Abstract
Purpose: Retinal organoids generated from human pluripotent stem cells exhibit considerable variability during differentiation. Our goals are to assess developmental maturity of the neural retina in vitro and design improved protocols based on objective criteria.
Methods: We performed transcriptome analyses of developing retinal organoids from human embryonic and induced pluripotent stem cell lines and utilized multiple bioinformatic tools for comparative analysis. Immunohistochemistry, immunoblotting and electron microscopy were employed for validation.
Results: We show that the developmental variability in organoids was reflected in gene expression profiles and could be evaluated by molecular staging with the human fetal and adult retinal transcriptome data. We also demonstrate that the addition of 9-cis retinal, instead of the widely used all-trans retinoic acid, accelerated rod photoreceptor differentiation in organoid cultures, with higher rhodopsin expression and more mature mitochondrial morphology evident by day 120.
Conclusion: Our studies provide an objective transcriptome-based modality for determining the differentiation state of retinal organoids and for comparisons across different stem cell lines and platforms, which should facilitate disease modeling and evaluation of therapies in vitro.
Introduction
Human development requires stringent and coordinated control of gene expression, signaling pathways, and cellular interactions that result in the generation of distinct cell types and tissues with complex morphological and functional phenotypes [1,2]. However, most of our current knowledge of the fundamental molecular events underlying cell-type specification and tissue differentiation has been derived from model organisms. Despite studies using human preimplantation embryos [3,4] and fetal tissue [5-7], the complexities of human organogenesis are poorly understood [8,9]. Pioneering advances in the generation of human embryonic stem cells (hESCs) [10] and induced pluripotent stem cells (iPSCs) [11], together with the development of three-dimensional (3D) organoid cultures [12-14], have revolutionized the studies of human development, facilitated individualized disease modeling, and rejuvenated the field of regenerative medicine [15-18].
Retinogenesis begins with specification of the forebrain neuroectoderm, and patterning of the early eye field is governed by finely tuned regulatory networks of signaling pathways and transcription factors [19]. Distinct morphological changes in the rostral neuroectoderm involve lateral expansion of bilateral eye fields to produce the optic vesicles, which invaginate and become the optic cups [20]. Landmark studies using fetal and neonatal tissues have provided unique insights, distinct from model organisms, into human retinal development [21-24]. By providing appropriate systemic and exogenous cues, human pluripotent stem cells can be directed to self-organize into 3D optic vesicle or optic cup structures [13,14]. Retinal organoids mimic early eye field development, with VSX2+ (also called CHX10) multipotent retinal progenitor cells differentiating into a polarized and laminated architecture harboring all types of retinal neurons and the Müller glia [25]. Rod and cone photoreceptors in organoid culture express opsin and other phototransduction genes [26-28], and develop rudimentary outer segment-like structures at late stages of differentiation [29,30]. However, current methods for characterizing retinal organoids have largely relied upon expression of select cell-type specific markers and histology, which provide limited information about the precise differentiation status. Although live imaging modalities have been employed recently for characterization and developmental staging [31,32], we still lack molecular insights into retinal organoid differentiation and maturation on a global scale, and how different experimental conditions (e.g., cell lines and protocols) could impact organoid cultures.
In this study, we performed transcriptome profiling of developing retinal organoids generated from hESCs and hiPSCs, using modifications of a widely used protocol [29]. Comparative transcriptome analyses with gene profiles of human fetal and adult retina revealed the molecular stages of retinal organoids and demonstrated their differentiation status and cellular composition more accurately. We also identified a specific role of 9-cis retinal (9CRAL) in expediting rod photoreceptor differentiation compared to the currently used all-trans retinoic acid (ATRA). Thus, these studies established a transcriptome-based molecular staging system using human fetal and adult data, enabling direct comparison of organoids under different experimental conditions for disease modeling and evaluation of therapies.
Methods
Maintenance and differentiation of human pluripotent stem cells
CRX-GFP H9 is a subclone of the H9 human ESC line, carrying a green fluorescent protein (GFP) gene under the control of the cone‐rod homeobox (CRX; Gene ID: 1406, associated OMIM numbers: 120970, 613829, 268000) promoter as previously reported [26]. The human iPSC lines PEN8E and NEI377 were reprogrammed from skin biopsies using integration-free Sendai virus carrying the four Yamanaka factors, as described [33], and their genome integrity and pluripotency have been evaluated [33]. The ESP1 and ESP2 lines were reprogrammed by Oct4, Klf4, Sox2, c-Myc, and Lin-28 mRNAs [34] and the Sendai virus (Ctrl1 FiPS4F1, Spanish National Stem Cell Bank, Valencia, Spain), respectively. The H9, PEN8E, and NEI377 cell lines were maintained in Essential 8 medium (E8; ThermoFisher Scientific, Waltham, MA) under hypoxia (5% O2), and the ESP1 and ESP2 lines in mTeSR1® (Stem Cell Technologies, Vancouver, Canada) under normoxia (20% O2). All lines were sustained on BD MatrigelTM human embryonic stem cell-qualified Basement Membrane Matrix (Corning, New York, NY)-coated plates. The PSCs were passaged every 3–4 days at 70–80% confluency using the EDTA-based protocol [35].
To start differentiation, all lines were detached and dissociated into small clumps with the EDTA dissociation protocol [35], and cultured in polyHEMA-coated or Ultra Low Attachment Culture Dishes (Corning) in E8 with 10 μM Y-27632 (Tocris) to form embryoid bodies (EBs). Neural-induction medium (NIM) consisting of Dulbecco's Modified Eagle's medium (DMEM/Ham's F-12 Nutrient Mix (F-12) (1:1), 1% N2 supplement (ThermoFisher Scientific), 1X Minimum essential medium (MEM) non-essential amino acids (NEAA; MilliporeSigma), and 2 µg/ml heparin (MilliporeSigma) was added at differentiation day (D)1 and D2 to reach a final ratio of 1:3 and 1:1 NIM:E8, respectively. Starting at D3, EBs were cultured in 100% NIM for an additional 4 days. EBs were collected at D7 and plated onto BD MatrigelTM Growth Factor Reduced (GFR) Basement Membrane Matrix (Corning)-coated plates. On D16, the medium was changed to photoreceptor induction medium (PIM) consisting of DMEM/F-12 (3:1) with 2% B27 without vitamin A, 1% antibiotic-antimycotic solution (ThermoFisher Scientific), and 1X NEAA. Medium was changed daily.
Upon the appearance of optic vesicles (OVs; typically, D21–28) with neuroepithelium morphology, the regions were excised with tungsten needles under magnification and transferred to suspension culture in polyHEMA-coated or Ultra Low Attachment 10 cm2 Culture Dishes (Corning) in PIM. After D42, PIM was supplemented with 10% fetal bovine serum (FBS; ThermoFisher Scientific), 100 mM taurine (MilliporeSigma), and 2 mM GlutaMAX (ThermoFisher Scientific). Starting at D63, the PIM was supplemented with 1 µM retinoid (MilliporeSigma) three times a week and switched to 0.5 µM at D92. The 1% N2 supplement was included starting at D92. The medium for the H9 and PEN8E-derived organoids was supplemented with 20 ng/ml insulin-like growth factor 1 (IGF1; ThermoFisher Scientific) and 55 nM beta-mercaptoethanol (2-ME; ThermoFisher Scientific) from dissection until the end of differentiation. 9-cis retinal (9CRAL; MilliporeSigma) was added at the described concentrations and corresponding time periods during the media change. Organoids for the data set PEN8E_2 were cultured with 10 ng/ml IGF1 from D42 to D200, and ATRA (MilliporeSigma) was supplemented to the PIM at 1 µM 5 times per week and at 0.5 µM twice a week for D63–92 and D92–180, respectively. The ESP-derived organoid cultures were maintained in the same manner as the PEN8E_2 organoids, except no IGF1 was used. For all experiments directly comparing 9CRAL and ATRA, media changes were performed in the dark under dim red light to reduce isomerization of the retinoids. Retinal organoids were collected in the dark and processed in the light as described for all other experiments.
Immunofluorescence
hPSC-derived retinal organoids were collected throughout the differentiation process, fixed, and immunostained as previously described [36]. Briefly, the organoids were fixed in 2% or 4% paraformaldehyde in 1X PBS (D-PBS, 1X, 138 mM NaCl, 2.6 mM KCl, 8 mM Na2HPO4-7H2O, 1.5 mM KH2PO4, pH 7.4) for 1 h at room temperature. Fixation was followed by cryoprotection in a sucrose gradient, 10–30%. Retinal tissues were then embedded in Shandon M1 embedding matrix and frozen on dry ice. Ten-micron sections were obtained using a Leica cryostat (Leica Biosystems, Buffalo Grove, IL) at −14 °C. The sections were placed on Superfrost Plus slides (Fisher Scientific, Hampton, NH) and stored at −20 °C until use. Specific antibodies and concentrations are summarized in Appendix 1.
