Summary
The intestinal epithelium forms a barrier between the microbiota and the rest of the body. In addition, beyond acting as a physical barrier, the function of intestinal epithelial cells (IECs) in sensing and responding to microbial signals is increasingly appreciated and likely has numerous implications for the vast network of immune cells within and below the intestinal epithelium. IECs also respond to factors produced by immune cells, and these can regulate IEC barrier function, proliferation and differentiation, as well as influence the composition of the microbiota. The mechanisms involved in IEC–microbe–immune interactions, however, are not fully characterized. In this review, we explore the ability of IECs to direct intestinal homeostasis by orchestrating communication between intestinal microbes and mucosal innate and adaptive immune cells during physiological and inflammatory conditions. We focus primarily on the most recent findings and call attention to the numerous remaining unknowns regarding the complex crosstalk between IECs, the microbiota and intestinal immune cells.
Keywords: bacterial, epithelial cell, gut, microbiota, mucosal immunology
Introduction
The mucosal surface of the gastrointestinal tract consists of a single layer of intestinal epithelial cells (IECs) that provide an interface for immune cells to detect and respond to environmental substances. These include food components and pathogenic or commensal microbial species of Archaea, bacteria, fungi, viruses and parasites, with around 1011 bacteria colonizing the human gastrointestinal tract.1 This creates an enormous source of potential immune stimuli; however, under homeostatic conditions the immune cells in and underlying the mucosa develop and function in a controlled manner, balancing inflammatory and regulatory responses to prevent overreaction to innocuous luminal antigens. During pathogenic infection, immune cells are mobilized to fight and clear invading microbes. Although the mechanisms that regulate intestinal immune responses during health and disease are still being elucidated, dialogue between intestinal microbes, IECs and innate and adaptive immune cells is increasingly appreciated to play a major role.
The IEC monolayer is composed of a number of cell types that differentiate from epithelial stem cells residing in the crypts. IEC types include goblet cells that produce mucin glycoproteins and form mucus, absorptive enterocytes, enteroendocrine cells, Paneth cells at the bottom of intestinal crypts that secrete antimicrobial peptides (AMPs), microfold (M) cells involved in antigen capture and presentation to immune cells, and tuft cells that promote type 2 immunity to intestinal parasites.2, 3 Single‐cell RNA sequencing has further defined the behaviour and characteristics of each IEC cell type,4 and a recent study identified two subtypes of tuft cells that change in frequency during helminth infection.5 Together, IECs form the boundary between the internal body and the outside environment, and studies in germ‐free mice have demonstrated that microbial colonization of the intestinal lumen influences IEC metabolism, proliferation, survival, barrier function and communication with immune cells.6 IECs are the main cell type in direct contact with stimuli from the luminal microbiota and are critical players in microbe–host interactions. As such, in addition to epithelial cell‐mediated defence mechanisms, IECs also coordinate the development and maturation of downstream immune responses from immune cells residing in the lamina propria and underlying lymphoid tissues. These immune cells help to contain microbes at the mucosa and maintain intestinal homeostasis.
Although much is known about the immune cell populations in the gut, less is known about the mechanisms by which IECs regulate the development and maturation of immune cells during homeostasis and how this is disrupted during different disease states. In addition, the stimuli from commensal bacterial species recognized by IECs and the receptors and signalling pathways involved are not thoroughly understood. In this review, we describe the role of IECs as important communication hubs and modulators that shape and coordinate the activity of both microbes and immune cells. We place special emphasis on the most recent findings and highlight the many open questions regarding the complex network of interactions between IECs, the microbiota and intestinal immune cells.
Microbiota–IEC crosstalk
The intestinal epithelium is a highly dynamic tissue that provides both physical and chemical barriers to protect the intestinal mucosa and peripheral organs from commensal microbes or invading pathogenic microorganisms. In addition to forming a barrier, IECs also detect a myriad of signals from intestinal microbes, allowing fine tuning of IEC proliferation and homeostatic functions (Fig. 1). Likewise, IEC programmes can influence the composition of the intestinal microbiota in a number of ways.
Figure 1.
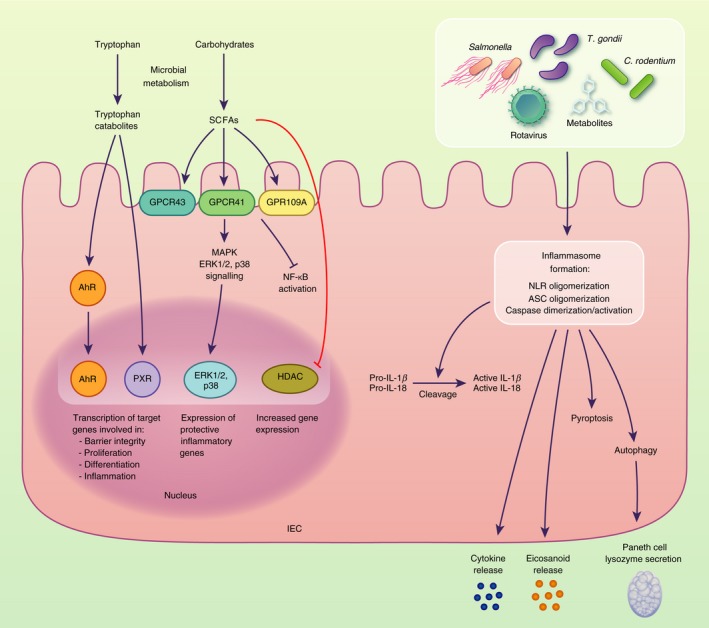
Intestinal epithelial cells (IECs) sense microbial stimuli through a number of different mechanisms that regulate IEC gene transcription and inflammatory responses. For example, tryptophan catabolites and short‐chain fatty acids (SCFAs) produced as a result of microbial metabolism trigger the activation of AhR, PXR, ERK1/2 and p38 that directly regulate the expression of target genes. The inflammasome complexes in IECs reported to respond to microbial stimuli include NLRP3, NAIP‐NLRC4, NLRP6 and NLRP9b, which trigger cell death pathways and the release of inflammatory cytokines and mediators.
Microbial regulation of IEC growth and function
Intestinal epithelial cells possess a number of mechanisms to sense and respond to the presence and activity of intestinal microbes. IECs express pattern recognition receptors (PRRs) to specifically detect molecular patterns from commensal and pathogenic gut microbes, and these have been extensively described in previous reviews.7, 8, 9 Following the detection of intestinal microbes, IECs enhance various components of the intestinal barrier to protect underlying host tissues from bacterial infiltration. These include AMP production, mucus secretion, tight junction integrity, and IEC growth and differentiation. IECs secrete a range of AMPs, many through PRR/MyD88‐dependent mechanisms, that accumulate in the mucus layer and possess broad antimicrobial activities.10, 11 Indeed, during Citrobacter rodentium infection, MyD88 signalling solely in IECs was recently shown to be sufficient to enhance IEC barrier integrity and increase production of RegIIIγ and immunomodulatory acute‐phase protein serum amyloid A1 (SAA1).12 Goblet cells secrete mucin glycoproteins to create the viscous mucus layer, and the importance of mucus in protection against invading microbes was recently highlighted in a study showing that the discontinuous mucus layer in the mouse caecum and corresponding uncovered areas of the epithelium form hotspots for Salmonella infection.13 A number of bacterial species have been shown to modulate mucin secretion by goblet cells. For example, commensal Ruminococcus gnavus 14 and Lactobacillus rhamnosus 15 stimulate the production of mucins, while pathogenic microbes including adherent and invasive Escherichia coli promote a less effective mucus barrier.16 In a recent study, Amuc_1100, a membrane protein from commensal Akkermansia muciniphila, was shown to interact with the PRR Toll‐like receptor 2 (TLR2) to increase intestinal barrier function, namely mucus thickness and tight junction protein (TJP) expression.17
Although PRR‐mediated mechanisms of sensing microbial products are the most extensively studied, IECs also use a number of other pathways. For example, inflammasomes have been shown to play an important role in IEC‐sensing of microbial stimuli and damage‐associated molecular patterns and in triggering protective barrier responses.18, 19, 20, 21 The NAIP‐NLRC4 inflammasome has recently been implicated in the IEC response to Salmonella infection in vivo, enabling pro‐inflammatory programmes that result in production of cytokines and the hormone‐like eicosanoid prostaglandin E2, as well as lytic cell death and the expulsion of infected IECs.22 The autophagy pathway has also been shown to be critical for maintaining intestinal epithelial integrity in response to microbes, and a recent study demonstrated that release of lysozyme by Paneth cells during bacterial infection is mediated through an autophagy‐based alternative secretion pathway.23 Although mechanisms of microbial modulation of and sensing by IECs continue to be uncovered, many pathways remain incompletely characterized.
