Abstract
The gastrointestinal tract harbors most of the microbiota associated with humans. In recent years, there has been a surge of interest in assessing the relationships between the gut microbiota and several gut alterations, including colorectal cancer. Changes in the gut microbiota in patients suffering colorectal cancer suggest a possible role of host-microbe interactions in the origin and development of this malignancy and, at the same time, open the door for novel ways of preventing, diagnosing, or treating this disease. In this review we survey current knowledge on the healthy microbiome of the gut and how it is altered in colorectal cancer and other related disease conditions. In describing past studies we will critically assess technical limitations of different approaches and point to existing challenges in microbiome research. We will have a special focus on host-microbiome interaction mechanisms that may be important to explain how dysbiosis can lead to chronic inflammation and drive processes that influence carcinogenesis and tumor progression in colon cancer. Finally, we will discuss the potential of recent developments of novel microbiota-based therapeutics and diagnostic tools for colorectal cancer.
Keywords: Gut microbiome, Colorectal cancer, Colon, Microbiota
Abbreviations: T, tissue; F, feces; NA, not available; SCFA, short-chain fatty acids; 16S, 16S ribosomal RNA sequencing and/or pyrosequencing; WGS, whole-genome shotgun sequencing; qPCR, quantitative PCR
1. Introduction
The gut comprises an abundant and diverse community of microorganisms (archaea, fungi, protozoa and viruses), which is collectively referred to as the “gut microbiome” (Costello et al., 2012). A typical gut microbiome may comprise trillions of microbial cells from over several hundreds of different species, which genomes can entail, globally, over three million genes (Sender et al., 2016). This complex ecosystem is not a passive colonizer of our gut, but rather interacts with the host in many ways, contributing to various processes such as nutritional absorption, metabolism, immunity, tissue development, and carcinogenesis (Bosch and McFall-Ngai, 2011; Dzutsev et al., 2015). Alterations in the composition of the gut microbiome - a state known as dysbiosis - have been associated with a growing number of prevalent human diseases, including cancer (Schwabe and Jobin, 2013). Indeed, it has been estimated that more than 20% of the cancer burden worldwide is attributable to known intestinal infectious agents that are often normal residents of the intestinal microbiota (Zur Hausen, 2009). In particular, dysbiosis-related inflammation and the biosynthesis of chemical carcinogens (e.g. acetaldehyde, N-nitroso compounds) by microbes are among several possible mechanisms through which the microbiota may have a role in carcinogenesis (Arthur et al., 2012; Kostic et al., 2013; Rubinstein et al., 2013; Sears et al., 2008). The colon is the most heavily colonized section of the digestive tract, and it has been estimated that this organ contains approximately 70% of the estimated human microbiome (Sekirov et al., 2010). The colon is also the section of the digestive tract that is more prone to develop cancer, with cancer incidence being 12-fold higher in the colon as compared to the small intestine (Gagnière et al., 2016). In addition, known risk factors in colorectal cancer (CRC), such as dietary habits and life-style (Moskal et al., 2016; Torre et al., 2015), are known to modulate the gut microbiota. Thus, a plausible hypothesis is that certain colonic microbes or alterations of the typical resident colonic flora may create a microenvironment that is more favorable to tumor development. This hypothesis is supported by findings from several studies that explored the gut microbiota associated with individuals with CRC (Gagnière et al., 2016; Kostic et al., 2013; Wang et al., 2012; Weir et al., 2013). In this regard, a growing number of studies report specific alterations in the gut microbiome associated with CRC and explored its value for CRC screening (Wong et al., 2017a). This is particularly important for CRC, as an early diagnosis of the tumor (in stages 0, I or II) is associated to 80% survival rate over five years, which is reduced to only 10% in later diagnosis (stage IV). In this context, the detection of specific microbiome alterations has emerged as a promising strategy for CRC diagnostics (Wong et al., 2017a; Zeller et al., 2014a). Finally, some bacterial genera have been hypothesized to be protective against CRC (Appleyard et al., 2011). This protective phenotype may be mediated through metabolite production, induction of immunological tolerance, or an ability to outcompete pathogenic bacteria or fungi (Coker et al., 2018a; Zhu et al., 2011). In the long run, a better knowledge of the relationships between the microbiota and the origin and progression of CRC may open novel opportunities for the development of therapies targeting the microbiome. In this regard the development and use of prebiotics, probiotics, specific antibiotics, phage therapies, or the transplantation of whole microbiomes may bring new tools for the prevention and treatment of CRC (Kelly, 2013; Lynch and Pedersen, 2016; Schmidt et al., 2018). In this review we provide an overview of how the gut microbiome is studied and survey recent research directed towards unveiling possible relationships between gut microbiota and the origin and development of CRC. We next provide an overview of potential applications related to microbiome-based diagnostic and therapeutic tools, which are still in very early-development stages. Finally, we discuss the current limitations and future potential of this field of research.
2. The gut microbiome and approaches to assess it
Metagenomic studies have most commonly used one of two main approaches to assess the composition of microbiomes: whole-genome shotgun (WGS) sequencing, and 16S ribosomal RNA amplicon sequencing. In both cases, DNA sequences of microbes present in a given sample are read using next generation sequencing technologies, and compared to a database of known sequences to determine the presence and abundance of particular taxa. Depending on the sequencing and analysis approach, a resolution at the strain, species, genus, or higher taxonomic levels can be achieved, often depending on the specific clades. In addition, WGS can provide information on the genes encoded by the strains present in the sample, at least the most abundant ones. This information can be used to reconstruct potential metabolic capacities of a given microbial ecosystem.
WGS is performed by randomly fragmenting all the DNA into small segments multiple times, so as to determine the sequences of millions of fragments in parallel. Then they are assembled into longer fragments (contigs) corresponding to chromosomal regions (perhaps full genomes for the most abundant species) by piecing together the overlapping ends (Anderson, 1981). Finally, these contigs are annotated and analyzed in terms of their taxonomic origin and functional capabilities. However, the most commonly used technique in metagenomic studies these days is the targeted sequencing of the 16S rRNA gene, which is present in all bacterial and archaeal genomes. This gene has highly conserved regions that allow for the use of universal primer sequences to bind to and specifically amplify them. Between the highly conserved regions of this 16S gene, there are nine highly variable regions (named V1 through V9), so mapping the read sequences to a database of known 16S rRNA gene sequences allows for taxonomic identification of the bacteria present in a sample (Weisburg et al., 1991). Focusing on just one or two of these segments of this particular gene highlights the advantage of the 16S sequencing technique: the vast reduction in cost and data management which enables cost-effective, large-scale studies. The primary disadvantage of 16S sequencing however is the lack of taxonomic resolution; while the variable regions of the gene are particular to different organisms, finding differences within this section of a few hundred base pairs versus differences across the entire genome can often limit identification to only the genus level. In contrast, WGS allows for more accurate detection of species, or even strains, and diversity within samples, as well as identification of the coding potential of the genome, which can only be indirectly inferred in the case of 16S sequencing, by extrapolation from known genomes (Ranjan et al., 2016). Further details on the methodologies used in metagenomics analyses can be found elsewhere (Mallick et al., 2017; Song et al., 2018).
Both 16S and WGS approaches have been extensively used to study the human gut microbiome, perhaps the most intensively studied niche so far, with two main large international consortia playing a major role in driving the field: the Metagenomics of the Human Intestinal Tract (MetaHIT) (Li et al., 2014), and the Human Microbiome Project (HMP) (Human Microbiome Project Consortium, 2012; Schmidt et al., 2018). One of the earliest large-scale analyses used WGS to provide a catalogue of bacterial genes present in the gut microbiome based on an analysis of 124 European individuals (Qin et al., 2010). They also catalogued between 1000 and 1150 prevalent bacterial species, of which each individual carried at least 160. In this early work, clustering analyses already showed distinct microbial compositions for individuals having specific gut alterations such as Crohn's disease or ulcerative colitis. Later analyses showed that healthy individuals also clustered into different microbiome composition profiles, which were dubbed “enterotypes” driven by the abundances of one or a few different organisms (Arumugam et al., 2011). The enterotype concept has been somewhat controversial in the field, with some authors suggesting that instead of placing samples into discrete clusters, a gradient of abundances should be recognized to better compare samples (Jeffery et al., 2012), while others prefer the cluster-centric approach, with the use of carefully defined boundaries (Costea et al., 2018). Moreover, it is as yet not entirely clear whether enterotypes are determined by extrinsic factors such as diet, or immune state, represent intrinsically different ecological optima, or both (Costea et al., 2018). Therefore, whatever the enterotype philosophy, samples may also be stratified, either instead by or alongside, other factors like medication (Forslund et al., 2015), age (Jeffery et al., 2016), lifestyle (Barton et al., 2018) or diet (Zhernakova et al., 2016). Similarly, there has been a major interest in finding what microbiome differences may associate with the presence (or the risk of developing it in the future) of a particular disease. Such studies compare microbiome compositions between donors having a particular condition, and a control group, to then find significant differences. Such studies have been dubbed Metagenome-Wide (or Microbiome-Wide) Association Studies (MWAS) in analogy to Genome-Wide Association Studies (GWAS) (Gilbert et al., 2016; Wang and Jia, 2016). Finding such associations is the first step not only to discover potential new mechanisms underlying the origin, progression, or the effects of the disease, but also for the development of future diagnostic and therapeutic tools based on the monitoring or modulation, respectively, of key elements of the microbiota (Schmidt et al., 2018). Particular differences found between CRC and control samples, and their relevance for understanding, detecting, and treating the disease are discussed in subsequent sections of this review.
The deep understanding of the functional roles of the gut microbiota and its interactions with the human host is a further step, which is needed to enable the application of microbiome knowledge to the clinics. In this line, the use of metatranscriptomics and metabolomics approaches can be complementary to metagenomics approaches. Metatranscriptomics is the sequencing-based analysis of expressed transcripts in a sample, which provides information on what genes are active at the time of the experiment. Metatranscriptomics can help to elucidate biological functions underlying microbial dysbiosis associated to multiple diseases. Some metatranscriptomic studies have been conducted to explore the functional role of the human gut microbiota (Franzosa et al., 2014; Ranjan et al., 2018) and its relationship with disorders such as inflammatory bowel disease (IBD) (Schirmer et al., 2018). However, in CRC there is an obvious gap, with, to the best of our knowledge, no exhaustive metatranscriptomics studies available in CRC patients. The scarcity of metatranscriptomic studies is likely related to specific limitations that complicate its use, such as the low stability of RNA, the need to deplete ribosomal RNA, and the complexity of downstream data analyses (Aguiar-Pulido et al., 2016).
Metabolomics is the study of small molecules or metabolites present in biological specimens. Metabolites and other chemical compounds are the ultimate link mediating host-pathogen interactions, and therefore the metabolome may provide important mechanistic insights into cancer-related processes (Armitage and Barbas, 2014). Metabolites of microbial or host origin are released in the gut and affect the tumor microenvironment. The main final aim of metabolomics research in CRC is to apply this knowledge to i) improving understanding of CRC etiology – for individual/population risk stratification, and effective cancer prevention policies, ii) describing metabolite profiles capable of predicting patients with CRC at early stages of tumorigenesis, even distinguishing solitary adenoma from disease-free controls (diagnosis biomarkers) (Farshidfar et al., 2016; Tan et al., 2013; Uchiyama et al., 2017), and iii) finding prognosis, survival and recurrence biomarkers to improve intervention and treatment strategies in patients with different molecular CRC subtypes (Liesenfeld et al., 2015; Qiu et al., 2014). In addition, metabolomic analyses can serve as orthogonal lines of evidence to confirm mechanistic hypotheses generated in MWASs.
3. Relationships between gut microbiome and colorectal cancer
CRC is a complex malignant disease whose multi-stage development involves numerous factors, including genetic and environmental risk factors. It has been shown that an accumulation of genetic and epigenetic alterations in proto-oncogenes, tumor suppressor genes, and/or DNA repair genes, leads to transformation of the normal colonic epithelium into tumoral cells (Fleming et al., 2012). The majority of CRC cases occur sporadically and less than 20% of CRC cases are hereditary (Carethers and Jung, 2015). Smoking, alcohol consumption, obesity and diabetes are all known factors involved in this disease. Dietary factors such as a diet rich in processed foods, animal fat and red meat coupled with a low intake of fibre and fruits have also been validated as an important risk factor for the development of sporadic CRC (Moskal et al., 2016). More recently, an increasing number of studies report alterations of the gut microbiota in CRC samples, suggesting that gut microbiota may be an essential contributing factor to the initiation and development of this cancer.
The gut microbiota is involved in the maintenance of mucosal homeostasis and epithelial barrier function. In a healthy situation, the intestinal barrier is efficiently compartmentalizing bacteria to the lumen, but perturbations in gut barrier function can lead to increased “intestinal permeability”, which has been shown to be associated with a variety of gastrointestinal disorders and diseases, including inflammatory bowel disease (IBD), irritable bowel syndrome (IBS), celiac disease, and CRC development (Arrieta et al., 2006; Bischoff et al., 2014; Owyang and Wu, 2014; Sattar et al., 1985). In addition, the collective activities of gut microbes and particularly their metabolic products can have a major influence on the immune response, and be a source of chronic inflammation. Microbiota and their metabolites, such as short chain fatty acids (e.g. butyrate) might induce dysfunction in the gut epithelial barrier, thereby activating proinflammatory mediators such as cytokines, interleukin-6 (IL-6), and tumor necrosis factor-α (TNF-α), that damage epithelial cells and their junctions (Wu et al., 2009; Yoshioka et al., 2009). Some specific bacterial species can trigger inflammatory responses or produce toxins that directly damage gut cells. For instance, Bacteroides fragilis and Enterococcus faecalis produce enterotoxins (i.e. fragylisin) and reactive oxygen species that cause oxidative DNA damage, induce inflammation, and damage the epithelial barrier (Goodwin et al., 2011; Wu et al., 2009). In addition, B. fragilis activates β-catenin nuclear signalling and induces cellular proliferation (Wu et al., 2003). Similarly Fusobacterium nucleatum can induce inflammatory changes by directly adhering to and invading colonic epithelial cells via the FadA surface protein, which interacts with E-cadherin to mediate changes in β-catenin and Wnt signalling (Kostic et al., 2013; Rubinstein et al., 2013). As we will see below, some of these bacterial species have been shown to have significantly different abundances in CRC samples as compared to healthy controls.