Immunoblots
The protocol for immunoblot analysis was as previously described [37]. Individual organoids were lysed in 50 μl of 1% Triton X-100 (MilliporeSigma) in 1X PBS (ThermoFisher Scientific) supplemented with 1X protease inhibitors (Roche, Basel, Switzerland). Gentle trituration was then used to dissociate the organoid until a homogenous solution was obtained. The samples were then incubated on a nutator at 4 °C for 1 h and centrifuged at 4 °C at 4,000 ×g for 5 min. The protein concentration was measured via the Pierce bicinchoninic acid (BCA) protein assay (ThermoFisher). Approximately 15 μg supernatant protein was diluted 4:1 in reducing 4X Laemmeli buffer for 1 h at room temperature and separated at 100 V for 1 h on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels, which were then transferred to polyvinylidene fluoride (PVDF) membranes at 100 V for 1 h. After blocking in 5% milk in 1X Tris-Buffered Saline, 0.1% Tween® 20 Detergent (TBST; Santa Cruz Biotech, Dallas, TX) for 1 h at room temperature, the blots were incubated in an antibody cocktail (1:1,000 1D4, 3A6, and 4D2, a generous gift from Dr. Robert Molday, Vancouver, Canada) overnight in 1% milk in 1X TBST. The following morning, the membranes were washed in 1X TBST and shaken at room temperature four times for 10 min each. The membranes were then incubated in appropriate secondary species immunoglobulin G (IgG) conjugated to horseradish peroxidase (1:10,000) in 1X TBST shaking at room temperature for 1.5 h. The membranes were then washed again four times in 1X TBST for 10 min each. Before imaging, the membranes were exposed to SuperSignal® West Pico enhanced chemiluminescence (ECL) solution (ThermoFisher) for 5 min, and chemiluminescence was captured using a Bio-Rad ChemiDoc™ touch (Bio-Rad, Hercules, CA). Membranes were next dried overnight at room temperature to remove antibody binding to the protein. Dry membranes were reactivated in 100% methanol and incubated with gamma-tubulin (1:1,000, ab11317, Abcam, Cambridge, United Kingdom) overnight as described above. Images were analyzed in Image Lab (Bio-Rad) and exported to Adobe Photoshop (San Jose, CA) for figure generation.
Transmission electron microscopy
Retinal organoids were processed for transmission electron microscopy (TEM) analysis as previously described [33]. Briefly, the organoids were initially fixed in 4% formaldehyde and 2% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4; Tousimis, Rockville, MD) for 2 h, washed in cacodylate buffer three times, and then fixed in osmium tetroxide (1% v/v in 0.1 M cacodylate buffer; Electron Microscope Science, Hatfield, PA) for 1 h in a room temperature. The organoids were washed in the same buffer three times, followed by acetate buffer (0.1 M, pH 4.2), and en-bloc staining in uranyl acetate (0.5% w/v; Electron Microscope Science) in acetate buffer for 1 h. The samples were dehydrated in ethanol solution (35%, 50%, 75%, 95%, and 100%), followed by propylene oxide. Subsequently, the samples were infiltrated in a mixture of propylene oxide and epoxy resin (1:1) overnight, embedded in a pure epoxy resin in a flat mold, and cured in a 55 °C oven for 48 h. Thin sections (70 to 80 nm) were made with an ultramicrotome (UC 7) and diamond knife (Diatome, Hatfield, PA), attached on 200-mesh copper grid, and counter-stained in aqueous solution of uranyl acetate (0.5% w/v) followed by lead citrate solutions. The thin sections were stabilized with carbon evaporation under a vacuum evaporator before the EM examination. The digital images were taken in the electron microscope (H7650)-equipped digital camera (AMT, Rosemont, IL).
RNA-seq alignment and quantitation
High-quality total RNA (100 ng, RIN >7) from at least two independent replicates for hPSCs at various differentiation time points were subjected to mRNA directional library construction as described previously [26,38]. Paired-end sequencing was performed to a length of 125 bases using HiSeq2500 (Illumina, San Diego, CA). Genome reference sequence GRCm38.p7 and Ensembl v82 annotation was used for alignment and quantitation. Quality control, sequence alignment, transcript, and gene-level quantification of primary RNA sequence (RNA-seq) data were accomplished using an established bioinformatics pipeline [39] Appendix 2. The transcriptome of adult human retina samples was obtained from NEI Commons (Brooks, MJ and Swaroop, A).
Differential expression and clustering analysis
Data generated for the five different cell lines, transcriptome comparison and 9CRAL/ATRA analysis were analyzed separately. For each data set, genes were kept for further analysis only if there were ≥5 count per million (CPM) in all replicates of at least one group of the data sets. The data were subjected to trimmed mean of log expression ratios values (TMM) normalization with edgeR v3.20.9 [40,41], and principle component analysis (PCA) and Pearson correlation were performed with normalized CPM (log2) values. Differential expression analysis was performed using limma v3.34.9 [42], and genes that had ≥2-fold change at least between two time points in either transcriptome of comparison and a false discovery rate (FDR) ≤0.01 were considered statistically significantly differentially expressed.
Gene expression clustering was performed on gene-wise Z-scores using AP (apcluster v1.4.7 package) [43,44] with the k-means option, where k=12 by choosing the corSimMat function as the similarity measure. Determination of the cluster number k was accomplished by empirical observation of the heatmap produced from the PCA projections of the genes-wise z-score.
Gene Ontology analysis
GO enrichment analysis was performed using clusterProfiler v3.6.0 [45]. To reduce redundancy associated with GO analysis, we performed Wang semantic similarity comparisons of the enriched GO terms using GOSemSim v2.4.1 [46] and semantic similarities clustered with AP. The term with the highest level in each cluster was taken as the enriched GO term. Heatmaps of GO term gene lists were generated using the log2 CPM values. The GO enrichment ratio was calculated with reference to the number of pathway genes found overlapping with the analysis.
Time-course stage comparison of retinal organoids with human fetal retina
Open-ended dynamic time warping (OE-DTW) analysis was used to compare the maturation state of the different retina organoid time-course stage expression data to the human fetal retina (GSE104827) and the human adult retina, as previously described [47]. Retina organoid time-course stages were defined as Stage 1: 25–36 days, Stage 2: 50–75 days, Stage 3: 80–90 days, Stage 4: 105–125 days, Stage 5: 145–172 days, and Stage 6: 186–205. Mean gene expression values at each stage were used for OE-DTW analysis. The gene set used for the OE-DTW analysis consisted of 118 well-known retina-centric transcription factors and cell-type markers (see Appendix 3 and Appendix 4). Co-inertia analysis was performed using MADE4 v1.52.0 package in R.
Results
Comparable morphology between hESC- and hiPSC-derived retinal organoids
Two human pluripotent stem cell lines, H9 (hESC) and PEN8E (hiPSC), were differentiated into retinal organoids using a widely used robust protocol [29], with minor modifications based on our mouse organoid differentiation method that included 9CRAL [36]. Both lines could generate phase-bright neural retina with cone and rod photoreceptors of comparable morphology (Figure 1A–C). S opsin (OPN1SW) was polarized to the apical side of the organoids (outer surface of organoid, exposed to media) as early as D90, with a further increase in expression from D120 to D200. L/M cone photoreceptors (labeled with L/M opsin, OPN1L/MW) were barely evident at D120 but increased at D200 (Figure 1C, left). Rhodopsin (RHO) immunostaining was observed at the apical side as early as D120 in the H9- and PEN8E-derived organoids, and elongated as the photoreceptors matured (Figure 1C, right). Retinal ganglion cells (RGCs) were evident at early stages, but could not be maintained through the end of differentiation, while all other neural retina cell types showed similar morphology and development in the H9- and PEN8E-organoids (Appendix 5).
Figure 1.