Microbial metabolites produced by bacterial fermentation of dietary components are also important signals detected by IECs. For example, tryptophan catabolites, detected by pregnane X receptor (PXR)24 and the aryl hydrocarbon receptor (AhR),25, 26 drive a multitude of anti‐inflammatory and protective barrier functions. IEC AhR sensing of dietary components and tryptophan catabolites contributes to the maintenance of intestinal barrier integrity by inducing IEC differentiation from crypt stem cells26 and mitigating inflammatory responses.27 PXR was recently shown to respond to indole 3‐propionic acid, a tryptophan metabolite produced by commensal Clostridium sporogenes, and mice deficient for PXR exhibited increased epithelial inflammatory injury and decreased tight junction protein expression. In contrast, germ‐free mice colonized with C. sporogenes and dosed with l‐tryptophan exhibited decreased intestinal permeability and increased expression of detoxifying PXR target genes.24 In addition to serving as a major energy source for enterocytes, microbiota‐derived short‐chain fatty acids (SCFAs) have also been implicated in the regulation of most IEC functions including cell turnover,28 tight junction protein expression,29 and inflammasome‐ or hypoxia‐inducible factor (HIF)‐mediated epithelial integrity.30, 31 SCFAs can directly influence gene transcription by binding to and inhibiting histone deacetylases (HDAC) or through binding to the metabolite‐sensing receptors GPR41, GPR43 and GPR109A.32 Indeed, a recent study showed that optimal expression of AMPs requires IEC‐sensing of SCFAs via GPR43. Using Gpr43 −/− mice and enteroids, investigators observed that the AMPs RegIIIγ and β‐defensins 1, 3 and 4 were reduced in the absence of GPR43 or downstream mammalian target of rapamycin (mTOR) and signal transducer and activator of transcription 3 (STAT3) activation.33
Microbes also induce a number of non‐barrier functions in IECs, including changes in metabolism and the biosynthesis of signalling molecules. For example, early during Citrobacter rodentium infection, IECs have been shown to exhibit changes in cholesterol and carbon metabolic pathways, suggesting that IEC metabolism is reprogrammed to meet increased cellular energetic demands during tissue repair.34 Some enterochromaffin cells, a subtype of enteroendocrine cell, have been shown to secrete serotonin (5‐hydroxytryptamine, 5‐HT) in response to mechanosensing via the mechanotransducer Piezo2,35 and 5‐HT is an important regulator of enteric nervous system development, and gastrointestinal tract motility and inflammation.36 In addition to mechanosensing, a recent study demonstrated that several metabolites from a consortium of commensal spore‐forming bacteria (predominantly Clostridial species) promote 5‐HT biosynthesis by colonic enterochromaffin cells in colonized mice.37 In response to microbes, IECs also secrete a number of cytokines and effector molecules including interleukin‐25 (IL‐25) and SAA.38, 39 These effectors regulate the development and function of intestinal immune cells, as described in the next section of this review. Collectively, these recent findings indicate that a broad range of IEC functions are affected by sensing of intestinal microbes (Table 1); however, it is worth noting that many of these studies were performed in the context of pathogenic microbial infection. Further studies are required to identify additional stimuli from commensal microbes and characterize commensurate IEC responses at steady state.
Table 1.
Gut microbial stimuli that interact with intestinal epithelial cells (IECs)
| IEC sensor/signalling pathway | Microbial stimuli | Microbial species used | In vivo/in vitro and study details | IEC response | References |
|---|---|---|---|---|---|
| Toll‐like receptor 9, nuclear factor‐κB | Unmethylated CpG bacterial DNA1 | Citrobacter rodentium (DBS100), Salmonella typhimurium (ATCC 14028), Helicobacter pylori (PMSS1) | In vivo, Tlr9 −/− mice | Decreases intestinal inflammation and damage following bacterial challenge | 140, 141, 142, 143 1 |
| Caspase‐3/7‐mediated apoptosis | Enterotoxins (TcdA and TcdB) | Clostridium difficile (VPI10463) | In vivo and in vitro intestinal organoids; Casp3/7 IEC‐KO mice | Restricts C. difficile growth in vivo | 144 |
| NAIP/NLRC4 inflammasome |
Flagellin1
Unknown2 |
Salmonella Typhimurium, Citrobacter rodentium 2 | In vivo, Casp1 −/−, Casp8 −/−, Nlrc4 −/− | Protects against enteric pathogen invasion; expulsion of pyroptotic IECs and release of eicosanoid and interleukin‐18 (IL‐18) | 21, 22, 145 1 |
| Toll‐like receptor 4, peroxisome proliferator‐activated receptor (PPAR) | Free fatty acids1 | Commensal gut microbes | In vivo, Tlr4 IEC‐KO | Prevents development of metabolic syndrome; regulates expression of lysozyme and PPAR‐controlled genes | 58, 146, 147, 1 |
| P2X7R/NLRP3 inflammasome | Ligands include extracellular ATP and K+ 1 | Toxoplasma gondii | In vitro, FHs 74 Int cells | IL‐1β secretion and inhibition of parasitic proliferation | 18, 148, 149, 1 |
| NLRP6 inflammasome | Unknown | Citrobacter rodentium | In vivo, Nlrp6 −/−, Asc −/−, Casp1/11 −/− | Orchestrates goblet cell mucin granule exocytosis | 19 |
| Nlrp9b inflammasome | dsRNA1 | Rotavirus EW | In vivo, Nlrp9b −/−, Nlrp9b IEC‐KO | Restricts rotavirus infection by IL‐18 production and pyroptosis | 20 |
| Aryl hydrocarbon receptor | Tryptophan indole derivatives | Lactobacilli, Clostridiales members | In vivo, Ahr −/− | IL‐22 production; resistance to enteric pathogens; maintenance of intestinal homeostasis and barrier functions | 25, 26, 27 |
| Receptors GPR41, GPR43 and GPR109; HDAC inhibition; mTOR, STAT3, ERK and MAPK signalling | short‐chain fatty acids | Various microbes including Bacteroides spp. | In vivo, GPR41 −/−, GPR43 −/−, GPR109 −/−, in vitro murine intestinal organoids | Protective inflammatory responses during pathogen infection; secretion of AMPs, chemokines and cytokines; controls IEC turnover and barrier functions; RALDH1 expression and vitamin A metabolism | 28, 29, 30, 31, 33, 150, 151, 152 |
| MyD88 signalling | Various TLR ligands | Citrobacter rodentium | In vivo, MyD88 −/− | Secretion of AMPs, control of bacterial infiltration, enhanced barrier integrity | 12 |
| GPCR and ERK/MAPK signalling | Pili, novel 3000 MW molecule | Lactobacillus rhamnosus (CNCM I‐3690), Ruminococcus gnavus (E1) | In vivo, in vitro HT29‐MTX cells | Expression of glycoroteins and mucus production by goblet cells; cytoprotective responses | 14, 15 |
| Various cellular stresses including nutrient deprivation, infection with microbes1 | Autophagy | Helicobacter hepaticus, Salmonella Typhimurium, Pasteurellaceae family | In vivo, Atg161 −/− , in vitro Atg161 −/− organoids | Control inflammation‐induced apoptosis, necroptosis and maintains intestinal barrier, lysozyme secretion by Paneth cells, promotes bacterial clearance | 23, 43, 45, 153, 154, 155 1 |
| Cellular forces | Mechanosensors/mechanotransducer Piezo2 | Clostridial species | In vivo, in vitro | Serotonin release by enterochromaffin cells | 35, 37 |
| Peptidoglycan components; muramyl dipeptide1 | Nod2 | Bacteroides vulgatus, Enterococcus faecium | In vivo, Nod2 −/− and in vitro | Restriction of bacterial growth or dissemination, expression of inflammatory genes, goblet cell function | 56, 156, 157, 1, 158 |
| Pregnane X receptor (PXR) | Indole 3‐propionic acid | Clostridium sporogenes | In vivo, Nr1i2 −/−, Nr1i2 −/−, Tlr4 −/−, Pxr −/− | Regulation of intestinal permeability and intestinal inflammation, defence against intracellular pathogens | 24, 159 |
1A finding from a different or additional study.
2While type 3 secretion system components expressed by C. rodentium are thought to provide the stimulus triggering NLRC4 inflammasome formation in vivo, this has yet to be demonstrated.
Influence of IECs on diversity and function of the intestinal microbiota
Although the effects of IEC–microbe crosstalk on IECs are beginning to be elucidated, the effects of this interaction on the gut microbiota are substantially less characterized. Still, numerous recent studies have indicated that IECs also have an impact on the microbial populations residing in the gut. Autophagy is particularly well studied in maintaining the function of Paneth cells and protecting against pathogenic bacteria.40, 41, 42, 43, 44 Recently, disruption of IEC autophagy has also been shown to dramatically alter the composition of the gut microbiota and reduce intestinal microbial alpha‐diversity in mice.45 Another recent study showed that serotonin production by enterochromaffin cells modulates gut microbial composition,46 and AMPs secreted by IECs have been broadly reported to influence the composition of intestinal gut microbes.47, 48, 49 The NLRP6‐inflammasome is also highly expressed by IECs, and previous studies have shown that NLRP6 helps to maintain eubiosis of the intestinal microbiota.50, 51 However, recent studies of Nlrp6 −/− and Asc −/− mice co‐housed with wild‐type littermates report that the NLRP6 inflammasome does not affect gut microbial diversity,52, 53 highlighting that non‐genetic confounding factors may impact in vivo studies investigating causal relationships between host gene deficiencies and alterations in the microbiota.54 Indeed, whereas previous studies eliminated a role for nucleotide‐binding oligomerization domain‐containing protein 1 (NOD1) and NOD2 in shaping microbiota composition based on polymerase chain reaction for 10 targeted bacterial groups in co‐housed littermates of different genotypes,55 NOD2 signalling in IECs was recently strongly implicated in specifically controlling the colonization and growth of commensal Bacteroides vulgatus.56 In this recent study, although wild‐type animals co‐housed with Nod2 −/− mice acquired the overabundance of B. vulgatus characteristic of knockout mice, this was diminished upon re‐separation. Given these conflicting observations, a strong case has been made for using crosses and littermate controls as a superior alternative (or addition) to co‐housing.57 Still, the effects of IEC PRRs on the composition of the intestinal microbiota remain contentious and, importantly, the mechanisms behind many of the microbiota alterations observed have not been fully uncovered.