A growing body of evidence indicates that CRC arises from a stepwise disturbance of the composition of the gut microbiota, induced by food components or diet, plus genetic alterations in oncogenes and tumor-suppressor genes (Fig. 1). Several microbes have been found to be differentially enriched in tumor versus normal tissues or in fecal samples from patients with CRC versus healthy control subjects. Altered taxa in CRC are different depending on whether the samples are obtained directly from mucosal or from fecal samples (Flemer et al., 2017; Gao et al., 2017). However, despite the fact that the fecal microbiome is only a proxy for the gut microbiome, this non-invasive approach has provided crucial information of the changes in the gut ecosystem associated with CRC, and is the most commonly used sampling method in gut microbiome studies. Numerous studies comparing fecal microbiota from CRC patients versus controls have shown that despite having overall similar microbiomes, sets of significantly enriched and depleted microorganisms can be identified that differentiate CRC and control populations (Table 1, Fig. 1). Microbiome in CRC patients is often enriched in pro-inflammatory opportunistic pathogens and microbes associated with metabolic disorders and depleted in butyrate-producing bacteria, which have been shown to be pivotal for the preservation of intestinal homeostasis (Gao et al., 2015; Marchesi et al., 2011). Some bacteria such as Streptococcus gallolyticus (in the past Streptococcus bovis), F. nucleatum, Escherichia coli, B. fragilis and E. faecalis, have high prevalence in CRC patients as compared to the normal population, whereas genera such as Roseburia, Clostridium, Faecalibacterium and Bifidobacterium are generally depleted in CRC patients (Feng et al., 2015; Gagnière et al., 2016; Gao et al., 2015; Shang and Liu, 2018; Wang et al., 2012; Yu et al., 2017; Zhang et al., 2019). Although there is significant interest in identifying specific oncomicrobes, no single species has been found to be universally present among all individuals with CRC and there is significant variation in microbial composition between individuals (Sears and Garrett, 2014). This suggests that different combinations of microorganisms - also known as co-abundance groups - may act synergistically. In addition, changes in both harmful and protective bacterial populations are probably responsible for colon tumor initiation and/or progression.
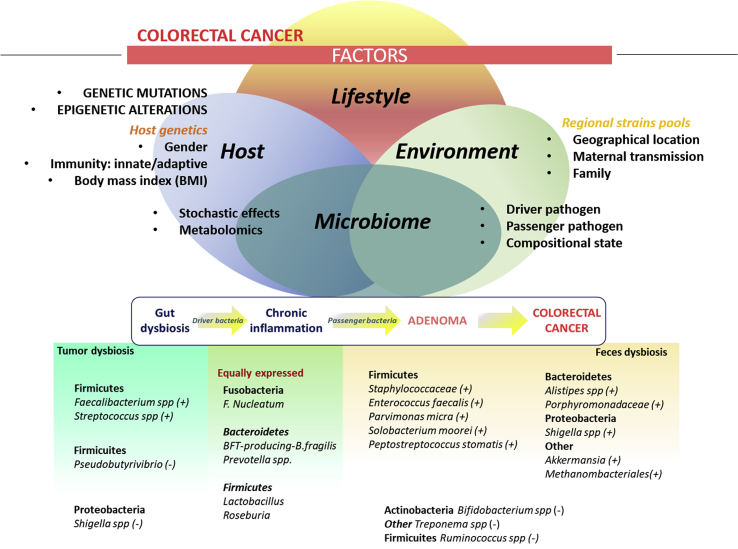
Fig. 1.
Microbiome dysbiosis in CRC. This figure summarizes factors influencing the gut microbiome, a model for dysbiosis in carcinogenesis process of CRC, and a description of the bacteria species accounting for dysbiosis in CRC. Upper part: factors shaping the gut microbiome: host (genetics and non-genetics), lifestyle (diet, exercise, sleeping time, etc), and environmental (pool of colonizing strains) factors. All those factors may have an influence in the microbiome, which in turn may influence the state of health/disease of individuals. Other factors influencing the state of gut microbiome include stochastic effects, presence of driver/passenger species, its compositional state and metabolomics. Middle part: The multi-step process of carcinogenesis and the influence of the microbiome. As suggested by several authors, imbalances in the normal content of the gut microbiome (gut dysbiosis) lead to colonization of driver bacteria that induce a chronic inflammation of the gut epithelia. This inflammation changes the microenvironment and biofilm and allow a new colonization by passenger bacteria, which may contribute to carcinogenesis process from adenomatosis to tumor formation. Bottom: Bacterial species enriched/depleted in CRC both tumor and feces samples as compared to control samples.
Table 1.
Gut microbiota identified in CRC patients and healthy individuals, classified in bacteria enriched, depleted or being controversial in CRC. Columns indicate, in this order: phyla of the bacteria, genus of the bacteria, species of the bacteria identified if having this level of resolution, type of sample studied (feces or tissue), description of the mechanism through which the described bacteria exert their function if known, and methodology used to assess the microbiome.
| Phyla | Genus | Species | Sample | Mechanism/Function | Method | References | |
|---|---|---|---|---|---|---|---|
| Enriched in CRC | |||||||
| ACTINOBACTERIA | |||||||
| Collinsella | --- | T | NA | 16S | Marchesi et al. (2011) | ||
| Slackia | --- | T | Anti-oxidant potential | 16S | Marchesi et al. (2011) | ||
| BACTEROIDETES | |||||||
| Alistipes | A. finegoldii* | F | Inflammatory | 16S, WGS | (Baxter et al., 2014; Dai et al., 2018a; Feng et al., 2015) | ||
| Bacteroides | B. fragilis* | F, T | Inflammatory, enterotoxigenic (fragilisin) | 16S, WGS | (Dai et al., 2018a; Drewes et al., 2017; Hale et al., 2018; Wang et al., 2012) | ||
| Porphyromonas | P. asaccharolytica* | F, T | Inflammatory | 16S, WGS, qPCR | (Ahn et al., 2013; Chen et al., 2012; Dai et al., 2018a; Wang et al., 2012) | ||
| Prevotella | P.intermedia* | F, T | Inflammatory | 16S, WGS, qPCR | (Dai et al., 2018a; Flemer et al., 2017; Sobhani et al., 2011) | ||
| EURYARCHAEOTA | |||||||
| Methanobrevibacter | – | F, T | NA/methane producer | 16S, qPCR | Mira-Pascual et al. (2015) | ||
| FIRMICUITES | |||||||
| Enterococcus | E. faecalis | F, T | Inflammatory, oxidative stress | 16S, qPCR | (Wang et al., 2012; Zhou et al., 2016) | ||
| Gemella | --- | F, T | NA | 16S | (Allali et al., 2015; Chen et al., 2012) | ||
| Mogibacterium | – | F, T | NA | 16S | Chen et al. (2012) | ||
| Parvimonas | P. micra* | F, T | Inflammatory, Immune response | 16S, WGS, qPCR | (Allali et al., 2015; Dai et al., 2018a; Drewes et al., 2017; Flemer et al., 2018; Yu et al., 2017) | ||
| Peptostreptococcus | P. stomatis*, P. anaerobious | F, T | Oxidative stress | 16S, WGS, qPCR | (Chen et al., 2012; Drewes et al., 2017; Tsoi et al., 2017; Wang et al., 2012) | ||
| Staphylococcus | --- | F | NA | 16S | Wu et al. (2013) | ||
| Solobacterium | S. moorei | F | NA | WGS, qPCR | Yu et al. (2017) | ||
| Streptococcus | S. gallolyticus* (previously S.bovis biotypeI) | F, T | Inflammatory | 16S, qPCR | (Allali et al., 2015; Boleij and Tjalsma, 2013; Klein et al., 1977; Shen et al., 2010; Wang et al., 2012) | ||
| FUSOBACTERIA | |||||||
| Fusobacterium | F. nucleatum* | F, T | Inflammatory, butyrate producer | 16S, WGS, qPCR | (Dai et al., 2018a; Drewes et al., 2017; Hamada et al., 2018; Kostic et al., 2013; Liang et al., 2017; Marchesi et al., 2011; Sinha et al., 2016; Wong et al., 2017a; Zhernakova et al., 2016) | ||
| PROTEOBACTERIA | |||||||
| Escherichia | --- | F, T | Genotoxin (colibactin), DNA mismatch repair, DNA damage checkpoint | 16S | (Nougayrède et al., 2006; Swidsinski et al., 1998; Taieb et al., 2016; Wang et al., 2012) | ||
| Helicobacter | H. pylori | T | Inflammatory | 16S | (Grahn et al., 2005; Kamada et al., 2013) | ||
| Klebsiella | – | F, T | Alter the cell biology of the host | 16S | (Chen et al., 2012; Wang et al., 2012) | ||
| SYNERGISTETES | |||||||
| Thermanaerovibrio | T. acidaminovorans* | F | NA | WGS | Dai et al. (2018a) | ||
| VERRUCOMICROBIA | |||||||
| Akkermansia | A. muciniphila | F, T | Immune modulatory (involved in PD-1 blockade efficacy) | 16S | (Mira-Pascual et al., 2015; Routy et al., 2018; Weir et al., 2013) | ||
| Depleted in CRC | |||||||
| ACTINOBACTERIA | |||||||
| Bifidobacterium | Several | F, T | Immune modulatory, anti-inflammatory, butyrate production | 16S, WGS, qPCR | (Candela et al., 2011; Chen et al., 2012; Feng et al., 2015; Gueimonde et al., 2007) | ||
| BACTEROIDETES | |||||||
| Bacteroides | B. vulgatus, B. uniformin | F | Inflammatory | 16S | Wang et al. (2012) | ||
| FIRMICUITES | |||||||
| Anaerostipes | – | T | Butyrate producer | 16S | Chen et al. (2012) | ||
| Clostridium | C. butyricum | F | Secondary bile acids producer, apoptosis of CRC cells, inhibition of tumorigenesis (mice) | WGS | Dai et al. (2018a) | ||
| Eubacterium | E. ventriosum | F, T | Inflammatory, butyrate producer, DNA damage | 16S, WGS, | (Chen et al., 2012; Wu et al., 2013; Yu et al., 2017) | ||
| Faecalibacterium | F. prausnitzii | F, T | Anti-inflammatory, butyrate producer | 16S, qPCR | (Balamurugan et al., 2008; Chen et al., 2012; Marchesi et al., 2011; Shah et al., 2018) | ||
| Lactobacillus | – | F, T | Immune modulatory (activation T-cells), mucus barrier maintenance | 16S | Chen et al. (2012) | ||
| Roseburia | – | F, T | Anti-inflammatory, butyrate producer | 16S | (Marchesi et al., 2011; Meehan and Beiko, 2014; Wang et al., 2012) | ||
| Ruminococcus | R. gnavus | T | SCFA producer, secondary bile acid producer | 16S | (Chen et al., 2012; Richard et al., 2018) | ||
| PROTEOBACTERIA | |||||||
| Citrobacter | – | T | Inflammatory | 16S | Marchesi et al. (2011) | ||
| Cronobacter | – | T | Inflammatory | 16S | Marchesi et al. (2011) | ||
| Kluyvera | – | T | Inflammatory | 16S | Marchesi et al. (2011) | ||
| Salmonella | – | T | Inflammatory | 16S | Marchesi et al. (2011) | ||
| Serratia | --- | T | Inflammatory | 16S | Marchesi et al. (2011) | ||
| SPIROCHAETES | |||||||
| Treponema | --- | F | NA | 16S | Zhu et al. (2014) | ||
| Enriched in CRC feces/Depleted in CRC tumor tissue | |||||||
| PROTEOBACTERIA | |||||||
| Shigella | --- | F, T | Inflammatory | 16S | (Marchesi et al., 2011; Tjalsma et al., 2012; Wang et al., 2012) | ||
Different hypotheses have been put forward to explain the role of microbial unbalance in carcinogenesis. Some authors propose that some types of dysbiotic gut microbiota originate a functional imbalance that triggers sustained pro-inflammatory responses and epithelial cell transformation, leading to cancer (Hajishengallis et al., 2012; Lamont and Hajishengallis, 2015; Maloy and Powrie, 2011). A related hypothesis is the ‘driver-passenger theory’ (Tjalsma et al., 2012). According to this model indigenous intestinal bacteria (defined as driver bacteria) trigger the DNA damage in epithelial cells, which in turn contributes to cancer initiation. In a second step, ongoing tumorigenesis alters the surrounding microenvironment, favoring the proliferation of opportunistic bacteria (termed bacterial passengers). Thus, this model proposes that disease progression causes changes in the microenvironment as a result of the growing tumor, and bacteria are replaced by others which show a competitive advantage in the tumor microenvironment and are capable of nurturing tumor progression. This scenario suggests that bacterial drivers and passengers have distinct temporal associations with CRC and may have separate roles in pathogenesis.