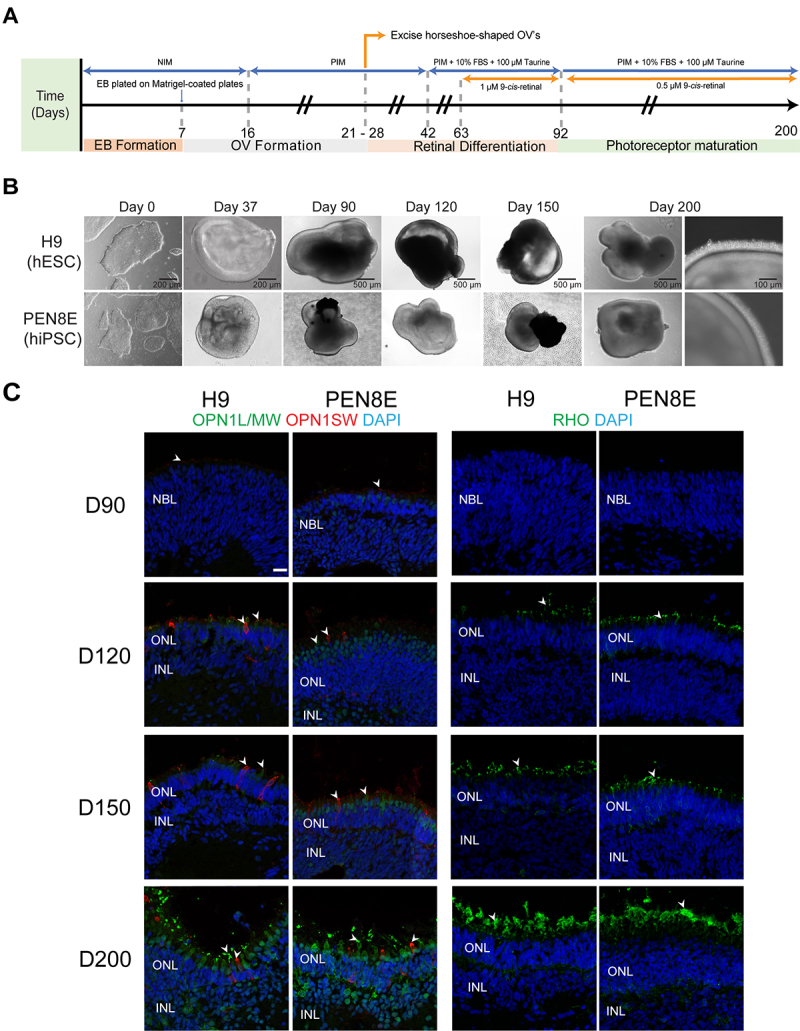
Differentiation of human pluripotent stem cells into retinal organoids. A: Differentiation protocol used in this study, modified from [29,33]. Numbers under the arrow indicate the differentiation day. NIM: neural induction medium; PIM: photoreceptor induction medium; EB: embryoid body; OV: optic vesicle; FBS: fetal bovine serum. B: Representative bright-field images of human embryonic stem cells (hESCs; H9) and human induced pluripotent stem cells (hiPSCs; PEN8E), and of differentiating organoids (from D37 to D200). C: Immunohistochemistry analysis of H9- and PEN8E-derived retinal organoids using marker antibodies for cones (OPN1L/MW, OPN1SW) and rods (RHO). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, blue). Arrowheads indicate relevant staining of each marker. Scale bar: 20 μm. D: differentiation day; NBL: neuroblastic layer; ONL: outer nuclear layer; INL, inner nuclear layer; OPL: outer plexiform layer; IPL: inner plexiform layer.
Differentially expressed gene clusters during organoid development
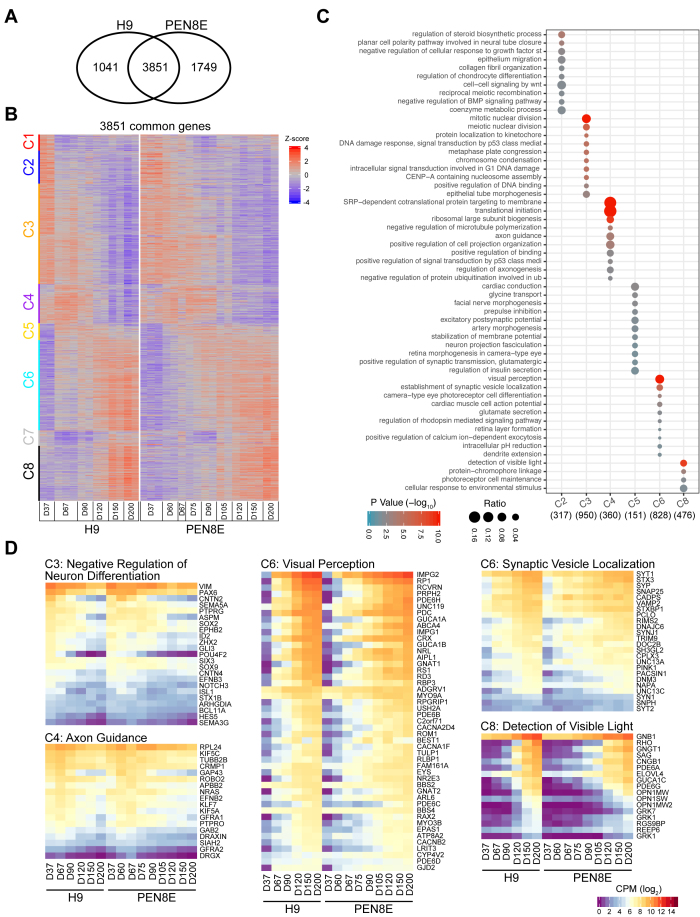
We then performed RNA-seq analysis of the developing retinal organoids to decipher the major differentiation stages in vitro. The H9- and PEN8E-derived retinal organoids showed a substantial overlap of a total of 3,851 differentially expressed (DE) genes that exhibited statistically significant changes in expression during differentiation (Figure 2A). The 1,041 DE genes unique to H9 and the 1,749 genes unique to PEN8E showed the same general trend in both data sets (Appendix 6). We noted that many of these genes were statistically significantly differentially expressed in both data sets under a less stringent cutoff of 1.5-fold and 5% FDR (data not shown). AP clustering analysis of the common DE genes between the two datasets yielded eight clusters (Cs) with either monotonically increasing or decreasing expression (Figure 2B). GO Biologic Process enrichment analysis uncovered key biologic pathways and genes that could be associated with specific stages of retinal development, and identified statistically significantly enriched terms for all clusters except C1 and C7 (Figure 2C). The C2 through C4 genes displayed a gradual decrease throughout differentiation, and were associated with negative regulation of neuronal differentiation, mitotic cell division, and neurodevelopmental processes and signaling (Figure 2D, Appendix 7). C4 also included axon guidance genes, which likely reflected the loss of ganglion cells in the organoid cultures. In contrast, the C5, C6, and C8 genes exhibited progressively increasing expression during organoid development, and included genes associated with neuronal or retinal differentiation and functions, including visual perception, synaptogenesis, and phototransduction.
Figure 2.
Comparative transcriptome analysis of H9 (hESC)- and PEN8E (hiPSC)-derived retinal organoids. A: Venn diagram showing differentially expressed (DE) genes during organoid development from the H9 and PEN8E lines. The vast majority of the DE genes are common (3851), with 1,041 and 1,749 unique to the H9 and PEN8E cell lines, respectively. B: Heatmap of 3,851 common DE genes, sorted and clustered based on row-wise z-score expression values in H9. Eight clusters are evident. C: Reduced-redundancy Gene Ontology (GO) Biologic Process enrichment analysis of genes in each cluster. Clusters 1 and 7 did not detect any statistically significantly enriched biologic processes. D: Heatmaps of GO cluster-related genes and their expression patterns in H9 and PEN8E retinal organoids. Expression count per million (CPM; log2) values were used for analysis; color scale shown at the bottom right.