Microbial gene expression is also influenced by IECs through several mechanisms. For instance, a recent study using IEC‐specific TLR4 knockout (TLR4IEC‐KO) mice demonstrated that TLR4 influences the composition and function of intestinal microbes, including the expression of microbial genes involved in the metabolism of lipids, amino acids and nucleotides.58 TLR4IEC‐KO mice developed metabolic syndrome, and lysozyme and genes regulated by peroxisome proliferator‐activated receptors were down‐regulated, suggesting a mechanism by which intestinal TLR4 may influence the microbiota. In another study, attaching and effacing enterohaemorrhagic E. coli was shown to require mechanosensing of IECs to express the locus of enterocyte effacement that encodes its type 3 secretion system, and this was responsible for forming lesions in the gastrointestinal tract.59 In addition, a recent study reported that miRNA is released by IECs into the intestinal lumen where it enters bacterial species such as Fusobacterium nucleatum and E. coli and regulates their gene expression and growth.60
The main nutrient source for gut microbes is typically diet‐derived components including polysaccharides or glycans. However, some gut microbes can also use host glycans on mucin proteins and the surface of IECs, providing an alternative energy source when dietary glycans are reduced.61, 62, 63 For example, several commensal Clostridiales members use the mucin‐associated sugars fucose and sialic acid as energy sources, promoting their colonization of the gut.27 Glycans are also ligands for bacterial attachment, and some gut microbial species such as Ruminococcus gnavus are hypothesized to target mucin glycans to assist their spread and persistence in niches in the intestinal lumen.64 Together, these studies demonstrate the multitude of interactions between microbes and IECs that can trigger various IEC programmes and shape the microbial ecosystem in the gut.
IEC–immune cell crosstalk
Intestinal epithelial cells possess a number of independent barrier functions to control and/or kill gut microbes, they also mediate crosstalk between the microbiota and intraepithelial and subepithelial immune cells by responding to microbial metabolites and coordinating immune responses. This is achieved by a number of known and unknown mechanisms including the secretion of chemokines, cytokines and other immunomodulatory molecules (Fig. 2), as well as the transport of microbial antigens and metabolites to underlying immune cells in the lamina propria. Reciprocally, intestinal immune cells support a number of important IEC functions (Fig. 3).
Figure 2.
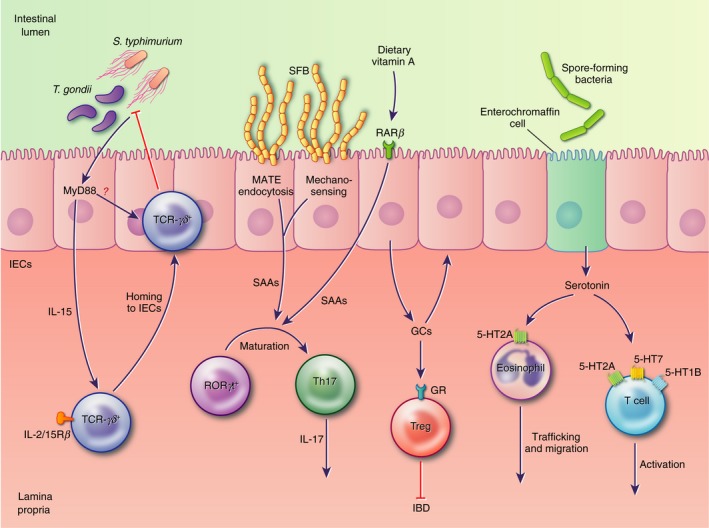
In response to microbial stimuli, intestinal epithelial cells (IECs) secrete factors that modulate various immune cell functions. In the small intestine these include interleukin‐15 (IL‐15), required for the recruitment of protective T‐cell receptor (TCR) ‐γδ + intraepiethlial lymphocytes (IELs) to the epithelial layer, and serum amyloids, which induce the differentiation of IL‐17‐secreting T helper type 17 cells. In the small and large intestine, glucocorticoids and serotonin promote anti‐inflammatory responses by immune cell populations, including lymphocytes and eosinophils, modulating inflammation and the development of disease pathology.
Figure 3.
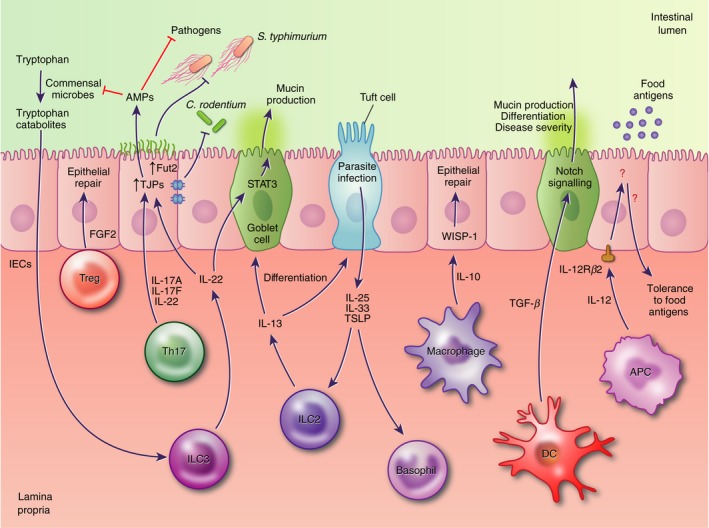
Immune cells contribute to the regulation of intestinal epithelial cell (IEC) differentiation and barrier function. For example, type 3 innate lymphoid cells (ILC3) secretion of interleukin‐22 (IL‐22) regulates IEC secretion of antimicrobial peptides (AMPs) and mucins, tight junction formation, and surface protein glycosylation, assisting in resistance to pathogenic microbes. Tolerance to food antigens is reported to involve IEC responsiveness to IL‐12; however, the subsequent IEC signalling pathways and immune cell types that mediate this response are not currently known.
IEC secretion of immunomodulatory molecules
Among the immunomodulatory molecules that are produced by IECs, thymic stromal lymphopoietin, transforming growth factor (TGF)‐β, retinoic acid and IL‐10 have been shown to impact a broad range of immune cells and have each earned their own detailed reviews.65, 66, 67, 68 In addition to these well‐described modulators of immune cell function, IEC production of IL‐15 has recently been shown to be required for the homing of protective T‐cell receptor‐γδ‐positive (TCR‐γδ +) intraepiethlial lymphocytes (IELs) to the epithelium of the small intestine.69 The TCR‐γδ+ IEL surveillance behaviour, antimicrobial responses and protection against pathogens such as Salmonella Typhimurium and Toxoplasma gondii are dependent on MyD88 signalling in IECs;70, 71 however, the mechanisms of IEC–IEL communication required for these functions are still unknown. In response to colonization by adherent microbes, IECs secrete SAAs, which promotes the functional maturation of retinoic acid‐related orphan receptor γt‐positive (RORγt+) T cells to IL‐17‐secreting T helper type 17 (Th17) cells.72, 73 This has been hypothesized to occur via mechanosensing of microbial contact, and a recent study has shown that in the case of segmented filamentous bacteria (SFB), the transfer of SFB antigens through IECs via microbial adhesion‐triggered endocytosis (MATE) plays a pivotal role.74 Another recent study shows that epithelial sensing of dietary vitamin A through retinoic acid receptor β is also required for IEC expression of SAAs.75
Perhaps less appreciated, glucocorticoids (GCs) and neurotransmitters are also abundantly produced by epithelial cells in the gut. GCs are well‐known for their general anti‐inflammatory effects, but beyond their production in adrenal glands, crypt IECs have been shown to release GCs in response to anti‐CD3‐mediated T‐cell activation, and IEC synthesis of GCs has been shown to control local inflammation and disease severity in a 2,4,6‐trinitrobenzene sulphonic acid colitis model.76, 77 As almost all vertebrate cells express glucocorticoid receptors (GRs) the effects of GCs are pleiotropic; however, T‐cell‐specific responses to GCs have been shown to be involved in T‐cell homeostasis, and regulatory T (Treg) cell‐specific GR deficiency was recently shown to impair Treg cell capacity to prevent the induction of disease in a mouse model of inflammatory bowel disease.78, 79 In addition, a recent study of mice with diminished GR responses revealed an interferon‐specific gene signature in the gut that was abrogated by antibiotic treatment, indicating a role for the microbiota.80 While information regarding intestinal production of GCs continues to emerge, the stimuli involved and immune cell effects have yet to be fully elucidated.