4. Microbiome-enabled early diagnostics of CRC
The transition from normal mucosa to CRC is a multi-step process, from the development of pre-neoplastic lesions to the adenomatous polyps. The adenoma-carcinoma sequence may take at least 10 years to develop, which makes CRC an ideal malignancy to be screened. Screening can prevent CRC by the detection and removal of precancerous growths, or by enabling the detection of cancer at an early stage, when treatment has a higher success rate. Guided by these promising facts, several countries have adopted population-wide screening and prevention programs aimed at improving CRC survival. The main screening strategies used worldwide are the fecal occult blood test (FOBT) and the fecal immunochemical test (FIT), which are generally coupled to a subsequent colonoscopy when the test is positive and other parameters recommend it (Navarro et al., 2017). A broad meta-analysis showed that FOBT and FIT reduced the mortality of CRC by 14% and 59%, respectively, as compared with no screening in average-risk populations (Zhang et al., 2017a). This study also concluded that colonoscopy is the most effective examination to reduce CRC mortality. However, colonoscopy requires previous bowel preparation and sedation, and it is an expensive, time-consuming, and invasive procedure that exposes patients to serious medical complications (Rutter et al., 2014). These limitations prevent the application of colonoscopy on population-wide screenings and highlight the need for efficient pre-colonoscopy tests. FOBT and FIT share the advantage of being non-invasive, stool-based techniques but they have a moderate sensitivity (69–86%) to detect CRC (Lee et al., 2014; Robertson et al., 2017). Thus, despite the undeniable advances for CRC prevention brought about by the current screening techniques, research for more accurate diagnostic and prognostic biomarkers is still needed. Most current research is directed towards finding additional criteria, such as risk factors and other biomarkers to be considered by the decision algorithms used to derive persons with a positive FOBT or FIT test to colonoscopy. In this sense, some molecular biomarkers related to the processes underlying carcinogenesis in CRC are being considered. These include circulating tumor cells, cell-free DNA, microRNAs as well as metabolites from plasma samples (Coghlin and Murray, 2015).
Given the growing evidence of the existence of microbiome alterations associated to CRC, and the likely involvement of the microbiota in the origin and progression of cancer discussed above, microbial markers have recently emerged as a promising additional factor to be considered in early screenings. In this context, over the last years, many studies have shown the possibility of using different microbiome signatures (especially in stool) to differentiate between patients with CRC and control individuals, and to some extent also from advanced adenomas (Konstantinov et al., 2013). A general observation from studies comparing CRC and control microbiomes is that the overall microbial composition does not seem to differ extensively, but differences rather involve abundances of some key organisms (Zeller et al., 2014). Accordingly, an effort in many such studies is to narrow down whole microbiome signatures to differences in specific species or sets of species that could act as indicators. This could significantly reduce the cost and time of diagnostic efforts by enabling the development of specific tests for the presence and abundance of those indicative species, without the need to assess the whole microbiome. For instance, as mentioned above, F. nucleatum has consistently been found associated with CRC development and progression, being enriched both in feces of patients with adenoma and CRC as compared to control individuals, as well as in tumoral tissue in comparison with surrounding normal tissue (Castellarin et al., 2012; Flanagan et al., 2014; Kostic et al., 2012; Yu et al., 2017). Consistently, a recent meta-analysis concluded that this sole species could be used as a biomarker for a non-invasive screening in CRC and colorectal adenoma (Zhang et al., 2019). Additional efforts have been conducted to identify bacterial markers to predict the risk of developing CRC in a more efficient way (Ai et al., 2017; Rezasoltani et al., 2018; Zeller et al., 2014), with a claim of similar accuracy to the standard FOBT (Zeller et al., 2014). However, it is still unclear whether such studies are extrapolable to other populations, as host genetics, and environmental and lifestyle factors are known to play an important role in the composition of the gut microbiota. Additionally, differences between studies, including the quality of samples, or the experimental protocols and bioinformatics tools used can have an influence. To overcome this potential lack of reproducibility and to obtain more consistent outcomes, two recent meta-analyses have been performed, which gathered 16S sequencing (Shah et al., 2018) or shotgun metagenomic data (Dai et al., 2018a) from different populations. In both of them, the authors identified microbial markers consistently enriched or depleted in CRC, regardless of differences in population and technologies applied between studies. Specifically, one of these studies (Dai et al., 2018a) found seven bacterial markers that were enriched in CRC patients and had a good a performance in differentially classifying CRC from controls across four ethnic cohorts from three different countries. A broader meta-analysis collecting 28 gut microbiome datasets showed that some studies claiming to present disease links actually shared some microbiome signatures, suggesting that some reported connections may actually be non-specific (Duvallet et al., 2017). Nevertheless, these authors did show that CRC patients could be clustered separately from controls based on the microbiome in all four CRC studies included, and that there was significant agreement in the observed microbial shifts in three out of the four. In any case, as in any other disease association study, the potential for overlap and uncertainty in markers identified by different studies requires further exploration. Hence, despite the great advances disentangling the bacterial signatures underlying CRC, the use of microbiome biomarkers for early detection of CRC is still not being used in clinical routine.
In addition to the use of the microbiome, some studies have explored the metabolome, mostly in feces, to find CRC-associated signatures [reviewed in (Zhang et al., 2017b)]. Some fecal metabolites (short-chain fatty acids such as acetate and butyrate, fructose, linoleic acid and nicotinic acid among others) have been suggested as potential diagnostic markers of CRC since they have been found at lower levels in fecal metabolome of CRC patients in comparison to healthy individuals (Monleón et al., 2009; Phua et al., 2014). Some other studies claim the possibility of discriminating between CRC patients and tumor-free controls using a panel of urinary metabolites (Cheng et al., 2012) or differentially expressed serum metabolites with a sensitivity of 0.981 and a specificity of 1.000 (Li et al., 2013). However, a prospective cohort study of patients with CRC could distinguish early-stage patients from more advanced stages of the disease but not intermediate stages based on the urinary metabolome (Liesenfeld et al., 2015). As in the case of the microbiome, extrinsic and genetic factors may affect the metabolome, and this technique also suffers from similar reproducibility and standardization problems (Villéger et al., 2018). Thus metabolic profiling has the potential of being used as a biomarker for CRC diagnosis, but still more extended and comparable studies are needed to obtain more clear and robust conclusions. Overall, the final purpose of all this emerging research is integrating information on genomics, epigenetics together with microbiome, metabolomics and lifestyle and environmental factors to construct new risk models which are more robust and precise, thereby enabling a better prediction on CRC initiation, progression as well as therapeutic strategies.
5. The gut microbiome as a therapeutic target in CRC
Apart from the effects of human host genetics (Hall et al., 2017; Kurilshikov et al., 2017), the composition of the gut microbiome is influenced by many host extrinsic factors, including diet (Singh et al., 2017), medications (Becattini et al., 2016; Le Bastard et al., 2018; Maier et al., 2018), and other lifestyle components, such as exercise, smoking, and sleep cycles (Biedermann et al., 2013; Claus et al., 2016; Das et al., 2018; Monda et al., 2017). In addition, research leading to the design of pre- and probiotics able to modulate the gut microbiome, the transplantation of fecal microbiota from healthy donors, or the use of phage therapy, among other strategies, has intensified in recent years (Zitvogel et al., 2018). This modulability of the gut microbiota opens the possibility of novel therapeutic approaches to prevent or treat diseases that are originated or influenced by alterations in the microbiota. Most clinical and pre-clinical studies assessing microbiome-modulation treatments suggest that these therapies may have an advantage over synthetic drugs, at least in terms of their potential side-effects (Juul et al., 2018; Lynch and Pedersen, 2016). Given the potential relationships of the gut microbiota with the origin and development of CRC discussed above, there is an increasing interest in exploring microbiome-related therapies for aiding in the prevention and treatment of this cancer. So far, strategies under development for CRC include fecal microbiota-transplantation (FMT), use of pre-/probiotics and diet, and phage therapy.
FMT was first described in 1958 (Eiseman et al., 1958), when it was used to treat Clostridium difficile infection (CDI), and it was shown to help restore a beneficial microbial composition. Later on, several studies showed that FMT was an effective treatment in more than 80% of patients with CDI (Bakken et al., 2011; Lee et al., 2016; van Nood et al., 2013). More recently, FMT has been re-evaluated as a promising therapeutic method to treat other disorders involving gut dysbiosis, such as CRC, ulcerative colitis, IBD, IBS, metabolic syndrome, obesity, types 1 and 2 diabetes, atopy, multiple sclerosis, autism (Filip et al., 2018). Some case reports have shown favorable outcomes, yet demonstrating these effects on a larger scale has proved difficult. In a single-arm open-label study of patients with IBS, FMT proved to be safe and relatively effective (Mizuno et al., 2017), however further studies are needed to further develop this approach.
To date there are no conclusive data from clinical trials in humans using FMT to treat CRC, but two recent studies using mouse models have shown its potential. The first one demonstrated that the gut microbiome of wild-type mice, when transplanted, conferred traits that promote host fitness and limit inflammation of induced neoplastic development (Rosshart et al., 2017). A second study showed that fecal microbiota from patients with CRC promoted tumorigenesis in germ-free mice and mice supplied with a carcinogen (Wong et al., 2017b). Overall, FMT stands out as a promising strategy in the treatment of CRC patients, although there are still open questions to be discussed and potential risks that need a deeper revision. For the use of FMT in this and other diseases, the debate is still open with respect to factors such as optimal storage conditions of stool, standardization of processes, selection of donors and recipients, as well as establishment of a proper cycle of FMT administration, dose, and frequency. There is at least one clinical study showing no significant difference in clinical resolution of patients with CDI comparing both frozen and fresh FMT (Lee et al., 2016). Safety and efficacy of FMT in CDI has been proven, although larger long-term follow-up studies are needed to identify potential long-term adverse effects. Potential risks of FMT include transmission of pathogens, particularly to immunocompromised patients, transmission of recessive elements silent in healthy donors (Sbahi and Di Palma, 2016), and transmission of other factors accounting for chronic diseases i.e. although controversial, a case report suggested transmission of obesity to a patient (Alang and Kelly, 2015). Among future desirable developments it is the combination of FMT with fecal DNA testing for accuracy in CRC screening, as well as a transition from whole microbiome transplant to more precise combinations of microbes (Dai et al., 2018b).
A more direct strategy is to specifically target bacteria associated to CRC development (i.e. F. nucleatum, B. fragilis, or E. coli). Antibiotics have been successfully used to modulate the microbiome and indirectly affect CRC progression. For instance, one study used antibiotics to treat mice xenografted with CRC (Bullman et al., 2017), and showed that the treatment reduced both the load of F. nucleatum and the overall growth of the tumor. However, the use of antibiotics generally have broad effects on the gut microbiota, often leading to dysbiosis and facilitating the acquisition of drug resistance. Therefore, there is a need for new products that more specifically target bacteria associated to CRC. In this context, some molecules have shown promising effects in CRC and other diseases. For instance, a glycopolymer, antagonist of the E. coli virulence factor FimH, showed a reduction in the adherence of E. coli to the intestinal epithelia, and therefore its invasiveness in a mouse model of Crohn's disease (Khan et al., 2017). Also, the exposure to a low dose of a recombinant BFT-2 enterotoxin (a major virulence factor of B. fragilis) decreased the formation of tumors in a CRC mouse model (Lv et al., 2017). Some authors have highlighted the importance of exploring the interaction between gut bacteria and the viral component, and its potential modulation via phage therapy. Phage therapy is a matter of current interest for its potential advantages in fighting antibiotic-resistant bacteria and its potential application the clinics (Mirzaei and Maurice, 2017).
Diet has been proposed as one of the most influential factors shaping the human gut microbiome (Singh et al., 2017). In particular, current data shows that gut microbiota changes rapidly in response to dietary changes. The observation that diet can modulate host-microbe interactions, suggests a promising therapeutic approach, with effect on immune response and metabolic pathways. Studies on mouse models have shown that alterations of the microbial community induced by deoxycholic acid (a secondary bile acid increased by western-type diet) promoted intestinal carcinogenesis (Cao et al., 2017), and that supplementation with high dietary fiber and butyrate-producing bacteria can significantly reduce colon tumor growth (Donohoe et al., 2014). Two further studies performed in pre-clinical tumor models showed that gut microbiome may influence the response of two different cancer immunotherapy agents (anti-CTLA4, and anti-PD-L1), by augmenting activation of dendritic and anti-tumor T cell responses (West and Powrie, 2015). In particular, the authors found that immunologic stimulation by Bacteroides spp. and Bifidobacterium spp. respectively, have a profound effect on therapy efficacy. Moreover, it has been reported that imbalance of dietary sphingolipids can have a major impact on the therapeutic efficacy of chemotherapy and radiation (Camp et al., 2017). In this context, several authors have suggested the need of developing more personalized methods to study dietary interventions which might increase the efficiency of CRC treatments, as for other diseases dietary interventions are already under clinical trials (Zeevi et al., 2015).