Transcriptomes of organoids from different iPSCs and protocols
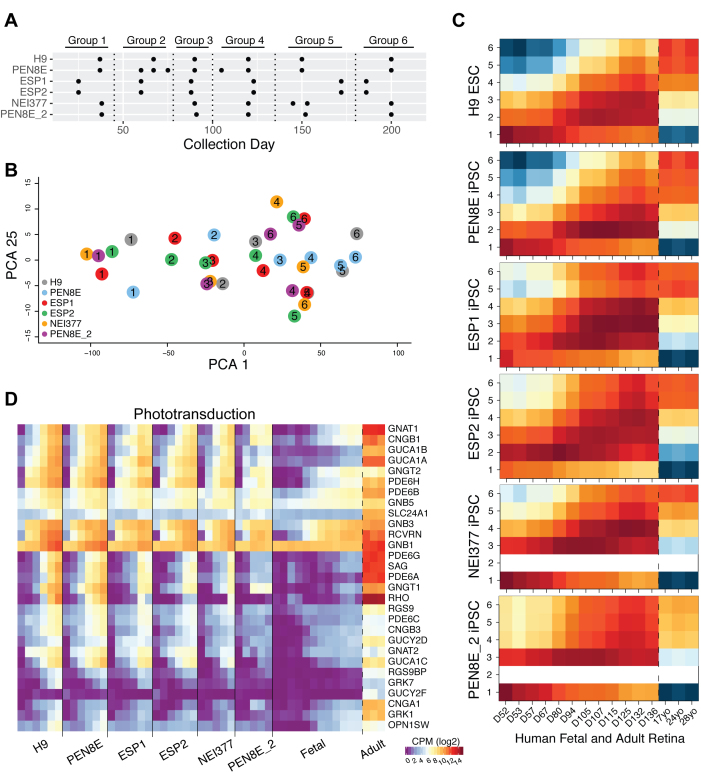
To evaluate the impact of iPSC lines or modifications in the differentiation protocol, we compared the expression of the 3,851 DE genes with organoids produced from different hiPSC lines (ESP1, ESP2, and NEI377) and protocols (PEN8E_2). PCA revealed a similar temporal progression in all organoid transcriptomes with increments on the differentiation days, as evident from PC1 (Appendix 4). As the harvesting time points varied among individuals and laboratories, we organized the organoid samples into six groups, for ease of comparison, based on the collection day (chronological age) and state of differentiation (Figure 3A) and performed PCA (Figure 3B) using a selection of established, retina-centric genes (Appendix 3). Although all organoids clustered together at an early stage of differentiation (group 1), the H9- and PEN8E-derived organoids (which included 9CRAL supplementation) showed accelerated differentiation from group 3 and onward, compared to the ESP, NEI377, and PEN8E_2 organoids (which used ATRA as described by [29]). In concordance, the ESP, NEI377, and PEN8E_2 organoids displayed lower expression of genes associated with phototransduction and outer segments compared to the H9- and PEN8E-derived organoids (Appendix 4). The NEI377 and PEN8E_2 organoids were generated by an identical protocol, and they displayed a similar developmental pattern, except group 6 of PEN8E_2, which is likely due to iPSC line differences.
Figure 3.
Comparative transcriptome analysis of hPSC-derived organoids and human retinal samples. A: Table showing organoid collection groups (Group 1: D25–37, Group 2: D50–75, Group 3: D80–90, Group 4: D105–125, Group 5: D145–172, Group 6: D185–200). B: Principal component analysis (PCA) plot of grouped human pluripotent stem cell (hPSC)-derived organoid samples based on retinal gene expression. PC1 shows the highest variation percentage and relates to time progression, while PC25 shows a non-statistically significant principle component used for ease of visualization. C: Dynamic time warping analysis showing the local cost matrix (LCM) for all hPSC-derived organoid groups and human fetal (D52–136) and adult (17, 24, and 28 years) samples. Warmer colors indicate lower levels of global dissimilarity (lowest: red, medium: orange), whereas cold colors (yellow and blue) represent higher levels of dissimilarity between samples. D: Heatmap demonstrating mean expression count per million (CPM; log2) value expression profiles of phototransduction across all hPSC-derived organoids and human retinal samples.
Molecular staging of retinal organoids based on developing human retina in vivo
To determine the maturity of organoid cultures using objective parameters, we performed OE-DTW analysis to align the time-series expression data between the organoids and the human fetal and adult retina gene profiles (Figure 3C) using a set of retina-centric genes (Appendix 3). Across all stem cell lines, group 1 organoids (D25–37) matched with the earliest human fetal time points (D52–67). Subsequently, the distinct PSC organoids showed differences in their highest correlation to corresponding human retinal stages. While organoids derived from ESP1, ESP2, NEI377, and PEN8E_2 lines revealed higher correlation with fetal retina samples; groups 4–6 H9 and PEN8E organoids were more concordant with the late fetal and adult retina (Figure 3C). For example, the group 4 H9 and PEN8E-derived organoids (D145–172) were highly correlated with adult samples (17, 24, and 28 years of age), whereas the ESP1, ESP2, NEI377, and PEN8E_2 organoids corresponded with late fetal samples (D105–136) at a similar stage (Figure 3C).
To confirm the result of the DTW analysis, we compared the expression of mature retinal genes associated with phototransduction and synaptic function between organoids and in vivo retinas. The late stage (group 5 and 6) H9 and PEN8E organoids showed similar expression of phototransduction genes as that of the adult retina (Figure 3D). However, most genes from “Transmission across chemical synapses” pathways were downregulated in the organoid cultures, suggesting the retinal organoids were not yet fully mature (Appendix 4).
Accelerated rod photoreceptor differentiation with 9CRAL
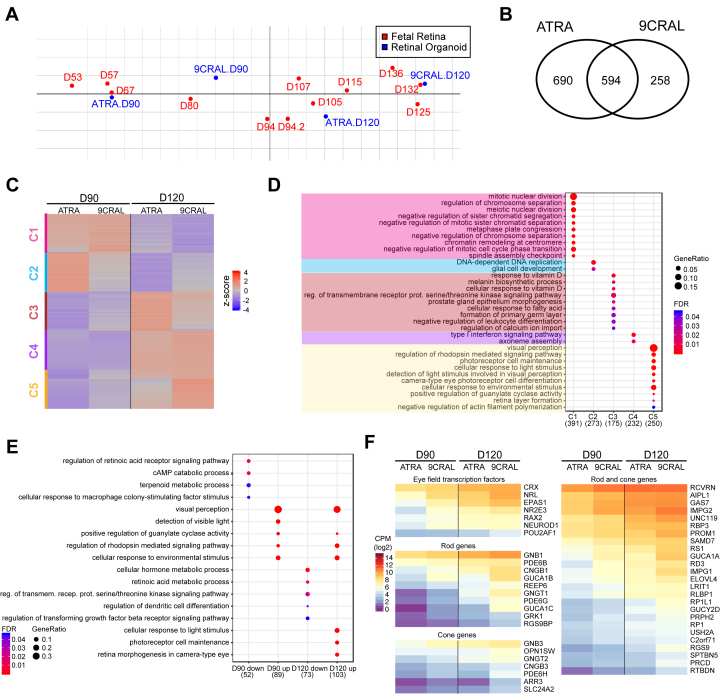
Much like its physiologically relevant isomer 11-cis retinal, 9CRAL can also bind opsin to form functioning rhodopsin, and retinoic acid (RA) can be generated as an oxidation product. Therefore, we hypothesized that replacing RA with 9CRAL in the modified protocol would result in improved rod differentiation. To evaluate this hypothesis, we performed a direct comparison of the D90 and D120 H9 organoids cultured with ATRA or 9CRAL from D64 onward. We then applied coinertia analysis to the transcriptomes of the ATRA- and 9CRAL-supplemented organoids and the developing human fetal retina (D53 to D136). The D90 and D120 of the 9CRAL organoid transcriptomes corresponded with the D80–94 and D125–136 human fetal retina gene profiles, respectively. However, the D90 and D120 ATRA transcriptomes matched with earlier fetal retina time points (D57–67 and D105–115, respectively; Figure 4A). Thus, treatment with 9CRAL expedited retinal development in organoid cultures and showed closer temporal transcriptome dynamics to the human fetal retina. The DE genes between D90 and D120 in the ATRA (1284 genes) and 9CRAL (852 genes) data presented an overlap of 594 genes (Figure 4B), which could, in turn, be grouped into five clusters with hierarchical clustering analysis (Figure 4C). Redundancy-reduced GO analysis of the common DE genes revealed a monotonic decrease in the C1 and C2 genes involved in the cell cycle, suggesting neural progenitor cells exit the cell cycle and become mature in retinal organoids (Figure 4D). The C3 through C5 genes were involved in energy metabolism and visual perception, which further characterized their progressive increase from D90 to D120. Although the ATRA and 9CRAL organoids showed comparable developmental patterns, changes in the DE genes during development varied between the two groups (Figure 4C). Day-matched comparison of DE genes revealed lower expression of genes involved in regulation of the RA signaling pathway, RA metabolic processing, and energy metabolism, whereas higher expression of visual perception and function genes was evident in the D90 and D120 9CRAL-treated organoids (Figure 4E). The heatmap of early eye field transcription factors and photoreceptor genes consistently showed expedited photoreceptor differentiation in the 9CRAL organoids, as revealed by the higher expression of these genes, compared to the ATRA organoids (Figure 4F).
Figure 4.
