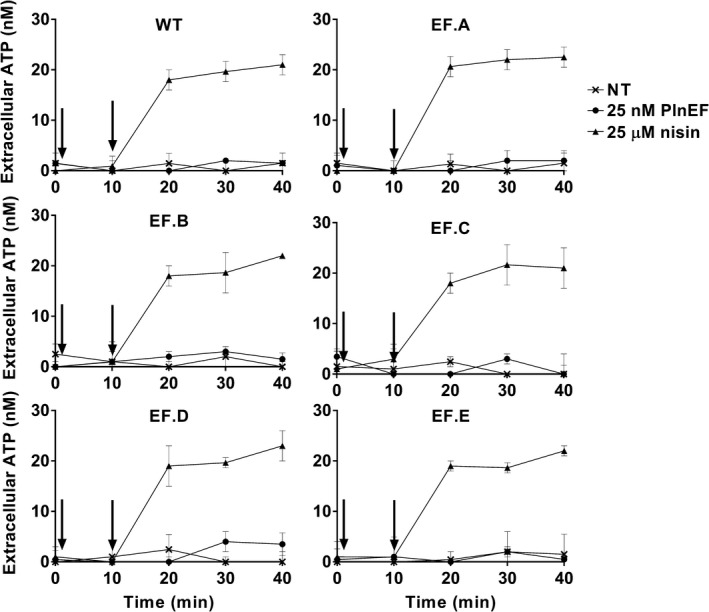
Effects of plantaricin EF (PlnEF) on extracellular concentrations of ATP. WT and PlnEF‐resistant isolates of LP965 were initially energized with 10 mM glucose (arrow at 1 min). At 10 min (second arrow), 25 nM of PlnEF, 25 µM nisin, or water (NT) was added to each culture. The avg ± SD of n = 3 replicates is shown
Figure A3.
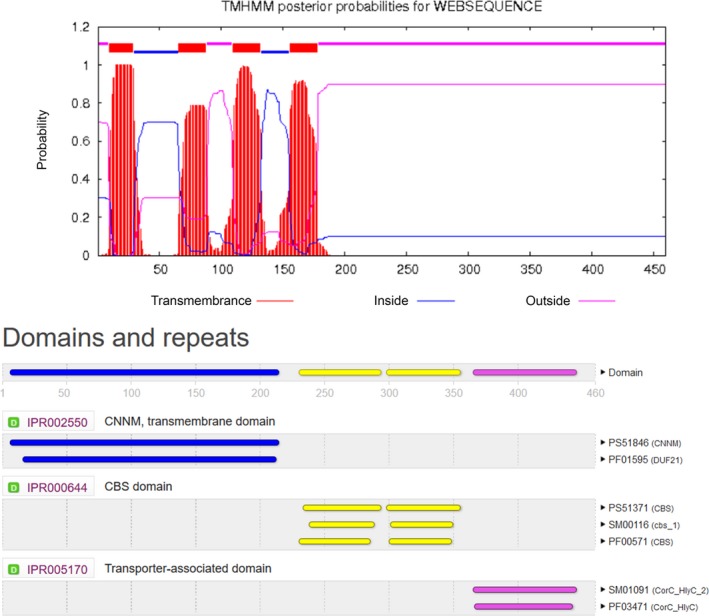
LP965 CorC domain annotation. LP965 CorC amino acid sequnce was submitted to the TMHMM server (top) and the InterPro domain annotation server (bottom). The alphanumeric strings in purple are the InterPro domain identifiers. The alphanumeric strings in black are the domain identifier links to other databases
Figure A4.