Another line of current research includes the use of probiotics. Probiotics help maintain healthy microbiota states by regulating pathogenic bacteria and immune system response, which in turn may reduce blood cholesterol, colitis, and prevent CRC (Raman et al., 2013). Different probiotics can inhibit CRC by different mechanisms: releasing detoxifying agents, anti-inflammatory factors, anti-cancer compounds (anti-angiogenesis, promoting anti-PDL1 drugs), and short-chain fatty acids (SCFA) that improve the intestinal barrier function (Lin et al., 2018). So far, several lines of evidence support a protective role of probiotics against CRC. For instance, it has been shown that the butyrate producing species Clostridium butyricum and Bacillus subtilis may have an anti-tumor effect in a CRC mouse model (Chen et al., 2015). Interestingly, another probiotic, Lactobacillus casei (strain BL23) not only inhibited CRC in a mouse model, but also, re-established the disrupted gut dysbiosis in CRC (Lenoir et al., 2016), whereas L. casei (strain ATCC 334) has been reported to produce a molecule: ferrichrome, which has been shown to inhibit progression of colon cancer by means of apoptosis mediated through the c-Jun N-terminal kinase pathway (Konishi et al., 2016). Another L. casei strain (variety rhamnosus, Lcr35) prevented induced intestinal mucositis in CRC-bearing mice (Chang et al., 2018). Recent clinical studies found out that Bifidobacterium probiotics could restore the equilibrium of gut dysbiosis and reduce small intestinal bacterial overgrowth induced in patients with CRC (Liang et al., 2016). Other authors suggest to focus on strategies that promote growth of positive bacteria to increase antitumor immunity, for instance, by use of prebiotics highly associated with diet. Also, the synergistic combination of prebiotics and probiotics in the form of dietary supplements or ingredients has been formulated, being called synbiotics. These include for instance OAT fiber/L. Plantarum and FOS/L. sporogens (Pandey et al., 2015). More recently, some authors highlighted the benefits of creating new generation probiotics for the treatment and prevention of CRC using genetic engineering techniques. For instance, increasing the potency of a well-documented probiotic species such as Bifidobacterium animalis subsp. Lactis (main source of butyrate: an anti-inflammatory and anti-tumor molecule), with the ability to produce mycosporin-like amino acids which exert prebiotic effects and is able to modulate host immunity by regulating cytokine production and the proliferation and differentiation of intestinal epithelial cells, macrophages and lymphocytes (Bozkurt et al., 2019). To date, 24 clinical trials of pro-/synbiotics therapies have been published and shown a favorable benefit for CRC patients (Ding et al., 2018). As previously mentioned, gut microbiome has a natural anticancer effect through the stimulation of the immune system. Therefore, it has been suggested that regulation of patients’ immune systems through the microbiota is one of the key mechanisms of tumor immunotherapy. It has been suggested that gut microbiota activate natural anticancer immune responses through three main mechanisms: microbial or tumoral antigens activate T-cell response which in turn may activate tumor-specific immune responses (Zitvogel et al., 2018). Among the species identified with known anti-tumoral response through activation of immune system (T-cells) there are: Bifidobacteria spp. (Matson et al., 2018; Sivan et al., 2015), Akkermansia muciniphilia (Routy et al., 2018), Enterococcus hirae (Daillère et al., 2016), Bacteroides spp (Vétizou et al., 2015). Alternative mechanisms include the activation of pattern recognition receptors that mediate pro-immune or anti-inflammatory effects, and, release of small metabolites that mediate systemic effects in the host (such as polyamines, vitamin and desaminotyrosine, among others (Ding et al., 2018)). Indeed, several studies showed a relation between the gut microbiota and the efficacy and toxicity of chemotherapies and immunotherapies (Ding et al., 2018; Iida et al., 2013; Pouncey et al., 2018). Taken altogether, these data highlight the interest of investigating the effect of new therapeutic strategies for the treatment of different types of cancer, including CRC, that benefit from the combined strategy of chemotherapy and/or immunotherapy with adjuvant treatments targeting the gut microbiota based on more personalized medicine, such as diet, probiotics and prebiotics. In this respect, recent studies have shown that oral therapy with bifidobacteria combined with PD-L1 inhibitors had a synergistic effect inhibiting almost completely the tumor growth, as compared with the individual effect of the drugs (Ding et al., 2018; Lee and Le, 2016; Routy et al., 2018).
6. Current challenges
As discussed in the previous sections, many studies have demonstrated the association between the microbiome composition and cancer, particularly CRC. Still the field faces many serious limitations that need to be considered. A recurrent limitation is that the non-bacterial components of the microbiome (i.e. fungi, protozoans, viruses) have been barely studied, mainly due to either their lower abundances or the technical difficulties in targeting them. In this regard, some promising fields which are growing rapidly are the study of the gut virome and mycobiome (i.e. the viral, and fungal components of the microbiome, respectively) in relation to CRC pathology. For example, a recent MWAS explored the virome community in fecal samples from CRC patients and control individuals using a WGS approach (Nakatsu et al., 2018). The authors reported a significant increase in the diversity of the bacteriophage virome in CRC patients and identified a set of viral markers (e.g. Orthobunyavirus, Inovirus and Tunalikevirus) that discriminated between CRC patients and healthy subjects and improved the performance of FOBT or FIT tests for detection of CRC. Gut virome dysbiosis was associated with CRC stage (early-vs late) and the prognosis of the disease, but it could not differentiate patients with colorectal adenomas from control individuals.
Similarly, the mycobiome has been the target of very few studies in CRC. Whereas it has been estimated that fungi are orders of magnitude less abundant than bacteria, they are ubiquitous and play key roles in the maintenance of equilibrium at the microbiome (Huffnagle and Noverr, 2013). A recent fecal metagenomics study compared the mycobiome in CRC and control individuals (Coker et al., 2018a). Principal component analyses showed different clusters of fungal components for controls, early-stage CRC and late-stage CRC patients, indicating that mycobiome profiles were stage-specific. In addition, the Basidiomycota:Ascomycota ratio, which is an indicator of fungal dysbiosis, was higher in CRC patients, which were enriched in Malasseziomycetes and depleted in Saccharomycetes and Pneumocystidomycetes. At the species level, a combination of 14 fungal biomarkers were able to distinguish CRC patients from controls, and controls from early-stage CRC, both findings being validated in two independent cohorts. However, there is still very limited information about the involvement of the fungal community in carcinogenic processes, particularly in CRC. Improvement of standardization and methodologies to study the mycobiome would contribute to shedding new light on the etiopathology of the disease and to refining of diagnosis and prognosis biomarkers.
Related to the limitation of under-studied taxonomic groups, there is the limited taxonomic resolution achieved in the studied ones, which currently stays at the genus or higher levels in most studies. This lack of resolution limits our understanding of the functional impact of the microbiome, as the coding and functional potential can vary greatly among species of the same microbial genus, and even among strains of the same species. Connected to this is the currently limited use of complementary techniques such as metatranscriptomics, metabolomics and metaproteomics (Mallick et al., 2017), which have the potential to help us provide a mechanistic understanding of the roles of the microbiome.
Sample collection is another challenging step in microbiome studies. Many studies on the gut microbiome, including those related to CRC disease, are conducted using fecal samples, since it is an easy and non-invasive procedure. While fecal samples represent a powerful strategy for finding biomarkers with diagnosis and prognosis purposes, tissue samples from colonic mucosa are more valuable to disentangle the physiopathology of CRC disease. In fact, there is strong evidence pointing to different microbiome profiles between mucosal and fecal samples (reviewed in (Yu et al., 2018)). A recent prospective study using paired fecal and mucosal samples from patients with CRC confirmed not only that the fecal microbiota did not totally reflect the underlying mucosal microbiota in CRC, but also that the colonic microbiota was different between patients with distal (including rectal) tumors and proximal tumors (Flemer et al., 2017). Accordingly, further studies should consider the most relevant sample collection procedure according to the objectives, and a better definition of subphenotypes and categorization of CRC tumors. Beyond sample collection issues, there are serious challenges regarding heterogeneity and limitations in study design and analysis. As mentioned above, broadening the scope of a study by including additional metadata can reveal potentially specific links to be confounding factors, but often few such data is collected or stored alongside the sequences. Additionally, new statistical methods for analyzing microbiome compositional data are indispensable (Dai et al., 2018b). For instance, current analyses treat every taxon independently, while we know of ecological correlations between taxa. Technical variation must also be considered in any omics study and batch effect correction is often necessary for comparisons between or even within studies (Costea et al., 2017; Duvallet et al., 2017). Furthermore, compositional data are generally analyzed with a focus on relative abundances of taxa within each sample. However, this approach does not account for the effects of changes in total abundances of taxa, which can be linked to various physiological factors or metabolite concentrations. A recent technique called quantitative microbiome profiling (QMP) addresses this issue by essentially sampling an equivalent number of sequences from each sample, and correcting the counts of taxa by both the number of copies of the 16S rRNA gene known to be in that organism and the number of cells from that sample, producing the total microbial load (Vandeputte et al., 2017). QMP has shown that alterations in taxa that were previously associated with Crohn's disease are mostly driven by differences in microbial load. This technique can also reveal potential artefacts, as in the same dataset they showed that Bacteroides was found at higher relative abundance in Crohn's samples, but there was no significant difference in microbial load. The reverse was true for Prevotella, wherein relative abundances were not significantly different between controls and Crohn's samples, but QMP revealed a significantly lower microbial load of Prevotella in Crohn's. QMP is an example of a straightforward technique that can help to mitigate some of the technical variation that occurs in microbiome studies.
Another limitation, shared by many GWASs and MWASs is the potential lack of representativity of existing datasets, which are mostly focused on population of the “westernized” world. A recent study revealed the limited scope of existing data (Pasolli et al., 2019). By combining 50 publicly available datasets collectively representing 9500 WGS samples from various body sites, they more than doubled the current public repository of microbial genomes associated with the human microbiome. Many of these new genomes were found in “non-westernized” populations, hinting at our lack of a global understanding of the human microbiome. Another study supporting this idea made available the microbiome of an uncontacted Amerindian population in Brazil (Clemente et al., 2015). The authors suggest the need for extensive characterization of the function of the microbiome and resistome in remote “non-westernized" populations before globalization of modern practices affects potentially beneficial bacteria harbored in the human body. The study of the microbiome in more diverse populations would enable the generation of worldwide databases which include epidemiological and host-genetic information, for further comprehensive studies.
7. Future perspectives: from bench to bedside
Despite the many limitations mentioned in the previous section, the good news is that many of them will be solved or attenuated by continuous improvements and cost reductions in sequencing technologies. Considering this, it is reasonable to predict that the near future will witness an increased focus on less covered taxonomic groups (i.e fungi, viruses) alongside bacteria, will provide increased resolution (i.e. at the species and strain level), and will additionally target less westernized populations (i.e African, Native American, etc). In addition improvements in study design, analytical methods, and increased awareness of interactions and confounding variables will help us to better disentangle relationships between microbes and origin and progression of diseases, including CRC. Similarly, we expect an increasing use, in combination with metagenomics, of complementary approaches such as metabolomics and metatranscriptomics, which will help us moving from descriptive studies to those able to derive mechanistic models of how microbes and hosts interact.
In this respect, it is also reasonable to predict a trend from generalized studies to those focusing on more personalized patterns. The analysis of longitudinal studies compiling multi-dimensional data about microbiome, diet, metabolic parameters and considering potential underlying disorders, will help to develop personalized diets and lifestyle recommendations to fine-tune the microbiota composition towards a healthy state. Also, we could hypothesize that once the composition of the microbiome of a given patient is characterized, he or she could benefit from personalized pre- and probiotics, or from a tailored phage therapy to restore a desired bacterial equilibrium (Paule et al., 2018). Furthermore, development of new culturing technologies may help to study for instance difficult-to-culture gut microorganisms in tissue engineering organoids (Williamson et al., 2018). This will constitute an ideal manipulable and fully-controlled platform to study mechanisms of host-pathogen interactions (Arnold et al., 2016). Moreover, it would pave the way for the testing and development of personalized therapies, drug screening, or pre-clinical studies for newly developed drugs.
Funding
This work was supported by grants from COST (European Cooperation in Science and Technology), COST Action CA17118; from the Spanish Ministry of Economy, Industry, and Competitiveness (MEIC) for the EMBL partnership, and grants ‘Centro de Excelencia Severo Ochoa’ SEV-2012-0208, and BFU2015-67107 cofounded by European Regional Development Fund; from the CERCA Programme/Generalitat de Catalunya; from the Catalan Research Agency (AGAUR) SGR857, and grants from the European Union’s Horizon 2020 research and innovation programme under the grant agreement ERC-2016-724173, and the Marie Sklodowska-Curie grant agreement No H2020-MSCA-ITN-2014-642095; and from Instituto Nacional de Bioinformatica (INB, grant PT17/0009/0023 - ISCIII-SGEFI/ERDF).
Declarations of interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mam.2019.05.001.
Contributor Information
Ester Saus, Email: ester.saus@crg.eu.
Susana Iraola-Guzmán, Email: susana.iraola@crg.eu.
Jesse R. Willis, Email: jesseR.Willis@crg.eu.
Anna Brunet-Vega, Email: ABrunetV@tauli.cat.