Transcriptome analysis of D90 and D120 organoids treated with ATRA or 9CRAL. A: Coinertia analysis projecting ordinations of maximum covariation of D90 and D120 all-trans retinoic acid (ATRA) and 9-cis retinal (9CRAL) organoids with human fetal transcriptome data. B: Venn diagram revealing differentially expressed (DE) genes during differentiation between ATRA and 9CRAL organoids. C: Heatmap of 594 common DE genes, sorted and clustered based on row-wise z-score expression values in H9. Five clusters are evident. D: Reduced-redundant Gene Ontology (GO) Biologic Process enrichment analysis of genes in each cluster. E: GO analysis of DE genes comparing 9CRAL to ATRA at D90 and D120 (day-wise comparison). F: Heatmaps showing expression profiles of genes encoding eye field transcription factors and photoreceptor genes.
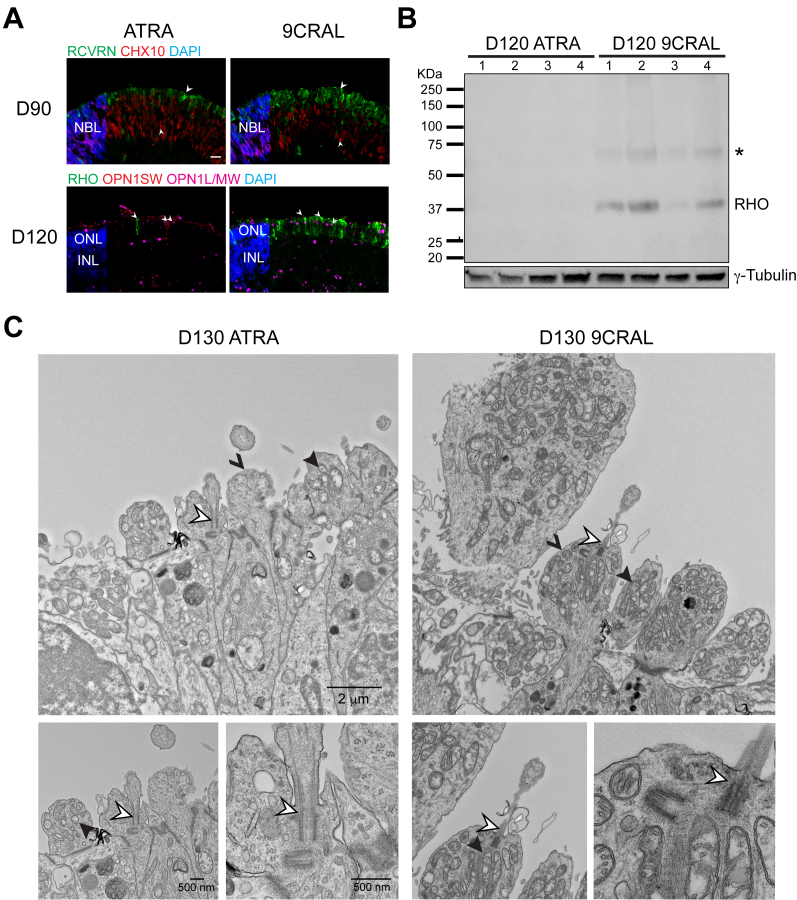
To validate the findings that 9CRAL accelerated photoreceptor differentiation, we performed immunohistochemistry on the D90 and D120 organoids. The D90 9CRAL organoids showed lower expression of retinal progenitor cell marker VSX2 (also called CHX10) and higher expression of a pan-photoreceptor marker (recoverin, RCVRN) compared to the ATRA organoids (Figure 5A, upper panel). At D120, the 9CRAL-treated organoids showed distinctively more RHO+ cells, which were barely detected in the ATRA organoids (Figure 5A, bottom panel), consistent with the higher rhodopsin expression in 9CRAL (14.0±9.00 CPM) compared to the ATRA (3.8±1.7 CPM) RNA-seq data. No marked difference was observed in the expression or morphology of the L/M-opsin and S-opsin+ cells (see Figure 5A, bottom panel), as well as other neural retina cell types (Appendix 8). In concordance, immunoblot analysis demonstrated increased rhodopsin expression in the 9CRAL organoids compared to the ATRA organoids at D120 (Figure 5B). Ultrastructural analysis of D130 retinal organoids using transmission electron microscopy (TEM) showed an advanced stage of maturation of the 9CRAL photoreceptors compared to the ATRA photoreceptors (Figure 5C). The ellipsoid (apical) side of the photoreceptor inner segments in the 9CRAL organoids contained a higher number of mitochondria with typical morphology, which could explain the divergent expression of genes involved in energy metabolism between the 9CRAL and ATRA organoids. No differences were evident in the morphology of the photoreceptor connecting cilia between the two groups. Taken together, these data demonstrate that 9CRAL supplementation expedited differentiation and maturation of rod photoreceptors in retinal organoids.
Figure 5.
9CRAL-expedited photoreceptor development. A: Representative images of immunostained sections of H9 retinal organoids supplemented with either all-trans retinoic acid (ATRA; left) or 9-cis retinal (9CRAL; right). D90 (top) 10 μm sections were immunolabeled for pan-photoreceptor marker recoverin (green) and retinal progenitor cell marker CHX10 (red). D120 sections (bottom) were immunolabeled for rod photoreceptor marker rhodopsin (green), cone photoreceptor marker OPN1SW (red), and L/M cone photoreceptor marker OPN1L/MW (magenta). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, blue). Arrowheads indicate relevant staining of a specific marker. Scale bar, 10 μm. B: Immunoblot showing rhodopsin expression in H9 ATRA and 9CRAL organoids at D120 (individual replicates are shown). The asterisk denotes a second band which is likely to be dimerization of rhodopsin. γ-tubulin is included as the protein loading control for protein amounts. C: Transmission electron microscopy of longitudinal sections at D130, showing inner segments and cilia. Hollow, solid, and v-shaped arrowheads indicate the relevant structure of the photoreceptor cilium, mitochondria, and inner segments, respectively. Magnifications (scale bars) in the right panel are the same as shown in the left.
Summary statement
Three-dimensional organoids derived from human pluripotent stem cells have been extensively applied for investigating organogenesis, modeling diseases, and developing therapies. However, substantial variations within organoids pose challenges for comparison among different cultures and studies. We generated transcriptomes of multiple distinct retinal organoids and compared them to human fetal and adult retina gene profiles for molecular staging of differentiation state of the cultures. The analysis revealed the advantage of using 9-cis retinal, instead of the widely used all-trans retinoic acid, in facilitating rod photoreceptor differentiation. Thus, a transcriptome-based comparison can provide an objective method for uncovering the maturity of organoid cultures across different lines and in various study platforms.
Discussion
The generation of in vitro 3D organoids from pluripotent stem cells has permitted rapid advances in our understanding of human organogenesis, disease mechanisms, and therapeutic interventions [16,18]. Relatively easy access and promising applications of cell replacement therapy to alleviate blinding diseases have prompted an explosion of studies on human retinal organoids, which exhibit appropriate stratified architecture and differentiation of all relevant cell types [26,29-31,48-51]. However, a lack of appropriate photoreceptor outer segments and synaptic connectivity, and the loss of RGCs after prolonged cultures, has hampered progress. The variability in temporal differentiation, depending on protocols and iPSC lines, also demands objective criteria for staging of human retinal organoids. Live imaging and reporter quantification assays have been used to characterize organoid development and staging [31,52]. More recently, retinal organoids from 16 hPSC lines were examined using multiple structural criteria that were then employed for developing a staging system [32]. In comparison, the present molecular staging provides a more objective measure of organoid maturity based on molecular staging with human retinal development. The present molecular staging profiles are concordant with a recent report describing similar temporal gene expression between in vivo human retina and in vitro cone-rich retinal organoid transcriptome data [53].
Human iPSCs harbor epigenetic memory of their somatic tissue of origin, which appears to favor their subsequent differentiation toward the lineage related to the donor cells and restrict other cell fates [54]. The presence of epigenetic signatures of donor tissues depends on the reprogramming methods and passages of iPSCs [55]; therefore, the differentiation capacity and therapeutic potential of hESCs and hiPSCs are not easily comparable. Interneurons in mouse retinal organoids differentiated from iPSCs cannot be well differentiated due to epigenetic markers of fibroblasts [5], but this phenotype could be alleviated in cultures with additional nutrients [36,39], suggesting that culture conditions and cell lines impact the epigenome in iPSCs. In this report, we differentiated genetically unmatched hESC and hiPSC lines reprogrammed from fibroblasts into retinal organoids, and the data showed a similar differentiation capacity of hESCs and hiPSCs in retinal lineages. The present results are in agreement with a recent study showing equivalent gene expression and neuronal differentiation potential between hESCs and iPSCs [56].