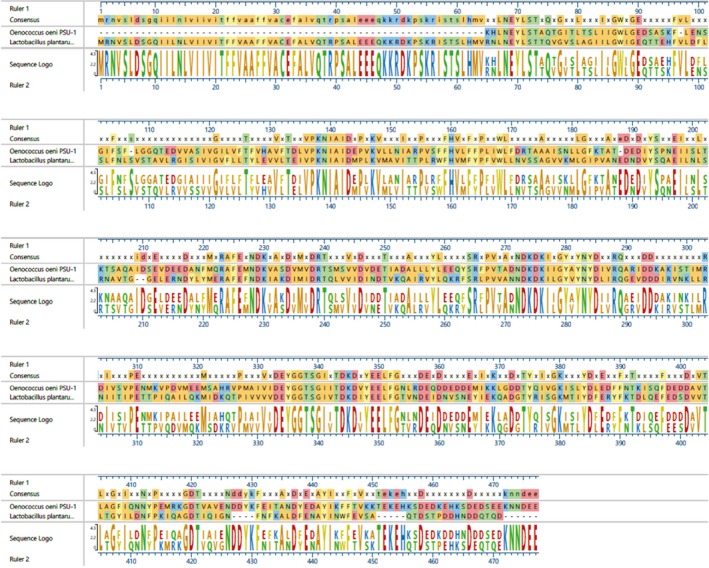
Alignment of CorC amino acid sequences. Amino acid sequences from LP965 and Oenococcus oeni were aligned using MUSCLE. Due to gaps in the alignment, the mutated glycine residue in LP965 (334) in this figure is in position 336
Figure A5.
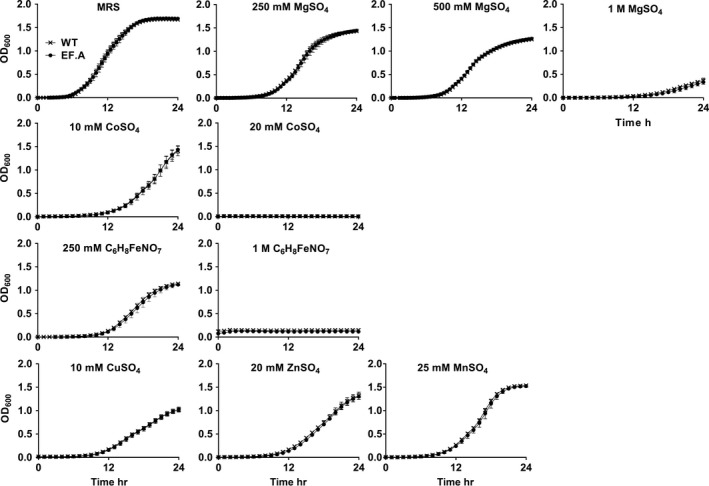
WT and EF.A metal sensitivity. WT and EF.A LP965 were grown in MRS or MRS supplemented with the indicated growth‐inhibiting concentration of metal salt
Heeney DD, Yarov‐Yarovoy V, Marco ML. Sensitivity to the two peptide bacteriocin plantaricin EF is dependent on CorC, a membrane‐bound, magnesium/cobalt efflux protein. MicrobiologyOpen. 2019;8:e827 10.1002/mbo3.827
DATA ACCESSIBILITY
LP965 WT sequence data was deposited into the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) with accession numbers CP023490 to CP023495. The EF.A PlnEF‐resistant strain was deposited in NCBI with accession numbers CP026505 to CP026510.
REFERENCES
- Acedo, Z. J. , Chiorean, S. , Vederas, J. C. , & van Belkum, M. J. (2018). The expanding structural variety among bacteriocins from gram‐positive bacteria. FEMS Microbiology Reviews, 42(6), 805–828. 10.1093/femsre/fuy033 [DOI] [PubMed] [Google Scholar]
- Akanuma, G. , Kobayashi, A. , Suzuki, S. , Kawamura, F. , Shiwa, Y. , Watanabe, S. , … Ishizuka, M. (2014). Defect in the formation of 70S ribosomes caused by lack of ribosomal protein L34 can be suppressed by magnesium. Journal of Bacteriology, 196(22), 3820–3830. 10.1128/JB.01896-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford, R. F. , Leaver‐Fay, A. , Jeliazkov, J. R. , O’Meara, M. J. , DiMaio, F. P. , Park, H. , … Gray, J. J. (2017). The Rosetta all‐atom energy function for macromolecular modeling and design. Journal of Chemical Theory and Computation, 13(6), 3031–3048. 10.1021/acs.jctc.7b00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderssen, E. L. , Diep, D. B. , Nes, I. F. , Eijsink, V. G. H. , & Nissen‐Meyer, J. (1998). Antagonistic activity of Lactobacillus plantarum C11: Two new two‐ peptide bacteriocins, plantaricins EF and JK, and the induction factor plantaricin A. Applied and Environmental Microbiology, 64(6), 2269–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitano, J. , Redder, P. , Guimarães, V. A. , & Linder, P. (2016). An essential factor for high Mg2+ tolerance of Staphylococcus aureus . Frontiers in Microbiology, 7, 1888–1899. 10.3389/fmicb.2016.01888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukrust, T. W. , Brurberg, M. B. , & Nes, I. F. (1995). Transformation of Lactobacillus by electroporation. In Electroporation Protocols for Microorganisms (pp. 201–208). New Jersey: Humana Press; 10.1385/0-89603-310-4:201 [DOI] [PubMed] [Google Scholar]
- Aziz, R. K. , Bartels, D. , Best, A. A. , DeJongh, M. , Disz, T. , Edwards, R. A. , … Zagnitko, O. (2008). The RAST Server: Rapid annotations using subsystems technology. BMC Genomics, 9(1), 75–79. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barwinska‐Sendra, A. , & Waldron, K. J. (2017). The role of intermetal competition and mis‐metalation in metal toxicity. Advances in Microbial Physiology, 70(70), 315–379. 10.1016/bs.ampbs.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Baykov, A. A. , Tuominen, H. K. , & Lahti, R. (2011). The CBS domain: A protein module with an emerging prominent role in regulation. ACS Chemical Biology, 6(11), 1156–1163. 10.1021/cb200231c [DOI] [PubMed] [Google Scholar]