Toni Gabaldón, Email: toni.gabaldon.bcn@gmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Aguiar-Pulido V., Huang W., Suarez-Ulloa V., Cickovski T., Mathee K., Narasimhan G. Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis. Evol. Bioinform. Online. 2016;12:5–16. doi: 10.4137/EBO.S36436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J., Sinha R., Pei Z., Dominianni C., Wu J., Shi J., Goedert J.J., Hayes R.B., Yang L. Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013;105:1907–1911. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai L., Tian H., Chen Z., Chen H., Xu J., Fang J.-Y. Systematic evaluation of supervised classifiers for fecal microbiota-based prediction of colorectal cancer. Oncotarget. 2017;8:9546–9556. doi: 10.18632/oncotarget.14488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alang N., Kelly C.R. Weight gain after fecal microbiota transplantation. Open Forum Infect Dis. 2015;2:ofv004. doi: 10.1093/ofid/ofv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allali I., Delgado S., Marron P.I., Astudillo A., Yeh J.J., Ghazal H., Amzazi S., Keku T., Azcarate-Peril M.A. Gut microbiome compositional and functional differences between tumor and non-tumor adjacent tissues from cohorts from the US and Spain. Gut Microb. 2015;6:161–172. doi: 10.1080/19490976.2015.1039223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S. Shotgun DNA sequencing using cloned DNase I-generated fragments. Nucleic Acids Res. 1981;9:3015–3027. doi: 10.1093/nar/9.13.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleyard C.B., Cruz M.L., Isidro A.A., Arthur J.C., Jobin C., De Simone C. Pretreatment with the probiotic VSL#3 delays transition from inflammation to dysplasia in a rat model of colitis-associated cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;301:G1004–G1013. doi: 10.1152/ajpgi.00167.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage E.G., Barbas C. Metabolomics in cancer biomarker discovery: current trends and future perspectives. J. Pharm. Biomed. Anal. 2014;87:1–11. doi: 10.1016/j.jpba.2013.08.041. [DOI] [PubMed] [Google Scholar]
- Arnold J.W., Roach J., Azcarate-Peril M.A. Emerging technologies for gut microbiome research. Trends Microbiol. 2016;24:887–901. doi: 10.1016/j.tim.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta M.C., Bistritz L., Meddings J.B. Alterations in intestinal permeability. Gut. 2006;55:1512–1520. doi: 10.1136/gut.2005.085373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur J.C., Perez-Chanona E., Mühlbauer M., Tomkovich S., Uronis J.M., Fan T.-J., Campbell B.J., Abujamel T., Dogan B., Rogers A.B., Rhodes J.M., Stintzi A., Simpson K.W., Hansen J.J., Keku T.O., Fodor A.A., Jobin C. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R., Fernandes G.R., Tap J., Bruls T., Batto J.-M., Bertalan M., Borruel N., Casellas F., Fernandez L., Gautier L., Hansen T., Hattori M., Hayashi T., Kleerebezem M., Kurokawa K., Leclerc M., Levenez F., Manichanh C., Nielsen H.B., Nielsen T., Pons N., Poulain J., Qin J., Sicheritz-Ponten T., Tims S., Torrents D., Ugarte E., Zoetendal E.G., Wang J., Guarner F., Pedersen O., de Vos W.M., Brunak S., Doré J., MetaHIT Consortium, Antolín M., Artiguenave F., Blottiere H.M., Almeida M., Brechot C., Cara C., Chervaux C., Cultrone A., Delorme C., Denariaz G., Dervyn R., Foerstner K.U., Friss C., van de Guchte M., Guedon E., Haimet F., Huber W., van Hylckama-Vlieg J., Jamet A., Juste C., Kaci G., Knol J., Lakhdari O., Layec S., Le Roux K., Maguin E., Mérieux A., Melo Minardi R., M’rini C., Muller J., Oozeer R., Parkhill J., Renault P., Rescigno M., Sanchez N., Sunagawa S., Torrejon A., Turner K., Vandemeulebrouck G., Varela E., Winogradsky Y., Zeller G., Weissenbach J., Ehrlich S.D., Bork P. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken J.S., Borody T., Brandt L.J., Brill J.V., Demarco D.C., Franzos M.A., Kelly C., Khoruts A., Louie T., Martinelli L.P., Moore T.A., Russell G., Surawicz C., Fecal microbiota transplantation workgroup treating Clostridium difficile infection with fecal microbiota transplantation. Clin. Gastroenterol. Hepatol. 2011;9:1044–1049. doi: 10.1016/j.cgh.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamurugan R., Rajendiran E., George S., Samuel G.V., Ramakrishna B.S. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J. Gastroenterol. Hepatol. 2008;23:1298–1303. doi: 10.1111/j.1440-1746.2008.05490.x. [DOI] [PubMed] [Google Scholar]
- Barton W., Penney N.C., Cronin O., Garcia-Perez I., Molloy M.G., Holmes E., Shanahan F., Cotter P.D., O'Sullivan O. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut. 2018;67:625–633. doi: 10.1136/gutjnl-2016-313627. [DOI] [PubMed] [Google Scholar]
- Baxter N.T., Zackular J.P., Chen G.Y., Schloss P.D. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome. 2014;2:20. doi: 10.1186/2049-2618-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becattini S., Taur Y., Pamer E.G. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol. Med. 2016;22:458–478. doi: 10.1016/j.molmed.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann L., Zeitz J., Mwinyi J., Sutter-Minder E., Rehman A., Ott S.J., Steurer-Stey C., Frei A., Frei P., Scharl M., Loessner M.J., Vavricka S.R., Fried M., Schreiber S., Schuppler M., Rogler G. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059260. e59260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff S.C., Barbara G., Buurman W., Ockhuizen T., Schulzke J.-D., Serino M., Tilg H., Watson A., Wells J.M. Intestinal permeability--a new target for disease prevention and therapy. BMC Gastroenterol. 2014;14:189. doi: 10.1186/s12876-014-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boleij A., Tjalsma H. The itinerary of Streptococcus gallolyticus infection in patients with colonic malignant disease. Lancet Infect. Dis. 2013;13:719–724. doi: 10.1016/S1473-3099(13)70107-5. [DOI] [PubMed] [Google Scholar]
- Bosch T.C.G., McFall-Ngai M.J. Metaorganisms as the new frontier. Zoology (Jena) 2011;114:185–190. doi: 10.1016/j.zool.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt H.S., Quigley E.M., Kara B. Bifidobacterium animalis subspecies lactis engineered to produce mycosporin-like amino acids in colorectal cancer prevention. SAGE Open Med. 2019;7 doi: 10.1177/2050312119825784. 2050312119825784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullman S., Pedamallu C.S., Sicinska E., Clancy T.E., Zhang X., Cai D., Neuberg D., Huang K., Guevara F., Nelson T., Chipashvili O., Hagan T., Walker M., Ramachandran A., Diosdado B., Serna G., Mulet N., Landolfi S., Ramon Y Cajal S., Fasani R., Aguirre A.J., Ng K., Élez E., Ogino S., Tabernero J., Fuchs C.S., Hahn W.C., Nuciforo P., Meyerson M. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp E.R., Patterson L.D., Kester M., Voelkel-Johnson C. Therapeutic implications of bioactive sphingolipids: a focus on colorectal cancer. Cancer Biol. Ther. 2017;18:640–650. doi: 10.1080/15384047.2017.1345396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela M., Guidotti M., Fabbri A., Brigidi P., Franceschi C., Fiorentini C. Human intestinal microbiota: cross-talk with the host and its potential role in colorectal cancer. Crit. Rev. Microbiol. 2011;37:1–14. doi: 10.3109/1040841X.2010.501760. [DOI] [PubMed] [Google Scholar]
- Cao H., Xu M., Dong W., Deng B., Wang Sinan, Zhang Y., Wang Shan, Luo S., Wang W., Qi Y., Gao J., Cao X., Yan F., Wang B. Secondary bile acid-induced dysbiosis promotes intestinal carcinogenesis. Int. J. Cancer. 2017;140:2545–2556. doi: 10.1002/ijc.30643. [DOI] [PubMed] [Google Scholar]
- Carethers J.M., Jung B.H. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology. 2015;149:1177–1190. doi: 10.1053/j.gastro.2015.06.047. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin M., Warren R.L., Freeman J.D., Dreolini L., Krzywinski M., Strauss J., Barnes R., Watson P., Allen-Vercoe E., Moore R.A., Holt R.A. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-W., Liu C.-Y., Lee H.-C., Huang Y.-H., Li L.-H., Chiau J.-S.C., Wang T.-E., Chu C.-H., Shih S.-C., Tsai T.-H., Chen Y.-J. Lactobacillus casei variety rhamnosus probiotic preventively attenuates 5-fluorouracil/oxaliplatin-induced intestinal injury in a syngeneic colorectal cancer model. Front. Microbiol. 2018;9:983. doi: 10.3389/fmicb.2018.00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Liu F., Ling Z., Tong X., Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.-F., Ai L.-Y., Wang J.-L., Ren L.-L., Yu Y.-N., Xu J., Chen H.-Y., Yu J., Li M., Qin W.-X., Ma X., Shen N., Chen Y.-X., Hong J., Fang J.-Y. Probiotics Clostridium butyricum and Bacillus subtilis ameliorate intestinal tumorigenesis. Future Microbiol. 2015;10:1433–1445. doi: 10.2217/fmb.15.66. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Xie G., Chen T., Qiu Y., Zou X., Zheng M., Tan B., Feng B., Dong T., He P., Zhao L., Zhao A., Xu L.X., Zhang Y., Jia W. Distinct urinary metabolic profile of human colorectal cancer. J. Proteome Res. 2012;11:1354–1363. doi: 10.1021/pr201001a. [DOI] [PubMed] [Google Scholar]
- Claus S.P., Guillou H., Ellero-Simatos S. The gut microbiota: a major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes. 2016;2:16003. doi: 10.1038/npjbiofilms.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente J.C., Pehrsson E.C., Blaser M.J., Sandhu K., Gao Z., Wang B., Magris M., Hidalgo G., Contreras M., Noya-Alarcón Ó., Lander O., McDonald J., Cox M., Walter J., Oh P.L., Ruiz J.F., Rodriguez S., Shen N., Song S.J., Metcalf J., Knight R., Dantas G., Dominguez-Bello M.G. The microbiome of uncontacted Amerindians. Sci Adv. 2015;1 doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlin C., Murray G.I. Biomarkers of colorectal cancer: recent advances and future challenges. Proteonomics Clin. Appl. 2015;9:64–71. doi: 10.1002/prca.201400082. [DOI] [PubMed] [Google Scholar]
- Coker O.O., Nakatsu G., Dai R.Z., Wu W.K.K., Wong S.H., Ng S.C., Chan F.K.L., Sung J.J.Y., Yu J. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. 2018 doi: 10.1136/gutjnl-2018-317178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costea P.I., Zeller G., Sunagawa S., Pelletier E., Alberti A., Levenez F., Tramontano M., Driessen M., Hercog R., Jung F.-E., Kultima J.R., Hayward M.R., Coelho L.P., Allen-Vercoe E., Bertrand L., Blaut M., Brown J.R.M., Carton T., Cools-Portier S., Daigneault M., Derrien M., Druesne A., de Vos W.M., Finlay B.B., Flint H.J., Guarner F., Hattori M., Heilig H., Luna R.A., van Hylckama Vlieg J., Junick J., Klymiuk I., Langella P., Le Chatelier E., Mai V., Manichanh C., Martin J.C., Mery C., Morita H., O'Toole P.W., Orvain C., Patil K.R., Penders J., Persson S., Pons N., Popova M., Salonen A., Saulnier D., Scott K.P., Singh B., Slezak K., Veiga P., Versalovic J., Zhao L., Zoetendal E.G., Ehrlich S.D., Dore J., Bork P. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 2017;35:1069–1076. doi: 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- Costea P.I., Hildebrand F., Arumugam M., Bäckhed F., Blaser M.J., Bushman F.D., de Vos W.M., Ehrlich S.D., Fraser C.M., Hattori M., Huttenhower C., Jeffery I.B., Knights D., Lewis J.D., Ley R.E., Ochman H., O'Toole P.W., Quince C., Relman D.A., Shanahan F., Sunagawa S., Wang J., Weinstock G.M., Wu G.D., Zeller G., Zhao L., Raes J., Knight R., Bork P. Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 2018;3:8–16. doi: 10.1038/s41564-017-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello E.K., Stagaman K., Dethlefsen L., Bohannan B.J.M., Relman D.A. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Zhenwei, Coker O.O., Nakatsu G., Wu W.K.K., Zhao L., Chen Z., Chan F.K.L., Kristiansen K., Sung J.J.Y., Wong S.H., Yu J. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. 2018;6:70. doi: 10.1186/s40168-018-0451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Zhujiang, Zhang J., Wu Q., Chen J., Liu J., Wang L., Chen C., Xu J., Zhang H., Shi C., Li Z., Fang H., Lin C., Tang D., Wang D. The role of microbiota in the development of colorectal cancer. Int. J. Cancer. 2018 doi: 10.1002/ijc.32017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daillère R., Vétizou M., Waldschmitt N., Yamazaki T., Isnard C., Poirier-Colame V., Duong C.P.M., Flament C., Lepage P., Roberti M.P., Routy B., Jacquelot N., Apetoh L., Becharef S., Rusakiewicz S., Langella P., Sokol H., Kroemer G., Enot D., Roux A., Eggermont A., Tartour E., Johannes L., Woerther P.-L., Chachaty E., Soria J.-C., Golden E., Formenti S., Plebanski M., Madondo M., Rosenstiel P., Raoult D., Cattoir V., Boneca I.G., Chamaillard M., Zitvogel L. Enterococcus hirae and barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity. 2016;45:931–943. doi: 10.1016/j.immuni.2016.09.009. [DOI] [PubMed] [Google Scholar]