Human organoid cultures manifest substantial variability that may result from multiple factors, including intrinsic genetic variations and the epigenome state of the iPSC lines, reprogramming method, and differentiation protocol. A recent report used a TaqMan array-based analysis of key marker genes to demonstrate differences in retinal organoids [57]. The present comprehensive transcriptome-based molecular staging method utilizes global gene expression profiles and can robustly capture organoid variability and developmental status by comparing them to in vivo retinal transcriptome data. The gene profiles of ESP1, ESP2, NEI377, and PEN8E_2 showed broadly similar gene profiles and developmental trajectories, suggesting a minimal impact of cell-line intrinsic and protocol variations. However, the H9 and PEN8E lines clearly exhibited a more mature developmental trajectory and transcriptomes, resulting from the use of 9CRAL in the organoid cultures.
The studies reported here demonstrated accelerated rod photoreceptor differentiation and detection of rhodopsin protein with immunoblot analysis in 9CRAL-supplemented organoids as early as D120. We suggest that 9CRAL is driving more cells toward a photoreceptor cell fate. In the experimental protocol, all media changes were performed in the dark under dim red light to avoid isomerization of 9CRAL to all-trans retinal; therefore, at least some of the 9CRAL is likely oxidized into 9CRA inside the cells. 9-cis retinoic acid (9CRA) is a potent agonist for retinoid X receptors (RXRs) and retinoic acid receptors (RARs), while ATRA binds only to RARs [58-60]. Retinoic acid has been shown to promote photoreceptor development [61-63] and induce the expression of rod differentiation factor NRL [64]. We should note that retinoid-related orphan receptor beta (RORβ) regulates rod development by activating NRL [65]. In general, the interplay of retinoic acid receptors with other nuclear receptors has a substantial impact on transcriptional regulation of genes involved in photoreceptor development [66]. Thus, expedited rod differentiation by 9CRAL could result from more potent activation of retinoic acid receptors that may induce rod genes through a NRL-regulated gene network [67]. Nevertheless, further investigations are needed to elucidate underlying mechanisms of expediated rod photoreceptor differentiation via 9CRAL. We note that transcriptome profiling of retinal organoids derived from mouse pluripotent stem cells was recently used to identify missing regulatory signals and produce a more mature neural retina in vitro using docosahexaenoic acid (DHA) and fibroblast growth factor 1 (FGF1) [68].
Acknowledgments
We are grateful to Drs. Samuel G. Jacobson and Brian Brooks for skin biopsy samples that were used for generating fibroblasts and subsequently iPSC lines at the Stem Cell Core facility of National Heart, Lung and Blood Institute. We thank Comparative Cytogenetics Core Facility of National Cancer Institute for karyotyping assay. We acknowledge Anupam Mondal, Ben Fadl, Lina Zelinger and Samantha Papal for insightful discussions and constructive comments, and Jacob Nellissery and John Wilson for technical maintenance. This research was supported by Intramural Research Program of the NEI (ZIAEY000450 and ZIAEY000456). Y.K.A. was partially supported by Department of Science and Technology (DST) INSPIRE Faculty grant (IFA13-LSBM-90). Stem cell work in Spain was supported by Spanish Ministry of Science, Innovation and Universities, Instituto de Salud Carlos III (ISCIII)-European Regional Developmental Fund (FEDER) PI16/00409, CP18/00033 and ISCIII-FEDER Platform for Proteomics, Genotyping and Cell Lines, PRB3, PT17/0019/0024. The EM work was funded by FNLCR Contract HHSN261200800001E. The bioinformatic analyses used the high-performance computational capabilities of the Biowulf Linux cluster at NIH. Data availability statement: All raw and processed data are available through Gene Expression Omnibus with accession GSE129104 and at https://neicommons.nei.nih.gov.
Appendix 1. Antibody information for immunostaining.
To access the data, click or select the words “Appendix 1.”
Appendix 2. Sample sequencing depth and transcriptome alignment statistics.
To access the data, click or select the words “Appendix 2.”
Appendix 3. Retina-centric genes used for DTW analysis.
To access the data, click or select the words “Appendix 3.”
Appendix 4. Comparative transcriptome analysis of retinal organoids and human retinal samples (relates to Figure 3).
To access the data, click or select the words “Appendix 4.” (A) PCA plot of the all hPSC-derived organoid samples using CPM (log2) gene expression values. PC1 shows the highest variation percentage and relates to time progression. (B) Heatmap showing CPM (log2) gene expression values of well-known retinal-centric genes (Appendix 3) across all hPSC-derived organoid samples. (C) Heatmaps demonstrating expression profiles from all hPSC-derived organoids and human retinal samples of the Reactome pathway Synaptic Transmission Across Chemical Synapse. Heatmap uses the same CPM scale as in B.
Appendix 5. Immunohistochemistry images showing differentiation of retinal cell types in H9 and PEN8E retinal organoids (relates to Figure 1).
To access the data, click or select the words “Appendix 5.” (A) Retinal ganglion cells, photoreceptors and horizontal/amacrine cells are shown by brain-specific homeobox/POU domain protein 3A (BRN3A, green), cone-rod homeobox (CRX, red) and Calbindin (CALB, red), respectively. (B) Ceh-10 Homeodomain-Containing Homolog (CHX10, green, left panel), Protein kinase C alpha (PKCα, red, left panel), cellular retinaldehyde-binding protein (CRALBP, green, middle panel), Bassoon (red, right panel) showing bipolar cells, rod bipolar cells, rod photoreceptors, Müller glia and ribbon synapses, respectively. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, blue). The arrowheads indicate relevant staining of each marker. Scale bars: 100 μm (A); 50 μm (B).
Appendix 6. Genes uniquely expressed in H9 or PEN8E organoids (relates to Figure 2).
To access the data, click or select the words “Appendix 6.” (A) Clustered heatmaps of differentially expressed genes uniquely expressed in H9 (1041) or PEN8E (1749) organoids. Unique gene sets are shown across all samples. Interestingly, those specifically expressed in H9 or PEN8E show a stronger trend of upregulation/downregulation (yellow rectangle) to hESC or hiPSC, respectively. (B) GO Biological Process enrichment plot of H9 and PEN8E unique genes based on trend of expression. No enrichment was found for H9 unique upregulated genes.
Appendix 7. Heatmaps of CPM (log2) gene expression values from selected enriched GO Biological Process terms within clusters C2, C3, and C5 from Figure 2.
To access the data, click or select the words “Appendix 7.”
Appendix 8. Immunohistochemistry images of H9 retinal organoids treated with all-trans retinoic acid (ARTA, left) or 9-cis retinal (9CRAL) (relates to Figure 5).
To access the data, click or select the words “Appendix 8.”Rod photoreceptor, bipolar cells, horizonal/amacrine cells, and Müller glia are indicated by Rhodopsin (RHO, green), Protein kinase C alpha (PKCa, red, upper panel), Calbindin (CALB, red, middle panel) and cellular retinaldehyde-binding protein (CRALBP, red, bottom panel), respectively. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, blue). The arrowheads indicate relevant staining of each marker. Scale bars: 50 μm.