- Bender, B. J. , Cisneros, A. , Duran, A. M. , Finn, J. A. , Fu, D. , Lokits, A. D. , … Moretti, R. (2016). Protocols for molecular modeling with Rosetta3 and RosettaScripts. Biochemistry, 55(34), 4748–4763. 10.1021/acs.biochem.6b00444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breukink, E. , Wiedemann, I. , van Kraaij, C. , Kuipers, O. P. , Sahl, H. G. , & de Kruijff, B. (1999). Use of the cell wall precursor lipid II by a pore‐forming peptide antibiotic. Science, 286(5448), 2361–2364. [DOI] [PubMed] [Google Scholar]
- Chikindas, M. L. , Weeks, R. , Drider, D. , Chistyakov, V. A. , & Dicks, L. M. (2018). Functions and emerging applications of bacteriocins. Current Opinion in Biotechnology, 49, 23–28. 10.1016/j.copbio.2017.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway, P. , Tyka, M. D. , DiMaio, F. , Konerding, D. E. , & Baker, D. (2014). Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Science, 23(1), 47–55. 10.1002/pro.2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter, P. D. , Hill, C. , & Ross, P. R. (2005). Bacteriocins: Developing innate immunity for food. NatureReviews Microbiology, 3(10), 777–788. 10.1038/nrmicro1273 [DOI] [PubMed] [Google Scholar]
- Darling, A. E. , Mau, B. , & Perna, N. T. (2010). ProgressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE, 5(6), 1–18. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep, D. B. , Skaugen, M. , Salehian, Z. , Holo, H. , & Nes, I. F. (2007). Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proceedings of the National Academy of Sciences of the USA, 104(7), 2384–2389. 10.1073/pnas.0608775104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5), 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekblad, B. , Nissen‐Meyer, J. , & Kristensen, T. (2017). Whole‐genome sequencing of mutants with increased resistance against the two‐peptide bacteriocin plantaricin JK reveals a putative receptor and potential docking site. PLoS ONE, 12(9), 1–12. 10.1371/journal.pone.0185279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimland, N. , Rogne, P. , Fimland, G. , Nissen‐Meyer, J. , & Kristiansen, P. E. (2008). Three‐dimensional structure of the two peptides that constitute the two‐peptide bacteriocin plantaricin EF. Biochimica Et Biophysica Acta ‐ Proteins and Proteomics, 1784(11), 1711–1719. 10.1016/j.bbapap.2008.05.003 [DOI] [PubMed] [Google Scholar]
- Finn, R. D. , Attwood, T. K. , Babbitt, P. C. , Bateman, A. , Bork, P. , Bridge, A. J. , … Mitchell, A. L. (2017). InterPro in 2017‐beyond protein family and domain annotations. Nucleic Acids Research, 45(D1), D190–D199. 10.1093/nar/gkw1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharsallaoui, A. , Oulahal, N. , Joly, C. , & Degraeve, P. (2016). Nisin as a food preservative: Part 1: Physicochemical properties, antimicrobial activity, and main uses. Critical Reviews in Food Science and Nutrition, 56(8), 1262–1274. 10.1080/10408398.2013.763765 [DOI] [PubMed] [Google Scholar]
- Gibson, M. M. , Bagga, D. A. , Miller, C. G. , & Maguire, M. E. (1991). Magnesium transport in Salmonella typhimurium: The influence of new mutations conferring Co2+ resistance on the CorA Mg2+ transport system. Molecular Microbiology, 5(11), 2753–2762. 10.1111/j.1365-2958.1991.tb01984.x [DOI] [PubMed] [Google Scholar]
- Groisman, E. A. , Hollands, K. , Kriner, M. A. , Lee, E.‐J. , Park, S.‐Y. , & Pontes, M. H. (2013). Bacterial Mg2+ homeostasis, transport, and virulence. Annual Review of Genetics, 47, 625–646. 10.1146/annurev-genet-051313-051025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot, M. N. , Klaassens, E. , de Vos, W. M. , Delcour, J. , Hols, P. , & Kleerebezem, M. (2005). Genome-based in silico detection of putative manganese transport systems in Lactobacillus plantarum and their genetic analysis. Microbiology, 151(4), 1229–1238. 10.1099/mic.0.27375-0. [DOI] [PubMed] [Google Scholar]