- Das B., Ghosh T.S., Kedia S., Rampal R., Saxena S., Bag S., Mitra R., Dayal M., Mehta O., Surendranath A., Travis S.P.L., Tripathi P., Nair G.B., Ahuja V. Analysis of the gut microbiome of rural and urban healthy Indians living in sea level and high altitude areas. Sci. Rep. 2018;8:10104. doi: 10.1038/s41598-018-28550-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C., Tang W., Fan X., Wu G. Intestinal microbiota: a novel perspective in colorectal cancer biotherapeutics. OncoTargets Ther. 2018;11:4797–4810. doi: 10.2147/OTT.S170626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe D.R., Holley D., Collins L.B., Montgomery S.A., Whitmore A.C., Hillhouse A., Curry K.P., Renner S.W., Greenwalt A., Ryan E.P., Godfrey V., Heise M.T., Threadgill D.S., Han A., Swenberg J.A., Threadgill D.W., Bultman S.J. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014;4:1387–1397. doi: 10.1158/2159-8290.CD-14-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes J.L., White J.R., Dejea C.M., Fathi P., Iyadorai T., Vadivelu J., Roslani A.C., Wick E.C., Mongodin E.F., Loke M.F., Thulasi K., Gan H.M., Goh K.L., Chong H.Y., Kumar S., Wanyiri J.W., Sears C.L. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes. 2017;3:34. doi: 10.1038/s41522-017-0040-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvallet C., Gibbons S.M., Gurry T., Irizarry R.A., Alm E.J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017;8:1784. doi: 10.1038/s41467-017-01973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzutsev A., Goldszmid R.S., Viaud S., Zitvogel L., Trinchieri G. The role of the microbiota in inflammation, carcinogenesis, and cancer therapy. Eur. J. Immunol. 2015;45:17–31. doi: 10.1002/eji.201444972. [DOI] [PubMed] [Google Scholar]
- Eiseman B., Silen W., Bascom G.S., Kauvar A.J. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery. 1958;44:854–859. [PubMed] [Google Scholar]
- Farshidfar F., Weljie A.M., Kopciuk K.A., Hilsden R., McGregor S.E., Buie W.D., MacLean A., Vogel H.J., Bathe O.F. A validated metabolomic signature for colorectal cancer: exploration of the clinical value of metabolomics. Br. J. Canc. 2016;115:848–857. doi: 10.1038/bjc.2016.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q., Liang S., Jia H., Stadlmayr A., Tang L., Lan Z., Zhang D., Xia H., Xu Xiaoying, Jie Z., Su L., Li Xiaoping, Li Xin, Li J., Xiao L., Huber-Schönauer U., Niederseer D., Xu Xun, Al-Aama J.Y., Yang H., Wang Jian, Kristiansen K., Arumugam M., Tilg H., Datz C., Wang Jun. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- Filip M., Tzaneva V., Dumitrascu D.L. Fecal transplantation: digestive and extradigestive clinical applications. Clujul Med. 2018;91:259–265. doi: 10.15386/cjmed-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan L., Schmid J., Ebert M., Soucek P., Kunicka T., Liska V., Bruha J., Neary P., Dezeeuw N., Tommasino M., Jenab M., Prehn J.H.M., Hughes D.J. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33:1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- Flemer B., Lynch D.B., Brown J.M.R., Jeffery I.B., Ryan F.J., Claesson M.J., O'Riordain M., Shanahan F., O'Toole P.W. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66:633–643. doi: 10.1136/gutjnl-2015-309595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemer B., Warren R.D., Barrett M.P., Cisek K., Das A., Jeffery I.B., Hurley E., O'Riordain M., Shanahan F., O'Toole P.W. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018;67:1454–1463. doi: 10.1136/gutjnl-2017-314814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming M., Ravula S., Tatishchev S.F., Wang H.L. Colorectal carcinoma: pathologic aspects. J. Gastrointest. Oncol. 2012;3:153–173. doi: 10.3978/j.issn.2078-6891.2012.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K., Hildebrand F., Nielsen T., Falony G., Le Chatelier E., Sunagawa S., Prifti E., Vieira-Silva S., Gudmundsdottir V., Pedersen H.K., Arumugam M., Kristiansen K., Voigt A.Y., Vestergaard H., Hercog R., Costea P.I., Kultima J.R., Li J., Jørgensen T., Levenez F., Dore J., MetaHIT consortium, Nielsen H.B., Brunak S., Raes J., Hansen T., Wang J., Ehrlich S.D., Bork P., Pedersen O. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa E.A., Morgan X.C., Segata N., Waldron L., Reyes J., Earl A.M., Giannoukos G., Boylan M.R., Ciulla D., Gevers D., Izard J., Garrett W.S., Chan A.T., Huttenhower C. Relating the metatranscriptome and metagenome of the human gut. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E2329–E2338. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnière J., Raisch J., Veziant J., Barnich N., Bonnet R., Buc E., Bringer M.-A., Pezet D., Bonnet M. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016;22:501–518. doi: 10.3748/wjg.v22.i2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Guo B., Gao R., Zhu Q., Qin H. Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 2015;6:20. doi: 10.3389/fmicb.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R., Kong C., Huang L., Li H., Qu X., Liu Z., Lan P., Wang J., Qin H. Mucosa-associated microbiota signature in colorectal cancer. Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:2073–2083. doi: 10.1007/s10096-017-3026-4. [DOI] [PubMed] [Google Scholar]
- Gilbert J.A., Quinn R.A., Debelius J., Xu Z.Z., Morton J., Garg N., Jansson J.K., Dorrestein P.C., Knight R. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature. 2016;535:94–103. doi: 10.1038/nature18850. [DOI] [PubMed] [Google Scholar]
- Goodwin A.C., Destefano Shields C.E., Wu S., Huso D.L., Wu X., Murray-Stewart T.R., Hacker-Prietz A., Rabizadeh S., Woster P.M., Sears C.L., Casero R.A. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahn N., Hmani-Aifa M., Fransén K., Söderkvist P., Monstein H.-J. Molecular identification of Helicobacter DNA present in human colorectal adenocarcinomas by 16S rDNA PCR amplification and pyrosequencing analysis. J. Med. Microbiol. 2005;54:1031–1035. doi: 10.1099/jmm.0.46122-0. [DOI] [PubMed] [Google Scholar]
- Gueimonde M., Ouwehand A., Huhtinen H., Salminen E., Salminen S. Qualitative and quantitative analyses of the bifidobacterial microbiota in the colonic mucosa of patients with colorectal cancer, diverticulitis and inflammatory bowel disease. World J. Gastroenterol. 2007;13:3985–3989. doi: 10.3748/wjg.v13.i29.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G., Darveau R.P., Curtis M.A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale V.L., Jeraldo P., Chen J., Mundy M., Yao J., Priya S., Keeney G., Lyke K., Ridlon J., White B.A., French A.J., Thibodeau S.N., Diener C., Resendis-Antonio O., Gransee J., Dutta T., Petterson X.-M., Sung J., Blekhman R., Boardman L., Larson D., Nelson H., Chia N. Distinct microbes, metabolites, and ecologies define the microbiome in deficient and proficient mismatch repair colorectal cancers. Genome Med. 2018;10:78. doi: 10.1186/s13073-018-0586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A.B., Tolonen A.C., Xavier R.J. Human genetic variation and the gut microbiome in disease. Nat. Rev. Genet. 2017;18:690–699. doi: 10.1038/nrg.2017.63. [DOI] [PubMed] [Google Scholar]
- Hamada T., Zhang X., Mima K., Bullman S., Sukawa Y., Nowak J.A., Kosumi K., Masugi Y., Twombly T.S., Cao Y., Song M., Liu L., da Silva A., Shi Y., Gu M., Li W., Koh H., Nosho K., Inamura K., Keum N., Wu K., Meyerhardt J.A., Kostic A.D., Huttenhower C., Garrett W.S., Meyerson M., Giovannucci E.L., Chan A.T., Fuchs C.S., Nishihara R., Giannakis M., Ogino S. Fusobacterium nucleatum in colorectal cancer relates to immune response differentially by tumor microsatellite instability status. Cancer Immunol Res. 2018;6:1327–1336. doi: 10.1158/2326-6066.CIR-18-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle G.B., Noverr M.C. The emerging world of the fungal microbiome. Trends Microbiol. 2013;21:334–341. doi: 10.1016/j.tim.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida N., Dzutsev A., Stewart C.A., Smith L., Bouladoux N., Weingarten R.A., Molina D.A., Salcedo R., Back T., Cramer S., Dai R.-M., Kiu H., Cardone M., Naik S., Patri A.K., Wang E., Marincola F.M., Frank K.M., Belkaid Y., Trinchieri G., Goldszmid R.S. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery I.B., Claesson M.J., O'Toole P.W., Shanahan F. Categorization of the gut microbiota: enterotypes or gradients? Nat. Rev. Microbiol. 2012;10:591–592. doi: 10.1038/nrmicro2859. [DOI] [PubMed] [Google Scholar]
- Jeffery I.B., Lynch D.B., O'Toole P.W. Composition and temporal stability of the gut microbiota in older persons. ISME J. 2016;10:170–182. doi: 10.1038/ismej.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juul F.E., Garborg K., Bretthauer M., Skudal H., Øines M.N., Wiig H., Rose Ø., Seip B., Lamont J.T., Midtvedt T., Valeur J., Kalager M., Holme Ø., Helsingen L., Løberg M., Adami H.-O. Fecal microbiota transplantation for primary Clostridium difficile infection. N. Engl. J. Med. 2018;378:2535–2536. doi: 10.1056/NEJMc1803103. [DOI] [PubMed] [Google Scholar]
- Kamada N., Seo S.-U., Chen G.Y., Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- Kelly C.P. Fecal microbiota transplantation--an old therapy comes of age. N. Engl. J. Med. 2013;368:474–475. doi: 10.1056/NEJMe1214816. [DOI] [PubMed] [Google Scholar]
- Khan A.A., Khan Z., Malik A., Kalam M.A., Cash P., Ashraf M.T., Alshamsan A. Colorectal cancer-inflammatory bowel disease nexus and felony of Escherichia coli. Life Sci. 2017;180:60–67. doi: 10.1016/j.lfs.2017.05.016. [DOI] [PubMed] [Google Scholar]
- Klein R.S., Recco R.A., Catalano M.T., Edberg S.C., Casey J.I., Steigbigel N.H. Association of Streptococcus bovis with carcinoma of the colon. N. Engl. J. Med. 1977;297:800–802. doi: 10.1056/NEJM197710132971503. [DOI] [PubMed] [Google Scholar]
- Konishi H., Fujiya M., Tanaka H., Ueno N., Moriichi K., Sasajima J., Ikuta K., Akutsu H., Tanabe H., Kohgo Y. Probiotic-derived ferrichrome inhibits colon cancer progression via JNK-mediated apoptosis. Nat. Commun. 2016;7:12365. doi: 10.1038/ncomms12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinov S.R., Kuipers E.J., Peppelenbosch M.P. Functional genomic analyses of the gut microbiota for CRC screening. Nat. Rev. Gastroenterol. Hepatol. 2013;10:741–745. doi: 10.1038/nrgastro.2013.178. [DOI] [PubMed] [Google Scholar]
- Kostic A.D., Gevers D., Pedamallu C.S., Michaud M., Duke F., Earl A.M., Ojesina A.I., Jung J., Bass A.J., Tabernero J., Baselga J., Liu C., Shivdasani R.A., Ogino S., Birren B.W., Huttenhower C., Garrett W.S., Meyerson M. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A.D., Chun E., Robertson L., Glickman J.N., Gallini C.A., Michaud M., Clancy T.E., Chung D.C., Lochhead P., Hold G.L., El-Omar E.M., Brenner D., Fuchs C.S., Meyerson M., Garrett W.S. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurilshikov A., Wijmenga C., Fu J., Zhernakova A. Host genetics and gut microbiome: challenges and perspectives. Trends Immunol. 2017;38:633–647. doi: 10.1016/j.it.2017.06.003. [DOI] [PubMed] [Google Scholar]
- Lamont R.J., Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol. Med. 2015;21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bastard Q., Al-Ghalith G.A., Grégoire M., Chapelet G., Javaudin F., Dailly E., Batard E., Knights D., Montassier E. Systematic review: human gut dysbiosis induced by non-antibiotic prescription medications. Aliment. Pharmacol. Ther. 2018;47:332–345. doi: 10.1111/apt.14451. [DOI] [PubMed] [Google Scholar]
- Lee V., Le D.T. Efficacy of PD-1 blockade in tumors with MMR deficiency. Immunotherapy. 2016;8:1–3. doi: 10.2217/imt.15.97. [DOI] [PubMed] [Google Scholar]
- Lee J.K., Liles E.G., Bent S., Levin T.R., Corley D.A. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Ann. Intern. Med. 2014;160:171. doi: 10.7326/M13-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.H., Steiner T., Petrof E.O., Smieja M., Roscoe D., Nematallah A., Weese J.S., Collins S., Moayyedi P., Crowther M., Ropeleski M.J., Jayaratne P., Higgins D., Li Y., Rau N.V., Kim P.T. Frozen vs fresh fecal microbiota transplantation and clinical resolution of diarrhea in patients with recurrent Clostridium difficile infection: a randomized clinical trial. J. Am. Med. Assoc. 2016;315:142–149. doi: 10.1001/jama.2015.18098. [DOI] [PubMed] [Google Scholar]
- Lenoir M., Del Carmen S., Cortes-Perez N.G., Lozano-Ojalvo D., Muñoz-Provencio D., Chain F., Langella P., de Moreno de LeBlanc A., LeBlanc J.G., Bermúdez-Humarán L.G. Lactobacillus casei BL23 regulates Treg and Th17 T-cell populations and reduces DMH-associated colorectal cancer. J. Gastroenterol. 2016;51:862–873. doi: 10.1007/s00535-015-1158-9. [DOI] [PubMed] [Google Scholar]