Similarly, although a monoamine neurotransmitter, serotonin is primarily produced in the intestines by enterochromaffin cells. As discussed earlier, a recent study using germ‐free mice colonized with spore‐forming bacteria identified a role for metabolites from commensal microbes in promoting serotonin biosynthesis by colonic enterochromaffin cells.37 Although the effects of serotonin on intestinal immune cells have not been completely characterized, most immune cells express the serotonin transporter (SERT), and there is evidence that functions as diverse as T‐cell activation, eosinophil trafficking and tumour necrosis factor‐α‐mediated inflammation are modulated by serotonin.81, 82, 83
IEC transport of microbial antigens and metabolites
An important mechanism by which intestinal epithelial cells direct adaptive immune responses to gut microbes is by antigen sampling and presentation to immune cells underlying the epithelium. Specialized M cells are concentrated in the follicle‐associated epithelium that overlies the luminal surface of Peyer's patches and isolated lymphoid follicles of the small intestine. M cells directly take up antigens and intact microorganisms from the intestinal lumen and transport them in a unidirectional way for presentation to resident immune cells. Antigen sampling by M cells is likely to be the key initiator of intestinal IgA responses to commensal bacteria as mice with impaired M cell differentiation display decreased faecal secretory IgA.84
In addition to M cells, goblet cells contribute to antigen sampling by forming goblet cell‐associated antigen passages (GAPs) to deliver intestinal lumen antigens to CD103+ dendritic cells in the lamina propria.85 Regulation of GAPs may constitute a dynamic means of modulating intestinal immune responses. While small intestine goblet cells form GAPs in response to acetylcholine, colonic goblet cell sensing of commensal microbes via MyD88 decreases their acetylcholine responsiveness and formation of GAPs to limit inflammatory immune responses to commensals.86 Timed control of GAPs during the pre‐weaning phase has been implicated in Treg cell‐mediated tolerance towards commensal bacteria,87 and during Salmonella infection IL‐1β inhibits GAP formation, leading to decreased bacterial dissemination.88
Enterocytes also participate in antigen presentation by several processes. These include presentation of lipid antigens to natural killer T cells via expression of CD1d, and IEC CD1d expression has been shown to suppress pro‐inflammatory natural killer T‐cell functions thereby reducing intestinal inflammation.89 In addition, MHC class II has been shown to be constituitively expressed by IECs in the upper villi of the small intestine, and surface expression appears to be increased in patients with inflammatory bowel disease and in response to interferon‐γ.90, 91, 92 Reciprocally, IEC antigen presentation was shown to promote interferon‐γ secretion by CD4+ T cells in co‐cultures of normal T cells with IECs from patients with inflammatory bowel disease;93 however, more recent studies suggest that interferon‐γ‐induced MHC class II expression on IECs plays a more anti‐inflammatory role by promoting a tolerogenic ratio of Treg cells to effector CD4+ T cells.94, 95 Still, the role of IEC antigen presentation in shaping intestinal immunity has not been thoroughly explored, and the intimate contact between the epithelium and commensal microbes provides ample opportunity for IECs to curate intestinal T‐cell responses.
Immune cell contributions to IEC differentiation and function
In addition to IECs regulating immune cell functions, several intestinal immune cell types influence IEC homeostasis and inflammatory responses (Fig. 3). For example, in response to microbial metabolites such as tryptophan catabolites, type 3 innate lymphoid cells (ILC3s) produce cytokines that regulate barrier functions of IECs.25 ILC3s secrete IL‐22, which promotes IEC homeostasis and repair, and can induce AMPs to control the growth of both pathogenic and commensal microbes.96, 97, 98 IL‐22 also affects the glycosylation of IEC surface proteins by inducing fucosyltransferase 2 (Fut2) expression, thereby enhancing host protection against Salmonella Typhimurium.99 Mucin production by IECs is also increased by IL‐22 through the activation of signal transducer and activator of transcription STAT3,100 and tight junction proteins such as claudin‐2 have recently been shown to be up‐regulated by IL‐22, inducing diarrhoea and facilitating clearance of Citrobacter rodentium in a mouse model of enteric infection.101
Beyond ILC3s and IL‐22, some other lymphoid cells also contribute to IEC responses. During parasitic infection, IECs secrete a number of cytokines that promote the expansion and activation of group 2 innate lymphoid cells (ILC2s) and basophils, including IL‐33 and thymic stromal lymphopoietin (TSLP), and IL‐25 produced by tuft cells.2, 5, 102, 103 Reciprocally, activated ILC2s secrete IL‐13, which promotes tuft and goblet cell differentiation and parasite clearance.3, 104 The signature cytokines secreted by Th17 cells (IL‐17A, IL‐17F and IL‐22) can also induce IEC‐mediated AMP secretion and reinforce IEC tight junctions.105, 106, 107, 108 In addition, production of fibroblast growth factor 2 (FGF2) by Treg cells has recently been shown to synergize with IL‐17 to enhance mechanisms of intestinal epithelial repair.109 IEC responsiveness to tumour necrosis factor also promotes mucosal repair and healing in individuals with Crohn's disease, human cells and mouse models.110
Myeloid cells also play key roles in IEC differentiation and function. For instance, perturbations to macrophage–IEC interactions lead to aberrant differentiation of IEC subtypes. Using CSF1R blockade to deplete macrophages that localize to the intestinal crypt epithelium, a recent study found that absence of macrophages results in reduced Lgr5+ intestinal stem cells, lysozyme‐expressing Paneth cells and Peyer's patch M cells, and increased goblet cell density.111 Macrophages have also been shown to be the likely source of IL‐10 in a colon biopsy‐induced injury model, and in this model, macrophage IL‐10 induced epithelial synthesis of the pro‐repair WNT1‐inducible signalling protein 1 to mediate IEC proliferation and mucosal wound healing.112 In DCs, transforming growth factor‐β signalling has been suggested to control goblet cell numbers, mucus production and disease severity in dextran sulphate sodium colitis via Notch signalling, although the effects of DC dysfunction and involvement of other immune cell types were not fully investigated in this study.113 More recently, IL‐12 responsiveness via IL‐12Rβ2 on IECs has been shown to play a protective role in food allergy; however, the precise mechanism of protection is once again unknown.114
Immune cell–microbiota crosstalk
Due to limited direct contact, most immune cell–microbiota communication is likely mediated, at least to some extent, by IECs; however, the contributions of IECs to many microbiota–immune cell interactions have yet to be fully realized. Nevertheless, a growing body of work has revealed the importance of commensal microbes for the proper development and function of immune cells (Fig. 4), and immune cells reciprocally shape the microbial habitat and microbiota diversity.
Figure 4.
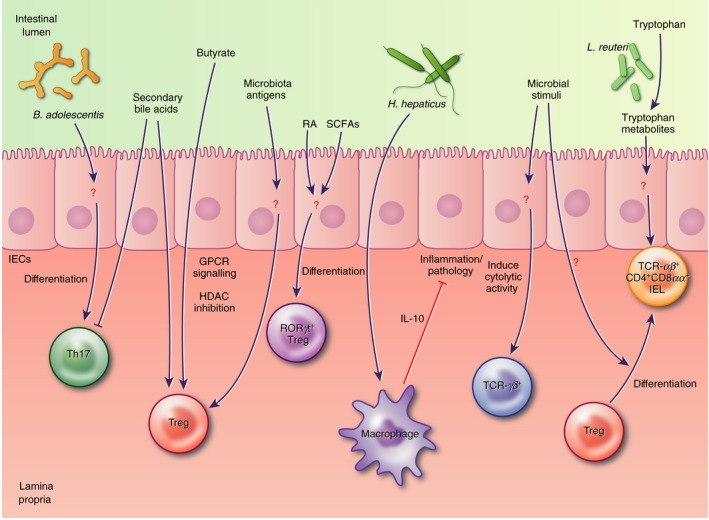
Microbial modulation of intestinal immune cells is reported to involve both direct interaction of lymphocytes and antigen‐presenting cells (APCs) with microbial stimuli, as well as relatively uncharacterized indirect interactions via intestinal epithelial cells (IECs). These interactions involve many subsets of intraepithelial and lamina propria T cells and microbial metabolites like short‐chain fatty acids, tryptophan catabolites and secondary bile acids, as well as currently undefined microbial antigens.
Microbiota modulation of intestinal lymphocytes
As mentioned earlier, the proper development of IL‐17‐secreting Th17 cells requires SAA production by IECs in response to microbial adhesion and specifically MATE in response to SFB adhesion. The human symbiont Bifidobacterium adolescentis, which closely associates with the gut epithelium, is also reported to induce Th17 cells in the murine intestine with a transcriptional programme distinct from SFB, suggesting that Th17 accumulation can also be promoted by another mechanism.115 Although precise roles for IECs have not been completely defined, roles for commensal microbial metabolites and antigens also continue to emerge for the generation and function of Treg cells. In three seminal studies, commensal‐derived butyrate was shown to drive induction of peripheral Treg cells in the colon.116, 117, 118 A later study also showed a role for recognition of antigens from commensal microbes in intestinal Treg cell differentiation. Transfer of naive transgenic T cells specific for commensal antigens into mice with a normal microbiota resulted in robust Foxp3 induction in these cells.119 At weaning, the intestinal microbiota induces a vigorous immune response associated with the generation of RORγt+ Treg cells in an SCFA and retinoic acid‐dependent manner, and inhibition of this response leads to later immunopathologies including colitis.120
RORγt+ Treg cells specific for Helicobacter hepaticus have also been shown to mediate tolerance to this commensal pathobiont,121 and a polysaccharide from the same species induces anti‐inflammatory IL‐10 secretion in intestinal macrophages.122 However, Helicobacter specificity itself does not dictate an anti‐inflammatory programme. A recent study demonstrated that the same Helicobacter‐specific T cells differentiate to Treg cells during homeostasis and effector T cells during colitis.123 Helicobacter bilis colonization, on the other hand, has previously been shown to induce persistent immune reactivity to other commensal bacteria.124 Collectively, these studies suggest the importance of antigen‐independent contextual cues during T‐cell activation in the gut for determining T‐cell fates. Indeed, two secondary bile acids, generated by commensal bacteria transformation of primary bile acids, were recently shown to inhibit Th17 differentiation and promote Treg cell induction.125 Identifying the full spectrum of contextual cues will be integral for understanding how intestinal T cells are programmed.