- Hayward, A. C. , & Davis, G. H. G. (1956). The isolation and classification of Lactobacillus strains from Italian saliva. British Dentistry Journal, 101, 43–46. [Google Scholar]
- Heeney, D. D. , Zhai, Z. , Bendiks, Z. , Barouei, J. , Martinic, A. , Slupsky, C. , & Marco, M. L. (2018). Lactobacillus plantarum bacteriocin is associated with intestinal and systemic improvements in diet‐induced obese mice and maintains epithelial barrier integrity in vitro. Gut Microbes, 1–16, 10.1080/19490976.2018.1534513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata, Y. , Funato, Y. , Takano, Y. , & Miki, H. (2014). Mg2+‐dependent interactions of ATP with the cystathionine‐β‐synthase (CBS) domains of a magnesium transporter. The Journal of Biological Chemistry, 289(21), 14731–14739. 10.1074/jbc.M114.551176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisetty, H. , Ovchinnikov, S. , & Baker, D. (2013). Assessing the utility of coevolution‐based residue‐residue contact predictions in a sequence‐and structure‐rich era. Proceedings of the National Academy of Sciences USA, 39(12), 15674–15679. 10.1073/pnas.1314045110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatib, F. , Cooper, S. , Tyka, M. D. , Xu, K. , Makedon, I. , Popovic, Z. , … Players, F. (2011). Algorithm discovery by protein folding game players. Proceedings of the National Academy of Sciences of the USA, 108(47), 18949–18953. 10.1073/pnas.1115898108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjos, M. , Oppegard, C. , Diep, D. B. , Nes, I. F. , Veening, J. W. , Nissen‐Meyer, J. , & Kristensen, T. (2014). Sensitivity to the two‐peptide bacteriocin lactococcin G is dependent on UppP, an enzyme involved in cell‐wall synthesis. Molecular Microbiology, 92(6), 1177–1187. 10.1111/mmi.12632 [DOI] [PubMed] [Google Scholar]
- Kjos, M. , Snipen, L. , Salehian, Z. , Nes, I. F. , & Diep, D. B. (2010). The Abi proteins and their involvement in bacteriocin self‐immunity. Journal of Bacteriology, 192(8), 2068–2076. 10.1128/JB.01553-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem, M. , Boekhorst, J. , van Kranenburg, R. , Molenaar, D. , Kuipers, O. P. , Leer, R. , … Siezen, R. J. (2003). Complete genome sequence of Lactobacillus plantarum WCFS1. Proceedings of the National Academy of Sciences USA, 100(4), 1990–1995. 10.1073/pnas.0337704100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, A. , Larsson, B. , Von Heijne, G. , & Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. Journal of Molecular Biology, 305(3), 567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kyriakou, P. K. , Ekblad, B. , Kristiansen, P. E. , & Kaznessis, Y. N. (2016). Interactions of a class IIb bacteriocin with a model lipid bilayer, investigated through molecular dynamics simulations. Biochimica Et Biophysica Acta ‐ Biomembranes, 1858(4), 824–835. 10.1016/j.bbamem.2016.01.005 [DOI] [PubMed] [Google Scholar]
- Leloup, L. , Ehrlich, S. D. , Zagorec, M. , & Morel‐Deville, F. (1997). Single‐crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Applied and Environmental Microbiology, 63(6), 2117–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod, R. A. , & Snell, E. E. (1947). Some mineral requirements of the lactic acid bacteria. Journal of Biological Chemistry, 170, 351–357. [Google Scholar]