References
- 1.Lagha M, Bothma JP, Levine M. Mechanisms of transcriptional precision in animal development. Trends Genet. 2012;28:409–16. doi: 10.1016/j.tig.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parikshak NN, Gandal MJ, Geschwind DH. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat Rev Genet. 2015;16:441–58. doi: 10.1038/nrg3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma H, Marti-Gutierrez N, Park SW, Wu J, Lee Y, Suzuki K, Koski A, Ji D, Hayama T, Ahmed R, Darby H, Van Dyken C, Li Y, Kang E, Park AR, Kim D, Kim ST, Gong J, Gu Y, Xu X, Battaglia D, Krieg SA, Lee DM, Wu DH, Wolf DP, Heitner SB, Belmonte JCI, Amato P, Kim JS, Kaul S, Mitalipov S. Correction of a pathogenic gene mutation in human embryos. Nature. 2017;548:413–9. doi: 10.1038/nature23305. [DOI] [PubMed] [Google Scholar]
- 4.Fogarty NME, McCarthy A, Snijders KE, Powell BE, Kubikova N, Blakeley P, Lea R, Elder K, Wamaitha SE, Kim D, Maciulyte V, Kleinjung J, Kim JS, Wells D, Vallier L, Bertero A, Turner JMA, Niakan KK. Genome editing reveals a role for OCT4 in human embryogenesis. Nature. 2017;550:67–73. doi: 10.1038/nature24033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiler D, Chen X, Hazen J, Kupriyanov S, Carroll PA, Qu C, Xu B, Johnson D, Griffiths L, Frase S, Rodriguez AR, Martin G, Zhang J, Jeon J, Fan Y, Finkelstein D, Eisenman RN, Baldwin K, Dyer MA. Quantification of Retinogenesis in 3D Cultures Reveals Epigenetic Memory and Higher Efficiency in iPSCs Derived from Rod Photoreceptors. Cell Stem Cell. 2015;17:101–15. doi: 10.1016/j.stem.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerrard DT, Berry AA, Jennings RE, Piper Hanley K, Bobola N, Hanley NA. An integrative transcriptomic atlas of organogenesis in human embryos. eLife. 2016;5:e15657. doi: 10.7554/eLife.15657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belle M, Godefroy D, Couly G, Malone SA, Collier F, Giacobini P, Chedotal A. Tridimensional Visualization and Analysis of Early Human Development. Cell. 2017;169:161–73. doi: 10.1016/j.cell.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Rossant J. Mouse and human blastocyst-derived stem cells: vive les differences. Development. 2015;142:9–12. doi: 10.1242/dev.115451. [DOI] [PubMed] [Google Scholar]
- 9.Ortega NM, Winblad N, Plaza Reyes A, Lanner F. Functional genetics of early human development. Curr Opin Genet Dev. 2018;52:1–6. doi: 10.1016/j.gde.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 13.Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, Sasai Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–85. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Phillips MJ, Wallace KA, Dickerson SJ, Miller MJ, Verhoeven AD, Martin JM, Wright LS, Shen W, Capowski EE, Percin EF, Perez ET, Zhong X, Canto-Soler MV, Gamm DM. Blood-derived human iPS cells generate optic vesicle-like structures with the capacity to form retinal laminae and develop synapses. Invest Ophthalmol Vis Sci. 2012;53:2007–19. doi: 10.1167/iovs.11-9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bermingham-McDonogh O, Corwin JT, Hauswirth WW, Heller S, Reed R, Reh TA. Regenerative medicine for the special senses: restoring the inputs. J Neurosci. 2012;32:14053–7. doi: 10.1523/JNEUROSCI.3336-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasai Y. Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell. 2013;12:520–30. doi: 10.1016/j.stem.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Ader M, Tanaka EM. Modeling human development in 3D culture. Curr Opin Cell Biol. 2014;31:23–8. doi: 10.1016/j.ceb.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Clevers H. Modeling Development and Disease with Organoids. Cell. 2016;165:1586–97. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 19.Centanin L, Wittbrodt J. Retinal neurogenesis. Development. 2014;141:241–4. doi: 10.1242/dev.083642. [DOI] [PubMed] [Google Scholar]
- 20.Sasai Y. Grow your own eye: biologists have coaxed cells to form a retina, a step toward growing replacement organs outside the body. Sci Am. 2012;307:44–9. [PubMed] [Google Scholar]
- 21.Hendrickson A, Drucker D. The development of parafoveal and mid-peripheral human retina. Behav Brain Res. 1992;49:21–31. doi: 10.1016/s0166-4328(05)80191-3. [DOI] [PubMed] [Google Scholar]
- 22.Xiao M, Hendrickson A. Spatial and temporal expression of short, long/medium, or both opsins in human fetal cones. J Comp Neurol. 2000;425:545–59. [PubMed] [Google Scholar]
- 23.Hendrickson A, Possin D, Vajzovic L, Toth CA. Histologic development of the human fovea from midgestation to maturity. Am J Ophthalmol. 2012;154:767–78. doi: 10.1016/j.ajo.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendrickson A. Development of Retinal Layers in Prenatal Human Retina. Am J Ophthalmol. 2016;161:29–35. doi: 10.1016/j.ajo.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips MJ, Perez ET, Martin JM, Reshel ST, Wallace KA, Capowski EE, Singh R, Wright LS, Clark EM, Barney PM, Stewart R, Dickerson SJ, Miller MJ, Percin EF, Thomson JA, Gamm DM. Modeling human retinal development with patient-specific induced pluripotent stem cells reveals multiple roles for visual system homeobox 2. Stem Cells. 2014;32:1480–92. doi: 10.1002/stem.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaewkhaw R, Kaya KD, Brooks M, Homma K, Zou J, Chaitankar V, Rao M, Swaroop A. Transcriptome Dynamics of Developing Photoreceptors in Three-Dimensional Retina Cultures Recapitulates Temporal Sequence of Human Cone and Rod Differentiation Revealing Cell Surface Markers and Gene Networks. Stem Cells. 2015;33:3504–18. doi: 10.1002/stem.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welby E, Lakowski J, Di Foggia V, Budinger D, Gonzalez-Cordero A, Lun ATL, Epstein M, Patel A, Cuevas E, Kruczek K, Naeem A, Minneci F, Hubank M, Jones DT, Marioni JC, Ali RR, Sowden JC. Isolation and Comparative Transcriptome Analysis of Human Fetal and iPSC-Derived Cone Photoreceptor Cells. Stem Cell Reports. 2017;9:1898–915. doi: 10.1016/j.stemcr.2017.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eldred KC, Hadyniak SE, Hussey KA, Brenerman B, Zhang PW, Chamling X, Sluch VM, Welsbie DS, Hattar S, Taylor J, Wahlin K, Zack DJ, Johnston RJ., Jr Thyroid hormone signaling specifies cone subtypes in human retinal organoids. Science. 2018;362 doi: 10.1126/science.aau6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong X, Gutierrez C, Xue T, Hampton C, Vergara MN, Cao LH, Peters A, Park TS, Zambidis ET, Meyer JS, Gamm DM, Yau KW, Canto-Soler MV. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun. 2014;5:4047. doi: 10.1038/ncomms5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wahlin KJ, Maruotti JA, Sripathi SR, Ball J, Angueyra JM, Kim C, Grebe R, Li W, Jones BW, Zack DJ. Photoreceptor Outer Segment-like Structures in Long-Term 3D Retinas from Human Pluripotent Stem Cells. Sci Rep. 2017;7:766. doi: 10.1038/s41598-017-00774-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Browne AW, Arnesano C, Harutyunyan N, Khuu T, Martinez JC, Pollack HA, Koos DS, Lee TC, Fraser SE, Moats RA, Aparicio JG, Cobrinik D. Structural and Functional Characterization of Human Stem-Cell-Derived Retinal Organoids by Live Imaging. Invest Ophthalmol Vis Sci. 2017;58:3311–8. doi: 10.1167/iovs.16-20796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capowski EE, Samimi K, Mayerl SJ, Phillips MJ, Pinilla I, Howden SE, Saha J, Jansen AD, Edwards KL, Jager LD, Barlow K, Valiauga R, Erlichman Z, Hagstrom A, Sinha D, Sluch VM, Chamling X, Zack DJ, Skala MC, Gamm DM. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development. 2019;146 doi: 10.1242/dev.171686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimada H, Lu Q, Insinna-Kettenhofen C, Nagashima K, English MA, Semler EM, Mahgerefteh J, Cideciyan AV, Li T, Brooks BP, Gunay-Aygun M, Jacobson SG, Cogliati T, Westlake CJ, Swaroop A. In Vitro Modeling Using Ciliopathy-Patient-Derived Cells Reveals Distinct Cilia Dysfunctions Caused by CEP290 Mutations. Cell Reports. 2017;20:384–96. doi: 10.1016/j.celrep.2017.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Artero Castro A, Leon M, Luna-Pelaez N, Martin Bernal A, Del Buey Furio V, Erceg S, Lukovic D. Generation of a human iPSC line by mRNA reprogramming. Stem Cell Res (Amst) 2018;28:157–60. doi: 10.1016/j.scr.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 35.Beers J, Gulbranson DR, George N, Siniscalchi LI, Jones J, Thomson JA, Chen G. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat Protoc. 2012;7:2029–40. doi: 10.1038/nprot.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DiStefano T, Chen HY, Panebianco C, Kaya KD, Brooks MJ, Gieser L, Morgan NY, Pohida T, Swaroop A. Accelerated and Improved Differentiation of Retinal Organoids from Pluripotent Stem Cells in Rotating-Wall Vessel Bioreactors. Stem Cell Reports. 2018;10:300–13. doi: 10.1016/j.stemcr.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelley RA, Al-Ubaidi MR, Naash MI. Retbindin is an extracellular riboflavin-binding protein found at the photoreceptor/retinal pigment epithelium interface. J Biol Chem. 2015;290:5041–52. doi: 10.1074/jbc.M114.624189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brooks MJ, Rajasimha HK, Swaroop A. Retinal transcriptome profiling by directional next-generation sequencing using 100 ng of total RNA. Methods Mol Biol. 2012;884:319–34. doi: 10.1007/978-1-61779-848-1_23. [DOI] [PubMed] [Google Scholar]