In addition to conventional T cells, IELs have proved to be markedly influenced by the commensal microbiota. For example, TCR‐αβ + IELs are almost absent in germ‐free mice,126, 127 and TCR‐γδ + IELs have impaired cytolytic activity.128 The mechanisms of this control are still under investigation, but they likely involve transmission of signals through the IECs. The gut microbiota is also an important factor in the generation of TCR‐αβ + CD4+ CD8αα + IELs. In a recent study, introduction of tryptophan‐metabolizing Lactobacillus reuteri in mice given a diet rich in tryptophan was sufficient to induce TCRαβ + CD4+ CD8αα + IEL differentiation.129 Another study has demonstrated microbiota‐dependent conversion of lamina propria Foxp3+ Treg cells into TCRαβ + CD4+ CD8αα + IELs upon homing to the intestinal epithelium.130 The ability of epithelial cells and microbial metabolites to contribute to the induction of this IEL subset is also still being elucidated.
Immune cell effects on the intestinal microbiota
Although historically met with scepticism and comparatively understudied, the influences of intestinal immune cells on the microbiota are also gaining appreciation. Evidence that the adaptive immune system shapes microbial composition and diversity in the gut has been provided using sequencing of bacteria in multiple intestinal loci in Rag‐deficient mice that lack B and T cells.131 However, while ILCs are present in Rag‐deficient mice, there is evidence that their number and function are altered,132 complicating the conclusions that can be drawn from these animals about the role of B and T cells. Further studies have identified an important role for polyreactive IgA in facilitating the induction of bacteria‐specific IgA, and differences in these significantly influence colonization by commensal microbes.133 Indeed, Bacteroides fragilis has now been shown to permit binding of IgA to facilitate its ability to occupy a privileged intestinal niche in close proximity to IECs.134 Very recently, an important role was identified for commensal‐specific IgG that results from epithelial disruption in the gut. Responsiveness to these IgGs in intestinal macrophages via activating FcγRs drives intestinal inflammation and colitis.135 Although the effects of these IgGs on microbiota composition have not yet been characterized, future studies may define functions for both intestinal IgA and IgG in modulating commensal microbial communities.
Immune cells in the gut are tasked with maintaining a balance of physiological inflammation and tolerance. The resulting intestinal immune cell programmes regulate the microbial ecosystem in the gut in a manner that allows for beneficial colonization and deters invasive pathogenic infection. For example, Foxp3+ Treg cells have been shown to support microbiota diversity both by suppressing inflammation and facilitating IgA selection in Peyer's patches.136 Conversely, a lack of peripheral Treg cells leads to increased type 2 immune responses and disruption of microbial niches for IEC border‐dwelling bacteria,137 highlighting the importance of these T cells in shaping the intestinal microbial environment. In addition to composition and diversity, the evolution of commensal bacterial species has also been shown to be influenced by host adaptive immunity. In the intestines of Rag‐deficient mice, the rate and predictability of E. coli adaptation are altered in comparison with wild‐type hosts.138 Taken together, these studies bring new insight into the intimate interdependence of the intestinal microbiota and immune system and open additional questions about the mechanisms involved and contribution of IECs.
Conclusion
Due to the anatomical location of IECs between the intestinal microbiota and the host intestinal tissues, it is reasonable to predict that IECs play an important role in controlling the interaction between the luminal microbiota and underlying immune cells. Indeed, recent literature has highlighted the ability of IECs to contribute to shaping both host intestinal immunity and gut microbial composition. However, despite recent progress in the field, several challenges remain to be addressed and overcome.
Demonstrating that IEC‐secreted factors are induced in response to microbe‐derived signals, and the effects of these factors on immune cells, has proved difficult. Most IEC‐derived cytokines are also produced by other cell types, therefore IEC involvement in vivo is usually inferred but not definitively demonstrated. Knockout mice for certain receptors or effector molecules expressed by IECs have yielded further insight into the roles of IECs as direct sensors of microbial signals; however, few studies have employed IEC‐specific genetic ablation in vivo. Studying the impact of microbe–IEC signalling on the function of immune cell subsets is also limited due to the difficulty in isolating and manipulating these cell types; the lifespan of IECs is extremely short as they are renewed every 2–6 days.139 Although in vitro models have provided valuable insight into IEC signalling pathways and production of effectors, they remain unable to recapitulate the complexity of the intestinal environment, and interpretation of these studies is consequently limited. By further elucidating the mechanisms involved in microbe–immune crosstalk at the intestinal epithelium, we can better understand the role of IECs in regulating host immunity during homeostasis as well as during states of dysbiosis and disease.
Disclosures
VAP is supported by a Royal Society and Wellcome Trust Sir Henry Dale Fellowship. The authors declare they have no conflicts of interest.
References
- 1. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 2016; 14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. von Moltke J, Ji M, Liang HE, Locksley RM. Tuft‐cell‐derived IL‐25 regulates an intestinal ILC2‐epithelial response circuit. Nature 2016; 529:221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV et al Tuft cells, taste‐chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 2016; 351:1329–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parikh K, Antanaviciute A, Fawkner‐Corbett D, Jagielowicz M, Aulicino A, Lagerholm C et al Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 2019; 567:49–55. [DOI] [PubMed] [Google Scholar]
- 5. Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C et al A single‐cell survey of the small intestinal epithelium. Nature 2017; 551:333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol 2007; 19:59–69. [DOI] [PubMed] [Google Scholar]
- 7. Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature 2016; 535:65–74. [DOI] [PubMed] [Google Scholar]
- 8. Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host‐microbial symbiosis. Nat Immunol 2013; 14:668–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Artis D. Epithelial‐cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol 2008; 8:411–20. [DOI] [PubMed] [Google Scholar]
- 10. Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host–microbial interface. Proc Natl Acad Sci USA 2008; 105:20858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fahlgren A, Hammarstrom S, Danielsson A, Hammarstrom ML. Increased expression of antimicrobial peptides and lysozyme in colonic epithelial cells of patients with ulcerative colitis. Clin Exp Immunol 2003; 131:90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Friedrich C, Mamareli P, Thiemann S, Kruse F, Wang Z, Holzmann B et al MyD88 signaling in dendritic cells and the intestinal epithelium controls immunity against intestinal infection with C. rodentium . PLoS Pathog 2017; 13:e1006357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Furter M, Sellin ME, Hansson GC, Hardt WD. Mucus architecture and near‐surface swimming affect distinct Salmonella typhimurium infection patterns along the murine intestinal tract. Cell Rep 2019; 27:2665–78 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Graziani F, Pujol A, Nicoletti C, Dou S, Maresca M, Giardina T et al Ruminococcus gnavus E1 modulates mucin expression and intestinal glycosylation. J Appl Microbiol 2016; 120:1403–17. [DOI] [PubMed] [Google Scholar]
- 15. Martin R, Chamignon C, Mhedbi‐Hajri N, Chain F, Derrien M, Escribano‐Vazquez U et al The potential probiotic Lactobacillus rhamnosus CNCM I‐3690 strain protects the intestinal barrier by stimulating both mucus production and cytoprotective response. Sci Rep 2019; 9:5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gibold L, Garenaux E, Dalmasso G, Gallucci C, Cia D, Mottet‐Auselo B et al The Vat‐AIEC protease promotes crossing of the intestinal mucus layer by Crohn's disease‐associated Escherichia coli . Cell Microbiol 2016; 18:617–31. [DOI] [PubMed] [Google Scholar]
- 17. Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L et al A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 2017; 23:107–13. [DOI] [PubMed] [Google Scholar]
- 18. Quan JH, Huang R, Wang Z, Huang S, Choi IW, Zhou Y et al P2X7 receptor mediates NLRP3‐dependent IL‐1β secretion and parasite proliferation in Toxoplasma gondii‐infected human small intestinal epithelial cells. Parasit Vectors 2018; 11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wlodarska M, Thaiss CA, Nowarski R, Henao‐Mejia J, Zhang JP, Brown EM et al NLRP6 inflammasome orchestrates the colonic host‐microbial interface by regulating goblet cell mucus secretion. Cell 2014; 156:1045–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu S, Ding S, Wang P, Wei Z, Pan W, Palm NW et al Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature 2017; 546:667–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nordlander S, Pott J, Maloy KJ. NLRC4 expression in intestinal epithelial cells mediates protection against an enteric pathogen. Mucosal Immunol 2014; 7:775–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rauch I, Deets KA, Ji DX, von Moltke J, Tenthorey JL, Lee AY et al NAIP‐NLRC4 inflammasomes coordinate intestinal epithelial cell expulsion with eicosanoid and IL‐18 release via activation of Caspase‐1 and ‐8. Immunity 2017; 46:649–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bel S, Pendse M, Wang Y, Li Y, Ruhn KA, Hassell B et al Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 2017; 357:1047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP et al Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll‐like receptor 4. Immunity 2014; 41:296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G et al Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin‐22. Immunity 2013; 39:372–85. [DOI] [PubMed] [Google Scholar]