- Marco, M. L. , Bongers, R. S. , de Vos, W. M. , & Kleerebezem, M. (2007). Spatial and temporal expression of Lactobacillus plantarum genes in the gastrointestinal tracts of mice. Applied and Environmental Microbiology, 73(1), 124–132. 10.1128/AEM.01475-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies, D. , Dalmas, O. , Borgnia, M. J. , Dominik, P. K. , Merk, A. , Rao, P. , … Subramaniam, S. (2016). Cryo‐EM structures of the magnesium channel CorA reveal symmetry break upon gating. Cell, 164(4), 747–756. 10.1016/j.cell.2015.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze, A. , Boel, G. , Zuniga, M. , Bourand, A. , Loux, V. , Yebra, M. J. , … Deutscher, J. (2010). Complete genome sequence of the probiotic Lactobacillus casei strain BL23. Journal of Bacteriology, 192(10), 2647–2648. 10.1128/JB.00076-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, S. , Coffey, A. , Hill, C. , Fitzgerald, G. F. , McAuliffe, O. , & Ross, R. P. (2005). Insertional inactivation of determinants for Mg2+ and Co2+ transport as a tool for screening recombinant Lactococcus species clones. Applied and Environmental Microbiology, 71(8), 4897–4901. 10.1128/AEM.71.8.4897-4901.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll, G. N. , van den Akker, E. , Hauge, H. H. , Nissen‐Meyer, J. , Nes, I. F. , Konings, W. N. , & Driessen, A. J. (1999). Complementary and overlapping selectivity of the two‐peptide bacteriocins plantaricin EF and JK. Journal of Bacteriology, 181(16), 4848–4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moomaw, A. S. , & Maguire, M. E. (2008). The unique nature of Mg2+ channels. Physiology, 23(5), 275–285. 10.1152/physiol.00019.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen‐Meyer, J. , Oppegård, C. , Rogne, P. , Haugen, H. S. , & Kristiansen, P. E. (2010). Structure and mode‐of‐action of the two‐peptide (class‐IIb) bacteriocins. Probiotics and Antimicrobial Proteins, 2(1), 52–60. 10.1007/s12602-009-9021-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivón, L. G. , Moretti, R. , & Baker, D. (2013). A pareto‐optimal refinement method for protein design scaffolds. PLoS ONE, 8(4), 1–10. 10.1371/journal.pone.0059004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppegård, C. , Kjos, M. , Veening, J. , Nissen‐Meyer, J. , & Kristensen, T. (2016). A putative amino acid transporter determines sensitivity to the two‐peptide bacteriocin plantaricin JK. MicrobiologyOpen, 5(4), 700–708. 10.1002/mbo3.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov, K. V. , Kristiansen, P. E. , Straume, D. , Jensen, M. S. , Aleksandrzak‐Piekarczyk, T. , Nes, I. F. , & Diep, D. B. (2017). The leaderless bacteriocin Enterocin K1 is highly potent against Enterococcus faecium: A study on structure, target spectrum and receptor. Frontiers in Microbiology, 8(774), 1–12. 10.3389/fmicb.2017.00774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, R. H. , Zendo, T. , & Sonomoto, K. (2014). Novel bacteriocins from lactic acid bacteria (LAB): Various structures and applications. Microbial Cell Factories, 13(S1), 1–13. 10.1186/1475-2859-13-S1-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E. F. , Goddard, T. D. , Huang, C. C. , Couch, G. S. , Greenblatt, D. M. , Meng, E. C. , & Ferrin, T. E. (2004). UCSF Chimera: A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Ramnath, M. , Beukes, M. , Tamura, K. , & Hastings, J. W. (2000). Absence of a putative mannose‐specific phosphotransferase system enzyme IIAB component in a leucocin a‐resistant strain of Listeria monocytogenes, as shown by two‐dimensional sodium dodecyl sulfate‐polyacrylamide gel electrophoresis. Applied and Environmental Microbiology, 66(7), 3098–3101. 10.1128/AEM.66.7.3098-3101.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault, P. , Corthier, G. , Goupil, N. , Delorme, C. , & Ehrlich, S. D. (1996). Plasmid vectors for gram‐positive bacteria switching from high to low copy number. Gene, 183(1–2), 175–182. 10.1016/S0378-1119(96)00554-9 [DOI] [PubMed] [Google Scholar]