- Li Fang, Qin X., Chen H., Qiu L., Guo Y., Liu H., Chen G., Song G., Wang X., Li Fenjie, Guo S., Wang B., Li Z. Lipid profiling for early diagnosis and progression of colorectal cancer using direct-infusion electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 2013;27:24–34. doi: 10.1002/rcm.6420. [DOI] [PubMed] [Google Scholar]
- Li J., Jia H., Cai X., Zhong H., Feng Q., Sunagawa S., Arumugam M., Kultima J.R., Prifti E., Nielsen T., Juncker A.S., Manichanh C., Chen B., Zhang W., Levenez F., Wang Juan, Xu X., Xiao L., Liang S., Zhang D., Zhang Z., Chen W., Zhao H., Al-Aama J.Y., Edris S., Yang H., Wang Jian, Hansen T., Nielsen H.B., Brunak S., Kristiansen K., Guarner F., Pedersen O., Doré J., Ehrlich S.D., MetaHIT Consortium, Bork P., Wang Jun, MetaHIT Consortium An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- Liang S., Xu L., Zhang D., Wu Z. Effect of probiotics on small intestinal bacterial overgrowth in patients with gastric and colorectal cancer. Turk. J. Gastroenterol. 2016;27:227–232. doi: 10.5152/tjg.2016.15375. [DOI] [PubMed] [Google Scholar]
- Liang Q., Chiu J., Chen Y., Huang Y., Higashimori A., Fang J., Brim H., Ashktorab H., Ng S.C., Ng S.S.M., Zheng S., Chan F.K.L., Sung J.J.Y., Yu J. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin. Cancer Res. 2017;23:2061–2070. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- Liesenfeld D.B., Habermann N., Toth R., Owen R.W., Frei E., Staffa J., Schrotz-King P., Klika K.D., Ulrich C.M. Changes in urinary metabolic profiles of colorectal cancer patients enrolled in a prospective cohort study (ColoCare) Metabolomics. 2015;11:998–1012. doi: 10.1007/s11306-014-0758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Cai X., Zhang J., Wang W., Sheng Q., Hua H., Zhou X. Role of gut microbiota in the development and treatment of colorectal cancer. 2018. Digestion 1–7. [DOI] [PubMed]
- Lv Y., Ye T., Wang H.-P., Zhao J.-Y., Chen W.-J., Wang X., Shen C.-X., Wu Y.-B., Cai Y.-K. Suppression of colorectal tumorigenesis by recombinant Bacteroides fragilis enterotoxin-2 in vivo. World J. Gastroenterol. 2017;23:603–613. doi: 10.3748/wjg.v23.i4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch S.V., Pedersen O. The human intestinal microbiome in health and disease. N. Engl. J. Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- Maier L., Pruteanu M., Kuhn M., Zeller G., Telzerow A., Anderson E.E., Brochado A.R., Fernandez K.C., Dose H., Mori H., Patil K.R., Bork P., Typas A. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick H., Ma S., Franzosa E.A., Vatanen T., Morgan X.C., Huttenhower C. Experimental design and quantitative analysis of microbial community multiomics. Genome Biol. 2017;18:228. doi: 10.1186/s13059-017-1359-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy K.J., Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- Marchesi J.R., Dutilh B.E., Hall N., Peters W.H.M., Roelofs R., Boleij A., Tjalsma H. Towards the human colorectal cancer microbiome. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020447. e20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M.-L., Luke J.J., Gajewski T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan C.J., Beiko R.G. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol Evol. 2014;6:703–713. doi: 10.1093/gbe/evu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira-Pascual L., Cabrera-Rubio R., Ocon S., Costales P., Parra A., Suarez A., Moris F., Rodrigo L., Mira A., Collado M.C. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J. Gastroenterol. 2015;50:167–179. doi: 10.1007/s00535-014-0963-x. [DOI] [PubMed] [Google Scholar]
- Mirzaei M.K., Maurice C.F. Ménage à trois in the human gut: interactions between host, bacteria and phages. Nat. Rev. Microbiol. 2017;15:397–408. doi: 10.1038/nrmicro.2017.30. [DOI] [PubMed] [Google Scholar]
- Mizuno S., Masaoka T., Naganuma M., Kishimoto T., Kitazawa M., Kurokawa S., Nakashima M., Takeshita K., Suda W., Mimura M., Hattori M., Kanai T. Bifidobacterium-rich fecal donor may Be a positive predictor for successful fecal microbiota transplantation in patients with irritable bowel syndrome. Digestion. 2017;96:29–38. doi: 10.1159/000471919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monda V., Villano I., Messina A., Valenzano A., Esposito T., Moscatelli F., Viggiano A., Cibelli G., Chieffi S., Monda M., Messina G. Exercise modifies the gut microbiota with positive health effects. Oxid Med Cell Longev. 2017;2017:3831972. doi: 10.1155/2017/3831972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monleón D., Morales J.M., Barrasa A., López J.A., Vázquez C., Celda B. Metabolite profiling of fecal water extracts from human colorectal cancer. NMR Biomed. 2009;22:342–348. doi: 10.1002/nbm.1345. [DOI] [PubMed] [Google Scholar]
- Moskal A., Freisling H., Byrnes G., Assi N., Fahey M.T., Jenab M., Ferrari P., Tjønneland A., Petersen K.E., Dahm C.C., Hansen C.P., Affret A., Boutron-Ruault M.-C., Cadeau C., Kühn T., Katzke V., Iqbal K., Boeing H., Trichopoulou A., Bamia C., Naska A., Masala G., de Magistris M.S., Sieri S., Tumino R., Sacerdote C., Peeters P.H., Bueno-de-Mesquita B.H., Engeset D., Licaj I., Skeie G., Ardanaz E., Buckland G., Castaño J.M.H., Quirós J.R., Amiano P., Molina-Portillo E., Winkvist A., Myte R., Ericson U., Sonestedt E., Perez-Cornago A., Wareham N., Khaw K.-T., Huybrechts I., Tsilidis K.K., Ward H., Gunter M.J., Slimani N. Main nutrient patterns and colorectal cancer risk in the European Prospective Investigation into Cancer and Nutrition study. Br. J. Canc. 2016;115:1430–1440. doi: 10.1038/bjc.2016.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu G., Zhou H., Wu W.K.K., Wong S.H., Coker O.O., Dai Z., Li X., Szeto C.-H., Sugimura N., Lam T.Y.-T., Yu A.C.-S., Wang X., Chen Z., Wong M.C.-S., Ng S.C., Chan M.T.V., Chan P.K.S., Chan F.K.L., Sung J.J.-Y., Yu J. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology. 2018;155:529–541. doi: 10.1053/j.gastro.2018.04.018. e5. [DOI] [PubMed] [Google Scholar]
- Navarro M., Nicolas A., Ferrandez A., Lanas A. Colorectal cancer population screening programs worldwide in 2016: an update. World J. Gastroenterol. 2017;23:3632–3642. doi: 10.3748/wjg.v23.i20.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougayrède J.-P., Homburg S., Taieb F., Boury M., Brzuszkiewicz E., Gottschalk G., Buchrieser C., Hacker J., Dobrindt U., Oswald E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- Owyang C., Wu G.D. The gut microbiome in health and disease. Gastroenterology. 2014;146:1433–1436. doi: 10.1053/j.gastro.2014.03.032. [DOI] [PubMed] [Google Scholar]
- Pandey K.R., Naik S.R., Vakil B.V. Probiotics, prebiotics and synbiotics- a review. J. Food Sci. Technol. 2015;52:7577–7587. doi: 10.1007/s13197-015-1921-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasolli E., Asnicar F., Manara S., Zolfo M., Karcher N., Armanini F., Beghini F., Manghi P., Tett A., Ghensi P., Collado M.C., Rice B.L., DuLong C., Morgan X.C., Golden C.D., Quince C., Huttenhower C., Segata N. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176:649–662. doi: 10.1016/j.cell.2019.01.001. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paule A., Frezza D., Edeas M. Microbiota and phage therapy: future challenges in medicine. Med. Sci. 2018;6 doi: 10.3390/medsci6040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phua L.C., Chue X.P., Koh P.K., Cheah P.Y., Ho H.K., Chan E.C.Y. Non-invasive fecal metabonomic detection of colorectal cancer. Cancer Biol. Ther. 2014;15:389–397. doi: 10.4161/cbt.27625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouncey A.L., Scott A.J., Alexander J.L., Marchesi J., Kinross J. Gut microbiota, chemotherapy and the host: the influence of the gut microbiota on cancer treatment. Ecancermedicalscience. 2018;12:868. doi: 10.3332/ecancer.2018.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D.R., Li J., Xu J., Li Shaochuan, Li D., Cao J., Wang B., Liang H., Zheng H., Xie Y., Tap J., Lepage P., Bertalan M., Batto J.-M., Hansen T., Le Paslier D., Linneberg A., Nielsen H.B., Pelletier E., Renault P., Sicheritz-Ponten T., Turner K., Zhu H., Yu C., Li Shengting, Jian M., Zhou Y., Li Y., Zhang X., Li Songgang, Qin N., Yang H., Wang Jian, Brunak S., Doré J., Guarner F., Kristiansen K., Pedersen O., Parkhill J., Weissenbach J., MetaHIT Consortium, Bork P., Ehrlich S.D., Wang Jun. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Cai G., Zhou B., Li D., Zhao A., Xie G., Li H., Cai S., Xie D., Huang C., Ge W., Zhou Z., Xu L.X., Jia Weiping, Zheng S., Yen Y., Jia Wei. A distinct metabolic signature of human colorectal cancer with prognostic potential. Clin. Cancer Res. 2014;20:2136–2146. doi: 10.1158/1078-0432.CCR-13-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M., Ambalam P., Kondepudi K.K., Pithva S., Kothari C., Patel A.T., Purama R.K., Dave J.M., Vyas B.R.M. Potential of probiotics, prebiotics and synbiotics for management of colorectal cancer. Gut Microb. 2013;4:181–192. doi: 10.4161/gmic.23919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan R., Rani A., Metwally A., McGee H.S., Perkins D.L. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016;469:967–977. doi: 10.1016/j.bbrc.2015.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan R., Rani A., Finn P.W., Perkins D.L. Multiomic strategies reveal diversity and important functional aspects of human gut microbiome. BioMed Res. Int. 2018:6074918. doi: 10.1155/2018/6074918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezasoltani S., Sharafkhah M., Asadzadeh Aghdaei H., Nazemalhosseini Mojarad E., Dabiri H., Akhavan Sepahi A., Modarressi M.H., Feizabadi M.M., Zali M.R. Applying simple linear combination, multiple logistic and factor analysis methods for candidate fecal bacteria as novel biomarkers for early detection of adenomatous polyps and colon cancer. J. Microbiol. Methods. 2018;155:82–88. doi: 10.1016/j.mimet.2018.11.007. [DOI] [PubMed] [Google Scholar]
- Richard M.L., Liguori G., Lamas B., Brandi G., da Costa G., Hoffmann T.W., Pierluigi Di Simone M., Calabrese C., Poggioli G., Langella P., Campieri M., Sokol H. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microb. 2018;9:131–142. doi: 10.1080/19490976.2017.1379637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D.J., Lee J.K., Boland C.R., Dominitz J.A., Giardiello F.M., Johnson D.A., Kaltenbach T., Lieberman D., Levin T.R., Rex D.K. Recommendations on fecal immunochemical testing to screen for colorectal neoplasia: a consensus statement by the US Multi-Society Task Force on colorectal cancer. Gastrointest. Endosc. 2017;85:2–21. doi: 10.1016/j.gie.2016.09.025. e3. [DOI] [PubMed] [Google Scholar]
- Rosshart S.P., Vassallo B.G., Angeletti D., Hutchinson D.S., Morgan A.P., Takeda K., Hickman H.D., McCulloch J.A., Badger J.H., Ajami N.J., Trinchieri G., Pardo-Manuel de Villena F., Yewdell J.W., Rehermann B. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell. 2017;171:1015–1028. doi: 10.1016/j.cell.2017.09.016. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillère R., Fluckiger A., Messaoudene M., Rauber C., Roberti M.P., Fidelle M., Flament C., Poirier-Colame V., Opolon P., Klein C., Iribarren K., Mondragón L., Jacquelot N., Qu B., Ferrere G., Clémenson C., Mezquita L., Masip J.R., Naltet C., Brosseau S., Kaderbhai C., Richard C., Rizvi H., Levenez F., Galleron N., Quinquis B., Pons N., Ryffel B., Minard-Colin V., Gonin P., Soria J.-C., Deutsch E., Loriot Y., Ghiringhelli F., Zalcman G., Goldwasser F., Escudier B., Hellmann M.D., Eggermont A., Raoult D., Albiges L., Kroemer G., Zitvogel L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- Rubinstein M.R., Wang X., Liu W., Hao Y., Cai G., Han Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M.D., Nickerson C., Rees C.J., Patnick J., Blanks R.G. Risk factors for adverse events related to polypectomy in the English Bowel Cancer Screening Programme. Endoscopy. 2014;46:90–97. doi: 10.1055/s-0033-1344987. [DOI] [PubMed] [Google Scholar]
- Sattar M.A., White A.G., Eklof B., Fenech F.F. Takayasu's disease in Arabs. Postgrad. Med. 1985;61:387–390. doi: 10.1136/pgmj.61.715.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbahi H., Di Palma J.A. Faecal microbiota transplantation: applications and limitations in treating gastrointestinal disorders. BMJ Open Gastroenterol. 2016;3 doi: 10.1136/bmjgast-2016-000087. e000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M., Franzosa E.A., Lloyd-Price J., McIver L.J., Schwager R., Poon T.W., Ananthakrishnan A.N., Andrews E., Barron G., Lake K., Prasad M., Sauk J., Stevens B., Wilson R.G., Braun J., Denson L.A., Kugathasan S., McGovern D.P.B., Vlamakis H., Xavier R.J., Huttenhower C. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat. Microbiol. 2018;3:337–346. doi: 10.1038/s41564-017-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.S.B., Raes J., Bork P. The human gut microbiome: from association to modulation. Cell. 2018;172:1198–1215. doi: 10.1016/j.cell.2018.02.044. [DOI] [PubMed] [Google Scholar]