- 26. Metidji A, Omenetti S, Crotta S, Li Y, Nye E, Ross E et al The environmental sensor AHR protects from inflammatory damage by maintaining intestinal stem cell homeostasis and barrier integrity. Immunity 2018; 49:353–62 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wlodarska M, Luo C, Kolde R, d'Hennezel E, Annand JW, Heim CE et al Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe 2017; 22:25–37 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park JH, Kotani T, Konno T, Setiawan J, Kitamura Y, Imada S et al Promotion of intestinal epithelial cell turnover by commensal bacteria: role of short‐chain fatty acids. PLoS One 2016; 11:e0156334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zheng L, Kelly CJ, Battista KD, Schaefer R, Lanis JM, Alexeev EE et al Microbial‐derived butyrate promotes epithelial barrier function through IL‐10 receptor‐dependent repression of claudin‐2. J Immunol 2017; 199:2976–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S et al Metabolite‐sensing receptors GPR43 and GPR109A facilitate dietary fibre‐induced gut homeostasis through regulation of the inflammasome. Nat Commun 2015; 6:6734. [DOI] [PubMed] [Google Scholar]
- 31. Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ et al Crosstalk between microbiota‐derived short‐chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 2015; 17:662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016; 16:341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao Y, Chen F, Wu W, Sun M, Bilotta AJ, Yao S et al GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol 2018; 11:752–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hopkins EGD, Roumeliotis TI, Mullineaux‐Sanders C, Choudhary JS, Frankel G. Intestinal epithelial cells and the microbiome undergo swift reprogramming at the inception of colonic Citrobacter rodentium infection. MBio 2019; 10:e00062‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alcaino C, Knutson KR, Treichel AJ, Yildiz G, Strege PR, Linden DR et al A population of gut epithelial enterochromaffin cells is mechanosensitive and requires Piezo2 to convert force into serotonin release. Proc Natl Acad Sci USA 2018; 115:E7632–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Terry N, Margolis KG. Serotonergic mechanisms regulating the GI tract: experimental evidence and therapeutic relevance. Handb Exp Pharmacol 2017; 239:319–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L et al Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015; 161:264–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang G, Liu J, Wu L, Fan Y, Sun L, Qian F et al Elevated expression of serum amyloid A 3 protects colon epithelium against acute injury through TLR2‐dependent induction of neutrophil IL‐22 expression in a mouse model of colitis. Front Immunol 2018; 9:1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schneider C, O'Leary CE, von Moltke J, Liang HE, Ang QY, Turnbaugh PJ et al A metabolite‐triggered tuft cell‐ILC2 circuit drives small intestinal remodeling. Cell 2018; 174:271–84 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC et al Autophagosome‐independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 2008; 4:458–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Castillo EF, Dekonenko A, Arko‐Mensah J, Mandell MA, Dupont N, Jiang S et al Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci USA 2012; 109:E3168–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Watson RO, Manzanillo PS, Cox JS, Extracellular M. Tuberculosis DNA targets bacteria for autophagy by activating the host DNA‐sensing pathway. Cell 2012; 150:803–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Conway KL, Kuballa P, Song JH, Patel KK, Castoreno AB, Yilmaz OH et al Atg16l1 is required for autophagy in intestinal epithelial cells and protection of mice from Salmonella infection. Gastroenterology 2013; 145:1347–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Benjamin JL, Sumpter R Jr, Levine B, Hooper LV. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe 2013; 13:723–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang L, Liu C, Zhao W, He C, Ding J, Dai R et al Impaired autophagy in intestinal epithelial cells alters gut microbiota and host immune responses. Appl Environ Microbiol 2018; 84:e00880‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kwon YH, Wang H, Denou E, Ghia JE, Rossi L, Fontes ME et al Modulation of gut microbiota composition by serotonin signaling influences intestinal immune response and susceptibility to colitis. Cell Mol Gastroenterol Hepatol 2019; 7:709–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muniz LR, Knosp C, Yeretssian G. Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front Immunol 2012; 3:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol 2011; 9:356–68. [DOI] [PubMed] [Google Scholar]
- 49. Salzman NH, Hung K, Haribhai D, Chu H, Karlsson‐Sjoberg J, Amir E et al Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol 2010; 11:76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Elinav E, Strowig T, Kau AL, Henao‐Mejia J, Thaiss CA, Booth CJ et al NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011; 145:745–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Levy M, Thaiss CA, Zeevi D, Dohnalova L, Zilberman‐Schapira G, Mahdi JA et al Microbiota‐modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 2015; 163:1428–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mamantopoulos M, Ronchi F, Van Hauwermeiren F, Vieira‐Silva S, Yilmaz B, Martens L et al Nlrp6‐ and ASC‐dependent inflammasomes do not shape the commensal gut microbiota composition. Immunity 2017; 47:339–48 e4. [DOI] [PubMed] [Google Scholar]
- 53. Lemire P, Robertson SJ, Maughan H, Tattoli I, Streutker CJ, Platnich JM et al The NLR protein NLRP6 does not impact gut microbiota composition. Cell Rep 2017; 21:3653–61. [DOI] [PubMed] [Google Scholar]
- 54. Stappenbeck TS, Virgin HW. Accounting for reciprocal host–microbiome interactions in experimental science. Nature 2016; 534:191–9. [DOI] [PubMed] [Google Scholar]
- 55. Robertson SJ, Zhou JY, Geddes K, Rubino SJ, Cho JH, Girardin SE et al Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes 2013; 4:222–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ramanan D, Tang MS, Bowcutt R, Loke P, Cadwell K. Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus . Immunity 2014; 41:311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Robertson SJ, Lemire P, Maughan H, Goethel A, Turpin W, Bedrani L et al Comparison of Co‐housing and littermate methods for microbiota standardization in mouse models. Cell Rep 2019; 27:1910–9 e2. [DOI] [PubMed] [Google Scholar]
- 58. Lu P, Sodhi CP, Yamaguchi Y, Jia H, Prindle T Jr, Fulton WB et al Intestinal epithelial Toll‐like receptor 4 prevents metabolic syndrome by regulating interactions between microbes and intestinal epithelial cells in mice. Mucosal Immunol 2018; 11:727–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Islam MS, Krachler AM. Mechanosensing regulates virulence in Escherichia coli O157:H7. Gut Microbes 2016; 7:63–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L et al The host shapes the gut microbiota via fecal MicroRNA. Cell Host Microbe 2016; 19:32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pudlo NA, Urs K, Kumar SS, German JB, Mills DA, Martens EC. Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. MBio 2015; 6:e01282‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012; 3:289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Goto Y, Uematsu S, Kiyono H. Epithelial glycosylation in gut homeostasis and inflammation. Nat Immunol 2016; 17:1244–51. [DOI] [PubMed] [Google Scholar]
- 64. Owen CD, Tailford LE, Monaco S, Suligoj T, Vaux L, Lallement R et al Unravelling the specificity and mechanism of sialic acid recognition by the gut symbiont Ruminococcus gnavus . Nat Commun 2017; 8:2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ihara S, Hirata Y, Koike K. TGF‐β in inflammatory bowel disease: a key regulator of immune cells, epithelium, and the intestinal microbiota. J Gastroenterol 2017; 52:777–87. [DOI] [PubMed] [Google Scholar]