- 39.Chen HY, Kaya KD, Dong L, Swaroop A. Three-dimensional retinal organoids from mouse pluripotent stem cells mimic in vivo development with enhanced stratification and rod photoreceptor differentiation. Mol Vis. 2016;22:1077–94. [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–97. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bodenhofer U, Kothmeier A, Hochreiter S. APCluster: an R package for affinity propagation clustering. Bioinformatics. 2011;27:2463–4. doi: 10.1093/bioinformatics/btr406. [DOI] [PubMed] [Google Scholar]
- 44.Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–6. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 45.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu G, Li F, Qin Y, Bo X, Wu Y, Wang S. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010;26:976–8. doi: 10.1093/bioinformatics/btq064. [DOI] [PubMed] [Google Scholar]
- 47.Hoshino A, Ratnapriya R, Brooks MJ, Chaitankar V, Wilken MS, Zhang C, Starostik MR, Gieser L, La Torre A, Nishio M, Bates O, Walton A, Bermingham-McDonogh O, Glass IA, Wong ROL, Swaroop A, Reh TA. Molecular Anatomy of the Developing Human Retina. Dev Cell. 2017;43:763–79. doi: 10.1016/j.devcel.2017.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reichman S, Slembrouck A, Gagliardi G, Chaffiol A, Terray A, Nanteau C, Potey A, Belle M, Rabesandratana O, Duebel J, Orieux G, Nandrot EF, Sahel JA, Goureau O. Generation of Storable Retinal Organoids and Retinal Pigmented Epithelium from Adherent Human iPS Cells in Xeno-Free and Feeder-Free Conditions. Stem Cells. 2017;35:1176–88. doi: 10.1002/stem.2586. [DOI] [PubMed] [Google Scholar]
- 49.Deng WL, Gao ML, Lei XL, Lv JN, Zhao H, He KW, Xia XX, Li LY, Chen YC, Li YP, Pan D, Xue T, Jin ZB. Gene Correction Reverses Ciliopathy and Photoreceptor Loss in iPSC-Derived Retinal Organoids from Retinitis Pigmentosa Patients. Stem Cell Reports. 2018;10:1267–81. doi: 10.1016/j.stemcr.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fligor CM, Langer KB, Sridhar A, Ren Y, Shields PK, Edler MC, Ohlemacher SK, Sluch VM, Zack DJ, Zhang C, Suter DM, Meyer JS. Three-Dimensional Retinal Organoids Facilitate the Investigation of Retinal Ganglion Cell Development, Organization and Neurite Outgrowth from Human Pluripotent Stem Cells. Sci Rep. 2018;8:14520. doi: 10.1038/s41598-018-32871-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hallam D, Hilgen G, Dorgau B, Zhu L, Yu M, Bojic S, Hewitt P, Schmitt M, Uteng M, Kustermann S, Steel D, Nicholds M, Thomas R, Treumann A, Porter A, Sernagor E, Armstrong L, Lako M. Human-Induced Pluripotent Stem Cells Generate Light Responsive Retinal Organoids with Variable and Nutrient-Dependent Efficiency. Stem Cells. 2018;36:1535–51. doi: 10.1002/stem.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vergara MN, Flores-Bellver M, Aparicio-Domingo S, McNally M, Wahlin KJ, Saxena MT, Mumm JS, Canto-Soler MV. Three-dimensional automated reporter quantification (3D-ARQ) technology enables quantitative screening in retinal organoids. Development. 2017;144:3698–705. doi: 10.1242/dev.146290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim S, Lowe A, Dharmat R, Lee S, Owen LA, Wang J, Shakoor A, Li Y, Morgan DJ, Hejazi AA, Cvekl A, DeAngelis MM, Zhou ZJ, Chen R, Liu W. Generation, transcriptome profiling, and functional validation of cone-rich human retinal organoids. Proc Natl Acad Sci USA. 2019 doi: 10.1073/pnas.1901572116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L, Hiler D, Xu B, AlDiri I, Chen X, Zhou X, Griffiths L, Valentine M, Shirinifard A, Sablauer A, Thiagarajan S, Barabas ME, Zhang J, Johnson D, Frase S, Dyer MA. Retinal Cell Type DNA Methylation and Histone Modifications Predict Reprogramming Efficiency and Retinogenesis in 3D Organoid Cultures. Cell Reports. 2018;22:2601–14. doi: 10.1016/j.celrep.2018.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 56.Marei HE, Althani A, Lashen S, Cenciarelli C, Hasan A. Genetically unmatched human iPSC and ESC exhibit equivalent gene expression and neuronal differentiation potential. Sci Rep. 2017;7:17504. doi: 10.1038/s41598-017-17882-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mellough CB, Collin J, Queen R, Hilgen G, Dorgau B, Zerti D, Felemban M, White K, Sernagor E, Lako M. Systematic Comparison of Retinal Organoid Differentiation from Human Pluripotent Stem Cells Reveals Stage Specific, Cell Line, and Methodological Differences. Stem Cells Transl Med. 2019 doi: 10.1002/sctm.18-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adamson PC, Widemann BC, Reaman GH, Seibel NL, Murphy RF, Gillespie AF, Balis FM. A phase I trial and pharmacokinetic study of 9-cis-retinoic acid (ALRT1057) in pediatric patients with refractory cancer: a joint Pediatric Oncology Branch, National Cancer Institute, and Children’s Cancer Group study. Clin Cancer Res. 2001;7:3034–9. [PubMed] [Google Scholar]
- 59.Heyman RA, Mangelsdorf DJ, Dyck JA, Stein RB, Eichele G, Evans RM, Thaller C. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 60.Allenby G, Bocquel MT, Saunders M, Kazmer S, Speck J, Rosenberger M, Lovey A, Kastner P, Grippo JF, Chambon P. Retinoic acid receptors and retinoid X receptors: interactions with endogenous retinoic acids. Proc Natl Acad Sci USA. 1993;90:30–4. doi: 10.1073/pnas.90.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hyatt GA, Schmitt EA, Fadool JM, Dowling JE. Retinoic acid alters photoreceptor development in vivo. Proc Natl Acad Sci USA. 1996;93:13298–303. doi: 10.1073/pnas.93.23.13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kelley MW, Williams RC, Turner JK, Creech-Kraft JM, Reh TA. Retinoic acid promotes rod photoreceptor differentiation in rat retina in vivo. Neuroreport. 1999;10:2389–94. doi: 10.1097/00001756-199908020-00031. [DOI] [PubMed] [Google Scholar]
- 63.Cvekl A, Wang WL. Retinoic acid signaling in mammalian eye development. Exp Eye Res. 2009;89:280–91. doi: 10.1016/j.exer.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khanna H, Akimoto M, Siffroi-Fernandez S, Friedman JS, Hicks D, Swaroop A. Retinoic acid regulates the expression of photoreceptor transcription factor NRL. J Biol Chem. 2006;281:27327–34. doi: 10.1074/jbc.M605500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jia L, Oh EC, Ng L, Srinivas M, Brooks M, Swaroop A, Forrest D. Retinoid-related orphan nuclear receptor RORbeta is an early-acting factor in rod photoreceptor development. Proc Natl Acad Sci USA. 2009;106:17534–9. doi: 10.1073/pnas.0902425106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Forrest D, Swaroop A. Minireview: the role of nuclear receptors in photoreceptor differentiation and disease. Mol Endocrinol. 2012;26:905–15. doi: 10.1210/me.2012-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim JW, Yang HJ, Brooks MJ, Zelinger L, Karakulah G, Gotoh N, Boleda A, Gieser L, Giuste F, Whitaker DT, Walton A, Villasmil R, Barb JJ, Munson PJ, Kaya KD, Chaitankar V, Cogliati T, Swaroop A. NRL-Regulated Transcriptome Dynamics of Developing Rod Photoreceptors. Cell Reports. 2016;17:2460–73. doi: 10.1016/j.celrep.2016.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brooks MJ, Chen HY, Kelley RA, Mondal AK, Li T, Chaitankar V, Swaroop A. Improved retinal organoid differentiation by modulating signaling pathways revealed by comparative transcriptome analyses with development in vivo. Stem Cell Reports. 2019 doi: 10.1016/j.stemcr.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]