- Rohl, C. A. , Strauss, C. E. M. , Misura, K. M. S. , & Baker, D. (2004). Protein structure prediction using Rosetta. Methods in Enzymology, 383, 66–93. 10.1016/S0076-6879(04)83004-0 [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , & Russell, D. W. (2006). Purification of nucleic acids by extraction with phenol:chloroform. Cold Spring Harbor Protocols, 2006(1), 4455–4567. 10.1101/pdb.prot4455 [DOI] [PubMed] [Google Scholar]
- Sherwood, I. R. (1939). The relation of certain lactic acid bacteria to open texture in cheddar cheese. Journal of Dairy Science, 10(2), 326–335. 10.1017/S0022029900002946 [DOI] [Google Scholar]
- Shin, J.‐H. , Wakeman, C. A. , Goodson, J. R. , Rodionov, D. A. , Freedman, B. G. , Senger, R. S. , & Winkler, W. C. (2014). Transport of magnesium by a bacterial Nramp‐related gene. PLoS Genetics., 10(6), 1–12. 10.1371/journal.pgen.1004429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachon, S. , Lee, B. , & Marco, M. L. (2014). Diet alters probiotic Lactobacillus persistence and function in the intestine. Environmental Microbiology, 16(9), 2915–2926. 10.1111/1462-2920.12297 [DOI] [PubMed] [Google Scholar]
- Taylor, R. G. , Walker, D. C. , & McInnes, R. R. (1993). E. coli host strains significantly affect the quality of small‐scale plasmid DNA preparations used for sequencing. Nucleic Acids Research, 21(7), 1677–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyka, M. D. , Keedy, D. A. , André, I. , DiMaio, F. , Song, Y. , Richardson, D. C. , … Baker, D. (2011). Alternate states of proteins revealed by detailed energy landscape mapping. Journal of Molecular Biology, 405(2), 607–618. 10.1016/j.jmb.2010.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccaro, B. J. , Lancaster, W. A. , Thorgersen, M. P. , Zane, G. M. , Younkin, A. D. , Kazakov, A. E. , … Adams, M. W. W. (2016). Novel metal cation resistance systems from mutant fitness analysis of denitrifying Pseudomonas stutzeri . Applied and Environmental Microbiology, 82(19), 6046–6056. 10.1128/AEM.01845-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, C. J. , Guinane, C. M. , Hill, C. , Ross, R. P. , O’Toole, P. W. , & Cotter, P. D. (2015). In silico identification of bacteriocin gene clusters in the gastrointestinal tract, based on the Human Microbiome Project’s reference genome database. BMC Microbiology, 15(183), 1–11. 10.1186/s12866-015-0515-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker, D. L. , Hughes, J. E. , Steele, J. L. , & Broadbent, J. R. (2015). High efficiency electrotransformation of Lactobacillus casei . FEMS Microbiology Letters, 362(2), 1–6. 10.1093/femsle/fnu033 [DOI] [PubMed] [Google Scholar]
- Yin, X. , Heeney, D. , Srisengfa, Y. , Golomb, B. , Griffey, S. , & Marco, M. (2017). Bacteriocin biosynthesis contributes to the anti‐inflammatory capacities of probiotic Lactobacillus plantarum . Beneficial Microbes, 9(2), 333–344. 10.3920/BM2017.0096 [DOI] [PubMed] [Google Scholar]
- Zhang, N. , Ren, X. , Zhu, D. , Li, D. , & Wang, D. (2010). Crystallization and preliminary crystallographic studies of CorC, a magnesium‐ion transporter. Acta Crystallographica, 66(6), 681–683. 10.1107/S1744309110012613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J. , Gänzle, M. G. , Lin, X. B. , Ruan, L. , & Sun, M. (2015). Diversity and dynamics of bacteriocins from human microbiome. Environmental Microbiology, 17(6), 2133–2143. 10.1111/1462-2920.12662 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
LP965 WT sequence data was deposited into the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) with accession numbers CP023490 to CP023495. The EF.A PlnEF‐resistant strain was deposited in NCBI with accession numbers CP026505 to CP026510.