- 66. Tsilingiri K, Fornasa G, Rescigno M. Thymic stromal lymphopoietin: to cut a long story short. Cell Mol Gastroenterol Hepatol 2017; 3:174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Oliveira LM, Teixeira FME, Sato MN. Impact of retinoic acid on immune cells and inflammatory diseases. Mediators Inflamm 2018; 2018:3067126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Andrews C, McLean MH, Durum SK. Cytokine tuning of intestinal epithelial function. Front Immunol 2018; 9:1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hu MD, Ethridge AD, Lipstein R, Kumar S, Wang Y, Jabri B, et al Epithelial IL‐15 is a critical regulator of γδ intraepithelial lymphocyte motility within the intestinal mucosa. J Immunol 2018; 201:747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ismail AS, Severson KM, Vaishnava S, Behrendt CL, Yu X, Benjamin JL et al γδ intraepithelial lymphocytes are essential mediators of host‐microbial homeostasis at the intestinal mucosal surface. Proc Natl Acad Sci USA 2011; 108:8743–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hoytema van Konijnenburg DP, Reis BS, Pedicord VA, Farache J, Victora GD, Mucida D. Intestinal epithelial and intraepithelial T cell crosstalk mediates a dynamic response to infection. Cell 2017; 171:783–94 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S et al Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 2015; 163:367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ et al An IL‐23R/IL‐22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell 2015; 163:381–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ladinsky MS, Araujo LP, Zhang X, Veltri J, Galan‐Diez M, Soualhi S et al Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science 2019; 363:eaat4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gattu S, Bang YJ, Pendse M, Dende C, Chara AL, Harris TA et al Epithelial retinoic acid receptor β regulates serum amyloid A expression and vitamin A‐dependent intestinal immunity. Proc Natl Acad Sci USA 2019; 116:10911–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cima I, Corazza N, Dick B, Fuhrer A, Herren S, Jakob S et al Intestinal epithelial cells synthesize glucocorticoids and regulate T cell activation. J Exp Med 2004; 200:1635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Coste A, Dubuquoy L, Barnouin R, Annicotte JS, Magnier B, Notti M et al LRH‐1‐mediated glucocorticoid synthesis in enterocytes protects against inflammatory bowel disease. Proc Natl Acad Sci USA 2007; 104:13098–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pazirandeh A, Xue Y, Prestegaard T, Jondal M, Okret S. Effects of altered glucocorticoid sensitivity in the T cell lineage on thymocyte and T cell homeostasis. FASEB J 2002; 16:727–9. [DOI] [PubMed] [Google Scholar]
- 79. Rocamora‐Reverte L, Tuzlak S, von Raffay L, Tisch M, Fiegl H, Drach M et al Glucocorticoid receptor‐deficient Foxp3+ regulatory T cells fail to control experimental inflammatory bowel disease. Front Immunol 2019; 10:472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ballegeer M, Van Looveren K, Timmermans S, Eggermont M, Vandevyver S, Thery F et al Glucocorticoid receptor dimers control intestinal STAT1 and TNF‐induced inflammation in mice. J Clin Invest 2018; 128:3265–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Leon‐Ponte M, Ahern GP, O'Connell PJ. Serotonin provides an accessory signal to enhance T‐cell activation by signaling through the 5‐HT7 receptor. Blood 2007; 109:3139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kang BN, Ha SG, Bahaie NS, Hosseinkhani MR, Ge XN, Blumenthal MN et al Regulation of serotonin‐induced trafficking and migration of eosinophils. PLoS One 2013; 8:e54840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nau F Jr, Yu B, Martin D, Nichols CD. Serotonin 5‐HT2A receptor activation blocks TNF‐α mediated inflammation in vivo . PLoS One 2013; 8:e75426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rios D, Wood MB, Li J, Chassaing B, Gewirtz AT, Williams IR. Antigen sampling by intestinal M cells is the principal pathway initiating mucosal IgA production to commensal enteric bacteria. Mucosal Immunol 2016; 9:907–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, Knoop KA et al Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature 2012; 483:345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Knoop KA, McDonald KG, McCrate S, McDole JR, Newberry RD. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol 2015; 8:198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Knoop KA, Gustafsson JK, McDonald KG, Kulkarni DH, Coughlin PE, McCrate S et al Microbial antigen encounter during a preweaning interval is critical for tolerance to gut bacteria. Sci Immunol 2017; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kulkarni DH, McDonald KG, Knoop KA, Gustafsson JK, Kozlowski KM, Hunstad DA et al Goblet cell associated antigen passages are inhibited during Salmonella typhimurium infection to prevent pathogen dissemination and limit responses to dietary antigens. Mucosal Immunol 2018; 11:1103–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Olszak T, Neves JF, Dowds CM, Baker K, Glickman J, Davidson NO et al Protective mucosal immunity mediated by epithelial CD1d and IL‐10. Nature 2014; 509:497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lin XP, Almqvist N, Telemo E. Human small intestinal epithelial cells constitutively express the key elements for antigen processing and the production of exosomes. Blood Cells Mol Dis 2005; 35:122–8. [DOI] [PubMed] [Google Scholar]
- 91. Colgan SP, Parkos CA, Matthews JB, D'Andrea L, Awtrey CS, Lichtman AH et al Interferon‐γ induces a cell surface phenotype switch on T84 intestinal epithelial cells. Am J Physiol 1994; 267:C402–10. [DOI] [PubMed] [Google Scholar]
- 92. Bar F, Sina C, Hundorfean G, Pagel R, Lehnert H, Fellermann K et al Inflammatory bowel diseases influence major histocompatibility complex class I (MHC I) and II compartments in intestinal epithelial cells. Clin Exp Immunol 2013; 172:280–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Dotan I, Allez M, Nakazawa A, Brimnes J, Schulder‐Katz M, Mayer L. Intestinal epithelial cells from inflammatory bowel disease patients preferentially stimulate CD4+ T cells to proliferate and secrete interferon‐γ . Am J Physiol Gastrointest Liver Physiol 2007; 292:G1630–40. [DOI] [PubMed] [Google Scholar]
- 94. Westendorf AM, Fleissner D, Groebe L, Jung S, Gruber AD, Hansen W et al CD4+ Foxp3+ regulatory T cell expansion induced by antigen‐driven interaction with intestinal epithelial cells independent of local dendritic cells. Gut 2009; 58:211–9. [DOI] [PubMed] [Google Scholar]
- 95. Thelemann C, Eren RO, Coutaz M, Brasseit J, Bouzourene H, Rosa M et al Interferon‐γ induces expression of MHC class II on intestinal epithelial cells and protects mice from colitis. PLoS One 2014; 9:e86844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lindemans CA, Calafiore M, Mertelsmann AM, O'Connor MH, Dudakov JA, Jenq RR et al Interleukin‐22 promotes intestinal‐stem‐cell‐mediated epithelial regeneration. Nature 2015; 528:560–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ngo VL, Abo H, Maxim E, Harusato A, Geem D, Medina‐Contreras O et al A cytokine network involving IL‐36γ, IL‐23, and IL‐22 promotes antimicrobial defense and recovery from intestinal barrier damage. Proc Natl Acad Sci USA 2018; 115:E5076–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hammer AM, Morris NL, Cannon AR, Khan OM, Gagnon RC, Movtchan NV et al Interleukin‐22 prevents microbial dysbiosis and promotes intestinal barrier regeneration following acute injury. Shock 2017; 48:657–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A et al Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 2014; 345:1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK et al IL‐22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest 2008; 118:534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Tsai PY, Zhang B, He WQ, Zha JM, Odenwald MA, Singh G et al IL‐22 upregulates epithelial claudin‐2 to drive diarrhea and enteric pathogen clearance. Cell Host Microbe 2017; 21:671–81 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V et al Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 2016; 529:226–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Camelo A, Rosignoli G, Ohne Y, Stewart RA, Overed‐Sayer C, Sleeman MA et al IL‐33, IL‐25, and TSLP induce a distinct phenotypic and activation profile in human type 2 innate lymphoid cells. Blood Adv 2017; 1:577–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Artis D, Wang ML, Keilbaugh SA, He W, Brenes M, Swain GP et al RELMβ/FIZZ2 is a goblet cell‐specific immune‐effector molecule in the gastrointestinal tract. Proc Natl Acad Sci USA 2004; 101:13596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q et al Interleukin‐22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med 2008; 14:282–9. [DOI] [PubMed] [Google Scholar]
- 106. Dixon BR, Radin JN, Piazuelo MB, Contreras DC, Algood HM. IL‐17a and IL‐22 induce expression of antimicrobials in gastrointestinal epithelial cells and may contribute to epithelial cell defense against Helicobacter pylori . PLoS One 2016; 11:e0148514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Maxwell JR, Zhang Y, Brown WA, Smith CL, Byrne FR, Fiorino M et al Differential roles for interleukin‐23 and interleukin‐17 in intestinal immunoregulation. Immunity 2015; 43:739–50. [DOI] [PubMed] [Google Scholar]
- 108. Lee JS, Tato CM, Joyce‐Shaikh B, Gulen MF, Cayatte C, Chen Y et al Interleukin‐23‐independent IL‐17 production regulates intestinal epithelial permeability. Immunity 2015; 43:727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Song X, Dai D, He X, Zhu S, Yao Y, Gao H et al Growth factor FGF2 cooperates with interleukin‐17 to repair intestinal epithelial damage. Immunity 2015; 43:488–501. [DOI] [PubMed] [Google Scholar]
- 110. Bradford EM, Ryu SH, Singh AP, Lee G, Goretsky T, Sinh P et al Epithelial TNF receptor signaling promotes mucosal repair in inflammatory bowel disease. J Immunol 2017; 199:1886–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Sehgal A, Donaldson DS, Pridans C, Sauter KA, Hume DA, Mabbott NA. The role of CSF1R‐dependent macrophages in control of the intestinal stem‐cell niche. Nat Commun 2018; 9:1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Quiros M, Nishio H, Neumann PA, Siuda D, Brazil JC, Azcutia V et al Macrophage‐derived IL‐10 mediates mucosal repair by epithelial WISP‐1 signaling. J Clin Invest 2017; 127:3510–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ihara S, Hirata Y, Serizawa T, Suzuki N, Sakitani K, Kinoshita H et al TGF‐β signaling in dendritic cells governs colonic homeostasis by controlling epithelial differentiation and the luminal microbiota. J Immunol 2016; 196:4603–13. [DOI] [PubMed] [Google Scholar]