- Schwabe R.F., Jobin C. The microbiome and cancer. Nat. Rev. Canc. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears C.L., Garrett W.S. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears C.L., Islam S., Saha A., Arjumand M., Alam N.H., Faruque A.S.G., Salam M.A., Shin J., Hecht D., Weintraub A., Sack R.B., Qadri F. Association of enterotoxigenic Bacteroides fragilis infection with inflammatory diarrhea. Clin. Infect. Dis. 2008;47:797–803. doi: 10.1086/591130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I., Russell S.L., Antunes L.C.M., Finlay B.B. Gut microbiota in health and disease. Physiol. Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Sender R., Fuchs S., Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14 doi: 10.1371/journal.pbio.1002533. e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M.S., DeSantis T.Z., Weinmaier T., McMurdie P.J., Cope J.L., Altrichter A., Yamal J.-M., Hollister E.B. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut. 2018;67:882–891. doi: 10.1136/gutjnl-2016-313189. [DOI] [PubMed] [Google Scholar]
- Shang F.-M., Liu H.-L. Fusobacterium nucleatum and colorectal cancer: a review. World J. Gastrointest. Oncol. 2018;10:71–81. doi: 10.4251/wjgo.v10.i3.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X.J., Rawls J.F., Randall T., Burcal L., Mpande C.N., Jenkins N., Jovov B., Abdo Z., Sandler R.S., Keku T.O. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microb. 2010;1:138–147. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.K., Chang H.-W., Yan D., Lee K.M., Ucmak D., Wong K., Abrouk M., Farahnik B., Nakamura M., Zhu T.H., Bhutani T., Liao W. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017;15:73. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R., Ahn J., Sampson J.N., Shi J., Yu G., Xiong X., Hayes R.B., Goedert J.J. Fecal microbiota, fecal metabolome, and colorectal cancer interrelations. PLoS One. 2016;11 doi: 10.1371/journal.pone.0152126. e0152126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan A., Corrales L., Hubert N., Williams J.B., Aquino-Michaels K., Earley Z.M., Benyamin F.W., Lei Y.M., Jabri B., Alegre M.-L., Chang E.B., Gajewski T.F. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhani I., Tap J., Roudot-Thoraval F., Roperch J.P., Letulle S., Langella P., Corthier G., Tran Van Nhieu J., Furet J.P. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016393. e16393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E.-J., Lee E.-S., Nam Y.-D. Progress of analytical tools and techniques for human gut microbiome research. J. Microbiol. 2018;56:693–705. doi: 10.1007/s12275-018-8238-5. [DOI] [PubMed] [Google Scholar]
- Swidsinski A., Khilkin M., Kerjaschki D., Schreiber S., Ortner M., Weber J., Lochs H. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology. 1998;115:281–286. doi: 10.1016/s0016-5085(98)70194-5. [DOI] [PubMed] [Google Scholar]
- Taieb F., Petit C., Nougayrède J.-P., Oswald E. The enterobacterial genotoxins: cytolethal distending toxin and colibactin. EcoSal Plus. 2016;7 doi: 10.1128/ecosalplus.ESP-0008-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B., Qiu Y., Zou X., Chen T., Xie G., Cheng Y., Dong T., Zhao L., Feng B., Hu X., Xu L.X., Zhao A., Zhang M., Cai G., Cai S., Zhou Z., Zheng M., Zhang Y., Jia W. Metabonomics identifies serum metabolite markers of colorectal cancer. J. Proteome Res. 2013;12:3000–3009. doi: 10.1021/pr400337b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalsma H., Boleij A., Marchesi J.R., Dutilh B.E. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat. Rev. Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Tsoi H., Chu E.S.H., Zhang X., Sheng J., Nakatsu G., Ng S.C., Chan A.W.H., Chan F.K.L., Sung J.J.Y., Yu J. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology. 2017;152:1419–1433. doi: 10.1053/j.gastro.2017.01.009. e5. [DOI] [PubMed] [Google Scholar]
- Uchiyama K., Yagi N., Mizushima K., Higashimura Y., Hirai Y., Okayama T., Yoshida N., Katada K., Kamada K., Handa O., Ishikawa T., Takagi T., Konishi H., Kuriu Y., Nakanishi M., Otsuji E., Itoh Y., Naito Y. Serum metabolomics analysis for early detection of colorectal cancer. J. Gastroenterol. 2017;52:677–694. doi: 10.1007/s00535-016-1261-6. [DOI] [PubMed] [Google Scholar]
- van Nood E., Vrieze A., Nieuwdorp M., Fuentes S., Zoetendal E.G., de Vos W.M., Visser C.E., Kuijper E.J., Bartelsman J.F.W.M., Tijssen J.G.P., Speelman P., Dijkgraaf M.G.W., Keller J.J. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Vandeputte D., Kathagen G., D’hoe K., Vieira-Silva S., Valles-Colomer M., Sabino J., Wang J., Tito R.Y., De Commer L., Darzi Y., Vermeire S., Falony G., Raes J. Quantitative microbiome profiling links gut community variation to microbial load. Nature. 2017;551:507–511. doi: 10.1038/nature24460. [DOI] [PubMed] [Google Scholar]
- Vétizou M., Pitt J.M., Daillère R., Lepage P., Waldschmitt N., Flament C., Rusakiewicz S., Routy B., Roberti M.P., Duong C.P.M., Poirier-Colame V., Roux A., Becharef S., Formenti S., Golden E., Cording S., Eberl G., Schlitzer A., Ginhoux F., Mani S., Yamazaki T., Jacquelot N., Enot D.P., Bérard M., Nigou J., Opolon P., Eggermont A., Woerther P.-L., Chachaty E., Chaput N., Robert C., Mateus C., Kroemer G., Raoult D., Boneca I.G., Carbonnel F., Chamaillard M., Zitvogel L. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villéger R., Lopès A., Veziant J., Gagnière J., Barnich N., Billard E., Boucher D., Bonnet M. Microbial markers in colorectal cancer detection and/or prognosis. World J. Gastroenterol. 2018;24:2327–2347. doi: 10.3748/wjg.v24.i22.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Jia H. Metagenome-wide association studies: fine-mining the microbiome. Nat. Rev. Microbiol. 2016;14:508–522. doi: 10.1038/nrmicro.2016.83. [DOI] [PubMed] [Google Scholar]
- Wang T., Cai G., Qiu Y., Fei N., Zhang M., Pang X., Jia W., Cai S., Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir T.L., Manter D.K., Sheflin A.M., Barnett B.A., Heuberger A.L., Ryan E.P. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070803. e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West N.R., Powrie F. Immunotherapy not working? Check your microbiota. Cancer Cell. 2015;28:687–689. doi: 10.1016/j.ccell.2015.11.010. [DOI] [PubMed] [Google Scholar]
- Williamson I.A., Arnold J.W., Samsa L.A., Gaynor L., DiSalvo M., Cocchiaro J.L., Carroll I., Azcarate-Peril M.A., Rawls J.F., Allbritton N.L., Magness S.T. A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Cell. Mol. Gastroenterol. Hepatol. 2018;6:301–319. doi: 10.1016/j.jcmgh.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.H., Kwong T.N.Y., Chow T.-C., Luk A.K.C., Dai R.Z.W., Nakatsu G., Lam T.Y.T., Zhang L., Wu J.C.Y., Chan F.K.L., Ng S.S.M., Wong M.C.S., Ng S.C., Wu W.K.K., Yu J., Sung J.J.Y. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2017;66:1441–1448. doi: 10.1136/gutjnl-2016-312766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.H., Zhao L., Zhang X., Nakatsu G., Han J., Xu W., Xiao X., Kwong T.N.Y., Tsoi H., Wu W.K.K., Zeng B., Chan F.K.L., Sung J.J.Y., Wei H., Yu J. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology. 2017;153:1621–1633. doi: 10.1053/j.gastro.2017.08.022. e6. [DOI] [PubMed] [Google Scholar]
- Wu S., Morin P.J., Maouyo D., Sears C.L. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology. 2003;124:392–400. doi: 10.1053/gast.2003.50047. [DOI] [PubMed] [Google Scholar]
- Wu S., Rhee K.-J., Albesiano E., Rabizadeh S., Wu X., Yen H.-R., Huso D.L., Brancati F.L., Wick E., McAllister F., Housseau F., Pardoll D.M., Sears C.L. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Yang X., Zhang R., Li J., Xiao X., Hu Y., Chen Y., Yang F., Lu N., Wang Z., Luan C., Liu Y., Wang B., Xiang C., Wang Y., Zhao F., Gao G.F., Wang S., Li L., Zhang H., Zhu B. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb. Ecol. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- Yoshioka N., Taniguchi Y., Yoshida A., Nakata K., Nishizawa T., Inagawa H., Kohchi C., Soma G.-I. Intestinal macrophages involved in the homeostasis of the intestine have the potential for responding to LPS. Anticancer Res. 2009;29:4861–4865. [PubMed] [Google Scholar]
- Yu J., Feng Q., Wong S.H., Zhang D., Liang Q.Y., Qin Y., Tang L., Zhao H., Stenvang J., Li Y., Wang X., Xu Xiaoqiang, Chen N., Wu W.K.K., Al-Aama J., Nielsen H.J., Kiilerich P., Jensen B.A.H., Yau T.O., Lan Z., Jia H., Li J., Xiao L., Lam T.Y.T., Ng S.C., Cheng A.S.-L., Wong V.W.-S., Chan F.K.L., Xu Xun, Yang H., Madsen L., Datz C., Tilg H., Wang Jian, Brünner N., Kristiansen K., Arumugam M., Sung J.J.-Y., Wang Jun. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66:70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- Yu L.C.-H., Wei S.-C., Ni Y.-H. Impact of microbiota in colorectal carcinogenesis: lessons from experimental models. Intest Res. 2018;16:346–357. doi: 10.5217/ir.2018.16.3.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi D., Korem T., Zmora N., Israeli D., Rothschild D., Weinberger A., Ben-Yacov O., Lador D., Avnit-Sagi T., Lotan-Pompan M., Suez J., Mahdi J.A., Matot E., Malka G., Kosower N., Rein M., Zilberman-Schapira G., Dohnalová L., Pevsner-Fischer M., Bikovsky R., Halpern Z., Elinav E., Segal E. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Zeller G., Tap J., Voigt A.Y., Sunagawa S., Kultima J.R., Costea P.I., Amiot A., Böhm J., Brunetti F., Habermann N., Hercog R., Koch M., Luciani A., Mende D.R., Schneider M.A., Schrotz-King P., Tournigand C., Tran Van Nhieu J., Yamada T., Zimmermann J., Benes V., Kloor M., Ulrich C.M., von Knebel Doeberitz M., Sobhani I., Bork P. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014;10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Cheng Z., Ma Y., He C., Lu Y., Zhao Y., Chang X., Zhang Y., Bai Y., Cheng N. Effectiveness of screening modalities in colorectal cancer: a network meta-analysis. Clin. Colorectal Cancer. 2017;16:252–263. doi: 10.1016/j.clcc.2017.03.018. [DOI] [PubMed] [Google Scholar]
- Zhang F., Zhang Y., Zhao W., Deng K., Wang Z., Yang C., Ma L., Openkova M.S., Hou Y., Li K. Metabolomics for biomarker discovery in the diagnosis, prognosis, survival and recurrence of colorectal cancer: a systematic review. Oncotarget. 2017;8:35460–35472. doi: 10.18632/oncotarget.16727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhu X., Cao Y., Fang J.-Y., Hong J., Chen H. Fecal Fusobacterium nucleatum for the diagnosis of colorectal tumor: a systematic review and meta-analysis. Cancer Med. 2019;8:480–491. doi: 10.1002/cam4.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A., Kurilshikov A., Bonder M.J., Tigchelaar E.F., Schirmer M., Vatanen T., Mujagic Z., Vila A.V., Falony G., Vieira-Silva S., Wang J., Imhann F., Brandsma E., Jankipersadsing S.A., Joossens M., Cenit M.C., Deelen P., Swertz M.A., LifeLines cohort study, Weersma R.K., Feskens E.J.M., Netea M.G., Gevers D., Jonkers D., Franke L., Aulchenko Y.S., Huttenhower C., Raes J., Hofker M.H., Xavier R.J., Wijmenga C., Fu J. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Youlian, He H., Xu H., Li Y., Li Z., Du Y., He J., Zhou Yongjian, Wang H., Nie Y. Association of oncogenic bacteria with colorectal cancer in South China. Oncotarget. 2016;7:80794–80802. doi: 10.18632/oncotarget.13094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Michelle Luo T., Jobin C., Young H.A. Gut microbiota and probiotics in colon tumorigenesis. Cancer Lett. 2011;309:119–127. doi: 10.1016/j.canlet.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Jin Z., Wu W., Gao R., Guo B., Gao Z., Yang Y., Qin H. Analysis of the intestinal lumen microbiota in an animal model of colorectal cancer. PLoS One. 2014;9 doi: 10.1371/journal.pone.0090849. e90849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L., Ma Y., Raoult D., Kroemer G., Gajewski T.F. The microbiome in cancer immunotherapy: diagnostic tools and therapeutic strategies. Science. 2018;359:1366–1370. doi: 10.1126/science.aar6918. [DOI] [PubMed] [Google Scholar]
- Zur Hausen H. The search for infectious causes of human cancers: where and why. Virology. 2009;392:1–10. doi: 10.1016/j.virol.2009.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.