- 114. Regoli M, Man A, Gicheva N, Dumont A, Ivory K, Pacini A et al Morphological and functional characterization of IL‐12Rβ2 chain on intestinal epithelial cells: implications for local and systemic immunoregulation. Front Immunol 2018; 9:1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Tan TG, Sefik E, Geva‐Zatorsky N, Kua L, Naskar D, Teng F et al Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci USA 2016; 113:E8141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM et al The microbial metabolites, short‐chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013; 341:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H et al Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013; 500:232–6. [DOI] [PubMed] [Google Scholar]
- 118. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P et al Metabolites produced by commensal bacteria promote peripheral regulatory T‐cell generation. Nature 2013; 504:451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Nutsch K, Chai JN, Ai TL, Russler‐Germain E, Feehley T, Nagler CR et al Rapid and efficient generation of regulatory T cells to commensal antigens in the periphery. Cell Rep 2016; 17:206–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Dejardin F et al A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity 2019; 50:1276–88 e5. [DOI] [PubMed] [Google Scholar]
- 121. Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ et al c‐MAF‐dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 2018; 554:373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Danne C, Ryzhakov G, Martinez‐Lopez M, Ilott NE, Franchini F, Cuskin F et al A large polysaccharide produced by Helicobacter hepaticus induces an anti‐inflammatory gene signature in macrophages. Cell Host Microbe 2017; 22:733–45 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Chai JN, Peng Y, Rengarajan S, Solomon BD, Ai TL, Shen Z et al Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci Immunol 2017; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Jergens AE, Wilson‐Welder JH, Dorn A, Henderson A, Liu Z, Evans RB et al Helicobacter bilis triggers persistent immune reactivity to antigens derived from the commensal bacteria in gnotobiotic C3H/HeN mice. Gut 2007; 56:934–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Hang S, Paik D, Devlin AS, Jamma T, Lu J, Ha S et al Bile acid metabolites control Th17 and Treg cell differentiation. bioRxiv 2018; 465344 10.1101/465344 [DOI] [Google Scholar]
- 126. Umesaki Y, Setoyama H, Matsumoto S, Okada Y. Expansion of αβ T‐cell receptor‐bearing intestinal intraepithelial lymphocytes after microbial colonization in germ‐free mice and its independence from thymus. Immunology 1993; 79:32–7. [PMC free article] [PubMed] [Google Scholar]
- 127. Imaoka A, Matsumoto S, Setoyama H, Okada Y, Umesaki Y. Proliferative recruitment of intestinal intraepithelial lymphocytes after microbial colonization of germ‐free mice. Eur J Immunol 1996; 26:945–8. [DOI] [PubMed] [Google Scholar]
- 128. Kawaguchi‐Miyashita M, Shimizu K, Nanno M, Shimada S, Watanabe T, Koga Y et al Development and cytolytic function of intestinal intraepithelial T lymphocytes in antigen‐minimized mice. Immunology 1996; 89:268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Cervantes‐Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J et al Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα + T cells. Science 2017; 357:806–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Sujino T, London M, Hoytema van Konijnenburg DP, Rendon T, Buch T, Silva HM et al Tissue adaptation of regulatory and intraepithelial CD4+ T cells controls gut inflammation. Science 2016; 352:1581–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Zhang H, Sparks JB, Karyala SV, Settlage R, Luo XM. Host adaptive immunity alters gut microbiota. ISME J 2015; 9:770–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Korn LL, Thomas HL, Hubbeling HG, Spencer SP, Sinha R, Simkins HM et al Conventional CD4+ T cells regulate IL‐22‐producing intestinal innate lymphoid cells. Mucosal Immunol 2014; 7:1045–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Fransen F, Zagato E, Mazzini E, Fosso B, Manzari C, El Aidy S et al BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen‐specific IgA and microbiota diversity. Immunity 2015; 43:527–40. [DOI] [PubMed] [Google Scholar]
- 134. Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC et al Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 2018; 360:795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Castro‐Dopico T, Dennison TW, Ferdinand JR, Mathews RJ, Fleming A, Clift D et al Anti‐commensal IgG drives intestinal inflammation and type 17 immunity in ulcerative colitis. Immunity 2019; 50:1099–114 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, Doi Y et al Foxp3+ T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 2014; 41:152–65. [DOI] [PubMed] [Google Scholar]
- 137. Campbell C, Dikiy S, Bhattarai SK, Chinen T, Matheis F, Calafiore M et al Extrathymically generated regulatory T cells establish a niche for intestinal border‐dwelling bacteria and affect physiologic metabolite balance. Immunity 2018; 48:1245–57 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Barroso‐Batista J, Demengeot J, Gordo I. Adaptive immunity increases the pace and predictability of evolutionary change in commensal gut bacteria. Nat Commun 2015; 6:8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Mayhew TM, Myklebust R, Whybrow A, Jenkins R. Epithelial integrity, cell death and cell loss in mammalian small intestine. Histol Histopathol 1999; 14:257–67. [DOI] [PubMed] [Google Scholar]
- 140. Yang H, Yu HB, Bhinder G, Ryz NR, Lee J, Yang H et al TLR9 limits enteric antimicrobial responses and promotes microbiota‐based colonisation resistance during Citrobacter rodentium infection. Cell Microbiol 2019; 21:e13026. [DOI] [PubMed] [Google Scholar]
- 141. Li Y, Liu M, Zuo Z, Liu J, Yu X, Guan Y et al TLR9 regulates the NF‐κB‐NLRP3‐IL‐1β pathway negatively in salmonella‐induced NKG2D‐mediated intestinal inflammation. J Immunol 2017; 199:761–73. [DOI] [PubMed] [Google Scholar]
- 142. Varga MG, Shaffer CL, Sierra JC, Suarez G, Piazuelo MB, Whitaker ME et al Pathogenic Helicobacter pylori strains translocate DNA and activate TLR9 via the cancer‐associated cag type IV secretion system. Oncogene 2016; 35:6262–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H et al A toll‐like receptor recognizes bacterial DNA. Nature 2000; 408:740–5. [DOI] [PubMed] [Google Scholar]
- 144. Saavedra PHV, Huang L, Ghazavi F, Kourula S, Vanden Berghe T, Takahashi N et al Apoptosis of intestinal epithelial cells restricts Clostridium difficile infection in a model of pseudomembranous colitis. Nat Commun 2018; 9:4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H et al The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 2011; 477:596–600. [DOI] [PubMed] [Google Scholar]
- 146. Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 links innate immunity and fatty acid‐induced insulin resistance. J Clin Invest 2006; 116:3015–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS et al Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator‐activated receptors α and γ . Proc Natl Acad Sci USA 1997; 94:4318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Murakami T, Ockinger J, Yu J, Byles V, McColl A, Hofer AM et al Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc Natl Acad Sci USA 2012; 109:11282–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Solle M, Labasi J, Perregaux DG, Stam E, Petrushova N, Koller BH et al Altered cytokine production in mice lacking P2X(7) receptors. J Biol Chem 2001; 276:125–32. [DOI] [PubMed] [Google Scholar]
- 150. Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short‐chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013; 145:396–406 e1–10. [DOI] [PubMed] [Google Scholar]
- 151. Goverse G, Molenaar R, Macia L, Tan J, Erkelens MN, Konijn T et al Diet‐derived short chain fatty acids stimulate intestinal epithelial cells to induce mucosal tolerogenic dendritic cells. J Immunol 2017; 198:2172–81. [DOI] [PubMed] [Google Scholar]
- 152. Koh A, De Vadder F, Kovatcheva‐Datchary P, Backhed F. From dietary fiber to host physiology: short‐chain fatty acids as key bacterial metabolites. Cell 2016; 165:1332–45. [DOI] [PubMed] [Google Scholar]
- 153. Pott J, Kabat AM, Maloy KJ. Intestinal epithelial cell autophagy is required to protect against TNF‐induced apoptosis during chronic colitis in mice. Cell Host Microbe 2018; 23:191–202 e4. [DOI] [PubMed] [Google Scholar]
- 154. Matsuzawa‐Ishimoto Y, Shono Y, Gomez LE, Hubbard‐Lucey VM, Cammer M, Neil J et al Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med 2017; 214:3687–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Casanova JE. Bacterial autophagy: offense and defense at the host‐pathogen interface. Cell Mol Gastroenterol Hepatol 2017; 4:237–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Pedicord VA, Lockhart AAK, Rangan KJ, Craig JW, Loschko J, Rogoz A et al Exploiting a host–commensal interaction to promote intestinal barrier function and enteric pathogen tolerance. Sci Immunol 2016; 1: eaai7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J et al Host recognition of bacterial muramyl dipeptide mediated through NOD2. implications for Crohn's disease. J Biol Chem 2003; 278:5509–12. [DOI] [PubMed] [Google Scholar]
- 158. Kim B, Wang YC, Hespen CW, Espinosa J, Salje J, Rangan KJ et al Enterococcus faecium secreted antigen A generates muropeptides to enhance host immunity and limit bacterial pathogenesis. Elife 2019; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Qiu Z, Cervantes JL, Cicek BB, Mukherjee S, Venkatesh M, Maher LA et al Pregnane X receptor regulates pathogen‐induced inflammation and host defense against an intracellular bacterial infection through toll‐like receptor 4. Sci Rep 2016; 6:31936. [DOI] [PMC free article] [PubMed] [Google Scholar]


